Allogeneic CAR T–cell therapies are being developed for hematologic malignancies. The authors implement a Cas12a chRDNA platform to generate allogeneic immune-cloaked BCMA-specific CAR T cells with resistance to host response–mediated rejection for evaluation in multiple myeloma.
Abstract
Allogeneic chimeric antigen receptor (CAR) T cell therapies hold the potential to overcome many of the challenges associated with patient-derived (autologous) CAR T cells. Key considerations in the development of allogeneic CAR T cell therapies include prevention of graft-vs-host disease (GvHD) and suppression of allograft rejection. Here, we describe preclinical data supporting the ongoing first-in-human clinical study, the CaMMouflage trial (NCT05722418), evaluating CB-011 in patients with relapsed/refractory multiple myeloma. CB-011 is a hypoimmunogenic, allogeneic anti–B-cell maturation antigen (BCMA) CAR T cell therapy candidate. CB-011 cells feature 4 genomic alterations and were engineered from healthy donor–derived T cells using a Cas12a CRISPR hybrid RNA–DNA (chRDNA) genome-editing technology platform. To address allograft rejection, CAR T cells were engineered to prevent endogenous HLA class I complex expression and overexpress a single-chain polyprotein complex composed of beta-2 microglobulin (B2M) tethered to HLA-E. In addition, T-cell receptor (TCR) expression was disrupted at the TCR alpha constant locus in combination with the site-specific insertion of a humanized BCMA-specific CAR. CB-011 cells exhibited robust plasmablast cytotoxicity in vitro in a mixed lymphocyte reaction in cell cocultures derived from patients with multiple myeloma. In addition, CB-011 cells demonstrated suppressed recognition by and cytotoxicity from HLA-mismatched T cells. CB-011 cells were protected from natural killer cell–mediated cytotoxicity in vitro and in vivo due to endogenous promoter-driven expression of B2M–HLA-E. Potent antitumor efficacy, when combined with an immune-cloaking armoring strategy to dampen allograft rejection, offers optimized therapeutic potential in multiple myeloma.
Introduction
Chimeric antigen receptor (CAR) T cell therapeutics show promise for improving the lives of patients with cancer. This is particularly evident for hematological malignancies, where several investigational and approved CAR T cell therapies show antitumor activity against B-cell lymphomas, leukemias, and multiple myeloma (1–7). Despite recent advances, autologous CAR T cell therapies have several limitations preventing universal access for patients such as high cost, lengthy manufacturing times, bridging therapy requirements, and product variability (8, 9). Allogeneic CAR T cell therapies aim to overcome these limitations; however, allogeneic CAR T cells must overcome different obstacles, including graft-vs-host disease (GvHD) and allograft rejection (9). We describe an allogeneic hypoimmune anti–B-cell maturation antigen (BCMA) CAR T cell therapeutic candidate (CB-011) engineered and optimized for safety and efficacy using a next-generation Cas12a CRISPR hybrid RNA–DNA (chRDNA) genome-editing platform. CB-011 is being evaluated for the treatment of relapsed/refractory multiple myeloma in the ongoing first-in-human CaMMouflage phase 1 clinical trial, which opened in 2023 (NCT05722418; ref. 10).
GvHD occurs when donor immune cells recognize and attack host tissue through immune cell–mediated responses. Disrupting the donor's native T-cell receptor (TCR) expression at the TRAC locus via genome editing largely prevents GvHD by eliminating nonspecific alloreactivity (2–4, 11). Allograft rejection is more difficult to overcome as it is caused by cell-extrinsic mechanisms, such as HLA mismatch and innate immune cell–mediated rejection. To evade allograft rejection, some allogeneic CAR T cell therapies use additional chemotherapeutic and/or biologic agents to suppress the host immune system, in addition to the standard lymphodepletion regimen (12). Genome-editing approaches were developed to allow allogeneic CAR T cells to evade elements of the host immune response to delay immune-mediated rejection. Specifically, blocking endogenous HLA expression through disruption of the beta-2 microglobulin (B2M) gene helps prevent allograft rejection due to HLA mismatch (13, 14). However, this strategy could leave engineered CAR T cells prone to clearance by natural killer (NK) cells through the “missing-self” response (15–17). Cell-surface overexpression of HLA-E has been shown to suppress the missing-self response in stem and bladder cancer cells (17, 18). A similar approach can be implemented in CAR T cells through genome editing and has been explored in vitro and in peripheral blood mononuclear cell (PBMC)–engrafted xenotransplantation models (3). Our studies extend this allogeneic CAR T-cell approach to characterize function, molecular responses, and NK-cell dynamics in vivo and demonstrate antitumor efficacy in murine xenograft models of multiple myeloma.
CRISPR-based genome editing has greatly enhanced the capabilities of engineering and producing improved CAR T cell therapies through targeted gene knockout (KO) and site-specific transgene knockin (KI; ref. 4). Although CRISPR–Cas9 is the gold standard for genome editing, a large collection of alternative CRISPR–Cas systems have distinct and potentially beneficial characteristics for CAR T cell engineering (19). A class 2 Cas enzyme, Cas12a, offers unique properties for genome editing in eukaryotic cells compared with Cas9, such as the recognition of a thymine-rich protospacer adjacent motif (PAM) and an increased sensitivity to mismatch between guide RNA and target DNA (20, 21). In addition, recent advancements involving the incorporation of DNA base(s) into the guide of CRISPR systems, termed chRDNA (21–24), demonstrated increased specificity, which may translate to improved safety for genome-editing applications for malignancies such as multiple myeloma.
Multiple myeloma is the second most common hematological malignancy with diverse treatment options, including proteasome inhibitors, immunomodulatory agents, and mAbs (25). However, because of disease relapse and other risks, alternative therapies must be considered. BCMA is highly expressed in multiple myeloma, establishing it as a promising therapeutic target (25). Although several autologous anti-BCMA CAR T cell therapies have been approved or are being investigated in clinical trials (26–28), allogeneic CAR T cell therapy approaches may enable a broader patient population to benefit from cell therapy. In addition, in the context of multiple myeloma, the standard use of immunostimulatory agents may negatively impact treatment with allogeneic cell therapies and as such, an allogeneic anti-BCMA CAR T cell product with immune-evasive properties may be a better therapeutic option (29, 30).
Materials and Methods
chRDNA screening and genome editing
Oligonucleotides used in this study are summarized in Supplementary Table S1 and were synthesized by Integrated DNA Technologies, Inc. with standard desalting.
T-cell handling and genome editing were carried out as previously described with minor modifications (22). Cas12a guide nucleoprotein complexes (NPCs) were formed by heating 250 pmol of guide to 95°C for 2 minutes and then allowing it to cool to room temperature. Guides were then mixed with 80 pmol of Cas12a protein (1:3 Cas12a:guides) in C12 buffer (60 mmol/L Tris-Acetate pH7.9, 150 mmol/L potassium acetate, 30 mmol/L magnesium acetate) in a final volume of 2.5 μL and incubated at 37°C for 10 minutes. NPCs were kept at 4°C or frozen at −20°C until transfection.
For transfection, T cells were isolated from cryopreserved PBMCs (STEMCELL Technologies) using RoboSep-S (STEMCELL Technologies) and the EasySep Human T cell Isolation Kit (STEMCELL Technologies) and activated for 3 days in the presence of anti–CD3/CD28 Dynabeads (Gibco) in ImmunoCult-XF T Cell Expansion Medium (STEMCELL Technologies) supplemented with 5% CTS immune cell serum replacement (Gibco) and rhIL-2. Beads were removed and cells were expanded for 24 hours with rhIL-2 before nucleofection. T cells were nucleofected using a 96-well Shuttle System (Lonza) with the prepared Cas12a chRDNA complexes. 48 hours after transfection cell lysate was prepared using 50 μL of QuickExtract DNA Extraction Solution (Lucigen) per 2e5 cells transfected and following the manufacturer's protocol. Genomic lysate was used for next-generation sequencing (NGS) analysis of the targeted region using the NextSeq platform (Illumina), and >10,000 reads were obtained for each sample. For a list of reagents with catalog numbers, see Supplementary Table S2.
Cas12a protein expression and purification
The Cas12a protein expression plasmid was generated at Caribou Biosciences, Inc. The AsCas12a sequence was codon optimized and synthesized by Twist Biosciences (South San Francisco), and then cloned into a recombinant protein expression plasmid backbone that was constructed from DNA fragments synthesized by Genscript Biotech. The recombinant Acidaminococcus sp. Cas12a protein, containing a cleavable N-terminal His6 tag and a C-terminal Simian Virus 40 nuclear localization signal, was expressed in Escherichia coli BL21 (DE3; New England Biolabs). Cells were lysed and soluble Cas12a protein was purified by immobilized metal affinity chromatography (Ni Sepharose 6 Fast Flow, Cytiva), followed by gel filtration (G-25 fine, Cytiva). After enzymatic cleavage of the affinity tag, the protein was further purified by cation exchange (SP HP, Cytiva) and size exclusion (Superdex 200, Cytiva) chromatography. Purified protein was concentrated at 50 mg/mL in buffer (25 mmol/L Tris:HCl pH 7.5, 10% glycerol, 0.1 mmol/L EDTA, 300 mmol/L NaCl, and 1 mmol/L TCEP).
CAR T cell production
CB-011 cells were generated from healthy donor–derived PBMCs. T cells were isolated from cryopreserved PBMCs (STEMCELL Technologies) using RoboSep-S (STEMCELL Technologies) and the EasySep Human T-cell Isolation Kit (STEMCELL Technologies) and activated for 3 days in the presence of anti–CD3/CD28 Dynabeads (Gibco) along with recombinant human interleukin-2 (rhIL-2; 100 U/mL). Beads were removed and cells were expanded for 24 hours with rhIL-2 before nucleofection and transduction. T cells were nucleofected using a 96-well Shuttle System (Lonza) with the prepared Cas12a chRDNA complexes and subsequently transduced with recombinant adeno-associated virus 6 (Signagen) engineered to deliver the CB-011 CAR transgene and the B2M–HLA-E fusion transgene. Cells were cultured for an additional 8 days in ImmunoCult-XF T Cell Expansion Medium (STEMCELL Technologies) supplemented with 5% CTS immune cell serum replacement (Gibco) and rhIL-2.
Human tumor cell lines and recombinant cell lines
Source information for tumor cell lines is provided in Supplementary Table S3. K562 cells were maintained in IMDM (Gibco) supplemented with 10% FBS (Corning) and 1× antibiotic–antimycotic (Gibco). K562 cells were transduced to express BCMA by lentiviral transduction and dilution cloning was performed to obtain a clonal cell population. The BCMA expression plasmid was cloned at Caribou Biosciences, Inc. The nucleic acid sequence encoding amino acids 1–184 of full-length BCMA/TNFRSF17 was codon optimized. The codon-optimized sequence was synthesized by Twist Biosciences (South San Francisco) and cloned using standard Gibson cloning into a lentiviral backbone. Recombinant BCMA-expressing cells were maintained in selection with 3 μg/mL puromycin (Gibco). MM.1S, NCI-H929, Daudi, U226B1, Ramos, and RPMI-8226 were maintained in RPMI-1640 (Gibco) supplemented with 10% FBS (Corning) and 1× antibiotic–antimycotic (Gibco). MM.1S, Daudi, and NCI-H929 were modified to express luciferase and GFP by lentiviral transduction. The luciferase and eGFP expression plasmid was cloned at Caribou Biosciences, Inc. The full-length firefly luciferase and eGFP coding sequences were codon optimized. The luciferase coding sequence was cloned into a lentiviral expression vector with the eGFP sequence directly 3′ of the luciferase coding sequence resulting in a single-chain polyprotein upon translation. Dilution cloning was performed to obtain a clonal cell population and recombinant GFP-luciferase expressing cells were maintained in 3 μg/mL blasticidin S HCl (Gibco). Where indicated, cells were treated with 0 to 1 μmol/L of the gamma-secretase inhibitor RO4929097 (Selleck Chemicals). All cell lines were tested for Mycoplasma contamination. The cells were maintained in culture for a maximum of three weeks, with approximately 6–8 passages conducted during this period.
