Acute lymphoblastic leukemia (ALL) is the most common malignancy among children in industrialized countries, with a peak incidence between 2 and 5 years of age.1 The early onset of this cancer and heterogeneity in incidence by race and ethnicity implicates the influence of inherited genetic susceptibility in which evidence from genome-wide association studies (GWAS) of childhood ALL have identified several genomic regions associated with risk.2 To date, the identification of risk loci has been driven by studies conducted in populations of Hispanic or European ancestry, with a paucity of genome-wide studies performed in Asian populations.3 Pursuit of potential population-specific loci through genome-wide assessment and characterization of known loci across diverse populations is important to advance our understanding of inherited genetic variation in the risk childhood ALL.
Our previous study of targeted loci conducted within the Tokyo Children Cancer Study Group (TCCSG) showed that risk associations for single nucleotide polymorphism (SNP) in ARID5B, IKZF1 and PIP4K2A transfer to the Japanese population.4 As a next step, this current study included two independent GWAS series assembled through TCCSG and the Japanese Pediatric Leukemia/Lymphoma Study Group (JPLSG), including a total of 1,088 cases and 5,315 controls, in the first comprehensive evaluation of genetic variation in the risk of childhood ALL in Japanese. The first series (TCCSG GWAS) comprised patients from the TCCSG clinical network,4, 5 and included childhood ALL patients diagnosed at age 19 years or younger prior to 2012 (N=621) from outpatient clinic visits between 2013 and 2015 through a convenience sampling approach. Controls comprised adult participants from the Nagahama Prospective Cohort for Comprehensive Human Bioscience (the Nagahama Study) (N=1,846) and the Hospital-based Epidemiologic Research Program at Aichi Cancer Center (HERPACC) Study (N=2,170).6, 7 The second series (JPLSG GWAS) comprised childhood B-cell precursor (BCP) ALL patients (N=572) aged 1 to 19 years newly diagnosed between 2012 and 2018 through the nationwide ALL-B12 clinical study (registry: UMIN000009339).5, 8 Controls comprised a subset of participants from the Nagahama Study (N=1,924). DNA were extracted from saliva samples (TCCSG) or peripheral blood at remission (JPLSG) and were genotyped with the Illumina HumanCoreExome and OmniExpress microarrays, respectively. Institutional review board approvals were obtained from St. Luke’s International University and the major collaborating centers.
We performed quality control (QC) steps separately for the TCCSG and JPLSG cases and controls followed by additional QC filters after merging the case-control series for the TCCSG and JPLSG GWAS separately. After sample and SNP exclusions based on a standard QC approach (Online Supplementary Figure S1), a total of 258,069 and 481,270 directly genotyped SNP was available for the TCCSG GWAS (540 cases and 3,714 controls) and JPLSG GWAS (548 cases and 1,601 controls), respectively. Genome-wide SNP imputation was performed using ShapeIT2 and Minimac4 with an in-house Japanese haplotype reference panel. After post-imputation QC, bi-allelic loci shared between the TCCSG and JPLSG case-control series resulted in data for a total of 6,446,781 SNP for both GWAS.
