Abstract
Microsatellite stable colorectal cancers (MSS-CRC) are resistant to anti-PD-1/PD-L1 therapy but the combination of immune checkpoints inhibitors (ICI) could be a clue to reverse resistance. Our aim was to evaluate ex vivo the capacity of the combination of atezolizumab (anti-PD-L1) and tiragolumab (anti-TIGIT) to reactivate the immune response of tumor infiltrating lymphocytes (TILs) in MSS-CRC. We analysed CRC tumor tissue and the associated blood sample in parallel. For each patient sample, extensive immunomonitoring and cytokine production were tested. We generated an ex vivo assay to study immune reactivity following immune stimulation with checkpoint inhibitors of tumor cell suspensions. Three microsatellite instable (MSI) and 13 MSS-CRC tumors were analysed. To generalize our observations, bioinformatics analyses were performed on public data of single cell RNA sequencing of CRC TILs and RNA sequencing data of TCGA. Atezolizumab alone could only reactivate T cells from MSI tumors. Atezolizumab and tiragolumab reactivated T cells in 46% of MSS-CRC samples. Reactivation by ICK was observed in patients with higher baseline frequency of Th1 and Tc1 cells, and was also associated with higher baseline T cell polyfunctionality and higher CD96 expression. We showed that a high frequency of CD96 expression on T cells could be a surrogate marker of atezolizumab and tiragolumab efficacy. Together these data suggest that the association of atezolizumab and tiragolumab could restore function of CD4 and CD8 TILs in MSS-CRC and could be tested in a clinical trial in colorectal cancer patients with MSS status.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00262-022-03182-9.
Keywords: Colorectal cancer, Anti-PDL1, Anti-TIGIT, CD8 T cells, TNFα
Background
T cell immune response is essential to shape tumor growth in colorectal cancer (CRC). The presence of T cells in the tumor bed is one of the strongest prognostic parameters in localized colorectal cancer. T-immune infiltration can be evaluated by different analytical methods, of which the most widely used is the Immunoscore®, an immune-histological assessment. This method, which determines CD3 and CD8 infiltration in CRC, can predict patient survival and seems to be better able to predict clinical prognosis than the TNM classification [1]. In addition, the immune infiltrate also seems to be associated with better outcome and response to chemotherapy in a metastatic setting, also in CRC [2, 3].
Despite the strong prognostic role of immune infiltrate in localized and metastatic CRC (mCRC), immune checkpoint inhibitors (ICI) aimed at reinvigorating intratumoral CD8 T cells are ineffective as monotherapy in most mCRC. Clinical studies underline that only tumors with microsatellite instability (MSI), which account for only 4% of mCRC, respond to anti-PD-1/PD-L1 antibody when used as monotherapy. This is explained by the fact that only MSI harbours a high number of mutations resulting in a high level of neoantigens that may favour induction of specific CD8 antitumor immune response. Despite the inefficiency of ICI monotherapies in microsatellite stable (MSS) tumors, some previous reports have shown that cytotoxic antitumor specific CD8 T cells are present at the tumor site in either primary tumor or metastases [4, 5]. These cells are able to recognize and kill autologous cancer cells, albeit at a lower frequency than in highly immunogenic tumors like melanoma. Therefore, three hypotheses can be proposed to explain the lack of effectiveness of ICI in MSS-CRC. First, the lack of ICI efficacy may rely on the absence of CD8 recruitment, or the absence of an expansion of the specific CD8 T immune response. Second, the accumulation of immunosuppressive cells may blunt the reactivation of CD8 T cells. The third possible hypothesis could be that CD8 T cells infiltrating MSS-CRC are in the terminal exhausted stage, which could impede their function and promote resistance to reactivation by classical ICI such as anti-PD-1/PD-L1.
During the exhaustion program, CD8 T cells will express other various immune checkpoints on their surface in addition to PD-1, such as Tim3, LAG3 or TIGIT. In addition to increase immune checkpoints expression, cytotoxic function and cytokine production both decrease in exhausted T cells, and eventually, these cells become resistant to anti-PD-1 when they complete the exhausted transcriptional program [6, 7]. TIGIT is a member of the adhesion molecules called nectins, of which CD96 and CD226 are also members. CD226 and TIGIT bind to CD112 and CD155, while CD96 binds to CD155 and CD111. CD96 and TIGIT are classically known as inhibitor receptors, which blunt T or NK cell function, while CD226 is an activating receptor. Previous studies in preclinical models have underlined that anti-TIGIT alone or in combination with anti-PD-L1 can be synergistic in the CT26 mouse colorectal cancer model [7]. Similarly, in the same model, we previously reported the existence of synergy between anti-PD-1/anti-TIGIT plus radiotherapy [8].
A recent report underlined the ability of ex vivo assays to predict response to ICI using “in sitro” (in situ/in vitro) assays [9, 10]. Using multiplexed functional immunology analyses, the authors determined that the tumor microenvironment can respond to ex vivo stimulation with various ICIs. Such assays predicted the response to anti-PD-1 monoclonal antibodies in this study. In the present study, also using an ex vivo assay, we investigated whether the combination of an anti-PD-L1 (Atezolizumab) plus an anti-TIGIT (Tiragolumab) can restore CD8 and CD4 T-cell function in TIL suspensions isolated from either primary tumors or liver metastases of MSS or MSI-CRC. We also investigated predictive biomarkers associated with ex vivo response to anti-PD-L1 (Atezolizumab) and anti-TIGIT (Tiragolumab) therapy.
Materials and methods
Study participants
This study included consecutive stage III and IV colorectal cancer treated by surgery for primary tumor or liver metastases at the Georges Francois Leclerc Center between April 2019 and May 2020. Written informed consent was obtained from all patients before enrolment. The hospital institutional review board approved the study in accordance with the principles of Good Clinical Practice, the Declaration of Helsinki, and other applicable local regulations. This study falls within the scope of the biobanking authorisation registered under the registration number AC-2014-2260.
MSI status was determined using a panel of 6 genes (BAT26, BAT40, BAT25, NR21, NR22 and NR27) by Gold Fast PCR and fragment analysis via an ABI3130 sequencer. Tumours with two or more unstable markers were considered to have a MSI status, whereas those with no unstable markers were considered as MSS.
Tumor dissociation
Fresh tumor tissues were collected on the day of surgery for each patient. Tumors were mechanically and enzymatically dissociated using a human tumor dissociation kit, according to the manufacturer's instructions (130-095-929, Miltenyi Biotec). In brief, tumors were cut into small pieces and transferred into gentleMACS C tubes containing the enzyme mix. The dissociation was performed using the gentleMACS Octo Dissociator with heaters and with the human tumor dissociation 37C_h_TDK_1 program. Samples were homogenized before being applied to a MACS SmartStrainer 70 µM (130-110-916, Miltenyi Biotec) placed on a 50 mL tube. Filters were washed with 20 mL serum-free RPMI (L0500-500, Dutscher) and then centrifuged at 300 g for 7 min. After complete aspiration of the supernatant, tumor cell suspensions were resuspended in RPMI and counted with trypan blue to remove dead cells. Then, 6⋅106 cells were put to one side for cytometry analyses and the rest of the cells were frozen in a solution of 50% Foetal Bovine Serum (FBS, Dutscher), 40% RPMI and 10% DMSO (P60-36720100, Dutscher) until further use.
Peripheral blood mononuclear cell isolation
Blood samples were also collected on the day of tumor resection for each patient. After recovering 1 mL of whole blood for cytometry analyses, peripheral blood mononuclear cells (PBMC) were isolated from the remaining whole blood by density gradient centrifugation (Lymphocyte Separation Medium, CMSMSL0101, Eurobio) with SepMate tubes (85460, Stemcell Technologies). Whole blood was transferred into Sepmate tubes at a rate of 17 mL of whole blood per tube and then centrifuged at 1200g for 10 min with an acceleration of 5 and the brake off. After removing as much plasma as possible, the phase containing the enriched PBMCs could be recovered. After washing with 45 mL PBS, centrifugation of 300g for 7 min was carried out and the PBMC pellet was resuspended in 5 mL PBS 1× for counting. Then, a final wash with 10 mL of PBS 1× was performed before cryopreservation, which consists in freezing at a rate of 8⋅106 cells per cryotube in a solution of 50% FBS, 40% RPMI and 10% DMSO until further use.
Ex vivo tumor assay restimulation
Tumor cell suspensions for each patient were thawed and diluted in RPMI1640 containing 10% FBS. After removing dead cells using the Dead Cell Removal Kit (Miltenyi Biotec), live tumor cell suspensions were labelled with 1 µM of CellTrace Violet (CTV; C34571, ThermoFisher Scientific) following the manufacturer's instructions. Then, cells were resuspended in the AIM V medium (12055091, Fisher Scientific) at a rate of 5.105 cells per well in a 96-well round-bottom culture plate, and stimulated with coated anti-CD3 antibody (10 ng/mL, clone OKT3, BE0001-2, BioXcell), or 5 µg/mL of anti-CD3 and anti-CD28 (clone CD28-2; 302933, BioLegend) antibodies for the positive control, in the presence or absence of 10 µg/mL of antibodies blocking immune checkpoint receptors (anti-PDL1 (clone 6E11) and anti-TIGIT (clone 10A7) provided by Roche Institute). After 4 days of culture at 37 °C with 5% CO2, 100 µL of supernatant were removed for each well and frozen at − 20 °C and replaced by 100 µL of Brefeldin A (420601, BioLegend) 2× solution for 20 h.
