Abstract
Up‐to‐date knowledge of gut microbial taxa associated with ischemic heart disease (IHD). Microbial metabolites for mechanistic dissection of IHD pathology. Microbiome‐based therapies in IHD prevention and treatment.
Ischemic heart disease (IHD) has caused a major burden on public health due to its high morbidity and mortality [1, 2]. IHD is commonly also referred to as coronary heart disease (CHD) or coronary artery disease, meaning a heart problem characterized by narrowed or blocked coronary arteries with reduced blood flow to the heart muscle (https://www.cdc.gov/heartdisease/coronary_ad.htm). Depending on the clinical manifestations of the disease, IHD can be classified into stable or chronic IHD and acute coronary syndrome reflecting an imbalance between myocardial oxygen demand and supply (https://www.nhs.uk/conditions/coronary-heart-disease). Patients who suffer from IHD are frequently accompanied by common risk factors for many years before overt IHD, including obesity, type 2 diabetes (T2D), and metabolic syndrome [3], calling for more specific and effective strategies for IHD prevention and intervention.
From the outcome of recent epidemiological, physiological, integrated omics‐based studies, followed by the findings from both animal and cellular investigations, it shows that a great proportion of the links between the environmental influences and human IHD may be contributed by microbial communities (termed gut microbiome) [4]. It has been revealed that the collection of all intestinal bacterial genes has more than an order of magnitude higher gene numbers than the human genome [4]. The total amount of gut bacteria exceeds 1014 microorganisms, whereas the gut virus has even more orders of magnitude higher quantity than that of bacteria [5]. The gut microbes, including bacteria, archea, virus, and unicellular eukaryotes, may collectively provide a repository of information characterizing the IHD development [6]. In the meanwhile, the various enzymes encoded by gut microbes may participate in pathways of producing numerous metabolites, which may via the blood circulation impact systemic and myocardial metabolism that are associated with IHD [7]. Therefore, it is reasonable to believe the strong involvement of gut microbes in the IHD development.
In the past two decades, rapid development in next‐generation sequencing technology and bioinformatic databases and tools allowed us to gain deeper knowledge of the relationships between gut microbial compositions, functional potentials, and host phenotypes, which has greatly sped up the field from cohort‐based towards personalized understanding [8]. Yet, the gap still remains between basic science and clinical translation. For instance, specific taxa is lacking for the precise diagnosis of IHD [9], the causality of microbiome on IHD is poorly understood [10], and microbiome‐based therapy in patients has not yielded satisfying efficiency [11]. Therefore, it is of great importance to explore additional mechanistic involvement of gut microbiota in the IHD development, either based on observational findings from populational studies or evidence from experimental validations. To determine the impact of gut microbiota on IHD development, the microbiota‐related metabolites may play an important and nonignorable role by mediating the alterations of microbial functionalities on host phenotypic changes.
In this review, we summarize recent advances in the IHD‐linked alterations in microbiome, not only with a focus on the taxa of bacteria (Figure 1 and Supporting Information: Table 1), but also with a particular emphasis on the microbiota‐related metabolites that regulate the initiation, escalation, and onset of IHD (Figure 2 and Table 1). We also provide insights into the updates and perspectives of microbiome‐based therapies against IHD development (Figure 3). In addition to that, we point out the gut virome/phageome as an emerging possibility in interfering the gut bacterial structure or function, thereby complementing therapeutic strategy on IHD (Figure 2).
Figure 1.
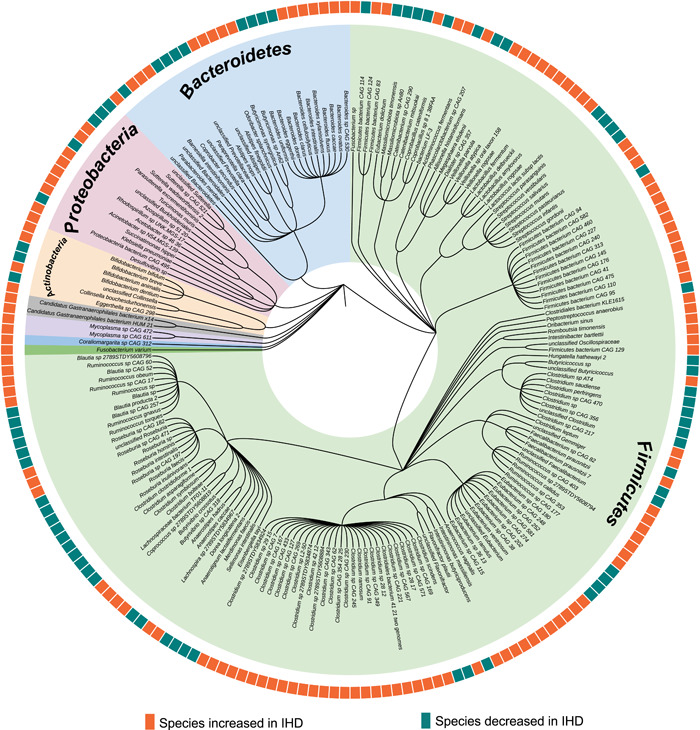
Phylogenetic tree of bacterial species associated with ischemic heart disease (IHD). This phylogenetic tree summarizes the numerous reported gut bacterial species associated with IHD. Background colors in the inner ring indicate all species that belong to the same phylum. The outer rings indicate the enrichment status of each listed bacterial species in IHD cases compared with healthy microbiota. Orange color denotes bacterial species increased in IHD cases, whereas green color denotes bacterial species decreased in IHD cases. This phylogenetic tree was created with iTOL (v6, https://itol.embl.de/).
Figure 2.
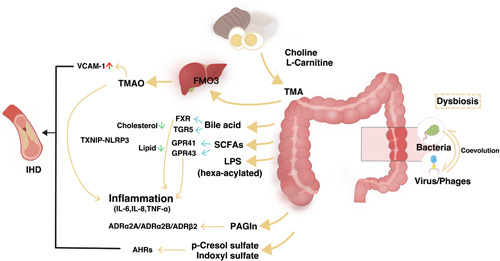
Gut‐heart axis: the potential mechanisms. An overview of some of the microbially produced compounds affecting the well‐being of cardiometabolic homeostasis during the dysbiosis associated with ischemic heart disease (IHD). Dietary l‐carnitine is metabolized by gut microbiota producing trimethylamine (TMA), which is subsequently N‐oxidized by liver flavin‐containing monooxygenases (FMOs) and producing trimethylamine N‐oxide (TMAO). TMAO has been recognized as an important contributor to atherosclerotic consequences. Secondary bile acids (BAs), synthesized from primary BAs by the intestinal microbiota, act through farnesoid X receptor (FXR) and Takeda G‐protein‐coupled receptor‐5 (TGR5) (also known as G protein‐coupled BA receptor 1 [Gpbar1]) receptors to reduce inflammation, thereby counteracting atherosclerosis. Short‐chain fatty acids (SCFAs) produced by gut microbiota through G‐protein receptor (GPR) 41/43 regulating systematic inflammation. Lipopolysaccharides (LPS), a class of pro‐inflammatory compounds, act on Toll‐like receptors, thereby activating atherosclerogenesis. Adrenoceptors have been identified as binding receptors for microbiota‐synthesized phenylacetylglutamine (PAGln). The Aryl hydrocarbon receptors (AHRs) plays crucial roles in mediating impact of two uremic toxins, p‐cresol sulfate and indoxyl sulfate, on cardiovascular system. IL, interleukin; NLRP3, NLR family pyrin domain containing 3; TNF‐α, tumor necrosis factor‐α; TXNIP, thioredoxin interacting protein; VCAM‐1, vascular cell adhesion molecule 1.
Table 1.
Bacterial producers of microbiota‐related metabolites and their signaling pathways involved in IHD.
| Human gut bacterial producers [18, 19, 20, 21, 22, 23] | Gut microbiota‐related metabolites | Potential receptors and molecular pathways linking gut microbial metabolites to IHD |
|---|---|---|
| p_Actinobacteria|c_Not assigned|o_Coriobacteriales|f_Coriobacteriaceae|g_Collinsella|s_Collinsella aerofaciens | Trimethylamine N‐oxide | Receptors: TAAR5 [24]; PERK [25], and so on |
| p_Bacteroidetes|c_Bacteroidia|o_Bacteroidales|f_ Bacteroidaceae|g_Bacteroides|s_Bacteroides caccae | ||
| p_Bacteroidetes|c_Bacteroidia|o_Bacteroidales|f_ Bacteroidaceae|g_Bacteroides|s_Bacteroides ovatus | Pathways: Atherosclerosis [26]; Thrombosis [27]; Inflammatory regulation [28]; Cardiorenal fibrosis [29], and so on | |
| p_Bacteroidetes|c_Bacteroidia|o_Bacteroidales|f_ Bacteroidaceae|g_Bacteroides|s_Bacteroides thetaiotaomicron | ||
| p_Firmicutes|c_Clostridia|o_Clostridiales|f_ Clostridiaceae|g_Clostridium|s_Clostridium asparagiforme | ||
| p_Firmicutes|c_Clostridia|o_Clostridiales|f_ Clostridiaceae|g_Clostridium|s_Clostridium hathewayi | ||
| p_Firmicutes|c_Clostridia|o_Clostridiales|f_ Clostridiaceae|g_Clostridium|s_Clostridium sporogenes | ||
| p_Firmicutes|c_Clostridia|o_Clostridiales|f_Eubacteriaceae|g_Eubacterium|s_Eubacterium rectale | ||
| p_Firmicutes|c_Clostridia|o_Clostridiales|f_Not assigned|g_Anaerococcus|s_Anaerococcus hydrogenalis | ||
| p_Proteobacteria|c_Gammaproteobacteria|o_Enterobacteriales|f_Enterobacteriaceae|g_Edwardsiella|s_Edwardsiella tarda | ||
| p_Proteobacteria|c_Gammaproteobacteria|o_Enterobacteriales|f_Enterobacteriaceae|g_Escherichia|s_Escherichia fergusonii | ||
| p_Proteobacteria|c_Gammaproteobacteria|o_Enterobacteriales|f_Enterobacteriaceae|g_Proteus|s_Proteus penneri | ||
| p_Proteobacteria|c_Gammaproteobacteria|o_Enterobacteriales|f_Enterobacteriaceae|g_Providencia|s_Providencia rettgeri | ||
| p_Bacteroidetes|c_Bacteroidia|o_Bacteroidales|f_ Bacteroidaceae|g_Bacteroides|s_Bacteroides fragilis | Short‐chain fatty acids | Receptors: GPR41/43 [30]; Olfr78 [31], and so on |
| p_Bacteroidetes|c_Bacteroidia|o_Bacteroidales|f_ Bacteroidaceae|g_Bacteroides|s_Bacteroides thetaiotaomicron | Pathways: Inflammatory modulation [32]; Myocardial regeneration [33]; Blood pressure homeostasis [34], and so on | |
| p_Firmicutes|c_Negativicutes|o_Selenomonadales|f_Acidaminococcaceae|g_Phascolarctobacterium|s_Phascolarctobacterium succinatutens | ||
| p_Firmicutes|c_Negativicutes|o_Selenomonadales|f_Veillonellaceae|g_ Dialister|s_Dialister succinatiphilus | ||
| p_Firmicutes|c_Negativicutes|o_Selenomonadales|f_Veillonellaceae|g_ Veillonella|s_Veillonella parvula | ||
| p_Firmicutes|c_Negativicutes|o_Selenomonadales|f_Veillonellaceae|g_Megasphaera|s_Megasphaera elsdenii | ||
| p_Firmicutes|c_Negativicutes|o_Selenomonadales|f_Veillonellaceae|g_Selenomonas|s_Selenomonas ruminantium | ||
| p_Actinobacteria|c_Not assigned|o_Bifidobacteriales|f_Bifidobacteriaceae|g_Bifidobacterium|s_Bifidobacterium adolescentis | Bile acids | Receptors: TGR5 [35]/FXR [36]/LXR [37], and so on |
| p_Actinobacteria|c_Not assigned|o_Bifidobacteriales|f_Bifidobacteriaceae|g_Bifidobacterium|s_Bifidobacterium bifidum | ||
| p_Actinobacteria|c_Not assigned|o_Bifidobacteriales|f_Bifidobacteriaceae|g_Bifidobacterium|s_Bifidobacterium longum | ||
| Pathways: Lipid metabolism regulation [38]; Glucose/Insulin homeostasis [39]; Immunity [40], and so on | ||
| p_Bacteroidetes|c_Bacteroidia|o_Bacteroidales|f_ Bacteroidaceae|g_Bacteroides|s_Bacteroides fragilis | ||
| p_Bacteroidetes|c_Bacteroidia|o_Bacteroidales|f_ Bacteroidaceae|g_Bacteroides|s_Bacteroides vulgatus | ||
| p_Firmicutes|c_Bacilli|o_Bacillales|f_Listeriaceae|g_Listeria|s_Listeria monocytogenes | ||
| p_Firmicutes|c_Bacilli|o_Lactobacillales|f_Lactobacillaceae|g_Lactobacillus|s_Lactobacillus acidophilus | ||
| p_Firmicutes|c_Bacilli|o_Lactobacillales|f_Lactobacillaceae|g_Lactobacillus|s_Lactobacillus johnsonii | ||
| p_Firmicutes|c_Bacilli|o_Lactobacillales|f_Lactobacillaceae|g_Lactobacillus|s_Lactobacillus plantarum | ||
| p_Firmicutes|c_Clostridia|o_Clostridiales|f_ Clostridiaceae|g_Clostridium|s_Clostridium perfringens | ||
| p_Bacteroidetes|c_Bacteroidia|o_Bacteroidales|f_ Bacteroidaceae|g_Bacteroides|s_Bacteroides caccae | Lipopolysaccharide | Receptors: TLR2/4 [41]/9 [42], and so on |
| p_Bacteroidetes|c_Bacteroidia|o_Bacteroidales|f_ Bacteroidaceae|g_Bacteroides|s_Bacteroides dorei | ||
| p_Bacteroidetes|c_Bacteroidia|o_Bacteroidales|f_ Bacteroidaceae|g_Bacteroides|s_Bacteroides fragilis | Pathways: Proinflammation [42]; Vascularly functional homeostasis, and so on | |
| p_Bacteroidetes|c_Bacteroidia|o_Bacteroidales|f_ Bacteroidaceae|g_Bacteroides|s_Bacteroides ovatus | ||
| p_Bacteroidetes|c_Bacteroidia|o_Bacteroidales|f_ Bacteroidaceae|g_Bacteroides|s_Bacteroides uniformis | ||
| p_Bacteroidetes|c_Bacteroidia|o_Bacteroidales|f_ Bacteroidaceae|g_Bacteroides|s_Bacteroides vulgatus | ||
| p_Bacteroidetes|c_Bacteroidia|o_Bacteroidales|f_Prevotellaceae|g_Prevotella|s_Prevotella copri | ||
| p_Actinobacteria|c_Not assigned|o_Coriobacteriales|f_ Coriobacteriaceae|g_Collinsella|s_ Collinsella intestinalis | Phenylacetylglutamine | Receptors: ADRA2A/ADRA2B/ADRB2 [43], and so on |
| p_Actinobacteria|c_Not assigned|o_Coriobacteriales|f_ Coriobacteriaceae|g_Collinsella|s_Collinsella aerofaciens | ||
| p_Bacteroidetes|c_Bacteroidia|o_Bacteroidales|f_ Bacteroidaceae|g_Bacteroides|s_Bacteroides caccae | ||
