Abstract
RNA turnover plays critical roles in the regulation of gene expression and allows cells to respond rapidly to environmental changes. In bacteria, the mechanisms of RNA turnover have been extensively studied in the models Escherichia coli and Bacillus subtilis, but not much is known in other bacteria. Cyanobacteria are a diverse group of photosynthetic organisms that have great potential for the sustainable production of valuable products using CO2 and solar energy. A better understanding of the regulation of RNA decay is important for both basic and applied studies of cyanobacteria. Genomic analysis shows that cyanobacteria have more than 10 ribonucleases and related proteins in common with E. coli and B. subtilis, and only a limited number of them have been experimentally investigated. In this review, we summarize the current knowledge about these RNA‐turnover‐related proteins in cyanobacteria. Although many of them are biochemically similar to their counterparts in E. coli and B. subtilis, they appear to have distinct cellular functions, suggesting a different mechanism of RNA turnover regulation in cyanobacteria. The identification of new players involved in the regulation of RNA turnover and the elucidation of their biological functions are among the future challenges in this field.
Keywords: cyanobacteria, RNA maturation, RNA metabolism, RNA turnover, ribonucleases
INTRODUCTION
“Life is short, and art is long”—Hippocrates
Gene expression is regulated at multiple levels, and the stability of RNA is a key regulatory step for gene expression. As intermediates of genetic information decoding process, the relative amounts of RNAs reflect the balance between transcription and degradation. Compared to proteins, the half‐lives of RNAs, in particular, those of messenger RNAs (mRNAs), are very short. In Escherichia coli, for example, the half‐life of mRNA averages 6.8 min, ranging from less than a minute to more than 10 min 1 , 2 , while a median half‐life of 2.4 min was determined for mRNAs from the cyanobacterium Prochlorococcus sp. MED4 3 . The rapid turnover of RNA constitutes an important mechanism of gene regulation and enables a cell to respond in a timely manner to changes in the environment. Beyond their role as carriers of genetic information, RNAs can be a structural component, such as the rRNAs in the ribosome; function as guide RNAs such as CRISPR RNAs (crRNAs); or have regulatory functions, such as riboswitches, trans‐acting small RNAs (sRNAs), or antisense RNAs. Therefore, studying the process of RNA decay is essential for our general understanding of gene regulation.
Bacterial RNA degradation has been extensively studied in the gram‐negative model E. coli, and to a lesser extent, in the gram‐positive model Bacillus subtilis. RNA decay requires a large number of proteins, which can be categorized into three groups: endoribonucleases, exoribonucleases, and auxiliary proteins 4 , 5 (Table 1). In E. coli, the endoribonucleases include RNase E, which cleaves internally in most RNA species; RNases Z and P, which generate the mature ends of transfer RNAs (tRNAs); RNase III, which cleaves at specific sites within double‐stranded RNAs (dsRNAs); and RNase H, which cuts the RNA strand in RNA–DNA duplexes. The exoribonucleases include PNPase, RNase R, and RNase II, which contribute to the degradation of bulk RNA; RNase D and PH, which mainly act on tRNA precursors; and Orn, which specifically degrades oligoribonucleotides. All these exoribonucleases degrade single‐stranded RNAs (ssRNAs) from 3ʹ to 5ʹ, with PNPase and RNase PH degrading RNA by phosphorolysis, and the others by hydrolysis. The major auxiliary proteins include the DEAD‐box RNA helicases (CsdA, RhlB, DbpA, RhlE, and SrmB), the RNA pyrophosphohydrolase RppH, and the sRNA‐binding proteins (Hfq and ProQ). These proteins do not cleave or degrade RNAs directly but can alter the rate of RNA turnover catalyzed by ribonucleases.
Table 1.
Genes encoding ribonucleases and related proteins for RNA processing or degradation in the γ‐proteobacterium Escherichia coli, the firmicute Bacillus subtilis, the unicellular cyanobacterium Synechocystis PCC 6803, and the filamentous cyanobacterium Anabaena PCC 7120.
| Genes from different bacteria | ||||||
|---|---|---|---|---|---|---|
| Protein | Activity and known functions | Escherichia coli | Bacillus subtilis | Synechocystis PCC 6803 | Anabaena PCC 7120 | |
| Endoribonucleases | ||||||
| RNase E/G | Cleaving ssRNA, participating in bulk RNA degradation | rne; rng | / | slr1129 (rne) | alr4331 (rne) | |
| RNase Y | Cleaving ssRNA, participating in bulk RNA degradation | / | rny | / | / | |
| RNase III | Cleaving dsRNAs, participating rRNA maturation and bulk RNA degradation | rnc | rnc | slr1646; slr0346 | alr0280; all4107 | |
| Mini‐III | Cleaving dsRNAs, mainly participating in 23S rRNA maturation | / | mrnC | slr0954 | alr1158 | |
| RNase HI | Cleaving the RNA strand in RNA–DNA hybrids | rnhA | / | slr0080 | alr0142 | |
| RNase HII | Cleaving the RNA strand in RNA–DNA hybrids | rnhB | rnhB | slr1130 | alr4332 | |
| RNase HIII | Cleaving the RNA strand in RNA–DNA hybrids | / | rnhC | / | / | |
| YbeY | Cleaving ssRNAs, mainly participating in 16S rRNA maturation | ybeY | ybeY | slr0053 | all0271 | |
| RNase P | Cleaving tRNA precursors to produce the mature 5ʹ ends | rnpA, rnpB | rnpA, rnpB | slr1469 (rnpA), rnpB | alr3413 (rnpA), rnpB | |
| RNase Z/BN | Cleaving tRNA precursors to produce the 3ʹ end for CAA addition | rbn | rnz | slr0050 | alr5152 | |
| RNase M5 | Cleaving specifically at a double‐strand region in 5S rRNA precursor to produce mature 3ʹ and 5ʹ ends | / | rnmV | / | / | |
| Cas6 | Cleaving specifically within the CRISPR repeats for the maturation of crRNAs | cas6 | cas6 | slr7014 (cas6‐1); slr7068 (cas6‐2a) | alr1482; alr1566 | |
| Exoribonucleases | ||||||
| PNPase | Degrading ssRNAs from 3ʹ by phosphorolysis, polyadenylating ssRNAs | pnp | pnp | sll1043 | all4396 | |
| RNase J | Hydrolyzing ssRNAs from 5ʹ end or cleaving it at internal sites | / | rnj1; rnj2 | slr0551 (rnj) | all3678 (rnj) | |
| RNase R/II | Hydrolyzing ssRNAs from 3ʹ end, participating in bulk RNA degradation | rnb; rnr | rnr | sll1290; sll1910 | all4450; alr1240 | |
| Orn | Oligoribonuclease, hydrolyzing short oligoribonucleotides into single ribonucleotides | b4162 | / | / | / | |
| nanoRNase | Oligoribonuclease, hydrolyzing short oligoribonucleotides into single ribonucleotides | / | nrnA; nrnB | / | / | |
| RNase PH | Degrading ssRNAs from 3ʹ end by phosphorolysis, mainly producing mature tRNA 3ʹ ends | rph | rph | / | alr0069 | |
| RNase D | Hydrolyzing ssRNAs from 3ʹ end, participating in tRNA maturation | rnd | / | sll0320 | all1697; all3791 | |
| RNase T | Hydrolyzing ssRNAs from 3ʹ end, mainly participating in the maturation of tRNAs, 5S RNA and 23S rRNA | rnt | / | / | / | |
| Auxiliary proteins | ||||||
| RNA helicase | Unwinding secondary structures of double‐strand RNA segments | rhlE; rhlB; deaD/csdA; dbpA; srmB | cshA; cshB; deaD/yxiN; and yfmL | slr0083 (crhR) | alr1223 (crhB); alr4718 (crhC) | |
| Hfq | Binding to sRNAs and mRNAs, facilitating the base pairing between sRNAs and their mRNA targets | hfq | hfq | ssr3341 (hfq) | asl2047 (hfq) | |
| ProQ | Binding mainly to mRNAs, acting as an RNA chaperone | proQ | / | / | / | |
| RppH | RNA 5ʹ pyrophosphohydrolase, converting 5ʹ triphosphate to 5ʹ monophosphate | rppH | rppH | / | / | |
crRNA, CRISPR RNA; dsRNA, double‐stranded RNA; mRNA, messenger RNA; sRNA, small RNA; ssRNA, single‐stranded RNA; tRNA, transfer RNA.
Tremendous insights have been gained into the bacterial RNA turnover process during the last 40 years 5 , 6 . It is now well recognized that the RNA degradation process in bacteria takes several steps (Figure 1). In E. coli, the primary transcripts are first cleaved internally mainly by the endoribonuclease RNase E. Before RNase E cleavage, most primary transcripts, which are 5ʹ‐triphosphorylated, need to be converted into the RNase E preferred 5ʹ‐monophosphorylated form by the RNA pyrophosphohydrolase RppH. The fragments generated by RNase E are further trimmed from the 3ʹ end to the 5ʹ end by various exoribonucleases, such as PNPase, RNase II, and RNase R and turned into 2‐ to 5‐nt oligoribonucleotide end products (also known as nanoRNAs). These short oligoribonucleotides are eventually converted into monoribonucleotides by the 3ʹ–5ʹ oligoribonuclease Orn, completing the degradation process. No 5ʹ–3ʹ exoribonuclease has been found in E. coli thus far. The auxiliary proteins, Hfq, ProQ, and DEAD‐box RNA helicases, are involved in the degradation of specific RNAs through their RNA binding and unwinding activities.
Figure 1.
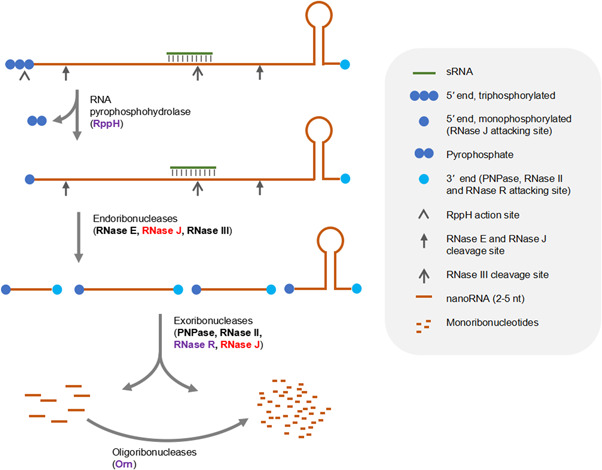
The principal pathway of messenger RNA (mRNA) degradation in bacteria. The major steps include the following: RNA phosphohydrolase converts the 5ʹ end of the primary transcripts from triphosphate to monophosphate; endoribonucleases internally cleave the transcripts into intermediate fragments; exoribonucleases degrade the intermediates into monoribonucleotides and generate the end products of 2‐ to 5‐nt oligoribonucleotides (nanoRNAs); and oligoribonucleases hydrolyze the oligoribonucleotides into monoribonucleotides, finishing the degradation process. Note that many transcripts in the triphosphorylated form can also be substrates of endoribonucleases and some transcripts may also be substrates of exoribonucleases before endoribonucleolytic cleavage. The major Escherichia coli and cyanobacterial enzymes involved in the degradation process are shown. The enzymes present in both E. coli and cyanobacteria (RNase E, RNase III, PNPase, and RNase II) are in black, those currently discovered in E. coli only (RppH, RNase R, and Orn) are in purple, and the one present in cyanobacteria but not in E. coli (RNase J) is in red. Note that RNase J acts as both an endoribonuclease and a 5ʹ−3ʹ exoribonuclease.
As different ribonucleases participate in the degradation of one RNA, bacteria have evolved various mechanisms to coordinate their activities for efficient turnover of cellular RNAs. One of the most important mechanisms is the formation of the RNA degradosome, a protein complex whose components are mostly RNA‐degrading enzymes 7 . The core components of the RNA degradosome in E. coli are the endoribonuclease RNase E, the exoribonuclease PNPase, the RNA helicase RhlB, and the glycolytic enzyme enolase. RNase E acts as the scaffold to recruit other degradosomal components. It is believed that within the degradosome, the RNA fragment produced by RNase E cleavage can be quickly captured and degraded by PNPase. RhlB is able to unwind RNA duplexes to facilitate the degradation of structured substrates by RNase E and PNPase. The coupled activities of RNase E, PNPase, and RhlB ensure efficient substrate degradation and prevent the accumulation of the intermediate products. Enolase does not directly act on RNAs but can modulate the activity of the degradosome under specific conditions 8 . When the RNA degradosome is disrupted, cell growth and cellular RNA metabolism are significantly affected 9 , 10 .
Compared to E. coli, B. subtilis presents important differences in RNA degradation systems 5 . A key difference is that B. subtilis does not have an ortholog of E. coli RNase E; instead, it has two ribonucleases that are absent in E. coli: RNase Y and RNase J. The endoribonuclease RNase Y is among the most important ribonucleases that control the abundance of bulk mRNAs in B. subtilis 11 . It shows no detectable similarity to E. coli RNase E in primary sequence but has many properties remarkably similar to those of RNase E. RNase Y is able to cut the same substrates in the same way as RNase E, and the activity of both enzymes is stimulated by the presence of a 5ʹ‐monophosphate group in the substrates 12 . RNase Y is also the ribonuclease that initiates RNA cleavage in vivo 13 . Last, RNase Y interacts with other proteins, such as RNA helicase and glycolytic enzyme, forming a complex that is functionally equivalent to the E. coli RNA degradosome 14 . RNase J is unique in that it acts as both a 5ʹ–3ʹ exoribonuclease and an endoribonuclease, and it also globally regulates RNA metabolism in B. subtilis 15 , 16 . More details regarding RNase J will be described below.
Our major understanding of bacterial RNA metabolism and ribonucleases has been obtained mainly by studies in E. coli and B. subtilis. In recent years, research interests have been extended to the RNA metabolism of other bacterial species, such as Caulobacter, Helicobacter, Staphylococcus and Mycobacterium 17 , 18 , 19 , 20 . Based on these studies, it is well recognized that different bacteria, even closely related species, may use different sets of enzymes for RNA turnover. The functional importance of the same enzyme can also vary greatly in different hosts. Thus, in addition to studying model organisms, it is very necessary to investigate the ribonucleases in species of interest, so that we can have a better understanding of RNA metabolism and gene regulation in bacteria.
Cyanobacterial enzymes for RNA maturation and degradation
Cyanobacteria diverged from E. coli and B. subtilis over 3 billion years ago 21 . They belong to a unique bacterial phylum that comprises species with great morphological, ecological, and genetic diversity. Cyanobacteria are the only prokaryotes that carry out oxygenic photosynthesis, and many of them can also fix atmospheric nitrogen, thus contributing greatly to the carbon and nitrogen cycles in the biosphere 22 , 23 . Therefore, cyanobacteria are excellent model organisms for studying photosynthesis and nitrogen fixation. In addition, they are being developed into efficient hosts for the renewable production of valuable products using solar energy and CO2 24 . In contrast to the importance of cyanobacteria in basic and applied studies, our understanding of ribonucleases and RNA metabolism in cyanobacteria remains very limited. Here we will summarize the current knowledge about ribonucleases and other relevant proteins in cyanobacteria and present our understanding of their roles in gene regulation at the posttranscriptional level in these organisms.
