Abstract
Glycosylation is a hallmark of cancer biology, and altered glycosylation influences multiple facets of melanoma growth and progression. To identify glycosyltransferases, glycans, and glycoproteins essential for melanoma maintenance, we conducted an in vivo growth screen with a pooled shRNA library of glycosyltransferases, lectin microarray profiling of benign nevi and melanoma patient samples, and mass spectrometry-based glycoproteomics. We found that α−2,3 sialyltransferases ST3GAL1 and ST3GAL2 and corresponding α−2,3-linked sialosides are upregulated in melanoma compared to nevi and are essential for melanoma growth in vivo and in vitro. Glycoproteomics revealed that glycoprotein targets of ST3GAL1 and ST3GAL2 are enriched in transmembrane proteins involved in growth signaling, including the amino acid transporter Solute Carrier Family 3 Member 2 (SLC3A2/CD98hc). CD98hc suppression mimicked the effect of ST3GAL1 and ST3GAL2 silencing, inhibiting melanoma cell proliferation. We found that both CD98hc protein stability and its pro-survival effect in melanoma are dependent upon α−2,3 sialylation mediated by ST3GAL1 and ST3GAL2. In summary, our studies reveal that α−2,3-sialosides functionally contribute to melanoma maintenance, supporting ST3GAL1 and ST3GAL2 as novel therapeutic targets in these tumors.
Keywords: Melanoma; glycosyltransferase; α-2,3 sialylation; ST3GAL1; ST3GAL2; cell growth
Introduction
Melanoma is the most aggressive skin cancer, the incidence of which has increased rapidly over the past decades. Early staged disease can be cured by surgery; however, despite the success of targeted and immune therapies, more than 50% of patients with melanoma metastasis still succumb to their disease. Thus, identifying novel drivers of melanoma progression and maintenance is essential to developing more effective treatments for advanced disease. Aberrant epigenetic, post-transcriptional, and post-translational programs such as glycosylation may provide unique intervention points. Melanoma originates from pigment-producing melanocytes, neural crest-derived cells found in the skin, eye, and other tissues throughout the body1, 2. Mutations in BRAF initiate the formation of nevi, benign melanocytic lesions. While most nevi have very low levels of proliferation, histopathological data shows that approximately one-third of melanomas arise from melanocytic nevi3. Transformation of nevi to melanoma in situ occurs through crosstalk between mutated cancer cells and their immediate microenvironment4, 5. As primary tumor cells override tumor-suppressing barriers via oncogenic adaptations, invasive melanoma cells grow into the dermis and access systemic circulation1. Cells surviving in circulation can extravasate, survive and colonize regional and distant organs2, 6.
Altered glycosylation is a hallmark of cancer biology7–9 that influences multiple facets of tumor growth and progression10. Recent findings of altered glycosylation in melanoma include upregulation of branching and bisecting N-linked glycans and truncated O-linked glycans contributing to the invasive and metastatic properties of melanoma11–14. In our previous work, we analyzed patient samples and identified core fucosylation, incorporated by α−1,6 fucosyltransferase FUT8, as a driver of melanoma metastasis15. However, current knowledge of glycans and their role in promoting melanoma growth and maintenance remains incomplete.
Herein, we conducted the first in vivo functional screen of glycosyltransferases in melanoma using a pooled library of doxycycline-inducible shRNAs. This analysis revealed a consistent depletion of melanoma cells carrying shRNAs targeting the family of α−2,3-sialyltransferases during in vivo growth. Concordantly, lectin array profiling of nevi vs. melanoma patient samples revealed enhanced α−2,3-sialylation, and in silico analyses of clinical transcriptomic datasets identified ST3GAL1 and ST3GAL2 as consistently upregulated in melanoma vs. nevi. Lectin pull-downs followed by mass-spectrometry analysis identified sialylated proteins responsible for promoting melanoma growth, including the amino acid transporter Solute Carrier Family 3 Member 2 (SLC3A2, CD98hc). Overall, our work sheds light on the role of glycosylation in melanoma biology and opens a novel path to developing glycan-based therapeutics to treat melanoma patients.
Results
Integrated in vivo shRNA functional screen and multi-omics analyses identify glycogenes and glycans essential for melanoma growth.
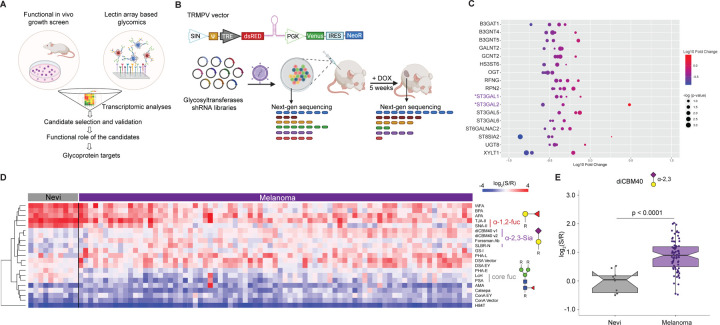
To identify glycogenes and their respective glycan epitopes required for melanoma cell survival, we utilized a multi-omics approach, integrating a functional in vivo growth screen of glycogenes with high throughput glycomic profiling of clinical melanoma samples (Fig. 1A). To identify glycogenes essential for melanoma growth, we designed a pooled library of doxycycline-inducible small hairpin RNA (shRNA) targeting glycan biosynthetic enzymes16. Our retrovirus-based shRNA library targets 199 glycosyltransferase genes, representing the majority of the 214 annotated in the human genome17 and contains six shRNAs/gene. These pooled libraries were divided into sub-libraries of 100 shRNAs to get sufficient coverage during next-generation sequencing (NGS) of xenograft tumors (Fig. 1B). Sub-libraries were used to transduce MeWo melanoma cells implanted subcutaneously in immunocompromised mice. To identify glycogenes essential for melanoma growth in vivo, we performed NGS of tumor cells before implantation and at 5 weeks post-injection. Our analysis revealed multiple consistently depleted shRNA (average fold change <0.6, t-test p<0.05, at least 3/6 shRNA read counts in tumors vs. baseline, Fig. S1A–B). The following glycogenes were found to be depleted: B3GAT1, B3GNT4, B3GNT5, GALNT2, GCNT2, HS3ST6, OGT, RFNG, RPN2, ST3GAL1, ST3GAL2, ST3GAL5, ST3GAL6, ST6GALNAC2, ST8SIA2, UGT8 and XYLT1 (Fig. 1C, Fig S1B). OGT and RPN2 are known to be essential for cell viability18–20, confirming the ability of our growth screen to identify essential genes. Of note, multiple members of the α−2,3 sialyltransferase family (ST3GAL1/2/5/6) showed depleted shRNAs in tumors, indicating a potential role in maintaining melanoma growth (Fig. 1C, Fig. S1B).
Fig. 1. In vivo functional genetic screen and multi-omics approach identify essential glycogenes and glycans for melanoma growth.
(A) Schematic illustration of our approach to identifying glycosylation enzymes involved in melanoma growth and their targets. (B) Schematic representation of dual-color TRINE vector that enables Tet-regulated shRNA expression to suppress glycosyltransferases involved in cell proliferation and survival. The in vivo growth screen schema is also presented. (C) Bubble plot representing log 10-fold change of the depleted or enriched shRNA in the MeWo cells transduced with glycosyltransferase shRNA libraries in tumors (5 weeks post-injection) relative to baseline (before injection). Glycosyltransferases corresponding to α−2,3 sialylation are highlighted in purple. (D) Heatmap of ratiometric lectin microarray data for nevi (n=10) and melanoma FFPE tissues (n=79); Only lectins showing significant differences between the 2 groups are shown (student’s t-test (two-tailed), p < 0.05). Pink, log2(S/R) > log2(Smedian/Rmedian); blue, log2(Smedian/Rmedian) > log2(S/R). Lectins corresponding to α−2,3 sialosides are highlighted in purple. The complete heatmap is given in Fig. S2A. (E) Whisker plot showing significantly increased diCBM40 binding in melanoma compared to nevi. Significance was determined using Wilcoxon’s t-test.
To complement our functional in vivo screens and identify glycosylation signatures associated to melanoma growth and progression, we analyzed melanocytic nevi (n=18) and melanoma (primary, n=10; metastatic, n=61) FFPE patient samples using lectin microarrays. Melanocytic nevi are benign lesions unlikely to progress to melanoma; however, accumulating evidence suggests they are melanoma precursors2, 3, 21. The inclusion of nevi thus allowed the identification of glycan changes happening during melanomagenesis. Formalin-fixed paraffin-embedded (FFPE) melanoma tissue samples were processed and analyzed using our dual-color lectin microarray technology15, 22. Lectin microarrays utilize carbohydrate-binding proteins with well-defined specificities23 to detect glycan changes at different stages of melanoma progression. Heatmaps (Figures 1D and S2A) show differences in lectin binding between nevi and melanomas (including primary, lymph node metastases, subcutaneous metastases, and brain metastases). In keeping with our earlier work, we see a loss of a-1,2 fucose (lectins: TJA-II, SNA-II)24 in primary melanoma compared to nevi that is even more striking in the metastasis (Fig. 1D). Interestingly, core fucose (lectins: LcH, PSA) is high in nevi and lower in primary melanoma but then appears to be regained in metastatic melanoma where we have shown it contributes to invasion15. We also observed a significant increase in binding to lectins diCBM4025 and SLBR-N26, which recognize α−2,3-sialosides, in both primary and metastatic melanoma compared to nevi (Fig. 1D–E and S2B). Combined with the results of our functional selection experiment, our data points to a role for members of the sialyltransferase family and α−2,3-sialosides in melanoma growth and survival.