| p_Bacteroidetes|c_Bacteroidia|o_Bacteroidales|f_ Bacteroidaceae|g_Bacteroides|s_Bacteroides cellulosilyticus | Pathways: Atherosclerosis [43]; Thrombosis [43], and so on | |
| p_Bacteroidetes|c_Bacteroidia|o_Bacteroidales|f_ Bacteroidaceae|g_Bacteroides|s_Bacteroides ovatus | ||
| p_Bacteroidetes|c_Bacteroidia|o_Bacteroidales|f_ Bacteroidaceae|g_Bacteroides|s_Bacteroides thetaiotaomicron | ||
| p_Bacteroidetes|c_Bacteroidia|o_Bacteroidales|f_ Bacteroidaceae|g_Bacteroides|s_Bacteroides uniformis | ||
| p_Bacteroidetes|c_Bacteroidia|o_Bacteroidales|f_ Rikenellaceae|g_ Alistipes|s_Alistipes indistinctus | ||
| p_Firmicutes|c_Bacilli|o_Bacillales|f_ Staphylococcaceae|g_Staphylococcus|s_Staphylococcus aureus | ||
| p_Firmicutes|c_Clostridia|o_Clostridiales|f_ Clostridiaceae|g_Clostridium|s_Clostridium asparagiforme | ||
| p_Firmicutes|c_Clostridia|o_Clostridiales|f_ Clostridiaceae|g_Clostridium|s_Clostridium hathewayi | ||
| p_Bacteroidetes|c_Bacteroidia|o_Bacteroidales|f_Muribaculaceae|g_Barnesiella|s_uncultured Barnesiella sp. | p‐Cresol sulfate, Indoxyl sulfate | Receptors: AHRs [44], and so on |
| p_Bacteroidetes|c_Bacteroidia|o_Bacteroidales|f_Muribaculaceae|g_Duncaniella|s_uncaniella dubosii | Pathways: Kidney loss of function [45]; ROS production [46], proinflammation [47], and so on | |
| p_Bacteroidetes|c_Bacteroidia|o_Bacteroidales|f_Muribaculaceae|g_Muribaculum|s_uncultured Muribaculum sp. | ||
| p_Bacteroidetes|c_Bacteroidia|o_Bacteroidales|f_Muribaculaceae|g_Unknown|s_uncultured Muribaculaceae bacterium sp. | ||
| p_Bacteroidetes|c_Bacteroidia|o_Bacteroidales|f_Odoribacteraceae|g_Culturomica|s_Culturomica sp. | ||
| p_Firmicutes|c_Clostridia|o_Clostridiales incertae sedis|f_Clostridiales family XIII Incertae Sedis|g_Aminipila|s_Aminipila butyrica | ||
| p_Firmicutes|c_Clostridia|o_Clostridiales|f_Lachnospiraceae|g_Anaerobium|s_Anaerobium sp. | ||
| p_Proteobacteria|c_Betaproteobacteria|o_Burkholderiales|f_Sutterellaceae|g_Turicimonas|s_Turicimonas muris |
Abbreviations: ADR, adrenoceptor; AHR, acryl hydrocarbon receptor; c_, class; f_, family; FXR, farnesoid X receptor; g_, genuss; GPR41/43, G‐protein‐coupled receptor 41/43; IHD, ischemic heart disease; LXR, liver X receptor; o_, order; Olfr78, olfactory receptor 78; p_, phylum; PERK, protein kinase R (PKR)‐like endoplasmic reticulum kinase; ROS, reactive oxygen species; s_, species; TAAR5, trace amine‐associated receptor 5; TGR5, G‐protein‐coupled bile acid receptor 1; TLR2/4/9, Toll‐like receptor 2/4/8.
Figure 3.
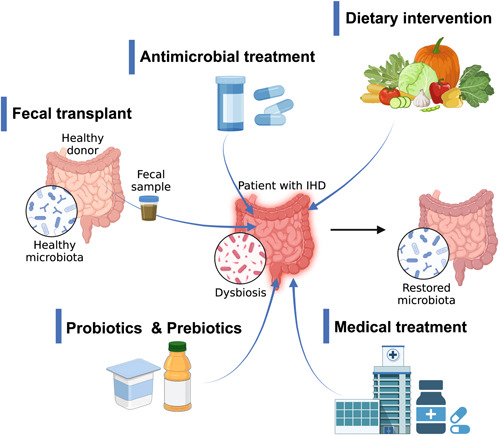
Gut microbiome‐targeted interventions in humans with ischemic heart disease (IHD). For general restoration of microbial composition and functions, there are interests in testing the microbiome‐targeted interventions on the disrupted microbiome of IHD patients. Various approaches include fecal microbiota transplantation either in heterologous or autologous manner; antibiotics treatment aiming at restructuring the gut microbiome; individualized nutrition to change the gut microbiome and metabolism; use of probiotics and prebiotics or combination of various probiotics strains and prebiotics; and finally, an emerging potential frontier of using drugs targeting specific IHD‐related microbial metabolites or pathways. Illustration was created with biorender (https://biorender.com/).
GUT BACTERIAL CHANGES ASSOCIATED WITH IHD DEVELOPMENT
Patients with different types of IHD are often found to be associated with gut dysbiosis at multiple resolutions (Figure 1). However, due to the fact that patients with atherosclerosis are frequently found to have prior clinically silent metabolic dysregulations for many years, it is therefore a major challenge to delineate the putative impact of gut bacterial imbalance on early‐stage metabolic dysfunction from IHD onset. In addition to that, most of the IHD patients are always heavily medicated on its various comorbidities, such as obesity, T2D, and so on, making it even more challenging to decode the gut microbial alterations directly linked to IHD itself without accounting for the dysbiosis induced by its premorbidities and comorbidities. In 2017, Cui et al. [12] found that the abundance of phylum Bacteroidetes and Proteobacteria in patients with IHD were lower than that in the controls without adjusting for any medications or comorbidities. In addition, the class Bacteroidia, belonging to phylum Bacteroidetes, was significantly decreased in the IHD patient group compared with the control group. In contrast, phylum Firmicutes and Fusobacteria was higher than that in the controls [12]. At the family level, in 2019, Liu et al. [13] reported that Lachnospiraceae and Ruminococcaceae decreased significantly in IHD patients. Meanwhile, they found that Proteobacteria phylotypes such as Streptococcus, Haemophilus, and Granulicatella increased higher in the group with more severe heart disease by multiple comparisons among its subgroups [13]. In addition, they found that the abundance of the co‐abundance group 17, which contained several Gram‐negative bacteria, such as Veillonella, Haemophilus, and Klebsiella, increased with IHD severity [13]. According to the previous findings, these bacteria trigger the innate immune response via lipopolysaccharide (LPS) production and elicit a subsequent inflammatory reaction [14]. At the genus level, Jie et al. [9] found that there was a relative reduction in Bacteroides and Prevotella, and enrichment in Streptococcus and Escherichia in gut bacteriome of patients with atherosclerotic cardiovascular disease (ACVD). The abundance of Enterobacteriaceae and the bacteria that are often found in the oral cavity, such as Streptococcus spp., Lactobacillus salivarius, Solobacterium moorei, and Atopobium parvulum, were also higher in patients with ACVD than in healthy controls. In contrast, butyrate‐producing bacteria including Roseburia intestinalis and Faecalibacterium prausnitzii were depleted in the ACVD gut bacteriome. It is worthwhile to note that the gut microbiota also showed differences in network structure between ACVD and healthy individuals. For instance, it was found that ACVD microbiome is characterized by a negative correlation between ACVD‐depleted commensals Bacteroides spp. and aerobes Streptococcus spp., which is intriguingly absent in normal control gut bacteriome. These results demonstrated profound imbalances in the composition and inter‐species relationship in the gut microbiome of ACVD patients as compared with healthy controls [9].
Of special interest is the impact of major disease confounders on IHD microbiome analysis, including promorbidities, comorbidities, and multidrug interventions, which have gained attention. A recent work focused on characterizing the altered microbial features along the nature history of IHD, including disease initiation and escalation, while accounting for the effects of medication and lifestyle, on different IHD stages. It was reported long before the early clinical manifestation of IHD, the major microbial and metabolic alterations had already begun. The researchers additionally by using machine learning algorithms identified deconfounded IHD‐specific microbiome and metabolome features, which likely provide better capacity in IHD subgroup classification than that of the conventional IHD biomarkers. The IHD‐specific bacterial features are composed of 23 species including Acinetobacter, Turcimonas, and Acetobacter depleted in IHD patients, and 8 species enriched in IHD which contains 2 species in Burkholderiales order [10]. One of the two species in Burkholderiales order was reported as a possible cause of endocarditis [15]. This work highlighted the importance of accounting for interactions by confounders when analyzing microbiome data in complex noncommunicable diseases. In another Israeli cohort, Yeela et al. [16] reported that 20 bacterial genomes significantly enriched in either the patients with acute coronary diseases (ACS) or the control individuals by adjusting the confounders including clinical parameters and multidrug usage. They found that butyrate‐producing bacteria such as Clostridium, Anaerostipes hadrus, Streptococcus thermophilus, and Blautia decreased, whereas the abundance of Odoribacter splanchnicus and Escherichia coli increased in ACS patients. In addition to known bacterial features, they found a previously unknown bacterial species of the Clostridiaceae family that was depleted in ACS [16]. Interestingly, researchers from both groups found that butyrate producers decreased in IHD patients, which may lead to the reduced production potential of short‐chain fatty acids (SCFAs). As for acute myocardial infarction (AMI), Han et al. [17] recruited 30 in‐hospital AMI patients in China and found bacteria belonging to the phyla Actinobacteria, Cyanobacteria, Proteobacteria, and Verrucomicrobia are enriched in AMI gut microbiome, whereas the phyla Fusobacteria and Tenericutes decreased. In addition, they reported that the patients who suffered the AMI caused by left anterior descending coronary stenosis are characterized by enriched Ruminantium group, Comamonadaceae, Comamonas, and unknown species belonging to the MollicutesRF9 order [17]. This study, although without considering the confounding effects of polypharmacy and lifestyle in a relatively small cohort, points to the potential for gut microbial involvement in AMI caused by coronary branch vessel stenosis.
MICROBIOTA‐RELATED METABOLITES FUEL THE MECHANISTIC UNDERSTANDING OF GUT BACTERIA IN IHD
Vast studies have revealed that the compositional and structural aberrancies in gut microbiota characterize IHD patients. However, the underlying mechanistic information of microbiota‐IHD associations remains unsystematically reviewed. In fact, the gut microbiome, as a genetic repository, is an immense factory that can release or synthesize overwhelming numbers of chemicals needed for the communication between gut commensals and host (Table 1). In the following section, we selectively summarize the IHD‐specific bacterial messengers and their impacts on IHD pathophysiology (Figure 2).
Production of trimethylamine‐N‐oxide (TMAO)
Dietary factors such as choline and carnitine are closely related to TMAO, which is proved as an independent risk factor for IHD [48, 49, 50]. TMAO comes from many sources, such as egg, fish, red meat, and so on [51]. In 2013, Hazen and colleagues [48] found that TMAO was an independent risk factor for IHD and subsequent experiments demonstrated that TMAO levels were associated with death and nonfatal myocardial infarction [48, 52, 53]. The precursor of TMAO is trimethylamine (TMA), which is produced by intestinal microorganisms from nutrients containing l‐carnitine or phosphatidylcholine [50]. TMA produced by intestinal microorganisms can enter the host circulation and reach hepatocytes. In the liver, kidney, and other tissues, TMA is metabolized by flavin‐containing monooxygenase (FMO), which is encoded by FMO gene [54, 55]. Higher production of TMAO will affect lipid metabolism and reduce cholesterol clearance by inhibiting the synthesis of bile acids (BAs) [56, 57]. This may be because TMAO induces the expression of two scavenging receptors (CD36 and scavenger receptor A) on the cell surface which lead to inhibit the reverse transport of cholesterol and the accumulation of cholesterol in macrophages [58]. Moreover, TMAO can also induce calcium release and platelet hyperreactivity, thereby affecting the IHD development [59]. TMAO can upregulate inflammatory factors such as tumor necrosis factor α (TNF‐α), interleukin (IL)‐6, IL‐18 through the activation of TXNIP‐NLRP3 [60]. It can boost the expression of vascular cell adhesion molecule 1 and monocyte adhesion, which can lead to plaque development [61].
