Abstract
To date, immunotherapy has opened a new chapter in the treatment of lung cancer. Precise biomarkers can help to screen subpopulations of lung cancer to provide the best treatment. Multiple studies suggest that specific gene mutations may be predictive markers in guiding non-small cell lung cancer (NSCLC) immune checkpoint inhibitor (ICI) treatment. A published immunotherapy cohort with mutational and survival data for 350 NSCLC patients was used. First, the mutational data of the immunotherapy cohort were used to identify gene mutations related to the prognosis of ICI therapy. The immunotherapy cohort and TCGA-NSCLC cohort were further studied to elucidate the relationships between specific gene mutations and tumor immunogenicity, antitumor immune response capabilities, and immune cell and mutation counts in the DNA damage response (DDR) pathway. In the immunotherapy cohort (N = 350), ZFHX3 mutations were an independent predictive biomarker for NSCLC patients receiving ICI treatment. Significant differences were observed between ZFHX3-mutant (ZFHX3-MT) and ZFHX3-wild type (ZFHX3-WT) patients regarding the overall survival (OS) time (P < 0.001, HR = 0.26, 95% Cl 0.17–0.41). ZFHX3-MT is significantly associated with higher tumor mutation burden (TMB) and neoantigen load (NAL), and ZFHX3-MT positively correlates with known immunotherapy response biomarkers, including T-cell infiltration, immune-related gene expression, and mutation counts in the DDR pathway in NSCLC. ZFHX3-MT is closely related to longer OS in NSCLC patients treated with ICIs, suggesting that ZFHX3 mutations be used as a novel predictive marker in guiding NSCLC ICI treatment.
Electronic supplementary material
The online version of this article (10.1007/s00262-020-02668-8) contains supplementary material, which is available to authorized users.
Keywords: Non-small cell lung cancer, ZFHX3, Immune checkpoint inhibitor, Biomarker
Introduction
Lung cancer ranks first in the incidence and mortality of malignant tumors worldwide. Non-small cell lung cancer (NSCLC) accounts for approximately 80–85% of lung cancer, and more than 50% of patients have distant metastasis at the time of diagnosis. The 5-year survival rate in patients with NSCLC is less than 15–20% [1]. In recent years, with the discovery of immune checkpoint molecules, including programmed cell death protein-1 (PD-1), its ligand PD-L1, and cytotoxic T-lymphocyte-associated protein 4 (CTLA-4), immunotherapy has become one of the most promising treatment strategies for lung cancer. In advanced NSCLC, the response rate of treatment with immune checkpoint inhibitor (ICIs) is approximately 17–21%, and some patients have a very long-lasting response [2]. However, many patients do not benefit from ICIs, and immunotherapy lacks precise biomarkers to predict efficacy [3, 4]. Therefore, identifying biomarkers to screen dominant populations for ICI efficacy is particularly important.
In recent years, studies have shown that PD-L1 expression, tumor mutation burden (TMB), microsatellite instability, mismatch repair gene deficiency, special gene mutations, tumor immune microenvironment, gene expression profiles (GEPs), and antigen presentation defects may serve as predictive markers for the efficacy of ICIs [5]. However, these markers also have certain limitations, so precise predictive markers for ICI treatment still need to be explored [6, 7].
Studies have shown a correlation between specific gene mutations and the sensitivity of ICIs [8–10]. The loss of TP53 function is associated with increased PD-L1 expression and increased TMB [11–13]. In NSCLC, EGFR mutations may cause the upregulation of PD-L1 expression [14]. In multiple cancers, TET1 mutations were strongly associated with longer progression-free survival (PFS) and improved overall survival (OS) in patients receiving ICI treatment [10]. In addition, mutations in the DNA damage response and repair (DDR) pathway can increase tumor immunogenicity by accumulating incorrect DNA damage responses to increase the efficacy of ICI treatment [15, 16]. The above results suggest that gene mutations may be a novel predictive biomarker for ICIs in NSCLC.
In this study, we used an NSCLC immunotherapy cohort (reported by Samstein et al. [17]) with mutational and clinical data to further evaluate the association between specific gene mutations and the efficacy of ICIs in NSCLC. The results suggest that ZFHX3 mutations can be used as an independent predictive biomarker for NSCLC patients receiving ICIs. In addition, ZFHX3 mutations are strongly associated with improved OS, enhanced tumor immunogenicity, activated antitumor immunity, T-cell infiltration, immune-related gene expression, and mutation counts in the DDR pathway.
Materials and methods
Clinical samples and cancer cell lines
To assess the relationship between gene mutations and ICI efficacy in NSCLC patients, we collected an anti-PD-(L)1 monotherapy or in combination with anti-CTLA-4 NSCLC clinical cohort (N = 350). The R package “TCGAbiolinks” [18] was used to download The Cancer Genome Atlas (TCGA)-Lung Adenocarcinoma (LUAD) and TCGA-Lung Squamous Cell Carcinoma (LUSC) cohorts, including mRNA expression profiling data, somatic mutation data and patient prognosis information from TCGA. Then, the TCGA-LUAD and TCGA-LUSC cohorts were combined into the TCGA-NSCLC cohort for subsequent analysis. The unit of gene expression was pan-cancer normalized log2 (fragments per kilobase of exon model per million mapped fragments [FPKM] + 1). In addition, we used cBioPortal (https://www.cbioportal.org) to download the survival data (disease-free survival, DFS) of the TCGA-LUAD and TCGA-LUSC patients [19]. We downloaded data for 67 NSCLC cell lines with whole-exome sequencing (WES) and drug sensitivity data from the Genomics of Drug Sensitivity in Cancer (GDSC) database. We downloaded an independent cohort including 75 patients with NSCLC treated with nivolumab plus ipilimumab as part of the CheckMate-012 study [20].
Identification of survival-related gene mutations and establishment of prognostic gene mutations
The mutational data of the immunotherapy cohort were used to identify specific mutated genes related to the prognosis of ICIs. Univariate and multivariate Cox regression model analyses and Kaplan–Meier analysis were used to evaluate the predictive function of specific mutated genes in ICI treatment. The detailed analysis process is shown in Supplementary Fig. S1.
Correlation analysis of tumor immunogenicity and immune characteristics
We used the CIBERSORT web portal (https://cibersort.stanford.edu/) to evaluate the 22 immune cell infiltration status of the TCGA-NSCLC cohort [21]. The immune-associated gene list, neoantigen load (NAL) data, immune-related genes, and their functional classifications were from Thorsson et al. [22], and the expression levels of these genes were quantified as log2 (FPKM + 1). We took the somatic called variants in the TCGA-NSCLC cohort as the raw mutation count. In addition, we used 38 Mb as the estimate of the exome size [23].
Pathway enrichment analysis and gene sets related to the DDR pathway
For gene annotation enrichment analysis, the clusterProfiler R package was used, and P < 0.05 indicated a significant difference for Gene Ontology (GO) terms, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways and Reactome pathways [24]. The gene set enrichment analysis (GSEA) gene set and the DDR pathway gene set were obtained from the Broad Institute Molecular Signatures Database (MSigDB) [25]. The details of DDR-related gene sets are shown in Supplementary Table. S1.
Statistical analysis
The association between ZFHX3 status and TMB, NAL, immune gene expression, and immune cells was examined using the Mann–Whitney U test. Assessment of the association between the top 20 mutated genes’ rate and ZFHX3 status was performed with Fisher’s exact test and the Chi-square test. The DFS and OS probabilities of ZFHX3-mutant (ZFHX3-MT) and ZFHX3-wild type (ZFHX3-WT) patients were analyzed by the Kaplan–Meier method, the log-rank test, and univariate and multivariate Cox proportional hazard regression analyses. P < 0.05 was considered statistically significant, and all statistical tests were two-sided. R software (version 3.6) was used for statistical analysis. The R package ComplexHeatmap was employed to visualize the mutational landscape and create a heatmap [26].
