Abstract
mRNA translation relies on identifying translation initiation sites (TISs) in mRNAs. Alternative TISs are prevalent across plant transcriptomes, but the mechanisms for their recognition are unclear. Using ribosome profiling and machine learning, we developed models for predicting alternative TISs in the tomato (Solanum lycopersicum). Distinct feature sets were predictive of AUG and nonAUG TISs in 5′ untranslated regions and coding sequences, including a novel CU-rich sequence that promoted plant TIS activity, a translational enhancer found across dicots and monocots, and humans and viruses. Our results elucidate the mechanistic and evolutionary basis of TIS recognition, whereby cis-regulatory RNA signatures affect start site selection. The TIS prediction model provides global estimates of TISs to discover neglected protein-coding genes across plant genomes. The prevalence of cis-regulatory signatures across plant species, humans, and viruses suggests their broad and critical roles in reprogramming the translational landscape.
Translation initiation is the first stage of protein synthesis and is also rate limiting, as it includes the selection of the translation initiation site (TIS) in the mRNA. The choice of TIS determines the coding sequence (CDS) of mRNA and ensures the accurate and timely production of a desired protein. This mechanism enables plants to rapidly respond to developmental cues and environmental stress (Merchante et al. 2017; Urquidi Camacho et al. 2020; Fang and Liu 2023). Advanced high-throughput computational and experimental workflows can be used to annotate protein-coding genes, decode plant genomes, and identify the genetic basis of phenotypic diversity among plant species. However, the current criteria for identifying protein-coding genes, which include the presence of an AUG initiation codon, a minimum open reading frame (ORF) length of 100 amino acids, and a (most likely) single ORF in eukaryotic mRNA, limit the identification of genes with small or nonAUG-initiated ORFs and may not fully capture the complexity of plant genomes (Yandell and Ence 2012; Kearse and Wilusz 2017; Hsu and Benfey 2018). Ribosome profiling, which allows the global mapping of TISs in vivo, has revealed numerous unannotated TISs in mRNAs in plants (Juntawong et al. 2014; Hsu et al. 2016; Willems et al. 2017; Wu et al. 2019; Li and Liu 2020). These alternative TISs (i.e., the AUG and nonAUG TISs that differ from the annotated AUG sites) mainly located at AUG and near-cognate codons (i.e., codons with one base difference from AUG) direct the translation of uncharacterized ORFs encoding novel peptides/proteins or different protein isoforms. These peptides/proteins play crucial roles in stress and other physiological responses in plants (Hanada et al. 2013; Juntawong et al. 2014; Tavormina et al. 2015; Hellens et al. 2016; Hsu et al. 2016; Willems et al. 2017; van der Horst et al. 2019; Li and Liu 2020). For example, multiple Arabidopsis (Arabidopsis thaliana) small ORFs initiated at AUG encode hormone-like peptides that regulate morphogenic development and salinity stress tolerance (Hanada et al. 2013; Nakaminami et al. 2018). The tomato (Solanum lycopersicum) valyl-tRNA synthetase gene encodes both mitochondrial and cytosolic proteins via an upstream ACG and annotated AUG initiation site, respectively (Li and Liu 2020). Whereas previous annotations of protein-coding genes overlooked alternative TIS-initiated ORFs, ribosome profiling studies have revealed unexpected proteome diversity in plants (Hanada et al. 2013; Juntawong et al. 2014; Tavormina et al. 2015; Hellens et al. 2016; Hsu et al. 2016; Willems et al. 2017; van der Horst et al. 2019; Li and Liu 2020). Thus, it is crucial to elucidate the general principles of plant TIS recognition to decode plant genome sequences (Kress et al. 2022).
How do plant ribosomes recognize start sites for protein synthesis? Although thousands of unannotated AUGs and near-cognate TISs are present in the 5′ untranslated regions (UTRs) and major CDSs of plant mRNAs, ribosomes do not initiate protein synthesis at every triplet they encounter (Hanada et al. 2013; Juntawong et al. 2014; Hsu et al. 2016; Willems et al. 2017; van der Horst et al. 2019; Li and Liu 2020), highlighting the need to understand how start codons and a subset of triplets are selected in mRNA. These mechanisms depend on the sequence context and cis-regulatory RNA elements surrounding the start codon (Hinnebusch 2017; Kearse and Wilusz 2017; Orr et al. 2020). For example, Kozak sequences and specific TIS-flanking nucleotides in the –2, –4, and +5 positions (where +1 refers to A of the AUG start site) can enhance the efficiency of translation initiation (Kozak 1984; Noderer et al. 2014). Nevertheless, they do not account for all AUG- and nonAUG TIS activities. Kozak motifs are commonly found in 5′ UTR–nonAUG TISs (TISs located in 5′ UTRs and using a nonAUG codon) and CDS-AUGs TISs (located in CDSs and using an AUG codon) but not in 5′ UTR–AUG TISs (located in 5′ UTRs and using an AUG codon) in both mammals and plants (Spealman et al. 2018; Benitez-Cantos et al. 2020; Li and Liu 2020). Thus, how plant ribosomes recognize different types of start codons in mRNA is not fully understood. Several questions remain, including which sequence features determine alternative TISs in plants, how these determinants jointly and distinctly regulate AUG and nonAUG TIS selection, the similarity of these features across plant species and eukaryotes, and how much of plant genomes truly encodes proteins.
Machine learning (ML) offers one way to uncover the protein-coding genes of plant genomes and understand the mechanisms underlying plant AUG and nonAUG TIS recognition. ML can systematically identify RNA cis-regulatory codes of plant alternative TISs and provide more accurate TIS annotations. ML frameworks, which build mathematical models and identify patterns in large data sets, have successfully elucidated complex gene regulatory networks and predicted phenotypes in plants. For example, ML-based predictions of dynamic gene expression responses and identification of novel regulators have revealed the transcriptional regulatory network architectures of plant stress responses (Ma et al. 2014; Wu et al. 2021; Wang et al. 2022). ML strategies have also helped characterize novel cis-regulatory DNA elements and their combined effects in regulating transcription and associating genetic variation with differential gene expression and phenotypic diversity (Azodi et al. 2020a,b). Therefore, integrating high-throughput translation initiation sequencing data sets from different plant species using ML techniques may facilitate TIS/ORF annotations and help decipher the underlying TIS-determining principles across plants (Willems et al. 2017; Li and Liu 2020).
Here, we combined ML-based, computational, and experimental techniques to investigate how plant ribosomes recognize different types of TISs along mRNA sequences, including those initiated at both canonical AUG and nonAUG codons, and explored uncharacterized TIS-initiated ORFs in the tomato. We built ML models using TIS-flanking mRNA sequence and TIS codon usage and characterized common or species-specific sequence features. We explored their conserved regulatory role across dicot and monocot plants, viruses, and humans.
Results
ML enables cross-species TIS predictions in plants
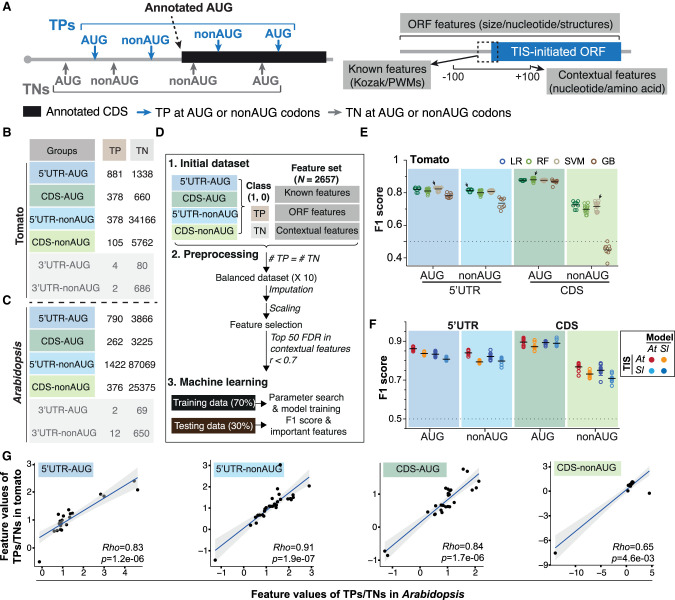
To comprehensively and precisely build models that predict TISs in plant mRNAs, we used tomato ribosome profiling data sets (Supplemental Fig. S1A; Willems et al. 2017; Li and Liu 2020) to globally profile experimentally supported alternative AUG and near-cognate TISs (referred to as true-positive [TP] TISs) and implemented an ML workflow to distinguish these TP TISs from AUG and near-cognate triplets with no significant translation initiation signals (true-negative [TN] TISs) (Fig. 1A–D). Bioinformatics and statistical analyses identified TPs (see the Supplemental Methods) and categorized them into six groups based on the locations of initiation codons and their sequences: 5′ UTR–AUG, 5′ UTR–nonAUG, CDS-AUG, CDS-nonAUG, 3′ UTR–AUG, and 3′ UTR–nonAUG (Fig. 1A, left). This allows us to explore the common and distinct regulatory mechanisms between AUG and nonAUG TIS recognition. We identified several hundred to thousands of TP TISs in the 5′ UTR and CDS but not the 3′ UTR (Fig. 1B,C; Supplemental Fig. S1). Therefore, we focused on the TP and TN TISs in the 5′ UTR and CDS for further analysis using the ML workflow illustrated in Figure 1D. The ML workflow includes the collection and selection of features comprising known (such as Kozak motifs) (Kozak 1984, 1989), ORF (such as mononucleotide contents within ORFs and ORF sizes), and contextual features (nucleotide/amino acid frequency of k-mers around TISs) (Fig. 1A, right) and their employment for the generation of TIS prediction models (see the Supplemental Methods).
Figure 1.
