Abstract
Myeloid-derived suppressor cells (MDSC) populate the peripheral blood and contribute to immune regulation in cancer. However, there is limited knowledge on the myeloid cell types with proinflammatory capacities that may serve as opponents of MDSC. In the circulation of cancer patients, a monocyte subpopulation was identified with a specific immunophenotype and transcriptomic signature. They were predominantly CD14+CD33hiCD16−/+HLA-DR+/hi cells that typically expressed CD66b. In accordance with the transcriptomics data, NALP3, LOX-1 and PAI-1 levels were also significantly upregulated. The CD66b+ monocytes displayed high phagocytic activity, matrix adhesion and migration, and provided costimulation for T cell proliferation and IFN-γ secretion; thus, they did not suppress T cell responses. Irrespective of clinical stage, they were identified in various cancers. In conclusion, the CD66b+ monocytes represent a novel myeloid subpopulation which is devoid of immune regulatory influences of cancer and displays enhanced proinflammatory capacities.
Electronic supplementary material
The online version of this article (10.1007/s00262-020-02656-y) contains supplementary material, which is available to authorized users.
Keywords: Myeloid-derived suppressor cells (MDSC), Monocyte, PMN-MDSC, Transcriptomics, Monocyte subtypes, Neutrophil
Introduction
Monocytes are bone marrow-derived mononuclear phagocytes which constitute the primary reserve for professional antigen-presenting cells (APC) [1, 2]. They patrol throughout the body, respond to various stimuli and differentiate into functionally distinct subsets of macrophages or dendritic cells (DC) [3]. These cells not only take place in the first line of defense against infections but also contribute to tissue repair and hemostasis. Monocytes and monocyte-derived cells ingest, process and present antigens to T cells; therefore, they constitute a link between innate and adaptive immunity [4–6].
Based on the expression of CD14 and CD16 markers, the human monocytes are divided into three major subsets. The CD14++CD16− classical monocytes are the major subtype in the peripheral blood; whereas, CD16 expression determines the two non-classical subtypes (CD14++CD16+ and CD14+CD16++) that increase in inflammatory disorders such as autoimmunity and cancer [7–9]. Nevertheless, according to the pathophysiological circumstances, monocytes frequently undergo functional and phenotypic adjustments; however, whether the variations and discrepancies amongst the monocytes could represent distinct subsets remains disputed [10, 11]. Inflammation alters the steady-state dynamics of myelopoiesis; critically, continuum of perpetual stimulation in chronic inflammatory diseases not only promotes the expansion of myeloid cells but also perturbs their differentiation and functional competence [12]. In cancer, these newly generated myeloid cells are characterized with immunosuppressive activities and defined as the myeloid-derived suppressor cells (MDSC) [13]. In humans, CD33 and low levels of HLA-DR are the hallmarks of MDSC and according to CD14, CD15 or CD66b expression, they can be further characterized as polymorphonuclear (PMN-MDSC), monocytic (M-MDSC) and early-stage (e-MDSC) subsets [14–17]. In response to the cancer-associated inflammation and the factors derived from tumor microenvironment, functional impairment or acquisition of suppressive properties have been reported almost in every type of myeloid cells [18–20]. Nevertheless, phenotypic and functional heterogeneity observed amongst individual cells of a specific myeloid type limits the consensus-building for the determination of novel subsets [21].
In this study, CD33hiCD14+ monocytes were profiled according to the CD66b expression. The CD66b+ monocytes constituted a subpopulation in the peripheral blood of cancer patients. In contrast to the previously defined myeloid-derived cells that emerge in cancer, CD66b+ monocytes possessed proinflammatory signatures, displayed increased functional competence, and did not interfere with T cell responses. In conclusion, CD66b marks a new monocyte subset that retains immune-stimulatory and proinflammatory capacities.
Materials and methods
Patients and healthy controls
Peripheral blood was obtained from newly diagnosed, treatment-naïve breast cancer (BC) patients [n = 66 (females), median age 54 (min 32–max 83)] or colorectal cancer (CRC) patients [n = 70 (32 females, 38 males), median age 62 (min 32–max 89)] (Supplementary Table 1). Samples from representative patients with small cell lung cancer, gastric cancer, and pancreatic cancer were also used. Blood collected from healthy volunteers [n = 48 (22 females, 26 males), median age 48 (min 24–max 62)] were used as controls. Overall survival was defined as time from diagnosis to death from colorectal cancer. This study was approved by Hacettepe University and University of Health Sciences local ethics committees and conducted in agreement with the guiding principles of the declaration of Helsinki and the good clinical practice. Informed consent was obtained from the participants and all methods were performed in accordance with the relevant guidelines and regulations.
Blood collection and processing
Blood samples were collected in trisodium citrate (3.2%) tubes (Vacutest) and processed within 2 h. For density gradient separation of peripheral blood mononuclear cells (PBMC), samples were diluted (1:1 with 1 × phosphate-buffered saline (PBS)), layered over 1.077 g/mL Ficoll-Hypaque (Sigma-Aldrich), and centrifuged at 400× g for 30 min. without brake and acceleration. Total PBMC was collected, washed, counted and used in further experiments.
Flow cytometry
Various panels of monoclonal antibodies (mAb) were used for immunophenotyping (Supplementary Table 2). Cells were suspended in FACS buffer (CellWASH, BD Biosciences) and incubated for 20 min. at 4 °C with appropriate combinations of mAb and Fc Block (BD). The samples were run on a FACSAria II cell sorter (BD) and the data were analyzed using FlowJo software v10.0.7 (Treestar). Percentage of the positive cells and median fluorescence intensity (MFI) values were calculated considering isotype-matched antibody and fluorescence minus one (FMO) controls.
Cell sorting and purification
CD14+ and CD66b+ populations amongst PBMC were isolated with anti-CD14 and anti-CD66b magnetic-activated cell sorting (MACS) immunobeads, respectively, according to the manufacturer’s instructions (Miltenyi Biotec). Then, the isolation of CD14+CD66b− and CD14+CD66b+ monocyte subpopulations, and CD14−CD66b+ PMN-MDSC was continued with fluorescence-activated cell sorting (FACS). Briefly, for the purification of CD66b+ monocytes and PMN-MDSC, freshly collected PBMC were resuspended in MACS buffer (1 × PBS/2 mM EDTA/0.5% BSA) and incubated with anti-CD66b immunobeads, applied onto MACS-LS columns (Miltenyi Biotec) and the immobilized cells were positively selected. This CD66b-enriched fraction was further labeled with anti-CD14 and anti-CD66b antibodies (BioLegend); CD14+CD66b+ and CD14−CD66b+ populations were purified by FACS. The PBMC collected in the flow-through of CD66b MACS were incubated with anti-CD14 magnetic beads and CD66b− monocytes were enriched by CD14 positive selection. Then, these cells were labeled with anti-CD14 and anti-CD66b antibodies and the CD14+CD66b− population were further purified by FACS. The purity of sorted cells was assessed by flow cytometric immunophenotyping (Supplementary Fig. 1).
