ABSTRACT
The ectopic gut colonization by orally derived pathobionts has been implicated in the pathogenesis of various gastrointestinal diseases, including inflammatory bowel disease (IBD). For example, gut colonization by orally derived Klebsiella spp. has been linked to IBD in mice and humans. However, the mechanisms whereby oral pathobionts colonize extra-oral niches, such as the gut mucosa, remain largely unknown. Here, we performed a high-density transposon (Tn) screening to identify genes required for the adaptation of an oral Klebsiella strain to different mucosal sites – the oral and gut mucosae – at the steady state and during inflammation. We find that K. aerogenes, an oral pathobiont associated with both oral and gut inflammation in mice, harbors a newly identified genomic locus named “locus of colonization in the inflamed gut (LIG)” that encodes genes related to iron acquisition (Sit and Chu) and host adhesion (chaperon usher pili [CUP] system). The LIG locus is highly conserved among K. aerogenes strains, and these genes are also present in several other Klebsiella species. The Tn screening revealed that the LIG locus is required for the adaptation of K. aerogenes in its ectopic niche. In particular, we determined K. aerogenes employs a CUP system (CUP1) present in the LIG locus for colonization in the inflamed gut, but not in the oral mucosa. Thus, oral pathobionts likely exploit distinct adaptation mechanisms in their ectopically colonized intestinal niche compared to their native niche.
KEYWORDS: Klebsiella, oral microbiota, pathobionts, periodontitis, inflammatory bowel disease, gut microbiota, Chaperon usher pili
Introduction
The oral cavity nourishes a large number of microorganisms referred to as the oral microbiota.1 The oral microbiota is the second most diverse and abundant microbial community, next to the gastrointestinal (GI) tract, harboring over 770 species of bacteria.2 The symbiotic oral microbiota potentiates oral mucosal immunity and resists colonization and expansion of pathogenic microorganisms causing oral disease through host immunity and/or direct competition.3,4 On the other hand, during oral disease, oral pathobionts, such as Porphyromonas gingivalis, Tannerella forsythia, Fusobacterium nucleatum, and Aggregatibacter actinomycetemcomitans, and species of Treponema and Prevotella, are expanded and cause various oral pathologies, such as oral mucosal inflammation and alveolar bone resorption.5–7
Emerging evidence supports a pathogenic link between oral bacteria and extra-oral conditions, such as GI diseases. The accumulating body of literature demonstrated that oral-associated bacteria are enriched in the gut mucosa of patients with inflammatory bowel disease (IBD) and colorectal cancer (CRC).8,9 Beyond the clinical correlation, preclinical studies have confirmed that gut colonization by oral-associated bacteria drives GI pathologies. For example, gut colonization by P. gingivalis and F. nucleatum aggravates the severity of dextran sulfate sodium (DSS) – induced colitis in mice.10–12 Also, Klebsiella spp. present in the saliva of healthy individuals and IBD patients can ectopically colonize the gut and induce a pathogenic Th1 response, leading to colitis in mice.13 In this regard, it is noteworthy that oral symbiotic bacteria are generally nonpathogenic, and their ectopic gut colonization does not cause inflammation.14 However, specific oral bacteria, particularly oral pathobionts associated with periodontal disease, increase the risk of GI disease through ectopic gut colonization. In this context, we have found that oral inflammation is a curial trigger for the expansion of oral pathobionts in the oral cavity and subsequent ectopic gut colonization by them. In a mouse periodontitis model, inflammasome-activating pathogenic Klebsiella and Enterobacter spp. accumulated in the oral cavity. The expanded Klebsiella and Enterobacter spp. translocated to the lower digestive tract and increased the susceptibility to DSS-induced colitis.14 In humans, there is accumulating evidence that IBD patients have a higher prevalence of periodontitis and gingivitis,15,16 and, consistent with preclinical findings, periodontal inflammation has been reported to be associated with worse clinical outcomes in IBD.17 Of note, consistent with animal studies, periodontitis is associated with increased gut colonization by an oral pathobiont Haemophilus parainfluenzae in patients with Crohn’s disease (CD).18 Thus, oral pathobionts, expanded during oral inflammation, contribute to the pathogenesis of GI diseases through ectopic colonization in the gut mucosa.
However, the mechanisms by which oral pathobionts thrive in the inflamed oral mucosa and subsequently re-adapt to the gut environment remain largely unclear. Given the difference in the microenvironment between the oral and gut mucosae, oral pathobionts likely utilize different strategies for colonization in their native niche (i.e., oral mucosa) and ectopically colonizing niche (i.e., gut mucosa). Here, we used the orally derived K. aerogenes (Ka), the most predominant Klebsiella spp. found in the inflamed oral cavity of periodontitis mice.14 Notably, the mono-association of Ka in germ-free (GF) mice can induce periodontitis (Figure S1). Hence, Ka represents an oral pathobiont capable of instigating periodontitis and, when ectopically colonizing the gut, also contributing to colitis.14 Leveraging this colitogenic oral pathobiont strain, we sought to identify genes exploited by Ka to adapt to the oral and gut mucosae using a transposon – insertion site sequencing (Tn-Seq) analysis.19–21
Results
Characterization of the K. aerogenes SK431 strain
We first determined the genomic sequence of SK431 and compared the genetic characteristics of this strain with the reference Ka strain KCTC 2190 and 17 other Ka strains identified in the NCBI GenBank database (Figure 1) by Roary.22 Ka strain SK431 showed high evolutionary relationships to AR0161, a poorly described Ka strain, among the different Ka strains studied (Figure 1a and Figure S2), as 94% of SK431 gene orthologues were found in AR0161. The putative virulence genes of Ka strains were predicted by homology with the entries in the virulence factor data base (VFDB)23 and compared with related Enterobacteriaceae pathogens (Figure 1b). The Ka SK431 and other 18 reference Ka strains have similar sets of putative virulence genes (Figure 1b). All Ka strains have common systems associated with iron acquisition, including Sit (inner membrane-located iron/manganese transporter), TonB-dependent enterobactin (Ent/Fep), salmochelin (Sal), aerobactin (Aer), and heme-iron acquisition (Chu), suggesting conservation of efficient iron acquisition of Ka in host-associated environments.24 Also, all Ka strains harbor type VI secretion systems (T6SS), but lack type III secretion system (T3SS) and type IV secretion system (T4SS), which are involved in competition with other bacteria and host immune invasion or intoxication, respectively.25 The Ka SK431 also harbors strain-specific competition factors, including a group A colicin (A-Col) and contact-dependent growth inhibition (Cdi) systems. Ka SK431 genome harbors several putative adhesion factors, including orthologues of known pili (Fim, CfA, Escherichia coli common pilus [Ecp]), and conserved but uncharacterized chaperon usher (CU) pili systems (named CUP1 to CUP6 here, Figure 1b). Notably, Sit, Chu, and CUP1 operons are located within a single locus of the SK431 genome (Figure 1c). Other Ka strains, including AR0161, also carry this locus in their genomes, whereas the CUP operon is separated from Sit and Chu in other Klebsiella species (Figure 1c), suggesting an evolutionary relationship of this putative virulence locus in the genus Klebsiella, as well as providing a specific advantage for colonizing a Ka niche. As this newly identified genetic locus is composed of genes that are required for the colonization in the inflamed gut (see below), we named it the “locus of colonization in the inflamed gut” (LIG). These findings suggest that Ka SK431 LIG, and other virulence genes conserved in Ka strains, may be responsible for the colonization and virulence of this oral pathobiont at specific sites.
Figure 1.