The unicellular model cyanobacterium Synechocystis PCC 6803 and the filamentous model cyanobacterium Anabaena (also called Nostoc) PCC 7120 were the first cyanobacterial strains to have sequenced genomes 25 , 26 . By genomic analysis, in both strains, we identified 16 proteins homologous to the known proteins involved in RNA turnover in E. coli and B. subtilis (Table 1). Eleven of these proteins are universally present in E. coli, B. subtilis, and cyanobacteria: PNPase, RNase PH, RNase Z, RNase III, RNase H, RNase II/R, RNase P, YbeY, RNA helicases, Cas6, and Hfq. Three are present in cyanobacteria and E. coli but are missing in B. subtilis: RNase E, RNase HI, and RNase D. Mini‐III and RNase J are present only in cyanobacteria and B. subtilis. Some proteins (RNase E, PNPase, RNase Z, YbeY, Mini‐III, and RNase J) are encoded by single genes in each cyanobacterial genome, while others (RNase D, RNase II/R and RNase III and RNase helicase) are encoded by more than one gene. Several proteins present in E. coli or B. subtilis do not have homologs in cyanobacteria, including RNase T, Orn, RNase Y, RNase HIII, RNase M5, NrnA/NrnB, ProQ, and RppH. Taken together, cyanobacteria contain a set of ribonucleases and related proteins that are different from those used by the model bacteria E. coli and B. subtilis, implying certain unique characteristics in their RNA metabolism regulation.
To date, only a handful of proteins involved in RNA turnover has been experimentally investigated in cyanobacteria. Some of the known proteins, such as RNase P, RNase Z, RNase PH, RNase D, and Cas6, mainly participate in the maturation of stable RNAs (i.e., tRNAs, rRNAs, and crRNAs) and are functionally conserved in diverse bacteria, while the others have more important roles in mRNA turnover. In this review, we will focus on the latter.
ENDORIBONUCLEASES
RNase E, a key endoribonuclease in cyanobacteria
At least one of the three ribonucleases, RNase E, RNase Y, and RNase J, was found when a representative set of 1535 bacterial genomes was scanned for these genes 27 . In E. coli, RNase E is the most important ribonuclease for bulk RNA turnover. Its orthologs are widely distributed in Proteobacteria, Actinobacteria, Bacteroidetes, Chlamydiae, Cyanobacteria, Firmicutes, and plant chloroplasts 28 . The 1061‐aa E. coli RNase E can be divided into two distinct parts: the conserved N‐terminal half (NTH) and the highly divergent C‐terminal half (CTH) 29 . The crystal structure of the NTH shows that it consists of a large domain and a small domain 30 . The large domain encompasses the subdomains S1, 5ʹ‐sensor, RNase H, DNase I, and Zn‐link, with the first four conferring substrate binding and cleavage activity and the Zn‐link mediating the tetramerization of RNase E. The small domain also plays a role in tetramerization. In contrast to the compact structure of the NTH, the CTH is highly disordered. CTH is not critical for RNase E activity in vitro, as RNase E without this region still cleaves the substrates efficiently 31 . However, it is required for normal RNase E function in vivo. CTH is the scaffold of the RNA degradosome 32 , and it also contains a cell membrane‐targeting sequence that is responsible for the membrane localization of RNase E 33 .
Cyanobacterial RNase E proteins also contain a catalytic N‐terminal region and a disordered C‐terminal region 34 , 35 . The N‐terminal region is similar to the NTH of E. coli RNase E, with the subdomains S1, 5ʹ‐sensor, RNase H, DNase I, and Zn‐link, but without the small domain found in E. coli RNase E. The C‐terminal region is also highly disordered, with no particular sequence similarity with the CTH of E. coli RNase E.
E. coli RNase E preferentially cleaves within AU‐rich regions of single‐stranded RNAs 36 , 37 , 38 . In the closely related Salmonella enterica, a uridine two nucleotides downstream of the cleavage sites was identified as an important recognition determinant for RNase E 39 . For most substrates, cleavage is efficient only when the 5ʹ end is monophosphorylated 40 , while for certain substrates, the cleavage efficiency is not affected by the 5ʹ‐end phosphorylation state 41 . The presence of both the 5ʹ‐sensing (i.e., 5ʹ‐monophosphate dependent) pathway and the direct entry (i.e., 5ʹ‐monophosphate independent) pathway of RNase E cleavage is supported by in vivo evidence 42 . The similarity between cyanobacterial RNase E and E. coli RNase E in the catalytic region suggests that they have similar catalytic activities. Indeed, cyanobacterial RNase E and its E. coli counterpart have the same substrate preference and cleavage pattern for all the tested substrates 34 , 35 . Additionally, the cyanobacterial rne gene can complement the rne mutant of E. coli, indicating that cyanobacterial RNase E and E. coli RNase E have similar activity on the cellular RNA substrates 34 .
The C‐terminal regions of cyanobacterial RNase E are much shorter and show no detectable sequence similarity to that of E. coli RNase E 35 . The sequences of the C‐terminal regions of all cyanobacterial RNase E proteins are also quite divergent; however, a careful alignment revealed four conserved subregions (C1, C2, C3, and C4) and three highly variable subregions (V1, V2, and V3) (Figure 2A). Three of the four conserved subregions can be detected universally in cyanobacterial homologs, but we noticed the absence of C1 in marine picocyanobacteria (Prochlorococcus and marine Synechococcus). As the E. coli CTH is the scaffold of the RNA degradosome, the C‐terminal region of cyanobacterial RNase E could have a similar role in vivo. Indeed, cyanobacterial RNase E was found to interact with PNPase via the Arg‐rich C4 subregion, forming a complex resembling the E. coli RNA degradosome 35 . The cyanobacterial degradosome is distinct from the E. coli degradosome in composition and the way of assembly (see more details below). The functions of the other subregions are currently unknown but we noticed that C2 is almost as Arg‐rich as C4. Moreover, for two residues in the variable subregion V2 of the Synechocystis PCC 6803 enzyme, K494, and K512, cross‐linking to an RNA substrate was observed 43 , pointing at possible functions in the positioning of the substrate.
Figure 2.
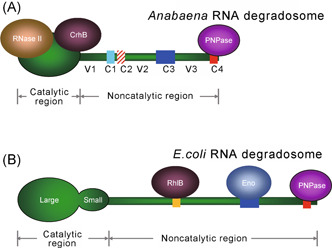
Schematic representation of the assembly of the Anabaena and the Escherichia coli RNA degradosome. (A) Anabaena RNA degradosome; (B) Escherichia coli RNA degradosome. RNase E forms tetramers; here, only one of them (in green) is shown for simplicity. The components of the degradosome are shown relative to the positions of the RNase E proteins. Four conserved subregions (C1, C2, C3 and C4) and three variable subregions (V1, V2 and V3) have been identified in the noncatalytic region of Anabaena RNase E 35 . Note that the E. coli RNase E catalytic region is composed of a large domain that is equivalent to the catalytic region of Anabaena RNase E and a small domain that has no counterpart in Anabaena RNase E.
E. coli RNase E is localized to the cell membrane via a 15‐residue membrane‐targeting motif forming an amphipathic α‐helix 33 . Disruption of its membrane localization by removing the targeting motif resulted in slow growth, indicating abnormal cellular RNA metabolism 33 . In the sequences of cyanobacterial RNase E, no membrane‐targeting motif can be detected. It has been shown that the RNase E of Anabaena PCC 7120 is in fact a cytoplasmic protein 44 . In the proteobacterium Caulobacter crescentus, which is evolutionally far from cyanobacteria but close to E. coli, RNase E also displays cytoplasmic localization 45 . Apparently, RNase E has evolved specific subcellular localization in different cellular contexts. Another potentially highly interesting interaction between RNase E and RNA has recently been reported, showing that the enzyme could be modified by the addition of a short RNA chain (called RNAylation) 46 . This observation requires further work, but if confirmed, might have far‐reaching implications.
The physiological function of RNase E has been experimentally investigated in a few cyanobacterial species. The rne gene was found to be indispensable in Synechococcus PCC 7002 and Synechocystis PCC 6803 47 , 48 , and our attempt to inactivate the rne gene in Anabaena was also unsuccessful. Thus, the rne gene is essential in cyanobacteria. In Anabaena PCC 7120, overexpressing either the full‐length or the catalytic region of RNase E led to severe growth inhibition and changes in cell morphology 49 . In an rne partial mutant of Synechocystis PCC 6803, more than 2000 genes showed altered expression 48 . These studies indicate a global role of RNase E in cyanobacteria.
An experimental approach to characterize the targetome of an essential RNase requires the construction of a strain expressing a temperature‐sensitive enzyme. Then, transcriptomes are analyzed by RNA‐seq before and after transfer of the mutant strain to the nonpermissive temperature and compared with each other and with a wild‐type control. This approach is called “transient inactivation of an essential ribonuclease followed by RNA‐seq” (TIER‐seq) 39 and has been applied recently to the transcriptome‐wide identification of RNase‐E‐dependent cleavage sites in Synechocystis PCC 6803 50 . One challenge in engineering a temperature‐sensitive RNase E in this cyanobacterium was that the introduction of the amino acid substitution G63S or I65F, which corresponds to G66S or L68F that renders the E. coli enzyme temperature‐sensitive 51 , was not sufficient. Instead, one of three additional mutations (V94A, V297A, or G281E) was needed to obtain viable mutant strains. Using one of such mutant strains for TIER‐seq analysis, 1472 RNase‐E‐dependent cleavage sites were mapped transcriptome‐wide in Synechocystis PCC 6803 50 . Careful analysis of these sites revealed enrichment for adenine residues at positions ‐4 and ‐3 upstream and uridine residues immediately downstream of the cleavage site, especially at position +2. These conserved sites could form an AU clamp, which is a possible signal for positioning the actual cleavage site 50 . Moreover, the uridine preferred at the +2 position in an AU‐rich RNA sequence stretch matches the target preference identified for the RNase E of Salmonella enterica 39 . Given the ability of RNase E to cleave potentially thousands of sites, it is important to understand how the specificity and target recognition of RNase E is modulated. Specific proteins mediating target selection were identified in E. coli, such as Hfq (see below) or the RNase adaptor protein RapZ, which binds to the sRNA GlmZ 52 , 53 , 54 . RapZ senses the cell envelope precursor glucosamine‐6‐phosphate 55 and has been found to boost RNase E activity in an intriguing way, through interaction with its catalytic domain 56 . Potential homologs of RapZ exist in several cyanobacteria (e.g., NJM11451, REJ55829, and WP_071838815, all with >60% similar residues), but none of these proteins has been functionally characterized thus far.
Interestingly, in addition to its role in bulk RNA metabolism, Synechocystis RNase E also participates in the maturation of crRNAs 43 . In most CRISPR‐Cas systems, specialized endoribonucleases, often belonging to the Cas6 family, mediate the maturation of crRNAs. In the case of the type III‐Bv CRISPR system in Synechocystis PCC 6803, RNase E was found to recognize the stem‐loop region of the precursor crRNA, followed by cleavage within the downstream single‐stranded region to produce the mature crRNA 43 . This is one of the few known cases where crRNA is not matured by a Cas protein.
Transcripts encoding photosynthesis proteins are a major target of RNase E in Synechocystis PCC 6803 50 . The relation between RNase E and some of these transcripts has been studied in more detail, such as for psaL, which encodes the photosystem I reaction center protein XI, or the psbA2 and psbA3 genes that encode the photosystem II reaction center protein D1 (Figure 3). The almost identical psbA2 and psbA3 transcripts are not stable in the dark due to the presence of an AU‐box within their 5ʹ UTRs. As RNase E could efficiently cleave within the AU‐box in vitro, it was thus suggested that it is responsible for the rapid degradation of those AU‐box containing transcripts in the dark 57 . This discovery raised the question of how the activity of RNase E on these transcripts would be prevented under other conditions. A potential answer lies in the fact that the AU‐box overlaps with the region for the initiation of translation. Thus, interacting ribosomes on these mRNAs that are strongly translated in the light would protect against degradation. This model was further refined by the discovery of weak promoters in Synechocystis PCC 6803 on the reverse strand that give rise to cis‐antisense RNAs called PsbA2R and PsbA3R. Since these antisense RNAs are coregulated with the psbA2/3 mRNAs, they protect the RNase E target sites not covered by ribosomal subunits 58 . An interplay between RNase E and an mRNA modulated by a noncoding RNA was also discovered for psaL. This gene encodes the photosystem I reaction center protein XI, which is involved in the trimerization of photosystem I. Under high light conditions, the monomeric status is the preferred configuration; hence, psaL expression needs to be reduced. Under this condition, transcription of the sRNA PsrR1 is stimulated. PsrR1 interacts with a target region in the psaL mRNA; upon binding, RNase E is recruited to this site, which then cleaves at a site seven nucleotides into the coding sequence 59 .
Figure 3.
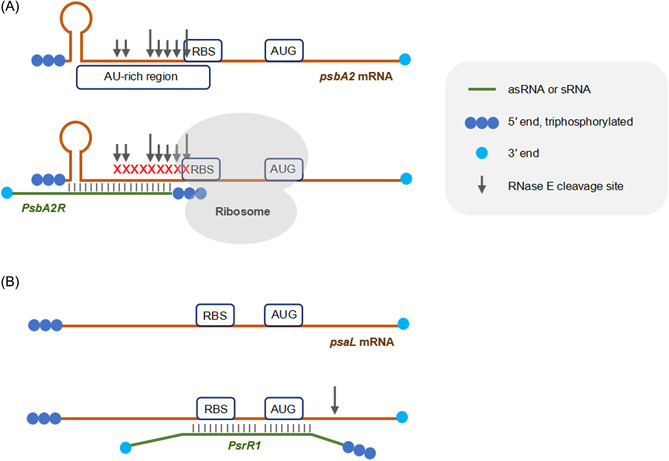
RNase E cleavage of messenger RNAs (mRNAs) encoding photosynthetic proteins in Synechocystis PCC 6803. (A) Upper panel: The psbA2 mRNA has a 49‐nt 5ʹ UTR that, in the dark, is sensitive to RNase E cleavage due to multiple RNase E cleavage sites within an AU‐rich sequence that extends into the likely ribosome binding site (RBS) 43 , 50 , 57 . Hence, rapidly interacting ribosomes under conditions such as high light, when this mRNA is strongly translated, can protect against degradation (lower panel). In addition, the antisense RNA (asRNA) PsbA2R is coregulated with the mRNA from a transcriptional start site leading to an overlap with the first 19 nucleotides of the psbA2 mRNA, hence protecting these sites against RNase E cleavage 58 , as indicated by the red crosses. A short stem‐loop near the mRNA 5ʹ end might be relevant for recognition. Note that in vivo additional ribonucleases are likely involved 50 . Note that the gene psbA3, which is almost identical to psbA2, is regulated in the same way 57 , 58 . (B) Upper panel: The psaL gene is preceded by a 55‐nt 5ʹ UTR that is not targeted by RNase E under most growth conditions. However, under high light, transcription of the gene for the sRNA PsrR1, which is located elsewhere in the genome, is stimulated. Except for two mismatches, PsrR1 interacts over 22 consecutive nucleotides with the psaL mRNA (lower panel). This interaction overlaps the 3ʹ end of the 5ʹ UTR, including the ribosome binding site, the start codon, and one additional nucleotide of the second codon. This interaction leads to conditional recruitment of RNase E, which then cleaves a single‐bond seven nucleotides into the coding sequence, effectively decapitating the psaL mRNA 59 .