Identification of prediction models and the associated features that predict plant alternative translation initiation sites (TISs). (A) Illustration of the identified true-positive (TP; blue) and true-negative (TN; gray) TISs that were categorized based on the location of initiation codons (i.e., 5′ UTR, CDS, or 3′ UTR) and their sequences in a transcript. The “known” features include the Kozak motif and position weight matrices (PWMs) generated from short sequences centered on TISs (Joshi et al. 1997; Reuter et al. 2016). The “ORF” features consist of various nucleotide compositions/sizes of the alternative TIS-initiated open reading frames (ORFs) and the annotated ORFs and also the RNA structures/nucleotide compositions of their surrounding regions. The “contextual” features based on k-mer enrichment analyses (k = 1–3) consist of the nucleotide and amino acid sequence contexts in the 200-bp region centered on the TISs (see the Supplemental Methods). Gray line indicates mRNAs; black boxes, annotated AUG TIS-initiated ORFs; and blue boxes, alternative TIS-initiated ORFs. (B,C) Numbers of the identified TP and TN TISs, categorized as described in A, in the tomato (B) and Arabidopsis (C). (D) Machine-learning (ML) workflow used to identify prediction models and the features that were informative for predicting TISs (see the Supplemental Methods). The Pearson correlation coefficient (r) and the false-discovery rate (FDR; determined by Wilcoxon rank-sum test) represent the correlation and the statistical significance of differences for the feature values between TP and TN TIS sets. “Top 50 FDR in contextual features” represents that the 50 contextual features with smallest FDRs were selected for further analysis (see the Supplemental Methods). The F1 score is the harmonic mean of precision and recall, which ranges from zero to one with one indicating a perfect model. (E) The prediction performance (represented as F1 scores) when all features (i.e., the known/ORF/contextual ones) were applied in predicting the four tomato TIS groups. A circle indicates the performance of a model with a given combination of model parameters in a randomly balanced TP and TN data set. A black line indicates the mean of the F1 scores for a given ML algorithm. An arrow indicates the best model (i.e., for the ML algorithm with highest mean performance, the model with highest F1 score). A dashed line indicates the baseline performance expected by random guessing. (F) The cross-species and within-species prediction performance (shown as F1 score) when using the best model built in one species to predict the TISs in another species (light blue and orange) and to predict the TISs within the same species (red and dark blue). Results are shown for the four TIS groups in the tomato and Arabidopsis. (G) The mean of the fold changes for the feature values between TP and TN TIS sets among 10 randomly balanced data sets. Rho indicates Spearman's rank correlation coefficient. The black line indicates the fitted linear regression line, and the gray area indicates the 95% confidence interval. To exclude the possibility of bias arising from 5′ UTR lengths, the lengths of 5′ UTR for genes with and without 5′ UTR–AUG and 5′ UTR–nonAUG are shown in Supplemental Figure S14.
The F1 scores from the model using the features from all categories ranged from 0.7 to 0.9 with the highest and lowest F1 scores observed for the 5′ UTR–AUG and CDS-nonAUG groups, respectively, in the tomato (Fig. 1E, arrows; Supplemental Fig. S2A; Supplemental Table S1). Similar results were also observed in Arabidopsis (Fig. 1A,C; Supplemental Fig. S1F, arrows). Furthermore, the model using the features from all categories outperformed those using the known, ORF, or contextual categories for TIS prediction (Supplemental Figs. S1F, S2A; Supplemental Table S1). Therefore, in addition to using features with known biological functions (e.g., Kozak score), using a combination of unexplored features (e.g., ORF and contextual features) and features with known biological functions is important for TIS prediction.
To assess the generality of TIS recognition mechanisms across plants, we explored whether the models generated from tomato could be used for predictions in Arabidopsis. The best models from tomato predicted the four types of Arabidopsis TISs with F1 scores ranging from 0.73 to 0.87 (Fig. 1F, orange; Supplemental Fig. S2B; Supplemental Table S1), showing the robustness of the established ML workflow in identifying TIS prediction models across plants. We observed similar patterns using Arabidopsis models to predict tomato TISs (Fig. 1F, light blue). The feature enrichment values (ratio of TP to TN feature values) showed significant positive correlations in the tomato and Arabidopsis for all groups tested (Fig. 1G). These results suggested that information gained from one species-based model could be useful for predicting TISs in other plant species.
Shared and distinct nearby sequence contexts and codon usage bias are associated with plant AUG and nonAUG TIS predictions
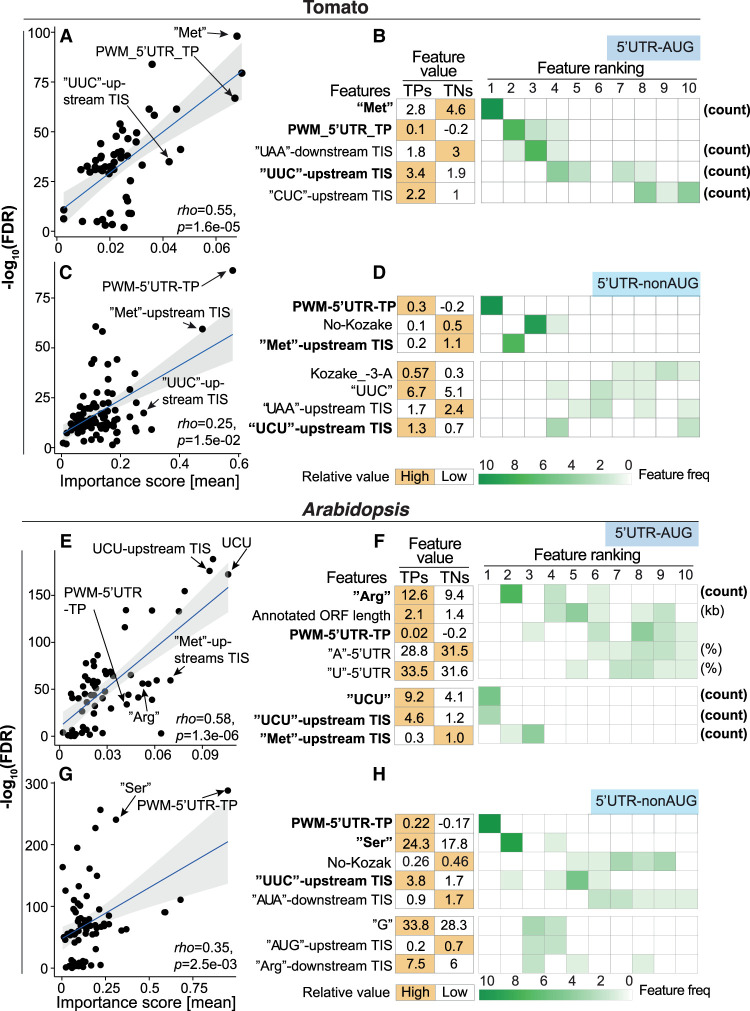
We asked to what extent each feature contributed to the performance of the TIS prediction model in plants. We identified the top features that contributed most to model performance for each TIS group in the tomato (Fig. 2A–D; Supplemental Fig. S3A). The known PWM–5′ UTR–TP feature, representing the nucleotide compositions of the flanking regions (see the Supplemental Methods; Noderer et al. 2014; Reuter et al. 2016; de Arce et al. 2018; Li and Liu 2020), substantially contributed to the accuracy of the model, especially for predicting tomato 5′ UTR–nonAUG TISs (Fig. 2C,D), as supported by its high feature importance score and significant enrichment (false-discovery rate [FDR]) (Fig. 2C), its top feature ranking and higher feature occurrence in 10 randomly balanced TP/TN data sets (Fig. 2D, right), and the differential feature values between TPs and TNs (Fig. 2D, left).
Figure 2.
The features that are most informative for predicting plant 5′ UTR TISs. (A) Comparison of the importance scores derived from the model and the statistical significance of differences (−log10(FDR), determined by a Wilcoxon signed-rank test with Bonferroni correction) between tomato 5′ UTR–AUG TPs and TNs for the features used in the best model. Rho indicates Spearman's rank correlation coefficient. The black line indicates the fitted linear regression line, and the gray area indicates the 95% confidence level interval. (B) The means of the feature values in the tomato 5′ UTR–AUG TP and TN data sets (right) and the frequency of features identified in 10 randomly balanced data sets (left) for the feature elimination–determined top 10 features (ranked using their importance) (see Supplemental Fig. S3A). The rank and frequency indicate the importance of a given feature in the prediction model and their robustness using 10 randomly balanced data sets. The features with a frequency greater than seven within the top 10 are shown. Orange indicates the TIS group with the higher feature value. (C–H) As described in A,B, but for the tomato 5′ UTR–nonAUG TIS group (C,D), the Arabidopsis 5′ UTR–AUG TIS group (E,F), and Arabidopsis 5′ UTR–nonAUG TIS group (G,H). To exclude the possibility of bias arising from random down-sampling, the correlation between two different strategies of random sampling is shown in Supplemental Figure S15.
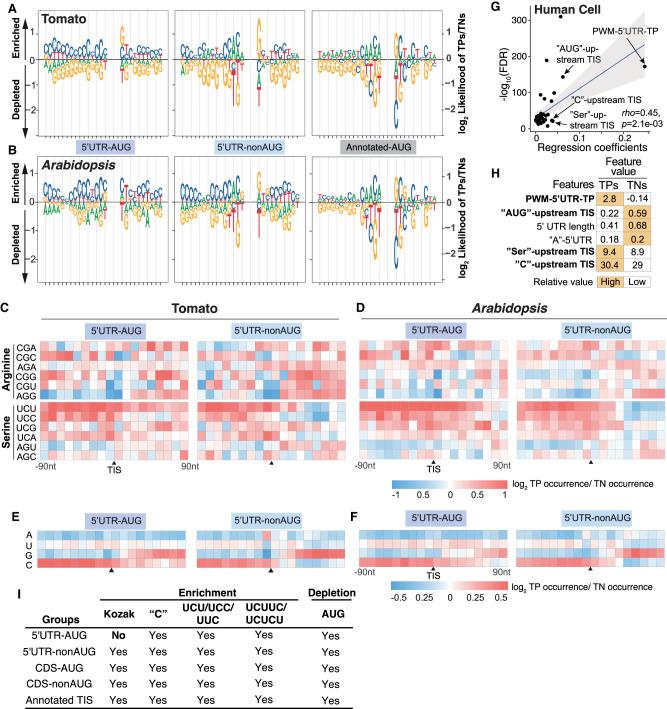
Analysis of the nucleotide compositions of the TIS-flanking regions showed that all tomato TIS groups have a high frequency of C nucleotides in nearby flanking regions (Fig. 3A; Supplemental Fig. S4A). On the other hand, we found that the 5′ UTR–nonAUG and CDS-AUG TPs, but not 5′ UTR–AUG TPs, tended to have A's at positions –4 to –1 and G and C at positions +4 and +5, respectively (Fig. 3A,I; Supplemental Fig. S4A), a pattern similar to the Kozak motif (Kozak 1984, 1989). This Kozak-motif pattern was also observed in annotated AUG TPs (Fig. 3A; Supplemental Fig. S4) and is in line with previously reported sequence requirements for mammalian TISs at AUG and nonAUG codons (Noderer et al. 2014; de Arce et al. 2018). These observations suggest that 5′ UTR–AUG, 5′ UTR-nonAUG, and CDS-AUG TISs have both shared and distinct sequence context dependencies for TIS recognition in the tomato.
