Abstract
The co‐occurrence of plasmid‐mediated multidrug resistance and hypervirulence in epidemic carbapenem‐resistant Klebsiella pneumoniae has emerged as a global public health issue. In this study, an ST23 carbapenem‐resistant hypervirulent K. pneumoniae (CR‐HvKP) strain VH1‐2 was identified from cucumber in China and harbored a novel hybrid plasmid pVH1‐2‐VIR. The plasmid pVH1‐2‐VIR carrying both virulence and multidrug‐resistance (MDR) genes was likely generated through the recombination of a virulence plasmid and an IncFIIK conjugative MDR plasmid in clinical ST23 18622 isolated from a sputum sample. The plasmid pVH1‐2‐VIR exhibited the capacity for transfer to the clinical ST11 carbapenem‐resistant K. pneumoniae (CRKP) strain via conjugation assay. Acquisition of pVH1‐2‐VIR plasmid directly converted a CRKP into CR‐HvKP strain characterized by hypermucoviscosity, heightened virulence for Galleria mellonella larvae, and increased colonization ability in the mouse intestine. The emergence of such a hybrid plasmid may expedite the spread of CR‐HvKP strains, posing a significant risk to human health.
Keywords: conjugative virulence plasmid, CR‐HvKP, hybrid plasmid, Klebsiella pneumoniae, ST23
Impact statement
An ST23 carbapenem‐resistant and hypervirulent K. pneumoniae (CR‐HvKP) strain VH1‐2 was detected from cucumber in China. VH1‐2 was observed to possess a hybrid virulence plasmid, pVH1‐2‐VIR, which may have arisen from the recombination of a virulence plasmid and an IncFIIK conjugative MDR plasmid in clinical ST23 18622 from a sputum sample. The hypervirulent plasmid pVH1‐2‐VIR was proficient in conjugative transfer to clinical ST11 CRKP, leading to an escalation in virulence and colonization levels. The appearance of such plasmids can be worrisome, which necessitates vigilant observation of their prevalence in the food chain.
INTRODUCTION
Klebsiella pneumoniae is a significant constituent of the human gut microbiota, yet it also serves as a prominent causative agent of invasive nosocomial infections including pyogenic liver abscesses that pose a substantial clinical treatment across the globe, particularly in Asia 1 . Clinical K. pneumoniae strains have undergone a transformation into hypervirulent K. pneumoniae (HvKP) (initially detected in Taiwan, China in 2002) and carbapenem‐resistant K. pneumoniae (CRKP), both of which are currently prevalent in Asia 2 , 3 , 4 . Infection by HvKP strains is the cause of significant life‐threatening liver abscesses or pneumonia thereby resulting in a high morbidity and mortality 5 . CRKP strains are responsible for the majority of (70%–90%) clinical carbapenem‐resistant Enterobacteriaceae (CRE) infections in several countries including China 6 . This has resulted in untreatable or difficult‐to‐treat infections and poses a pressing public health concern globally. Fortunately, most HvKP strains as of now are susceptible to antibiotic therapy.
A significant threat to animal and human health is the acquisition of a carbapenemase‐encoding plasmid by an HvKP strain, which results in a carbapenem‐resistant HvKP (CR‐HvKP) 7 , 8 . A fatal outbreak caused by ST11 CR‐HvKP strains carrying a pLVPK‐like hypervirulence plasmid was reported in China 3 and subsequent CR‐HvKP infections have been documented 9 , 10 . It was recently discovered that the integration of carbapenemase gene bla KPC‐2 into an HvKP virulence plasmid, along with the capsular polysaccharide (CPS) regulator rmpA2, had resulted in the emergence of hyper‐resistance and hyper‐virulence phenotypes 8 . Fortunately, this plasmid was determined to be nontransferable even though it was mobilizable. A recent discovery in China revealed the insertion of a transposon element carryingbla CTX‐M‐24 into a pLVPK‐like hypervirulence plasmid in an ST23 CR‐HvKP 11 . However, the nonconjugative characteristic of pLVPK‐like virulence plasmids has constrained the dissemination levels of CR‐HvKP. The greatest concern, however, is a hybrid virulence plasmid that is conjugative. Moreover, composite pLVPK and IncFIB plasmid has been detected in a clinical Klebsiella variicola isolate, suggesting a potential for increased dissemination of CR‐HvKP 12 . All previously reported CR‐HvKP strains have been isolated solely from hospitalized patients with no instances from animals, environment, or retail food.
The current safety status of the food chain has attracted significant public attention due to contamination risks and its potential to act as a reservoir for antibiotic resistance genes (ARGs). Numerous studies have reported a high prevalence of antibiotic‐resistant bacteria in the global food chain, particularly in retail 13 , 14 , 15 . CRE are also frequently detected in retail meat with some new isolates carrying both bla NDM type carbapenemases and the mobilized colistin resistance gene mcr‐1 16 . The safety of fresh vegetables has also become a growing concern due to the rising number of foodborne disease outbreaks linked to contaminated vegetables. These infections are often caused by bacteria exhibiting hyper‐resistance and hyper‐virulence phenotypes, such as Shiga toxin‐producing Escherichia coli (STEC), which encoded ESBL in sprouts in Germany in 2011 17 . In our previous study, two CRKP isolates were found in ready‐to‐eat vegetables but the virulence potential of these isolates is currently unknown 18 .
The current study presents novel findings on the identification of a conjugative virulence plasmid (undergone hybridization) with a conjugative IncFII MDR plasmid (isolated from cucumber). Acquisition of this virulence plasmid was observed to understand the level of virulence and intestinal colonization.
RESULTS
Genome sequence analysis of K. pneumoniae strains VH1‐2 and 18622
Between May and November 2017, the CR‐HvKP ST23 strains 18622 and VH1‐2 were obtained from an inpatient sputum sample and cucumber, respectively, in the Qingdao region of Shandong province, China. These strains exhibited highly similar pulsed‐field gel electrophoresis (PFGE) patterns (Figure S1). ARG and virulence gene analysis indicated a significant degree of relatedness between strain 18622 and VH1‐2 (Table S1). Altogether, these findings indicate that VH1‐2 and 18622 are phylogenetically closely related.
