Abstract
Ubiquitination is one of the most common post-translational modifications in eukaryotic cells. Depending on the architecture of polyubiquitin chains, substrate proteins can meet different cellular fates, but our understanding of how chain linkage controls protein fate remains limited. UBL-UBA shuttle proteins, such as UBQLN2, bind to ubiquitinated proteins and to the proteasome or other protein quality control machinery elements and play a role in substrate fate determination. Under physiological conditions, UBQLN2 forms biomolecular condensates through phase separation, a physicochemical phenomenon in which multivalent interactions drive the formation of a macromolecule-rich dense phase. Ubiquitin and polyubiquitin chains modulate UBQLN2’s phase separation in a linkage-dependent manner, suggesting a possible link to substrate fate determination, but polyubiquitinated substrates have not been examined directly. Using sedimentation assays and microscopy we show that polyubiquitinated substrates induce UBQLN2 phase separation and incorporate into the resulting condensates. This substrate effect is strongest with K63-linked substrates, intermediate with mixed-linkage substrates, and weakest with K48-linked substrates. Proteasomes can be recruited to these condensates, but proteasome activity towards K63-linked and mixed linkage substrates is inhibited in condensates. Substrates are also protected from deubiquitinases by UBQLN2-induced phase separation. Our results imply that phase separation can act as a regulatory switch that controls the fate of ubiquitinated substrates in a chain-linkage dependent manner, thus serving as an interpreter of the ubiquitin code.
Keywords: Condensates, Deubiquitination, Phase Separation, Protein Degradation, Protein Quality Control, Proteasome, Ubiquitin
Classification: Biological Sciences: Biochemistry
Introduction
Eukaryotic proteins can be post-translationally modified in many ways, with ubiquitination one of the most common modifications (1). Ubiquitin (Ub) is typically attached to the amino group of a lysine on a substrate via an isopeptide bond, with additional Ubs attached to one or more lysines of the first Ub to form a polyubiquitin (polyUb) chain. The seven lysines and the N-terminal amine within Ub give rise to diverse polyUb chain architectures, which can lead to different fates within the cell (2–4). For example, K48-linked chains and some mixed linkage chains (K11/K48 and K48/K63) target proteins to the proteasome for degradation, whereas K63-linked chains are involved in DNA damage repair, and M1-linked chains are involved in immune response. These different polyUb chains make up the Ub code. However, our understanding of how these different chain types signal for different protein fates remains limited, especially since chain types with different in vivo fates (i.e. K48 and K63) can both be used to signal proteasomal degradation in vitro (5).
An additional layer of regulatory complexity comes from a family of UBL-UBA shuttle proteins, which bind to the proteasome or other protein quality control machinery elements (via the UBL domain) and to ubiquitinated proteins (via one or more UBA domains) (6). In some cases, shuttle proteins facilitate protein degradation, but in other cases, they seem to be protective and actually prevent degradation (7–11).
We and others have recently shown that some of these shuttle proteins such as Rad23B, UBQLN2, and its homologs UBQLN1, UBQLN4 and yeast Dsk2 can undergo phase separation, a phenomenon in which proteins self-assemble via multiple interactions to form biomolecular condensates (membrane-free compartments) in the cell or liquid droplets in vitro (12–17). UBQLN2 forms stress-induced condensates in cells and can be recruited to stress granules, a type of stress-induced biomolecular condensate hypothesized to form via phase separation of RNA-binding proteins and RNA (14, 15). UBQLN2 mutations linked to frontotemporal dementia and amyotrophic lateral sclerosis affect phase separation behavior of UBQLN2 both in cells and in vitro (18–21). Importantly, Ub also affects UBQLN2’s propensity to phase separate, where monoUb drives disassembly of UBQLN2 droplets in vitro in a concentration-dependent manner (15). Recent experiments with multiple polyUb linkage types revealed that length of the chain, linkage type, Ub-Ub spacing, and concentration are all important variables that modulate UBQLN2 phase separation (22, 23). Generally, K48-linked polyUb chains inhibit phase separation while K63-linked polyUb chains promote phase separation. In these situations, the valency of the polyUb chain and the arrangement of the Ub units can enable it to act as a scaffold and bring multiple shuttle proteins together to promote phase separation.
Although experiments with unanchored polyUb chains have revealed some of the rules that control phase separation of UBQLN2, we do not yet know how polyubiquitinated substrates behave in the presence of UBQLN2, or how substrate localization to phase separated condensates affects substrate fate (24). Using model substrates in an in vitro reconstituted system, we show that UBQLN2 and ubiquitinated substrates can reciprocally induce one another’s phase separation and that phase separation can protect substrates from both proteasomal degradation and deubiquitination.
Results
To determine how substrate ubiquitination affects UBQLN2 phase separation, we ubiquitinated a model substrate consisting of R-Neh2Dual, an artificial degron derived from the N-terminus of Nrf2, followed by superfolder GFP (25) (sGFP; Figure 1A). The R-Neh2Dual degron can be ubiquitinated with a Ubr1 E3 ligase system, generating K48-linked chains; an Rsp5 system, generating K63-linked chains; or a Cul3/Rbx1/Keap1 system, generating branched ubiquitin chains containing K48, K63, and other linkages (26–29). We hereafter refer to these substrates as K63-linked, K48-linked or mixed. Mass spectrometry showed that the substrates were principally ubiquitinated on lysines within the degron region, although some substrates were also ubiquitinated within GFP, particularly in the mixed linkage substrate (Supplemental Figure S1). All substrates were highly ubiquitinated to a similar degree (estimated ≥15 Ub/substrate), with the bulk of the ubiquitinated substrate running at the top of the gel (Figure 1B).
Figure 1.
Ubiquitinated substrates phase separate and sediment UBQLN2 in a linkage-dependent manner. A) Fluorescence microscopy of 10 μM UBQLN2 (1% labeled with Alexa Fluor 647) incubated with or without 500 nM ubiquitinated (or not Ub’ed) substrate for 1 hr at 30 °C. B) Quantification of droplet size from images in E (K63: n=125, Mixed: 161, K48: 102 droplets) and statistics applied with Welch’s two-tailed t-test (****, p < 0.0001). C) Sedimentation assays use R-Neh2Dual-sGFP substrate ubiquitinated with K63-linked (Rsp5), K48-linked (Ubr1), or mixed linkage chains (Keap1/Cul3/Rbx1). After mixing substrate with UBQLN2 and incubating at 30 °C to allow phase separation to occur, condensates are pelleted (P) by centrifugation and separated from soluble (S) proteins before gel analysis. D) 10 μM UBQLN2 (1% labeled with Alexa Fluor 647) was incubated ± 1 μM ubiquitinated substrate for 1 hr; soluble (S) and pelleted (P) proteins were separated by centrifugation. E) UBQLN2 sedimentation as a function of substrate concentration. F) Quantification of replicate data (n=4–6) from E.
