Abstract
Objectives
We aim to estimate geographic variability in total numbers of infections and infection fatality ratios (IFR; the number of deaths caused by an infection per 1,000 infected people) when the availability and quality of data on disease burden are limited during an epidemic.
Methods
We develop a noncentral hypergeometric framework that accounts for differential probabilities of positive tests and reflects the fact that symptomatic people are more likely to seek testing. We demonstrate the robustness, accuracy, and precision of this framework, and apply it to the United States (U.S.) COVID-19 pandemic to estimate county-level SARS-CoV-2 IFRs.
Results
The estimators for the numbers of infections and IFRs showed high accuracy and precision; for instance, when applied to simulated validation data sets, across counties, Pearson correlation coefficients between estimator means and true values were 0.996 and 0.928, respectively, and they showed strong robustness to model misspecification. Applying the county-level estimators to the real, unsimulated COVID-19 data spanning April 1, 2020 to September 30, 2020 from across the U.S., we found that IFRs varied from 0 to 44.69, with a standard deviation of 3.55 and a median of 2.14.
Conclusions
The proposed estimation framework can be used to identify geographic variation in IFRs across settings.
Keywords: Infection fatality ratio, Infection fatality rate, Noncentral hypergeometric distribution, COVID-19, SARS-CoV-2
1. Introduction
During the course of an epidemic, accurate estimation of the infection fatality ratio (“IFR”) – the number of deaths caused by an infection per 1,000 infected people – is crucial for planning and developing risk mitigation strategies. IFRs may vary by age distribution of the population, distribution of infection across demographic groups, prevalence of underlying health conditions, access to healthcare resources, and other factors (Shen et al., 2021; Wood et al., 2021). Therefore, a flexible framework for estimating IFRs among different populations that vary across geographic regions is essential.
Accurately ascertaining both components of the IFR, the number of deaths and the number of infections, can be challenging (Dana Flanders & Kleinbaum, 1995). While the number of deaths may be under- or over-reported in some settings (Aron et al., 2020), even more problematic are potential inaccuracies in ascertaining the number of infections. While case mortality data may provide an upper bound for the IFR, the total number of infections is likely to be dramatically underestimated because infected people may not be tested or report illness. This issue was well-documented during the COVID-19 pandemic (Campbell et al., 2020; Orin & Topol, 2020; Rubin, 2020). In addition, the availability and uptake of testing, as well as the reliability of testing data, may also vary over time and geography. For example, during the COVID-19 pandemic, the number of SARS-CoV-2 nucleic acid amplification tests (NAATs) administered was not consistently recorded for all counties (The Atlantic, 2021).
Here we develop an inferential framework for estimating area-level variation in IFRs and the total numbers of infections to address common data limitations. We utilize a non-central hypergoemetric model to derive estimators, and validate these estimators with simulations. To demonstrate their application, we use the estimators to assess geographic patterns in the number of SARS-CoV-2 infections and IFRs across the U.S. during the COVID-19 pandemic, finding novel patterns.
2. Methods
2.1. Overview of estimators
2.1.1. Definitions
Our goal is to estimate, over a specified period of time, the total numbers of infections and IFRs for a pathogen within discrete geographic subregions contained within a larger region. For example, the period of time may be a month or year; the subregions may be states or provinces; and the larger region may be the country or continent containing the subregions.
We suppose that infections with the pathogen can be detected reliably with testing, but that there is a bias toward testing infected individuals. Over the period of time, across geographic subregion j, let Tj denote the number of tests for the pathogen that are administered, Dj denote the number of deaths caused by the pathogen, and Cj denote the number of observed disease cases (i.e., positive tests). Let Pj be the population size of the subregion, assumed effectively constant. Let the number of infections (both observed and unobserved) be denoted ιj. Define ωj as the odds ratio of testing an infected vs. an uninfected individual. Let φj be the IFR in subregion j. Across the entire region, as opposed to the subregions, we will denote the total numbers of tests, deaths, and observed cases, and the overall population, number of infections, odds ratio, and IFR without subscripts (i.e., T, D, C, P, ι, ω, and φ, respectively). We denote estimators for parameters with “hat” notation; e.g., an estimator for ω will be denoted by .
2.1.2. General Assumptions
To derive an estimator for the number of infections and IFR in each subregion j, we make three assumptions:
-
1.
Odds ratios (ω and ωj): Although it is not necessary to assume actual values of ω and ωj, it is necessary to assume that the odds ratio of testing an infected vs. an uninfected person is constant across geographic subregions, so that for all subregions i and j, ω = ωi = ωj.
-
2.
Global IFR (φ): To estimate geographic variation in IFR, in our framework it is necessary to assume that the baseline IFR (φ) for the entire region under consideration is known.
-
3.
Observed quantities: we assume that the population (Pj) and numbers of deaths (Dj), observed cases (Cj), and tests (Tj) in each subregion are known, as well as P, D, C, and T for the entire region.