In vitro cytotoxicity, cytokine secretion, and proliferation assays
Target cells were labeled with CellTrace Violet (CTV; Thermo Fisher Scientific) and were cocultured with CB-011 or double KO (DKO) T cells (engineered with a TRAC and B2M DKO) at different effector to target (E:T) ratios (10:1 to 0:1) for 48 hours. Live target cell counts were determined by flow cytometry using a Sartorius, iQue 3, and percentage-specific lysis calculated using the formula: 100×[1− (live target cells in test sample/live target cells in control sample)]. AUC was calculated to represent overall specific lysis against each target cell line. Where indicated, 0.1 μmol/L RO4929097 was added to the coculture.
For multiple myeloma bone marrow mononuclear cell (BMNC) samples (Discovery Life Sciences), in addition to propidium iodide (PI) staining (BioLegend), samples were stained with human anti-CD138 and human anti-CD38. The E:T ratio was subsequently adjusted to reflect the actual number of CD138+CD38+ cells that were plated. For multiple myeloma PBMC samples, in addition to a Live/Dead Fixable Near-IR Dead cell staining (Thermo Fisher Scientific), samples were stained with human anti-CD38, human anti-CD27, human anti-CD56, human anti-CD19, human anti-CD14, human anti-CD16, and human anti-CD3. Live multiple myeloma plasma cells were identified as CD3−CD14−CD16−CD19−CD56+/−CD38+CD27+/−. A list of human-reactive mAbs is provided in Supplementary Table S4. The E:T ratio was subsequently adjusted to reflect the actual number of multiple myeloma plasma cells that were plated. CountBright Beads (Invitrogen) were added to coculture samples before acquisition and used to calculate cell counts. Samples were acquired using a LSRFortessa X-20 flow cytometer (BD Biosciences). Data were analyzed using FlowJo v.10.
An in vitro cytotoxicity assay was also used to evaluate the protection conferred by the immune-cloaking armoring strategy of CB-011 against PBMC-derived CD8+ T cells. Briefly, HLA-mismatched PBMCs were purchased (STEMCELL Technologies) and CD8+ T cells isolated using RoboSep-S (STEMCELL Technologies) and the EasySep Human CD8 T-cell Isolation Kit (STEMCELL Technologies). CB-011 or CAR KI-only T cells (identically engineered CAR T cells without the B2M KO or B2M–HLA-E fusion) were labeled with CTV and were cocultured with PBMC-derived CD8+ T cells at different E:T ratios (10:1 to 0:1) for 48 hours. Cell counts were performed, and specific cell lysis was calculated as described above.
For cytokine secretion, cells were cocultured at a 1:1 E:T ratio for 24 hours. Effector- and target-only controls were used to measure background cytokine detection. Supernatant was collected and cytokines were multiplexed using a human ProcartaPlex immunoassay kit (PPX-08, Thermo Fisher Scientific). Data collection and analysis were performed using the Magpix instrument (Luminex Corporation).
Antigen-dependent proliferation of CB-011 cells was evaluated in vitro using BCMA+ cell lines; BCMA− K562 cells served as the control. Target cells were cocultured at a 1:1 ratio with CB-011 cells or DKO T cells labeled with CTV in the absence of rhIL-2. Proliferation was measured by reduction in CTV fluorescence 72 hours after initiation of the coculture using flow cytometry (Intellicyt iQue Screener Plus). Gating for CTV reduction was based on CTV− target cells at the same time point. Fold dye dilution was calculated using the geometric mean of CTV using the formula: CTV geometric mean0h/CTV geometric mean72h. Source information for primary cell samples is provided in Supplementary Table S5.
Serial rechallenge assay
Serial rechallenge with tumor cells was assayed at different E:T ratios (2:1 to 0:1). Luciferase+ MM.1S, Daudi, or NCI-H929 target cells were added to CB-011, CAR KI, CAR KI B2M KO, or DKO T cells every 3 to 4 days. Tumor cell–only controls were included in triplicate to measure spontaneous cell death. Luciferase activity was measured before adding new target cells using Promega One-GLO reagent (Promega Corp.) according to the manufacturer's instructions. After reading the luminescence with a SpectraMax i3x plate reader (Molecular Devices, LLC), specific lysis was calculated in relative luminometer units (RLU) using the formula: 100×(1− [RLU in test sample/RLU of spontaneous death sample]). The AUC was calculated to represent overall specific lysis against BCMA+ target. Concurrently, during the serial rechallenge assay, we assessed the proliferative capacity of CB-011 or DKO cells in response to repeated antigen challenges. For each round of the assay, a fraction of each of the cocultured wells was chosen for assessment of Ki-67 staining to measure proliferation.
Cells from these selected wells were harvested and stained with near-IR viability dye, followed by surface staining with primary conjugated antibodies as detailed in the flow cytometry analysis section. After surface staining, the cells were washed with Cell Staining Buffer (BioLegend), followed by the addition of ice-cold 70% ethanol (Thermo Fisher Scientific) for one hour at −20°C. Subsequently, the cells were washed in Cell Staining Buffer and stained with an anti–Ki-67 for 30 minutes at room temperature (Supplementary Table S4). Flow cytometry was used to analyze and quantify the percentage of Ki-67+ CB-011 or DKO T cells.
NK-cell evasion assay
NK-92 cells (ATCC) were cultured in minimum essential medium alpha (MEM alpha, Corning) supplemented with 12.5% heat-inactivated FBS (Corning), 12.5% heat-inactivated horse serum (Sigma-Aldrich), 1X l-glutamine (GlutaMAX, Gibco), 1X antibiotic–antimycotic (Gibco), and 5 ng/mL rhIL-2 (Peprotech). Cell seeding was performed at a density of 3×105 cells/mL, and the culture medium was refreshed every 2 to 3 days. NK-92 cultures were maintained for a maximum of three weeks, with a total of 6–9 passages. NK-92 cells were tested for Mycoplasma contamination. CAR T cells were thawed and rested for 72 hours in ImmunoCult T-cell expansion medium (STEMCELL Technologies) supplemented with 5% CTS immune cell serum replacement (Gibco) and 100IU rhIL-2 (PeproTech). CAR T cells were labeled with CTV (Thermo Fisher Scientific) and incubated with NK-92 cells at a 1:1 E:T ratio for ≤72 hours. At each time point, coculture samples were analyzed via flow cytometry for expression of CD56, HLA-E, and HLA-ABC and labeling of CTV. Information about the antibodies used can be found in Supplementary Table S4). CountBright Beads (Invitrogen) were added to coculture samples before acquisition and used to calculate cell counts. Counts of HLA-E+ and HLA-ABC+ CAR T cells were compared and normalized with a 0 hours time point.
Flow cytometry analysis
Cells were washed with FACS buffer (PBS, 1% BSA, and 1 mmol/L EDTA) and incubated with near-IR viability staining solution (Invitrogen, L10119) for 10 minutes at 4°C. Cells were washed and incubated with Human TruStain FcX (BioLegend) and primary conjugated antibodies for 20 minutes at 4°C. For staining mouse xenograft tissues, samples were first mechanically digested and incubated with ammonium chloride solution (STEMCELL Technologies) to lyse red blood cells. Cells were stained as previously described with the addition of Mouse BD Fc Block (BD Biosciences). Surface expression of the BCMA CAR was detected using phycoerythrin (PE)-conjugated recombinant human BCMA protein (Creative Biomart). A list of human-reactive mAbs is provided in Supplementary Table S4. Samples were acquired using the LSRFortessa X-20 flow cytometer (BD Biosciences). Data were analyzed using FlowJo v.10.
Primary NK-engrafted NOG-tg (hu-IL15) mouse model
NK cells were isolated from a fresh leukopak (AllCells) using a human NK-cell isolation kit (Miltenyi Biotec) in conjunction with the QuadroMACS Separator (Miltenyi Biotec), MACS MultiStand (Miltenyi Biotec), and LS Columns (Miltenyi Biotec). The isolation process followed the manufacturer's recommended protocol. NK cells were activated and expanded using a human NK-cell activation/expansion kit (Miltenyi Biotech, Inc.). NK cells were maintained in NK expansion media (1:2 F12/DMEM (Gibco), 20% heat-inactivated human AB serum (Innovative Research Inc.), 20 mg/L l-ascorbic acid Sigma-Aldrich), 5 ng/mL sodium selenite (Sigma-Aldrich), 50 μmol/L ethanolamine (Sigma-Aldrich), and 1× antibiotic–antimycotic (Gibco) supplemented with 100 IU/mL rhIL-15 and 100 IU/mL rhIL-2 (Peprotech). All mice were group housed with environmental enrichment; irradiated feed and sterilized water were provided ad libitum. Female 7-week-old NOD.Cg-Prkdcscid Il2rgtm1Sug Tg(CMV-IL2/IL15)1–1Jic/JicTac (NOG-Tg; Hu-IL15) mice (Taconic Biosciences) were acclimated for 7 days, randomized according to body weight, and engrafted with day 9 postactivation/expansion NK cells (5×105 NK cells intravenously/mouse). One day later (day 0), a total of 1×107 CAR+ CB-011 or CAR KI B2M KO T cells were administered intravenously. rhIL-2 was dosed intraperitoneally 3×/wk (10,000 IU/dose). Spleens were harvested on days 4 and 7.
All animal experiments were performed unblinded according to national ethical guidelines with guidance and approval from the Institutional Animal Care and Use Committee of Explora Biolabs/Charles River Laboratories. For all studies, euthanasia was conducted in compliance with the current requirements of the Guide for the Care and Use of Laboratory Animals, 8th Edition, and the American Veterinary Medical Association Guidelines on Euthanasia.
Orthotopic tumor xenograft mouse models
Female 7-week-old NOD-scid IL2Rgammanull (NSG) mice (The Jackson Laboratory) were acclimated for 7 days, randomized according to body weight (minimally established models) or tumor volume (well-established models), and were injected with tumor cells at approximately 8 weeks of age. Tumor cells were washed twice in prewarmed PBS, centrifuging each time at 300 × g for 7 minutes. Viability was determined after washing, and cells were prepared for tumor engraftment in a final volume of 200 μL/animal. Tumor engraftment by tail vein injection was performed. CAR T cells were washed once in prewarmed PBS and centrifuged at 400 × g for 5 minutes. Viability was determined after washing, and cells were prepared in a final volume of 200 μL/animal. Vehicle was used as a negative control. Body weight was measured after CAR T cell administration. Animals were euthanized after losing >20% of their body weight compared with day 0 or after developing paralysis. Bioluminescence imaging was performed using an IVIS Spectrum system (PerkinElmer). Tumor-bearing mice received 150 mg/kg luciferin (PerkinElmer) intraperitoneally 10 minutes before imaging. Mice were anesthetized with 2% isoflurane and imaged without an emission filter (840 nm, total light output – open filter) for 5 minutes.