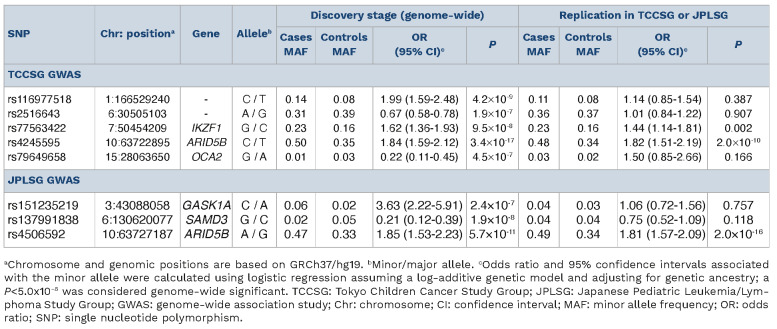
Patients included in the TCCSG and JPLSG series comprised predominately of B-cell ALL (TCCSG, 93%; JPLSG, 100%), greater numbers of males (TCCSG, 52%; JPLSG, 58%) than females, and showed the majority to be between 1 and 6 years of age (TCCSG, 69%; JPLSG, 57%). We first performed a discovery analysis in the TCCSG series and observed a novel association represented by SNP rs116977518 (odds ratio [OR] =1.99, P=4.2x10-9) at 1q24.1 (intergenic, proximity to FMO8P) and an association at a known region represented by rs4245595 (OR=1.84, P=3.4x10-17) located at 10q21.2 (ARID5B) (Table 1; Online Supplementary Figure S2A). An association with the previously identified IKZF1 region was also found (rs77563422, OR=1.62, P=9.5x10-8). Only SNP in ARID5B (rs4245595, OR=1.82, P=2.0x10-10) and IKZF1 (rs77563422, OR=1.44, P=0.002) replicated in the JPLSG series (Table 1). Confirmation is still necessary for the putative risk locus at 1q24.1. This locus is located adjacent to the FMO8P and FMO9P pseudogenes, and contains expression quantitative trait loci (eQTL) in blood for the deoxyuridine triphosphatase pseudogene 6 (DUTP6) gene as documented in the Genotype-Tissue Expression (GTEx) portal. In a gene expression profiling study of tonsil squamous cell carcinoma, DUTP6, along with other pseudogenes and small nuclear RNA, were found to be upregulated in blood mononuclear cells of patients compared to controls.9 Interestingly, the leading SNP in this region, rs116977518, is rare or not present in most other racial and ethnic populations.
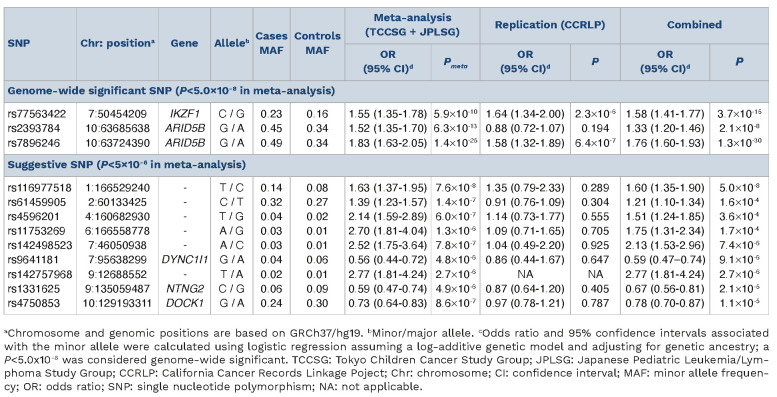
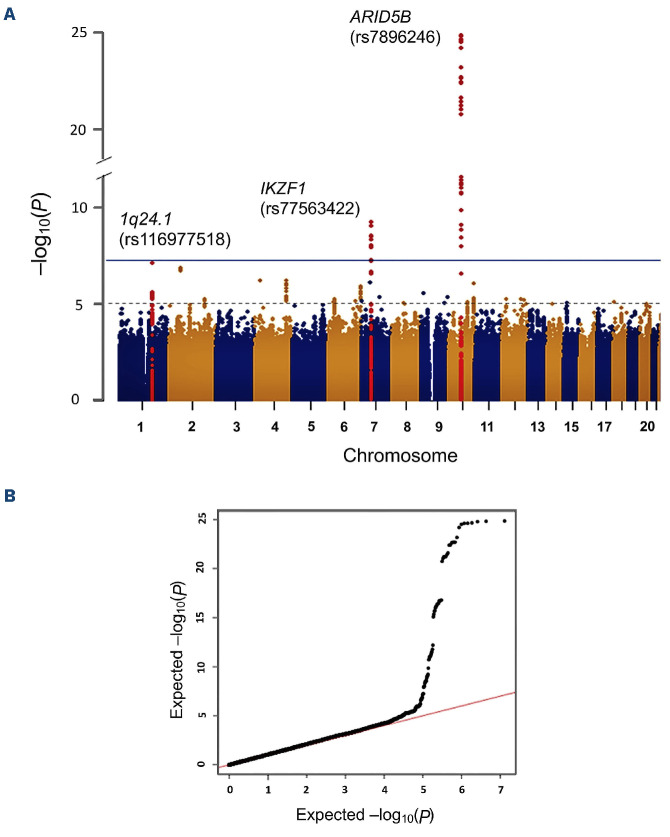
Next, we performed a discovery analysis in the JPLSG GWAS, and observed an association with the known ARID5B region (rs4506592, OR=1.85, P=5.7x10-1 1), along with another region at 6q23.1 in the sterile α motif domain containing 3 (SAMD3) gene (rs137991838, OR=0.21, P=1.9x10-8) (Table 1; Online Supplementary Figure S2B). The novel SAMD3 SNP association did not appear to replicate in the overall TCCSG case-control series, but limiting to only B-cell ALL showed a reduced risk (rs137991838, OR=0.67, P=0.046). The SAMD3 gene exhibits the highest expression levels in lymphoid tissues and blood.10 It belongs to the sterile α motif (SAM) domain superfamily in which the characteristic SAM domain suggests involvement in diverse protein-protein interactions important in assembly, regulation, and localization of functional elements.11 The leading SNP, rs137991838, is unique to the Japanese population and resides within a region that contain eQTL for SAMD3 in lymphoblastoid cell lines according to GTEx and RegulomeDB. Chromosomal aberrations of the 6q23 region are known to be common across a diverse range of tumor types, including hematologic malignancies.12 In