Abstract
Global efforts to eradicate malaria are threatened by multiple factors, particularly the emergence of antimalarial drug resistant strains of Plasmodium falciparum. Heat shock proteins (HSPs), particularly P. falciparum HSPs (PfHSPs), represent promising drug targets due to their essential roles in parasite survival and virulence across the various life cycle stages. Despite structural similarities between human and malarial HSPs posing challenges, there is substantial evidence for subtle differences that could be exploited for selective drug targeting. This review provides an update on the potential of targeting various PfHSP families (particularly PfHSP40, PfHSP70, and PfHSP90) and their interactions within PfHSP complexes as a strategy to develop new antimalarial drugs. In addition, the need for a deeper understanding of the role of HSP complexes at the host–parasite interface is highlighted, especially heterologous partnerships between human and malarial HSPs, as this opens novel opportunities for targeting protein–protein interactions crucial for malaria parasite survival and pathogenesis.
Keywords: Plasmodium falciparum, Malaria, Antimalarial drugs, Molecular chaperones, Heat shock proteins, Protein folding
Introduction
The unicellular protozoan parasite, Plasmodium falciparum, invades human cells and transforms them into vehicles of pathology, thereby causing the most virulent form of human malaria. Over the 2000–2015 period, annual deaths due to malaria declined from nearly 900,000 to just over 500,000; however, since then malaria mortalities have been increasing, with the COVID-19 pandemic contributing to this trend by disrupting essential malaria services.1 There are a number of factors that are threatening efforts to eliminate and ultimately eradicate malaria, particularly the emergence of antimalarial drug resistant in P. falciparum strains.2 Resistance to artemisinin, the recommended first-line treatment for malaria, has been reported to be linked to mutations in the gene encoding P. falciparum Kelch13, with certain mutations (e.g. K395T) causing a reduction in levels of this protein.3, 4, 5, 6 Such P. falciparum Kelch13 mutations are reported to be associated with upregulation of protein folding and turnover, enabling mutant parasites to readily remove the damaged proteins that result from artemisinin treatment, leading to parasites' survival and ultimately drug resistance.4 This emerging drug resistance has highlighted: (i) the scarcity of validated antimalarial drug targets; (ii) the need to focus on drug discovery strategies that develop synergistic drugs acting on distinct and complementary targets; and (iii) the identification of a target, or combinations of targets, that can act across the entire parasite life cycle.7 The survival, establishment, and resulting pathology of the malaria parasite throughout the various stages of its life cycle rely on highly evolved interactions between the human host and the parasite. Elucidating the structural properties of these host–parasite protein–protein interfaces are critical to the development of novel antimalarial drugs. Key drug discovery steps include (i) the identification of a target parasite protein or (preferably) protein complex required for the growth or survival of the parasite; (ii) the identification of a readily accessible chemical series of modulators of the parasite target; and (iii) the design of modulators with preferential affinity for the parasite target over its human homologs. Heat shock proteins (HSPs) have been shown to meet the threshold criteria to be considered bona fide drug targets, especially those that serve as guardians of proteostasis through their roles as molecular chaperones.8 This review provides an update on the potential of targeting P. falciparum HSP (PfHSP) families, especially PfHSP40s, PfHSP70s, and PfHSP90s and their interactions within homologous and heterologous complexes (with human HSPs), as novel strategies to develop new antimalarial drugs.
Malarial HSPs and their complexes as drug targets
There is substantial evidence that PfHSPs are not only genuine drug targets but that they also carry highly druggable structural features for antimalarial drug discovery.9, 10, 11 HSPs were discovered on the basis that their expression was significantly upregulated in response to physical, chemical, or physiological stress factors, and they were named according to their apparent molecular weight; for example, HSP70 (HSP70) is a 70 kDa protein. The major families of HSPs (HSP10, HSP40, HSP60, HSP70, HSP90, and HSP100) have now been shown to play essential roles in cell survival not only under stressful conditions but also under normal physiology, with many of them demonstrated to be molecular chaperones with both inducible and constitutive isoforms.12 They are highly conserved from prokaryotes to eukaryotes, and in eukaryotes, they are found in all of the major subcellular compartments. The various chaperone families and associated regulatory cochaperones often work together to perform specific tasks. For example, HSP40 (also called J domain protein or JDP)13 and HSP70 bind to each other and to misfolded proteins to limit aggregation and promote folding or degradation. Likewise, the HSP10–HSP60 system and HSP70–HSP90 systems work together to stabilize “client” proteins and promote their function, while the HSP100 family is involved in thermal tolerance, unfolding, and degradation of aggregated proteins.12 Together, this network of chaperones is important for protein homeostasis (proteostasis), especially in response to stress.14, 15 Consistent with this role, most PfHSPs have been found to be essential for malaria parasite growth, survival, viability, and differentiation at all stages of its life cycle.16, 17 Many PfHSPs are also essential for the parasite’s pathogenesis and virulence, and a number form unique complexes at the host–parasite interface.11 Furthermore, unlike many other antimalarial drug targets studied to date, modulation of PfHSPs could have much broader effects on the survival of the parasite across all stages of the life cycle.
The key challenge in targeting PfHSPs (such as PfHSP40s, PfHSP70s, and PfHSP90s) as an antimalarial strategy, is the relatively high structural and sequence identity with their human orthologs. However, a number of reports have highlighted that there are subtle and potentially significant structural and functional differences that might be exploited (Figure 1).8, 18, 19, 20 Indeed, over the last decade these subtle differences have begun to be interrogated, resulting in a growing list of modulators of PfHSP40s, PfHSP70s, and PfHSP90s that are potential candidates for antimalarial drug development. Table 1 provides detailed key information on those small molecule compounds that have been experimentally characterized for their biochemical and biological effects. The ensuing narrative provides a critical appraisal of these potential antimalarial drug candidates, as well as highlighting other chemical entities emerging from virtual and experimental compound database screening strategies.
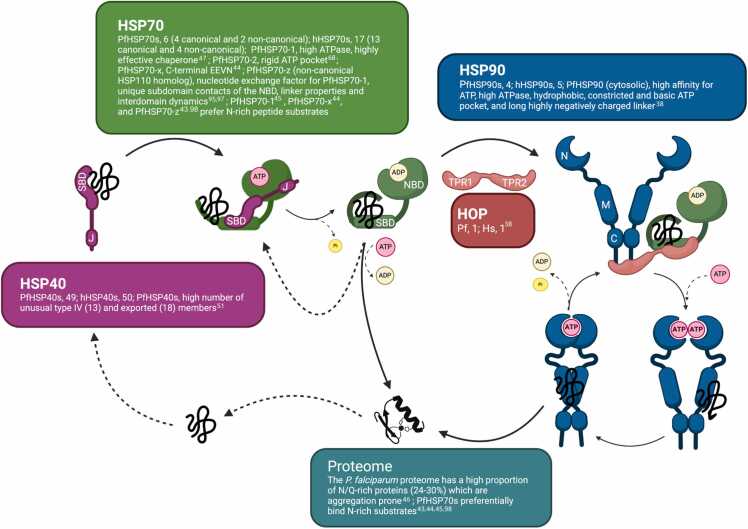
Fig. 1.
HSP40s, HSP70s, and HSP90s of the malaria parasite and humans form similar chaperone networks but have different intrinsic properties. The number, structure, and function of the malarial HSPs and the malarial proteome show significant differences compared to the human system (the unique differences reported for PfHSPs compared to human HSPs are described in colored boxes; human HSP40, hHSP40; human HSP70, hHSP70; human HSP90, hHSP90). The graphic illustrates the pathway of a newly synthesized or unfolded/misfolded protein (black coil) through a chaperone network until it is folded (black helix/sheet). The protein substrate is bound by HSP40 (purple; substrate binding domain, SBD, and J domain, J), which associates with ATP-bound HSP70 (green; SBD, and nucleotide-binding domain, NBD), hands over the substrate, stimulating ATP hydrolysis, thereby locking the substrate in the SBD of HSP70. Upon nucleotide exchange, the substrate can be released from HSP70 to fold into the native state, or the HSP70−substrate complex can associate with HSP90 (blue; N-terminal domain, N, middle domain, M, and C-terminal domain, C) with the assistance of HOP (HSP70−HSP90 organizing protein; red; tetratricopeptide domain 1 and 2, TPR1, and TPR2), resulting in another ATP-driven chaperone cycle, before the substrate is released to fold into the native state. This graphic was created with BioRender (biorender.com). Abbreviations used: HSP, heat shock protein; PfHSP, Plasmodium falciparum HSPs; Hs, Homo sapiens; Pf, Plasmodium falciparum.