Transcriptional profiling of in vivo models
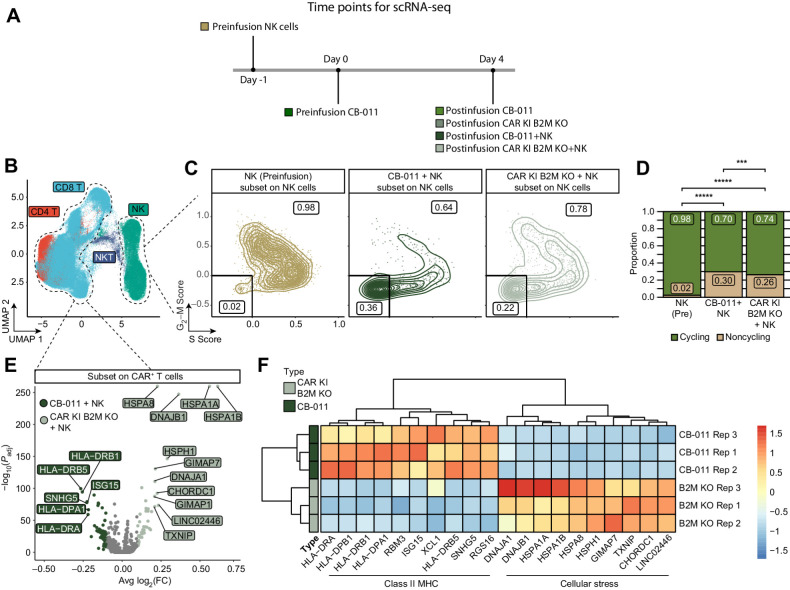
For profiling of NK cells and T cells following infusion in vivo, 6 different cohorts were established in NOG-Tg (Hu-IL15) mice according to the protocol described above: (i) 2 cohorts of CB-011 cells without NK cells (CB-011 only, n = 2 mice/cohort), (ii) 2 cohorts of CB-011 cells with NK cells (CB-011+NK, n = 3 mice/cohort), (iii) 1 cohort of B2M KO CAR T cells without NK (B2M KO only, n = 2 mice), and (iv) 1 cohort of B2M KO CAR T cells with NK cells (B2M KO+NK, n = 3 mice). In addition, NK and CB-011 cells were isolated for single-cell RNA sequencing (scRNA-seq; NK intraperitoneally and CB-011 intraperitoneally, n = 1/group) before infusion. NK and CAR T cells from either the CB-011 or the CAR KI B2M KO cohorts were isolated from the spleens of NOG-Tg (Hu-IL15 mice) mice 4 days post CAR T cell infusion by FACS using a human CD45-specific antibody. Cells were resuspended in PBS + 0.4% BSA at a concentration of a total of 1×106 cells/mL and scRNA-seq libraries were generated.
Histology and imaging of murine specimens
Tissues were extracted from NSG mice treated with a single bolus of vehicle, a total of 3×107 WT unmodified T cells, or 3×107 CB-011 cells, and immediately fixed in 10% neutral-buffered formalin for 24 hours before extensive wash in water. Femur samples were incubated in 10% di- and tetra-sodium EDTA until complete decalcification, followed by serial ethanol dehydration and paraffin embedding. All other tissues taken for histopathology were placed in 70% ethanol until paraffin embedding following formalin fixation. For characterization of human CD45+ cell presence in sectioned tissues, IHC staining was performed using a Leica BOND automated immunostainer and a rabbit anti-human CD45 (clone EP322Y, Abcam).
IHC image analysis
Where appropriate, a sample with no primary antibody was used as a negative control. Whole-slide images were generated using a Panoramic SCAN (3D Histech) and ImageDx was used to generate AI-powered quantitative data. CD45+ cells were identified and quantitated within stained tissue sections as the number of positive cells within the total image analysis area for each sample. An algorithm using automated identification of tissue areas on whole-slide images followed by segmentation of cell nuclei and classification of positive cells was visualized by overlaid masks. The number of positive cells was counted and measured as the total amount of positive cells, total positive cell area, and percentage of positive cells within the total image analysis area for each sample. The intensity measurement was a mean intensity value of all positive cells. Data are reported per tissue.
SITE-seq assay and in cell validation
SITE-seq assay was performed as described previously with minor modifications (22). Unmodified human T cells were isolated from cryopreserved PBMCs (STEMCELL Technologies) using RoboSep-S (STEMCELL Technologies) and the EasySep Human T-cell Isolation Kit (STEMCELL Technologies) and activated for 3 days in the presence of anti–CD3/CD28 Dynabeads (Gibco) along with rhIL-2 (100 U/mL). Beads were removed and cells were expanded for 24 hours with rhIL-2 before nucleofection, and high–molecular weight genomic DNA (gDNA) was extracted from the human primary T cells using the Blood and Cell Culture DNA Maxi Kit (Qiagen), according to the manufacturer's protocol. NPCs were formulated with the TRAC and B2M chRDNAs, and 10 μg of gDNA was digested with both 16 and 128 nmol/L of each NPC in a total volume of 50 μL at 37°C for 4 hours. Library preparation and sequencing were done as previously described (31) with an additional deoxyadenosine (dA)-tailing step post-NPC cleavage using 5 μL of End Repair Reaction Buffer components of the NEBNext Ultra End Repair/dA-Tailing Module (New England BioLabs). Samples were incubated at 20°C for 30 minutes and then 65°C for 30 minutes, and the standard procedure was resumed. NGS was performed using the NextSeq platform (Illumina), and approximately 3 million reads were obtained for each sample. Any sites recovered by the SITE-seq assay without off-target motifs located within 1 nucleotide of a cut-site were considered false positives and discarded. A full list of recovered biochemical off targets is shown in Supplementary Table S6.
To measure indel frequencies at off-target sites recovered using the SITE-seq assay, T cells were transfected as previously described with either TRAC or B2M chRDNA NPCs, and editing rates were determined for each off-target site recovered at the 128 nmol/L concentration. Mutant reads were defined as any nonreference variant calls within 20 base pairs (bp) of the cut-site. Sites that had low sequencing coverage (<1,000 reads in the combined, Cas12a NPC–treated samples or <200 reads in the reference samples) or >2% variant calls in the reference samples were discarded. Sites were tallied as cellular off-targets if they accumulated >0.1% mutant reads in the combined, Cas12a NPC–treated samples.
Processing of scRNA-seq data
Construction of scRNA-seq libraries was performed using the Chromium Next GEM Single Cell 5′ HT Reagent Kit v2 (10x Genomics). For each sample, 10,000 cells were targeted for capture based on a loading concentration of 1,000 cells/μL. Following gene expression library construction, the samples were multiplexed on the same lane of a NextSeq 2000 platform (Illumina) and sequenced to a target depth of 20,000 to 30,000 reads/cell.
The Cell Ranger Single-Cell Software Suite (version 7.0.1) was used to perform sample demultiplexing, barcode assignments, and gene alignments (10x Genomics). Raw base BCL files from the NextSeq 2000 were demultiplexed using the Cell Ranger mkfastq pipeline into sample-specific FASTQ files. FASTQ files were processed individually using the Cell Ranger count pipeline with prebuilt GRCh38 human reference genome provided by Cell Ranger (10x Genomics). Aligned reads were filtered for valid cell barcodes and unique molecular identifiers (UMI). For samples that contained CAR T cells, an additional alignment to a custom build of GRCh38 containing the coding sequence of the BCMA-CAR was performed and the CAR status of individual cells was added to the metadata of the sample. For downstream analysis, gene expression levels were determined using the pre-built GRCh38 reference genome alignment.
The resulting cell-gene count matrix was imported into Seurat for quality control (QC) filtering and analysis (32). The following QC filtering criteria were applied to the samples: Library complexity below an outlier cutoff value [defined on a per-sample basis as a value below quartile 1 (Q10 – 1.5×interquartile range (IQR) of all cells in the sample] or a percentage of mitochondrial gene expression above an outlier cutoff value (defined on a per-sample basis as a value above Q3 + 1.5×IQR) or 15%, whichever was lower. Outlier genes were excluded from downstream analysis if they did not contain ≥1 UMI count in ≥3 cells in each sample. Each sample was processed individually following the standard Seurat R functions according to the recommended workflow tutorial for normalizing with SCTransform. Doublet cells were identified as the majority consensus (4/5) of 5 different doublet detection algorithms and removed (33–35). Following data normalization and doublet removal, all samples were integrated by condition with the Seurat IntegrateData workflow using “SCT” as the normalization method and “rpca” as the reduction method. A small percentage of contaminating mouse cells was removed on the basis of low library complexity and percent mitochondrial gene expression. The final dataset contained 125,750 single cells combined across all the conditions described above.
Following integration, the Seurat object was subjected to principal component analysis, clustering, and uniform manifold projection (UMAP) analyses on the “integrated” assay. The first 30 principal components were used as input to both the RunUMAP() and FindNeighbors() functions of Seurat. Individual cell clusters were used to identify individual cell types based on canonical marker gene expression from the RNA assay. In addition, to aid in visualization, the RNA assay was used as input to the MAGIC package for expression level visualization (36). These imputed RNA values were used for visualization of the various marker genes and feature UMAP plots.
For NK cell– and CAR+ T cell–subset analysis, the clusters identified at a resolution of 0.4 were used. Marker genes for each cluster were determined by using the FindMarkers() function in Seurat with the “MAST” test statistic (37). To remove the effect of unequal sample sizes when determining the proportion of each condition in the clusters, the cell counts for each condition were weighted by using inverse probability weighting. For cell-cycle analysis of NK cells, each cell was scored using the CellCycleScoring() function in Seurat for 2 sets of genes associated with either the S phase or G2–M phase of the cell cycle as previously described (38). The heat map visualization of the differential gene expression between CAR+ T cells from either the CB-011 or B2M-KO conditions was performed using the R package pheatmap (version 1.0.12).
Droplet digital PCR–based quantification of CB-011 balanced translocations
Genomic DNA was extracted from CB-011 CAR T cells, on the Qiacube Connect (Qiagen), using the DNeasy Blood and Tissue Kit from Qiagen. Genomic DNA was quantified using the Qubit and normalized by concentration. Primers and probes were ordered from Integrated DNA Technologies. The sequence of each primer and probe set is listed in Supplementary Table S7. The droplet digital PCR (ddPCR) supermix for probes (No UTP; Bio-Rad) was aliquoted in a 96-well plate, then the quantified and normalized gDNA was added to the appropriate wells. The completed PCR plate was sealed with a heat sealer, vortexed, and centrifuged briefly before performing droplet generation using a Bio-Rad QX200. Once droplets were generated, the PCR plate was immediately put on a Bio-Rad C1000 thermal cycler, and the appropriate program was performed. After thermal cycling, ddPCR reactions were analyzed on the QX200 droplet reader to count the presence or absence of the targeted translocation and/or RPP30 in each droplet. Each well of the plate was manually gated, and populations were appropriately grouped.
Statistical analysis
Data were analyzed using two-way ANOVA with Sidak or Tukey corrections, an unpaired t test with a Welch correction, Fisher exact test, or Wilcoxon test with Bonferroni corrected P values. In animal studies, Kaplan–Meier survival curves and the 95% confidence interval for fractional survival at any time (Mantel–Cox log-rank test) were developed. Statistical analyses were performed using GraphPad Prism v7.0d and are denoted in each figure legend.