Ubiquitinated substrates condense with UBQLN2 in a linkage-specific manner
We first assessed the effect of ubiquitinated substrates on UBQLN2 phase separation via fluorescence microscopy (Figure 1A). Under conditions where full-length UBQLN2 exhibited little phase separation (10 μM UBQLN2, low salt, Figure 1A), K63-linked and mixed substrates (500 nM), but not K48-linked substrates, substantially promoted UBQLN2 phase separation, (Figure 1A,B). Both UBQLN2 and polyubiquitinated substrates were localized into the same droplets, as previously observed for UBQLN2 and polyubiquitin chains (22).
We next quantified the effect of ubiquitinated substrates on UBQLN2 phase separation using sedimentation assays, in which components of the dense phase were pelleted upon centrifugation (Figure 1C). Consistent with the microscopy data (Figure 1A,B), both K63-linked and mixed substrates (1 μM) induced robust phase separation of UBQLN2 (10 μM) while K48-linked substrates induced limited phase separation of UBQLN2 (Figure 1D). Similar results were obtained with 1 μM UBQLN2 (Supplemental Figure S2), in line with physiological concentrations (15). Phase separation was reciprocal (Figure 1D), such that ubiquitinated substrates co-sedimented with UBQLN2. There was a marked preference for longer chains in the condensates, as the highest molecular weight substrates were overrepresented in the pelleted fractions and underrepresented in the soluble fractions.
UBQLN2 sedimentation also depended on substrate concentration (Figure 1E,F). For K63-linked substrates, there was an increase in sedimentation with increasing substrate concentration. For K48-linked and, to a lesser extent, mixed substrates, biphasic behavior was observed, with an initial increase in sedimentation at lower substrate concentrations followed by a decrease at higher concentrations. Similar results were observed using enhanced GFP (eGFP) instead of sGFP (Supplemental Figure S3). These results agree with previous results using unanchored polyubiquitin chains, where K63-linked chains promoted UBQLN2 phase separation over a much broader concentration range than K48-linked chains (22). The biphasic behavior is consistent with re-entrant phase separation behavior stemming from heterotypic interactions between UBQLN2 and polyUb chains (22).
Proteasomes localize into ubiquitinated substrate/UBQLN2 condensates
We next asked whether proteasome would follow the substrate into phase-separated condensates using sedimentation assays (Figure 2, Supplemental Figure S4). Incubating TagRFP-T-labeled proteasome (Methods) with Alexa647-labeled UBQLN2 and ubiquitinated substrates in the presence of proteasome peptidase inhibitors revealed that the proteasome significantly sedimented in the presence of UBQLN2 and both K63-linked and mixed-linkage substrates. There was also a trend towards increased proteasome sedimentation in the presence of the K48-linked substrate, although this was not statistically significant (p = 0.12), perhaps because under these conditions only some UBQLN2 phase separated (Figure 1). Interestingly, the proteasome’s deubiquitinase (DUB) activity, which was especially noticeable for the K63-linked substrate (30), was inhibited in the presence of UBQLN2 (compare substrate + UBQLN + proteasome to substrate + proteasome in Figure 2A). These data suggest that, even though both the substrate and proteasome are concentrated in the condensate pellet fraction, condensation may protect the substrate from proteasomal activity instead of enhancing it. Microscopy experiments confirmed co-localization of UBQLN2, proteasome and substrates (Figure 2D). Small proteasome puncta were observed in condensates regardless of linkage type (Supplemental Figure S5).
Figure 2.
Proteasome is recruited to phase separated UBQLN2-substrate condensates. A) 100 nM TagRFP-T-Rpn6 containing proteasome and 500 nM K48-linked or K63-linked or 250 nM mixed linkage substrate were incubated with 10 μM UBQLN2 (1% Alexa 647 labeled) and 100 μM proteasome inhibitor cocktail (epoxomicin, MG132, bortezomib) for 1 hr as indicated. Individual channels are shown in Supplemental Figure S4. Soluble (S) and pelleted (P) proteins were separated by centrifugation and imaged using fluorescence. B, C) Quantification of replicate data from A. * indicates p≤0.05 relative to proteasome alone (B) or UBQLN2 alone (C) (two-tailed Welch’s t-test). Error bars are SEM from 3–4 measurements. D) Fluorescence microscopy of 10 μM UBQLN2, 100 nM proteasome, 500 nM ubiquitinated sGFP substrates, proteasome inhibitor cocktail, 1x deg buffer, and 1 hr at 30 °C. Scale bar 10 μm.