ST3GAL1 and ST3GAL2 display higher transcriptional and protein levels in melanoma compared to nevi
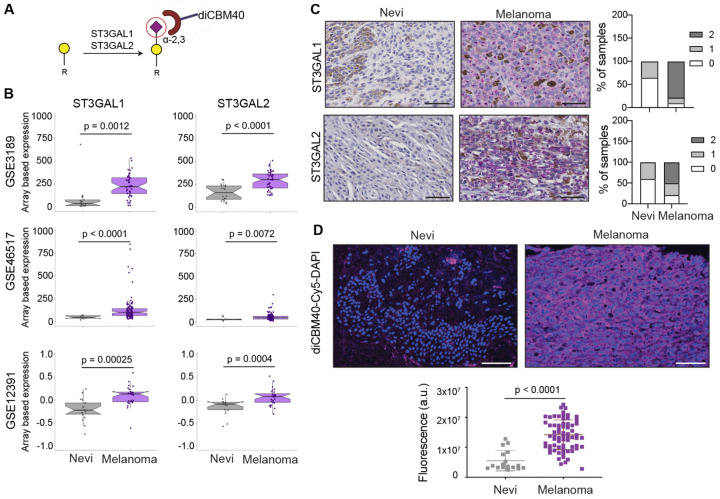
Our in vivo growth screens and glycomic analysis of benign and melanoma patient samples revealed multiple candidate regulators of melanoma growth. To narrow the list, we further filtered candidates by transcriptomics analysis of melanoma patient cohorts with the expectation that essential genes would show transcriptional upregulation in melanoma (Fig. 1A). We observed that most of the glycogene candidates from our in vivo growth screens did not show significant transcript differences in benign vs. melanoma samples (Fig. 1A, Fig. S3A–C). While ST3GAL1 and ST3GAL2 transcripts were significantly upregulated in melanoma compared to benign nevi (Fig. 2B) in multiple datasets. These analyses suggest that ST3GAL1 and ST3GAL2 upregulation is associated with neoplastic transformation and could contribute to melanomagenesis. ST3GAL1 and ST3GAL2 are sialyltransferases that catalyze the incorporation of α−2,3 sialic acids onto Galβ1,3GalNAc acceptors on glycoproteins and glycolipids (Fig. 2A)27. ST3GAL1 transfers sialic acid onto O-linked glycans (e.g., mucins)28, while ST3GAL2 can biosynthesize both O-glycans and glycolipids29, 30. Immunohistochemistry (IHC) analysis of an independent cohort of nevi and melanoma tissue microarray confirmed higher ST3GAL1 and ST3GAL2 protein in melanoma as demonstrated by perinuclear granular staining (Fig. 2C, Fig. S4A). In concordance with this, we observed significantly increased staining by diCBM40, a lectin that binds α−2,3-linked sialosides, in melanoma vs. nevi25 (p<0.001; Fig. 2D). We confirmed the specificity of diCBM40 with α−2,3-specific neuraminidase (NEB, P0743), which diminished the staining from this lectin (Fig. S4B). Our analyses suggest that transcriptional upregulation of ST3GAL1 and ST3GAL2 in melanoma results in increased sialyltransferase activity and higher levels of α−2,3-sialylated proteins. We postulated that ST3GAL1 and ST3GAL2 upregulation in melanoma is pro-tumorigenic and/or positively selected during tumorigenesis to support survival and growth during transformation.
Fig. 2. ST3GAL1, ST3GAL2, and α−2,3-sialosides are upregulated in melanoma relative to nevi.
(A) α−2,3 sialylated glycans generated by ST3GAL1 and ST3GAL2 and recognized by diCBM40 lectin. (B) Whisker plot illustrating significant upregulation of ST3GAL1 and ST3GAL2 mRNA expression in melanoma samples compared to nevi in multiple datasets: GSE318981, GSE4651782, GSE1239183. Two-tailed unpaired t-test. (C) Representative images of IHC staining with ST3GAL1 and ST3GAL2 antibodies in 15 nevi and 50 melanoma samples show a perinuclear staining pattern (Fast red counterstaining). IHC score was calculated by combining the signal intensity and percentage of positive cells within the section. The histogram shows the distribution of ST3GAL1 and ST3GAL2 IHC scores in nevi and melanoma samples. Scale bar, 10 μm. (D) Representative images of diCBM40 lectin fluorescence microscopy of nevi and melanoma FFPE tissues (n = 17 for nevi and 68 for melanomas). diCBM40-Alexa 647 (magenta) and DAPI-stained sections of TMA. Scale bar, 100 μm. Dot plots represent the average fluorescence intensity of five fields per image, two-tailed unpaired t-test.
ST3GAL1 and ST3GAL2 are essential for melanoma proliferation in vitro
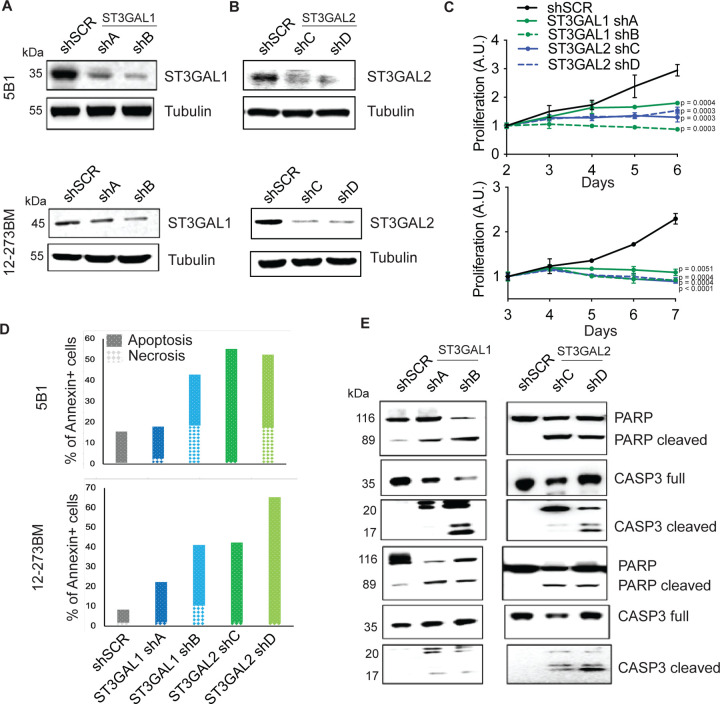
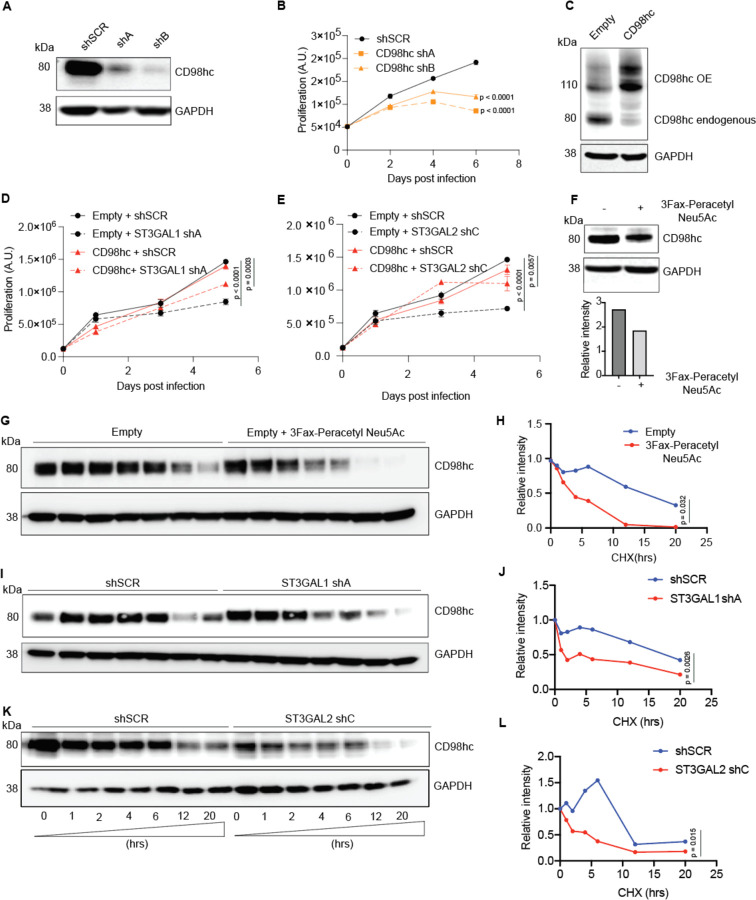
To further investigate the role of α−2,3-sialylation in growth, we silenced ST3GAL1 or ST3GAL2 in a human metastatic melanoma cell line 131/4–5B1 (hereafter, 5B1)31 and a patient-derived short-term culture, 12–273BM32. In brief, 5B1 and 12–273BM cells were stably transduced with lentivirus carrying one of two independent short hairpin RNAs (shRNAs) targeting ST3GAL1 (shA or shB) or ST3GAL2 (shC or shD) or a non-targeting, scrambled control (shSCR). We confirmed ST3GAL1/2 knockdown by qRT-PCR and Western blot (WB) (Fig. 3A–B, Figure S5A–B). Silencing of ST3GAL1 or ST3GAL2 substantially decreased binding by diCBM40, implying a concomitant reduction in α−2,3-linked sialosides (Fig. S5C). In both cases, silencing these genes also substantially decreased melanoma cell proliferation (Fig. 3C). No impact on cell growth was observed in normal melanocytes (NHEM) or in non-melanoma cell lines derived from the human embryonic kidney (HEK293) and lung adenocarcinoma (A549, Fig. S5D–E). Knockout of these sialyltransferases in mouse models have been generated by others and are viable33, 34, arguing that the anti-proliferative effect observed is not general to all cell types and may be specific to melanoma. To further characterize this growth defect, we investigated the effects of ST3GAL1 and ST3GAL2 knockdown on apoptosis. Flow cytometry analysis for Annexin V- and PI positive cells (Fig. 3D), together with WB for cleaved PARP and caspase-3 (Fig. 3E), all classical apoptosis markers, demonstrate that melanoma cells in which ST3GAL1 or ST3GAL2 are knocked down indeed undergo apoptosis. Moreover, simultaneous incubation with pan-caspase inhibitor Q-VD-OPh impaired melanoma cell death (Fig. S5F), further supporting caspase-dependent apoptosis in ST3GAL2 silenced cells.
Fig. 3. ST3GAL1 and ST3GAL2 are essential for melanoma proliferation in vitro.
ST3GAL1 (A) and ST3GAL2 (B) protein levels in 5B1 and 12–273BM cells stably transduced with non-targeting scrambled control shRNA (shSCR), ST3GAL1 shRNAs (shA and shB) and ST3GAL2 shRNAs (shC and shD) were assessed by Western blotting. Western blot images are representative of three independent experiments. (C) Relative growth curves of 5B1 and 12–273BM cells stably transduced with non-targeting scrambled control shRNA (shSCR), ST3GAL1 shRNAs (shA and shB), and ST3GAL2 shRNAs (shC and shD). The data shown are representative of three independent experiments. Two-tailed unpaired t-tests and p-values are shown in the figures. (D) The percentage of melanoma cells positive for Annexin V only (early apoptosis) or PI only (necrosis), Experiment was performed in duplicates. (E) Representative Western blots for caspase 3 and PARP on lysates from 5B1 and 12–273BM cells with shRNA against ST3GAL1 and ST3GAL2.
