Summary
In this work, we examine the use of environment-sensitive fluorescent dyes in fluorescence lifetime imaging microscopy (FLIM) biosensors. We screened merocyanine dyes to find an optimal combination of environment-induced lifetime changes, photostability, and brightness at wavelengths suitable for live-cell imaging. FLIM was used to monitor a biosensor reporting conformational changes of endogenous Cdc42 in living cells. The ability to quantify activity using phasor analysis of a single fluorophore (e.g., rather than ratio imaging) eliminated potential artifacts. We leveraged these properties to determine specific concentrations of activated Cdc42 across the cell.
Keywords: biosensor, Cdc42, FLIM, solvatochromic, dye, merocyanine, phasor, fluorescence, cell
Graphical abstract

Highlights
-
•
Characterize solvent-dependent fluorescence lifetimes to generate live-cell biosensors
-
•
Dye-protein conjugate changes lifetime upon binding activated Cdc42
-
•
Quantifies the concentration of a specific Cdc42 conformation in cells
-
•
FLIM biosensor used in cells to report localized Cdc42 activity during cell protrusion
Motivation
Fluorescent biosensors that read out protein activity as changes in the fluorescence lifetime of incorporated dyes can avoid artifacts associated with other designs and can be used to quantify the intracellular concentration of specific conformations. Here, we identify dyes suitable for live-cell imaging that undergo solvent-dependent changes in lifetime and construct a biosensor reporting the conformation of endogenous Cdc42 in living cells.
Mehl et al. construct biosensors based on dyes whose fluorescence lifetimes respond to their solvent environment. Lifetime changes of a biosensor reflect the changing level and location of Cdc42 activation during cell protrusion and enable quantitation of the activated concentration in cells.
Introduction
Fluorescent biosensors have been a powerful tool for untangling signaling behavior in living cells.1,2,3,4,5,6 Most often, they have been based on fluorescence resonance energy transfer (FRET), with readouts quantified using ratiometric fluorescence intensity measurements. Although ratiometric imaging can achieve fast acquisition with relatively high signal-to-noise ratio, fluorescence lifetime imaging microscopy (FLIM) can provide important advantages over FRET and other biosensor approaches.7,8,9 Unlike some ratiometric biosensors (e.g., those using two fluorophores on separate protein chains), single-dye FLIM biosensors do not need corrections for varying biosensor concentrations across the cell. In FRET biosensors, two fluorophores can bleach at different rates, complicating quantitation of photobleaching effects. It is difficult to distinguish between true FRET changes (reflecting actual biological interactions or conformational changes) and artifactual changes (due to incomplete folding, variable expression, or photobleaching) because all these changes manifest as alterations in fluorescence intensity at similar wavelengths. Using a single fluorophore on the biosensor simplifies multiplexed imaging.10,11,12,13
FLIM was previously explored for intracellular biosensor imaging, including studies of NAD(P)H,14 calcium,15,16 sodium,17 and protein conformation. For the latter, FLIM was used largely to image FRET,8,9,18,19,20,21 but FLIM biosensors based on single dyes have the potential for brighter emission than FRET because the dyes are directly excited.22,23,24 Environment-sensing dyes have been monitored in cells using fluorescence intensity changes,22,23,24,25,26 but biosensors based on the FLIM of such dyes remain largely unexplored.27,28
For biosensors, environment-sensing dyes have been attached to target proteins,6,29 where they are affected by conformational changes,30,31,32 or to “affinity reagents” (ARs) that bind selectively to the activated conformation of the target, causing a spectral change in the attached dye when the AR binds its target.22,24 ARs have been based on proteins, peptides, or even small molecules22,24,33 (Figure 1A). They can report the activation of endogenous, unmodified targets.
Figure 1.
Dye-based biosensors
(A) The dye-based Cdc42 biosensor consisted of an AR that specifically binds to the active conformation of Cdc42, with a covalently attached environment-sensing dye (red). Upon Cdc42 activation, the AR bound to Cdc42, altering the dye’s local environment and affecting optical properties, including brightness and fluorescence lifetime.
(B) Parent dyes (left) and derivatives with a cysteine-reactive linker (middle). Pht, phthalimide; BA, barbituric acid; TBA, thiobarbituric acid; SO, benzothiophene. n indicates the number of double bonds as shown in the structure at top. mero81 and mero77 are based on the red-shifted fluorophore.
(C) Phase lifetime measurements of parent merocyanine dyes in DMSO, methanol, and butanol (n = 3, data shown as mean ± SD).
Here, we make a FLIM-based biosensor by attaching a merocyanine dye to an AR that selectively binds the activated conformation of Cdc42. Such a biosensor was previously constructed using dyes undergoing intensity changes,24 and we here take advantage of the previous biochemical characterization of that biosensor, constructing it again but with dyes for FLIM readouts. Dyes were screened for optimal fluorescence and solvent-sensitive lifetime, and the dye with the best combination of brightness and lifetime response to Cdc42 binding was used in the final FLIM biosensor.23,24,25,26 The final FLIM biosensor provided sensitive readouts of Cdc42 activity in living cells. By adapting an approach used for calcium biosensors, we were able to determine the actual concentration of activated species, not just the relative distribution of activity. In summary, our results identify dyes suitable for FLIM biosensors and show how they can be used to quantify the subcellular concentrations of activated Cdc42. This can be a paradigm for the application of the dyes to other proteins.
Results
Merocyanine dyes
Merocyanine dyes can show changes in quantum yield, extinction coefficient, excitation maxima, and/or emission maxima when placed in different solvent environments.23,26,34,35,36,37,38 Dyes selected for photostability, brightness, and emission greater than 595 nm in DMSO23 were tested for FLIM changes that could be used in biosensors. Merocyanine dyes are characterized by electron donor and acceptor moieties linked together through conjugation. An indolenine donor can be combined with different strong electron acceptors to produce merocyanines with a useful compromise between brightness, photostability, and environment-sensitive fluorescence.23,25,26,36,37 Here, we linked indolenine to phthalimide (Pht), barbituric acid (BA), thiobarbituric acid (TBA), and benzothiophene (SO) (Figure 1B). Simple versions of the dyes, without reactive side chains, were assessed for FLIM and other fluorescence properties. Useful fluorophores were then derivatized with a cysteine-selective linker for attachment to the AR (Figure 1B). We examined four dyes with known favorable fluorescence properties, previously used in biosensors but not for FLIM, and one red-shifted dye synthesized for this study, potentially useful for multiplexed imaging. The dyes were screened for the sensitivity of their fluorescence lifetimes to environment. Figure 1C shows the measured phase lifetimes, τp, for each of the parent dyes prior to attachment of the cysteine-reactive linker. Lifetime was measured in three solvents with different polarity and hydrogen-bonding strengths.39 The parent merocyanine dyes included mero50, mero51, mero89, and mero88,23 all of which contain an indolenine donor but different acceptors (Pht, BA, TBA, and SO). The previously unpublished dyes, mero81 and mero77 (Figure 1B), had extended polyene systems that produced red-shifted absorption and emission spectra valuable for multiplexed imaging applications (Figure 2). They were shown to have photostability greater than Cy5, a well-known dye used for live-cell imaging (Figure S1A).
Figure 2.
Photophysical properties of mero77 and mero81
(A) The excitation and emission spectra of mero77 dissolved in DMSO.