Substrates in UBQLN2 condensates can be protected from degradation
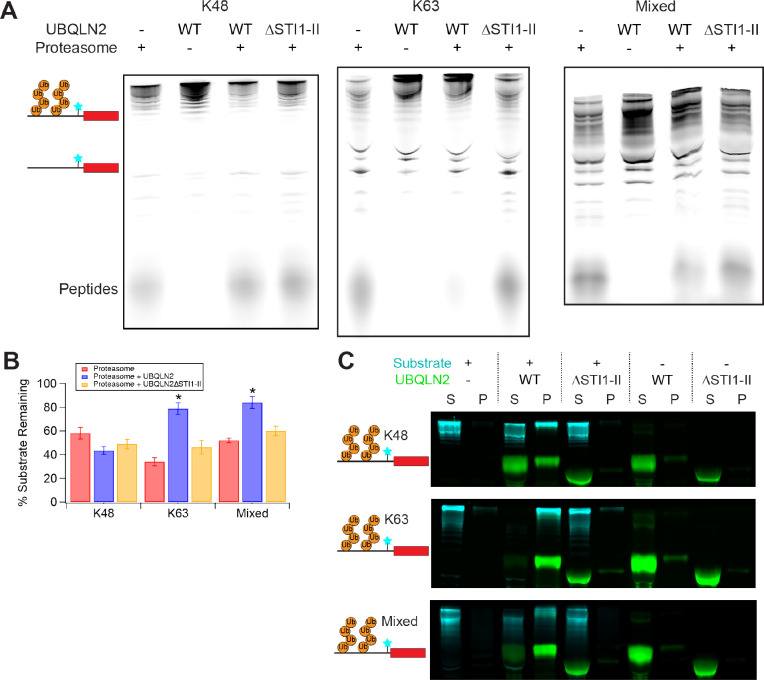
Does proteasomal recruitment to UBQLN2-substrate condensates have functional consequences? To test whether proteasome activity is enhanced (as has been shown for Rad23 in mammalian cells (12, 13)) or repressed, we examined the degradation of substrates in the presence of UBQLN2 condensates. We pre-incubated substrate and UBQLN2 to allow phase separation to occur, and then added proteasome to initiate the reaction. Since sGFP is quite stable, and, depending on ubiquitination state, is not always fully degraded by the proteasome (31, 32), we used a Cy5-labeled model substrate containing the same degron followed by two domains – the unstructured activation domain from the p160 transcriptional co-activator for thyroid hormone and retinoid receptors (ACTR) (33) followed by folded E. coli dihydrofolate reductase (DHFR) (34). Although DHFR is stably folded, the proteasome can easily unfold and degrade the entire substrate in the absence of UBQLN2 (Figure 3), as indicated by the Cy5-labeled peptides that appear at the bottom of the gel (34). UBQLN2 protected both K63-linked and mixed substrates, but not K48-linked substrates, against proteasomal degradation (Figure 3A,B). One potential explanation would be that UBQLN2 binds ubiquitinated substrates and shields them from the proteasome. To decouple binding to ubiquitinated substrates from phase separation, we used a UBQLN2 deletion construct, the ΔSTI1-II mutant, which exhibits greatly reduced phase separation propensity (15, 17), but unaltered ability to bind to ubiquitin (Supplemental Figure S7, Supplemental Table 1). The presence of the ΔSTI1-II mutant failed to protect substrates from degradation, indicating that ubiquitin binding is not sufficient for protection, and that phase separation is required. Crucially, under the same conditions as degradation occurs, the DHFR-containing substrate, like the sGFP substrate, reciprocally phase separates with UBQLN2, but not with the ΔSTI1-II mutant (Figure 3C).
Figure 3. UBQLN2-induced phase separation inhibits proteasomal degradation in a polyUb-linkage dependent manner.
A) Degradation of 250 nM ubiquitinated R-Neh2Dual-ACTR-DHFR substrate (Cy5-labeled N-terminal to DHFR) by 100 nM 26S proteasome with or without 10 μM UBQLN2 (1% DyLight-488 labeled) at 30 °C for 30’. ΔSTI1-II is a UBQLN2 variant that does not phase separate on its own. B) Quantification of replicate data (% substrate remaining, quantifying total amount of full-length and ubiquitinated substrate) from A. Error bars are SEM from 3–4 measurements. * indicates p≤0.01 for proteasome + UBQLN2 relative to both proteasome alone and proteasome + UBQLN2ΔSTI1-II (two-tailed Welch’s t-test). Quantified regions of example gels are shown in Supplemental Figure S6. C) Sedimentation assays under the same condition as assays in A (but without proteasome) confirm reciprocal co-sedimentation of ubiquitinated substrates with WT but not ΔSTI1-II UBQLN2.
Substrates in UBQLN2 condensates are protected from deubiquitination
Despite proteasomal recruitment to condensates, K63-linked and mixed substrates, but not K48-linked substrates, were protected from degradation and deubiquitination by the proteasome. To determine whether such protection was a general feature of UBQLN2-mediated condensates, we analyzed the ability of UBQLN2 to protect polyubiquitinated substrates from deubiquitination by DUBs. Ubiquitinated substrates and UBQLN2 were pre-incubated to allow condensate formation, and then different DUBs were added. We tested three DUBs: oTUB1, which is specific for K48-linked chains; AMSH, which is specific for K63-linked chains; and vOTU, which has no linkage specificity (35). Under conditions similar to those in which K63-linked and mixed substrates were protected from proteasomal degradation by UBQLN2, all three substrates (K48, K63 and mixed-linkage) were protected from DUB activity in the presence of WT UBQLN2 but not UBQLN2 ΔSTI1-II (Figure 4A,B). We therefore conclude that UBQLN2-mediated phase separation protects substrates from DUBs. Strikingly, K48-linked substrates show some protection from DUBs under the same conditions in which there is no protection from proteasomal degradation, indicating that there can be differential regulation of different enzymatic activities within condensates.
Figure 4. Phase separation protects polyubiquitin and polyubiquitinated substrates from DUB activity.
A) Deubiquitination of 250 nM ubiquitinated (Ub +) R-Neh2Dual-ACTR-DHFR substrate (Cy5-labeled N-terminal to DHFR) by oTUB1, AMSH, or vOTU with or without 10 μM UBQLN2. Nonubiquitinated substrate (Ub -) is shown as a size reference. B) Quantification of replicate data (% highly ubiquitinated substrate remaining) from A. Error bars are SEM from 3–4 measurements. * indicates p<0.05 for DUB + UBQLN2 relative to both DUB alone and DUB + UBQLN2ΔSTI1-II (two-tailed welch’s t-test). Quantified regions of example gels are shown in Supplemental Figure S6. C) The linkage-independent DUB vOTU (100 nM AF488-labeled vOTU) colocalizes with 50 μM UBQLN2 in the presence of unlabeled 50 μM K63-Ub4. Scale bar = 10 μm. D) 25 μM Ub (6.25 μM Ub4) of K63-Ub4 or K48-Ub4 was incubated with 50 nM vOTU in the presence of 60 μM UBQLN2 or UBQLN2 ΔSTI1-II at 37 °C and time points were run on an SDS-PAGE gel. E) Quantification of DUB assays from D. The Ub4 band volumes were quantified as single-exponential fits (lines). Error bars are SD from n=3 DUB assay experiments. F) 100 nM each K48-linked R-Neh2Dual-ACTR-DHFR and K63-linked R-Neh2Dual-sGFP were incubated with 20 nM vOTU in the presence of 1 μM UBQLN2 or UBQLN2 ΔSTI1-II at 30 °C and time points were run on an SDS-PAGE gel and detected via fluorescence. Arrow indicates highly ubiquitinated K63-linked substrate that persists in the presence of UBQLN2.