Identification of α−2,3-sialylated glycoproteins in melanoma reveals novel regulators of viability and maintenance
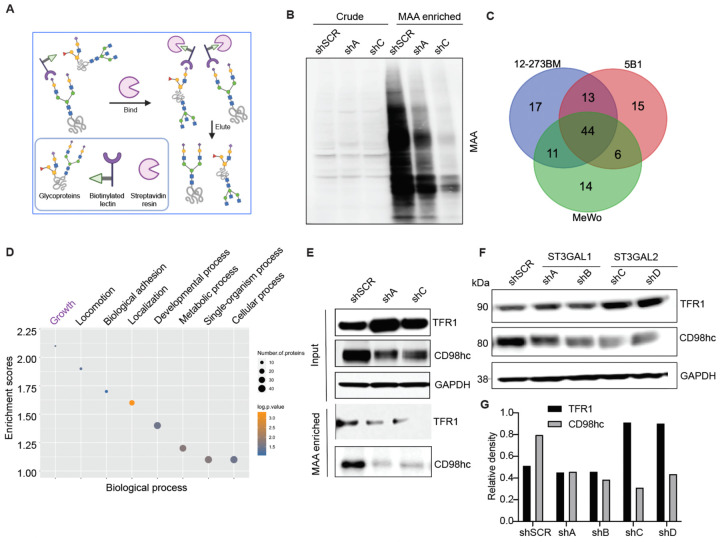
To identify α−2,3-sialylated glycoproteins that could mediate the role of ST3GAL1 and ST3GAL2 in melanoma proliferation, we performed proteomic analysis of α−2,3-sialylated membrane proteins. We collected membrane extracts from three melanoma cell lines, 5B1 (BRAF mutant), 12–273BM (NRAS mutant), and MeWo (NF1 mutant/BRAF wild type/NRAS wild type), which represent the major melanoma genotypes, to identify shared sialylated glycoproteins. The α−2,3-sialylated glycoproteins were enriched using lectin chromatography with MAA (Fig. 4A). We confirmed α−2,3-sialylated glycoprotein enrichment by silver staining and MAA lectin blot (Fig. S6), and samples were analyzed by liquid chromatography coupled with mass spectrometry (LC-MS). As expected, less MAA-bound protein was observed from ST3GAL1, and ST3GAL2 silenced cells (Fig. 4B). Mass spectrometry data were processed by median filtering. Contaminant proteins such as ribosomal proteins, tubulins, and heat shock proteins are known to be non-glycosylated and were removed from the analysis. We identified 85, 78, and 75 candidate proteins in the MAA enrichment of 12–273BM, 5B1, and MeWo, respectively, and 44 out of the total proteins were common to all (Fig. 4C). Gene ontology analysis (DAVID) of shared sialylated proteins revealed enrichment in growth, locomotion, and adhesion factors (Fig. 4D). Proteins bound by MAA within the cell growth category include Transferrin receptor 1 (TFR1), Solute carrier family three member 2 SLC3A2 (also known as CD98 heavy chain (hc)), Apolipoprotein E (ApoE), Solute carrier family one member 4 (SLC1A4), Serpin Family H Member 1 (SERPINH1), and Galectin-3 binding protein (LGALS3BP) (Table S1). We investigated the potential association of these genes with the survival of melanoma cells using the Dependency Map (DepMap)35–37, which catalogs genetic dependencies in cancer cells by pooled RNAi or CRISPR screens. We observed that many melanoma cell lines and cells of other cancer types are frequently dependent on TFR1 and SLC3A2 for survival (Fig. S7 A–C). 32
Fig. 4. Identification of α−2,3-sialylated glycoproteins in melanoma reveals regulators of melanoma growth.
(A) Schematic illustration of the experimental approach showing affinity enrichment of α−2,3 sialylated proteins by MAA lectin affinity chromatography. (B) MAA affinity chromatography of whole-cell lysates of 5B1 cells transfected with NTC or ST3GAL1/ST3GAL2 shRNA followed by lectin blot with MAA lectin. An equal concentration of crude protein and an equal volume of MAA-enriched fractions was loaded for MAA lectin blot. (C) A number of proteins were identified by mass spectrometry analysis of the MAA-enriched fractions from 5B1, 12–273BN, and MeWo cell lines. (D) Gene ontology enrichment analysis (biological processes category) of α−2,3 sialylated proteins common to the three cell lines. Also, see Table S1 (E) Western blot analysis with a-TFR1 or a-CD98hc antibodies of MAA-pulldown and corresponding input from lysates of 5B1 cells transfected with NTC or ST3GAL1/ST3GAL2 shRNA. (F) Western blot analysis of TFR1 and CD98hc in lysates from 5B1 cells transfected with NTC or ST3GAL1 or ST3GAL2 shRNAs. (G) Densitometric analysis of CD98hc on lysates from 5B1 cells transfected with NTC or ST3GAL1 or ST3GAL2 shRNAs. The graph is representative of three replicates. Experiments in B, E, and F were performed in triplicate, and representative images were shown.
To validate our proteomic analysis, we further examined the glycosylation state of TFR1 and CD98hc. MAA lectin enrichment followed by Western blot showed reduced TFR1 and CD98hc levels in shA (shST3GAL1) and shC (shST3GAL2) compared with shSCR-transduced 5B1 cells (Fig. 4E), consistent with lower α−2,3-sialylation on those proteins. Interestingly, silencing of ST3GAL1/2 also led to reduced total CD98hc protein levels (Fig. 4E,F). In contrast, TFR1 levels did not change in ST3GAL1-silenced cells and were found to increase in shST3GAL2-transduced cells (Fig. 4F). Overall, our analysis suggests CD98hc and TFR1 are α−2,3-sialylated by ST3GAL1 and ST3GAL2 and that CD98hc levels may be influenced by sialylation.
α−2,3-sialylation of SLC3A2 (CD98hc) is required for its stability and anti-proliferative effect
CD98 heavy chain (CD98hc), encoded by SLC3A2, consists of a transmembrane domain, a heavily glycosylated extracellular domain, and a cytoplasmic tail38, 39. SLC3A2 is then disulfide-linked with the non-glycosylated light chains (CD98lc) encoded by either LAT-1 or LAT-240. CD98 participates in multiple biological functions, including amplifying integrin signaling41 and mediating amino acid transport42, which ultimately impact cell proliferation and survival. Moreover, altered SLC3A2 expression has been associated with poor prognosis in several types of cancer43–46. A recent study showed that SLC3A2 was highly expressed in metastatic melanoma tissue compared to benign melanocytic nevi, and higher SLC3A2 mRNA levels were strongly correlated with lower survival rates47, proposing SLC3A2 as a prognostic biomarker for melanoma. We found that silencing of CD98hc suppresses melanoma cell proliferation (Fig. 5A–B) and has no effect on other cell types, such as HEK 293T (Fig. S8), mimicking the effects of ST3GAL1 or ST3GAL2 silencing on those cells. We also found that CD98hc overexpression partially rescues the antiproliferative effect of ST3GAL1- or ST3GAL2 silencing (Fig. 5C–E; Fig. S9), suggesting that CD98hc may be a critical mediator of ST3GAL1/2 effects on melanoma growth.
Fig. 5. α−2,3 sialylation of SLC3A2 (CD98hc) is required for its stability and anti-proliferative effect.
(A) Western blot for CD98hc on lysates from 5B1 cells transfected with NTC or CD98hc shRNAs. (B) Relative growth curves of 5B1 cells stably transduced with non-targeting control shRNA (shSCR) and CD98hc shRNAs (shA and shB). The data shown are representative of two independent experiments. Two-tailed unpaired t-tests and p-values are shown in the figures. (C) Western blot of CD98hc in 5B1 cells stably overexpressing CD98hc or control vector. (D) Cell proliferation assay on 5B1 melanoma cells stably overexpressing CD98hc or empty vector and transduced with non-targeting control shSCR or shST3GAL1. (E) Cell proliferation assay on 5B1 melanoma cells stably overexpressing CD98hc or empty vector and transduced with non-targeting control shSCR or shST3GAL2. (F) Western blot of CD98hc in 5B1 cells treated with or without 200 uM 3Fax-peracetylNeu5Ac. Densitometric analysis is shown below. (G–H) Western blot analysis of CD98hc protein in 5B1 cells. 3Fax-peracetylNeu5Ac treated 5B1 cells were further treated with 10 μM CHX at indicated intervals and analyzed by western blot analysis. The intensity of CD98hc protein was quantified using a densitometer. Western blot analysis of SLC3A2 protein in 5B1 cells silenced for ST3GAL1 (I–J) or ST3GAL2 (K–L) and treated with 10 μM cycloheximide (CHX) at indicated intervals and analyzed by western blot analysis. Paired t-test is shown. Western blot images are representative of 2 independent experiments.
We next sought to determine whether α−2,3-sialylation affects CD98hc protein stability. We used 3Fax-Peracetyl Neu5Ac, a cell-permeable sialic acid analog inhibitor of sialyltransferases, to assess the effect of blocking sialylation on CD98hc stability. As expected, 3Fax-Peracetyl Neu5Ac treatment reduced sialic acid-specific MAL-2 lectin binding while increasing Galβ1–3 GalNAc specific PNA lectin binding (Fig. S10A–B). Indeed P-3FAX-Neu5Ac treatment mimicked the effect of ST3GAL1 or ST3GAL2 silencing by reducing CD98hc levels (Fig. 4F, 5F). Moreover, in the presence of protein synthesis inhibitor cycloheximide (CHX), the turnover rate for de-sialylated CD98hc was much faster than for sialylated CD98hc (Fig. 5G–H). Similarly, ST3GAL1 or ST3GAL2 silencing resulted in more rapid CD98hc degradation in the presence of CHX (Fig. 5I–L). Collectively our data indicates that α−2,3-sialylation supports the protein stability of CD98hc, an amino acid transporter required for melanoma cell survival (Fig. 6).
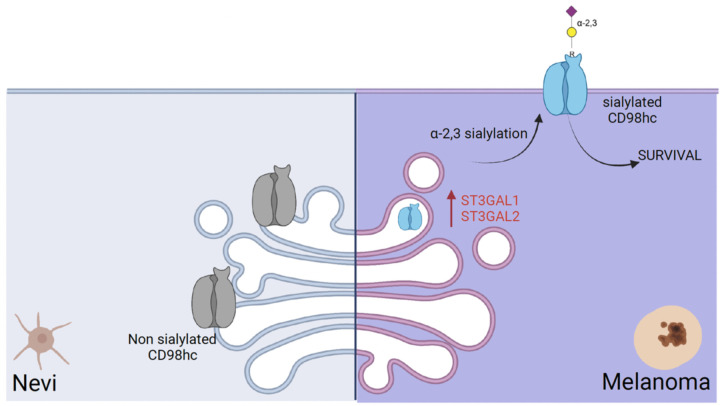
Fig. 6.
A schematic model suggests that α−2,3 sialylated transmembrane glycoprotein is required for melanoma cell survival.
Discussion
Aberrant glycosylation influences multiple facets of malignant transformation and tumor progression10. In our previous work, we obtained glycan profiles of matched primary and metastatic melanomas using clinical specimens and identified core fucosylation generated by FUT8 as a molecular driver of melanoma metastasis15. However, little is known about glycans’ role in melanoma genesis and maintenance. This study provides experimental and clinical evidence of the role of α−2,3 sialyltransferases in melanoma cell growth. Further, we identify α−2,3 sialylation of solute transporter protein CD98hc as a key regulator of melanoma growth.