(B–G) Emission spectra (B) and normalized emission spectra (C) of mero81, excitation spectra (D) and normalized excitation spectra (E) of mero81, absorption spectra (F) and normalized absorption spectra (G) of mero81.
(H) Molar extinction coefficients of mero81 at λmax.
(I) Solvents used in (B)–(G).
All parent dyes had lifetimes in the nanosecond range, suitable for current lifetime instrumentation.40 The shortest lifetimes were observed when the parent dyes were measured in the strongly polar, hydrogen-bonding solvent DMSO. In contrast, the longest lifetimes were observed in the least polar hydrogen-bonding solvent, butanol. Response to environment can be seen as resulting from differential solvent interaction with specific resonance structures41,42,43,44 or electron distributions.34,35,36,37
In vitro assay of biosensor response
The solvent dependence of dye lifetimes was promising, but it was hard to predict how a given dye would change FLIM when attached to an AR and exposed to a given protein (see discussion). Therefore, each dye was tested in an actual Cdc42 biosensor. A water-soluble, cysteine-reactive side chain was attached to each dye for covalent coupling to the single cysteine of the AR. This was based on the approach previously used to attach intensiometric dyes to the same cysteine.24 The AR was based on the Cdc42-binding domain (CBD) of Wiskott-Aldrich syndrome protein.23,24 It has been shown that this domain binds selectively to the activated, GTP-bound conformation of Cdc42.45 In vitro assays were performed to measure the difference in lifetime between bound and unbound biosensors. The final products were named mero-CBD biosensors,23 with the number denoting the dye (e.g., mero87-CBD for the AR CBD labeled with dye mero87).
The phasor approach is a powerful way to visualize lifetime changes without the aid of complex fitting routines.13,46,47 Figure 3A shows a phasor plot of mero87-CBD in the presence and absence of a constitutively active Cdc42 mutant (Q61L). The free and bound forms of the biosensor had unique phasor locations within the phasor plot, defining characteristic lifetimes for each state. Utilizing the property of linear combination, a phasor that contains a portion of free and bound biosensors will be located somewhere along a line connecting the two distinct states. We observed that upon biosensor binding to Cdc42, the lifetime of all tested biosensors increased (Figure 3B). Additionally, the phasors did not lie on the “universal circle,”8,13,47 indicating complex decay kinetics.
Figure 3.
In vitro assay for lifetime changes associated with Cdc42 binding and measurement of endogenous Cdc42 activation
(A) Phasor histogram of mero87-CBD biosensor (400 nM) in the presence and absence of 12 μM constitutively active Cdc42 mutant (Q61L). Excess Cdc42 was used to shift the equilibrium of the biosensor to the bound form. G = M∗cosφ; S = M∗sinφ, where M is the modulation and φ is the phase shift in the frequency domain measurements.8,13,47
(B) Bar graph displaying phase lifetimes13 of biosensors made with the indicated dyes (n = 3, data shown as mean ± SD).
(C–E) Intensity (C), activated Cdc42 concentration (D), and phasor histogram (E) for cells injected with mero87-CBD (12 cells) or mero87-mCBD (AR mutated to abrogate Cdc42 binding; 7 cells). The phasor histograms are labeled with the locations of free and bound biosensors (solid red circles). The hash marks indicate 10% increments of protein activation. Scale bar: 10 μm.
All Cdc42 biosensors exhibited an appreciable change in lifetime. Mero87-CBD showed the largest difference between the free and bound conformations. Although dye mero81 performed the best in solvent tests (Figure 1C), the biosensor derived from it (through reaction with mero77, its reactive derivative) showed a relatively weak response to Cdc42 binding. This again showed that response to proteins is complex and that dye selection for a given biosensor cannot be based simply on the performance of free dye in solution. We chose to pursue mero87-CBD because of its strong response to Cdc42. Changes in pH over the physiological range minimally affected the fluorescence of the attached dye (Figure S1B). The small changes observed outside this range were likely due to effects on the AR, not the dye, as there were no dye functional groups with pKa near this range.
Fluorescence lifetime imaging in living cells
NIH 3T3 mouse embryonic fibroblasts were microinjected with mero87-CBD and imaged on a confocal FLIM microscope. FLIM allowed us to visualize both the distribution of the biosensor (Figure 3C) and the absolute concentration of active Cdc42 (Figure 3D; STAR Methods). We used phasor locations of the free and bound biosensors from the in vitro assay (pf and pb, respectively, solid circles; Figure 3E, column 3); phasor locations determined in cells were comparable (Figures 4A and 4B; data are available at Zenodo, https://doi.org/10.5281/zenodo.10654587). The increase in emission intensity produced by binding active Cdc42 resulted in a nonlinear relationship between the fraction of activated protein (hash marks, increments of 10%) and the fractional contribution of each species to the overall lifetime (see discussion). Cells loaded with the wild-type biosensor (Figures 3C–3E, columns 1 and 2) had phasors that displayed a smear along the line connecting the two states, indicating a distribution of Cdc42 activity within the cells. The majority of pixels ranged between 0% and 10% of what would be observed for a fully bound biosensor, as indicated by the first two hash marks. This was consistent with previous biochemical studies.48,49 It is noteworthy that this relatively small change in binding produced a readily discernable change in lifetime shown by the phasor plot (see discussion). In the negative control cells where a mutation of the AR abolished the interaction with Cdc42 (Figures 3C–3E, column 3), the measured lifetimes were bunched near the free form of the biosensor, signifying negligible Cdc42 activation.
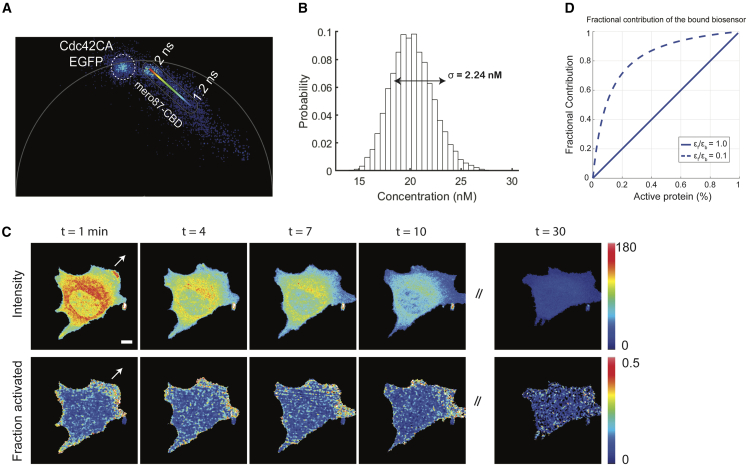
Figure 4.
Phasor analysis and FLIM imaging of mero87-CBD in a migrating mouse embryonic fibroblast (MEF) cell
(A) The phasor plot of mero87-CBD co-expressed with constitutively active Cdc42 in living cells showed that bounded biosensors could reach an extreme (2 ns) similar to that seen in vitro (Figure 3B).
(B) The slight discrepancy between the in vitro and live-cell measurements of phasor locations causes negligible changes in quantified concentrations. The simulation used the KD of mero87-CBD binding to Cdc42 with the assumption that the activation level was at 10%.
(C) Time course of the distribution (top) and fraction activated (bottom) of mero87-CBD in a migrating MEF cell. The cell was continuously imaged until it was completely photobleached at 30 min.