a genome-wide SNP meta-analysis of the TCCSG and JPLSG GWAS combined, three SNP representing regions with genome-wide significant associations included rs77563422 (IKZF1, OR=1.55, P=5.9x10-10), and two uncorrelated SNP in ARID5B separated by about 38 kb (r2=0.07), rs2393784 (OR=1.52, P=6.3x10-13) and rs7896246 (OR=1.83, P=1.4x10-25) (Table 2; Figure 1). Replication opportunities of discovery results were pursued within cases (N=318) and controls (N=5,107) of East Asian ancestry from the California Cancer Records Linkage Project (CCRLP), a study based on the birth population of California previously reported.13 The associations were confirmed in this CCRLP replication series except for rs2393784 in ARID5B (Table 2). Conditional analysis of the two ARID5B SNP showed attenuation in effect size for both loci, but evidence of independent associations remained (rs2393784, OR=1.22, P=2.1x10-3; rs7896246, OR=1.69, P=1.6x10-16). rs2393784 is located about 38 kb upstream in intron 2, a SNP in LD (rs6479778) has been shown associated with both ALL relapse and disease risk in a US population.14 ARID5B SNP associations represent some of the most consistently observed in childhood ALL susceptibility, all of which suggest a role for variation in intronic regions and thus, mechanisms that involve gene regulation through affecting RNA splicing, transcription factor binding, and other processes. In a UK study, fine-mapping in high-hyperdiploid ALL cases and controls identified two plausibly casual SNP in LD, one of which is the same top hit identified in the current study (rs7896246).15
Table 1.
Results of top single nucleotide polymorphisms (P<1.0×10-6) associated with childhood acute lymphoblastic leukemia risk observed in the TCCSG and JPLSG genome-wide association analyses.
Table 2.
Meta-analysis of TCCSG and JPLSG genome-wide association analysis results and independent replication in CCRLP.
The association between IKZF1 and ALL risk has been confirmed repeatedly for rs4132601 and rs11978267 in populations of European, Hispanic, and African ancestry, but has been less clear for East Asians.3 For both SNP, East Asians exhibit among the lowest allele frequencies (MAF~0.08), and previous studies in this population may have been hampered by statistical power. ALL associations replicated for both rs4132601 and rs11978267 (P<0.01). We also identified a genome-wide significant region in IKZF1 (rs77563422), which is uncorrelated with the known risk locus (r2<0.01), and results conditioning on the presence of rs4132601 resulted in a stronger effect size and significance for both variants (rs77563422, OR=1.61, P=2.6x10-11; rs4132601, OR=1.49, P=5.5x10-5). SNP rs77563422 is rare in populations of European ancestry and is located in a different intronic region about 16 kb upstream of the other known variants.