An intriguing mechanism was discovered for the upregulation of RNase E expression in marine picocyanobacteria. During lytic infection of Prochlorococcus MED4 with the T7‐like cyanophage P‐SSP7, rne, the gene encoding RNase E was one of very few upregulated host genes. This result was interpreted as a mechanism by which the phage could benefit from the stimulated degradation of host transcripts for use as substrates for phage deoxynucleotide synthesis 60 . To achieve upregulation, an alternative transcription start site was identified that was only activated during infection and from which an mRNA was derived that lacked the 5ʹ UTR, thereby bypassing the normally tight control of RNase E expression 61 .
Most cyanobacteria having more than one RNase III gene
RNase III family proteins are endoribonucleases that specifically cleave dsRNAs. They are widely distributed in Bacteria, Archaea, and Eukarya. Based on their sequence features, bacterial RNase III family proteins can be grouped into two categories. The first category includes the canonical homodimeric RNase III enzymes, with each subunit containing one RNase III domain and a dsRNA binding domain (dsRBD). The second category corresponds to the Mini‐III enzymes, which also act as a dimer, but each subunit consists of a single RNase III domain only 62 . Here, we refer to the enzymes in these two groups as RNase III and Mini‐III, respectively.
E. coli and B. subtilis each have only one gene encoding RNase III. In contrast, different cyanobacterial species usually have 1 to 3 RNase III‐encoding genes. For instance, two RNase III family proteins (A0061 and A2542) are present in Synechococcus PCC 7002, and both can cleave many substrates of E. coli RNase III, although they do not always cleave at the same sites as E. coli RNase III 47 . A0061 was shown to be involved in the maturation of tRNAs and rRNAs 63 . Note that E. coli RNase III also plays an important role in rRNA maturation 64 , 65 . Thus, maturing stable RNAs may be a common function of bacterial RNase III proteins. Disruption of the gene encoding A2542, the other RNase III family protein in Synechococcus PCC 7002, did not affect rRNA maturation but greatly increased the copy number of the endogenous plasmid pAQ3 63 . The mechanism of plasmid copy number control by A2542 remains unclear. Anabaena PCC 7120 also has two RNase III family proteins. One of them (Alr0280) could cleave an artificial dsRNA, while the other (All4107) could not 66 . We noticed that the start codon of all4107 was misidentified in the original annotation, and the recombinant All4107 prepared by Gao et al. 66 , which is based on this incorrect annotation, lacks the first 40 residues. This is probably the reason why All4107 was inactive in their studies.
Although RNase III family proteins participate in the maturation and degradation of many cellular RNAs, they are not required for the viability of E. coli and many other bacteria 67 , 68 , 69 . One noticeable exception is B. subtilis RNase III, which is essential for cell growth 70 . It is now clear that the essentiality of B. subtilis RNase III results from its activity toward the toxin genes borne by two prophages and not from its influence on the metabolism of other cellular RNAs 71 , 72 . In cyanobacteria, RNase III proteins are likely also dispensable. Loss of each of the two RNase III proteins, or of both, in Synechococcus PCC 7002 did not significantly affect cell growth, although it led to altered expression of hundreds of genes 47 , 63 . In Anabaena PCC 7120, at least one of the RNase III‐encoding genes, alr0280, could be inactivated; however, the phenotype of the mutant was not mentioned 66 . A significant share of the bacterial transcriptome consists of overlapping antisense transcripts (for review, see reference 73 ). In cyanobacteria, some antisense transcripts were demonstrated to control the level of the respective overlapping mRNA in a codegradation mechanism, such as for the isiA gene in Synechocystis PCC 6803 under iron starvation 74 . RNase III was found to be the enzyme responsible for such codegradation in bacteria 75 . In cyanobacteria, this was clearly demonstrated for an antisense RNA called as_glpX that is transcribed in Anabaena PCC 7120 within the gene glpX encoding sedoheptulose‐1,7‐bisphosphatase/fructose‐1,6‐bisphosphatase (SBPase). The as_glpX transcript is specifically transcribed in heterocysts and the RNase III Alr0280 was demonstrated to mediate the codegradation of as_glpX and its cognate mRNA 76 . The outcome of this codegradation is a reduced amount of SBPase contributing to the shutdown of the Calvin cycle specifically in those cells that are undergoing differentiation into heterocysts. RNase III‐encoding genes are found in all sequenced cyanobacterial species, and such a wide distribution suggests that they may be required for the acclimation of cyanobacteria to fluctuating environments.
The activity of E. coli RNase III was shown to be regulated by the O‐acetyl‐ADP‐ribose deacetylase YmdB 77 , 78 . YmdB homologs are present in a few cyanobacteria, including Synechocystis PCC 6803, where it is fused as a C‐terminal domain to a YfbK‐domain‐containing protein, encoded by slr7060 on plasmid pSYSA. It would be interesting to test whether RNase III proteins could be regulated by YmdB homologs in these cyanobacterial species.
Although absent in E. coli, Mini‐III family proteins are also widely distributed. Mini‐III was originally identified and characterized in B. subtilis 79 . It mainly participates in the maturation of the 23 S rRNA precursor 79 , 80 . Genes encoding a Mini‐III family protein are present in every sequenced cyanobacterial species. Mini‐III in Synechococcus PCC 7002 was found to mature 23S rRNA 63 . Likewise, in the plant chloroplast, Mini‐III is involved in the maturation of 23S rRNA, 16S rRNA, and 4.5S rRNA 81 . Thus, maturation of rRNAs (particularly 23S rRNA) is likely a function common to Mini‐III family proteins. B. subtilis Mini‐III seems to have much more stringent substrate specificity than canonical RNase III, as it has been shown to cut dsRNA preferentially at the ACC^U site 82 . The substrate specificity of Mini‐III proteins from cyanobacteria and other bacteria has not yet been determined.
YbeY, one of the most conserved bacterial endoribonucleases
The endoribonuclease YbeY, which cleaves both single and double‐stranded RNA substrates, is one of the most conserved proteins in bacteria 83 . In E. coli, Sinorhizobium meliloti and many other bacteria, YbeY participates in the maturation of ribosomal RNAs (particularly 16S rRNA) 84 , 85 , 86 , 87 . In addition, YbeY was also shown to be involved in sRNA‐mediated mRNA degradation, probably by directly cleaving the sRNA‐mRNA duplex 88 , 89 . YbeY‐encoding gene is present in every sequenced cyanobacterial genome, and the high conservation of YbeY in bacteria suggests that this protein may also function in rRNA mutation and posttranscriptional gene regulation in cyanobacteria.
RNase H family proteins
RNase H family proteins are the primary enzymes responsible for recognizing and cleaving RNA–DNA hybrids. Such hybrids are prevalent in cells and can compromise genome integrity if not eliminated. RNA–DNA hybrids exist in several forms in vivo, including the R‐loop, which occurs during transcription when the nascent RNA strand bases pair with its template DNA; covalently incorporated single ribonucleotides or short polymers of RNA due to DNA replication error; and the oligoribonucleotides used to prime the synthesis of Okazaki fragments 90 , 91 , 92 , 93 . RNase H family proteins, which specifically remove these RNA–DNA hybrids, are widely present in bacteria and can be classified into two groups based on sequence similarity: class one, which is represented by RNase HI, and class two, which consists of RNase HII and RNase HIII 94 . RNase HI cleaves within RNA–DNA hybrids and such substrates should have four or more consecutive ribonucleoside monophosphates (rNMPs). RNase HII has been shown to cleave 5ʹ to single embedded rNMPs or 3ʹ to the first rNMP at an RNA‐DNA junction when multiple rNMPs are present 95 , 96 . It should be noted that the genes encoding RNase HII in many cyanobacteria occur in a conserved dicistron together with the gene encoding RNase E.
Although RNase HIII is more closely related to RNase HII in sequence, its activity is very similar to that of RNase HI, cleaving RNA–DNA hybrids with three or more consecutive rNMPs 97 . As RNase HI and RNase HIII have similar activities, most bacteria have either RNases HI and HII, or HII and HIII 98 . For example, E. coli has RNase HI (encoded by rnhA) and RNases HII (encoded by rnhB), and B. subtilis has RNase HII and RNase HIII (encoded by rnhC). In E. coli, RNase HI mainly removes R‐loops by cleaving the RNA strand in the RNA–DNA hybrids, generating RNA primers that could either be used by DNA polymerase I (Pol I) for the synthesis of Okazaki fragments or be degraded by Pol I 99 , 100 . RNase HII is involved in removing the misincorporated rNMPs in genomic DNA 92 , 93 , 101 . While inactivation of rnhB did not result in an obvious growth defect, inactivation of rnhA led to slow growth in E. coli 93 , 102 . Furthermore, the inactivation of both genes resulted in much poorer growth and impaired chromosome replication and segregation 103 , suggesting the critical roles of these two RNase H proteins in vivo. An RNase HI and an RNase HII are encoded by each sequenced cyanobacterial genome, and they both show high sequence similarities to their E. coli counterparts. It is likely that they also play important roles in eliminating R‐loops and misincorporated rNMPs from genomic DNA.
EXORIBONUCLEASES
RNase J and its enigmatic role in cyanobacteria
RNase J was originally identified in B. subtilis 104 in which two RNase J family proteins exist, RNase J1 (encoded by rnjA) and RNase J2 (encoded by rnjB), which can form heterodimers in vivo 105 . RNase J1 acts as both an endoribonuclease and an exoribonuclease; however, structural analyses suggest that it mainly acts as an exoribonuclease 106 , 107 . This enzyme is the first 5ʹ–3ʹ exoribonuclease identified in bacteria 15 . RNase J2 has much lower activity than RNase J1, although the two proteins show high sequence similarity 105 . Both rnjA and rnjB can be deleted in B. subtilis but the consequences are quite different. While the deletion of rnjB did not affect growth, that of rnjA led to severe growth defects, indicating that RNase J1 is much more important than RNase J2 in vivo 16 . A recent study showed that RNase J could resolve the stalled RNA polymerase (RNAP) complex from DNA by degrading the nascent RNA in the complex and finally colliding the RNAP off DNA, thereby playing an important role in preventing transcription–replication collisions 108 .
RNase J family proteins are present in many archaeal and bacterial species 104 . However, the number of RNase J homologs and their importance in different organisms may vary significantly. For instance, two RNase J homologs are present in the pathogen Staphylococcus aureus, and although they also seem to function as a complex in vivo, deletion of either of them could greatly affect growth 109 , 110 . The only RNase J homolog in the gastric pathogen Helicobacter pylori is essential for viability, and it was shown to associate with the DEAD‐box RNA helicase RhpA, forming a minimal RNA degradosome 111 . A single RNase J‐encoding gene, rnj, is present in every sequenced cyanobacterial genome. Gene expression of rnj in Synechocystis PCC 6803 was found to respond to a variety of environmental stresses such as iron starvation 112 , or to the absence of the RNA helicase CrhR 113 . The gene (ID slr0551, cf. Table 1) has a very long 5ʹ UTR of 455 nt 114 , which is also detectable as a separate transcript in the cell and may function as an sRNA (called NC‐117 115 ). To date, limited investigation of the function of RNase J has been carried out. RNase J from Synechocystis PCC 6803 has catalytic properties very similar to those of B. subtilis RNase J1: as an exoribonuclease that degrades RNAs whose 5ʹ end is not triphosphorylated, and as an endoribonuclease that cuts substrates at sites where B. subtilis RNase J1 does 48 . Attempts to inactivate rnj in several cyanobacterial species have been unsuccessful, implying that RNase J is essential in cyanobacteria 47 , 48 , 116 . In a partially segregated rnj mutant of Synechocystis PCC 6803, the maturation of the CRISPR3 crRNA was affected, but unlike RNase E, there was no evidence provided that RNase J could directly act on the precursor crRNA 48 . In this partial mutant, only 180 genes, most of which are located on endogenous plasmids, had altered expression 48 . The small number of genes affected may be due to the insufficient depletion of RNase J in this strain. Given that RNase J is essential in cyanobacteria, it is expected to have a global role in gene regulation.
Existing knowledge of RNase J in cyanobacteria suggests that it is a conserved and essential enzyme, but its true functionality has remained unknown thus far. In contrast, the functions of RNase J in plant chloroplasts are understood much better. Plant RNase J is nucleus‐encoded but localized in chloroplasts. Its catalytic properties are similar to those of B. subtilis RNase J1 117 . An Arabidopsis mutant without RNase J could not form normal chloroplasts, resulting in aberrant embryo development 118 , 119 . Massive accumulation of antisense RNAs was observed when RNase J was depleted from the chloroplast 120 , and it is likely that RNase J plays a key role in 5ʹ end maturation of chloroplast mRNAs 121 . Because of the cyanobacterial origin of chloroplasts, knowledge of chloroplast RNase J could provide valuable insights into the functions of RNase J in cyanobacteria.
RNase II/R
Two RNB family 3ʹ−5ʹ exoribonucleases are present in E. coli: RNase II (encoded by rnb) and RNase R (encoded by rnr). RNase II only degrades the 3ʹ‐single‐stranded region of substrates 122 , while RNase R, which has RNA helicase activity, can efficiently degrade structured RNAs that have a single‐stranded tail 123 , 124 . The two enzymes have different cellular substrates in E. coli. RNase II is mainly involved in the degradation of mRNAs 125 , although it was also shown to protect rpsO mRNA from attack by other exoribonucleases by removing its poly(A) tail appended by poly(A) polymerase 126 . In contrast, RNase R plays an important role in the degradation of rRNAs and mRNAs with complicated structures 123 , 127 . B. subtilis has only one RNB family protein, RNase R, which, similar to E. coli RNase R, is capable of degrading structured RNAs 128 .