Table 1.
Experimentally characterized inhibitors of PfHSP90, PfHSP70, PfHSP40, and their complexes.
| PfHSPa | Inhibitor | Biochemical activityb | Biological activity | Ref.c |
|---|---|---|---|---|
| PfHSP90 | Beta carboline alkaloids (Harmine analogs; 17A and 21A) | Moderate affinity but greater affinity for the ATP-binding site of PfHSP90 than human HSP90 | Inhibit in vitro growth of parasite-infected erythrocytes; reduce parasitemia and extend survival of parasite-infected mice in combination with DHA | 21, 22 |
| 2-Aminobenzamides (SNX-0723) |
High affinity for the ATP-binding site of PfHSP90 | Inhibit in vitro growth of parasite-infected erythrocytes and parasite-infected liver cells; inhibit parasite growth in liver cells synergistically with PIK-75 (phosphatidylinositol 3-kinase inhibitor); inhibit in vivo growth of parasite-infected mice | 23 | |
| Amino-alcohol carbazoles (N-CBZ; compound 5B) | Moderate affinity binding to a PfHSP90-specific hydrophobic extension of the ATP-binding site; no binding to human HSP90 | Inhibit in vitro growth of parasite-infected erythrocytes | 24 | |
| Pyrimidinediones (compound FM2, which is MMV Malaria Box compound MMV019066) | Moderate affinity binding to PfHSP90 (Surface Plasmon Spectroscopy), and no binding to human HSP90 (competition binding assays with geldanamycin); inhibit ATPase activity of PfHSP90 | Inhibit in vitro growth of parasite-infected erythrocytes; high selectivity index from in vitro cytotoxicity assays with mammalian cells | 25, 26 | |
| Terpenoids (IMA; UAA) | Bind to PfHSP90 with moderate affinity (binding site not experimentally determined); inhibit ATPase activity and protein aggregation suppression activity of PfHSP90 | Inhibit in vitro growth of parasite-infected erythrocytes; inhibit in vivo growth of parasite-infected mice | 27 | |
| PfHSP70 | Polymyxin B | Bind to PfHSP70-1 (primarily ATPase domain) with high affinity and to PfHSP70-z with moderate-to-high affinity; inhibit ATPase activity and protein aggregation suppression activity of PfHSP70-1 and PfHSP70-z; disrupt the interaction of PfHSP70-1 with PfHSP70-z and PfHOP | Not determined | 28 |
| (−)-Epigallocatechin-3-gallate | Bind to PfHSP70-1 (primarily ATPase domain) and PfHSP70-z with high affinity; inhibit ATPase activity and protein aggregation suppression activity of PfHSP70-1 and PfHSP70-z; disrupt the interaction of PfHSP70-1 with PfHSP70-z and PfHOP | Inhibit in vitro growth of parasite-infected erythrocytes | 29 | |
| Terpenoids (IMA; UAA) | Bind to PfHSP70-1 (affinity and binding site not experimentally determined). IMA Inhibits ATPase activity and protein aggregation suppression activity of PfHSP70-1; UAA inhibits protein aggregation suppression activity of PfHSP70-1 | Inhibit in vitro growth of parasite-infected erythrocytes; inhibit in vivo growth of parasite-infected mice | 30 | |
| Malonganenones A, B, and C (MalA, B, and C) | MalA–C inhibit the protein aggregation suppression activity of PfHSP70-1; MalA also inhibits the protein aggregation suppression activity of PfHSP70-x; MalA does not inhibit the basal ATPase activity of PfHSP70-1, PfHSP70-x, and human HSP70, but does inhibit the HSP40-stimulated ATPase activity of PfHSP70-1 and PfHSP70-x, but not human HSP70 | MalA and C inhibit in vitro growth of parasite-infected erythrocytes | 31, 32 | |
| 1,4 Naphthoquinones (lapachol) | Preferentially inhibit protein aggregation suppression activity of PfHSP70-1 compared to a “human-like” HSP70 control; inhibit the basal and HSP40-stimulated ATPase activity of PfHSP70-x but not that of PfHSP70-1 and human HSP70 | Inhibit in vitro growth of parasite-infected erythrocytes | 31, 32 | |
| Pyrimidinones (DMT002264) | Inhibit PfHSP40-stimulated ATPase activity of PfHSP70-1 (single turnover assay); inhibit HSP40-stimulated ATPase activity of human HSP70 (steady state assay) | Inhibit in vitro growth of parasite-infected erythrocytes | 33 | |
| Quinoline–pyrimidine hybrids (compound 7a) | Bind to PfHSP70-1 and PfHSP70-z with high affinity (binding site not experimentally determined) | Inhibit in vitro growth of parasite-infected erythrocytes | 34 | |
| 2-Phenylthynesulfonamide | Bind with high affinity to PfHSP70-1 (primarily the substrate binding domain) and with moderate affinity to PfHOP; disrupt the interaction of PfHSP70-1 with PfHOP; inhibit protein aggregation suppression activity of PfHSP70-1 | Inhibit in vitro growth of parasite-infected erythrocytes | 35 | |
| PfHSP40 | Chalcones (C86) | A pan-inhibitor of all classes of human HSP40, binding specifically to the J domain; inhibited the PFE0055c-stimulated ATPase activity of PfHSP70-x but not its basal ATPase activity | Not determined | 36, 37 |
Abbreviations used: DHA, dihydroartemisinin; HSP, heat shock protein; IMA, iso-mukaadial acetate; PfHSP, Plasmodium falciparum heat shock protein; UAA, ursolic acid acetate.
The title “PfHSP” refers to the PfHSP family that is the primary target of the indicated inhibitors.
High affinity indicates submicromolar or nanomolar affinity, while moderate affinity indicates micromolar affinity.
The title “Ref.” refers to reference citations.
PfHSP90
Since cytoplasmic human HSP90 is a specialized chaperone with many oncogenic client proteins, it has been extensively studied as an anticancer drug target, with many of the resulting inhibitors and drug candidates being readily accessible starting points for PfHSP90-based antimalarial drug discovery. Furthermore, while there is a 64% identity between cytosolic PfHSP90 and cytoplasmic human HSP90 (HSP90β), PfHSP90 has a number of distinct biochemical and structural differences (Figure 1),38 which have already been investigated but could be further explored. Inhibitors of HSP90 typically bind to the highly conserved ATP-binding site in the N-terminus, and this appears to be the case for many of the PfHSP90 inhibitors (harmine analogs, 2-aminobenzamides, amino-alcohol carbazoles, pyrimidinediones and terpenoids; Table 1). Harmine, a β-carboline alkaloid, has been found to bind PfHSP90 about four-fold tighter than to human HSP90 (Table 1).21 More recent efforts reported the synthesis and screening of ~40 new analogs of harmine, revealing compounds 17A and 21A22 that inhibit PfHSP90 two-fold better than harmine (~20 µM). These compounds also reduce parasitemia and extend the survival of mice infected with Plasmodium berghei when combined with dihydroartemisinin (Table 1). While promising, it is likely that greater selectivity for PfHSP90 over human HSP90 is needed. One important reason is that numerous HSP90 inhibitors have failed in oncology clinical trials, often because of unacceptable toxicity.39 The toxicity was particularly pronounced for those inhibitors that targeted the N-terminus since they were pan-inhibitors, capable of binding to all human Hsp90 isoforms, which ultimately triggered the observed toxicity. Nevertheless, recent efforts have revived interest in HSP90 inhibitors.39 Thus, it will be important to exploit the subtle structural differences between PfHSP90 and human HSP90,40 including the differences between their heterocomplexes with clients and cochaperones.38 For example, the glycine-rich hinge loop (GHL) motif of the ATP-binding pocket of PfHSP90 forms a unique hydrophobic extension that could potentially be leveraged by analogs of some of the known HSP90 inhibitors.41 Many of the known human HSP90 inhibitors, such as PU-H7121 and geldanamycin,42 have nanomolar activity against parasite growth, providing multiple, strong starting points. Other recent efforts have taken a comparative approach to develop new chemotypes that are selective for PfHSP90. For example, Derbyshire’s group screened PfHSP90 and human HSP90 in vitro,23 revealing an aminobenzamide (SNX-0723) as a promising lead, with high binding affinity for the PfHSP90 ATP pocket (low nM range), and capable of dual-stage parasite growth inhibition (Table 1). Another complementary approach has been taken by Picard and colleagues, who docked known antimalarials to PfHSP90 to show that the carbazole, N-CBZ, might achieve its antiparasitic activity, in part, through inhibiting the chaperone.24 Interestingly, while N-CBZ bound to PfHSP90 with moderate affinity (low μM range), it bound to the unique hydrophobic extension of the ATP pocket formed by the GHL motif, and was not able to bind to the ATP pocket of human HSP90 (Table 1).