We conceptualize the process by which people are tested with a noncentral hypergeometric model, wherein each test is administered to either a sick or healthy person with some bias toward administering tests to symptomatic people (e.g., because sick people are more likely to seek testing (Campbell et al., 2020); quantified by ω). For estimation, we utilize an approximation for the mean of the Wallenius’ noncentral hypergeometric distribution (Equation 16 in reference Fog, 2008):
| (1) |
and
| (2) |
The noncentral hypergeometric process is appropriate because it results from randomly picking balls without replacement from an urn, where there are two types of balls (in this case sick and healthy people from a finite population) and there is a bias toward picking one type of ball (sick people). Although Expressions (1) and (2) are close approximations, for clarity, in the following we will refer to them as equalities.
2.1.3. Estimators
We develop a moments estimator for ιj based on Expressions (1) and (2). Because by Assumptions 2 and 3 the region-wide IFR (φ) and the total number of deaths (D) are known, a moments estimator for ι is given by
| (3) |
(By definition, φ ≡ 1000 ⋅ D/ι.) A moments estimator for the odds ratio (ω) then follows by plugging C, , T, and P into Expression (1) and solving for ω; that is,
| (4) |
It follows from Assumption 1 that a moments estimator for ιj can be found by substituting for ωj in Expression (2) and solving for ιj; that is, solving the following equation for :
| (5) |
Although there is no general closed form solution for , the appropriate root can easily be found numerically with the constraint that . A moments estimator for the IFR in subregion j is then given by
| (6) |
To estimate the variance of these estimators, a parametric bootstrap can be utilized: that is, parametric bootstrap resamples using the Wallenius’ noncentral hypergeometric model can be generated using the parameter estimates, and estimates of variance and confidence intervals calculated via usual bootstrap methods.
2.2. Validation and application to COVID-19
We now use data from the COVID-19 pandemic to validate and apply these estimators. We utilize the observed numbers of COVID-19 cases and deaths, and the numbers of SARS-CoV-2 NAAT tests from the U.S. between April 1, 2020 and September 30, 2020 as the basis for an extensive numerical analysis assessing the robustness, accuracy, and precision of the estimators. We then estimate the numbers of SARS-CoV-2 infections and IFRs across counties and states in the U.S. during this same period.
2.2.1. Data
For validation and application analyses, we utilize publicly-available case and mortality data from the COVID-19 Data Repository by the Center for Systems Science and Engineering (CSSE) at Johns Hopkins University (Dong et al., 2020). These case and mortality data, which are at a daily temporal resolution, are shifted by 14 days to account for the fact that COVID-19 deaths result from SARS-CoV-2 infections acquired approximately two weeks prior to death rather than at the time of death. We utilize NAAT testing data from The U.S. Covid Atlas (Kolak et al., 2021).
2.2.2. Definitions
For estimation purposes, we divide the period between April 1, 2020 and September 30, 2020 into seven-day intervals, hereafter referred to as “weeks.” Following the terminology developed above, we treat the entire U.S. as the “region,” and counties and states as “subregions” where the number of infections and IFRs are to be estimated. In the U.S., each state contains multiple counties, so we make two sets of estimates, one for each state and one for each county.
County- and state-level variables are denoted with the subscripts c and s respectively (following the notation developed above, country-wide variables lack a subscript). For instance the number of observed cases in state j will be denoted Csj and the number of cases across the entire U.S. will be denoted C. Where distinguishing between individual counties and states is unnecessary, we omit the j subscript for clarity; for instance denoting Csj by Cs.
2.2.3. COVID Case Study Assumptions
To apply the estimators to the COVID-19 pandemic, we further specify the above assumptions.
-
1.1
Odds ratios (ω, ωc, ωs): Following Assumption 1 (above) here we assume that for a given week in the pandemic, the odds ratio of testing an individual infected vs. uninfected with SARS-CoV-2 is the same for all counties, states, and the entire U.S; that is, for any county i and state j, ω = ωci = ωsj. However, the odds ratios may change across weeks.
-
2.1
Regional-wide IFR (φ): Following Assumption 2 (above), we assume that the SARS-CoV-2 IFR across the entire U.S. was five deaths per 1,000 infections from April 1, 2020 to September 30, 2020, a widely agreed upon value (O’Driscoll et al., 2021).