Since different chain linkages have different abilities to induce phase separation, it might be possible to regulate DUB activity via differential phase separation. To explore this possibility, we used unanchored tetraubiquitin (Ub4), as K63-linked Ub4 strongly induces UBQLN2 condensation while K48-linked Ub4 represses UBQLN2 condensation (22). The linkage-independent DUB vOTU was recruited to condensates formed by K63-linked Ub4 and UBQLN2 (Figure 4C). The presence of WT UBQLN2 protected K63-linked Ub4 chains from disassembly by vOTU, but had no effect on K48-linked Ub4 (Figure 4D,E). Neither substrate was protected in the presence of the non-phase-separating ΔSTI1-II mutant, revealing that phase separation can act as a switch which converts a promiscuous DUB into a linkage-specific one. To further explore this possibility, we treated a mixture of K63-linked DHFR-containing substrate and K48-linked GFP-containing substrate with vOTU in the presence or absence of a physiological concentration (1 μM) of UBQLN2. Strikingly, while there was no substantial effect of UBQLN2 on deubiquitination of the K48-linked substrate, some of the highly ubiquitinated K63-linked substrate was protected from deubiquitination in the presence of UBQLN2 but not the ΔSTI1-II mutant (Figure 4F). Thus, in a complex cellular environment UBQLN2-dependent phase separation might protect a subset of ubiquitinated proteins, but this is likely to be dependent on the substrate and cellular context.
Discussion
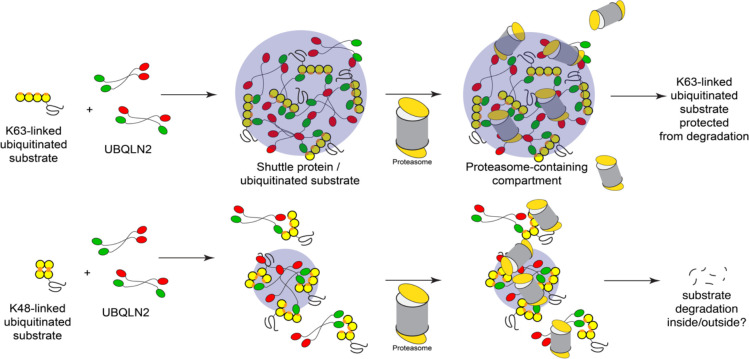
Our results indicate that ubiquitinated substrates can induce phase separation of UBQLN2, and are themselves recruited to the UBQLN2 condensates (Figure 5). Consistent with previous results with unanchored polyUb chains (22), extended K63-linked substrates are the most potent drivers of phase separation, while compact K48-linked substrates induce some phase separation at lower substrate concentrations but then disfavor phase separation as the substrate concentration is increased. Branched chains containing multiple K48 and K63 linkages have intermediate properties. We observe a strong preference for longer chains in condensates, particularly for K63 and mixed-linkage chains. Conceptually, longer extended ubiquitin chains would be expected to serve as better hubs or platforms for phase separation to occur, providing increased valency and multiple interaction surfaces (36), both via the UBA domain of UBQLN2 and via other weaker interactions. The substrate itself may be able to influence the phase separation process. However, the polyUb chains seem to be the major driver, as sGFP, eGFP and DHFR-containing substrates all showed qualitatively similar patterns of polyUb linkage-dependent phase separation. Similarly, although we cannot rule out a role for the location of the ubiquitin modifications on the substrate in the phase separation process, there was no obvious correlation between predominant ubiquitination sites in the substrate and phase separation behavior. Moreover, although GFP can be directly ubiquitinated on surface lysines, the DHFR-containing substrate only contains lysines in the R-Neh2Dual degron, suggesting the site of ubiquitination is not controlling phase separation.
Figure 5. Ubiquitinated substrates condense with UBQLN2 to form membraneless compartments capable of recruiting protein quality control machinery.
Our data show that ubiquitinated substrates can induce condensate formation with UBQLN2 as well as also be recruited into UBQLN2-substrate condensates, with K63-linked substrates showing greater propensity for this behavior over K48-linked substrates at similar substrate concentrations. Proteasomes can be recruited into these condensates although a proportion remains soluble and excluded from the condensate population. K63-linked ubiquitinated substrates are protected from degradation while K48-linked ubiquitinated substrates can still be degraded.
We find that UBQLN2-ubiquitinated substrate condensates, but not UBQLN2 or substrates on their own, recruit the proteasome. Interestingly, there appears to be some degree of internal structure to the condensates, with proteasome forming puncta within the larger UBQLN2-substrate droplets (Supplemental Figure S5); this seems to be a pattern regardless of linkage type (K48 or K63). These results suggest the possibility for similar multicomponent condensates, potentially with internal organization, to form within the cell, although the condensates observed here in vitro are micron-sized, which are significantly larger than those observed in cells. Cellular condensates could also include other protein quality control components. For example, under various conditions UBQLN has been shown to interact with p62 and, separately, with various chaperones and other protein quality control components in nuclear condensates (37–39). In yeast, the UBQLN2 homolog Dsk2 interacts with both proteasomes and another shuttle factor Rad23 in nuclear condensates upon prolonged cell growth in a ubiquitin-dependent process (16).
Our data suggest that the proteasome is hindered from degrading substrates in the UBQLN2-substrate-proteasome condensates we form in vitro, at least for the K63-linked and mixed substates (Figure 5). There is a trend towards improved degradation of K48-linked substrates (p=0.052), but since these substrates incompletely induce phase separation, it is unclear whether this potential increase in proteasome activity is due to phase separation. Our results suggest an additional reason that, although K63- and K48-linked chains both lead to degradation in vitro, K63-linked chains are generally not used as proteasomal targeting signals in vivo (30, 40). In cells, additional components could potentially further modulate the fates of ubiquitinated substrates within condensates. For example, recruitment of ubiquitination machinery to condensates may also promote ubiquitination of substrates (41). Ubiquitination can be regulated within UBQLN2 condensates as UBQLN2 interacts with E3 ligases such as E6AP (42).
We suspect that the network architectures of UBQLN2-substrate phase separated condensates, governed by the interactions between UBQLN2 and ubiquitinated substrates with different polyUb chain linkages, can determine whether a substrate is protected or not from degradation or deubiquitination. Inclusion/exclusion of components may be regulated by the conformational entropy of disordered segments in proteins (43). Similarly, the differences in conformational ensembles of polyUb chains may play a role in regulating internal network architecture of condensates (23) and consequently, their activity. Intriguingly, our data show that although all polyUb chain linkages are protected from DUB activity, under the same conditions only some linkages are protected from proteasomal degradation even though both DUBs and the proteasome can be recruited to condensates. Although the exact mechanism of this differential protection remains unclear (i.e. whether degradation is occurring inside or outside of condensates), it points to the ability of phase separation to serve as a regulatory mechanism to control the fate of ubiquitinated substrates. This work adds to a growing body of literature that shows that phase separation has functional consequences, with condensates regulating the activity of multiple processes, including ubiquitination (41), immune system activation (44–46), and signaling in development (46, 47).