Whole genome sequencing (WGS) analysis of the genomes of these two strains revealed the presence of three and four plasmid types in VH1‐2 and 18622, respectively (Figure 1A). Isolate 18622 contained bla KPC‐2 on an F35:A‐:B1 type plasmid pCRKP18622‐KPC (134,794 bp, Genbank Accession No. CP027866), which carried aac(6′)‐Ib, bla TEM‐1B, bla CTX‐M‐14, aac(6′)‐Ib‐cr, cmlA1, and iucABCD‐iutA. Notably, isolate VH1‐2 from cucumber contained a highly similar bla KPC‐2‐bearing F35:A‐:B1 type plasmid. The backbone of both bla KPC‐2‐bearing plasmids was organized similar to plasmid p2246‐CTXM (Genbank Accession No. KX646543) carrying bla CTX‐M‐14 from Shigella boydii. The primary component of the multidrug‐resistant (MDR) region on pCRKP18622‐KPC was bla KPC‐2 and bla CTX‐M‐14. The bla KPC‐2 was located in the ΔTn6296 that inserted at a site downstream of ISKpn26 in both strains 18622 and VH1‐2 and differed from the site in p112298‐KPC (Genbank Accession No. NZ_KP987215). The bla CTX‐M‐14 transposition unit was flanked by 5 bp direct repeats (DRs) at both ends, located downstream of bla KPC‐2 and inserted on a reverse ΔTn1721 on plasmid pCRKP18622‐KPC. However, the bla CTX‐M‐14 transposition unit was truncated and further disrupted by ΔIn153 and IS26 elements on plasmid pVH1‐2‐KPC, resulting in the minor differences between pVH1‐2‐KPC and pCRKP18622‐KPC (Figure 1B).
Figure 1.
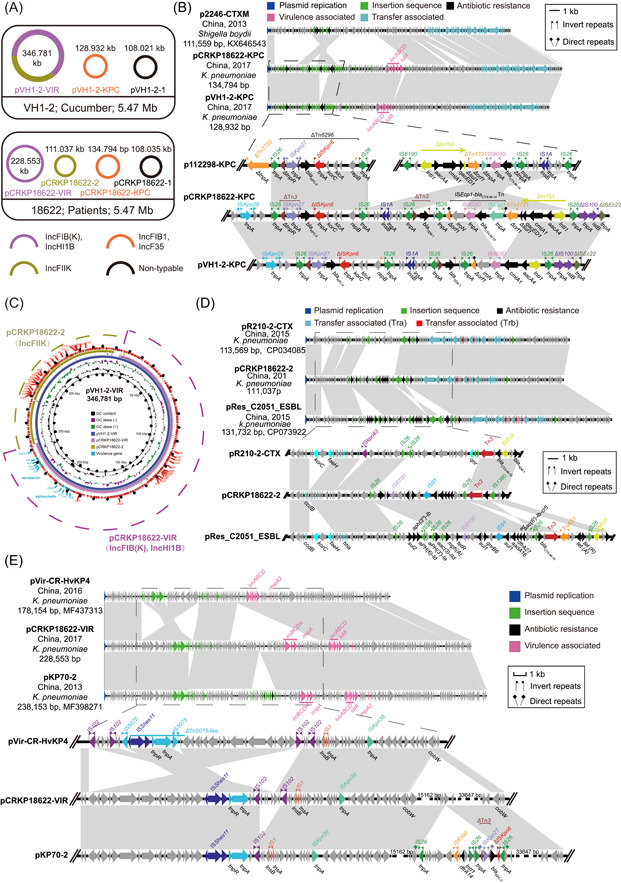
Genome sequence analysis of Klebsiella pneumoniae strains VH1‐2 and 18622. (A) Species, sources, and chromosome sizes of the CRKP isolates. (B) Comparison of plasmids pCRKP18622‐KPC, pVH1‐2‐KPC, and p112298‐KPC. (C) Circular comparison of plasmid pVH1‐2‐VIR, pCRKP18622‐VIR, and pCRKP18622‐2. The plasmid pVH1‐2‐VIR located at the innermost circle was used as the reference plasmid to perform sequence alignment with BLASTn by the BRIG software. Genetic regions associated with virulence are highlighted in cyan. (D) Linear alignment of complete plasmid sequences of pR210‐2‐CTX, pCRKP18622‐2, and pRes_C2051_ESBL. Genes are denoted by arrows and colored based on function classification. Shaded regions denote shared regions of homology (95% nucleotide identity). (E) Linear alignment of complete plasmid sequences of pVir‐CR‐HvKP4, pCRKP18622‐VIR, and pKP70‐2.
Numerous virulence genes were identified on the plasmids in both strains and VH1‐2 possessed rmpA, iroBCDN, iucABCD‐iutA, and rmpA2 on a 346,781 bp IncHI1B‐FIIK2‐FIB(K) plasmid designated as pVH1‐2‐VIR, which also carried ARGs for β‐lactams (bla CTX‐M‐15, bla TEM‐1C), fluoroquinolones (qnrB6, aac(6′)‐Ib‐crcr), aminoglycosides (aadA16), rifampicin (arr‐3), sulfonamides (sul1), macrolide (mph(A)), and trimethoprim (dfrA27). Interestingly, the virulence plasmid pVH1‐2‐VIR covered all the regions of both the IncFIIK resistance plasmid (pCRKP18622‐2) and the IncFIB(K)‐IncHI1B virulence plasmid (pCRKP18622‐VIR) and carried the ARGs aadA16, bla CTX‐M‐15, bla TEM‐1C, aac(6′)‐Ib‐cr, qnrB6, mph(A), arr‐3, sul1, dfrA27 as well as virulence genes iucABCD‐iutA, iroBCDN, rmpA, and rmpA2, respectively (Table S1). Moreover, sequence comparisons indicated that the virulence plasmid pVH1‐2‐VIR was formed by the fusion of pCRKP18622‐2 and pCRKP18622‐VIR carried by strain 18622 (Figure 1C).
The backbone of plasmids pR210‐2‐CTX (Genbank Accession No. CP034085) and pRes‐C2051‐ESBL (Genbank Accession No. CP073922) from K. pneumoniae in the GenBank database were similar to pCRKP18622‐2, especially for pRes‐C2051‐ESBL. Compared with pCRKP18622‐2, the plasmid pRes‐C2051‐ESBL was inserted with a 5036 bp hp‐korc‐hp‐fesH‐hp fragment and a 9221 bp MDR region mediated by IS26 in two identical directions. In particular, a 10,971 bp tetracycline resistance gene region flanked by TnAS1 and IS26 in two opposite directions was inserted in plasmid pRes‐C2051‐ESBL (Figure 1D). The conjugative transfer regions including tra and trb genes were identified in the plasmid pCRKP18622‐2 and pVH1‐2‐VIR (Figure 1C,D).