-
3.1
Observed quantities: Following Assumption 3 (above), we assume that the population (Pc, Ps, P), and the numbers of COVID-19 deaths (Dc, Ds, D) and cases (Cc, Cs, C) in each county, state, and across the entire U.S. are known. Likewise, we assume that the number of SARS-CoV-2 NAAT tests in each state (Ts) and consequently across the entire U.S. (T) is known. However, we assume that the number of tests in some counties (Tc) for some weeks is unknown. The latter assumption reflects the actual situation where, in the U.S., the number of SARS-CoV-2 NAATs in some counties was not consistently recorded during 2020, despite the fact that it was recorded in other counties and in aggregate for each state. For instance, reliable testing data are unavailable for all counties in the state of Washington, but they are available for all counties in neighboring states (Holmgren et al., 2020; The Atlantic, 2021)
-
3.2Number of tests in each county (Tc): Heuristically, having data on testing is needed, because otherwise it is impossible to know whether observing few cases in a county is indicative of low infection rates or just a limited testing effort, and vice versa for high rates. Because the number of SARS-CoV-2 NAATs in some counties in some weeks is unknown, it is necessary to estimate it. To do so, we assume that the number of tests administered Tcj in county j can be predicted by some subset of the population size Pcj, number of observed cases Ccj, and number of COVID-19 deaths Dcj. Specifically, we start with the following full model:
(7) where ϵj is a normal random variable with mean 0 (log transformations were implemented to improve the linearity of empirical relationships). This linear model was strongly-supported by regression diagnostics. For each week, we fit this model using the counties where NAAT testing data are available, and all-subsets model selection with 51-fold cross validation, omitting the counties for each state and the District of Columbia separately in each fold (Fig. S1A). The selected models consistently had cross-validation coefficients of determination in the range 0.8–0.95, giving us high confidence in their predictive power (Fig. S1B). For each time point, we then use the fitted model to interpolate the number of NAATs in the counties where testing data were unavailable.
2.2.4. Estimates
Following the sequence in which the estimators in Section 2.1.3 are developed, we first estimate ι and ω at the country level, and then ιc, ιs, φc, and φs at the county and state levels (Table 1; Supplementary Information, Section 1.1). As an added step specific to COVID-19, we initially estimate Tc in counties where testing data is unavailable. To calculate the uncertainty of these point estimates, we use a parametric bootstrap. Specifically, using the point estimates for the odds ratio and the number of infections, for each time point and region, we generate 100 parametric bootstrap resamples using the Wallenius’ noncentral hypergeometric model in Expressions (1) and (2). Dividing the observed mortality by the resulting bootstrap estimates of the number of infections yields an estimate of the sampling distribution of the estimator for the IFR .
Table 1.
Estimators used for finding unobserved quantities (see Table S1). Bold numbers indicate the order of calculation. The estimators for the total number of infections at the state and county levels [(7) and (10); and ] are defined implicitly and are found by solving the given equations numerically. These estimators and are based on the approximation for the mean of the Wallenius’ noncentral hypergeometric distribution given in Expression (1). Time (t), state (j), and county (i) subscripts are omitted for readability, but all estimators are time-, state-, and county-specific.
| Region | Infection fatality ratio | Infections | Tests | Odds ratio |
|---|---|---|---|---|
| Country | φ = 5 (observed) | (4) | (3) | (5) |
| State | (8) | (7) | (2) | (6) |
| County | (11) | (10) | (1) (linear model) | (9) |
2.2.5. Validation: performance when assumptions are met
Under scenarios with set numbers of infections, we generated simulated COVID-19 case data from April 1, 2020 to September 30, 2020 (Supplementary Information, Section 1.2). Using the simulated values of the known quantities in the original data set (black quantities in Table S1) and the estimation procedure described above, we found estimates of all of the unknown quantities (red quantities in Table S1), and also the country-wide IFR, φ. Comparing estimates from 100 iterations of the sampling process to the known values yielded measures of the performance of the estimators.
2.2.6. Validation: performance when assumptions are violated
We performed simulations to assess the performance of the estimators when (a) odds ratios varied spatially in addition to temporally, a violation of Assumption 1.1, and (b) the region-wide IFR differed from five deaths per 1,000 infections, a violation of Assumption 2.1. Specifically, to simulate data from a model where Assumption 1.1 was violated, we multiplied the odds ratio for each state by a factor of (a) 0.9 to 1.1, which equaled 0.896 + 0.004 ⋅ S and (b) 0.5 to 1.5, which equaled 0.482 + 0.018 ⋅ S, where S is the numeric FIPS code of the state (a pseudo-random factor). To simulate data from a model where Assumption 2.1 was violated, we substituted a value of 0.005 and 0.5 for 0.0196 in Step 3, above, giving an overall infection fatality ratio of approximately 0.21 and 18 deaths per 1,000 infections rather than 5, respectively. In addition, for the real (unsimulated) case data, we compared the summed county-level infection estimates within each state to the state-level infection estimates to check for consistency.
2.2.7. Validation: comparison to published seroprevalence IFR estimates
We compared the IFR estimates from the hypergeometric estimators to those from seroprevalence studies summarized in reference (Ioannidis, 2021). We matched the geographic regions (e.g., counties containing cities where seroprevalence estimates are available) and time periods as closely as possible, calculating estimates based on the number of infections and deaths integrated over three months before the date of the seroprevalence estimate. All seroprevalence IFR estimates considered were from between April and August 2020.