Results
ZFHX3 gene mutation is an independent predictive biomarker for the treatment response of NSCLC patients receiving ICIs
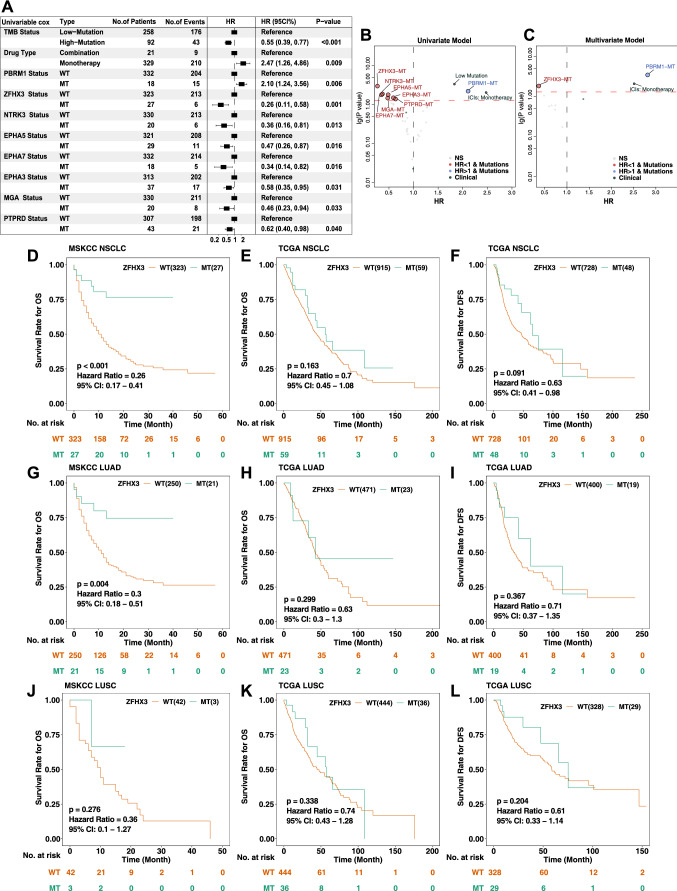
We used cBioPortal to collect a published cohort study of NSCLC immunotherapy patients (Samstein et al.), including 350 patients receiving ICIs (anti-PD-(L)1 monotherapy or in combination with anti-CTLA-4) with available mutational and clinical data to further explore the relationship between gene mutations and the prognosis of ICIs (Supplementary Fig. S1). Gene mutations (mutation frequency > 5% cases) were analyzed by a univariate Cox regression model. The analysis showed that the mutation counts, the types of immunotherapy drugs, and specific gene mutations (PBRM1, ZFHX3, NTRK3, EPHA5, EPHA7, EPHA3, MGA, and PTPRD) were related to the prognosis of immunotherapy, and the differences were significant (Fig. 1a, P < 0.05). Among them, low mutation counts, PBRM1 mutations, and immune monotherapy were associated with a worse prognosis of immunotherapy; on the contrary, other gene mutations, including ZFHX3, NTRK3, EPHA5, EPHA7, EPHA3, and MGA, were associated with a better prognosis of immunotherapy (Fig. 1b). Statistically significant (P < 0.05) indicators in univariate Cox analysis were introduced into the multivariate Cox regression model, and the results showed that ZFHX3 mutations were associated with a better prognosis for immunotherapy and that PBRM1 mutations were associated with a worse prognosis for immunotherapy (Fig. 1c; Supplementary Fig. 2a). Survival analysis showed that the OS of patients with ZFHX3-MT NSCLC was significantly longer than that of patients with ZFHX3-WT (P < 0.001, HR = 0.26, 95% Cl 0.17 − 0.41; Fig. 1d). In addition, subgroup analysis showed that patients with ZFHX3-MT LUAD had a longer OS after ICI treatment than patients with ZFHX3-WT (Fig. 1g, P = 0.004, HR = 0.3, 95% Cl 0.18–0.51). To further validate the predictive function of ZFHX3-MT and TMB on OS benefit, Kaplan–Meier curves were used to investigating the prognostic impact of ZFHX3 status and TMB status in the ICI-treated cohort from Samstein et al. (N = 350). In patients with known TMB status (according to a median of TMB; N = 350); 92 of them were TMB-High, while 27 were ZFHX3-MT, and 18 patients were both TMB-high and ZFHX3-MT. Notably, in patients with TMB-Low, ZFHX3-MT could equal to that of TMB-High patients (Group 3 vs Group 2: p = 0.6027; Supplementary Fig. 2b). As expected, the longest OS of patients with ZFHX3-MT and TMB-High was observed (Supplementary Fig. 2c, Group 3 vs Group 1: p = 4e-04; Group 3 vs Group 2: p = 0.0046; Group 3 vs Group 4: p = 0.0327). The OS benefit from ICI treatment was worst in the ZFHX3-WT and TMB-Low compared with other groups. (Supplementary Fig. 2c). In a validation cohort (N = 75; reported by Hellmann et. al [20]), the PFS benefit from ICI treatment was more prominent in the ZFHX3-MT group than that in the ZFHX3-WT group (Supplementary Fig. 2d, median PFS, not reached in the ZFHX3-MT group versus 7.8 months in the ZFHX3-WT group). However, there was no numerically significant PFS benefit (p = 0.08, HR = 0.21 [95%Cl, 0.08 to 0.64]), probably due to the limited sample size of the ZFHX3-MT group (N = 4). Therefore, ZFHX3 mutations can be used as an independent predictive biomarker for NSCLC patients receiving ICI treatment. To confirm that the OS benefit from ICI treatment in patients with ZFHX3-MT was not simply attributed to its general prognostic impact, we further evaluated the survival differences between ZFHX3-MT and ZFHX3-WT patients in the non-ICI-treated cohort (TCGA cohort). The relationship between ZFHX3 mutations and the OS and DFS of TCGA-NSCLC/LUAD/LUSC was further explored. There was no significant difference in the prognosis of LUAD/LUSC (Fig. 1e–l).
Fig. 1.