The gut microbial composition is a major factor that impacts TMAO production. Gwen's study identified a total of 102 genomes from 36 species classified as Firmicutes, Proteobacteria, and Actinobacteria [51] that influenced the production of TMA. Another study found that eight species representing phylum Firmicutes and Proteobacteria, and six genera consume more than 60% of choline presented in the media, which subsequently led to a remarkable production of TMA [18, 62]. Additional experiments have also expanded the TMA‐producing bacteria to Ruminococcus [63]. In addition, the taxonomic identification of TMA‐producing gut bacteria, biosynthetic genes, and gene clusters (BGCs) responsible for the production TMA have been reported. Two dominant TMA synthesis pathways have been extensively studied; these are as follows: (1) using choline as a substrate via the choline TMA‐lyase (CutC) and its activator CutD [64]; (2) acting on carnitine through a two‐component Rieske‐type oxygenase/reductase (CntA/B) [65]. In addition, the enzyme complex YeaW/X has also been shown to take part in the TMA synthesis [66]. The γ‐butyrobetaine (γBB)‐specific BGCs are six adjacent genes consisting of one acyl‐CoA dehydrogenase (gbuA), two acyl‐CoA transferases (gbuB, gbuC), a ubiquinone oxidoreductase (gbuD), a betaine/carnitine/choline transporter (gbuE), and one acyl‐CoA thioester hydrolase (gbuF), among which four genes were identified as necessary and sufficient for TMA production in the non‐native E. coli host: gbuA, gbuB, gbuC, and gbuE [67].
Although TMAO is the most widely studied independent risk factor related to IHD, more microbiota‐dependent risk factors have been found. For instance, trimethyllysine (TML), a precursor of the synthesis of carnitine, which can be metabolized to proatherogenic TMA, is a strong predictor of incident IHD, independent of TMAO [68]. Another proatherogenic agent, γBB, which is an intermediate in gut microbial transformation of carnitine to TMA, was also found to be closely related to the risk of IHD in clinical cohort (n = 2918). Furthermore, N,N,N‐trimethyl‐5‐aminovaleric acid (TMAVA), which was derived from TML through the gut microbial metabolism, was elevated with gradually increased risk of cardiac mortality and transplantation in a prospective heart failure cohort (n = 1647) [69]. Zhao et al. [69] found that TMAVA increased significantly, especially in patients with hypertension, which may lead to cardiac hypertrophy. In addition, they supplemented mice on a high‐fat diet for 12 weeks. They discovered that heart weight was increased in the TMAVA‐treated mice, compared with the untreated controls, which suggested that TMAVA aggravates cardiac hypertrophy and dysfunction induced by heart failure. They also found that TMAVA treatment leads to myocardial lipid accumulation and carnitine reduction in plasma and myocardium. They supposed that TMAVA functions through γBB hydroxylase (BBOX) by mice experimenting with BBOX deficiency [69].
In conclusion, TMAO and other related precursors in its synthetic pathway, play an important role in the occurrence and development of IHD, and relevant pathways remain to be further explored.
Synthesis of SCFAs
SCFAs including acetate, propionate, and butyrate are fermented from monosaccharides and are the main bacterial products [70, 71]. Acetate and propionate are mostly produced by the phylum Bacteroidetes, whereas butyrate is mainly produced by the phylum Bacteroidetes and Firmicutes [72, 73]. Research by Jie et al. [9] showed that the gut microbiome of IHD patients is characterized by a reduction in Roseburia and Eubacterium, two known producers of butyrate. Consistently, they found the functional potential for butyrate production reduced in IHD patients [9]. SCFAs have positive effects including regulating intestinal pH, decreasing body weight, improving insulin sensitivity, and promoting intestinal motility [73, 74]. The SCFAs can also reduce blood lipid levels by transferring cholesterol to the liver and blocking cholesterol synthesis [75]. SCFAs are transported by specific monocarboxylate transporters through the intestine into the blood. SCFAs work by acting as a ligand for the G‐protein‐coupled receptors (GPR43 and GPR41) [76, 77]. These receptors play a vital role in the regulation of energy consumption and expenditure, and immune response. Furthermore, another research reported that there was a strong negative correlation between butyrate‐producing genes and C‐reactive protein (CRP) levels [78], which has been reported to be closely related to the occurrence of IHD.
The gut SCFA‐producing bacteria have been shown to be less abundant in certain IHD and hypertension patients [78, 79]. Both SCFA‐producing bacterial features and SCFAs are considered as a protective element in IHD development. Thus, targeted strategies enriching the SCFAs and its producers are potential therapeutic means for IHD preventions.
BA modulation
Gut microbiota is one of the main contributors in regulating circulating BAs [80, 81]. Primary BAs are synthesized by the oxidation of cholesterol in the liver and secreted into the intestine as taurine‐ or glycine‐conjugated forms at C24 to dissolve lipids for absorption through the rate‐limiting enzyme cholesterol 7‐α‐hydroxylase (CYP7A1) [82]. Primary BAs (cholic acid and chenodeoxycholic acid) are converted into secondary BAs (deoxycholic acid, lithocholic acid [LCA], ursodeoxycholic acid [UDCA], and so on) through microbial dehydroxylation [83]. About 95% of BAs are reabsorbed and recycled from the intestine, except for LCA and UDCA [84]. BAs can act as ligands activating nuclear receptor farnesoid X receptor and Takeda G‐protein‐coupled receptor‐5 (TGR5) [82, 85]. Through activating the two receptors, BAs can reduce the serum cholesterol level [86]. Moreover, upon activation of TGR5, BAs can also protect LPS‐induced inflammation [87]. More specifically related to IHD development, the study led by Mayerhofer et al. [88] demonstrated that BAs reduce heart rate and regulate vascular tension via regulating channel conductance and calcium dynamics. Moreover, they found that the primary to secondary BAs ratio is positively correlated with the level of circulating cholesterol in patients with heart failure and IHD development [88]. Although most studies focus on describing the associations between gut microbes and circulating BAs, a deeper understanding towards the regulator of gut microbiota responsible for BAs metabolism is sparse. As a notable example, Wang et al. [89] demonstrated the gut bacterial structural variations (SVs) greatly determine the BAs metabolism. Systematically characterizing two types of SVs, deletion and variable SVs, in the human gut microbiome from two cohorts consisting of 1437 participants, and associating the SVs profile to circulating BAs, allowed the investigators to identify the genetic regions in specific bacterial genomes that are responsible for BAs regulation. More interestingly, such a strategy also identifies putative regions encoding BA‐metabolizing enzymes, although experimental evidence is still lacking due to the big challenge in isolating bacterial strains carrying the identified regions.
We assume that if the gut microbial features (taxas, functional potentials, and SVs), which is related to BAs metabolism, are in a state of imbalance, IHD is developed. Therefore, the BA‐relevant bacterial functions, receptors, and pathways need to be explored and may be targeted for therapeutic intervention of IHD.
LPS and immune regulation
The depletion of butyrate‐producing bacteria may not only cause reduction of butyrate but also lead to intestinal mucosal barrier dysfunction and increase the passive leakage of microbial toxins, such as LPS and other receptors of the innate immune system, leading to inflammation [90, 91]. Recently, Awoyemi et al. [92] reported that increasing levels of LPS‐binding protein associated with high risk of IHD. Intestinal leakage may also lead to the translocation of LPS [93]. Several studies have reported that hexa‐acylated LPS but not penta‐acylated LPS can lead to systematic inflammation [94, 95]. Therefore, we highlight that hexa‐acylated LPS may be a potential target IHD treatment.
Gut microbiota can lead to IHD development via regulating our immune system. IHD is a chronic inflammatory disease, whereas AMI is suspected to be associated with acute inflammation [96, 97]. In our body, oxidized low density lipoprotein (oxLDL) can promote atherosclerosis and inflammation by activating endothelial cells, macrophages, and T cells. Macrophages can promote generation of inflammatory factors (TNF‐α, IL‐6, IL‐18, and IL‐37) by devouring oxLDL and leading to IHD development as a consequence [98, 99]. The composition of gut microbiota can strongly influence body's immune system. The study by Mikelsaar et al. [100] reported that the quantity of Lactobacillus reuteri, which exists in the intestine, is associated with high levels of white blood cells. Furthermore, Low Oscillibacter, Faecalibacterium, and Ruminococcus are correlated with high CRP level [101, 102]. Besides, germ‐free mouse models showed that the development of T cells is directly influenced by gut microbiota, particularly the differentiation of T helper 17 cells (Th17) [103, 104, 105]. The research by Gil‐Cruz et al. [60] reported that myocarditis may depend on specific Th17 cells derived from gut microbiota. They additionally found that Bacteroides thetaiotaomicron and B. faecis can promote inflammatory myocardiopathy [60]. Furthermore, butyrate produced by gut bacteria promotes the forkhead box P3 (Foxp3+) regulatory T cell induction [106], as well as acts on the GPR43 and GPR41 for affecting immune system [76, 107].
Phenylacetylglutamine (PAGln) production
PAGln, a product from microbial fermentation of dietary phenylalanine followed by conjugation to glutamine, has been reported to be associated with IHDs and major adverse cardiovascular events independently [108]. Among patients with carotid plaque, plasma level of PAGln was significantly lower in protected phenotype rather than other more severe phenotype. Therefore, it is considered that lower PAGln may contribute to plaque stability in carotid atherosclerosis [109]. As for patients with IHD, Liu et al. [110] reported an independent association between plasma PAGln levels and the coronary atherosclerotic burden. Patients with the higher PAGln levels had higher risks of obstructive IHD and higher coronary lesion complexity [110]. Mechanically, genetic engineering studies followed by microbial transplantation showed that PAGln contribute to the thrombosis potential by accelerating platelet clot formation, calcium release, and responsiveness to multiple agonists. By using multiple genetic and pharmacological screening, PAGln was found to interact with G‐protein‐coupled receptors, in particular adrenergic receptors (ADRs), including α2A, α2B, and β2‐ADRs, which highly present on human platelets. ADRs are crucial for cardiovascular functions and closely related to cardiovascular events [111] and platelet activity [112]. Selective ADR inhibitors can reduce the platelet hyperreactivity induced by PAGln and the acceleration rate of thrombosis in vivo [43]. Similar to TMAO, PAGln and PAGln‐releasing gut microbes appear to be the other potential targets for treating IHD in future efforts.
Other microbiota‐related metabolites
Among the well‐known microbiota‐related metabolites, two protein‐bound uremic toxins, p‐cresol sulfate (PCS) and indoxyl sulfate (IS), are associated with cardiovascular events [113, 114, 115] and cardiovascular stiffening [116] in patients with chronic kidney disease (CKD). Indeed, CKD patients are often found to display a substantial increase in cardiovascular disease [117]. In the rat CKD model, etiological evidence has been described that PCS and IS may, via activating the coagulation and pro‐inflammatory pathways, contribute to the onset and development of calcification in the vessel wall [118] (Table 1). However, it is important to note that conflicting results exist for the absence in associations between PCS, IS, and cardiovascular outcomes in patients undergoing hemodialysis [119]. Caution is required in the causal interpretation of PCS and IS in IHD development.
MICROBIOTA‐BASED THERAPY OF IHD
Therapy targeting gut microbiota
Considering the high mortality and morality of IHD in modern society, clinical translation of the identified IHD‐specific microbiota‐dependent targets is urgently needed. In response to the arising evidence indicating gut microbiota plays a crucial role in IHD, more and more attention has been paid to the therapeutic strategies targeting gut microbial modulation. In the following section, we will highlight several tools and strategies to modulate gut microbial community and their potential in IHD intervention (Figure 3).
Fecal microbiota transplantation (FMT)
FMT allows for the nutritional enrichment or depletion to the host microbiota, inhibits the growth of pathogenic bacteria, and regulates the host's immune system by transplanting and recolonizing live functional bacterial community from healthy donors into the patient's gastrointestinal tract [120]. FMT is the most fundamental intervention for intestinal microbiota and it is also an established and widely accepted method for the treatment of recurrent Clostridium difficile infection [121]. Although it has been shown that obese individuals who receive FMT from lean donors gained enhanced insulin sensitivity and improved phenotypic parameters related to metabolic syndromes [122], the outcomes are highly varied among studies. For instance, the TMAO levels in individuals with metabolic syndrome are unexpectedly not associated with FMT from a single vegan donor, whereas the gut microbial composition of recipients changes towards that of vegan's, pointing to the importance of big sample size and prolonged follow‐up periods in FMT to get desired effects. In addition to the unremarkable changes in TMAO production upon FMT, recent study also reported that transplanting drug‐resistant E. coli led to the death of one patient [123], raising the safety concerns of FMT for clinical use. It must be mentioned that challenges in FMT still exists, including the knowledge about optimal conditions for anaerobic handling of donor stools, the incompatibility between recipients and donors, as well as the instability in the survival and recolonization of donor bacteria in recipients’ intestinal tract.
Antibiotics
Broad‐spectrum antibiotics are commonly used in early experiments targeting the intestinal microbiota for IHD. In 2004, the study by Conraads et al. [124] evidenced that broad‐spectrum antibiotics reduces the biomarker of systematic inflammation in patients with heart failure, but not specifically focused on clinical symptoms. Galla et al. [125] found that minocycline and vancomycin intervention remarkably increased systolic blood pressure in salt‐sensitive rats and decreased systolic blood pressure in spontaneously hypertensive rats. Rune et al. [126] showed in ApoE‐deficient mice, ampicillin intervention could reduce blood low‐density lipoprotein and very low‐density lipoprotein levels. It is interesting to see in the recent study, the oral administration of broad‐spectrum antibiotics increased the mortality of myocardial infarction murine model [127]. This is contrary to the previously reported results that oral vancomycin or a mixture of streptomycin, neomycin, polymyxin B, and bacitracin can reduce myocardial infarction size and improve cardiac function [128, 129]. In trials on patients, outcomes also vary, where some studies showed beneficial effects of antibiotics on IHD [130], whereas others did not. The 10‐year follow‐up data from the Claricor trial showed an increase in cardiovascular death in patients with stable CHD treated with clarithromycin [131]. Therefore, treating IHD with antibiotics remains controversial. Considering these safety problems and the lack of reliable clinical consequences in many trials, antibiotics should be used with caution in future studies aiming at re‐structuring intestinal microbiome of IHD.
