Abstract
Non-keratinizing nasopharyngeal carcinoma (NPC) is a malignancy with a poor prognosis for relapsing patients and those with metastatic disease. Here, we identify a novel disease mechanism of NPC which may be its Achilles’ heel that makes it susceptible to immunotherapy. CD137 is a potent costimulatory receptor on activated T cells, and CD137 agonists strongly enhance anti-tumor immune responses. A negative feedback mechanism prevents overstimulation by transferring CD137 from T cells to CD137 ligand (CD137L)-expressing antigen presenting cells (APC) during cognate interaction, upon which the CD137-CD137L complex is internalized and degraded. We found ectopic expression of CD137 on 42 of 122 (34.4%) NPC cases, and that CD137 is induced by the Epstein-Barr virus latent membrane protein (LMP) 1. CD137 expression enables NPC to hijack the inbuilt negative feedback mechanism to downregulate the costimulatory CD137L on APC, facilitating its escape from immune surveillance. Further, the ectopically expressed CD137 signals into NPC cells via the p38-MAPK pathway, and induces the expression of IL-6, IL-8 and Laminin γ2. As much as ectopic CD137 expression may support the growth and spread of NPC, it may be a target for its immunotherapeutic elimination. Natural killer cells that express a CD137-specific chimeric antigen receptor induce death in CD137+ NPC cells, in vitro, and in vivo in a murine xenograft model. These data identify a novel immune escape mechanism of NPC, and lay the foundation for an urgently needed immunotherapeutic approach for NPC.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00262-022-03183-8.
Keywords: Nasopharyngeal carcinoma, Immune evasion, Trogocytosis, CD137, IL-6, IL-8
Introduction
Non-keratinizing nasopharyngeal carcinoma (NPC) is a common cancer in Southeast Asia with an annual incidence as high as 25–30 per 100,000, especially in ethnic groups from Southern China [1]. In 2018, 129,000 cases were diagnosed worldwide [2]. If NPC can be detected at stage I or II, treatment by a combined chemotherapy and radiation therapy results in > 80% 5-year survival rates. However, many patients are diagnosed with stage IV or V NPC, which have 50–60% 5-year survival rates. The situation is especially dire for the 20% of patients who develop metastatic disease as their median overall survival is only 12–15 months. In relapsed NPC, chemotherapy and radiation therapy are merely palliative with very low survival rates [3]. This resulted in 73,000 deaths from NPC [2, 4, 5].
NPC is an Epstein-Barr virus (EBV) driven malignancy that is typically associated with a rich lymphocytic infiltration, indicating a T cell inflamed phenotype and tumor immune evasion dominated by immunosuppressive tumor microenvironment. PD-L1 is frequently expressed in NPC, and promotes immune tolerance through suppression of T cell activation. However, anti-PD1 monoclonal antibody therapy display modest activity in NPC [6].
EBV uses several mechanisms to escape immune surveillance, e.g., it interferes with the MHC class I and class II antigen presentation pathways to avoid recognition and subsequent elimination by T cells. For example, the EBV proteins BGLF5 and BILF1 reduce surface levels of MHC class I which diminishes recognition by CD8+ T cells [7]. The latent EBV protein EBNA1 contains an internal glycine-alanine repeat domain (GAr) which decreases the rate of translation of EBNA1 mRNA and proteasomal degradation to peptides that could be displayed on MHC [7]. EBV-encoded microRNAs (miRNA) control gene expression of MHC class II and of several lysosomal enzymes that are important for in MHC class II peptide processing [8]. Further, EBV encodes vIL-10, a viral homolog of IL-10, that downregulates the costimulatory molecules CD86 and CD80 on EBV-infected monocytes [7]. Another immune evasion strategy that is employed by EBV in Hodgkin Lymphoma is the ectopic expression of CD137 (TNFRSF9, 4-1BB) [9].
The costimulatory molecule CD137 is a main driver for cellular immune responses. Crosslinking of CD137 strongly enhances proliferation, IFN-γ secretion and the cytolytic activity of T cells. CD137 costimulation enables the immune system to eliminate tumors as shown in a wide plethora of murine tumor models [10–12]. Antibodies against human CD137 are currently being evaluated for tumor immunotherapy in human patients [13]. Agonistic anti-CD137 antibodies also strongly enhance anti-virus immune responses [14–16]. The potent immune-stimulatory activity of CD137 became further evident by the inclusion of the cytoplasmic CD137 signaling domain into the second generation of CARs that greatly enhanced their efficacy in augmenting CAR-mediated T cell responses by reducing exhaustion and enhancing the persistence of CAR T cells [17, 18].
Since CD137 signaling is a main driver of cellular immune responses, and since cellular immune responses are responsible for the elimination of intracellular pathogens, it was quite unexpected to discover that EBV induces CD137 in infected NKT cells and Hodgkin Lymphoma [9, 19]. The main advantage EBV gains from inducing CD137 expression may be the ability to usurp a negative feedback mechanism for cellular immune responses which in turn facilitates its escape from immune surveillance. Negative feedback loops prevent an overshooting of immune reactions which otherwise could lead to tissue damage. A negative feedback mechanism is especially important for such a powerful costimulatory molecule as CD137. During cognate interactions when T cells get stimulated through CD137 by antigen presenting cells (APC), CD137 gets transferred by trogocytosis to the CD137L-expressing APC where it forms a complex with CD137L which then gets internalized [9]. This removes CD137 from the T cells preventing their further costimulation through CD137. It also removes CD137L from the APC, removing their ability to costimulate additional T cells [20–22].
Here, we show ectopic expression of CD137 in NPC, which is driven by LMP1, an EBV oncogenic protein expressed in type 2 latency states. NPC cells transfer CD137 to CD137L expressing APC where it leads to a downregulation of CD137L and a lower costimulatory capacity of APC. Further, CD137 signaling into NPC induces expression of genes that are associated with tumor progression and metastasis and poor patient prognosis. However, ectopic CD137 expression is also an Achilles heel for immunotherapy of NPC as it can be targeted by CARs.
Materials and methods
Cells
The nasopharyngeal carcinoma (NPC) cell lines C666, HK-1 and HONE-1 were acquired from Dr Paul Macary and Dr Zhang Yongliang (National University of Singapore). The DG-75 cell line was purchased from the German Collection of Microorganisms and Cell Cultures (DSMZ, Braunschweig, Germany). The cells were cultured in RPMI 1640 (Sigma, St. Louis, MO, USA) with 10% fetal bovine serum (Biowest, Kansas City, MO, USA) in a humidified chamber at 37 °C and 5% CO2. CD137-expressing stable cell lines were generated by lentiviral transduction (Ho et al., 2013). Cells were selected with 2–10 μg/ml blasticidin followed by Fluorescent Activated Cell Sorting on a SY3200 (Flow Cytometry Laboratory, LSI, Singapore) using PE-conjugated anti-CD137 (clone 4B4-1, BD Pharmingen, San Jose, USA).
Immunohistochemistry
Tissue arrays of 135 NPC cases were acquired from US Biomax (NPC1506, MD, USA). The tissues were deparaffinised in Clear-Rite (Thermo-Fisher Scientific, Waltham, USA) and hydrated with an alcohol gradient before epitope retrieval in Accel-EDTA antigen retrieval solution (GBI Labs, Bothwell, WA) for 20 min at 100 °C. The tissue was blocked with 5% non-fat milk for 30 min, Klear dual enzyme block (GBI Labs, Bothwell, WA) for 10 min and TNB blocking buffer for 30 min (Perkin Elmer, Waltham, USA). Mouse anti-CD137 primary antibody (clone BBK-2, Thermo-Fisher Scientific, Waltham, USA) or isotype (mouse IgG1 κ, clone MOPC-21, Sigma-Aldrich) was added followed by anti-mouse horseradish peroxidase (HRP) polymer (GBI Labs). Signal amplification was performed with biotin tyramide working solution and horseradish peroxidase-streptavidin (HRP-SA) (Perkin Elmer, Waltham, USA). Slides were developed with DAB (GBI labs) and counterstained with haematoxylin solution (Thermo-Fisher Scientific, Waltham, USA). Slides were imaged with a Leica DM2000 microscope.