(D) Fractional contribution changes upon activation for a dye-based biosensor: brightness ratios of the free to the bound form are 1.0 (solid blue) or 0.1 (dashed blue). Dye-based FLIM biosensors are most sensitive to activation changes when protein activation levels are low. Scale bar: 10 μm.
Knowledge of active protein concentrations provides information valuable for the understanding and quantitative modeling of biochemical regulatory cycles. The spatial distribution of active Cdc42 concentrations across individual cells was calculated by analyzing phasors as described in the STAR Methods section (Equation 5). Shown in Figure 3D are the pseudo-colored, mapped concentrations of active Cdc42. Endogenous Cdc42 activation was observed near protrusions, consistent with previous results24,50 (Figures 3D and 4C).
Discussion
Here, we evaluated a series of bright dyes that showed solvent-induced changes in lifetime, promising for FLIM biosensors. Although one dye was clearly superior for monitoring Cdc42, the others provide a tool chest of dyes that could prove to be more useful for other targeted proteins. The response of the dyes to solvent changes did not indicate the extent of their response to our specific target, Cdc42. Interaction of dyes with proteins is complex and multifactorial, so this was not surprising. The dyes were marginally soluble in water (just enough to enable protein labeling when low concentrations of DMSO were included in buffers). This suggested that they interacted with protein surfaces rather than extended from the proteins, completely solvated by buffer. When the dye on AR was exposed to target protein, this could have changed interactions with specific residues or the ability of specific atoms in the dyes to undergo hydrogen bonding with water or protein side chains. Previous studies have shown that hydrogen bonding to specific atoms on the dye can have a strong effect and that general solvent effects play a role as well.37,38 The rigidity and length of the linker will be important in drawing the maximum response from each dye.
In the FLIM biosensors, only one fluorophore was needed, providing advantages. They did not show the complex photobleaching behavior encountered due to differential photobleaching of two fluorophores in FRET biosensors. In the future, simultaneous imaging of multiple biosensors can be simplified because each biosensor uses less of the spectrum.
The biosensors showed the greatest change in overall FLIM (or phasor location) at lower activation levels (Figures 4D; data available at Zenodo, https://doi.org/10.5281/zenodo.10654587). This feature will likely be especially valuable for GTPases, which can initiate cell behaviors when less than 10% of the protein population is active.48,49 The biosensor design used here responds to endogenous protein, reducing artifacts that come from overexpression or substituting target protein with modified analogs. Combining reduced perturbation with the ability to quantify low activated concentrations bodes well for the study of low-abundance targets or phenomena driven by low activation levels. We showed that the mero87-CBD biosensor could not only report the relative subcellular distribution of activity but could quantify the actual concentrations of activated species at different locations. We hope that the dyes described here can be used to provide quantitative readouts of active species for a range of protein targets.
Limitations of the study
One of the more difficult challenges in using dye-protein conjugates as biosensors is delivery of the biosensors to the interior of cells. A number of methods can accomplish this, including microinjection,22,24 electroporation,51 bead loading,52 and cell-penetrating peptides.53 However, these can be substantially more cumbersome than genetic encoding. More recently, biosensors have been generated by genetically encoding the protein portion and using membrane-permeable dyes to covalently attach dyes to proteins within live cells. This has been accomplished using unnatural amino acids for site-specific labeling26,54 or through attachment of fluorophores to enzyme fragments that catalyze attachment (e.g., SNAP-tag55 and HaloTag56). Labeling of unnatural amino acids with merocyanine dyes in living cells has already been used to generate biosensors.26
Another issue with dye-labeled biosensors is the propensity of some dyes to cause aggregation within cells or the cells’ recognition of the biosensor as a foreign body. Both result in packaging of the biosensors into vesicles, leading to punctate fluorescence superimposed on the signal being studied (see Figures 3C–3E and S1C). The impact of this depends on the kinetics of vesicle formation. In some cases, no puncta are seen for hours, if at all. For other biosensors, puncta form within minutes. Our control studies (Figure S1C) showed that, for the biosensor mero87-CBD (Figures 3C–3E), the presence of the dye increased the cell’s propensity to produce vesicles, albeit not to levels that prohibited our experiments regarding motility signaling. Finally, the merocyanine dyes here show a bi-exponential decay, which may present difficulties for some types of FLIM analysis. This may be due to the exposure of the dye to multiple environments or different solvent effects. Further study could improve this property.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| 2,3,3-Trimethylindolenine | TCI America | T0766 |
| N-(5-Anilino-2,4-pentadienylidene)aniline hydrochloride | TCI America | G0201 |
| Benzo[b]thiophen-3(2H)-one 1,1-dioxide | Ambeed Inc | A532355 |
| 2,3,3-trimethyl-1-(3-((3-sulfopropyl)amino)propyl)indolinium bromide | MacNevin et al.23 | N/A |
| 1,2,3,3-Tetramethylindolium iodide | Wang et al.57 | N/A |
| Dulbecco’s Modified Eagle’s Medium | Cellgro | 15-013-CV |
| Fetal Bovine Serum | Hyclone | SH30088.03 |
| GlutaMax | GIBCO | 35050061 |
| Hams/F12 | Caisson Labs | HFL12 |
| HEPES | GIBCO | 15630106 |
| Deposited data | ||
| Spectra of dyes and intermediates | This paper | In Zenodo: 10.5281/zenodo.10654587 |
| Experimental models: Cell lines | ||
| NIH 3T3 mouse embryo fibroblast | ClonTech | 630914 |
| HeLa cells | American Type Culture Collection (ATCC) | CCL-2; RRID: CVCL_0030 |
| Recombinant DNA | ||
| Cdc42 binding domain (CBD) of Wiskott-Aldrich Syndrome Protein (WASP), (Nalbant et al.24) | Provided by Dr. Gary Bokoch, Scripps Research Institute | N/A |
| Cdc42 Q61L (Nalbant et al.24) | Provided by Dr. Gary Bokoch, Scripps Research Institute | N/A |
| Software and algorithms | ||
| MATLAB 2016, 2020, 2022 | The MathWorks, Inc. | https://ww2.mathworks.cn/en/ |
| Code for FLIM analysis | This paper | In Zenodo: https://doi.org/10.5281/zenodo.10654587 |
Resource availability
Lead contact
Further information and requests for resources should be directed to and will be fulfilled by the lead contact, Klaus M. Hahn (khahn@med.unc.edu)
Materials availability
Plasmids in this study are available by request to lead contact, Dr. Klaus M. Hahn (khahn@med.unc.edu).
Data and code availability
-
•
Raw data reported in this paper is available from the lead contact upon request. Spectra of dyes and intermediates can be found in Figures S2 and S3 and at Zenodo: (https://doi.org/10.5281/zenodo.10654587).
-
•
Codes used for FLIM analysis and to generate Figures 4B and 4D can be found at Zenodo: (https://doi.org/10.5281/zenodo.10654587). These are also listed in the KRT and are available from the lead contact upon request.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contactupon request
Experimental model and study participant details
Cell lines
NIH 3T3 mouse embryo fibroblasts (MEF) were obtained from Clontech (RRID: CVCL_0594), authenticated by Short Tandem Repeat (STR) profile. HeLa cells were purchased from the American Type Culture Collection (ATCC, https://www.atcc.org/). All cells were tested for mycoplasma by polymerase chain reaction (PCR). MEF and HeLa cells were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% fetal bovine serum and 1X GlutaMax. Before imaging, cells were re-plated on fibronectin-coated coverslips and allowed to attach for at least 2 h. Before imaging, the media was replaced with Hams/F12 medium supplemented with 5% Fetal Bovine Serum and 10 mM HEPES.