Figure 3.
The C/U nucleotide compositions and the flanking sequences of 5′ UTR TISs. (A,B) Sequence logo plots showing the differential enrichment of A/U/C/G nucleotides between TPs and TNs in the regions 15-bp upstream of and 13-bp downstream from the TISs, represented as the log2 ratio of the site frequencies between TPs and TNs, for the 5′ UTR TISs and annotated AUG sites in tomato (A) and Arabidopsis (B). (C,D) Enrichment of sites with the indicated 3-mer sequences in the tomato (C) and Arabidopsis (D) TIS groups, represented as the log2 ratio of the site frequencies between TPs and TNs in the 180-bp region centered on tomato TISs with a 10-bp window. (E,F) As described in C,D, but for the A, U, C, and G mononucleotides. (G) As described in Figure 2A, but shown for the regression coefficients (x-axis) for the features used in the best linear regression model of predicting human TISs. (H) As described in Figure 2B, but for the top six features with highest regression coefficients in human TIS prediction model. (I) Summary of the ML-revealed features of the Kozak motif (Kozak), the mononucleotide C content (“C”), and CU-rich tracts for their importance across different TIS groups.
The tomato model performances were slightly worse for the nonAUG groups relative to AUG groups regardless of TIS location (Fig. 1E; Supplemental Table S1). Not every near-cognate codon serves as a TIS with equal activity, with CUG, ACG, and GUG generally being the most efficient (Kearse and Wilusz 2017; de Arce et al. 2018; Li and Liu 2020). Hence, we attempted to increase prediction accuracy by adding the feature TIS codon usage bias (i.e., enrichment values of each near-cognate codon among TP TISs) (Supplemental Fig. S1D). In general, models with TIS codon usage bias information slightly outperformed those without this feature (Supplemental Fig. S5A), and the rate of TN → TP misclassification (i.e., a TN TIS inaccurately classified as a TP TIS) decreased by 6% in the tomato (Supplemental Fig. S5B, light brown). The major codons contributing to misclassification were AAG and AGG (Supplemental Fig. S5C, arrows), suggesting that, in addition to the flanking sequences context, the codon preference of TISs themselves is also important for tomato nonAUG prediction.
We observed similar patterns when examining the prediction models of Arabidopsis TISs (Figs. 2E–H, 3B,I; Supplemental Figs. S3B,G–J, S4B, and S5), reflecting the shared sequence features between the tomato and Arabidopsis (Fig. 1G) and suggesting these features are evolutionarily conserved across plants.
CU-rich tracts are important for plant TIS predictions
We identified that the UUC and UCU short sequences, as well as the amino acids encoded by CU-rich codons such as Ser, were important for model accuracy (Fig. 2B,D,F,H). To determine whether the amino acids themselves or the encoded codons are crucial, we examined the codon usages for those amino acids. We found that the codons enriched in TP groups were biased toward C and U, regardless of the types of amino acids in both the tomato and Arabidopsis (Fig. 3C,D; Supplemental Fig. S6A–F). We further dissect codon sequences into mononucleotides and found the C/U enrichments were more obvious in four groups across species (Fig. 3E,F,I; Supplemental Fig. S6G,H). Similar C/U enrichment was also identified when we examined the TIS-predictive features from human cells (Fig. 3G,H; Reuter et al. 2016). These results suggest that CU-rich nucleotide tracts, regardless of the amino acids they encode, are important for alternative TIS prediction in all TIS groups and are a shared cis-regulatory RNA signature across eukaryotes, at least in plants and humans.
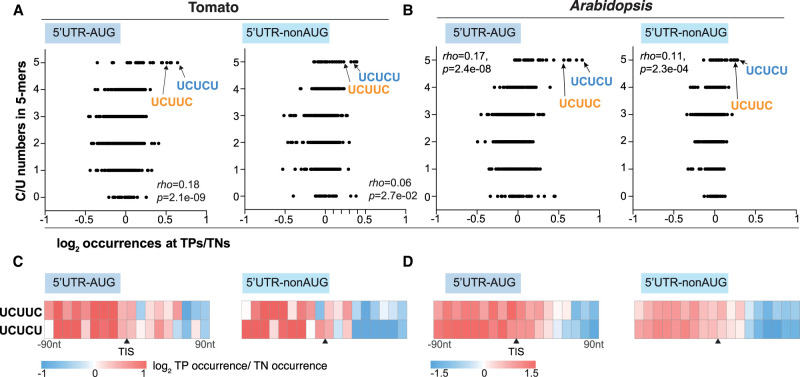
We asked whether longer specific sequences also regulate TIS selection. We conducted k-mer enrichment analyses, which started from any k-mers with length or four or more and successfully narrowed down to around 111–153 putative cis-elements, with the predominant size of 5–7 nt (Supplemental Fig. S6I) and being predictive of TISs in Arabidopsis (Supplemental Fig. S6J). Using the combined set of these cis-regulatory RNA elements (n = 444) (Supplemental Fig. S6I), we developed a TIS prediction model with F1/AUROC scores of 0.74/0.78 (Supplemental Fig. S6P,Q) with the top motifs of UCUUC and UCUCU (Supplemental Fig. S6R). The “UCUUC” and “UCUCU” motifs were significantly enriched and prevalent in the upstream regions of four TIS groups in the tomato (Fig. 4; Supplemental. Fig. S6K–M), highlighting their significance in AUG and nonAUG TIS activity. There was also a positive correlation between the number of C/Us and the enrichment of sequence occurrence of all 5-mers for all tomato TIS groups (Fig. 4A; Supplemental Fig. S6N). Similar results were also observed in Arabidopsis (Fig. 4B,D; Supplemental Fig. S6). These results suggest that both CU-rich nucleotide tracts and specific CU-related putative cis-elements are associated with alternative TIS selection in plants.
Figure 4.
Characterization of putative RNA cis-elements predictive of plant TISs. (A,B) Relationship between the number of C/Us (y-axis) and the enrichment of sequence occurrence (x-axis) for all 5-mers in the tomato (A) and Arabidopsis (B) 5′ UTR–AUG and 5′ UTR–nonAUG TIS groups. The enrichment is represented as the log2 ratio of median sequence occurrence between TPs and TNs in the 200-bp regions centered on TIS sites. Rho indicates Spearman's rank correlation coefficient. (C,D) As described in Figure 3C, but for the enrichment of putative RNA cis-elements in the tomato (C) and Arabidopsis (D) 5′ UTR TIS groups. The putative “UCUUC” and “UCUCU” elements with the highest importance score and with the highest enrichment, which are indicated by arrows and highlighted in blue and orange in C,D, are shown.
Note that the perfect AUROC score of annotated TIS prediction (Supplemental Fig. S7A) indicated a bias in our TN collection strategy toward AUG triplets in 5′ UTRs. We thus examine feature similarities between the annotated and alternative TIS groups, focusing on the important features from alternative TIS prediction model (Figs. 2–4). Similar patterns of sequences enrichments of the UCU, UCC, UUC, C mononucleotides, and UCUUC/UCUCU, as well as the depletion of AUG, were observed across annotated and alternative TIS groups (Figs. 3C–F, 4C,D; Supplemental Figs. S6A–L; S7B–I). In contrast, the well-known Kozak motif was not enriched in the 5′ UTR–AUG group but was present in the rest of the alternative and annotated TIS groups (Fig. 3A,B; Supplemental Fig. S4A,B). The principal component analyses and pairwise comparison of the feature correlations showed that the annotated AUG TP TISs were closest to the CDS-AUG TP TISs (rho = 0.97), followed by 5′ UTR–nonAUG (rho = 0.95) and then 5′ UTR–ATG (rho = 0.94) in the tomato and Arabidopsis (Supplemental Fig. S7J–M). These findings underscore the similarity of most features among TIS groups while highlighting the unique presence of the Kozak motif. Our ML models effectively predicted the four groups—5′ UTR–AUG, 5′ UTR–nonAUG, CDS-AUG, and CDS-nonAUG—whereas the CDS-nonAUG group had a relatively lower score. Despite some sequence similarities in the CDS-nonAUG group (Fig. 3I), we prioritized the first three TIS groups for discussion.
