Abstract
Chronic liver disease stands as a significant global health problem with an estimated 2 million annual deaths across the globe. Combining the use of next-generation sequencing technologies with evolving knowledge in the interpretation of genetic variation across the human genome is propelling our understanding, diagnosis, and management of both rare and common liver diseases. Here, we review the contribution of risk and protective alleles to common forms of liver disease, the rising number of monogenic diseases affecting the liver, and the role of somatic genetic variants in the onset and progression of oncological and non-oncological liver diseases. The incorporation of genomic information in the diagnosis and management of patients with liver disease is driving the beginning of a new era of genomics-informed clinical hepatology practice, facilitating personalized medicine, and improving patient care.
Liver disease constitutes a substantial global health burden, causing 2 million deaths each year worldwide. 1 Despite advancements in the treatment of viral hepatitis, morbidity and mortality from liver diseases continue to rise, largely due to the global obesity epidemic and the increasing incidence of metabolic dysfunction–associated steatotic liver disease (MASLD). 1 Investigation of the genetic underpinnings of chronic liver disease (CLD) has (i) identified risk and protective alleles for a variety of common liver diseases, such as MASLD; (ii) uncovered novel monogenic diseases 2; and (iii) expanded our understanding of the contribution of somatic genetic variants to oncological and non-oncological liver disease. The intersection of genes, genomes, and liver disease is propelling our understanding of liver biology, and in turn, improving patient care. This underscores the need for incorporating training and multidisciplinary discussions on the clinical utility of genomic analysis among hepatologists and other providers caring for patients with liver disease.
RISK AND PROTECTIVE ALLELES FOR LIVER DISEASE
Similarly to other complex traits, which are influenced by interactions between common genetic variants and the environment, MASLD has also been shown to have a heritable component.3–5 The genetic drivers of MASLD in the population are primarily composed of common variants with relatively small effect sizes, which have been uncovered by genome-wide association studies (Figure 1). To date, the most well-validated single nucleotide polymorphism driving hepatic steatosis is the p.I148M variant (rs738409) in PNPLA3. 6 This gene is upregulated after carbohydrate feeding and associates with lipid droplets.7,8 Studies have shown that p.I148M results in triglyceride accumulation.9,10 It has also been shown that p.I148M is associated with an increased risk of liver fibrosis and HCC.11–14 While Hispanics have a relatively higher incidence of MASLD, African Americans have a lower incidence, mirroring the prevalence of the p.I148M genotype in these populations.15–17 Another common variant associated with MASLD is p.P446L (rs1260326) in GCKR, which encodes a regulator of glucokinase. 18 This variant is thought to alter the response of glucokinase to fructose-6-phosphate, which results in continuous glucose uptake by the liver and increased de novo lipogenesis.19,20 A less common variant, p.E167K (rs58542926) in TM6SF2 has also repeatedly been associated with an increased risk of MASLD. 21 While its exact function is still poorly understood, it has been shown to play a role in triglyceride secretion. 22 In addition, an individual’s increase in body mass index (BMI) has been shown to enhance the penetrance of a number of these risk variants, most notably p.I148M in PNPLA3. 12 Furthermore, genetic variants in several other genes involved in metabolic function, such as MBOAT7, GPAM, and APOE, have been associated with MASLD.23–26
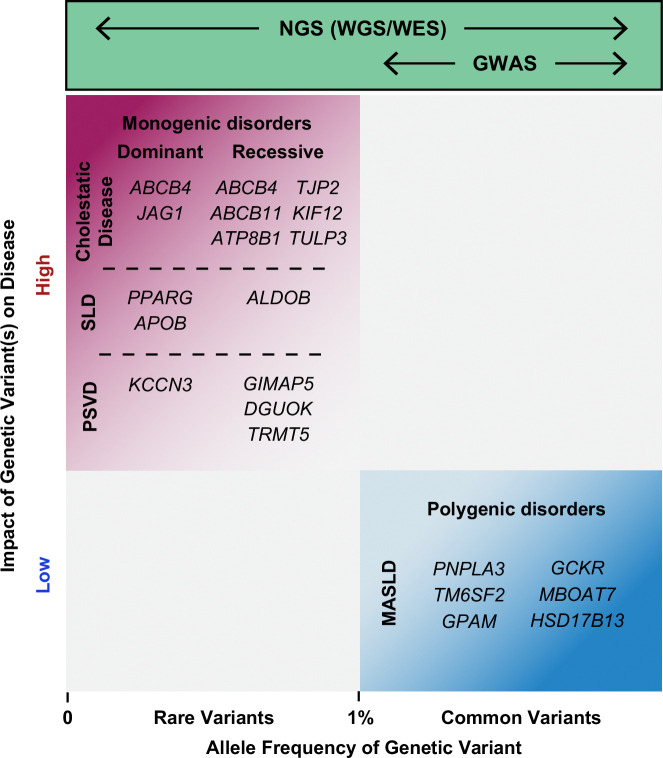
FIGURE 1.
Schematic representation of the contribution of rare (minor allele frequency less than or equal to 1%) genetic variants to monogenic liver diseases, and more frequent genetic variants as risk or protective alleles to common (polygenic) forms of liver disease. Illustrative examples are mentioned. NGS technologies can be used to identify variants across the allele frequency spectrum, while conventional GWAS using genotype arrays only identify common variants associated with disease. However, GWAS are increasingly using WES and WGS data, allowing the identification of both rare and common genetic variants that confer risk for or protection from disease. Abbreviations: GWAS, genome-wide association study; MASLD, metabolic dysfunction–associated steatotic liver disease; NGS, next-generation sequencing; PSVD, porto-sinusoidal vascular disease; SLD, steatotic liver disease; WES, whole-exome sequencing; WGS, whole-genome sequencing.
Recent genome-wide association studies using whole-exome sequencing (WES) data from larger cohorts have confirmed the role of rare variants in APOB, ABCB4, SLC30A10, and TM6SF2 as risk alleles for liver disease. 27 Moreover, these studies have also uncovered genetic variants in HSD17B13 and CIDEB, which provide protection against MASLD and liver disease in general.27,28 Both HSD17B13 and CIDEB encode proteins which associate with lipid droplets. Functional studies have demonstrated how loss of function of both of these proteins alters the dynamics of lipid droplet accumulation.27,29 The characterization of these genes involved in hepatic lipid homeostasis allows for accurate stratification of each patient’s risk of developing liver disease and for the development of novel therapeutic targets. Small-interfering RNAs targeting PNPLA3 and HSD17B13 are 2 illustrative examples of genetic medicine approaches currently being evaluated in clinical trials. 30
The association of several genetic variants with MASLD has also encouraged the development of polygenic risk scores.31–33 These represent a weighted sum of well-characterized disease risk alleles carried by an individual, in hopes of using them to better stratify individual disease risk and therefore more accurately inform disease management. While polygenic risk scores have been proposed to outperform any individual risk allele in predicting the risk of MASLD progression, robust data on their performance in the clinical setting are largely lacking. In addition, their efficacy in predicting long-term outcomes has yet to be evaluated.33,34 In an attempt to incorporate genetic information into the stratification of HCC risk, variants impacting hepatic lipid accumulation and the Wnt/β-catenin pathway have recently been proposed to slightly enhance the performance of conventional clinical scores. 33 Additional studies with larger and more ethnically diverse cohorts are required to fully understand the interplay between risk and protective alleles, alongside environmental risk factors, to inform accurate disease risk stratification and appropriate management.
“Take-to-clinic message [1]”: While polygenic risk scores for the management of MASLD are still not prime time for routine clinical practice, individuals who are overweight (BMI >25), obese (BMI >30), or morbidly obese (BMI >35) and carry homozygous risk allele p.I148M in PNPLA3 have a considerably higher risk to progress to cirrhosis and develop HCC. Thus, clinicians may consider determining their patients’ PNPLA3 p.I148M genotype to inform personalized counseling on lifestyle modifications and weight loss more effectively, particularly in individuals at higher risk of CLD progression and complications (Table 1, Figure 2).35–37
TABLE 1.
Summary of 6 considerations for translating recent genomics-based knowledge in liver disease to clinical practice.
| Clinical presentation | Genomics-based knowledge | Management considerations |
|---|---|---|
| “Take-to-clinic” messages | ||
| [1] MASLD | • Individuals who are overweight, obese, or morbidly obese and harbor homozygous p.I148M in PNPLA3 have a higher risk of progression to cirrhosis and develop HCC | • Consider obtaining PNPLA3 p.I148M genotype to inform personalized counseling about lifestyle modifications/weight loss |
| [2] Unexplained liver disease | • Up to 30% of adults with liver disease of unclear etiology despite a comprehensive workup harbor a monogenic liver disease | • Consider genomic analysis in patients with idiopathic liver disease despite a comprehensive workup |
| [3] Hepatic steatosis in lean individuals | • Lean individuals with hepatic steatosis, transaminase elevation, and no alcohol overuse or visceral adiposity are more likely to have an underlying monogenic liver disease | • Consider genomic analysis in patients who meet these criteria |
| [4] Idiopathic cholestasis | • Nearly half of adults with idiopathic cholestasis appear to have an underlying monogenic liver disease | • Consider genomic analysis (WES or TGS panel) in adults with idiopathic cholestasis |
| [5] PSVD | • Novel genetic causes of PSVD are being discovered in children and adults | • Consider genomic analysis in patients with PSVD of unknown cause |
| [6] HCA | • HCAs with CTNNB1 exon 3-mutations show a higher risk for malignant transformation | • Consider tumor molecular profiling (when a biopsy is performed) for optimal patient management |
Abbreviations: HCA, hepatocellular adenoma; MASLD, metabolic dysfunction–associated steatotic liver disease; PSVD, porto-sinusoidal vascular disease; TGS, targeted gene sequencing; WES, whole-exome sequencing.