Method details
Dye synthesis and protein conjugation
Dyes were synthesized as previously described,48,49 except for the dyes mero81 and mero77, whose synthesis is described below (Scheme: Synthesis of merocyanine dyes). Production of the WASP domain used as the affinity reagent, and its conjugation with dyes, was as described in MacNevin et al.,23 a modification of the procedure in Nalbant et al.24
2,3,3-Trimethylindolenine and N-(5-anilino-2,4-pentadienylidene)aniline hydrochloride were purchased from TCI America. Benzo[b]thiophen-3(2H)-one 1,1-dioxide was purchased from Ambeed Inc. All other reagents were purchased from Sigma-Aldrich. The following compounds were prepared according to published procedures: 1,2,3,3-tetramethylindolium iodide,57 and 2,3,3-trimethyl-1-(3-((3-sulfopropyl)amino)propyl)indolinium bromide.23 Reactions were run using anhydrous solvents. All operations with dyes were performed under dim light. UV-Visible spectra were recorded on a Hewlett-Packard 8453 diode array spectrophotometer or Agilent Cary 60 UV-Vis spectrophotometer. Fluorescence spectra were obtained on a Spex Fluorolog 2 spectrofluorometer. NMR spectra were taken on a Inova-400 or Bruker-500 or Bruker-700 or Bruker-850 spectrometer using deuterated solvents purchased from Cambridge Isotope Laboratories. Reference peaks of 7.26 ppm (CDCl3), 2.50 ppm and (DMSO-d6) were used for 1H NMR spectra. Reference peaks of 77.2 ppm (CDCl3), and 39.5 ppm (DMSO-d6) were used for 13C NMR spectra. High resolution mass spectra were obtained on a Thermo Scientific Q Exactive HF-X mass spectrometer with direct infusion. The synthesis is shown in Scheme: Synthesis of merocyanine dyes. NMR spectra and high resolution mass spectra of compounds S2, mero81, S3 and meo77 are available at Zenodo (https://doi.org/10.5281/zenodo.10654587) and in supplemental information (Figures S2 and S3).
Scheme. Synthesis of merocyanine dyes
N-((1E,3E,5E)-5-(1,1-dioxido-3-oxobenzo(b)thiophen-2(3H)-ylidene)penta-1,3-dien-1-yl)-N-phenylacetamide (S2)
A mixture of the benzo[b]thiophen-3(2H)-one 1,1-dioxide (2.73 g, 15.0 mmol) and N-(5-anilino-2,4-pentadienylidene)aniline hydrochloride (5.98 g, 21.0 mmol) were diluted in acetic anhydride (25 mL). The mixture was heated to reflux under argon for 2 h and cooled to room temperature. The resulting precipitate was filtered, washed with ethyl acetate, and dried under vacuum. The product was re-crystalized from isopropanol to give 4.16 g dark brown solid (73% yield). 1H NMR (CDCl3, 400 MHz) δ ppm 8.21 (d, J = 16.0 Hz, 1H), 8.07 (d, J = 8.0 Hz, 1H), 7.99 (d, J = 8.0 Hz, 1H), 7.89 (t, J = 8.0 Hz, 1H), 7.81 (t, J = 8.0 Hz, 1H), 7.70–7.60 (m, 4H), 7.28–7.14 (m, 3H), 6.86 (t, J = 12.0 Hz, 1H), 5.41 (t, J = 12.0 Hz, 1H), 2.02 (s, 3H); 13C NMR (CDCl3, 100 MHz) δ ppm 178.6, 169.4, 152.8, 144.1, 143.4, 140.8, 138.3, 136.0, 134.0, 133.2, 130.8, 130.0, 128.4, 128.1, 124.6, 122.3, 121.2, 113.3, 23.5; HRMS (positive mode) obsd 380.0946, calcd 380.0951 [(M + H)+, M = C21H17NO4S].
Mero81
1,2,3,3-Tetramethylindolium iodide (0.15 g, 0.50 mmol), compound S2 (0.21 g, 0.55 mmol), and sodium acetate (0.053 g, 0.65 mmol) were dissolved in methanol (2.0 mL) and heated to reflux for 10 min. The reaction mixture was cooled to room temperature, concentrated via rotary evaporation, and purified by silica gel chromatography using dichloromethane and ethyl acetate. The product was isolated as 0.136 g blue-olive solid (65% yield). 1H NMR (CDCl3, 400 MHz) δ ppm 8.00 (d, J = 8.0 Hz, 1H), 7.94 (d, J = 8.0 Hz, 1H), 7.81–7.66 (m, 3H), 7.48–7.39 (m, 1H), 7.28–7.18 (m, 3H), 7.03 (t, J = 8.0 Hz, 1H), 6.92 (t, J = 12.0 Hz,1H), 6.82 (d, J = 8.0 Hz, 1H), 6.38 (t, J = 12.0 Hz, 1H), 5.64 (d, J = 12.0 Hz, 1H), 3.31 (s, 3H), 1.63 (s, 6H); 13C NMR (CDCl3, 100 MHz) δ ppm 177.9, 165.9, 156.6, 145.3, 143.9, 143.9, 143.8, 139.8, 134.9, 133.9, 133.6, 128.3, 124.6, 124.1, 123.4, 122.4, 122.0, 120.8, 119.3, 108.0, 99.1, 47.4, 29.9, 28.4; HRMS (positive mode) obsd 418.1478, calcd 418.1471 [(M + H)+, M = C25H23NO3S].
3-((tert-Butoxycarbonyl)(3-((E)-2-((2E,4E,6Z)-6-(1,1-dioxido-3-oxobenzo[b]thiophen-2(3H)-ylidene)hexa-2,4-dien-1-ylidene)-3,3-dimethylindolin-1-yl)propyl)amino)propane-1-sulfonic acid (S3)
2,3,3-Trimethyl-1-(3-((3-sulfopropyl)amino)propyl)indolinium bromide (0.420 g, 1.00 mmol), and di-tert-butyl dicarbonate (1.09 g, 5.00 mmol) were mixed with water (0.2 mL). The resulting mixture was sonicated for a minute and then heated at 80°C under reduced pressure in a rotary evaporator for 30 min. The residue obtained was mixed with S2 (0.100 g, 0.264 mmol), sodium acetate (0.082 g, 1.00 mmol) and methanol (5.0 mL) under argon. The reaction mixture was stirred at 80°C under argon for 30 min. The solvent was removed under reduced pressure. The residue was purified on a flash chromatography using methanol/dichloromethane mixture (1:9) as an eluent. The product was isolated as a dark blue solid (105 mg, 58% yield). E/Z isomerism in tertiary amide caused peak doubling in 1H NMR and 13C NMR. 1H NMR (DMSO-d6, 850 MHz) δ ppm 8.03 (d, J = 7.6 Hz, 1H), 7.93–7.89 (m, 2H), 7.87–7.81 (m, 2H), 7.73 (t, J = 12.8 Hz, 1H), 7.55 (d, J = 13.6 Hz, 1H), 7.50 (d, J = 7.3 Hz, 1H), 7.33 (t, J = 7.7 Hz, 1H), 7.20 (d, J = 7.9 Hz, 1H), 7.12 (t, J = 7.4 Hz, 1H), 6.60 (t, J = 12.8 Hz, 1H), 6.51 (t, J = 12.8 Hz, 1H), 6.07 (d, J = 13.6 Hz, 1H), 3.96 (br s, 2H), 3.22–3.27 (br m, 4H), 2.38 (t, J = 7.4 Hz, 2H), 1.90 (br s, 2H), 1.79 (br s, 2H), 1.62 (s, 6H), 1.41 & 1.27 (2 s, 9H); 13C NMR (DMSO-d6, 213.7 MHz) δ ppm 175.6, 168.3, 158.4, 154.4, 149.5, 143.1, 142.5, 141.9, 140.2, 135.0, 134.0, 133.0, 128.2, 124.0, 123.2, 122.2, 120.4, 119.1, 116.6, 109.6, 100.6, 78.4, 49.0, 47.8, 46.1, 44.4, 44.2, 40.6, 28.0, 27.5, 26.2, 25.6, 24.9, 24.5; HRMS (negative mode) obsd 681.2316, calcd 681.2310 [(M-H)-, M = C35H42N2O8S2].