The plant CU-rich tracts function as translation enhancers to promote initiation activity
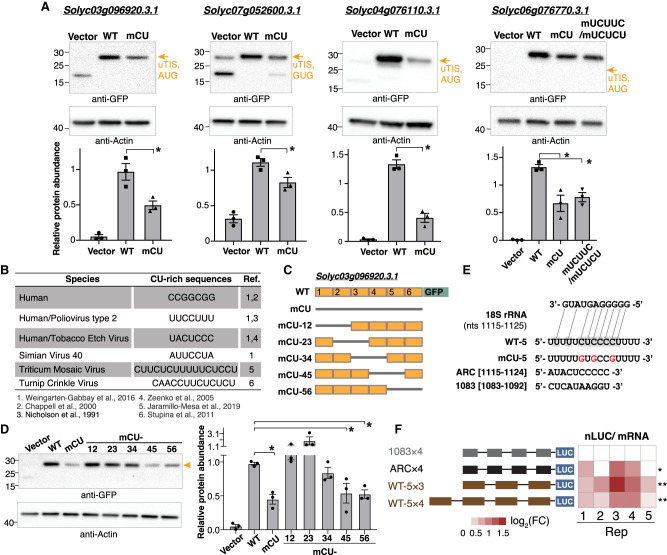
To validate the regulatory roles of the CU-related features in recognizing TISs, we mutated the CU-rich nucleotide tracts in the upstream regions of alternative AUG and nonAUG TISs selected from tomato. In all five TISs examined, mutating CU-rich tracts led to much lower protein abundance (wild-type sequences [WT] vs. CU-tract mutation [mCU]) (Fig. 5A; Supplemental Fig. S8A–E), whereas steady-state mRNA levels remained comparable (Supplemental Fig. S8F–I), indicating that the CU-rich tracts generally promote the efficiency of protein synthesis. Mutating “UCUUC” and “UCUCU” in the 5′ UTR–AUG TIS of Solyc06g076770.3.1 also decreased protein abundance but not mRNA abundance (WT vs. UCUUC/UCUCU mutations [mUCUUC/mUCUCU]) (Fig. 5A; Supplemental Fig. S8H). Mutating the CU-tract or “UCUUC”/”UCUCU” in the 5′ UTR–nonAUG TIS of Solyc06g009750.3.1 did not change translation efficiency (Supplemental Fig. S8F,G), implying other factors influence TIS activity.
Figure 5.
Mutation of CU-rich sequences on plant cellular mRNAs attenuated the TIS activities. (A) Immunoblotting analyses of proteins with translation initiated from the TISs indicated in Supplemental Figure S8 (orange arrows) and with translation driven by the upstream 100-nt wild-type (WT) sequence or sequences with mutations of CU tracts (mCU) or UCUUC/UCUCU (mUCUUC/mUCUCU) sites. Proteins were expressed in Nicotiana benthamiana (tobacco) leaves using the Agrobacterium-mediated transient expression system. Vector indicates tobacco leaves infiltrated with agrobacteria containing the expression vector (i.e., the GFP or FLAG-containing plasmid without a target gene sequence). (Bottom) The protein abundance of the reporter genes relative to actin for three biological repeats (dots) and the corresponding means and standard errors are shown. (B) The known CU-rich sequences found on human transcripts and on plant and animal viral mRNAs promote translation efficiency as reported in the indicated literature. (C) Illustration of swapping the WT (orange box) and the CU-mutated (mCU; gray line) sequences for the TIS-upstream region of a 5′ UTR–AUG TIS of Solyc03g096920.3.1 examined in A. The TIS-upstream region was divided into six subregions (indicated in Supplemental Fig. S8A) to generate distinct mCU mutants with indicated CU-mutated regions fused with the GFP reporter gene as described in A. (D) As described in A, but for the protein of the mCU mutants as indicated in C. (E) The putative binding site on WT sequences (shaded), but not on CU-mutated ones, to the plant 18S rRNA (sold and dashed lines). The sequences of the plant 18S rRNA at positions nt 1115 to 1125 and the WT sequence or sequence with mutations of CT tracts (red) in the fifth region indicated in C are shown. (*) P-values < 0.05, which is derived from one-way ANOVA test, representing the significant difference between WT and the samples with CU sequence mutations. (F) Heatmap shows the nLUC/ mRNA ratio for five biological replicates: 1083 × 4 and ARC × 4 denote negative and positive controls with four copies of 1083 and ARC fragments indicated in E. WT-5 × 3 and WT-5 × 4 denote vectors with three and four copies of the WT-5 indicated in E. (nLUC) Nano luciferase. The color keys represent log2 fold change (FC) relative to the negative control. (*) P-value < 0.05, (**) P-value < 0.01; determined by one-tailed Student's t-test.
CU-rich elements enhance TIS activity in an internal ribosome entry site (IRES)–like manner in human, human viral, and plant viral mRNAs (Fig. 5B; Nicholson et al. 1991; Chappell et al. 2000; Zeenko and Gallie 2005; Stupina et al. 2011; Weingarten-Gabbay et al. 2016; Jaramillo-Mesa et al. 2019), whereas their roles in plants are largely unexplored. Focusing on the orthologous gene pairs between Arabidopsis and human, the CU contents of the annotated TP TISs were higher than those without initiation signals (Supplemental Fig. S9), suggesting the similarity of the translation initiation regulation in both species. To characterize the CU-rich elements in plant mRNAs, we swapped the TIS upstream regions between WT and mCU mutants from the 5′ to 3′ end in the 5′ UTR–AUG TIS of Solyc03g096920.3.1 (Fig. 5C). The protein signals of the GFP reporter were significantly weaker for mCU-45 and mCU-56 than for the WT and were most similar to that of mCU (Fig. 5D); however, their mRNA abundances were comparable (Supplemental Fig. S8I). The WT fifth regions contained a CU-rich site that can base pair with a purine-rich (especially G) region of plant 18S rRNAs (Fig. 5E), a highly conserved region across rice (Oryza sativa), tobacco (Nicotiana tabacum), maize (Zea mays), and wheat (Triticum aestivum) that efficiently enhances translation (Akbergenov et al. 2004). These results provide experimental evidence that CU-rich elements enhance translation initiation in plant mRNAs.
Furthermore, we asked whether the insertion of repeats of CU-rich tracts can enhance initiation activities. We inserted three and four copies of the WT-5 sequences upstream of the nano-luciferase (nLUC) reporter (WT-5 × 3 and WT-5 × 4) (Fig. 5E,F, brown) and measured the nLUC/mRNA ratio (representing translation initiation activity). We used published sequences as negative (four copies of 1083; 1083 × 4) and positive (four copies of ARC; ARC × 4) controls (Akbergenov et al. 2004) and calculated the fold changes (FCs) between the negative control and positive control, as well as the WT-5 repeats. In general, we observed an increase of translation initiation activity (Fig. 5F). These results, together with our findings from the ML-characterized importance of CU-rich features across species (Figs. 2–4), as well as the experimentally supported role of specific CU-rich elements in translation initiation (Fig. 5), collectively pointed a conserved translational strategy of CU-rich tracts and specific CU-related cis-elements in regulating AUG and nonAUG TIS recognition across plants and even in humans and viruses.
TIS prediction models discover plant TISs
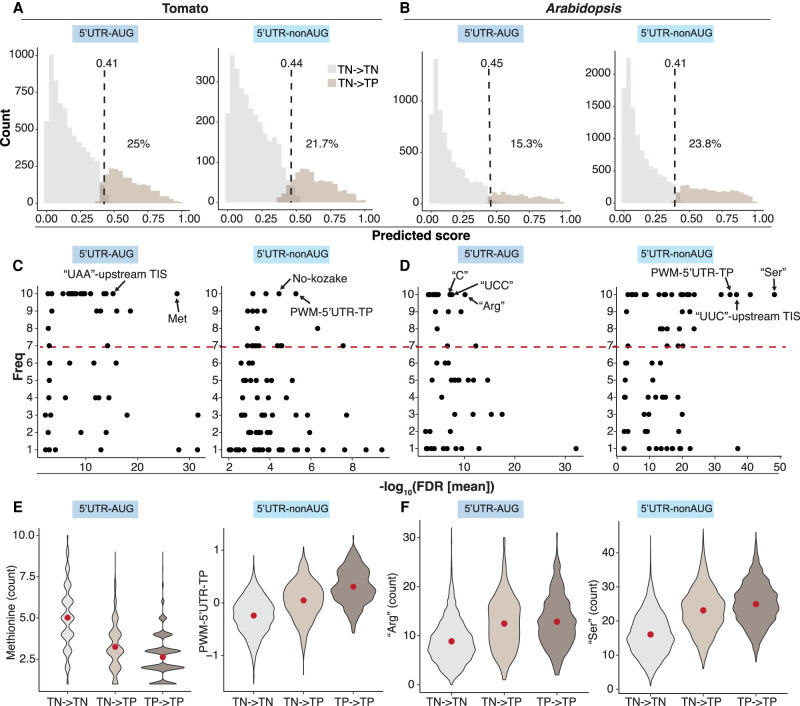
Although our plant TIS prediction models performed well (Fig. 1E; Supplemental Fig. S1F), ∼9%–25% of TNs were misclassified as TPs (i.e., TNs with a high TP prediction score and classified as TPs; TNs → TPs) in each group (Fig. 6A,B, light brown; Supplemental Fig. S10A,B). To explore the possibility that misclassified TNs function as initiation sites, we compared the feature values between correctly predicted TNs (true TNs; TNs → TNs) and incorrectly predicted TNs (misclassified TNs; TNs → TPs) (Fig. 6A,B; Supplemental Fig. S10A,B). The features that likely contributed to misclassification (i.e., with significant enrichment [FDR] and high frequency in 10 randomly balanced TP/TN data sets) (Fig. 6C,D, arrows; Supplemental Fig. S10C,D) were those with high importance scores in the built prediction models (Fig. 2; Supplemental Fig. S3; Cusack et al. 2021). For example, in the tomato 5′ UTR–AUG group, the number of Met residues was significantly different between mispredicted TNs (TNs → TPs) and true TNs (TNs → TNs) (Fig. 6C,E), and this feature also had a high importance score in the corresponding model (Fig. 2B, “Met”). Likewise, in the Arabidopsis 5′ UTR–nonAUG group, the number of Ser residues (which reflects CU nucleotide enrichment) was significantly mispredicted, and this feature also had a high importance score in the corresponding model (Figs. 6D,F, 2H, “Ser”).
Figure 6.