3. Results
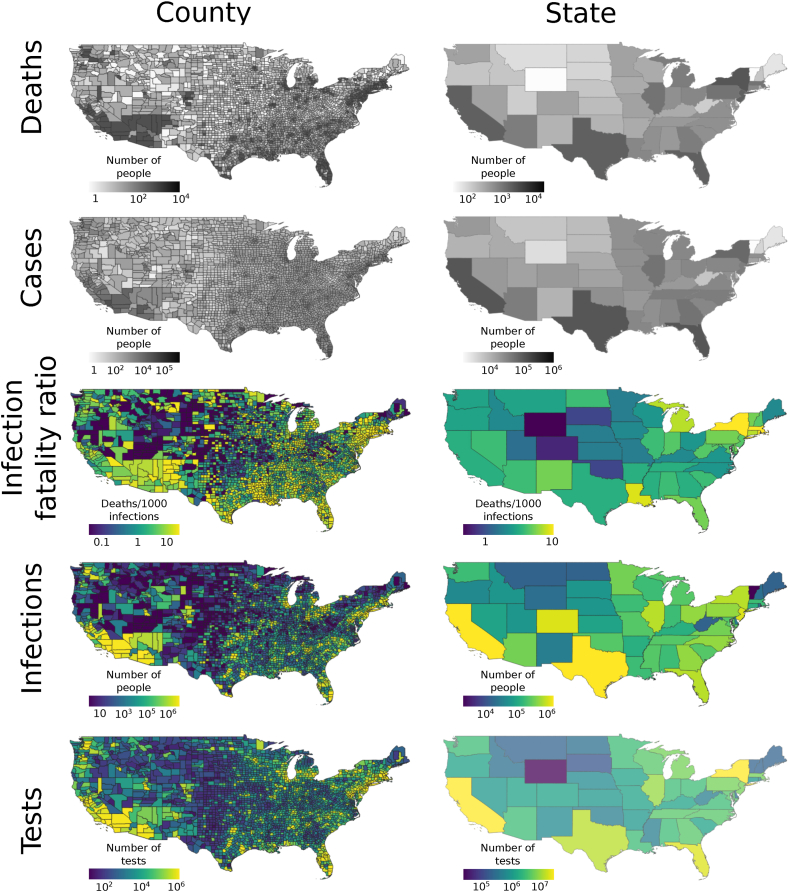
Applying the county-level estimators to the real, unsimulated, COVID-19 data spanning April 1, 2020 to September 30, 2020 from across the U.S., we found that IFRs varied across counties from 0 to 44.69, with a mean of 3.15 and a median of 2.14 (standard deviation 3.55). IFRs were the lowest in the Intermountain West, High Plains, and New England, and the highest in Arizona, Western New Mexico, and scattered other regions of the U.S. The number of SARS-CoV-2 infections (not corrected for population) was highest in the Southwest and scattered other parts of the U.S. County-level IFRs showed a relatively low degree of spatial autocorrelation. Similar trends were evident at the state level, with lower spatial grain (Fig. 1). All of the estimators had low variance (Fig. S2).
Fig. 1.
Maps showing estimated and observed values of the number of COVID-19 deaths and cases, and SARS-CoV-2 IFRs, number of infections, and number of NAATs in the U.S. over the period spanning March 1, 2020 to October 31, 2020. Estimated quantities are shown in color, while observed quantities are shown in gray. The number of SARS-CoV-2 NAATs was estimated in some counties, while observed in others.
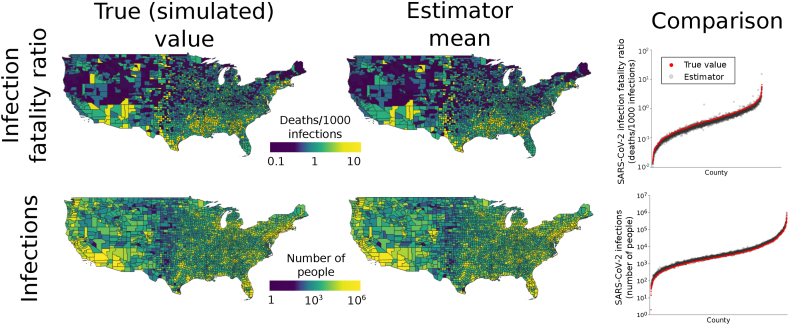
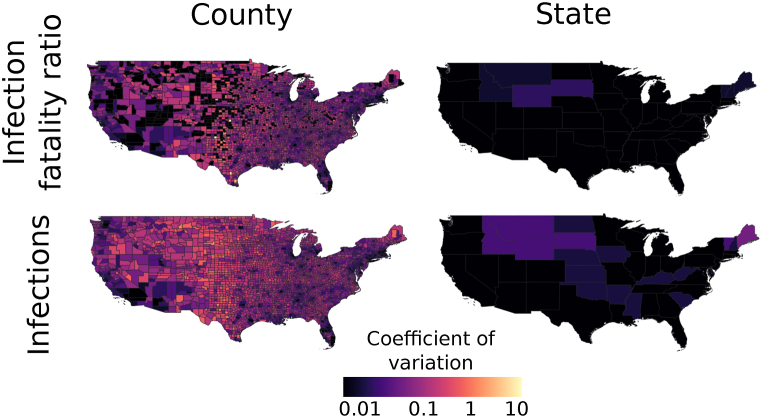
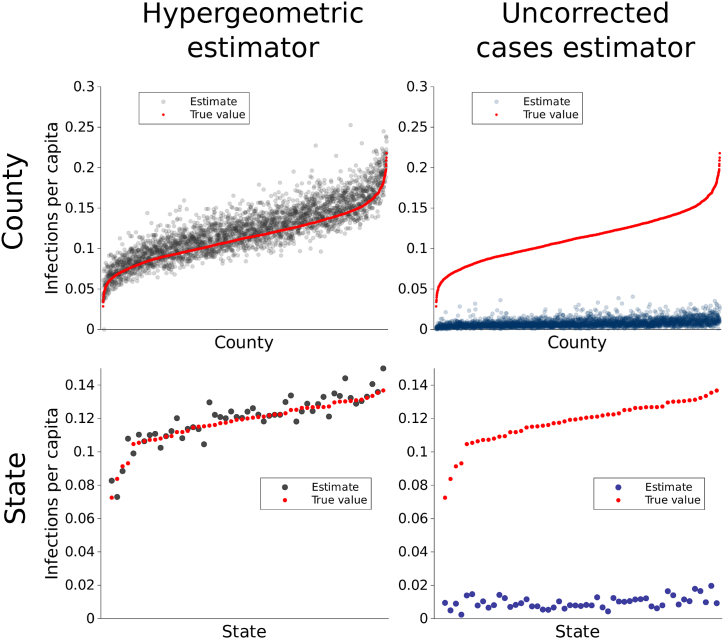
In our validation, we found that the mean of the county-level estimates of IFRs and number of SARS-CoV-2 infections had high accuracy (Fig. 2; Pearson correlation coefficients for estimator means and true values 0.927 and 0.996 respectively). Likewise, at the state level, the mean of the estimates for IFRs and number of infections had high accuracy and precision (Fig. S3; Pearson correlation coefficients for estimator means and true values 0.991 and 0.996, respectively). Moreover, the estimators had coefficients of variation below 1 for almost all counties and states (Fig. 3; for example, for IFR, 99.7%, 96.7%, and 40.1% of counties had coefficients of variation below 1, 0.5, and 0.1, respectively). In contrast, estimates of the number of infections and IFR obtained simply by using the raw numbers of cases and case fatality ratios, respectively, were biased by up to several orders of magnitude and had low precision (Fig. S4). These results also held for per capita infection estimates (Fig. 4), indicating that the performance was not driven by varying populations across counties and states. The estimator was robust against deviations from Assumption 1: when odds ratios varied spatially, performance remained good (Figs. S5A–B, S6A-B). When