Multiplex immunohistochemistry
Multiplex immunohistochemistry was performed on an NPC tissue array according to manufacturer’s protocol for opal multiplex immunofluorescent system (Perkin Elmer, Waltham, USA) and for PanPath (PanPath B.V., Amsterdam, The Netherlands). The tissues were probed with EBER oligo probe (PanPath B.V., Amsterdam, The Netherlands) followed by HRP-SA (Perkin Elmer). Primary antibodies anti-CD137 (clone BBK-2, Thermo-Fisher Scientific, Waltham, USA), rabbit anti-CD3 (Dako, Glostrup, Denmark) or mouse pan-cytokeratin (Abcam, Cambridge, UK) were used followed by anti-mouse or anti-rabbit HRP polymers, respectively (GBI Labs, Bothell, USA). The tissues were developed with 4-color Opal IHC kit (Perkin Elmer) and nuclei staining was performed with DAPI. Tissue sections were imaged on Vectra Imaging system, and analyzed with inForm (Perkin Elmer).
Transfection
NPC cell lines were transfected with LMP1 or GFP expression vectors generated using Axygen midiprep kit (Corning, NY, USA) according to Amaxa 4D-Nucleofector Basic Protocol for Primary Mammalian Epithelial Cells (Program #EL-110, Lonza, Basel, Switzerland). Cells were incubated in a humidified chamber at 37 °C, 5% CO2 for 48 h and analyzed by flow cytometry. The plasmid pVAX-GFP was obtained from Dr. Dario Campana, National University of Singapore) and p1990 SV40-LMP1 was obtained from Dr. Bill Sugden (Addgene plasmid # 26654).
CD137-directed trogocytosis of CD137L on DG-75
DG-75 cells were labeled with CellTrace Violet (Thermo-Fisher Scientific) and added to wells. Parental and CD137-expressing NPC cells were added to wells or 8.0 μm transwell inserts (Corning, NY, USA) at a 1:1 ratio, and incubated in a humidified chamber at 37 °C, 5% CO2 for 20 min. Cells were collected and incubated with FcR blocker (Miltenyi Biotec, Bergisch Gladbach, Germany) before labeling with primary antibodies against CD137L (PerCP efluor 701-conjugated, clone 4H3, eBioscience, San Jose, USA) or CD137 (PE-conjugated, clone 4B4-1, BD Pharmingen, San Jose, USA). Cells were analyzed on Fortessa Analyser, and data were interpreted using FlowJo.
Flow cytometry
Cells were labeled with PE-conjugated mouse primary antibodies for human CD137 (clone 4B4-1, BD Pharmingen, San Jose, USA) or with APC-conjugated anti-human CD137L antibodies (eBioscience, CA, USA) or mouse IgG1κ (clone MOPC-21, eBioscience, San Diego, USA) for 30 min in the dark at 4 °C. Cells were analyzed on a Fortessa Analyser (BD Biosciences) and data were interpreted using FlowJo (FlowJo, LLC, Ashland, USA). Recombinant Anti-LAMC2 antibody (clone EPR23654-127) was from Abcam (Cambridge, UK).
Stimulation of CD137 on NPC cell lines
Wells were coated with PBS (negative control) or 100 µl of 5 µg/ml rhCD137L for 3 h at 37˚C or overnight at 4˚C. The wells were then washed with sterile PBS and seeded with 5 × 104 or 105 C666-CD137 or HK-1-CD137 cells. Cells were incubated for 24 h for intracellular staining or 72 h for cytokine quantification. When indicated, VX-702 (SelleckChem) was used at a final concentration of 10 µM.
Enzyme linked immunosorbent assay (ELISA)
Levels of IL-6 and IL-8 in supernatants were measured using the respective Human ELISA kits (Invitrogen, San Diego, USA).
RNA extraction and sequencing
Cells were treated with immobilized rhCD137L or BSA (control) for 24 h, washed and harvested by trypsinization. Cells were then pelleted, snap-frozen with dry ice and stored at − 80 °C. RNA extraction was performed using the RNeasy RNA extraction kit (74104, QIAGEN). RNA was eluted using ddH2O and stored at − 80 °C before being sent for sequencing at NovogeneAIT.
RNA Sequencing Analysis: Raw RNA data, Fasta files were processed to bam files by STAR RNA-seq Aligner [23] mapped to human and EBV genome, grch38-ebv genome with indices created using samtools (1.11; Danecek) [23, 24]. Duplicates were marked using picard tools (2.23.8, Broad institute). Read summarization was conducted using subread::featurecounts (2.0.1; Yang L) packages [25]. Quality control was conducted using fastqc (0.11.9; Babraham Bioinformatics) and Multiqc (1.9; ewels). R (4.0.5; R Foundation for Statistical Computing, Vienna, Austria) was used for the differential gene expression analysis and gene set enrichment analysis using packages: Deseq2 (1.28.1) and fgsea (1.14.0), respectively.
Raw fastq files were mapped to the combined human GRCh38 and EBV genomes by STAR Aligner [23] Mapping parameters were as per ENCODE RNA-Seq parameters (https://github.com/ENCODE-DCC/long-rna-seq-pipeline/blob/master/DAC/STAR_RSEM.sh), with the exception of applying “—outFilterMultimapNmax 1”, to exclude any multimapping to both the human and EBV genomes. Duplicates were marked using picard tools (2.23.8, Broad institute). Gene expression features were quantified using the featureCounts function from Subread (v1.6.4) [25]. Quality control was performed using fastqc (0.11.9; Babraham Bioinformatics) and Multiqc (1.9; ewels).
Further downstream processing was performed in R (4.0.5; R Foundation for Statistical Computing, Vienna, Austria). Differential gene expression analysis was performed by DESeq2 (1.28.1) using it’s default parameters; [26], while gene set enrichment was performed with Fgsea (1.14.0). GO gene sets from downloaded from the Molecular Signatures Database v7.0 (Broad Institute) [27].
Sample preparation for quantitative proteomics analysis
Untreated and rhCD137L-treated NPC cells were prepared in biological triplicates and harvested by standard protocols as cell pellets. Cells were resuspended in lysis buffer containing 8 M urea in 50 mM HEPES, pH 8.0 and lysed using probe sonication with 5 s pulse and 5 s pause cycles over 45 s on ice. Ice-cold acetone was added at 4:1 lysate volume ratio, incubated at − 20 °C for 30 min, followed by centrifugation at 20,000×g at 4 °C for 5 min to precipitate proteins. The supernatant was removed and residual acetone was air-dried before the precipitated proteins were resuspended in 100 mM triethylammonium bicarbonate (TEAB), pH 8.5. Total protein content from each sample was measured using standard bicinchoninic acid (BCA) assay (Thermo Scientific, USA).
An aliquot of cell lysate containing 100 µg of protein was taken from each sample and added with a final concentration of 10 mM tris(2-carboxyethyl)phosphine (TCEP) and 55 mM 2-chloroacetamide (CAA) before incubation in the dark for 30 min for reduction and alkylation. Proteins were digested using LysC endopeptidase (1:50 ratio of LysC:protein) at 37 °C for 4 h and further digested with trypsin (1:100 ratio of trypsin:protein) at 37 °C overnight. Samples were acidified with a final concentration of 1% (v/v) trifluoroacetic acid (TFA) and centrifuged at 20,000g for 5 min to remove any precipitate. Peptides were desalted using Waters Oasis HLB cartridges with 30 mg sorbent weight (Waters, Milford, MA). Briefly, following activation with 500 µl of 100% acetonitrile and equilibration with 500 µl of 0.5% (v/v) acetic acid in water, samples were loaded onto the column and washed with 500 µl of 0.5% (v/v) acetic acid in water. Peptides were eluted with 500 µl of 65% acetonitrile in 0.5% (v/v) acetic acid. Peptide amount in each sample was quantified using quantitative fluorometric peptide assay (Thermo Scientific) according to manufacturer’s protocol before an aliquot containing 25 µg of peptides were taken from each sample and dried using centrifugal evaporator.