mero77
A solution of compound S3 (48 mg, 0.070 mmol) in dichloromethane (3.0 mL) was treated with trifluoroacetic acid (1.0 mL) and stirred under argon for 2 h. Completion of Boc deprotection was confirmed by TLC analysis. Then, the reaction mixture was concentrated under reduced pressure and the residue was thoroughly washed with diethyl ether. The resulting residue was mixed with iodoacetic anhydride (50 mg, 0.14 mmol), THF (1.0 mL) and N,N-diisopropylethyl amine (0.33 mmol, 0.057 mL) and stirred under argon for 1 h. The solvent was removed by rotary evaporation under reduced pressure. The residue was purified on a flash chromatography using methanol/dichloromethane mixture (3:17) as an eluent. The product was isolated as a dark blue solid (33 mg, 54% yield). E/Z isomerism in tertiary amide caused peak doubling in 1H NMR and 13C NMR. 1H NMR (DMSO-d6, 500 MHz) δ 8.02 (d, J = 7.3 Hz, 1H), 7.92–7.79 (m, 4H), 7.71 (t, J = 12.0 Hz, 1H), 7.54–7.49 (m, 2H), 7.36–7.31 (m, 1H), 7.24–7.20 (m, 1H), 7.12 (t, J = 7.6 Hz, 1H), 6.97 (t, J = 6.3 Hz, 0.5H), 6.60–6.46 (m, 1.5H), 6.17–6.06 (m, 1H), 5.25 (m, 0.5H), 4.47–4.31 (m, 1.5H), 4.00–3.90 (m, 2H), 3.48–3.36 (m, 4H), 2.46–2.36 (m, 2H), 1.92–1.74 (m, 4H), 1.62 (s, 6H); 13C NMR (DMSO-d6, 176 MHz) δ 175.5, 168.1, 167.4, 166.0, 165.6, 158.4, 149.6, 143.0, 142.5, 141.9, 140.1, 136.6, 135.1, 133.9, 132.7, 128.2, 124.2, 123.2, 122.2, 120.4, 118.9, 116.5, 109.6, 100.8, 48.6, 47.83, 47.77, 46.4, 42.8, 42.6, 42.0, 27.5, 27.4, 24.9, 24.8, 24.6, 23.5, −0.5, −4.4; HRMS (negative mode) obsd 749.0860, calcd 749.0858 [(M)-, M = C32H34IN2O7S2].
Lifetime measurements
Lifetimes in vitro and in live cells were collected on a modified laser scanning confocal microscope (Olympus FV1000) equipped with a UPLFL40X, 1.30 NA objective. The merocyanine dyes were excited at 594 nm using a pulsed supercontinuum laser source (Fianium) at 20 MHz in combination with an acousto-optic tunable filter (AOTF) (Crystal Technologies). The images were obtained with 256 x 256, 20 μs/pixel, 1.660 s/frame acquisition settings. Lifetime images were averaged over 35 frames for a total acquisition period of 58 s. The emission was detected with an external GaAsP photomultiplier detector (Hamamatsu: H722P-40). An emission bandpass filter (Semrock: FF-02-628/40-25) was placed before the PMT to reject unwanted excitation light. A digital frequency domain approach was used to acquire lifetime measurements (ISS: A320 FastFlim box).23 Calibration of the system was performed by measuring Texas red, which has a known lifetime of 4.2 ns in water. Processing was done with custom scripts developed in MATLAB.
The fluorescence decay of each merocyanine dye was acquired with a time-correlated single photon counting fluorometer based on an ISS Koala automated sample compartment, supercontinuum laser source (Fianium), Hamamatsu photomultiplier tube, and Becker & Hickl SPC-144 board. For each acquisition, the bin time was 24.43 ps and points were integrated over 60 s. Depending on the spectral properties of each merocyanine dye the Fianium laser was tuned to either 570 nm or 600 nm while emission was collected using a 600/25 nm or 625/25 nm bandpass filter placed in the front of the detector. The instrument response was calibrated by measuring the instantaneous response from an aqueous dispersion of glycogen. Experiments for Figures 4A, 4B, 4D and S1C were carried out with a Leica SP8X Falcon microscope, with excitation at 561 nm and emission collected between 570 and 640 nm.
Photobleaching of Cy5 and mero77
Photobleaching curves were measured as previously described.25 In brief, 1 μM dye was dissolved in 1% low-melting point agarose and sandwiched between two glass coverslips. Samples were imaged on a home-built inverted microscope equipped with a 100x, NA1.50 oil objective (UPLAPO100XOHR, Olympus) and a scientific CMOS (sCMOS, Photometrics 95B) camera. Samples were excited with a mercury lamp (U-LH100HG, Olympus) filtered by HQ630/45M (Chroma) and the fluorescence was collected through an HQ700/75M (Chroma) emission filter.
Calculation of active Cdc42 concentration
The following is based on a similar derivation previously published for the calcium sensor, CG5N.58 The following description has been included for completeness. The free, and bound, forms of the biosensors have unique locations within the phasor plot, defining characteristic lifetimes for both states. Utilizing the property of linear combinations, a phasor which contains a portion of free and bound biosensor, will be located somewhere along a line connecting the two states. This phasor position is given by the following equation.
| Equation 1 |
and correspond to the relative brightness of the free () and bound () states of the biosensor, respectively. The asterisk designates that Cdc42 is in its active, GTP-bound conformation. With knowledge of these phasor locations along with the relative brightness of the two states, the fraction of bound biosensor, can be calculated.
| Equation 2 |
To determine the concentration of active Cdc42, the dissociation constant, must be determined. Mathematically, this is presented through the law of mass action.
| Equation 3 |
When Equation 3 is inserted into Equation 2 and after some algebraic reorganization, the concentration of active Cdc42 is given by Equation 5. It is proportional to the dissociation constant and dependent on the fraction of bound biosensor.
| Equation 4 |
| Equation 5 |
The dissociation constant, and ratio of free to bound brightness were previously published for mero87-CBD; 186 ± 40 nM and 1/8.9 respectively.23 Pseudo-colored images of activated Cdc42 concentration were generated by first determining the fraction of bound biosensor for every pixel. This was achieved by projecting the measured phasor p for each pixel on the line connecting pf and pb (Equation 2). After this, the concentration was determined using Equation 5, with knowledge of the dissociation constant. Step-by-step instructions for use of the software are shown below.