PfHSP70 (canonical) and PfHSP40
Similar possibilities and challenges exist for the other families of HSPs. A substantial number of HSP70 inhibitors have been identified that target the N-terminal ATP pocket, the C-terminal substrate binding domain, and the interface with cochaperones, and this is reflected in the array of inhibitors emerging against PfHSP70 (polymyxin B, (−)-epigallocatechin-3-gallate, terpenoids, malonganenones, naphthoquinones, pyrimidinones, quinoline–pyrimidine hybrids, phenylthynesulfonamide, and chalcones; Table 1). The P. falciparum proteome has a high proportion of N/Q-rich proteins (24–30%) which are aggregation-prone, and the evidence suggests that PfHSP70s have evolved to specifically recognize N-rich peptides, leading to the proposal that they protect N-rich malarial proteins from misfolding and aggregation (Figure 1).43, 44, 45, 46 Furthermore, certain PfHSP70s have distinct biochemical properties that suggest they are more effective chaperones than human HSP70s (e.g. PfHSP70-1; Figure 1),47 while others have structure–function features that distinguish them from human HSP70s (e.g. PfHSP70-x C-terminal EEVN; Figure 1).44 PfHSP70-1 is an abundant chaperone of the parasite cytosol that is required for survival.10 Known inhibitors of PfHSP70-1 include polymyxin B and (−)-epigallocatechin-3-gallate, both of which bind with high affinity (submicromolar) to the full-length protein and a nucleotide-binding domain (NBD) construct, suggesting that the primary bind site is the N-terminal ATPase domain (Table 1).28, 29 Similarly, the phytocompound iso-mukaadial acetate inhibits the basal ATPase activity of PfHSP70-1,30 while malonganenones, including malonganenone A, B, and C, and 1,4 naphthoquinones, particularly lapachol and its derivatives, have been reported to inhibit the protein aggregation suppression activity of PfHSP70-1 (Table 1).31 These studies and others have provided multiple starting points for selective inhibitors. Unfortunately, compared to HSP90 inhibitors, less effort has been devoted to exploiting the biochemical and structural differences between PfHSP70-1 and human HSP70. Deeper research is needed to develop highly selective inhibitors against PfHSP70s, and to show structure–activity relationships between PfHSP70 inhibition and parasite growth inhibition for a series of chemotypes for each scaffold. Achieving this selectivity is going to be critical for patient safety, as clinical trials of HSP70 inhibitors have revealed significant liver toxicity.48
Given the high conservation between HSP70s across humans and parasites,49 it is important to consider other approaches to achieving selectivity. An essential component of the HSP70 chaperone cycle is its interaction with HSP40 (Figure 1); a key protein–protein interaction (PPI) that is essential for optimal activation of the HSP70 ATPase and chaperone activity.50 In the parasite, many of the exported PfHSP40s are expressed during the early stages of the asexual phase of the malaria parasite life cycle, and they are vital for malaria protein export and survival during febrile episodes.51, 52, 53 Some of these exported PfHSP40s have been confirmed to functionally interact with the only exported PfHSP70, PfHSP70-x. For example, Daniyan et al.54 and Dutta et al.36 found compelling evidence that the exported type II PfHSP40s, PFA0660w and PFE0055c have a functional association with PfHSP70-x. These studies aligned with other reports that identified a complex of these proteins (so-called J-Dots) in the cytosol of P. falciparum-infected erythrocytes, which appeared to be involved in the trafficking of the key virulence factor P. falciparum erythrocyte membrane protein 1.55, 56, 57, 58 These observations have focused attention on the PPIs between HSP70s and HSP40s as potential targets. This possibility has been explored using the pyrimidinone-based inhibitors of HSP70, MAL3-39, and DMT002264. These compounds bind at the interface of the HSP70–HSP40 contact and they were found to inhibit the HSP40-stimulated ATPase activity of human HSP70.59 Interestingly, however, only DMT002264 was able to also inhibit the PfHSP40-stimulated ATPase activity of PfHSP70-1 (Table 1).33 Likewise, malonganenone A was shown to inhibit the aggregation suppression activity of exported PfHSP70-x and disrupt its functional interaction with HSP40 (Table 1).32 Furthermore, recently the chalcone C86 (a pan-inhibitor of human HSP40s binding specifically to the signature J domain),37 was reported to inhibit the functional interaction of PFE0055c with PfHsp70-x (Table 1).36 These observations suggest that structural differences between the PfHSP70–PfHSP40 and human HSP70–HSP40 interfaces might be exploited. To explore this idea, key differences in the PPIs between P. falciparum and human cochaperone–chaperone complexes have been revealed based on structural analyses.36, 53, 60, 61, 62, 63 Furthermore, molecular docking studies have compared PfHSP70–PfHSP40 complexes to their human HSP70–HSP40 equivalents, and used these complexes to screen drug repurposing small molecule libraries to identify compounds with preferential binding to the malarial over the human system.64 As expected, the J domain–NBD interfaces of PfHSP70-x–PFE0055c and human HSP70 (HSPA1A)-DNAJA1 both involved multiple topologically equivalent hydrogen bond interactions between highly conserved positively charged J domain helix II residues and NBD negatively charged residues (Figure 2).64 However, the interfaces were not identical, reflecting differences in the nature and number of contacts, with the malarial J domain–NBD interface containing a greater number of hydrogen bonds and an electrostatic energy of interaction that suggested a higher binding affinity.64 Virtual screening studies identified five drug-like compounds that were potential preferential modulators of the malarial system: three binding at the interface of the PfHSP70–PfHSP40 complex (MBX 1641, PfHSP70-x–PFE0055c; and zoliflodacin and itraconazole, PfHSP70-2-PfSec63); and two binding to the J domain of the PfHSP40s (ezetimibe and a benzodiazepinone, PFE0055c J domain). Interestingly, itraconazole and ezetimibe have both been approved by the Food and Drug Administration (FDA, USA), and shown to inhibit the growth of the malaria parasite.64 However, further experimental biophysical, biochemical, and medicinal chemistry efforts on these candidate compounds are required to validate their potential as specific modulators of the PfHSP70–PfHSP40 system, as well as their potential for development into antimalarial drugs.
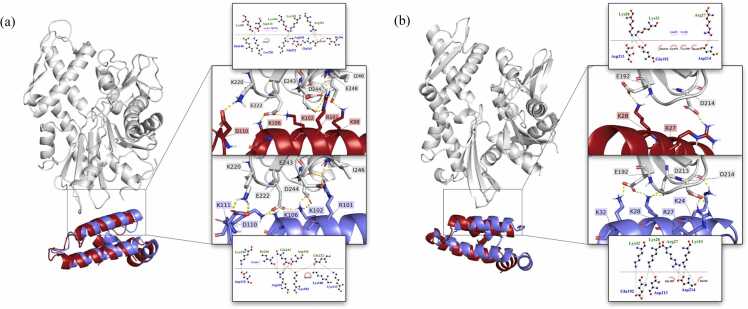
Fig. 2.
The J domain−NBD interfaces of PfHSP70-x–PFE0055c and human HSP70 (HSPA1A)-DNAJA1 are similar but not identical. The predicted three-dimensional structures of the complexes were generated using HADDOCK (J domain; red helices)65 and ClusPro (J domain; blue helixes).66 (a) PfHsp70-x–PFE00055c complex (b) human HSP70-DNAJA1 complex. The complexes were graphically rendered using PyMol 2.5.2 (PyMOL Molecular Graphics System, Version 2.0 Schrödinger, LLC). The insets on the right-hand side of each protein structure highlight the major interacting residues at the interface (yellow dotted lines for hydrogen bonds) and associated contact analyses (LigPlot+67). For the contact analyses, the HADDOCK complex J domain bonds are shown with red lines, the ClusPro complex J domain bonds are shown with blue lines, NBD bonds are shown with gray lines, nitrogen, and oxygen are highlighted with blue and red colors dots, respectively, hydrogen bonds are shown by green dashed lines with the length of the bond printed in the middle, salt bridges shown by red dashed lines and hydrophobic contacts between proteins are indicated by the brick-red spoked arcs for the NBD and pink spoked arcs for the J domain. Abbreviations used: HSP, heat shock protein; NBD, nucleotide-binding domain; PfHSP, Plasmodium falciparum HSP.