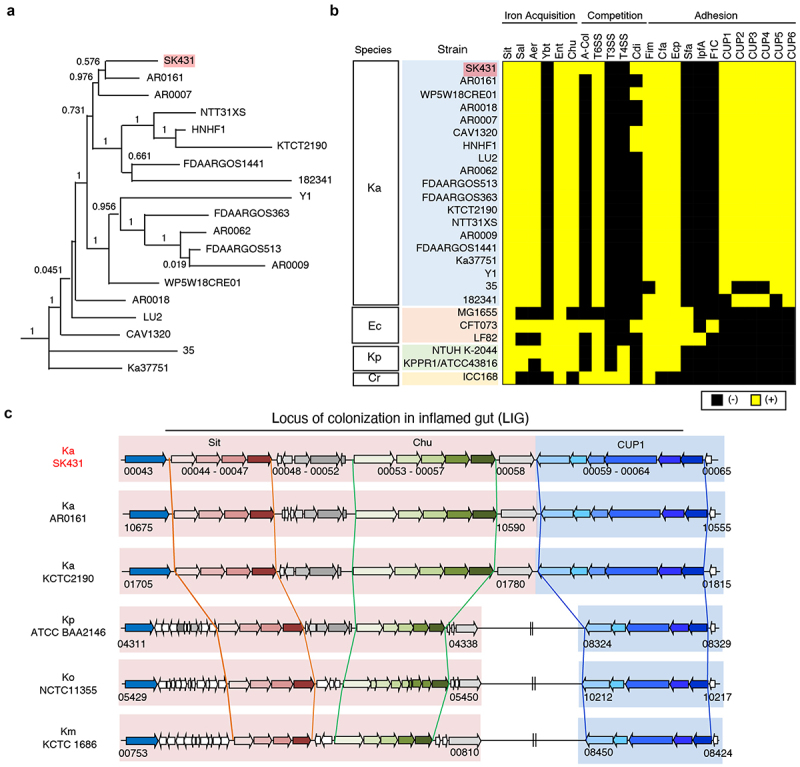
Characterization of K. aerogenes SK431 genome. (a) Phylogenetic tree of Ka SK431 and 18 reference Ka strains. (b) Presence of virulence-associated genes in indicated strains of Ka; Escherichia coli (Ec); K. pneumoniae (Kp); Citrobacter rodentium (Cr). The presence and absence of operon orthologues are indicated by yellow and black, respectively. (c) Schematic representation of the “locus of colonization in the inflamed gut” (LIG) virulence factors in SK431 and other Klebsiella strains. Syntenic regions and gene orthologues are indicated by the same arrows and background colors. The genes of uncharacterized proteins are shown in white arrows. Direction of arrows indicates the transcriptional direction. The locus numbers in gene-annotated genomes (GenBank accession numbers, CP028951, NC015663, 000364385, 900478285, 000240325 for Ka AR0161, Ka KCTC2190, Kp ATCC BA2146, Ko NTCC 11,355, Km KCTC1686, respectively) are indicated below the respective first and last genes of each syntenic region.
Identification of K. aerogenes SK431 genes required for the colonization in the oral and gut mucosae
To identify the genes required for the colonization of the oral pathobiont Ka SK431 in its natural (oral) and ectopic (gut) niches, both in the steady state and during inflammation, we generated a SK431 mutant library by Tn insertion,26 comprising ~ 63,000 independent mutants. To identify genes responsible for the colonization in the healthy and inflamed oral environments, we inoculated the SK431 mutant library into dental ligatures of the oral cavity14 (Figure 2a). The SK431 mutant strains were then harvested from the inserted ligatures on day 1 (before the development of periodontitis) and day 14 (after the development of periodontitis). To mimic ectopic gut colonization by the oral pathobiont, we also performed orogastric inoculation of the SK431 mutant library (Figure 2a). In the gut environment, the SK431 mutant strains were harvested from feces. DNA was extracted from recovered bacteria (outputs) and the initial inoculum (input; IN) and subjected to Illumina sequencing (Figure 2a). The outputs included ligature 1-day (control, non-inflamed oral cavity; nOC), ligature 14 days (periodontitis, inflamed oral cavity; iOC), feces of water-treated mice (control, non-inflamed gut; nGT), and feces of DSS-treated mice (colitis, inflamed gut; iGT) samples (Figure 2A). Illumina sequencing analysis showed that the library contained multiple Tn inserts in individual genes of Ka SK431 and no obvious locus-based bias on the Ka reference genome (Figure 2b). The overall Tn insertion frequencies in particular genes of the outputs (i.e., nOC, iOC, nGT, iGT) were lower than the IN, indicating that these genes were required for optimal colonization at the periodontal site or in the gut (Table S1). Next, we performed a coverage analysis of the fold difference between two groups to identify genes required to colonize the tested in vivo conditions (Figure 2c, and results are summarized in Figure 2d). We compared the IN and output samples to identify the genes required for in vivo colonization. Numbers of genes are enriched in Group A (IN) compared to Group B (output), indicating that mutants of these genes have reduced fitness in the output groups (i.e., genes are required for in vivo adaptation). Notably, the number of required genes was higher in the inflamed gut compared to the input (IN/iGT) than in the other conditions (IN/nOC, IN/iOC, and IN/nGT), although the level of requirement was mild to moderate (the majority of genes showed 10- to 25-fold enrichment in IN) (Figure 2c, d). These results suggested that the pathobiont K. aerogenes SK431 requires more genes to adapt to the ectopic niche (gut) compared to its natural niche (oral), particularly during inflammation.
Figure 2.
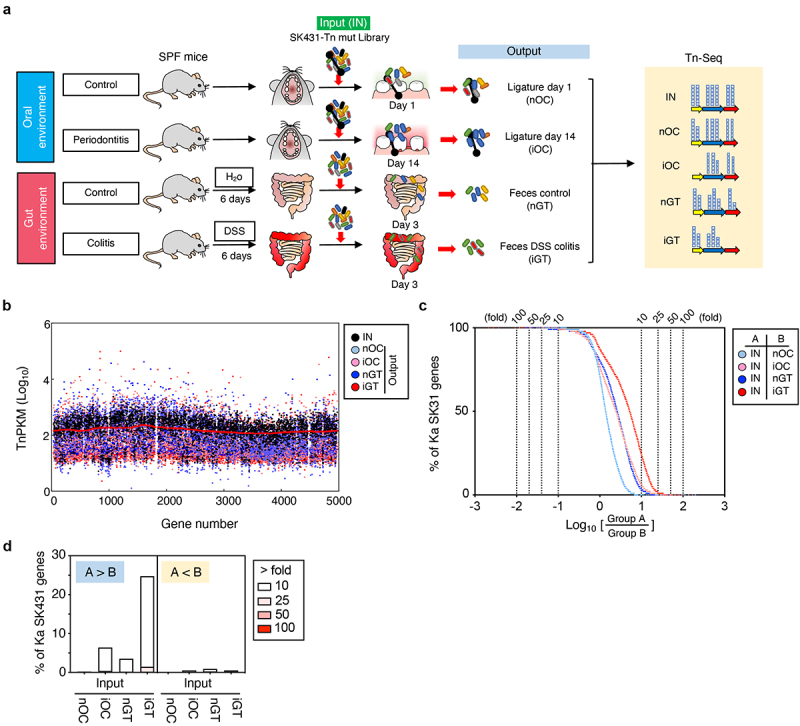
Experimental design and overview of genes required for the colonization in the steady state and the inflamed oral and gut mucosae. (a) A transposon (tn) mutant (mut) library of Ka SK431 was inoculated into specific pathogen-free (SPF) C57BL/6 mice by soaked ligature and orogastric gavage for the oral and gut sites, respectively (input: IN). Ligature and feces were collected from the healthy control mice and the inflamed (periodontitis or colitis) mice on indicated days after inoculation (output). DNA was isolated from the input and output samples and sequenced by Illumina NovaSeq. The resulting reads were mapped to the Ka SK431 genome, and the abundance of reads at each insertion site from all output samples were compared to those for the input samples to determine a fold-change value for each gene. (b) Genome-wide distribution and frequency of mutations in the library. Individual tn densities per kilobase gene per million reads (TnPKM) are shown as dots on the map of Ka KCTC2190 complete genome, instead of incomplete SK431 genome. Red line, 3,000 gene moving average of IN. (c) Number of genes that were required for the colonization in the indicated locations and conditions. The log ratio of the average TnPKM of indicated group a per group B for individual genes is shown on the X-axis. The accumulation curves of the number of the gene from the top (rich in group B) to the bottom (rich in group A) are shown on the Y-axis. (d) Graph summarizing the results of (C).
Identification of metabolic pathways required for colonization in the oral and gut mucosae
To identify the metabolic pathways that are required for colonization in the healthy and inflamed oral and gut environments, we first obtained the Tn densities per kilobase gene per million reads (TnPKM) sum for the entire metabolic pathways and compared to those of paired groups by the signal-to-noise ratio (SNR) (IN vs iOC [Figure 3, open bars] and IN vs iGT [Figure 3, filled bars]). Pathways required for nitrogen metabolism and quinone cofactors synthesis were significantly enriched in the input compared to iOC, suggesting that these pathways are required for colonization of Ka SK431 in the inflamed oral mucosa (Figure 3). Likewise, pathways associated with triacylglycerol metabolism, DNA metabolism, inorganic sulfur assimilation, sulfur metabolism, proline and 4-hydroxyproline metabolism, branched-chain amino acids (AA) metabolism, glycogen metabolism, pyridoxine, capsular and extracellular polysaccharides synthesis, cell wall and capsule synthesis, phosphorus metabolism, iron acquisition and metabolism, and cation transports were significantly enriched in the input compared to iGT (Figure 3), indicating the importance of these pathways in the ectopic gut colonization of Ka SK431, especially in the inflamed condition.
Figure 3.