To identify glycogenes and their glycan epitopes required for melanoma survival, we utilized a multi-omics approach, including an unbiased analysis of glycogenes using a functional in vivo growth screen. The decision to conduct the screen in vivo was based on the fact that in vivo models provide a more physiologic context for studying requirements for melanoma growth than in vitro systems. Our in vivo shRNA library consisting of relatively small shRNA pools (100 different shRNAs/sub-library), allowed us to examine the role of glycosyltransferases with high resolution. Supporting the value of this strategy, our screen identified glycogenes like OGT and RPN2 previously implicated in cancer cell survival18, 20. In addition, our analysis identified multiple members of the sialyltransferases family48, some of which are known to play an essential role in cancer cell survival, immune regulation, and angiogenesis (Fig. 1B)49, 50.
Most studies of glycan alterations in melanoma have been limited to in vitro assays on cell lines11, 12, 51–53. Our parallel high-throughput lectin profiling of benign nevi and clinical melanoma samples addresses this prior limitation. We observed increasing levels of α−2,3-sialosides (diCBM40, SLBR-N) in primary and metastatic melanoma compared to benign nevi. This could be due to the enhanced activity of sialyltransferases, including ST3GAL1, which has been associated with melanoma migration and invasion12. Only a handful of studies have examined ST3GAL1 function in human cancer. ST3GAL1 is overexpressed in ovarian54 and breast cancers55, and associated with poor prognosis in renal cancer56 and glioblastoma57. Induced expression of ST3GAL1 in mouse mammary cells promotes tumorigenesis58. ST3GAL2 is even less studied in cancer, although recently, overexpression of both the enzyme and a corresponding sialylated glycolipid SSEA-4 were associated with chemotherapy-resistant breast and ovarian cancers, correlating with poor outcomes59.
Our two parallel approaches (i.e., genetic screens and lectin profiling) provided multiple interesting candidates with a possible role in melanoma growth. We applied an additional filter based on transcriptomics data analysis to identify the most robust candidate glycogenes potentially modulating melanoma cell survival. Altogether, our transcriptomics, lectin fluorescence, and immunohistochemistry validation analysis in independent cohorts support a role for ST3GAL1 and ST3GAL2 generated α−2,3-sialosides in melanoma survival and growth. ST3GAL1 is partially essential in various cancer cell types with a median essentiality score of −0.2 as observed by DepMaP CRISPR perturbation data. While ST3GAL1 and ST3GAL2 are essential genes for melanoma cells, their silencing does not abrogate the survival of normal melanocytes such as NHEM-A or cell lines from other cancer types (Fig. S5D). These results support the value of an integrated approach to identifying specific glycogenes and corresponding glycan changes contributing to melanoma transformation. Moreover, our data suggest that ST3GAL1 and ST3GAL2 upregulation in melanoma is either directly pro-oncogenic or an event positively selected for during tumorigenesis to support survival or growth during neoplastic transformation.
Glycans can influence cancer cells properties by altering the glycosylation of adhesion molecules, membrane receptors, etc. To dissect the role of α−2,3 sialylation on growth-related programs of melanoma, we identified sialylated proteins and the mechanisms by which α−2,3 sialylation impacts them. MAA lectin-based glycoproteomic analysis of melanoma cell lines identified a set of α−2,3 sialylated membrane proteins, enriched in factors involved in cell growth, among other biological processes. These include TFR1 or transferrin receptor, a membrane glycoprotein which can import iron by binding a plasma glycoprotein, transferrin (TF)60, and involved in tumor progression and abundantly expressed in liver, breast, lung, and colon cancer cells61–64; ApoE which suppresses melanoma progression and metastasis65, 66; SLC1A4, a glutamine transporter which promotes apoptosis in Hepatocellular Carcinoma67 and decreases growth of prostate cancer cells68; SERPINH1, a serine proteinase inhibitor which contributes to gastric cancer progression69; and LGALS3BP, a galectin binding protein which is associated with poor prognosis in several human cancers70, 71. Some of these proteins have been previously shown to be glycosylated72. We focused our studies on SLC3A2 (CD98 heavy chain), a type II transmembrane protein that covalently links to one of several L-type amino acids (AA) transporters (light chains) to form large, functionally heterodimeric neutral amino acid transport systems73, 74. CD98hc plays a central role in AA and glucose cellular nutrition, redox homeostasis, and nucleotide availability, all key for cell proliferation75. In addition to playing a role in amino acid transport, CD98hc has been shown to associate with β1 and β3 integrins and regulate integrin signaling, which is in turn involved in cell adhesion, fusion, proliferation, and growth76, 77. Multiple reports suggest that overexpression of CD98hc stimulates FAK and AKT phosphorylation77–79, and deletion of CD98hc impairs integrin signaling41.
We found that CD98hc stability is dependent upon ST3GAL1/2-mediated sialylation. Indeed, desialylated CD98hc degraded much faster than sialylated CD98hc. Moreover, CD98hc silencing reduced melanoma cell proliferation, and CD98hc ectopic overexpression rescued the antiproliferative effect of ST3GAL1 or ST3GAL2 depletion (Fig. 5D–E). As CD98hc interacts with integrins and activates PI3K/AKT signaling, our data suggest that α−2,3 sialylation by ST3GAL1 and ST3GAL2 contributes to survival by enhancing CD98hc stability in melanoma cells. A possibility is that CD98hc sialylation influences protein export and membrane localization and, indirectly, protein stability. The rapid degradation of de-sialylated CD98hc may be a consequence of improper folding, inefficient binding to a chaperon, or changes in post-translational modifications resulting in protein cleavage or degradation. In fact, LAT1/CD98hc has been found among the multiple transmembrane protein clients of the CRBN-HSP90 co-chaperon complex80.
Overall, our work reveals ST3GAL1 and ST3GAL2 as robust pro-survival genes in melanoma, highlighting an essential role of α−2,3 sialylated glycans in melanoma biology and pathogenesis. Moreover, our data support the function of α−2,3 sialylated glycans in the modulation of the activity of CD98hc, an α−2,3 sialylated transmembrane glycoprotein required for melanoma survival (Fig. 6). Our findings may have therapeutic applications in melanoma by targeting melanoma-specific sialylated glycopeptide/s of CD98hc.
Methods
Cell Lines and Cell Culture
131/4–5B1 (hereafter 5B1) melanoma cells, A549 non-small cell lung cancer (NSCLC) cell line, and HEK293T cells (for lentivirus production) and were grown in DMEM with 10% FBS, 1 mmol/L sodium pyruvate, 4 mmol/L L-glutamine, 25 mmol/L D-glucose, 100 units/mL penicillin, and 100 μg/mL streptomycin. MeWo was cultured in EMEM with 10% FBS, 1 mmol/L sodium pyruvate, 4 mmol/L L-glutamine, and 25 mmol/L D-glucose. Melanocytes NHEM was purchased from Promocell, cultured in Melanocyte growth media (Promocell), and dissociated for harvest using Detach Kit (Promocell). Cell lines were maintained in a 5% CO2 incubator at 37°C and were routinely tested for Mycoplasma contamination. Low-passage melanoma STCs, derived in the Osman laboratory as describe32, were grown in Dulbecco’s Modified Eagle Medium (DMEM) with 10% FBS, 1 mmol/L sodium pyruvate, 4 mmol/L L-glutamine, 25 mmol/L D-glucose, 1% nonessential amino acids (NEAA).
Clinical Specimens
Human melanoma specimens (nevi, primary, metastatic) were collected at the time of surgery (Table S2). Approval to collect specimens was granted by New York University Institutional Review Board protocol number i10362, “Development of an NYU Interdisciplinary Melanoma Cooperative Group: A clinicopathological database.” Informed consent was obtained from all subjects included.
Mice
NOD/SCID/IL2γR−/− female mice (Jackson labs, Cat# 05557) were used for in vivo studies. Experiments were conducted following protocols approved by the NYU Institutional Animal Care Use Committee (IACUC) (protocol number S16–00051).
Generation of custom shRNA library of glycosyltransferases and in vivo shRNA mediated growth screen.
To identify glycosyltransferases whose silencing can confer growth inhibition in melanoma, we built a custom shRNA library targeting 199 glycosyltransferases (5–6 shRNAs, a total of 1169 shRNAs) using miR30-adapted sequences (5–6 shRNAs per target) by PCR-cloning a pool of oligonucleotides synthesized on 55k customized arrays (Agilent Technologies) using a well-established system84–87. The library was sub-cloned in TRMPV-Neo using 12 independent sub-pools, each consisting of 100 shRNAs, to ensure that shRNA representation was not lost after grafting the tumor cells in vivo. Sequence verification was performed by random screening and Sanger sequencing of individual clones from each subpooled library, as well as Illumina sequencing of the final library. We checked that library purity (percentage of non-mutated shRNAs) is high, around 90%. We found that all the shRNAs are represented in the corresponding libraries, and no cross-contamination was found.
In Vivo shRNA Mediated Screen and HiSeq
Each pool of the library was transduced into MeWo melanoma cells stably expressing rtTA (Addgene #26429) using conditions that predominantly lead to a single retroviral integration and represent each shRNA in a calculated number of 9,600 cells (a total of 1.2 million cells at infection, 8% transduction efficiency, in technical triplicates). For viral library production, GP2 cells were plated on 100 mm tissue culture plates precoated with collagen at about 50% confluency 24 hours before transfection. Transfection was performed with a total of 12 μg of library and 8 μg of VSV-G using Lipofectamine 2000 (Invitrogen) overnight, and media was exchanged for DMEM with 10% Tet-free FBS (Clontech). Supernatants were collected and 0.45 μm filtered at 48 and 72 hours post-transfection and immediately used for infection of MeWo-rtTA target cells in the presence of 8 μg/ml of polybrene (Fisher). Transduced cells were selected for ten days using 600 μg/ml G418 (Invitrogen); at each passage, more than ten million cells were maintained to preserve library representation throughout the experiment. Before induction, time point zero (T0) samples were obtained (one million cells per replicate), and cells were subsequently cultured with 600 μg/ml G418 and 2 μg/ml doxycycline to induce shRNA expression. We combined two sub-pools of transduced MeWo cells in equal numbers before injecting them subcutaneously. Briefly, cells were resuspended in sterile PBS at a concentration of 2 × 106 cells per 150 ml, aliquoted into Eppendorf tubes (150 ml) and maintained on ice until injection. Immediately before injection, cell aliquots were mixed with 150 ml Matrigel (Becton Dickinson). Cell/Matrigel (1:1) suspensions were injected subcutaneously in the right flank of NOD/Shi-scid/IL-2Rgamma null (NSG) 8-week-old female mice (n=10 for each group). Each sub-pool of cells was injected in mice in triplicates. Primary tumors were resected five weeks post-injection. Genomic DNA from 5 weeks post-injection tumors was isolated by two rounds of phenol extraction using PhaseLock tubes (5 prime) followed by isopropanol precipitation. Deep-sequencing template libraries were generated by PCR amplification of shRNA guide strands86. Libraries were analyzed on an Illumina HiSeq at a final concentration of 10 pM; 18 nucleotides of the guide strand were sequenced using a custom primer (miR30 EcoRISeq, TAGCCCCTTGAATTCCGAGGCAGTAGGCA). Sequence processing was performed using a customized Galaxy platform88. For each shRNA and timepoint, the number of matching reads was normalized to the total number of library-specific reads per lane. For scoring of enrichment/depletion of each shRNA, an average value for each timepoint divided by the respective input was averaged, and log10 fold change was calculated. We applied various cutoffs in a sequential manner to identify shRNAs depleted or enriched, as shown in fig S1A.