Data availability statement
The data generated in this study can be found in the article and its Supplementary Materials or are available upon reasonable request from the corresponding author. Raw scRNA-seq data are available through the Gene Expression Omnibus (accession number GSE249316).
Results
Next-generation Cas12a chRDNA genome-editing platform enables “immune cloaking” of allogeneic anti-BCMA CAR T cells
To identify suitable target sequences in the TRAC and B2M genes, all Acidaminococcus sp. Cas12a PAM sites (5′TTTV) in the coding regions of each gene were identified in the hg38 genomic sequence of the TRAC and B2M genes using in-house sequence visualization software. A library of all-RNA CRISPR RNA (crRNA) guides were designed comprising a spacer complementary to the 20 nucleotides downstream of each identified PAM sequence appended to the 20 nucleotide Cas12a repeat sequence and produced via chemical synthesis. Each crRNA was complexed with recombinantly expressed Cas12a and transfected into primary T cells isolated from PBMCs. Two days after transfection, the genome editing efficiency of each guide was determined by NGS and the highest efficiency guide for each gene was selected for subsequent chRDNA development (Fig. 1A and B; Supplementary Fig. S1A; ref. 22).
Figure 1.
Cas12a chRDNA guides generate high-precision genome editing for the development of CB-011 cells. chRDNA guides included DNA bases to confer high target specificity. Cas12a chRDNA guides were designed to target the TRAC and B2M genes. A and B, The chRDNA design process started with gene-tiling to identify optimal all-RNA CRISPR guides (crRNA). Once optimal all-RNA guides were identified (A and B, left), iterative DNA position screening proceeded for a given target sequence. Individual chRDNAs were tested with a single DNA base at each position in the 20-nucleotide spacer and evaluated for on-target editing activity in T cells (A and B, middle). The positions where DNA bases did not reduce editing efficiencies were combined in subsequent chRDNA guide designs to identify optimal chRDNAs with multiple DNA bases in the sequence (A and B, right). C and D, Schematic representations of the insertion of the anti-BCMA CAR into TRAC exon 1 and the B2M–HLA-E fusion into B2M exon 2 using Cas12a chRDNA guides. E, Dot plots of BCMA-CAR transgene insertion with AAV MOI titration. F, Dot plots of B2M–HLA-E fusion protein transgene insertion with AAV MOI titration. G, Representative dot plot of the drug product achieving >50% of intended gene edits. A–C, Each point represents a biological replicate, bars show the mean of biological replicates. E–G, Representative data from three independent experiments. APC, allophycocyanin; BGH, bovine growth hormone; MOI, multiplicity of infection; scFv, single-chain variable fragment; SS, secretion signal; SSC-A, side scatter area; TM, transmembrane.
To understand the capacity of Cas12a to support activity with chRDNA guides, a screening process was developed involving the generation of 20 chRDNA guides containing a DNA base at each position along the spacer. For each chRDNA guide, the editing rates were tested for selected TRAC and B2M targets in T cells. DNA incorporation produced a wide range of editing efficiencies between the 2 gene targets, where the TRAC target showed more variable efficiencies compared with the B2M target (Fig. 1A and B). For the TRAC target, DNA incorporation within the seed region had the most deleterious impact to editing activity, save for the first nucleotide position in the spacer, where DNA incorporation at positions 5 and 6 showed <20% editing activity. Although editing rates for the B2M target were largely unaffected by DNA base incorporation, the design with a DNA base at position 5 showed an approximately 30% reduction in editing rate.
We combined the individual DNA-tolerant positions identified in our single position screen into chRDNA designs incorporating multiple DNA residues. For each target, 7 chRDNAs with between 2 and 3 DNA bases in the spacer were generated and editing in T cells was evaluated compared with an all-RNA crRNA control. Many designs had editing efficiencies equivalent to an all-RNA crRNA, with chRDNA designs with DNA bases at positions 1, 11, and 20 in the spacer being among the top designs emerging from the screens. A suitable pair of targets in the TRAC and B2M genes was identified that showed approximately 90% indel formation (Fig. 1A and B).
To understand the off-target profile of these reagents in primary T cells, all biochemical off-target sites from genomic DNA extracted from primary T cells using the SITE-seq assay were identified (31). Subsequently, T cells were transfected with Cas12a programmed with the TRAC or B2M chRDNA and targeted amplicon sequencing was performed at both on-target and off-target sites [identified at the highest digestion concentration (128 nmol/L) from the SITE-seq assay]. No off-target editing in cells was observed above the limit of detection (0.1%) across the evaluated off-target sites, confirming the specificity of the chRDNA guides (Supplementary Fig. S1B, Supplementary Table S6).
The identified Cas12a chRDNA guides were used to engineer primary T cells in which a transgene encoding a second-generation CAR was site-specifically inserted into exon 1 of the TRAC locus, and a transgene encoding a B2M–HLA-E fusion protein was inserted into exon 2 of the B2M locus (Fig. 1C and D). The resulting cells lacked expression of a TCR and canonical HLA class I complexes on the T-cell surface (Supplementary Fig. S1E)
High-efficiency gene editing at intended KI loci is a prerequisite for high-efficiency transgene insertion when using homology-directed repair at a double-strand break. Having optimized Cas12a chRDNA guides for specific gene editing at the TRAC and B2M loci, we optimized transgene insertion through multiplicity of infection (MOI) titration of recombinant adeno-associated virus (rAAV)–mediated transgene delivery. CAR and B2M–HLA-E transgene cassettes were designed to be flanked with homology arms complementary to between the edited loci. In addition to phenotypic characterization of KO/KI efficiency the intended Cas12a chRDNA cut-site to yield site-specific transgene insertion (Fig. 1C and D; Supplementary Fig. S1C and S1D, Supplementary Table S8). A delivery MOI of a total of 2×105 rAAV genomes per cell yielded high-efficiency transgene insertion (Fig. 1E and F), resulting in a product that can achieve >50% homogeneity for all 4 of the intended gene edits (Fig. 1G; Supplementary Fig. S1F). Gene editing at multiple loci can lead to translocation events by flow cytometry, we assessed the potential for predictable, balanced translocations between the edited TRAC and B2M loci by ddPCR. The observed balanced translocation frequencies ranged from 0.02% to 0.05% (Supplementary Table S9).
Cas12a chRDNA precision genome–edited CB-011 cells demonstrated BCMA-targeted antitumor activity
We sought to evaluate the BCMA-specific antitumor efficacy of the immune-cloaked CAR T cells (CB-011 cells). CB-011 cells showed BCMA-dependent cytotoxic activity, highlighted by low activity against the K562 cell line (which does not express BCMA) and potent activity against the K562-BCMA cell line (engineered to overexpress recombinant BCMA; Fig. 2A and B). CB-011 cells were cocultured with transformed BCMA+ cell lines, including the multiple myeloma cell lines MM.1S and NCI-H929. Dose-dependent–specific lysis of multiple myeloma cell lines was observed at increasing E:T ratios. Significant BCMA-targeted cytotoxicity was observed when compared with control genome-edited DKO T cells, as summarized by calculating the AUC from dose-titration experiments (Fig. 2A and B). BCMA-targeted cytotoxic activity was accompanied by secretion of the Th1 cytokines IL2, IFNγ, and TNFα, as well as the cytolytic molecules granzyme A and B and perforin (Fig. 2C). In the absence of cytokine or antigen-mediated stimulation (T cells only or K562 coculture conditions), neither control T cells nor CB-011 cells underwent cell division as demonstrated by a lack of dilution of a cell proliferation tracker dye (Fig. 2D). Conversely, CB-011 cells proliferated in the presence of BCMA+ cells, including the multiple myeloma cell lines MM.1S and NCI-H929 (Fig. 2D and E). In serial rechallenge assays, CB-011 cells maintained cytotoxic potential following multiple rounds of multiple myeloma cell line–mediated stimulation (Fig. 2F and G), and the combined gene-editing approach had no negative impact on CB-011 cell fitness, as demonstrated in serial rechallenge assays comparing CB-011 CAR T cells with unarmored CAR T cells (Supplementary Fig. S2). Serial rechallenge assays were further used to examine the proliferative potential of CB-011 cells following multiple rounds of stimulation. CB-011 cells maintained expression of the proliferative marker Ki-67 when repeatedly challenged with BCMA-expressing myeloma cell lines, consistent with sustained cytotoxic activity (Fig. 2H).
Figure 2.
CB-011 cells exhibit antitumor activity, cytokine expression, and proliferation in response to BCMA-expressing tumor cells in vitro. A, T-cell cytotoxicity assay at a range of E:T ratios of CB-011 cells (green lines) or negative T-cell control DKO cells (black lines) against BCMA− cell line K562 and BCMA+ cell lines K562-BCMA, MM.1S, NCI-H929, and Daudi. B, The AUC of data in A [x-axis (E:T ratios) vs. y-axis (percentage of specific cell lysis)]. Cumulative data from three independent experiments are presented. Error bars representing SEM. Statistical analysis using a two-way ANOVA with Sidak multiple comparisons was performed (C) Secretion of IL2, IFNγ, TNFα, Perforin, Granzyme A, and Granzyme B by CB-011 cells (green bars) or DKO cells (black bars) 24 hours after coculture with target cells was quantified by Luminex multiplex immunoassay. Two-way ANOVA with Sidak correction was performed. D, CB-011 cells and target cell lines were incubated at a 1:1 E:T ratio and proliferation was monitored by CTV dye dilution at 72 hours. E, Proliferation of DKO T cells or CB-011 cells at 72 hours. F, Serial rechallenge assay demonstrating T-cell cytotoxicity data following an 8-round rechallenge assay at a range of E:T ratios. CB-011 cells (green lines) or negative T-cell control DKO cells (black lines) in coculture with the BMCA+ cell lines MM.1S, NCI-H929, and Daudi. Each symbol depicted in the key below the plots represents the data point for each round of restimulation (R = round). Within the plots the symbols are connected by either a green line, indicating CB-011, or a black line, indicating DKO. G, Cumulative AUC value representing the sum of the AUC data calculated following each of round of serial rechallenge. H, The percentage of proliferating (Ki67+) CB-011 or DKO T cells during the first 5 rounds of rechallenge. A, C–G, Representative experiments from at least 3 independent experiments. Mean ± SEM of three replicates per data point shown. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant.
CB-011 cells demonstrate BCMA-targeted antitumor activity toward BMNCs derived from patients with multiple myeloma
BMNCs derived from patients with multiple myeloma were cocultured with CB-011 cells. Specific lysis of the myeloma plasma cells was determined by flow cytometry–mediated gating of the CD38+CD138+ population. In this population, BCMA expression was consistent, although lower than in the myeloma MM.1S cell line. Consistently, we observed dose-dependent cytotoxicity of CB-011 cells toward myeloma plasma cells across 3 separate patient-derived samples (Fig. 3) and toward myeloma plasma cells found in peripheral blood (Supplementary Fig. S3). Because the density of cell surface BCMA can be modulated by gamma secretase cleavage activity, we examined the effect of gamma secretase inhibition on CB-011 cytotoxic activity. Gamma secretase inhibition led to an increase in BCMA cell surface expression across all tested BCMA-expressing cell lines. CB-011 displayed comparable high-level cytotoxic activity in the presence and absence of gamma secretase activity, suggesting that BCMA expression, within the range shown experimentally, does not affect CB-011–induced cytotoxicity (Supplementary Fig. S4).
Figure 3.