Figure 1.
Results of meta-analysis combining the TCCSG and JPLSG genome-wide association analysis results. (A) The Manhattan plot shows the -log10 (P value) of the logistic regression analysis plotted against the chromosomal position (GRCh37/ hg19). The blue line indicates the genome-wide significant P value threshold of 5x10-8, and the dotted line was the threshold (1x10-6) for considering results as suggestive. (B) A quantile-quantile plot of the observed versus expected distribution of the -log10 (P value) from the analysis. TCCSG: Tokyo Children Cancer Study Group; JPLSG: Japanese Pediatric Leukemia/Lymphoma Study Group.
We were able to confirm associations for known risk loci representing ARID5B, IKZF1, DDC, CEBPE, PIP4K2A, GATA3, IKZF3, and 8q24.21, with some showing a different leading SNP in Japanese (Online Supplementary Table S1). There are several reasons why certain associations may not have been detected, including insufficient statistical power due to lower allele frequencies and/or effect sizes, unavailable SNP data in sufficient LD with the causal locus, and analyses without similar subtype specificity as the original study. An overall limitation of the current study included limited access to molecular subtype data for this analysis. Notably, it is possible that the recruitment strategy of the TCCSG series may have over-represented patients with higher survival probabilities and specific molecular subtype profiles which could have affected replication attempts for loci that show subtype-specificity. In addition, the CCRLP replication population represented a broadly defined group of cases and controls of East Asian ancestry, and differing genetic substructure between Japanese and others of East Asian origins needs consideration in interpreting the failure to replicate.
In this first case-control GWAS effort in Japanese, we confirmed the strong ALL risk associations with ARID5B and IKZF1 variation, and we report two putative ALL risk associations suggesting a role for the 1q24.1 region and SAMD3, but confirmation is necessary. Together with also characterizing the effects of known risk loci in Japanese, we expect this study to aid efforts in understanding the heritability of childhood ALL in this population, a key step for elucidating the causes of this devastating disease.
Supplementary Material
Acknowledgments
We would like to thank the patients and families participating in this study, and staff of the collaborating hospitals for their various contributions. This study made use of data from the 1000 Genomes Project (http://www.internationalgenome.org/data) and the International HapMap Project.
Funding Statement
Funding: This work was support by funding from St. Luke’s Life Science Institute (Tokyo, Japan), Japan Society for the Promotion of Science (JSPS) KAKENHI grant number 26253041), Japan Agency for Medical Research and Development (grant numbers 15km0305013h0101, 16km0405107h0004, 21kk0305014), the Children’s Cancer Association of Japan, and the Japan Leukemia Research Fund. The HERPACC Study was supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Science, Sports, Culture and Technology of Japan Priority Areas of Cancer (17015018), Innovative Areas (221S0001), and JSPS KAKENHI grants (JP16H06277 and 22H04923 [CoBiA], JP26253041, JP20K10463) and Grant-in-Aid for the Third Term Comprehensive 10-year Strategy for Cancer Control from the Ministry of Health, Labour and Welfare of Japan. The work pertaining to the CCRLP replication series was supported by grants from the US National Institutes of Health (R01CA155461, R35GM142783). The biospecimens and/or data used in CCRLP were obtained from the California Biobank Program (SIS request#26), Section 6555(b), 17 CCR. The collection of cancer incidence data used in the CCRLP study was supported by the California Department of Public Health pursuant to California Health and Safety Code Section 103885, Centers for Disease Control and Prevention’s (CDC) National Program of Cancer Registries, under cooperative agreement 5NU58DP003862-04/DP003862, the National Cancer Institute’s Surveillance, Epidemiology and End Results Program under contract HHSN261201000140C awarded to the Cancer Prevention Institute of California, contract HHSN261201000035C awarded to the University of Southern California, and contract HHSN261201000034C awarded to the Public Health Institute. The ideas and opinions expressed herein are those of the author(s) and do not necessarily reflect the opinions of the State of California, Department of Public Health, the National Institutes of Health, and the Centers for Disease Control and Prevention or their Contractors and Subcontractors. This study used birth data obtained from the State of California Center for Health Statistics and Informatics. The California Department of Public Health is not responsible for the analyses, interpretations, or conclusions drawn by the authors regarding the birth data used in this publication.