The features of misclassified TN TISs. (A,B) Prediction score distribution of the TN TISs for the 5′ UTR–AUG and 5′ UTR–nonAUG TIS groups in tomato (A) and Arabidopsis (B). The mean threshold (dashed lines) for the classification of TN → TN (the TN TISs predicted as TNs; gray) and TN → TP (the TN TISs predicted as TPs; light brown) and the mean percentage (%) of the TN TISs misclassified as TP (TN → TP group) derived from the best models in the 10 randomly balanced data sets (as indicated in Fig. 1) are shown. (C,D) Dot plots show the frequency (y-axis) of a given feature used for TIS prediction in 10 randomly balanced data sets and the feature enrichment (FDR; x-axis) between the TN → TN and TN → TP groups for the TIS groups indicated in A,B. The red line represents the threshold (frequency of seven or more) of important features as indicated in Figure 2. (E,F) Violin plots show the feature value distributions for the features of methionine counts or PWM–5′ UTR–TP and “Arg” or “Ser” that were most enriched in C,D for the TN → TN (gray) and TN → TP (light brown) groups, indicated in A and the TP → TP (the TP TISs predicted as TPs, dark brown) group. The red dot represents the median value.
To explore the potential of the TIS prediction models to identify alternative TISs in plant mRNAs, we assessed whether the mispredicted TNs with high prediction scores function as TISs in vivo (for the details of selected tomato TISs, see Supplemental Figs. S8, S11). Immunoblotting detected proteins corresponding to the expected sizes for these misclassified AUG and nonAUG TIS-initiated ORFs (vector vs. WT) (Fig. 5A; Supplemental Figs. S8, S11A,B). In addition, the mispredicted CDS-AUG TIS site of Solyc11g039830.2.1 (encoding glycyl-tRNA synthetase) could potentially generate a protein isoform with distinct N termini, likely affecting mitochondrion targeting signals (Supplemental Fig. S11C).
We further applied this pipeline to the noncoding RNA (ncRNA) genes annotated in Araport11 (n = 5178). We identified three novel TISs, including the one encoding a novel small ORF (Supplemental Fig. S12A; Hsu et al. 2016), and they all had CU-rich tracts in their upstream regions (Supplemental Fig. S12, gray boxes). These findings suggested that alternative TISs can be located in genes with known functions and the ncRNAs and direct the translation of novel polypeptides or protein isoforms that diversify the proteome. Thus, TIS prediction models can help identify potential TISs, even without experimental evidence.
Predicting TISs with conserved features in monocots and dicots via transfer learning
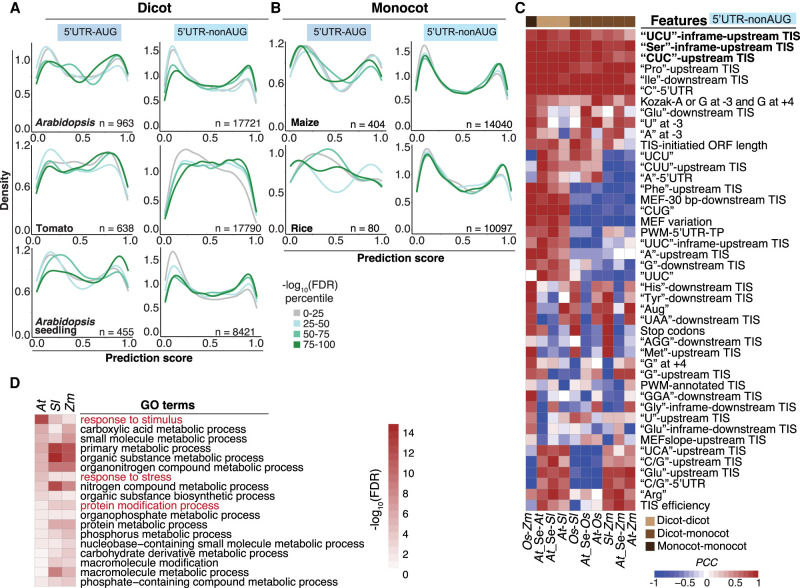
To explore whether our TIS prediction models can be applied to different tissues or even monocots, we generated TIS lists from the cycloheximide-treated ribosome profiling data sets of different plant species and tissues (Lei et al. 2015; Willems et al. 2017; Li and Liu 2020; Yang et al. 2021) using RiboTISH software (Zhang et al. 2017a). We grouped the RiboTISH-reported TISs based on FDR values and examined their prediction scores generated by our best tomato TIS prediction model (Fig. 1E). Compared with TISs with lower FDRs (Fig. 7A, gray and light green lines, top and middle panels; Supplemental Fig. S13), TISs with higher FDRs had higher prediction scores, especially in 5′ UTR–nonAUG, in Arabidopsis (suspension cells) and tomato (leaves) (Fig. 7A, lime green and dark green lines, top and middle panels; Supplemental Fig. S12). Similarly, among the potential TISs in Arabidopsis seedlings identified by RiboTISH, their FDRs and predicted scores from our model were positively correlated (Fig. 7A, bottom). In the monocot maize and rice, we also observed this correlation in the 5′ UTR–nonAUG TIS groups (Fig. 7B, left). Note that the small sample sizes of the RiboTISH-reported 5′ UTR–AUG and CDS-AUG groups might influence the results (Fig. 7A,B; Supplemental Fig. S13). In addition, once the LTM- and Harringtonine-based Ribo-seq experiments in rice and maize are available, they will be valuable to examine the performance of our TIS prediction model between monocots and dicots. Together, these results show the feasibility and flexibility of our analytical pipelines and the utility of the TIS prediction model for identifying TISs from different plant tissues, as well as within and across species.
Figure 7.
Prediction of monocot and dicot TISs and their characteristics. (A) Distribution of the TIS prediction scores generated by the tomato best models for the 5′ UTR–AUG and 5′ UTR–nonAUG TIS groups identified by the RiboTISH algorism and with RiboTISH-reported FDRs (FDR percentile in which the 0–25 category includes TISs with lowest FDR values) using ribosome profiling data sets generated from dicot plants including Arabidopsis (suspension cells), tomato (leaves) and Arabidopsis seedings. (B) As indicated in A, but for the data sets generated from monocot plants including maize and rice. (C) Pairwise Pearson correlation coefficient of the features among dicot and monocot plants for the 5′ UTR–nonAUG TISs identified by the RiboTISH algorism. Monocots: (At) Arabidopsis, (Sl) tomato, and (At-Se) Arabidopsis seedlings; monocots: (Zm) maize and (Os) rice. (D) GO biological process terms significantly enriched (FDR < 0.05) in genes that have RiboTISH-reported TISs with the top 10% highest prediction scores and with the top 25% highest CG contents in the upstream 100-bp regions.
Next, we analyzed the pairwise Pearson correlation coefficient (PCC) to identify conserved features in monocots and dicots. Features linked to “UCU,” “Ser,” and “CUC” content had strong correlations (PCC > 0.8), indicating CU tracts are key for TIS prediction (Fig. 7C, bold). The functions of genes with alternative TISs with the top 10% highest prediction scores and with frequent CU tracts (top 25% in the highest-CU contents in its upstream 100-bp region) were associated with abiotic/biotic stress responses in all species examined, except for genes from the rice data set, in which no Gene Ontology (GO) terms were enriched (Fig. 7D). Given the known role of CU-rich tracts in plant virus TIS activity (Fig. 5B; Stupina et al. 2011; Jaramillo-Mesa et al. 2019), their validated function in promoting plant translation initiation (Fig. 5), and their association with stress-related plant genes (Fig. 7), CU tracts are likely important for triggering plant immunity and viral pathogenesis in both plants and plant viruses.
Discussion
Gene annotation is critical for inferring gene family structure, evolution, and function and for decoding genomes from sequences to phenotypes, as it provides physical and biological contexts to an assembled genome sequence (Kress et al. 2022). The prevalence of unannotated protein-coding regions and the lack of a workflow for revising protein-coding gene annotations of existing plant genome references have limited fundamental discovery. Here, we addressed these issues by focusing on the agriculturally important tomato crop. Exploiting an ML pipeline, we discovered known Kozak motifs, novel CU-rich sequences, and the codons of TISs themselves with joint and distinct influences on alternative AUG and nonAUG TIS recognition, the observations also found in a model species, Arabidopsis (Figs. 1–4). We identified CU-rich elements in plant mRNAs that promote translation initiation (Fig. 5) and were evolutionarily conversed for accurate TIS prediction within and across plant species, as well as in humans and viruses (Figs. 1–3, 7). Lastly, our models revealed hidden TISs based on mRNA sequences across monocots and dicots, thereby improving gene annotation in plants (Figs. 6, 7). The translation of small or nonAUG TIS-initiated ORFs can expand proteome diversity and produce proteins with varied functions (Figs. 5, 6; Supplemental Fig. S8; Hanada et al. 2013; Willems et al. 2017; Wu et al. 2019; Li and Liu 2020). Alternatively, some noncanonical ORFs act as cis-regulatory units to interfere with the translation of the main ORF in a transcript (Tanaka et al. 2016). Thus, the coding regions or peptide compositions of these noncanonical ORFs may not be critical or conserved across plants (von Arnim et al. 2014). Additionally, because unannotated TIS-initiated ORFs tend to be short, the use of computational approaches becomes less effective owing to low conservation scores (Couso 2015). The read coverage of Ribo-seq data sets, the abundance of translated transcripts, and the mRNA features or structures triggering ribosome pausing (Richter and Coller 2015; Merchante et al. 2017) might affect the phasing patterns of genes revealed via CHX-based Ribo-seq data sets. These highlight the limitation of relying on sequence conservation and 3-nt periodicity in revealing protein-coding regions for ORF annotation. Thus, complementary to comparative-genomic approaches and the CHX-based ribosome profiling analyses that have successfully characterized some alternative AUG and nonAUG TIS-initiated ORFs that are conserved across eudicot plant species (Hsu et al. 2016; van der Horst et al. 2019), our ML pipeline together with LTM-supported TIS profiles serves as an alternative and complementary method to generate global estimates of the TISs used in vivo, providing protein-coding information for genomic sequences, especially in information-poor crop species. We should note that our approach and data set are limited in identifying TISs that are specifically sensitive to the LTM chemical, which may not provide a comprehensive profile of every TIS on the genome. In addition, the LTM-induced ribosome stalling near a given initiation site might form a block to pause scanning ribosomes along mRNAs and lead to the bias in identifying its further upstream TISs (Lee et al. 2012; Gao et al. 2015) that might noise TIS prediction models and informative features found. The extent to which the LTM-revealed TISs reflect translation events on genomes remains to be addressed. Thus, the additional experimental validation on the ML-derived novel TISs and TIS features (Fig. 5), the ribosome profiling with distinct chemical treatments and time-course design, and the high-throughput and high-sensitive proteomics approaches such as peptidomes and N-terminal proteomics will facilitate profiling TISs on genome and elucidating plant TIS recognition mechanisms (Lee et al. 2012; Stern-Ginossar et al. 2012; Fields et al. 2015; Gao et al. 2015; Willems et al. 2021; Fan et al. 2023).