the global IFR differed from five deaths per 1,000 infections, the estimator became biased, as would be expected. However, it continued to correctly estimate the relative values of the county-level IFRs; that is, to indicate area-level variation in IFRs (Figs. S5C–D, S6C-D). Using the observed (non-simulated) numbers of cases, the summed county level estimates of the number of infections within each state matched the number of infections directly estimated for those states (Fig. S7). At both the state level and county level, seroprevalence IFR estimates matched the IFR estimates from the hypergeometric estimator (Fig. S8), further indicating its validity.
Fig. 2.
Performance of county-level estimators. Maps in the first column show true (simulated) SARS-CoV-2 IFRs and numbers of infections. All values were conditioned on observed numbers of COVID-19 deaths. Counties with zero COVID-19 deaths are omitted from the SARS-CoV-2 IFR estimation results because they have ratios of zero. The second column gives the means of the estimates over 100 simulated data sets for each county. The close match between true values and estimates indicates that the estimators have high accuracy. The accuracy is further illustrated by the graphs in the third column, which plot the true values in each county compared to the mean of the estimator. In each plot, counties (x-axis) are ordered by increasing value.
Fig. 3.
The estimators for IFRs and the number of infections have high precision. At both the county and state levels, the estimators have low coefficients of variation, generally below 0.1 and often below 0.01.
Fig. 4.
Performance of county-level estimators for SARS-CoV-2 infections with population taken into account. The hypergeometric infections estimator per capita had low bias and high precision (column 1), displaying superior performance to a naive estimator that used the uncorrected number of COVID-19 cases (column 2). On the x-axis, counties and states are ordered by increasing number of SARS-CoV-2 infections, shown in red.
4. Discussion
We present an inferential framework for estimating the geographic variation in the total number of infections of a pathogen and its IFR. We apply this framework to SARS-CoV-2 and the COVID-19 pandemic, finding that it is robust and performs accurately and precisely. The approach relies on a noncentral hypergeometric model that accounts for differential probabilities of positive tests (i.e. because sick individuals are more likely to seek testing than healthy individuals (Campbell et al., 2020)), which we show is well-suited for estimating IFRs.
