Abstract
RNA therapeutics are an emerging, powerful class of drugs with potential applications in a wide range of disorders. A central challenge in their development is the lack of clear pharmacokinetic (PK)-pharmacodynamic relationship, in part due to the significant delay between the kinetics of RNA delivery and the onset of pharmacologic response. To bridge this gap, we have developed a physiologically based PK/pharmacodynamic model for systemically administered mRNA-containing lipid nanoparticles (LNPs) in mice. This model accounts for the physiologic determinants of mRNA delivery, active targeting in the vasculature, and differential transgene expression based on nanoparticle coating. The model was able to well-characterize the blood and tissue PKs of LNPs, as well as the kinetics of tissue luciferase expression measured by ex vivo activity in organ homogenates and bioluminescence imaging in intact organs. The predictive capabilities of the model were validated using a formulation targeted to intercellular adhesion molecule-1 and the model predicted nanoparticle delivery and luciferase expression within a 2-fold error for all organs. This modeling platform represents an initial strategy that can be expanded upon and utilized to predict the in vivo behavior of RNA-containing LNPs developed for an array of conditions and across species.
Keywords: MT: Delivery Strategies, mRNA, lipid nanoparticles, nucleic acid delivery, drug targeting, pharmacokinetics, pharmacodynamics, physiologically based pharmacokinetics
Graphical abstract

Glassman and colleagues have evaluated how the spatiotemporal disconnect between LNP delivery and transgene expression poses challenges in prediction of PK/PD relationships. A physiologically based PK/PD model was developed to bridge this gap, which was used to predict the in vivo behavior of both untargeted and targeted LNPs.
Introduction
RNA-based therapeutics have gained prominence in recent years, including regulatory approvals of two small interfering RNA drugs—patisiran and givosiran1,2—and U.S. Food and Drug Administration approvals for two mRNA-based vaccines against coronavirus disease 2019 (COVID-19).3,4 As use of these next-generation therapeutics expands in clinical pharmacotherapy, it will be increasingly important to develop quantifiable, mechanism-based relationships between pharmacokinetics (PK) and pharmacodynamics (PD). This will facilitate dose selection for obtaining the desired efficacy-safety profile in the target patient population. Often, RNA is encapsulated within lipid nanoparticles (LNPs) for delivery, as in the COVID-19 vaccines and patisiran. Therefore, a thorough understanding of LNP biodistribution and PK would have utility in (1) characterizing distribution of nucleic acid therapeutics encapsulated in LNP and (2) anticipating off-target toxicities related to high local concentrations of components of the delivery system. Determining the PK/PD parameters in healthy organisms is the first, basal phase that should be followed by systematic studies recapitulating these parameters in animal models of human pathology intended to be managed by specific formulations.
A challenge in the field of gene delivery is the spatiotemporal disconnect between level of tissue uptake of the delivery vehicle PK/biodistribution and transgene expression (often reported by luciferase activity).5 In many cases, transgene expression is the sole readout provided to describe distribution of mRNA-LNP in the body. While useful for characterizing the functional activity of the cargo RNA, this metric does not necessarily reflect the biodistribution of the formulation, as different tissues and cell types will likely express deliver nucleic acids with different efficiencies, due to local changes in endocytosis, endosomal escape, translation machinery, nucleic acid degradation, and transgene-encoded protein stability. Further, approaches based on detection of activity of the transgene or other reporter signal in vivo do not provide readouts in blood. Therefore, the key information on the PK is missing.
It is appreciated that conjugation of affinity ligands (e.g., monoclonal antibodies [mAbs] and fragments) toward a cell surface molecule of interest to a drug delivery system can enhance accumulation in a desired tissue, cell, or subcellular compartment.6 However, it is less clear whether these changes in cellular and subcellular addressing of targeted LNPs may also lead to changes in processing and expression of the transgene. In this study, we compare LNPs coated with immunoglobulins with differing specificities (Table S1) to evaluate the role of active targeting, not only on particle biodistribution, but also on transgene expression. It is only within the last few years that antibody targeting has begun to be evaluated for mRNA-LNP delivery.7,8,9,10 Mechanism-based modeling, coupled with well-designed experiments, may be uniquely positioned to provide insights into these so far unanswered questions and bridge the gap of knowledge.
Physiologically based PK (PBPK) models are a gold standard approach for prediction of blood and tissue PK of therapeutic agents. While these models are regularly used in clinical development of small molecule drugs11,12 and have been gaining prominence in development of biologics,13 there is a relative paucity of PBPK models for nanoparticle-based drug delivery systems. PBPK models integrate knowledge of physiology (tissue volumes, blood flow rates, and organization of the circulatory system) with mechanistic determinants of drug disposition. The use of this framework will facilitate description of mechanistic determinants of LNP distribution and clearance and, due to its basis in physiology, will provide an optimal platform for extrapolating results obtained in preclinical species to predict PK in patient populations.
In this report, we have developed and validated a PBPK model to characterize the delivery of a targeted mRNA delivery system. This model incorporated mechanistic determinants of LNP tissue uptake, clearance, and transgene expression. Critically, the PBPK framework was able to simultaneously characterize blood and tissue PK and the kinetics of transgene expression, despite a significant disconnect between these two processes. Simulations with this model were able to predict the behavior of an LNP targeted to a distinct target from that used for model development, demonstrating the overall utility of this modeling framework. The proposed model structure is expected to have utility in development of future LNP therapeutics.
Results
Nanoparticle properties
We have previously reported a detailed description of the physicochemical properties of LNP used in this study.5 Briefly, conjugation of IgG or mAb to LNP led to an increase in size from 82.5 to 100 nm and polydispersity index (0.06–0.2). Conjugation also resulted in modest changes in zeta potential (−6.5 to −4.1 mV) and approximately 80 antibody molecules/LNP.
Bioluminescence imaging of intact organs: Bare vs. IgG LNPs
Studies were carried out to compare the kinetics of luciferase activity following administration of bare and IgG-coated LNPs. This was necessary, as LNPs rapidly shed their pegylated (PEG) lipid after in vivo administration due to the short length of the lipids.14 The PEG lipid used for conjugation of the targeting ligand has a longer carbon tail and remains associated with the LNP. Similar to luciferase activity in organ homogenates, bioluminescence imaging (BLI) of intact organs showed maximal luciferase activity in liver, with peak expression at 4.5 h after injection of LNPs (Figure S1). Because units of activity and the kinetics of signal decay were distinct between BLI of intact organs and luciferase activity in organ homogenates, individual parameter sets were estimated for each assay.