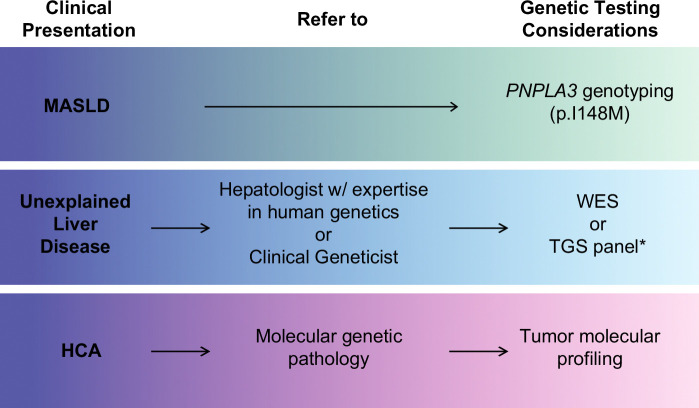
FIGURE 2.
Framework for considering and incorporating genetic testing into the evaluation and management of patients with liver disease. Hepatologists may consider obtaining the PNPLA3 p.I148M genotype for individuals with MASLD to assess their risk for progression to advanced liver disease. In cases of unexplained liver disease despite a thorough workup, genomic analysis is recommended as the next step, and referral to a hepatologist with expertise in human genetics or a clinical geneticist may be beneficial. *If there is strong suspicion for a specific group of genetic liver diseases (eg, cholestasis, iron overload, cystic liver/kidney disease, etc.), TGS for a relevant gene panel may be considered. Otherwise, unbiased WES should be considered. For patients diagnosed with HCA, referral to molecular genetic pathology for liver tumor molecular profiling is advised to guide further management. Abbreviations: HCA, hepatocellular adenoma; MASLD, metabolic dysfunction–associated steatotic liver disease; TGS, targeted gene sequencing; WES, whole-exome sequencing.
A genomic approach applied to autoimmune liver diseases, namely autoimmune hepatitis (AIH), primary biliary cholangitis (PBC), and primary sclerosing cholangitis (PSC), has revealed insights into their underlying genetic determinants. 38 AIH is thought to be caused by loss of tolerance to liver antigens, while PBC and PSC represent the most relevant immune-mediated biliary diseases.39–41 Similarly to other autoimmune conditions, these diseases are thought to exhibit complex gene-gene and gene-environment interactions. A Danish study has estimated the pairwise concordance rate of AIH between monozygotic twins to be about 8.7%, suggesting relatively low heritability. 42 On the other hand, studies in monozygotic twins have found the pairwise concordance rate of PBC to be among the highest across all autoimmune diseases at 63%. 43 First-degree relatives of patients with PSC have also been found to harbor an increased risk of PSC. 44 In line with these findings, genome-wide association studies have identified susceptibility loci associated with these conditions, many of which are pleiotropic across other autoimmune diseases. 45 These include stronger associations with human leukocyte antigen (class I and II HLA genes) as compared with non-HLA loci.45–50 These findings help to advance our understanding of these diseases, particularly those that diverge from the typical demographic profile for autoimmune diseases. This is the case for PSC, given its male predominance, as well as its relative lack of responsiveness to immunosuppression. 47 Moreover, large studies in certain ethnic groups, such as PSC in non-European individuals, are needed. 45 More detailed discussion of non-HLA susceptibility loci in AIH, PBC, and PSC can be found elsewhere.51–53
THE EVOLVING UNDERSTANDING OF MONOGENIC LIVER DISEASES
Despite the early characterization of various monogenic liver diseases, our understanding continues to evolve. The implication of the HFE gene in the pathogenesis of hereditary hemochromatosis (HH) paved the way for understanding the physiology of iron metabolism, leading to the discovery of other genes associated with HH, such as HJV, TFR2, and HAMP, and SLC40A1.54–59 It has been reported that iron overload–related disease in individuals with p.C282Y homozygosity is more common in males than females, attributed largely to the physiologic loss of excess iron due to menstrual bleeding. 60 Our knowledge about the contribution of the p.C282Y and p.H63D HFE genotypes to disease burden has also continued to expand. While these genetic variants were initially described as causal for HH, large-scale genome sequencing studies have revealed that the frequency of these variants in the population is higher than expected, suggesting they more likely act as risk alleles contributing to the development of HH.61,62
Wilson disease is caused by biallelic variants affecting the gene ATP7B, encoding transmembrane copper-transporting ATPase 2. ATP7B is crucial for both facilitating the transport of copper into bile and its incorporation into the copper transport protein ceruloplasmin. 63 As copper is metabolized by the liver, the disease results in the accumulation of copper in hepatocytes and an excess of free serum copper in the bloodstream, presumably leading to pathologic accumulation in other tissues such as the brain. 63 Though a wide spectrum of genetic variants have been associated with the disease, attempts toward establishing genotype-phenotype correlations have been largely elusive. 63 Prevalence of the disease had previously been estimated to be 1/30,000 individuals, but a study based on genetic case confirmation estimated this number to be higher at 1/7000. 64 Previous underestimates may result from a lack of consideration of this disease in differential diagnosis, as well as limitations in the adequate interpretation of ATP7B variants. One such example is the rare synonymous variant p.Phe764Phe (c.2292C>T, rs372979339) in ATP7B, which has been shown to cause skipping of exon 8, thought to result in functionally inactive protein.65–67 Moreover, genetic testing should be performed to confirm the diagnosis of Wilson disease, as there are other rare genetic diseases that can mimic the clinical presentation of Wilson disease, such as MPV17-hepatocerebral mitochondrial DNA depletion syndrome 6, also known as Navajo neurohepatopathy. 68
Alpha-1-antitrypsin (AAT) deficiency is a monogenic disease affecting the liver and lungs. Most individuals carry 2 copies of the normal PiM allele of the gene SERPINA1 encoding AAT, which is synthesized by the liver and secreted into the bloodstream in homeostasis. In AAT deficiency, commonly the result of a homozygous p.Glu342Lys substitution (PiZ allele), an altered protein is produced and abnormally accumulates as AAT aggregates in hepatocytes, leading to liver toxicity. Subsequently, this results in AAT deficiency in circulation, and is therefore unable to perform its primary role in protecting the lung tissue from destruction by neutrophil elastase. 69 While the PiZZ genotype accounts for the majority of cases with severe disease, a milder form of the disease is seen in individuals who harbor p.Glu264Val (PiS allele). 69 The highest prevalence of the PiZZ genotype is detected in individuals of European descent, with 1/2000 Europeans affected with the disease; 1/25 individuals of European descent have the PiMZ genotype and appear to be at risk for milder liver disease. 69 This risk is compounded by the presence of coexisting conditions such as MASLD or alcohol use.69–71 Liver disease manifestations of AAT deficiency can be highly variable in their presentation in both children and adults.72–74 This disorder remains underdiagnosed, highlighting the importance of screening those for which there is high clinical suspicion of the disease.69,75 The gold standard test is either AAT protein phenotyping or genotyping performed by an experienced reference lab. 73 While no therapies are currently available for the disease, a promising phase 2 trial using an RNA interference approach demonstrated efficacy in significantly reducing both liver AAT accumulation and liver enzyme measurements in those with the PiZZ genotype. 76
Cystic fibrosis (CF) is caused by biallelic variants in the gene CFTR, encoding a chloride channel on the apical membrane of epithelial cells, leading to disease manifestations in the lungs, pancreas, gastrointestinal, and hepatobiliary tracts. CFTR is expressed throughout the biliary tract, and CF-related liver disease is thought to be caused by impaired bile flow, leading to retention of toxic bile acids and peribiliary fibrosis. 77 The F508del allele is the most common variant associated with the disease worldwide.77,78 The characterization of CF-causing variants into 6 variant classes and their association with genotype-phenotype correlations at the population level have been well described elsewhere.78,79 While numerous patients with CF have evidence of hepatobiliary disease, this is not clinically significant for most patients. 77 Progression to cirrhosis with portal hypertension is only seen in a minority of individuals with CF. 80 Disease manifestations in other organs, implications of genetic heterogeneity on management, and advancements in novel treatments have been comprehensively reviewed by others.77,79
RARE GENETIC VARIANTS UNDERLYING LIVER DISEASE PATHOGENESIS
While CLD can result from a variety of etiologies, up to 30% of the patients with cirrhosis and more than 10% of those under consideration for liver transplant have advanced liver disease of an unknown cause. 81 Implementing genomic analysis in cases of undiagnosed liver disease has been fruitful in the discovery of novel genetic liver diseases82–89 expediting the diagnosis of pediatric liver diseases, and in some instances, limiting invasive liver biopsies. Furthermore, genomic analysis provides an actionable diagnosis for up to 30% of adults with unexplained liver disease despite a comprehensive workup.4,5 This enables a definitive diagnosis, and therefore targeted treatment and management, while also facilitating the characterization of a broader phenotypic disease spectrum, which may not have been recognized in the absence of a molecular understanding of the disease pathogenesis.4,5,90 These advances are largely due to the application of next-generation sequencing (NGS), which has transformed our ability to rapidly and effectively establish a link between rare genomic variation and disease (Figure 1). WES allows for the putative detection of any genetic variant within protein-coding regions, as well as adjacent intronic splice-site regions for nearly all the 20,000 human protein-coding genes. Given that about 85% of disease-causing variants are predicted to be in coding regions, which only represent ~1% of the entire genome, WES provides a balance between sequencing and storage cost, time of analysis, and scope of interpretable genetic information. 91 On the other hand, whole-genome sequencing, another application of NGS, which examines nucleotides across the entire (coding and noncoding) genome, provides a much larger amount of genetic information. However, it presents with increased costs and larger challenges in interpreting sequencing data as compared with WES. Thus, our incomplete understanding of linking genetic variation in noncoding regions of the human genome with disease has largely hindered the adoption of whole-genome sequencing in most clinical settings.92–95