Results of Cox proportional hazard regression analysis for the ICI-treated NSCLC cohort (Samstein et al. N = 350) and survival curves for patients with NSCLC stratified by ZFHX3 status. a Forest plots showing the loge hazard ratio (95% confidence interval). Cox p values less than 0.05 are shown. Bubble plot showing the result of univariate b and multivariate c Cox proportional hazard regression analysis between ZFHX3-MT and ZFHX3-WT tumors. d Kaplan–Meier estimates of OS in the ICI-treated NSCLC cohort comparing patients with ZFHX3-MT with their respective counterparts without ZFHX3-MT. Patients (NSCLC) who harbored ZFHX3 mutations showed a better prognosis for ICI-based immunotherapy (P < 0.001, log-rank test). e Kaplan–Meier estimates of OS in the TCGA-NSCLC cohort comparing patients with ZFHX3-MT with their respective counterparts without ZFHX3-MT. f Kaplan–Meier estimates of DFS in the TCGA-NSCLC cohort comparing patients with ZFHX3-MT with their respective counterparts without ZFHX3-MT.g Kaplan–Meier estimates of OS in the ICI-treated LUAD cohort comparing patients with ZFHX3-MT with their respective counterparts without ZFHX3-MT. Patients (LUAD) who harbored ZFHX3 mutations showed a better prognosis for ICI-based immunotherapy. h Kaplan–Meier estimates of OS comparing patients in the TCGA-LUAD cohort with ZFHX3-MT with their respective counterparts without ZFHX3-MT. i Kaplan–Meier estimates of DFS in the TCGA-LUAD cohort comparing patients with ZFHX3-MT with their respective counterparts without ZFHX3-MT. j Kaplan–Meier estimates of OS in the ICI-treated LUSC cohort comparing patients with ZFHX3-MT with their respective counterparts without ZFHX3-MT. k Kaplan–Meier estimates of OS in the TCGA-LUSC cohort comparing patients with ZFHX3-MT with their respective counterparts without ZFHX3-MT. l Kaplan–Meier estimates of DFS in the TCGA-LUSC cohort comparing patients with ZFHX3-MT with their respective counterparts without ZFHX3-MT
Correlation of ZFHX3 mutations with other gene mutations and clinical characteristics
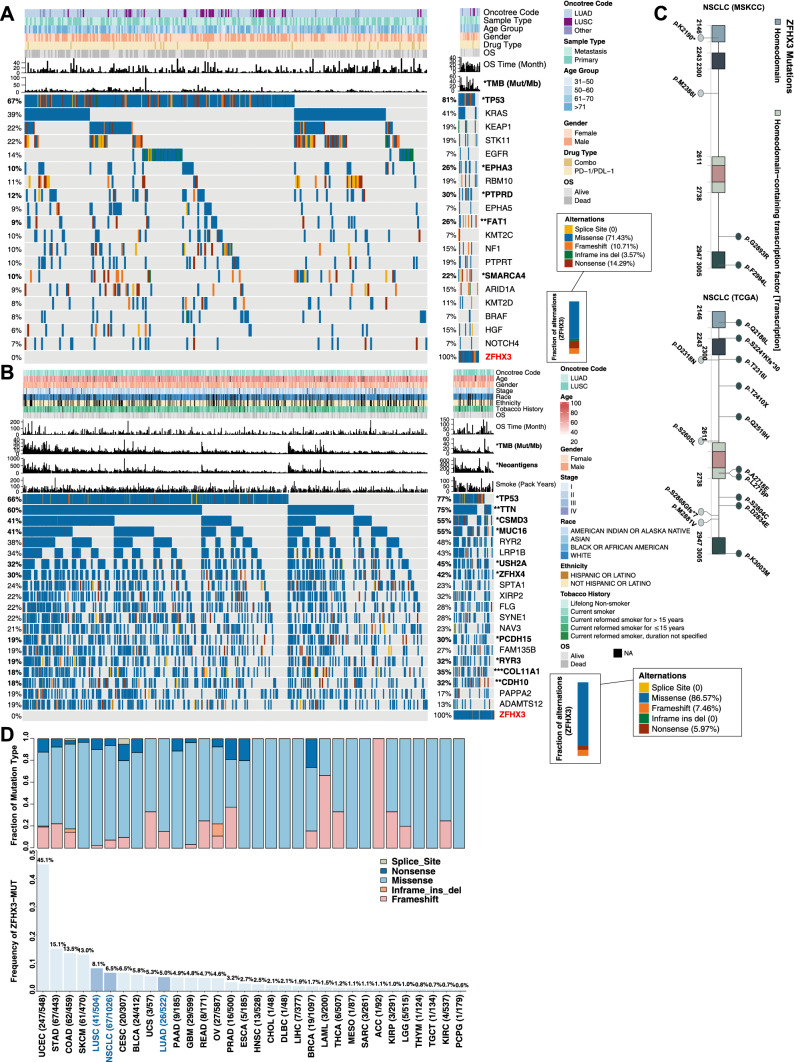
To further explore the clinical characteristics of patients with ZFHX3 mutations, we further analyzed the relationships between clinical characteristics (such as age, sex, immunotherapy drug type, smoking history, and clinical stage) and ZFHX3 mutations, and the results showed that there was no statistical difference in the correlations between these clinical characteristics and ZFHX3 mutations (Fig. 2a–b). In contrast, patients with ZFHX3 mutations had higher TMB. The types of ZFHX3 mutations in the immunotherapy and TCGA-NSCLC cohorts were mostly missense mutations (71.43% and 86.57%), nonsense mutations (14.29% and 5.97%), and frameshift mutations (10.71% and 7.46%). In contrast, there was no splice site in the ZFHX3 mutation type. The mutational landscape of the immunotherapy cohort is shown in Fig. 2a. In the immunotherapy cohort, the mutation frequency of genes in ZFHX3-MT patients was usually higher than that in ZFHX3-WT patients, such as TP53 (81% vs 67%), PTPRD (30% vs 12%), EPHA3 (26% vs 10%), and SMARCA4 (22% vs 10%) mutations. In the TCGA-NSCLC cohort (Fig. 2b), patients with ZFHX3-MT were often accompanied by other genetic mutations, including TP53 mutations (77% vs 66%), TTN mutations (75% vs 60%), and CSMD3 mutations (55% vs 41%), MUC16 mutations (55% vs 41%), etc. For detailed results, see Supplementary Table S2. Figure 2c shows the mutation sites of the ZFHX3 gene, including p.K2190, p.M2386I, p.G2893R, p.F2994L, p.Q2186L, p.T2316I, p.D2318N, p.M2881V, etc. The somatic mutation sites of the ZFHX3 gene were more evenly distributed and did not include any annotated functional hotspot mutations from 3D Hotspots (https://www.3dhotspots.org). In the TCGA database cohort of 33 tumor sample types, the statistical analysis of the frequency of ZFHX3 gene mutations (Fig. 2d) showed that the mutation rates of ZFHX3 in NSCLC, LUAD, and LUSC were 6.5% (67/1026), 5.0% (26/522), and 8.1% (41/504), respectively. Among these 33 cancer types, the average mutation rate of ZFHX3 is 5.2%, of which 27 mutation rates are higher than 1%. The gastrointestinal tract and urogenital system (including UCEC, STAD, COAD, etc.) are among the organs with the highest mutation rates.
Fig. 2.
Mutational landscape, clinical information of NSCLC patients, and the characteristics of ZFHX3 mutations in patients (NSCLC and each cancer type in TCGA). a Top 20 frequently mutated genes in NSCLC in the Samstein cohort (ICI-treated). The genes were ranked by the mutation frequency in NSCLC patients. The alteration type, ZFHX3 status, sex, histological subtype, OS status, OS time, treatment type, and age group are annotated. Significantly different genes are highlighted in bold (significance was calculated using Fisher’s exact test). b Top 20 frequently mutated genes in NSCLC in the TCGA-NSCLC cohort. The genes were ranked by the mutation frequency in NSCLC patients. The alteration type, survival status, survival time, ZFHX3 status, histological subtype, clinical stage, age, race, sex, tobacco smoking history, and number of pack-years smoked are annotated. c Lollipop plot shows the distribution of ZFHX3 mutations in the ICI-treated cohort (left panel) and TCGA-NSCLC cohort. d The proportion of ZFHX3-MT tumors identified for each cancer type in TCGA. The numbers above the barplot indicate the alteration frequency, and the numbers close to the cancer names indicate the number of ZFHX3-MT patients and the total number of patients. The fractions of mutation types of ZFHX3-MT tumors identified for each cancer type in TCGA (top panel)
Associations of ZFHX3 mutations with enhanced immunogenicity and activated antitumor immunity
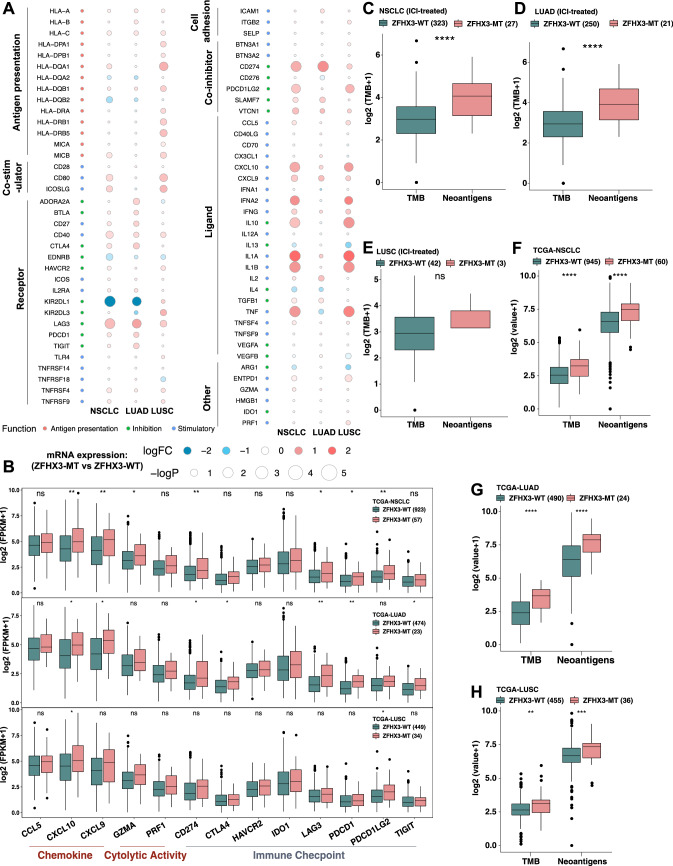
To investigate the immune characteristics of ZFHX3-MT NSCLC, we compared the differences in the expression patterns of immune-related genes between ZFHX3-MT and ZFHX3-WT tumors (Fig. 3a). The results showed that the expression of antigen-presentation-related molecules, stimulating immune-related ligands and receptors and cosuppressor molecules in patients with ZFHX3-MT were usually upregulated. Figure 3b shows that the expression of chemokines such as CXCL10 (FPKM: 72.8 vs 40.0) and CXCL9 (FPKM: 53.1 vs 35.9) in the ZFHX3-MT group was significantly upregulated. ZFHX3-MT tumors had a higher expression of mRNAs related to cytolytic activity, such as GZMA (FPKM: 16.0 vs 13.3), than ZFHX3-WT tumors. In addition, the ZFHX3-MT group had significantly increased immune checkpoint gene profiles, such as CD274, LAG3, PDCD1, PDCD1LG2, and TIGIT.