CB-011 cells exhibit in vitro antitumor activity against BMNCs from patients with multiple myeloma. A, T-cell cytotoxicity assay at a range of E:T ratios of CB-011 cells (green lines) or negative T-cell control DKO cells (black lines) against BMNCs from samples from patients with multiple myeloma. B, The AUC of data in A [x-axis (E:T ratios) vs. y-axis (percentage of specific cell lysis)]. C, Dot plot representing the gating strategy to identify multiple myeloma cells in the BMNC samples. D, Histogram showing expression of BCMA in BMNC samples (BCMA FMO in gray, samples in black). E, BCMA density on BMNCs from samples from patients with multiple myeloma and MM.1S as a reference control. Three sets of BMNCs from patients with multiple myeloma was used with technical triplicates. Error bars indicate SEM. FMO, fluorescence minus one; MM, multiple myeloma.
CB-011 cell treatment confers a significant survival advantage in murine xenograft models of multiple myeloma
Multiple myeloma xenograft models were established in mice over 21 days following which animals were treated with vehicle or a single dose of CB-011 cells. Bioluminescence intensity per animal was used to track tumor burden of the luciferase expressing multiple myeloma cell line, MM.1S (Fig. 4A and B). Despite high tumor burden at the time of treatment, CB-011 cells significantly reduced tumor levels and significantly prolonged survival in multiple myeloma xenograft animals (Fig. 4C). A single dose of CB-011 cells administered when tumor burden was comparatively low also conferred significant prolongation of survival in several xenograft models of multiple myeloma (Supplementary Figs. S5 and S6). No evidence of GvHD was observed over 74 days in NSG mice treated with a high dose of CB-011 cells, as indicated by limited infiltration of CB-011 cells noted in the bone marrow, lungs, lymph nodes, or spleen and maintenance of body weight and survival (Fig. 4D, E, G; Supplementary Tables S10 and S11). Consistent with this, in-life observations of CB-011–treated animals were normal, whereas animals treated with unedited T cells developed disease characteristics consistent with GvHD, such as dull fur, hunching, skin lesions, and diarrhea (Fig. 4F).
Figure 4.
BCMA-targeted CAR T cells engineered with an immune-cloaking armoring strategy induced long-term survival in a disseminated xenograft model of multiple myeloma and do not induce GvHD. Human MM.1S-GFP-Luc+ tumor cells were engrafted intravenously in NSG mice on day 0, and a single bolus dose of vehicle (A) or a total of 1×107 CB-011 cells (B) was administered intravenously on day 21 (9 mice per group). Bioluminescence imaging was performed using an IVIS Spectrum system. A and B, Lines represent individual animal bioluminescent intensity for each group. C, Kaplan–Meier survival plot represents the percentage of survival for each group after tumor engraftment. Median survival, Vehicle, 15 days; CB-011 (1×107 CAR+ T cells/animal), 90 days (P < 0.0001, Mantel–Cox test). Experiment representative of three independent experiments. NSG mice (6 mice per group) were treated on day 0 with a single bolus of vehicle, 3×107 WT unmodified T cells, or 3×107 CB-011 cells. Animal body weight was obtained by an electronic balance. Scheduled takedowns at day 35 eliminated some of the mice from subsequent body weight analyses. Signs of acute GvHD were observed in WT T cell–dosed animals, but not in vehicle or CB-011 cell–dosed cohorts. D, Example images of hCD45 staining (shown at 20×) from day 74 samples in vehicle (top row), WT unmodified T cell (middle row), and CB-011 cell (bottom row) groups. Quantification of the percentage of hCD45 staining from samples collected at days 35 and 74, respectively, are shown. E, The percentage of body weight decline in the WT T cell–dosed animals is shown. Lines represent individual animal body weight percentage to change after treatment. F, Clinical signs of GvHD, including hunching, skin lesions, and dull fur, were observed in the WT T cell–dosed animals but not in vehicle or CB-011 cell–dosed cohorts. G, Kaplan–Meier survival plot representing the percentage of survival for each group. Median survival, vehicle, undefined; WT unmodified T cells, 63 days; and CB-011, undefined. Experiment representative of two independent experiments. WT, wild-type.
Immune-cloaking strategy protects CB-011 cells from CD8+ T cell– and NK cell–mediated cytotoxicity in vitro
The immune-cloaking strategy was designed to protect CB-011 cells from allogeneic graft rejection in the following 2 ways: suppression of endogenous HLA class I complex expression through the abrogation of B2M expression, and suppression of “missing-self”–mediated activation of NK cells through overexpression of a B2M–HLA-E fusion polyprotein. Non–immune-cloaked CAR KI-only (CAR KI) T cells were compared with CB-011 cells to functionally characterize suppression of HLA class I–mismatched CAR T cells. Fully HLA class I–mismatched, PBMC-derived CD8+ T cells from 3 separate donors were cocultured with either CB-011 or CAR KI CAR T cells at increasing CD8+ to CAR T cell ratios and cytotoxic activity across the dose titration was summarized by calculating the AUC (Fig. 5A). CAR T cells lacking an immune-cloaking armoring strategy were highly susceptible to mismatched T cell–mediated cytotoxicity. Conversely, T cell–mediated cytotoxicity was significantly reduced toward CB-011 cells. To address the second element of the immune-cloaking strategy, CB-011 cells or CAR T cells without a B2M–HLA-E fusion (CAR KI B2M KO) were cocultured with the transformed NK-cell line, NK-92. In the absence of NK-92 cells, the B2M KO population (HLA-ABC−) grew as expected over 3 days (D3 vs. D0). In contrast, the presence of NK-92 cells led to a significant reduction in the B2M KO population, whereas the B2M–HLA-E+ CB-011 cell population was enriched following 3 days of coculture with NK-92 cells (Fig. 5B). These results were confirmed by plotting the dynamics of CB-011 cells as a function of the B2M KO CAR T cells (HLA-ABC−; Fig. 5C; Supplementary Fig. S7).
Figure 5.
Insertion of a B2M–HLA-E peptide fusion in the B2M locus of CB-011 cells protected against CD8+ T-cell and NK cell–mediated lysis. A, T-cell cytotoxicity assay at a range of E:T ratios of wild-type CD8+ T cells derived from 3 different healthy donors against CB-011 or CAR KI T cells. The AUC of data in A [x-axis (E:T ratios) vs. y-axis (percentage of specific cell lysis)]. Mean values ± SEM from 3 replicates. B, Ratio of HLA-ABC− CB-011 or CAR KI B2M KO T cells at day 3 over day 0 with or without NK-92 cells. Two-way ANOVA with Tukey correction was performed. Representative results show mean values ± SEM from 3 replicates. C, Ratio of HLA-E+ to HLA-ABC− CB-011 cells at different time points in cultures with or without NK-92 cells. Two-way ANOVA with Tukey correction was performed. Representative results show mean values ± SEM from 3 replicates. D, Scheme of in vivo study design. E, Count of NK cells in spleen at day 4 after engraftment. The unpaired t test with Welch correction was performed. F, The percentage of NKG2A+ or NKG2C+ NK cells in spleen at day 4 after engraftment. G, Fold change of HLA-ABC− CB-011 or CAR KI B2M KO T-cell count in presence of NK cells at day 4 in NOG-Tg (Hu-IL15) mice. The unpaired t test was performed. H, The percentage of HLA-E+ CB-011 cells at days 4 and 7 in NOG-Tg (Hu-IL15) mice. Two-way ANOVA with Sidak correction was performed. E–H, 5 mice per group. Representative results show mean values ± SEM. D3/D0, day 3 versus 0.
Immune-cloaking strategy protects CB-011 cells from primary NK cell–mediated rejection in vivo
The resistance of CB-011 cells to NK cell–cytotoxic activity was demonstrated in vivo by examining CB-011 cell dynamics in immunodeficient NOG-Tg (Hu-IL15) mice in the presence of PBMC-derived primary NK cells. NK cells were expanded in vitro before intravenous engraftment (Fig. 5D). Levels of NK cells in mice treated with either CB-011 cells or control CAR T cells were similar (Fig. 5E) and immunophenotyping of the inhibitory or activating receptors (NKG2A or NKG2C) showed that isolated NK-cell populations predominantly expressed the inhibitory receptor NKG2A (Fig. 5F). In the absence of NK cells, the abundance of B2M KO (HLA-ABC−) cells was the same at day 4 in mice treated with CB-011 or control CAR T cells; however, a significant reduction was observed in this population in NK-engrafted mice treated with control CAR T cells. Conversely, this population was protected from NK cells in CB-011–treated mice (Fig. 5G). Stabilization of the B2M KO population of CB-011 cells was accompanied by an enrichment of the B2M–HLA-E+ population in the presence of NK cells (Fig. 5H; Supplementary Fig. S7).
Immune-cloaking armoring strategy reduced NK-cell proliferation and T-cell stress signatures
A comparison of the transcriptional profiles of CB-011 cells versus unarmored CAR T cells isolated from the spleens of CAR T/NK-cell co-infused animals highlighted differential proliferative states and gene expression profiles (Fig. 6A). Following dimensionality reduction and clustering, NK and CD4/8 T cells were identified on the basis of known marker genes (Supplementary Fig. S8A and S8B). The NK- and T-cell compartments were investigated for differential gene expression profiles between CB-011 cells and unarmored CAR T cells. Of note, the NK-cell compartment demonstrated higher scoring for proliferative markers and genes associated with cytotoxicity when co-infused with unarmored CAR T cells, consistent with increased NK-cell activation (Fig. 6C and D; Supplementary Fig. S8C–S8E). In the T-cell compartment, differential gene expression analysis highlighted that unarmored CAR T cells, which do not express the B2M–HLA-E fusion, demonstrated transcriptional upregulation of genes associated with cellular stress, which may correlate with their reduced inhibition of NK-cell activation and thus T cell–directed, NK-cell cytotoxicity (Fig. 6E and F).
Figure 6.
CB-011 immune-cloaking armoring strategy reduced NK-cell proliferation and CAR+ T cell stress signatures. A, Scheme of in vivo experiment. B, UMAP plots colored by cell type identity. C, Scatter plots demonstrating single-cell module scores for a set of genes associated with either S phase (S.Score) or G2–M phase (G2–M.Score) of the cell cycle, subset to only NK cells from either of the in vivo datasets (CB011 + NK In Vivo or CAR KI B2M KO + NK In Vivo, representative plot for 1 replicate each is shown) or the NK cells before injection, but after in vitro activation (NK pre). Cells negative for both scores were considered non-cycling and cells single or double positive were considered cycling. The proportion of each of those populations is indicated on the scatter plot. D, Bar plots demonstrating the proportion of cycling and noncycling NK cells in the indicated conditions (CB-011 and CAR KI B2M KO represent NK cells from in vivo co-injection with T cells; NK pre represent NK cells before injection). Data visualized the combined cells from all replicate samples. The CB-011 immune-cloaking armoring strategy significantly reduced the proportion of cycling NK cells in vivo relative to CAR KI B2M KO cells and NK pre cells. Significance determined by the Fisher's exact test. ***, P < 0.001; *****, P < 1 × 10−15. E, Volcano plot of differentially expressed genes between CB-011 or CAR KI B2M KO CAR+ T cells following co-injection with NK cells in vivo. Genes enriched in CB-011 CAR+ T cells had a negative mean log2(FC) and genes enriched in CAR KI B2M-KO CAR+ T cells had a positive mean log2(FC). Significance was determined with a Wilcoxon test with Bonferroni corrected P values. F, Heat map of the top 10 differentially expressed genes from the analysis in (E). The expression of each gene was averaged across all CAR+ T cells in one lot replicate of CB-011 and CAR KI B2M KO. CAR KI B2M KO CAR+ T cells showed significant upregulation of stress-response gene signatures indicative of diminished fitness when challenged by the presence of NK cells. Spleens from three mice per group were processed for transcriptional profiling. FC, fold change.