Distinct from previously outlined diseases with well-characterized pathophysiology, the study of patients who do not fit the typical diagnostic framework is expanding the spectrum of liver-related Mendelian diseases. 96 An illustrative example is atypical presentations of common diseases such as steatotic liver disease (SLD). 97 SLD in lean patients (BMI <25) is estimated to occur in about 10%–20% of the population.98,99 Patients with associated visceral adiposity and insulin resistance make up the majority of these cases and are thought to have similar drivers of disease as nonlean patients with MASLD. 3 However, lean patients with SLD without visceral adiposity or alcohol overuse appear to be more enriched with monogenic disorders as the primary drivers of disease. 3 Our group has previously identified several cases of SLD with monogenic causes among individuals with biopsy-proven SLD. This includes cases of familial lipodystrophy type 3 caused by PPARG deficiency, familial hypobetalipoproteinemia caused by APOB deficiency, and hereditary fructose intolerance caused by ALDOB deficiency, among others.3,5,90 These genetic diagnoses allowed for targeted treatment in certain situations. This also highlights cases in which clinicians should consider a genetic diagnosis, particularly in the presence of an atypical phenotype or multisystemic involvement.5,100,101
“Take-to-clinic message [2]”: Genomic analysis should be considered in adults with unexplained liver disease despite a comprehensive workup, especially if they also have any of the following features: 40 years of age or younger, multisystemic disease, congenital malformations, positive family history, or being offspring of a consanguineous union (Table 1, Figure 2).100,102 This approach has been successful in uncovering a variety of known genetic liver diseases, some of them primarily described in pediatric patients and not traditionally included in the differential diagnosis in adults.
“Take-to-clinic message [3]”: Genomic analysis should be considered in adults who are lean, with no visceral adiposity or alcohol overuse, and present with unexplained hepatic steatosis and transaminase elevation (Table 1, Figure 2).
Progressive familial intrahepatic cholestasis (PFIC) types 1, 2, and 3, which consist of monogenic disorders characterized initially in pediatric patients and attributed to rare biallelic variants in genes ATP8B1, ABCB11, and ABCB4, respectively, have played an integral role in our understanding of the genetic underpinnings of hepatobiliary pathology. These 3 genes encode proteins that are crucial for the appropriate composition of phospholipids and bile acids in bile. Alterations in these proteins are associated with varying disease onset and severity of cholestatic liver disease. ABCB4, which encodes for MDR3 protein, has been implicated in heterogeneous cholestatic diseases of varying severity, including PFIC3, intrahepatic cholestasis of pregnancy, and low phospholipid–associated cholestasis.4,5,103,104 However, disease presentation can blur the lines of these conditions, subverting conventional thought on disease inheritance. While less severe disease such as intrahepatic cholestasis of pregnancy or low phospholipid–associated cholestasis is classically thought to result from rare heterozygous loss-of-function variants in ABCB4, PFIC3, which can progress to end-stage liver disease, requires recessive inheritance. 105 However, a number of studies have highlighted cases of patients with cryptogenic cirrhosis found to have only a single damaging allele.106,107 While genetic modifiers and environmental interactions could be contributors to this clinical variation, additional studies are required to gain a clearer understanding of genetic and environmental contributors in these cases. Furthermore, novel monogenic causes of low and high gamma-glutamyl transferase cholestasis continue to be described, the scope of which has been outlined in a recent comprehensive review. 108
Heterogeneity in cholestatic syndromes also extends to Alagille syndrome (ALGS), an autosomal dominant condition caused primarily by rare heterozygous loss-of-function variants in JAG1, or in a smaller proportion of patients due to rare heterozygous variants in NOTCH2. ALGS has long been known to show clinical variability, as patients within the same family often have varying clinical phenotypes. 109 Many patients receive a diagnosis purely based on the presence of classic diagnostic criteria. 109 However, genetic studies continue to reveal disease-causing JAG1 variants in adult patients who many times do not meet the classic ALGS criteria, but rather, present with atypical disease, which would have been overlooked without a molecular diagnosis. This includes patients with predominantly renal and vascular involvement.4,110–112 It has also been proposed the role of genetic modifiers contributing to the clinical heterogeneity in this syndrome. 113 Overall, these findings suggest that ALGS is likely underdiagnosed in adults, highlighting the need for increased awareness in the clinical setting.4,112,114
“Take-to-clinic message [4]”: Nearly half of the adults with idiopathic cholestasis despite a comprehensive workup who underwent genomic analysis were found to have an underlying genetic cause for their disease, including patients diagnosed with ALGS and MDR3 deficiency in adulthood.4,5 Thus, genomic analysis should be considered in the evaluation of these patients (Table 1, Figure 2).
Insights into porto-sinusoidal vascular diseases (PSVDs), a clinically heterogeneous group of disorders that coalesce into a common phenotype of noncirrhotic portal hypertension, have been further elucidated by the recent discovery of several monogenic diseases underlying their pathogenesis. 115 These include rare biallelic variants in DGUOK, GIMAP5, and TRMT5, as well as heterozygous variants in KCNN3, FOPV (C4ORF54), and FCHSD1.82,83,116–119 GIMAP5, KCNN3, and FOPV have been suggested to contribute to maintaining the integrity of the liver vasculature, while DGUOK and TRMT5 play a role in mitochondrial DNA maintenance. While these discoveries continue to enhance our understanding of PSVD, further research is required to disentangle the mechanisms among these heterogeneous disorders, for which we expect to illuminate innovative therapeutics.
“Take-to-clinic message [5]”: Genetic causes should be investigated in individuals with features of porto-sinusoidal vascular disease of unknown cause (Table 1, Figure 2).
SOMATIC VARIANTS IN HEALTHY AND CIRRHOTIC LIVERS
The application of NGS to germline and tissue DNA from the same individual has also allowed the effective identification of somatic variants in non-oncological liver diseases. Recent studies have highlighted age-related increases in somatic variation in non-diseased liver, confirming previously observed findings. Despite the heterogeneity in variant burden detected in healthy individuals, cirrhotic livers were shown to have a significantly increased burden of somatic variants.120–122 Furthermore, while a moderate number of genetic variants were found in healthy livers, structural variants, including chromothripsis, were much more common in cirrhotic compared to healthy livers. In patients with MASLD, an excess of somatic variants was observed in genes that protect hepatocytes from lipotoxicity. 120 These genes include FOXO1, a key insulin-signaling transcription factor, CIDEB, highlighted previously as implicated in germline protection from MASLD, and GPAM, which is thought to be a regulator of lipid processing and protecting hepatocytes from lipotoxicity.27,123–125 Interestingly, none of FOXO1, CIDEB, and GPAM were found in excess in tissue samples from HCC. 120 The implication of genes in germline and somatic protection raises unique opportunities for exploring innovative therapeutic targets.
These findings raise further questions regarding the role of somatic variants in liver disease physiology. In addressing this question, one study used WES to profile nonmalignant diseased liver tissue and replicated previous findings of high mutational burden in tissue samples with severe disease. 126 Interestingly, further investigation in these samples with ultradeep sequencing of a target set of HCC-associated genes revealed recurrent variants in PKD1, PPARGC1B, KMT2D, and ARID1A, which were not found using standard coverage WES. 126 Subsequent murine validation revealed that loss of Pkd1, Kmt2d, and Arid1a was sufficient to promote clonal expansion and increase hepatocyte fitness. 126 In a separate study, this group leveraged a novel mouse platform for lineage tracing of somatic clones to identify genes that confer protection from steatosis, including Tbx3, Bcl6, and Smyd2. 127 While these studies have begun to uncover the contribution of somatic variants to liver disease pathophysiology, future studies will elucidate the scope of their contribution to progression and protection from CLD.