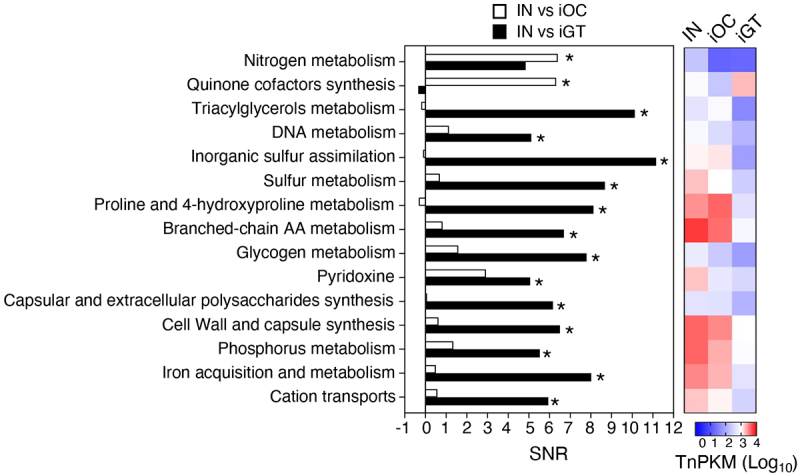
Functional pathways required for the colonization in the oral and gut mucosae. The genes were grouped in functional categories and an SNR analysis was performed using input/output ratios of TnPKM sums (left). The heatmap (right) shows the transposon mutant gene abundance (log10 averaged TnPKM, n = 4 to 6 mice) in the input (IN), the inflamed oral cavity (iOC), and the inflamed gut (iGT). The positive values of SNR indicate more in input. *, statistically significant TnPKM ratio (fold differences > 10, absolute SNR (|SNR|) > 5, p < 0.05, FDR < 0.05). AA, amino acid; TnPKM, transposon (tn) densities per kilobase per million mapped reads; SNR, signal-to-noise ratio; FDR, false discovery rate.
Identification of virulence factors required for the colonization in the oral and gut mucosae
Ka SK431 is an oral pathobiont that can ectopically colonize the gut, causing colitis.14 Like other bacterial strains belonging to the Enterobacteriaceae family, Ka SK431 harbors various virulence-related factors (Figure 1b). However, it remains uncertain which virulence factors are required to colonize the original niche (i.e., the oral mucosa) and the ectopic niche (i.e., the gut mucosa). To identify virulence genes required for mucosal site-specific adaptation, we analyzed the relative abundance of mutants in virulence genes between the input and outputs. We found that several genes were required to colonize the oral and gut mucosae (Figure 4). For example, putative competition factors (T6SS1 and T6SS2) and adhesion factors (Fim, Cfa, CUP2, CUP5, and CUP 6) are required for the colonization in the inflamed oral cavity (iOC) (Figure 4, right). Likewise, genes related to iron acquisition (Sit, Sal, Ent, Chu), competition (T6SS1 and T6SS2), and adhesion (Fim, Cfa, Ecp, CUP1, CUP3, CUP5, and CUP6) were necessary for the colonization in the inflamed gut (iGT). Notably, most of these genes are essential only in the inflamed condition, not in the steady state, in neither the oral nor the gut mucosae, suggesting that Ka SK431 uses virulence factors to adapt to the inflamed mucosal environment. Also, these results showed that Ka SK431 uses more complex virulence mechanisms to adapt to a distant mucosal site – the gut.
Figure 4.
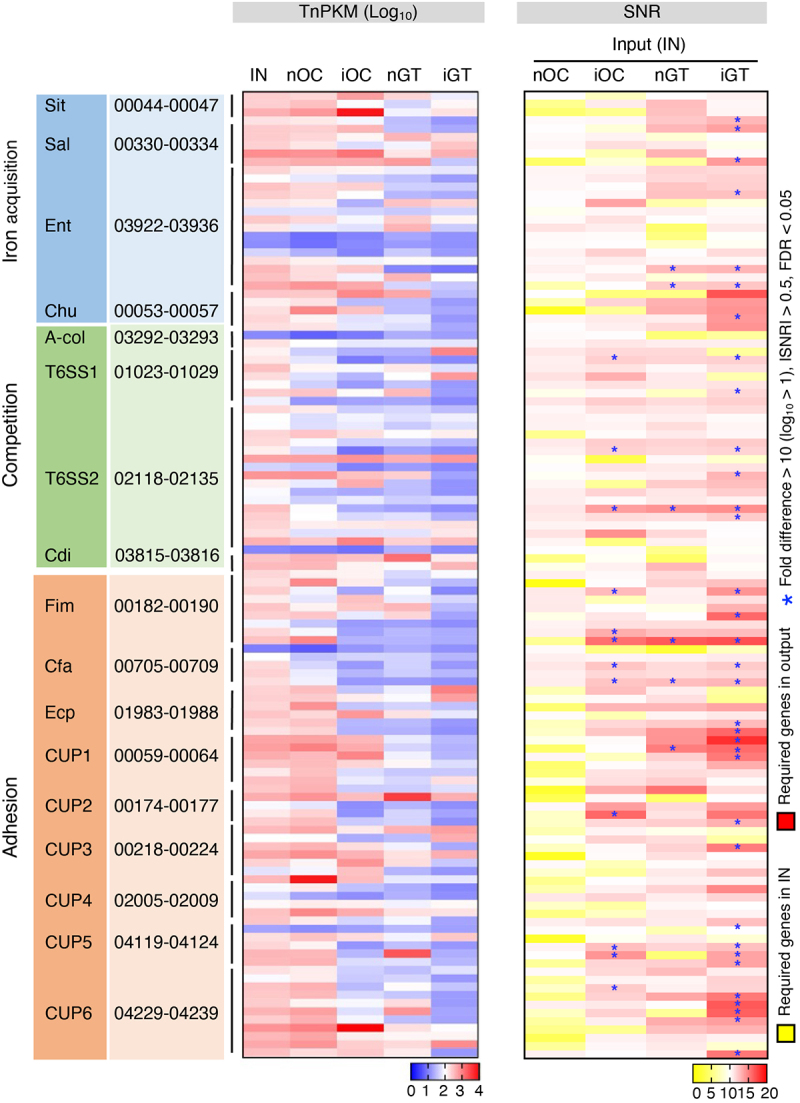
Virulence genes required by K. aerogenes SK431 for colonization in differential mucosal sites. The relative tn abundance (log10TnPKM, left) and signal-to-noise (input to output) ratio (SNR, right) of virulence gene mutants of indicated groups are depicted in heatmaps. The gene IDs of each operon are listed. T6SS1 and T6SS2 represent two core operons of T6SS. [Left; TnPKM]: higher gene abundance is indicated in red, while lower abundance is shown in blue. [Right; SNR]: genes required in input (IN) condition are indicated in yellow. Genes required in output conditions are shown in red. *, genes with significant differences (fold differences > 10, |SNR| > 0.5, FDR < 0.05, p < 0.05).
K. aerogenes uses CUP1-mediated binding to colonize the inflamed gut mucosa
Next, we attempted to validate the requirement of genes required for ectopic gut colonization by Ka SK431. To this end, we focused on the importance of CUP-mediated adhesion, as Tn-Seq data showed that CUP systems are required for ectopic gut colonization by Ka SK431 (Figure 4). In particular, we focused on the CUP1 system, which is located in the LIG and appears to be essential for colonization in the inflamed gut based on the Input/iGT ratio (Figure 4). To confirm the extent to which CUP1 contributes to the colonization of K. aerogenes in the gut mucosa, we first generated an isogenic mutant for the CUP1 operon and assessed the adhesiveness of the mutant strain to human colonic epithelial cell line T84 cells (Figure 5). As a control, the strain lacking CUP2, which is dispensable for gut colonization (Figure 4), was used. As expected, the Ka SK431 CUP1, but not CUP2, mutant strain showed a significant defect in the adhesion to the colonic epithelium cell line (Figure 5a). Next, we examined the importance of CUP1 in Ka fitness in the inflamed gut. To this end, WT and mutant (CUP1 or CUP2) Ka SK431 strains were co-inoculated into the oral cavity or the gut of SPF mice – both steady state and inflamed conditions – and the in vivo fitness of these strains in the different mucosal environments was assessed. WT and CUP1 mutant strains colonized equally in the oral cavity both in the steady state and inflammation (Figure 5b). In contrast, the CUP1 mutant exhibited a significant defect in its fitness in the inflamed gut (Figure 5b). CUP2 is not required for the fitness of Ka SK431 in both the oral and gut environments even in the presence of inflammation (Figure 5b). These results suggest that CUP1-mediated adhesion to the colonic epithelium is crucial for the adaptation of K. aerogenes to the gut environment, particularly in the presence of inflammation.
Figure 5.