RNA sequencing (RNA-seq) analysis
Total RNA was isolated from highly purified CD66b− and CD66b+ monocyte subpopulations according to the manufacturer’s instructions (RNeasy RNA isolation kit, QIAGEN). The RNA samples were pooled in three groups as healthy controls (n = 9), breast cancer patients (n = 12), and colorectal cancer patients (n = 12). Gene expression levels of over 2 × 104 human RefSeq genes were measured by Ampliseq Human Gene Expression Panel (Thermo Fisher Scientific) on an Ion Torrent next-generation sequencing platform (Ion S5, Thermo Fisher Scientific). Heat maps showing hierarchical clustering and differentially expressed genes were generated with integrated differential expression and pathway (iDEP) software [22]. Briefly, the genes with read counts per million < 0.5 were excluded by pre-processing on iDEP. Heat map data were centered by subtracting the average expression level for each gene. For cluster analysis and principal component analysis, started-log transformation, which is equivalent to the logCPM offered in edgeR, was applied to read counts. Differentially expressed genes (DEG) between CD66b+ and CD66b− monocyte subpopulations were determined according to two criteria: i. A fold change > 4 where adjusted p value is ≤ 0.01, and ii. No significant difference (a fold change < 2 where adjusted p value is ≤ 0.05) in the gene expression levels between CD66b+ monocytes obtained from CRC and BC patients. Out of 24 DEG that were determined according to these stringent criteria given above, six genes (OLR1, CCL2, DND1, CCL7, NLRP3, and SERPINE1) were identified with an adjusted p value < 0.001 and marked on volcano plots. Search tool for the retrieval of interacting genes/proteins (STRING) adjusted to high-level of confidence was used to construct protein–protein interaction network [23]. Kyoto encyclopedia of genes and genomes (KEGG) pathway enrichment analyses were performed by manual examination of gene lists. Percentile enrichment was calculated by normalization of total read counts of each KEGG gene set. Gene set enrichment analysis (GSEA) was used for further assessment of enriched specific biological pathways or signatures. The most variable genes were surveyed in the collection of molecular signatures database (MsigDB) by GSEA 3.0 [24] (Supplementary Table 3).
Microscopy
Purified cell populations were cytocentrifuged onto glass slides (50× g for 3 min), air-dried, and fixed in 4% paraformaldehyde for 20 min for immunofluorescence or in methanol for 5 min for May-Grünwald Giemsa (Merck) staining. Paraformaldehyde-fixed cells were permeabilized with 0.1% Triton X-100 (Sigma-Aldrich) in PBS and blocked with 1% BSA (Sigma-Aldrich). Then, the slides were incubated with anti-human CD14 (EPR3653; 1:400), CD66b (G10F5; 1:400), NALP3 (polyclonal; 1:400), LOX-1 (EPR4025, 1:400), PAI-1 (polyclonal, 1:400) primary antibodies, followed by Alexa488- or Alexa555-conjugated secondary antibodies and DAPI (Abcam) staining. Where needed, FITC-conjugated CD14 (M5E2; 1:200) or FITC-conjugated CD66b (G10F5; 1:200) antibodies were also used. The slides were mounted and representative micrographs were taken and processed by ImageJ (NIH).
Stimulation of monocytes
CD66b− monocytes from the healthy individuals were purified by CD14 MACS and cultured (5 × 105 cells/well in 96-well tissue culture plate) in complete RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS) or 10% patient serum, L-glutamine (2 mM), penicillin (100 U/mL) and streptomycin (100 μg/mL) at 37 °C in a humidified 5% CO2 incubator. The monocytes were stimulated with recombinant cytokines IFN-γ (50 ng/mL), IL-6 (10 ng/mL), G-CSF (100 ng/mL) (R&D Systems) or their combinations for 48 h. Then, the cells were harvested and CD66b, CD80, CD86, and HLA-DR levels on CD14+ cells were analyzed by flow cytometry.
Analysis of monocytes’ co-stimulatory influence on T cell responses
Monocytes were depleted from healthy donors’ PBMC using CD14 MACS (Miltenyi Biotec); then, the monocyte-depleted PBMC were labelled with carboxyfluorescein succinimidyl ester (CFSE, 5 µM) (Invitrogen) according to the manufacturer’s protocol. Purified CD66b+ and CD66b− monocytes from cancer patients or CD66b− monocytes from healthy controls were co-cultured with the CFSE-labelled monocyte-depleted allogeneic PBMC (105 cells/200 µL in U-bottom 96-well plates) at different ratios for 72 h in the presence of anti-CD3 mAb (25 ng/mL; HIT3a; BioLegend) which mimics the first signal for T cell activation. CD8+ and CD4+ T cells were gated and proliferation was assessed according to CFSE fluorescence dilution by flow cytometry. Monocyte-derived CD1a+CD83+ DC were also used in the co-cultures as technical positive controls for maximal T cell costimulation. For the generation of DC, purified CD14+ monocytes (5 × 105 cells/mL) were cultured in complete RPMI 1640 medium supplemented with 100 ng/mL granulocyte–macrophage colony-stimulating factor (GM-CSF) and 30 ng/mL IL-4 for 24 h; followed by an additional 48 h incubation with 50 ng/mL GM-CSF and 60 ng/mL IL-4. On the 5th day, DC maturation was induced by lipopolysaccharide (LPS, 50 ng/mL) for 48 h.
In addition to T cell proliferation, supernatants were collected from the co-cultures and the levels of IL-1α, IL-1β, IL-2, IL-4, IL-6, IL-10, IL-12, IL-17A, IFN-γ, TNF-α, and GM-CSF were determined by an ELISA array system (QIAGEN). Expression of CD69, CD137, and CD25 activation markers on CD8+ and CD4+ T cells was also determined in the co-cultures at 24 h by flow cytometry (Supplementary Table 4).
Determination of ROS and NO production
PBMC fraction was labeled with anti-CD14 and anti-CD66b antibodies (BioLegend), resuspended in serum-free RPMI 1640 medium, and incubated either with the reactive oxygen species (ROS) indicator, 5-(and-6)-carboxy-2′,7′-dichlorofluorescein diacetate (1 µM, H2-DCF-DA; Sigma-Aldrich), or with the nitric oxide (NO) indicator, 4,5-diaminofluorescein-diacetate (2 µM, DAF-2DA; Sigma-Aldrich), for 25 min. at room temperature. Maximal ROS and NO production was induced with phorbol 12-myristate 13-acetate (PMA, 800 nM; Sigma-Aldrich) and the subpopulations of interest were gated by flow cytometry and H2-DCF-DA or DAF-2DA fluorescence was simultaneously analyzed. ROS or NO production capacity index was calculated by normalization of MFI from PMA-stimulated cells to unstimulated cells.
Latex bead capture assay
PBMC in RPMI 1640 medium containing 5% FBS were incubated with 4% fluorescent-tagged carboxylate-modified polystyrene latex beads (2 μm, Sigma-Aldrich) for 4 h at 37 °C. After incubation, unbound beads were washed away and the cells were stained with anti-CD14 and anti-CD66b mAb (BioLegend). CD14+CD66b− and CD14−CD66b+ monocyte subpopulations and PMN-MDSC were gated and percentage of the cells that captured latex beads was determined by flow cytometry.