Each sequenced cyanobacterial genome contains two or three RNB family exoribonucleases. In Synechocystis PCC 6803, one of the RNB family proteins, Sll1290, has the same substrate specificity as that of E. coli RNase II and only degrades ssRNAs while producing 4‐nt end products 129 . Whereas E. coli RNase II prefers polyadenylated substrates, Sll1290 does not have such a preference, in line with the fact that cyanobacterial mRNAs do not have a homogenous poly(A) tail 130 . Another cyanobacterial RNB family protein, Alr1240, from Anabaena PCC 7120 also has properties similar to those of E. coli RNase II, as it can only degrade unstructured substrates 44 . It is still unclear whether RNase R‐like RNB family exoribonucleases exist in cyanobacteria.
RNB family proteins are dispensable in E. coli and B. subtilis as their activities can be replaced by other exoribonucleases 125 , 128 , 131 . In contrast, the essentiality of RNB family proteins in cyanobacteria seems to be species dependent. In Synechocystis PCC 6803, sll1290 is indispensable 129 , while the other RNB protein‐encoding gene sll1910 could be inactivated, and the resulting strain showed resistance to the herbicide acetazolamide 132 . Similarly, only one of the two RNB‐family genes in Synechococcus PCC 7002 could be inactivated and the mutant strain grew only slightly slower than wild type 47 . In contrast, each of the RNB‐family protein‐encoding genes in Synechococcus PCC 7942 could be inactivated by transposon mutation 116 .
Recently, the Anabaena PCC 7120 RNB family protein Alr1240 was shown to form a complex with RNase E, and these two proteins colocalized in the cytoplasm 44 . Alr1240 enhanced substrate cleavage activities by RNase E in vitro 44 . These results suggest that Alr1240 and RNase E act together in vivo and that Alr1240 may have a regulatory role in the activity of RNase E.
PNPase
PNPase, encoded by the pnp gene, is an exoribonuclease that catalyzes the phosphorolysis of RNAs from 3ʹ‐ends, and it can also catalyze the reverse reaction that adds heteropolymeric tails to RNA 3ʹ ends 133 . This protein is widely distributed in bacteria, chloroplasts and mitochondria, but is missing in Archaea 134 . PNPase processively degrades RNAs with 10‐ to 12‐nt long single‐stranded 3ʹ ends. Similar to RNase II, the action of PNPase is easily stopped by stable secondary structures in RNAs 135 , 136 . Nevertheless, PNPase can cooperate with the RNA helicase RhlB, either by directly forming the PNPase‐RhlB complex or coexisting in the RNA degradosome, to degrade structured RNAs 137 , 138 .
Together with RNase II and RNase R, PNPase plays a global role in exoribonucleic degradation of cellular RNAs in E. coli, and its depletion could influence the expression of more than 1000 genes 139 , 140 , 141 , 142 . Although dispensable for growth under standard conditions, it is essential for growth at low temperatures in E. coli 143 , 144 . Such a function could be attributed to the role of PNPase in rRNA maturation and ribosome biogenesis, as defects in ribosome biogenesis are often associated with cold sensitivity 145 , 146 , 147 , 148 , 149 . PNPase also selectively degrades the mRNAs of cold shock proteins after the low‐temperature acclimation phase to allow cells to resume growth 150 . In many other bacteria, including B. subtilis, PNPase was also shown to be necessary for survival at low temperatures 144 , 151 , 152 , suggesting that it has a common function in these organisms.
Cyanobacterial PNPases have not yet been biochemically investigated. However, they are close to E. coli PNPase in sequence (e.g., Anabaena PNPase shares 48% identities and 64% similarities with its counterpart in E. coli), and such a high similarity suggests conserved catalytic properties. Cyanobacterial PNPases also interact with RNase E in vivo within the RNA degradosome 33 . Inactivation of the pnp gene in Synechocystis PCC 6803 and Synechococcus PCC 7002 was not successful 47 , 130 , suggesting that PNPase has much more important physiological roles in cyanobacteria than in E. coli. Considering that cyanobacteria do not possess the homologs of poly(A) polymerase, which is the major enzyme for RNA polyadenylation in E. coli 153 , PNPase is likely the only enzyme that adds heterogeneous poly(A)‐rich tails to RNA molecules important for efficient degradation 130 .
AUXILIARY PROTEINS
DEAD box RNA helicases
DEAD‐box RNA helicases are the major RNA helicases in bacteria. They facilitate the degradation of structured RNAs by unwinding double‐stranded regions, particularly under low‐temperature conditions. In E. coli, the DEAD‐box helicases RhlB and CsdA are found in the RNA degradosome 154 , 155 . In addition to their roles in RNA degradation, DEAD‐box helicases also participate in other biological processes, such as ribosome biogenesis and the regulation of translational initiation 156 .
Many bacteria have multiple DEAD‐box helicases. For instance, E. coli and B. subtilis have five and four DEAD‐box proteins, respectively. Cyanobacteria usually have 1 to 3 genes encoding DEAD‐box helicases, except that Synechococcus PCC 7942 and a few other species have no such genes 157 . According to their sequence similarity, cyanobacterial DEAD‐box helicases have been classified into three groups: CsdA‐like, RhlE‐like, and CrhR‐like 157 .
The only DEAD‐box RNA helicase in Synechocystis PCC 6803, CrhR, has been extensively studied in recent years. CrhR is a bidirectional, ATP‐stimulated RNA helicase that unwinds the RNA duplex from either 5ʹ–3ʹ or 3ʹ–5ʹ; additionally, it also catalyzes the formation of RNA duplexes by annealing two complementary RNA strands 158 . By combining unwinding and annealing activities, CrhR is able to catalyze RNA strand exchange 158 . Temperature is the environmental factor that significantly influences the expression of crhR. When transferred from standard growth temperature (30°C or 34°C) to a low temperature (24°C or 20°C), both the crhR transcript and the CrhR protein accumulate greatly in cells 159 , 160 . At 30°C, CrhR could be degraded rapidly by an unknown protease in vivo 161 .
At 30°C or 34°C, the crhR mutant of Synechocystis PCC 6803 did not show altered growth, and it only had a small number of genes with altered expression 113 , 159 , 162 , consistent with the low level of expression of crhR under these conditions. However, the mutant showed a retarded growth at 24°C and lost viability at 20°C 159 , 163 , 164 . One conclusion from these studies was that under low temperature, CrhR is required to maintain efficient photosynthesis. Consistent with this conclusion, RNA coimmunoprecipitation experiments with extracts from Synechocystis PCC 6803 strains expressing FLAG‐tagged CrhR yielded a striking bias toward photosynthesis‐associated and redox‐controlled transcripts 165 . In addition, the transcription and mRNA stability of crhR were also shown to be correlated with the redox state of the electron transport chain between Q A in photosystem II and Q O in cyt b6f 166 . While the plastoquinone pool was in the reduced state in wild type, it was in the oxidized state in the crhR mutant. Additionally, the amount of PSI trimers, the transcript levels of psaA and psaB, the PSII activity, and the carbon fixation rate all decreased in the mutant 163 , 164 . Analysis of intracellular structures by scanning electron microscopy (SEM) indicated that the architecture of ribosomes, carboxysomes, and thylakoid membranes were all highly disorganized 164 . How CrhR depletion leads to these physiological and structural changes in the cells remains unknown. A further interaction between CrhR and RNA metabolism was found when RNase E was identified as the endoribonuclease involved in the autoregulation of crhR expression and operon discoordination in the rimO‐crhR operon 165 , 167 .
Anabaena PCC 7120 has two RNA helicases: CrhB (CrhR‐like) and CrhC (RhlE‐like). The enzymatic properties of CrhC have been characterized in vitro. Unlike the bidirectional CrhR, ChrC only unwinds the RNA duplex from 5ʹ to 3ʹ; it also lacks annealing activity 168 . Nevertheless, the expression of crhC is also cold inducible. The crhC transcript was highly unstable at higher temperatures and it was detectable only when the cells were grown at temperatures lower than 25°C 169 , 170 . CrhC was shown to be localized to the cytoplasmic membrane, although it had no discernable membrane‐targeting motif 171 . CrhC may function with other proteins in vivo, as a coimmunoprecipitation assay showed that CrhC coprecipitated with several other proteins, and one of them was further shown to interact with CrhC directly 168 . However, the identities of the coprecipitated proteins were not further determined.
Another Anabaena helicase, CrhB, shows constant expression levels in a wide range of temperatures (at least from 20°C to 44°C), and under various stress conditions, including darkness, nitrogen starvation, and salt stress 169 . The constitutively expressed CrhB and the cold‐inducible ChrC may have distinct biological functions. A recent study showed that CrhB could interact with RNase E, and thus it may be one component of the cyanobacterial RNA degradosome 172 . The exact roles of CrhB and CrhC in RNA turnover in Anabaena PCC 7120 remain to be discovered.
Hfq, not binding RNA in cyanobacteria
Hfq is one of the most important RNA chaperones in bacteria. Although Hfq does not degrade RNA, it can bind to small RNAs (sRNAs) and promote annealing between sRNAs and their target mRNAs, thereby regulating the stability and translation of these mRNAs 173 , 174 . In fact, Hfq is now recognized as a global regulator that controls cell fitness under various conditions 175 , 176 . Another RNA chaperone of similar functional relevance as Hfq in the Enterobacteriaceae, which was more recently recognized, is ProQ 177 . For a review on the differences and similarities between Hfq and ProQ and the comparison to other enterobacterial RNA‐binding proteins, see reference 178 .
Hfq homologs are present in most cyanobacteria, except in obligate symbionts and some Prochlorococcus strains 179 . Within the Prochlorococcus group, an interesting discrepancy was observed. While strains such as MIT9313, which represent a more deeply rooted subclade, possess an hfq gene, other strains with a more streamlined genome, such as MED4, lack an hfq homolog, although the genomic region flanking hfq is otherwise conserved 180 . Although cyanobacterial and E. coli Hfq proteins share a low similarity in primary sequence, they are strikingly similar in tertiary structure 181 . However, genetic and biochemical assays indicated that cyanobacterial Hfq proteins are unlikely to be related to RNA metabolism. First, the cyanobacterial Hfq lacks the key residues involved in RNA binding found in the Hfq proteins of other bacteria, and it has very low affinity to RNA; second, the cyanobacterial hfq gene could not complement the E. coli hfq mutant; and third, inactivation of cyanobacterial hfq did not influence cell growth 181 . In fact, it is now clear that Hfq in cyanobacteria functions as a regulator of cell motility instead of being an RNA chaperone. Inactivation of hfq in Synechocystis PCC 6803 led to the loss of cell motility and pili formation 182 . Hfq, together with PilB and EbsA, was recently shown to form a tripartite complex that regulates the biogenesis of type IV pili 183 .
Although cyanobacterial Hfq does not function as the sRNA‐binding RNA chaperone, sRNAs are prevalent in cyanobacteria, and several of them were shown to regulate gene expression at the posttranscriptional level 59 , 114 , 184 , 185 , 186 , 187 , 188 . The efficient regulation mediated by these sRNAs suggests that non‐Hfq‐type RNA chaperones exist in cyanobacteria.
Novel RNA‐binding protein candidates in cyanobacteria
Gradient profiling by sequencing (Grad‐seq) analyses are an approach to identify unknown RNA chaperones. Following fractionation of whole‐cell lysates on sucrose density gradients by ultracentrifugation, colocalizing proteins and RNA molecules are determined by mass spectrometry and RNA sequencing 177 . This approach led to the discovery of ProQ as a novel major RNA chaperone in Salmonella enterica 177 and the involvement of exoribonuclease in the stabilization and activation of sRNAs in the gram‐positive pathogen Streptococcus pneumonia 189 . For cyanobacteria, the first such global analysis of stable RNA‐protein complexes has been presented recently, focusing on Synechocystis PCC 6803 190 . The stability of such complexes during cell lysis and fractionation was inferred from the colocalization of known RNA‐protein complexes involved in transcription, RNA metabolism, and translation initiation (Figure 4). The data showed the occurrence of a larger number of RNAs in the fractions containing higher molecular weight complexes, suggesting their binding to cognate proteins (Group 1 and Group 2 RNAs in Figure 4). Following hierarchical clustering, the prediction of RNA‐binding protein candidates using RNApred 191 and considering phylogenetic conservation in 57 and synteny in 34 diverse cyanobacteria, a short list of previously uncharacterized protein candidates for the interaction with sRNAs was obtained 190 . Among them were cyanobacterial homologs of KhpA/B proteins, which are also considered as sRNA chaperones in other bacteria 192 .
Figure 4.
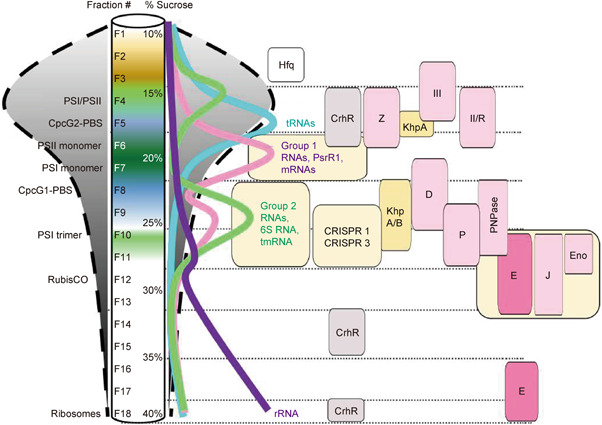
Grad‐seq analysis aids the analysis of ribonucleases, auxiliary proteins, and ribonucleoprotein complexes. A typical sedimentation profile obtained in the analysis of the cyanobacterium Synechocystis PCC 6803 is shown 190 . The positions of several major macromolecular complexes as determined by mass spectrometry are given to the left, the respective fraction numbers and sucrose percentages are indicated along the gradient. The different colors result from the native pigmentation of protein–pigment complexes involved in photosynthesis. The distribution of distinct groups of RNAs is sketched by the colored lines to the right of the gradient. The positions of abundant RNA–protein complexes, such as two of the three CRISPR complexes 193 and noncoding RNA–ribonucleoprotein complexes containing 6S RNA or transfer‐messenger RNA (tmRNA) are shown. Note that characterized regulatory small RNAs such as PsrR1 59 peaked in fraction 7 (F7) together with the bulk of mRNAs, but there were secondary peaks in mRNA abundance in other fractions (for details, see Riediger et al. 190 ). Several proteins and RNA‐protein complexes involved in RNA metabolism, such as RNase D (D), RNase J (J), RNase E (E), RNase P (P), PNPase, enolase (Eno), and CrhR occur in the higher molecular fractions, indicating their association with larger complexes. Most RNAs were detected in overlapping fractions as well, indicating their likely direct association with such complexes. The striking overlap in the in‐gradient distribution of PNPase, enolase, RNase E and J, consistent with their possible colocalization into degradosomes is boxed.