SOMATIC VARIANTS UNDERLYING MOLECULAR SUBTYPES OF HEPATOCELLULAR ADENOMAS
Hepatocellular adenomas (HCA) are rare benign liver tumors classically associated with excessive hormonal exposure, such as the use of oral contraceptives or anabolic steroids, as well as other conditions such as glycogen storage diseases and metabolic syndrome. Advances in NGS have facilitated the classification of HCA subtypes based on underlying genetic architecture, namely (i) HNF1A-inactivating HCA, (ii) CTNNB1-activating HCA, (iii) inflammatory HCA resulting from genetic alterations in IL6/JAK/STAT signaling pathway, and (iv) Sonic Hedgehog HCA, along with an unclassified group encompassing the remaining cases. 128
HCAs with biallelic loss of HNF1A typically show marked steatosis with minimal cytological abnormalities or inflammatory infiltrates.129,130 While most of these cases are found to be due to somatic biallelic inactivating variants, studies have noted up to 10% of cases demonstrating one inactivating allele of germline origin. As germline HNF1A variants are known to cause maturity-onset diabetes of the young type 3, it is important to consider the risk for the development of HCA in these patients.131–133 HCAs due to alterations in CTNNB1 typically lead to overactivation of the Wnt/β-catenin pathway. Subtypes of CTNNB1-activating HCA have also been identified, namely those with activating mutations in exon 3, or in exons 7/8. These subtypes are primarily distinguished by their different risk for malignant potential. HCAs with CTNNB1 exon 3 mutation(s) demonstrate higher potential for malignant transformation as compared with HCAs with CTNNB1 mutation(s) in exons 7/8 or other HCA subtypes. Thus, HCAs with mutation(s) in exon 3 are more likely to warrant surgical management. 128 Inflammatory HCA is characterized by variants in a variety of genes, including IL6ST, FRK, STAT3, GNAS, and JAK1, comprising up to 50% of cases and showing a strong association with estrogen exposure. These genes are involved in the IL6/JAK/STAT pathway, leading to its activation and an inflammatory infiltrate within tumors.134,135 Sonic Hedgehog HCA accounts for about 4%–5% of cases, resulting from somatic fusion events between the genes INHBE and GLI1. GLI1 encodes a transcription factor in the Hedgehog pathway, and INHBE acts as the driver of GLI1 activity in these HCAs. 128 Studies have also highlighted the association between this type of HCA and estrogen exposure, obesity, and an increased risk of bleeding complications. 136 These findings underscore the clinical importance of identifying these genetic subtypes, particularly in informing diagnostic and management strategies.
“Take-to-clinic message [6]”:In cases where a biopsy of hepatocellular adenoma is obtained, tissue molecular profiling as described above should be considered. Genetic information will assist in disease subtype classification, determination of association with estrogen exposure, and assessment of risk for malignant transformation, with a direct impact on optimal patient care (Table 1, Figure 2).
SOMATIC VARIANTS UNDERLYING HCC
HCC is the most common type of primary liver cancer in adulthood, with its development often arising in background livers with chronic hepatitis B or cirrhosis.137,138 Similarly to CLD, the global burden of HCC remains a public health challenge with a projection of over 1 million cases annually by 2025, largely driven by the obesity epidemic and MASLD. 139 Over the past decade, numerous studies have utilized NGS to investigate the genetic architecture of HCC. These studies have demonstrated that HCC is a highly heterogeneous malignancy with an average of 40–60 coding variants per tumor, including 2–6 of which are located in putative driver genes. 122 Mutations in the TERT promoter are the most common driver, detected in early low-grade dysplastic nodules, and increasing in prevalence as the dysplastic nodules progress to HCC. 122 Recurrent somatic mutations have also been identified in CTNNB1, TP53, ARID1A, ARID2, RB1, AXIN1, and NFE2L2, demonstrating the role of the Wnt/β-catenin pathway, cell cycle regulators, and chromatin remodelers in driving the progression to HCC. 140
The heterogeneity of genetic variants observed in liver tumors introduces unique challenges in therapeutic development in HCC, limiting a treatment approach toward a single specific target. In addition, while both in vivo mouse and in vitro cell culture models can provide important insights into disease mechanism(s), both have limitations in their ability to model the complex biology contributing to the onset and development of human HCC. Despite these challenges, which also exist across other cancer types, an increasing number of studies in various cancers have demonstrated that genomic analysis of tumors can improve patient outcomes.68,141–143 Thus, while the translation of HCC genomics into clinical decision-making in hepatology is lagging, continued efforts toward its future incorporation into patient care are needed.
A VISION FOR A (NEAR) FUTURE OF GENOMICS-INFORMED HEPATOLOGY CLINICAL PRACTICE
The wide range of recent discoveries highlighting the contribution of germline genetic variants to liver disease underscores the importance of continued investigation in this realm. Whether related to common or rare diseases, these findings offer valuable insights into our understanding of disease pathogenesis, with some of these observations being ready for translation to the clinic (Table 1). 102 Continued innovations, such as the release of the first human pangenome reference, are likely to enhance our understanding of ancestry-specific genetic factors and their impact on disease. 144 Furthermore, studies incorporating large population databases, such as the UK Biobank and the more diverse US-based All of Us cohort, will continue to facilitate advances in precision medicine.145,146
As NGS becomes an affordable tool in the research and clinical armamentarium, we anticipate that it will enable further discoveries across the spectrum of liver diseases. Beyond improving diagnosis and our understanding of disease pathogenesis, the next step is translating these findings toward optimal patient management and new effective therapeutic interventions. 27 Advances in understanding the diverse liver cell types and their respective gene expression profiles at the single cell level, in addition to breakthroughs in modeling the liver’s microenvironment using humanized liver cell types within a mouse model and development of liver organoid model systems, allow for unprecedented approaches for the study of human liver physiology.147–151 These approaches can be applied toward modeling patient’s disease, therapeutic screening, and propelling the field of regenerative medicine.152–154
In the clinic, a key step in translating these findings into improvements in patient care is adopting a multidisciplinary, team-based approach. In many cases, discussion and collaboration with other specialists about what genetic test to order and how to properly interpret its results is recommended (Figure 2). Numerous institutions host dedicated sessions for the discussion of oncology cases known as “tumor boards,” drawing additional insights from imaging, pathology, and genetics to optimize patient care. We envision the development of similar forums for the discussion of genetic disease cases with significant liver involvement, many of which benefit from the participation of members across different fields to provide expertise in achieving a proper diagnosis and to guide appropriate management. Accordingly, some institutions have already begun to establish these forums (eg, Hepatology Genome Rounds). 102 This provides a crucial step forward in accelerating the translation of new knowledge generated through research endeavors toward our goals of precision medicine and excellent patient care in hepatology.
AUTHOR CONTRIBUTIONS
All authors conceptualized, wrote, reviewed, and approved the final manuscript.
FUNDING INFORMATION
Silvia Vilarinho is supported by the NIH/NIDDK (R01 DK131033) and Doris Duke Charitable Foundation Grant #2019081. Shanin Chowdhury is supported by the National Institute of General Medical Sciences of the National Institutes of Health under award 1T32GM136651-01. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
CONFLICTS OF INTEREST
Silvia Vilarinho served as a consultant to Albireo Pharma, Inc., and receives research grant support from Moderna Therapeutics, Inc. The remaining authors have no conflicts to report.
Footnotes
Abbreviations: AAT, alpha-1-antitrypsin; AIH, autoimmune hepatitis; ALGS, Alagille syndrome; BMI, body mass index; CF, cystic fibrosis; CLD, chronic liver disease; HCA, hepatocellular adenoma; HH, hereditary hemochromatosis; MASLD, metabolic dysfunction–associated steatotic liver disease; NGS, next-generation sequencing; PBC, primary biliary cholangitis; PFIC, progressive familial intrahepatic cholestasis; PSC, primary sclerosing cholangitis; PSVD, porto-sinusoidal vascular disease; SLD, steatotic liver disease; WES, whole-exome sequencing.
Chigoziri Konkwo and Shanin Chowdhury contributed equally.
Contributor Information
Chigoziri Konkwo, Email: chigoziri.konkwo@yale.edu.
Shanin Chowdhury, Email: shanin.chowdhury@yale.edu.
Silvia Vilarinho, Email: silvia.vilarinho@yale.edu.