Analysis of migration capacity
Purified cells were resuspended in RPMI 1640 medium with 1% FBS, and seeded (105 cells/well) onto polycarbonate 24-well transwell inserts (pore size, 5 μm; Corning-Costar). The lower chambers were filled with complete medium. After 3 h of incubation, the upper surface of the inserts was swabbed and the cells that migrated and adhered underneath the inserts were fixed with methanol, stained with Giemsa, and counted under a light microscope.
Adhesion assay
Adhesion kinetics of the cells were examined on a xCELLigence real-time cell analysis platform (ACEA Biosciences). E-plates (ACEA Biosciences) were coated with the extracellular matrix components, laminin, fibronectin (Biological Industries), matrigel, and collagen type 1 (Corning-Costar), according to the manufacturers’ recommendations. Following a background impedance reading with complete medium, the purified myeloid cell subpopulations (105 cells/200 μL) were seeded. The plates were placed into xCELLigence station and impedance was measured for 8 h.
Quantification and statistical analysis
Graphical output and statistical analysis of significance were performed with Prism 6 software (GraphPad). Statistical analyses were carried out by one-way analysis of variance (ANOVA) with post-hoc analyses or Student’s paired or unpaired t test where appropriate. Differences were considered as statistically significant when p value was ≤ 0.05. Otherwise noted, the data are shown as the mean ± standard error of the mean (SEM).
Results
CD66b+ monocytes emerge in the circulation of cancer patients and are not related to MDSC
MDSC have been described as immature myeloid regulatory cells that emerge within the peripheral blood mononuclear cells (PBMC) of cancer patients [13]. Either M-MDSC or PMN-MDSC subsets are universally recognized as HLA-DR−/lo leukocytes where the latter bears CD15 and CD66b granulocyte-specific surface antigens [21]. Moreover, the monocytes have been renowned as CD66b-negative cells [25]. Nevertheless, when the HLA-DR gating was omitted, a subpopulation of CD11b+CD33hi cells (i.e., CD14+ monocytes) that moderately expressed CD66b was identified together with the CD14−CD33moCD66bhi PMN-MDSC. The CD66b+ monocytes appeared as negligible events when the HLA-DR−/lo cells were gated with the common immunophenotyping strategy used for M-MDSC (Fig. 1a). Fluorescence minus one (FMO) and isotype-matched Ig controls indicated a specific labeling for CD66b on these CD14+CD33hi cells (Fig. 1b). Immunophenotyping was also confirmed with two different anti-human CD66b monoclonal antibody clones (Supplementary Fig. 2a and b). Immunofluorescence imaging on the purified CD66b+ or CD14+ leukocytes from the PBMC fraction (< 1.077 g/mL) of breast cancer (BC) or colorectal cancer (CRC) patients validated the coexistence of CD14 and CD66b (Supplementary Fig. 3 and Fig. 1c). In contrast to CD66b− monocytes and CD66b+ PMN-MDSC, the CD66b+ monocytes displayed various nuclear morphologies: mostly bi-lobed, irregular with prominent folds or delicate convolutions (Fig. 1d). They were CD11bhi and negative for the non-PMN granulocyte marker, CD125. Minor and variable percentages of CD66b+ monocytes were HLA-DR−/lo (15.95 ± 11.30%), CD15mo (13.77 ± 19.98%), CD16mo (23.11 ± 13.32%), and CD16hi (9.55 ± 5.33%). When CD66b+ monocytes were back-gated, 8.70 ± 5.15% of M-MDSC, 25.71 ± 15.27% of CD14+CD16mo intermediate monocytes, and 16.27 ± 11.86% of CD14+CD16hi non-classical monocytes found to be positive for CD66b (Fig. 1e). To test whether the expression of CD66b was inducible, monocytes from healthy donors were stimulated with inflammatory factors (IFN-γ, IL-6, G-CSF or the combinations of these cytokines) and patient sera; however, CD66b was not upregulated on the CD14+ monocytes, ex vivo (Supplementary Fig. 4).
Fig. 1.
Identification of CD66b+ monocytes by immunophenotyping. a Flow cytometry gating strategy commonly (yellow gates) used for detection of MDSC and the alternative gating strategy (purple gates) used to identify CD66b+ monocyte subpopulation in a cancer patient. b Fluorescence minus one (FMO) analysis for CD66b staining. c The presence of CD66b+CD14+ subpopulation amongst the PBMCs from breast cancer (BC) and colorectal cancer (CRC) patients was confirmed by immunofluorescence staining of the cells purified with CD66b or CD14 MACS (Supplementary Fig. 3) (scale bar, 10 μm). d May-Grünwald Giemsa staining on cytospin preparations of CD66b− and CD66b+ monocytes and PMN-MDSC from cancer patients (scale bar, 10 μm). e Distribution of CD66b+ monocytes amongst the myeloid cell populations according to CD11b, CD125, CD15, HLA-DR, CD14, CD16 expression levels. (CD14+CD33hiCD66b+ monocytes, red; CD66b+CD14+CD33hi monocytes, green; CD14−CD33moCD66b+ PMN-MDSC, gray)
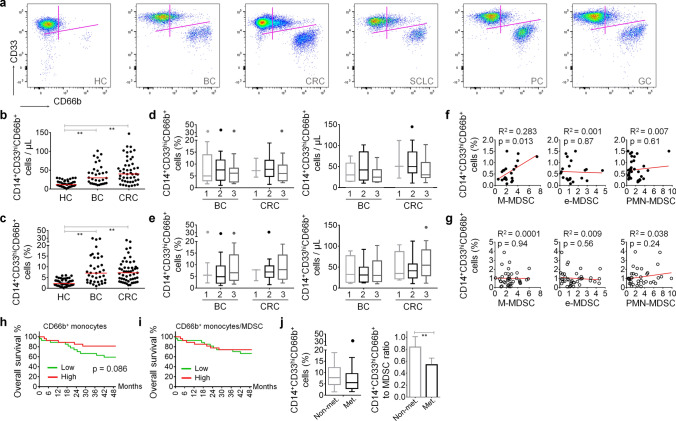
These CD14+CD33hiCD66b+ cells were barely detectable in healthy subjects (median 2.7%; range 0.2–6.03% of CD14+ monocytes); whereas, they constituted a notable population in the cancer patients’ circulation (median 8.45%; range 1.06–29.5% of CD14+ monocytes) including breast, colorectal, lung, pancreatic, and gastric cancers (Fig. 2a). Both absolute count and percentage of these cells were augmented in BC and CRC patients (Fig. 2b and c); intriguingly, no significant difference was observed when the data were stratified according to the histopathological or clinical grades (Fig. 2d and e). In general, the percentage of CD66b+ monocytes was not significantly correlated with that of MDSC subsets (Fig. 2f and g) or conventional monocyte subtypes (Supplementary Fig. 5). Only in CRC, the percentage of CD14+CD33hiHLA-DR−/loCD66b− M-MDSC was significantly correlated with CD66b+ monocytes (Fig. 2f and g). Albeit not reaching to the level of statistical significance, a trend was observed between the percentages of CD66b+ monocytes and CD14+CD16mo or CD14+CD16hi monocytes in BC, but not in CRC (Supplementary Fig. 5).
Fig. 2.