This figure and the figure legend are adapted with permission from the supplementary materials of a previous publication.64
Another member of the HSP70 family, the P. falciparum HSP70-2 (PfHSP70-2), an essential endoplasmic reticulum (ER)-resident chaperone,17 has recently gained attention as a possible target for antimalarial drug discovery.68, 69 PfHSP70-2 provides cytoprotection to the malaria parasite, particularly during the physiological and environmental changes it experiences during transmission from the vector to the host.70 PfHSP70-2 is also known as P. falciparum immunoglobulin binding protein; and P. falciparum glucose-regulated protein 78 kDa.71, 72, 73 The human ortholog of PfHSP70-2 is GRP78 which resides in the lumen of the ER, binds to newly formed polypeptides translocating through the ER Sec complex and assists folding into the native state.74, 75 During stress conditions, normal ER functions are disturbed leading to the accumulation of misfolded proteins, which triggers the onset of the unfolded protein response (UPR) pathway to restore normal ER functioning. The UPR, in turn, upregulates the expression of ER chaperones to degrade misfolded proteins.76 In addition, the UPR attenuates translation and selectively degrades mRNAs to further reduce the folding load.77, 78 GRP78 plays a very important role in maintaining ER homeostasis by facilitating correct protein folding, and reducing physiological as well as pathological harm that may lead to misfolded protein accumulation and ER stress.79 GRP78 interacts with its partner ER-resident HSP40, Sec63, which is involved in protein translocation into the ER.75, 80 PfHSP70-2 has been found to regulate eIF2-α-mediated arrest of protein translation, an important event during the schizont and gametocyte stages.77, 81, 82, 83 PfHsp70-2 was reported to be involved in the oxidative folding of ER proteins in association with ER-resident folding catalysts, a PfHSP40 (Pfj2), and a protein disulfide isomerase (PfPDI-8).84 PfHSP70-2 has also been reported to be present in ER-like structures in P. falciparum72 and the Maurer’s clefts.85 As in the human system, PfHSP70-2 is proposed to interact with the ER-resident PfHSP40 homologous to Sec63 (PfSec63), and through its association with PfSec63 and the PfSec complex, to play a role in protein translocation into the ER.11, 80, 86, 87, 88 Similarly to PfHSP70-x, PfHSP70-2 also interacts with exported virulence proteins such as P. falciparum erythrocyte membrane protein 1.87, 89, 90 Very limited information about small molecule inhibitors of PfHSP70-2 is available.10 Using in vitro binding assays, four commercially available small molecule inhibitors with broad specificity to the HSP70 family members viz., gilvocarcin V, apoptozole, MKT-077, and VER-15500891, 92, 93, 94 were found to interact with PfHSP70-2.68 All these small molecules exhibited very little difference in their binding affinities to PfHSP70-2 and GRP78, with the exception of VER-155008 which showed three-fold lesser binding affinity to PfHSP70-2 as compared to GRP78. PfHSP70-2 has a somewhat compact ATP binding pocket (Figure 1), leading to the proposal that this results in a rigidity that may be responsible for the lower binding affinity to VER-155008.68 Interestingly, the rigidity of the ATP pocket of PfHSP70-2 also accounts for the low affinity for ADP/ATP, which is potentially advantageous in competitive inhibitor screening. Recently, eight potential nucleoside inhibitors (HP2, HP3, HP5, HP10, HP19, HP20, HP22, and HP23/VER-155008) binding to the ATP binding domain of PfHSP70-2 were identified using a crystal-ligand soaking-based assay.69 As already alluded to, virtual screening studies identified two drug-like compounds that were potential preferential modulators of PfHSP70-2-PfSec63 over human GRP78-Sec63 (zoliflodacin and itraconazole).64 Building on these recent findings, further experimental studies are required to validate and develop these novel compounds into derivatives with proven specificity for the malarial chaperones and potential candidate antimalarial drugs.
PfHSP70 (noncanonical)
The malaria parasite does not appear to have the typical array of nucleotide exchange factors (NEFs) as found in the human system (e.g. BAG1-6 and HSPBP1).95 However, a number of reports have proposed that the cytosolic noncanonical PfHSP70, PfHSP70-z (homolog of cytosolic human HSP110) may serve as a NEF for cytosolic PfHSP70-1.96, 97, 98 Similarly, the ER-resident noncanonical PFHSP70, PfHSP70-y (homolog of human GRP170), may also serve as an NEF for PfHSP70-2. There are very few studies on inhibitors of HSP110/GRP170 in general,99 and no such studies have been carried out on the noncanonical PfHSP70s (PfHSP70-z and PfHSP70-y). Given that they are essential to survival of the malaria parasite,17, 43 and have unique structural features compared to their human homologs (e.g. for PfHSP70-z, differences in subdomain contacts of the NBD, linker properties and interdomain dynamics; Figure 1),95 PfHSP70-z and PfHSP70-y and their complexes with partners PfHSP70s represent novel targets for future small molecule inhibitor screening studies.
HSP complexes at the host–parasite interface are also drug targets
Interestingly, there is compelling evidence to suggest that it is imperative to more deeply explore the HSP complexes at the host–parasite interface.11, 63, 100 Comprehensive studies conducted over 20 years ago have already established that full-length and functional human HSP70/HSP90 organizing protein (HOP), HSP70 and HSP90 were present in the infected erythrocyte cytosol, potentially as both the free form and together in a highly complexed state.101 These findings have been validated by more recent proteomics analyses of knob-associated protein complexes.102 Furthermore, human HOP, HSP40s, HSP70s, and HSP90s have been found by proteomics studies on malaria parasite-infected erythrocytes, to be relatively enriched in the erythrocyte cytosol.63, 103 With this backdrop, it is not surprising that reports are emerging to suggest that exported PfHSP40s are not only capable of forming homologous partnerships (e.g. PFE0055c and PA0660w with PfHsp70-x) but also heterologous pairings with human HSP70 (e.g. PFB0090c/KAHSP40104; PFE0055c105; PFA0660w106; PF11_0034/eCiJp107). Structural bioinformatics analysis of the J domains of these PfHSP40s suggests that the nature of their interactions with human HSP70 may well be quite different (Figure 3). PFE0055c and PFB0090c/KAHSP40 have canonical J domains with the key helix II motifs required for the initial interaction with HSP70 (RK26-27 and HPD33-35; Escherichia coli DnaJ residue numbering) and would be predicted to optimally stimulate HSP70 chaperone activity. On the other hand, PFA0660w has a disruptive substitution of one of the key residues (L in the RK motif) and PF11_0034/eCiJp has disruptive substitutions of all the motif residues, apart from one (D in the HPD motif), and so would be predicted to suboptimally stimulate, not stimulate, or inhibit HSP70 chaperone activity. Given the enrichment of human HSPs in the infected erythrocyte cytosol, and the large number (18) of exported PfHSP40s,36 probably operating at lower levels in a catalytic capacity, heterologous HSP70–HSP40 partnerships may well be the predominant scenario (especially human HSP70–PfHSP40 partnerships). While the evidence is limited,44 the existence of heterologous complexes of PfHSP70-x with human HOP and HSP90 is also a possibility. These potential heterologous complexes would have different PPI interfaces compared to homologous complexes, given that PfHSP70-x has a C-terminal EEVN while human Hsp70 has an EEVD. These potential scenarios suggest that the door is wide open with respect to potential PPI interfaces that could be important drug targets for future antimalarial drug development.
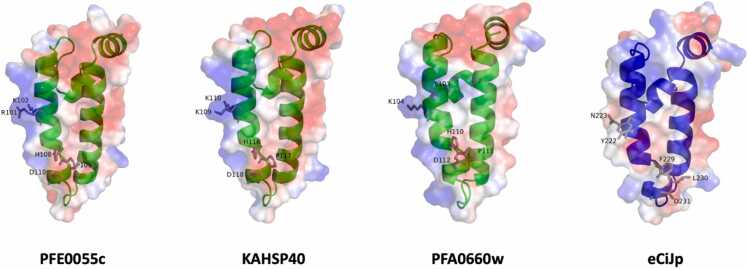
Fig. 3.