Most studies utilize an averaged IFR, despite known geographic and temporal variation in this parameter. The problematic nature of variation in IFR estimates is well-documented in the literature; for example, multiple systematic reviews have highlighted differences in SARS-CoV-2 IFRs across time and geographic settings (Ioannidis, 2021; O'Driscoll et al., 2021; Verity et al., 2020). Although in limited settings, comprehensive case reporting and tracking enable more accurate estimation of the IFR, most estimates are based on a snapshot of seroprevalence data, which may not be representative of the population of interest and is subject to information delays and issues around accuracy of testing. (Levin et al., 2020; Takahashi et al., 2020).
Our framework helps to address the challenges of IFR estimation posed by incompletely observed data. There are sparse examples of other approaches to address this issue: for example, Reich et al. (Reich et al., 2012) propose using a log-linear model to estimate the relative case fatality ratio, and this approach has been applied to the SARS-CoV-2 epidemic to develop a corrected estimator of case fatality rate that accounts for time lag and imperfect reporting of deaths and recoveries (Angelopoulos et al., 2020). Similarly, Brazeau et al. (Brazeau et al., 2022) used serologic test sensitivity and specificity alongside a Bayesian framework to account for uncertainties in the parameters for determining IFR, including consideration of delay-distributions from time from infection to seroconversion, time to death, and time to seroreversion (i.e., antibody waning), and showed improvements in state-level estimates of SARS-CoV-2 infections in the U.S.
Our approach differs in that we use data on testing effort, where reliably reported, to address incompletely observed seroprevalence data, which are widely used to estimate IFR. Most seroprevalence data are limited geographically, may have non-representative samples, or fail to account for potential delays from onset to seroconversion. Therefore, our framework can be seen as complementary to the methods described above.
The number of excess deaths is another frequently used metric of the burden of mortality, particularly with regard to the COVID-19 pandemic. Typically defined as the difference between the observed number of deaths and the expected number of deaths across a given time period (Centers for Disease Control and Prevention, 2021), it includes changes in mortality that may directly or indirectly be attributable to COVID-19 (such as from delayed care or behavioral health crises). While useful in some settings, excess death estimates are highly sensitive to the reference time period used and negative estimates often occur in younger age strata (Woolf et al., 2021; Excess Deaths Associated with COVID). Our proposed method allows for more detailed area-level estimates of mortality directly attributable to an infection.
As with all statistical tools, the accuracy and precision of our method are limited by the quality of the data that are available. An advantage of our approach is that it allows investigators to rely on the best available data and estimate quantities where data may be lacking. In the application to COVID-19, our estimates use data on NAAT (also known as molecular or PCR tests) and do not include data from antigen tests including home tests, which did not receive FDA approval until late 2020. The NAAT are the most reliable for detecting infections and were the most widely used during the study period.
Our approach is also limited by a number of assumptions, though our numerical analyses show that the performance of the approach is robust to many violations of these assumptions. However, as with any inferential approach, sufficiently extreme violations of assumptions will reduce the performance of the method, and in these situations, it may be necessary to modify the method. Specifically, Assumption 1 – that the odds testing an infected vs. an uninfected individual is the same across geographic regions – can be relaxed in numerous ways. While some geographic or temporal structure must be assumed to estimate the odds ratios, which are latent variables, this structure does not need to be geographically homogeneous. With additional covariates (e.g., data on demographics, health care access), it may also be possible to further infer specific aspects of the odds ratios. Second, regarding Assumption 2, while an overall IFR must be assumed, this value can in principle be any number between 0 and 1 dependent on the disease, time, location, and other factors. Moreover, when the aim is to rank IFRs in subregions relative to each other instead of estimate their precise values, the assumed overall IFR may not matter. And third, an added degree of complexity specific to COVID-19 in the U.S. is imparted here because the number of NAATs in each state, but not every county, are known. In a more typical scenario, when testing effort is recorded for each subregion, one could directly make estimates for a single class of subregions (e.g., counties) from a superregion (e.g., country). This would omit the need for our Assumption 3.2. It should be possible to adapt our approach for application at different geographic and temporal scales, and it may be particularly useful in settings in which resources to carry out large, representative seroprevalence studies are unavailable.
Local estimates are likely to be more accurate and useful for policy planning purposes than generalized or country-level estimates. The importance of flexible estimates is highlighted by the work of Solis et al. (Solís et al., 2021), who examined how COVID-related models performed differently in different states and the impact on morbidity with respect to state-level policy decisions driven by a lack of fit. Our approach can serve as an extension to other methods for estimation of epidemic burden to allow for greater precision around geographic and temporal population subgroups.