PK: Untargeted LNPs
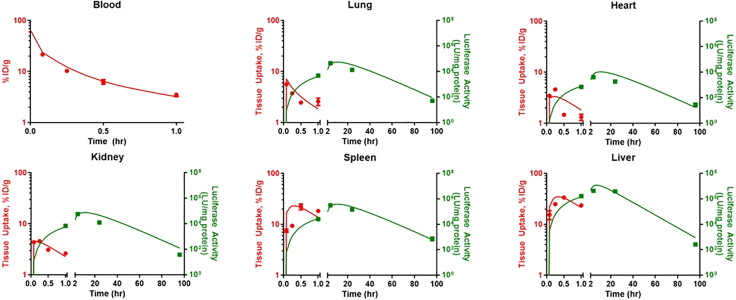
The PBPK model (Figure 1) was fit to blood and tissue concentration vs. time data for both bare and IgG-coated LNP to obtain parameter estimates for tissue-specific uptake clearances (CLups) and for the LNP degradation rate (kdeg). For both types of particles, the model was able to well-characterize the observed data (Figures 2 and 3; Table S2) and all parameters were estimated with good confidence (Table 1).
Figure 1.
Structure of the PBPK model of LNPs
(A) Whole body structure of the PBPK model. Venous blood flow is depicted in blue, arterial blood in red. The relative thickness of arrows to explicitly depicted organs is representative of relative blood flow. (B) Tissue-level model structure. LNPs in the tissue vasculature are able to enter the extravascular space either by non-specific uptake (CLup) or via binding to target (KD), followed by internalization (kint). Internalized LNPs were assumed to be eliminated in a first-order process (kdeg). (C) Luciferase expression model. LNP degradation was assumed to drive the release of mRNA into the cytosol. mRNA elimination was assumed to be via a first-order process (kmRNA). Intact mRNA was assumed to drive production of transgene in a tissue-specific manner (Stissue) and luciferase signal was assumed to decay in a first-order manner (kluc).
Figure 2.
Model-estimated blood and tissue PK and kinetics of luciferase activity (luciferase assay in organ homogenates) following administration of bare LNPs
Symbols represent observed data and lines represent model fitted profiles. PK measurements are depicted in red and luciferase activity is depicted in green. Data depicted as mean ± SEM. n = 3 mice/time point.
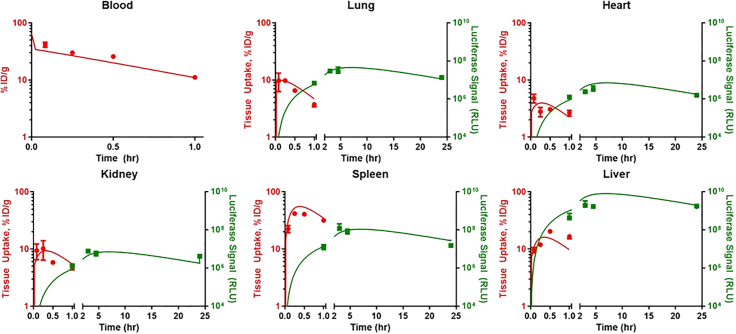
Figure 3.
Model-estimated blood and tissue PK and kinetics of luciferase activity (BLI of intact organs) following administration of control IgG-LNPs
Symbols represent observed data and lines represent model fitted profiles. PK measurements are depicted in red and luciferase activity is depicted in green. Data depicted as mean ± SEM. n = 3 mice/time point.
Table 1.
Estimated PK parameters for untargeted LNPs
| Parameter | Bare LNPsa | IgG LNPsa |
|---|---|---|
| CLlung (mL/h) | 0.134 (16.0%) | 0.272 (15.8%) |
| CLheart (mL/h) | 0.0999 (12.3%) | 0.0976 (14.1%) |
| CLkidney (mL/h) | 0.428 (12.7%) | 0.836 (12.8%) |
| CLspleen (mL/h) | 0.656 (10.4%) | 1.41 (11.5%) |
| CLliver (mL/h) | 16.3 (9.66%) | 5.64 (13.1%) |
| CLblood (mL/h) | 1.96 (19.1%) | 5.32 (19.6%) |
| kdeg (h−1) | 1.00 (6.38%) | 1.21 (8.18%) |
CLorgan, organ uptake clearance; kdeg, intracellular first-order LNP kdeg constant.
Parameter estimates represented as mean (% coefficient of variation, calculated as relative standard error).
Luciferase activity: Bare LNPs
Recognizing that there are two major approaches used to measure luciferase in animals (luciferase activity in organ homogenates, BLI of intact organs), we elected to fit the model to luciferase expression data obtained via both types of assay. The model was able to well-characterize both the kinetics and amplitude of luciferase activity, regardless of assay format used for detection (Figures 2 and S2; Tables 2 and S2). These results demonstrate the flexibility of this model structure in characterizing transgene expression data. Additionally, the ability of the model to effectively capture the temporal disconnect between PK and luciferase expression highlights the utility of PBPK modeling in gaining a mechanistic understanding of the in vivo performance of gene delivery strategies.
Table 2.
Estimated luciferase expression parameters
| Parameter | Bare LNPsa | Bare LNPsa | IgG LNPsa | PECAM LNPsa |
|---|---|---|---|---|
| Assay format | Homogenateb | BLIb | BLIb | Homogenateb |
| Slung (Signal/h) | 2.57 × 105 (12.2%) | 2.05 × 109 (5.53%) | 1.31 × 108 (9.67%) | 1.27 × 105 (7.08%) |
| Sheart (Signal/h) | 6.70 × 104 (10.6%) | 2.04 × 108 (5.53%) | 5.73 × 107 (10.5%) | 2.68 × 104 (8.78%) |
| Skidney (Signal/h) | 1.12 × 105 (10.5%) | 1.40 × 108 (5.53%) | 6.74 × 106 (9.64%) | 3.49 × 104 (11.1%) |
| Sspleen (Signal/h) | 3.82 × 105 (9.07%) | 3.27 × 108 (5.53%) | 6.74 × 107 (9.88%) | 4.53 × 105 (9.47%) |
| Sliver (Signal/h) | 8.64 × 105 (8.74%) | 1.77 × 109 (7.02%) | 1.20 × 109 (7.66%) | 3.40 × 104 (24.6%) |
| kluc (h−1) | 0.0940 (1.20%) | 0.250 (4.25%) | 0.250 (Fixed) | 0.0940 (Fixed) |
| kluc, liver (h−1) | 0.247 (7.02%) | 0.258 (8.57%) | 0.258 (Fixed) | 0.224 (Fixed) |
Sorgan, slope relating intracellular mRNA to the production rate of luciferase; kluc, first-order rate constant describing the loss of luciferase signal.