Two different gene products were detected for RNase II/R (II/R) and RNase III (III) in the lighter fractions, while Mini‐III was not detected at all. The strong correlation between RNase Z (Z) and the bulk of tRNAs is consistent with the role of this enzyme in tRNA maturation. Note that Hfq was found only in very light fractions, consistent with its non‐RNA binding character in cyanobacteria. Candidates for alternative RNA chaperones are the KhpA/B homologs Slr0287 and Slr1472. See Table 1 for the gene IDs of all other mentioned proteins. The entire data set is available at https://sunshine.biologie.uni-freiburg.de/GradSeqExplorer/. Reprinted in modified form with permission from Riediger et al. 190 .
RNA DEGRADOSOME
As the degradation of most RNA species involves several steps catalyzed by different ribonucleases, the rapid turnover of cellular RNAs requires the highly coordinated actions of various ribonucleases 5 . The formation of the RNA degradosome is one of the most important mechanisms for the coordinated actions of RNases found in bacteria. As mentioned earlier, the RNA degradosome of E. coli is mainly composed of RNase E, PNPase, RhlB, and enolase (Figure 2B). RNase E as the core of the RNA degradosome recruits other degradosomal components via its C‐terminal noncatalytic region 194 . Another RNase E‐based degradosome identified in the α‐proteobacterium C. crescentus is similar to that of E. coli, consisting of RNase E, PNPase, a DEAD‐box helicase, and the Krebs cycle enzyme aconitase 17 . In B. subtilis, the RNA degradosome is likely composed of RNase Y, RNase J1, RNase J2, and some other proteins 14 . The orthologs of RNase E are universally present in cyanobacteria, while the orthologs of RNase Y are absent (Table 1). The noncatalytic regions of cyanobacterial RNase E proteins are much shorter than those of E. coli RNase E, and their primary sequences show no detectable similarity, making it impossible to predict whether an E. coli‐like RNA degradosome also exists in cyanobacteria. However, recent works have clearly shown that an RNase E‐based RNA degradosome is present in cyanobacteria (Figure 2A). PNPase was the first to be copurified with the noncatalytic region of RNase E from Anabaena PCC 7120 cells, and it interacts with a highly conserved nonapeptide at the very end of the C‐terminus of RNase E within subregion C4 35 . A further copurification assay using the full‐length RNase E identified another RNase E‐associated ribonuclease, RNase II 44 . Fluorescence resonance energy transfer (FRET) analysis using a strain in which RNase E and RNase II were tagged with BFP and CFP respectively revealed that RNase E and RNase II form a compact complex in vivo 44 . In vitro assays showed that RNase E binds to RNase II at a stoichiometry of 1:1 via the catalytic region 44 . As RNase E interacts with PNPase and RNase II via its noncatalytic region and catalytic region, respectively, the three ribonucleases are likely to form one complex in vivo. Note that the RNase E‐based RNA degradosomes of other bacteria only have one exoribonuclease, PNPase; the benefit of two exoribonucleases coexisting in the degradosome remains unclear. The RNA helicase CrhB was recently shown to interact with the catalytic region of RNase E, suggesting that it might be another component of the cyanobacterial RNA degradosome 172 . The same study also showed that the enolase also interacted with the catalytic region of RNase E, but such an interaction needs to be supported by more evidence.
Taken together, these discoveries clearly show that an RNase E‐based RNA degradosome exists in Anabaena PCC 7120. It is very likely that similar RNA degradosomes also exist in other cyanobacteria, as RNase E (particularly, the catalytic region and the PNPase‐interacting motif), PNPase, and RNase II are all highly conserved in cyanobacteria, and CrhB orthologs are also present in most cyanobacterial species. Moreover, these proteins showed overlapping fractionation patterns in the Grad‐seq analysis of Synechocystis PCC 6803 (Figure 4). Note that cyanobacterial RNase E has several other conserved motifs of unknown function in the noncatalytic region, and it is possible that the functions of these motifs may be related to interactions with some other unknown degradosomal components or with RNA substrates.
The RNA degradosomes from E. coli, B. subtilis, and Anabaena all contain at least one endoribonuclease, one exoribonuclease, and an RNA helicase, indicating that the concerted actions of these ribonucleases and the helicase are important for the efficient turnover of cellular RNAs. PNPase is present in all known RNase E‐based RNA degradosomes and the PNPase‐binding sites in the RNase E proteins of E. coli, C. crescentus, and Anabaena PCC 7120 have been determined 17 , 35 , 195 . Despite the fact that these sites are quite divergent in their amino acid sequences, they are all located at the very end of RNase E, implying that the interaction between PNPase and the very end of RNase E is required for efficient cooperation between PNPase and RNase E.
In different organisms, the importance of RNA degradosome may vary significantly. Deletion of the noncatalytic region of E. coli RNase E, which is the scaffold for degradosome assembly, did not lose viability under lab conditions, although it significantly increased the stability of cellular mRNAs 9 . Additionally, among all E. coli degradosomal components, only RNase E is indispensable 10 , 151 , 196 , 197 , 198 . The deletion of RhlB or PNPase only influences the stability of certain RNA species 10 . In B. subtilis, all the ribonucleases in the degradosome are dispensable, although the deletion of RNase Y or RNase J1 severely impairs cell growth 16 , 151 , 199 . Among the known degradosomal components in cyanobacteria, RNase E is essential for cell viability 47 , 48 , a characteristic similar to that of E. coli. PNPase could not be inactivated in Synechocystis PCC 6803 and Synechococcus PCC 7002 47 , 130 , and our attempt to inactivate the PNPase‐encoding gene in Anabaena PCC 7120 also failed (unpublished), indicating that PNPase is essential in cyanobacteria. This result contrasts with the finding that deletion of E. coli pnp only slightly affects cell growth 148 , 200 . The importance of RNase II and CrhB in cyanobacteria remains unclear, but they may be less important than RNase E and PNPase, as their orthologs were shown to be dispensable in some unicellular cyanobacteria 130 , 159 .
PERSPECTIVES
More than 10 RNA turnover‐related enzymes, including ribonucleases and RNA helicases, have been identified in cyanobacteria by genome analysis. Cyanobacteria are evolutionarily distant from E. coli and B. subtilis 21 . Because both E. coli and B. subtilis have some species‐specific ribonucleases, it is anticipated that cyanobacteria also have such unique enzymes that will need to be discovered experimentally. According to the general principle of RNA degradation in bacteria, the enzymes for at least two activities remain to be identified in cyanobacteria: oligoribonuclease and RNA pyrophosphohydrolase.
Oligoribonuclease. Bacterial RNA degradation starts with internal cleavage, and the generated intermediates are further degraded by exoribonucleolytic enzymes from either the 3ʹ or 5ʹ end. However, most exoribonucleases are not able to fully degrade the substrates as their activities can only turn the RNA fragments into oligoribonucleotides of 2–5 nt. These oligoribonucleotides, also known as nanoRNAs, are toxic at least due to their ability to alter global gene expression by priming transcription initiation 201 . Therefore, these oligoribonucleotides need to be converted into single ribonucleotides by oligoribonucleases. The first bacterial oligoribonuclease, Orn, was discovered in E. coli 202 . Orn is required for cell viability, and its depletion leads to a dramatic accumulation of oligoribonucleotides with a length of 2–5 nt 203 . B. subtilis has no Orn homologs; however, it contains two distinct oligoribonucleases (NrnA and NrnB) that can complement the E. coli orn mutant well 204 , 205 . The homologs of all these known oligoribonucleases are missing in cyanobacteria, indicating that cyanobacteria may use a different type of enzyme for the degradation of oligoribonucleotides.
RNA pyrophosphohydrolase. Bacterial primary transcripts, which have a triphosphate 5ʹ end, are not efficiently recognized by decay‐initiating enzymes, such as E. coli RNase E and B. subtilis RNase Y. Therefore, these RNAs generally need to be converted into the 5ʹ monophosphorylated form before degradation. This conversion requires the activity of RNA pyrophosphohydrolase. The first RNA pyrophosphohydrolase, RppH, was discovered in E. coli 206 . RppH belongs to the Nudix hydrolase family protein. B. subtilis has no close homologs of E. coli RppH, but its genome encodes several proteins of the Nudix hydrolase family. One of the Nudix hydrolases, YtkD, was found to have the RNA pyrophosphohydrolase activity and hence was reannotated as B. subtilis RppH (BsRppH) 207 . The close homologs of E. coli RppH and B. subtilis RppH are all absent in cyanobacteria. However, each sequenced cyanobacterium has multiple Nudix hydrolase family proteins, and it is worth testing whether some of them have RNA pyrophosphohydrolase activity.
Although ribonucleases of the same type from different bacteria usually have similar catalytic properties, their cellular substrates are not the same; thus, they can have very different impacts on the growth and physiology in vivo. For instance, PNPase is not essential and only important for the growth of E. coli at lower temperatures 148 , it is however obligately required for cell viability under normal conditions in cyanobacteria 47 , 130 . To understand how ribonucleases function in cyanobacteria, it is necessary to investigate the phenotypes of their mutation (or conditional mutation) and overexpression strains and to determine their cellular substrates.
Cyanobacteria have evolved a unique set of RNA‐degrading enzymes. Our understanding of how these enzymes function in cyanobacteria is still very preliminary. In addition, cyanobacteria may contain proteins that regulate the activities of RNA degradation enzymes. These regulatory proteins, such as RraA and RraB, have been found to have important functions in E. coli 208 , 209 . Continuing to uncover their regulation, cellular substrates, physiological functions, and cooperation mechanisms will help us develop a better understanding of RNA metabolism in cyanobacteria.
ACKNOWLEDGMENTS
This study is supported by the National Natural Science Foundation of China (grant No. 32070037), the Featured Institute Service Project from the Institute of Hydrobiology, the Chinese Academy of Sciences (grant No. Y85Z061601), and the German Science Foundation (DFG) research training group BioInMe 322977937/GRK2344 and grant HE 2544/14‐2 (to Wolfgang R. Hess).
Zhang J‐Y, Hess WR, Zhang C‐C. “Life is short, and art is long”: RNA degradation in cyanobacteria and model bacteria. mLife. 2022;1:21–39. 10.1002/mlf2.12015
Edited by Li Huang, Institute of Microbiology, Chinese Academy of Sciences
Contributor Information
Wolfgang R. Hess, Email: wolfgang.hess@biologie.uni-freiburg.de.
Cheng‐Cai Zhang, Email: cczhang@ihb.ac.cn.
REFERENCES
- 1. Bernstein JA, Khodursky AB, Lin PH, Lin‐Chao S, Cohen SN. Global analysis of mRNA decay and abundance in Escherichia coli at single‐gene resolution using two‐color fluorescent DNA microarrays. Proc Natl Acad Sci USA. 2002;99:9697–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Selinger DW, Saxena RM, Cheung KJ, Church GM, Rosenow C. Global RNA half‐life analysis in Escherichia coli reveals positional patterns of transcript degradation. Genome Res. 2003;13:216–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Steglich C, Lindell D, Futschik M, Rector T, Steen R, Chisholm SW. Short RNA half‐lives in the slow‐growing marine cyanobacterium Prochlorococcus . Genome Biol. 2010;11:R54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mohanty BK, Kushner SR. Regulation of mRNA Decay in Bacteria. Annu Rev Microbiol. 2016;70:25–44. [DOI] [PubMed] [Google Scholar]
- 5. Bechhofer DH, Deutscher MP. Bacterial ribonucleases and their roles in RNA metabolism. Crit Rev Biochem Mol Biol. 2019;54:242–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hui MP, Foley PL, Belasco JG. Messenger RNA degradation in bacterial cells. Annu Rev Genet. 2014;48:537–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Carpousis AJ. The RNA degradosome of Escherichia coli: an mRNA‐degrading machine assembled on RNase E. Annu Rev Microbiol. 2007;61:71–87. [DOI] [PubMed] [Google Scholar]
- 8. Murashko ON, Lin‐Chao S. Escherichia coli responds to environmental changes using enolasic degradosomes and stabilized DicF sRNA to alter cellular morphology. Proc Natl Acad Sci USA. 2017;114:E8025–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lopez PJ, Marchand I, Joyce SA, Dreyfus M. The C‐terminal half of RNase E, which organizes the Escherichia coli degradosome, participates in mRNA degradation but not rRNA processing in vivo . Mol Microbiol. 1999;33:188–99. [DOI] [PubMed] [Google Scholar]
- 10. Bernstein JA, Lin PH, Cohen SN, Lin‐Chao S. Global analysis of Escherichia coli RNA degradosome function using DNA microarrays. Proc Natl Acad Sci USA. 2004;101:2758–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Durand S, Gilet L, Bessières P, Nicolas P, Condon C. Three essential ribonucleases‐RNase Y, J1, and III‐control the abundance of a majority of Bacillus subtilis mRNAs. PLoS Genet. 2012;8:e10025‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lehnik‐Habrink M, Newman J, Rothe FM, Solovyova AS, Rodrigues C, Herzberg C, et al. RNase Y in Bacillus subtilis: a natively disordered protein that is the functional equivalent of RNase E from Escherichia coli. J Bacteriol. 2011;193:5431–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Laalami S, Zig L, Putzer H. Initiation of mRNA decay in bacteria. Cell Mol Life Sci. 2014;71:1799–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Commichau FM, Rothe FM, Herzberg C, Wagner E, Hellwig D, Lehnik‐Habrink M, et al. Novel activities of glycolytic enzymes in Bacillus subtilis: interactions with essential proteins involved in mRNA processing. Mol Cell Proteomics. 2009;8:1350–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mathy N, Bénard L, Pellegrini O, Daou R, Wen T, Condon C. 5ʹ‐to‐3ʹ exoribonuclease activity in bacteria: role of RNase J1 in rRNA maturation and 5ʹ stability of mRNA. Cell. 2007;129:681–92. [DOI] [PubMed] [Google Scholar]