Cytometry analysis
Lymphoid and myeloid population identification
Antibodies for lymphoid cell analysis: multicolour flow cytometry was performed using Beckman Coulter's custom design service and its dry coating technology, custom tubes containing anti-CD56-ECD (clone N901), anti-CD45-PECy5.5 (clone J.33), anti-PD-1-APC (clone PD1.3), anti-CD3-AA750 (Clone UCTH1), anti-CD45RA-PacBlue (clone 2H4LDH11LDB9), anti-CD8-KromeOrange (Clone B9.11) and a mortality marker DRAQ7 were produced. The following liquid antibodies were added to the custom tubes: anti-DNAM-1-FITC (BioLegend, clone TX25), anti-CD96-PE (BioLegend, clone NK92.39), anti-TIGIT-PECy7 (BioLegend, clone A15153G), anti-CCR7-BV605 (BioLegend, clone G043H7), anti-Tim-3-BV650 (BioLegend, clone F38-E2E2) and anti-CD4-BV785 (BioLegend, clone OKT4).
Antibodies for myeloid cell analysis: multicolour flow cytometry was also performed using Beckman Coulter's custom design service and its dry coating technology, custom tubes containing anti-CD11b-FITC (clone Bear1), anti-HLA-DR-ECD (clone Immu-357), anti-PD-L1-APC (clone PDL1.3.1), anti-CD15-PacBlue (clone 80H5), anti-CD14-KromeOrange (clone RMO52) and a mortality marker DRAQ7 were produced. The following liquid antibodies were added to the custom tubes: anti-CD111-PE (BioLegend, clone R1.302), anti-CD155-PerCPCy5.5 (BioLegend, clone SKIL4), anti-CD112-PC7 (BioLegend, clone TX31), anti-CD163-APCCy7 (BioLegend, clone GHI/61), anti-CD3-BV605 (BioLegend, clone OKT3), anti-CD19-BV605 (BioLegend, clone HIB19), anti-CD20-BV605 (BioLegend, clone 2H7), anti-CD56-BV605 (BioLegend, clone HCD56), anti-Galectin-9-Biotin (Miltenyi Biotec, clone RG9-35.7), anti-Streptavidin-BV650 (BioLegend) and anti-CD45-BV785 (BioLegend, clone HI30).
Staining protocol: 100 μL of total heparinized blood or 1.106 cells of tumor cell suspension was added to each DURAClone tube containing liquid antibodies, vortexed immediately for 15 s and incubated for 15 min at room temperature in the dark. Two millilitres of red blood lysis solution (VersaLyse solution, A09777, Beckman Coulter) containing 50 μL of the fixative agent IOTest 3 Fixative solution (A07800, Beckman Coulter) was added, inverted and incubated for 15 min in the dark. After centrifugation and washing with 3 mL of PBS 1×, cells were resuspended in 150 µL PBS 1× before acquisition on a CytoFLEX cytometer (Beckman Coulter). The gating strategies are described in Supplementary Figs. 1 and 2.
Lymphocyte function analysis
Using Beckman Coulter's custom design service and its dry coating technology, custom tubes containing anti-IFNγ-FITC (clone 45.15), anti-TNFα-PE (clone IPM2 (188)), anti-IL-4-PECy7 (clone MP4-25D2), anti-Foxp3-AF647 (Clone 259D), anti-IL-17A-AF700 (Clone BL168), anti-CD3-AA750 (clone UCHT1), anti-CD4-PacBlue (clone 13B8.2) and anti-CD8-KromeOrange (clone B9.11) were produced. Liquid antibodies were also used: anti-GranzymeB-PECy5.5 (ThermoFisher, clone GB11), anti-IL-2-BV650 (BioLegend, clone MQ1-17H12) and anti-CD45-BV785 (BioLegend, clone HI30).
Staining procedure: 1.106 cells of tumor cell suspension in 100 µL of RPMI1640 were stained with anti-CD45 mAb (clone HI30, BioLegend) in the dark for 15 min at RT. The cells were then washed twice and the pellet was resuspended with 50 µL of total heparinized blood. This solution was transferred into a DURactive 1 tube (C11101, Beckman Coulter) for 3 h at 37 °C in the dark. After activation, 25 μL of PerFix-NC R1 buffer (PerFix-NC kit, B31168, Beckman Coulter) was added on vortex and incubated for 15 min at room temperature. Then, 2 mL of PBS 1× was added, and after centrifugation, the pellet was resuspended in 25 μL of FBS (Dutscher) and 300 μL of PerFix-NC R2 buffer was added. A 325 μL aliquot was transferred to a DURAClone tube containing the liquid antibody, vortexed immediately for 15 s and incubated for 1 h at room temperature in the dark. PBS 1× (3 mL) was added to the tubes, incubated for 5 min at room temperature in the dark before centrifugation for 5 min at 500 g. After supernatant removal, the cells were resuspended in 3 mL of 1× PerFix-NC R3 buffer before another 5 min centrifugation at 500 g. The pellet was dried and resuspended in 150 μL of 1× R3 buffer. Acquisition was done on a CytoFLEX cytometer. The gating strategy is described in Supplementary Fig. 3.
Analysis post re-stimulation with anti-PD-L1 and anti-TIGIT blocking antibodies
After 5 days of culture, tumor cell suspensions were harvested and washed with 2 mL of PBS 1×. After centrifugation, the pellets were resuspended with 50 µL of FBS and fixed with addition of 12.5 µL of PerFix-NC R1 buffer (PerFix-NC kit, B31168, Beckman Coulter) on vortex and incubated 15 min at room temperature. After incubation, 150 μL of PerFix-NC R2 buffer and the following fluorochrome-conjugated mAbs (Beckman Coulter) were added: anti-CD3 (clone UCHT1), anti-CD4 (clone 13B8.2), anti-CD8 (clone B9.11), anti-IFNγ (45.15) and anti TNFα (clone IPM2 (188)) and incubated for 1 h at room temperature in the dark. PBS 1× (3 mL) was added to the tubes, incubated for 5 min at room temperature in the dark before centrifugation for 5 min at 500g. After supernatant removal, the cells were resuspended in 3 mL of 1× PerFix-NC R3 buffer before another 5 min centrifugation at 500g. The pellet was dried and resuspended in 150 μL of 1× R3 buffer. Acquisition was done on a CytoFLEX cytometer. The gating strategy is described in Supplementary Fig. 4.
CTVlo T cells were counted as proliferated cells. To further evaluate the proliferation of T cells, the mitotic index was calculated by dividing mitotic events by the absolute number of precursor cells based on the number of cells in each mitotic division. We counted the number of divided cells up to the fifth mitotic division based on the fluorescence intensity of CTV.
All cytometry analyses were done with Kaluza 1.3 software (Beckman Coulter).
Bioinformatic analysis
Single cell RNAseq analysis was based on tumor data from 5 patients with colorectal cancer taken from Qian et al. [11]. Processed count data were downloaded from the ArrayExpress database at EMBL-EBI (http://www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-8107.
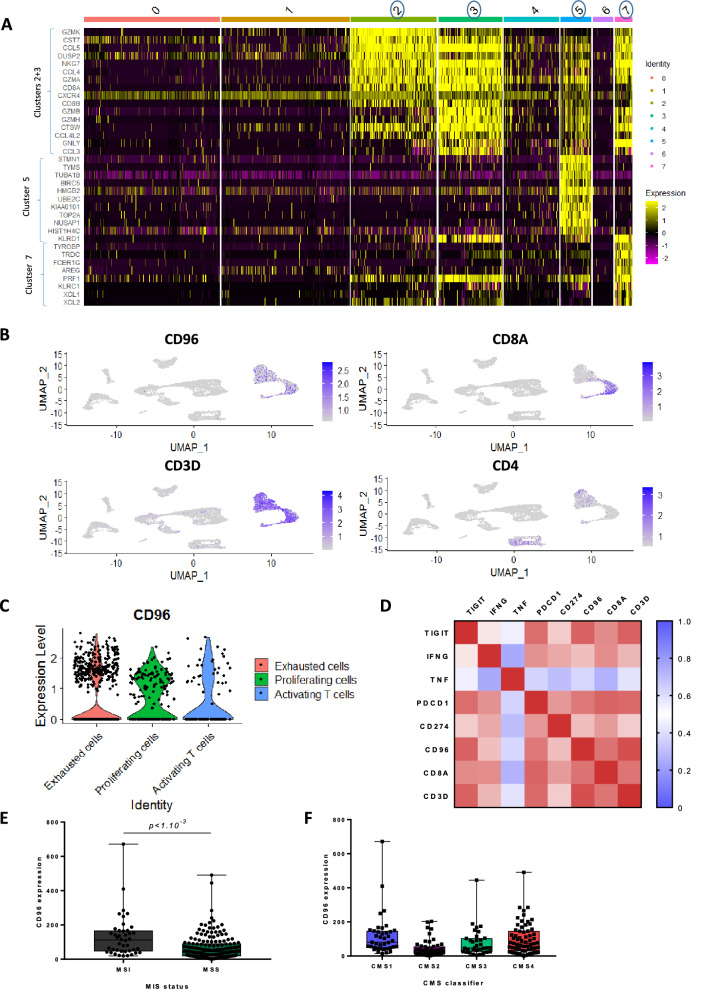
All variably expressed genes were used to construct principal components (PCs) and PCs covering the highest variance in the dataset were selected. Clusters were calculated by the FindClusters function from the Seurat library [12] and visualized using the t-SNE dimensional reduction method. Gene characteristics of each cluster were selected using the FindMarkers function. Annotation of the resulting clusters to cell types was based on the expression of marker genes. First, only clusters expressing following genes were kept for further analysis: CD96, IFNG, TNF, PDCD1, CD274, HAVCR2, CD101, SLAMF6, TCF7. A new clustering was performed on these cell subsets, leading to 7 clusters. Among these clusters we kept 3 clusters expressing CD8 cells, and annotated among them exhausted, proliferating and activating cells (Fig. 5A, B). The selection of the 3 clusters expressing CD8 was based on heatmap provided in Fig. 5A. This heatmap shows expression for given cells and features; only features most representative of each cluster are showed. These markers defining clusters were obtained via differential expression, by comparing expression of markers from cells of one cluster vs all other cells. Markers characteristics of each cluster help labelling each cluster and led to the identification of clusters 2, 3, 5 and 7 as expressing CD8. We chose to group clusters 2 and 3 because cells from these 2 clusters had similar profiles (Fig. 5A).