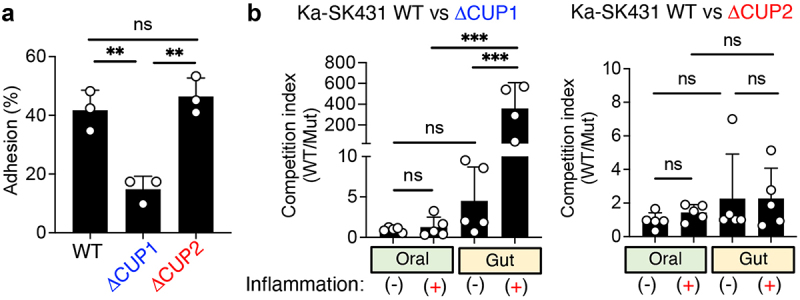
CUP1 genes are required for the ectopic colonization of K. aerogenes in the inflamed gut. (A) Ka SK431 WT, CUP1, or CUFP2 strains were co-cultured with human colonic epithelial cell line T84 for 3 hours. T84 cells were then washed, and adherent bacteria were cultured on LB plates. The percentages of adherent bacteria relative to inoculum are shown. (B) For the oral niche, WT and mutant (CUP1 or CUP2) Ka SK431 strains were co-inoculated into SPF C57BL/6 mice by soaked ligature. Inserted ligatures were harvested on day 1 (inflammation-) or day 14 (inflammation+). CFUs of colonized strains were measured by plating in selective media. For the gut niche, WT and mutant Ka SK431 strains were co-inoculated into SPF B6 mice (naïve and DSS day 5) by oral gavage. Inoculated bacterial strains were then harvested 3 days post-challenge. The competition indexes (Wt/mutant) are shown. Results are the mean ± SD. ns; not significant, **; p < 0.01, ***; p < 0.001 by 1-way ANOVA with Bonferroni post hoc test.
Discussion
In the current study, we have demonstrated that the oral pathobiont Ka SK431 exploits distinct virulence and metabolic strategies to establish colonization depending on the environment in which it resides – the oral or gut mucosa. Ka strains, including oral pathobiont strain SK431, harbor several putative virulence genes associated with iron acquisition (Sit, Sal, Aer, Ent, and Chu), competition with other bacteria (T6SS), and adhesion to the host epithelium (Fim, Cfa, Ecp, and CUP1–6) (Figure 1b). Notably, some of these virulence genes (Sit, Chu, and CUP1) are present within a single locus (Figure 1c). More importantly, the locus, namely the LIG, was found to be important for the adaptation of orally derived Ka SK431 in the ectopic gut niche, particularly during inflammation.
Adhesion to host tissues is a crucial first step for bacteria pathogens to establish infection and elicit disease responses. Fimbriae and pili are non-flagellar filamentous appendages present on the bacterial cell surface that are known to play an important role in the adhesion to host epithelial cells.27,28 Klebsiella spp. are known to employ fimbriae to colonize the mucosal tissues and cause inflammation. For example, K. pneumoniae forms a biofilm using type 1 and type 3 fimbriae to establish colonization in the bladder during urinary tract infections.29,30 In this study, we found that Ka strains harbor various types of adhesion molecules, including Fim, Cfa, Ecp, and CUP systems 1–6. Although Fim, CFA, and Ecp are highly conserved in various bacterial species belonging to the family of Enterobacteriaceae, such as E. coli, CUP systems are present only in Ka and selected strains of other Klebsiella (Figure 1b, c). In particular, CUP1 is located in the LIG (Figure 1c). Thus, CUP1 may be involved in ectopic gut colonization by orally derived Ka. Consistent with this notion, we found that CUP1 is required for the adhesion of oral Ka SK431 to the colonic epithelial cells (Figure 5a). Moreover, CUP1 is essential for the fitness of oral Ka SK431 in the inflamed gut environment (Figure 5b). Of note, CUP1 is not required for colonization in the oral mucosa, while other fimbrial genes, Fim and Cfa, are required for colonization in both the oral and gut mucosae. These results suggest that the CUP systems may be the central adhesion mechanism for Ka utilized in the ectopic niche. In this regard, CUP1 is present in the IBD-associated K2 clade K. pneumoniae strains (e.g., Kp KSB1_4E, Kp FDAARGOS_156)31 (Figure S3). This fact indicates that CUP1-mediated adhesion of Klebsiella strains (some probably orally derived) to colonic epithelial cells may be a crucial process in developing pathobiont-driven inflammation in IBD. Nonetheless, it remains possible that not all strains of Klebsiella harboring the CUP1 system can serve as colitogenic pathobionts through ectopic gut colonization. As shown in Figure 1, the Ka SK431 strain harbors the A-Col competition factor, while other analyzed K. aerogenes strains lack this factor. It is conceivable that, in addition to CUP1, A-Col is necessary for gut ectopic colonization by K aerogenes. If this were the case, only K. aerogenes strains that possess both CUP1 and A-Col, such as strain SK431, could serve as ‘colitogenic’ oral pathobionts. Future studies with other Klebsiella strains carrying CUP1 but naturally lacking A-Col (such as Ka AR0161), and/or a Ka SK431 mutant deleted in such factor, will test this hypothesis.
The Chu and Sit operons residing in the LIG contribute to iron metabolism in Ka and promote fitness in the inflammatory microenvironment, particularly in the gut mucosa. Iron is an essential nutrient for both the host and most microbes. During inflammation, however, the host produces several iron-binding proteins, such as transferrin, lactoferrin, and lipocalin-2, and scavenges available iron to limit the excessive growth of bacteria.32 To overcome this host-mediated iron limitation, pathobionts have evolved strategies to efficiently acquire iron in limited-iron environments. The Chu system is utilized for heme uptake, transport, utilization, and degradation, thereby allowing bacteria to grow in iron-poor, heme-rich environments, such as damaged mucosal tissues.33,34 The Sit system exists in most pathogenic enterobacteria, such as E. coli, Salmonella,35 Shigella,36,37 and Yersinia,38 but is generally absent in nonpathogenic bacteria. Genes encoded in the Sit operon are responsible for ferrous iron transport in low-oxygen environments. In addition to the Chu and Sit, Ka strains also have multiple siderophores (e.g., enterobactin, salmochelin, and aerobactin) in non-LIG genetic regions (Figure 1b). These siderophores are conserved in other Klebsiella spp. and are known to contribute to their in vivo growth and virulence.39–41 Enterobactin is known to show a greater affinity to iron than host molecules such as transferrin and lactoferrin.42 Also, salmochelin can counteract host-mediated iron starvation during inflammation.43–45 Interestingly, these iron acquisition genes are not required for the colonization in the oral mucosa even during inflammation (Figure 4).
In addition to the LIG virulence, the Tn-Seq data exhibited that Ka SK431 may use T6SSs to colonize the inflamed gut. T6SSs were first identified in V. cholerae as a syringe-like apparatus anchored within the bacterial cell membrane that serves to inject various effector molecules and toxins into target cells.46,47 T6SSs target both other bacteria and eukaryotic cells, suggesting a dual role in bacterial competition and pathogenesis. Based on our bioinformatics analysis, Ka SK431 harbors putative T6SS gene clusters within 2 loci. In K. pneumoniae, the T6SS is required for competition within the same bacterial species (i.e., intra-species competition), between different bacterial species (i.e., inter-species competition), and with fungi (i.e., inter-kingdom competition).48 Further studies are needed to unravel the mechanisms by which the oral pathobiont Ka SK431 exploits its T6SSs to compete with gut resident microbes to establish ectopic colonization in the gut. Clearly, Ka SK431 requires multiple virulence mechanisms to adapt to its ectopic microenvironment – the gut mucosa. Notably, not all functional genes required for gut adaptation of Ka SK431 are essential to establish the colonization in its native niche (i.e., the oral mucosa). These results suggest that oral pathobionts use complex virulence mechanisms to adapt to ectopic mucosal sites, such as the GI tract.