Probiotics and prebiotics
Probiotics can functionally and compositionally interfere with or modulate intestinal microbiota, subsequently activating the immune system and conferring a health benefit [132]. Common probiotics, which have been widely used in clinical practice include Lactobacillus and Bifidobacterium [133, 134]. Prebiotics, which can stimulate the activity of probiotics, are substrates selectively utilized by the host microorganisms. Most prebiotics are carbohydrates, which can induce the increase in SCFAs and improve metabolic health [134, 135]. Animal models have already suggested that some probiotics and prebiotics such as Lactobacilli and inulin can slow down atherosclerosis. Rats treated with Lactobacillus plantarum 299v before coronary artery ligation reduced myocardial infarction area and improved heart function [129]. Mohania et al. [136] found that deposition of cholesterol and TAGs in liver and aorta were significantly reduced in rats fed with probiotic dahi. Another study found that obese volunteers who received 20 g/day of inulin‐propionate ester have reduced pro‐inflammatory interleukin‐8 levels compared with those who received cellulose, whereas inulin had no impact on the systemic inflammatory markers [137]. These observations suggested that probiotics and prebiotics may have therapeutic capacity of reducing hyperlipidemia and diet‐induced hypercholesterolemia. In patients with chronic systolic heart failure that was submitted to a 3‐month daily oral supplementation of Saccharomyces boulardii (1 g per day) present an improvement in left ventricular ejection fraction and a reduction on left atrial diameter [138]. Except for the relatively small sample size, the beneficial enlightenment of probiotics on IHD represents a promising therapeutic measure for preventing IHD and its related complications.
Controlling for diet and medication
Genetic factors and environment are both play a great role in the composition of gut microbiota. Dietary details in published studies are commonly lacking or ignored. In the human cohort, dietary records are difficult to access and highly individualized. One recent study found associations between the composition of gut microbiota and red wine, salt intake. Alcohol consumption frequency itself was robustly associated with differences in the distribution of several microbiota such as Firmicutes and Actinobacteria. In addition, the consumption of wine and beer or cider was strongly associated with differences in gut microbial composition [139]. Several studies have examined the impacts of diet on intestinal flora and disease by giving mice a high‐salt diet (HSD). The composition of gut microbiota was changed in the HSD mice. Erwinia, Christensenellaceae and Corynebacteriaceae, increased in HSD mice. In contrast, the quantity of Anaerostipes reduced in the HSD mice [140]. In subsequent studies, Bier et al. found seven unique taxa that were significantly associated with blood pressure [141]. There was a significant difference in fecal acetic acid, as well as propionic and isobutyric acids, but not in the butyric acid composition between HSD mice and normal‐diet ones [142]. Dietary control is a very practical way to adjust intestinal microbiota. For example, reducing red meat intake is a feasible and effective method to control TMAO level [143], high‐fiber diet for a short term significantly alters the gut microbiome and reduces constipation.
Medication is another crucial but unignorable confounder of gut microbiota. A recent study has shown that 19 drug groups and their metabolism were associated to the composition of intestinal microbiome [144]. Some antihyperglycemic drugs such as metformin, likely through restructuring the gut microbiome at multi‐taxonomic levels, as well as regulating various microbial functions, particularly microbial genes encoding metalloproteins or metal transporters, to impact human glucose homeostasis [145]. Statins are widely prescribed in clinic combined with aspirin for synergistically lowering blood atherosclerotic lipoproteins [145], it has been found that, although without accounting for other potential confounders, nor inferring the causality behind the observational outcomes, statin therapy is associated with lower prevalence of Bact2 enterotype [146], which was considered as a pro‐inflammatory gut microbial community type. Despite the findings indicating statins are possible targets for designing drug‐based gut microbial modulations, longitudinal, double‐blinded, randomized, placebo‐control study design adjusting for any potential confounders is required to further elucidate the causal relationship between statin therapy and lower Bact2 prevalence. This strategy is also recommended to be acknowledged for observational microbiome studies in cross‐sectional cohorts for a better clarification on the nature of microbiome‐disease links.
GUT VIROME: AN “EMERGING STAR” FOR UNDERSTANDING AND TREATING IHD
In the past few years, IHD has been proven to be associated with a variety of viruses such as hepatitis virus [147, 148, 149], human immunodeficiency virus [150], and periodontal viruses such as cytomegalovirus and Epstein‐Barr virus [151]. It remains to be explored whether gut virome alterations contribute to the development of IHD. As an important part of the gut microbiome, the function of the virome has gained more and more attention in recent years. The human gut virome is also known as the phageome, as phages make up the vast majority of it [152]. Roughly, there are 109 virus‐like particles (VLPs) per gram of feces, which is an order of magnitude higher than the total number of bacterial cells [153]. Although the advance of next‐generation sequencing technology enables us to further obtain the relevant sequencing reads of enterovirus [154], many challenges still exist. First, it must be mentioned that the gut virome is highly specific and dynamic among individuals, with scant overlap between healthy subjects [154]. Second, as phages rely on bacteria as their host to grow and function, so most enterovirus groups cannot be evaluated by conventional laboratory methodologies except for high‐throughput isolation of single phage species/strains, which is extremely time‐consuming and labor‐intensive. Currently, the mainstream strategy for profiling gut virome are VLPs DNA sequencing or whole‐community metagenome sequencing that require complex data processing and computational resources, while present viral genome database only covers a small proportion of existing viruses in human intestine [152, 155]. These challenges need to be addressed before a high‐resolution snapshot of gut virome is defined.
In recent years, research examining compositions of intestinal phages in specific type of IHD is gradually emerging despite the technical challenges in profiling the gut virome. A compositional analysis of gut virome where viral sequences were profiled in metegenomes of patients with CHD has resulted in Virgaviridae and Microviridae as the two dominant types of viruses in the enteric virome of CHD subjects [156]. Compared with the gut virome of healthy individuals, CHD gut virome is characterized by enriched Virgaviridae but reduced Microviridae; however, the underlying mechnisms remained to be explored. Additionally, Jie et al. [9] have identified a panel of differential bacteriophages that are specifically altered in ACVD, but displaying non‐significance in patients with rheumatoid arthritis, T2D, and obesity. Of interest, the known hosts for ACVD‐specific bacteriophages are dominant by Enterobacteriaceae at family level or Streptococcus at genus level. In another study of individuals with hypertension, a risk factor for IHD [157], it was found a panel consisting of 32 viruses displayed high discriminative power than that of gut bacteriome for the differentiation between people with hypertension and healthy individuals and prehypertention group.
Compared with the sparse knowledge of gut virome functionality from a genetic point of view, the importance of gut virome from an evolutionary perspective appears to be better clarified. Bacteria–phages coevolution, the reciprocal evolution between bacterial hosts and the phages that live on or infect them, is an important driver of ecological and evolutionary processes in microbial communities. There is growing evidence from both laboratory and natural populations that coevolution can maintain phenotypic and genetic diversity, increase the rate of bacterial and phage evolution and divergence, affect community structure, and shape the evolution of ecologically relevant bacterial traits [158]. In the transkingdom interactions, the specific type of interaction between bacteria and phage can be quickly reflected in host immunity and infectious phenotype [159]. This directly or indirectly promotes genetic and phenotypic divergence, competitions, and cooperations [160, 161]. Indeed, in the transkingdom interaction analysis between gut virome and bacteriome in participants with hypertension, it was shown that hypertension group has higher number of linkages between viruses and bacteria in comparison with the healthy controls and prehypertension subjects. Gut phage is also considered a potential therapeutic target due to its lytic interaction with bacteria. In a recent study, Yi et al. [162] investigated the therapeutic effects of bacteriophages that target cytolytic E. faecalis by using humanized mice that were colonized with bacteria from the faeces of patients with alcoholic hepatitis. They showed that these phages decrease cytolysin in the liver and abolish ethanol‐induced liver disease in humanized mice [162]. Torben et al. [163] found that faecal virome transplantation cases showed significant weight loss in murine models. They supposed that faecal virome transplantation can ameliorate obesity and diabetes by changing the gut microbiota [163]. Considering IHD is closely related to infection and immunity, it is reasonable to hypothesize gut virome may play an important inducing role in the occurrence of IHD, and whether bacteriophages are potent in treating IHD by infecting IHD‐enriched specific pathogens remains to be further explored.
FUTURE PERSPECTIVES
Despite the immense alterations in taxonomic and functional potentials of gut microbiome in IHD, major reports on IHD microbiome studies appear to be observational, association‐based, and lacking specificity. With the focus on the influence of the gut microbiome on the overall functional readouts of IHD, much still needs to be learned. It is of great importance to decipher and annotate hundreds of yet non‐annotated chemicals in the metabolome of various biological fluids as well as their origins (solely host, microbial, and dietary origin, or combined origin). As most studies are based on cross‐sectional cohorts, sparse information of the microbial dynamics in IHD is given, either relevant bioinformatic algorithms predicting the short‐ or long‐term dynamics of gut microbiome, or longitudinal data is required to fill this gap. Last but not least, little of the novel knowledge is validated or has maturated to gain potential in being translated to guide clinical practice of IHD intervention. Future, with the progress in sequencing‐based and culture‐based gut microorganism surveys combined with mechanistic exploitations of the gut bacteriome, phageome, and virome, our knowledge towards the interactions within the global gut microbial system will be exponentially expanded.
CONCLUSIONS
As discussed in various literatures linking gut microbial features and IHD, human gut microbiota‐related metabolites appear as major mediators. In this review, we have not only summarized the various gut microbial taxa that are linked to multiple IHD stages, but also highlighted the up‐to‐date knowledges of microbial metabolites and their potential roles in mediating the impact of gut microbial alterations on progression of IHDs. Last but not least, we highlighted the gut virome as an additional dimension for mechanistic dissections and understandings of IHD.
AUTHOR CONTRIBUTIONS
Yong Fan and Hanbin Cui contributed to the overall conceptualization. Yong Fan and Jiajun Ying contributed equally to the writing and discussion of the main content of this manuscript. All authors have read and approved the final manuscript.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflict of interest.
Supporting information
Supporting information.
ACKNOWLEDGMENTS
The authors would like to thank the reviewers and editors for valuable discussion and critical reading. Hanbin Cui is supported by key research and development project of Zhejiang Province, China (Grant No. 2021C03096), and major project of science and technology innovation 2025 in Ningbo, China (Grant number 2021Z134).
Fan, Yong , Ying Jiajun, Ma Hongchuang, and Cui Hanbin. 2023. “Microbiota‐related metabolites fueling the understanding of ischemic heart disease.” iMeta 2, e94. 10.1002/imt2.94
Yong Fan and Jiajun Ying contributed equally to this work.
DATA AVAILABILITY STATEMENT
This manuscript does not generate any code or data.