Fig. 5.
CD96 expression in lymphoid cells characterized a particular CRC subtype. A Heatmap representing gene expression matrix among the 7 clusters of cells expressing CD96, IFNG, TNF, PDCD1, CD274, HAVCR2, CD101, SLAMF6, TCF7. Gene expression levels are represented by colors. Only genes most characteristics of each cluster were kept for visualization. B Umap visualization of cells from tumor data from 5 patients with colorectal cancer analysed using single cell RNAseq. Each dot corresponds to a single cell. The 4 panels respectively highlight expression of CD96, CD8A, CD3D and CD4 genes among clusters. C Violin plots showing expression level of CD96 in exhausted, proliferating and activated T cell clusters based on single cell RNAseq analysis. D RNAseq expression correlation matrix involving genes used for the metagene estimation, namely TIGIT, IFNG, TNF, PDCD1, CD274, CD8A, CD3D, and CD96. E, F Boxplots showing the RNAseq expression of CD96 given the MSI status (E) and the CMS classification (F)
RNAseq COAD and READ TCGA datasets were downloaded using TCGA2stat R package [13]. MSS status was obtained through the TRONCO R package [14]. CMS classification was obtained using the CMScallerR package [15]. All these data were available for 211 patients.
Statistical analysis
Statistical analyses were performed using Prism GraphPad software [not significant (ns), *p < 0.05; **p < 0.01; ***p < 0.001; and ****p < 0.0001]. Results are shown as the mean ± SD. Datasets were compared using an unpaired Mann–Whitney–Wilcoxon test. No statistical corrections were performed.
Results
Double blockade of PD-L1 and TIGIT restores tumor infiltrating lymphocyte function in some cancer patients with MSS CRC
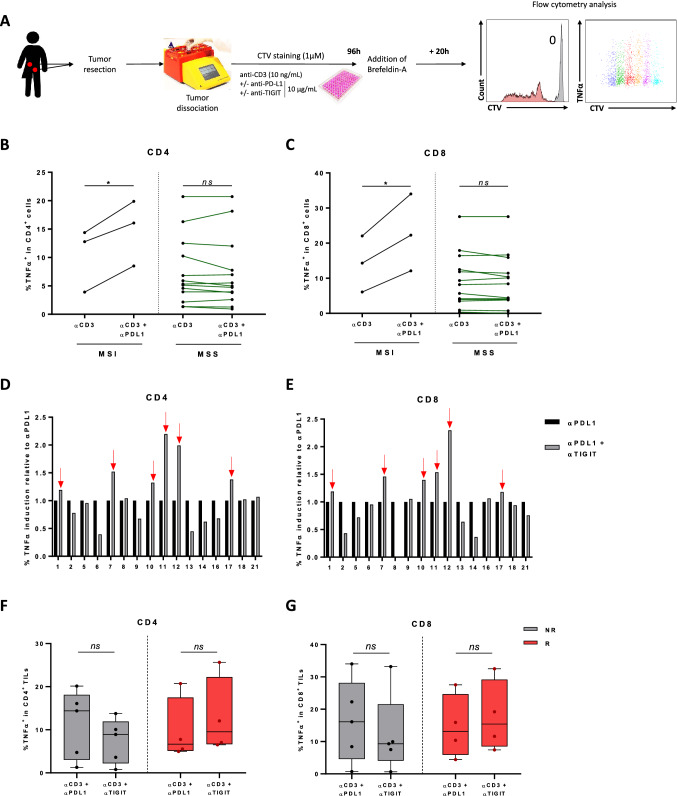
We included 21 patients with CRC. We obtained enough material from only 16 patients for TIL analysis. In the cohort, 3 patients had a tumor with MSI status, and 13 had MSS status. For 13 patients, the tumour sample came from a primary tumor, and for 3 patients, from liver metastasis. To test the ability of monoclonal antibodies targeting immune checkpoints to restore T cell function, we stimulated tumor cell suspensions with anti-CD3 in the presence or absence of atezolizumab and/or tiragolumab for 5 days (Fig. 1A). When we looked at the effect of atezolizumab ex vivo, we found that it enhanced TNFα production in CD8 and CD4 T cells only in MSI tumors (Fig. 1B, C). This suggests the clinical relevance of our test. Then, we investigated whether tiragolumab could improve the effector functions of CD8 and CD4 TILs. From our ex vivo test, we selected as responders to tiragolumab those patients in whom the combination of tiragolumab and atezolizumab enhanced TNFα production in CD4 or CD8 T cells compared to the control group treated with atezolizumab alone (Fig. 1D, E, Supplementary Fig. 5A). We observed that 6 patients were responders to combotherapy and that these patients all had MSS tumors. Ex vivo, tiragolumab did not induce a significant increase in IFNγ production in CD4 and CD8 T cells (Supplementary Fig. 5B). Tiragolumab alone had a moderate effect on TNFα production in CD8 and CD4 T cells, with a tendency to increase it only in patients sensitive to combotherapy with a rather lower relative intensity (Fig. 1F, G).
Fig. 1.
Double blockade of PD-L1 and TIGIT restores tumor infiltrating lymphocyte function in some cancer patients with MSS CRC. A Design of the experiment conducted with tumor samples (n = 16 patients with colorectal cancer) to test the efficacy of a combination of anti-PDL1 and anti-TIGIT immunotherapies. B, C Box plots showing the percentage of TNFα expression in CD4 (B) and CD8 (C) TILs according to MSS or MSI tumor status and comparing the effect of the anti-CD3 plus anti-PD-L1 condition versus the anti-CD3 alone condition. D, E The percentage induction of TNFα expression of anti-PDL1 and anti-TIGIT blockade compared to anti-PDL1 alone in CD4 (D) and CD8 (E) TILs are depicted. Patients are classified as responders when induction is greater than control and are marked with a red arrow. F, G Box plots showing the percentage of TNFα expression in CD4 (F) and CD8 (G) TILs according to responder (R) or non-responder (NR) status and comparing the effect of anti-TIGIT versus anti-PD-L1 condition. Statistical difference was determined by a Mann–Whitney test. ns not significant and *p < 0.05
Taken together, these data suggest that atezolizumab can induce activation of an immune response in TILs only in MSI tumors, while tiragolumab alone or atezolizumab plus tiragolumab combotherapy is able to induce reactivation of CD8 and CD4 TILs in some colorectal cancer patients with MSS status.
Association between baseline lymphoid cell number and response to combo-immunotherapy
Using exhaustive immunomonitoring of TILs or peripheral blood, we searched for predictive biomarkers associated with response to atezolizumab plus tiragolumab combotherapy. Before looking for these predictive biomarkers of response to the immunotherapy combination, we described the proportions of the different cell populations found in the blood and tumor of colon cancer patients (Supplementary Fig. 6A and B). The tumors analysed show a different and variable level of immune cell infiltration, indicating a different recruitment capacity depending on the tumor (Supplementary Fig. 6A). We also showed that in the tumors studied the infiltrate is largely composed of lymphocytes, in particular CD3+, CD4+ and CD8+ T cells, and that myeloid cells are very rarely present in the tumor whatever the subtype (CD14+, CD15+ or gMDSC) compared with the patients’ blood (Supplementary Fig. 6B).
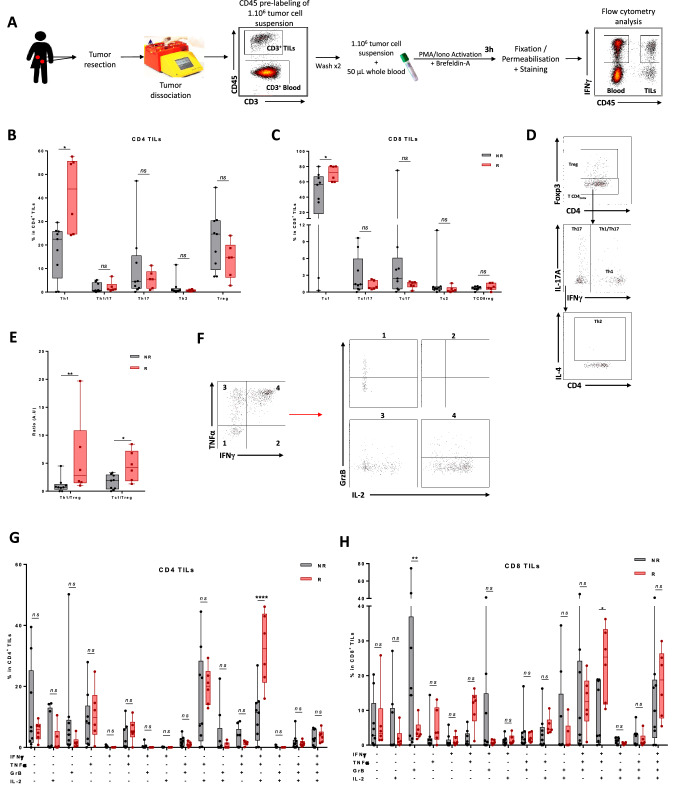
We then analyzed the function of intratumoral lymphocytes (Fig. 2A) and we observed that at baseline, a high number of Th1 was associated with a better response to the combination of atezolizumab and tiragolumab (Fig. 2B, D). Similarly, a high percentage of Tc1 cells was associated with a better response to combotherapy (Fig. 2C, D). We also noted that Th1/Treg and Tc1/Treg ratio is increased in responder patients which confirms that the presence of high levels of cytotoxic T cells and low levels of immunosuppressive cells are associated with response to combination therapy (Fig. 2E). When we looked at the production of Th1 and Tc1 effector molecules (IFNγ, TNFα, IL-2 and granzyme B), we observed that the responders were enriched in polyfunctional CD4 T cells which expressed the 3 cytokines related to a Th1 signature (Fig. 2F, G). For CD8 T cells, we observed a similar accumulation of polyfonctional T cells expressing the 3 classical Tc1 cytokines (Fig. 2F, H). In contrast, we found no association between the frequency of CD4 or CD8 subpopulations or cytokine production in the patients’ blood, and response to the combination of atezolizumab and tiragolumab (Supplementary Fig. 7A and B). Furthermore, when looking at the expression of granzyme B by CD4 and CD8 TILs, we can see that non-responders express more granzyme B than responders (Supplementary Fig. 7C). This cytokine is used to identify exhausted lymphocytes, thus revealing that non-responders have more exhausted lymphocytes and thus their intra-tumor immune response is less cytotoxic than responders to dual immunotherapy.