CD66b+ monocytes are increased in cancer patients. a Representative flow cytometry scatter plots showing CD66b+ subpopulation of CD33hi monocytes and CD66b+CD33mo PMN-MDSC in healthy controls (HC), breast cancer (BC), colorectal cancer (CRC), small cell lung cancer (SCLC), pancreatic cancer (PC), and gastric cancer (GC) patients’ circulation. b Absolute count of CD66b+ monocytes and (c) their percentage in CD14+ monocytic cells in healthy subjects (n = 44) and in cancer patients (CRC, n = 51; BC, n = 37). b, c Red lines represent the median values. Distribution of absolute count and percentage data according to the (d) histopathological and (e) clinical stages of BC and CRC patients. Correlation analyses of CD66b+ monocytes with MDSC subtypes that emerge in (f) BC and (g) CRC peripheral blood. Kaplan–Meier curves for the CRC patients followed-up for 48 months are given according to (h) the percentage of CD14+CD33hiCD66b+ monocytes and (i) their proportion to MDSC. (j) The distribution of CD14+CD33hiCD66b+ cells (left panel) and their proportion to MDSC (right panel) in CRC patients with or without metastasis are given. Data were obtained from at least three independent experiments and are shown as mean ± SEM (Student’s t-test; *p < 0.05, **p < 0.01)
Even though the clinical follow-up of the patients enrolled to this study was limited to ~ 48 months, high levels of CD14+CD33hiCD66b+ cells tended to associate with longer overall survival in CRC but it did not reach the level of statistical significance (Fig. 2h). On the other hand, when the increment in MDSC population was considered as a factor related to disease progression, the positive tendency of CD66b+ monocytes on the survival was completely lost (Fig. 2h). Accordingly, not the amount of CD66b+ monocytes but their proportion to MDSC was significantly decreased in the patients with metastatic disease (Fig. 2j).
Collectively, in the peripheral blood of cancer patients, a subset of CD14+CD33hi monocytes was identified with CD66b expression. These cells were essentially HLA-DRhiCD16−; however, CD66b was also detected on minor populations which displayed surface antigens previously associated with archetypal sub-types of monocytic cells. Certain inflammatory factors did not induce CD66b on healthy monocytes and CD66b+ monocytes did not show strong correlation with MDSC or conventional monocyte subtypes. Moreover, high levels of CD66b+ monocytes were associated with overall survival.
CD66b+ monocytes constitute a distinct subpopulation with proinflammatory profile
Next, we performed a flow cytometry-based surface antigen screening array and a whole genome mRNA transcriptomics analysis to better define the CD66b+ monocytes. Critically, out of 80 surface antigens tested, which are associated with hematological lineages, 33 were differentially expressed by the CD66b+ monocytes when compared to the PMN-MDSC which are also identified with CD66b marker. Twenty-three markers were expressed at different levels on the CD66b+ monocytes when compared to the CD66b− monocytes (Fig. 3a). There were only 7 molecules (CD275, CD276, CD278, CD284, CD68, CCRL2, and CD66b) that could be useful to discriminate the CD66b+ monocytes from both the CD66b− conventional monocytes and the CD66b+ PMN-MDSC (Supplementary Fig. 6). Nevertheless, CD66b displayed the most distinctive expression pattern and the other six molecules’ expression has been previously shown to be altered in response to inflammatory mediators [26–30].
Fig. 3.
Inflammatory signatures are prominent in CD66b+ monocytes. a Flow cytometry-based array for 80 molecules expressed in CD66b+ and CD66b− monocytes, and PMN-MDSC. MFI values from specific subpopulations were normalized to those of from total events counted. b RNA-seq and hierarchical clustering analyses were performed on purified cells, and expression data are presented for each monocyte subpopulation. c Gene set enrichment heatmaps for monocyte subpopulations, (d) network analysis, (e) percent enrichment of the pathways, and (f) volcano plots showing differentially expressed genes in CD66b+ monocytes compared to that of CD66b− in BC and CRC patients. The data from CD66b− monocytes are available in Supplementary Fig. 9. Please note the similar pattern plotted between CD66b+ monocytes from BC and CRC
Two different strategies were used to determine the differentially expressed genes (DEGs) between CD14+CD66b+ and CD14+CD66b− monocyte subpopulations in BC and CRC. When statistical criteria on the transcripts were set to at least twofold difference and adjusted p value was ≤ 0.05; 2087 upregulated, 68 downregulated genes were identified in BC, and 349 upregulated, 276 downregulated genes were identified in CRC (Supplementary Table 5). The number of DEGs overlapped both in BC and CRC was 219 (208 upregulated and 11 downregulated). Thus, the transcriptomic analysis revealed a large difference between BC- and CRC-derived monocytes (Fig. 3b–f). For transcriptomics, RNA samples of CD14+CD66b+ and CD14+CD66b− monocyte subpopulations isolated from different patients were pooled; thus, we preferred to apply more stringent criteria for identifying the signature genes amongst DEGs with a fold change > 4 and adjusted p value < 0.01. Accordingly, the hierarchical clustered heatmaps obtained from RNA-Seq analyses of CD14+CD66b+ and CD14+CD66b− monocyte subpopulations showed a set of genes (24 DEGs with adjusted p value < 0.05 and fold change > 4) strongly upregulated in CD66b+ monocytes from both CRC and especially BC patients (Supplementary Table 6). The transcriptomic profile of the CD66b− monocytes was very close to that of healthy control subjects (Fig. 3b and Supplementary Fig. 7). The Inflammation-associated gene set enrichment demonstrated increased expression of certain cytokines and costimulatory molecules including the members of chemokine, interleukin (IL), interferon (IFN), and tumor necrosis factor (TNF) families. Critically, many transcription factors related to cell cycle, differentiation and development, and epigenetic regulation processes were explicitly augmented in CD14+CD66b+ subpopulation (Fig. 3c). The network biology integration of the genes that were significantly upregulated in the CD66b+ monocytes indicated an increase in the cellular activities related to cell migration and adhesion, detection of chemical stimuli, and expression of factors related to IFN family, cAMP signaling, chromosomal stability, ribosomal biogenesis, and cell division (Fig. 3d). The genes responsible for extracellular matrix (ECM) interaction and cell adhesion, and inflammatory responses were upregulated in the CD14+CD66b+ cells. Nevertheless, expression of the genes employed in basal cellular functions, metabolism and maintenance showed a decline (Fig. 3e). The gene set enrichment analysis (GSEA) revealed that the over-represented genes in CD66b+ monocytes were not compatible with those previously reported macrophage or monocyte gene sets (nominal p value < 0.05) (Supplementary Fig. 8). As signature genes (fold change > 4, and adjusted p value ≤ 0.001), oxidized low-density lipoprotein receptor 1 (OLR1), CCL2, CCL7, nod-like receptor family pyrin domain containing 3 (NLRP3), DND microRNA-mediated repression inhibitor 1 (DND1), and serine protease inhibitor 1 (SERPINE1) were explicitly and differentially expressed in the CD66b+ monocytes (Supplementary Table 6). These genes were at similar levels in the CD66b+ subpopulations both from BC and CRC (Fig. 3f). High-level expression of LOX-1, NALP3, and PAI-1, which are the respective proteins encoded by OLR1, NLRP3, and SERPINE1, was also determined in the CD14+CD66b+ subpopulation (Supplementary Fig. 10).