Methods
Constructs & Strains
Constructs expressing UBQLN2 and UBQLN2ΔSTI1-II (deletion of residues 379–462) were described previously (17). Constructs expressing His-SUMO-R-Neh2Dual-sGFP, His-SUMO-R-Neh2Dual-eGFP were cloned into previously existing His-SUMO-R-Neh2Dual constructs (26) using Gibson assembly and verified using Sanger sequencing. A construct expressing His-SUMO-R-Neh2Dual-ACTR-DHFR (which is lysine-free except for the degron region and contains a single cysteine between ACTR and DHFR) was described previously (34). An S. cerevisiae strain with TagRFP-T-Rpn6 (yDAK56) and a 3X-Flag-tag on Rpn11 was created using CRISPR as described previously (48).
Protein Purification & Labeling
UBQLN2 proteins were expressed, purified and, as indicated, labeled with Alexa Fluor 647, Alexa Fluor 555, or DyLight-488 as described previously (15). Substrates were purified by Ni-NTA chromatography and SUMO protease cleavage as described previously, and R-Neh2Dual-ACTR-DHFR was labeled with sulfo-cyanine5 maleimide (Lumiprobe) on a single unique cysteine immediately N-terminal to DHFR, and repurified by gel filtration as described previously (26, 34). 26S Proteasome was purified from S. cerevisiae via a 3X-FLAG tag on Rpn11 as described previously (26). AMSH*, oTUB1* and vOTU were purified as described previously (32). vOTU was labeled with Alexa Fluor 555 and repurified by gel filtration. Ub4 was synthesized and purified as described previously (22). Substrate, UBQLN2 and DUB protein sequences are given in Supplemental Table S2
Ubiquitination
Substrates were ubiquitinated largely as described previously (26). To generate K63-linkages, substrates (5 μM) were incubated with mammalian E1 (166 nM), UbcH7 (E2, 2.9 μM), and Rsp5 (E3, 2.9 μM) ligases in 25 mM Tris-Cl pH 7.5, 50 mM NaCl, 4 mM MgCl2, 1.33 mg mL−1 ubiquitin, 4 mM ATP, and 1 μM DTT. To generate K48-linkages, substrates (5 μM) were incubated with mammalian E1 (100 nM), Ubc2 (E2, 12 μM), and Ubr1 (E3, 400 nM) ligases in 50 mM HEPES pH 7.5, 150 mM NaCl, 10 mM MgCl2, 1.33 mg mL−1 ubiquitin, and 5 mM ATP. To generate mixed linkages, substrates (4 uM) were incubated with mammalian E1 (E1, 130 nM), UbcH5 (E2, 4 μM), Cul3/Rbx1 (E3, 4 μM) and Keap1 C151S (4 μM) in 45 mM Tris-Cl pH 8.0, 100 mM NaCl, 10 mM MgCl2, 0.73 mg mL−1 ubiquitin, 2 mg mL−1 ovalbumin and 5 mM ATP. All reactions took place in the dark at 30°C for 1.5–2 hours. Polyubiquitinated substrates were purified in 1x degradation assay buffer (50 mM Tris-Cl pH 7.5, 5 mM MgCl2, 5% glycerol) by spin size exclusion using Sephadex G75. Concentrations were determined using the fluorescence intensity of the final sample.
Mass spectrometry
On-column digests of ubiquitination reactions were performed using the Protifi S-Trap™ mini kit (K02-Mini-10) following the manufacturer’s protocol in preparation for peptide analysis by LC-MS. The proteases trypsin and Lys-C (Promega, V5073) were used to digest the LtEc proteins overnight at 37°C after reduction with 5 mM TCEP at 55°C and alkylation with 20 mM iodoacetamide at room temperature. Peptides were separated on an Agilent InfinityLab Poroshell 120 EC-C18 (50 × 2.1 mm, 2.7 μm particle, # 699775–902) column maintained at 40°C on a SCIEX Exion LC (Framingham, MA). The HPLC solvents used were (A) water with 0.1% formic acid and (B) acetonitrile with 0.1% formic acid at a flowrate of 0.4 mL/min. Gradient elution consisted of 100% A hold for 5 minutes, followed by two gradient steps (0–40% B from 5–60 min and 40–95% B from 60–75 min) with a final hold at 95% B from 70–90 minutes. Eluted peptides were analyzed by data dependent acquisition (DDA) on a SCIEX 5600+ TripleTOF mass spectrometer calibrated with the SCIEX APCI positive calibrant solution prior to each analysis. Positive ESI precursor ions for DDA were determined under high sensitivity mode TOF from 200–1250 Da with the following source parameters: DP = 100 V, CE = 10, GAS1 = GAS2 = 60 psi, CUR = 30 psi, ISFV = 5500 V, and a source temperature of 500°C. DDA fragmentation experiments selected the ten most abundant ions from survey scan criteria between 400–1250 m/z, with charge states between 2+ to 5+, using dynamic background selection and rolling collision energy (CE). Selected target ions were excluded for 7 sec (2 cycles). Spectra were analyzed for ubiquitin modification using ProteinPilot (SCIEX, v. 5.0.2), with a custom database consisting of the proteins known to be present in the ubiquitination reactions (E1, E2, E3 enzymes, substrate and ubiquitin). A custom modification to tyrosine (loss of OH4) was used to enable detection of the GFP chromophore derived from TYG; false positives containing this modification on other peptides were discarded. Distinct peptides at an FDR of 1% (or, for Keap1 ubiquitination, at a confidence level of 99%) were used to assess locations of ubiquitin modification, with manual assignments of ambiguous peptides based on theoretical confidence levels and the typical absence of a cleavage event after a ubiquitinated lysine. Data is deposited as MassIVE MSV000093780.