REFERENCES
- 1. Benjamin, Emelia J. , Blaha Michael J., Chiuve Stephanie E., Cushman Mary, Das Sandeep R., Deo Rajat, de Ferranti Sarah D., et al. 2017. “Heart Disease and Stroke Statistics‐2017 Update: A Report From the American Heart Association.” Circulation 135: e146–e603. 10.1161/cir.0000000000000485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mogensen, Ulrik M. , Gong Jianjian, Jhund Pardeep S., Shen Li, Køber Lars, Desai Akshay S., Lefkowitz Martin P., et al. 2018. “Effect of Sacubitril/Valsartan on Recurrent Events in the Prospective Comparison of ARNI with ACEI to Determine Impact on Global Mortality and Morbidity in Heart Failure Trial (PARADIGM‐HF): Effect of Sacubitril/Valsartan on Recurrent Events.” Eur J Heart Fail 20: 760–68. 10.1002/ejhf.1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Crowe, Francesca , Zemedikun Dawit T., Okoth Kelvin, Adderley Nicola Jaime, Rudge Gavin, Sheldon Mark, Nirantharakumar Krishnarajah, and Marshall Tom. 2020. “Comorbidity Phenotypes and Risk of Mortality in Patients with Ischaemic Heart Disease in the UK.” Heart 106: 810–16. 10.1136/heartjnl-2019-316091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Piccioni, Andrea , de Cunzo Tommaso, Valletta Federico, Covino Marcello, Rinninella Emanuele, Raoul Pauline, Zanza Christian, Mele Maria Cristina, and Franceschi Francesco. 2021. “Gut Microbiota and Environment in Coronary Artery Disease.” International Journal of Environmental Research and Public Health 18: 4242. 10.3390/ijerph18084242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sender, Ron , Fuchs Shai, and Milo Ron. 2016. “Revised Estimates for the Number of Human and Bacteria Cells in the Body.” PLoS Biol 14: e1002533. 10.1371/journal.pbio.1002533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fan, Yong , and Pedersen Oluf. 2021. “Gut Microbiota in Human Metabolic Health and Disease.” Nature Reviews Microbiology 19: 55–71. 10.1038/s41579-020-0433-9 [DOI] [PubMed] [Google Scholar]
- 7. Almeida, Alexandre , Nayfach Stephen, Boland Miguel, Strozzi Francesco, Beracochea Martin, Shi Zhou Jason, Pollard Katherine S., et al. 2021. “A Unified Catalog of 204,938 Reference Genomes from the Human Gut Microbiome.” Nature Biotechnology 39: 105–14. 10.1038/s41587-020-0603-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pedersen, Helle K. , Gudmundsdottir Valborg, Nielsen Henrik B., Hyotylainen Tuulia, Nielsen Trine, Jensen Benjamin A. H., Forslund Kristoffer, et al. 2016. “Human Gut Microbes Impact Host Serum Metabolome and Insulin Sensitivity.” Nature 535: 376–81. 10.1038/nature18646 [DOI] [PubMed] [Google Scholar]
- 9. Jie, Zhuye , Xia Huihua, Zhong Shi‐Long, Feng Qiang, Li Shenghui, Liang Suisha, Zhong Huanzi, et al. 2017. “The Gut Microbiome in Atherosclerotic Cardiovascular Disease.” Nature Communications 8: 845. 10.1038/s41467-017-00900-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fromentin, Sebastien , Forslund Sofia K., Chechi Kanta, Aron‐Wisnewsky Judith, Chakaroun Rima, Nielsen Trine, Tremaroli Valentina, et al. 2022. “Microbiome and Metabolome Features of the Cardiometabolic Disease Spectrum.” Nature Medicine 28: 303–14. 10.1038/s41591-022-01688-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schmidt, Thomas S. B. , Li Simone S., Maistrenko Oleksandr M., Akanni Wasiu, Coelho Luis Pedro, Dolai Sibasish, Fullam Anthony, et al. 2022. “Drivers and Determinants of Strain Dynamics Following Fecal Microbiota Transplantation.” Nature Medicine 28: 1902–12. 10.1038/s41591-022-01913-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cui, Li , Zhao Tingting, Hu Haibing, Zhang Wen, and Hua Xiuguo. 2017. “Association Study of Gut Flora in Coronary Heart Disease Through High‐Throughput Sequencing.” BioMed Research International 2017: 3796359. 10.1155/2017/3796359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liu, Honghong , Chen Xi, Hu Xiaomin, Niu Haitao, Tian Ran, Wang Hui, Pang Haiyu, et al. 2019. “Alterations in the Gut Microbiome and Metabolism with Coronary Artery Disease Severity.” Microbiome 7: 68. 10.1186/s40168-019-0683-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cybulsky, M. I. , Chan M. K., and Movat H. Z.. 1988. “Acute Inflammation and Microthrombosis Induced by Endotoxin, Interleukin‐1, and Tumor Necrosis Factor and Their Implication in Gram‐Negative Infection.” Laboratory Investigation 58: 365–78. [PubMed] [Google Scholar]
- 15. Velusamy, Ragani , and Muhi Stephen. 2020. “Melioidosis and the Heart: A Systematic Review.” Tropical Medicine and Infectious Disease 5: 121. 10.3390/tropicalmed5030121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Talmor‐Barkan, Yeela , Bar Noam, Shaul Aviv A., Shahaf Nir, Godneva Anastasia, Bussi Yuval, Lotan‐Pompan Maya, et al. 2022. “Metabolomic and Microbiome Profiling Reveals Personalized Risk Factors for Coronary Artery Disease.” Nature Medicine 28: 295–302. 10.1038/s41591-022-01686-6 [DOI] [PubMed] [Google Scholar]
- 17. Han, Ying , Gong Zhaowei, Sun Guizhi, Xu Jing, Qi Changlu, Sun Weiju, Jiang Huijie, Cao Peigang, and Ju Hong. 2021. “Dysbiosis Of Gut Microbiota In Patients With Acute Myocardial Infarction.” Frontiers in Microbiology 12: 680101. 10.3389/fmicb.2021.680101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Romano, Kymberleigh A. , Vivas Eugenio I., Amador‐Noguez Daniel, and Rey Federico E.. 2015. “Intestinal Microbiota Composition Modulates Choline Bioavailability From Diet and Accumulation Of the Proatherogenic Metabolite Trimethylamine‐N‐Oxide.” mBio 6: e02481. 10.1128/mBio.02481-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Reichardt, Nicole , Duncan Sylvia H., Young Pauline, Belenguer Alvaro, McWilliam Leitch Carol, Scott Karen P., Flint Harry J., and Louis Petra. 2014. “Phylogenetic Distribution Of Three Pathways for Propionate Production Within the Human Gut Microbiota.” The ISME Journal 8: 1323–35. 10.1038/ismej.2014.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ridlon, Jason M. , Kang Dae‐Joong, and Hylemon Phillip B.. 2006. “Bile Salt Biotransformations by Human Intestinal Bacteria.” Journal of Lipid Research 47: 241–59. 10.1194/jlr.R500013-JLR200 [DOI] [PubMed] [Google Scholar]
- 21. d'Hennezel, Eva , Abubucker Sahar, Murphy Leon O., and Cullen Thomas W.. 2017. “Total Lipopolysaccharide From the Human Gut Microbiome Silences Toll‐Like Receptor Signaling.” mSystems 2. 10.1128/mSystems.00046-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhu, Yijun , Dwidar Mohammed, Nemet Ina, Buffa Jennifer A., Sangwan Naseer, Li Xinmin S., Anderson James T., et al. 2023. “Two Distinct Gut Microbial Pathways Contribute to Meta‐Organismal Production of Phenylacetylglutamine with Links to Cardiovascular Disease.” Cell Host & Microbe 31: 18–32.e9. 10.1016/j.chom.2022.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bermudez‐Martin, Patricia , Becker Jérôme A. J., Caramello Nicolas, Fernandez Sebastian P., Costa‐Campos Renan, Canaguier Juliette, Barbosa Susana, et al. 2021. “The Microbial Metabolite P‐Cresol Induces Autistic‐Like Behaviors in Mice By Remodeling the Gut Microbiota.” Microbiome 9: 157. 10.1186/s40168-021-01103-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wallrabenstein, Ivonne , Kuklan Jonas, Weber Lea, Zborala Sandra, Werner Markus, Altmüller Janine, Becker Christian, et al. 2013. “Human Trace Amine‐Associated Receptor TAAR5 Can be Activated by Trimethylamine.” PLoS One 8: e54950. 10.1371/journal.pone.0054950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chen, Sifan , Henderson Ayana, Petriello Michael C., Romano Kymberleigh A., Gearing Mary, Miao Ji, Schell Mareike, et al. 2019. “Trimethylamine N‐Oxide Binds and Activates PERK to Promote Metabolic Dysfunction.” Cell Metabolism 30: 1141–1151.e1145. 10.1016/j.cmet.2019.08.021 [DOI] [PubMed] [Google Scholar]
- 26. Wang, BingYu , Qiu Jun, Lian JiangFang, Yang Xi, and Zhou JianQing. 2021. “Gut Metabolite Trimethylamine‐N‐Oxide in Atherosclerosis: From Mechanism to Therapy.” Frontiers in Cardiovascular Medicine 8: 723886. 10.3389/fcvm.2021.723886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhu, Weifei , Gregory Jill C., Org Elin, Buffa Jennifer A., Gupta Nilaksh, Wang Zeneng, Li Lin, et al. 2016. “Gut Microbial Metabolite TMAO Enhances Platelet Hyperreactivity and Thrombosis Risk.” Cell 165: 111–24. 10.1016/j.cell.2016.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yang, Shengjie , Li Xinye, Yang Fan, Zhao Ran, Pan Xiandu, Liang Jiaqi, Tian Li, et al. 2019. “Gut Microbiota‐Dependent Marker TMAO in Promoting Cardiovascular Disease: Inflammation Mechanism, Clinical Prognostic, and Potential as a Therapeutic Target.” Frontiers in Pharmacology 10: 1360. 10.3389/fphar.2019.01360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zou, Deling , Li Yanyu, and Sun Guangping. 2021. “Attenuation of Circulating Trimethylamine N‐Oxide Prevents the Progression of Cardiac and Renal Dysfunction in a Rat Model of Chronic Cardiorenal Syndrome.” Frontiers in Pharmacology 12: 751380. 10.3389/fphar.2021.751380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kim, Myung H. , Kang Seung G., Park Jeong H., Yanagisawa Masashi, and Kim Chang H.. 2013. “Short‐Chain Fatty Acids Activate GPR41 and GPR43 on Intestinal Epithelial Cells to Promote Inflammatory Responses in Mice.” Gastroenterology 145: 396–406.e391‐310. 10.1053/j.gastro.2013.04.056 [DOI] [PubMed] [Google Scholar]
- 31. Pluznick, Jennifer L. , Protzko Ryan J., Gevorgyan Haykanush, Peterlin Zita, Sipos Arnold, Han Jinah, Brunet Isabelle, et al. 2013. “Olfactory Receptor Responding to Gut Microbiota‐Derived Signals Plays a Role in Renin Secretion and Blood Pressure Regulation.” Proceedings of the National Academy of Sciences of the United States of America 110: 4410–15. 10.1073/pnas.1215927110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Vinolo, Marco A. R. , Rodrigues Hosana G., Nachbar Renato T., and Curi Rui. 2011. “Regulation of Inflammation by Short Chain Fatty Acids.” Nutrients 3: 858–76. 10.3390/nu3100858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bartolomaeus, Hendrik , Balogh András, Yakoub Mina, Homann Susanne, Markó Lajos, Höges Sascha, Tsvetkov Dmitry, et al. 2019. “Short‐Chain Fatty Acid Propionate Protects From Hypertensive Cardiovascular Damage.” Circulation 139: 1407–21. 10.1161/circulationaha.118.036652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pluznick, Jennifer L . 2017. “Microbial Short‐Chain Fatty Acids and Blood Pressure Regulation.” Current Hypertension Reports 19: 25. 10.1007/s11906-017-0722-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kawamata, Yuji , Fujii Ryo, Hosoya Masaki, Harada Masataka, Yoshida Hiromi, Miwa Masanori, Fukusumi Shoji, et al. 2003. “A G Protein‐Coupled Receptor Responsive to Bile Acids.” Journal of Biological Chemistry 278: 9435–40. 10.1074/jbc.M209706200 [DOI] [PubMed] [Google Scholar]
- 36. Wang, Haibo , Chen Jasmine, Hollister Kevin, Sowers Lawrence C., and Forman Barry M.. 1999. “Endogenous Bile Acids are Ligands for the Nuclear Receptor FXR/BAR.” Molecular Cell 3: 543–53. 10.1016/s1097-2765(00)80348-2 [DOI] [PubMed] [Google Scholar]
- 37. Uppal, Hirdesh , Saini Simrat P. S., Moschetta Antonio, Mu Ying, Zhou Jie, Gong Haibiao, Zhai Yonggong, et al. 2007. “Activation of LXRs Prevents Bile Acid Toxicity and Cholestasis in Female Mice.” Hepatology 45: 422–32. 10.1002/hep.21494 [DOI] [PubMed] [Google Scholar]
- 38. Qi, Yunpeng , Jiang Changtao, Cheng Jie, Krausz Kristopher W., Li Tiangang, Ferrell Jessica M., Gonzalez Frank J., and Chiang John Y. L.. 2015. “Bile Acid Signaling in Lipid Metabolism: Metabolomic and Lipidomic Analysis of Lipid and Bile Acid Markers Linked to Anti‐Obesity and Anti‐Diabetes in Mice.” Biochimica et Biophysica Acta (BBA) ‐ Molecular and Cell Biology of Lipids 1851: 19–29. 10.1016/j.bbalip.2014.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ahmad, Tiara R. , and Haeusler Rebecca A.. 2019. “Bile Acids in Glucose Metabolism and Insulin Signalling ‐ Mechanisms and Research Needs.” Nature Reviews Endocrinology 15: 701–12. 10.1038/s41574-019-0266-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chen, Mei L. , Takeda Kiyoshi, and Sundrud Mark S.. 2019. “Emerging Roles of Bile Acids in Mucosal Immunity and Inflammation.” Mucosal Immunology 12: 851–61. 10.1038/s41385-019-0162-4 [DOI] [PubMed] [Google Scholar]
- 41. Takeuchi, Osamu , Hoshino Katsuaki, Kawai Taro, Sanjo Hideki, Takada Haruhiko, Ogawa Tomohiko, Takeda Kiyoshi, and Akira Shizuo. 1999. “Differential Roles of TLR2 and TLR4 in Recognition of Gram‐Negative and Gram‐Positive Bacterial Cell Wall Components.” Immunity 11: 443–51. 10.1016/s1074-7613(00)80119-3 [DOI] [PubMed] [Google Scholar]