In summary, CD66b+ monocytes that expand in cancer were distinguished as a distinct subpopulation with potential influences on the inflammatory responses.
T cell costimulation capacity and myeloid functions are pronounced in CD66b+ monocytes
In cancer, the myeloid cell compartment becomes functionally hampered [13]. Not only the specific subsets of myeloid regulatory cells emerge but also the overall magnitude of myeloid cells’ inflammatory capacities are reduced [12]. Thus, the influence of CD66b+ monocyte-derived costimulatory signals on T cell reactions was evaluated. For this purpose, the monocytes were co-cultured with T cells in the presence of anti-CD3 mAb that mimics the signal-1 produced by T cell receptor (TCR) complex. The early T cell activation markers CD69, CD137, and CD25 were induced at similar levels upon co-culturing with CD66b+ or CD66b− monocytes (Supplementary Table 4). Nevertheless, the CD66b− monocytes from cancer patients either were inefficient at providing costimulation for the effector responses, or harbored immunosuppressive populations; however, the costimulation by CD66b+ monocytes, which was compatible with that of healthy monocytes, maintained the T cell proliferation and differentiation (Fig. 4a–e). T cell proliferation in the presence of CD66b+ monocytes from either BC or CRC patients (at 0.125:1 monocyte:T cell ratio; range, 46.78–72.7%) was approximately 1.5-fold higher than that of observed with the CD66b− monocytes (range 31.43–57.53%). Nevertheless, they were not as proficient as the monocyte-derived DC at providing costimulatory signals for T cell proliferation (Fig. 4a). In comparison with the patient-derived CD66b− monocytes, under the influence of CD66b+ monocytes, secretion of proinflammatory cytokines such as IL-1β, IL-6, and IFN-γ was not suppressed (Fig. 4b) and both CD4+ and CD8+ T cells were essentially proliferated (Fig. 4d and e). Accordingly, T cell-derived IFN-γ levels were comparable to or even higher (in BC) than that of produced in the presence of healthy donor CD66b− monocytes (Fig. 4c). Next, the functions directly related to myeloid cells were evaluated. There was no significant difference in NO or ROS production capacities between CD66b+ and CD66b− monocyte subpopulations (Fig. 4f). Nevertheless, the CD66b+ monocytes were able to capture high amount of latex beads (Fig. 4g), displayed enhanced migratory capacity (Fig. 4h), and efficiently adhered to the extracellular matrix such as Matrigel, fibronectin, and collagen (Fig. 4i).
Fig. 4.
Functional evaluation of CD66b+ monocytes in BC and CRC patients. a Influence of CD66b+ or CD66b− monocytes from healthy controls (HC), breast cancer (BC) or colorectal cancer (CRC) patients on the proliferation of T cells amongst monocyte-depleted PBMC stimulated with anti-CD3 monoclonal antibody (the first activation signal for T cells). This setup enables to monitor costimulation (the second signals) provided by the monocytes. Monocyte-derived DC were used as positive controls that provide proficient costimulation for T cell proliferation. Representative histograms showing CFSE dilution in the T cells proliferated under the influence of monocyte subpopulations are given in the right panels. b Graphical output of inflammatory cytokine ELISArray and (c) IFN-γ ELISA performed with the supernatants collected from the co-cultures of anti-CD3-stimulated PBMC and CD66b− or CD66b+ monocytes. Change in the proliferation of CD4+ and CD8+ T cells stimulated with anti-CD3 and the monocyte subpopulations from (d) BC and (e) CRC patients is shown. CD66b+ and CD66b− monocytes and PMN-MDSC from cancer patients were evaluated for (f) nitric oxide (NO) and reactive oxygen species (ROS) production (upper panels, representative flow cytometry overlay histograms of PMA-induced cells), (g) latex-bead capture (right panel, representative flow cytometry overlay histograms). h Percentage of myeloid cells migrated towards high-FBS gradient were assayed with transwell chambers with 5-μm pore-size inserts. i Adhesion kinetics of CD66b+ or CD66b− monocyte subpopulations, and PMN-MDSC onto extracellular matrix components were measured on a xCELLigence real-time cell analysis platform. Data were obtained from at least three independent experiments and are shown as mean ± SEM (Student’s t test; *p < 0.05, **p < 0.01)
All in all, the CD66b+ monocytes were not of the myeloid regulatory cells which emerge in cancer; in contrast, they represented a functionally distinct subpopulation which provokes inflammatory responses.
Discussion
The monocyte lineage retains a high capacity to adapt microenvironmental changes and differentiate into functional subsets of macrophages or dendritic cells [3]. Monocytes represent one of the most plastic leukocyte types that circulate without full maturation or terminal differentiation [4]. Accordingly, both phenotypic and functional heterogeneity are observed amongst the circulating monocytes in the context of various diseases [7–9]. Nevertheless, to date, only limited number of monocyte subtypes has been described under inflammatory or pathological circumstances [10, 16, 31–34]. Here, in the cancer patients’ circulation, a unique subpopulation of monocytes was identified to carry CD66b, a marker classically associated with the granulocyte lineage. Even though the loss-of-functional competence in the myeloid cells is a common facet in cancer, the CD66b+ monocytes retained proinflammatory capacity and functionally contributed to the immune reactions, ex vivo.
Ever since the initial description of the phagocytic cells, our knowledge on the monocytes became stronger through new concepts and changing perspectives regarding origin, morphology, molecular signatures, and functional differentiation. Under physiological conditions, the circulating monocytes are characterized according to the differential expression of the lipopolysaccharide (LPS) co-receptor CD14 and the type III Fcγ receptor CD16 [25]. Accordingly, classical monocytes, non-classical monocytes, and intermediate monocytes are three well-defined subtypes; while distinctions amongst them are clearly established, recent studies pointed out further heterogeneity in the monocyte lineage both in human and mouse [35, 36]. Even though numerous surface molecules have been identified on the peripheral blood monocytes, only a few has been reported to be associated with specific subpopulations that are distinguished with functional discrepancies [6, 30]. Especially, the pathological conditions, which induce emergency monopoiesis and alter hematopoietic signaling pathways, result in the egress of monocyte populations with distinct features that are not commonly observed in healthy individuals [5]. In cancer, chronic inflammation and tumor-derived factors not only hamper anti-tumor capacities of myeloid cells but also lead to accumulation of monocyte-related M-MDSC and e-MDSC populations [13]. In addition, circulating FcεRI+ monocytes [32], CD64hiCD33hiCD13loCSF1R+ proliferating monocytes [33], M1 monocytes [10], TIE2-expressing monocytes [37] as well as precursors of fibrocytes [31] and osteoclasts [34] have been detected in various diseases. In mouse, CD209ahiMHCIIhi monocytes [38], Ceacam1+Msr1+Ly6C−F4/80−Mac1+ monocytes that share granulocyte characteristics [39], and the neutrophil-like Ly6Chi monocytes [40] have been previously reported. Recently, monocyte-like precursors of granulocytes (MLPGs) were identified in human cancers that commit to PMN-MDSC, a similar phenomenon previously reported in the tumor-bearing mice [41, 42].