Parameter estimates represented as mean (% coefficient of variation calculated as relative standard error).
Homogenate represents ‘luciferase activity in organ homogenates’ and BLI represents ‘BLI in intact organs.’
Luciferase activity: Control IgG-LNPs
Data were obtained via imaging for control IgG-LNP that were then used to fit the model to evaluate any differences between bare and control IgG-IgG LNP with respect to transgene expression. Loss of luciferase signal was assumed to occur at the same rate as following administration of bare LNP (e.g., decay of signal is independent of LNP formulation). The model was able to well characterize the observed data (Figure 3; Table S2) and all parameters were estimated with good confidence (Table 1). Model-estimated parameters for production of luciferase signal were 3- to 17-fold lower than those estimated for bare LNPs in a similar assay (BLI of intact organs) (Figure S2). This suggests that conjugation of immunoglobulins via 1,2-distearoyl-sn-glycero-3-phosphoethanolamine (DSPE)-PEG to the surface of LNPs, even in the absence of active targeting, reduces the efficiency of transgene expression. These results highlight the need for direct studies into the mechanism of this observation, considering mechanisms, such as (1) inhibition of endosomal escape of mRNA, (2) shielding the LNP surface from opsonization by serum proteins that could enhance optimal gene delivery, (3) interactions with plasma components that may fortuitously alter LNP behavior and tissue uptake (e.g., Apo-E), and/or (4) cognizant redirection of LNPs to different cell types enabled by conjugation of PEG and affinity ligands providing targeting.
In vivo behavior: Platelet endothelial cell adhesion molecule-targeted LNPs
Data for blood and tissue PK and luciferase expression following platelet endothelial cell adhesion molecule (PECAM)-targeted LNP administration were used to evaluate the PBPK framework for targeted LNP delivery. PK data were used to obtain estimates of the concentration of PECAM accessible to LNP in tissue vasculature, as well as binding affinity and internalization rate. The final model provided good agreement between observed and model-fitted data (Figure 4; Table S2), with a minimal number of estimated parameters (Table 3). To evaluate whether targeting LNPs to a specific epitope had an impact on transgene expression, a comparison was made between a priori prediction of luciferase expression kinetics using values obtained for IgG-coated LNPs and estimating unique parameters for PECAM-targeted LNPs. In all organs except for liver, a priori simulations significantly underpredicted luciferase activity at all time points (Figure 4), indicating that specific delivery to endothelial cells via PECAM binding led to enhanced mRNA delivery and translation. In contrast, parameter estimates obtained by fitting to the observed data (Table 2) allowed for good characterization of the observed luciferase kinetics (Figure 4). In sum, these results suggest that PECAM targeting not only enhances particle accumulation in target tissues (PK), but also leads to a greater than anticipated increase in transgene expression in target tissues relative to IgG-coated particles (luciferase expression).
Figure 4.
Model-fitted PK and luciferase expression (luciferase activity in organ homogenates) following administration of PECAM-targeted LNPs
PK measurements are depicted in red and luciferase activity is depicted in green. Solid red lines represent model-fitted PK profiles and solid green lines represent model-fitted luciferase expression. Data depicted as mean ± SEM. n = 3 mice/time point.
Table 3.
Estimated PK parameters for PECAM-targeted LNPs
| Parameter | Estimatea |
|---|---|
| PECAMlung (μg/mL) | 16.4 (7.83%) |
| PECAMheart (μg/mL) | 1.21 (14.8%) |
| PECAMkidney (μg/mL) | 2.56 (10.4%) |
| PECAMspleen (μg/mL) | 2.98 (12.7%) |
| PECAMliver (μg/mL) | 15.2 (2.90%) |
| CLPECAM (mL/h) | 0.366 (14.9%) |
| KD (μg/mL) | 0.00152 (51.3%) |
PECAMorgan, organ-specific binding capacity for PECAM-targeted LNPs; kPECAM, first-order internalization rate of PECAM; KD, equilibrium dissociation constant between PECAM and PECAM-targeted LNPs.
Parameter estimates represented as mean (% coefficient of variation calculated as relative standard error).
Model validation: Intracellular adhesion molecule-targeted LNPs
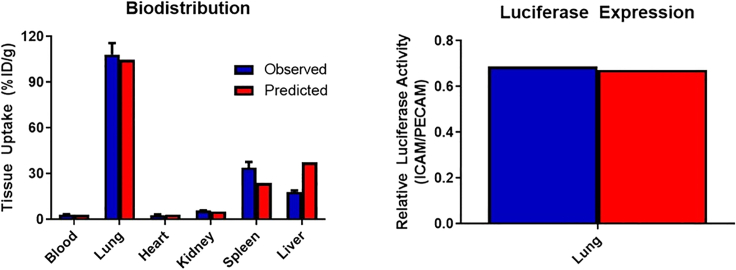
As the ultimate goal of PBPK model building is to develop a predictive framework, we performed simulations, with no additional fitting of parameters, to describe the biodistribution and luciferase expression at a single time point following the administration of intracellular adhesion molecule (ICAM)-targeted LNPs. To account for differences between ICAM and PECAM expression, relative estimates of accessible target were obtained from our recent publication.15 These values were then scaled to relevant values for LNPs based on the estimate of LNP-accessible PECAM obtained in this study. All other parameters were fixed to those values used for PECAM-targeted LNPs. Both tissue biodistribution and luciferase expression were well-predicted by the model (Figure 5). Predicted values for biodistribution at 30 min were all within 2-fold of observed values (mean percent prediction error [%PE], 37.0%), with the best predictions being in kidney, lung, and heart. Data for luciferase expression was expressed relative to PECAM-targeted LNP due to inter-assay variability in absolute signal obtained from the luciferase assay. Simulation of luciferase expression in the lung was in good agreement with observed data (%PE, 17.7%).