Lectin Microarray Printing, Hybridization and Analysis
Lectins were purchased from E.Y. Laboratories or Vector Laboratories with the following exceptions: recombinant CVN, SVN and Griffithsin were gifts from B. O’Keefe (NCI, Frederick, MD); TJA-I and TJA-II were from NorthStar Bioproducts. Printing, hybridization, and data analysis were performed as previously described89. Our data were normally distributed as determined by the Lilliefors test in MATLAB. Lectins were excluded from the analysis if they did not meet our minimum threshold for activity.
FFPE Nevi, Primary, and Metastatic Melanoma Sample Deparaffinization, Protein Extraction and Labeling
Formalin-fixed paraffin-embedded (FFPE) tissues from benign nevi (n= 10) and patients with primary melanoma (n = 18), LN metastasis (n = 22), subcutaneous metastasis (n = 21) brain metastasis (n = 18) were analyzed on our lectin microarrays. In brief, melanoma samples were fixed in 10% neutral buffered formalin. To analyze tumor samples, hematoxylin and eosin-stained slides were reviewed to ensure enough viable tumors. Unstained cut sections mounted on slides were macro dissected to remove contaminating normal cells, and two 20 μm sections of each FFPE tissue were scraped into a 1.5-ml microcentrifuge tube. One mL xylene and 200 ml ethanol were added to a microcentrifuge tube, incubated for 15 min at room temperature, centrifuged at 10,000 × g for 2 min, and the supernatant was removed. The deparaffinized tissue pellets were then rehydrated with a graded series of ethanol. One mL of 95% ethanol was added to each tissue pellet, incubated for 10 min at room temperature, and centrifuged at 10,000 × g for 2 min. This was followed by rehydration with 70% ethanol, spun down to remove excess supernatant, and allowed samples to dry at room temperature. The rehydrated tumor tissue and cell sections were re-suspended in 200 ml 10 mM sodium citrate buffer (pH 6.0) and incubated at 95° C for 60 min. The supernatant was removed, and the tissue was washed with PBS. Pellet was solubilized with 250 ml (100 ml for smaller pellets) Cy buffer (0.1 M NaHCO3, pH=9.3) containing 0.5% NP40 (Nonidet P-40). The sample was sonicated gently (70% power, total time 1 min [10 s on, 10 s off]) and incubated on ice for 60 min. The sample was centrifuged at 12,000 × g for 5 min at 4° C. Supernatant was collected for analysis. Total protein concentration was quantified by BCA assay. Samples were labeled with NHS-Cy3 or -Cy5 and analyzed on our lectin microarrays in dual color using standard protocols89. The reference sample was a mixture of 3 nevi, 6 primary, and 6 metastatic samples. Normalized data were subjected to hierarchical clustering using the Pearson correlation coefficient (R) as the distance metric and average linkage analysis to generate heat maps.
RNA Extraction and Real-Time qPCR
Total RNA was extracted from samples (miRNeasy mini kit; Qiagen), quantified using Nanodrop 8000, and stored at −80° C. 500 ng of RNA were reverse transcribed using TaqMan RT reagents (Applied Biosystems) with random hexamers following manufacturer’s recommendations. Transcripts were quantified by real-time qPCR using Power SYBR Green PCR MasterMix (Applied Biosystems) in StepOne System (ABI). Primers were designed by using PrimerSelect. Some qPCR primers were purchased from Origene (Table S3). Cycle threshold values were normalized to those of the housekeeping genes GAPDH. The average for three biological replicates was plotted as relative transcript abundance.
Plasmids
pLKO.1 plasmids targeting human ST3GAL1, ST3GAL2, CD98hc and a non-targeting control were purchased from Sigma. DOX inducible PLKO plasmids were subcloned using shRNA and scramble sequences, as shown in table S3. CD98hc overexpression plasmid pCMV6-Myc-DDK was purchased from Origene.
Viral Production
4×106 HEK293T cells were seeded per 10 cm tissue culture dish and incubated overnight at 37°C and 5% CO2 for 16–20 hr. After seeding, HEK293T was co-transfected with lentiviral expression constructs (12 μg), viral packaging plasmid (psPAX2, 8 μg), and viral envelope plasmid (pMD2.G, 4 μg) using Lipofectamine 2000 (Invitrogen) following manufacturer’s recommendations. Viral supernatant was collected and 0.45 μm filtered at 72 hr post-transfection and stored at 4° C for short-term use (1–5 days) or −20 C for long-term storage (5–30 days).
Viral Transduction
Target cells were seeded and incubated overnight prior to infection. The medium was replaced with 1:2 diluted viral supernatant supplemented with eight μg/mL polybrene and incubated for 6–8 hr, followed by replacement with growth medium. Control and shRNA cells were selected using puromycin (2 μg/mL) or neomycin (500 μg/mL) prior to use in experiments.
In Vitro Proliferation Assay
Cells were seeded at 5000 cells/well in 96-well plates. Cell proliferation was measured using CellTiter-Glo 2.0 Luminescent Cell Viability Assay (Promega) following the manufacturer’s instructions.
Caspase activity assay
Cells were treated with or without a pan-caspase inhibitor (10 nM, QVD) 24 hr post-lentiviral infection. Cells were then rinsed 1x with PBS and resuspended in 1X lysis buffer (20 mM HEPES/NaOH, pH 7.2, 10% sucrose, 150 mM NaCl, ten mM DTT, five mM EDTA, 1% Igepal CA-630, 0.1% CHAPS, and 1X EDTA-free Complete protease inhibitor mixture (Roche)). Lysates were cleared by centrifugation at 16,000xg for 5 min, and supernatants were quantified by the Lowry method (Bio-Rad). Assays were performed in triplicate using 25 μg of protein in lysis buffer supplemented with 25 μM of the fluorogenic substrate Ac-DEVD-afc. Plates were read in a fluorimeter using a 400 nm excitation filter and a 505 nm emission filter.
Immunohistochemistry (IHC)
Unconjugated anti-human ST3GAL1 and ST3GAL2, raised against human ST3GAL1 (Sigma HPA040466) and ST3GAL2 (Abcam ab96028), were used for IHC. Antibody optimization was performed on formalin-fixed, paraffin-embedded 4-micron composite tissue microarrays (TissueArray.Com) containing normal and tumor: skin, stomach, prostate, liver, and pancreas. IHC parameters were validated on formalin-fixed, paraffinembedded 4-micron human hippocampus with cells clearly expressing cytoplasmic-membrane staining considered positive. Chromogenic IHC was performed on a Ventana Medical Systems Discovery XT instrument with online deparaffinization using Ventana’s reagents and detection kits unless otherwise noted (Ventana Medical Systems Tucson, AZ USA). AMIGO2 was antigen-retrieved in Ventana Cell Conditioner 1 (Tris-BorateEDTA) for 20 min. Antibody was diluted 1:50 in PBS and incubated for 6 hr at room temperature. Primary antibody was detected with goat anti-mouse, horseradish peroxidase-conjugated multimer incubated for 8 min. The complex was visualized with alpha-naphthol pyronin incubated for 8 min. Slides were washed in distilled water, counterstained with Hematoxylin, dehydrated, and mounted with permanent media. Negative controls were incubated with diluent instead of primary antibody. The staining score was performed in a blinded manner. The IHC score was calculated using staining intensity (0, 1, and 2) × percentage of positive cells.
Lectin Fluorescence Microscopy
Lectin fluorescence was performed on 4 mm formalin-fixed, paraffin-embedded tissue sections using diCBM40-Cy5 lectin. Both lectins and secondary antibodies were incubated for 1 hr each. DAPI staining was used to determine the nuclei morphology. Fluorescent slides were stored at −20° C until imaging. Stained slides were imaged by Hamamatsu fluorescent slide scanner, and images were extracted using NDPIS software. Lectin fluorescence microscopy for cultured cells was performed as previously described90.
Western Blotting
Cells were lysed in cold RIPA buffer supplemented with protease inhibitors. Equal amounts of protein were resolved by 4–12% Bis-Tris/PAGE, transferred onto PVDF membranes, and blocked in buffer [5% (wt/vol) milk, TBST (TBS, pH 7.4, 0.05% Tween-20), 1 hr, room temperature]. Primary antibodies were diluted in blocking buffer, as follows: ST3GAL1 (1:1000, Sigma HPA040466), ST3GAL2 (1:1000, ab96028), CD98hc (1:5000, Abcam ab244356), TFR1 1:5000, Abcam ab269513), PAPR (1:4000, Cell signaling, 9542), Caspase-3 Antibody (1:4000, Cell signaling, #9662), Cleaved-caspase3 (1:2000, Cell signaling, #9661), (GAPDH (1:5,000; Abcam, ab8245); α-Tubulin (1:5000; Sigma, T9026). Secondary antibodies were a-mouse or α-rabbit-HRP (1:5,000; BioRad). Blots were developed using Clarity Western ECL Blotting Substrate (BioRad) and imaged in the LICOR Odyssey Fc imaging system.
MAA Chromatography
For lectin enrichment assays, cells were lysed with lysis buffer containing 1% Nonidet P-40. 1000 mg of lysate was mixed with 100 μl of biotinylated MAA lectin, and a volume made up to 1000 ml with PBS containing 1mM MnCl2, 1mM MgCl2, and 1mM CaCl2 and incubated with rotation at 4° C overnight. Then, 60 μl of a 1:1 suspension of agarose coupled streptavidin was added, and incubation was continued for 4 hr. The beads were washed five times with PBST (Tween, 0.05%) buffer and subsequently extracted with SDS-PAGE sample buffer at 95° C for 5 min. The samples were separated by 4–12% Bis-Tris/PAGE and subjected to immunoblotting with α-CD98hc and α-TFR1 antibodies.
GO Analysis
GeneOntology Enrichment Pathway analysis was performed using DAVID to determine biological process categories enriched in MAA-enriched alpha 2,3 sialylated glycoproteins. The input gene lists were generated from overlapping α−2,3 sialylated proteins in 3 melanoma cell types consisting of 44 proteins.
Data mining of human transcriptomics datasets
The log fold changes in gene expression between melanoma or nevi were calculated in the following three Affymetrix transcriptomic datasets: Talantov et al.81 (GSE3189; 18 nevi and 45 melanomas), Kabbarah et al.82 (GSE46517; 9 nevi and 104 melanomas) and Scatolini et al.83 (GSE12391; 18 nevi and 28 melanomas). Statistical comparisons were made by Student’s t-testing. Significant differences in gene expression were considered when p < 0.05.