Fig. 3.
ZFHX3 mutations were associated with enhanced tumor immunogenicity and activated antitumor immunity. a Bubble plot depicting the mean differences in immune-related gene mRNA expression between ZFHX3-MT and ZFHX3-WT tumors in the TCGA-NSCLC/LUAD/LUSC cohort. The x-axis of the bubble plot indicates different histological subtypes, and the y-axis indicates tumor-infiltrating leukocytes, immune signatures, or gene names. The size of the circle represents the difference (-log10(p-value)) of each indicated immune signature or immune-related gene between ZFHX3-MT and ZFHX3-WT tumors in each cancer type. Red indicates upregulation, while blue indicates downregulation. b The expression levels of immune-related genes, such as chemokines, cytolytic activity-associated genes and immune checkpoints in ZFHX3-MT tumors versus ZFHX3-WT tumors (TCGA-NSCLC, LUAD and LUSC). Comparison of TMB between ZFHX3-MT and ZFHX3-WT tumors in Samstein’s NSCLC (c), LUAD (d) and LUSC (e) cohorts. Comparison of TMB and NAL between ZFHX3-MT and ZFHX3-WT tumors in the TCGA-NSCLC (f), LUAD (g) and LUSC (h) cohorts. (b–h *, P < 0.05; **, P < 0.01; ***, P < 0.001, ****, P < 0.0001, Mann–Whitney U test)
We compared tumor immunogenicity between ZFHX3-MT and ZFHX3-WT tumors. The TMB of ZFHX3-MT tumors in the immunotherapy cohort (Fig. 3c, P < 0.0001) and TCGA-NSCLC cohort (Fig. 3f, P < 0.0001) was significantly higher than that of ZFHX3-WT tumors. The NAL of ZFHX3-MT tumors in the TCGA-NSCLC cohort was also significantly higher (Fig. 3f, P < 0.0001), indicating that ZFHX3-MT is associated with increased tumor immunogenicity. Subgroup analysis showed that ZFHX3-MT in LUAD is associated with increased TMB (Fig. 3d: P < 0.0001; Fig. 3g: P < 0.0001) and NAL (Fig. 3g: P < 0.0001). In addition, ZFHX3-MT in TCGA-LUSC has a higher TMB (P < 0.01) and NAL (P < 0.001).
Patterns of immune cells and transcriptome traits based on MET status
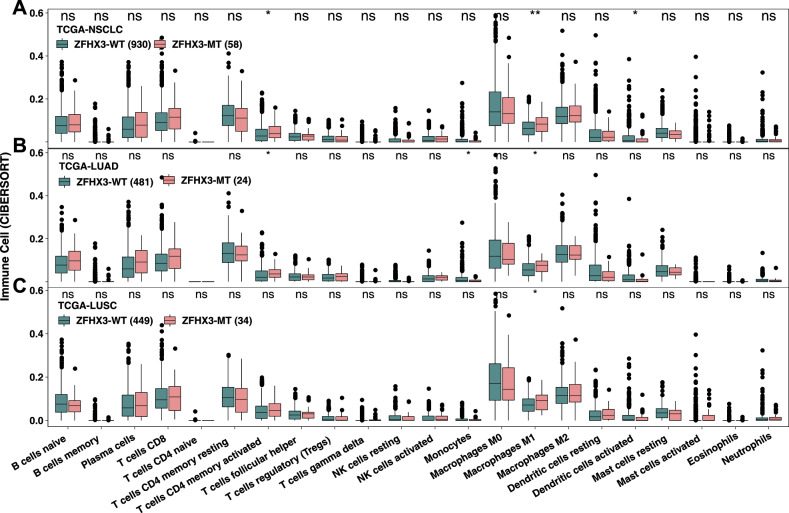
To further study the tumor immune microenvironment of ZFHX3-MT NSCLC, we used the CIBERSORT algorithm to estimate the TCGA-NSCLC immune cell infiltration status and compared the differences in immune cell infiltration patterns between ZFHX3-MT and ZFHX3-WT tumors (Fig. 4). The results showed that activated CD4 memory T cells (P < 0.05), M1 macrophages (P < 0.01), and activated dendritic cells (DCs) (P < 0.05) were more abundant in ZFHX3-MT tumors. This finding indicates that immune-activated cells were significantly richer in the ZFHX3-MT group. Subgroup analysis showed that in TCGA-LUAD, activated CD4 memory T cells (P < 0.05), monocytes (P < 0.05), and M1 macrophages (P < 0.05) were significantly higher in ZFHX3-MT tumors than in ZFHX3-WT tumors.
Fig. 4.
Comparison of immune cells between ZFHX3-MT and ZFHX3-WT tumors in the TCGA-NSCLC (a), LUAD (b), and LUSC (c) cohorts. Gene expression profiles were prepared using standard annotation files, and data were uploaded to the CIBERSORT web portal (https://cibersort.stanford.edu/), with the algorithm run using the LM22 signature and 1,000 permutations
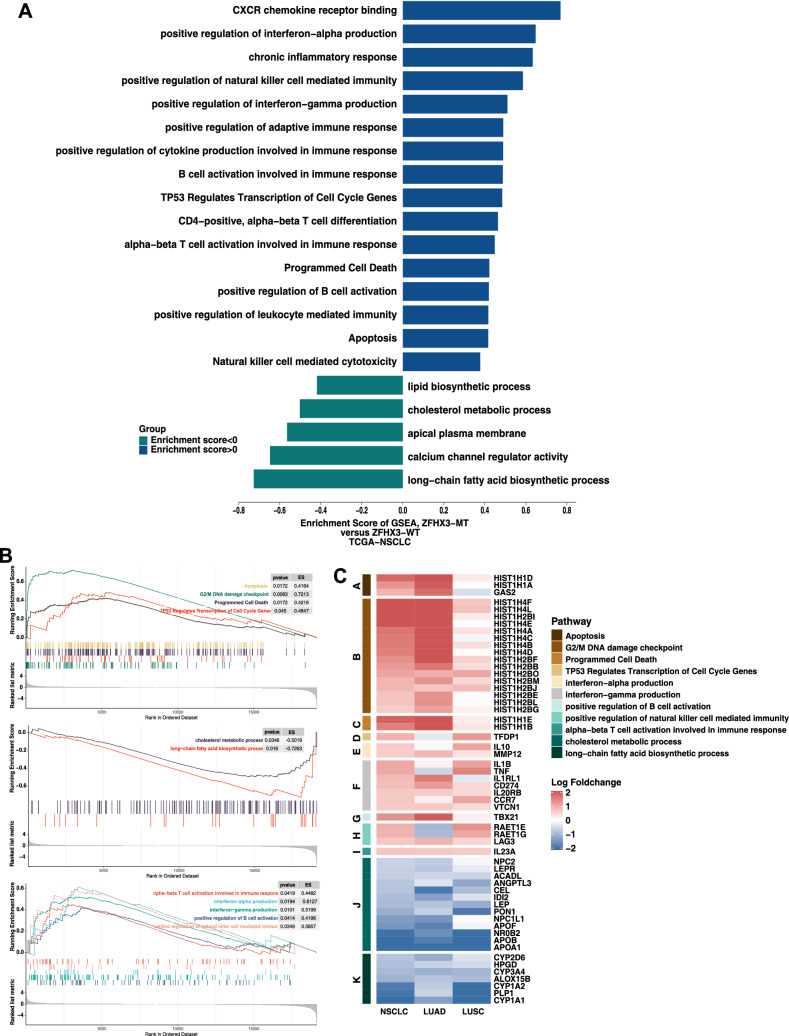
Further analysis of the differences in potential biological mechanisms between ZFHX3-MT and ZFHX3-WT tumors (Fig. 5) and the results of GSEA (Fig. 5a–b) showed that the immune response and cell cycle-related pathways in the ZFHX3-MT group were significantly upregulated, such as B-cell activation involved in the immune response, positive regulation of interferon − gamma production, and positive regulation of natural killer cell-mediated immunity. In addition, metabolic-related pathways were significantly downregulated in the ZFHX3-MT group (Fig. 5b), such as cholesterol metabolic process and long-chain fatty acids. The differential expression profile of core genes in some pathways is shown in Fig. 5c, and genes related to immune response and cell cycle pathways were significantly upregulated in the ZFHX3-MT group. In contrast, genes related to metabolic pathways were significantly downregulated in the ZFHX3-MT group.