Fig. 2.
Association between baseline lymphoid cell number and response to combo-immunotherapy. A Fresh tumor tissues and associated blood samples for each patient (n = 16) were activated, stained and then analysed by flow cytometry. B The frequency of Th1 (CD4+ Foxp3− IFNγ+ IL-17A−), Th17 (CD4+ Foxp3− IFNγ− IL-17A+), Th1-Th17 (CD4+ Foxp3− IFNγ+ IL-17A+), Th2 (CD4+ Foxp3− IFNγ− IL-17A− IL-4+) and Treg (CD4+ Foxp3+) cells in CD4+ TILs (CD45+ CD3+ CD4+) according to responder versus non-responder status is depicted. C The frequency of Tc1 (CD8+ Foxp3− IFNγ+ IL-17A−), Tc17 (CD8+ Foxp3− IFNγ− IL-17A+), Tc1-Tc17 (CD8+ Foxp3− IFNγ+ IL-17A+), Tc2 (CD8+ Foxp3− IFNγ− IL-17A− IL-4+) and T CD8reg (CD8+ Foxp3+) cells in CD8+ TILs (CD45+ CD3+ CD8+) according to responder (R) versus non-responder (NR) status is depicted. D Representative dot plots of helper T and CD8 T subtype function analysis shown in B and C. E The Th1-Treg and Tc1-Treg ratio is plotted against responder (R) and non-responder (NR) status. F Representative dot plots of CD4 T and CD8 T function analysis shown in G and H. G, H Box plots showing the different combinations of expression of the 4 cytokines IFNγ, TNFα, Granzyme B (GrB) and IL-2 in CD4 (CD45+ CD3+ CD4+) (G) and CD8 (CD45+ CD3+ CD8+) (H) TILs according to responder (R) versus non-responder (NR) status. Statistical difference was determined by a Mann–Whitney test. ns not significant, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001
Together, these data suggest that the presence of polyfunctional Th1 and Tc1 cells at the tumor site is associated with better efficacy of atezolizumab and tiragolumab combotherapy.
Association between baseline nectin expression and response to combo-immunotherapy
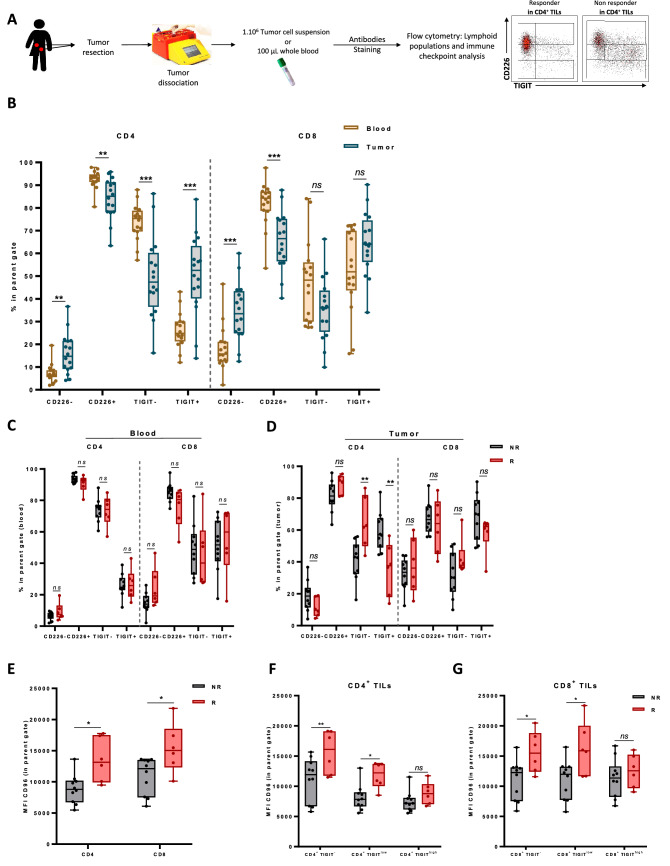
We further analysed the relationship between the expression of nectin receptors on T cells and the response to the combination of atezolizumab and tiragolumab (Fig. 3A). Different expression profiles of CD226 and TIGIT were observed between CD4 T and CD8 T both in the blood and in the tumor (Supplementary Fig. 8A and B), with higher CD226 expression in CD4 T compared to CD8 T cells, and higher TIGIT expression in CD8 T cells compared to CD4 T cells. When we compared the expression of these two markers between blood and tumor in each lymphoid subpopulation, we observed that CD226 is expressed at higher levels in the blood by CD4 and CD8 T cells, whereas TIGIT is expressed more at the tumor site, suggesting a more exhausted phenotype of lymphocytes in the tumor (Fig. 3B). However, high expressions of TIGIT and CD226 in blood and tumor by both CD4 and CD8 T cells were not associated with response to the combination of atezolizumab and tiragolumab. (Fig. 3C, D and Supplementary Fig. 8C and D). When we focused on the expression on CD96 on lymphocytes, we found a high expression of CD96 on the surface of the T cells (100% of the cells are CD96+). We therefore looked for differences in the median fluorescence index (MFI) of this marker. Surprisingly, the responders to the combination of atezolizumab and tiragolumab expressed higher CD96 levels on both CD8 and CD4 cells in the tumor but not in the blood (Fig. 3E and Supplementary Fig. 9). We could observe that, contrary to what we would expect, the expression of TIGIT is less important in the responders (Fig. 3D). However, when we studied the fluorescence intensity of the CD96 marker on the populations expressing more or less TIGIT, we found that in the responders, the TIGITlow and TIGIT− cells are those where the fluorescence intensity of CD96 is the most important contrary to the TIGIThigh cells (Fig. 3F, G).
Fig. 3.
Association between baseline nectin expression and response to combo-immunotherapy. A Fresh tumor tissues and associated blood samples for each patient (n = 16) were stained and then analysed by flow cytometry. B Box plots showing the frequency of CD226+, CD226−, TIGIT+ and TIGIT− populations within CD4 T cells and CD8 T cells in patients’ blood and tumor. C, D Box plots showing the frequency of CD226+, CD226−, TIGIT+ and TIGIT− populations within CD4 T cells and CD8 T cells in patients’ blood (C) and tumor (D) according to responder (R) or non-responder (NR) status. E Box plots showing the median fluorescence intensity (MFI) of CD96 marker in CD4 TILs and CD8 TILs in patient tumors according to responder (R) or non-responder (NR) status. F, G Box plots showing the median fluorescence intensity (MFI) of CD96 marker in each cell subtype expressing or not TIGIT: TIGIT−, TIGITlow and TIGIThigh in CD4 TILs (F) and CD8 TILs (G) in patient tumors according to responder (R) or non responder (NR) status. Statistical difference was determined by a Mann–Whitney test. ns not significant, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001
Overall, these data highlight that TILs express a higher level of TIGIT and that high CD96 expression in TIGIT−/low TILs is associated with a better response to atezolizumab and tiragolumab combotherapy.
Association between baseline myeloid cell number and response to combo-immunotherapy
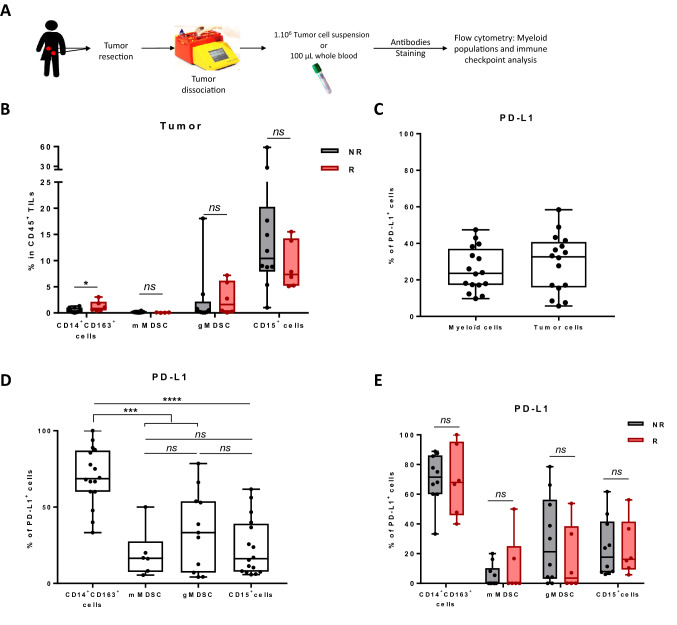
We then analysed the link between the myeloid infiltrate and the response to the combination of atezolizumab and tiragolumab (Fig. 4A). The most frequent myeloid cells in the tumor were CD15+ neutrophils (Fig. 4B). We observed that CD14+ CD163+ macrophages were present in higher numbers in responders compared to non-responders (Fig. 4B). PD-L1 is expressed almost as much on myeloid cells as on tumor cells (Fig. 4C). When we looked at the level of PD-L1 expression on the different myeloid cell subtypes, we observed that macrophages expressed a significantly higher level of PD-L1 than the other myeloid cell subsets (Fig. 4D). However, PD-L1 expression on myeloid cells, regardless of subtype, was not associated with response to the combination of atezolizumab and tiragolumab (Fig. 4E). Concerning the expression of TIGIT ligands, only high CD155 in macrophage was associated with response to combotherapy (Supplementary Fig. 10). For the expression of the CD96 ligand, called CD111, its expression was not found to be associated with response to the combination of atezolizumab and tiragolumab (Supplementary Fig. 10).