Supplementary Material
Acknowledgments:
We thank the NYU School of Medicine Histopathology and IHC Core Laboratories for tissue processing and histology and the NYULMC Proteomics Resource Center for mass spectrometry data. We thank Garrett Cooper (Interdisciplinary Melanoma Cooperative Group, NYU Langone Health) for providing clinical data for melanoma cases. This work was funded by the NIH/NCI R01 CA202027, U54 CA263001. P.A. has been supported by a career development grant Harry J Lloyd charitable trust.
References:
- 1.Damsky WE, Rosenbaum LE, Bosenberg M. Decoding melanoma metastasis. Cancers (Basel). 2010;3(1):126–63. Epub 2010/01/01. doi: 10.3390/cancers3010126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shain AH, Bastian BC. From melanocytes to melanomas. Nat Rev Cancer. 2016;16(6):345–58. Epub 20160429. doi: 10.1038/nrc.2016.37. [DOI] [PubMed] [Google Scholar]
- 3.Damsky WE, Bosenberg M. Melanocytic nevi and melanoma: unraveling a complex relationship. Oncogene. 2017;36(42):5771–92. Epub 20170612. doi: 10.1038/onc.2017.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Turner N, Ware O, Bosenberg M. Genetics of metastasis: melanoma and other cancers. Clin Exp Metastasis. 2018;35(5–6):379–91. Epub 2018/05/04. doi: 10.1007/s10585-018-9893-y. [DOI] [PubMed] [Google Scholar]
- 5.Schadendorf D, van Akkooi ACJ, Berking C, Griewank KG, Gutzmer R, Hauschild A, Stang A, Roesch A, Ugurel S. Melanoma. The Lancet. 2018;392(10151):971–84. doi: 10.1016/s0140-6736(18)31559-9. [DOI] [PubMed] [Google Scholar]
- 6.Elder DE. Melanoma progression. Pathology. 2016;48(2):147–54. Epub 2016/03/30. doi: 10.1016/j.pathol.2015.12.002. [DOI] [PubMed] [Google Scholar]
- 7.Peixoto A, Relvas-Santos M, Azevedo R, Santos LL, Ferreira JA. Protein Glycosylation and Tumor Microenvironment Alterations Driving Cancer Hallmarks. Front Oncol. 2019;9:380. Epub 2019/06/04. doi: 10.3389/fonc.2019.00380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rodrigues JG, Balmana M, Macedo JA, Pocas J, Fernandes A, de-Freitas-Junior JCM, Pinho SS, Gomes J, Magalhaes A, Gomes C, Mereiter S, Reis CA. Glycosylation in cancer: Selected roles in tumour progression, immune modulation and metastasis. Cell Immunol. 2018;333:46–57. Epub 2018/03/27. doi: 10.1016/j.cellimm.2018.03.007. [DOI] [PubMed] [Google Scholar]
- 9.Munkley J, Elliott DJ. Hallmarks of glycosylation in cancer. Oncotarget. 2016;7(23):35478–89. Epub 2016/03/24. doi: 10.18632/oncotarget.8155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pinho SS, Reis CA. Glycosylation in cancer: mechanisms and clinical implications. Nat Rev Cancer. 2015;15(9):540–55. Epub 20150820. doi: 10.1038/nrc3982. [DOI] [PubMed] [Google Scholar]
- 11.Pochec E, Zabczynska M, Bubka M, Homa J, Litynska A. beta1,6-branched complex-type N-glycans affect FAK signaling in metastatic melanoma cells. Cancer Invest. 2016;34(1):45–56. Epub 2016/01/09. doi: 10.3109/07357907.2015.1102928. [DOI] [PubMed] [Google Scholar]
- 12.De Vellis C, Pietrobono S, Stecca B. The Role of Glycosylation in Melanoma Progression. Cells. 2021;10(8). Epub 2021/08/28. doi: 10.3390/cells10082136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sobiepanek A, Paone A, Cutruzzola F, Kobiela T. Biophysical characterization of melanoma cell phenotype markers during metastatic progression. Eur Biophys J. 2021;50(3–4):523–42. Epub 2021/03/18. doi: 10.1007/s00249-021-01514-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tang L, Chen X, Zhang X, Guo Y, Su J, Zhang J, Peng C, Chen X. N-Glycosylation in progression of skin cancer. Med Oncol. 2019;36(6):50. Epub 2019/05/01. doi: 10.1007/s12032-019-1270-4. [DOI] [PubMed] [Google Scholar]
- 15.Agrawal P, Fontanals-Cirera B, Sokolova E, Jacob S, Vaiana CA, Argibay D, Davalos V, McDermott M, Nayak S, Darvishian F, Castillo M, Ueberheide B, Osman I, Fenyo D, Mahal LK, Hernando E. A Systems Biology Approach Identifies FUT8 as a Driver of Melanoma Metastasis. Cancer Cell. 2017;31(6):804–19 e7. Epub 2017/06/14. doi: 10.1016/j.ccell.2017.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zuber J, McJunkin K, Fellmann C, Dow LE, Taylor MJ, Hannon GJ, Lowe SW. Toolkit for evaluating genes required for proliferation and survival using tetracycline-regulated RNAi. Nat Biotechnol. 2011;29(1):79–83. Epub 20101205. doi: 10.1038/nbt.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Joshi HJ, Hansen L, Narimatsu Y, Freeze HH, Henrissat B, Bennett E, Wandall HH, Clausen H, Schjoldager KT. Glycosyltransferase genes that cause monogenic congenital disorders of glycosylation are distinct from glycosyltransferase genes associated with complex diseases. Glycobiology. 2018;28(5):284–94. doi: 10.1093/glycob/cwy015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang L, Chen S, Zhang Z, Zhang J, Mao S, Zheng J, Xuan Y, Liu M, Cai K, Zhang W, Guo Y, Zhai W, Yao X. Suppressed OGT expression inhibits cell proliferation while inducing cell apoptosis in bladder cancer. BMC Cancer. 2018;18(1):1141. Epub 20181120. doi: 10.1186/s12885-018-5033-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li H, Al-Japairai K, Tao Y, Xiang Z. RPN2 promotes colorectal cancer cell proliferation through modulating the glycosylation status of EGFR. Oncotarget. 2017;8(42):72633–51. Epub 20170807. doi: 10.18632/oncotarget.20005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tominaga N, Hagiwara K, Kosaka N, Honma K, Nakagama H, Ochiya T. RPN2-mediated glycosylation of tetraspanin CD63 regulates breast cancer cell malignancy. Mol Cancer. 2014;13:134. Epub 20140531. doi: 10.1186/1476-4598-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elder DE. Precursors to melanoma and their mimics: nevi of special sites. Mod Pathol. 2006;19 Suppl 2:S4–20. doi: 10.1038/modpathol.3800515. [DOI] [PubMed] [Google Scholar]
- 22.Pilobello KT, Krishnamoorthy L, Slawek D, Mahal LK. Development of a lectin microarray for the rapid analysis of protein glycopatterns. Chembiochem. 2005;6(6):985–9. Epub 2005/03/31. doi: 10.1002/cbic.200400403. [DOI] [PubMed] [Google Scholar]
- 23.Bojar D, Meche L, Meng G, Eng W, Smith DF, Cummings RD, Mahal LK. A Useful Guide to Lectin Binding: Machine-Learning Directed Annotation of 57 Unique Lectin Specificities. bioRxiv. 2021. doi: 10.1101/2021.08.31.458439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bojar D, Meche L, Meng G, Eng W, Smith DF, Cummings RD, Mahal LK. A Useful Guide to Lectin Binding: Machine-Learning Directed Annotation of 57 Unique Lectin Specificities. ACS Chem Biol. 2022;17(11):2993–3012. Epub 20220127. doi: 10.1021/acschembio.1c00689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ribeiro JP, Pau W, Pifferi C, Renaudet O, Varrot A, Mahal LK, Imberty A. Characterization of a high-affinity sialic acid-specific CBM40 from Clostridium perfringens and engineering of a divalent form. Biochem J. 2016;473(14):2109–18. Epub 20160517. doi: 10.1042/BCJ20160340. [DOI] [PubMed] [Google Scholar]
- 26.Bensing BA, Li Q, Park D, Lebrilla CB, Sullam PM. Streptococcal Siglec-like adhesins recognize different subsets of human plasma glycoproteins: implications for infective endocarditis. Glycobiology. 2018;28(8):601–11. doi: 10.1093/glycob/cwy052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harduin-Lepers A, Vallejo-Ruiz V, Krzewinski-Recchi MA, Samyn-Petit B, Julien S, Delannoy P. The human sialyltransferase family. Biochimie. 2001;83(8):727–37. [DOI] [PubMed] [Google Scholar]
- 28.Priatel JJ, Chui D, Hiraoka N, Simmons CJ, Richardson KB, Page DM, Fukuda M, Varki NM, Marth JD. The ST3Gal-I sialyltransferase controls CD8+ T lymphocyte homeostasis by modulating O-glycan biosynthesis. Immunity. 2000;12(3):273–83. [DOI] [PubMed] [Google Scholar]
- 29.Chandrasekaran EV, Xue J, Xia J, Locke RD, Patil SA, Neelamegham S, Matta KL. Mammalian sialyltransferase ST3Gal-II: its exchange sialylation catalytic properties allow labeling of sialyl residues in mucintype sialylated glycoproteins and specific gangliosides. Biochemistry. 2011;50(44):9475–87. doi: 10.1021/bi200301w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sturgill ER, Aoki K, Lopez PH, Colacurcio D, Vajn K, Lorenzini I, Majic S, Yang WH, Heffer M, Tiemeyer M, Marth JD, Schnaar RL. Biosynthesis of the major brain gangliosides GD1a and GT1b. Glycobiology. 2012;22(10):1289–301. doi: 10.1093/glycob/cws103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cruz-Munoz W, Man S, Xu P, Kerbel RS. Development of a preclinical model of spontaneous human melanoma central nervous system metastasis. Cancer research. 2008;68(12):4500–5. doi: 10.1158/0008-5472.CAN-08-0041. [DOI] [PubMed] [Google Scholar]