- 42. Burgueño, Joan F. , Barba Albert, Eyre Elena, Romero Carolina, Neunlist Michel, and Fernández Ester. 2016. “TLR2 and TLR9 Modulate Enteric Nervous System Inflammatory Responses to Lipopolysaccharide.” Journal of Neuroinflammation 13: 187. 10.1186/s12974-016-0653-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nemet, Ina , Saha Prasenjit P., Gupta Nilaksh, Zhu Weifei, Romano Kymberleigh A., Skye Sarah M., Cajka Tomas, et al. 2020. “A Cardiovascular Disease‐Linked Gut Microbial Metabolite Acts via Adrenergic Receptors.” Cell 180: 862–877.e822. 10.1016/j.cell.2020.02.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Berg, Anders H. , Kumar Sanjeev, and Karumanchi S. Ananth. 2022. “Indoxyl Sulfate in Uremia: An Old Idea With Updated Concepts.” Journal of Clinical Investigation 132: e155860. 10.1172/jci155860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Campillo, Sofía , Bohorquez Lourdes, Gutiérrez‐Calabrés Elena, García‐Ayuso Diego, Miguel Verónica, Griera Mercedes, Calle Yolanda, et al. 2022. “Indoxyl Sulfate‐ and P‐Cresol‐Induced Monocyte Adhesion and Migration Is Mediated By Integrin‐Linked Kinase‐Dependent Podosome Formation.” Experimental and Molecular Medicine 54: 226–38. 10.1038/s12276-022-00738-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Liu, Wen‐Chih , Tomino Yasuhiko, and Lu Kuo‐Cheng. 2018. “Impacts Of Indoxyl Sulfate and P‐Cresol Sulfate on Chronic Kidney Disease and Mitigating Effects of AST‐120.” Toxins 10: 367. 10.3390/toxins10090367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Poveda, Jonay , Sanchez‐Niño Maria D., Glorieux Griet, Sanz Ana B., Egido Jesús, Vanholder Raymond, and Ortiz Alberto. 2014. “P‐Cresyl Sulphate Has Pro‐Inflammatory and Cytotoxic Actions on Human Proximal Tubular Epithelial Cells.” Nephrology Dialysis Transplantation 29: 56–64. 10.1093/ndt/gft367 [DOI] [PubMed] [Google Scholar]
- 48. Tang, W. H. Wilson , Wang Zeneng, Levison Bruce S., Koeth Robert A., Britt Earl B., Fu Xiaoming, Wu Yuping, and Hazen Stanley L.. 2013. “Intestinal Microbial Metabolism of Phosphatidylcholine and Cardiovascular Risk.” The New England Journal of Medicine 368: 1575–84. 10.1056/NEJMoa1109400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Koeth, Robert A. , Wang Zeneng, Levison Bruce S., Buffa Jennifer A., Org Elin, Sheehy Brendan T., Britt Earl B., et al. 2013. “Intestinal Microbiota Metabolism of L‐Carnitine, a Nutrient In Red Meat, Promotes Atherosclerosis.” Nature Medicine 19: 576–85. 10.1038/nm.3145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wang, Zeneng , Klipfell Elizabeth, Bennett Brian J., Koeth Robert, Levison Bruce S., Dugar Brandon, Feldstein Ariel E., et al. 2011. “Gut Flora Metabolism of Phosphatidylcholine Promotes Cardiovascular Disease.” Nature 472: 57–63. 10.1038/nature09922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Falony, Gwen , Vieira‐Silva Sara, and Raes Jeroen. 2015. “Microbiology Meets Big Data: The Case of Gut Microbiota‐Derived Trimethylamine.” Annual Review of Microbiology 69: 305–21. 10.1146/annurev-micro-091014-104422 [DOI] [PubMed] [Google Scholar]
- 52. Tang, W. H. Wilson , and Hazen Stanley L.. 2014. “The Contributory Role of Gut Microbiota in Cardiovascular Disease.” Journal of Clinical Investigation 124: 4204–11. 10.1172/jci72331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tang, W. H. Wilson , Wang Zeneng, Kennedy David J., Wu Yuping, Buffa Jennifer A., Agatisa‐Boyle Brendan, Li Xinmin S., Levison Bruce S., and Hazen Stanley L.. 2015. “Gut Microbiota‐Dependent Trimethylamine N‐Oxide (TMAO) Pathway Contributes to Both Development of Renal Insufficiency and Mortality Risk in Chronic Kidney Disease.” Circulation Research 116: 448–55. 10.1161/circresaha.116.305360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zeisel, Steven H. , and Warrier Manya. 2017. “Trimethylamine N‐Oxide, the Microbiome, and Heart and Kidney Disease.” Annual Review of Nutrition 37: 157–81. 10.1146/annurev-nutr-071816-064732 [DOI] [PubMed] [Google Scholar]
- 55. Phillips, Ian R. , Dolphin Colin T., Clair Philippe, Hadley Mark R., Hutt Andrew J., McCombie Richard R., Smith Robert L., and Shephard Elizabeth A.. 1995. “The Molecular Biology of the Flavin‐Containing Monooxygenases of Man.” Chemico‐Biological Interactions 96: 17–32. 10.1016/0009-2797(94)03580-2 [DOI] [PubMed] [Google Scholar]
- 56. Lu, Yingchang , Feskens Edith J. M., Boer Jolanda M. A., and Müller Michael. 2010. “The Potential Influence of Genetic Variants in Genes Along Bile Acid and Bile Metabolic Pathway On Blood Cholesterol Levels in the Population.” Atherosclerosis 210: 14–27. 10.1016/j.atherosclerosis.2009.10.035 [DOI] [PubMed] [Google Scholar]
- 57. Allayee, Hooman , and Hazen Stanley L.. 2015. “Contribution of Gut Bacteria to Lipid Levels: Another Metabolic Role for Microbes? Circulation Research 117: 750–54. 10.1161/circresaha.115.307409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Geng, Jin , Yang Chenchen, Wang Bingjian, Zhang Xiwen, Hu Tingting, Gu Yang, and Li Ju. 2018. “Trimethylamine N‐Oxide Promotes Atherosclerosis Via CD36‐dependent MAPK/JNK Pathway.” Biomedicine & Pharmacotherapy 97: 941–47. 10.1016/j.biopha.2017.11.016 [DOI] [PubMed] [Google Scholar]
- 59. Al‐Obaide, Mohammed , Singh Ruchi, Datta Palika, Rewers‐Felkins Kathy, Salguero Maria, Al‐Obaidi Ibtisam, Kottapalli Kameswara, and Vasylyeva Tetyana. 2017. “Gut Microbiota‐Dependent Trimethylamine‐N‐oxide and Serum Biomarkers in Patients with T2DM and Advanced CKD.” Journal of Clinical Medicine 6: 86. 10.3390/jcm6090086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Gil‐Cruz, Cristina , Perez‐Shibayama Christian, De Martin Angelina, Ronchi Francesca, van der Borght Katrien, Niederer Rebekka, Onder Lucas, et al. 2019. “Microbiota‐Derived Peptide Mimics Drive Lethal Inflammatory Cardiomyopathy.” Science 366: 881–86. 10.1126/science.aav3487 [DOI] [PubMed] [Google Scholar]
- 61. Garber, Ken . 2015. “Drugging the Gut Microbiome.” Nature Biotechnology 33: 228–31. 10.1038/nbt.3161 [DOI] [PubMed] [Google Scholar]
- 62. Liu, Tianxing , Niu Haitao, and Zhang Shuyang. 2015. “Intestinal Microbiota Metabolism and Atherosclerosis.” Chinese Medical Journal 128: 2805–11. 10.4103/0366-6999.167362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wang, Zeneng , Roberts Adam B., Buffa Jennifer A., Levison Bruce S., Zhu Weifei, Org Elin, Gu Xiaodong, et al. 2015. “Non‐Lethal Inhibition of Gut Microbial Trimethylamine Production for the Treatment of Atherosclerosis.” Cell 163: 1585–95. 10.1016/j.cell.2015.11.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Craciun, Smaranda , and Balskus Emily P.. 2012. “Microbial Conversion of Choline to Trimethylamine Requires a Glycyl Radical Enzyme.” Proceedings of the National Academy of Sciences of the United States of America 109: 21307–12. 10.1073/pnas.1215689109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zhu, Yijun , Jameson Eleanor, Crosatti Marialuisa, Schäfer Hendrik, Rajakumar Kumar, Bugg Timothy D. H., and Chen Yin. 2014. “Carnitine Metabolism To Trimethylamine By an Unusual Rieske‐Type Oxygenase From Human Microbiota.” Proceedings of the National Academy of Sciences of the United States of America 111: 4268–73. 10.1073/pnas.1316569111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Koeth, Robert A. , Levison Bruce S., Culley Miranda K., Buffa Jennifer A., Wang Zeneng, Gregory Jill C., Org Elin, et al. 2014. “γ‐Butyrobetaine is a Proatherogenic Intermediate in Gut Microbial Metabolism of L‐Carnitine to TMAO.” Cell Metabolism 20: 799–812. 10.1016/j.cmet.2014.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Buffa, Jennifer A. , Romano Kymberleigh A., Copeland Matthew F., Cody David B., Zhu Weifei, Galvez Rachel, Fu Xiaoming, et al. 2022. “The Microbial Gbu Gene Cluster Links Cardiovascular Disease Risk Associated With Red Meat Consumption to Microbiota L‐Carnitine Catabolism.” Nature Microbiology 7: 73–86. 10.1038/s41564-021-01010-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Li, Xinmin S. , Wang Zeneng, Cajka Tomas, Buffa Jennifer A., Nemet Ina, Hurd Alex G., Gu Xiaodong, et al. 2018. “Untargeted Metabolomics Identifies Trimethyllysine, a TMAO‐producing Nutrient Precursor, As a Predictor of Incident Cardiovascular Disease Risk.” JCI Insight 3. 10.1172/jci.insight.99096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Zhao, Mingming , Wei Haoran, Li Chenze, Zhan Rui, Liu Changjie, Gao Jianing, Yi Yaodong, et al. 2022. “Gut Microbiota Production of trimethyl‐5‐aminovaleric Acid Reduces Fatty Acid Oxidation and Accelerates Cardiac Hypertrophy.” Nature Communications 13: 1757. 10.1038/s41467-022-29060-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Canfora, Emanuel E. , Jocken Johan W., and Blaak Ellen E.. 2015. “Short‐Chain Fatty Acids in Control of Body Weight and Insulin Sensitivity.” Nature Reviews Endocrinology 11: 577–91. 10.1038/nrendo.2015.128 [DOI] [PubMed] [Google Scholar]
- 71. Macfarlane, George T. , and Macfarlane Sandra. 2011. “Fermentation in the Human Large Intestine: Its Physiologic Consequences and the Potential Contribution of Prebiotics.” Journal of Clinical Gastroenterology 45(Suppl): S120–127. 10.1097/MCG.0b013e31822fecfe [DOI] [PubMed] [Google Scholar]
- 72. Macfarlane, Sandra , and Macfarlane George T.. 2003. “Regulation of Short‐Chain Fatty Acid Production.” Proceedings of the Nutrition Society 62: 67–72. 10.1079/pns2002207 [DOI] [PubMed] [Google Scholar]
- 73. Lin, Hua V. , Frassetto Andrea, Kowalik Edward J. Jr., Nawrocki Andrea R., Lu Mofei M., Kosinski Jennifer R., Hubert James A., et al. 2012. “Butyrate and Propionate Protect Against Diet‐Induced Obesity and Regulate Gut Hormones Via Free Fatty Acid Receptor 3‐independent Mechanisms.” PLoS One 7: e35240. 10.1371/journal.pone.0035240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Musso, Giovanni , Gambino Roberto, and Cassader Maurizio. 2011. “Interactions Between Gut Microbiota and Host Metabolism Predisposing to Obesity and Diabetes.” Annual Review of Medicine 62: 361–80. 10.1146/annurev-med-012510-175505 [DOI] [PubMed] [Google Scholar]
- 75. De Preter, Vicky , Coopmans Tamara, Rutgeerts Paul, and Verbeke Kristin. 2006. “Influence of Long‐Term Administration of Lactulose and Saccharomyces boulardii on the Colonic Generation of Phenolic Compounds in Healthy Human Subjects.” Journal of the American College of Nutrition 25: 541–49. 10.1080/07315724.2006.10719570 [DOI] [PubMed] [Google Scholar]
- 76. Pluznick, Jennifer . 2014. “A Novel SCFA Receptor, the Microbiota, and Blood Pressure Regulation.” Gut Microbes 5: 202–7. 10.4161/gmic.27492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Ichimura, Atsuhiko , Hasegawa Sae, Kasubuchi Mayu, and Kimura Ikuo. 2014. “Free Fatty Acid Receptors as Therapeutic Targets for the Treatment of Diabetes.” Frontiers in Pharmacology 5: 236. 10.3389/fphar.2014.00236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Karlsson, Fredrik H. , Fåk Frida, Nookaew Intawat, Tremaroli Valentina, Fagerberg Björn, Petranovic Dina, Bäckhed Fredrik, and Nielsen Jens. 2012. “Symptomatic Atherosclerosis is Associated with an Altered Gut Metagenome.” Nature Communication 3: 1245. 10.1038/ncomms2266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Yang, Tao , Santisteban Monica M., Rodriguez Vermali, Li Eric, Ahmari Niousha, Carvajal Jessica M., Zadeh Mojgan, et al. 2015. “Gut Dysbiosis is Linked to Hypertension.” Hypertension 65: 1331–40. 10.1161/hypertensionaha.115.05315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Jones, Brian V. , Begley Máire, Hill Colin, Gahan Cormac G. M., and Marchesi Julian R.. 2008. “Functional and Comparative Metagenomic Analysis of Bile Salt Hydrolase Activity in the Human Gut Microbiome.” Proceedings of the Natliona Academy of Sciences of the United States of America 105: 13580–85. 10.1073/pnas.0804437105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Wahlström, Annika , Sayin Sama I., Marschall Hanns‐Ulrich, and Bäckhed Fredrik. 2016. “Intestinal Crosstalk Between Bile Acids and Microbiota and its Impact on Host Metabolism.” Cell Metabolism 24: 41–50. 10.1016/j.cmet.2016.05.005 [DOI] [PubMed] [Google Scholar]
- 82. Ferrell, Jessica M. , Boehme Shannon, Li Feng, and Chiang John Y. L.. 2016. “Cholesterol 7α‐hydroxylase‐deficient Mice are Protected from High‐Fat/High‐Cholesterol Diet‐Induced Metabolic Disorders.” Journal of Lipid Research 57: 1144–54. 10.1194/jlr.M064709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Kumar, Purnima S. , Mason Matthew R., Brooker Michael R., and O'Brien Kelly. 2012. “Pyrosequencing Reveals Unique Microbial Signatures Associated with Healthy and Failing Dental Implants.” Journal of Clinical Periodontology 39: 425–33. 10.1111/j.1600-051X.2012.01856.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Chiang, John Y. L . 2009. “Bile Acids: Regulation of Synthesis.” Journal of Lipid Research 50: 1955–66. 10.1194/jlr.R900010-JLR200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Hodge, R. J. , and Nunez D. J.. 2016. “Therapeutic Potential of Takeda‐G‐protein‐receptor‐5 (TGR5) Agonists. Hope or Hype? Diabetes, Obesity and Metabolism 18: 439–43. 10.1111/dom.12636 [DOI] [PubMed] [Google Scholar]