Fig. 5.
Transcriptome biological function traits of ZFHX3-MT and ZFHX3-WT tumors in the TCGA-NSCLC cohort. a Differences in pathway activities scored by GSEA between ZFHX3-MT and ZFHX3-WT tumors in the TCGA-NSCLC cohort. Enrichment results with significant associations between ZFHX3-MT and ZFHX3-WT tumors are shown. The blue bar indicates that the enrichment score (ES) of the pathway is more than 0, while the green bar indicates that the ES of the pathway is less than 0. b GSEA of hallmark gene sets downloaded from MSigDB. All transcripts were ranked by the log2 (fold change) between ZFHX3-MT and ZFHX3-WT tumors in the TCGA-NSCLC cohort. Each run was performed with 1000 permutations. Enrichment results with significant associations between ZFHX3-MT and ZFHX3-WT tumors are shown. c Heatmap depicting the mean differences in the enrichment results with significant associations between ZFHX3-MT and ZFHX3-WT tumors in the TCGA-NSCLC, LUAD, and LUSC cohorts. The x-axis of the heatmap indicates different histological subtypes, and the y-axis indicates gene names and pathway signatures between ZFHX3-MT and ZFHX3-WT tumors in TCGA-NSCLC, LUAD, and LUSC. Red indicates upregulation, while blue indicates downregulation
Correlation between ZFHX3-MT and DDR pathway mutation characteristics
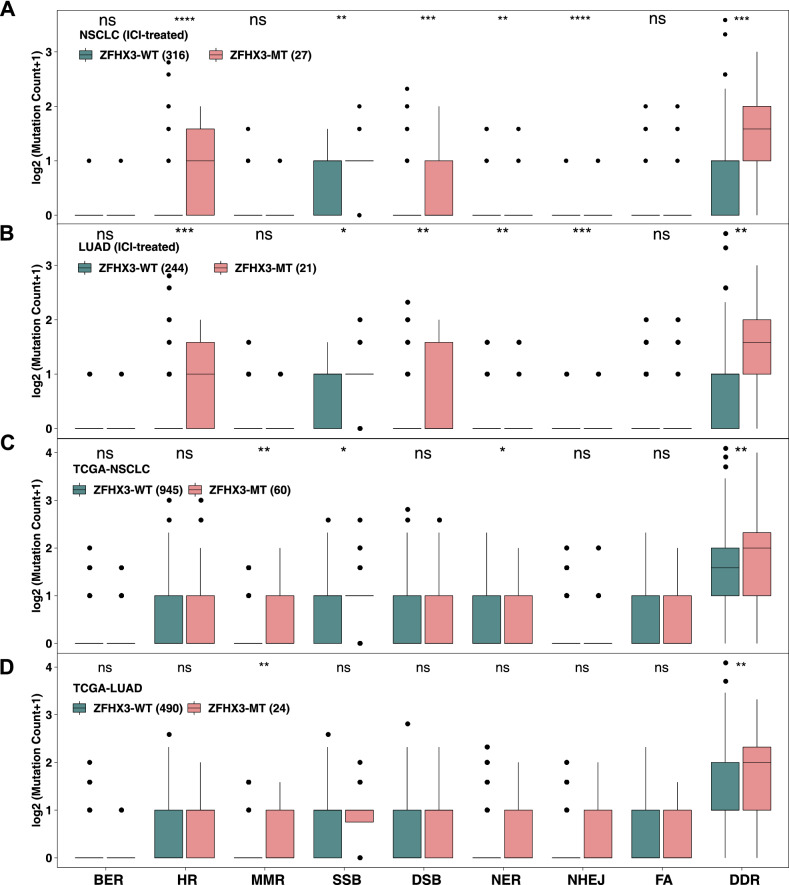
In recent years, many studies have shown that genetic mutations in the DDR pathway are related to the efficacy of ICIs [15, 16, 27], so we used the DDR gene set (Supplementary Table S1) from MSigDB to compare the differences in the number of DDR pathway mutations between ZFHX3-MT and ZFHX3-WT tumors (Fig. 6). In the immunotherapy cohort, ZFHX3-MT tumors had a significantly increased number of DDR pathway mutations (including homologous recombination (HR), single-strand breaks (SSB), DSB (double-strand breaks), NER (nucleotide excision repair), NHEJ (non-homologous end joining), etc.). In the TCGA-NSCLC cohort, the number of DDR pathway mutations in ZFHX3-MT tumors was also greater. Subgroup analysis shows that ZFHX3-MT LUAD usually also has more DDR pathway mutations. In contrast, there was no difference in the number of mutations in the DDR pathway between ZFHX3-WT and ZFHX3-MT LUSC (Supplementary Fig. S3).
Fig. 6.
Comparison of DNA damage-related gene set alterations between ZFHX3-MT and ZFHX3-WT tumors in cell lines from the ICI-treated NSCLC (a), ICI-treated LUAD (b), TCGA-NSCLC (c), and TCGA-LUAD (d) cohorts. (*, P < 0.05; **, P < 0.01; ***, P < 0.001, ****, P < 0.0001, Mann–Whitney U test)
Discussion
In recent years, ICIs represented by PD-1/PD-L1 inhibitors have become one of the important options for the treatment of patients with advanced NSCLC. However, ICI treatment is not effective for all patients with NSCLC, and the key to improving efficacy is to screen dominant populations. Studies have shown that specific mutated genes, such as alterations in the DDR pathway (including POLE, POLD1, MLH1, etc.) [28, 29], interferon signal pathway gene mutations [30, 31], driver gene mutations (KRAS, BRAF, ALK, EGFR, etc.) [32, 33], and other gene mutations (such as, SERPINB) [8], are related to the prognosis of ICI treatment. Therefore, we tried to explore the correlation between gene mutations and the prognosis of NSCLC immunotherapy. In this study, we systematically collected and consolidated a large amount of genomic and clinical data to evaluate the predictive function of mutations in key genes involving in NSCLC receiving ICI treatment (Supplementary Fig. S1). And we found that mutation in ZFHX3, a transcription factor that encoded four homeobox domains and 23 zinc fingers [34], was predictive of improved overall survival (OS) to ICI treatment in NSCLC. ZFHX3-MT was strongly associated with better OS, increased tumor immunogenicity, antitumor immune response ability, and the number of DDR pathway mutations. In addition, in the GDSC database, there was no significant difference in the sensitivity of common chemotherapy drugs between ZFHX3-MT and ZFHX3-WT NSCLC, LUAD, and LUSC cell lines (Supplementary Fig. S4), which suggests that ZFHX3-MT may have no significant predictive significance for chemotherapy in patients with lung cancer.