In human IBD, accumulating evidence indicates oral bacteria, likely oral pathobionts, colonize the gut mucosa.8,9 However, it remains unclear whether these bacteria play a causal role in triggering inflammation or colonize secondary to inflamed tissues, exacerbating inflammation further. In this regard, it has been reported that ectopic gut colonization by some oral pathobionts does not lead to the development of colitis in normal mice, while these pathobionts trigger colitis in genetically susceptible mice (i.e., IL-10-deficient mice) or exacerbates DSS-induced colitis.13,14 For example, we reported that mice with periodontitis do not develop colitis.14 Similarly, gut colonization of wild-type GF mice with oral pathobionts, including Ka SK431, does not induce colitis.14 These results suggest that periodontitis alone does not trigger gastrointestinal pathology in normal (i.e., WT) mice, despite promoting oral pathobiont expansion and their natural gut translocation. On the other hand, gut colonization by oral pathobionts induces colitis in IL-10-deficient IBD-prone mice or exacerbates DSS-induced colitis.14 This finding is consistent with human oral pathobionts.13 Gut colonization of the K. pneumoniae strain Kp-2H7, isolated from a CD patient’s saliva, induces colitis in IL-10-deficient mice but not in WT mice. Thus, in human IBD, it is conceivable that ectopic gut colonization by oral pathobionts might cause inflammation in a subset of patients with genetic susceptibilities (e.g., mutations in IL-10 signaling-related pathway genes, such as IL10 and IL10R). However, even in patients without genetic susceptibility related to responses to oral pathobionts, ectopic gut colonization by oral pathobionts can exacerbate inflammation caused by different factors. In this study, we used normal WT mice for oral and gut colonization by an oral pathobiont Ka SK431, and therefore employed DSS administration to induce colitis. This condition may not fully replicate what occurs in human IBD patients with genetic susceptibility. However, the approach used in this study can mimic the setting of colitis exacerbation by oral pathobionts, which may also occur in some IBD patients. Hence, the knowledge obtained through this study may, at least in part, translate to the understanding of the mechanisms underlying the oral-gut axis in human IBD. This study successfully identified genes required for an oral pathobiont Ka SK431 to adapt to the inflamed gut mucosa. Notably, these genes, including CUP1, are not necessary for colonization in the inflamed oral mucosa. Thus, the genes identified in this study are not merely factors involved in tolerating the inflammatory environment; instead, they are required for site-specific adaptation.
In this study, we used Ka as a model oral pathobiont, as we and others reported Klebsiella spp. reside in the oral cavity in humans and mice, particularly during periodontal disease,13,14,49,50 and, more importantly, orally derived Klebsiella can serve as colitogenic pathobionts through ectopic gut colonization.13,14 However, Enterobacteriaceae such as Klebsiella spp. are not considered classical oral pathobionts of humans and may be enriched only in a subset of patients with oral diseases. In this context, it would be interesting to examine if other major oral pathobionts use similar adaptation mechanisms in their ectopic niche and cause gut pathologies. In this regard, the genus Haemophilus, a major constituent of the oral resident microbiota and a known contributor to oral disease,51,52 utilizes iron/heme acquisition for growth53 and CUP for mucosal colonization.54–56 Thus, it is possible that the ectopic colonization mechanisms identified in this study may also be used in a wider range of oral pathobionts. Collectively, the current study provides valuable insights, as a proof of concept, into the possible site-specific adaptation mechanisms used by oral pathobionts.
Methods
Bacteria and mice
The oral pathobiont Ka SK43114 was cultured in Luria Broth (LB) medium and stored at −80°C in 25 (v/v) % glycerol until use. Specific pathogen-free (SPF) and germ-free (GF) C57BL/6 (B6) mice (age 8–12 weeks, female and male) were obtained from The Jackson Laboratory (Bar Harbor, Maine) and bred under SPF and GF conditions in our mouse facility at the University of Michigan. All SPF mice were regularly maintained on conventional rodent chow (Laboratory Rodent Diet 5001, LabDiet, St. Louis, MO). The gut microbiotas of SPF mice were normalized by exchanging their bedding 2–3 times a week for 2 weeks before use. All GF mice were housed in flexible film isolators, provided with distilled water ad libitum, and fed Autoclavable Rodent Breeder Diet 5013 (LabDiet). Their GF status was checked weekly by aerobic and anaerobic culture. The absence of microbiota was verified by microscopic analysis of stained cecal contents, which detects any unculturable contamination. All animal study protocols were approved by the Institutional Animal Care & Use Committee at the University of Michigan.
Transposon mutant library generation and inoculation
The transposon mutant library was generated by the conjugative transfer of the suicide plasmid pSAM_Cam, carrying a mariner-based transposon flanking the kanamycin resistant (KanR) cassette,19,20 from the donor strain E. coli S17–1 (KanR, ampicillin sensitive [AmpS]) to the recipient Ka strain SK431 (Ampicillin resistant [AmpR], Kanamycin sensitive [KanS]). A library of random transposon mutants was created by mating a mid-log phase culture of E. coli S17–1 with a pre-heat shock (20 min, 42 ºC) mid-log phase culture of Ka SK431 at a 1:1 ratio. Mating mixtures were centrifuged (4,500 rpm, 10 min), washed with PBS, spread onto 0.22 µm filter discs (MilliporeSigma, Burlington, MA), plated on LB agar plates with 250 µM IPTG (Thermo Fisher Scientific, Waltham, MA), and incubated at 37°C for 5 h. Then, the filter discs were transferred to tubes and rinsed with 1 mL PBS. The bacterial suspensions were plated onto LB agar plus 50 µg/mL Kan and 100 µg/mL Amp and incubated at 37°C overnight to isolate the SK431 mutants. The mutants were flooded with PBS and harvested using a cell spreader. Multiple conjugation experiments were conducted, and mutants from multiple plates were pooled. The pooled stock solution, containing ~ 63,000 SK431 mutant colonies, was stored in 25% glycerol at −80°C until needed. To prepare the inoculum (i.e., input), an aliquot of the frozen stock was thawed at 4°C and cultured in LB medium at 37°C with shaking until the culture reached an OD600 0.6–0.8.
Experimental periodontitis model in mice
Periodontitis was induced by ligature placement.57 For the Ka-induced periodontitis model, Ka SK431 (109 CFU) was inoculated into GF C57BL/6 mice. After 1 week, Ka SK431 mice were subjected to ligature silk suture thread (SUT-15-1; Roboz Surgical Instrument Co., Gaithersburg, MD) for 3 hours or 14 days and inserted between the first and second maxillary molars on the contralateral right and left sides (i.e., ligature placement) in GF B6 mice. Then mice were euthanized after the treatment of ligature insertion. Maxilla tissues were used for histological evaluation of periodontitis. Tissue biopsies from the maxillae were fixed in 4% paraformaldehyde. Alveolar bone loss was measured by micro – computed tomography (micro-CT) and visualized by MicroView Standard software (Parallax Innovations, 2.5.0–3139), and sections were stained with hematoxylin and eosin (HE) for histological analysis. Histological scores were assigned blindly by a trained pathologist who graded the severity of periodontitis from 0 to 4 as described.57–60
For Tn-Seq analysis, silk suture thread was soaked with the 109 CFU of Ka SK431 WT strain or SK431 Tn mutant library for 3 hours, and then inserted into SPF mice. The oral ligatures remained in place for 1 day (control group) or 14 days (inflamed group). Mice with periodontitis (14 day-ligature) or without periodontitis (1-day ligature) were euthanized, and the oral ligatures were removed. The ligatures were suspended in PBS, plated onto LB agar plus 50 µg/mL Kan and 100 µg/mL Amp, and incubated at 37°C overnight to harvest SK431 mutants colonized in the oral cavity (i.e., oral ligatures). Bacterial colonies were flooded with PBS and harvested using a cell spreader for bacterial DNA extraction.
Experimental colitis model in mice
A dextran sulfate sodium (DSS) – induced colitis model was used in mice. For the Tn-Seq analysis, colitis was induced in SPF mice by administering 2 (w/v) % DSS for 6 days. On day 6 post DSS treatment, the SK431 Tn mutant library (1 × 109 CFU/mouse) was administered by oral gavage. Colonized SK431 mutants were harvested from feces at 3 days post-challenge. Harvested feces were plated on LB agar plus 50 µg/mL Kan and 100 µg/mL Amp and incubated at 37°C overnight. SK431 mutants were flooded with PBS and collected using a cell spreader and used for bacterial DNA extraction. The presence of inflammation was confirmed by measuring fecal lipocalin 2 (Lcn2) levels.61
Genome analysis of K. aerogenes SK431
DNA was isolated from an overnight culture of Ka SK431 and subjected to random sequencing by Illumina MiSeq as described.62 The resulting paired-end sequences were assembled to contigs by SPAdes.63 The genes of SK431 and other reference genomes, whose complete sequences were obtained from NCBI GenBank, were annotated by Prokka.64 Putative virulence genes were predicted by BLASTP with virulence factor database (VFDB) entries.23 Orthologue gene groups and the phylogenetic tree distances were determined by roary.22 The Newick file of the resulting phylogenetic tree was visualized by TreeGraph 2.65 To compare Tn mutation frequencies at pathway level among groups, total TnPKM of pathways were calculated based on the subsystem categories of RAST.66 Synteny maps were obtained using SyntTax.67
Mutant library sequencing and bioinformatic analyses
Bacterial DNA was purified using the DNeasy Blood & Tissue Kit (Qiagen Sciences, Germantown, MD) from the pathogen library grown in vitro (input), and mutants recovered from selective LB plates from the ligature (oral) and fecal (gut) samples (output).68 Preparation of the libraries for Illumina sequencing was performed following a published protocol.69 Briefly, the NEB Ultra II FS kit (NEB #E6177L, New England Biolabs, Ipswich, MA) was used for enzymatic fragmentation of genomic DNA, end-repair of fragments, and ligation of the adaptor. Then, the transposon – genome junctions were enriched by PCR and indexed using NEBNext Multiplex Oligos (NEB #E7600S, New England Biolabs). The libraries were quantified by the University of Michigan Advanced Genomics Core using the Agilent TapeStation system (Agilent, Santa Clara, CA) and by quantitative PCR (qPCR) using a KAPA Library Quantification Kit for Illumina sequencing platforms (Kapa Biosystems, Roche, Wilmington, MA). Samples were multiplexed and sequenced with a 15% spike-in of PhiX DNA in an Illumina NovaSeq platform (Illumina, San Diego, CA) using a 200-cycle single-end read at the same facility. Tn sequences were mapped to the genome of SK43114 using Bowtie 270 and annotated/counted by HTSeq).71 Required genes were defined as the relative Tn abundance per kilobase gene length per million reads (TnPKM) in the output divided by the input. Total frequencies of the pathways were calculated based on the subsystem categories of RAST66 using signal-to-noise ratios (SNR) between two groups as determined by Morpheus (https://software.broadinstitute.org/morpheus/).