In summary, we propose versatile inference methods that leverage the best available data to yield comprehensive estimates that allow for variance in IFRs across geographic regions or time intervals. We show in validation models that our method can be used to accurately and precisely estimate SARS-CoV-2 IFR at the county level in the U.S.
CRediT authorship contribution statement
Joshua Ladau: Writing – review & editing, Writing – original draft, Visualization, Validation, Software, Project administration, Methodology, Investigation, Formal analysis, Data curation, Conceptualization. Eoin L. Brodie: Writing – review & editing, Writing – original draft, Project administration, Funding acquisition, Conceptualization. Nicola Falco: Writing – review & editing, Writing – original draft, Conceptualization. Ishan Bansal: Writing – review & editing, Writing – original draft, Investigation, Conceptualization. Elijah B. Hoffman: Writing – review & editing, Writing – original draft, Investigation, Conceptualization. Marcin P. Joachimiak: Writing – review & editing, Writing – original draft, Investigation, Conceptualization. Ana M. Mora: Writing – review & editing, Writing – original draft, Methodology. Angelica M. Walker: Writing – review & editing, Writing – original draft, Investigation, Conceptualization. Haruko M. Wainwright: Writing – review & editing, Writing – original draft, Conceptualization. Yulun Wu: Writing – review & editing, Writing – original draft, Validation, Methodology, Investigation, Formal analysis. Mirko Pavicic: Writing – review & editing, Writing – original draft, Investigation. Daniel Jacobson: Writing – review & editing, Writing – original draft, Validation, Supervision, Project administration, Methodology, Investigation, Funding acquisition, Data curation, Conceptualization. Matthias Hess: Writing – review & editing, Writing – original draft, Supervision, Project administration, Investigation, Conceptualization. James B. Brown: Writing – review & editing, Writing – original draft, Visualization, Validation, Supervision, Project administration, Methodology, Investigation, Funding acquisition, Formal analysis, Data curation, Conceptualization. Katrina Abuabara: Writing – review & editing, Writing – original draft, Visualization, Supervision, Project administration, Methodology, Investigation, Funding acquisition, Data curation, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
K.A. and J.L. were supported by a grant from the Benioff Center for Microbiome Medicine. This research used resources of the Oak Ridge Leadership Computing Facility, which is a DOE Office of Science User Facility supported under Contract DE-AC05-00OR22725. This manuscript has been coauthored by UT-Battelle, LLC under contract no. DE-AC05-00OR22725 with the U.S. Department of Energy. The United States Government retains and the publisher, by accepting the article for publication, acknowledges that the United States Government retains a nonexclusive, paid-up, irrevocable, world-wide license to publish or reproduce the published form of this manuscript, or allow others to do so, for United States Government purposes. The Department of Energy will provide public access to these results of federally sponsored research in accordance with the DOE Public Access Plan (http://energy.gov/downloads/doe-public-access-plan, last accessed September 16, 2020). Work at Oak Ridge and Lawrence Berkeley National Laboratories was supported by the DOE Office of Science through the National Virtual Biotechnology Laboratory, a consortium of DOE national laboratories focused on response to COVID-19, with funding provided by the Coronavirus CARES Act, and was facilitated by previous breakthroughs obtained through the Laboratory Directed Research and Development Programs of the Lawrence Berkeley and Oak Ridge National Laboratories. M.P.J. was supported by a grant from the Laboratory Directed Research and Development (LDRD) Program of Lawrence Berkeley National Laboratory under U.S. Department of Energy Contract No. DE-AC02-05CH11231. Oak Ridge National Laboratory would also like to acknowledge funding from the U.S. National Science Foundation (EF-2133763).
Handling Editor: Dr Daihai He
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.idm.2024.02.009.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Angelopoulos A.N., Pathak R., Varma R., Jordan M.I. On Identifying and Mitigating Bias in the Estimation of the COVID-19 Case Fatality Rate. Harvard Data Science Review. 2020 doi: 10.1162/99608f92.f01ee285. Special Issue 1. [DOI] [Google Scholar]
- Aron, J., Muellbauer, J., Giattino, C., & Ritchie, H. (2020). A pandemic primer on excess mortality statistics and their comparability across countries. Our World in Data. https://ourworldindata.org/covid-excess-mortality
- Brazeau N.F., Verity R., Jenks S., Han F., Whittaker C., Peter W.…Schnekenberg R.P. Estimating the COVID-19 infection fatality ratio accounting for seroreversion using statistical modelling. Communications Medicine. 2022;2(1):54. doi: 10.1038/s43856-022-00106-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell H., Valpine P.de, Maxwell L., Valentijn MT. de J., Debray T., Jänisch T., Gustafson P. 2020. Bayesian adjustment for preferential testing in estimating the COVID-19 infection fatality rate: Theory and methods.https://arxiv.org/abs/2005.08459v2 [Google Scholar]
- Centers for Disease Control and Prevention . 2021. Notes from the Field: Update on Excess Deaths Associated with the COVID-19 Pandemic — United States, January 26, 2020–February 27, 2021.https://www.cdc.gov/mmwr/volumes/70/wr/mm7015a4.htm [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dana Flanders W., Kleinbaum D.G. Basic Models for Disease Occurrence in Epidemiology. International Journal of Epidemiology. 1995;24(1):1–7. doi: 10.1093/ije/24.1.1. [DOI] [PubMed] [Google Scholar]
- Dong E., Du H., Gardner L. An interactive web-based dashboard to track COVID-19 in real time. The Lancet Infectious Diseases. 2020;20(5):533–534. doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excess Deaths Associated with COVID-19. https://www.cdc.gov/nchs/nvss/vsrr/covid19/excess_deaths.htm. Accessed: 2021-09-02.