- 86. Lefebvre, Philippe , Cariou Bertrand, Lien Fleur, Kuipers Folkert, and Staels Bart. 2009. “Role of Bile Acids and Bile Acid Receptors in Metabolic Regulation.” Physiological Reviews 89: 147–91. 10.1152/physrev.00010.2008 [DOI] [PubMed] [Google Scholar]
- 87. Wang, Yan‐Dong , Chen Wei‐Dong, Yu Donna, Forman Barry M., and Huang Wendong. 2011. “The G‐Protein‐coupled Bile Acid Receptor, Gpbar1 (TGR5), Negatively Regulates Hepatic Inflammatory Response Through Antagonizing Nuclear Factor Kappa Light‐Chain Enhancer Of Activated B Cells (NF‐κB) in Mice.” Hepatology 54: 1421–32. 10.1002/hep.24525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Mayerhofer, Cristiane C. K. , Ueland Thor, Broch Kaspar, Vincent Royce P., Cross Gemma F., Dahl Christen P., Aukrust Pål, et al. 2017. “Increased Secondary/Primary Bile Acid Ratio in Chronic Heart Failure.” Journal of Cardiac Failure 23: 666–71. 10.1016/j.cardfail.2017.06.007 [DOI] [PubMed] [Google Scholar]
- 89. Wang, Daoming , Doestzada Marwah, Chen Lianmin, Andreu‐Sánchez Sergio, van den Munckhof Inge C. L., Augustijn Hannah E., Koehorst Martijn, et al. 2021. “Characterization Of Gut Microbial Structural Variations as Determinants of Human Bile Acid Metabolism.” Cell Host & Microbe 29: 1802–1814.e1805. 10.1016/j.chom.2021.11.003 [DOI] [PubMed] [Google Scholar]
- 90. Cani, Patrice D. , Amar Jacques, Iglesias Miguel A., Poggi Marjorie, Knauf Claude, Bastelica Delphine, Neyrinck Audrey M., et al. 2007. “Metabolic Endotoxemia Initiates Obesity and Insulin Resistance.” Diabetes 56: 1761–72. 10.2337/db06-1491 [DOI] [PubMed] [Google Scholar]
- 91. Brandsma, Eelke , Kloosterhuis Niels J., Koster Mirjam, Dekker Daphne C., Gijbels Marion J. J., van der Velden Saskia, Ríos‐Morales Melany, et al. 2019. “A Proinflammatory Gut Microbiota Increases Systemic Inflammation and Accelerates Atherosclerosis.” Circular Research 124: 94–100. 10.1161/circresaha.118.313234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Awoyemi, Ayodeji , Trøseid Marius, Arnesen Harald, Solheim Svein, and Seljeflot Ingebjørg. 2019. “Effects of Dietary Intervention and n‐3 PUFA Supplementation on Markers of Gut‐Related Inflammation and Their Association with Cardiovascular Events in a High‐Risk Population.” Atherosclerosis 286: 53–59. 10.1016/j.atherosclerosis.2019.05.004 [DOI] [PubMed] [Google Scholar]
- 93. Sandek, Anja , Bauditz Juergen, Swidsinski Alexander, Buhner Sabine, Weber‐Eibel Jutta, von Haehling Stephan, Schroedl Wieland, et al. 2007. “Altered Intestinal Function in Patients with Chronic Heart Failure.” Journal of the American College of Cardiology 50: 1561–69. 10.1016/j.jacc.2007.07.016 [DOI] [PubMed] [Google Scholar]
- 94. Brix, Susanne , Eriksen Carsten, Larsen Jeppe Madura, and Bisgaard Hans. 2015. “Metagenomic Heterogeneity Explains Dual Immune Effects of Endotoxins.” Journal of Allergy and Clinical Immunology 135: 277–80. 10.1016/j.jaci.2014.09.036 [DOI] [PubMed] [Google Scholar]
- 95. Vatanen, Tommi , Kostic Aleksandar D., d'Hennezel Eva, Siljander Heli, Franzosa Eric A., Yassour Moran, Kolde Raivo, et al. 2016. “Variation in Microbiome LPS Immunogenicity Contributes to Autoimmunity in Humans.” Cell 165: 842–53. 10.1016/j.cell.2016.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Mailer, Reiner K. W. , Gisterå Anton, Polyzos Konstantinos A., Ketelhuth Daniel F. J., and Hansson Göran K.. 2017. “Hypercholesterolemia Induces Differentiation of Regulatory T Cells in the Liver.” Circular Research 120: 1740–53. 10.1161/circresaha.116.310054 [DOI] [PubMed] [Google Scholar]
- 97. Frostegård, Johan . 2013. “Immunity, Atherosclerosis and Cardiovascular Disease.” BMC Medicine 11: 117. 10.1186/1741-7015-11-117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Kazemian, Negin , Mahmoudi Morteza, Halperin Frank, Wu Joseph C., and Pakpour Sepideh. 2020. “Gut Microbiota and Cardiovascular Disease: Opportunities and Challenges.” Microbiome 8: 36. 10.1186/s40168-020-00821-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Korporaal, Suzanne J. A. , Van Eck Miranda, Adelmeijer Jelle, Ijsseldijk Martin, Out Ruud, Lisman Ton, Lenting Peter J., Van Berkel Theo J. C., and Akkerman Jan‐Willem N.. 2007. “Platelet Activation by Oxidized Low Density Lipoprotein is Mediated by CD36 and Scavenger Receptor‐A.” Arteriosclerosis, Thrombosis, and Vascular Biology 27: 2476–83. 10.1161/atvbaha.107.150698 [DOI] [PubMed] [Google Scholar]
- 100. Mikelsaar, Marika , Stsepetova Jelena, Hütt Pirje, Kolk Helgi, Sepp Epp, Lõivukene Krista, Zilmer Kersti, and Zilmer Mihkel. 2010. “Intestinal Lactobacillus sp. is Associated with Some Cellular and Metabolic Characteristics of Blood in Elderly People.” Anaerobe 16: 240–46. 10.1016/j.anaerobe.2010.03.001 [DOI] [PubMed] [Google Scholar]
- 101. Claesson, Marcus J. , Jeffery Ian B., Conde Susana, Power Susan E., O'Connor Eibhlís M., Cusack Siobhán, Harris Hugh M. B., et al. 2012. “Gut Microbiota Composition Correlates With Diet and Health in the Elderly.” Nature 488: 178–84. 10.1038/nature11319 [DOI] [PubMed] [Google Scholar]
- 102. Martínez, Inés , Lattimer James M., Hubach Kelcie L., Case Jennifer A., Yang Junyi, Weber Casey G., Louk Julie A., et al. 2013. “Gut Microbiome Composition is Linked to Whole Grain‐Induced Immunological Improvements.” The ISMEJ Journal 7: 269–80. 10.1038/ismej.2012.104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Tlaskalová‐Hogenová, Helena , Štěpánková Renata, Kozáková Hana, Hudcovic Tomáš, Vannucci Luca, Tučková Ludmila, Rossmann Pavel, et al. 2011. “The Role of Gut Microbiota (Commensal Bacteria) and the Mucosal Barrier in the Pathogenesis of Inflammatory and Autoimmune Diseases and Cancer: Contribution of Germ‐Free and Gnotobiotic Animal Models of Human Diseases.” Cellular and Molecular Immunology 8: 110–20. 10.1038/cmi.2010.67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Littman, Dan R. , and Rudensky Alexander Y.. 2010. “Th17 and Regulatory T Cells in Mediating and Restraining Inflammation.” Cell 140: 845–58. 10.1016/j.cell.2010.02.021 [DOI] [PubMed] [Google Scholar]
- 105. van den Munckhof, I. C. L. , Kurilshikov A., Ter Horst R., Riksen N. P., Joosten L. A. B., Zhernakova A., Fu J., et al. 2018. “Role of Gut Microbiota in Chronic Low‐Grade Inflammation as Potential Driver for Atherosclerotic Cardiovascular Disease: A Systematic Review Of Human Studies: Impact of Gut Microbiota on Low‐Grade Inflammation.” Obesity Reviews 19: 1719–34. 10.1111/obr.12750 [DOI] [PubMed] [Google Scholar]
- 106. Furusawa, Yukihiro , Obata Yuuki, Fukuda Shinji, Endo Takaho A., Nakato Gaku, Takahashi Daisuke, Nakanishi Yumiko, et al. 2013. “Commensal Microbe‐Derived Butyrate Induces the Differentiation of Colonic Regulatory T Cells.” Nature 504: 446–50. 10.1038/nature12721 [DOI] [PubMed] [Google Scholar]
- 107. Maslowski, Kendle M. , Vieira Angelica T., Ng Aylwin, Kranich Jan, Sierro Frederic, Yu Di, Schilter Heidi C., et al. 2009. “Regulation of Inflammatory Responses by Gut Microbiota and Chemoattractant Receptor GPR43.” Nature 461: 1282–86. 10.1038/nature08530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Poesen, Ruben , Claes Kathleen, Evenepoel Pieter, de Loor Henriette, Augustijns Patrick, Kuypers Dirk, and Meijers Björn. 2016. “Microbiota‐Derived Phenylacetylglutamine Associates with Overall Mortality and Cardiovascular Disease in Patients With CKD.” Journal of the American Society of Nephrology 27: 3479–87. 10.1681/asn.2015121302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Bogiatzi, Chrysi , Gloor Gregory, Allen‐Vercoe Emma, Reid Gregor, Wong Ruth G., Urquhart Bradley L., Dinculescu Vincent, et al. 2018. “Metabolic Products of the Intestinal Microbiome and Extremes of Atherosclerosis.” Atherosclerosis 273: 91–97. 10.1016/j.atherosclerosis.2018.04.015 [DOI] [PubMed] [Google Scholar]
- 110. Liu, Yang , Liu Shaoyan, Zhao Zhizhuang, Song Xiang, Qu Haixian, and Liu Hongbin. 2021. “Phenylacetylglutamine is Associated with the Degree of Coronary Atherosclerotic Severity Assessed by Coronary Computed Tomographic Angiography in Patients with Suspected Coronary Artery Disease.” Atherosclerosis 333: 75–82. 10.1016/j.atherosclerosis.2021.08.029 [DOI] [PubMed] [Google Scholar]
- 111. Wang, Jialu , Gareri Clarice, and Rockman Howard A.. 2018. “G‐Protein‐Coupled Receptors in Heart Disease.” Circular Research 123: 716–35. 10.1161/circresaha.118.311403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Offermanns, Stefan . 2006. “Activation of Platelet Function Through G Protein‐Coupled Receptors.” Circulation Research 99: 1293–304. 10.1161/01.Res.0000251742.71301.16 [DOI] [PubMed] [Google Scholar]
- 113. Meijers, Björn K. I. , Claes Kathleen, Bammens Bert, de Loor Henriette, Viaene Liesbeth, Verbeke Kristin, Kuypers Dirk, Vanrenterghem Yves, and Evenepoel Pieter. 2010. “P‐Cresol and Cardiovascular Risk in Mild‐To‐Moderate Kidney Disease.” Clinical Journal of the American Society of Nephrology 5: 1182–89. 10.2215/cjn.07971109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Lin Cheng‐Jui, Wu Chih‐Jen, Pan Chi‐Feng, Chen Yi‐Chou, Sun Fang‐Ju, Chen Han‐Hsiang. 2010. “Serum Protein‐Bound Uraemic Toxins and Clinical Outcomes In Haemodialysis Patients.” Nephrology Dialysis Transplantion 25: 3693–700. 10.1093/ndt/gfq251 [DOI] [PubMed] [Google Scholar]
- 115. Barreto, Fellype C. , Barreto Daniela V., Liabeuf Sophie, Meert Natalie, Glorieux Griet, Temmar Mohammed, Choukroun Gabriel, Vanholder Raymond, and Massy Ziad A.. 2009. “Serum Indoxyl Sulfate is Associated with Vascular Disease and Mortality in Chronic Kidney Disease Patients.” Clinical Journal of the American Society of Nephrology 4: 1551–58. 10.2215/cjn.03980609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Rossi, Megan , Campbell Katrina L., Johnson David W., Stanton Tony, Vesey David A., Coombes Jeff S., Weston Kassia S., et al. 2014. “Protein‐Bound Uremic Toxins, Inflammation and Oxidative Stress: A Cross‐Sectional Study in Stage 3‐4 Chronic Kidney Disease.” Archives of Medical Research 45: 309–17. 10.1016/j.arcmed.2014.04.002 [DOI] [PubMed] [Google Scholar]
- 117. Wen, Chi P. , Cheng Ting Y. D., Tsai Min K., Chang Yen C., Chan Hui T., Tsai Shan P., Chiang Po H., et al. 2008. “All‐Cause Mortality Attributable to Chronic Kidney Disease: A Prospective Cohort Study Based on 462 293 Adults in Taiwan.” The Lancet 371: 2173–82. 10.1016/s0140-6736(08)60952-6 [DOI] [PubMed] [Google Scholar]
- 118. Opdebeeck, Britt , Maudsley Stuart, Azmi Abdelkrim, De Maré Annelies, De Leger Wout, Meijers Bjorn, Verhulst Anja, et al. 2019. “Indoxyl Sulfate and P‐Cresyl Sulfate Promote Vascular Calcification and Associate With Glucose Intolerance.” Journal of the American Society of Nephrology 30: 751–66. 10.1681/asn.2018060609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Shafi, Tariq , Sirich Tammy L., Meyer Timothy W., Hostetter Thomas H., Plummer Natalie S., Hwang Seungyoung, Melamed Michal L., et al. 2017. “Results of the HEMO Study Suggest that P‐Cresol Sulfate and Indoxyl Sulfate are not Associated with Cardiovascular Outcomes.” Kidney International 92: 1484–92. 10.1016/j.kint.2017.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Gallo, Antonella , Passaro Giovanna, Gasbarrini Antonio, Landolfi Raffaele, and Montalto Massimo. 2016. “Modulation of Microbiota as Treatment for Intestinal Inflammatory Disorders: An Uptodate.” World Journal of Gastroenterology 22: 7186–202. 10.3748/wjg.v22.i32.7186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Khan, Muhammad Y. , Dirweesh Ahmed, Khurshid Talal, and Siddiqui Waqas J.. 2018. “Comparing Fecal Microbiota Transplantation to Standard‐of‐Care Treatment for Recurrent Clostridium difficile Infection: A systematic reviewSystematic Review and meta‐analysisMeta‐Analysis.” The European Journal of Gastroenterology & Hepatology 30: 1309–17. 10.1097/meg.0000000000001243 [DOI] [PubMed] [Google Scholar]
- 122. Kootte, Ruud S. , Levin Evgeni, Salojärvi Jarkko, Smits Loek P., Hartstra Annick V., Udayappan Shanti D., Hermes Gerben, et al. 2017. “Improvement of Insulin Sensitivity After Lean Donor Feces in Metabolic Syndrome is Driven by Baseline Intestinal Microbiota Composition.” Cell Metabolism 26: 611–619.e6. 10.1016/j.cmet.2017.09.008 [DOI] [PubMed] [Google Scholar]
- 123. DeFilipp, Zachariah , Bloom Patricia P., Torres Soto Mariam, Mansour Michael K., Sater Mohamad R. A., Huntley Miriam H., Turbett Sarah, et al. 2019. ““Drug‐Resistant E. coli Bacteremia Transmitted by Fecal Microbiota Transplant.” The New England Journal of Medicine 381: 2043–50. 10.1056/NEJMoa1910437 [DOI] [PubMed] [Google Scholar]