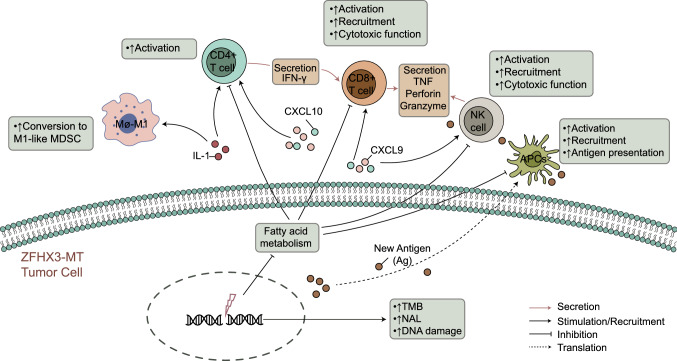
Zinc finger homeobox 3 (ZFHX3) was initially identified as ATBF1, a transcription factor that encoded four homeobox domains and 23 zinc fingers; ZFHX3 was shown to be involved in suppressing alpha-fetoprotein transcription [34, 35]. It was identified as a candidate tumor suppressor gene for prostate, breast, and gastric cancers, which acted by inducing cell cycle arrest [36–38]. ZFHX3 is frequently mutated in metastatic or high-grade human cancers, and many of the mutations are frameshifting and thus function inactivating [34, 35, 39]. Although these studies indicate an important role of ZFHX3 in cancers, it is unknown what ZFHX3 mutations exert function in NSCLC patients receiving ICI treatment. Furthermore, we have summarized the possible mechanisms underlying the improved efficacy and prognosis in ZFHX3-MT NSCLC receiving ICIs (Fig. 7).
Fig. 7.
The possible mechanism underlying the improved efficacy and prognosis in ZFHX3-MT NSCLC receiving ICIs
The immunogenicity of the tumor is the basis for the initiation of the antitumor immune response, and higher frequency somatic mutations may cause tumor cells to produce more new antigens to enhance the immune killing abilities of T cells to tumor cells [40]. In addition, the immunogenicity of the tumor is also affected by factors in the TME, such as the efficiency of antigen processing and presentation of DCs, the most powerful antigen-presenting cells in the body [41]. The TME is closely related to the efficacy of ICIs in patients with lung cancer. For example, CD4 + T cells are associated with a better efficacy of immunotherapy [42]. Therefore, higher TMB, NAL, DCs, and CD4 + T cells may be why ICIs are more effective in ZFHX3-MT patients than in ZFHX3-WT patients.
In recent years, studies have shown that GEPs can be used as a novel potential predictor of the efficacy of immunotherapy [43, 44]. Jiang et al. [44] showed that a T-cell inflamed GEP is related to the clinical benefit of patients receiving ICI treatment. In addition, they evaluated CD8A, CD8B, GZMA, GZMB, and PRF1 expression levels to evaluate tumor cytotoxic T lymphocyte (CTL) infiltration levels, and found that the survival time of immunotherapy patients was significantly prolonged in the high-infiltration CTL groups. In addition, CD8 + T cells can be recruited to enhance immune infiltration and antitumor immunity by chemokines such as CXCL9 and CXCL10 [45]. Therefore, chemokines (such as CXCL10 and CCL5) and molecules related to cytolytic activity (such as GZMA) that are highly expressed at the mRNA level may be one of the reasons why the efficacy of ICIs is better in ZFHX3-MT patients than in ZFHX3-WT patients.
The DDR pathway is critical for maintaining genomic integrity. Alterations in the DDR pathway increase genomic instability and are associated with higher TMB. The previous studies have shown that mutations in the DDR pathway may serve as a potential predictive biomarker for ICI treatment and improve the clinical results of ICI treatment [15, 16]. Therefore, more DDR pathway mutations may be one of the reasons why ICIs are more effective in ZFHX3-MT patients than in ZFHX3-WT patients.
Studies have shown that IFN-γ can reduce the infiltration of Tregs, thereby enhancing the antitumor immune effect [46]. In addition, the expression profile of IFN-γ-related GEP can predict the clinical outcome of PD-1 treatment [43]. Studies have shown that cholesterol can be combined with the TCRβ transmembrane region or that disrupting the TCR signaling pathway further inhibits the antitumor activity of T cells [47, 48]. Cholesterol affects immune cells and promotes the metastasis and recurrence of breast cancer [49]. Ma et al. found that high cholesterol will promote the expression of T-cell immune checkpoints, making T cells more easily inhibited, thus losing antitumor function [50]. GSEA showed that immune response-related pathways (including interferon-gamma production, alpha–beta T-cell activation involved in the immune response, etc.) are significantly upregulated in ZFHX3-MT patients. In contrast, the cholesterol metabolic process was significantly downregulated in patients with ZFHX3-MT. Further comparison of core gene expression in these pathways revealed that the expression of core genes in the interferon-gamma production-related pathway in ZFHX3-MT patients was significantly upregulated, and core gene expression of the cholesterol metabolic process was significantly downregulated. Therefore, the GSEA results and gene expression profile also provide evidence that ZFHX3-MT patients can benefit more from ICI treatment than ZFHX3-WT patients.
We found that, in LUSC, there was no significant difference between ZFHX3-MT and ZFHX3-WT tumors in immune cell infiltration patterns, immune-related gene profiles, and number of mutations in the DDR pathway. Lambrechts et al. suggest that there are differences between stromal cell marker genes and tumor stromal cell subtypes in LUAD and LUSC tumors; moreover, the low expression of CD8 + T-cell cluster genes in LUSC is associated with worse survival prognosis. Therefore, these findings suggest that the presence of the TME between LUAD and LUSC is different [51].
This study explored the association between the prognosis of NSCLC immunotherapy and specific mutated genes and tried to clarify the possible mechanism of ZFHX3-MT as an independent predictive marker of the prognosis of NSCLC immunotherapy. However, there are still some limitations. First, this study included only one NSCLC immunotherapy cohort, and there was bias in screening biomarkers for the prognosis of NSCLC immunotherapy. Second, the immunotherapy cohort used targeted sequencing (the Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets [MSK-IMPACT] panel) to detect gene mutations. Finally, our analysis discussed only the two most important subtypes of NSCLC; the remaining subtypes were not discussed. Therefore, more research involving a large number of samples and diverse ethnic groups is needed for analysis and validation.
Conclusions
The study found that ZFHX3 mutations are independent predictors of the prognosis of NSCLC immunotherapy. ZFHX3-MT is associated with longer OS after immunotherapy, and ZFHX3-MT is positively correlated with known predictive markers of immunotherapy, including TMB, NAL, immune-related genes, immune cells, and the number of DDR pathway mutations. Therefore, ZFHX3 mutations can be used as a novel potential predictive marker to guide NSCLC ICI treatment. A series of prospective clinical studies and molecular mechanism explorations are still needed in the future.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary file1 (PDF 101 kb)
Fig. S1. Flowchart of data processing of the TCGA-NSCLC/LUAD/LUSC dataset and the ICI-treated NSCLC cohort (Samstein et al.)
Supplementary file2 (PDF 318 kb)
Fig. S2. a Results of Multivariable Cox proportional hazards regression analysis for the ICI-treated NSCLC cohort (Samstein et al., N=350); b–c Kaplan-Meier curves comparing the OS in the TMB-High and ZFHX3-MT, TMB-High and ZFHX3-WT, TMB-Low and ZFHX3-MT, TMB-Low and ZFHX3-WT groups in the ICI-treated NSCLC cohort (Samstein et al.); d Kaplan-Meier curves investigating the prognostic impact of ZFHX3-MT in the independent-ICI-treated cohort from Hellmann et al (N=75)
Supplementary file3 (PDF 289 kb)
Fig. S3. Comparison of DNA damage-related gene set alterations between ZFHX3-MT and ZFHX3-WT tumors in cell lines from the ICI-treated LUSC and TCGA-LUSC cohorts. (*, P<0.05; **, P<0.01; ***, P<0.001, ****, P<0.0001, Mann-Whitney U test)
Supplementary file4 (PDF 288 kb)
Fig. S4. Comparison of the drug sensitivity between the ZFHX3-MT and ZFHX3-WT cell lines from the GDSC-NSCLC (a), LUAD (b) and LUSC (c) cell lines. (*, P<0.05; **, P<0.01; ***, P<0.001, ****, P<0.0001, Mann-Whitney U test)
Supplementary file5 (PDF 63 kb)
Table S1. List of genes included in DNA damage response (DDR) gene set used for Comparison Analysis (MsiDB)
Supplementary file6 (PDF 24 kb)
Table S2. Comparison of gene mutations (top20) between the ZFHX3-MT and ZFHX3-WT tumors in the ICI-treated cohort (Samstein et.al) and the TCGA-NSCLC cohort
Acknowledgements
We would like to thank the biotrainee, Dr. Jianming Zeng (University of Macau), XiaoYa HuaTu, Lianchuan Biotechnology Limited (subsidiary of LC Sciences, Hangzhou, China), and Haplox Biotechnology Co., Ltd.