Generation of mutant Ka strains
The isogenic deletion mutants were constructed using the λ-Red recombinase system as previously described.72 Briefly, deletion cassettes were amplified by PCR with specific primers (Table S2) using the plasmid pKD4 (Kanamycin resistance cassette). The resulting PCR products were purified using a QIAquick PCR purification kit (Qiagen Sciences, Germantown, MD). Cultures of SK431 strain containing the pKD47 (λ-Red recombinase, Spectinomycin resistant) plasmid were induced with 1 mM L-Arabinose (final concentration) and then individually transformed with the purified PCR products using a MicroPluser Electroporator (BioRad, Hercules, CA). Mutants were selected onto LB agar plates containing kanamycin and streptomycin.
Bacteria adhesion assay
T84 cells (human colonic epithelial cells, ATCC CCL-248) were purchased from the American Type Culture Collection (ATCC) and cultured in Ham’s F-12 nutrient mixture + DMEM (1:1) supplemented with 10% FBS and antibiotics (penicillin-streptomycin). T84 cells were seeded onto 24-well plates (2 × 105 cells/well in 500 μl) and cultured for 3 weeks. Polarized cells were then washed three times with ice-cold PBS, and then added DMEM media without antibiotics. SK431 and mutant strains (2 × 106 cells/well) were then added to the T84 cell culture. Cell culture plates were then centrifuged (900 g for 10 min) to promote bacterial attachment to the cells, then incubated for 3 h at 37°C with 20% O2 and 5% CO2. After 3 h, cells were washed thoroughly to remove unattached bacteria, and the adhered bacteria were then quantified by culturing on LB agar plates.
Competitive colonization of Ka WT/mutant strains in vivo
For the oral colonization, SPF B6 mice were inserted with oral ligatures (soaked with the SK431 WT and a mutant with a 1:1 ratio [1 × 109 CFU each]) for 1 day (no inflammation) or 14 days (inflamed), respectively. After the indicated days, ligatures were harvested and resuspended in sterile PBS. For the gut colonization, SPF B6 mice were treated with 2% DSS or regular water (control) for 6 days. At day 6 post-DSS treatment, Ka SK431 strains (WT and a mutant with a 1:1 ratio [1 × 109 CFU each]) were inoculated. Three days after bacterial inoculation, feces were collected and resuspended in sterile PBS. Bacterial loads were determined by plating the suspended bacterial material on LB agar plates containing the appropriate antibiotics.
Quantification and statistical analysis
Statistical analyses were performed using GraphPad Prism 10 software (GraphPad Software, San Diego, CA). Differences between two groups were evaluated using a 2-tailed unpaired Student t test and Mann – Whitney U test for parametric and non-parametric datasets (determined by Shapiro-Wilk test and F test), respectively. Differences among multiple groups were evaluated using the Dunnett test or the One-Way ANOVA with Bonferroni post-hoc test. Statistical significance in highly repetitive comparison between two groups was evaluated by false positive discovery rate (FDR) analysis using Morpheus. Differences at p < 0.05 and FDR < 0.05 were considered significant.
Supplementary Material
Acknowledgments
The authors wish to thank the University of Michigan Center for Gastrointestinal Research (NIH 5P30DK034933), and the Host Microbiome Initiative, the Germ-Free Mouse Facility, the Advanced Genomics Core for technical assistance, and the In-Vivo Animal for histological evaluation, all at the University of Michigan. This work was supported by the National Institutes of Health grants DK119219 (to N.K.), JSPS KAKENHI JP23H00404 (to N.K.), K99AI159620 (to G. C.-F.), National Natural Science Foundation of China grant 82070546 (to Y.G.), Crohn’s & Colitis Foundation Research Fellowship Award and Career Development Award (to. Y.G., Y.K., and K.S.), the Office of the Assistant Secretary of Defense for Health Affairs endorsed by the Department of Defense through the Peer-Reviewed Cancer Research Program under Award No. W81XWH2010547 (to S.K.).
Funding Statement
The work was supported by the Crohn’s and Colitis Foundation Japan Society for the Promotion of Science [JP23H00404]; National Institutes of Health [DK119219]; National Institutes of Health [DK034933]; National Natural Science Foundation of China [82070546].
Disclosure statement
The authors declare that the research was conducted without any commercial or financial relationships that could be construed as a potential conflict of interest
Author contributions
Y.G. and N.K. conceived and designed the experiments. Y.G. performed experiments with help from S.K., G.C.-F., Y.K., D.W., K.S., G.N., and C.J.A. N.I. performed bioinformatic analysis of Tn-seq data. Y.G. and N.K. wrote the manuscript with contributions from all authors.
Data availability statement
The data that support the findings of this study are available from the corresponding author, NK, upon reasonable request. The Klebsiella aerogenes SK431 genomic sequence, Tn-Seq data used in this study are available from the BioProject website (https://www.ncbi.nlm.nih.gov/bioproject/), accession numbers PRJNA763502.