Sedimentation assays
UBQLN2 was incubated with polyubiquitinated substrates in 1X degradation assay buffer containing 2 mM DTT, 1 mg mL−1 BSA, and 1% DMSO in Eppendorf Protein Lo-Bind tubes for 1 hour at 30°C. For titration experiments, substrates were serially diluted in 1x degradation assay buffer prior to the addition of UBQLN2. For experiments including TagRFP-T-Rpn6 proteasome, reaction mixture also contained 1 mM ATP, 10 mM creatine phosphate, 0.1 mg mL−1 creatine phosphokinase and a proteasome inhibitor cocktail consisting of 100 μM each epoxomicin, bortezomib, and MG132.
Following incubation, reactions were spun at 15871 × g for 5 minutes at room temperature to separate soluble and phase-separated proteins. Pellets were solubilized in 2X SDS-PAGE sample buffer (49). Proteins were run on 9.25% tris-tricine gels, scanned on a Typhoon FLA 9500 imager (Cytiva) with voltage adjusted to assure all samples were within the linear range of the detector, and relative amounts of UBQLN2, substrate, and proteasome present in soluble and pelleted fractions were determined using ImageQuant.
Microscopy
Samples were prepared on ice to contain either 10 μM UBQLN2 (spiked with 20 nM UBQLN2 labelled with Alexa Fluor 647) and/or 500 nM of sGFP substrates in 1x degradation assay buffer. Samples with 100 nM TagRFP-T-Rpn6 containing proteasome were also prepared with 100 μM proteasome inhibitor cocktail. Samples were added to Eisco Labs Microscope Slides, with Single Concavity, and covered with MatTek coverslips that had been coated with 5% bovine serum albumin (BSA) to minimize changes due to surface interactions, and incubated coverslip-side down at 30°C for 1 hour. Samples of deubiquitinase vOTU contained 50 μM UBQLN2 (spiked with Alexa Fluor 647-labelled UBQLN2), 50 μM Ub of K63-Ub4 (spiked with Alexa Fluor 488-labeled K63-Ub4), 20 mM HEPES, 200 mM NaCl, 1 mM DTT, pH 7, and were incubated at 30 °C and imaged at indicated time points. Phase separation was imaged on an ONI Nanoimager (Oxford Nanoimaging Ltd, Oxford, UK) equipped with a Hamamatsu sCMOS ORCA flash 4.0 V3 camera using an Olympus 100×/1.4 N.A. objective. Images were prepared using Fiji (Schindelin et al, 2012) and FigureJ plugin.
Microscopy analysis
For puncta count, image background subtraction was performed using a rolling ball radius of 50 pixels. Images were subsequently thresholded using default settings. Puncta were analyzed using a minimum size of 10 pixel^2 and a circularity range of 0.3 – 1.0 to eliminate edge artifacts.
Degradation assays
Cy5-labeled polyubiquitinated R-Neh2Dual-ACTR-DHFR (250 nM) was preincubated with 10 μM UBQLN2 in 1x degradation assay buffer containing 1 mM ATP, 10 mM creatine phosphate, 0.1 mg mL−1 creatine phosphokinase, 2 mM DTT, 1 mg mL−1 BSA, and 1% DMSO for 1 hour at 30°C. 100 nM 26S proteasome was then added, and the reaction was incubated for anther 30 minutes. Reactions were stopped by the addition of an equal volume of SDS-PAGE sample buffer (100 mM Tris pH 6.8, 24% glycerol, 8% SDS, 0.2 mg mL−1 coomassie blue R250, 0.2 M DTT). Samples were run on 9.25% tris-tricine gels, scanned on a Typhoon FLA 9500 imager with voltage adjusted to assure all samples were within the linear range of the detector, and relative amounts of substrate remaining in each reaction were determined using ImageQuant.
Deubiquitinase assays
Using polyubiquitinated substrates:
Cy5-labeled polyubiquitinated R-Neh2Dual-ACTR-DHFR (250 nM) was preincubated with 10 μM UBQLN2 in 1x degradation assay buffer containing 2 mM DTT, 1 mg mL−1 BSA, and 1% DMSO for 1 hour at 30°C. For K63-linked substrates, 1 μM AMSH or 40 nM vOTU was added and the reactions were incubated for 15 or 10 min, respectively. For K48-linked substrates, 1 μM OTUB1 or 200 nM vOTU was added and the reactions were incubated for 20 or 10 minutes, respectively. For mixed linkage substrates, 200 nM vOTU was added and the reaction was incubated for 30 minutes. Incubation times and DUB concentrations were chosen in control experiments to ensure partial deubiquitination in the absence of UBQLN2. DUB reactions were stopped by the addition of an equal volume of SDS-PAGE sample buffer. Samples were run on 9.25% tris-tricine gels, scanned on a Typhoon FLA 9500 imager with voltage adjusted to assure all samples were within the linear range of the detector, and relative amounts of highly ubiquitinated substrate remaining in each reaction were determined using ImageQuant.
Using free polyUbiquitin chains:
Samples were prepared on ice to contain 50 nM vOTU, 30 μM of K48-Ub4 or K63-Ub4, with and without 60 μM of full-length UBQLN2 or UBQLN2 ΔSTI1-II in 20 mM NaP, 500 mM NaCl, pH 6.8. Samples were aliquoted into separated microfuge tubes and incubated at 37 °C. At indicated time points, the reactions were stopped by the addition of an equal volume of SDS-PAGE sample buffer. Samples were run on 4–20% gradient gels (BioRad), stained with Coomassie Blue, destained, imaged on Gel Doc EZ imager (BioRad) and bands intensities were quantified with Image Lab (BioRad).
Nuclear Magnetic Resonance (NMR) Spectroscopy
Protein solutions of 50 μM 15N UBQLN2 or UBQLN2 ΔSTI1-II were prepared in 20 mM NaPhosphate buffer (pH 6.8) with 0.5 mM EDTA, 0.02 % NaN3, and 5 % D2O. NMR data (either standard 1H-15N SOFAST-HMQC or 1H-15N TROSY-HSQC experiments) were acquired at 25°C using a Bruker Avance III 800 MHz spectrometer equipped with TCI cryoprobe. NMR data were subsequently processed using NMRPipe (50) and analyzed using CCPNMR 2.5.2 (51). UBA peak assignments were transferred by visual inspection using previously-determined UBQLN2 450–624 assignments (15). To determine Ub binding affinity , we prepared separate NMR samples with known amounts of unlabeled Ub (up to a stoichiometry of ~2:1 Ub:UBQLN2). Binding was monitored as a function of different ligand:protein ratios. Assuming the fast-exchange limit, the chemical shift perturbation (CSP) for each backbone amide was calculated as , where is the CSP in the fully-bound state, and [PL] and [Pt] represent the ligand-bound and the total UBQLN2 protein concentrations, respectively. The CSP is quantified as where and are the differences in 1H and 15N chemical shifts in ppm for amide resonances compared between the NMR spectrum at a specific titration point and the ligand-free NMR spectrum. Titration data for each amide resonance were fit to the single-site binding model with an in-house MATLAB program: . [Lt] is the total ligand (Ub) concentration and is the binding affinity. Reported values are averages of residue-specific values with the error reflecting the standard deviation of these values.