Discussion
Dominant drivers of allograft rejection for adoptive cell therapies (ACT) include both CD8+ T and NK cells, with contributions by myeloid-derived phagocytic cells. To address both GvHD and allograft rejection dynamics after ACT, we generated CB-011, an allogeneic CAR T cell therapy with genome-edited TCR (TRAC gene) and HLA class I (B2M gene), with insertion of a BCMA-specific CAR and a B2M–HLA-E fusion at these loci, respectively. To achieve this, we developed a Cas12a chRDNA genome-editing platform. Previously, we demonstrated that the incorporation of DNA bases into the guide of the Cas9 nuclease enhances the specificity of genome editing (22). We extended these findings to Cas12a and developed a chRDNA screening pipeline to identify TRAC- and B2M-targeting chRDNAs with impeccable specificity. The positions within Cas12a guides tolerant to DNA substitution appear to be dependent on the target DNA sequence, as demonstrated by the B2M target, which was more permissible of DNA substitutions in the spacer compared with the TRAC target. Using optimized chRDNA guides, 4 genome edits provided CB-011 cells with immune-evasive properties and specific targeting of multiple myeloma cells, functionally addressing both allograft rejection by the host lymphoid compartment and GvHD.
BCMA targeting with autologous CAR T cell therapies has demonstrated a substantial impact on disease progression in relapsed/refractory multiple myeloma (26, 27, 39). Despite this success, a variety of challenges remain such as general inaccessibility, long manufacturing delays necessitating bridging therapy for waiting patients, and T-cell exhaustion resulting in manufacturing failures, among other causes (40–43). The alternative approach of using allogeneic CAR T cells, such as CB-011, addresses some of the challenges and limitations of autologous CAR T cell therapy. Given the 3–4-week reconstitution window for both CD8+ T cells and NK cells from bone marrow after lymphodepletion (44–46), the hypoimmune strategy in CB-011 cells may enable enhanced circulation time and therapeutic effect.
Immune-cloaking armoring strategies like those used here have been previously shown to enable CAR T cells to survive NK cell–mediated destruction in vivo (3, 17, 47–51). In this study, CB-011 cells demonstrated increased resistance to HLA class I–mismatched T cell– and NK cell–mediated cytotoxicity in vitro when compared with unarmored CAR T cells. We extended these observations by demonstrating that HLA-E+ CB-011 cells were enriched, and NK-cell proliferation was reduced when the “missing-self” HLA antigen was reconstituted in a mouse model of NKG2A+ NK cell immune surveillance. In addition, CB-011 cells exhibited limited molecular stress signatures when compared with non–immune-cloaked CAR T cells. NK-cell phenotypes are an important determinant of NK cell–mediated innate immune responses and regulatory roles (52). Expression of the B2M–HLA-E fusion protein by CB-011 cells likely confers protection through the high-affinity interaction of HLA-E with the inhibitory NK-cell receptor NKG2A. However, HLA-E may interact with NKG2C in an axis that could lead to activated NK-cell profiles (53). A study of NK-cell phenotypes from patients with relapsed/refractory multiple myeloma showed that clinical responses to daratumumab could be associated with NK-cell characteristics. In that study, the authors note that in patients with relapsed/refractory multiple myeloma, the proportion of NKG2A+ NK cells was approximately 2-fold that of NKG2C+ NK cells (54). Although the interplay between NK-cell phenotype and functional fate is complex, these data suggest that the relapsed/refractory multiple myeloma setting may be amenable to NK-cell suppression through the HLA-E/NKG2A signaling pathway.
The use of immunostimulatory agents is foundational in the treatment of multiple myeloma at all stages of disease, including maintenance treatment after autologous stem cell transplant (55). However, in the context of allogeneic cell therapies, immunostimulatory agents may enhance the propensity for allograft rejection (29, 56). As such, an immune-cloaking armoring strategy such as that implemented for CB-011 cells may be particularly impactful in treating patients with relapsed/refractory multiple myeloma.
Alternative strategies to enhance the expansion and persistence of allogeneic CAR T cells in this setting are being explored, including gene-editing approaches that allow for the use of deeper lymphodepletion regimens (57, 58). The evaluation of ALLO-715, an allogeneic anti-BCMA CAR T cell therapy candidate engineered with a CD52 KO, in conjunction with a lymphodepletion regimen, including fludarabine, cyclophosphamide, and anti-CD52, has demonstrated the safety and tolerability of such an approach, and there is a measurable overall response rate in the initial dose-escalation study.
Clinical development of allogeneic CAR T cell therapies may require a balance between enabling robust persistence of allogeneic cells through the depth of depletion of a patient's immune cell repertoire versus enabling increased resistance of allogeneic cells to host response-mediated rejection. CB-011 was developed to enhance resistance to allogeneic response-mediated clearance and is being evaluated for the treatment of relapsed/refractory multiple myeloma in the ongoing first-in-human CaMMouflage phase 1 clinical trial (NCT05722418; ref. 10). To our knowledge, this is the first clinical evaluation of an allogeneic CAR T cell therapy with an immune-cloaking strategy, including both B2M KO and expression of a B2M–HLA-E transgene. Other hypoimmune approaches have not included the B2M–HLA-E KI in CAR T cells (14), which may have yielded resistance to patient-derived T-cell rejection at the expense of limited protection from patient NK cell–mediated rejection. CaMMouflage aims to evaluate the safety, tolerability, pharmacokinetics, and efficacy of CB-011 in patients with relapsed/refractory multiple myeloma. A major determinant will be whether increased resistance to lymphoid compartment rejection will promote longer CB-011 CAR T cell residence time in circulation to enable durable antitumor activity.
Supplementary Material
Supplementary Tables S1 through S11
chRDNA-mediated gene editing did not result in any off-target editing above the limit of detection.
In vitro serial rechallenge assay demonstrates long-term cytotoxicity activity of CB-011 against BCMA+ tumor cells.
CB-011 cells exhibit in vitro antitumor activity against PBMCs from patients with multiple myeloma.
Gamma-secretase inhibitor RO4929097 increases BCMA expression on-target cell lines but does not improve CB-011 antitumor efficacy.
Antitumor activity of CB-011 CAR-T cells in a xenograft model of multiple myeloma.
Antitumor activity of CB-011 CAR-T cells in a xenograft model of multiple myeloma.
CB-011 immune-cloaking armoring strategy suppresses cytotoxic T and NK cell–mediated allograft rejection.
CB-011 immune-cloaking armoring strategy reduced NK cell proliferation and CAR+ T cell stress signatures.
Acknowledgments
This study was funded by Caribou Biosciences, Inc. We thank Peter Lauer (Caribou Biosciences, Inc) for direct technical assistance. This study was funded by Caribou Biosciences, Inc. Medical writing and editorial support were provided by Steven Walker of Red Nucleus and were funded by Caribou Biosciences, Inc.
Footnotes
Note: Supplementary data for this article are available at Cancer Immunology Research Online (http://cancerimmunolres.aacrjournals.org/).
Authors' Disclosures
P.D. Donohoue reports US patents 1051946, US 9688972, US 9580701, US 9868962, US 9650617, US 9771601, US 10988781, and US 11459588 issued and licensed. S. Gradia reports personal fees from Caribou Biosciences during the conduct of the study and personal fees from Caribou Biosciences outside the submitted work. S.B. Kanner reports non-financial support from Red Nucleus during the conduct of the study. No disclosures were reported by the other authors.
Authors' Contributions
É Degagné: Conceptualization, data curation, formal analysis, supervision, investigation, writing–review and editing. P.D. Donohoue: Data curation, methodology, writing–review and editing. S. Roy: Investigation. J. Scherer: Investigation. T.W. Fowler: Data curation, supervision, writing–review and editing. R.T. Davis: Conceptualization, formal analysis, supervision, investigation, visualization, writing–review and editing. G.A. Reyes: Investigation. G. Kwong: Investigation, writing–review and editing. M. Stanaway: Investigation. V. Larroca Vicena: Investigation. D. Mutha: Investigation. R. Guo: Investigation. L. Edwards: Investigation. B. Schilling: Investigation. M. Shaw: Investigation. S.C. Smith: Resources, investigation. B. Kohrs: Resources, investigation, writing–review and editing. H.J. Kufeldt: Investigation. G. Churchward: Investigation. F. Ruan: Investigation. D.B. Nyer: Investigation. K. McSweeney: Investigation. M.J. Irby: Investigation. C.K. Fuller: Data curation, formal analysis, methodology. L. Banh: Investigation. M.S. Toh: Investigation. M. Thompson: Data curation, supervision. A.L.G. Owen: Data curation, software, formal analysis. Z. An: Supervision. S. Gradia: Supervision. J. Skoble: Supervision. M. Bryan: Project administration. E. Garner: Supervision, writing–original draft. S.B. Kanner: Supervision, writing–review and editing.