Different ML pipelines have been used to predict TISs in human and mouse cells (Ingolia et al. 2011; Reuter et al. 2016; Zhang et al. 2017b). These studies revealed significant sequence features that provided information for predicting TISs (Reuter et al. 2016; Zhang et al. 2017b). In addition to known biological features such as the Kozak sequence and the sequences nearby flanking TISs, several important contextual features also contribute to TIS recognition mechanisms. For example, the number of upstream AUG/Ser residues contributed to the models with high importance scores in both humans and mice (Fig. 3G,H; Reuter et al. 2016; Zhang et al. 2017b). CU-rich tracts regulate TIS activities in humans and plant and human viruses (Figs. 3, 5C; Stupina et al. 2011; Weingarten-Gabbay et al. 2016; Jaramillo-Mesa et al. 2019). Thus, given the similarity of the contextual features required for TIS prediction/activities in humans, mice, viruses, and plants (Figs. 2, 3, 5), it is likely that these features are biologically meaningful for the regulation of TIS selection across different kingdoms rather than being merely the result of sequencing or experimental bias. The conservation of TIS recognition mechanisms further indicates their critical and universal roles in controlling mRNA translation, which is indispensable for all organisms. The codon bias of TIS sites was also an important feature for plant TIS prediction (Supplemental Fig. S5), which is in line with findings for mammalian TIS prediction (Zhang et al. 2017b) and the observation that AAG and AGG are generally poor start codons, as revealed using in vitro/in vivo reporter assay systems (Kearse and Wilusz 2017). These observations show that both contextual features and TIS codon preference play important roles in TIS recognition.
The polypyrimidine CU-tract element enhances translation initiation of preferred start sites via cap-independent and IRES-mediated translation in plant viruses. These CU-tract regions provide complementary interactions between the CU-rich regions of viral mRNAs and the conserved regions of 18S rRNAs within 40S small ribosomes, as reported for tobacco etch virus, blackcurrant reversion virus, turnip crinkle virus, and triticum mosaic virus (Zeenko and Gallie 2005; Karetnikov and Lehto 2007; Stupina et al. 2011; Jaramillo-Mesa et al. 2019). A systemic high-throughput screen of IRES elements in humans and human viruses revealed short motif sequences, including the known viral “UUCCUUU” and “UACUCC” IRES elements, and novel short C/U-related sequences (such as “CCCUCUU” and “UUCCUU) that can base pair with 18S rRNAs within a scanning ribosome to enhance cap-independent translation (Weingarten-Gabbay et al. 2016). However, much less evidence is available for their regulatory roles in plant mRNA translation, with only a single report showing that a 100-bp CU-rich region within the 5′ UTR of OsMac1 in rice promotes translation initiation (Mutsuro-Aoki et al. 2021). Base-paring interaction between plant mRNAs and plant 18S RNAs is critical for translation initiation efficiency, as observed in the short 5′ UTRs of Arabidopsis ribosomal protein S18C and a plant translation enhancer element with the active plant ribosomal 18S RNA complementary sequences (Akbergenov et al. 2004; Vanderhaeghen et al. 2006). In line with these findings for individual genes, our studies systemically and experimentally characterized the broad influence of CU-rich sequences in plant TIS recognition across different plant species, likely via interaction with plant 18S RNAs, and highlighted a conserved translational strategy that modulated initiation site recognition in different species (Figs. 3, 5, 7). Genes with alternative TISs were associated with abiotic/biotic stress responses (Fig. 7D). Although it is proposed that base-pairing interaction between the polypyrimidine CU-tract on mRNAs and the purine-rich regions of 18S rRNAs slows down scanning ribosomes and facilitates ribosome positioning on preferred TISs along a transcript with multiple AUG codons (Akbergenov et al. 2004; Weingarten-Gabbay et al. 2016), the mechanisms underlying these actions, particularly regarding the structural interaction among the CU-rich motifs, rRNAs, and the preferred TISs, as well as whether the plant CU-rich motifs function similarly to IRESs as shown in humans and viruses (Akbergenov et al. 2004; Weingarten-Gabbay et al. 2016; Jaramillo-Mesa et al. 2019) remain largely unclear and require further investigation. Together, it will be interesting to investigate how plants and plant viruses use these CU-rich tracts for protein synthesis and which translated proteins result in plant immunity.
Kozak sequences and the sequences nearby flanking TISs are well-known cis-regulatory signatures that enhance initiation at either AUG or nonAUG start codons (Kozak 1984, 1989). Here, we showed that Kozak motifs are preferentially associated with 5′ UTR–nonAUG and CDS-AUG but not with 5′ UTR–AUG TISs (Fig. 3; Supplemental Fig. S4). How can ribosomes recognize these cis-regulatory sequences when scanning mRNAs? Multiple trans-acting factors including (but not limited to) eukaryotic translation initiation factors (eIFs) play vital roles in selectively regulating TIS efficiency (Roy and von Arnim 2013; Kearse and Wilusz 2017; Fang and Liu 2023). How different sets of trans-regulatory factors and cis-regulatory elements work together coordinately to determine initiation sites and start protein synthesis, especially when plants are exposed to different stress conditions, will be an interesting topic for further research.
Our TIS prediction models identified informative cis-regulatory features in the tomato and Arabidopsis, revealing the mechanistic basis of alternative TIS recognition across dicot and monocot plants. The evolutionary similarities of plant TIS recognition principles highlight the feasibility of applying TIS prediction models to crop species with little experimentally derived gene information. Integrating these prediction models into existing bioinformatics tools would leverage the power of protein-coding gene annotation pipelines across diverse plant species. Moreover, these cis-regulatory features will facilitate efforts to reveal the associations between genetic variation, gene expression, and phenotypic diversity, as well as ultimately bridge genotype to phenotype in plants. Lastly, our approaches and findings are a key initial first step, but not the only one, to profile all protein products from genomes and elucidate the mechanistic basis of selecting alternative TISs in plants. When the initiation step is completed, how will the elongation and termination steps involve protein products? How the general activation and condition-specific regulation can jointly orchestrate the TIS selections for dynamic regulation of gene expression in plants remains largely unclear and will be worth studying in the future.
Methods
Identification of putative cis-elements for predicting TISs
To search for the putative cis-regulatory elements associated with Arabidopsis AUG/nonAUG TIS activities, an enrichment-based k-mer (oligomer with the length of k)-finding pipeline was used as described previously (Liu et al. 2018). Briefly, all possible k-mer sequences (k ≥ 4) were examined for significant enrichment in the 200-bp transcript regions centered on a given TIS between the TP and TN TISs. For the shorter k-mer sequences that were perfectly matched to the longer k-mer sequences, only the one with higher enrichment significance (i.e., the lower P-value) was referred to as a putative cis-regulatory element in the downstream TIS prediction analyses.
Construction and evaluation of predictive ML models
An ML pipeline described previously (Uygun et al. 2019; Wu et al. 2021) was retrieved from GitHub (https://github.com/ShiuLab/ML-Pipeline; https://github.com/azodichr/ML-Pipeline/tree/master/Workshop). Briefly, we used scikit-learn (v0.24.2) in Python (v3.7.0) to train and test the models. For each TIS group, we split balanced data into training (70%) and testing (30%) sets and tested four classification methods: random forest (RF), support vector machine (SVM), logistic regression (LR), and gradient boost (GB). We used 10-fold internal cross-validation to select the optimized hyperparameters. The parameters used to train the models for the identification of the best model among different ML algorisms are as follows: (1) RF—“max_depth” = [3, 5, 10], “max_features” = [0.1, 0.25, 0.5, 0.75, “sqrt”, “log2”, None], “n_estimators” = [100,500,1000]; (2) GB—“max_depth” = [3, 5, 10], “max_features” = [0.1, 0.5, sqrt, log2, None], “n_estimators” = [100, 500, 1000], “learning_rate” = [0.001, 0.01, 0.1, 0.5, 1]; (3) LR—“C” = [0.01, 0.1, 0.5, 1, 10, 50, 100], “intercept_scaling” = [0.1, 0.5, 1, 2, 5, 10], “penalty” = [“l1”,“l2”]; and (4) SVM—“kernel” = [“linear”], “C” = [0.01, 0.1, 0.5, 1, 10, 50, 100]. The F1 score was used to select the best model for each TIS group. Note that to have TIS-predictive features representative in a given species, only the features used in the best model with a frequency of seven or more in the 10 randomly balanced data sets were included in Figure 1G. To comprehensively reveal the features contributing to the best model of predicting TISs, the features used at least in one of the 10 randomly balanced data sets were included in Figure 2, A, C, E, and G, and Supplemental Figure S3, C, E, G, and I. The importance scores infer the importance of each feature in a given model. For the RF and GB methods, this score represents the Gini index, whereas for LR and SVM, it is the coefficient.
Generation of the candidate TIS-initiated protein expression constructs with mutations and expression tags
The best models generated (Fig. 1E; Supplemental Fig. S1F) were used to compute the possibility of a given triplet being classified as a TP. The triplets with the prediction scores higher than the threshold of classifying TP/TNs used in the best models (Fig. 6A; Supplemental Fig. S10) were selected for further functional validation.