The exported PfHSP40s reported to interact with human HSP70. Three-dimensional models of the J domains for the malarial proteins PFE0055c (PF3D7_0501100), KAHSP40 (PFB0090c/PF3D7_0201800), PFA0660w (PF3D7_0113700), and eCiJp (PF11_0034/PF3D7_1102200) are shown. The J domains of type II (green cartoons; PFE0055c, KAHSP40, and PFA0660w) and type IV (blue cartoon; eCiJp) PfHSP40s are shown. The conserved RK and HPD motifs are shown as gray sticks. The positive charge is shown on blue colored surface, the negative charge is shown on red colored surface, and neutral potentials are shown on white colored surface. The surface electrostatic potential was calculated by APBS. The PFA0660w J domain was obtained from the Protein Data Bank (PDB code 6RZY), and homology models were prepared for the other J domains using SWISS-MODEL (6RZY was the template for PFE0055c and KAHSP40; 4J7Z was the template for eCiJp).108 The models were graphically rendered using PyMol 2.5.2 (PyMOL Molecular Graphics System, Version 2.0 Schrödinger, LLC). Abbreviations used: HSP, heat shock protein; PfHSP, Plasmodium falciparum HSP.
Conclusion
There is growing evidence that targeting chaperone–cochaperone network of PfHSPs offers a promising avenue for antimalarial drug development. PfHSPs play pivotal roles in parasite survival and pathology, are important for progression through the various stages of the parasite life cycle, and are incorporated into complexes at the host–parasite interface. Despite challenges posed by structural similarities between human and malarial HSPs, recent studies have unveiled subtle differences that could be exploited to design selective modulators targeting PfHSPs (Fig. 1, Fig. 2, Fig. 3). While inhibitors have been identified with higher affinity for PfHSP90 over human HSP90, challenges will be faced with off-target effects and toxicity when progressing candidate drugs through preclinical and clinical trials, and with resistance when the drugs are released as a treatment. Studies on model organisms have shown that Hsp90 has a global function as an evolutionary capacitator, suggesting that PfHSP90 may also enable the malaria parasite to tolerate mutations for deleterious phenotypes when its expression is high, while also harboring mutations that could promote the development of antimalarial drug resistance when its levels are low or inhibited.109 Lessons need to be learned from the excellent progress made in addressing these challenges for HSP90-based anticancer drugs.110 Promising results have been seen with the use of highly specific Hsp90 inhibitors applied in combination therapies at well-calibrated low-level doses.110 Furthermore, PfHSP90-based combination therapies could be explored with inhibitors of the other PfHSPs. Indeed, investigations into PfHSP70s have identified potential inhibitors, highlighting promising initial steps in targeting these proteins. Exploring PPIs between PfHSP70s and PfHSP40s, essential for parasite protein export and survival, presents another intriguing avenue for drug development, with virtual screening identifying potential drug-like candidates from drug repurposing small molecule libraries. Additionally, understanding the roles of HSP complexes at the host–parasite interface, especially the potential for heterologous partnerships between human and malarial HSPs, opens new possibilities for targeting PPIs crucial for malaria parasite survival. Further research and validation of potential modulators identified through computational screening are necessary next steps toward the development of specific antimalarial drugs targeting HSPs.
Author contributions
All authors listed have made a direct, and intellectual contribution to the work and approved the manuscript for publication. The contributions made by the authors were considered equal, leading to the collective decision to list the authors in alphabetical order.
Funding and support
This work was supported by the Higher Colleges of Technology, UAE (Interdisciplinary Research Grant number 213471).
Declarations of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Data availability statement
No data was used for the research described in the article.
References
- 1.WHO. World Malaria Report. Geneva: World Health Organization; 2022. https://www.who.int/teams/global-malaria-programme/reports. Accessed July 17, 2023.
- 2.Balikagala B., Fukuda N., Ikeda M., et al. Evidence of artemisinin-resistant malaria in Africa. N Engl J Med. 2021;385:1163–1171. doi: 10.1056/NEJMoa2101746. [DOI] [PubMed] [Google Scholar]
- 3.Mok S., Ashley E.A., Ferreira P.E., et al. Population transcriptomics of human malaria parasites reveals the mechanism of artemisinin resistance. Science. 2015;347:431–435. doi: 10.1126/science.1260403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mok S., Stokes B.H., Gnädig N.F., et al. Artemisinin-resistant K13 mutations rewire Plasmodium falciparum's intra-erythrocytic metabolic program to enhance survival. Nat Commun. 2021;12:530. doi: 10.1038/s41467-020-20805-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Siddiqui F.A., Liang X., Cui L. Plasmodium falciparum resistance to ACTs: emergence, mechanisms, and outlook. Int J Parasitol Drugs Drug Resist. 2021;16:102–118. doi: 10.1016/j.ijpddr.2021.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paloque L., Coppée R., Stokes B.H., et al. Mutation in the Plasmodium falciparum BTB/POZ domain of K13 protein confers artemisinin resistance. Antimicrob Agents Chemother. 2022;66 doi: 10.1128/AAC.01320-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shibeshi M.A., Kifle Z.D., Atnafie S.A. Antimalarial drug resistance and novel targets for antimalarial drug discovery. Infect Drug Resist. 2020;13:4047–4060. doi: 10.2147/IDR.S279433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Edkins A.L., Blatch G.L. In: Drug Discovery in Africa. Chibale K., Davies-Coleman M.T., Masimirembwa C., editors. Springer-Verlag; Berlin: 2012. Targeting conserved pathways as a strategy for novel drug development: disabling the cellular stress response; pp. 101–126. [Google Scholar]
- 9.Pesce E.-R., Blatch G.L., Edkins A.L. In: McAlpine S.R., Edkins A.L., editors. Vol. 19. Springer International Publishing; Switzerland: 2016. Hsp40 co-chaperones as drug targets: current status on development of inhibitors; pp. 163–195. (Heat Shock Protein Inhibitors: Success Stories - Topics in Medicinal Chemistry). [Google Scholar]
- 10.Barth J., Schach T., Przyborski J.M. HSP70 and their co-chaperones in the human malaria parasite P. falciparum and their potential as drug targets. Front Mol Biosci. 2022;9 doi: 10.3389/fmolb.2022.968248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blatch G.L. Plasmodium falciparum molecular chaperones: guardians of the malaria parasite proteome and renovators of the host proteome. Front Cell Dev Biol. 2022;10 doi: 10.3389/fcell.2022.921739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edkins A.L., Boshoff A. General structural and functional features of molecular chaperones. Adv Exp Med Biol. 2021;1340:11–73. doi: 10.1007/978-3-030-78397-6_2. [DOI] [PubMed] [Google Scholar]
- 13.Kampinga H.H., Andreasson C., Barducci A., et al. Function, evolution, and structure of J-domain proteins. Cell Stress Chaperones. 2019;24:7–15. doi: 10.1007/s12192-018-0948-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Malinverni D., Zamuner S., Rebeaud M.E., Barducci A., Nillegoda N.B., De Los Rios P. Data-driven large-scale genomic analysis reveals an intricate phylogenetic and functional landscape in J-domain proteins. Proc Natl Acad Sci USA. 2023;120 doi: 10.1073/pnas.2218217120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang R., Malinverni D., Cyr D.M., Rios P.L., Nillegoda N.B. J-domain protein chaperone circuits in proteostasis and disease. Trends Cell Biol. 2023;33:30–47. doi: 10.1016/j.tcb.2022.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shonhai A., G. Maier A., M. Przyborski J., L. Blatch G. Intracellular protozoan parasites of humans: the role of molecular chaperones in development and pathogenesis. Protein Pept Lett. 2011;18:143–157. doi: 10.2174/092986611794475002. [DOI] [PubMed] [Google Scholar]