REFERENCES
- 1. Devarbhavi H, Asrani SK, Arab JP, Nartey YA, Pose E, Kamath PS. Global burden of liver disease: 2023 update. J Hepatol. 2023;79:516–537. [DOI] [PubMed] [Google Scholar]
- 2. Zheng M, Allington G, Vilarinho S. Genomic medicine for liver disease. Hepatology. 2022;76:860–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Vilarinho S, Ajmera V, Zheng M, Loomba R. Emerging role of genomic analysis in clinical evaluation of lean individuals with NAFLD. Hepatology. 2021;74:2241–2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zheng M, Hakim A, Konkwo C, Deaton AM, Ward LD, Silveira MG, et al. Advancing diagnosis and management of liver disease in adults through exome sequencing. EBioMed. 2023;95:104747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hakim A, Zhang X, DeLisle A, Oral EA, Dykas D, Drzewiecki K, et al. Clinical utility of genomic analysis in adults with idiopathic liver disease. J Hepatol. 2019;70:1214–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Romeo S, Kozlitina J, Xing C, Pertsemlidis A, Cox D, Pennacchio LA, et al. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. 2008;40:1461–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Huang Y, He S, Li JZ, Seo YK, Osborne TF, Cohen JC, et al. A feed-forward loop amplifies nutritional regulation of PNPLA3. Proc Natl Acad Sci USA. 2010;107:7892–7897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ruhanen H, Perttilä J, Hölttä-Vuori M, Zhou Y, Yki-Järvinen H, Ikonen E, et al. PNPLA3 mediates hepatocyte triacylglycerol remodeling [S]. J Lipid Res. 2014;55:739–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pingitore P, Pirazzi C, Mancina RM, Motta BM, Indiveri C, Pujia A, et al. Recombinant PNPLA3 protein shows triglyceride hydrolase activity and its I148M mutation results in loss of function. Biochim Biophys Acta Mol Cell Biol Lipids. 2014;1841:574–580. [DOI] [PubMed] [Google Scholar]
- 10. He S, McPhaul C, Li JZ, Garuti R, Kinch L, Grishin NV, et al. A sequence variation (I148M) in PNPLA3 associated with nonalcoholic fatty liver disease disrupts triglyceride hydrolysis. J Biol Chem. 2010;285:6706–6715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Trepo E, Guyot E, Ganne-Carrie N, Degre D, Gustot T, Franchimont D, et al. PNPLA3(rs738409 C>G) is a common risk variant associated with hepatocellular carcinoma in alcoholic cirrhosis. Hepatology. 2012;55:1307–1308. [DOI] [PubMed] [Google Scholar]
- 12. Stender S, Kozlitina J, Nordestgaard BG, Tybjærg-Hansen A, Hobbs HH, Cohen JC. Adiposity amplifies the genetic risk of fatty liver disease conferred by multiple loci. Nat Genet. 2017;49:842–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen VL, Oliveri A, Miller MJ, Wijarnpreecha K, Du X, Chen Y, et al. PNPLA3 genotype and diabetes identify patients with nonalcoholic fatty liver disease at high risk of incident cirrhosis. Gastroenterology. 2023;164:966–977.e917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Guyot E, Sutton A, Rufat P, Laguillier C, Mansouri A, Moreau R, et al. PNPLA3 rs738409, hepatocellular carcinoma occurrence and risk model prediction in patients with cirrhosis. J Hepatol. 2013;58:312–318. [DOI] [PubMed] [Google Scholar]
- 15. Samji NS, Snell PD, Singal AK, Satapathy SK. Racial disparities in diagnosis and prognosis of nonalcoholic fatty liver disease. Clin Liver Dis. 2020;16:66–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rich NE, Oji S, Mufti AR, Browning JD, Parikh ND, Odewole M, et al. Racial and ethnic disparities in nonalcoholic fatty liver disease prevalence, severity, and outcomes in the United States: A systematic review and meta-analysis. Clin Gastroenterol Hepatol. 2018;16:198–210.e192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kallwitz ER, Tayo BO, Kuniholm MH, Cai J, Daviglus M, Cooper RS, et al. American ancestry is a risk factor for suspected nonalcoholic fatty liver disease in Hispanic/Latino adults. Clin Gastroenterol Hepatol. 2019;17:2301–2309. [DOI] [PubMed] [Google Scholar]
- 18. Santoro N, Zhang CK, Zhao H, Pakstis AJ, Kim G, Kursawe R, et al. Variant in the glucokinase regulatory protein (GCKR) gene is associated with fatty liver in obese children and adolescents. Hepatology. 2012;55:781–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Beer NL, Tribble ND, McCulloch LJ, Roos C, Johnson PR, Orho-Melander M, et al. The P446L variant in GCKR associated with fasting plasma glucose and triglyceride levels exerts its effect through increased glucokinase activity in liver. Hum Mol Genet. 2009;18:4081–4088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Valenti L, Alisi A, Nobili V. Unraveling the genetics of fatty liver in obese children: Additive effect of P446LGCKRand I148MPNPLA3 polymorphisms. Hepatology. 2012;55:661–663. [DOI] [PubMed] [Google Scholar]
- 21. Kozlitina J, Smagris E, Stender S, Nordestgaard BG, Zhou HH, Tybjærg-Hansen A, et al. Exome-wide association study identifies a TM6SF2 variant that confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. 2014;46:352–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mahdessian H, Taxiarchis A, Popov S, Silveira A, Franco-Cereceda A, Hamsten A, et al. TM6SF2 is a regulator of liver fat metabolism influencing triglyceride secretion and hepatic lipid droplet content. Proc Natl Acad Sci USA. 2014;111:8913–8918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chen Y, Du X, Kuppa A, Feitosa MF, Bielak LF, O’Connell JR, et al. Genome-wide association meta-analysis identifies 17 loci associated with nonalcoholic fatty liver disease. Nat Genet. 2023;55:1640–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hakim A, Moll M, Brancale J, Liu J, Lasky-Su JA, Silverman EK, et al. Genetic variation in the mitochondrial glycerol-3-phosphate acyltransferase is associated with liver injury. Hepatology. 2021;74:3394–3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jamialahmadi O, Mancina RM, Ciociola E, Tavaglione F, Luukkonen PK, Baselli G, et al. Exome-wide association study on alanine aminotransferase identifies sequence variants in the GPAM and APOE associated with fatty liver disease. Gastroenterology. 2021;160:1634–1646.e1637. [DOI] [PubMed] [Google Scholar]
- 26. Vujkovic M, Ramdas S, Lorenz KM, Guo X, Darlay R, Cordell HJ, et al. A multiancestry genome-wide association study of unexplained chronic ALT elevation as a proxy for nonalcoholic fatty liver disease with histological and radiological validation. Nat Genet. 2022;54:761–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Verweij N, Haas ME, Nielsen JB, Sosina OA, Kim M, Akbari P, et al. Germline mutations in CIDEB and protection against liver disease. N Engl J Med. 2022;387:332–344. [DOI] [PubMed] [Google Scholar]
- 28. Abul-Husn NS, Cheng X, Li AH, Xin Y, Schurmann C, Stevis P, et al. A protein-truncating HSD17B13 variant and protection from chronic liver disease. N Engl J Med. 2018;378:1096–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ma Y, Belyaeva OV, Brown PM, Fujita K, Valles K, Karki S, et al. 17-beta hydroxysteroid dehydrogenase 13 is a hepatic retinol dehydrogenase associated with histological features of nonalcoholic fatty liver disease. Hepatology. 2019;69:1504–1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lindén D, Romeo S. Therapeutic opportunities for the treatment of NASH with genetically validated targets. J Hepatol. 2023;79:1056–1064. [DOI] [PubMed] [Google Scholar]
- 31. Khera AV, Chaffin M, Aragam KG, Haas ME, Roselli C, Choi SH, et al. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat Genet. 2018;50:1219–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Torkamani A, Wineinger NE, Topol EJ. The personal and clinical utility of polygenic risk scores. Nat Rev Genet. 2018;19:581–590. [DOI] [PubMed] [Google Scholar]
- 33. Nahon P, Bamba-Funck J, Layese R, Trépo E, Zucman-Rossi J, Cagnot C, et al. Integrating genetic variants into clinical models for hepatocellular carcinoma risk stratification in cirrhosis. J Hepatol. 2023;78:584–595. [DOI] [PubMed] [Google Scholar]
- 34. Trépo E, Valenti L. Update on NAFLD genetics: From new variants to the clinic. J Hepatol. 2020;72:1196–1209. [DOI] [PubMed] [Google Scholar]
- 35. Shen J, Wong GL-H, Chan HL-Y, Chan RS-M, Chan H-Y, Chu WC-W, et al. PNPLA3 gene polymorphism and response to lifestyle modification in patients with nonalcoholic fatty liver disease. J Gastroenterol Hepatol. 2015;30:139–146. [DOI] [PubMed] [Google Scholar]
- 36. Krawczyk M, Stachowska E, Milkiewicz P, Lammert F, Milkiewicz M. Reduction of caloric intake might override the prosteatotic effects of the PNPLA3 p.I148M and TM6SF2 p.E167K variants in patients with fatty liver: Ultrasound-based prospective study. Digestion. 2016;93:139–148. [DOI] [PubMed] [Google Scholar]