An identical multidrug resistance region (MRR) of 29,479 bp containing bla CTX‐M‐15, aac (6′)‐Ib‐cr and qnrB6 as well as another six ARGs were found in both pVH1‐2‐VIR and pCRKP18622‐2 and both harbored virulence genes iucABCD‐iutA, iroBCDN, rmpA, and rmpA2. In the GenBank database, there was no highly similar plasmid to pVH1‐2‐VIR whereas pKP70‐2 (Genbank Accession No. MF398271) from a clinical CR‐HvKP isolate in China showed the highest similarity to pCRKP18622‐VIR in this study. The minor differences between these two plasmids were two unique regions and a segment of 3645 bp franked by two IS102 in the same orientation was inserted in pCRKP18622‐VIR. Furthermore, an MDR region of 14,780 bp flanked by two IS26 elements containing 8 bp DRs (CTAAAATT) in the same orientation was inserted between the backbone region in pKP70‐2, suggesting that the insertion of this MDR region into the virulence plasmid pCRKP18622‐VIR was mediated by IS26 to form pKP70‐2 (Figure 1E).
Prevalence of pVH‐2‐VIR‐like plasmids
To investigate the prevalence of such virulence plasmid, we retrospectively screened 45 clinical CRKP strains collected from Qingdao, Beijing, and Kunming in China in 2017–2018. All the 45 clinical CRKP strains that carried bla KPC‐2 and 23 (51.11%) possessed the virulence plasmids harboring rmpA and rmpA2. Both the CRKP isolates (1742 and 1743) from Beijing were CR‐HvKP, possessing rmpA and rmpA2, and 21/45 (46.67%) clinical CRKP strains were classified as CR‐HvKP. WGS analysis indicated that five different ST types were present in the 23 clinical CR‐HvKP strains, among which ST11 (10 strains) and ST23 (10 strains) were the most prevalent (Figure 2A). We also found that all ST11 and ST1764 CR‐HvKP strains lacked the clbA‐S virulence gene cluster, which was found in all examined ST23 and ST65 clinical CR‐HvKP strains. Of note, all 23 CR‐HvKP strains carried the gene cluster iucABCD‐iutA, which was also located in the virulence plasmid pVH1‐2‐VIR harboring rmpA2, and genes ybtA/E/P/Q/S/T/U/X were found in 42 clinical CRKP strains (Figure 2A).
Figure 2.
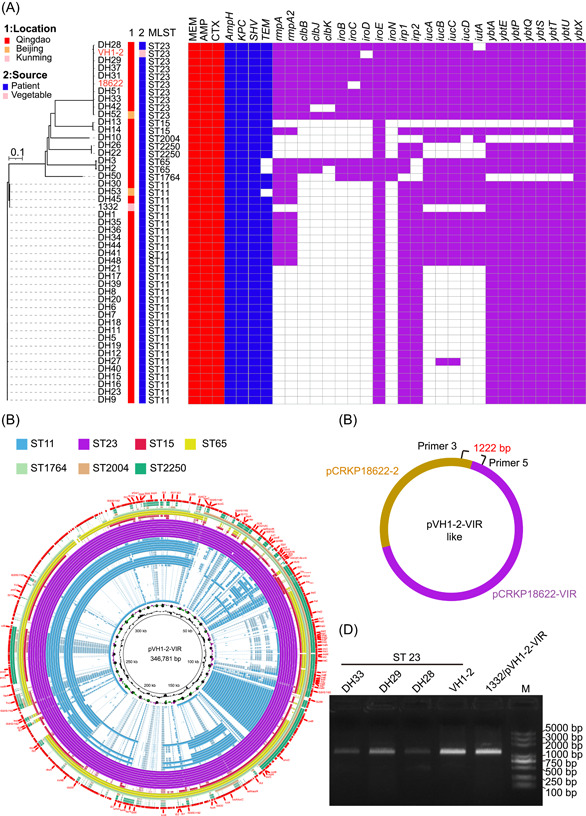
Prevalence of pVH1‐2‐VIR‐like plasmids. (A) Phylogenetic structures, MLST, antibiotic resistance phenotypes, ARGs, and virulence genes of CRKP isolates. Isolate location and sources are indicated. Red and blue squares represent positivity for resistance phenotypes and resistance genes, respectively. Purple squares represent positivity for virulence genes. AMP, ampicillin; CTX, cefotaxime; MEM, meropenem. (B) Sequence alignment of virulence plasmids harboring rmpA and rmpA2 in the 23 clinical CR‐HvKP strains of different STs used in this study. The 346.781 kb virulence plasmid pVH1‐2‐VIR found in this study is used as a reference and genetic regions associated with virulence are highlighted in green. (C) Schematic depicting for investigating potential pVH1‐2‐VIR‐like plasmids. Primers 3 and 5 were used to amplify hybrid regions. (D) PCR confirmed the presence of the pVH1‐2‐VIR‐like plasmids in CRKP isolates. CRKP, carbapenem‐resistant K. pneumoniae.
Owing to its hybrid nature encompassing both the virulence plasmid pCRKP18622‐VIR and resistance plasmid pCRKP18622‐2, the 346.781 kb virulence plasmid pVH1‐2‐VIR was aligned to pVH1‐2‐VIR in 47 CR‐HvKP strains of varying STs. A retrospective analysis of clinical records revealed that the plasmid contigs within the ST23 strains covered nearly all regions of pVH1‐2‐VIR (Figure 2B) indicating that CR‐HvKP from patients exhibited similarity with those from ready‐to‐eat vegetables.
To further investigate the prevalence of the pVH1‐2‐VIR‐like plasmid, two pairs of primers were designed to target the hybrid region for PCR amplification and sequencing (Figure 2C). The results of the PCR amplification and sequencing confirmed that four ST23 isolates from hospital patients in Qingdao, Shandong province, China (DH33, DH29, DH28, and VH1‐2) were positive for the hybrid region (Figure 2D). This indicated that the occurrence of pVH1‐2‐VIR‐like virulence plasmids was not limited to VH1‐2 but was also present in other clinical ST23 CRKP strains.
The pVH1‐2‐VIR plasmid contributes to hypervirulence in clinical ST11 CRKP
Given that ST11 has been the predominant CRKP type in China and the majority of ST11 CRKP strains are not hypervirulent, conjugation experiments were conducted to assess the potential hazard of the pVH1‐2‐VIR plasmid using a clinical ST11 strain as the recipient. The results of the conjugation experiments demonstrated that the pVH1‐2‐VIR plasmid was transmissible to ST11 CRKP1332 from VH1‐2 at a conjugative frequency of 2.2 ± 0.56 × 10−6. Of note, the transconjugant 1332 demonstrated resistance to amikacin while remaining susceptible to rifampicin whereas VH1‐2 exhibited resistance to rifampicin but remained susceptible to amikacin. MIC tests demonstrated that the acquisition of pVH1‐2‐VIR resulted in resistance to amikacin and rifampicin, indicating the successful transfer of pVH1‐2‐VIR into strain 1332. Additionally, both 1332/pVH1‐2‐VIR and 1332 exhibited comparable susceptibilities to meropenem, rifampicin, fosfomycin, gentamicin, ciprofloxacin, tetracycline, and doxycycline (Tables S2 and S3).