- 32.Kleffman K, Levinson G, Rose IVL, Blumenberg LM, Shadaloey SAA, Dhabaria A, Wong E, Galan-Echevarria F, Karz A, Argibay D, Von Itter R, Floristan A, Baptiste G, Eskow NM, Tranos JA, Chen J, Vega YSdMEC, Call M, Rogers R, Jour G, Wadghiri YZ, Osman I, Li YM, Mathews P, DeMattos RB, Ueberheide B, Ruggles KV, Liddelow SA, Schneider RJ, Hernando E. Melanoma-Secreted Amyloid Beta Suppresses Neuroinflammation and Promotes Brain Metastasis. Cancer Discov. 2022;12(5):1314–35. Epub 2022/03/10. doi: 10.1158/2159-8290.CD-21-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Orr SL, Le D, Long JM, Sobieszczuk P, Ma B, Tian H, Fang X, Paulson JC, Marth JD, Varki N. A phenotype survey of 36 mutant mouse strains with gene-targeted defects in glycosyltransferases or glycan-binding proteins. Glycobiology. 2013;23(3):363–80. doi: 10.1093/glycob/cws150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lopez PH, Aja S, Aoki K, Seldin MM, Lei X, Ronnett GV, Wong GW, Schnaar RL. Mice lacking sialyltransferase ST3Gal-II develop late-onset obesity and insulin resistance. Glycobiology. 2017;27(2):129–39. doi: 10.1093/glycob/cww098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, Smith I, Tothova Z, Wilen C, Orchard R, Virgin HW, Listgarten J, Root DE. Optimized sgRNA design to maximize activity and minimize offtarget effects of CRISPR-Cas9. Nat Biotechnol. 2016;34(2):184–91. Epub 20160118. doi: 10.1038/nbt.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meyers RM, Bryan JG, McFarland JM, Weir BA, Sizemore AE, Xu H, Dharia NV, Montgomery PG, Cowley GS, Pantel S, Goodale A, Lee Y, Ali LD, Jiang G, Lubonja R, Harrington WF, Strickland M, Wu T, Hawes DC, Zhivich VA, Wyatt MR, Kalani Z, Chang JJ, Okamoto M, Stegmaier K, Golub TR, Boehm JS, Vazquez F, Root DE, Hahn WC, Tsherniak A. Computational correction of copy number effect improves specificity of CRISPR-Cas9 essentiality screens in cancer cells. Nat Genet. 2017;49(12):1779–84. Epub 20171030. doi: 10.1038/ng.3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsherniak A, Vazquez F, Montgomery PG, Weir BA, Kryukov G, Cowley GS, Gill S, Harrington WF, Pantel S, Krill-Burger JM, Meyers RM, Ali L, Goodale A, Lee Y, Jiang G, Hsiao J, Gerath WFJ, Howell S, Merkel E, Ghandi M, Garraway LA, Root DE, Golub TR, Boehm JS, Hahn WC. Defining a Cancer Dependency Map. Cell. 2017;170(3):564–76 e16. doi: 10.1016/j.cell.2017.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cantor JM, Ginsberg MH. CD98 at the crossroads of adaptive immunity and cancer. J Cell Sci. 2012;125(Pt 6):1373–82. Epub 2012/04/14. doi: 10.1242/jcs.096040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deuschle FC, Schiefner A, Skerra A. Structural differences between the ectodomains of murine and human CD98hc. Proteins. 2019;87(8):693–8. Epub 2019/04/09. doi: 10.1002/prot.25686. [DOI] [PubMed] [Google Scholar]
- 40.Scalise M, Galluccio M, Console L, Pochini L, Indiveri C. The Human SLC7A5 (LAT1): The Intriguing Histidine/Large Neutral Amino Acid Transporter and Its Relevance to Human Health. Front Chem. 2018;6:243. Epub 2018/07/11. doi: 10.3389/fchem.2018.00243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Feral CC, Nishiya N, Fenczik CA, Stuhlmann H, Slepak M, Ginsberg MH. CD98hc (SLC3A2) mediates integrin signaling. Proc Natl Acad Sci U S A. 2005;102(2):355–60. Epub 20041229. doi: 10.1073/pnas.0404852102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yanagida O, Kanai Y, Chairoungdua A, Kim DK, Segawa H, Nii T, Cha SH, Matsuo H, Fukushima J-i, Fukasawa Y, Tani Y, Taketani Y, Uchino H, Kim JY, Inatomi J, Okayasu I, Miyamoto K-i, Takeda E, Goya T, Endou H. Human L-type amino acid transporter 1 (LAT1): characterization of function and expression in tumor cell lines. Biochimica et Biophysica Acta (BBA) - Biomembranes. 2001;1514(2):291–302. doi: 10.1016/s0005-2736(01)00384-4. [DOI] [PubMed] [Google Scholar]
- 43.El Ansari R, Craze ML, Diez-Rodriguez M, Nolan CC, Ellis IO, Rakha EA, Green AR. The multifunctional solute carrier 3A2 (SLC3A2) confers a poor prognosis in the highly proliferative breast cancer subtypes. Br J Cancer. 2018;118(8):1115–22. Epub 2018/03/17. doi: 10.1038/s41416-018-0038-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kaira K, Oriuchi N, Imai H, Shimizu K, Yanagitani N, Sunaga N, Hisada T, Kawashima O, Kamide Y, Ishizuka T, Kanai Y, Nakajima T, Mori M. CD98 expression is associated with poor prognosis in resected non-small-cell lung cancer with lymph node metastases. Ann Surg Oncol. 2009;16(12):3473–81. Epub 2009/09/25. doi: 10.1245/s10434-009-0685-0. [DOI] [PubMed] [Google Scholar]
- 45.Toyoda M, Kaira K, Shino M, Sakakura K, Takahashi K, Takayasu Y, Tominaga H, Oriuchi N, Nikkuni O, Suzuki M, Iijima M, Tsukamoto N, Nagamori S, Kanai Y, Oyama T, Chikamatsu K. CD98 as a novel prognostic indicator for patients with stage III/IV hypopharyngeal squamous cell carcinoma. Head Neck. 2015;37(11):1569–74. Epub 2014/06/11. doi: 10.1002/hed.23797. [DOI] [PubMed] [Google Scholar]
- 46.Alfarsi LH, El-Ansari R, Craze ML, Masisi BK, Mohammed OJ, Ellis IO, Rakha EA, Green AR. Co-Expression Effect of SLC7A5/SLC3A2 to Predict Response to Endocrine Therapy in Oestrogen-Receptor-Positive Breast Cancer. Int J Mol Sci. 2020;21(4). Epub 2020/02/26. doi: 10.3390/ijms21041407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Theodosakis N, Micevic G, Sharma R, Baras AS, Lazova R, Bosenberg MW, Rodic N. Integrative discovery of CD98 as a melanoma biomarker. Pigment Cell Melanoma Res. 2016;29(3):385–7. Epub 2016/02/07. doi: 10.1111/pcmr.12464. [DOI] [PubMed] [Google Scholar]
- 48.Harduin-Lepers A, Vallejo-Ruiz V, Krzewinski-Recchi M-A, Samyn-Petit B, Julien S, Delannoy P. The human sialyltransferase family. Biochimie. 2001;83(8):727–37. doi: 10.1016/s0300-9084(01)01301-3. [DOI] [PubMed] [Google Scholar]
- 49.Dobie C, Skropeta D. Insights into the role of sialylation in cancer progression and metastasis. Br J Cancer. 2021;124(1):76–90. Epub 20201104. doi: 10.1038/s41416-020-01126-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rodrigues E, Macauley MS. Hypersialylation in Cancer: Modulation of Inflammation and Therapeutic Opportunities. Cancers (Basel). 2018;10(6). Epub 20180618. doi: 10.3390/cancers10060207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Krishnan V, Bane SM, Kawle PD, Naresh KN, Kalraiya RD. Altered melanoma cell surface glycosylation mediates organ specific adhesion and metastasis via lectin receptors on the lung vascular endothelium. Clin Exp Metastasis. 2005;22(1):11–24. Epub 2005/09/01. doi: 10.1007/s10585-005-2036-2. [DOI] [PubMed] [Google Scholar]
- 52.Laidler P, Litynska A, Hoja-Lukowicz D, Labedz M, Przybylo M, Ciolczyk-Wierzbicka D, Pochec E, Trebacz E, Kremser E. Characterization of glycosylation and adherent properties of melanoma cell lines. Cancer Immunol Immunother. 2006;55(1):112–8. Epub 2005/08/03. doi: 10.1007/s00262-005-0019-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kinoshita M, Mitsui Y, Kakoi N, Yamada K, Hayakawa T, Kakehi K. Common glycoproteins expressing polylactosamine-type glycans on matched patient primary and metastatic melanoma cells show different glycan profiles. J Proteome Res. 2014;13(2):1021–33. Epub 2013/12/21. doi: 10.1021/pr401015b. [DOI] [PubMed] [Google Scholar]
- 54.Wu X, Zhao J, Ruan Y, Sun L, Xu C, Jiang H. Sialyltransferase ST3GAL1 promotes cell migration, invasion, and TGF-beta1-induced EMT and confers paclitaxel resistance in ovarian cancer. Cell Death Dis. 2018;9(11):1102. doi: 10.1038/s41419-018-1101-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Burchell J, Poulsom R, Hanby A, Whitehouse C, Cooper L, Clausen H, Miles D, Taylor-Papadimitriou J. An alpha2,3 sialyltransferase (ST3Gal I) is elevated in primary breast carcinomas. Glycobiology. 1999;9(12):1307–11. [DOI] [PubMed] [Google Scholar]
- 56.Bai Q, Liu L, Xia Y, Long Q, Wang J, Xu J, Guo J. Prognostic significance of ST3GAL-1 expression in patients with clear cell renal cell carcinoma. BMC Cancer. 2015;15:880. doi: 10.1186/s12885-015-1906-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chong YK, Sandanaraj E, Koh LW, Thangaveloo M, Tan MS, Koh GR, Toh TB, Lim GG, Holbrook JD, Kon OL, Nadarajah M, Ng I, Ng WH, Tan NS, Lim KL, Tang C, Ang BT. ST3GAL1-Associated Transcriptomic Program in Glioblastoma Tumor Growth, Invasion, and Prognosis. J Natl Cancer Inst. 2016;108(2). doi: 10.1093/jnci/djv326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Picco G, Julien S, Brockhausen I, Beatson R, Antonopoulos A, Haslam S, Mandel U, Dell A, Pinder S, Taylor-Papadimitriou J, Burchell J. Over-expression of ST3Gal-I promotes mammary tumorigenesis. Glycobiology. 2010;20(10):1241–50. doi: 10.1093/glycob/cwq085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Aloia A, Petrova E, Tomiuk S, Bissels U, Deas O, Saini M, Zickgraf FM, Wagner S, Spaich S, Sutterlin M, Schneeweiss A, Reitberger M, Ruberg S, Gerstmayer B, Agorku D, Knobel S, Terranegra A, Falleni M, Soldati L, Sprick MR, Trumpp A, Judde JG, Bosio A, Cairo S, Hardt O. The sialyl-glycolipid stage-specific embryonic antigen 4 marks a subpopulation of chemotherapy-resistant breast cancer cells with mesenchymal features. Breast Cancer Res. 2015;17(1):146. doi: 10.1186/s13058-015-0652-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lopez A, Cacoub P, Macdougall IC, Peyrin-Biroulet L. Iron deficiency anaemia. Lancet. 2016;387(10021):907–16. Epub 20150824. doi: 10.1016/S0140-6736(15)60865-0. [DOI] [PubMed] [Google Scholar]