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/19490976.2024.2333463
References
- 1.Deo PN, Deshmukh R.. Oral microbiome: unveiling the fundamentals. J Oral Maxillofac Pathol. 2019;23(1):122–16. doi: 10.4103/jomfp.JOMFP_304_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Escapa IF, Chen T, Huang Y, Gajare P, Dewhirst FE, Lemon KP, Xu J.. New insights into human nostril microbiome from the expanded human oral microbiome database (eHOMD): a resource for the microbiome of the human aerodigestive tract. mSystems. 2018;3(6). doi: 10.1128/mSystems.00187-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Devine DA, Marsh PD, Meade J. Modulation of host responses by oral commensal bacteria. J Oral Microbiol. 2015;7(1):26941. doi: 10.3402/jom.v7.26941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zenobia C, Herpoldt K-L, Freire M. Is the oral microbiome a source to enhance mucosal immunity against infectious diseases? NPJ Vaccines. 2021;6(1). doi: 10.1038/s41541-021-00341-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perez-Chaparro PJ, Gonçalves C, Figueiredo LC, Faveri M, Lobão E, Tamashiro N, Duarte P, Feres M. Newly identified pathogens associated with periodontitis: a systematic review. J Dent Res. 2014;93(9):846–858. doi: 10.1177/0022034514542468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bodet C, Chandad F, Grenier D. Potentiel pathogénique de Porphyromonas gingivalis, Treponema denticola et Tannerella forsythia, le complexe bactérien rouge associé à la parodontite. Pathol Biol. 2007;55(3–4):154–162. doi: 10.1016/j.patbio.2006.07.045. [DOI] [PubMed] [Google Scholar]
- 7.Krishnan K, Chen T, Paster B. A practical guide to the oral microbiome and its relation to health and disease. Oral Dis. 2017;23(3):276–286. doi: 10.1111/odi.12509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yachida S, Mizutani S, Shiroma H, Shiba S, Nakajima T, Sakamoto T, Watanabe H, Masuda K, Nishimoto Y, Kubo M. et al. Metagenomic and metabolomic analyses reveal distinct stage-specific phenotypes of the gut microbiota in colorectal cancer. Nat Med. 2019;25(6):968–976. doi: 10.1038/s41591-019-0458-7. [DOI] [PubMed] [Google Scholar]
- 9.Gevers D, Kugathasan S, Denson L, Vázquez-Baeza Y, Van Treuren W, Ren B, Schwager E, Knights D, Song S, Yassour M. et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe. 2014;15(3):382–392. doi: 10.1016/j.chom.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsuzuno T, Takahashi N, Yamada‐Hara M, Yokoji‐Takeuchi M, Sulijaya B, Aoki‐Nonaka Y, Matsugishi A, Katakura K, Tabeta K, Yamazaki K. et al. Ingestion of porphyromonas gingivalis exacerbates colitis via intestinal epithelial barrier disruption in mice. J Periodontal Res. 2021;56(2):275–288. doi: 10.1111/jre.12816. [DOI] [PubMed] [Google Scholar]
- 11.Liu L, Liang L, Liang H, Wang M, Lu B, Xue M, Deng J, Chen Y. Fusobacterium nucleatum aggravates the progression of colitis by regulating M1 macrophage polarization via AKT2 pathway. Front Immunol. 2019;10:1324. doi: 10.3389/fimmu.2019.01324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Su W, Chen Y, Cao P, Chen Y, Guo Y, Wang S, Dong W. Fusobacterium nucleatum promotes the development of ulcerative colitis by inducing the autophagic cell death of intestinal epithelial. Front Cell Infect Microbiol. 2020;10:594806. doi: 10.3389/fcimb.2020.594806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Atarashi K, Suda W, Luo C, Kawaguchi T, Motoo I, Narushima S, Kiguchi Y, Yasuma K, Watanabe E, Tanoue T. et al. Ectopic colonization of oral bacteria in the intestine drives TH1 cell induction and inflammation. Science. 2017;358(6361):359–365. doi: 10.1126/science.aan4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kitamoto S, Nagao-Kitamoto H, Jiao Y, Gillilland MG, Hayashi A, Imai J, Sugihara K, Miyoshi M, Brazil JC, Kuffa P. et al. The intermucosal connection between the mouth and gut in commensal pathobiont-driven colitis. Cell. 2020;182(2):447–462.e414. doi: 10.1016/j.cell.2020.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Papageorgiou SN, Hagner M, Nogueira AV, Franke A, Jager A, Deschner J. Inflammatory bowel disease and oral health: systematic review and a meta-analysis. J Clin Periodontol. 2017;44(4):382–393. doi: 10.1111/jcpe.12698. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Y, Qiao D, Chen R, Zhu F, Gong J, Yan F. The association between periodontitis and inflammatory bowel disease: a systematic review and meta-analysis. Biomed Res Int. 2021;2021:6692420. doi: 10.1155/2021/6692420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Imai J, Ichikawa H, Kitamoto S, Golob JL, Kaneko M, Nagata J, Takahashi M, Gillilland MG, Tanaka R, Nagao-Kitamoto H. et al. A potential pathogenic association between periodontal disease and Crohn’s disease. JCI Insight. 2021;6(23). doi: 10.1172/jci.insight.148543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sohn J, Li L, Zhang L, Genco RJ, Falkner KL, Tettelin H, Rowsam AM, Smiraglia DJ, Novak JM, Diaz PI. et al. Periodontal disease is associated with increased gut colonization of pathogenic Haemophilus parainfluenzae in patients with Crohn’s disease. Cell Rep. 2023;42(2):112120. doi: 10.1016/j.celrep.2023.112120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Caballero-Flores G, Pickard JM, Fukuda S, Inohara N, Nunez G. An enteric pathogen subverts colonization resistance by evading competition for amino acids in the gut. Cell Host Microbe. 2020;28(4):526–533 e525. doi: 10.1016/j.chom.2020.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bachman MA, Breen P, Deornellas V, Mu Q, Zhao L, Wu W, Cavalcoli JD, Mobley HLT. Genome-wide identification of Klebsiella pneumoniae fitness genes during lung infection. mBio. 2015;6(3):e00775. doi: 10.1128/mBio.00775-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shea AE, Marzoa J, Himpsl SD, Smith SN, Zhao L, Tran L, Mobley HLT. Escherichia coli CFT073 fitness factors during urinary tract infection: identification using an ordered transposon library. Appl Environ Microbiol. 2020;86(13). doi: 10.1128/AEM.00691-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MTG, Fookes M, Falush D, Keane JA, Parkhill J. et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31(22):3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen L. VFDB: a reference database for bacterial virulence factors. Nucleic Acids Res. 2004;33:D325–D328. Database issue. doi: 10.1093/nar/gki008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ellermann M, Arthur JC. Siderophore-mediated iron acquisition and modulation of host-bacterial interactions. Free Radical Biol Med. 2017;105:68–78. doi: 10.1016/j.freeradbiomed.2016.10.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hsieh P-F, Lu Y-R, Lin T-L, Lai L-Y, Wang J-T. Klebsiella pneumoniaeType VI secretion system contributes to bacterial competition, cell invasion, type-1 fimbriae expression, and in vivo colonization. J Infect Dis. 2019;219(4):637–647. doi: 10.1093/infdis/jiy534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Opijnen T, Bodi KL, Camilli A. Tn-seq: high-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nat Methods. 2009;6(10):767–772. doi: 10.1038/nmeth.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schembri MA, Blom J, Krogfelt KA, Klemm P. Capsule and fimbria interaction in Klebsiella pneumoniae. Infect Immun. 2005;73(8):4626–4633. doi: 10.1128/IAI.73.8.4626-4633.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shanmugasundarasamy T, Karaiyagowder Govindarajan D, Kandaswamy K. A review on pilus assembly mechanisms in gram-positive and gram-negative bacteria. Cell Surf. 2022;8:100077. doi: 10.1016/j.tcsw.2022.100077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stahlhut SG, Struve C, Krogfelt KA, Reisner A. Biofilm formation of Klebsiella pneumoniae on urethral catheters requires either type 1 or type 3 fimbriae. FEMS Immunol Med Microbiol. 2012;65(2):350–359. doi: 10.1111/j.1574-695X.2012.00965.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Subashchandrabose S, Mobley HLT, Mulvey MA, Stapleton AE, Klumpp DJ. Virulence and fitness determinants of uropathogenic Escherichia coli. Microbiol Spectr. 2015;3(4). doi: 10.1128/microbiolspec.UTI-0015-2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Federici S, Kredo-Russo S, Valdés-Mas R, Kviatcovsky D, Weinstock E, Matiuhin Y, Silberberg Y, Atarashi K, Furuichi M, Oka A. et al. Targeted suppression of human IBD-associated gut microbiota commensals by phage consortia for treatment of intestinal inflammation. Cell. 2022;185(16):2879–2898 e2824. doi: 10.1016/j.cell.2022.07.003. [DOI] [PubMed] [Google Scholar]
- 32.Cassat JE, Skaar EP. Iron in infection and immunity. Cell Host Microbe. 2013;13(5):509–519. doi: 10.1016/j.chom.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nagy G, Dobrindt U, Kupfer M, Emody L, Karch H, Hacker J. Expression of hemin receptor molecule ChuA is influenced by RfaH in uropathogenic Escherichia coli strain 536. Infect Immun. 2001;69(3):1924–1928. doi: 10.1128/IAI.69.3.1924-1928.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Torres AG, Payne SM. Haem iron-transport system in enterohaemorrhagic Escherichia coli O157: H7. Mol Microbiol. 1997;23(4):825–833. doi: 10.1046/j.1365-2958.1997.2641628.x. [DOI] [PubMed] [Google Scholar]
- 35.Janakiraman A, Slauch JM. The putative iron transport system SitABCD encoded on SPI1 is required for full virulence of salmonella typhimurium. Mol Microbiol. 2000;35(5):1146–1155. doi: 10.1046/j.1365-2958.2000.01783.x. [DOI] [PubMed] [Google Scholar]