- 16. Figaro S, Durand S, Gilet L, Cayet N, Sachse M, Condon C. Bacillus subtilis mutants with knockouts of the genes encoding ribonucleases RNase Y and RNase J1 are viable, with major defects in cell morphology, sporulation, and competence. J Bacteriol. 2013;195:2340–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hardwick SW, Chan VS, Broadhurst RW, Luisi BF. An RNA degradosome assembly in Caulobacter crescentus . Nucleic Acids Res. 2011;39:1449–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Płociński P, Macios M, Houghton J, Niemiec E, Płocińska R, Brzostek A, et al. Proteomic and transcriptomic experiments reveal an essential role of RNA degradosome complexes in shaping the transcriptome of Mycobacterium tuberculosis . Nucleic Acids Res. 2019;47:5892–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tejada‐Arranz A, Galtier E, El Mortaji L, Turlin E, Ershov D, De Reuse H. The RNase J‐based RNA degradosome is compartmentalized in the gastric pathogen Helicobacter pylori . mBio. 2020;11:e01173‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Raj R, Nadig S, Patel T, Gopal B. Structural and biochemical characteristics of two Staphylococcus epidermidis RNase J paralogs RNase J1 and RNase J2. J Biol Chem. 2020;295:16863–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dickerson RE. Cytochrome c and the evolution of energy metabolism. Sci Am. 1980;242:137–53. [PubMed] [Google Scholar]
- 22. Garcia‐Pichel F. Solar ultraviolet and the evolutionary history of cyanobacteria. Orig Life Evol Biosph. 1998;28:321–47. [DOI] [PubMed] [Google Scholar]
- 23. Rasmussen B, Fletcher IR, Brocks JJ, Kilburn MR. Reassessing the first appearance of eukaryotes and cyanobacteria. Nature. 2008;455:1101–4. [DOI] [PubMed] [Google Scholar]
- 24. Hagemann M, Hess WR. Systems and synthetic biology for the biotechnological application of cyanobacteria. Curr Opin Biotechnol. 2018;49:94–9. [DOI] [PubMed] [Google Scholar]
- 25. Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, et al. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein‐coding regions. DNA Res. 1996;3:109–36. [DOI] [PubMed] [Google Scholar]
- 26. Kaneko T. Complete genomic sequence of the filamentous nitrogen‐fixing cyanobacterium Anabaena sp. strain PCC 7120. DNA Res. 2001;8:227–53. [DOI] [PubMed] [Google Scholar]
- 27. Tejada‐Arranz A, de Crécy‐Lagard V, de Reuse H. Bacterial RNA degradosomes: molecular machines under tight control. Trends Biochem Sci. 2020;45:42–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Aït‐Bara S, Carpousis AJ. RNA degradosomes in bacteria and chloroplasts: classification, distribution and evolution of RNase E homologs. Mol Microbiol. 2015;97:1021–35. [DOI] [PubMed] [Google Scholar]
- 29. Cohen SN, McDowall KJ. RNase E: still a wonderfully mysterious enzyme. Mol Microbiol. 1997;23:1099–106. [DOI] [PubMed] [Google Scholar]
- 30. Callaghan AJ, Marcaida MJ, Stead JA, McDowall KJ, Scott WG, Luisi BF. Structure of Escherichia coli RNase E catalytic domain and implications for RNA turnover. Nature. 2005;437:1187–91. [DOI] [PubMed] [Google Scholar]
- 31. McDowall KJ, Cohen SN. The N‐terminal domain of the rne gene product has RNase E activity and is non‐overlapping with the arginine‐rich RNA‐binding site. J Mol Biol. 1996;255:349–55. [DOI] [PubMed] [Google Scholar]
- 32. Vanzo NF, Li YS, Py B, Blum E, Higgins CF, Raynal LC, et al. Ribonuclease E organizes the protein interactions in the Escherichia coli RNA degradosome. Genes Dev. 1998;12:2770–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Khemici V, Poljak L, Luisi BF, Carpousis AJ. The RNase E of Escherichia coli is a membrane‐binding protein. Mol Microbiol. 2008;70:799–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kaberdin VR, Miczak A, Jakobsen JS, Lin‐Chao S, McDowall KJ, von Gabain A. The endoribonucleolytic N‐terminal half of Escherichia coli RNase E is evolutionarily conserved in Synechocystis sp. and other bacteria but not the C‐terminal half, which is sufficient for degradosome assembly. Proc Natl Acad Sci USA. 1998;95:11637–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhang JY, Deng XM, Li FP, Wang L, Huang QY, Zhang CC, et al. RNase E forms a complex with polynucleotide phosphorylase in cyanobacteria via a cyanobacterial‐specific nonapeptide in the noncatalytic region. RNA. 2014;20:568–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. McDowall KJ, Lin‐Chao S, Cohen SN. A+U content rather than a particular nucleotide order determines the specificity of RNase E cleavage. J Biol Chem. 1994;269:10790–6. [PubMed] [Google Scholar]
- 37. Lin‐Chao S, Wong TT, McDowall KJ, Cohen SN. Effects of nucleotide sequence on the specificity of rne‐dependent and RNase E‐mediated cleavages of RNA I encoded by the pBR322 plasmid. J Biol Chem. 1994;269:10797–803. [PubMed] [Google Scholar]
- 38. Kaberdin VR. Probing the substrate specificity of Escherichia coli RNase E using a novel oligonucleotide‐based assay. Nucleic Acids Res. 2003;31:4710–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chao Y, Li L, Girodat D, Forstner KU, Said N, Corcoran C, et al. In vivo cleavage map illuminates the central role of RNase E in coding and non‐coding RNA pathways. Mol Cell. 2017;65:39–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mackie GA. Ribonuclease E is a 5ʹ‐end‐dependent endonuclease. Nature. 1998;395:720–3. [DOI] [PubMed] [Google Scholar]
- 41. Kime L, Jourdan SS, Stead JA, Hidalgo‐Sastre A, McDowall KJ. Rapid cleavage of RNA by RNase E in the absence of 5ʹ monophosphate stimulation. Mol Microbiol. 2010;76:590–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Anupama K, Leela JK, Gowrishankar J. Two pathways for RNase E action in Escherichia coli in vivo and bypass of its essentiality in mutants defective for Rho‐dependent transcription termination. Mol Microbiol. 2011;82:1330–48. [DOI] [PubMed] [Google Scholar]
- 43. Behler J, Sharma K, Reimann V, Wilde A, Urlaub H, Hess WR. The host‐encoded RNase E endonuclease as the crRNA maturation enzyme in a CRISPR‐Cas subtype III‐Bv system. Nat Microbiol. 2018;3:367–77. [DOI] [PubMed] [Google Scholar]
- 44. Zhou C, Zhang J, Hu X, Li C, Wang L, Huang Q, et al. RNase II binds to RNase E and modulates its endoribonucleolytic activity in the cyanobacterium Anabaena PCC 7120. Nucleic Acids Res. 2020;48:3922–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bayas CA, Wang J, Lee MK, Schrader JM, Shapiro L, Moerner WE. Spatial organization and dynamics of RNase E and ribosomes in Caulobacter crescentus . Proc Natl Acad Sci USA. 2018;115:E3712–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Höfer K, Schauerte M, Grawenhoff J, Wulf A, Welp LM, Billau FA, et al. Viral ADP‐ribosyltransferases attach RNA chains to host proteins. bioRxiv. 2021. 10.1101/2021.06.04.446905. [DOI] [Google Scholar]
- 47. Cameron JC, Gordon GC, Pfleger BF. Genetic and genomic analysis of RNases in model cyanobacteria. Photosynth Res. 2015;126:171–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cavaiuolo M, Chagneau C, Laalami S, Putzer H. Impact of RNase E and RNase J on global mRNA metabolism in the cyanobacterium Synechocystis PCC6803. Front Microbiol. 2020;11:1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yan H, Cheng Y, Wang L, Chen W. Function analysis of RNase E in the filamentous cyanobacterium Anabaena sp. PCC 7120. Res Microbiol. 2020;171:194–202. [DOI] [PubMed] [Google Scholar]
- 50. Hoffmann UA, Heyl F, Rogh SN, Wallner T, Backofen R, Hess WR, et al. Transcriptome‐wide in vivo mapping of cleavage sites for the compact cyanobacterial ribonuclease E reveals insights into its function and substrate recognition. Nucleic Acids Res. 2021;49:13075–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. McDowall KJ, Hernandez RG, Lin‐Chao S, Cohen SN. The ams‐1 and rne‐3071 temperature‐sensitive mutations in the ams gene are in close proximity to each other and cause substitutions within a domain that resembles a product of the Escherichia coli mre locus. J Bacteriol. 1993;175:4245–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Moll I, Afonyushkin T, Vytvytska O, Kaberdin VR, Bläsi U. Coincident Hfq binding and RNase E cleavage sites on mRNA and small regulatory RNAs. RNA. 2003;9:1308–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Göpel Y, Papenfort K, Reichenbach B, Vogel J, Görke B. Targeted decay of a regulatory small RNA by an adaptor protein for RNase E and counteraction by an anti‐adaptor RNA. Genes Dev. 2013;27:552–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Göpel Y, Khan MA, Görke B. Ménage à trois: post‐transcriptional control of the key enzyme for cell envelope synthesis by a base‐pairing small RNA, an RNase adaptor protein, and a small RNA mimic. RNA Biol. 2014;11:433–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Khan MA, Görke B. A multifunctional small RNA binding protein for sensing and signaling cell envelope precursor availability in bacteria. Microb Cell. 2020;7:139–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Durica‐Mitic S, Göpel Y, Amman F, Görke B. Adaptor protein RapZ activates endoribonuclease RNase E by protein‐protein interaction to cleave a small regulatory RNA. RNA. 2020;26:1198–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Horie Y, Ito Y, Ono M, Moriwaki N, Kato H, Hamakubo Y, et al. Dark‐induced mRNA instability involves RNase E/G‐type endoribonuclease cleavage at the AU‐box and SD sequences in cyanobacteria. Mol Genet Genomics. 2007;278:331–46. [DOI] [PubMed] [Google Scholar]
- 58. Sakurai I, Stazic D, Eisenhut M, Vuorio E, Steglich C, Hess WR, et al. Positive regulation of psbA gene expression by cis‐encoded antisense RNAs in Synechocystis sp. PCC 6803. Plant Physiol. 2012;160:1000–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Georg J, Dienst D, Schürgers N, Wallner T, Kopp D, Stazic D, et al. The small regulatory RNA SyR1/PsrR1 controls photosynthetic functions in cyanobacteria. Plant Cell. 2014;26:3661–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lindell D, Jaffe JD, Coleman ML, Futschik ME, Axmann IM, Rector T, et al. Genome‐wide expression dynamics of a marine virus and host reveal features of co‐evolution. Nature. 2007;449:83–6. [DOI] [PubMed] [Google Scholar]
- 61. Stazic D, Pekarski I, Kopf M, Lindell D, Steglich C. A novel strategy for exploitation of host RNase E activity by a marine cyanophage. Genetics. 2016;203:1149–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Court DL, Gan J, Liang YH, Shaw GX, Tropea JE, Costantino N, et al. RNase III: genetics and function; structure and mechanism. Annu Rev Genet. 2013;47:405–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Gordon GC, Cameron JC, Pfleger BF. Distinct and redundant functions of three homologs of RNase III in the cyanobacterium Synechococcus sp. strain PCC 7002. Nucleic Acids Res. 2018;46:1984–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Young RA, Steitz JA. Complementary sequences 1700 nucleotides apart form a ribonuclease III cleavage site in Escherichia coli ribosomal precursor RNA. Proc Natl Acad Sci USA. 1978;75:3593–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Bubunenko M, Court DL, Al Refaii A, Saxena S, Korepanov A, Friedman DI, et al. Nus transcription elongation factors and RNase III modulate small ribosome subunit biogenesis in Escherichia coli . Mol Microbiol. 2013;87:382–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Gao Y, Gong Y, Xu X. RNase III‐dependent down‐regulation of ftsH by an artificial internal sense RNA in Anabaena sp. PCC 7120. FEMS Microbiol Lett. 2013;344:130–7. [DOI] [PubMed] [Google Scholar]
- 67. Kindler P, Keil TU, Hofschneider PH. Isolation and characterization of a ribonuclease 3 deficient mutant of Escherichia coli . Mol Gen Genet. 1973;126:53–9. [DOI] [PubMed] [Google Scholar]
- 68. Price B, Adamidis T, Kong R, Champness WA. Streptomyces coelicolor antibiotic regulatory gene, absB, encodes an RNase III homolog. J Bacteriol. 1999;181:6142–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Viegas SC, Pfeiffer V, Sittka A, Silva IJ, Vogel J, Arraiano CM. Characterization of the role of ribonucleases in Salmonella small RNA decay. Nucleic Acids Res. 2007;35:7651–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Herskovitz MA, Bechhofer DH. Endoribonuclease RNase III is essential in Bacillus subtilis . Mol Microbiol. 2000;38:1027–33. [DOI] [PubMed] [Google Scholar]
- 71. Durand S, Gilet L, Condon C. The essential function of B. subtilis RNase III is to silence foreign toxin genes. PLoS Genet. 2012;8:e1003181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Commichau FM, Stülke J. A mystery unraveled: essentiality of RNase III in Bacillus subtilis is caused by resident prophages. PLoS Genet. 2012;8:e1003199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Georg J, Hess WR. Widespread antisense transcription in prokaryotes. Microbiol Spectr. 2018;8:56470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Dühring U, Axmann IM, Hess WR, Wilde A. An internal antisense RNA regulates expression of the photosynthesis gene isiA . Proc Natl Acad Sci USA. 2006;103:7054–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Lasa I, Toledo‐Arana A, Gingeras TR. An effort to make sense of antisense transcription in bacteria. RNA Biol. 2012;9:1039–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Olmedo‐Verd E, Brenes‐Álvarez M, Vioque A, Muro‐Pastor AM. A heterocyst‐specific antisense RNA contributes to metabolic reprogramming in Nostoc sp. PCC 7120. Plant Cell Physiol. 2019;60:1646–55. [DOI] [PubMed] [Google Scholar]
- 77. Kim KS, Manasherob R, Cohen SN. YmdB: a stress‐responsive ribonuclease‐binding regulator of E. coli RNase III activity. Genes Dev. 2008;22:3497–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Kim KS, Manasherob R, Cohen SN. Identification of macrodomain proteins as novel O‐acetyl‐ADP‐ribose deacetylases. J Biol Chem. 2011;286:13261–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Redko Y, Bechhofer DH. Condon C. Mini‐III, an unusual member of the RNase III family of enzymes, catalyses 23S ribosomal RNA maturation in B. subtilis . Mol Microbiol. 2008;68:1096–106. [DOI] [PubMed] [Google Scholar]
- 80. Redko Y, Condon C. Ribosomal protein L3 bound to 23S precursor rRNA stimulates its maturation by Mini‐III ribonuclease. Mol Microbiol. 2009;71:1145–54. [DOI] [PubMed] [Google Scholar]