Procedures for calculating protein concentrations in single cells.
1. Introduction
IFLI_Viewer is a GUI developed in Dr. Enrico Gratton’s Laboratory at the University of California, Irvine. The main functions include decode.fli FLIM data, phasor analysis, and FRET-FLIM analysis. The most recent version is maintained at the Laboratory for Fluorescence Dynamics, named as Globus for Images. This document gives step-by-step instructions for generating a protein concentration map in single cells using IFLI_Viewer.
2. Prerequisite
-
•
MATLAB (tested on 2016, 2020, 2022)
-
•
Phasor locations of the active and inactive species, obtained from in vitro data
-
•
FLIM data (.fli format) of single cells
-
•
The cell mask (binary image) generated by ImageJ/Fiji or MovieThresh (see Hahn lab)
3. Step-by-step instructions
3.1 Open IFLI View and load .fli data
-
•
Open MATLAB and change the current directory to the directory where IFLI_Viewer is located
-
•
Type in IFLI_View in the command window to run the GUI
-
•
Go to File>Open IFLI File to open an FLIM dataset (∗.fli)
-
•
The intensity image will show up, along with the raw phasor plot
-
•
A cell mask will be applied to clean up the phasor. This can be done under Intensity Image Tools>Apply Mask
3.2 Calculate fraction bound
-
•
Step up phasor locations of active and inactive species
-
•
Click Calculation Image Tools > Calculate Linear Projection > Calculate Fraction Bound and type in the intensity ratio of the free and bound states
-
•
To adjust the colorbar of the Fraction Bound Image, click on the Fraction Bound Image window, then go to the MATLAB Command Window and type in caxis([0 0.2])
3.3 Calculate concentration
-
•
Click Calculation Image Tools > Calculate Linear Projection > Calculate Concentration and type in the intensity ratio of the free and bound states, as well as the KD
Quantification and statistical analysis
Both the in vitro and in vivo FLIM data were analyzed using home-built software written in MATLAB (see data and code availability). Statistical details can be found in the figure legends. Data (Figures 1C, and 3B) were shown as mean ± SD.
Acknowledgments
We thank Nanjing Genvivo Biotech Co., Ltd. (China), for help with initial syntheses. We gratefully acknowledge the NIH for their support (NIGMS R35 GM122596). B.L. acknowledges partial support from the National Key R&D Program of China (2022YFC3400600) and the National Natural Science Foundation of China (32371519). Some FLIM experiments were performed at the UNC Neuroscience Microscopy Core (RRID:SCR_019060), supported partly by funding from the NIH-NINDS Neuroscience Center Support Grant P30 NS045892 and the NIH/NICHD Intellectual and Developmental Disabilities Research Center Support Grant U54 HD079124. UNC core facilities supporting this work were funded by the National Science Foundation (grant no. CHE-1726291 for the Mass Spec facility), and the NIH (P30CA016086 for the NMR facility and S10OD032476 for upgrading the 500 MHz NMR spectrometer).
Author contributions
B.P.M. initiated the study together with K.M.H. and led the work involving live-cell imaging and FLIM characterization. P.V., C.J.M., and C.-W.H. synthesized and characterized the dyes. L.L. contributed to lifetime imaging in living cells. E.H. determined lifetimes in solvent. B.L. carried out imaging studies, phasor analysis, and quantitation of concentrations. E.G., B.L., and K.M.H. contributed intellectually and organized the studies. The manuscript was written by B.P.M., K.M.H., and B.L. with input from all authors.
Declaration of interests
The authors declare no competing interests.
Published: March 18, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.crmeth.2024.100734.
Contributor Information
Bei Liu, Email: beiliu@pku.edu.cn.
Klaus M. Hahn, Email: khahn@med.unc.edu.
Supplemental information
References
- 1.Greenwald E.C., Mehta S., Zhang J. Genetically encoded fluorescent biosensors illuminate the spatiotemporal regulation of signaling networks. Chem. Rev. 2018;118:11707–11794. doi: 10.1021/acs.chemrev.8b00333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Komatsu N., Aoki K., Yamada M., Yukinaga H., Fujita Y., Kamioka Y., Matsuda M. Development of an optimized backbone of FRET biosensors for kinases and GTPases. Mol. Biol. Cell. 2011;22:4647–4656. doi: 10.1091/mbc.E11-01-0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Machacek M., Hodgson L., Welch C., Elliott H., Pertz O., Nalbant P., Abell A., Johnson G.L., Hahn K.M., Danuser G. Coordination of Rho GTPase activities during cell protrusion. Nature. 2009;461:99–103. doi: 10.1038/nature08242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marston D.J., Vilela M., Huh J., Ren J., Azoitei M.L., Glekas G., Danuser G., Sondek J., Hahn K.M. Multiplexed GTPase and GEF biosensor imaging enables network connectivity analysis. Nat. Chem. Biol. 2020;16:826–833. doi: 10.1038/s41589-020-0542-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pertz O., Hodgson L., Klemke R.L., Hahn K.M. Spatiotemporal dynamics of RhoA activity in migrating cells. Nature. 2006;440:1069–1072. doi: 10.1038/nature04665. [DOI] [PubMed] [Google Scholar]
- 6.Yasuda R., Harvey C.D., Zhong H., Sobczyk A., van Aelst L., Svoboda K. Supersensitive Ras activation in dendrites and spines revealed by two-photon fluorescence lifetime imaging. Nat. Neurosci. 2006;9:283–291. doi: 10.1038/nn1635. [DOI] [PubMed] [Google Scholar]
- 7.Bastiaens P.I., Squire A. Fluorescence lifetime imaging microscopy: spatial resolution of biochemical processes in the cell. Trends Cell Biol. 1999;9:48–52. doi: 10.1016/s0962-8924(98)01410-x. [DOI] [PubMed] [Google Scholar]
- 8.Hinde E., Digman M.A., Welch C., Hahn K.M., Gratton E. Biosensor Förster resonance energy transfer detection by the phasor approach to fluorescence lifetime imaging microscopy. Microsc. Res. Tech. 2012;75:271–281. doi: 10.1002/jemt.21054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hinde E., Digman M.A., Hahn K.M., Gratton E. Millisecond spatiotemporal dynamics of FRET biosensors by the pair correlation function and the phasor approach to FLIM. Proc. Natl. Acad. Sci. USA. 2013;110:135–140. doi: 10.1073/pnas.1211882110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berney C., Danuser G. FRET or no FRET: a quantitative comparison. Biophys. J. 2003;84:3992–4010. doi: 10.1016/S0006-3495(03)75126-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Datta R., Heaster T.M., Sharick J.T., Gillette A.A., Skala M.C. Fluorescence lifetime imaging microscopy: fundamentals and advances in instrumentation, analysis, and applications. J. Biomed. Opt. 2020;25:1–43. doi: 10.1117/1.JBO.25.7.071203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elangovan M., Wallrabe H., Chen Y., Day R.N., Barroso M., Periasamy A. Characterization of one- and two-photon excitation fluorescence resonance energy transfer microscopy. Methods. 2003;29:58–73. doi: 10.1016/s1046-2023(02)00283-9. [DOI] [PubMed] [Google Scholar]