- 36.Runyen-Janecky LJ, Reeves SA, Gonzales EG, Payne SM. Contribution of the shigella flexneri sit, iuc, and feo iron acquisition systems to iron acquisition in vitro and in cultured cells. Infect Immun. 2003;71(4):1919–1928. doi: 10.1128/IAI.71.4.1919-1928.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fisher CR, Davies NM, Wyckoff EE, Feng Z, Oaks EV, Payne SM. Genetics and virulence association of the shigella flexneri sit iron transport system. Infect Immun. 2009;77(5):1992–1999. doi: 10.1128/IAI.00064-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fetherston JD, Mier I Jr., Truszczynska H, Perry RD, Payne SM. The yfe and Feo transporters are involved in microaerobic growth and virulence of Yersinia pestis in bubonic plague. Infect Immun. 2012;80(11):3880–3891. doi: 10.1128/IAI.00086-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Holden VI, Breen P, Houle S, Dozois CM, Bachman MA. Klebsiella pneumoniae Siderophores Induce Inflammation, bacterial dissemination, and HIF-1α stabilization during pneumonia. mBio. 2016;7(5). doi: 10.1128/mBio.01397-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Holden VI, Wright MS, Houle S, Collingwood A, Dozois CM, Adams MD, Bachman MA. Iron acquisition and siderophore release by carbapenem-resistant sequence type 258 Klebsiella pneumoniae. mSphere. 2018;3(2). doi: 10.1128/mSphere.00125-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Namikawa H, Niki M, Niki M, Oinuma K-I, Yamada K, Nakaie K, Tsubouchi T, Tochino Y, Takemoto Y, Kaneko Y. et al. Siderophore production as a biomarker for Klebsiella pneumoniae strains that cause sepsis: a pilot study. J Formos Med Assoc. 2021;121(4):848–855. doi: 10.1016/j.jfma.2021.06.027. [DOI] [PubMed] [Google Scholar]
- 42.Raymond KN, Dertz EA, Kim SS. Enterobactin: an archetype for microbial iron transport. Proc Natl Acad Sci U S A. 2003;100(7):3584–3588. doi: 10.1073/pnas.0630018100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Flo TH, Smith KD, Sato S, Rodriguez DJ, Holmes MA, Strong RK, Akira S, Aderem A. Lipocalin 2 mediates an innate immune response to bacterial infection by sequestrating iron. Nature. 2004;432(7019):917–921. doi: 10.1038/nature03104. [DOI] [PubMed] [Google Scholar]
- 44.Lam MMC, Wyres KL, Judd LM, Wick RR, Jenney A, Brisse S, Holt KE. Tracking key virulence loci encoding aerobactin and salmochelin siderophore synthesis in Klebsiella pneumoniae. Genome Med. 2018;10(1):77. doi: 10.1186/s13073-018-0587-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bachman MA, Miller VL, Weiser JN, Isberg RR. Mucosal lipocalin 2 has pro-inflammatory and iron-sequestering effects in response to bacterial enterobactin. PloS Pathog. 2009;5(10):e1000622. doi: 10.1371/journal.ppat.1000622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alteri CJ, Himpsl SD, Pickens SR, Lindner JR, Zora JS, Miller JE, Arno PD, Straight SW, Mobley HLT. et al. Multicellular bacteria deploy the type VI secretion system to preemptively strike neighboring cells. PloS Pathog. 2013;9(9):e1003608. doi: 10.1371/journal.ppat.1003608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alteri CJ, Mobley HLT, Kudva IT. The versatile type VI secretion system. Microbiol Spectr. 2016;4(2):337–356. doi: 10.1128/microbiolspec.VMBF-0026-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Storey D, McNally A, Åstrand M, Sa-Pessoa Graca Santos J, Rodriguez-Escudero I, Elmore B, Palacios L, Marshall H, Hobley L, Molina M. et al. Klebsiella pneumoniae type VI secretion system-mediated microbial competition is PhoPQ controlled and reactive oxygen species dependent. PloS Pathog. 2020;16(3):e1007969. doi: 10.1371/journal.ppat.1007969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goncalves MO, Coutinho-Filho WP, Pimenta FP, Pereira GA, Pereira JAA, Mattos-Guaraldi AL, Hirata R. Periodontal disease as reservoir for multi-resistant and hydrolytic enterobacterial species. Lett Appl Microbiol. 2007;44(5):488–494. doi: 10.1111/j.1472-765X.2007.02111.x. [DOI] [PubMed] [Google Scholar]
- 50.Baker JL, Hendrickson EL, Tang X, Lux R, He X, Edlund A, McLean JS, Shi W. Klebsiella and providencia emerge as lone survivors following long-term starvation of oral microbiota. Proc Natl Acad Sci U S A. 2019;116(17):8499–8504. doi: 10.1073/pnas.1820594116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Caselli E, Fabbri C, D’Accolti M, Soffritti I, Bassi C, Mazzacane S, Franchi M. Defining the oral microbiome by whole-genome sequencing and resistome analysis: the complexity of the healthy picture. BMC Microbiol. 2020;20(1):120. doi: 10.1186/s12866-020-01801-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu G, Luan Q, Chen F, Chen Z, Zhang Q, Yu X. Shift in the subgingival microbiome following scaling and root planing in generalized aggressive periodontitis. J Clin Periodontol. 2018;45(4):440–452. doi: 10.1111/jcpe.12862. [DOI] [PubMed] [Google Scholar]
- 53.Whitby PW, Seale TW, VanWagoner TM, Morton DJ, Stull TL. The iron/heme regulated genes of Haemophilus influenzae: comparative transcriptional profiling as a tool to define the species core modulon. BMC Genomics. 2009;10(1):6. doi: 10.1186/1471-2164-10-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.St Geme JW, Pinkner JS, Krasan GP, Heuser J, Bullitt E, Smith AL, Hultgren SJ. Haemophilus influenzae pili are composite structures assembled via the HifB chaperone. Proc Natl Acad Sci U S A. 1996;93(21):11913–11918. doi: 10.1073/pnas.93.21.11913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nuccio SP, Baumler AJ. Evolution of the chaperone/usher assembly pathway: fimbrial classification goes Greek. Microbiol Mol Biol Rev. 2007;71(4):551–575. doi: 10.1128/MMBR.00014-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Busch A, Waksman G. Chaperone–usher pathways: diversity and pilus assembly mechanism. Philos Trans R Soc Lond B Biol Sci. 2012;367(1592):1112–1122. doi: 10.1098/rstb.2011.0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Marchesan J, Girnary MS, Jing L, Miao MZ, Zhang S, Sun L, Morelli T, Schoenfisch MH, Inohara N, Offenbacher S. et al. An experimental murine model to study periodontitis. Nat Protoc. 2018;13(10):2247–2267. doi: 10.1038/s41596-018-0035-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Casili G, Ardizzone A, Lanza M, Gugliandolo E, Portelli M, Militi A, Cuzzocrea S, Esposito E, Paterniti I. Treatment with luteolin improves lipopolysaccharide-induced periodontal diseases in rats. Biomedicines. 2020;8(10):442. doi: 10.3390/biomedicines8100442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hiyari S, Wong RL, Yaghsezian A, Naghibi A, Tetradis S, Camargo PM, Pirih FQ. Ligature-induced peri-implantitis and periodontitis in mice. J Clin Periodontol. 2018;45(1):89–99. doi: 10.1111/jcpe.12817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Abe T, Hajishengallis G. Optimization of the ligature-induced periodontitis model in mice. J Immunol Methods. 2013;394(1–2):49–54. doi: 10.1016/j.jim.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chassaing B, Srinivasan G, Delgado MA, Young AN, Gewirtz AT, Vijay-Kumar M, Bereswill S. Fecal lipocalin 2, a sensitive and broadly dynamic non-invasive biomarker for intestinal inflammation. PloS One. 2012;7(9):e44328. doi: 10.1371/journal.pone.0044328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ohno M, Hasegawa M, Hayashi A, Caballero-Flores G, Alteri CJ, Lawley TD, Kamada N, Núñez G, Inohara N. et al. Lipopolysaccharide O structure of adherent and invasive Escherichia coli regulates intestinal inflammation via complement C3. PloS Pathog. 2020;16(10):e1008928. doi: 10.1371/journal.ppat.1008928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD. et al. Spades: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19(5):455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30(14):2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 65.Stöver BC, Müller KF. TreeGraph 2: combining and visualizing evidence from different phylogenetic analyses. BMC Bioinf. 2010;11(7). doi: 10.1186/1471-2105-11-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M. et al. The RAST server: rapid annotations using subsystems technology. BMC Genomics. 2008;9(1):75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Oberto J. SyntTax: a web server linking synteny to prokaryotic taxonomy. BMC Bioinf. 2013;14(4). doi: 10.1186/1471-2105-14-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nagao-Kitamoto H, Shreiner AB, Gillilland MG, Kitamoto S, Ishii C, Hirayama A, Kuffa P, El-Zaatari M, Grasberger H, Seekatz AM. et al. Functional characterization of inflammatory bowel disease–associated gut dysbiosis in gnotobiotic mice. Cell Mol Gastroenterol Hepatol. 2016;2(4):468–481. doi: 10.1016/j.jcmgh.2016.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Palani NP. Transposon insertion sequencing (tn-seq) library preparation protocol - includes UMI for PCR duplicate removal. 2019. [Google Scholar]
- 70.Langmead B, Salzberg SL. Fast gapped-read alignment with bowtie 2. Nat Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Anders S, Pyl PT, Huber W. Htseq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97(12):6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author, NK, upon reasonable request. The Klebsiella aerogenes SK431 genomic sequence, Tn-Seq data used in this study are available from the BioProject website (https://www.ncbi.nlm.nih.gov/bioproject/), accession numbers PRJNA763502.