Figure 5.
Model predicted biodistribution (30 min after injection) and luciferase activity in organ homogenates (5 h after injection) of ICAM-targeted LNPs
Predictions were made with no fitting of parameters and were able to describe both biodistribution and luciferase expression well. Data depicted as mean ± SEM. n = 3 mice/time point.
Discussion
LNP-mediated delivery of mRNA requires passage across many, not insignificant, barriers, including (1) delivery to the target tissue, (2) escape from the bloodstream for extravascular targets, (3) endocytosis by recipient cells, (4) endosomal escape of intact mRNA, and (5) translation of the message into protein. Engineering of the mRNA, nanoparticle, and nanoparticle coating creates a matrix of possible strategies to achieve optimal gene expression. Although discussion of specific engineering strategies for LNPs and/or targeting are beyond the scope of this paper, they are described in.16 Thorough studies of the kinetics of both delivery system in the blood and tissue distribution and transgene expression are necessary to identify steps where a given strategy has succeeded or failed. This permits the design of focused, hypothesis-driven experiments to optimize a delivery strategy.
Several approaches can be utilized to trace LNP biodistribution and transgene expression, each with their own strengths and weaknesses.17 Here, we utilized radioisotope labeling of the LNP itself to measure PK and tissue distribution. Unlike other methods that require labeling of the particle, radiotracing has no tissue background or permeation issues and features an unparalleled linear dynamic range; however, its level of spatial resolution does not afford cellular attribution of the signal. It is critical to consider that any form of particle labeling (radioisotope, fluorophore) may affect the particle properties and, therefore, biodistribution. In this study, we compared mRNA-driven luciferase expression via two commonly used methods: luciferase activity in organ homogenates and BLI of intact organs. While the absolute values obtained differ between these two methods, the expression patterns provided by the methods are comparable. However, there is a significant difference in the decay of luciferase signal obtained by the two methods. The modeling approach described here estimates that signal measured by BLI of intact organs decays approximately 2.5 times faster than that measured by luciferase activity in organ homogenates. This suggests that, while the results obtained from the two methods are relatively interchangeable at static time points, the kinetics of luciferase activity are likely to differ significantly. This is critical information for those pursuing model-based analyses of luciferase activity following mRNA delivery, as parameters related to expression kinetics will have to be estimated independently for different assay formats.
It should be noted that the method of tracing of LNP PK/biodistribution may impact the absolute magnitude of tissue uptake that is measured during these studies due to several factors, including signal interference due to tissue matrix, stability of the label for tracing on the LNP itself, and processing of the labeled component of the LNP following particle breakdown. Methods for tracing the LNP that have been reported in the literature include pre-labeling of specific components of the particle with radioisotopes (3H-cholesteryl hexadecyl ether, 14C phospholipid),18,19,20,21 radio-iodination of pre-formed particles (Parhiz et al. JCR. 2018),5,7 and liquid chromatography tandem mass spectrometry analysis for specific lipid components.22 With this in mind, the parameter values estimated with our PBPK model framework are likely unique to the tracing methodology used in this manuscript (radio-iodination of pre-formed particles). A comparison of the PK and biodistribution of PECAM-targeted LNPs when labeled with 125I vs. 3H is shown in Figure S3 to highlight the potential impact of labeling strategy on parameter estimates. However, the underlying assumptions and underlying physiological assumptions should transfer between labeling approaches. Therefore, we would stress the need for model validation and parameter re-estimation for alternative tracing methods. The need for this approach has been previously shown for mAbs labeled with isotopes that have different degrees of tissue retention.23 Future studies will be focused on the impact of LNP tracing methodology on PBPK model parameters. In this case, our expectation would be that both 125I and 3H would be delivered to tissues to a similar extent; however, their behavior would differ following LNP breakdown inside of cells. The hypothesis would be that 125I would not efficiently residualize within tissues and would be rapidly eliminated following LNP breakdown (as is the case with antibodies) and the 3H incorporated into cholesteryl hexadecyl ether would follow the disposition pattern of the cholesterol ether, which would likely involve hepatic accumulation, as this is the primary site of cholesterol elimination and processing. Additionally, normalization to the injected dose, rather than the amount of LNP found in the body at the first time point is critical. In our results, the percent of injected dose that was recovered in collected organs ranged from 54.6% (bare) to 84.2% (control IgG), with PECAM-targeted falling in the middle (72.4%). These differences may be attributed to rapid uptake and elimination of bare LNPs as well as potential uptake by organs that were not collected in the study. Highlighting these differences in recovery, even just 5 min after an intravenous injection, provides an impetus to ensure that the injected dose is well controlled and that concentrations are reported either as absolute values (e.g., mass mRNA/volume) or as a fraction of the injected dose.
Mechanism-based models, spanning a range of scales, from sub-cellular to whole body, are uniquely positioned to not only describe the behavior of current formulations, but also to serve as a platform to generate testable hypotheses about the properties of next-generation formulations. Use of a physiologically relevant framework to describe LNP PK allowed investigation of tissue-specific differences in LNP behavior (binding, uptake, and transgene expression). In the current work, we leveraged prior knowledge of tissue expression patterns of ICAMs vs. PECAMs and used that information to make accurate, a priori predictions of particle biodistribution and transgene expression in the primary target tissue. Critically, the model estimated that were substantial differences in target expression across organs of the body, likely related to differences in endothelial surface area between organs. This relationship has been previously measured experimentally in mice, where a direct correlation between PECAM expression (μg PECAM/g tissue) and endothelial surface area (cm2/g tissue) was observed in a subset of organs.24 These results provide a proof of principle for the use of this approach for prediction of LNPs directed against other targets and for scaling from mice to higher species.