- 37. Boeckmans J, Gatzios A, Schattenberg JM, Koek GH, Rodrigues RM, Vanhaecke T. PNPLA3 I148M and response to treatment for hepatic steatosis: A systematic review. Liver Int. 2023;43:975–988. [DOI] [PubMed] [Google Scholar]
- 38. Webb GJ, Hirschfield GM. Using GWAS to identify genetic predisposition in hepatic autoimmunity. J Autoimmun. 2016;66:25–39. [DOI] [PubMed] [Google Scholar]
- 39. Hirschfield GM, Karlsen TH, Lindor KD, Adams DH. Primary sclerosing cholangitis. Lancet. 2013;382:1587–1599. [DOI] [PubMed] [Google Scholar]
- 40. Kaplan MM, Gershwin ME. Primary biliary cirrhosis. N Engl J Med. 2005;353:1261–1273. [DOI] [PubMed] [Google Scholar]
- 41. Mieli-Vergani G, Vergani D, Czaja AJ, Manns MP, Krawitt EL, Vierling JM, et al. Autoimmune hepatitis. Nat Rev Dis Primers. 2018;4:18017. [DOI] [PubMed] [Google Scholar]
- 42. Grønbæk L, Vilstrup H, Pedersen L, Christensen K, Jepsen P. Family occurrence of autoimmune hepatitis: A Danish nationwide registry-based cohort study. J Hepatol. 2018;69:873–877. [DOI] [PubMed] [Google Scholar]
- 43. Selmi C, Mayo MJ, Bach N, Ishibashi H, Invernizzi P, Gish RG, et al. Primary biliary cirrhosis in monozygotic and dizygotic twins: Genetics, epigenetics, and environment. Gastroenterology. 2004;127:485–492. [DOI] [PubMed] [Google Scholar]
- 44. Bergquist A, Montgomery SM, Bahmanyar S, Olsson R, Danielsson Å, Lindgren S, et al. Increased risk of primary sclerosing cholangitis and ulcerative colitis in first-degree relatives of patients with primary sclerosing cholangitis. Clin Gastroenterol Hepatol. 2008;6:939–943. [DOI] [PubMed] [Google Scholar]
- 45. Ellinghaus D. How genetic risk contributes to autoimmune liver disease. Semin Immunopathol. 2022;44:397–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wang C, Zheng X, Tang R, Han C, Jiang Y, Wu J, et al. Fine mapping of the MHC region identifies major independent variants associated with Han Chinese primary biliary cholangitis. J Autoimmun. 2020;107:102372. [DOI] [PubMed] [Google Scholar]
- 47. Jiang X, Karlsen TH. Genetics of primary sclerosing cholangitis and pathophysiological implications. Nat Rev Gastroenterol Hepatol. 2017;14:279–295. [DOI] [PubMed] [Google Scholar]
- 48. Karlsen TH, Lammert F, Thompson RJ. Genetics of liver disease: From pathophysiology to clinical practice. J Hepatol. 2015;62:S6–S14. [DOI] [PubMed] [Google Scholar]
- 49. Qiu F, Tang R, Zuo X, Shi X, Wei Y, Zheng X, et al. A genome-wide association study identifies six novel risk loci for primary biliary cholangitis. Nat Commun. 2017;8:14828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Li Y, Sun Y, Liu Y, Wang B, Li J, Wang H, et al. Genome-wide meta-analysis identifies susceptibility loci for autoimmune hepatitis type 1. Hepatology. 2022;76:564–575. [DOI] [PubMed] [Google Scholar]
- 51. Engel B, Laschtowitz A, Janik MK, Junge N, Baumann U, Milkiewicz P, et al. Genetic aspects of adult and pediatric autoimmune hepatitis: A concise review. Eur J Med Genet. 2021;64:104214. [DOI] [PubMed] [Google Scholar]
- 52. Chung BK, Hirschfield GM. Immunogenetics in primary sclerosing cholangitis. Curr Opin Gastroenterol. 2017;33:93–98. [DOI] [PubMed] [Google Scholar]
- 53. Gerussi A, Carbone M, Corpechot C, Schramm C, Asselta R, Invernizzi P. The genetic architecture of primary biliary cholangitis. Eur J Med Genet. 2021;64:104292. [DOI] [PubMed] [Google Scholar]
- 54. Roetto A, Papanikolaou G, Politou M, Alberti F, Girelli D, Christakis J, et al. Mutant antimicrobial peptide hepcidin is associated with severe juvenile hemochromatosis. Nat Genet. 2003;33:21–22. [DOI] [PubMed] [Google Scholar]
- 55. Papanikolaou G, Samuels ME, Ludwig EH, MacDonald MLE, Franchini PL, Dubé M-P, et al. Mutations in HFE2 cause iron overload in chromosome 1q–linked juvenile hemochromatosis. Nat Genet. 2004;36:77–82. [DOI] [PubMed] [Google Scholar]
- 56. Montosi G, Donovan A, Totaro A, Garuti C, Pignatti E, Cassanelli S, et al. Autosomal-dominant hemochrom-atosis is associated with a mutation in the ferroportin (SLC11A3) gene. J Clin Invest. 2001;108:619–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Pietrangelo A, Montosi G, Totaro A, Garuti C, Conte D, Cassanelli S, et al. Hereditary hemochromatosis in adults without pathogenic mutations in the hemochromatosis gene. N Engl J Med. 1999;341:725–732. [DOI] [PubMed] [Google Scholar]
- 58. Fleming RE, Migas MC, Holden CC, Waheed A, Britton RS, Tomatsu S, et al. Transferrin receptor 2: Continued expression in mouse liver in the face of iron overload and in hereditary hemochromatosis. Proc Natl Acad Sci USA. 2000;97:2214–2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Camaschella C, Roetto A, Calì A, De Gobbi M, Garozzo G, Carella M, et al. The gene TFR2 is mutated in a new type of haemochromatosis mapping to 7q22. Nat Genet. 2000;25:14–15. [DOI] [PubMed] [Google Scholar]
- 60. Allen KJ, Gurrin LC, Constantine CC, Osborne NJ, Delatycki MB, Nicoll AJ, et al. Iron-overload–related disease in HFE hereditary hemochromatosis. N Engl J Med. 2008;358:221–230. [DOI] [PubMed] [Google Scholar]
- 61. Grosse SD, Gurrin LC, Bertalli NA, Allen KJ. Clinical penetrance in hereditary hemochromatosis: Estimates of the cumulative incidence of severe liver disease among HFE C282Y homozygotes. Genet Med. 2018;20:383–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Whitlock EP, Garlitz BA, Harris EL, Beil TL, Smith PR. Screening for hereditary hemochromatosis: A systematic review for the U.S. Preventive Services Task Force. Ann Intern Med. 2006;145:209–223. [DOI] [PubMed] [Google Scholar]
- 63. Członkowska A, Litwin T, Dusek P, Ferenci P, Lutsenko S, Medici V, et al. Wilson disease. Nat Rev Dis Primers. 2018;4:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Coffey AJ, Durkie M, Hague S, McLay K, Emmerson J, Lo C, et al. A genetic study of Wilson’s disease in the United Kingdom. Brain. 2013;136:1476–1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Espinós C, Ferenci P. Are the new genetic tools for diagnosis of Wilson disease helpful in clinical practice? JHEP Rep. 2020;2:100114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Panzer M, Viveiros A, Schaefer B, Baumgartner N, Seppi K, Djamshidian A, et al. Synonymous mutation in adenosine triphosphatase copper-transporting beta causes enhanced exon skipping in Wilson disease. Hepatol Commun. 2022;6:1611–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Xu WQ, Wang RM, Dong Y, Wu ZY. Pathogenicity of intronic and synonymous variants of ATP7B in Wilson disease. J Mol Diagn. 2023;25:57–67. [DOI] [PubMed] [Google Scholar]
- 68. Vilarinho S, Choi M, Jain D, Malhotra A, Kulkarni S, Pashankar D, et al. Individual exome analysis in diagnosis and management of paediatric liver failure of indeterminate aetiology. J Hepatol. 2014;61:1056–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Strnad P, McElvaney NG, Lomas DA. Alpha(1)-antitrypsin deficiency. N Engl J Med. 2020;382:1443–1455. [DOI] [PubMed] [Google Scholar]
- 70. Strnad P, Buch S, Hamesch K, Fischer J, Rosendahl J, Schmelz R, et al. Heterozygous carriage of the alpha1-antitrypsin Pi*Z variant increases the risk to develop liver cirrhosis. Gut. 2019;68:1099–1107. [DOI] [PubMed] [Google Scholar]
- 71. Hakim A, Moll M, Qiao D, Liu J, Lasky-Su JA, Silverman EK, et al. Heterozygosity of the alpha 1-antitrypsin Pi*Z allele and risk of liver disease. Hepatol Commun. 2021;5:1348–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Stockley RA, Turner AM. α-1-Antitrypsin deficiency: clinical variability, assessment, and treatment. Trends Mol Med. 2014;20:105–115. [DOI] [PubMed] [Google Scholar]
- 73. Patel D, McAllister SL, Teckman JH. Alpha-1 antitrypsin deficiency liver disease. Transl Gastroenterol Hepatol. 2021;6:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Fromme M, Schneider CV, Trautwein C, Brunetti-Pierri N, Strnad P. Alpha-1 antitrypsin deficiency: A re-surfacing adult liver disorder. J Hepatol. 2022;76:946–958. [DOI] [PubMed] [Google Scholar]
- 75. Greene CM, Marciniak SJ, Teckman J, Ferrarotti I, Brantly ML, Lomas DA, et al. α1-Antitrypsin deficiency. Nat Rev Dis Primers. 2016;2:16051. [DOI] [PubMed] [Google Scholar]
- 76. Strnad P, Mandorfer M, Choudhury G, Griffiths W, Trautwein C, Loomba R, et al. Fazirsiran for liver disease associated with alpha1-antitrypsin deficiency. N Engl J Med. 2022;387:514–524. [DOI] [PubMed] [Google Scholar]
- 77. Ooi CY, Durie PR. Cystic fibrosis from the gastroenterologist’s perspective. Nat Rev Gastroenterol Hepatol. 2016;13:175–185. [DOI] [PubMed] [Google Scholar]