The granulocyte-associated markers such as CD66b and especially CD15, which is preferred over CD66b, are frequently used to discriminate between M-MDSC and PMN-MDSC [21]. Currently, MDSC immunophenotyping relies on the presence or absence of certain surface molecules and should be followed by functional analyses [11]. Nonetheless, strict gating strategies might preclude new observations on the alterations in distinct cell populations [43]. In our study, the exclusion of HLA-DRmo/hi monocytes, which is a common immunophenotyping approach for M-MDSC detection, was one of the main factors reducing the percentage of CD66b+CD33hiCD14+ cells to a negligible level. In addition, the presence of distinct fractions amongst CD66b+ monocytes with various levels of CD16 or CD15, could be another reason for the ignorance of this subpopulation by other researchers. Even the diverse nuclear shape and other morphological features of CD66b+CD33hiCD14+ cells were indicative of a certain level of heterogeneity from the point of view of classical cytology. CD66b expression can be found on e-MDSC as well; however, they are distinguished as CD14 negative and CD33mo cells [21]. Alternatively, in severe infections, activated neutrophil granulocytes can express CD14 and HLA-DR, and loose CD15 and CD66b [44–46]. Nevertheless, in addition to the immunophenotyping arrays where many monocyte-associated surface molecules were identified, whole genome transcriptomics enabled the discrimination of CD66b+ monocytes from the conventional monocytes, PMN and MDSC. Moreover, healthy donor monocytes failed to upregulate CD66b upon stimulation with certain cytokines and patient sera. Therefore, CD66b, which is an important indicator for the granulocytes both in the peripheral blood and bone marrow, marked a subpopulation of monocytes in the cancer patients’ circulation.
Even though CD66b was the most distinctive surface molecule for this monocyte subpopulation, the expression of CD275, CD276, CD278, CD284, CD68, and CCRL2 was also remarkable. CD275 (B7-H2) and CD276 (B7-H3) are of the B7 family of immune regulatory ligands which could directly enhance or modulate T cell responses such as proliferation, differentiation and cytokine production [47]. CD68, CCRL2 (atypical chemokine receptor 5), and CD284 (Toll-like receptor 4) can indicate increased functional capacities such as phagocytosis, LPS sensing, and regulation of chemotaxis [29, 48–50]. Furthermore, CD66b+ monocytes displayed transcriptional signatures distinct from the previously reported monocyte subtypes. The most significant DEGs OLR1, CCL2, CCL7, NLRP3, and SERPINE1 were also indicating the migration, phagocytosis, and inflammatory potential of these cells [51–55]. Even though our results argue a common signature for CD66b+ monocytes, large transcriptomic differences were also noted between the monocytes from breast cancer and colorectal cancer patients that specify a disease-associated differentiation state. The pathway enrichment analyses on the CD66b+ monocytes highlighted their activities on many cellular processes, and indicated an increased capacity for migration, adhesion, and detection of chemical stimuli. In addition to the inflammatory mediators such as cytokines and chemokines, mRNA levels of numerous transcription factors were upregulated. Notably, the transcription factors UHRF1 and PHF5A are epigenetic regulators that bridge DNA methylation and chromatin modifications together with ZNF16, TFDP1, and E2F6 which are also implicated in cell cycle regulation [56–60]. The distinctive transcription factor signatures are essential for functional differentiation and subtype classification of the cells. Collectively, the CD66b+ monocytes that emerge in the cancer patients’ circulation are in an active state and display a proinflammatory profile.
The monocytes serve as the precursors of macrophages and DC, which are professional at perceiving extracellular stimuli, engulfing and processing antigens, migration, providing costimulatory molecules and T cell-polarizing cytokines [3, 5]. These processes should be fine-tuned for a successful anti-tumor immunity which is essentially mediated through Th1-oriented responses [61]. The CD66b+ monocytes both from breast cancer and colorectal cancer patients can be distinguished with two aspects: First, they did not fail to support both CD8+ cytotoxic T cell and CD4+ Th cell proliferation and to induce the secretion of key proinflammatory cytokines, such as IFN-γ. Second, the CD66b+ monocytes were distinctive with their improved abilities related to cell migration, adhesion, and phagocytosis. Nevertheless, whether these cells are in an interim functional state, display plasticity similar to that observed in the intermediate monocytes or represent a functionally differentiated population remain to be comprehensively investigated.
The importance of functional characterization over classical immunophenotyping, especially for myeloid regulatory cell subsets, has been highlighted [21]. Many discrepancies observed during functional assays can be attributed to diversity of the laboratory practices or the patient samples. In addition, the cells isolated according to specific markers may harbor various fractions of heterogeneous populations which may impede the consistency of functional assays [11]. In the laboratory practice, the CD66b+ monocytes might be contained amongst M-MDSC and PMN-MDSC isolated from the cancer patients, and may interfere or mask the immune suppression anticipated from these suppressive cells. The dysregulation in the myeloid cells is not only seen in cancer but also in many chronic inflammatory diseases such as infections and autoimmunity [7–9]. Therefore, the presence of CD66b+ monocytes under many pathological circumstances and their contribution to inflammatory course of the diseases need to be defined.
Conclusion
The CD66b+ monocytes were identified as a novel myeloid cell subpopulation that emerge in cancer patients’ circulation along with the MDSC. In contrast to the pro-tumor subversion observed in the myeloid cells, the CD66b+ monocytes displayed unique molecular and functional signatures of proinflammatory capacities that highlight their immune-provoking potential in cancer. A better understanding of this monocyte subpopulation might have important implications for cancer immunology and immunotherapy approaches.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This work was partially supported by The Scientific and Technological Research Council of Turkey, (TÜBİTAK; Project No. 115S636) and Hacettepe University Scientific Research and Coordination Unit, (Grant No. TSA-2018-17239), and covered under the European Cooperation in Science and Technology (COST-EU) Action BM1404 (Mye-EUNITER) (https://www.myeeuniter.eu). COST is supported by the EU Framework Program Horizon 2020. We acknowledge the technical support by Beren Karaosmanoglu, PhD. The authors thank all patients and nurses who contributed to the study, especially Nuraydın Sahin for providing blood samples.