The introduction of protein expression constructs into tobacco using an Agrobacterium-mediated transient expression system and the detection of expression were performed as described previously (Li and Liu 2020) with some minor modifications. Briefly, the 5′ UTR and CDS fragments of the gene with a given target TIS site ranging from the upstream 100 bp to the downstream 9 nt (including the target TIS site) were amplified by PCR and fused with a reporter gene encoding GFP, 10 × MYC, or 3 × FLAG, which was used previously (Li and Liu 2020; Chiu et al. 2022). All mutagenesis of the tested sites of genes was performed using synthetic primers listed in Supplemental Table S2.
Quantitative reverse-transcription PCR analyses
The total RNA purification from tobacco leaves, the cDNA preparation, and quantitative reverse-transcription PCR (qRT-PCR) analyses were described previously (Li and Liu 2020; Chiu et al. 2022). Ubiquitin 3 (Rotenberg et al. 2006) was applied as an internal control. The primers used are listed in Supplemental Table S2.
Detecting potential TISs using RiboTISH software and generating their prediction scores
The public CHX-treated ribosome-profiling data sets in Arabidopsis (suspension cells), tomato, maize, and rice were retrieved from the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) (under accession numbers GSE88790, GSE143311) (Willems et al. 2017; Li and Liu 2020) and NCBI Sequence Read Archive (SRA; https://www.ncbi.nlm.nih.gov/sra) database (under accession numbers PRJNA523300 and SRP052520) (Lei et al. 2015; Yang et al. 2021). To assess the model performance between different plant tissues in Arabidopsis, the CHX-treated ribosome profiling data sets of 6-d-old Arabidopsis seedlings were generated as described previously (Li and Liu 2020). The raw reads were trimmed and mapped as described in previous respective studies (Lei et al. 2015; Li and Liu 2020; Yang et al. 2021) and then input into a RiboTISH algorism (Zhang et al. 2017a) with default settings and the additional parameters of “- longest -alt” for the identification of the TISs with FDRs (i.e., the BH correction Q-value of frame test). The prediction scores for each RiboTISH-reported TIS were calculated in scikit-learn using the best model from Arabidopsis (suspension cells). The genome version used in this study was Zm-B73-REFERENCE-NAM-5.0 and IRGSP-1.0 for maize and rice, respectively.
GO analysis
GO term enrichment analysis was performed with the PANTHER database (Hanada et al. 2013) using the Fisher's exact test to calculate the degree of enrichment with FDR for a multiple testing adjustment. Significantly enriched GO terms (FDR < 0.05) were visualized as heatmaps.
Supplementary Material
Acknowledgments
We thank Mr. Te-Chang Hsu, Dr. Yao-Cheng Lin, and the AS-BCST Bioinformatics Core for high-performance computing services; Dr. Shin-Han Shiu and Dr. Ho-Ming Chen for critical reading; and Dr. Melissa Lehti-Shiu for English editing of this article. This research was financially supported by National Science and Technology Council grants MOST 110-2628-B-001-023, MOST 111-2311-B-001-005 and Academia Sinica grant AS-CDA-111-L06 to M.-J.L.
Author contributions: T.-Y.W. designed the research, performed bioinformatics analyses, and wrote the paper. K.-J.C. performed bioinformatics analyses. Y.-R.L. and J.-C.F. performed the experimental analyses. D.U. revised the paper. M.-J.L. conceived/designed the research, performed bioinformatics analyses, and wrote the paper.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.278100.123.
Freely available online through the Genome Research Open Access option.
Competing interest statement
The authors declare no competing interests.
References
- Akbergenov RZ, Zhanybekova SS, Kryldakov RV, Zhigailov A, Polimbetova NS, Hohn T, Iskakov BK. 2004. ARC-1, a sequence element complementary to an internal 18S rRNA segment, enhances translation efficiency in plants when present in the leader or intercistronic region of mRNAs. Nucleic Acids Res 32: 239–247. 10.1093/nar/gkh176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azodi CB, Pardo J, VanBuren R, de Los Campos G, Shiu SH. 2020a. Transcriptome-based prediction of complex traits in maize. Plant Cell 32: 139–151. 10.1105/tpc.19.00332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azodi CB, Tang J, Shiu SH. 2020b. Opening the black box: interpretable machine learning for geneticists. Trends Genet 36: 442–455. 10.1016/j.tig.2020.03.005 [DOI] [PubMed] [Google Scholar]
- Benitez-Cantos MS, Yordanova MM, O'Connor PBF, Zhdanov AV, Kovalchuk SI, Papkovsky DB, Andreev DE, Baranov PV. 2020. Translation initiation downstream from annotated start codons in human mRNAs coevolves with the Kozak context. Genome Res 30: 974–984. 10.1101/gr.257352.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappell SA, Edelman GM, Mauro VP. 2000. A 9-nt segment of a cellular mRNA can function as an internal ribosome entry site (IRES) and when present in linked multiple copies greatly enhances IRES activity. Proc Natl Acad Sci 97: 1536–1541. 10.1073/pnas.97.4.1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu CW, Li YR, Lin CY, Yeh HH, Liu MJ. 2022. Translation initiation landscape profiling reveals hidden open-reading frames required for the pathogenesis of tomato yellow leaf curl Thailand virus. Plant Cell 34: 1804–1821. 10.1093/plcell/koac019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couso JP. 2015. Finding smORFs: getting closer. Genome Biol 16: 189. 10.1186/s13059-015-0765-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusack SA, Wang P, Lotreck SG, Moore BM, Meng F, Conner JK, Krysan PJ, Lehti-Shiu MD, Shiu SH. 2021. Predictive models of genetic redundancy in Arabidopsis thaliana. Mol Biol Evol 38: 3397–3414. 10.1093/molbev/msab111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Arce AJD, Noderer WL, Wang CL. 2018. Complete motif analysis of sequence requirements for translation initiation at non-AUG start codons. Nucleic Acids Res 46: 985–994. 10.1093/nar/gkx1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan KT, Hsu CW, Chen YR. 2023. Mass spectrometry in the discovery of peptides involved in intercellular communication: from targeted to untargeted peptidomics approaches. Mass Spectrom Rev 42: 2404–2425. 10.1002/mas.21789 [DOI] [PubMed] [Google Scholar]
- Fang JC, Liu MJ. 2023. Translation initiation at AUG and non-AUG triplets in plants. Plant Sci 335: 111822. 10.1016/j.plantsci.2023.111822 [DOI] [PubMed] [Google Scholar]
- Fields AP, Rodriguez EH, Jovanovic M, Stern-Ginossar N, Haas BJ, Mertins P, Raychowdhury R, Hacohen N, Carr SA, Ingolia NT, et al. 2015. A regression-based analysis of ribosome-profiling data reveals a conserved complexity to mammalian translation. Mol Cell 60: 816–827. 10.1016/j.molcel.2015.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Wan J, Liu B, Ma M, Shen B, Qian SB. 2015. Quantitative profiling of initiating ribosomes in vivo. Nat Methods 12: 147–153. 10.1038/nmeth.3208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada K, Higuchi-Takeuchi M, Okamoto M, Yoshizumi T, Shimizu M, Nakaminami K, Nishi R, Ohashi C, Iida K, Tanaka M, et al. 2013. Small open reading frames associated with morphogenesis are hidden in plant genomes. Proc Natl Acad Sci 110: 2395–2400. 10.1073/pnas.1213958110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens RP, Brown CM, Chisnall MAW, Waterhouse PM, Macknight RC. 2016. The emerging world of small ORFs. Trends Plant Sci 21: 317–328. 10.1016/j.tplants.2015.11.005 [DOI] [PubMed] [Google Scholar]
- Hinnebusch AG. 2017. Structural insights into the mechanism of scanning and start codon recognition in eukaryotic translation initiation. Trends Biochem Sci 42: 589–611. 10.1016/j.tibs.2017.03.004 [DOI] [PubMed] [Google Scholar]
- Hsu PY, Benfey PN. 2018. Small but mighty: functional peptides encoded by small ORFs in plants. Proteomics 18: e1700038. 10.1002/pmic.201700038 [DOI] [PubMed] [Google Scholar]
- Hsu PY, Calviello L, Wu HL, Li FW, Rothfels CJ, Ohler U, Benfey PN. 2016. Super-resolution ribosome profiling reveals unannotated translation events in Arabidopsis. Proc Natl Acad Sci 113: E7126–E7135. 10.1073/pnas.1614788113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Lareau LF, Weissman JS. 2011. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147: 789–802. 10.1016/j.cell.2011.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaramillo-Mesa H, Gannon M, Holshbach E, Zhang J, Roberts R, Buettner M, Rakotondrafara AM. 2019. The triticum mosaic virus internal ribosome entry site relies on a picornavirus-like YX-AUG motif to designate the preferred translation initiation site and to likely target the 18S rRNA. J Virol 93: e01705-18. 10.1128/JVI.01705-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi CP, Zhou H, Huang X, Chiang VL. 1997. Context sequences of translation initiation codon in plants. Plant Mol Biol 35: 993–1001. 10.1023/A:1005816823636 [DOI] [PubMed] [Google Scholar]
- Juntawong P, Girke T, Bazin J, Bailey-Serres J. 2014. Translational dynamics revealed by genome-wide profiling of ribosome footprints in Arabidopsis. Proc Natl Acad Sci 111: E203–E212. 10.1073/pnas.1317811111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karetnikov A, Lehto K. 2007. The RNA2 5′ leader of blackcurrant reversion virus mediates efficient in vivo translation through an internal ribosomal entry site mechanism. J Gen Virol 88: 286–297. 10.1099/vir.0.82307-0 [DOI] [PubMed] [Google Scholar]
- Kearse MG, Wilusz JE. 2017. Non-AUG translation: a new start for protein synthesis in eukaryotes. Gene Dev 31: 1717–1731. 10.1101/gad.305250.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. 1984. Point mutations close to the AUG initiator codon affect the efficiency of translation of rat preproinsulin in vivo. Nature 308: 241–246. 10.1038/308241a0 [DOI] [PubMed] [Google Scholar]