References
- 1. Good Z, Spiegel JY, Sahaf B, Malipatlolla MB, Ehlinger ZJ, Kurra S, et al. Post-infusion CAR TReg cells identify patients resistant to CD19-CAR therapy. Nat Med 2022;28:1860–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hu X, Manner K, DeJesus R, White K, Gattis C, Ngo P, et al. Hypoimmune anti-CD19 chimeric antigen receptor T cells provide lasting tumor control in fully immunocompetent allogeneic humanized mice. Nat Commun 2023;14:2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jo S, Das S, Williams A, Chretien AS, Pagliardini T, Le Roy A, et al. Endowing universal CAR T cell with immune-evasive properties using TALEN-gene editing. Nat Commun 2022;13:3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lau E, Kwong G, Fowler TW, Sun BC, Donohoue PD, Davis RT, et al. Allogeneic chimeric antigen receptor T cells with CRISPR-disrupted programmed death-1 checkpoint exhibit enhanced functional fitness. Cytotherapy 2023;25:750–62. [DOI] [PubMed] [Google Scholar]
- 5. Locke FL, Ghobadi A, Jacobson CA, Miklos DB, Lekakis LJ, Oluwole OO, et al. Long-term safety and activity of axicabtagene ciloleucel in refractory large B-cell lymphoma (ZUMA-1): a single-arm, multicentre, phase 1–2 trial. Lancet Oncol 2019;20:31–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rodriguez-Marquez P, Calleja-Cervantes ME, Serrano G, Oliver-Caldes A, Palacios-Berraquero ML, Martin-Mallo A, et al. CAR density influences antitumoral efficacy of BCMA CAR T cells and correlates with clinical outcome. Sci Adv 2022;8:eabo0514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Scholler N, Perbost R, Locke FL, Jain MD, Turcan S, Danan C, et al. Tumor immune contexture is a determinant of anti-CD19 CAR T-cell efficacy in large B-cell lymphoma. Nat Med 2022;28:1872–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. de Lima Lopes G, Nahas GR. Chimeric antigen receptor T cells, a savior with a high price. Chin Clin Oncol 2018;7:21. [DOI] [PubMed] [Google Scholar]
- 9. Graham C, Jozwik A, Pepper A, Benjamin R. Allogeneic CAR T cells: more than ease of access? Cells 2018;7:155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Berdeja JG, Martin TG, Rossi A, Essell JH, Siegel DSD, Mailankody S, et al. A first-in-human phase 1, multicenter, open-label study of CB-011, a next-generation CRISPR-genome edited allogeneic anti-BCMA immune-cloaked CAR T cell therapy, in patients with relapsed/refractory multiple myeloma (CAMMOUFLAGE trial). J Clin Oncol 2023;41:TPS8063–TPS. [Google Scholar]
- 11. Eyquem J, Mansilla-Soto J, Giavridis T, van der Stegen SJ, Hamieh M, Cunanan KM, et al. Targeting a CAR to the TRAC locus with CRISPR/Cas9 enhances tumour rejection. Nature 2017;543:113–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Benjamin R, Graham C, Yallop D, Jozwik A, Mirci-Danicar OC, Lucchini G, et al. Genome-edited, donor-derived allogeneic anti-CD19 chimeric antigen receptor T cells in paediatric and adult B-cell acute lymphoblastic leukaemia: results of two phase 1 studies. Lancet 2020;396:1885–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lee J, Sheen JH, Lim O, Lee Y, Ryu J, Shin D, et al. Abrogation of HLA surface expression using CRISPR/Cas9 genome editing: a step toward universal T-cell therapy. Sci Rep 2020;10:17753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ren J, Liu X, Fang C, Jiang S, June CH, Zhao Y. Multiplex genome editing to generate universal CAR T Cells resistant to PD1 inhibition. Clin Cancer Res 2017;23:2255–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kagoya Y, Guo T, Yeung B, Saso K, Anczurowski M, Wang CH, et al. Genetic ablation of HLA class I, class II, and the T-cell receptor enables allogeneic T cells to be used for adoptive T-cell therapy. Cancer Immunol Res 2020;8:926–36. [DOI] [PubMed] [Google Scholar]
- 16. Karre K, Ljunggren HG, Piontek G, Kiessling R. Selective rejection of H-2–deficient lymphoma variants suggests alternative immune defence strategy. Nature 1986;319:675–8. [DOI] [PubMed] [Google Scholar]
- 17. Gornalusse GG, Hirata RK, Funk SE, Riolobos L, Lopes VS, Manske G, et al. HLA-E-expressing pluripotent stem cells escape allogeneic responses and lysis by NK cells. Nat Biotechnol 2017;35:765–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Salome B, Sfakianos JP, Ranti D, Daza J, Bieber C, Charap A, et al. NKG2A and HLA-E define an alternative immune checkpoint axis in bladder cancer. Cancer Cell 2022;40:1027–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Koonin EV, Makarova KS. Evolutionary plasticity and functional versatility of CRISPR systems. PLoS Biol 2022;20:e3001481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 2015;163:759–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kim H, Lee WJ, Oh Y, Kang SH, Hur JK, Lee H, et al. Enhancement of target specificity of CRISPR-Cas12a by using a chimeric DNA–RNA guide. Nucleic Acids Res 2020;48:8601–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Donohoue PD, Pacesa M, Lau E, Vidal B, Irby MJ, Nyer DB, et al. Conformational control of Cas9 by CRISPR hybrid RNA-DNA guides mitigates off-target activity in T cells. Mol Cell 2021;81:3637–49. [DOI] [PubMed] [Google Scholar]
- 23. Rueda FO, Bista M, Newton MD, Goeppert AU, Cuomo ME, Gordon E, et al. Mapping the sugar dependency for rational generation of a DNA-RNA hybrid-guided Cas9 endonuclease. Nat Commun 2017;8:1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yin H, Song CQ, Suresh S, Kwan SY, Wu Q, Walsh S, et al. Partial DNA-guided Cas9 enables genome editing with reduced off-target activity. Nat Chem Biol 2018;14:311–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kleber M, Ntanasis-Stathopoulos I, Terpos E. BCMA in multiple myeloma—a promising key to therapy. J Clin Med 2021;10:4088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Berdeja JG, Madduri D, Usmani SZ, Jakubowiak A, Agha M, Cohen AD, et al. Ciltacabtagene autoleucel, a B-cell maturation antigen-directed chimeric antigen receptor T-cell therapy in patients with relapsed or refractory multiple myeloma (CARTITUDE-1): a phase 1b/2 open-label study. Lancet 2021;398:314–24. [DOI] [PubMed] [Google Scholar]
- 27. Munshi NC, Anderson LD Jr, Shah N, Madduri D, Berdeja J, Lonial S, et al. Idecabtagene vicleucel in relapsed and refractory multiple myeloma. N Engl J Med 2021;384:705–16. [DOI] [PubMed] [Google Scholar]
- 28. Raje N, Berdeja J, Lin Y, Siegel D, Jagannath S, Madduri D, et al. Anti-BCMA CAR T cell therapy bb2121 in relapsed or refractory multiple myeloma. N Engl J Med 2019;380:1726–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lum EL, Huang E, Bunnapradist S, Pham T, Danovitch G. Acute kidney allograft rejection precipitated by lenalidomide treatment for multiple myeloma. Am J Kidney Dis 2017;69:701–4. [DOI] [PubMed] [Google Scholar]
- 30. Pazina T, AWt MF, Bernabei L, Dulaimi E, Kotcher R, Yam C, et al. Alterations of NK cell phenotype in the disease course of multiple myeloma. Cancers 2021;13:226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cameron P, Fuller CK, Donohoue PD, Jones BN, Thompson MS, Carter MM, et al. Mapping the genomic landscape of CRISPR-Cas9 cleavage. Nat Methods 2017;14:600–6. [DOI] [PubMed] [Google Scholar]
- 32. Hao Y, Hao S, Andersen-Nissen E, Mauck WM, 3rd, Zheng S, Butler A, et al. Integrated analysis of multimodal single-cell data. Cell 2021;184:3573–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bais AS, Kostka D. scds: computational annotation of doublets in single-cell RNA sequencing data. Bioinformatics 2020;36:1150–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. DePasquale EAK, Schnell DJ, Van Camp PJ, Valiente-Alandi I, Blaxall BC, Grimes HL, et al. DoubletDecon: deconvoluting doublets from single-cell RNA-sequencing data. Cell Rep 2019;29:1718–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. McGinnis CS, Murrow LM, Gartner ZJ. DoubletFinder: doublet detection in single-cell RNA sequencing data using artificial nearest neighbors. Cell Syst 2019;8:329–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. van Dijk D, Sharma R, Nainys J, Yim K, Kathail P, Carr AJ, et al. Recovering gene interactions from single-cell data using data diffusion. Cell 2018;174:716–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Finak G, McDavid A, Yajima M, Deng J, Gersuk V, Shalek AK, et al. MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol 2015;16:278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tirosh I, Izar B, Prakadan SM, Wadsworth MH, 2nd, Treacy D, Trombetta JJ, et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 2016;352:189–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. San-Miguel J, Dhakal B, Yong K, Spencer A, Anguille S, Mateos MV, et al. Cilta-cel or standard care in lenalidomide-refractory multiple myeloma. N Engl J Med 2023;389:335–47. [DOI] [PubMed] [Google Scholar]
- 40. Ahmed N, Shahzad M, Shippey E, Bansal R, Mushtaq MU, Mahmoudjafari Z, et al. Socioeconomic and racial disparity in chimeric antigen receptor T-cell therapy access. Transplant Cell Ther 2022;28:358–64. [DOI] [PubMed] [Google Scholar]
- 41. Al Hadidi S, Zangari M, van Rhee F. Chimeric antigen receptor T-cell therapy in multiple myeloma-challenges and potential solutions. JAMA Oncol 2022;8:823–4. [DOI] [PubMed] [Google Scholar]
- 42. Kourelis T, Bansal R, Patel KK, Berdeja JG, Raje NS, Alsina M, et al. Ethical challenges with CAR T slot allocation with idecabtagene vicleucel manufacturing access. J Clin Oncol 2022;40:. e20021 [Google Scholar]
- 43. Roddie C, O'Reilly M, Dias Alves Pinto J, Vispute K, Lowdell M. Manufacturing chimeric antigen receptor T cells: issues and challenges. Cytotherapy 2019;21:327–40. [DOI] [PubMed] [Google Scholar]
- 44. Strati P, Varma A, Adkins S, Nastoupil LJ, Westin J, Hagemeister FB, et al. Hematopoietic recovery and immune reconstitution after axicabtagene ciloleucel in patients with large B-cell lymphoma. Haematologica 2021;106:2667–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wang Y, Cao J, Zhiling Y, Qiao J, Li D, Li Z, et al. Kinetics of immune reconstitution after CD19 CAR T cell therapy in ALL patients. Blood 2019;134:1301. [Google Scholar]
- 46. Wang Y, Li H, Song X, Qi K, Cheng H, Cao J, et al. Kinetics of immune reconstitution after anti-CD19 chimeric antigen receptor T-cell therapy in relapsed or refractory acute lymphoblastic leukemia patients. Int J Lab Hematol 2021;43:250–8. [DOI] [PubMed] [Google Scholar]
- 47. Crew MD, Cannon MJ, Phanavanh B, Garcia-Borges CN. An HLA-E single chain trimer inhibits human NK cell reactivity towards porcine cells. Mol Immunol 2005;42:1205–14. [DOI] [PubMed] [Google Scholar]
- 48. Deuse T, Hu X, Gravina A, Wang D, Tediashvili G, De C, et al. Hypoimmunogenic derivatives of induced pluripotent stem cells evade immune rejection in fully immunocompetent allogeneic recipients. Nat Biotechnol 2019;37:252–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hoerster K, Uhrberg M, Wiek C, Horn PA, Hanenberg H, Heinrichs S. HLA class I knockout converts allogeneic primary NK cells into suitable effectors for "off-the-shelf" immunotherapy. Front Immunol 2020;11:586168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wang B, Iriguchi S, Waseda M, Ueda N, Ueda T, Xu H, et al. Generation of hypoimmunogenic T cells from genetically engineered allogeneic human induced pluripotent stem cells. Nat Biomed Eng 2021;5:429–40. [DOI] [PubMed] [Google Scholar]
- 51. Mo F, Watanabe N, McKenna MK, Hicks MJ, Srinivasan M, Gomes-Silva D, et al. Engineered off-the-shelf therapeutic T cells resist host immune rejection. Nat Biotechnol 2021;39:56–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Cooper MA, Fehniger TA, Caligiuri MA. The biology of human natural killer-cell subsets. Trends Immunol 2001;22:633–40. [DOI] [PubMed] [Google Scholar]
- 53. Lauterbach N, Wieten L, Popeijus HE, Voorter CE, Tilanus MG. HLA-E regulates NKG2C+ natural killer cell function through presentation of a restricted peptide repertoire. Hum Immunol 2015;76:578–86. [DOI] [PubMed] [Google Scholar]
- 54. Verkleij CPM, Frerichs KA, Broekmans MEC, Duetz C, O'Neill CA, Bruins WSC, et al. NK cell phenotype is associated with response and resistance to daratumumab in relapsed/refractory multiple myeloma. Hemasphere 2023;7:e881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Holstein SA, McCarthy PL. Immunomodulatory drugs in multiple myeloma: mechanisms of action and clinical experience. Drugs 2017;77:505–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Meyers DE, Adu-Gyamfi B, Segura AM, Buja LM, Mallidi HR, Frazier OH, et al. Fatal cardiac and renal allograft rejection with lenalidomide therapy for light-chain amyloidosis. Am J Transplant 2013;13:2730–3. [DOI] [PubMed] [Google Scholar]
- 57. Mailankody S, Matous JV, Chhabra S, Liedtke M, Sidana S, Oluwole OO, et al. Allogeneic BCMA-targeting CAR T cells in relapsed/refractory multiple myeloma: phase 1 UNIVERSAL trial interim results. Nat Med 2023;29:422–9. [DOI] [PubMed] [Google Scholar]
- 58. Sommer C, Boldajipour B, Kuo TC, Bentley T, Sutton J, Chen A, et al. Preclinical evaluation of allogeneic CAR T cells targeting BCMA for the treatment of multiple myeloma. Mol Ther 2019;27:1126–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Tables S1 through S11
chRDNA-mediated gene editing did not result in any off-target editing above the limit of detection.