Dried peptides were resuspended in 25 µl of 100 mM TEAB, pH 8.5 and added with 10 µl of the respective isobaric tandem mass tags (TMT) (Thermo Scientific). Reaction mixtures were incubated at 25 °C overnight before they were quenched by transferring them a tube containing 50 µl of 1 M ammonium formate, pH 10. Quenched samples were pooled, desalted, and step-fractionated using spin columns containing C18 reversed-phase beads (Reprosil-Pur Basic, Dr Maisch GmbH, Germany). Briefly, the beads were activated using 350 µl of 100% acetonitrile followed by equilibration with 350 µl of 10 mM ammonium formate, pH 10. Sample was then completed loaded onto the spin column, washed with 350 µl of 10 mM ammonium formate, pH 10 twice, and eluted sequentially with step-fractionation buffers containing 14%, 18%, 21%, 24%, 27%, 32%, and 60% acetonitrile in 10 mM ammonium formate, pH 10. Eluted fractions were dried by centrifugal evaporation and washed twice using 60% acetonitrile in 0.1% (v/v) formic acid to remove residual ammonium formate before tandem mass spectrometry.
Tandem mass spectrometry and data analysis
Dried fractions were resuspended in 2% (v/v) acetonitrile solution containing 0.06% (v/v) TFA and 0.5% (v/v) acetic acid and loaded onto an autosampler plate. Online chromatography was performed using EASY-nLC 1200 (ThermoScientific) in single-column setup using 0.1% formic acid in water and 0.1% formic acid in 95% acetonitrile as mobile phases. Samples were injected and separated on the reversed-phase C18 analytical column (EASY-Spray, 75 µm inner diameter × 50 cm, 2 µm particle size) (ThermoScientific) maintained at 50 °C using a 3–33% (v/v) acetonitrile gradient over 45 min, then ramped to 60% over the next 15 min, and finally to 90% within 5 min. The final gradient was maintained for 5 min to elute all remaining peptides. Total run duration for each sample was 70 min at a constant flow rate of 300 nl/min.
Data were acquired using an Orbitrap Fusion Lumos mass spectrometer (ThermoScientific) in data-dependent acquisition (DDA) mode. Samples were ionized using 2100 V and 300 °C at the nanospray source and positively-charged precursor MS1 signals between a mass-to-charge ratio range of 350–1550 m/z were detected using an Orbitrap analyzer set to 60,000 resolution, automatic gain control (AGC) target of 400,000 ions, and maximum injection time (IT) of 100 ms. Precursors with charges between + 2 to + 7 within a mass tolerance of ± 10 ppm were further fragmented using higher-energy collisional dissociation (HCD) at 42% normalized collision energy and their MS2 signals were analyzed by Orbitrap at a resolution of 50,000, an AGC target of 75,000, maximum IT of 86 ms, and fixed first mass scan starting from 110 m/z. Precursors used for MS2 scans were excluded for 70 s in order to avoid re-sampling of high abundance peptides. The MS1–MS2 cycles were repeated every 3 s until completion of the run.
Acquired data were analyzed using Proteome Discoverer (v2.4) (ThermoScientific). Raw mass spectra from all fractions were combined and searched against human primary protein sequences from SwissProt (retrieved 11 June 2019). Carbamidomethylation on Cys was set as the fixed modification and deamidation of asparagine and glutamine, acetylation on protein N-terminus and oxidation of Met methionine were set as dynamic modifications for the search. Trypsin/P was set as the digestion enzyme and was allowed up to three missed cleavage sites. Precursors and fragments were accepted if they had a mass error within 10 ppm and 0.06 Da, respectively. Reporter ions were quantitated using the TMT6 method. Peptides were matched to spectra at a false discovery rate (FDR) of 1% (strict) and 5% (relaxed) against the decoy database. The data were exported for differential analysis with a R-based script that was built upon the limma package from Bioconductor. Protein with differential expression were identified by comparing the treated samples to the negative control (PBS) with the cutoffs of log2 fold-change of 0.5 and adjusted p values of < 0.05 as significant hits.
Generation of anti-CD137 CAR-NK cells
KHYG-1 cells were kindly provided by Dr. Jeak Ling Ding (National University of Singapore). Anti-CD137 antibodies were isolated by phage display technology [28]. The anti-CD137-CAR gene was constructed by combining the anti-CD137 single chain fragment, the CD8 hinge and transmembrane domain, CD137 and CD3ζ intracellular domain. The anti-CD137-CAR gene was cloned into pLenti6-V5 D/TOPO vector and used to generate a 3rd generation lentiviruses together with pRSV-Rev, pMDLg/pRRE, and pMD2.G plasmids (Addgene). Anti-CD137-CAR NK cells were obtained by lentiviral spin-inoculation and sorting by flow cytometry.
Cell lysis assay
5 × 103 NPC cells were seeded on 96-well plate overnight before being co-cultured with KHYG-1 cells at an E:T = 10:1 ratio for 24 h. The supernatants were collected for the measurement of LDH activities by CyQUANT™ LDH Cytotoxicity Assay (Thermo Fisher Scientific). The percentage of cell lysis was calculated according to manufacturer’s protocol.
Xenograft studies
2 × 106 HK-1 and HK-1-CD137 cells were injected in the left and right flanks of NSG mice, respectively. When tumors reached a size of 100 mm3, the mice were divided into 2 groups, and injected intravenously with 4 × 106 control NK cells or anti‐CD137‐CAR‐NK cells. Tumor size and body weight of mice were monitored. When tumors reached a size of 2 cm3 for a single solid tumor or 3 cm3 for multiple tumors, mice were euthanized. These experiments were approved by the NUS IACUC under protocol 2020-00764.
Statistical analysis
Statistical significance was determined by Student t-test unless indicated otherwise. A p value of < 0.05 was considered as significant.
Results
Ectopic expression of CD137 in NPC
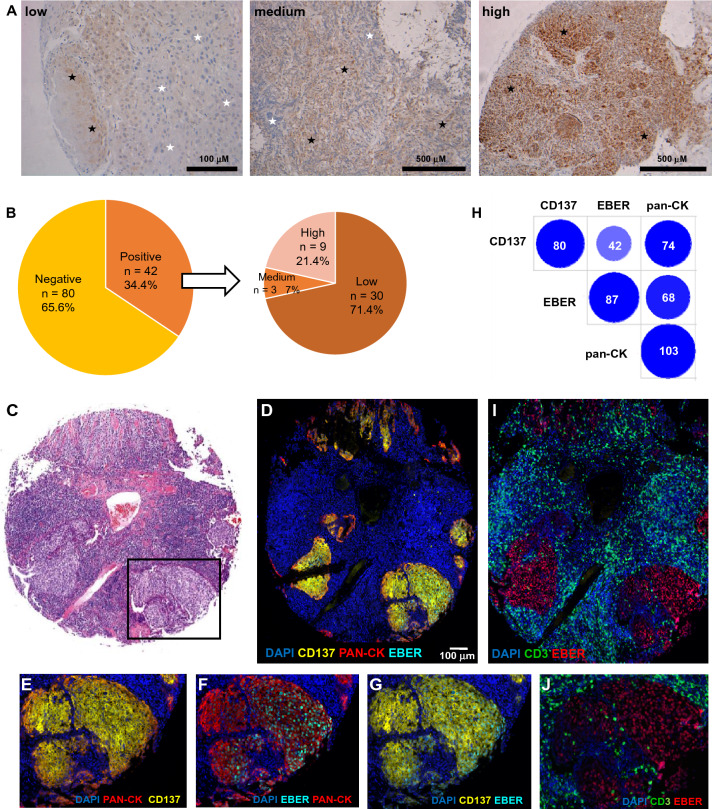
CD137 expression could be detected in NPC with areas of staining being heterogeneous in pattern and intensity, and intermixed with CD137-negative tissue (Fig. 1A). Of the 135 NPC cases on the tissue microarray, 122 cases were usable after the staining procedure and were investigated. CD137 expression was detected in 42 (34%) of them. Of the 42 CD137+ NPC cases, 30 (71.5%), 3 (7%) and 9 (21.5%) had low, medium and high expression, respectively (Fig. 1B).
Fig. 1.
Expression of CD137 in NPC. A Immunohistochemical staining for CD137 (brown) in NPC. Shown are representative cases for low, medium and high CD137 expression. Black and white stars indicate CD137‐positive and CD137‐negative areas on the images, respectively. B Pie chart illustrating the percentage and intensities of CD137 staining in NPC tissue array. CD137 positive cases were further categorized according to the intensity of CD137 staining. Actual number of cases are indicated in parentheses. C H&E staining by US Biomax. The area in the black box is shown enlarged in E, F, G and J. D–G and J–I Multiplex immunofluorescence staining with colors as indicated. H Graphic depiction of correlation of tissue cores with CD137, EBER and cytokeratin expression (≥ 5 cells). Intensity of color and size of circles represent the number of positive NPC cases, which are also stated within the circles
NPC is characterized by an extensive tumor stroma with an abundant presence of leukocytes, of which T cells and NK cells express CD137 upon activation. Further, vascular endothelial cells in tumors can also express CD137 [29]. Therefore, we employed multiplex immunofluorescence staining to unequivocally identify the CD137-expressing cells. Cytokeratin and CD3 were used as marker for epithelial cells and T cells, respectively, and staining for EBV-encoded small RNA (EBER) was employed to label EBV-infected cells.