- 124. Conraads, Viviane M. , Jorens Philippe G., De Clerck Luc S., Van Saene Hendrik K., Ieven Margaretha M., Bosmans Johan M., Schuerwegh Annemie, et al. 2004. “Selective Intestinal Decontamination in Advanced Chronic Heart Failure: A Pilot Trial.” European Journal of Heart Failure 6: 483–91. 10.1016/j.ejheart.2003.12.004 [DOI] [PubMed] [Google Scholar]
- 125. Galla, S. , Chakraborty S., Cheng X., Yeo J., Mell B., Zhang H., Mathew A. V., Vijay‐Kumar M., and Joe B.. 2018. “Disparate Effects of Antibiotics on Hypertension.” Physiological Genomics 50: 837–45. 10.1152/physiolgenomics.00073.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Rune, Ida , Rolin Bidda, Larsen Christian, Nielsen Dennis Sandris, Kanter Jenny E., Bornfeldt Karin E., Lykkesfeldt Jens, et al. 2016. “Modulating the Gut Microbiota Improves Glucose Tolerance, Lipoprotein Profile and Atherosclerotic Plaque Development in ApoE‐Deficient Mice.” PLoS One 11: e0146439. 10.1371/journal.pone.0146439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Tang, Tony W. H. , Chen Hung‐Chih, Chen Chen‐Yun, Yen Christopher Y. T., Lin Chen‐Ju, Prajnamitra Ray P., Chen Li‐Lun, et al. 2019. “Loss of Gut Microbiota Alters Immune System Composition and Cripples Postinfarction Cardiac Repair.” Circulation 139: 647–59. 10.1161/circulationaha.118.035235 [DOI] [PubMed] [Google Scholar]
- 128. Lam, Vy , Su Jidong, Hsu Anna, Gross Garrett J., Salzman Nita H., and Baker John E.. 2016. “Intestinal Microbial Metabolites are Linked to Severity of Myocardial Infarction in Rats.” PLoS One 11: e0160840. 10.1371/journal.pone.0160840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Lam, Vy , Su Jidong, Koprowski Stacy, Hsu Anna, Tweddell James S., Rafiee Parvaneh, Gross Garrett J., Salzman Nita H., and Baker John E.. 2012. “Intestinal Microbiota Determine Severity Of Myocardial Infarction in Rats.” Faseb j 26: 1727–35. 10.1096/fj.11-197921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Li, Zehua , Wu Zhiye, Yan Jianyun, Liu Hailin, Liu Qicai, Deng Yi, Ou Caiwen, and Chen Minsheng. 2019. “Gut Microbe‐Derived Metabolite Trimethylamine N‐Oxide Induces Cardiac Hypertrophy and Fibrosis.” Laboratory Investigation 99: 346–57. 10.1038/s41374-018-0091-y [DOI] [PubMed] [Google Scholar]
- 131. Winkel, Per , Hilden Jørgen, Hansen Jørgen Fischer, Kastrup Jens, Kolmos Hans Jørn, Kjøller Erik, Jensen Gorm Boje, et al. 2015. “Clarithromycin for Stable Coronary Heart Disease Increases All‐Cause and Cardiovascular Mortality and Cerebrovascular Morbidity Over 10 Years in the CLARICOR Randomised, Blinded Clinical Trial.” International Journal of Cardiology 182: 459–65. 10.1016/j.ijcard.2015.01.020 [DOI] [PubMed] [Google Scholar]
- 132. Hill, Colin , Guarner Francisco, Reid Gregor, Gibson Glenn R., Merenstein Daniel J., Pot Bruno, Morelli Lorenzo, et al. 2014. “The International Scientific Association for Probiotics and Prebiotics Consensus Statement on the Scope and Appropriate Use of the Term Probiotic.” Nature Reviews Gastroenterology & Hepatology 11: 506–14. 10.1038/nrgastro.2014.66 [DOI] [PubMed] [Google Scholar]
- 133. Patel, Rachna , and DuPont Herbert L.. 2015. “New Approaches for Bacteriotherapy: Prebiotics, New‐Generation Probiotics, and Synbiotics.” Clinical Infectious Diseases 60(Suppl 2): S108–121. 10.1093/cid/civ177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Markowiak, Paulina , and Śliżewska Katarzyna. 2017. “Effects of Probiotics, Prebiotics, and Synbiotics on Human Health.” Nutrients 9: 1021. 10.3390/nu9091021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Chambers, Edward S. , Preston Tom, Frost Gary, and Morrison Douglas J.. 2018. “Role of Gut Microbiota‐Generated Short‐Chain Fatty Acids in Metabolic and Cardiovascular Health.” Current Nutrition Reports 7: 198–206. 10.1007/s13668-018-0248-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Mohania, Dheeraj , Kansal Vinod K., Shah Dilip, Nagpal Ravinder, Kumar Manoj, Gautam Sanjeev K., Singh Birbal, and Behare Pradip V.. 2013. “Therapeutic Effect of Probiotic Dahi on Plasma, Aortic, and Hepatic Lipid Profile of Hypercholesterolemic Rats.” Journal of Cardiovascular Pharmacology and Therapeutics 18: 490–97. 10.1177/1074248413487431 [DOI] [PubMed] [Google Scholar]
- 137. Chambers, Edward S. , Byrne Claire S., Morrison Douglas J., Murphy Kevin G., Preston Tom, Tedford Catriona, Garcia‐Perez Isabel, et al 2019. “Dietary Supplementation with Inulin‐Propionate Ester or Inulin Improves Insulin Sensitivity in Adults with Overweight and Obesity with Distinct Effects on the Gut Microbiota, Plasma Metabolome and Systemic Inflammatory Responses: A Randomised Cross‐Over Trial.” Gut 68: 1430–38. 10.1136/gutjnl-2019-318424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Costanza, Annelise C. , Moscavitch Samuel D., Faria Neto Hugo C. C., and Mesquita Evandro T.. 2015. “Probiotic Therapy with Saccharomyces boulardii for Heart Failure Patients: A Randomized, Double‐Blind, Placebo‐Controlled Pilot Trial.” International Journal of Cardiology 179: 348–50. 10.1016/j.ijcard.2014.11.034 [DOI] [PubMed] [Google Scholar]
- 139. Vujkovic‐Cvijin, Ivan , Sklar Jack, Jiang Lingjing, Natarajan Loki, Knight Rob, and Belkaid Yasmine. 2020. “Host Variables Confound Gut Microbiota Studies of Human Disease.” Nature 587: 448–54. 10.1038/s41586-020-2881-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Miranda, Pedro M. , De Palma Giada, Serkis Viktoria, Lu Jun, Louis‐Auguste Marc P., McCarville Justin L., Verdu Elena F., Collins Stephen M., and Bercik Premysl. 2018. “High Salt Diet Exacerbates Colitis in Mice by Decreasing Lactobacillus Levels and Butyrate Production.” Microbiome 6: 57. 10.1186/s40168-018-0433-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Ariel, Bier , Braun Tzipi, Khasbab Rawan, Segni Ayelet Di, Grossman Ehud, Haberman Yael, and Leibowitz Avshalom. 2018. “A high salt diet modulates the gut microbiota and short chain fatty acids production in a salt-sensitive hypertension rat model.” Nutrients 10(9): 1154. 10.3390/nu10091154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Marques, Francine Z. , Nelson Erin, Chu Po‐Yin, Horlock Duncan, Fiedler April, Ziemann Mark, Tan Jian K., et al. 2017. “High‐Fiber Diet and Acetate Supplementation Change the Gut Microbiota and Prevent the Development of Hypertension and Heart Failure in Hypertensive Mice.” Circulation 135: 964–77. 10.1161/circulationaha.116.024545 [DOI] [PubMed] [Google Scholar]
- 143. Wang, Zeneng , Bergeron Nathalie, Levison Bruce S., Li Xinmin S., Chiu Sally, Jia Xun, Koeth Robert A., et al. 2019. “Impact of Chronic Dietary Red Meat, White Meat, or Non‐Meat Protein on Trimethylamine N‐Oxide Metabolism and Renal Excretion In Healthy Men and Women.” The European Heart Journal 40: 583–94. 10.1093/eurheartj/ehy799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Zhernakova, Alexandra , Kurilshikov Alexander, Bonder Marc Jan, Tigchelaar Ettje F., Schirmer Melanie, Vatanen Tommi, Mujagic Zlatan, et al. 2016. “Population‐Based Metagenomics Analysis Reveals Markers for Gut Microbiome Composition and Diversity.” Science 352: 565–69. 10.1126/science.aad3369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Wu, Hao , Esteve Eduardo, Tremaroli Valentina, Khan Muhammad Tanweer, Caesar Robert, Mannerås‐Holm Louise, Ståhlman Marcus, et al. 2017. “Metformin Alters the Gut Microbiome of Individuals with Treatment‐Naive Type 2 Diabetes, Contributing to the Therapeutic Effects of the Drug.” Nature Medicine 23: 850–58. 10.1038/nm.4345 [DOI] [PubMed] [Google Scholar]
- 146. Vieira‐Silva, Sara , Falony Gwen, Belda Eugeni, Nielsen Trine, Aron‐Wisnewsky Judith, Chakaroun Rima, Forslund Sofia K., et al. 2020. “Statin Therapy is Associated with Lower Prevalence of Gut Microbiota Dysbiosis.” Nature 581: 310–15. 10.1038/s41586-020-2269-x [DOI] [PubMed] [Google Scholar]
- 147. Auer, Johann , Berent Robert, and Eber Bernd. 2003. “Hepatitis A Virus Seropositivity and Coronary Artery Disease.” Circulation 108: e8. 10.1161/01.Cir.0000079069.35358.53 [DOI] [PubMed] [Google Scholar]
- 148. Tong, De‐Yan . 2005. “Hepatitis B Virus Infection and Coronary Atherosclerosis: Results from a Population with Relatively High Prevalence of Hepatitis B Virus.” World Journal of Gastroenterology 11: 1292–96. 10.3748/wjg.v11.i9.1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Wen, Dan , Du Xin, Dong Jian‐Zeng, and Ma Chang‐Sheng. 2019. “Hepatitis C Virus Infection and Risk of Coronary Artery Disease: A Meta‐Analysis.” European Journal of Internal Medicine 63: 69–73. 10.1016/j.ejim.2019.03.004 [DOI] [PubMed] [Google Scholar]
- 150. Hsue, Priscilla Y. , and Waters David D.. 2019. “HIV Infection and Coronary Heart Disease: Mechanisms and Management.” Nature Reviews Cardiology 16: 745–59. 10.1038/s41569-019-0219-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Ilango, Paavai , Mahendra Jaideep, Mahendra Little, Cherian Sanjay M., Kathaperumal Kumanan, Suresh Vasugi, Mahalingam Arulpari, and Abirami T. 2021. “Evidence Linking the Role of Periodontal Viruses in Coronary Artery Disease with and without Periodontitis.” Journal of Periodontology 92: 113–22. 10.1002/jper.19-0704 [DOI] [PubMed] [Google Scholar]
- 152. Shkoporov, Andrey N. , and Hill Colin. 2019. “Bacteriophages Of the Human Gut: The “Known Unknown” of the Microbiome.” Cell Host & Microbe 25: 195–209. 10.1016/j.chom.2019.01.017 [DOI] [PubMed] [Google Scholar]
- 153. Wylie, Kristine M. , Weinstock George M., and Storch Gregory A.. 2012. “Emerging View of the Human Virome.” Translational Research 160: 283–90. 10.1016/j.trsl.2012.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Manrique, Pilar , Bolduc Benjamin, Walk Seth T., van der Oost John, de Vos Willem M., and Young Mark J.. 2016. “Healthy Human Gut Phageome.” Proceedings of the National Academy of Sciences of the United States of America 113: 10400–10405. 10.1073/pnas.1601060113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Ansari, Mina H. , Ebrahimi Mehregan, Fattahi Mohammad R., Gardner Michael G., Safarpour Ali R., Faghihi Mohammad A., and Lankarani Kamran B.. 2020. “Viral Metagenomic Analysis of Fecal Samples Reveals an Enteric Virome Signature in Irritable Bowel Syndrome.” BMC Microbiology 20: 123. 10.1186/s12866-020-01817-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Guo, Lianghua , Hua Xiuguo, Zhang Wen, Yang Shixing, Shen Quan, Hu Haibing, Li Jingjiao, et al. 2017. “Viral Metagenomics Analysis of Feces From Coronary Heart Disease Patients Reveals the Genetic Diversity of the Microviridae.” Virologica Sinica 32: 130–38. 10.1007/s12250-016-3896-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Han, Maozhen , Yang Pengshuo, Zhong Chaofang, and Ning Kang. 2018. “The Human Gut Virome in Hypertension.” Frontiers in Microbiology 9: 3150. 10.3389/fmicb.2018.03150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Koskella, Britt , and Brockhurst Michael A.. 2014. “Bacteria‐Phage Coevolution as a Driver of Ecological and Evolutionary Processes in Microbial Communities.” FEMS Microbiology Reviews 38: 916–31. 10.1111/1574-6976.12072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Betts, Alex , Kaltz Oliver, and Hochberg Michael E.. 2014. “Contrasted Coevolutionary Dynamics Between a Bacterial Pathogen and Its Bacteriophages.” Proceedings of the National Academy of Sciences of the United States of America 111: 11109–14. 10.1073/pnas.1406763111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Paterson, Steve , Vogwill Tom, Buckling Angus, Benmayor Rebecca, Spiers Andrew J., Thomson Nicholas R., Quail Mike, et al. 2010. “Antagonistic Coevolution Accelerates Molecular Evolution.” Nature 464: 275–78. 10.1038/nature08798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Buckling, Angus , and Rainey Paul B.. 2002. “The Role of Parasites in Sympatric and Allopatric Host Diversification.” Nature 420: 496–99. 10.1038/nature01164 [DOI] [PubMed] [Google Scholar]
- 162. Duan, Yi , Llorente Cristina, Lang Sonja, Brandl Katharina, Chu Huikuan, Jiang Lu, White Richard C., et al. 2019. “Bacteriophage Targeting of Gut Bacterium Attenuates Alcoholic Liver Disease.” Nature 575: 505–11. 10.1038/s41586-019-1742-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Rasmussen, Torben S. , Mentzel Caroline M. J., Kot Witold, Castro‐Mejía Josué L., Zuffa Simone, Swann Jonathan R., Hansen Lars H., et al. 2020. “Faecal Virome Transplantation Decreases Symptoms of Type 2 Diabetes and Obesity in a Murine Model.” Gut 69: 2122–30. 10.1136/gutjnl-2019-320005 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information.
Data Availability Statement
This manuscript does not generate any code or data.