In vitro serial rechallenge assay demonstrates long-term cytotoxicity activity of CB-011 against BCMA+ tumor cells.
CB-011 cells exhibit in vitro antitumor activity against PBMCs from patients with multiple myeloma.
Gamma-secretase inhibitor RO4929097 increases BCMA expression on-target cell lines but does not improve CB-011 antitumor efficacy.
Antitumor activity of CB-011 CAR-T cells in a xenograft model of multiple myeloma.
Antitumor activity of CB-011 CAR-T cells in a xenograft model of multiple myeloma.
CB-011 immune-cloaking armoring strategy suppresses cytotoxic T and NK cell–mediated allograft rejection.
CB-011 immune-cloaking armoring strategy reduced NK cell proliferation and CAR+ T cell stress signatures.
Data Availability Statement
The data generated in this study can be found in the article and its Supplementary Materials or are available upon reasonable request from the corresponding author. Raw scRNA-seq data are available through the Gene Expression Omnibus (accession number GSE249316).



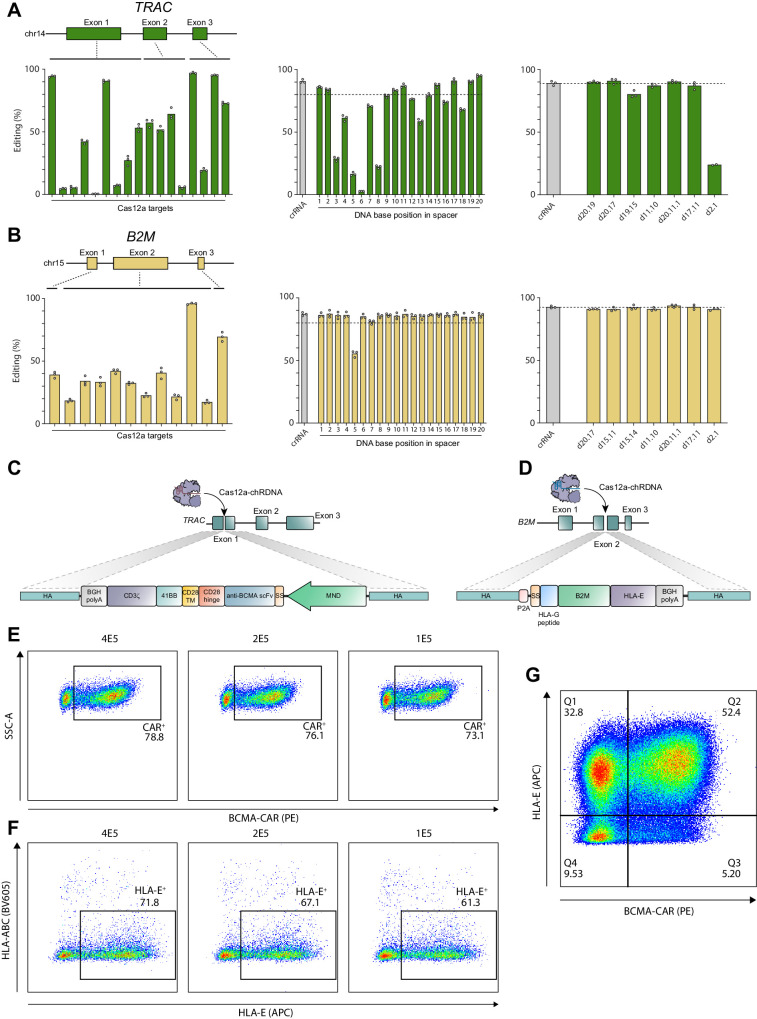
![Figure 2. CB-011 cells exhibit antitumor activity, cytokine expression, and proliferation in response to BCMA-expressing tumor cells in vitro. A, T-cell cytotoxicity assay at a range of E:T ratios of CB-011 cells (green lines) or negative T-cell control DKO cells (black lines) against BCMA− cell line K562 and BCMA+ cell lines K562-BCMA, MM.1S, NCI-H929, and Daudi. B, The AUC of data in A [x-axis (E:T ratios) vs. y-axis (percentage of specific cell lysis)]. Cumulative data from three independent experiments are presented. Error bars representing SEM. Statistical analysis using a two-way ANOVA with Sidak multiple comparisons was performed (C) Secretion of IL2, IFNγ, TNFα, Perforin, Granzyme A, and Granzyme B by CB-011 cells (green bars) or DKO cells (black bars) 24 hours after coculture with target cells was quantified by Luminex multiplex immunoassay. Two-way ANOVA with Sidak correction was performed. D, CB-011 cells and target cell lines were incubated at a 1:1 E:T ratio and proliferation was monitored by CTV dye dilution at 72 hours. E, Proliferation of DKO T cells or CB-011 cells at 72 hours. F, Serial rechallenge assay demonstrating T-cell cytotoxicity data following an 8-round rechallenge assay at a range of E:T ratios. CB-011 cells (green lines) or negative T-cell control DKO cells (black lines) in coculture with the BMCA+ cell lines MM.1S, NCI-H929, and Daudi. Each symbol depicted in the key below the plots represents the data point for each round of restimulation (R = round). Within the plots the symbols are connected by either a green line, indicating CB-011, or a black line, indicating DKO. G, Cumulative AUC value representing the sum of the AUC data calculated following each of round of serial rechallenge. H, The percentage of proliferating (Ki67+) CB-011 or DKO T cells during the first 5 rounds of rechallenge. A, C–G, Representative experiments from at least 3 independent experiments. Mean ± SEM of three replicates per data point shown. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/d56c/10985478/18c034e04de8/462fig2.jpg)
![Figure 3. CB-011 cells exhibit in vitro antitumor activity against BMNCs from patients with multiple myeloma. A, T-cell cytotoxicity assay at a range of E:T ratios of CB-011 cells (green lines) or negative T-cell control DKO cells (black lines) against BMNCs from samples from patients with multiple myeloma. B, The AUC of data in A [x-axis (E:T ratios) vs. y-axis (percentage of specific cell lysis)]. C, Dot plot representing the gating strategy to identify multiple myeloma cells in the BMNC samples. D, Histogram showing expression of BCMA in BMNC samples (BCMA FMO in gray, samples in black). E, BCMA density on BMNCs from samples from patients with multiple myeloma and MM.1S as a reference control. Three sets of BMNCs from patients with multiple myeloma was used with technical triplicates. Error bars indicate SEM. FMO, fluorescence minus one; MM, multiple myeloma.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/d56c/10985478/b54195dbe6f5/462fig3.jpg)
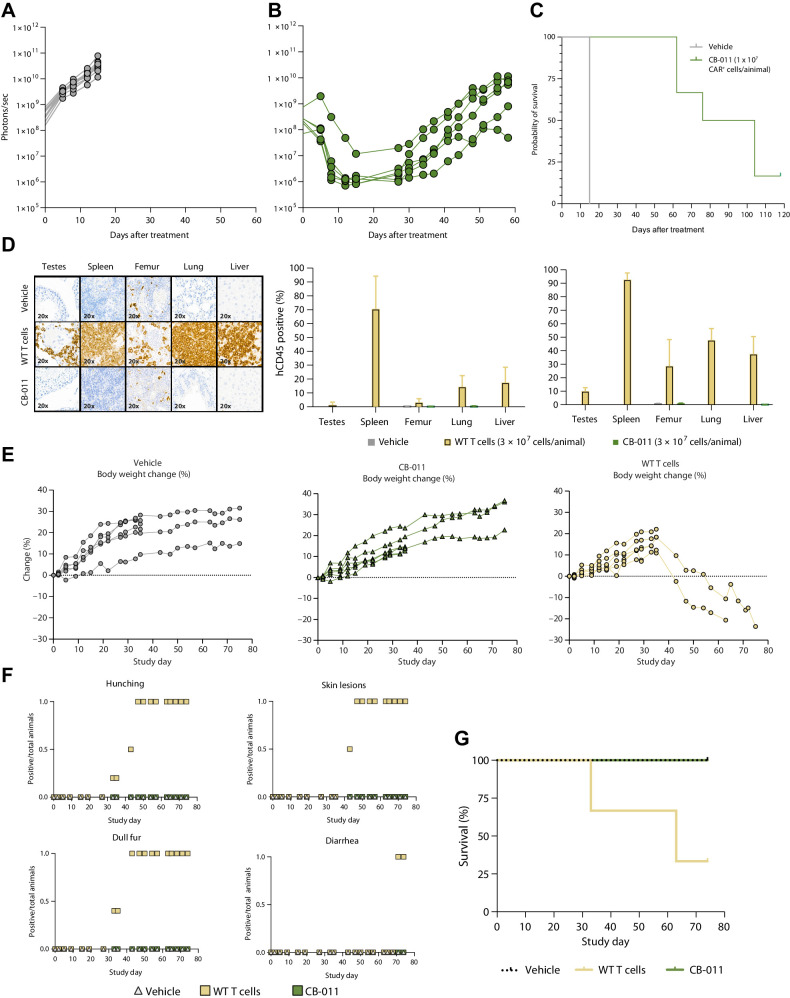
![Figure 5. Insertion of a B2M–HLA-E peptide fusion in the B2M locus of CB-011 cells protected against CD8+ T-cell and NK cell–mediated lysis. A, T-cell cytotoxicity assay at a range of E:T ratios of wild-type CD8+ T cells derived from 3 different healthy donors against CB-011 or CAR KI T cells. The AUC of data in A [x-axis (E:T ratios) vs. y-axis (percentage of specific cell lysis)]. Mean values ± SEM from 3 replicates. B, Ratio of HLA-ABC− CB-011 or CAR KI B2M KO T cells at day 3 over day 0 with or without NK-92 cells. Two-way ANOVA with Tukey correction was performed. Representative results show mean values ± SEM from 3 replicates. C, Ratio of HLA-E+ to HLA-ABC− CB-011 cells at different time points in cultures with or without NK-92 cells. Two-way ANOVA with Tukey correction was performed. Representative results show mean values ± SEM from 3 replicates. D, Scheme of in vivo study design. E, Count of NK cells in spleen at day 4 after engraftment. The unpaired t test with Welch correction was performed. F, The percentage of NKG2A+ or NKG2C+ NK cells in spleen at day 4 after engraftment. G, Fold change of HLA-ABC− CB-011 or CAR KI B2M KO T-cell count in presence of NK cells at day 4 in NOG-Tg (Hu-IL15) mice. The unpaired t test was performed. H, The percentage of HLA-E+ CB-011 cells at days 4 and 7 in NOG-Tg (Hu-IL15) mice. Two-way ANOVA with Sidak correction was performed. E–H, 5 mice per group. Representative results show mean values ± SEM. D3/D0, day 3 versus 0.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/d56c/10985478/2c72b35c7b56/462fig5.jpg)