Interestingly, the growth curves of the transconjugant 1332/pVH1‐2‐VIR and donor were indistinguishable (Figure 3A). This indicated that the introduction of pVH1‐2‐VIR plasmid did not alter the normal growth of donor 1332. Moreover, since plasmids can introduce fitness costs to their hosts, we performed a quantitative evaluation of the fitness costs linked to pVH1‐2‐VIR carriage. We carried out a competition between isogenic 1332/pVH1‐2‐VIR and 1332 clones. However, the outcomes of these assays indicated that pVH1‐2‐VIR introduced only minor fitness costs (Figure 3B).
Figure 3.
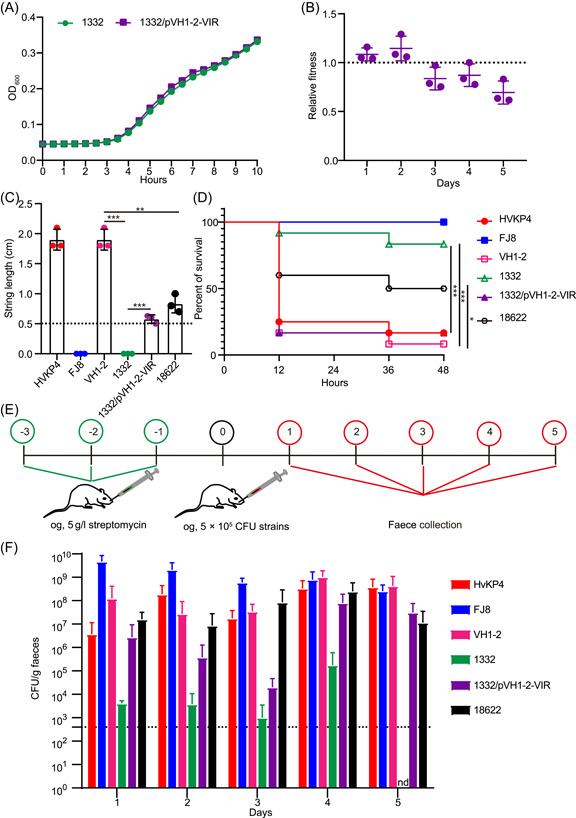
The pVH1‐2‐VIR plasmid contributes to hypervirulence to clinical ST11 CRKP. (A) Growth curve of 1332 and 1332/pVH1‐2‐VIR (n = 3). (B) Relative fitness of pVH1‐2‐VIR plasmid carrying clones compared with pVH1‐2‐VIR‐free clones obtained by competition assays. Bars represent normalized relative fitness after subtracting the effect of pVH1‐2‐VIR (n = 3). (C) String length observed in tested CRKP isolates (n = 3). (D) Virulence potential of CRKP isolates tested in a Galleria mellonella infection model. Survival of mice infected with 5 × 105 CFU of each Klebsiella pneumoniae at 48 h is shown (n = 12). (E) Experimental setup for the intestinal colonization assay. (F) Colonization levels of CRKP isolates in feces at Days 1 to 5 after infection (n = 6). *p < 0.05; **p < 0.01; ***p < 0.001. CFU, colony‐forming unit; CRKP, carbapenem‐resistant K. pneumoniae; nd, not detected.
The virulence potentials of strains 18622, VH1‐2, 1332, and 1332/pVH1‐2‐VIR were also assessed through viscous string assay and the G. mellonella larvae survival assay. Plasmid HVKP4 was used as a hyper‐virulence control and FJ8 as a low‐virulence control 18 . We found that both FJ8 and 1332 generated viscous string <0.5 cm, indicating a low‐mucoviscous phenotype. In contrast, HVKP4, 18622, VH1‐2, and 1332/pVH1‐2‐VIR exhibited a higher degree of mucoviscosity with strings >0.5 cm compared to the low‐virulence control FJ8 (Figure 3C). The virulence potential using G. mellonella larvae indicated that inoculation with 5 × 105 CFU of VH1‐2 strain resulted in a mortality of 91.67% at 48 h and this surpassed that of the hypervirulent control strain HVKP4 (83.33% mortality) and was significantly higher than the low‐virulence control strain FJ8 (0% mortality).
The inoculation of G. mellonella larvae with 5.0 × 105 CFU 1332/pVH1‐2‐VIR resulted in a mortality rate of 83.33% at 48 h and was comparable to that of HVKP4 but higher than that of 1332 (12.67%, p < 0.001). These findings provide further evidence that the acquisition of the pVH1‐2‐VIR plasmid by the ST11 CRKP strain 1332 facilitated its transformation into a hypervirulent strain. In contrast, strain 18622 displayed a lower degree of mucoviscosity (p < 0.01) and mortality (50%, p < 0.05) compared to VH1‐2, suggesting that its virulence level may be lower than that of the VH1‐2 strain (Figure 3D).
The process of gastrointestinal colonization plays a crucial role in the development of healthcare‐associated infections caused by K. pneumoniae strains 19 . To assess the colonization potential of CRKP strains, a streptomycin‐treated mouse model was utilized. Interestingly, despite the seemingly low virulence of strain FJ8, its colonization level was found to be significantly high in the mouse model. Specifically, infection with either 5 × 105 VH1‐2 or 1332/pVH1‐2‐VIR at 5 days postinfection (dpi) resulted in a colonization level of 4.28 × 108 and 3.12 × 107 CFU/g, respectively. However, CRKP strain 1332 was not detected in the feces at 5 dpi. The data indicated that the presence of a pVH1‐2‐VIR plasmid in 1332 resulted in an elevated level of colonization whereas 18622 demonstrated a slightly lower level of colonization compared to VH1‐2 (Figure 3E).