A challenge in developing a quantitative framework linking LNP PK and transgene expression is the significant disconnect between the kinetics of tissue uptake (within minutes) and peak expression (hours). One approach that would be capable of describing this lag would be a signal transduction model; however, these models are black boxes that do not explicitly describe specific mechanisms. Nonetheless, they have been used with great success to describe the PD of drugs affecting gene transcription, such as corticosteroids.25,26 To mechanistically account for processes related to intracellular processing of LNPs and mRNA, the model described here includes the following steps and assumptions: (1) breakdown of LNPs (loss of 125I signal) is correlated with endosomal escape of mRNA, assuming that loss of 125I from tissue would be the slowest possible rate of mRNA release, (2) mRNA breakdown in tissues occurs at a similar rate to estimates from in vitro assays, and (3) intact mRNA drives expression of the transgene. This relatively simple set of assumptions was able to accurately characterize the kinetics of luciferase expression following administration of LNP. Efforts to improve the predictive capacity of this model (by minimizing the number of estimated parameters) could focus on characterizing processes such as the kinetics and efficiency of internalization and endosomal escape and the efficiency of modified mRNA translation by different cell types. Additionally, further studies of a matrix of LNP parameters (size, zeta potential, etc.) would provide useful insights into the effects of these parameters on PK and transgene expression.
In the present model, first-order processes were assumed for all parameters related to PK and transgene expression. This was largely due to our observation that in vivo luciferase expression in mice scaled linearly with administered dose in our prior work.5 It is likely that, at certain doses, both PK processes (e.g., receptor-mediated uptake, reticuloendothelial system (RES) uptake) and transgene expression (mRNA translation) will be capacity limited. As, in our studies, we have not achieved the point of saturation of any processes included in the model we utilized first-order processes in the interest of model parsimony. As always, model structure is driven by data, and if saturation is achieved in any process, requisite updates to the model would need to be made to account for the relevant process using functions typical in the field (e.g., Michaelis-Menten or Hill-type functions).
The cellular destination of LNPs can have a significant impact on the efficacy of gene delivery, due to differences in endo-lysosomal trafficking between cell types and internalization pathways.27,28,29 To achieve the desired tissue and cellular specificity in vivo, antibodies (and fragments) are often conjugated to the surface of nanoparticles. This strategy, which can enhance accumulation by orders of magnitude in the desired tissue, also has the potential to interfere with the endosomal escape of mRNA that was optimized for an uncoated particle. Considering the typical clearance organ for untargeted LNP, the liver, our modeling results lead to the hypothesis that, irrespective of changes in tissue biodistribution, insertion of DSPE-PEG into the LNP membrane, followed by conjugation to control IgG, leads to a decrease in the efficiency of luciferase signal production, as suggested by differences in estimated values of the rate constant describing luciferase production in the liver (Sliver), which drives production of luciferase signal in the model proportionally to the amount of LNP taken up into the organ. For example, in the liver there was an approximately 7-fold lower amount of luciferase expression for control IgG vs. bare LNPs; however, this was associated with an approximately 2-fold lower exposure of LNPs in the organ. The remainder of the difference in expression would be accounted for by differences in Sliver.
Another potential explanation for differences in the efficiency of transgene production between bare vs. control IgG vs. PECAM-targeted mRNA-LNPs lies in the intracellular processing of the LNPs. We would anticipate that each formulation tested here would display differences in endocytic routes and processing. For example, unmodified LNPs will rapidly shed their PEG coating following injection, leading to apolipoprotein E (ApoE)-dependent uptake in hepatocytes in a low-density lipoprotein receptor-dependent process. Our strategy for conjugated antibodies to the surface of LNPs involved introduction of a PEG lipid with a longer acyl chain that would not be shed upon injection. This would likely prevent ApoE adsorption and would promote Fc-dependent uptake of LNPs by other cell types, likely macrophages. Finally, coating with anti-PECAM will promote delivery to endothelial cells using CAM-mediated endocytosis. All three endocytic routes are associated with distinct intracellular kinetics and environments, likely resulting in differential endosomal escape patterns of mRNA. This is a likely factor underlying the differences in efficiency of luciferase production for different LNP formulations in this study. To further validate this hypothesis, detailed studies of the intra-organ distribution of LNPs would be required, namely, to identify the cell types responsible for the uptake of LNPs within a given organ and the relative distribution of LNPs within the given cell types.
Many mRNA delivery strategies do not rely on intravascular administration, but rather injection either intramuscularly or subcutaneously, particularly in the area of vaccine development. While there are reports of the kinetics of transgene expression following extravascular administration of LNPs,8 data are lacking on the PK and biodistribution of the LNP itself. Future iterations of this model will include more tissues, particularly those relevant to extravascular administration, with the goal of describing PK following multiple, clinically relevant, routes of administration. This will have utility, not only in characterizing the behavior of LNPs and transgenes for vaccination, but also in evaluating the potential of LNP-based mRNA delivery for systemic expression following dosing via more convenient routes of administration than intravenous. These models will build on the existing framework to account for local expression at the site of injection and for any systemic exposure of LNP. We anticipate that routes of absorption into the circulation would be similar to those observed for large biologics, namely, drainage from the tissue interstitium into the lymphatics and passage through lymph nodes before entry into the blood at the thoracic duct. A recent publication has attempted to study the behavior of LNP following intramuscular injection in mice revealed that it takes several hours for LNP to reach maximal concentrations in plasma and that the kinetics of transgene expression in non-muscle tissue was delayed (peak in muscle, 6 h; peak in liver, 10 h).30
In conclusion, we have developed and validated a PBPK model for LNP-based delivery of mRNA, which was used to describe the PK and kinetics of transgene expression for untargeted vs. targeted LNPs and for measurement of transgene expression via two complementary methods (luciferase activity in organ homogenates and BLI of intact organs). This model was able to accurately capture the temporal disconnect between tissue uptake (minutes) and transgene expression (hours) by describing processes related to mRNA turnover and transgene signal production in sequence. Model validation was performed by simulating the biodistribution and peak transgene signal following administration of LNP against ICAM. Future iterations of this model will characterize the behavior of LNP following alternate routes of administration, the kinetics of therapeutic transgene expression, and in pathological conditions.
Materials and Methods
Experimental
Ethics statement
We followed the “Guide for the Care and Use of Laboratory Animals” by the Committee on Care of Laboratory Animal Resources Commission on Life Sciences, National Research Council for animal studies performed in this study. The animal facilities at the University of Pennsylvania are fully accredited by the American Association for Accreditation of Laboratory Animal Care. All studies were conducted under protocols approved by the University of Pennsylvania Institutional Animal Care and Use Committee.