Supplementary Material
Significance.
Covalent attachment of polyubiquitin chains to eukaryotic proteins is a common protein quality control mechanism. Ubiquitination often marks proteins for degradation by the proteasome, but can also result in non-degradative outcomes. Proteins, including UBQLN2, that bind both polyubiquitin and the proteasome can either enhance or inhibit degradation. The ALS-related UBQLN2 can form membraneless organelles in the cell and undergoes phase separation in vitro, but the effects of phase separation on substrate fate are unknown. Herein we show that UBQLN2 phase separation is modulated by polyubiquitinated substrates in a linkage-dependent fashion. Further, we show that functional outcomes, degradation or deubiquitination, are differentially affected by phase separation. Our results suggest that phase separation is a regulatory mechanism that can control protein fate.
Acknowledgements
The authors thank members of the Kraut and Castañeda labs, especially Dr. Edwin Ragwan, Dr. Billy Haws, Dr. Anitha Rajendran, Dr. Nirbhik Acharya, as well as Dr. Jeroen Roelofs and Dr. Jeremy Schmit for valuable discussions and Matt Schnell, Dr. Anthony Lagalante and Luke Lagalante for assistance with mass spectrometry experiments.
Funding
This material is based upon work supported by the National Science Foundation under Grant No. 1935596 to D.A.K (sedimentation assays, substrates, proteasome, DUBs). This work was supported by NIH R01GM136946 (UBQLN2 protein purifications, Ub4, NMR) to C.A.C.. Data collected on a Bruker 800 MHz NMR magnet (SU, SUNY-ESF) was supported by NIH shared instrumentation grant 1S10OD012254. Data collected on a SCIEX 5600+ was supported by a Major Research Instrumentation grant from the National Science Foundation (Grant No. CHE-2018399).
Data Availability
Mass spectrometry data for ubiquitinated substrates are deposited in MassIVE: ftp://massive.ucsd.edu/v06/MSV000093780/. All other data are available from corresponding authors upon request.
References
- 1.Finley D, Ulrich HD, Sommer T, Kaiser P (2012) The ubiquitin-proteasome system of Saccharomyces cerevisiae. Genetics 192(2):319–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akutsu M, Dikic I, Bremm A (2016) Ubiquitin chain diversity at a glance. J Cell Sci 129(5):875–880. [DOI] [PubMed] [Google Scholar]
- 3.Wang Y-S, Wu K-P, Jiang H-K, Kurkute P, Chen R-H (2020) Branched Ubiquitination: Detection Methods, Biological Functions and Chemical Synthesis. Molecules (Basel, Switzerland) 25(21):5200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kolla S, Ye M, Mark KG, Rapé M (2022) Assembly and function of branched ubiquitin chains. Trends Biochem Sci 47(9):759–771. [DOI] [PubMed] [Google Scholar]
- 5.Saeki Y et al. (2009) Lysine 63-linked polyubiquitin chain may serve as a targeting signal for the 26S proteasome. The EMBO Journal 28(4):359–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zientara-Rytter K, Subramani S (2019) The Roles of Ubiquitin-Binding Protein Shuttles in the Degradative Fate of Ubiquitinated Proteins in the Ubiquitin-Proteasome System and Autophagy. Cells 8(1):40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raasi S, Pickart CM (2003) Rad23 ubiquitin-associated domains (UBA) inhibit 26 S proteasome-catalyzed proteolysis by sequestering lysine 48-linked polyubiquitin chains. J Biol Chem 278(11):8951–8959. [DOI] [PubMed] [Google Scholar]
- 8.Lipinszki Z et al. (2011) Overexpression of Dsk2/dUbqln results in severe developmental defects and lethality in Drosophila melanogaster that can be rescued by overexpression of the p54/Rpn10/S5a proteasomal subunit. FEBS J 278(24):4833–4844. [DOI] [PubMed] [Google Scholar]
- 9.Livneh I, Cohen-Kaplan V, Cohen-Rosenzweig C, Avni N, Ciechanover A (2016) The life cycle of the 26S proteasome: from birth, through regulation and function, and onto its death. Cell Res 26(8):869–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsuchiya H et al. (2017) In Vivo Ubiquitin Linkage-type Analysis Reveals that the Cdc48-Rad23/Dsk2 Axis Contributes to K48-Linked Chain Specificity of the Proteasome. Mol Cell 66(4):488–502.e7. [DOI] [PubMed] [Google Scholar]
- 11.Jablonski AM et al. (2015) Loss of RAD-23 Protects Against Models of Motor Neuron Disease by Enhancing Mutant Protein Clearance. J Neurosci 35(42):14286–14306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yasuda S et al. (2020) Stress- and ubiquitylation-dependent phase separation of the proteasome. Nature 578(7794):296–300. [DOI] [PubMed] [Google Scholar]
- 13.Uriarte M et al. (2021) Starvation-induced proteasome assemblies in the nucleus link amino acid supply to apoptosis. Nat Commun 12(1):6984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alexander EJ et al. (2018) Ubiquilin 2 modulates ALS/FTD-linked FUS-RNA complex dynamics and stress granule formation. Proc Natl Acad Sci U S A 115(49):E11485–E11494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dao TP et al. (2018) Ubiquitin Modulates Liquid-Liquid Phase Separation of UBQLN2 via Disruption of Multivalent Interactions. Mol Cell 69(6):965–978.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Waite KA, Vontz G, Lee SY, Roelofs J (2024) Proteasome condensate formation is driven by multivalent interactions with shuttle factors and ubiquitin chains. Proc Natl Acad Sci U S A 121(10):e2310756121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dao TP, Rajendran A, Galagedera SKK, Haws W, Castañeda CA (2023) Short disordered termini and proline-rich domain are major regulators of UBQLN1/2/4 phase separation. Biophysical Journal [DOI] [PubMed] [Google Scholar]