Fig. 4.
Association between baseline myeloid cell number and response to combo-immunotherapy. A Fresh tumor tissues and associated blood samples for each patient (n = 16) were stained and then analysed by flow cytometry. B The frequency of macrophages (CD14+ CD163+), mMDSC (CD14+ CD163− HLA-DRlow), gMDSC (CD15+ CD14+) and granulocytes (CD15+) in tumor samples according to responder (R) or non-responder (NR) status is depicted. C Box plots showing PD-L1 expression on myeloid (CD11b+) and tumor cells. D Box plots showing PDL1 expression on different myeloid subtypes (macrophages (CD14+ CD163+), mMDSC (CD14+ CD163− HLA-DRlow), gMDSC (CD15+ CD14+) and granulocytes (CD15+)) in tumor samples. E Box plots showing PDL1 expression on different myeloid subtypes (macrophages (CD14+ CD163+), mMDSC (CD14+ CD163− HLA-DRlow), gMDSC (CD15+ CD14+) and granulocytes (CD15+)) in tumor samples according to responder (R) or non-responder (NR) status. Statistical difference was determined by a Mann–Whitney test. ns not significant, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001
CD96 expression in lymphoid cells characterized a particular CRC subtype
Because CD96 expression predicted response to the combination of atezolizumab and tiragolumab, we hypothesized that high expression of CD96 could be associated with a particular CRC subtype, which presents specific immune infiltrate. Based on public single cell RNA sequencing data from CRC tissue, we observed the presence of CD4 and CD8, which expressed high levels of CD96 mRNA. Interestingly, CD96 was more expressed in CD8 exhausted cluster than in other CD8 clusters (Fig. 5A–C). In the TCGA dataset, we observed a strong correlation between CD96/TIGIT/IFN/TNF/PD1/PDL1/CD8/CD3 suggesting that expression of all these genes are related to a same single pathway (Fig. 5D). CD96 expression was more frequent in MSI tumors (Wilcoxon test p < 1.10–3), (Fig. 5E), and in CMS1 (n = 39) and 4 (n = 64) tumors, but almost absent in CMS2 (n = 51) and 3 (n = 33) tumors (Fig. 5F) (Wilcoxon test p = 0.03). To address the clinical relevance of CD96 expression and to avoid bias due to the correlation between CD96 expression and lymphocyte accumulation, which is a well-known prognostic factor, we split the cohort according to the number of T cells present in each tumor. A metagene reflecting lymphoid cells was computed by averaging expression of TIGIT/IFN/TNF/PD1/PDL1/CD8/CD3 genes. While CD96 had no prognostic role in tumor poorly infiltrated with lymphoid cells (p = 0.8), CD96 was associated with poor OS in tumor with high lymphoid infiltration (HR = 2.24 [1.14; 4.50]; p = 0.02) (Supplementary Fig. 10A and B).
Together these data underline that CMS1 and 4 tumors are enriched in CD96 high exhausted T cells. Accumulation of these cells is connected with poor prognosis in patients untreated with immunotherapy.
Discussion
We report here the potential ability of an anti-PD-L1 and an anti-TIGIT to restore intratumoral CD4 and CD8 immune function in some colorectal cancers, independently of microsatellite status, in either primary tumor or liver metastasis. Immunotherapy in CRC patients with MSS status remains a challenge for clinical oncology. Despite the lack of efficacy of single targeting of the anti-PD-1/PD-L1 pathway, some preliminary results support the ability of an association of an anti-CTLA4 with an anti-PD-1/PD-L1, or a combination of chemo-immunotherapy with an anti-PD-L1/anti-CTLA4 to restore an immune response in situ and to induce some clinically relevant antitumor effects in a subset of patients [16, 17]. These data raise the hypothesis that a combination of immune checkpoints inhibitors could be of interest in MSS-CRC. However, the optimal combination of checkpoint inhibitors and the optimal biomarker remain to be determined.
The T cell immunoreceptor with Ig and ITIM domain (TIGIT) belongs to the PVR-nectin family and recognizes CD155 (poliovirus receptor, PVR), and CD112 [18, 19]. TIGIT is known as an inhibitory receptor on both T cells and NK cells [20, 21], and is highly expressed in exhausted T cells. Preclinical data underline the efficacy of combining immune checkpoints with an anti-TIGIT in various animal models [8, 22–25]. In addition, some preliminary clinical data show the efficacy of combination therapy comprising anti-PDL1 (atezolizumab) and anti-TIGIT (tiragolumab) in non-small cell lung cancer (NSCLC) [26]. In CRC, this strategy could also be relevant. In particular, we showed a high expression of TIGIT on both CD8 and CD4 T cells at the tumor site. In addition, TIGIT is more expressed at the tumor site than in the blood, suggesting that targeting TIGIT could induce preferentially reactivation of intratumoral T cells, thus limiting autoimmunity. We also observed a high expression of the TIGIT ligands (CD155 and CD122) and PD-L1 at the tumor site, suggesting the relevance of combotherapy targeting PDL1 and TIGIT in CRC. Surprisingly, responder patients express less TIGIT than non-responders on the surface of CD4 and CD8 cells. However, accumulation of TIGIT high cells may reflect terminal exhausted cells which might not be reactivable by immune checkpoints.
To assess the putative ability of anti-PD-L1 and anti-TIGIT to reactivate TILs, we decided to generate an ex vivo tumor cell suspension restimulation model with anti-PD-L1 and anti-TIGIT. Previous data have demonstrated in MSI CRC tumors, and also in several anti-PD-1 sensitive tumors, that tumor-derived CD8 T cells can be reinvigorated ex vivo with anti-PD-1 stimulation [27, 28]. More recently, Dubuisson et al. set up a functional dynamic multiplexed immunophenotyping assay, measuring up to 50 parameters after 3 days of ex vivo stimulation with various immune checkpoint blockers and linked ex vivo activation to in vivo response [9]. Similarly Voabil et al. report a patient-derived tumor fragment platform to dissect ex vivo effect of PD-1 blockade. These also reported that the capacity of immune cells to be reactivated ex vivo was predictive of a clinical response [10]. These data provide proof of concept that the use of ex vivo restimulation of TILs could be a relevant test for predicting the effectiveness of immune checkpoint blockers.
Based on this rationale, we observed that T cell reactivation using the combination of atezolizumab and tiragolumab could be achieved in 37.5% of patients with CRC. This reactivation occurred in tumors with stable microsatellite status, whereas reactivation with atezolizumab alone only had an impact in microsatellite instable tumors, as expected. Although it is described that liver metastases differ from primary lesions in terms of immune cell infiltration [29–31], we find a reactivation of T cells for primary lesions but also for liver metastases. Interestingly, we noted that some immune response patterns were associated with response to immune checkpoint blocker combotherapy. The presence of a high number of Tc1 and Th1 cells and the presence of polyfunctional cells was associated with a better response to immune checkpoint blockers. However, in responder patients these polyfunctional CD4 and CD8 cells show low granzyme B expression in contrast to non-responder patients. It has been shown in mice that terminally exhausted CD8 T cells are cells that express high levels of granzyme B in contrast to progenitor exhausted CD8 T cells that do not express granzyme B and express TNFα [32]. This highlights that responder patients have polyfunctional cells within their tumour that are progenitor exhausted and will therefore be able to induce a cytotoxic response against the tumour unlike non-responder patients who instead have terminally exhausted TILs revealing an inability to induce or restore an anti-tumour immune response. In the same way, PD-L1 expression by myeloid cells and especially macrophages, was a surrogate marker of response to immune checkpoint blockers in this cohort. Indeed, PD-L1 expression by myeloid cells is a strong surrogate marker of a pre-existing IFNγ-dependent immune response. Taken together, these findings confirm that the presence of type I effector cells and polyfunctional cells is associated with improved efficacy of immune checkpoint blockers.
We also observed an association between CD96 expression and response to checkpoint inhibitors. While CD96 and TIGIT are known to be two potential inhibitory receptors that bind to CD155, this result is rather surprising. Blocking CD96 in various murine cancer models reduced tumor growth and may be synergistic with anti-PD-L1 and anti-TIGIT blockade [33–35]. Most of these studies focused on the role of CD96 in NK in mouse models. The cytoplasmic tail of both human and mouse CD96 has an ITIM inhibitory motif, but human CD96 also contains an activating motif [36]. Thus, a discrepancy might be observed between mice and humans. Accordingly, another study that focused on human and mice CD8, suggested that CD96 had a co-stimulatory function on CD8 T cells. The crosslinking of CD96 on murine or human CD8 T cells induces activation, effector cytokine production, and proliferation [37]. Since our study shows that high CD96 expression is associated with an exhausted phenotype and a better ability of T cells to be reactivated, we hypothesize that CD96 might be a surrogate marker for reactivable exhausted T cells. Therefore, CD96 is found to be more expressed on TIGIT−/low cells, which might not be terminally exhausted and hence reactivable. We also show that CD14+ CD163+ macrophages are present in higher numbers in responders than in non-responders. This seems counterintuitive as these macrophages are frequently described as immunosuppressive cells in cancers. However, in colon cancer, myeloid cells express checkpoint ligands more than tumor cells. The presence of macrophages in responders can be explained by the fact that the interaction of a checkpoint with its ligand is more likely and therefore the use of immunotherapies is more effective.