Tumor cell areas were well distinguishable on the HE stain of a squamous cell carcinoma by the clear tumor/stroma boundary and the significantly lower rate of leukocyte infiltration (Fig. 1C). Cytokeratin (Pan-CK), CD137 and EBER staining coincided with these areas (Fig. 1D). Cytokeratin and CD137 co-localize, demonstrating CD137 expression on epithelial cells (Fig. 1E). EBER staining co-localized with cytokeratin (Fig. 1F) and with CD137 (Fig. 1G) proving that it is the malignant EBV-infected NPC cells that express CD137. Pan-CK, CD137 double-positive cells were found in 74 of 116 (63.8%) NPC cases. A similar number, 68 of 116 (58.6%), was found for Pan-CK, EBER double positive cells. CD137, EBER double positive cells could be detected in 42 of 116 (36.2%) of NPC cases (Fig. 1H).
T cells were present in 75/122 (61.5%) of cases, and often there was an abundance of T cells around the tumor area. However, very few T cells were present within the tumor (Fig. 1I, J). CD3 and CD137 staining occupied different tissue areas, i.e., tumor and peritumoral tissue, respectively, demonstrating that the vast majority of these T cells do not express CD137. EBER and CD3 do not colocalize, demonstrating that T cells are not EBV-infected (Fig. 1I, J).
LMP1 induces CD137 expression on NPC, which inhibits immune activity
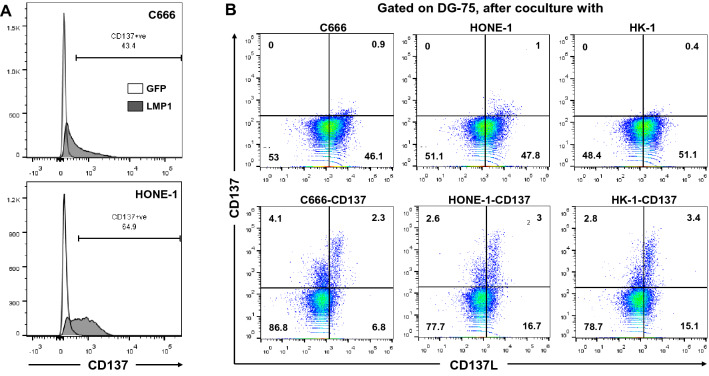
CD137 is induced by LMP1 in NKT cell lymphoma, Hodgkin Lymphoma (HL) and healthy T cells [19, 20, 30]. Since NPC is strongly associated with EBV, we speculated that LMP1 is also the factor that induces ectopic CD137 expression in NPC. Transfection of a LMP1 expression vector, induced CD137 expression in the two NPC cell lines C666 and HONE-1 (Fig. 2A) whereas transfection of GFP, as a negative control, did not induce CD137 expression.
Fig. 2.
LMP1-induced ectopic CD137 suppresses immune activity. A C666 and HONE-1 cells were transfected with an expression vector for GFP or LMP1, and expression of CD137 was quantified by flow cytometry 48 h later. B CD137-expressing NPC downregulate CD137L on APC. Control and CD137-expressing HK-1, HONE-1 and C666 cells were cocultured with CellTrace Violet-labeled DG-75 cells for 20 min. Cells were stained for CD137 and CD137L and analyzed by flow cytometry. Gating was performed on CellTrace Violet+ singlets. Numbers indicate percentages positive cells. Data are representative of at least two independent experiments
In order to address functional effects of ectopic CD137 expression, we established stable CD137-expressing lines for the NPC cell lines C666, HONE-1 and HK-1 (Supplementary Fig. 1). Similarly as HRS cells [9] and as in LMP1-transfected cells [20], NPC cells transferred CD137 to co-cultured CD137L-expressing APC, and led to a downregulation of CD137L on the APC within 20 min (Fig. 2B). Identical data have been obtained at the 1 and 2 h time points (not shown).
CD137 signal transduction in NPC cells
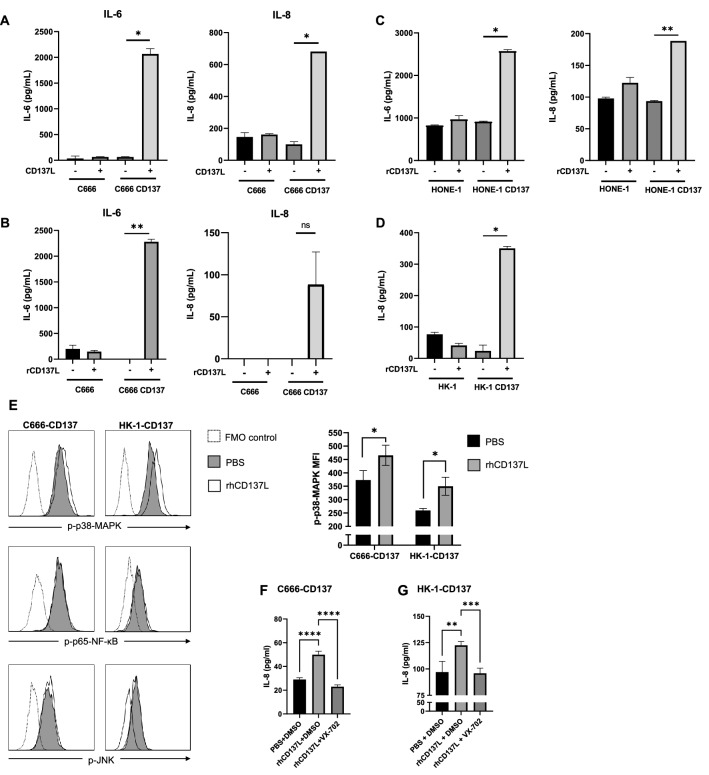
CD137 transmits signals not only into the cells, it is naturally expressed on, such as T cells, NK cells and endothelial cells but also into cells, where it is ectopically expressed, such as HRS cells [31]. Therefore, we investigated whether engagement of CD137 on NPC cells influences cellular functions. Treatment of control and CD137-expressing C666 cells with control and CD137L-expressing HEK293 cells demonstrated that CD137–CD137L interaction induces the secretion of IL-6 and IL-8 (Fig. 3A). We ruled out the confounding possibility of cytokine secretion by the HEK293 cells by replacing them with recombinant CD137L. Levels of IL-8 were negligible or below detection limit in the control conditions, and only CD137-expressing C666 cells released IL-6 and IL-8 upon activation by rCD137L (Fig. 3B). Identical data were obtained with HONE-1 cells for IL-6 and IL-8 (Fig. 3C), and with HK-1 cells for IL-8 (Fig. 3D).
Fig. 3.
CD137 signal transduction into NPC cells. A 105 C666 or C666-CD137 cells were cocultured at a 1:1 ratio with control or CD137L-expressing HEK-293T cells (CD137L). B–D 104 control or CD137-expressing C666 (B), HONE-1 (C) or HK-1 (D) cells were seeded on BSA or rCD137L coated plates. After 3 days, cytokine concentrations in the supernatant were measured by ELISA. *p < 0.05; **p < 0.01, ns, non-significant, by two-tailed paired t-test ND: non-detectable. The data are representative of at least two independent experiments with comparable results. E Wells were coated with 5 µg/ml rhCD137L or PBS before 105 C666-CD137 or HK-1-CD137 cells were added for 24 h, and analyzed assessed by flow cytometry. The histogram shows the signal gated on live single cells (F). Wells were coated with 5 µg/ml rhCD137L or PBS before 105 C666-CD137 or HK-1-CD137 cells were seeded and incubated for 72 h with either 10 mM VX-702 or an equal volume of DMSO control. IL-8 was measured in the supernatant. Unpaired T test (F) and one-way ANOVA followed by multiple comparison test (F–G) were used for statistical analysis. At least three replicates are performed. *p < 0.05, ****p < 0.0001
We assessed the phosphorylation of the transcription factors NF-κB p65, JNK, and p38-MAPK to identify the pathways triggered by CD137 activation in NPC. Engagement of CD137 on C666-CD137 and HK-1-CD137 cells by rhCD137L, increased the phosphorylation of p38-MAPK, while no significant changes of the phosphorylation status of NF-κB p65 and JNK could be detected (Fig. 3E). Inhibition of the p38-MAPK pathway with the p38-MAPK-specific inhibitor, VX-702, reversed the induction of IL-8 in these NPC cells, confirming the pivotal role of p38-MAPK in the CD137-induced activation of NPC cells (Fig. 3F).