- 81. Hotto AM, Castandet B, Gilet L, Higdon A, Condon C, Stern DB. Arabidopsis chloroplast mini‐ribonuclease III participates in rRNA maturation and intron recycling. Plant Cell. 2015;27:724–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Hotto AM, Castandet B, Gilet L, Higdon A, Condon C, Stern DB. Sequence‐specific cleavage of dsRNA by Mini‐III RNase. Nucleic Acids Res. 2015;43:2864–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Gil R, Silva FJ, Peretó J, Moya A. Determination of the core of a minimal bacterial gene set. Microbiol Mol Biol Rev. 2004;68:518–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Gil R, Silva FJ, Peretó J, Moya A. Role of Escherichia coli YbeY, a highly conserved protein, in rRNA processing. Mol Microbiol. 2010;78:506–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Davies BW, Köhrer C, Jacob AI, Simmons LA, Zhu J, Aleman LM, et al. The highly conserved bacterial RNase YbeY is essential in Vibrio cholerae, playing a critical role in virulence, stress regulation, and RNA processing. PLoS Pathog. 2014;10:e1004175‐18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Babu VMP, Sankari S, Budnick JA, Caswell CC, Walker GC. Sinorhizobium meliloti YbeY is a zinc‐dependent single‐strand specific endoribonuclease that plays an important role in 16S ribosomal RNA processing. Nucleic Acids Res. 2020;48:332–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Babu V, Sankari S, Budnick JA, Caswell CC, Walker GC. Endoribonuclease YbeY is essential for RNA processing and virulence in Pseudomonas aeruginosa . mBio. 2020;11:e00659‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Xia Y, Weng Y, Xu C, Wang D, Pan X, Tian Z, et al. Sinorhizobium meliloti YbeY is an endoribonuclease with unprecedented catalytic features, acting as silencing enzyme in riboregulation. Nucleic Acids Res. 2017;45:1371–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Saramago M, Peregrina A, Robledo M, Matos RG, Hilker R, Serrania J, et al. YbeY controls the type III and type VI secretion systems and biofilm formation through RetS in Pseudomonas aeruginosa . Appl Environ Microbiol. 2020;45:1371–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Thomas M, White RL, Davis RW. Hybridization of RNA to double‐stranded DNA: formation of R‐loops. Proc Natl Acad Sci USA. 1976;73:2294–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Rowen L, Kornberg A. Primase, the dnaG protein of Escherichia coli. An enzyme which starts DNA chains. J Biol Chem. 1978;253:758–64. [PubMed] [Google Scholar]
- 92. Rowen L, Kornberg A. Abundant ribonucleotide incorporation into DNA by yeast replicative polymerases. Proc Natl Acad Sci USA. 2010;107:4949–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Yao NY, Schroeder JW, Yurieva O, Simmons LA, O'Donnell ME. Cost of rNTP/dNTP pool imbalance at the replication fork. Proc Natl Acad Sci USA. 2013;110:12942–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Tadokoro T, Kanaya S. Ribonuclease H: molecular diversities, substrate binding domains, and catalytic mechanism of the prokaryotic enzymes. FEBS J. 2009;276:1482–93. [DOI] [PubMed] [Google Scholar]
- 95. Haruki M, Tsunaka Y, Morikawa M, Kanaya S. Cleavage of a DNA‐RNA‐DNA/DNA chimeric substrate containing a single ribonucleotide at the DNA‐RNA junction with prokaryotic RNases HII. FEBS Lett. 2002;531:204–8. [DOI] [PubMed] [Google Scholar]
- 96. Nowotny M, Gaidamakov SA, Crouch RJ, Yang W. Crystal structures of RNase H bound to an RNA/DNA hybrid: substrate specificity and metal‐dependent catalysis. Cell. 2005;121:1005–16. [DOI] [PubMed] [Google Scholar]
- 97. Ohtani N, Haruki M, Morikawa M, Crouch RJ, Itaya M, Kanaya S. Identification of the genes encoding mn2+‐dependent RNase HII and mg2+‐dependent RNase HIII from Bacillus subtilis: classification of RNases H into three families. Biochemistry. 1999;38:605–18. [DOI] [PubMed] [Google Scholar]
- 98. Ohtani N, Haruki M, Morikawa M, Kanaya S. Molecular diversities of RNases H. J Biosci Bioeng. 1999;88:12–9. [DOI] [PubMed] [Google Scholar]
- 99. Ogawa T, Pickett GG, Kogoma T, Kornberg A. RNase H confers specificity in the dnaA‐dependent initiation of replication at the unique origin of the Escherichia coli chromosome in vivo and in vitro . Proc Natl Acad Sci USA. 1984;81:1040–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Hong X, Cadwell GW, Kogoma T. Escherichia coli RecG and RecA proteins in R‐loop formation. EMBO J. 1995;14:2385–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. McDonald JP, Vaisman A, Kuban W, Goodman MF, Woodgate R. Mechanisms employed by Escherichia coli to prevent ribonucleotide incorporation into genomic DNA by Pol V. PLoS Genet. 2012;8:e1003030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Itaya M, Crouch RJ. A combination of RNase H (rnh) and recBCD or sbcB mutations in Escherichia coli K12 adversely affects growth. Mol Gen Genet. 1991;227:424–32. [DOI] [PubMed] [Google Scholar]
- 103. Kouzminova EA, Kadyrov FF, Kuzminov A. RNase HII saves rnhA mutant Escherichia coli from R‐loop‐associated chromosomal fragmentation. J Mol Biol. 2017;429:2873–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Kouzminova EA, Kadyrov FF, Kuzminov A. Ribonucleases J1 and J2: two novel endoribonucleases in B. subtilis with functional homology to E. coli RNase E. Nucleic Acids Res. 2005;33:2141–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Even S, Pellegrini O, Zig L, Labas V, Vinh J, Bréchemmier‐Baey D, et al. Bacillus subtilis ribonucleases J1 and J2 form a complex with altered enzyme behaviour. Mol Microbiol. 2010;75:489–98. [DOI] [PubMed] [Google Scholar]
- 106. Mathy N, Hébert A, Mervelet P, Bénard L, Dorléans A, Li de la Sierra‐Gallay I, et al. Molecular basis for the recognition and cleavage of RNA by the bifunctional 5ʹ‐3ʹ exo/endoribonuclease RNase. J. Structure. 2011;19:1252–61. [DOI] [PubMed] [Google Scholar]
- 107. Newman JA, Hewitt L, Rodrigues C, Solovyova A, Harwood CR, Lewis RJ. Unusual, dual endo‐ and exonuclease activity in the degradosome explained by crystal structure analysis of RNase J1. Structure. 2011;19:1241–51. [DOI] [PubMed] [Google Scholar]
- 108. Newman JA, Hewitt L, Rodrigues C, Solovyova A, Harwood CR, Lewis RJ. The torpedo effect in Bacillus subtilis: RNase J1 resolves stalled transcription complexes. EMBO J. 2020;39:e102500‐51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Redder P, Linder P. New range of vectors with a stringent 5‐fluoroorotic acid‐based counterselection system for generating mutants by allelic replacement in Staphylococcus aureus . Appl Environ Microbiol. 2012;78:3846–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Linder P, Lemeille S, Redder P. Transcriptome‐wide analyses of 5ʹ‐ends in RNase J mutants of a gram‐positive pathogen reveal a role in RNA maturation, regulation and degradation. PLoS Genet. 2014;10:e1004207‐3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Linder P, Lemeille S, Redder P. A minimal bacterial RNase J‐based degradosome is associated with translating ribosomes. Nucleic Acids Res. 2013;41:288–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Redko Y, Aubert S, Stachowicz A, Lenormand P, Namane A, Darfeuille F, et al. A minimal bacterial RNase J‐based degradosome is associated with translating ribosomes. Iron deprivation in Synechocystis: inference of pathways, non‐coding RNAs, and regulatory elements from comprehensive expression profiling. G3 (Bethesda). 2012;2:1475–95.23275872 [Google Scholar]
- 113. Georg J, Rosana ARR, Chamot D, Migur A, Hess WR, Owttrim GW. Inactivation of the RNA helicase CrhR impacts a specific subset of the transcriptome in the cyanobacterium Synechocystis sp. PCC 6803. RNA Biol. 2019;16:1205–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Kopf M, Klähn S, Scholz I, Matthiessen JK, Hess WR, Voß B. Comparative analysis of the primary transcriptome of Synechocystis sp. PCC 6803. DNA Res. 2014;21:527–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Bi Y, Pei G, Sun T, Chen Z, Chen L, Zhang W. Regulation mechanism mediated by trans‐encoded sRNA Nc117 in short chain alcohols tolerancein Synechocystis sp. PCC 6803. Front Microbiol. 2018;9:863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Bi Y, Pei G, Sun T, Chen Z, Chen L, Zhang W. The essential gene set of a photosynthetic organism. Proc Natl Acad Sci USA. 2015;112:E6634–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Halpert M, Liveanu V, Glaser F, Schuster G. The Arabidopsis chloroplast RNase J displays both exo‐ and robust endonucleolytic activities. Plant Mol Biol. 2019;99:17–29. [DOI] [PubMed] [Google Scholar]
- 118. Halpert M, Liveanu V, Glaser F, Schuster G. Identification of genes required for embryo development in Arabidopsis . Plant Physiol. 2004;135:1206–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Chen H, Zou W, Zhao J. Ribonuclease J is required for chloroplast and embryo development in Arabidopsis . J Exp Bot. 2015;66:2079–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Sharwood RE, Halpert M, Luro S, Schuster G, Stern DB. Chloroplast RNase J compensates for inefficient transcription termination by removal of antisense RNA. RNA. 2011;17:2165–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Luro S, Germain A, Sharwood RE, Stern DB. RNase J participates in a pentatricopeptide repeat protein‐mediated 5ʹ end maturation of chloroplast mRNAs. Nucleic Acids Res. 2013;41:9141–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Coburn GA, Mackie GA. Overexpression, purification, and properties of Escherichia coli ribonuclease II. J Biol Chem. 1996;271:1048–53. [DOI] [PubMed] [Google Scholar]
- 123. Cheng ZF, Deutscher MP. An important role for RNase R in mRNA decay. Mol Cell. 2005;17:313–8. [DOI] [PubMed] [Google Scholar]
- 124. Cheng ZF, Deutscher MP. Escherichia coli RNase R has dual activities, helicase and RNase. J Bacteriol. 2010;192:1344–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Cheng ZF, Zuo Y, Li Z, Rudd KE, Deutscher MP. The vacB gene required for virulence in Shigella flexneri and Escherichia coli encodes the exoribonuclease RNase R. J Biol Chem. 1998;273:14077–80. [DOI] [PubMed] [Google Scholar]
- 126. Marujo PE, Hajnsdorf E, Le Derout J, Andrade R, Arraiano CM, Régnier P. RNase II removes the oligo(A) tails that destabilize the rpsO mRNA of Escherichia coli . RNA. 2000;6:1185–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Cheng ZF, Deutscher MP. Purification and characterization of the Escherichia coli exoribonuclease RNase R. Comparison with RNase II. J Biol Chem. 2002;277:21624–9. [DOI] [PubMed] [Google Scholar]
- 128. Oussenko IA, Bechhofer DH. The yvaJ gene of Bacillus subtilis encodes a 3ʹ‐to‐5ʹ exoribonuclease and is not essential in a strain lacking polynucleotide phosphorylase. J Bacteriol. 2000;182:2639–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Matos RG, Fialho AM, Giloh M, Schuster G, Arraiano CM. The rnb gene of Synechocystis PCC6803 encodes a RNA hydrolase displaying RNase II and not RNase R enzymatic properties. PLoS One. 2012;7:e32690‐42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Rott R, Zipor G, Portnoy V, Liveanu V, Schuster G. RNA polyadenylation and degradation in cyanobacteria are similar to the chloroplast but different from Escherichia coli . J Biol Chem. 2003;278:15771–7. [DOI] [PubMed] [Google Scholar]
- 131. Oussenko IA, Abe T, Ujiie H, Muto A, Bechhofer DH. Participation of 3ʹ‐to‐5ʹ exoribonucleases in the turnover of Bacillus subtilis mRNA. J Bacteriol. 2005;187:2758–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Beuf L, Bédu S, Cami B, Joset F. A protein is involved in accessibility of the inhibitor acetazolamide to the carbonic anhydrase(s) in the cyanobacterium Synechocystis PCC 6803. Plant Mol Biol. 1995;27:779–88. [DOI] [PubMed] [Google Scholar]
- 133. Mohanty BK, Kushner SR. Bacterial/archaeal/organellar polyadenylation. Wiley Interdiscip Rev: RNA. 2011;2:256–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Lin‐Chao S, Chiou NT, Schuster G. The PNPase, exosome and RNA helicases as the building components of evolutionarily‐conserved RNA degradation machines. J Biomed Sci. 2007;14:523–32. [DOI] [PubMed] [Google Scholar]
- 135. Donovan WP, Kushner SR. Polynucleotide phosphorylase and ribonuclease II are required for cell viability and mRNA turnover in Escherichia coli K‐12. Proc Natl Acad Sci USA. 1986;83:120–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Coburn GA, Mackie GA. Reconstitution of the degradation of the mRNA for ribosomal protein S20 with purified enzymes. J Mol Biol. 1998;279:1061–74. [DOI] [PubMed] [Google Scholar]
- 137. Py B, Higgins CF, Krisch HM, Carpousis AJ. A DEAD‐box RNA helicase in the Escherichia coli RNA degradosome. Nature. 1996;381:169–72. [DOI] [PubMed] [Google Scholar]
- 138. Lin PH, Lin‐Chao S. RhlB helicase rather than enolase is the beta‐subunit of the Escherichia coli polynucleotide phosphorylase (PNPase)‐exoribonucleolytic complex. Proc Natl Acad Sci USA. 2005;102:16590–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Hajnsdorf E, Steier O, Coscoy L, Teysset L, Régnier P. Roles of RNase E. RNase II and PNPase in the degradation of the rpsO transcripts of Escherichia coli: stabilizing function of RNase II and evidence for efficient degradation in an ams pnp rnb mutant. EMBO J. 1994;13:3368–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Mohanty BK, Kushner SR. Genomic analysis in Escherichia coli demonstrates differential roles for polynucleotide phosphorylase and RNase II in mRNA abundance and decay. Mol Microbiol. 2003;50:645–58. [DOI] [PubMed] [Google Scholar]
- 141. Cheng ZF, Deutscher MP. Quality control of ribosomal RNA mediated by polynucleotide phosphorylase and RNase R. Proc Natl Acad Sci USA. 2003;100:6388–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Pobre V, Arraiano CM. Next generation sequencing analysis reveals that the ribonucleases RNase II, RNase R and PNPase affect bacterial motility and biofilm formation in E. coli . BMC Genomics. 2015;16:72–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Zangrossi S, Briani F, Ghisotti D, Regonesi ME, Tortora P, Dehò G. Transcriptional and post‐transcriptional control of polynucleotide phosphorylase during cold acclimation in Escherichia coli . Mol Microbiol. 2000;36:1470–80. [DOI] [PubMed] [Google Scholar]