- 13.Malacrida L., Ranjit S., Jameson D.M., Gratton E. The phasor plot: A universal circle to advance fluorescence lifetime analysis and interpretation. Annu. Rev. Biophys. 2021;50:575–593. doi: 10.1146/annurev-biophys-062920-063631. [DOI] [PubMed] [Google Scholar]
- 14.Blacker T.S., Mann Z.F., Gale J.E., Ziegler M., Bain A.J., Szabadkai G., Duchen M.R. Separating NADH and NADPH fluorescence in live cells and tissues using FLIM. Nat. Commun. 2014;5:3936. doi: 10.1038/ncomms4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herman B., Wodnicki P., Kwon S., Periasamy A., Gordon G.W., Mahajan N., Wang X.F. Recent developments in monitoring calcium and protein interactions in cells using fluorescence lifetime microscopy. J. Fluoresc. 1997;7:85–91. doi: 10.1007/BF02764581. [DOI] [Google Scholar]
- 16.Zheng K., Jensen T.P., Rusakov D.A. Monitoring intracellular nanomolar calcium using fluorescence lifetime imaging. Nat. Protoc. 2018;13:581–597. doi: 10.1038/nprot.2017.154. [DOI] [PubMed] [Google Scholar]
- 17.Despa S., Steels P., Ameloot M. Fluorescence lifetime microscopy of the sodium indicator sodium-binding benzofuran isophthalate in HeLa cells. Anal. Biochem. 2000;280:227–241. doi: 10.1006/abio.2000.4505. [DOI] [PubMed] [Google Scholar]
- 18.Berezin M.Y., Achilefu S. Fluorescence lifetime measurements and biological imaging. Chem. Rev. 2010;110:2641–2684. doi: 10.1021/cr900343z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ng T., Squire A., Hansra G., Bornancin F., Prevostel C., Hanby A., Harris W., Barnes D., Schmidt S., Mellor H., et al. Imaging protein kinase Calpha activation in cells. Science. 1999;283:2085–2089. doi: 10.1126/science.283.5410.2085. [DOI] [PubMed] [Google Scholar]
- 20.Orthaus S., Buschmann V., Bülter A., Fore S., König M., Erdmann R. 2009. Quantitative in Vivo Imaging of Molecular Distances Using FLIM-FRET.https://www.picoquant.com/images/uploads/page/files/7267/appnote_flim_fret.pdfhttps://www.picoquant.com/images/uploads/page/files/7267/appnote_flim_fret.pdf [Google Scholar]
- 21.Wallrabe H., Periasamy A. Imaging protein molecules using FRET and FLIM microscopy. Curr. Opin. Biotechnol. 2005;16:19–27. doi: 10.1016/j.copbio.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 22.Gulyani A., Vitriol E., Allen R., Wu J., Gremyachinskiy D., Lewis S., Dewar B., Graves L.M., Kay B.K., Kuhlman B., et al. A biosensor generated via high-throughput screening quantifies cell edge Src dynamics. Nat. Chem. Biol. 2011;7:437–444. doi: 10.1038/nchembio.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.MacNevin C.J., Gremyachinskiy D., Hsu C.-W., Li L., Rougie M., Davis T.T., Hahn K.M. Environment-sensing merocyanine dyes for live cell imaging applications. Bioconjug. Chem. 2013;24:215–223. doi: 10.1021/bc3005073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nalbant P., Hodgson L., Kraynov V., Toutchkine A., Hahn K.M. Activation of endogenous Cdc42 visualized in living cells. Science. 2004;305:1615–1619. doi: 10.1126/science.1100367. [DOI] [PubMed] [Google Scholar]
- 25.MacNevin C.J., Toutchkine A., Marston D.J., Hsu C.-W., Tsygankov D., Li L., Liu B., Qi T., Nguyen D.-V., Hahn K.M. Ratiometric imaging using a single dye enables simultaneous visualization of rac1 and cdc42 activation. J. Am. Chem. Soc. 2016;138:2571–2575. doi: 10.1021/jacs.5b09764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.MacNevin C.J., Watanabe T., Weitzman M., Gulyani A., Fuehrer S., Pinkin N.K., Tian X., Liu F., Jin J., Hahn K.M. Membrane-Permeant, Environment-Sensitive Dyes Generate Biosensors within Living Cells. J. Am. Chem. Soc. 2019;141:7275–7282. doi: 10.1021/jacs.8b09841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berezin M.Y., Akers W.J., Guo K., Fischer G.M., Daltrozzo E., Zumbusch A., Achilefu S. Long fluorescence lifetime molecular probes based on near infrared pyrrolopyrrole cyanine fluorophores for in vivo imaging. Biophys. J. 2009;97:L22–L24. doi: 10.1016/j.bpj.2009.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klymchenko A.S. Solvatochromic and Fluorogenic Dyes as Environment-Sensitive Probes: Design and Biological Applications. Acc. Chem. Res. 2017;50:366–375. doi: 10.1021/acs.accounts.6b00517. [DOI] [PubMed] [Google Scholar]
- 29.Yasuda R. Imaging spatiotemporal dynamics of neuronal signaling using fluorescence resonance energy transfer and fluorescence lifetime imaging microscopy. Curr. Opin. Neurobiol. 2006;16:551–561. doi: 10.1016/j.conb.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 30.Garrett S.C., Hodgson L., Rybin A., Toutchkine A., Hahn K.M., Lawrence D.S., Bresnick A.R. A biosensor of S100A4 metastasis factor activation: inhibitor screening and cellular activation dynamics. Biochemistry. 2008;47:986–996. doi: 10.1021/bi7021624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hahn K., DeBiasio R., Taylor D.L. Patterns of elevated free calcium and calmodulin activation in living cells. Nature. 1992;359:736–738. doi: 10.1038/359736a0. [DOI] [PubMed] [Google Scholar]
- 32.Nomanbhoy T.K., Leonard D.A., Manor D., Cerione R.A. Investigation of the GTP-binding/GTPase cycle of Cdc42Hs using extrinsic reporter group fluorescence. Biochemistry. 1996;35:4602–4608. doi: 10.1021/bi951743d. [DOI] [PubMed] [Google Scholar]
- 33.Konze K.D., Ma A., Li F., Barsyte-Lovejoy D., Parton T., Macnevin C.J., Liu F., Gao C., Huang X.-P., Kuznetsova E., et al. An orally bioavailable chemical probe of the Lysine Methyltransferases EZH2 and EZH1. ACS Chem. Biol. 2013;8:1324–1334. doi: 10.1021/cb400133j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Han W.-G., Liu T., Himo F., Toutchkine A., Bashford D., Hahn K.M., Noodleman L. A theoretical study of the UV/visible absorption and emission solvatochromic properties of solvent-sensitive dyes. ChemPhysChem. 2003;4:1084–1094. doi: 10.1002/cphc.200300801. [DOI] [PubMed] [Google Scholar]