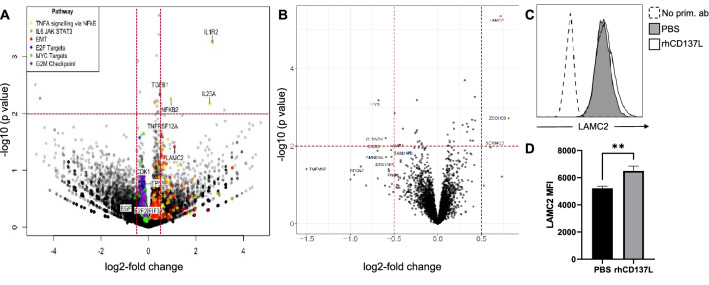
In order to identify additional genes that are regulated by CD137 signaling, we performed RNA sequencing and mass spectrometry analyses. C666-CD137, HK-1-CD137 and HONE-1-CD137 cells were treated with BSA (negative control) or rhCD137L for 24 h and sent for RNA-Seq. Differentially expressed genes common to all 3 cell lines were identified by comparing rhCD137L-treated cells with controls. Consistent with the role of CD137 as a driver of immune response, enrichment of genes associated with TNF signaling and the IL6/JAK/STAT3 pathway was observed, as well as upregulation of NFKB2, a key transcription factor of non-canonical NF-κB signaling. Expression of IL-1R2 and IL-23a was several times increased relative to control which was statistically highly significant (Fig. 4A). NF-kB2, TGFB1 and TNFRSF12A were also increased, although to more varying degrees in the three different NPC cell lines (Suppl. Figure 2 and Suppl. Table 1). These gene expression changes appeared to be most prominent in the EBV-positive C666 cell line, while the EBV-negative HK-1 and HONE-1 cell lines showed less prominent changes (Suppl. Figure 2). Genes associated with EMT, including TGFB1 and Laminin γ2 (LAMC2) were also observed to be upregulated.
Fig. 4.
A CD137-expressing C666, HONE-1 and HK-1 cells were seeded on BSA or rCD137L-coated plates for 24 h. RNA was extracted and sequenced. Depicted are differentially regulated genes vs statistical significance, colored by pathway association. B C666-CD137 cells were seeded for 24 h onto wells coated with either rhCD137L or BSA as a negative control. Protein was extracted and sequenced by mass spectroscopy. Depicted are differentially regulated proteins vs statistical significance. C, D CD137 stimulation enhances LAMC2 expression. 105 C666-CD137 cells were seeded onto wells coated with either rhCD137L or PBS control, and incubated for 24 h. C Cells were then stained intracellularly with primary anti-LAMC2 antibody and fluorochrome-conjugated secondary antibody. Live single cells are selected for expression analysis, n = 3. D Mean ± SD of LAMC2 MFI, **p < 0.01, unpaired Student’s t test
For C666-CD137 cells, we also analyzed global changes at the protein level in response to CD137 engagement. Cell lysates of C666-CD137 cells that had been treated with BSA (negative control) or rhCD137L were sequenced by mass spectrometry. Similar as at the mRNA level, we found a low induction of Laminin γ2 (Fig. 4B) which was further confirmed by flow cytometry (Fig. 4C).
Anti-CD137 CAR-NK cells specifically kill CD137-expressing NPC cells
The ectopic expression of CD137 could provide NPC with growth and selection advantages. However, it may make NPC susceptible to immunotherapy by targeting CD137. For proof of concept, we generated an anti-CD137 CAR that targets CD137 cell surface expression, and armed it on the NK cell line KHYG-1 (Suppl. Figure 3).
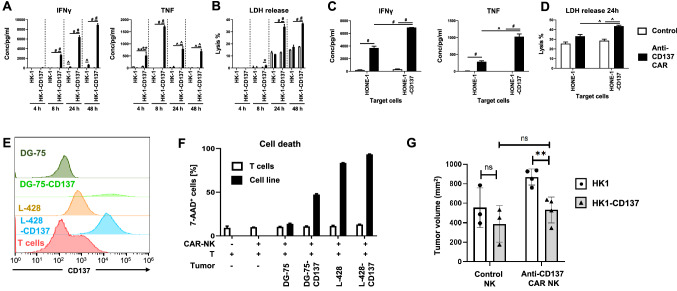
When anti-CD137 CAR-NK cells came in contact with CD137-expressing HK-1 cells, they became activated as evidenced by the continuous increase in IFNγ secretion from 4 to 48 h, and the increase in TNF secretion which peaked at 8 h. However, control NK cells without the anti-CD137 CAR were not activated and neither were the anti-CD137 CAR-NK cells by CD137-negative HK-1 cells (Fig. 5A). Activation of the anti-CD137 CAR-NK cells enabled them to specifically and time-dependently kill CD137-expressing HK-1 cells (Fig. 5B). Similar data were obtained with HONE-1 cells for IFNγ and TNF release (Fig. 5C) and cytotoxicity (Fig. 5D).
Fig. 5.
Anti-CD137-CAR NK cells are specifically activated by and kill CD137+ tumor cells. HK-1 cells were cocultured with control of anti-CD137-CAR-expressing NK cells at a T:E = 1:10 for indicated period of time. The supernatants were collected for the measurement of A IFNγ and TNF secretion or B the release of LDH. Another NPC cell line HONE-1 cells were cocultured with control of anti-CD137-CAR-expressing NK cells at an T:E = 1:10 for 24 h. The supernatants were collected for the measurement of C IFNγ and TNF secretion or D the release of LDH. E Activated T cells were isolated from anti-CD3 and anti-CD28-activated PBMCs and stained for CD137 expression along with DG-75, DG-75-CD137, L-428 and L428-CD137 cells. F Activated T cells were labeled by CellTrace Violet and cocultured with the other indicated target cells that were labeled by CFSE, in the presence of anti-CD137-CAR NK cells at a 1:5 ratio for 24 h. The degree of cell killing was measured by 7-AAD staining in respective gatings. Results are representative of at least 2 independent experiments. *p < 0.05, **p < 0.01, ^p < 0.001, #p < 0.0001 (unpaired two-tailed t test). G In vivo xenograft studies. Depicted are means ± SD of tumor volume of mice at day 17 after tumor implantation. Data are representative from three independent experiments with 3–5 biological replicates per condition per experiment. ns not significant, **p < 0.01, as determined by Student’s t-test
Since activated T cells also express CD137, we investigated whether anti-CD137 CAR-NK cells would induce death in effector T cells, and so reduce an anti-tumor T cell response. Therefore, we co-cultured CAR-NK cells with (1) peripheral T cells that were activated by anti-CD3 and anti-CD28 or (2) with a B cell line (DG-75, WT and transfected with CD137), or (3) a HL cell line (L-428, WT and transfected with CD137) to simulate strongly CD137-expressing tumors, and to evaluate whether preferentially effector T cells or CD137-expressing tumor cells would be killed by the CAR-NK cells (Fig. 5E). Cell lysis was induced in the CD137-expressing DG-75 and L-428 cells (CD137-moderate and CD137-high) but not in the control DG75 or in the moderately CD137-positive T cells (Fig. 5F), indicating that the comparatively low CD137 levels on T cells do not elicit killing by the anti-CD137 CAR-NK cells.
Based on these encouraging in vitro results, we tested the ability of the anti-CD137 CAR-NK cells to limit the growth of NPC in vivo in a murine xenograft. Control HK-1 and HK-1-CD137 cells were transferred to the left and right flanks of NSG mice, and once tumors had grown to a size of 100 mm3, mice were injected with either NK or anti-CD137 CAR NK cells. Treatment with anti-CD137 CAR NK cells resulted in a significantly smaller tumor size (863.6 vs 531.5 mm3), (Fig. 5G).