Author contributions
G.E. conceptualized the project and designed the experiments. U.H. and D.Y.-E. performed experiments. G.E., U.H., and D.Y.-E. interpreted data. U.H. performed bioinformatics analysis of RNA-seq data and contributed to preparation of the figures. E.Z.T performed assays related to transcriptomics. K.B.Y., E.H., and D.K. assisted with histopathological and clinical evaluation of the patients. G.E. and U.H. wrote the manuscript. All authors reviewed and approved the manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Nagl M, Kacani L, Mullauer B, et al. Phagocytosis and killing of bacteria by professional phagocytes and dendritic cells. Clin Vaccine Immunol. 2002;9:1165–1168. doi: 10.1128/cdli.9.6.1165-1168.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jakubzick CV, Randolph GJ, Henson PM. Monocyte differentiation and antigen-presenting functions. Nat Rev Immunol. 2017;17:349–362. doi: 10.1038/nri.2017.28. [DOI] [PubMed] [Google Scholar]
- 3.Boyette LB, MacEdo C, Hadi K, et al. Phenotype, function, and differentiation potential of human monocyte subsets. PLoS ONE. 2017;12:1–20. doi: 10.1371/journal.pone.0176460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ginhoux F, Jung S (2014) Monocytes and macrophages: developmental pathways and tissue homeostasis TL—14. Nat Rev Immunol 14 VN-r:392–404. 10.1038/nri3671 [DOI] [PubMed]
- 5.Guilliams M, Mildner A, Yona S. Review developmental and functional heterogeneity of monocytes. Immunity. 2018;49:595–613. doi: 10.1016/j.immuni.2018.10.005. [DOI] [PubMed] [Google Scholar]
- 6.Patel AA, Zhang Y, Fullerton JN, et al. The fate and lifespan of human monocyte subsets in steady state and systemic inflammation. J Exp Med. 2017;214:1913–1923. doi: 10.1084/jem.20170355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gordon S, Taylor PR. Monocyte and macrophage heterogeneity. Nat Rev Immunol. 2005;5:953–964. doi: 10.1038/nri1733. [DOI] [PubMed] [Google Scholar]
- 8.Ong SM, Hadadi E, Dang TM, et al. The pro-inflammatory phenotype of the human non-classical monocyte subset is attributed to senescence article. Cell Death Dis. 2018;9:1–12. doi: 10.1038/s41419-018-0327-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wildgruber M, Aschenbrenner T, Wendorff H, et al. The “Intermediate” CD14++CD16+ monocyte subset increases in severe peripheral artery disease in humans. Sci Rep. 2016;6:39483. doi: 10.1038/srep39483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marimuthu R, Francis H, Dervish S, et al. Characterization of human monocyte subsets by whole blood flow cytometry analysis. J Vis Exp. 2018;140:1–10. doi: 10.3791/57941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bruger AM, Dorhoi A, Esendagli G, et al. How to measure the immunosuppressive activity of MDSC: assays, problems and potential solutions. Cancer Immunol Immunother. 2019;68:631–644. doi: 10.1007/s00262-018-2170-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gabrilovich DI, Nagaraj S. Myeloid-derived suppressor cells as regulators of the immune system. Nat Rev Immunol. 2009;9:162–174. doi: 10.1038/nri2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gabrilovich DI. Myeloid-derived suppressor cells. Cancer Immunol Res. 2017;5:3–8. doi: 10.1158/2326-6066.CIR-16-0297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Onicescu G, Rosato A, Mandruzzato S, et al. A human promyelocytic-like population is responsible for the immune suppression mediated by myeloid-derived suppressor cells. Blood. 2011;118:2254–2265. doi: 10.1182/blood-2010-12-325753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brandau S, Trellakis S, Bruderek K, et al. Myeloid-derived suppressor cells in the peripheral blood of cancer patients contain a subset of immature neutrophils with impaired migratory properties. J Leukoc Biol. 2011;89:311–317. doi: 10.1189/jlb.0310162. [DOI] [PubMed] [Google Scholar]
- 16.Filipazzi P, Valenti R, Huber V, et al. Identification of a new subset of myeloid suppressor cells in peripheral blood of melanoma patients with modulation by a granulocyte-macrophage colony-stimulation factor-based antitumor vaccine. J Clin Oncol. 2007;25:2546–2553. doi: 10.1200/JCO.2006.08.5829. [DOI] [PubMed] [Google Scholar]
- 17.Kumar V, Patel S, Tcyganov E, Gabrilovich DI. The nature of myeloid-derived suppressor cells in the tumor microenvironment. Trends Immunol. 2016;37:208–220. doi: 10.1016/j.it.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Umansky V, Blattner C, Gebhardt C, Utikal J. The role of myeloid-derived suppressor cells (MDSC) in cancer progression. Vaccines. 2016;4:36. doi: 10.3390/vaccines4040036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moses K, Brandau S. Human neutrophils: their role in cancer and relation to myeloid-derived suppressor cells. Semin Immunol. 2016;28:187–196. doi: 10.1016/j.smim.2016.03.018. [DOI] [PubMed] [Google Scholar]
- 20.Qu P, Wang L, Lin PC. Expansion and functions of myeloid-derived suppressor cells in the tumor microenvironment. Cancer Lett. 2016;380:253–256. doi: 10.1016/j.canlet.2015.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bronte V, Brandau S, Chen S-H, et al. Recommendations for myeloid-derived suppressor cell nomenclature and characterization standards. Nat Commun. 2016;7:12150. doi: 10.1038/ncomms12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ge SX, Son EW, Yao R. iDEP: An integrated web application for differential expression and pathway analysis of RNA-Seq data. BMC Bioinform. 2018;19:1–24. doi: 10.1186/s12859-018-2486-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Szklarczyk D, Morris JH, Cook H, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ziegler-heitbrock L, Ancuta P, Crowe S, et al. Nomenclature of monocytes and dendritic cells in blood. Blood. 2010;116:5–7. doi: 10.1182/blood-2010-02-258558. [DOI] [PubMed] [Google Scholar]
- 26.Yoon BR, Yoo S, Choi Y, et al. Functional phenotype of synovial monocytes modulating inflammatory T-cell responses in rheumatoid arthritis ( RA ) PLoS ONE. 2014;9:e109775. doi: 10.1371/journal.pone.0109775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aicher A, Hayden-Ledbetter M, Brady WA, et al. Characterization of human inducible costimulator ligand expression and function. J Immunol. 2000;164:4689–4696. doi: 10.4049/jimmunol.164.9.4689. [DOI] [PubMed] [Google Scholar]
- 28.Ruth JH, Rottman JB, Kingsbury GA, et al. ICOS and B7 costimulatory molecule expression identifies activated cellular subsets in rheumatoid arthritis. Cytom Part A. 2007;71:317–326. doi: 10.1002/cyto.a.20383. [DOI] [PubMed] [Google Scholar]
- 29.Galligan CL, Matsuyama W, Matsukawa A, et al. Up-regulated expression and activation of the orphan chemokine receptor, CCRL2, in rheumatoid arthritis. Arthritis Rheum. 2004;50:1806–1814. doi: 10.1002/art.20275. [DOI] [PubMed] [Google Scholar]