DISCUSSION
The incidence of CR‐HvKP strains has been on the rise, particularly in China 12 , 20 . Previous reports have indicated that the carbapenemase gene bla KPC‐2 and virulence genes rmpA/rmpA2 were typically situated on separate plasmids in most of CR‐HvKP strains 3 , 21 . Plasmids carrying bla KPC‐2, for example pVH1‐2‐KPC, were frequently found to be transferable 6 , 22 . Whereas hypervirulence plasmids, for example pLVPK‐like plasmids, often lacked conjugative regions like tra 20 . These findings prompted the hypothesis that hypervirulent plasmids present in clinical HvKP strains lacked conjugative properties. Consequently, the formation of CR‐HvKP was believed to primarily occur via conjugative bla KPC‐2 plasmid acquisition from HvKP strains. Although pLVPK‐like virulence plasmids were initially deemed nonconjugative, a recent investigation revealed that virulence plasmids could mobilize independently or in conjunction with a helper plasmid at a lower rate 23 . Furthermore, the nonconjugative virulence plasmid could be supplied with transfer functions from a conjugative plasmid during conjugation and mobilize at a higher frequency 24 .
Previous reports have indicated that ARGs, such as bla CTX‐M 11 and bla KPC‐2 8 , are colocated on pLVPK‐like plasmids. However, these plasmids are devoid of conjugative regions. In contrast to previously identified virulence plasmids, a novel conjugative virulence plasmid was recently identified in K. variicola, which integrated a 100 kb virulence fragment into a conjugative IncFIB plasmid. This plasmid was capable of being transferred to various Klebsiella strains via conjugation but did not harbor any ARGs 12 . In scattered clinical CRKP strains, 11 virulence plasmids were identified that were potentially capable of conjugation and contained the transfer region of plasmid IncFII, that is, tra genes. In addition, most of these plasmids also harbored ARGs 25 , 26 while only two have been demonstrated to be transferable. Recently, a conjugative hybrid plasmid of 479 kb was reported in a clinical ST15 CR‐HvKP in China. This plasmid shared regions from a 290 kb virulence plasmid and a conjugative MDR plasmid (188 kb), both of which are present in another clinical ST15 CR‐HvKP 27 .
There have been recent increases in the incidence of foodborne illnesses caused by contaminated vegetables. This is compounded by the presence of hypervirulent MDR organisms in ready‐to‐eat vegetables and poses a significant food safety concern. The current study aims to characterize a CR‐HvKP strain associated with foodborne transmission. We identified an ST23 CR‐HvKP strain from cucumber that harbored a 347 kb conjugative hybrid virulence plasmid. This plasmid was likely formed through the recombination of a virulence plasmid (228 kb) and an IncFIIK conjugative MDR plasmid (111 kb) present in clinical ST23 CRKP strains. The virulence plasmid (pVH1‐2‐VIR) contained IncFIIK, IncFIB(K), and IncHI1B replicons as well as transfer regions tra and trb. These three types of Inc plasmids are predominantly distributed among Enterobacteriaceae, including E. coli, K. pneumoniae, Shigella flexneri, and Salmonella enterica Typhimurium 28 , 29 , 30 . Our conjugation assays demonstrated the transferability of the pVH1‐2‐VIR plasmid to the widely prevalent clinical ST11 CRKP strain. Our conjecture is that these virulence plasmids have the potential to directly disseminate among bacterial pathogens, especially Enterobacteriaceae, resulting in the simultaneous expression of phenotypic carbapenem resistance and high‐level virulence. This could pose a significant threat to human health through the food chain. To the best of our knowledge, this is the first documented report of this type of particularly hypervirulent MDR plasmid, particularly in the context of the food chain.
The ST23 clonal group is associated with hypervirulence and has been identified as a prevalent ST in clinical K. pneumoniae across several countries including China 31 . Examination of virulence genes in CR‐HvKP revealed that all ST23 CR‐HvKP strains contained nearly all virulence genes, resulting in heightened virulence compared to other STs. This finding further supports the notion that ST23 clonal group members are highly virulent. In addition, our findings indicated that CR‐HvKP strains (specifically ST23 VH1‐2) that possess virulence plasmids containing CPS regulatory genes, exhibit greater virulence than CRKP strains lacking such plasmids. This was indicated quantitively here using G. mellonella assays. The hypervirulent potential of the foodborne strain VH1‐2 and its virulence plasmid transconjugant was assessed using the string test, a widely used method for identifying HvKP strains 31 . The results confirmed that VH1‐2 and its transconjugants exhibited high levels of virulence and provided additional evidence that the acquisition of a conjugative hypervirulent plasmid by ST11 1332 contributed to the observed increase in virulence.
The cocarriage of ARGs and virulence factors within a singular virulence vector can facilitate the concurrent transmission of both traits, thereby promoting the rapid emergence and dissemination of multidrug‐resistant hypervirulent K. pneumoniae (MDR HvKP) strains 32 . Our results of colonization assay further confirmed that the acquisition of the virulence plasmid could augment mammalian colonization, thereby posing an unprecedented threat to human health via the food chain. Notably, VH1‐2 exhibited a greater degree of virulence and colonization relative to 18622, suggesting that the fusion of the two plasmids enhanced not only virulence but also bacterial colonization. However, the underlying mechanisms remain unclear and require further investigation.
In brief, this study delineated the emergence of an infrequent conjugative MDR hypervirulent plasmid in an ST23 CR‐HvKP strain isolated from fresh cucumber. This conjugative plasmid has the potential to directly disseminate into human CRKP and contribute to augmented virulence and colonization, thereby posing an unparalleled risk to human health through the food chain. The emergence of conjugative MDR hypervirulent plasmids in pathogens found in ready‐to‐eat food is a concerning trend and it is imperative to closely monitor the prevalence of such plasmids among clinical strains and the food chain.
MATERIALS AND METHODS
Bacterial strains antimicrobial susceptibility testing and PFGE typing
In 2017–2018, a total of 47 isolates were obtained from inpatient sputum and cucumber samples in Qingdao (n = 44), Beijing (n = 2), and Kunming (n = 1), China. The agar dilution method was used to determine the minimum inhibitory concentrations (MIC) for meropenem, cefotaxime, ampicillin, fosfomycin, amikacin, ciprofloxacin, gentamicin, tetracycline, trimethoprim/sulfamethoxazole, rifampicin, and florfenicol. The results were interpreted in accordance with the Clinical and Laboratory Standards Institute (CLSI) guidelines. Broth microdilution method was used to examine the MIC of tigecycline. E. coli ATCC 25922 was used as a quality control strain. Clonal relationships of VH1‐2 and 18622 were investigated using PFGE of XbaI‐digested genomic DNA as previously described 33 .