CD96 expression in the TCGA cohort is mainly observed in the CMS1 and 4 subtypes. CD96 expression is associated with poor prognosis in tumors highly infiltrated with lymphoid cells, suggesting that CRC invaded in large numbers by CD96+ cells is a particular subgroup of CRC, with a specific phenotype and prognosis. This subgroup is probably the most likely to respond to the combination of atezolizumab and tiragolumab.
Conclusions
To conclude, we observed that the association of atezolizumab and tiragolumab could restore the function of CD8 and CD4 TILs in an ex vivo restimulation system in a subset of CRC patients with stable microsatellite status. This event only occurs in patients with polyfunctional Tc1 and Th1 cells, thus suggesting that a pre-existing effector immune response is required for immune checkpoint blockers to be effective. High frequencies of CD96 expression on T cells and PD-L1 expression on myeloid cells could be surrogate markers of atezolizumab and tiragolumab efficacy. Clinical trials testing this combination in colorectal cancer with stable microsatellite status are warranted.
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary file2Supplementary figure 1: Gating strategy for “Lymphoid cell analysis” panel. The gating strategy was done on a tumour cell suspension sample and is performed in the same way on blood samples. The expression of CD226, TIGIT and PD1 was studied in the same way as on CD8 T cells on NK cells. Expression of naive/memory subpopulations dependent on CD45RA and CCR7 expression are studied in the same way as CD8 T cells on CD4 T cells, as well as the expression of CD226, TIGIT and PD1. The expression of CD96 is positive for all CD4 and CD8 T cells, so we analyzed the median intensity fluorenscence of this marker on our populations of interest. Supplementary figure 2: Gating strategy for “Myeloid cell analysis” panel. The gating strategy was done on a blood sample and is performed in the same way on tumour samples. The expression of PD-L1, CD111, CD112 and CD155 markers was studied in each of the following populations: granulocytes (CD15+), macrophages (CD14+ CD163+), monocytic MDSCs (CD14+ CD15- CD163- HLA-DRlow) and granulocytic MDSCs (CD14+ CD15+). Supplementary figure 3: Gating strategy for “Lymphocyte function analysis” panel. The gating strategy is developed on the CD3+ CD45- blood cell population. The same gating strategy was applied to the CD3+ CD45+ population corresponding to TILs. TILs was pre-labeled with an anti-CD45 antibody so that blood and TILs could be analyzed in the same tube. For the study of cytokine expression, we first analysed the expression of TNFα and IFNγ on conventional CD4 T cells (T CD4conv), regulatory CD4 T cells (Treg), conventional CD8 T cells (T CD8conv) and regulatory CD8 T cells (T CD8reg) and then the expression of Granzyme B and IL-2 on each subpopulation. Secondly, we analysed within the CD4conv T cells each of the following subpopulations: Th1 (CD4+ Foxp3- IFNγ+ IL-17A-), Th17 (CD4+ Foxp3- IFNγ- IL-17A+), Th1/17 (CD4+ Foxp3- IFNγ+ IL-17A+) and Th2 (CD4+ Foxp3- IFNγ- IL-17A- IL-4+) cells. We performed the same analysis within CD8conv T cells: Tc1, Tc17, Tc1/17 and Tc2 cells. For each of these populations, we studied the expression of the cytokines TNFα and IL-2 as shown for Th1 in this gating strategy. Supplementary figure 4: Gating strategy for “Ex vivo restimulation analysis”. Supplementary figure 5: Double blockade of PD-L1 and TIGIT restore tumor infiltration lymphocytes function in some cancer patients with MSS CRC. Tumor cell suspensions from 16 patients with cancer colorectal cancers were thawed and after removing dead cells were labelled with 1µM of CellTrace Violet. Then, the cells were plated at 5.105 cells per well and stimulated with anti-CD3 antibody (10 ng/mL) in the presence or absence of 10 µg/mL of anti-PD-L1 (clone 6E11) and/or anti-TIGIT (clone 10A7) during 5 days. Cells were also stimulated with 5 µg/mL of anti-CD3 and anti-CD28 as positive control experiments. 20 hours before the end of the culture, Brefeldin A was added to block transport processes during cell activation and allow the staining of intracellular cytokines. After 5 days of culture, to study the effect of immune checkpoints blockade antibodies on the proliferation and cytotoxic capacities of TILs, tumor cell suspensions were stained with anti-CD3, anti-CD4, anti-CD8, anti-IFNγ and anti-TNFα and analysed by flow cytometry. A. The percentage of TNFα expression in CD4 (left) and CD8 (right) is studied for the following 3 conditions: anti-CD3, anti-CD3 plus anti-PD-L1 and anti-CD3 plus anti-PD-L1 + anti-TIGIT. Patients classified as responders to double immunotherapy are shown in colour. B. The percentage of IFNγ expression in CD4 (left) and CD8 (right) comparing the anti-CD3 plus anti-TIGIT condition to anti-CD3 alone is depicted. Supplementary figure 6: Description of the cell composition in the blood and tumour of patients. Fresh tumor tissues and associated blood samples for each patient (n=16) were stained and then analysed by flow cytometry. A. Box plots showing the frequency of CD45+ and CD45- populations in patients’ blood and tumor. B. Box plots showing the frequency of CD3+, CD4+ and CD8+ T cells, NK and NKT cells, CD11b+; CD14+, CD15+ cells and gMDSC population in patients’ blood and tumor. Supplementary figure 7: Association between baseline lymphoid cell number and response to comboimmunotherapy. Fresh tumor tissues and associated blood samples for each patient were taken on the day of surgery. After tumor dissociation, 1.106 tumor cell suspension were stained with anti-CD45, resuspended in 50 µL of whole blood sample and then transferred to a tube containing PMA, Ionomycin and Brefeldin A for 3 hours. After activation, cells were fixed, permeabilized and stained with anti-IFNγ, anti-TNFα, anti-GranzymeB, anti-IL-4, anti-Foxp3, anti-IL-17A, anti-CD3, anti-CD4, anti-CD8 and anti-IL-2 antibodies and analysed by flow cytometry. A-B. Box plots showing the different combinations of expression of the 4 cytokines IFNγ, TNFα, Granzyme B (GrB) and IL-2 in CD4 (CD45- CD3+ CD4+) (A) and CD8 (CD45- CD3+ CD8+) (B) T cells in blood patients’ depending on the responder (R) versus non responder (NR) status. C. The frequency of Granzyme B (GrzB) expression according to responder (R) or non-responder status is depicted in tumor samples. Statistical difference is determined by a Mann–Whitney test. ns = no significant, * = p< 0.05 and ** = p< 0.01. Supplementary figure 8: Association between baseline nectin expression and response to comboimmunotherapy. Fresh tumor tissues and associated blood samples for each patient were taken on the day of surgery. After tumor dissociation, 1.106 tumor cell suspension and 100 µL of whole blood sample were stained with anti-DNAM-1 (CD226), anti-CD56, anti-CD96, anti-CD45, anti-TIGIT, anti-PD-1, anti-CD3, anti-CD45RA, anti-CD8, anti-CCR7, anti-Tim-3 and anti-CD4 antibodies and with a mortality marker DRAQ7 and then analysed by flow cytometry. A-B. Box plots showing the frequency of CD226+, CD226-, TIGIT+ and TIGIT- populations in the blood (A) and tumor (B) samples by comparing expression on CD4, CD8 and NK populations. C-D. Box plots showing the different combinations of expression of the 2 nectins CD226 and TIGIT within CD4 T cells and CD8 T cells in patient blood (C) and tumor (D) according to responder (R) or non responder (NR) status. Statistical difference is determined by a Mann–Whitney test. ns = no significant and * = p< 0.05, ** = p<0.01, *** = p<0.001 and **** = p<0.0001. Supplementary figure 9: Association between baseline nectin expression and response to comboimmunotherapy. Box plots showing the median fluorescence intensity (MFI) of CD96 marker in CD4 T cells and CD8 T cells in patient blood depending on the responder (R) or non responder (NR) status. Statistical difference is determined by a Mann–Whitney test. ns = no significant and * = p< 0.05. Supplementary figure 10: Association between baseline myeloid cell number and response to comboimmunotherapy. Fresh tumor tissues and associated blood samples for each patient were taken on the day of surgery. After tumor dissociation, 1.106 tumor cell suspension and 100 µL of whole blood sample were stained with anti-CD11b, anti-HLA-DR, anti-CD111, anti-CD155, anti-CD112, anti-PD-L1, anti-CD163, anti-CD15, anti-CD14, anti-CD3, anti-CD19, anti-CD20, anti-CD56, anti-Galectin-9 and anti-CD45 antibodies and with a mortality marker DRAQ7 and then analysed by flow cytometry. The frequency of CD111, CD112 and CD155 positive populations in macrophages (CD14+ CD163+), mMDSC (CD14+ CD163- HLA-DRlow), gMDSC (CD15+ CD14+) and granulocytes (CD15+) in tumor samples according to responder (R) or non-responder (NR) status is depicted. Statistical difference is determined by a Mann–Whitney test. ns = no significant and * = p< 0.05. Supplementary figure 11: CD96 expression in lymphoid cells characterized a particular CRC subtype. A-B. Kaplan-Meier curves for overall survival with patients stratified according to the level of CD96 RNAseq expression, respectively for patients with high (A) or low (B) level of the metagene. (PPTX 3848 kb)
Acknowledgements
This study was carried out in partnership with the Roche Institute, which supplied us with atezolizumab and tiragolumab. We would like to thank Dr. Laurent Arnould and his anatomopathology laboratory for their help in the recovery of the tumor samples. Flow cytometry analyses were performed on Beckman Coulter Cytoflex. Thanks to Olivier Jaen for his help in setting up the immunomonitoring panels.