- Fog A. Calculation Methods for Wallenius’ Noncentral Hypergeometric Distribution. Communications in Statistics - Simulation and Computation. 2008;37(2):258–273. doi: 10.1080/03610910701790269. [DOI] [Google Scholar]
- Holmgren A.J., Apathy N.C., Adler-Milstein J. Barriers to hospital electronic public health reporting and implications for the COVID-19 pandemic. Journal of the American Medical Informatics Association. 2020;27(8):1306–1309. doi: 10.1093/jamia/ocaa112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioannidis J.P.A. Infection fatality rate of COVID-19 inferred from seroprevalence data. Bulletin of the World Health Organization. 2021;99(1):19–33F. doi: 10.2471/BLT.20.265892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolak M., Lin Q., Halpern D., Paykin S., Martinez-Cardoso A., Li X. 2021. https://www.uscovidatlas.org (The US Covid Atlas). [Google Scholar]
- Levin A.T., Hanage W.P., Owusu-Boaitey N., Cochran K.B., Walsh S.P., Meyerowitz-Katz G. Assessing the age specificity of infection fatality rates for COVID-19: systematic review, meta-analysis, and public policy implications. European Journal of Epidemiology. 2020;35(12):1123–1138. doi: 10.1007/s10654-020-00698-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Driscoll M., Ribeiro Dos Santos G., Wang L., Cummings D.A.T., Azman A.S., Paireau J., Fontanet A., Simon C., Salje H. Age-specific mortality and immunity patterns of SARS-CoV-2. Nature. 2021;590(7844):140–145. doi: 10.1038/s41586-020-2918-0. [DOI] [PubMed] [Google Scholar]
- Orin D., Topol E. Prevalence of Asymptomatic SARS-CoV-2 Infection. Annals of Internal Medicine. 2020;173(5):362–367. doi: 10.7326/M20-3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich N.G., Lessler J., Cummings D.A.T., Brookmeyer R. Estimating Absolute and Relative Case Fatality Ratios from Infectious Disease Surveillance Data. Biometrics. 2012;68(2):598–606. doi: 10.1111/j.1541-0420.2011.01709.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin R. First it was masks; now some refuse testing for SARS-CoV-2. JAMA. 2020;324(20):2015–2016. doi: 10.1001/jama.2020.22003. [DOI] [PubMed] [Google Scholar]
- Shen C., VanGennep D., Siegenfeld A.F., Bar-Yam Y. Unraveling the flaws of estimates of the infection fatality rate for COVID-19. Journal of Travel Medicine. 2021;28(2) doi: 10.1093/jtm/taaa239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solís P., Dasarathy G., Pavan T., Drake A., Vora K.J., Sajja A., Raaman A., Praharaj S., Lattus R. Understanding the spatial patchwork of predictive modeling of first wave pandemic decisions by US governors. Geographical Review. 2021;111(4):592–615. doi: 10.1080/00167428.2021.1947139. [DOI] [Google Scholar]
- Takahashi S., Greenhouse B., Rodríguez-Barraquer I. Are Seroprevalence Estimates for Severe Acute Respiratory Syndrome Coronavirus 2 Biased? The Journal of Infectious Diseases. 2020;222(11):1772–1775. doi: 10.1093/infdis/jiaa523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Atlantic. (2021). The COVID Tracking Project. https://covidtracking.com
- Verity R., Okell L.C., Dorigatti I., Peter W., Whittaker C., Imai N.…Ferguson N.M. Estimates of the severity of coronavirus disease 2019: a model-based analysis. The Lancet Infectious Diseases. 2020;20(6):669–677. doi: 10.1016/S1473-3099(20)30243-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood S.N., Wit E.C., Fasiolo M., Green P.J. COVID-19 and the difficulty of inferring epidemiological parameters from clinical data. The Lancet Infectious Diseases. 2021;21(1):27–28. doi: 10.1016/S1473-3099(20)30437-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolf S.H., Chapman D.A., Sabo R.T., Zimmerman E.B. Excess Deaths From COVID-19 and Other Causes in the US, March 1, 2020, to January 2, 2021. JAMA. 2021;325(17):1786–1789. doi: 10.1001/jama.2021.5199. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.