WGS and phylogenetic analysis
WGS was conducted on 47 selected isolates using the Illumina MiSeq system (Illumina), and the paired‐end Illumina reads were assembled using SPAdes v3.6.2 34 . ARG was identified via ResFinder 3.1 (https://cge.cbs.dtu.dk/services/ResFinder/) while transposon and insertion sequence (IS) elements were predicted using ISfinder (https://www-is.biotoul.fr). The prediction of virulence genes was facilitated with the Virulence Factor Database (http://www.mgc.ac.cn/VFs/). The genome was annotated through the Prokaryotic Genome Annotation Pipeline server. The BLASTn algorithm, integrated in BLAST ring image generator (BRIG) Version 0.95 was employed for sequence comparisons and map generation 35 . In addition, EasyFig. 2.1 (BLASTn, default setting) was utilized for the linear comparison of plasmids 36 .
Identification of pVH1‐2‐VIR‐like plasmid
The CRKP isolates were screened for the presence of pVH1‐2‐VIR‐like hybrid plasmid using primers F3: 5′‐GCACTCCGGAATACATACTGATG‐3′, F5: 5′‐GCAAAAAGGGCGTGAACTTCAGG‐3′. The samples were run with the following PCR program: 95°C 3 min, 95°C 15 s–55°C 15 s–72°C 1 min for 30 cycles, 72°C 5 min. The presence of the targeted gene was confirmed by Sanger sequencing.
Conjugation experiments
The conjugation experiments were conducted following previously established protocols with minor modifications 37 . Specifically, the donor VH1‐2 (ST23, rifampicin‐resistant) and recipient K. pneumoniae 1332 (ST11, amikacin‐resistant) were each inoculated into 5 ml of LB broth at a concentration of 108 colony‐forming units (CFU)/ml and incubated at 37°C. After 6 h of liquid mating, the mixed suspension was subjected to serial dilutions and plated on selective media as follows: (1) LB agar supplemented with rifampicin (128 µg/ml) and amikacin (64 µg/ml) for the enumeration of transconjugants; (2) LB agar containing amikacin (64 µg/ml) for the calculation of recipient cells. The identity of transconjugants was confirmed by PCR assays and ERIC‐PCR 38 . Conjugative frequency was calculated as: γ = T/R, T and R represent the number of transconjugants and recipients per milliliter, respectively 39 . The experiment was performed in triplicate.
Growth curves
The bacterial cells of K. pneumoniae 1332 and 1332/pVH1‐2‐VIR were cultured overnight and subsequently adjusted to a concentration of 5 × 105 CFU/ml with LB broth. The cultures were then incubated at 37°C with shaking at 180 rpm. Growth curves were generated by measuring the optical density at 600 nm for a duration of 10 h utilizing an EnSight Multimode Microplate Reader (PerkinElmer). Prism 8 software (GraphPad) was employed to construct the growth curves.
Competition assay
The K. pneumoniae 1332 and 1332/pVH1‐2‐VIR strains were cultured overnight, washed, and resuspended in LB broth. Subsequently, the cultures were combined in a 1:1 ratio and inoculated at a 1:100 dilution. The resulting mixtures were incubated at 37°C for 1, 2, 3, 4, and 5 days. Following incubation, LB agar supplemented with or without rifampicin (128 µg/ml) was utilized to plate dilutions of the bacterial mixture, enabling differentiation between the rifampicin‐resistant 1332/pVH1‐2‐VIR and 1332 strains. Relative fitness was calculated as the ratio of Malthusian parameters W = log10(1332/pVH1‐2‐VIRend/1332/pVH1‐2‐VIRstart)/log10(1332end/1332start) as previously described 40 . Three independent competition experiments were performed to calculate the Malthusian parameters.
String tests
The methodology for the string test was executed in accordance with the previously published protocol 5 . All CRKP isolates were cultured on agar plates supplemented with 5% sheep blood and incubated at 37°C for one night. The hyper‐mucoviscous phenotype was identified by observing the formation of a viscous string measuring greater than 5 mm in length generated by pulling a single colony upwards using a standard inoculation loop.
The Galleria mellonella infection assay
The G. mellonella infection assay was conducted in accordance with the methodology outlined in a previous study 41 . Each group was comprised of a sample population of 12 larvae weighing 300 ± 50 mg. All CRKP isolates were cultured in LB broth and harvested during the exponential phase. Following a wash with sterile phosphate‐buffered saline (PBS), 10 μl of 5 × 105 CFU bacterial cells were administered via injection into the rear left proleg of the larvae. The survival rate of the G. mellonella larvae was monitored and recorded at regular 12 h intervals over a period of 48 h. Survival was analyzed using the Kaplan–Meier method and implemented with Prism 8.0 (GraphPad).
Mouse colonization assay
Female BALB/c SPF mice (8 weeks old, weighing 20 ± 2 g) were utilized for colonization assays. All mice were housed in the Laboratory Animal Center of Southern Medical University in Guangzhou, China. To overcome colonization resistance, the mice were pretreated with 200 μl streptomycin (5 g/l) via intragastric gavage for 3 consecutive days. The CRKP strains were experimentally introduced as previously described 42 . K. pneumoniae strains were cultured overnight, washed, and adjusted to a concentration of 5 × 107 CFU/ml with sterile PBS. The mice were subjected to oral gavage with 200 µl of 5 × 105 CFU bacterial cells following which their feces were collected on Days 1 to 5 post‐CRKP administration. The level of colonization was assessed by plating on LB agar with 100 μg/ml ampicillin and quantifying CRKP CFU in fecal samples. The colonization assay was conducted in accordance with the guidelines stipulated in the Guide for the Care and Use of Laboratory Animals at the Laboratory Animal Center of South China Agricultural University.
Statistical analysis
Statistical analysis was performed using the software GraphPad Prism version 8.0. The significance of differences (p < 0.05) was determined using the unpaired t test with a two‐tailed nonparametric analysis.
AUTHOR CONTRIBUTIONS
Gong Li: Investigation (lead); methodology (equal); software (equal); validation (equal); visualization (lead); writing—original draft (lead). Ling Jia: Data curation (equal); formal analysis (equal); methodology (equal); validation (supporting); visualization (equal). Lei Wan: Investigation (supporting). Lijuan Xia: Investigation (supporting); methodology (supporting). Ang Gao: Methodology (supporting). Runshi Yang: Investigation (supporting); validation (supporting); visualization (supporting). Ruanyang Sun: Methodology (supporting). Minge Wang: Data curation (supporting); formal analysis (supporting); validation (supporting); visualization (supporting). Juan Du: Formal analysis (supporting); validation (supporting). Xinlei Lian: Software (supporting). Rongmin Zhang: Software (supporting); visualization (supporting). Liangxing Fang: Writing—review and editing (supporting). Xiaoping Liao: Supervision (supporting). Yahong Liu: Funding acquisition (equal); project administration (equal); supervision (lead). Bao‐Tao Liu: Conceptualization (equal); data curation (equal); funding acquisition (supporting); methodology (supporting); project administration (supporting); resources (equal); validation (lead); writing—original draft (equal); writing—review and editing (equal). Jian Sun: Conceptualization (equal); funding acquisition (equal); project administration (equal); resources (supporting); validation (equal); writing—original draft (equal); writing—review and editing (lead).