Discussion
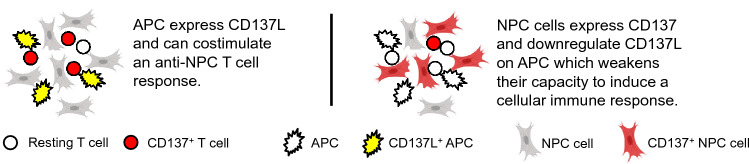
This study shows that in a significant subset of NPC, EBV through LMP-1, induces expression of CD137. Ectopic CD137 expression usurps a physiological negative feedback mechanism to quench T cell activity that helps EBV and NPC to evade immune surveillance. In addition, CD137 signals into NPC cells which promotes cancer progression and spread.
During an immune response, this negative feedback mechanism is very limited in scale since CD137 is expressed only transiently on antigen-activated T cells [32, 33], while the more strongly and permanently expressed ectopic CD137 on NPC cells is able to downregulate CD137L on APC more profoundly and for a prolonged time (Fig. 6). In this respect, the CD137-expressing NPC cells imitate regulatory T cells (Treg), since Treg can express CD137 at very high levels, and CD137 marks the most potent Treg, and is important for their suppressive activity [34, 35]. Treg use the negative CD137 feedback system to downregulate the immunostimulatory CD137L [22].
Fig. 6.
Schematic depiction of the effects of CD137 expression on NPC
There was an abundance of T cells in the NPC cores but the vast majority of T cells had not infiltrated the malignant tissue but were found around the tumor. Also, most of these T cells did not express CD137, indicating that effector T cells were not activated and were not participating in an anti-NPC T cell response. The absence of CD137 on T cells means also that CD137-expressing Treg may not play a major role in the tumor microenvironment of NPC. Since the CD137-expressing NPC cells already achieve CD137L downregulation through their own CD137 expression, CD137+ Treg may not be required for maintaining an immunosuppressive microenvironment in NPC. It would be interesting to investigate whether there is a correlation between CD137 levels in tumor tissues and prognosis of NPC patients. Unfortunately, we were not able to conduct such an analysis since we used a commercial tissue microarray which does not provide patient data.
This study shows that the ectopic expression of CD137 is not restricted to HL and NKTCL but also occurs in NPC. All these cancers are associated with EBV, and EBV induces CD137 via LMP1. This consistent downregulation of CD137L by EBV indicates the importance of CD137 for an effective anti-EBV immune response. Indeed, patients who lack CD137 develop EBV-related pathologies [36]. It seems that intracellular pathogens do not induce a large number of molecules for inhibiting a cellular immune response of which CD137 is just one of many. Saito et al., 2004 and Pichler et al., 2008 did a wider analysis and found that it is specifically CD137 that is induced by infection with M. bovis BCG and HTLV-1 [37, 38]. The explanation may be, that it is the CD137-CD137L interaction that most potently drives a cellular immune response, and hence its disruption is most important for viruses and cancers [39]. Of note, in a subgroup of rhabdomyosarcoma, which is not associated with viral infection, malignant cells expressing CD137 can be found, and there the inducer for CD137 is TNF [40].
CD137 stimulation has been reported to work through the NF-κB, JNK and p38-MAPK pathways [41–43]. Thus, we investigated the activation of transcription factors specific for each of these three pathways. Because NPC cells are in a permanent activation state to avoid programmed cell death, it is not surprising to see that many of these pathways are already constitutively activated in NPC cells. Since almost every C666-CD137 and HK-1-CD137 cell contained phosphorylated NF-κB, JNK, and p38-MAPK, even before CD137 stimulation, it was difficult to observe an increase. Nevertheless, CD137 engagement consistently enhanced the level of phosphorylated p38-MAPK in the two NPC cell lines, C666 and HK-1. Conversely, blocking the p38-MAPK pathway, reversed the CD137-mediated upregulation of IL-8.
The cytokines that are induced by CD137 signaling in NPC cells are known to contribute to NPC pathogenesis. IL-6 is expressed in clinical NPC specimens, and IL-6 concentrations are elevated in sera of NPC patients, and correlate with advanced disease and poor prognosis of NPC patients. Interestingly, IL-6 receptor too is overexpressed in NPC tissues which implies an auto- or paracrine stimulation that drives pathogenesis. Indeed, increased IL-6 receptor expression enhanced IL-6-induced growth and tumorigenicity of NPC cell lines in vivo in xenografts [44, 45]. Exogenous IL-6 enhanced proliferation, migration and invasion of NPC cell lines which was at least in part mediated by the IL-6-induced expression of MMP-2 and MMP-9 [46]. Intratumoral IL-8 levels in NPC correlate with poor prognosis and shorter overall survival of patients. IL-8 was shown to promote epithelial–mesenchymal transition and migration, invasion and metastasis of NPC cell lines in murine xenografts [47]. IL-8, alone [48] and in combination with MIF [49] or HGF [50], correlates significantly with angiogenesis in NPC. The expression of IL-8 receptor alpha in NPC also correlates significantly with a shorter overall survival of patients [51] enabling an auto- or paracrine loop, similarly as for IL-6.
There was a rather poor correlation of CD137 signaling-regulated genes identified by RNA sequencing and by mass spectrometry. Part of the reason was likely posttranscriptional control. However, Laminin γ2 was consistently upregulated in both experimental approaches, and was independently confirmed by flow cytometry. Laminins are major components of the basal lamina, which regulate adhesion, migration, differentiation and survival of healthy and malignant cells. Laminin γ2 is overexpressed by many solid cancers including in NPC [52]. It increases metastasis and EMT, and is associated with a poor prognosis. Laminin γ2 expression correlates with genes associated with the ATP-binding cassette (ABC) transporters, and it facilitates resistance to chemotherapy [53, 54].
The last decade has witnessed enormous progress in the treatment of some hematological malignancies, most notably of CD19+ B cell leukemia or Hodgkin lymphoma using novel immunotherapeutic approaches such as CARs or immune checkpoint inhibitors, respectively (Page et al., 2014; Salter et al., 2018). Unfortunately, these treatments exert limited therapeutic effect in most solid cancers. Among the malignancies for which more effective treatments are much required, is NPC.
The ectopic CD137 expression by NPC may be an Achilles’ heel that can be exploited for therapy, either by an antibody or a CAR-based approach. Indeed, anti-CD137 CAR-transfected NK cells killed CD137-expressing NPC cells specifically in vitro, and resulted in a reduced tumor mass of human xenografts in mice. Susceptibility to NK cell killing is not only dependent on levels of target expression but on several other factors as well, which likely accounts for the absence of a strict correlation of killing rates with CD137 levels on the cell lines and Treg. E.g. the level of CD137 on WT L-428 cells may already be saturating to induce effective killing, so that the higher CD137 levels on L-428-CD137 cells do not increase killing much further.
Killing CD137-expressing cells may also deplete activated T cells, NK cells and vascular endothelial cells, as these cells may also express CD137 upon activation. The transient depletion of activated T cells and NK cells may be acceptable for the benefit of a significant reduction in tumor mass and/or progression. It may even be advantageous for the anti-tumor immune response since it can lead to the elimination of Treg cells [55]. Indeed, ADCC-inducing anti-CD137 antibodies depleted Treg while having no noticeable effect on effector T cells [56]. The higher expression levels of CD137 on tumor cells than on T cells certainly contributed to the preferential killing of the tumor cells in Fig. 5F. In addition, different cell types have different levels of resistance against killing, mediated among others, by different levels of pro- and antiapoptotic proteins. Also, since the therapeutic effect is opposite to the potential side effect, we cannot discern whether [1] there was no side effect, or (2) whether the side effect reduced the actually larger therapeutic effect. The killing of CD137-expressing vascular endothelial cells may also be beneficial since CD137 is expressed by blood vessels at sites of inflammation, particularly in tumors [29], and could help in starving the tumor by reducing its vascularization. Adverse effects, however, would be expected if the patient would suffer from non-tumor related inflammation.