- 30.Gren ST, Rasmussen TB, Janciauskiene S. A single-cell gene-expression profile reveals inter-cellular heterogeneity within human monocyte subsets. PLoS ONE. 2015;10:1–20. doi: 10.1371/journal.pone.0144351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bucala R, Spiegel LA, Chesney J, et al. Circulating fibrocytes define a new leukocyte subpopulation that mediates tissue repair. Mol Med. 1994;1:71–81. doi: 10.1007/BF03403533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maurer D. Expression of functional high affinity immunoglobulin E receptors (Fc epsilon RI) on monocytes of atopic individuals. J Exp Med. 1994;179:745–750. doi: 10.1084/jem.179.2.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clanchy FIL. Detection and properties of the human proliferative monocyte subpopulation. J Leukoc Biol. 2006;79:757–766. doi: 10.1189/jlb.0905522. [DOI] [PubMed] [Google Scholar]
- 34.Komano Y, Nanki T, Hayashida K, et al. Identification of a human peripheral blood monocyte subset that differentiates into osteoclasts. Arthritis Res Ther. 2006;8:1–14. doi: 10.1186/ar2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Villani A-C, Satija R, Reynolds G, et al (2017) Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science 356: eaah4573. 10.1126/science.aah4573 [DOI] [PMC free article] [PubMed]
- 36.Mildner A, Schönheit J, Giladi A, et al. genomic characterization of murine monocytes reveals C/EBPβ transcription factor dependence of Ly6C− cells. Immunity. 2017;46:849–862.e7. doi: 10.1016/j.immuni.2017.04.018. [DOI] [PubMed] [Google Scholar]
- 37.Venneri MA, De Palma M, Ponzoni M, et al. Identification of proangiogenic TIE2-expressing monocytes (TEMs) in human peripheral blood and cancer. Blood. 2007;109:5276–5285. doi: 10.1182/blood-2006-10-053504. [DOI] [PubMed] [Google Scholar]
- 38.Menezes S, Melandri D, Anselmi G, et al. The heterogeneity of Ly6Chi monocytes controls their differentiation into iNOS+ macrophages or monocyte-derived dendritic cells. Immunity. 2016;45:1205–1218. doi: 10.1016/j.immuni.2016.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Satoh T, Nakagawa K, Sugihara F, et al. Identification of an atypical monocyte and committed progenitor involved in fibrosis. Nature. 2017;541:96–101. doi: 10.1038/nature20611. [DOI] [PubMed] [Google Scholar]
- 40.Yáñez A, Coetzee SG, Olsson A, et al. Granulocyte-monocyte progenitors and monocyte-dendritic cell progenitors independently produce functionally distinct monocytes. Immunity. 2017;47:890–902.e4. doi: 10.1016/j.immuni.2017.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Youn JI, Kumar V, Collazo M, et al. Epigenetic silencing of retinoblastoma gene regulates pathologic differentiation of myeloid cells in cancer. Nat Immunol. 2013;14:211–220. doi: 10.1038/ni.2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mastio J, Condamine T, Dominguez G, et al (2019) Identification of monocyte-like precursors of granulocytes in cancer as a mechanism for accumulation of PMN-MDSCs. J Exp Med jem.20181952. 10.1084/jem.20181952 [DOI] [PMC free article] [PubMed]
- 43.Mandruzzato S, Brandau S, Britten CM, et al. Toward harmonized phenotyping of human myeloid-derived suppressor cells by flow cytometry: results from an interim study. Cancer Immunol Immunother. 2016;65:161–169. doi: 10.1007/s00262-015-1782-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wright HL, Moots RJ, Bucknall RC, Edwards SW. Neutrophil function in inflammation and inflammatory diseases. Rheumatology. 2010;49:1618–1631. doi: 10.1093/rheumatology/keq045. [DOI] [PubMed] [Google Scholar]
- 45.Wittmann S, Rothe G, Schmitz G, Fröhlich D. Cytokine upregulation of surface antigens correlates to the priming of the neutrophil oxidative burst response. Cytom Part A. 2004;57:53–62. doi: 10.1002/cyto.a.10108. [DOI] [PubMed] [Google Scholar]
- 46.Wagner C, Deppisch R, Denefleh B, et al. Expression patterns of the lipopolysaccharide receptor CD14, and the FCgamma receptors CD16 and CD64 on polymorphonuclear neutrophils: data from patients with severe bacterial infections and lipopolysaccharide-exposed cells. Shock. 2003;19:5–12. doi: 10.1097/00024382-200301000-00002. [DOI] [PubMed] [Google Scholar]
- 47.Sharpe AH, Freeman GJ. The B7-CD28 superfamily. Nat Rev Immunol. 2002;2:116–126. doi: 10.1038/nri727. [DOI] [PubMed] [Google Scholar]
- 48.Chistiakov DA, Killingsworth MC, Myasoedova VA, et al. CD68/macrosialin: not just a histochemical marker. Lab Investig. 2017;97:4–13. doi: 10.1038/labinvest.2016.116. [DOI] [PubMed] [Google Scholar]
- 49.Sabroe I, Jones EC, Usher LR, et al. Toll-like receptor (TLR)2 and TLR4 in human peripheral blood granulocytes: a critical role for monocytes in leukocyte lipopolysaccharide responses. J Immunol. 2002;168:4701–4710. doi: 10.4049/jimmunol.168.9.4701. [DOI] [PubMed] [Google Scholar]
- 50.Sarmadi P, Tunali G, Esendagli-Yilmaz G, et al. CRAM-A indicates IFN-γ-associated inflammatory response in breast cancer. Mol Immunol. 2015;68:692–698. doi: 10.1016/j.molimm.2015.10.019. [DOI] [PubMed] [Google Scholar]
- 51.Hayashida K, Kume N, Minami M, Kita T. Lectin-like oxidized low-density lipoprotein receptor-1 (LOX-1) supports adhesion of leukocytes under both static and flow conditions. Circ J. 2002;66:432. [Google Scholar]
- 52.Kedde M, Strasser MJ, Boldajipour B, et al. RNA-binding protein Dnd1 inhibits MicroRNA access to target mRNA. Cell. 2007;131:1273–1286. doi: 10.1016/j.cell.2007.11.034. [DOI] [PubMed] [Google Scholar]
- 53.Ingersoll MA, Platt AM, Potteaux S, Randolph GJ. Monocyte trafficking in acute and chronic inflammation. Trends Immunol. 2011;32:470–477. doi: 10.1016/j.it.2011.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sha W, Mitoma H, Hanabuchi S, et al. Human NLRP3 inflammasome senses multiple types of bacterial RNAs. Proc Natl Acad Sci. 2014;111:16059–16064. doi: 10.1073/pnas.1412487111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pavón MA, Arroyo-Solera I, Téllez-Gabriel M, et al. Enhanced cell migration and apoptosis resistance may underlie the association between high SERPINE1 expression and poor outcome in head and neck carcinoma patients. Oncotarget. 2015;6:29016–29033. doi: 10.18632/oncotarget.5032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang Y, Venkatraj V, Fisher S, et al. Genomic cloning and chromosomal assignment of the E2F dimerization partner TFDP gene family. Genomics. 1997;39:95–98. doi: 10.1006/geno.1996.4473. [DOI] [PubMed] [Google Scholar]
- 57.Bostick M, Kim JK, Esteve P-O, et al. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science. 2007;317:1760–1765. doi: 10.1126/science.1147939. [DOI] [PubMed] [Google Scholar]
- 58.Li XB, Chen J, Deng MJ, et al. Zinc finger protein HZF1 promotes K562 cell proliferation by interacting with and inhibiting INCA1. Mol Med Rep. 2011;4:1131–1137. doi: 10.3892/mmr.2011.564. [DOI] [PubMed] [Google Scholar]
- 59.Teng T, Tsai JH, Puyang X, et al. Splicing modulators act at the branch point adenosine binding pocket defined by the PHF5A-SF3b complex. Nat Commun. 2017;8:1–16. doi: 10.1038/ncomms15522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Trimarchi JM, Fairchild B, Verona R, et al. E2F–6, a member of the E2F family that can behave as a transcriptional repressor. Proc Natl Acad Sci. 2002;95:2850–2855. doi: 10.1073/pnas.95.6.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nonaka K, Saio M, Suwa T, et al. Skewing the Th cell phenotype toward Th1 alters the maturation of tumor-infiltrating mononuclear phagocytes. J Leukoc Biol. 2008;84:679–688. doi: 10.1189/jlb.1107729. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.