Reagents
Radioactive isotope 125I was purchased from PerkinElmer (Wellesley, MA, USA). Whole molecule rat IgG was from Thermo Fisher Scientific (Waltham, MA, USA). Anti-mouse-PECAM-1/CD31 monoclonal was obtained from BioLegend (San Diego, CA, USA). N-succinimidyl S-acetylthioacetate (SATA) was purchased from Pierce Biotechnology (Rockford, IL, USA). All chemical reagents were purchased from Sigma Aldrich, unless stated otherwise.
Preparation of antibody-conjugated LNP-mRNA
mRNAs were produced, as described previously,31 using T7 RNA polymerase (Megascript, Ambion) on linearized plasmids encoding codon-optimized firefly luciferase (pTEV-Luc2-A101). To make modified nucleoside-containing mRNA, m1ψ-5′-triphosphate (TriLink) was incorporated instead of UTP. mRNAs contained 101 nucleotide-long poly(A) tails. Capping of the in vitro transcribed mRNAs was performed co-transcriptionally using the trinucleotide cap1 analog, CleanCap (TriLink). mRNA was purified by cellulose purification, as described.32
mRNAs were then encapsulated in LNPs using a self-assembly process in which an aqueous solution of mRNA at a pH of 4.0 is rapidly mixed with a solution of lipids dissolved in ethanol.22 LNPs used in this study were similar in composition to those described previously.22,33
LNPs were conjugated with mAb specific for PECAM-1 (MEC13.3, rat IgG2a), ICAM-1 (YN1, rat IgG2b), or pooled rat IgG (plasma purified). The LNP construct was modified with maleimide functioning groups (DSPE-PEG-mal) by a post-insertion technique with minor modifications.5 Targeting antibodies or control isotype-matched IgG was functionalized with SATA (Sigma-Aldrich) to introduce sulfhydryl groups allowing conjugation to maleimide. SATA was deprotected using 0.5 M hydroxylamine followed by removal of the unreacted components by G-25 Sephadex Quick Spin Protein columns (Roche Applied Science, Indianapolis, IN, USA). The reactive sulfhydryl group on the antibody was then conjugated to maleimide moieties using thioether conjugation chemistry.34 Purification was carried out using Sepharose CL-4B gel filtration columns (Sigma-Aldrich). mRNA content was calculated by performing a modified Quant-iT RiboGreen RNA assay (Invitrogen, Carlsbad, CA, USA). It was estimated that the final conjugation density was approximately 80 mAb/LNP.
LNPs were radiolabeled with 125I as previously described.5 Briefly, LNPs were incubated with Pierce (Rockford, IL, USA) Iodination Beads and Na125I for 15 min at room temperature. Following incubation, unreacted 125I was removed using desalting columns (G-25 Sephadex, Roche Applied Sciences, Indianapolis, IN, USA). For studies utilizing 3H labeling, LNPs were formulated by addition of a trace amount of 3H-cholesteryl hexadecyl ester to the ethanolic lipid mixture prior to the formulation of LNPs.
Animal studies
All animal experiments were carried out in male, 6- to 8-week-old C57BL/6 mice (The Jackson Laboratory, Bar Harbor, ME, USA). Injections were performed via the retro-orbital plexus via rapid bolus injection of 100 μL/mouse.
Biodistribution studies in mice
Radiolabeled mRNA-LNPs were administered at a dose of 8 μg mRNA/mouse by retro-orbital injection. Animals were sacrificed at specific time points after mRNA-LNP injection and their blood was collected from the inferior vena cava (n = 3 mice/time point). Organs (liver, spleen, lung, kidney, and heart) were harvested, rinsed with saline, blotted dry, and weighed. Tissue radioactivity measurement in organs and 100-μL samples of blood was performed in a gamma counter (Wallac 1470 Wizard gamma counter, Gaithersburg, MD, USA). Radioactivity values and weight of the samples were then used to calculate targeting parameters of nanoparticles, including tissue uptake as percent of injected dose per gram tissue.
Luciferase mRNA expression in mice: Luciferase activity in organ homogenates
A detailed description of the luciferase assay can be found in our prior publication describing the results used for modeling.5 Briefly, at select time points following IV injection of mRNA-LNPs, mice (n = 3/time point) were euthanized and organs (lungs, heart, liver, kidneys, and spleen) were collected, washed in PBS, and frozen at −80°C. Samples were then homogenized and processed to collect cell lysate and luciferase activity was measured in the supernatant using a Victor3 1420 Multilabel Plate Counter (PerkinElmer, Wellesley, MA, USA).
Luciferase mRNA expression in mice: BLI of intact organs
Mice were intravenously injected with unconjugated or antibody-conjugated LNP-luciferase mRNA formulations. At desired time points, mice (n = 3 mice/time point) were administered an intraperitoneal injection of D-luciferin at a dose of 150 mg/kg. After 5 min, the mice were euthanized; organs were quickly harvested, and placed on the imaging platform. BLI was performed as described previously5 using an IVIS Spectrum imaging system (Caliper Life Sciences, Waltham, MA, USA).
Organ luminescence was measured on the IVIS imaging system using an exposure time of 5 s or longer to ensure that the signal obtained was within operative detection range. Bioluminescence values were also quantified by measuring photon flux (photons/s) in the region of interest using LivingImage software provided by Caliper.
Theoretical
PK model structure and parameters
The PBPK model developed for LNP delivery was based on a published model for liposomal amphotericin B (AmBisome) developed by Kagan and colleagues.35 The macro-level PBPK model structure is depicted in Figure 1A. Briefly, key organs (blood, lungs, heart, kidneys, liver, and spleen) were linked in an anatomically relevant fashion with venous blood first passing through the lungs prior to distribution to the remaining organs. To achieve whole body mass balance, remainder compartments were made to account for the remaining organs that drain into the portal vein (stomach, small intestine, large intestine, and pancreas) and all other tissues. Outflow from both the spleen and portal vein remainder compartments first passed through the liver prior to returning to the venous blood. The sub-organ level (micro) model structure is shown in Figure 1B. Briefly, LNPs within the vascular space of each tissue could (1) return to venous blood at a rate consistent with tissue blood flow, (2) irreversibly leave the blood via a non-specific CLup, and (3) bind to target expressed on accessible cells and be internalized. Following internalization into cells, particle degradation was assumed to follow first-order kinetics. All physiologically relevant parameters (tissue volumes, blood flows, etc.) were obtained from the BioDMET database and scaled to a 25-g mouse (Table S3).