This study identifies a novel pathogenesis mechanism of NPC which is based on EBV-induced CD137 expression, and that helps NPC to evade immune surveillance and to gain growth advantages. However, this ectopic CD137 expression may serve as a target, and opens up the possibility of developing a novel immunotherapeutic treatment for NPC.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
This research was supported by a grant (NMRC/BnB/018b/2015) from the National Medical Research Council (NMRC), Singapore to H.S. and by A*STAR core funding and Singapore NRF under its NRF-SIS “SingMass” scheme to R.M.S. J.K.T. is supported by the Transition Award (TA20nov-0025) from NMRC, and K.S.L is supported by the Individual Research Grant (CIRG18nov-0045) from NMRC. We thank the LSI core facility under the leadership of Dr. Paul Hutchinson for excellent help with flow cytometry.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Mukul Prasad and Sashigala Ponnalagu have contributed equally to this work.
References
- 1.Smith C, Khanna R. A new approach for cellular immunotherapy of nasopharyngeal carcinoma. Oncoimmunology. 2012;1(8):1440–1442. doi: 10.4161/onci.21286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 3.Ma BB, Chan AT. Systemic treatment strategies and therapeutic monitoring for advanced nasopharyngeal carcinoma. Expert Rev Anticancer Ther. 2006;6(3):383–394. doi: 10.1586/14737140.6.3.383. [DOI] [PubMed] [Google Scholar]
- 4.Petersson F. Nasopharyngeal carcinoma: a review. Semin Diagn Pathol. 2015;32(1):54–73. doi: 10.1053/j.semdp.2015.02.021. [DOI] [PubMed] [Google Scholar]
- 5.Lee AW, Ma BB, Ng WT, Chan AT. Management of nasopharyngeal carcinoma: current practice and future perspective. J Clin Oncol: Off J Am Soc Clin Oncol. 2015;33(29):3356–3364. doi: 10.1200/JCO.2015.60.9347. [DOI] [PubMed] [Google Scholar]
- 6.Outh-Gauer S, Alt M, Le Tourneau C, Augustin J, Broudin C, Gasne C, et al. Immunotherapy in head and neck cancers: a new challenge for immunologists, pathologists and clinicians. Cancer Treat Rev. 2018;65:54–64. doi: 10.1016/j.ctrv.2018.02.008. [DOI] [PubMed] [Google Scholar]
- 7.Ressing ME, Horst D, Griffin BD, Tellam J, Zuo J, Khanna R, et al. Epstein-Barr virus evasion of CD8(+) and CD4(+) T cell immunity via concerted actions of multiple gene products. Semin Cancer Biol. 2008;18(6):397–408. doi: 10.1016/j.semcancer.2008.10.008. [DOI] [PubMed] [Google Scholar]
- 8.Tagawa T, Albanese M, Bouvet M, Moosmann A, Mautner J, Heissmeyer V, et al. Epstein-Barr viral miRNAs inhibit antiviral CD4+ T cell responses targeting IL-12 and peptide processing. J Exp Med. 2016;213(10):2065–2080. doi: 10.1084/jem.20160248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ho WT, Pang WL, Chong SM, Castella A, Al-Salam S, Tan TE, et al. Expression of CD137 on Hodgkin and Reed-Sternberg cells inhibits T-cell activation by eliminating CD137 ligand expression. Can Res. 2013;73(2):652–661. doi: 10.1158/0008-5472.CAN-12-3849. [DOI] [PubMed] [Google Scholar]
- 10.Thum E, Shao Z, Schwarz H. CD137, implications in immunity and potential for therapy. Front Biosci. 2009;14:4173–4188. doi: 10.2741/3521. [DOI] [PubMed] [Google Scholar]
- 11.Lee SW, Croft M. 4-1BB as a therapeutic target for human disease. In: Grewal IS, editor. Therapeutic targets of the TNF superfamily. New York: Springer; 2009. pp. 120–129. [DOI] [PubMed] [Google Scholar]
- 12.Wang S, Chen L. Immunobiology of cancer therapies targeting CD137 and B7-H1/PD-1 cosignal pathways. Curr Top Microbiol Immunol. 2011;344:245–267. doi: 10.1007/82_2010_81. [DOI] [PubMed] [Google Scholar]
- 13.Sanmamed MF, Etxeberria I, Otano I, Melero I. Twists and turns to translating 4-1BB cancer immunotherapy. Sci Transl Med. 2019;11(496):eaax4738. doi: 10.1126/scitranslmed.aax4738. [DOI] [PubMed] [Google Scholar]
- 14.Mbanwi AN, Watts TH. Costimulatory TNFR family members in control of viral infection: outstanding questions. Sem Immunol. 2014;26(3):210–219. doi: 10.1016/j.smim.2014.05.001. [DOI] [PubMed] [Google Scholar]
- 15.Lee S, Mittler RS, Moore ML. Targeting CD137 enhances vaccine-elicited anti-respiratory syncytial virus CD8+ T cell responses in aged mice. J Immunol. 2014;192(1):293–299. doi: 10.4049/jimmunol.1300453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang R, Freywald A, Chen Y, Xu J, Tan X, Xiang J. Transgenic 4-1BBL-engineered vaccine stimulates potent Gag-specific therapeutic and long-term immunity via increased priming of CD44(+)CD62L(high) IL-7R(+) CTLs with up- and downregulation of anti- and pro-apoptosis genes. Cell Mol Immunol. 2015;12(4):456–465. doi: 10.1038/cmi.2014.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Campana D, Schwarz H, Imai C. 4-1BB chimeric antigen receptors. Cancer J. 2014;20(2):134–140. doi: 10.1097/PPO.0000000000000028. [DOI] [PubMed] [Google Scholar]
- 18.Long AH, Haso WM, Shern JF, Wanhainen KM, Murgai M, Ingaramo M, et al. 4-1BB costimulation ameliorates T cell exhaustion induced by tonic signaling of chimeric antigen receptors. Nat Med. 2015;21(6):581–590. doi: 10.1038/nm.3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoshimori M, Imadome K, Komatsu H, Wang L, Saitoh Y, Yamaoka S, et al. CD137 expression is induced by Epstein-Barr virus infection through LMP1 in T or NK cells and mediates survival promoting signals. PLoS ONE. 2014;9(11):e112564. doi: 10.1371/journal.pone.0112564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu M, Wong HY, Lin JL, Moliner A, Schwarz H. Induction of CD137 expression by viral genes reduces T cell costimulation. J Cell Physiol. 2019;234(11):21076–21088. doi: 10.1002/jcp.28710. [DOI] [PubMed] [Google Scholar]
- 21.Shao Z, Harfuddin Z, Pang WL, Nickles E, Koh LK, Schwarz H. Trogocytic CD137 transfer causes an internalization of CD137 ligand on murine APCs leading to reduced T cell costimulation. J Leukoc Biol. 2015;97(5):909–919. doi: 10.1189/jlb.3A0213-079RRR. [DOI] [PubMed] [Google Scholar]
- 22.Luu K, Patwardhan MV, Zeng Q, Wickstrom SL, Lundqvist A, Schwarz H. Regulatory T cells inhibit T cell activity by downregulating CD137 ligand via CD137 trogocytosis. Cells. 2021;10(2):353. doi: 10.3390/cells10020353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liao Y, Smyth GK, Shi W. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 2013;41(10):e108. doi: 10.1093/nar/gkt214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liberzon A, Birger C, Thorvaldsdottir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1(6):417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rajendran S, Li Y, Ngoh E, Wong HY, Cheng MS, Wang CI, et al. Development of a bispecific antibody targeting CD30 and CD137 on Hodgkin and Reed-Sternberg cells. Front Oncol. 2019;9:945. doi: 10.3389/fonc.2019.00945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Broll K, Richter G, Pauly S, Hofstaedter F, Schwarz H. CD137 expression in tumor vessel walls. High correlation with malignant tumors. Am J Clin Pathol. 2001;115(4):543–549. doi: 10.1309/E343-KMYX-W3Y2-10KY. [DOI] [PubMed] [Google Scholar]
- 30.Aravinth SP, Rajendran S, Li Y, Wu M, Yi Wong AH, Schwarz H. Epstein-Barr virus-encoded LMP1 induces ectopic CD137 expression on Hodgkin and Reed-Sternberg cells via the PI3K-AKT-mTOR pathway. Leuk Lymphoma. 2019;60(11):2697–2704. doi: 10.1080/10428194.2019.1607330. [DOI] [PubMed] [Google Scholar]