Abbreviations
- CRC
ColoRectal Cancer
- CTLA-4
Cytotoxic T-Lymphocyte-Associated protein 4
- CTV
Cell Trace Violet
- FBS
Foetal Bovine Serum
- ICK
Immune ChecKpoint
- ICI
Immune Checkpoint Inhibitors
- IFNγ
InterFeroN gamma
- IL-2
InterLeukin-2
- LAG3
Lymphocyte-Activation Gene 3
- mAbs
Monoclonal antibodies
- mCRC
Metastatic ColoRectal Cancer
- MFI
Median Fluorescence Intensity
- MSS
MicroSatellite Stable
- MSI
MicroSatellite Instable
- NK
Natural Killer
- NSCLC
Non-Small Cell Lung Cancer
- PBMC
Peripheral Blood Mononuclear Cells
- PBS
Phosphate-Buffered Saline
- PD-1
Programmed cell Death protein 1
- PD-L1
Programmed cell Death protein Ligand 1
- TIGIT
T cell Immunoreceptor with Ig and ITIM domains
- Th1
T helper 1
- TILs
Tumor Infiltrating Lymphocytes
- TNFα
Tumor Necrosis Factor alpha
Author contributions
MT participated in the design of the study, the establishment of patient collection and draft the manuscript. LH performed patient collection, carried out the ex vivo and flow cytometry analysis and participated in analyse obtained data. EB and CT carried out all bioinformatics analysis. EL participated in the design of the study and helped to draft the manuscript. FG conceived of the study, and participated in its design and coordination and draft the manuscript. All authors read and approved the final manuscript.
Funding
This research was supported and granted by Genentech, Inc.
Declarations
Conflict of interest
The authors declare that there is no potential conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Pagès F, Mlecnik B, Marliot F, et al. International validation of the consensus Immunoscore for the classification of colon cancer: a prognostic and accuracy study. Lancet. 2018;391:2128–2139. doi: 10.1016/S0140-6736(18)30789-X. [DOI] [PubMed] [Google Scholar]
- 2.Reichling C, Taieb J, Derangere V, et al. Artificial intelligence-guided tissue analysis combined with immune infiltrate assessment predicts stage III colon cancer outcomes in PETACC08 study. Gut. 2020;69:681–690. doi: 10.1136/gutjnl-2019-319292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bindea G, Mlecnik B, Tosolini M, et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity. 2013;39:782–795. doi: 10.1016/j.immuni.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 4.Turcotte S, Gros A, Hogan K, et al. Phenotype and function of T cells infiltrating visceral metastases from gastrointestinal cancers and melanoma: implications for adoptive cell transfer therapy. J Immunol. 2013;191:2217–2225. doi: 10.4049/jimmunol.1300538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Robbins PF, Lu Y-C, El-Gamil M, et al. Mining exomic sequencing data to identify mutated antigens recognized by adoptively transferred tumor-reactive T cells. Nat Med. 2013;19:747–752. doi: 10.1038/nm.3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kong Y, Zhu L, Schell TD, et al. T-cell immunoglobulin and ITIM domain (TIGIT) associates with CD8+ T-cell exhaustion and poor clinical outcome in AML patients. Clin Cancer Res. 2016;22:3057–3066. doi: 10.1158/1078-0432.CCR-15-2626. [DOI] [PubMed] [Google Scholar]
- 7.Johnston RJ, Comps-Agrar L, Hackney J, et al. The immunoreceptor TIGIT regulates antitumor and antiviral CD8+ T cell effector function. Cancer Cell. 2014;26:923–937. doi: 10.1016/j.ccell.2014.10.018. [DOI] [PubMed] [Google Scholar]
- 8.Grapin M, Richard C, Limagne E, et al. Optimized fractionated radiotherapy with anti-PD-L1 and anti-TIGIT: a promising new combination. J Immunother Cancer. 2019 doi: 10.1186/s40425-019-0634-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dubuisson A, Fahrner J, Goubet A, et al. Immunodynamics of explanted human tumors for immuno-oncology. EMBO Mol Med. 2021 doi: 10.15252/emmm.202012850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Voabil P, de Bruijn M, Roelofsen LM, et al. An ex vivo tumor fragment platform to dissect response to PD-1 blockade in cancer. Nat Med. 2021;27:1250–1261. doi: 10.1038/s41591-021-01398-3. [DOI] [PubMed] [Google Scholar]
- 11.Qian J, Olbrecht S, Boeckx B, et al. A pan-cancer blueprint of the heterogeneous tumor microenvironment revealed by single-cell profiling. Cell Res. 2020;30:745–762. doi: 10.1038/s41422-020-0355-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stuart T, Butler A, Hoffman P, et al. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wan Y-W, Allen GI, Liu Z. TCGA2STAT: simple TCGA data access for integrated statistical analysis in R. Bioinformatics. 2016;32:952–954. doi: 10.1093/bioinformatics/btv677. [DOI] [PubMed] [Google Scholar]
- 14.Antoniotti M, Caravagna G, Sano LD et al (2021) TRONCO: TRONCO, an R package for TRanslational ONCOlogy. Bioconductor version: Release (3.12)
- 15.Eide PW, Bruun J, Lothe RA, Sveen A. CMScaller: an R package for consensus molecular subtyping of colorectal cancer pre-clinical models. Sci Rep. 2017 doi: 10.1038/s41598-017-16747-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chalabi M, Fanchi LF, Dijkstra KK, et al. Neoadjuvant immunotherapy leads to pathological responses in MMR-proficient and MMR-deficient early-stage colon cancers. Nat Med. 2020;26:566–576. doi: 10.1038/s41591-020-0805-8. [DOI] [PubMed] [Google Scholar]
- 17.Somaiah N, Conley AP, Lin HY, et al. A phase II multi-arm study of durvalumab and tremelimumab for advanced or metastatic sarcomas. JCO. 2020;38:11509–11509. doi: 10.1200/JCO.2020.38.15_suppl.11509. [DOI] [Google Scholar]
- 18.Yu X, Harden K, Gonzalez LC, et al. The surface protein TIGIT suppresses T cell activation by promoting the generation of mature immunoregulatory dendritic cells. Nat Immunol. 2009;10:48–57. doi: 10.1038/ni.1674. [DOI] [PubMed] [Google Scholar]
- 19.Manieri NA, Chiang EY, Grogan JL. TIGIT: a key inhibitor of the cancer immunity cycle. Trends Immunol. 2017;38:20–28. doi: 10.1016/j.it.2016.10.002. [DOI] [PubMed] [Google Scholar]
- 20.Joller N, Hafler JP, Brynedal B, et al. TIGIT has T cell intrinsic inhibitory functions. J Immunol. 2011;186:1338–1342. doi: 10.4049/jimmunol.1003081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stanietsky N, Simic H, Arapovic J, et al. The interaction of TIGIT with PVR and PVRL2 inhibits human NK cell cytotoxicity. PNAS. 2009;106:17858–17863. doi: 10.1073/pnas.0903474106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yeo J, Ko M, Lee D-H, et al. TIGIT/CD226 axis regulates anti-tumor immunity. Pharmaceuticals (Basel) 2021 doi: 10.3390/ph14030200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen F, Xu Y, Chen Y, Shan S. TIGIT enhances CD4+ regulatory T-cell response and mediates immune suppression in a murine ovarian cancer model. Cancer Med. 2020;9:3584–3591. doi: 10.1002/cam4.2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guillerey C, Harjunpää H, Carrié N, et al. TIGIT immune checkpoint blockade restores CD8+ T-cell immunity against multiple myeloma. Blood. 2018;132:1689–1694. doi: 10.1182/blood-2018-01-825265. [DOI] [PubMed] [Google Scholar]
- 25.Zhou X-M, Li W-Q, Wu Y-H, et al. Intrinsic expression of immune checkpoint molecule TIGIT could help tumor growth in vivo by suppressing the function of NK and CD8+ T cells. Front Immunol. 2018;9:2821. doi: 10.3389/fimmu.2018.02821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Phase 2 CITYSCAPE Trial Shows Promise for Tiragolumab in NSCLC. In: Cancer network. https://www.cancernetwork.com/view/phase-2-cityscape-trial-shows-promise-for-tiragolumab-in-nsclc. Accessed 22 Mar 2021
- 27.Kumar R, Yu F, Zhen Y-H, et al. PD-1 blockade restores impaired function of ex vivo expanded CD8+ T cells and enhances apoptosis in mismatch repair deficient EpCAM+PD-L1+ cancer cells. Onco Targets Ther. 2017;10:3453–3465. doi: 10.2147/OTT.S130131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pfannenstiel LW, Diaz-Montero CM, Tian YF, et al. Immune-checkpoint blockade opposes CD8+ T-cell suppression in human and murine cancer. Cancer Immunol Res. 2019;7:510–525. doi: 10.1158/2326-6066.CIR-18-0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Van den Eynde M, Mlecnik B, Bindea G, et al. The link between the multiverse of immune microenvironments in metastases and the survival of colorectal cancer patients. Cancer Cell. 2018;34:1012–1026.e3. doi: 10.1016/j.ccell.2018.11.003. [DOI] [PubMed] [Google Scholar]
- 30.Halama N, Spille A, Lerchl T, et al. Hepatic metastases of colorectal cancer are rather homogeneous but differ from primary lesions in terms of immune cell infiltration. OncoImmunology. 2013;2:e24116. doi: 10.4161/onci.24116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jakubowska K, Koda M, Kisielewski W, et al. Tumor-infiltrating lymphocytes in primary tumors of colorectal cancer and their metastases. Exp Ther Med. 2019 doi: 10.3892/etm.2019.8146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller BC, Sen DR, Al Abosy R, et al. Subsets of exhausted CD8+ T cells differentially mediate tumor control and respond to checkpoint blockade. Nat Immunol. 2019;20:326–336. doi: 10.1038/s41590-019-0312-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blake SJ, Stannard K, Liu J, et al. Suppression of metastases using a new lymphocyte checkpoint target for cancer immunotherapy. Cancer Discov. 2016;6:446–459. doi: 10.1158/2159-8290.CD-15-0944. [DOI] [PubMed] [Google Scholar]