Abbreviations
- CTLA-4
Cytotoxic T lymphocyte antigen 4
- GEP
Gene expression profile
- Mb
Megabase
- NSCLC
Non-small-cell lung cancer
- ORR
Objective response rate
- OS
Overall survival
- PD
Progression of disease
- PD-(L)1
Programmed cell death (ligand) 1
- PFS
Progression-free survival
- ICIs
Immune checkpoint inhibitors
- TCGA
The cancer genome atlas
- GSEA
Gene set enrichment
- TMB
Tumor mutational burden
- ZFHX3-MT
ZFHX3-mutant
- ZFHX3-WT
ZFHX3-wildtype
- MSK-IMPACT
The Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets
- NAL
Neoantigen loads
- LUAD
Lung adenocarcinoma
- LUSC
Lung squamous cell carcinoma
- DDR
DNA damage response
- GDSC
The genomics of drug sensitivity in cancer
- WES
Whole-exome sequencing
- CTL
Cytotoxic T lymphocyte
- FA
Fanconi anemia
- NHEJ
Non-homologous end joining
- BER
Base excision repair
- MMR
Mismatch repair
- NER
Nucleotide excision repair
- DSB
Double-strand breaks
- SSB
Single-strand breaks
- HR
Homologous recombination
Author contributions
Conceptualization, LLG, WLZ, and JZ; formal analysis, JXZ, NNZ, AQL, PL, and XC; software, JXZ, NNZ, AQL, and PL; supervision, LLG, WLZ, and JZ; visualization, JXZ, NNZ, AQL, and PL; writing–original draft, JXZ, NNZ, AQL, PL, XC, HJD, and SJK; writing–review & editing, JXZ, NNZ, AQL, PL, and XC.
Funding
This work was supported by grants from the National Natural Science Foundation of China (Nos. 81572244, 81602631, 81672267, 81772458, 81871859, and 81972809); Natural Science Foundation of Guangdong Province (key) (2015A030311028); The Clinical Research Initiative Project of Southern Medical University (LC2016ZD029 and LC2019ZD016); Major Basic Research Projects and Major Applied Research Projects of Educational Commission of Guangdong Province (2017KZDXM015).
Availability of data and material
All of the data we used in this study were publicly available as described in the Method section.
Compliance with ethical standards
Conflict of interests
The authors declare that they have no conflict interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Jiexia Zhang, Ningning Zhou, Anqi Lin, Peng Luo, and Xin Chen have contributed equally to this work.
Contributor Information
Linlang Guo, Email: linlangg@yahoo.com.
Weiliang Zhu, Email: duarion@126.com.
Jian Zhang, Email: blacktiger@139.com.
References
- 1.Fitzmaurice C, Allen C, Barber RM, et al. Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 32 cancer groups, 1990 to 2015. JAMA Oncol. 2017;3:524. doi: 10.1001/jamaoncol.2016.5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Garon EB, Rizvi NA, Hui R, et al. Pembrolizumab for the treatment of non–small-cell lung cancer. N Engl J Med. 2015;372:2018–2028. doi: 10.1056/NEJMoa1501824. [DOI] [PubMed] [Google Scholar]
- 3.Havel JJ, Chowell D, Chan TA. The evolving landscape of biomarkers for checkpoint inhibitor immunotherapy. Nat Rev Cancer. 2019;19:133–150. doi: 10.1038/s41568-019-0116-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zou W, Wolchok JD, Chen L. PD-L1 (B7–H1) and PD-1 pathway blockade for cancer therapy: Mechanisms, response biomarkers, and combinations. Sci Transl Med. 2016 doi: 10.1126/scitranslmed.aad7118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang S, He Z, Wang X, et al. Antigen presentation and tumor immunogenicity in cancer immunotherapy response prediction. Elife. 2019 doi: 10.7554/eLife.49020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boeri M, Milione M, Proto C, et al. Circulating miRNAs and PD-L1 tumor expression are associated with survival in advanced NSCLC patients treated with immunotherapy: A prospective study. Clin Cancer Res. 2019;25:2166–2173. doi: 10.1158/1078-0432.CCR-18-1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cristescu R, Mogg R, Ayers M, et al. Pan-tumor genomic biomarkers for PD-1 checkpoint blockade-based immunotherapy. Science. 2018 doi: 10.1126/science.aar3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Riaz N, Havel JJ, Kendall SM, et al. Recurrent SERPINB3 and SERPINB4 mutations in patients who respond to anti-CTLA4 immunotherapy. Nat Genet. 2016;48:1327–1329. doi: 10.1038/ng.3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang F, Zhao Q, Wang YN, et al. Evaluation of POLE and POLD1 mutations as biomarkers for immunotherapy outcomes across multiple cancer types. JAMA Oncol. 2019;5:1504–1506. doi: 10.1001/jamaoncol.2019.2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu HX, Chen YX, Wang ZX, et al. Alteration in TET1 as potential biomarker for immune checkpoint blockade in multiple cancers. J Immunother Cancer. 2019;7:264. doi: 10.1186/s40425-019-0737-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dong Z-Y, Zhong W-Z, Zhang X-C, et al. Potential predictive value of TP53 and KRAS mutation status for response to PD-1 blockade immunotherapy in lung adenocarcinoma. Clin Cancer Res. 2017;23:3012–3024. doi: 10.1158/1078-0432.CCR-16-2554. [DOI] [PubMed] [Google Scholar]
- 12.Ji M, Liu Y, Li Q, et al. PD-1/PD-L1 expression in non-small-cell lung cancer and its correlation with EGFR/KRAS mutations. Cancer Biol Ther. 2016;17:407–413. doi: 10.1080/15384047.2016.1156256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spigel DR, Schrock AB, Fabrizio D, et al. Total mutation burden (TMB) in lung cancer (LC) and relationship with response to PD-1/PD-L1 targeted therapies. J Clin Oncol. 2016;34:9017–9017. doi: 10.1200/jco.2016.34.15_suppl.9017. [DOI] [Google Scholar]
- 14.Lin A, Wei T, Meng H, et al. Role of the dynamic tumor microenvironment in controversies regarding immune checkpoint inhibitors for the treatment of non-small cell lung cancer (NSCLC) with EGFR mutations. Mol Cancer. 2019;18:139. doi: 10.1186/s12943-019-1062-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang J, Wang Z, Zhao J, et al. Co-mutations in DNA damage response pathways serve as potential biomarkers for immune checkpoint blockade. Cancer Res. 2018 doi: 10.1158/0008-5472.CAN-18-1814. [DOI] [PubMed] [Google Scholar]
- 16.Teo MY, Seier K, Ostrovnaya I, et al. Alterations in DNA damage response and repair genes as potential marker of clinical benefit from PD-1/PD-L1 blockade in advanced urothelial cancers. J Clin Oncol. 2018;36:1685–1694. doi: 10.1200/JCO.2017.75.7740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Samstein RM, Lee C-H, Shoushtari AN, et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat Genet. 2019;51:202–206. doi: 10.1038/s41588-018-0312-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Colaprico A, Silva TC, Olsen C, et al. TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016;44:e71–e71. doi: 10.1093/nar/gkv1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cerami E, Gao J, Dogrusoz U, et al. The cBio Cancer Genomics Portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hellmann MD, Nathanson T, Rizvi H, et al. Genomic features of response to combination immunotherapy in patients with advanced non-small-cell lung cancer. Cancer Cell. 2018;33:843–852.e4. doi: 10.1016/j.ccell.2018.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12:453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thorsson V, Gibbs DL, Brown SD, et al. The immune landscape of cancer. Immunity. 2018;48:812–830.e14. doi: 10.1016/j.immuni.2018.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chalmers ZR, Connelly CF, Fabrizio D, et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med. 2017;9:34. doi: 10.1186/s13073-017-0424-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu G, Wang L-G, Han Y, He Q-Y. cluster profiler: an R package for comparing biological themes among gene clusters. Omi A J Integr Biol. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32:2847–2849. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- 27.Luo P, Lin A, Li K, et al. DDR pathway alteration, tumor mutation burden, and cisplatin sensitivity in small cell lung cancer: difference detected by whole exome and targeted gene sequencing. J Thorac Oncol. 2019;14:e276–e279. doi: 10.1016/j.jtho.2019.08.2509. [DOI] [PubMed] [Google Scholar]