- 31.Rajendran S, Ho WT, Schwarz H. CD137 signaling in Hodgkin and Reed-Sternberg cell lines induces IL-13 secretion, immune deviation and enhanced growth. Oncoimmunology. 2016;5(6):e1160188. doi: 10.1080/2162402X.2016.1160188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wolfl M, Kuball J, Ho WY, Nguyen H, Manley TJ, Bleakley M, et al. Activation-induced expression of CD137 permits detection, isolation, and expansion of the full repertoire of CD8+ T cells responding to antigen without requiring knowledge of epitope specificities. Blood. 2007;110(1):201–210. doi: 10.1182/blood-2006-11-056168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ye Q, Song DG, Powell DJ., Jr Finding a needle in a haystack: Activation-induced CD137 expression accurately identifies naturally occurring tumor-reactive T cells in cancer patients. Oncoimmunology. 2013;2(12):e27184. doi: 10.4161/onci.27184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bacher P, Heinrich F, Stervbo U, Nienen M, Vahldieck M, Iwert C, et al. Regulatory T cell specificity directs tolerance versus allergy against aeroantigens in humans. Cell. 2016;167(4):1067–1078. doi: 10.1016/j.cell.2016.09.050. [DOI] [PubMed] [Google Scholar]
- 35.Kachapati K, Adams DE, Wu Y, Steward CA, Rainbow DB, Wicker LS, et al. The B10 Idd9.3 locus mediates accumulation of functionally superior CD137(+) regulatory T cells in the nonobese diabetic type 1 diabetes model. J Immunol. 2012;189(10):5001–5015. doi: 10.4049/jimmunol.1101013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luu K, Nickles E, Schwarz H. Destroy, what destroys you. Oncoimmunology. 2020;9(1):1685301. doi: 10.1080/2162402X.2019.1685301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saito K, Ohara N, Hotokezaka H, Fukumoto S, Yuasa K, Naito M, et al. Infection-induced up-regulation of the costimulatory molecule 4-1BB in osteoblastic cells and its inhibitory effect on M-CSF/RANKL-induced in vitro osteoclastogenesis. J Biol Chem. 2004;279(14):13555–13563. doi: 10.1074/jbc.M303791200. [DOI] [PubMed] [Google Scholar]
- 38.Pichler K, Kattan T, Gentzsch J, Kress AK, Taylor GP, Bangham CR, et al. Strong induction of 4-1BB, a growth and survival promoting costimulatory receptor, in HTLV-1-infected cultured and patients' T cells by the viral Tax oncoprotein. Blood. 2008;111(9):4741–4751. doi: 10.1182/blood-2007-10-115220. [DOI] [PubMed] [Google Scholar]
- 39.Dharmadhikari B, Wu M, Abdullah NS, Rajendran S, Ishak ND, Nickles E, et al. CD137 and CD137L signals are main drivers of type 1, cell-mediated immune responses. Oncoimmunology. 2016;5(4):e1113367. doi: 10.1080/2162402X.2015.1113367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee KY, Wong HY, Zeng Q, Le Lin J, Cheng MS, Kuick CH, et al. Ectopic CD137 expression by rhabdomyosarcoma provides selection advantages but allows immunotherapeutic targeting. Oncoimmunology. 2021;10(1):1877459. doi: 10.1080/2162402X.2021.1877459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arch RH, Thompson CB. 4-1BB and Ox40 are members of a tumor necrosis factor (TNF)-nerve growth factor receptor subfamily that bind TNF receptor-associated factors and activate nuclear factor kappaB. Mol Cell Biol. 1998;18(1):558–565. doi: 10.1128/MCB.18.1.558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cannons JL, Choi Y, Watts TH. Role of TNF receptor-associated factor 2 and p38 mitogen-activated protein kinase activation during 4-1BB-dependent immune response. J Immunol. 2000;165(11):6193–6204. doi: 10.4049/jimmunol.165.11.6193. [DOI] [PubMed] [Google Scholar]
- 43.Cannons JL, Hoeflich KP, Woodgett JR, Watts TH. Role of the stress kinase pathway in signaling via the T cell costimulatory receptor 4-1BB. J Immunol. 1999;163(6):2990–2998. [PubMed] [Google Scholar]
- 44.Chow KC, Chiou SH, Ho SP, Tsai MH, Chen CL, Wang LS, et al. The elevated serum interleukin-6 correlates with the increased serum butyrate level in patients with nasopharyngeal carcinoma. Oncol Rep. 2003;10(4):813–819. [PubMed] [Google Scholar]
- 45.Zhang G, Tsang CM, Deng W, Yip YL, Lui VW, Wong SC, et al. Enhanced IL-6/IL-6R signaling promotes growth and malignant properties in EBV-infected premalignant and cancerous nasopharyngeal epithelial cells. PLoS ONE. 2013;8(5):e62284. doi: 10.1371/journal.pone.0062284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sun W, Liu DB, Li WW, Zhang LL, Long GX, Wang JF, et al. Interleukin-6 promotes the migration and invasion of nasopharyngeal carcinoma cell lines and upregulates the expression of MMP-2 and MMP-9. Int J Oncol. 2014;44(5):1551–1560. doi: 10.3892/ijo.2014.2323. [DOI] [PubMed] [Google Scholar]
- 47.Li XJ, Peng LX, Shao JY, Lu WH, Zhang JX, Chen S, et al. As an independent unfavorable prognostic factor, IL-8 promotes metastasis of nasopharyngeal carcinoma through induction of epithelial–mesenchymal transition and activation of AKT signaling. Carcinogenesis. 2012;33(7):1302–1309. doi: 10.1093/carcin/bgs181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yoshizaki T, Horikawa T, Qing-Chun R, Wakisaka N, Takeshita H, Sheen TS, et al. Induction of interleukin-8 by Epstein-Barr virus latent membrane protein-1 and its correlation to angiogenesis in nasopharyngeal carcinoma. Clin Cancer Res: Off J Am Assoc Cancer Res. 2001;7(7):1946–1951. [PubMed] [Google Scholar]
- 49.Liao B, Zhong BL, Li Z, Tian XY, Li Y, Li B. Macrophage migration inhibitory factor contributes angiogenesis by up-regulating IL-8 and correlates with poor prognosis of patients with primary nasopharyngeal carcinoma. J Surg Oncol. 2010;102(7):844–851. doi: 10.1002/jso.21728. [DOI] [PubMed] [Google Scholar]
- 50.Xie LQ, Bian LJ, Li Z, Li Y, Liang YJ. Co-elevated expression of hepatocyte growth factor and Interleukin-8 contributes to poor prognosis of patients with primary nasopharyngeal carcinoma. Oncol Rep. 2010;23(1):141–150. [PubMed] [Google Scholar]
- 51.Horikawa T, Kaizaki Y, Kato H, Furukawa M, Yoshizaki T. Expression of interleukin-8 receptor A predicts poor outcome in patients with nasopharyngeal carcinoma. Laryngoscope. 2005;115(1):62–67. doi: 10.1097/01.mlg.0000150675.37860.f7. [DOI] [PubMed] [Google Scholar]
- 52.Garg M, Braunstein G, Koeffler HP. LAMC2 as a therapeutic target for cancers. Expert Opin Ther Targets. 2014;18(9):979–982. doi: 10.1517/14728222.2014.934814. [DOI] [PubMed] [Google Scholar]
- 53.Moon YW, Rao G, Kim JJ, Shim HS, Park KS, An SS, et al. LAMC2 enhances the metastatic potential of lung adenocarcinoma. Cell Death Differ. 2015;22(8):1341–1352. doi: 10.1038/cdd.2014.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Okada Y, Takahashi N, Takayama T, Goel A. LAMC2 promotes cancer progression and gemcitabine resistance through modulation of EMT and ATP-binding cassette transporters in pancreatic ductal adenocarcinoma. Carcinogenesis. 2021;42(4):546–556. doi: 10.1093/carcin/bgab011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Buchan SL, Dou L, Remer M, Booth SG, Dunn SN, Lai C, et al. Antibodies to costimulatory receptor 4-1BB enhance anti-tumor immunity via T regulatory cell depletion and promotion of CD8 T cell effector function. Immunity. 2018;49(5):958–970. doi: 10.1016/j.immuni.2018.09.014. [DOI] [PubMed] [Google Scholar]
- 56.Freeman ZT, Nirschl TR, Hovelson DH, Johnston RJ, Engelhardt JJ, Selby MJ, et al. A conserved intratumoral regulatory T cell signature identifies 4-1BB as a pan-cancer target. J Clin Investig. 2020;130(3):1405–1416. doi: 10.1172/JCI128672. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.