ETHICS STATEMENT
Animal experimentation was approved by the Ethics Committee of the Laboratory Animal Center of South China Agricultural University (Guangzhou, China), approval number 2022C16.
CONFLICT OF INTERESTS
The authors declare no conflict of interests.
Supporting information
Supporting information.
Supporting information.
ACKNOWLEDGMENTS
This study was supported in part by the Foundation for Innovative Research Groups of the National Natural Science Foundation of China (32121004), Local Innovative and Research Teams Project of Guangdong Pearl River Talents Program (2019BT02N054), Laboratory of Lingnan Modern Agriculture Project (NT2021006), Guangdong Major Project of Basic and Applied Basic Research (grant 2020B0301030007), Innovation Team Project of Guangdong University (2019KCXTD001), the 111 Project (grant D20008), Natural Science Foundation of Shandong Province of China (ZR2022MC001), and the Scientific and Technological Projects of Qingdao (19‐6‐1‐94‐nsh). The hyper‐virulence strain HVKP4 and low‐virulence strain FJ8 were kindly gifted by Professor Rong Zhang, Zhejiang University.
Li G, Jia L, Wan L, Xia L, Gao A, Yang R, et al. Acquisition of a novel conjugative multidrug‐resistant hypervirulent plasmid leads to hypervirulence in clinical carbapenem‐resistant Klebsiella pneumoniae strains. mLife. 2023;2:317–327. 10.1002/mlf2.12086
Edited by Liang Yang, Southern University of Science and Technology, China
Contributor Information
Bao‐Tao Liu, Email: liubaotao-1986@163.com.
Jian Sun, Email: jiansun@scau.edu.cn.
DATA AVAILABILITY
All sequencing data have been deposited in GenBank under BioProject accession number PRJNA885955.
REFERENCES
- 1. Meatherall BL, Gregson D, Ross T, Pitout JDD, Laupland KB. Incidence, risk factors, and outcomes of Klebsiella pneumoniae bacteremia. Am J Med. 2009;122:866–873. [DOI] [PubMed] [Google Scholar]
- 2. Liu YM, Li BB, Zhang YY, Zhang W, Shen H, Li H, et al. Clinical and molecular characteristics of emerging hypervirulent Klebsiella pneumoniae bloodstream infections in mainland China. Antimicrob Agents Chemother. 2014;58:5379–5385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gu D, Dong N, Zheng Z, Lin D, Huang M, Wang L, et al. A fatal outbreak of ST11 carbapenem‐resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect Dis. 2018;18:37–46. [DOI] [PubMed] [Google Scholar]
- 4. Lee CR, Lee JH, Park KS, Jeon JH, Kim YB, Cha CJ, et al. Antimicrobial resistance of hypervirulent Klebsiella pneumoniae: epidemiology, hypervirulence‐associated determinants, and resistance mechanisms. Front Cell Infect Microbiol. 2017;7:483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shon AS, Bajwa RPS, Russo TA. Hypervirulent (hypermucoviscous) Klebsiella pneumoniae: a new and dangerous breed. Virulence. 2013;4:107–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhang R, Liu L, Zhou H, Chan EW, Li J, Fang Y, et al. Nationwide surveillance of clinical carbapenem‐resistant Enterobacteriaceae (CRE) strains in China. EBioMedicine. 2017;19:98–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dong N, Sun Q, Huang Y, Shu L, Ye L, Zhang R, et al. Evolution of carbapenem‐resistant serotype K1 hypervirulent Klebsiella pneumoniae by acquisition of bla (VIM‐1)‐bearing plasmid. Antimicrob Agents Chemother. 2019;63:01056‐19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dong N, Lin D, Zhang R, Chan EW, Chen S. Carriage of bla KPC‐2 by a virulence plasmid in hypervirulent Klebsiella pneumoniae . J Antimicrob Chemoth. 2018;73:3317–3321. [DOI] [PubMed] [Google Scholar]
- 9. Du P, Zhang Y, Chen C. Emergence of carbapenem‐resistant hypervirulent Klebsiella pneumoniae . Lancet Infect Dis. 2018;18:23–24. [DOI] [PubMed] [Google Scholar]
- 10. Wong MHY, Shum HP, Chen JHK, Man MY, Wu A, Chan EWC, et al. Emergence of carbapenem‐resistant hypervirulent Klebsiella pneumoniae . Lancet Infect Dis. 2018;18:24. [DOI] [PubMed] [Google Scholar]
- 11. Shen D, Ma G, Li C, Jia X, Qin C, Yang T, et al. Emergence of a multidrug‐resistant hypervirulent Klebsiella pneumoniae sequence type 23 strain with a rare bla (CTX‐M‐24)‐harboring virulence plasmid. Antimicrob Agents Chemother. 2019;63:e02273‐18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yang X, Wai‐Chi Chan E, Zhang R, Chen S. A conjugative plasmid that augments virulence in Klebsiella pneumoniae . Nat Microbiol. 2019;4:2039–2043. [DOI] [PubMed] [Google Scholar]
- 13. Xie M, Lin D, Chen K, Chan EWC, Yao W, Chen S. Molecular characterization of Escherichia coli strains isolated from retail meat that harbor bla CTX‐M and fosA3 genes. Antimicrob Agents Chemother. 2016;60:2450–2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Casella T, Nogueira MCL, Saras E, Haenni M, Madec JY. High prevalence of ESBLs in retail chicken meat despite reduced use of antimicrobials in chicken production, France. Int J Food Microbiol. 2017;257:271–275. [DOI] [PubMed] [Google Scholar]
- 15. Hayashi W, Ohsaki Y, Taniguchi Y, Koide S, Kawamura K, Suzuki M, et al. High prevalence of blaCTX‐M‐14 among genetically diverse Escherichia coli recovered from retail raw chicken meat portions in Japan. Int J Food Microbiol. 2018;284:98–104. [DOI] [PubMed] [Google Scholar]
- 16. Zhang Q, Lv L, Huang X, Huang Y, Zhuang Z, Lu J, et al. Rapid increase in carbapenemase‐producing Enterobacteriaceae in retail meat driven by the spread of the bla (NDM‐5)‐carrying IncX3 plasmid in China from 2016 to 2018. Antimicrob Agents Chemother. 2019;63:e00573‐19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Buchholz U, Bernard H, Werber D, Böhmer MM, Remschmidt C, Wilking H, et al. German outbreak of Escherichia coli O104:H4 associated with sprouts. N Engl J Med. 2011;365:1763–1770. [DOI] [PubMed] [Google Scholar]