- 18.Riley JF, Fioramonti PJ, Rusnock AK, Hehnly H, Castañeda CA (2021) ALS-linked mutations impair UBQLN2 stress-induced biomolecular condensate assembly in cells. J Neurochem 159(1):145–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dao TP et al. (2019) ALS-Linked Mutations Affect UBQLN2 Oligomerization and Phase Separation in a Position- and Amino Acid-Dependent Manner. Structure 27(6):937–951.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sharkey LM et al. (2018) Mutant UBQLN2 promotes toxicity by modulating intrinsic self-assembly. Proc Natl Acad Sci U S A 115(44):E10495–E10504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Safren N et al. (2024) Pathogenic mutations in UBQLN2 exhibit diverse aggregation propensity and neurotoxicity. Sci Rep 14(1):6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dao TP et al. (2022) Mechanistic insights into enhancement or inhibition of phase separation by different polyubiquitin chains. EMBO Rep e55056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Galagedera SKK et al. (2023) Polyubiquitin ligand-induced phase transitions are optimized by spacing between ubiquitin units. Proc Natl Acad Sci U S A 120(42):e2306638120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dao TP, Castañeda CA (2020) Ubiquitin-Modulated Phase Separation of Shuttle Proteins: Does Condensate Formation Promote Protein Degradation? BioEssays : news and reviews in molecular, cellular and developmental biology 42(11):e2000036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pédelacq J-D, Cabantous S, Tran T, Terwilliger TC, Waldo GS (2005) Engineering and characterization of a superfolder green fluorescent protein. Nature Biotechnology 24(1):79–88. [DOI] [PubMed] [Google Scholar]
- 26.Hurley CM, Kraut DA (2021) Determination of Proteasomal Unfolding Ability. Methods Mol Biol 2365:217–244. [DOI] [PubMed] [Google Scholar]
- 27.Cundiff MD et al. (2019) Ubiquitin receptors are required for substrate-mediated activation of the proteasome’s unfolding ability. Sci Rep 9(1):14506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pan M et al. (2021) Structural insights into Ubr1-mediated N-degron polyubiquitination. Nature 600(7888):334–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim HC, Huibregtse JM (2009) Polyubiquitination by HECT E3s and the determinants of chain type specificity. Molecular and cellular biology 29(12):3307–3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jacobson AD et al. (2009) The lysine 48 and lysine 63 ubiquitin conjugates are processed differently by the 26 s proteasome. The Journal of biological chemistry 284(51):35485–35494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bragança CE, Kraut DA (2020) Mode of targeting to the proteasome determines GFP fate. J Biol Chem 295(47):15892–15901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reichard EL et al. (2016) Substrate Ubiquitination Controls the Unfolding Ability of the Proteasome. J Biol Chem 291(35):18547–18561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dogan J, Schmidt T, Mu X, Engström Å, Jemth P (2012) Fast association and slow transitions in the interaction between two intrinsically disordered protein domains. J Biol Chem 287(41):34316–34324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ragwan ER et al. (2024) Slippery sequences stall the 26S proteasome at multiple points along the translocation pathway. bioRxiv 10.1101/2024.03.14.584990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hospenthal MK, Mevissen TET, Komander D (2015) Deubiquitinase-based analysis of ubiquitin chain architecture using Ubiquitin Chain Restriction (UbiCRest). Nature Protocols 10(2):349–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pappu RV, Cohen SR, Dar F, Farag M, Kar M (2023) Phase Transitions of Associative Biomacromolecules. Chem Rev 123(14):8945–8987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hjerpe R et al. (2016) UBQLN2 Mediates Autophagy-Independent Protein Aggregate Clearance by the Proteasome. Cell 166(4):935–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fu A, Cohen-Kaplan V, Avni N, Livneh I, Ciechanover A (2021) p62-containing, proteolytically active nuclear condensates, increase the efficiency of the ubiquitin-proteasome system. Proc Natl Acad Sci U S A 118(33):e2107321118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mohan HM et al. (2022) RTL8 promotes nuclear localization of UBQLN2 to subnuclear compartments associated with protein quality control. Cellular and Molecular Life Sciences 79(3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nathan JA, Kim HT, Ting L, Gygi SP, Goldberg AL (2013) Why do cellular proteins linked to K63-polyubiquitin chains not associate with proteasomes? The EMBO Journal 32(4):552–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li J et al. (2023) Feedback regulation of ubiquitination and phase separation of HECT E3 ligases. Proc Natl Acad Sci U S A 120(33):e2302478120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Buel GR et al. (2023) E6AP AZUL interaction with UBQLN1/2 in cells, condensates, and an AlphaFold-NMR integrated structure. Structure S0969–2126(23)00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grigorev V, Wingreen NS, Zhang Y (2023) Conformational entropy of intrinsically disordered proteins bars intruders from biomolecular condensates. bioRxiv 10.1101/2023.03.03.531005 [DOI] [Google Scholar]
- 44.Du M, Ea CK, Fang Y, Chen ZJ (2022) Liquid phase separation of NEMO induced by polyubiquitin chains activates NF-κB. Mol Cell 82(13):2415–2426.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goel S et al. (2023) Linear ubiquitination induces NEMO phase separation to activate NF-κB signaling. Life Sci Alliance 6(4):e202201607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meyer HM, Hotta T, Malkovskiy A, Zheng Y, Ehrhardt DW (2023) Thermo-sensitive condensation of SUF4 tunes flowering time inArabidopsis thaliana. bioRxiv 10.1101/2023.11.01.565081 [DOI] [Google Scholar]
- 47.Powers SK et al. (2019) Nucleo-cytoplasmic Partitioning of ARF Proteins Controls Auxin Responses in Arabidopsis thaliana. Mol Cell 76(1):177–190.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cresti JR et al. (2021) Proteasomal conformation controls unfolding ability. Proc Natl Acad Sci U S A 118(25):e2101004118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang Z, Zhang G, Zhang H (2019) Protocol for analyzing protein liquid–liquid phase separation. Biophysics Reports 5(1):1–9. [Google Scholar]
- 50.Delaglio F et al. (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6(3):277–293. [DOI] [PubMed] [Google Scholar]
- 51.Vranken WF et al. (2005) The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins 59(4):687–696. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Mass spectrometry data for ubiquitinated substrates are deposited in MassIVE: ftp://massive.ucsd.edu/v06/MSV000093780/. All other data are available from corresponding authors upon request.