Data-sharing statement
The datasets generated and analyzed during the current study are not publicly available due to privacy and ethical restrictions, but are available from the corresponding author on reasonable request.
References
- 1.Hunger SP, Mullighan CG. Acute lymphoblastic leukemia in children. N Engl J Med. 2015;373(16):1541-1552. [DOI] [PubMed] [Google Scholar]
- 2.Moriyama T, Relling MV, Yang JJ. Inherited genetic variation in childhood acute lymphoblastic leukemia. Blood. 2015;125(26):3988-3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shi Y, Du M, Fang Y, et al. Identification of a novel susceptibility locus at 16q23.1 associated with childhood acute lymphoblastic leukemia in Han Chinese. Hum Mol Genet. 2016;25(13):2873-2880. [DOI] [PubMed] [Google Scholar]
- 4.Urayama KY, Takagi M, Kawaguchi T, et al. Regional evaluation of childhood acute lymphoblastic leukemia genetic susceptibility loci among Japanese. Sci Rep. 2018;8(1):789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kato M, Manabe A. Treatment and biology of pediatric acute lymphoblastic leukemia. Pediatr Int. 2018;60(1):4-12. [DOI] [PubMed] [Google Scholar]
- 6.Inoue M, Tajima K, Takezaki T, et al. Epidemiology of pancreatic cancer in Japan: a nested case-control study from the Hospital-based Epidemiologic Research Program at Aichi Cancer Center (HERPACC). Int J Epidemiol. 2003;32(2):257-262. [DOI] [PubMed] [Google Scholar]
- 7.Terao C, Ota M, Iwasaki T, et al. IgG4-related disease in the Japanese population: a genome-wide association study. Lancet Rheumatol. 2019;1(1):e14-e22. [DOI] [PubMed] [Google Scholar]
- 8.Koh K, Kato M, Saito AM, et al. Phase II/III study in children and adolescents with newly diagnosed B-cell precursor acute lymphoblastic leukemia: protocol for a nationwide multicenter trial in Japan. Jpn J Clin Oncol. 2018;48(7):684-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marcussen M, Sonderkaer M, Bodker JS, et al. Oral mucosa tissue gene expression profiling before, during, and after radiation therapy for tonsil squamous cell carcinoma. PLoS One. 2018;13(1):e0190709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Uhlen M, Fagerberg L, Hallstrom BM, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347(6220):1260419. [DOI] [PubMed] [Google Scholar]
- 11.Qiao F, Bowie JU. The many faces of SAM. Sci STKE. 2005;2005(286):re7. [DOI] [PubMed] [Google Scholar]
- 12.Wang DM, Miao KR, Fan L, et al. Intermediate prognosis of 6q deletion in chronic lymphocytic leukemia. Leuk Lymphoma. 2011;52(2):230-237. [DOI] [PubMed] [Google Scholar]
- 13.Jeon S, de Smith AJ, Li S, et al. Genome-wide trans-ethnic meta-analysis identifies novel susceptibility loci for childhood acute lymphoblastic leukemia. Leukemia. 2022;36(3):865-868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu H, Cheng C, Devidas M, et al. ARID5B genetic polymorphisms contribute to racial disparities in the incidence and treatment outcome of childhood acute lymphoblastic leukemia. J Clin Oncol. 2012;30(7):751-757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Studd JB, Vijayakrishnan J, Yang M, Migliorini G, Paulsson K, Houlston RS. Genetic and regulatory mechanism of susceptibility to high-hyperdiploid acute lymphoblastic leukaemia at 10p21.2. Nat Commun. 2017;8:14616. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and analyzed during the current study are not publicly available due to privacy and ethical restrictions, but are available from the corresponding author on reasonable request.