- 17.Zhang M., Wang C., Otto T.D., et al. Uncovering the essential genes of the human malaria parasite Plasmodium falciparum by saturation mutagenesis. Science. 2018;360:p.eaap7847. doi: 10.1126/science.aap7847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cooper R., Carucci D. Proteomic approaches to studying drug targets and resistance in Plasmodium. Curr Drug Targets Infect Disord. 2004;4:41–51. doi: 10.2174/1568005043480989. [DOI] [PubMed] [Google Scholar]
- 19.Pesce E.-R., Cockburn I.L., Goble J.L., Stephens L.L., Blatch G.L. Malaria heat shock proteins: drug targets that chaperone other drug targets. Infect Disord Drug Targets. 2010;10:147–157. doi: 10.2174/187152610791163417. [DOI] [PubMed] [Google Scholar]
- 20.Daniyan M.O. Heat shock proteins as targets for novel antimalarial drug discovery. Adv Exp Med Biol. 2021;1340:205–236. doi: 10.1007/978-3-030-78397-6_9. [DOI] [PubMed] [Google Scholar]
- 21.Shahinas D., Folefoc A., Taldone T., Chiosis G., Crandall I., Pillai D.R. A purine analog synergizes with chloroquine (CQ) by targeting Plasmodium falciparum Hsp90 (PfHsp90) PloS ONE. 2013;8 doi: 10.1371/journal.pone.0075446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bayih A.G., Folefoc A., Mohon A.N., et al. In vitro and in vivo anti-malarial activity of novel harmine-analog heat shock protein 90 inhibitors: a possible partner for artemisinin. Malaria J. 2016;15:579. doi: 10.1186/s12936-016-1625-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Posfai D., Eubanks A.L., Keim A.I., et al. Identification of Hsp90 inhibitors with anti-Plasmodium activity. Antimicrob Agents Chemother. 2018;62 doi: 10.1128/AAC.01799-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang T., Mäser P., Picard D. Inhibition of Plasmodium falciparum Hsp90 contributes to the antimalarial activities of aminoalcohol-carbazoles. J Med Chem. 2016;59:6344–6352. doi: 10.1021/acs.jmedchem.6b00591. [DOI] [PubMed] [Google Scholar]
- 25.Mafethe O., Ntseane T., Dongola T.H., Shonhai A., Gumede N.J., Mokoena F. Pharmacophore model-based virtual screening workflow for discovery of inhibitors targeting Plasmodium falciparum Hsp90. ACS Omega. 2023;8:38220–38232. doi: 10.1021/acsomega.3c04494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cowell A.N., Istvan E.S., Lukens A.K., et al. Mapping the malaria parasite druggable genome by using in vitro evolution and chemogenomics. Science. 2018;359:191–199. doi: 10.1126/science.aan4472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nndwammbi A.A.T., Dongola T.H., Shonhai A., Mokoena F., Pooe O.J., Simelane M.B.C. Ursolic acid acetate and iso-mukaadial acetate bind to Plasmodium falciparum Hsp90, abrogating its chaperone function in vitro. Naunyn-Schmiedeberg's Arch Pharmacol. 2024 doi: 10.1007/s00210-024-02944-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zininga T., Pooe O.J., Makhado P.B., et al. Polymyxin B inhibits the chaperone activity of Plasmodium falciparum Hsp70. Cell Stress Chaperones. 2017;22:707–715. doi: 10.1007/s12192-017-0797-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zininga T., Ramatsui L., Makhado P., et al. (−)-Epigallocatechin-3-Gallate inhibits the chaperone activity of Plasmodium falciparum Hsp70 chaperones and abrogates their association with functional partners. Molecules. 2017;22:2139. doi: 10.3390/molecules22122139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Salomane N., Pooe O.J., Simelane M.B.C. Iso-mukaadial acetate and ursolic acid acetate inhibit the chaperone activity of Plasmodium falciparum heat shock protein 70-1. Cell Stress Chaperones. 2021;26:685–693. doi: 10.1007/s12192-021-01212-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cockburn I.L., Pesce E.-R., Pryzborski J.M., et al. Screening for small molecule modulators of Hsp70 chaperone activity using protein aggregation suppression assays: inhibition of the plasmodial chaperone PfHsp70-1. Biol Chem. 2011;392:431–438. doi: 10.1515/BC.2011.040. [DOI] [PubMed] [Google Scholar]
- 32.Cockburn I.L., Boshoff A., Pesce E.-R., Blatch G.L. Selective modulation of plasmodial Hsp70s by small molecules with antimalarial activity. Biol Chem. 2014;395:1353–1362. doi: 10.1515/hsz-2014-0138. [DOI] [PubMed] [Google Scholar]
- 33.Botha M., Chiang A.N., Needham P.G., et al. Plasmodium falciparum encodes a single cytosolic type I Hsp40 that functionally interacts with Hsp70 and is upregulated by heat shock. Cell Stress Chaperones. 2011;16:389–401. doi: 10.1007/s12192-010-0250-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kayamba F., Malimabe T., Ademola I.K., et al. Design and synthesis of quinoline-pyrimidine inspired hybrids as potential plasmodial inhibitors. Eur J Med Chem. 2021;217 doi: 10.1016/j.ejmech.2021.113330. [DOI] [PubMed] [Google Scholar]
- 35.Muthelo T., Mulaudzi V., Netshishivhe M., et al. Inhibition of Plasmodium falciparum Hsp70-Hop partnership by 2-phenylthynesulfonamide. Front Mol Biosci. 2022;9 doi: 10.3389/fmolb.2022.947203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dutta T., Singh H., Gestwicki J.E., Blatch G.L. Exported plasmodial J domain protein, PFE0055c, and PfHsp70-x form a specific co-chaperone-chaperone partnership. Cell Stress Chaperones. 2021;26:355–366. doi: 10.1007/s12192-020-01181-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moses M.A., Kim Y.S., Rivera-Marquez G.M., et al. Targeting the Hsp40/Hsp70 chaperone axis as a novel strategy to treat castration-resistant prostate cancer. Cancer Res. 2018;78:4022–4035. doi: 10.1158/0008-5472.CAN-17-3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dutta T., Singh H., Edkins A.L., Blatch G.L. Hsp90 and associated co-chaperones of the malaria parasite. Biomolecules. 2022;12:1018. doi: 10.3390/biom12081018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sanchez J., Carter T.R., Cohen M.S., Blagg B.S. Old and new approaches to target the Hsp90 Chaperone. Curr Cancer Drug Targets. 2020;20:253–270. doi: 10.2174/1568009619666191202101330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stofberg M.L., Caillet C., de Villiers M., Zininga T. Inhibitors of the Plasmodium falciparum Hsp90 towards selective antimalarial drug design: the past, present and future. Cells. 2021;10:2849. doi: 10.3390/cells10112849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang T., Bisson W.H., Mäser P., Scapozza L., Picard D. Differences in conformational dynamics between Plasmodium falciparum and human Hsp90 orthologues enable the structure-based discovery of pathogen-selective inhibitors. Journal of medicinal chemistry. 2024;57:2524–2535. doi: 10.1021/jm401801t. [DOI] [PubMed] [Google Scholar]
- 42.Banumathy G., Singh V., Pavithra S.R., Tatu U. Heat shock protein 90 function is essential for Plasmodium falciparum growth in human erythrocytes. J Biol Chem. 2003;278:18336–18345. doi: 10.1074/jbc.M211309200. [DOI] [PubMed] [Google Scholar]
- 43.Muralidharan V., Oksman A., Pal P., Lindquist S., Goldberg D.E. Plasmodium falciparum heat shock protein 110 stabilizes the asparagine repeat-rich parasite proteome during malarial fevers. Nat Commun. 2012;3:1310. doi: 10.1038/ncomms2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mabate B., Zininga T., Ramatsui L., et al. Structural and biochemical characterization of Plasmodium falciparum Hsp70-x reveals functional versatility of its C-terminal EEVN motif. Proteins. 2018;86:1189–1201. doi: 10.1002/prot.25600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lebepe C.M., Matambanadzo P.R., Makhoba X.H., Achilonu I., Zininga T., Shonhai A. Comparative characterization of Plasmodium falciparum Hsp70-1 relative to E. coli DnaK reveals the functional specificity of the parasite chaperone. Biomolecules. 2020;10:856. doi: 10.3390/biom10060856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rajapandi T. Chaperoning of asparagine repeat-containing proteins in Plasmodium falciparum. J Parasit Dis. 2020;44:687–693. doi: 10.1007/s12639-020-01251-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Anas M., Shukla A., Tripathi A., et al. Structural-functional diversity of malaria parasite's PfHSP70-1 and PfHSP40 chaperone pair gives an edge over human orthologs in chaperone-assisted protein folding. Biochem J. 2020;477:3625–3643. doi: 10.1042/BCJ20200434. [DOI] [PubMed] [Google Scholar]