- 144. Zangrossi S, Briani F, Ghisotti D, Regonesi ME, Tortora P, Dehò G. Long‐term survival of Campylobacter jejuni at low temperatures is dependent on polynucleotide phosphorylase activity. Appl Environ Microbiol. 2009;75:7310–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Bryant RE, Sypherd PS. Genetic analysis of cold‐sensitive ribosome maturation mutants of Escherichia coli . J Bacteriol. 1974;117:1082–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Dammel CS, Noller HF. A cold‐sensitive mutation in 16S rRNA provides evidence for helical switching in ribosome assembly. Genes Dev. 1993;7:660–70. [DOI] [PubMed] [Google Scholar]
- 147. Zhou Z, Deutscher MP. An essential function for the phosphate‐dependent exoribonucleases RNase PH and polynucleotide phosphorylase. J Bacteriol. 1997;179:4391–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Awano N, Inouye M, Phadtare S. RNase activity of polynucleotide phosphorylase is critical at low temperature in Escherichia coli and is complemented by RNase II. J Bacteriol. 2008;190:5924–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Sulthana S, Deutscher MP. Multiple exoribonucleases catalyze maturation of the 3ʹ terminus of 16S ribosomal RNA (rRNA). J Biol Chem. 2013;288:12574–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Yamanaka K, Inouye M. Selective mRNA degradation by polynucleotide phosphorylase in cold shock adaptation in Escherichia coli . J Bacteriol. 2001;183:2808–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Luttinger A, Hahn J, Dubnau D. Polynucleotide phosphorylase is necessary for competence development in Bacillus subtilis . Mol Microbiol. 1996;19:343–56. [DOI] [PubMed] [Google Scholar]
- 152. Goverde RL, Huis in't Veld JH, Kusters JG, Mooi FR. The psychrotrophic bacterium Yersinia enterocolitica requires expression of pnp, the gene for polynucleotide phosphorylase, for growth at low temperature (5 degrees C). Mol Microbiol. 1998;28:555–69. [DOI] [PubMed] [Google Scholar]
- 153. Mohanty BK, Kushner SR. Polynucleotide phosphorylase functions both as a 3ʹ right‐arrow 5ʹ exonuclease and a poly(A) polymerase in Escherichia coli . Proc Natl Acad Sci USA. 2000;97:11966–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Miczak A, Kaberdin VR, Wei CL, Lin‐Chao S. Proteins associated with RNase E in a multicomponent ribonucleolytic complex. Proc Natl Acad Sci USA. 1996;93:3865–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Prud'homme‐Généreux A, Beran RK, Iost I, Ramey CS, Mackie GA, Simons RW. Physical and functional interactions among RNase E, polynucleotide phosphorylase and the cold‐shock protein, CsdA: evidence for a 'cold shock degradosome'. Mol Microbiol. 2004;54:1409–21. [DOI] [PubMed] [Google Scholar]
- 156. Redder P, Hausmann S, Khemici V, Yasrebi H, Linder P. Bacterial versatility requires DEAD‐box RNA helicases. FEMS Microbiol Rev. 2015;39:392–412. [DOI] [PubMed] [Google Scholar]
- 157. Whitford DS, Whitman BT, Owttrim GW. Genera specific distribution of DEAD‐box RNA helicases in cyanobacteria. Microb Genom. 2021;7:000517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Chamot D, Colvin KR, Kujat‐Choy SL, Owttrim GW. RNA structural rearrangement via unwinding and annealing by the cyanobacterial RNA helicase, CrhR. J Biol Chem. 2005;280:2036–44. [DOI] [PubMed] [Google Scholar]
- 159. Chamot D, Colvin KR, Kujat‐Choy SL, Owttrim GW. An RNA helicase, CrhR, regulates the low‐temperature‐inducible expression of heat‐shock genes groES, groEL1 and groEL2 in Synechocystis sp. PCC 6803. Microbiology. 2010;156:442–51. [DOI] [PubMed] [Google Scholar]
- 160. Rosana AR, Chamot D, Owttrim GW. Autoregulation of RNA helicase expression in response to temperature stress in Synechocystis sp. PCC 6803. PLoS One. 2012;7:e48683‐451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Tarassova OS, Chamot D, Owttrim GW. Conditional, temperature‐induced proteolytic regulation of cyanobacterial RNA helicase expression. J Bacteriol. 2014;196:1560–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Rowland JG, Simon WJ, Prakash JS, Slabas AR. Proteomics reveals a role for the RNA helicase crhR in the modulation of multiple metabolic pathways during cold acclimation of Synechocystis sp. PCC6803. J Proteome Res. 2011;10:3674–89. [DOI] [PubMed] [Google Scholar]
- 163. Rowland JG, Simon WJ, Prakash JS, Slabas AR. RNA helicase, CrhR is indispensable for the energy redistribution and the regulation of photosystem stoichiometry at low temperature in Synechocystis sp. PCC6803. Biochim Biophys Acta. 2012;1817:1525–36. [DOI] [PubMed] [Google Scholar]
- 164. Sireesha K, Radharani B, Krishna PS, Sreedhar N, Subramanyam R, Mohanty P, et al. Inactivation of a low temperature‐induced RNA helicase in Synechocystis sp. PCC 6803: physiological and morphological consequences. Plant Cell Physiol. 2012;53:646–58. [DOI] [PubMed] [Google Scholar]
- 165. Rosana AR, Ventakesh M, Chamot D, Patterson‐Fortin LM, Tarassova O, Espie GS, et al. The temperature‐regulated DEAD‐box RNA helicase CrhR interactome: autoregulation and photosynthesis‐related transcripts. J Exp Bot. 2021;72:7564–79. [DOI] [PubMed] [Google Scholar]
- 166. Kujat SL, Owttrim GW. Redox‐regulated RNA helicase expression. Plant Physiol. 2000;124:703–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Kujat SL, Owttrim GW. RNA helicase‐regulated processing of the Synechocystis rimO‐crhR operon results in differential cistron expression and accumulation of two sRNAs. J Biol Chem. 2020;295:6372–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Yu E, Owttrim GW. Characterization of the cold stress‐induced cyanobacterial DEAD‐box protein CrhC as an RNA helicase. Nucleic Acids Res. 2000;28:3926–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Chamot D, Magee WC, Yu E, Owttrim GW. A cold shock‐induced cyanobacterial RNA helicase. J Bacteriol. 1999;181:1728–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Chamot D, Owttrim GW. Regulation of cold shock‐induced RNA helicase gene expression in the cyanobacterium Anabaena sp. strain PCC 7120. J Bacteriol. 2000;182:1251–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. El‐Fahmawi B, Owttrim GW. Polar‐biased localization of the cold stress‐induced RNA helicase, CrhC, in the cyanobacterium Anabaena sp. strain PCC 7120. Mol Microbiol. 2003;50:1439–48. [DOI] [PubMed] [Google Scholar]
- 172. Yan H, Qin X, Wang L, Chen W. Both enolase and the DEAD‐Box RNA helicase CrhB can form complexes with RNase E in Anabaena sp. strain PCC 7120. Appl Environ Microbiol. 2020;86:e004251‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Vogel J, Luisi BF. Hfq and its constellation of RNA. Nat Rev Microbiol. 2011;9:578–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Santiago‐Frangos A, Woodson SA. Hfq chaperone brings speed dating to bacterial sRNA. Wiley Interdiscip Rev: RNA. 2018;9:e1475‐589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Valentin‐Hansen P, Eriksen M, Udesen C. The bacterial Sm‐like protein Hfq: a key player in RNA transactions. Mol Microbiol. 2004;51:1525–33. [DOI] [PubMed] [Google Scholar]
- 176. Kavita K, de Mets F, Gottesman S. New aspects of RNA‐based regulation by Hfq and its partner sRNAs. Curr Opin Microbiol. 2018;42:53–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Kavita K, de Mets F, Gottesman S. Grad‐seq guides the discovery of ProQ as a major small RNA‐binding protein. Proc Natl Acad Sci USA. 2016;113:11591–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Holmqvist E, Vogel J. RNA‐binding proteins in bacteria. Nat Rev Microbiol. 2018;16:601–15. [DOI] [PubMed] [Google Scholar]
- 179. Holmqvist E, Vogel J. Binding of the RNA chaperone Hfq to the type IV pilus base is crucial for its function in Synechocystis sp. PCC 6803. Mol Microbiol. 2014;92:840–52. [DOI] [PubMed] [Google Scholar]
- 180. Axmann IM, Kensche P, Vogel J, Kohl S, Herzel H, Hess WR. Identification of cyanobacterial non‐coding RNAs by comparative genome analysis. Genome Biol. 2005;6:R73–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Bøggild A, Overgaard M, Valentin‐Hansen P, Brodersen DE. Cyanobacteria contain a structural homologue of the Hfq protein with altered RNA‐binding properties. FEBS J. 2009;276:3904–15. [DOI] [PubMed] [Google Scholar]
- 182. Bøggild A, Overgaard M, Valentin‐Hansen P, Brodersen DE. The cyanobacterial homologue of the RNA chaperone Hfq is essential for motility of Synechocystis sp. PCC 6803. Microbiology. 2008;154:3134–43. [DOI] [PubMed] [Google Scholar]
- 183. Dienst D, Dühring U, Mollenkopf HJ, Vogel J, Golecki J, Hess WR, et al. A cyanobacterial component required for pilus biogenesis affects the exoproteome. mBio. 2021;12:e03674‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Yegorov Y, Sendersky E, Zilberman S, Nagar E, Waldman Ben‐Asher H, Shimoni E, et al. An experimentally anchored map of transcriptional start sites in the model cyanobacterium Synechocystis sp. PCC6803. Proc Natl Acad Sci USA. 2011;108:2124–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Mitschke J, Vioque A, Haas F, Hess WR, Muro‐Pastor AM. Dynamics of transcriptional start site selection during nitrogen stress‐induced cell differentiation in Anabaena sp. PCC7120. Proc Natl Acad Sci USA. 2011;108:20130–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Mitschke J, Vioque A, Haas F, Hess WR, Muro‐Pastor AM. The sRNA NsiR4 is involved in nitrogen assimilation control in cyanobacteria by targeting glutamine synthetase inactivating factor IF7. Proc Natl Acad Sci USA. 2015;112:E6243–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Álvarez‐Escribano I, Vioque A, Muro‐Pastor AM. NsrR1, a nitrogen stress‐repressed sRNA, contributes to the regulation of nblA in Nostoc sp. PCC 7120. Front Microbiol. 2018;9:2267–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Zhan J, Steglich C, Scholz I, Hess WR, Kirilovsky D. Inverse regulation of light harvesting and photoprotection is mediated by a 3ʹ‐end‐derived sRNA in cyanobacteria. Plant Cell. 2021;33:358–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Zhan J, Steglich C, Scholz I, Hess WR, Kirilovsky D. Grad‐seq in a Gram‐positive bacterium reveals exonucleolytic sRNA activation in competence control. EMBO J. 2020;39:e103852‐380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Riediger M, Spät P, Bilger R, Voigt K, Maček B, Hess WR. Analysis of a photosynthetic cyanobacterium rich in internal membrane systems via gradient profiling by sequencing (Grad‐seq). Plant Cell. 2021;33:248–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Kumar M, Gromiha MM, Raghava GP. SVM based prediction of RNA‐binding proteins using binding residues and evolutionary information. J Mol Recognit. 2011;24:303–13. [DOI] [PubMed] [Google Scholar]
- 192. Olejniczak M, Jiang X, Basczok MM, Storz G. KH domain proteins: another family of bacterial RNA matchmakers? Mol Microbiol. 2022;117:10–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Scholz I, Lange SJ, Hein S, Hess WR, Backofen R. CRISPR‐Cas systems in the cyanobacterium Synechocystis sp. PCC6803 exhibit distinct processing pathways involving at least two Cas6 and a Cmr2 protein. PLoS One. 2013;8:e56470‐269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Bandyra KJ, Bouvier M, Carpousis AJ, Luisi BF. The social fabric of the RNA degradosome. Biochim Biophys Acta. 2013;1829:514–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Callaghan AJ, Aurikko JP, Ilag LL, Günter Grossmann J, Chandran V, Kühnel K, et al. Studies of the RNA degradosome‐organizing domain of the Escherichia coli ribonuclease RNase E. J Mol Biol. 2004;340:965–79. [DOI] [PubMed] [Google Scholar]
- 196. Apirion D. Isolation, genetic mapping and some characterization of a mutation in Escherichia coli that affects the processing of ribonuleic acid. Genetics. 1978;90:659–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Ono M, Kuwano M. A conditional lethal mutation in an Escherichia coli strain with a longer chemical lifetime of messenger RNA. J Mol Biol. 1979;129:343–57. [DOI] [PubMed] [Google Scholar]
- 198. Khemici V, Poljak L, Toesca I, Carpousis AJ. Evidence in vivo that the DEAD‐box RNA helicase RhlB facilitates the degradation of ribosome‐free mRNA by RNase E. Proc Natl Acad Sci USA. 2005;102:6913–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Britton RA, Wen T, Schaefer L, Pellegrini O, Uicker WC, Mathy N, et al. Maturation of the 5' end of Bacillus subtilis 16S rRNA by the essential ribonuclease YkqC/RNase J1. Mol Microbiol. 2007;63:127–38. [DOI] [PubMed] [Google Scholar]
- 200. Wang W, Bechhofer DH. Properties of a Bacillus subtilis polynucleotide phosphorylase deletion strain. J Bacteriol. 1996;178:2375–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Goldman SR, Sharp JS, Vvedenskaya IO, Livny J, Dove SL, Nickels BE. NanoRNAs prime transcription initiation in vivo . Mol Cell. 2011;42:817–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. Niyogi SK, Datta AK. A novel oligoribonuclease of Escherichia coli. I. Isolation and properties. J Biol Chem. 1975;250:7307–12. [PubMed] [Google Scholar]
- 203. Ghosh S, Deutscher MP. Oligoribonuclease is an essential component of the mRNA decay pathway. Proc Natl Acad Sci USA. 1999;96:4372–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Mechold U, Fang G, Ngo S, Ogryzko V, Danchin A. YtqI from Bacillus subtilis has both oligoribonuclease and pAp‐phosphatase activity. Nucleic Acids Res. 2007;35:4552–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205. Fang M, Zeisberg WM, Condon C, Ogryzko V, Danchin A, Mechold U. Degradation of nanoRNA is performed by multiple redundant RNases in Bacillus subtilis . Nucleic Acids Res. 2009;37:5114–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206. Deana A, Celesnik H, Belasco JG. The bacterial enzyme RppH triggers messenger RNA degradation by 5' pyrophosphate removal. Nature. 2008;451:355–8. [DOI] [PubMed] [Google Scholar]
- 207. Richards J, Liu Q, Pellegrini O, Celesnik H, Yao S, Bechhofer DH, et al. An RNA pyrophosphohydrolase triggers 5'5'‐exonucleolytic degradation of mRNA in Bacillus subtilis . Mol Cell. 2011;43:940–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208. Lee K, Zhan X, Gao J, Qiu J, Feng Y, Meganathan R, et al. RraA. A protein inhibitor of RNase E activity that globally modulates RNA abundance in E. coli . Cell. 2003;114:623–34. [PubMed] [Google Scholar]
- 209. Gao J, Lee K, Zhao M, Qiu J, Zhan X, Saxena A, et al. Differential modulation of E. coli mRNA abundance by inhibitory proteins that alter the composition of the degradosome. Mol Microbiol. 2006;61:394–406. [DOI] [PubMed] [Google Scholar]