Luciferase expression model
The model structure for luciferase expression kinetics is depicted in Figure 1C. Briefly, the availability of mRNA for translation was assumed to be driven by the kinetics of LNP degradation. The turnover of mRNA was assumed to occur at a first-order rate, consistent with previously reported in vitro measurements of modified luciferase mRNA in cell culture (kRNA = 0.115 h−1).36 Intact mRNA was used to drive the expression of luciferase using a linear scalar (Sorgan) relating mRNA to the production of luciferase. Elimination of luciferase signal was assumed to follow first-order kinetics (kluc).
Model fitting
Data for fitting were obtained from our recent publication describing the kinetics of blood and tissue distribution of radiolabeled LNPs and the kinetics of tissue expression of luciferase for untargeted and PECAM-targeted LNPs.5 To estimate the kinetics of target-independent PK processes (e.g., non-specific cellular uptake, LNP degradation), tissue-specific CLup and a single, first-order LNP kdeg were estimated for bare and control IgG LNP. Simultaneously, parameters related to luciferase expression kinetics were estimated for bare LNPs. These included tissue-specific coefficients relating mRNA to production of luciferase signal (Sorgan) and first-order rate constants for the kinetics of loss of luciferase signal (kluc). Target-specific PK parameters were obtained by fixing CLup and kdeg to the values estimated for control IgG LNP and estimating parameters related to PECAM expression (Porgan), LNP-PECAM complex internalization (kPECAM), and the intracellular kdeg of PECAM-bound LNP (kdeg, PECAM). A comparison was made between a single luciferase production rate regardless of internalization pathway (simulation) and distinct rates for PECAM-mediated vs. non-specific endocytosis (e.g., Sorgan, nonspecific was set equal to that for IgG-coated LNPs and Sorgan, PECAM was estimated).
Model validation
To evaluate the predictive capacity of the PBPK model, biodistribution and luciferase expression following injection of ICAM-targeted LNPs were simulated using the model, with no additional fitting of parameters. Accessible ICAM concentrations in each tissue were fixed based on the relative expression of ICAM and PECAM estimated via PBPK of data from our recent publication.15 Model predictions were compared to observed data.
Software
All model fitting and simulation was performed using ADAPT 5 (BMSR) and the maximum likelihood estimator.
Data and code availability
All data supporting the findings described in this manuscript are available from the corresponding author upon reasonable request.
Acknowledgments
This research was funded by the following sources: National Institutes of Health (HL155106 [V.R.M., D.W.], HL126874 [V.R.M., D.W.], and R00HL153696 [P.M.G.]). Approved for public release; distribution is unlimited. This research has been funded in whole or part with federal funds by Defense Advanced Research Projects Agency Pandemic Prevention Platform (“P3”) under a grant from DARPA (Contract No. HR0011-17-2-0069) the content of the information does not necessarily reflect the position or the policy of the Government, and no official endorsement should be inferred.
Author contributions
H.P.: Conceptualization, methodology, formal analysis, investigation, writing – original draft, visualization; V.V.S.: Methodology, investigation; Q.L.: Investigation; A.Y.: Investigation; T.P.: Investigation; H.S.: Investigation; R.S.: Investigation; S.C.S.: Resources, writing – review and editing; B.L.M.: Resources, writing – review and editing; D.W.: Conceptualization, writing – original draft, supervision, funding acquisition; V.R.M.: Conceptualization, writing – original draft, supervision, funding acquisition; P.M.G.: Conceptualization, methodology, formal analysis, writing – original draft, visualization, funding acquisition.
Declaration of interests
H.P. and D.W. are scientific founders and hold equity in Capstan Therapeutics. S.C.S. and B.L.M. are employees and hold equity in Acuitas Therapeutics. H.P. and D.W. receive research support from BioNTech. H.P., V.V.S., D.W., and V.R.M. are named on patents that describe the use of nucleoside-modified mRNA and targeted LNP.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2024.102175.
Supplemental information
References
- 1.Adams D., Gonzalez-Duarte A., O'Riordan W.D., Yang C.C., Ueda M., Kristen A.V., Tournev I., Schmidt H.H., Coelho T., Berk J.L., et al. Patisiran, an RNAi Therapeutic, for Hereditary Transthyretin Amyloidosis. N. Engl. J. Med. 2018;379:11–21. doi: 10.1056/NEJMoa1716153. [DOI] [PubMed] [Google Scholar]
- 2.Balwani M., Sardh E., Ventura P., Peiró P.A., Rees D.C., Stölzel U., Bissell D.M., Bonkovsky H.L., Windyga J., Anderson K.E., et al. Phase 3 Trial of RNAi Therapeutic Givosiran for Acute Intermittent Porphyria. N. Engl. J. Med. 2020;382:2289–2301. doi: 10.1056/NEJMoa1913147. [DOI] [PubMed] [Google Scholar]