- 78. Castellani C, Cuppens H, Macek M, Jr, Cassiman JJ, Kerem E, Durie P, et al. Consensus on the use and interpretation of cystic fibrosis mutation analysis in clinical practice. J Cyst Fibros. 2008;7:179–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Bell SC, Mall MA, Gutierrez H, Macek M, Madge S, Davies JC, et al. The future of cystic fibrosis care: A global perspective. Lancet Respir Med. 2020;8:65–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Bartlett JR, Friedman KJ, Ling SC, Pace RG, Bell SC, Bourke B, et al. Genetic modifiers of liver disease in cystic fibrosis. JAMA. 2009;302:1076–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Czaja AJ. Cryptogenic chronic hepatitis and its changing guise in adults. Digest Dis Sci. 2011;56:3421–3438. [DOI] [PubMed] [Google Scholar]
- 82. Vilarinho S, Sari S, Yilmaz G, Stiegler AL, Boggon TJ, Jain D, et al. Recurrent recessive mutation in deoxyguanosine kinase causes idiopathic noncirrhotic portal hypertension. Hepatology. 2016;63:1977–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Drzewiecki K, Choi J, Brancale J, Leney-Greene MA, Sari S, Dalgiç B, et al. GIMAP5 maintains liver endothelial cell homeostasis and prevents portal hypertension. J Exp Med. 2021;218:e20201745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Ünlüsoy Aksu A, Das SK, Nelson-Williams C, Jain D, Özbay Hoşnut F, Evirgen Şahin G, et al. Recessive mutations in KIF12 cause high gamma-glutamyltransferase cholestasis. Hepatol Commun. 2019;3:471–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Gao E, Cheema H, Waheed N, Mushtaq I, Erden N, Nelson-Williams C, et al. Organic solute transporter alpha deficiency: A disorder with cholestasis, liver fibrosis, and congenital diarrhea. Hepatology. 2020;71:1879–1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Vilarinho S, Sari S, Mazzacuva F, Bilgüvar K, Esendagli-Yilmaz G, Jain D, et al. ACOX2 deficiency: A disorder of bile acid synthesis with transaminase elevation, liver fibrosis, ataxia, and cognitive impairment. Proc Natl Acad Sci USA. 2016;113:11289–11293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Moreno Traspas R, Teoh TS, Wong PM, Maier M, Chia CY, Lay K, et al. Loss of FOCAD, operating via the SKI messenger RNA surveillance pathway, causes a pediatric syndrome with liver cirrhosis. Nat Genet. 2022;54:1214–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Devane J, Ott E, Olinger EG, Epting D, Decker E, Friedrich A, et al. Progressive liver, kidney, and heart degeneration in children and adults affected by TULP3 mutations. Am J Hum Genet. 2022;109:928–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Maddirevula S, Alhebbi H, Alqahtani A, Algoufi T, Alsaif HS, Ibrahim N, et al. Identification of novel loci for pediatric cholestatic liver disease defined by KIF12, PPM1F, USP53, LSR, and WDR83OS pathogenic variants. Genet Med. 2019;21:1164–1172. [DOI] [PubMed] [Google Scholar]
- 90. Zheng M, Huang DQ, Konkwo C, Agrawal S, Khera AV, Loomba R, et al. Genomic analysis of lean individuals with NAFLD identifies monogenic disorders in a prospective cohort study. JHEP Rep. 2023;5:100692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Choi M, Scholl UI, Ji W, Liu T, Tikhonova IR, Zumbo P, et al. Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proc Natl Acad Sci USA. 2009;106:19096–19101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Wu AC, McMahon P, Lu C. Ending the diagnostic odyssey—Is whole-genome sequencing the answer? JAMA Pediatr. 2020;174:821–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Hayeems RZ, Bhawra J, Tsiplova K, Meyn MS, Monfared N, Bowdin S, et al. Care and cost consequences of pediatric whole genome sequencing compared to chromosome microarray. Eur J Hum Genet. 2017;25:1303–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Wade CH, Tarini BA, Wilfond BS. Growing up in the genomic era: Implications of whole-genome sequencing for children, families, and pediatric practice. Annu Rev Genomics Hum Genet. 2013;14:535–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. James KN, Clark MM, Camp B, Kint C, Schols P, Batalov S, et al. Partially automated whole-genome sequencing reanalysis of previously undiagnosed pediatric patients can efficiently yield new diagnoses. NPJ Genom Med. 2020;5:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Gao E, Hercun J, Heller T, Vilarinho S. Undiagnosed liver diseases. Transl Gastroenterol Hepatol. 2021;6:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Eslam M, George J. Genetic contributions to NAFLD: Leveraging shared genetics to uncover systems biology. Nat Rev Gastroenterol Hepatol. 2020;17:40–52. [DOI] [PubMed] [Google Scholar]
- 98. Hagström H, Nasr P, Ekstedt M, Hammar U, Stål P, Hultcrantz R, et al. Risk for development of severe liver disease in lean patients with nonalcoholic fatty liver disease: A long-term follow-up study. Hepatol Commun. 2018;2:48–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Leung JC-F, Loong TC-W, Wei JL, Wong GL-H, Chan AW-H, Choi PC-L, et al. Histological severity and clinical outcomes of nonalcoholic fatty liver disease in nonobese patients. Hepatology. 2017;65:54–64. [DOI] [PubMed] [Google Scholar]
- 100. Vilarinho S, Mistry PK. Exome sequencing in clinical hepatology. Hepatology. 2019;70:2185–2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Loomba R, Schork N, Chen CH, Bettencourt R, Bhatt A, Ang B, et al. Heritability of hepatic fibrosis and steatosis based on a prospective twin study. Gastroenterology. 2015;149:1784–1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Chung DH, Zheng M, Bale AE, Vilarinho S. Hepatology Genome Rounds: An interdisciplinary approach to integrate genomic data into clinical practice. J Hepatol. 2023;79:1065–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Ziol M, Barbu V, Rosmorduc O, Frassati-Biaggi A, Barget N, Hermelin B, et al. ABCB4 heterozygous gene mutations associated with fibrosing cholestatic liver disease in adults. Gastroenterology. 2008;135:131–141. [DOI] [PubMed] [Google Scholar]
- 104. Schatz SB, Jüngst C, Keitel-Anselmo V, Kubitz R, Becker C, Gerner P, et al. Phenotypic spectrum and diagnostic pitfalls of ABCB4 deficiency depending on age of onset. Hepatol Commun. 2018;2:504–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Reichert MC, Lammert F. ABCB4 gene aberrations in human liver disease: An evolving spectrum. Semin Liver Dis. 2018;38:299–307. [DOI] [PubMed] [Google Scholar]
- 106. Nayagam JS, Foskett P, Strautnieks S, Agarwal K, Miquel R, Joshi D, et al. Clinical phenotype of adult-onset liver disease in patients with variants in ABCB4, ABCB11, and ATP8B1. Hepatol Commun. 2022;6:2654–2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Avena A, Puggelli S, Morris M, Cerny A, Andrade AR, Pareti E, et al. ABCB4 variants in adult patients with cholestatic disease are frequent and underdiagnosed. Dig Liver Dis. 2021;53:329–344. [DOI] [PubMed] [Google Scholar]
- 108. Ibrahim SH, Kamath BM, Loomes KM, Karpen SJ. Cholestatic liver diseases of genetic etiology: Advances and controversies. Hepatology. 2022;75:1627–1646. [DOI] [PubMed] [Google Scholar]
- 109. Mitchell E, Gilbert M, Loomes KM. Alagille syndrome. Clin Liver Dis. 2018;22:625–641. [DOI] [PubMed] [Google Scholar]
- 110. Kamath BM, Spinner NB, Rosenblum ND. Renal involvement and the role of Notch signalling in Alagille syndrome. Nat Rev Nephrol. 2013;9:409–418. [DOI] [PubMed] [Google Scholar]
- 111. Kamath BM, Podkameni G, Hutchinson AL, Leonard LD, Gerfen J, Krantz ID, et al. Renal anomalies in Alagille syndrome: A disease-defining feature. Am J Med Genet Part A. 2012;158A:85–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Shrivastava R, Williams A, Mikhail A, Roberts D, Richards M, Aithal V. An unusual cause of hypertension and renal failure: A case series of a family with Alagille syndrome. Nephrol Dial Transplant. 2010;25:1501–1506. [DOI] [PubMed] [Google Scholar]
- 113. Tsai EA, Gilbert MA, Grochowski CM, Underkoffler LA, Meng H, Zhang X, et al. THBS2 is a candidate modifier of liver disease severity in Alagille Syndrome. Cell Mol Gastroenterol Hepatol. 2016;2:663–675.e662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Saleh M, Kamath BM, Chitayat D. Alagille syndrome: Clinical perspectives. Appl Clin Genet. 2016;9:75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Shalaby S, Ronzoni L, Hernandez-Gea V, Valenti L. The genetics of portal hypertension: Recent developments and the road ahead. Liver Int. 2023;43:2592–2603. [DOI] [PubMed] [Google Scholar]