- 35.Liu T., Han W.-G., Himo F., Ullmann G.M., Bashford D., Toutchkine A., Hahn K.M., Noodleman L. Density Functional Vertical Self-Consistent Reaction Field Theory for Solvatochromism Studies of Solvent-Sensitive Dyes. J. Phys. Chem. A. 2004;108:3545–3555. doi: 10.1021/jp031062p. [DOI] [Google Scholar]
- 36.Toutchkine A., Kraynov V., Hahn K. Solvent-sensitive dyes to report protein conformational changes in living cells. J. Am. Chem. Soc. 2003;125:4132–4145. doi: 10.1021/ja0290882. [DOI] [PubMed] [Google Scholar]
- 37.Toutchkine A., Nguyen D.-V., Hahn K.M. Merocyanine dyes with improved photostability. Org. Lett. 2007;9:2775–2777. doi: 10.1021/ol070780h. [DOI] [PubMed] [Google Scholar]
- 38.Toutchkine A., Han W.-G., Ullmann M., Liu T., Bashford D., Noodleman L., Hahn K.M. Experimental and DFT studies: novel structural modifications greatly enhance the solvent sensitivity of live cell imaging dyes. J. Phys. Chem. A. 2007;111:10849–10860. doi: 10.1021/jp073197r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hansen C.M. CRC Press; 2000. Hansen Solubility Parameters: A User’s Handbook. [Google Scholar]
- 40.Becker W. Fluorescence lifetime imaging--techniques and applications. J. Microsc. 2012;247:119–136. doi: 10.1111/j.1365-2818.2012.03618.x. [DOI] [PubMed] [Google Scholar]
- 41.Blanchard-Desce M., Wortmann R., Lebus S., Lehn J.-M., Krämer P. Intramolecular charge transfer in elongated donor-acceptor conjugated polyenes. Chem. Phys. Lett. 1995;243:526–532. doi: 10.1016/0009-2614(95)00895-B. [DOI] [Google Scholar]
- 42.Bublitz G.U., Ortiz R., Marder S.R., Boxer S.G. Stark spectroscopy of donor/acceptor substituted polyenes. J. Am. Chem. Soc. 1997;119:3365–3376. doi: 10.1021/ja9640814. [DOI] [Google Scholar]
- 43.Würthner F., Wortmann R., Matschiner R., Lukaszuk K., Meerholz K., DeNardin Y., Bittner R., Bräuchle C., Sens R. Merocyanine dyes in the cyanine limit: A new class of chromophores for photorefractive materials. Angew. Chem. Int. Ed. Engl. 1997;36:2765–2768. doi: 10.1002/anie.199727651. [DOI] [Google Scholar]
- 44.Würthner F., Wortmann R., Meerholz K. Chromophore design for photorefractive organic materials. ChemPhysChem. 2002;3:17–31. doi: 10.1002/1439-7641(20020118)3:1<17::AID-CPHC17>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 45.Abdul-Manan N., Aghazadeh B., Liu G.A., Majumdar A., Ouerfelli O., Siminovitch K.A., Rosen M.K. Structure of Cdc42 in complex with the GTPase-binding domain of the “Wiskott-Aldrich syndrome” protein. Nature. 1999;399:379–383. doi: 10.1038/20726. [DOI] [PubMed] [Google Scholar]
- 46.Colyer R.A., Lee C., Gratton E. A novel fluorescence lifetime imaging system that optimizes photon efficiency. Microsc. Res. Tech. 2008;71:201–213. doi: 10.1002/jemt.20540. [DOI] [PubMed] [Google Scholar]
- 47.Digman M.A., Caiolfa V.R., Zamai M., Gratton E. The phasor approach to fluorescence lifetime imaging analysis. Biophys. J. 2008;94:L14–L16. doi: 10.1529/biophysj.107.120154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Boulter E., Garcia-Mata R., Guilluy C., Dubash A., Rossi G., Brennwald P.J., Burridge K. Regulation of Rho GTPase crosstalk, degradation and activity by RhoGDI1. Nat. Cell Biol. 2010;12:477–483. doi: 10.1038/ncb2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jennings R.T., Knaus U.G. Rho family and Rap GTPase activation assays. Methods Mol. Biol. 2014;1124:79–88. doi: 10.1007/978-1-62703-845-4_6. [DOI] [PubMed] [Google Scholar]
- 50.Itoh R.E., Kurokawa K., Ohba Y., Yoshizaki H., Mochizuki N., Matsuda M. Activation of rac and cdc42 video imaged by fluorescent resonance energy transfer-based single-molecule probes in the membrane of living cells. Mol. Cell Biol. 2002;22:6582–6591. doi: 10.1128/MCB.22.18.6582-6591.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weaver J.C. Electroporation: a general phenomenon for manipulating cells and tissues. J. Cell. Biochem. 1993;51:426–435. doi: 10.1002/jcb.2400510407. [DOI] [PubMed] [Google Scholar]
- 52.McNeil P.L., Warder E. Glass beads load macromolecules into living cells. J. Cell Sci. 1987;88(Pt 5):669–678. doi: 10.1242/jcs.88.5.669. (Pt 5) [DOI] [PubMed] [Google Scholar]
- 53.Fonseca S.B., Pereira M.P., Kelley S.O. Recent advances in the use of cell-penetrating peptides for medical and biological applications. Adv. Drug Deliv. Rev. 2009;61:953–964. doi: 10.1016/j.addr.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 54.Lang K., Chin J.W. Cellular incorporation of unnatural amino acids and bioorthogonal labeling of proteins. Chem. Rev. 2014;114:4764–4806. doi: 10.1021/cr400355w. [DOI] [PubMed] [Google Scholar]
- 55.Keppler A., Gendreizig S., Gronemeyer T., Pick H., Vogel H., Johnsson K. A general method for the covalent labeling of fusion proteins with small molecules in vivo. Nat. Biotechnol. 2003;21:86–89. doi: 10.1038/nbt765. [DOI] [PubMed] [Google Scholar]
- 56.Los G.V., Encell L.P., McDougall M.G., Hartzell D.D., Karassina N., Zimprich C., Wood M.G., Learish R., Ohana R.F., Urh M., et al. HaloTag: a novel protein labeling technology for cell imaging and protein analysis. ACS Chem. Biol. 2008;3:373–382. doi: 10.1021/cb800025k. [DOI] [PubMed] [Google Scholar]
- 57.Wang J., Cao W.-F., Su J.-H., Tian H., Huang Y.-H., Sun Z.-R. Syntheses and nonlinear absorption of novel unsymmetrical cyanines. Dyes Pigments. 2003;57:171–179. doi: 10.1016/S0143-7208(03)00034-2. [DOI] [Google Scholar]
- 58.Celli A., Sanchez S., Behne M., Hazlett T., Gratton E., Mauro T. The epidermal Ca(2+) gradient: Measurement using the phasor representation of fluorescent lifetime imaging. Biophys. J. 2010;98:911–921. doi: 10.1016/j.bpj.2009.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Raw data reported in this paper is available from the lead contact upon request. Spectra of dyes and intermediates can be found in Figures S2 and S3 and at Zenodo: (https://doi.org/10.5281/zenodo.10654587).
-
•
Codes used for FLIM analysis and to generate Figures 4B and 4D can be found at Zenodo: (https://doi.org/10.5281/zenodo.10654587). These are also listed in the KRT and are available from the lead contact upon request.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contactupon request