- 61.Horniblow RD, Bedford M, Hollingworth R, Evans S, Sutton E, Lal N, Beggs A, Iqbal TH, Tselepis C. BRAF mutations are associated with increased iron regulatory protein-2 expression in colorectal tumorigenesis. Cancer Sci. 2017;108(6):1135–43. Epub 20170602. doi: 10.1111/cas.13234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kindrat I, Tryndyak V, de Conti A, Shpyleva S, Mudalige TK, Kobets T, Erstenyuk AM, Beland FA, Pogribny IP. MicroRNA-152-mediated dysregulation of hepatic transferrin receptor 1 in liver carcinogenesis. Oncotarget. 2016;7(2):1276–87. doi: 10.18632/oncotarget.6004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rychtarcikova Z, Lettlova S, Tomkova V, Korenkova V, Langerova L, Simonova E, Zjablovskaja P, Alberich-Jorda M, Neuzil J, Truksa J. Tumor-initiating cells of breast and prostate origin show alterations in the expression of genes related to iron metabolism. Oncotarget. 2017;8(4):6376–98. doi: 10.18632/oncotarget.14093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang B, Zhang J, Song F, Tian M, Shi B, Jiang H, Xu W, Wang H, Zhou M, Pan X, Gu J, Yang S, Jiang L, Li Z. EGFR regulates iron homeostasis to promote cancer growth through redistribution of transferrin receptor 1. Cancer Lett. 2016;381(2):331–40. Epub 20160811. doi: 10.1016/j.canlet.2016.08.006. [DOI] [PubMed] [Google Scholar]
- 65.Ostendorf BN, Bilanovic J, Adaku N, Tafreshian KN, Tavora B, Vaughan RD, Tavazoie SF. Common germline variants of the human APOE gene modulate melanoma progression and survival. Nat Med. 2020;26(7):1048–53. Epub 20200525. doi: 10.1038/s41591-020-0879-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pencheva N, Tran H, Buss C, Huh D, Drobnjak M, Busam K, Tavazoie SF. Convergent multi-miRNA targeting of ApoE drives LRP1/LRP8-dependent melanoma metastasis and angiogenesis. Cell. 2012;151(5):1068–82. Epub 20121108. doi: 10.1016/j.cell.2012.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Peng X, Chen R, Cai S, Lu S, Zhang Y. SLC1A4: A Powerful Prognostic Marker and Promising Therapeutic Target for HCC. Front Oncol. 2021;11:650355. Epub 20210311. doi: 10.3389/fonc.2021.650355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.White MA, Lin C, Rajapakshe K, Dong J, Shi Y, Tsouko E, Mukhopadhyay R, Jasso D, Dawood W, Coarfa C, Frigo DE. Glutamine Transporters Are Targets of Multiple Oncogenic Signaling Pathways in Prostate Cancer. Mol Cancer Res. 2017;15(8):1017–28. Epub 20170515. doi: 10.1158/1541-7786.MCR-16-0480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tian S, Peng P, Li J, Deng H, Zhan N, Zeng Z, Dong W. SERPINH1 regulates EMT and gastric cancer metastasis via the Wnt/beta-catenin signaling pathway. Aging (Albany NY). 2020;12(4):3574–93. Epub 20200224. doi: 10.18632/aging.102831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Capone E, Iacobelli S, Sala G. Role of galectin 3 binding protein in cancer progression: a potential novel therapeutic target. J Transl Med. 2021;19(1):405. Epub 20210926. doi: 10.1186/s12967-021-03085-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kimura R, Yoshimaru T, Matsushita Y, Matsuo T, Ono M, Park JH, Sasa M, Miyoshi Y, Nakamura Y, Katagiri T. The GALNT6LGALS3BP axis promotes breast cancer cell growth. Int J Oncol. 2020;56(2):581–95. Epub 20191213. doi: 10.3892/ijo.2019.4941. [DOI] [PubMed] [Google Scholar]
- 72.Hu Y, Shah P, Clark DJ, Ao M, Zhang H. Reanalysis of Global Proteomic and Phosphoproteomic Data Identified a Large Number of Glycopeptides. Anal Chem. 2018;90(13):8065–71. Epub 20180611. doi: 10.1021/acs.analchem.8b01137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Deves R, Boyd CA. Surface antigen CD98(4F2): not a single membrane protein, but a family of proteins with multiple functions. J Membr Biol. 2000;173(3):165–77. doi: 10.1007/s002320001017. [DOI] [PubMed] [Google Scholar]
- 74.Yan Y, Vasudevan S, Nguyen HT, Merlin D. Intestinal epithelial CD98: an oligomeric and multifunctional protein. Biochim Biophys Acta. 2008;1780(10):1087–92. Epub 20080624. doi: 10.1016/j.bbagen.2008.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cano-Crespo S, Chillaron J, Junza A, Fernandez-Miranda G, Garcia J, Polte C, RdlB L, Ignatova Z, Yanes O, Zorzano A, Stephan-Otto Attolini C, Palacin M. CD98hc (SLC3A2) sustains amino acid and nucleotide availability for cell cycle progression. Sci Rep. 2019;9(1):14065. Epub 20191001. doi: 10.1038/s41598-019-50547-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bajaj J, Konuma T, Lytle NK, Kwon HY, Ablack JN, Cantor JM, Rizzieri D, Chuah C, Oehler VG, Broome EH, Ball ED, van der Horst EH, Ginsberg MH, Reya T. CD98-Mediated Adhesive Signaling Enables the Establishment and Propagation of Acute Myelogenous Leukemia. Cancer Cell. 2016;30(5):792–805. Epub 20161027. doi: 10.1016/j.ccell.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cai S, Bulus N, Fonseca-Siesser PM, Chen D, Hanks SK, Pozzi A, Zent R. CD98 modulates integrin beta1 function in polarized epithelial cells. J Cell Sci. 2005;118(Pt 5):889–99. Epub 20050215. doi: 10.1242/jcs.01674. [DOI] [PubMed] [Google Scholar]
- 78.Fenczik CA, Sethi T, Ramos JW, Hughes PE, Ginsberg MH. Complementation of dominant suppression implicates CD98 in integrin activation. Nature. 1997;390(6655):81–5. doi: 10.1038/36349. [DOI] [PubMed] [Google Scholar]
- 79.Rintoul RC, Buttery RC, Mackinnon AC, Wong WS, Mosher D, Haslett C, Sethi T. Cross-linking CD98 promotes integrin-like signaling and anchorage-independent growth. Mol Biol Cell. 2002;13(8):2841–52. doi: 10.1091/mbc.01-11-0530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Heider M, Eichner R, Stroh J, Morath V, Kuisl A, Zecha J, Lawatscheck J, Baek K, Garz AK, Rudelius M, Deuschle FC, Keller U, Lemeer S, Verbeek M, Gotze KS, Skerra A, Weber WA, Buchner J, Schulman BA, Kuster B, Fernandez-Saiz V, Bassermann F. The IMiD target CRBN determines HSP90 activity toward transmembrane proteins essential in multiple myeloma. Mol Cell. 2021;81(6):1170–86 e10. Epub 20210210. doi: 10.1016/j.molcel.2020.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Talantov D, Mazumder A, Yu JX, Briggs T, Jiang Y, Backus J, Atkins D, Wang Y. Novel genes associated with malignant melanoma but not benign melanocytic lesions. Clin Cancer Res. 2005;11(20):7234–42. doi: 10.1158/1078-0432.CCR-05-0683. [DOI] [PubMed] [Google Scholar]
- 82.Kabbarah O, Nogueira C, Feng B, Nazarian RM, Bosenberg M, Wu M, Scott KL, Kwong LN, Xiao Y, Cordon-Cardo C, Granter SR, Ramaswamy S, Golub T, Duncan LM, Wagner SN, Brennan C, Chin L. Integrative genome comparison of primary and metastatic melanomas. PLoS One. 2010;5(5):e10770. Epub 20100524. doi: 10.1371/journal.pone.0010770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Scatolini M, Grand MM, Grosso E, Venesio T, Pisacane A, Balsamo A, Sirovich R, Risio M, Chiorino G. Altered molecular pathways in melanocytic lesions. Int J Cancer. 2010;126(8):1869–81. doi: 10.1002/ijc.24899. [DOI] [PubMed] [Google Scholar]
- 84.Fellmann C, Hoffmann T, Sridhar V, Hopfgartner B, Muhar M, Roth M, Lai Dan Y, Barbosa Inês AM, Kwon Jung S, Guan Y, Sinha N, Zuber J. An Optimized microRNA Backbone for Effective Single-Copy RNAi. Cell Reports. 2013;5(6):1704–13. doi: 10.1016/j.celrep.2013.11.020. [DOI] [PubMed] [Google Scholar]
- 85.Fellmann C, Zuber J, McJunkin K, Chang K, Malone Colin D, Dickins Ross A, Xu Q, Hengartner Michael O, Elledge Stephen J, Hannon Gregory J, Lowe Scott W. Functional Identification of Optimized RNAi Triggers Using a Massively Parallel Sensor Assay. Molecular Cell. 2011;41(6):733–46. doi: 10.1016/j.molcel.2011.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zuber J, McJunkin K, Fellmann C, Dow LE, Taylor MJ, Hannon GJ, Lowe SW. Toolkit for evaluating genes required for proliferation and survival using tetracycline-regulated RNAi. Nat Biotech. 2011;29(1):79–83. doi: http://www.nature.com/nbt/journal/v29/n1/abs/nbt.1720.html#supplementary-information. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, Magoon D, Qi J, Blatt K, Wunderlich M, Taylor MJ, Johns C, Chicas A, Mulloy JC, Kogan SC, Brown P, Valent P, Bradner JE, Lowe SW, Vakoc CR. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478(7370):524–8. doi: http://www.nature.com/nature/journal/v478/n7370/abs/nature10334.html#supplementary-information. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hillman-Jackson J, Clements D, Blankenberg D, Taylor J, Nekrutenko A, Team G. Using Galaxy to Perform Large-Scale Interactive Data Analyses. Current Protocols in Bioinformatics. 2012;38(1):10.5.1–.5.47. doi: doi: 10.1002/0471250953.bi1005s38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pilobello KT, Agrawal P, Rouse R, Mahal LK. Advances in lectin microarray technology: optimized protocols for piezoelectric print conditions. Curr Protoc Chem Biol. 2013;5(1):1–23. doi: 10.1002/9780470559277.ch120035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Agrawal P, Kurcon T, Pilobello KT, Rakus JF, Koppolu S, Liu Z, Batista BS, Eng WS, Hsu KL, Liang Y, Mahal LK. Mapping posttranscriptional regulation of the human glycome uncovers microRNA defining the glycocode. Proc Natl Acad Sci U S A. 2014;111(11):4338–43. Epub 20140303. doi: 10.1073/pnas.1321524111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.