- 18. Dong N, Liu L, Zhang R, Chen K, Xie M, Chan EWC, et al. An IncR plasmid harbored by a hypervirulent carbapenem‐resistant Klebsiella pneumoniae strain possesses five tandem repeats of the bla (KPC‐2)::NTE(KPC)‐Id fragment. Antimicrob Agents Chemother. 2019;63:e01775‐18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Osbelt L, Wende M, Almási É, Derksen E, Muthukumarasamy U, Lesker TR, et al. Klebsiella oxytoca causes colonization resistance against multidrug‐resistant K. pneumoniae in the gut via cooperative carbohydrate competition. Cell Host Microbe. 2021;29 11:1663–1679. [DOI] [PubMed] [Google Scholar]
- 20. Yang X, Dong N, Chan EWC, Zhang R, Chen S. Carbapenem resistance‐encoding and virulence‐encoding conjugative plasmids in Klebsiella pneumoniae . TIM. 2021;29:65–83. [DOI] [PubMed] [Google Scholar]
- 21. Zhao Q, Feng Y, Zong Z. Conjugation of a hybrid plasmid encoding hypervirulence and carbapenem resistance in Klebsiella pneumoniae of sequence type 592. Front Microbiol. 2022;13:852596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liu BT, Zhang XY, Wan SW, Hao JJ, Jiang RD, Song FJ. Characteristics of carbapenem‐resistant Enterobacteriaceae in ready‐to‐eat vegetables in China. Front Microbiol. 2018;9:1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Xu Y, Zhang J, Wang M, Liu M, Liu G, Qu H, et al. Mobilization of the nonconjugative virulence plasmid from hypervirulent Klebsiella pneumoniae . Genome Med. 2021;13:119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Xie M, Chen K, Ye L, Yang X, Xu Q, Yang C, et al. Conjugation of virulence plasmid in clinical Klebsiella pneumoniae strains through formation of a fusion plasmid. Adv Biosyst. 2020;4:1900239. [DOI] [PubMed] [Google Scholar]
- 25. Lam MMC, Wyres KL, Wick RR, Judd LM, Fostervold A, Holt KE, et al. Convergence of virulence and MDR in a single plasmid vector in MDR Klebsiella pneumoniae ST15. J Antimicrob Chemother. 2019;74:1218–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Turton J, Davies F, Turton J, Perry C, Payne Z, Pike R. Hybrid resistance and virulence plasmids in “High‐Risk” clones of Klebsiella pneumoniae, including those carrying bla (NDM‐5) . Microorganisms. 2019;7:326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li R, Cheng J, Dong H, Li L, Liu W, Zhang C, et al. Emergence of a novel conjugative hybrid virulence multidrug‐resistant plasmid in extensively drug‐resistant Klebsiella pneumoniae ST15. Int J Antimicro Ag. 2020;55:105952. [DOI] [PubMed] [Google Scholar]
- 28. de Toro M, Garcilláon‐Barcia MP, De La Cruz F. Plasmid diversity and adaptation analyzed by massive sequencing of Escherichia coli plasmids. Microbiol Spectrum. 2014;2:31. [DOI] [PubMed] [Google Scholar]
- 29. Liebert CA, Hall RM, Summers AO. Transposon Tn21, flagship of the floating genome. Microbiol Mol Biol Rev. 1999;63:507–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sherburne CK. The complete DNA sequence and analysis of R27, a large IncHI plasmid from Salmonella typhi that is temperature sensitive for transfer. Nucleic Acids Res. 2000;28:2177–2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Russo TA, Olson R, Fang CT, Stoesser N, Miller M, MacDonald U, et al. Identification of biomarkers for differentiation of hypervirulent Klebsiella pneumoniae from classical K. pneumoniae . J Clin Microbiol. 2018;56:e00776‐18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yang X, Dong N, Liu X, Yang C, Ye L, Chan EWC, et al. Co‐conjugation of virulence plasmid and KPC plasmid in a clinical Klebsiella pneumoniae strain. Front Microbiol. 2021;12:739461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gautom RK. Rapid pulsed‐field gel electrophoresis protocol for typing of Escherichia coli O157:H7 and other gram‐negative organisms in 1 day. J Clin Microbiol. 1997;35:2977–2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: a new genome assembly algorithm and its applications to single‐cell sequencing. J Comput Biol. 2012;19:455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Alikhan NF, Petty NK, Ben Zakour NL, Beatson SA. BLAST ring image generator (BRIG): simple prokaryote genome comparisons. BMC Genomics. 2011;12:402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sullivan MJ, Petty NK, Beatson SA. Easyfig: a genome comparison visualizer. Bioinformatics. 2011;27:1009–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Neil K, Allard N, Grenier F, Burrus V, Rodrigue S. Highly efficient gene transfer in the mouse gut microbiota is enabled by the Incl(2) conjugative plasmid TP114. Commun Biol. 2020;3:523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. de Bruijn FJ. Use of repetitive (repetitive extragenic palindromic and enterobacterial repetitive intergeneric consensus) sequences and the polymerase chain reaction to fingerprint the genomes of Rhizobium meliloti isolates and other soil bacteria. Appl Environ Microbiol. 1992;58:2180–2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gama JA, Zilhão R, Dionisio F. Conjugation efficiency depends on intra and intercellular interactions between distinct plasmids: plasmids promote the immigration of other plasmids but repress co‐colonizing plasmids. Plasmid. 2017;93:6–16. [DOI] [PubMed] [Google Scholar]
- 40. Hall JPJ, Wright RCT, Harrison E, Muddiman KJ, Wood AJ, Paterson S, et al. Plasmid fitness costs are caused by specific genetic conflicts enabling resolution by compensatory mutation. PLoS Biol. 2021;19:e3001225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. McLaughlin MM, Advincula MR, Malczynski M, Barajas G, Qi C, Scheetz MH. Quantifying the clinical virulence of Klebsiella pneumoniae producing carbapenemase Klebsiella pneumoniae with a Galleria mellonella model and a pilot study to translate to patient outcomes. BMC Infect Dis. 2014;14:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bohnhoff M, Drake BL, Miller CP. Effect of streptomycin on susceptibility of intestinal tract to experimental Salmonella infection. Exp Biol Med. 1954;86:132–137. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information.
Supporting information.
Data Availability Statement
All sequencing data have been deposited in GenBank under BioProject accession number PRJNA885955.