- 116. Warasnhe K, Özçay F, Aydin Hİ, Özgün G, Ceylaner S. A novel mutation in TRMT5 associated with idiopathic non-cirrhotic portal hypertension and hepatopulmonary syndrome: Case report of two siblings. Clin Res Hepatol Gastroenterol. 2022;46:101928. [DOI] [PubMed] [Google Scholar]
- 117. Koot BG, Alders M, Verheij J, Beuers U, Cobben JM. A de novo mutation in KCNN3 associated with autosomal dominant idiopathic non-cirrhotic portal hypertension. J Hepatol. 2016;64:974–977. [DOI] [PubMed] [Google Scholar]
- 118. Besmond C, Valla D, Hubert L, Poirier K, Grosse B, Guettier C, et al. Mutations in the novel gene FOPV are associated with familial autosomal dominant and non-familial obliterative portal venopathy. Liver Int. 2018;38:358–364. [DOI] [PubMed] [Google Scholar]
- 119. Shan J, Megarbane A, Chouchane A, Karthik D, Temanni R, Romero AR, et al. Genetic predisposition to porto-sinusoidal vascular disorder: A functional genomic-based, multigenerational family study. Hepatology. 2023;77:501–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Ng SWK, Rouhani FJ, Brunner SF, Brzozowska N, Aitken SJ, Yang M, et al. Convergent somatic mutations in metabolism genes in chronic liver disease. Nature. 2021;598:473–478. [DOI] [PubMed] [Google Scholar]
- 121. Brunner SF, Roberts ND, Wylie LA, Moore L, Aitken SJ, Davies SE, et al. Somatic mutations and clonal dynamics in healthy and cirrhotic human liver. Nature. 2019;574:538–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Müller M, Bird TG, Nault J-C. The landscape of gene mutations in cirrhosis and hepatocellular carcinoma. J Hepatol. 2020;72:990–1002. [DOI] [PubMed] [Google Scholar]
- 123. Lee S, Dong HH. FoxO integration of insulin signaling with glucose and lipid metabolism. J Endocrinol. 2017;233:R67–r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Sveinbjornsson G, Ulfarsson MO, Thorolfsdottir RB, Jonsson BA, Einarsson E, Gunnlaugsson G, et al. Multiomics study of nonalcoholic fatty liver disease. Nat Genet. 2022;54:1652–1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Mann JP, Hoare M. A minority of somatically mutated genes in pre-existing fatty liver disease have prognostic importance in the development of NAFLD. Liver Int. 2022;42:1823–1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Zhu M, Lu T, Jia Y, Luo X, Gopal P, Li L, et al. Somatic mutations increase hepatic clonal fitness and regeneration in chronic liver disease. Cell. 2019;177:608–621.e612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Wang Z, Zhu S, Jia Y, Wang Y, Kubota N, Fujiwara N, et al. Positive selection of somatically mutated clones identifies adaptive pathways in metabolic liver disease. Cell. 2023;186:1968–1984.e1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Nault JC, Paradis V, Ronot M, Zucman-Rossi J. Benign liver tumours: Understanding molecular physiology to adapt clinical management. Nat Rev Gastroenterol Hepatol. 2022;19:703–716. [DOI] [PubMed] [Google Scholar]
- 129. Bluteau O, Jeannot E, Bioulac-Sage P, Marqués JM, Blanc JF, Bui H, et al. Bi-allelic inactivation of TCF1 in hepatic adenomas. Nat Genet. 2002;32:312–315. [DOI] [PubMed] [Google Scholar]
- 130. Yamagata K, Oda N, Kaisaki PJ, Menzel S, Furuta H, Vaxillaire M, et al. Mutations in the hepatocyte nuclear factor-1alpha gene in maturity-onset diabetes of the young (MODY3). Nature. 1996;384:455–458. [DOI] [PubMed] [Google Scholar]
- 131. Bacq Y, Jacquemin E, Balabaud C, Jeannot E, Scotto B, Branchereau S, et al. Familial liver adenomatosis associated with hepatocyte nuclear factor 1alpha inactivation. Gastroenterology. 2003;125:1470–1475. [DOI] [PubMed] [Google Scholar]
- 132. Willson JS, Godwin TD, Wiggins GA, Guilford PJ, McCall JL. Primary hepatocellular neoplasms in a MODY3 family with a novel HNF1A germline mutation. J Hepatol. 2013;59:904–907. [DOI] [PubMed] [Google Scholar]
- 133. Fu J, Wang T, Zhai X, Xiao X. Primary hepatocellular adenoma due to biallelic HNF1A mutations and its co-occurrence with MODY 3: Case-report and review of the literature. Endocrine. 2020;67:544–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Pilati C, Amessou M, Bihl MP, Balabaud C, Nhieu JT, Paradis V, et al. Somatic mutations activating STAT3 in human inflammatory hepatocellular adenomas. J Exp Med. 2011;208:1359–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Pilati C, Letouzé E, Nault JC, Imbeaud S, Boulai A, Calderaro J, et al. Genomic profiling of hepatocellular adenomas reveals recurrent FRK-activating mutations and the mechanisms of malignant transformation. Cancer Cell. 2014;25:428–441. [DOI] [PubMed] [Google Scholar]
- 136. Nault JC, Couchy G, Balabaud C, Morcrette G, Caruso S, Blanc JF, et al. Molecular classification of hepatocellular adenoma associates with risk factors, bleeding, and malignant transformation. Gastroenterology. 2017;152:880–894.e886. [DOI] [PubMed] [Google Scholar]
- 137. El-Serag HB, Rudolph KL. Hepatocellular carcinoma: Epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–2576. [DOI] [PubMed] [Google Scholar]
- 138. Tarao K, Nozaki A, Ikeda T, Sato A, Komatsu H, Komatsu T, et al. Real impact of liver cirrhosis on the development of hepatocellular carcinoma in various liver diseases-meta-analytic assessment. Cancer Med. 2019;8:1054–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Llovet JM, Kelley RK, Villanueva A, Singal AG, Pikarsky E, Roayaie S, et al. Hepatocellular carcinoma. Nat Rev Dis Primers. 2021;7:6. [DOI] [PubMed] [Google Scholar]
- 140. Vilarinho S, Erson-Omay EZ, Mitchell-Richards K, Cha C, Nelson-Williams C, Harmanci AS, et al. Exome analysis of the evolutionary path of hepatocellular adenoma-carcinoma transition, vascular invasion and brain dissemination. J Hepatol. 2017;67:186–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Takeda M, Sakai K, Terashima M, Kaneda H, Hayashi H, Tanaka K, et al. Clinical application of amplicon-based next-generation sequencing to therapeutic decision making in lung cancer. Ann Oncol. 2015;26:2477–2482. [DOI] [PubMed] [Google Scholar]
- 142. Nakagawa H, Fujita M. Whole genome sequencing analysis for cancer genomics and precision medicine. Cancer Sci. 2018;109:513–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Iannelli F, Collino A, Sinha S, Radaelli E, Nicoli P, D’Antiga L, et al. Massive gene amplification drives paediatric hepatocellular carcinoma caused by bile salt export pump deficiency. Nat Commun. 2014;5:3850. [DOI] [PubMed] [Google Scholar]
- 144. Liao W-W, Asri M, Ebler J, Doerr D, Haukness M, Hickey G, et al. A draft human pangenome reference. Nature. 2023;617:312–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Bycroft C, Freeman C, Petkova D, Band G, Elliott LT, Sharp K, Motyer A, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Denny JC, Rutter JL, Goldstein DB, Philippakis A, Smoller JW, Jenkins G, et al. The “all of us” research program. N Engl J Med. 2019;381:668–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Kaffe E, Roulis M, Zhao J, Qu R, Sefik E, Mirza H, et al. Humanized mouse liver reveals endothelial control of essential hepatic metabolic functions. Cell. 2023;186:3793–3809.e3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Aizarani N, Saviano A, Sagar, Mailly L, Durand S, Herman JS, et al. A human liver cell atlas reveals heterogeneity and epithelial progenitors. Nature. 2019;572:199–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Brancale J, Vilarinho S. A single cell gene expression atlas of 28 human livers. J Hepatol. 2021;75:219–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Guan Y, Enejder A, Wang M, Fang Z, Cui L, Chen SY, et al. A human multi-lineage hepatic organoid model for liver fibrosis. Nat Commun. 2021;12:6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Jager M, Blokzijl F, Sasselli V, Boymans S, Janssen R, Besselink N, et al. Measuring mutation accumulation in single human adult stem cells by whole-genome sequencing of organoid cultures. Nat Protoc. 2018;13:59–78. [DOI] [PubMed] [Google Scholar]
- 152. Sampaziotis F, Muraro D, Tysoe OC, Sawiak S, Beach TE, Godfrey EM, et al. Cholangiocyte organoids can repair bile ducts after transplantation in the human liver. Science. 2021;371:839–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Watanabe S, Kobayashi S, Ogasawara N, Okamoto R, Nakamura T, Watanabe M, et al. Transplantation of intestinal organoids into a mouse model of colitis. Nat Protoc. 2022;17:649–671. [DOI] [PubMed] [Google Scholar]
- 154. Roper J, Tammela T, Cetinbas NM, Akkad A, Roghanian A, Rickelt S, et al. In vivo genome editing and organoid transplantation models of colorectal cancer and metastasis. Nat Biotechnol. 2017;35:569–576. [DOI] [PMC free article] [PubMed] [Google Scholar]