- 34.Chan CJ, Martinet L, Gilfillan S, et al. The receptors CD96 and CD226 oppose each other in the regulation of natural killer cell functions. Nat Immunol. 2014;15:431–438. doi: 10.1038/ni.2850. [DOI] [PubMed] [Google Scholar]
- 35.Hung AL, Maxwell R, Theodros D, et al. TIGIT and PD-1 dual checkpoint blockade enhances antitumor immunity and survival in GBM. Oncoimmunology. 2018 doi: 10.1080/2162402X.2018.1466769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meyer D, Seth S, Albrecht J, et al. CD96 interaction with CD155 via its first Ig-like domain is modulated by alternative splicing or mutations in distal Ig-like domains. J Biol Chem. 2009;284:2235–2244. doi: 10.1074/jbc.M807698200. [DOI] [PubMed] [Google Scholar]
- 37.Chiang EY, de Almeida PE, de Nagata DE, et al. CD96 functions as a co-stimulatory receptor to enhance CD8+ T cell activation and effector responses. Eur J Immunol. 2020;50:891–902. doi: 10.1002/eji.201948405. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary file2Supplementary figure 1: Gating strategy for “Lymphoid cell analysis” panel. The gating strategy was done on a tumour cell suspension sample and is performed in the same way on blood samples. The expression of CD226, TIGIT and PD1 was studied in the same way as on CD8 T cells on NK cells. Expression of naive/memory subpopulations dependent on CD45RA and CCR7 expression are studied in the same way as CD8 T cells on CD4 T cells, as well as the expression of CD226, TIGIT and PD1. The expression of CD96 is positive for all CD4 and CD8 T cells, so we analyzed the median intensity fluorenscence of this marker on our populations of interest. Supplementary figure 2: Gating strategy for “Myeloid cell analysis” panel. The gating strategy was done on a blood sample and is performed in the same way on tumour samples. The expression of PD-L1, CD111, CD112 and CD155 markers was studied in each of the following populations: granulocytes (CD15+), macrophages (CD14+ CD163+), monocytic MDSCs (CD14+ CD15- CD163- HLA-DRlow) and granulocytic MDSCs (CD14+ CD15+). Supplementary figure 3: Gating strategy for “Lymphocyte function analysis” panel. The gating strategy is developed on the CD3+ CD45- blood cell population. The same gating strategy was applied to the CD3+ CD45+ population corresponding to TILs. TILs was pre-labeled with an anti-CD45 antibody so that blood and TILs could be analyzed in the same tube. For the study of cytokine expression, we first analysed the expression of TNFα and IFNγ on conventional CD4 T cells (T CD4conv), regulatory CD4 T cells (Treg), conventional CD8 T cells (T CD8conv) and regulatory CD8 T cells (T CD8reg) and then the expression of Granzyme B and IL-2 on each subpopulation. Secondly, we analysed within the CD4conv T cells each of the following subpopulations: Th1 (CD4+ Foxp3- IFNγ+ IL-17A-), Th17 (CD4+ Foxp3- IFNγ- IL-17A+), Th1/17 (CD4+ Foxp3- IFNγ+ IL-17A+) and Th2 (CD4+ Foxp3- IFNγ- IL-17A- IL-4+) cells. We performed the same analysis within CD8conv T cells: Tc1, Tc17, Tc1/17 and Tc2 cells. For each of these populations, we studied the expression of the cytokines TNFα and IL-2 as shown for Th1 in this gating strategy. Supplementary figure 4: Gating strategy for “Ex vivo restimulation analysis”. Supplementary figure 5: Double blockade of PD-L1 and TIGIT restore tumor infiltration lymphocytes function in some cancer patients with MSS CRC. Tumor cell suspensions from 16 patients with cancer colorectal cancers were thawed and after removing dead cells were labelled with 1µM of CellTrace Violet. Then, the cells were plated at 5.105 cells per well and stimulated with anti-CD3 antibody (10 ng/mL) in the presence or absence of 10 µg/mL of anti-PD-L1 (clone 6E11) and/or anti-TIGIT (clone 10A7) during 5 days. Cells were also stimulated with 5 µg/mL of anti-CD3 and anti-CD28 as positive control experiments. 20 hours before the end of the culture, Brefeldin A was added to block transport processes during cell activation and allow the staining of intracellular cytokines. After 5 days of culture, to study the effect of immune checkpoints blockade antibodies on the proliferation and cytotoxic capacities of TILs, tumor cell suspensions were stained with anti-CD3, anti-CD4, anti-CD8, anti-IFNγ and anti-TNFα and analysed by flow cytometry. A. The percentage of TNFα expression in CD4 (left) and CD8 (right) is studied for the following 3 conditions: anti-CD3, anti-CD3 plus anti-PD-L1 and anti-CD3 plus anti-PD-L1 + anti-TIGIT. Patients classified as responders to double immunotherapy are shown in colour. B. The percentage of IFNγ expression in CD4 (left) and CD8 (right) comparing the anti-CD3 plus anti-TIGIT condition to anti-CD3 alone is depicted. Supplementary figure 6: Description of the cell composition in the blood and tumour of patients. Fresh tumor tissues and associated blood samples for each patient (n=16) were stained and then analysed by flow cytometry. A. Box plots showing the frequency of CD45+ and CD45- populations in patients’ blood and tumor. B. Box plots showing the frequency of CD3+, CD4+ and CD8+ T cells, NK and NKT cells, CD11b+; CD14+, CD15+ cells and gMDSC population in patients’ blood and tumor. Supplementary figure 7: Association between baseline lymphoid cell number and response to comboimmunotherapy. Fresh tumor tissues and associated blood samples for each patient were taken on the day of surgery. After tumor dissociation, 1.106 tumor cell suspension were stained with anti-CD45, resuspended in 50 µL of whole blood sample and then transferred to a tube containing PMA, Ionomycin and Brefeldin A for 3 hours. After activation, cells were fixed, permeabilized and stained with anti-IFNγ, anti-TNFα, anti-GranzymeB, anti-IL-4, anti-Foxp3, anti-IL-17A, anti-CD3, anti-CD4, anti-CD8 and anti-IL-2 antibodies and analysed by flow cytometry. A-B. Box plots showing the different combinations of expression of the 4 cytokines IFNγ, TNFα, Granzyme B (GrB) and IL-2 in CD4 (CD45- CD3+ CD4+) (A) and CD8 (CD45- CD3+ CD8+) (B) T cells in blood patients’ depending on the responder (R) versus non responder (NR) status. C. The frequency of Granzyme B (GrzB) expression according to responder (R) or non-responder status is depicted in tumor samples. Statistical difference is determined by a Mann–Whitney test. ns = no significant, * = p< 0.05 and ** = p< 0.01. Supplementary figure 8: Association between baseline nectin expression and response to comboimmunotherapy. Fresh tumor tissues and associated blood samples for each patient were taken on the day of surgery. After tumor dissociation, 1.106 tumor cell suspension and 100 µL of whole blood sample were stained with anti-DNAM-1 (CD226), anti-CD56, anti-CD96, anti-CD45, anti-TIGIT, anti-PD-1, anti-CD3, anti-CD45RA, anti-CD8, anti-CCR7, anti-Tim-3 and anti-CD4 antibodies and with a mortality marker DRAQ7 and then analysed by flow cytometry. A-B. Box plots showing the frequency of CD226+, CD226-, TIGIT+ and TIGIT- populations in the blood (A) and tumor (B) samples by comparing expression on CD4, CD8 and NK populations. C-D. Box plots showing the different combinations of expression of the 2 nectins CD226 and TIGIT within CD4 T cells and CD8 T cells in patient blood (C) and tumor (D) according to responder (R) or non responder (NR) status. Statistical difference is determined by a Mann–Whitney test. ns = no significant and * = p< 0.05, ** = p<0.01, *** = p<0.001 and **** = p<0.0001. Supplementary figure 9: Association between baseline nectin expression and response to comboimmunotherapy. Box plots showing the median fluorescence intensity (MFI) of CD96 marker in CD4 T cells and CD8 T cells in patient blood depending on the responder (R) or non responder (NR) status. Statistical difference is determined by a Mann–Whitney test. ns = no significant and * = p< 0.05. Supplementary figure 10: Association between baseline myeloid cell number and response to comboimmunotherapy. Fresh tumor tissues and associated blood samples for each patient were taken on the day of surgery. After tumor dissociation, 1.106 tumor cell suspension and 100 µL of whole blood sample were stained with anti-CD11b, anti-HLA-DR, anti-CD111, anti-CD155, anti-CD112, anti-PD-L1, anti-CD163, anti-CD15, anti-CD14, anti-CD3, anti-CD19, anti-CD20, anti-CD56, anti-Galectin-9 and anti-CD45 antibodies and with a mortality marker DRAQ7 and then analysed by flow cytometry. The frequency of CD111, CD112 and CD155 positive populations in macrophages (CD14+ CD163+), mMDSC (CD14+ CD163- HLA-DRlow), gMDSC (CD15+ CD14+) and granulocytes (CD15+) in tumor samples according to responder (R) or non-responder (NR) status is depicted. Statistical difference is determined by a Mann–Whitney test. ns = no significant and * = p< 0.05. Supplementary figure 11: CD96 expression in lymphoid cells characterized a particular CRC subtype. A-B. Kaplan-Meier curves for overall survival with patients stratified according to the level of CD96 RNAseq expression, respectively for patients with high (A) or low (B) level of the metagene. (PPTX 3848 kb)