- Kozak M. 1989. Context effects and inefficient initiation at non-AUG codons in eukaryotic cell-free translation systems. Mol Cell Biol 9: 5073–5080. 10.1128/mcb.9.11.5073-5080.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kress WJ, Soltis DE, Kersey PJ, Wegrzyn JL, Leebens-Mack JH, Gostel MR, Liu X, Soltis PS. 2022. Green plant genomes: what we know in an era of rapidly expanding opportunities. Proc Natl Acad Sci 119: e2115640118. 10.1073/pnas.2115640118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Liu B, Lee S, Huang SX, Shen B, Qian SB. 2012. Global mapping of translation initiation sites in mammalian cells at single-nucleotide resolution. Proc Natl Acad Sci 109: E2424–E2432. 10.1073/pnas.1207846109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei L, Shi J, Chen J, Zhang M, Sun S, Xie S, Li X, Zeng B, Peng L, Hauck A, et al. 2015. Ribosome profiling reveals dynamic translational landscape in maize seedlings under drought stress. Plant J 84: 1206–1218. 10.1111/tpj.13073 [DOI] [PubMed] [Google Scholar]
- Li YR, Liu MJ. 2020. Prevalence of alternative AUG and non-AUG translation initiators and their regulatory effects across plants. Genome Res 30: 1418–1433. 10.1101/gr.261834.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu MJ, Sugimoto K, Uygun S, Panchy N, Campbell MS, Yandell M, Howe GA, Shiu SH. 2018. Regulatory divergence in wound-responsive gene expression between domesticated and wild tomato. Plant Cell 30: 1445–1460. 10.1105/tpc.18.00194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C, Xin M, Feldmann KA, Wang X. 2014. Machine learning-based differential network analysis: a study of stress-responsive transcriptomes in Arabidopsis. Plant Cell 26: 520–537. 10.1105/tpc.113.121913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchante C, Stepanova AN, Alonso JM. 2017. Translation regulation in plants: an interesting past, an exciting present and a promising future. Plant J 90: 628–653. 10.1111/tpj.13520 [DOI] [PubMed] [Google Scholar]
- Mutsuro-Aoki H, Teramura H, Tamukai R, Fukui M, Kusano H, Schepetilnikov M, Ryabova LA, Shimada H. 2021. Dissection of a rice OsMac1 mRNA 5′ UTR to uncover regulatory elements that are responsible for its efficient translation. PLoS One 16: e0253488. 10.1371/journal.pone.0253488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaminami K, Okamoto M, Higuchi-Takeuchi M, Yoshizumi T, Yamaguchi Y, Fukao Y, Shimizu M, Ohashi C, Tanaka M, Matsui M, et al. 2018. Atpep3 is a hormone-like peptide that plays a role in the salinity stress tolerance of plants. Proc Natl Acad Sci 115: 5810–5815. 10.1073/pnas.1719491115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson R, Pelletier J, Le SY, Sonenberg N. 1991. Structural and functional analysis of the ribosome landing Pad of poliovirus type 2: in vivo translation studies. J Virol 65: 5886–5894. 10.1128/jvi.65.11.5886-5894.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noderer WL, Flockhart RJ, Bhaduri A, de Arce AJD, Zhang JJ, Khavari PA, Wang CL. 2014. Quantitative analysis of mammalian translation initiation sites by FACS-seq. Mol Syst Biol 10: 748. 10.15252/msb.20145136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr MW, Mao YH, Storz G, Qian SB. 2020. Alternative ORFs and small ORFs: shedding light on the dark proteome. Nucleic Acids Res 48: 1029–1042. 10.1093/nar/gkz734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter K, Biehl A, Koch L, Helms V. 2016. PreTIS: a tool to predict non-canonical 5′ UTR translational initiation sites in human and mouse. PLoS Comput Biol 12: e1005170. 10.1371/journal.pcbi.1005170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter JD, Coller J. 2015. Pausing on polyribosomes: make way for elongation in translational control. Cell 163: 292–300. 10.1016/j.cell.2015.09.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotenberg D, Thompson TS, German TL, Willis DK. 2006. Methods for effective real-time RT-PCR analysis of virus-induced gene silencing. J Virol Methods 138: 49–59. 10.1016/j.jviromet.2006.07.017 [DOI] [PubMed] [Google Scholar]
- Roy B, von Arnim AG. 2013. Translational regulation of cytoplasmic mRNAs. Arabidopsis Book 11: e0165. 10.1199/tab.0165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spealman P, Naik AW, May GE, Kuersten S, Freeberg L, Murphy RF, McManus J. 2018. Conserved non-AUG uORFs revealed by a novel regression analysis of ribosome profiling data. Genome Res 28: 214–222. 10.1101/gr.221507.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern-Ginossar N, Weisburd B, Michalski A, Le VT, Hein MY, Huang SX, Ma M, Shen B, Qian SB, Hengel H, et al. 2012. Decoding human cytomegalovirus. Science 338: 1088–1093. 10.1126/science.1227919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stupina VA, Yuan XF, Meskauskas A, Dinman JD, Simon AE. 2011. Ribosome binding to a 5′ translational enhancer is altered in the presence of the 3′ untranslated region in cap-independent translation of turnip crinkle virus. J Virol 85: 4638–4653. 10.1128/JVI.00005-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Sotta N, Yamazumi Y, Yamashita Y, Miwa K, Murota K, Chiba Y, Hirai MY, Akiyama T, Onouchi H, et al. 2016. The minimum open reading frame, AUG-stop, induces boron-dependent ribosome stalling and mRNA degradation. Plant Cell 28: 2830–2849. 10.1105/tpc.16.00481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavormina P, De Coninck B, Nikonorova N, De Smet I, Cammue BP. 2015. The plant peptidome: an expanding repertoire of structural features and biological functions. Plant Cell 27: 2095–2118. 10.1105/tpc.15.00440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urquidi Camacho RA, Lokdarshi A, von Arnim AG. 2020. Translational gene regulation in plants: a green new deal. Wiley Interdiscip Rev RNA 11: e1597. 10.1002/wrna.1597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uygun S, Azodi CB, Shiu SH. 2019. Cis-regulatory code for predicting plant cell-type transcriptional response to high salinity. Plant Physiol 181: 1739–1751. 10.1104/pp.19.00653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderhaeghen R, De Clercq R, Karimi M, Van Montagu M, Hilson P, Van Lijsebettens M. 2006. Leader sequence of a plant ribosomal protein gene with complementarity to the 18S rRNA triggers in vitro cap-independent translation. FEBS Lett 580: 2630–2636. 10.1016/j.febslet.2006.04.012 [DOI] [PubMed] [Google Scholar]
- van der Horst S, Snel B, Hanson J, Smeekens S. 2019. Novel pipeline identifies new upstream ORFs and non-AUG initiating main ORFs with conserved amino acid sequences in the 5′ leader of mRNAs in Arabidopsis thaliana. RNA 25: 292–304. 10.1261/rna.067983.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Arnim AG, Jia Q, Vaughn JN. 2014. Regulation of plant translation by upstream open reading frames. Plant Sci 214: 1–12. 10.1016/j.plantsci.2013.09.006 [DOI] [PubMed] [Google Scholar]
- Wang P, Schumacher AM, Shiu SH. 2022. Computational prediction of plant metabolic pathways. Curr Opin Plant Biol 66: 102171. 10.1016/j.pbi.2021.102171 [DOI] [PubMed] [Google Scholar]
- Weingarten-Gabbay S, Elias-Kirma S, Nir R, Gritsenko AA, Stern-Ginossar N, Yakhini Z, Weinberger A, Segal E. 2016. Comparative genetics: systematic discovery of cap-independent translation sequences in human and viral genomes. Science 351: eaad4939. 10.1126/science.aad4939 [DOI] [PubMed] [Google Scholar]
- Willems P, Ndah E, Jonckheere V, Stael S, Sticker A, Martens L, Van Breusegem F, Gevaert K, Van Damme P. 2017. N-terminal proteomics assisted profiling of the unexplored translation initiation landscape in Arabidopsis thaliana. Mol Cell Proteomics 16: 1064–1080. 10.1074/mcp.M116.066662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willems P, Ndah E, Jonckheere V, Van Breusegem F, Van Damme P. 2021. To new beginnings: riboproteogenomics discovery of N-terminal proteoforms in Arabidopsis thaliana. Front Plant Sci 12: 778804. 10.3389/fpls.2021.778804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu HL, Song G, Walley JW, Hsu PY. 2019. The tomato translational landscape revealed by transcriptome assembly and ribosome profiling. Plant Physiol 181: 367–380. 10.1104/pp.19.00541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TY, Goh H, Azodi CB, Krishnamoorthi S, Liu MJ, Urano D. 2021. Evolutionarily conserved hierarchical gene regulatory networks for plant salt stress response. Nat Plants 7: 787–799. 10.1038/s41477-021-00929-7 [DOI] [PubMed] [Google Scholar]
- Yandell M, Ence D. 2012. A beginner's guide to eukaryotic genome annotation. Nat Rev Genet 13: 329–342. 10.1038/nrg3174 [DOI] [PubMed] [Google Scholar]
- Yang X, Song B, Cui J, Wang L, Wang S, Luo L, Gao L, Mo B, Yu Y, Liu L. 2021. Comparative ribosome profiling reveals distinct translational landscapes of salt-sensitive and -tolerant rice. BMC Genomics 22: 612. 10.1186/s12864-021-07922-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeenko V, Gallie DR. 2005. Cap-independent translation of tobacco etch virus is conferred by an RNA pseudoknot in the 5′-leader. J Biol Chem 280: 26813–26824. 10.1074/jbc.M503576200 [DOI] [PubMed] [Google Scholar]
- Zhang P, He D, Xu Y, Hou J, Pan BF, Wang Y, Liu T, Davis CM, Ehli EA, Tan L, et al. 2017a. Genome-wide identification and differential analysis of translational initiation. Nat Commun 8: 1749. 10.1038/s41467-017-01981-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Hu H, Jiang T, Zhang L, Zeng J. 2017b. TITER: predicting translation initiation sites by deep learning. Bioinformatics 33: i234–i242. 10.1093/bioinformatics/btx247 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.