- 48.Koya K., Li Y., Wang H., et al. MKT-077, a novel rhodacyanine dye in clinical trials, exhibits anticarcinoma activity in preclinical studies based on selective mitochondrial accumulation. Cancer Res. 1996;56:538–543. [PubMed] [Google Scholar]
- 49.Shonhai A., Boshoff A., Blatch G.L. The structural and functional diversity of Hsp70 proteins from Plasmodium falciparum. Protein Sci. 2007;16:1803–1818. doi: 10.1110/ps.072918107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu Q., Liang C., Zhou L. Structural and functional analysis of the Hsp70/Hsp40 chaperone system. Protein Sci. 2020;29:378–390. doi: 10.1002/pro.3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dutta T., Pesce E.-R., Maier A., Blatch G.L. Role of the J domain protein family in the survival and pathogenesis of Plasmodium falciparum. Adv Exp Med Biol. 2021;1340:97–123. doi: 10.1007/978-3-030-78397-6_4. [DOI] [PubMed] [Google Scholar]
- 52.Bozdech Z., Llinás M., Pulliam B.L., Wong E.D., Zhu J., DeRisi J.L. The transcriptome of the intraerythrocytic developmental cycle of Plasmodium falciparum. PLoS Biol. 2003;1 doi: 10.1371/journal.pbio.0000005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Almaazmi S.Y., Singh H., Dutta T., Blatch G.L. Exported J domain proteins of the human malaria parasite. Front Mol Biosci. 2022;9 doi: 10.3389/fmolb.2022.978663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Daniyan M.O., Boshoff A., Prinsloo E., Pesce E.-R., Blatch G.L. The malarial exported PFA0660w is an Hsp40 co-chaperone of PfHsp70-x. PLoS ONE. 2016;11 doi: 10.1371/journal.pone.0148517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Külzer S., Rug M., Brinkmann K., et al. Parasite-encoded Hsp40 proteins define novel mobile structures in the cytosol of the P. falciparum-infected erythrocyte. Cell Microbiol. 2010;12:1398–1420. doi: 10.1111/j.1462-5822.2010.01477.x. [DOI] [PubMed] [Google Scholar]
- 56.Külzer S., Charnaud S., Dagan T., et al. Plasmodium falciparum-encoded exported hsp70/hsp40 chaperone/co-chaperone complexes within the host erythrocyte. Cell Microbiol. 2012;14:1784–1795. doi: 10.1111/j.1462-5822.2012.01840.x. [DOI] [PubMed] [Google Scholar]
- 57.Petersen W., Külzer S., Engels S., et al. J-dot targeting of an exported HSP40 in Plasmodium falciparum-infected erythrocytes. Int J Parasitol. 2016;46:519–525. doi: 10.1016/j.ijpara.2016.03.005. [DOI] [PubMed] [Google Scholar]
- 58.Behl A., Kumar V., Bisht A., Panda J.J., Hora R., Mishra P.C. Cholesterol bound Plasmodium falciparum co-chaperone 'PFA0660w' complexes with major virulence factor 'PfEMP1' via chaperone 'PfHsp70-x. Sci Rep. 2019;9:2664. doi: 10.1038/s41598-019-39217-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wisén S., Bertelsen E.B., Thompson A.D., et al. Binding of a small molecule at a protein-protein interface regulates the chaperone activity of hsp70-hsp40. ACS Chem Biol. 2010;5:611–622. doi: 10.1021/cb1000422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hatherley R., Blatch G.L., Bishop O.T. Plasmodium falciparum Hsp70-x: a heat shock protein at the host-parasite interface. J Biomol Struct Dyn. 2014;32:1766–1779. doi: 10.1080/07391102.2013.834849. [DOI] [PubMed] [Google Scholar]
- 61.Daniyan M.O., Blatch G.L. Plasmodial Hsp40s: new avenues for antimalarial drug discovery. Curr Pharm Des. 2017;23:4555–4570. doi: 10.2174/1381612823666170124142439. [DOI] [PubMed] [Google Scholar]
- 62.Day J., Passecker A., Beck H.P., Vakonakis I. The Plasmodium falciparum Hsp70-x chaperone assists the heat stress response of the malaria parasite. FASEB J. 2019;33:14611–14624. doi: 10.1096/fj.201901741R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Almaazmi S.Y., Kaur R.P., Singh H., Blatch G.L. The Plasmodium falciparum exported J domain proteins fine-tune human and malarial Hsp70s: pathological exploitation of proteostasis machinery. Front Mol Biosci. 2023;10 doi: 10.3389/fmolb.2023.1216192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Singh H., Almaazmi S.Y., Dutta T., Keyzers R.A., Blatch G.L. In silico identification of modulators of J domain protein-Hsp70 interactions in Plasmodium falciparum: a drug repurposing strategy against malaria. Front Mol Biosci. 2023;10:1158912. doi: 10.3389/fmolb.2023.1158912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.van Zundert G.C.P., Rodrigues J.P.G.L.M., Trellet M., et al. The HADDOCK2.2 Web Server: User-Friendly Integrative Modeling of Biomolecular Complexes. Journal of molecular biology. 2016;428:720–725. doi: 10.1016/j.jmb.2015.09.014. [DOI] [PubMed] [Google Scholar]
- 66.Kozakov D., Hall D.H., Xia B., et al. The ClusPro web server for protein–protein docking. Nat Protoc. 2017;12:255–278. doi: 10.1038/nprot.2016.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Laskowski R.A., Swindells M.B. LigPlot+: multiple ligand–protein interaction diagrams for drug discovery. J Chem Inf Model. 2011;51:2778–2786. doi: 10.1021/ci200227u. [DOI] [PubMed] [Google Scholar]
- 68.Chen Y., Murillo-Solano C., Kirkpatrick M.G., Antoshchenko T., Park H.W., Pizarro J.C. Repurposing drugs to target the malaria parasite unfolding protein response. Sci Rep. 2018;8:1–12. doi: 10.1038/s41598-018-28608-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mrozek A., Antoshchenko T., Chen Y., et al. A non-traditional crystal-based compound screening method targeting the ATP binding site of Plasmodium falciparum GRP78 for identification of novel nucleoside analogues. Front Mol Biosci. 2022;9 doi: 10.3389/fmolb.2022.956095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Przyborski J.M., Diehl M., Blatch G.L. Plasmodial HSP70s are functionally adapted to the malaria parasite life cycle. Front Mol Biosci. 2015;2:34. doi: 10.3389/fmolb.2015.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kumar N., Syin C.A., Carter R., Quakyi I., Miller L.H. Plasmodium falciparum gene encoding a protein similar to the 78-kDa rat glucose-regulated stress protein. Proc Natl Acad Sci USA. 1988;85:6277–6281. doi: 10.1073/pnas.85.17.6277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kumar N., Koski G., Harada M., Aikawa M., Zheng H. Induction and localization of Plasmodium falciparum stress proteins related to the heat shock protein 70 family. Mol Biochem Parasitol. 1991;48:47–58. doi: 10.1016/0166-6851(91)90163-z. [DOI] [PubMed] [Google Scholar]
- 73.Kumar N., Zheng H. Nucleotide sequence of a Plasmodium falciparum stress protein with similarity to mammalian 78-kDa glucose-regulated protein. Mol Biochem Parasitol. 1992;56:353–356. doi: 10.1016/0166-6851(92)90187-o. [DOI] [PubMed] [Google Scholar]
- 74.Anelli T., Sitia R. Protein quality control in the early secretory pathway. EMBO J. 2008;27:315–327. doi: 10.1038/sj.emboj.7601974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Melnyk A., Lang S., Sicking M., Zimmermann R., Jung M. Co-chaperones of the human endoplasmic reticulum: an update. Subcell Biochem. 2023;101:247–291. doi: 10.1007/978-3-031-14740-1_9. [DOI] [PubMed] [Google Scholar]