- 28.Le DT, Uram JN, Wang H, et al. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med. 2015;372:2509–2520. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rizvi NA, Hellmann MD, Snyder A, et al. Mutational landscape determines sensitivity to PD-1 blockade in non–small cell lung cancer. Science. 2015 doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao J, Shi LZ, Zhao H, et al. Loss of IFN-γ pathway genes in tumor cells as a mechanism of resistance to anti-CTLA-4 therapy. Cell. 2016;167:397–404.e9. doi: 10.1016/j.cell.2016.08.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zaretsky JM, Garcia-Diaz A, Shin DS, et al. Mutations associated with acquired resistance to PD-1 blockade in melanoma. N Engl J Med. 2016;375:819–829. doi: 10.1056/NEJMoa1604958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gainor JF, Shaw AT, Sequist LV, et al. EGFR mutations and ALK rearrangements are associated with low response rates to PD-1 pathway blockade in non-small cell lung cancer: A retrospective analysis. Clin Cancer Res. 2016;22:4585–4593. doi: 10.1158/1078-0432.CCR-15-3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peng W, Chen JQ, Liu C, et al. Loss of PTEN promotes resistance to T cell-mediated immunotherapy. Cancer Discov. 2016;6:202–216. doi: 10.1158/2159-8290.CD-15-0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu Q, Zhang B, Chen R, et al. ZFHX3 is indispensable for ERβ to inhibit cell proliferation via MYC downregulation in prostate cancer cells. Oncogenesis. 2019;8:28. doi: 10.1038/s41389-019-0138-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fu C, An N, Liu J, et al. The transcription factor ZFHX3 is crucial for the angiogenic function of hypoxia-inducible factor 1α in liver cancer cells. J Biol Chem. 2020;295:7060–7074. doi: 10.1074/jbc.RA119.012131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jung C-G, Kim H-J, Kawaguchi M, et al. Homeotic factor ATBF1 induces the cell cycle arrest associated with neuronal differentiation. Development. 2005;132:5137–5145. doi: 10.1242/dev.02098. [DOI] [PubMed] [Google Scholar]
- 37.Kataoka H, Miura Y, Joh T, et al. Alpha-fetoprotein producing gastric cancer lacks transcription factor ATBF1. Oncogene. 2001;20:869–873. doi: 10.1038/sj.onc.1204160. [DOI] [PubMed] [Google Scholar]
- 38.Zaw KTT, Sato N, Ikeda S, et al. Association of ZFHX3 gene variation with atrial fibrillation, cerebral infarction, and lung thromboembolism: An autopsy study. J Cardiol. 2017;70:180–184. doi: 10.1016/j.jjcc.2016.11.005. [DOI] [PubMed] [Google Scholar]
- 39.Grasso CS, Wu Y-M, Robinson DR, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487:239–243. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gubin MM, Artyomov MN, Mardis ER, Schreiber RD. Tumor neoantigens: building a framework for personalized cancer immunotherapy. J Clin Invest. 2015;125:3413–3421. doi: 10.1172/JCI80008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mellman I, Steinman RM. Dendritic Cells. Cell. 2001;106:255–258. doi: 10.1016/S0092-8674(01)00449-4. [DOI] [PubMed] [Google Scholar]
- 42.Spitzer MH, Carmi Y, Reticker-Flynn NE, et al. Systemic immunity is required for effective cancer immunotherapy. Cell. 2017;168:487–502.e15. doi: 10.1016/j.cell.2016.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ayers M, Lunceford J, Nebozhyn M, et al. IFN-γ–related mRNA profile predicts clinical response to PD-1 blockade. J Clin Invest. 2017;127:2930–2940. doi: 10.1172/JCI91190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jiang P, Gu S, Pan D, et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat Med. 2018;24:1550–1558. doi: 10.1038/s41591-018-0136-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peng D, Kryczek I, Nagarsheth N, et al. Epigenetic silencing of TH1-type chemokines shapes tumour immunity and immunotherapy. Nature. 2015;527:249–253. doi: 10.1038/nature15520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Overacre-Delgoffe AE, Chikina M, Dadey RE, et al. Interferon-γ drives treg fragility to promote anti-tumor immunity. Cell. 2017;169:1130–1141.e11. doi: 10.1016/j.cell.2017.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Swamy M, Beck-Garcia K, Beck-Garcia E, et al. A cholesterol-based allostery model of t cell receptor phosphorylation. Immunity. 2016;44:1091–1101. doi: 10.1016/j.immuni.2016.04.011. [DOI] [PubMed] [Google Scholar]
- 48.Wang F, Beck-García K, Zorzin C, et al. Inhibition of T cell receptor signaling by cholesterol sulfate, a naturally occurring derivative of membrane cholesterol. Nat Immunol. 2016;17:844–850. doi: 10.1038/ni.3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Baek AE, Yu YRA, He S, et al. The cholesterol metabolite 27 hydroxycholesterol facilitates breast cancer metastasis through its actions on immune cells. Nat Commun. 2017;8:864. doi: 10.1038/s41467-017-00910-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ma X, Bi E, Lu Y, et al. Cholesterol induces CD8+ T cell exhaustion in the tumor microenvironment. Cell Metab. 2019;30:143–156.e5. doi: 10.1016/j.cmet.2019.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lambrechts D, Wauters E, Boeckx B, et al. Phenotype molding of stromal cells in the lung tumor microenvironment. Nat Med. 2018;24:1277–1289. doi: 10.1038/s41591-018-0096-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary file1 (PDF 101 kb)
Fig. S1. Flowchart of data processing of the TCGA-NSCLC/LUAD/LUSC dataset and the ICI-treated NSCLC cohort (Samstein et al.)
Supplementary file2 (PDF 318 kb)
Fig. S2. a Results of Multivariable Cox proportional hazards regression analysis for the ICI-treated NSCLC cohort (Samstein et al., N=350); b–c Kaplan-Meier curves comparing the OS in the TMB-High and ZFHX3-MT, TMB-High and ZFHX3-WT, TMB-Low and ZFHX3-MT, TMB-Low and ZFHX3-WT groups in the ICI-treated NSCLC cohort (Samstein et al.); d Kaplan-Meier curves investigating the prognostic impact of ZFHX3-MT in the independent-ICI-treated cohort from Hellmann et al (N=75)
Supplementary file3 (PDF 289 kb)
Fig. S3. Comparison of DNA damage-related gene set alterations between ZFHX3-MT and ZFHX3-WT tumors in cell lines from the ICI-treated LUSC and TCGA-LUSC cohorts. (*, P<0.05; **, P<0.01; ***, P<0.001, ****, P<0.0001, Mann-Whitney U test)
Supplementary file4 (PDF 288 kb)
Fig. S4. Comparison of the drug sensitivity between the ZFHX3-MT and ZFHX3-WT cell lines from the GDSC-NSCLC (a), LUAD (b) and LUSC (c) cell lines. (*, P<0.05; **, P<0.01; ***, P<0.001, ****, P<0.0001, Mann-Whitney U test)
Supplementary file5 (PDF 63 kb)
Table S1. List of genes included in DNA damage response (DDR) gene set used for Comparison Analysis (MsiDB)
Supplementary file6 (PDF 24 kb)
Table S2. Comparison of gene mutations (top20) between the ZFHX3-MT and ZFHX3-WT tumors in the ICI-treated cohort (Samstein et.al) and the TCGA-NSCLC cohort
Data Availability Statement
All of the data we used in this study were publicly available as described in the Method section.