- 76.Gardner B.M., Pincus D., Gotthardt K., Gallagher C.M., Walter P. Endoplasmic reticulum stress sensing in the unfolded protein response. Cold Spring Harb Perspect Biol. 2013;5:a013169. doi: 10.1101/cshperspect.a013169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Narasimhan J., Joyce B.R., Naguleswaran A., et al. Translation regulation by eukaryotic initiation factor-2 kinases in the development of latent cysts in Toxoplasma gondii. J Biol Chem. 2008;283:16591–16601. doi: 10.1074/jbc.M800681200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang M., Joyce B.R., Sullivan W.J., Nussenzweig V. Translational control in Plasmodium and Toxoplasma parasites. Eukaryot Cell. 2013;12:161–167. doi: 10.1128/EC.00296-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ibrahim I.M., Abdelmalek D.H., Elfiky A.A. GRP78: A cell’s response to stress. Life Sci. 2019;226:156–163. doi: 10.1016/j.lfs.2019.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zimmermann R., Blatch G.L. A novel twist to protein secretion in eukaryotes. Trends Parasitol. 2009;25:147–150. doi: 10.1016/j.pt.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 81.Zhang M., Mishra S., Sakthivel R., et al. PK4, a eukaryotic initiation factor 2α(eIF2α) kinase, is essential for the development of the erythrocytic cycle of Plasmodium. Proc Natl Acad Sci USA. 2012;109:3956–3961. doi: 10.1073/pnas.1121567109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chaubey S., Grover M., Tatu U. Endoplasmic reticulum stress triggers gametocytogenesis in the malaria parasite. J Biol Chem. 2014;289:16662–16674. doi: 10.1074/jbc.M114.551549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Paloque L., Ramadani A.P., Mercereau-Puijalon O., Augereau J.M., Benoit-Vical F. Plasmodium falciparum: multifaceted resistance to artemisinins. Malaria J. 2016;15:149. doi: 10.1186/s12936-016-1206-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cobb D.W., Florentin A., Fierro M.A., Krakowiak M., Moore J.M., Muralidharan V. A redox-active crosslinker reveals an essential and inhibitable oxidative folding network in the endoplasmic reticulum of malaria parasites. PLoS pathogens. 2021;17 doi: 10.1371/journal.ppat.1009293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Vincensini L., Richert S., Blisnick T., et al. Proteomic analysis identifies novel proteins of the Maurer’s clefts, a secretory compartment delivering Plasmodium falciparum proteins to the surface of its host cell. Mol Cell Proteomics. 2005;4:582–593. doi: 10.1074/mcp.M400176-MCP200. [DOI] [PubMed] [Google Scholar]
- 86.Tuteja R. Unraveling the components of protein translocation pathway in human malaria parasite Plasmodium falciparum. Arch Biochem Biophys. 2007;467:249–260. doi: 10.1016/j.abb.2007.08.031. [DOI] [PubMed] [Google Scholar]
- 87.Cortés G.T., Wiser M.F., Gómez-Alegría C.J. Identification of Plasmodium falciparum HSP70-2 as a resident of the Plasmodium export compartment. Heliyon. 2020;6 doi: 10.1016/j.heliyon.2020.e04037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Shonhai A. The role of Hsp70s in the development and pathogenicity of Plasmodium falciparum. Adv Exp Med Biol. 2021;1340:75–95. doi: 10.1007/978-3-030-78397-6_3. [DOI] [PubMed] [Google Scholar]
- 89.Saridaki T., Sanchez C.P., Pfahler J., Lanzer M. A conditional export system provides new insights into protein export in Plasmodium falciparum-infected erythrocytes. Cell Microbiol. 2008;10:2483–2495. doi: 10.1111/j.1462-5822.2008.01223.x. [DOI] [PubMed] [Google Scholar]
- 90.Batinovic S., McHugh E., Chisholm S.A., et al. An exported protein-interacting complex involved in the trafficking of virulence determinants in Plasmodium-infected erythrocytes. Nat Commun. 2017;8:16044. doi: 10.1038/ncomms16044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Matsumoto A., Hanawalt P.C. Histone H3 and heat shock protein GRP78 are selectively cross-linked to DNA by photoactivated gilvocarcin V in human fibroblasts. Cancer Res. 2000;60:3921–3926. [PubMed] [Google Scholar]
- 92.Rousaki A., Miyata Y., Jinwal U.K., Dickey C.A., Gestwicki J.E., Zuiderweg E.R. Allosteric drugs: the interaction of antitumor compound MKT-077 with human Hsp70 chaperones. J Mol Biol. 2011;411:614–632. doi: 10.1016/j.jmb.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kim S.H., Kang J.G., Kim C.S., et al. The hsp70 inhibitor VER155008 induces paraptosis requiring de novo protein synthesis in anaplastic thyroid carcinoma cells. Biochem Biophys Res Commun. 2014;454:36–41. doi: 10.1016/j.bbrc.2014.10.060. [DOI] [PubMed] [Google Scholar]
- 94.Park S.H., Kim W.J., Li H., et al. Anti-leukemia activity of a Hsp70 inhibitor and its hybrid molecules. Sci Rep. 2017;7:3537. doi: 10.1038/s41598-017-03814-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tripathi A., Del Galdo S., Chandramouli B., Kumar N. Distinct dynamical features of plasmodial and human HSP70-HSP110 highlight the divergence in their chaperone-assisted protein folding. Biochim Biophys Acta Proteins Proteom. 2023;1871 doi: 10.1016/j.bbapap.2023.140942. [DOI] [PubMed] [Google Scholar]
- 96.Zininga T., Achilonu I., Hoppe H., Prinsloo E., Dirr H.W., Shonhai A. Overexpression, purification and characterisation of the Plasmodium falciparum Hsp70-z (PfHsp70-z) protein. PloS ONE. 2015;10 doi: 10.1371/journal.pone.0129445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zininga T., Achilonu I., Hoppe H., Prinsloo E., Dirr H.W., Shonhai A. Plasmodium falciparum Hsp70-z, an Hsp110 homologue, exhibits independent chaperone activity and interacts with Hsp70-1 in a nucleotide-dependent fashion. Cell Stress Chaperones. 2016;21:499–513. doi: 10.1007/s12192-016-0678-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chakafana G., Mudau P.T., Zininga T., Shonhai A. Characterisation of a unique linker segment of the Plasmodium falciparum cytosol localised Hsp110 chaperone. Int J Biol Macromol. 2021;180:272–285. doi: 10.1016/j.ijbiomac.2021.03.056. [DOI] [PubMed] [Google Scholar]
- 99.Hu L., Sun C., Kidd J.M., et al. A first-in-class inhibitor of Hsp110 molecular chaperones of pathogenic fungi. Nat Commun. 2023;14:2745. doi: 10.1038/s41467-023-38220-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Jonsdottir T.K., Gabriela M., Gilson M.P.R. The role of malaria parasite heat shock proteins in protein trafficking and remodelling of red blood cells. Adv Exp Med Biol. 2021;1340:141–167. doi: 10.1007/978-3-030-78397-6_6. [DOI] [PubMed] [Google Scholar]
- 101.Banumathy G., Singh V., Tatu U. Host chaperones are recruited in membrane-bound complexes by Plasmodium falciparum. J Biol Chem. 2002;277:3902–3912. doi: 10.1074/jbc.M110513200. [DOI] [PubMed] [Google Scholar]
- 102.Alampalli S.V., Grover M., Chandran S., Tatu U., Acharya P. Proteome and structural organization of the knob complex on the surface of the Plasmodium infected red blood cell. Proteomics Clin Appl. 2018;12 doi: 10.1002/prca.201600177. [DOI] [PubMed] [Google Scholar]
- 103.Siddiqui G., De Paoli A., MacRaild C.A., et al. A new mass spectral library for high-coverage and reproducible analysis of the Plasmodium falciparum-infected red blood cell proteome. GigaScience. 2022;11 doi: 10.1093/gigascience/giac008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Acharya P., Chaubey S., Grover M., Tatu U. An exported heat shock protein 40 associates with pathogenesis related knobs in Plasmodium falciparum infected erythrocytes. PloS ONE. 2012;7 doi: 10.1371/journal.pone.0044605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhang Q., Ma C., Oberli A., Zinz A., Engels S., Przyborski J.M. Proteomic analysis of exported chaperone/co-chaperone complexes of P. falciparum reveals an array of complex protein-protein interactions. Sci Rep. 2017;7:42188. doi: 10.1038/srep42188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Diehl M., Roling L., Rohland L., et al. Co-chaperone involvement in knob biogenesis implicates host-derived chaperones in malaria virulence. PLoS Pathog. 2021;17 doi: 10.1371/journal.ppat.1009969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sahu W., Bai T., Panda P.K., et al. Plasmodium falciparum HSP40 protein eCiJp traffics to the erythrocyte cytoskeleton and interacts with the human HSP70 chaperone HSPA1. FEBS Lett. 2022;596:95–111. doi: 10.1002/1873-3468.14255. [DOI] [PubMed] [Google Scholar]
- 108.Waterhouse A., Bertoni M., Bienert S., et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nuc Acids Res. 2018;46:W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Anas M., Kumari V., Gupta N., Dube A., Kumar N. Protein quality control machinery in intracellular protozoan parasites: hopes and challenges for therapeutic targeting. Cell Stress Chaperones. 2019;24:891–904. doi: 10.1007/s12192-019-01016-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lacey T., Lacey H. Linking hsp90's role as an evolutionary capacitator to the development of cancer. Cancer Treat Res Commun. 2021;28 doi: 10.1016/j.ctarc.2021.100400. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data was used for the research described in the article.