- 3.Oliver S.E., Gargano J.W., Marin M., Wallace M., Curran K.G., Chamberland M., McClung N., Campos-Outcalt D., Morgan R.L., Mbaeyi S., et al. The Advisory Committee on Immunization Practices' Interim Recommendation for Use of Pfizer-BioNTech COVID-19 Vaccine - United States, December 2020. MMWR Morb. Mortal. Wkly. Rep. 2020;69:1922–1924. doi: 10.15585/mmwr.mm6950e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oliver S.E., Gargano J.W., Marin M., Wallace M., Curran K.G., Chamberland M., McClung N., Campos-Outcalt D., Morgan R.L., Mbaeyi S., et al. The Advisory Committee on Immunization Practices' Interim Recommendation for Use of Moderna COVID-19 Vaccine - United States, December 2020. MMWR Morb. Mortal. Wkly. Rep. 2021;69:1653–1656. doi: 10.15585/mmwr.mm695152e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parhiz H., Shuvaev V.V., Pardi N., Khoshnejad M., Kiseleva R.Y., Brenner J.S., Uhler T., Tuyishime S., Mui B.L., Tam Y.K., et al. PECAM-1 directed re-targeting of exogenous mRNA providing two orders of magnitude enhancement of vascular delivery and expression in lungs independent of apolipoprotein E-mediated uptake. J. Control. Release. 2018;291:106–115. doi: 10.1016/j.jconrel.2018.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Glassman P.M., Myerson J.W., Ferguson L.T., Kiseleva R.Y., Shuvaev V.V., Brenner J.S., Muzykantov V.R. Targeting drug delivery in the vascular system: Focus on endothelium. Adv. Drug Deliv. Rev. 2020;157:96–117. doi: 10.1016/j.addr.2020.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marcos-Contreras O.A., Greineder C.F., Kiseleva R.Y., Parhiz H., Walsh L.R., Zuluaga-Ramirez V., Myerson J.W., Hood E.D., Villa C.H., Tombacz I., et al. Selective targeting of nanomedicine to inflamed cerebral vasculature to enhance the blood-brain barrier. Proc. Natl. Acad. Sci. USA. 2020;117:3405–3414. doi: 10.1073/pnas.1912012117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pardi N., Tuyishime S., Muramatsu H., Kariko K., Mui B.L., Tam Y.K., Madden T.D., Hope M.J., Weissman D. Expression kinetics of nucleoside-modified mRNA delivered in lipid nanoparticles to mice by various routes. J. Control. Release. 2015;217:345–351. doi: 10.1016/j.jconrel.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rosenblum D., Gutkin A., Kedmi R., Ramishetti S., Veiga N., Jacobi A.M., Schubert M.S., Friedmann-Morvinski D., Cohen Z.R., Behlke M.A., et al. CRISPR-Cas9 genome editing using targeted lipid nanoparticles for cancer therapy. Sci. Adv. 2020;6 doi: 10.1126/sciadv.abc9450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Q., Chan C., Peterson N., Hanna R.N., Alfaro A., Allen K.L., Wu H., Dall'Acqua W.F., Borrok M.J., Santos J.L. Engineering Caveolae-Targeted Lipid Nanoparticles To Deliver mRNA to the Lungs. ACS Chem. Biol. 2020;15:830–836. doi: 10.1021/acschembio.0c00003. [DOI] [PubMed] [Google Scholar]
- 11.Jones H.M., Chen Y., Gibson C., Heimbach T., Parrott N., Peters S.A., Snoeys J., Upreti V.V., Zheng M., Hall S.D. Physiologically based pharmacokinetic modeling in drug discovery and development: a pharmaceutical industry perspective. Clin. Pharmacol. Ther. 2015;97:247–262. doi: 10.1002/cpt.37. [DOI] [PubMed] [Google Scholar]
- 12.Jones H., Rowland-Yeo K. Basic concepts in physiologically based pharmacokinetic modeling in drug discovery and development. CPT Pharmacometrics Syst. Pharmacol. 2013;2 doi: 10.1038/psp.2013.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Glassman P.M., Balthasar J.P. Physiologically-based modeling of monoclonal antibody pharmacokinetics in drug discovery and development. Drug Metab. Pharmacokinet. 2019;34:3–13. doi: 10.1016/j.dmpk.2018.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heyes J., Palmer L., Bremner K., MacLachlan I. Cationic lipid saturation influences intracellular delivery of encapsulated nucleic acids. J. Control. Release. 2005;107:276–287. doi: 10.1016/j.jconrel.2005.06.014. [DOI] [PubMed] [Google Scholar]
- 15.Kiseleva R.Y., Glassman P.G., LeForte K.M., Walsh L.R., Villa C.H., Shuvaev V.V., Myerson J.W., Aprelev P.A., Marcos-Contreras O.A., Muzykantov V.R., Greineder C.F. Bivalent engagement of endothelial surface antigens is critical to prolonged surface targeting and protein delivery in vivo. FASEB J. 2020;34:11577–11593. doi: 10.1096/fj.201902515RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cullis P.R., Hope M.J. Lipid Nanoparticle Systems for Enabling Gene Therapies. Mol. Ther. 2017;25:1467–1475. doi: 10.1016/j.ymthe.2017.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hawthorne G., Henderson N., Hölttä M., Khan S., Lindqvist J., Wilson A. Overcoming analytical challenges to generate data critical to understanding lipid nanoparticle-delivered modified mRNA biodistribution. Bioanalysis. 2019;11:1993–2001. doi: 10.4155/bio-2019-0138. [DOI] [PubMed] [Google Scholar]
- 18.van der Meel R., Chen S., Zaifman J., Kulkarni J.A., Zhang X.R.S., Tam Y.K., Bally M.B., Schiffelers R.M., Ciufolini M.A., Cullis P.R., Tam Y.Y.C. Modular Lipid Nanoparticle Platform Technology for siRNA and Lipophilic Prodrug Delivery. Small. 2021;17 doi: 10.1002/smll.202103025. [DOI] [PubMed] [Google Scholar]
- 19.Chen S., Tam Y.Y.C., Lin P.J.C., Sung M.M.H., Tam Y.K., Cullis P.R. Influence of particle size on the in vivo potency of lipid nanoparticle formulations of siRNA. J. Control. Release. 2016;235:236–244. doi: 10.1016/j.jconrel.2016.05.059. [DOI] [PubMed] [Google Scholar]
- 20.Chen S., Tam Y.Y.C., Lin P.J.C., Leung A.K.K., Tam Y.K., Cullis P.R. Development of lipid nanoparticle formulations of siRNA for hepatocyte gene silencing following subcutaneous administration. J. Control. Release. 2014;196:106–112. doi: 10.1016/j.jconrel.2014.09.025. [DOI] [PubMed] [Google Scholar]
- 21.Mui B.L., Tam Y.K., Jayaraman M., Ansell S.M., Du X., Tam Y.Y.C., Lin P.J., Chen S., Narayanannair J.K., Rajeev K.G., et al. Influence of Polyethylene Glycol Lipid Desorption Rates on Pharmacokinetics and Pharmacodynamics of siRNA Lipid Nanoparticles. Mol. Ther. Nucleic Acids. 2013;2 doi: 10.1038/mtna.2013.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maier M.A., Jayaraman M., Matsuda S., Liu J., Barros S., Querbes W., Tam Y.K., Ansell S.M., Kumar V., Qin J., et al. Biodegradable lipids enabling rapidly eliminated lipid nanoparticles for systemic delivery of RNAi therapeutics. Mol. Ther. 2013;21:1570–1578. doi: 10.1038/mt.2013.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eigenmann M.J., Karlsen T.V., Krippendorff B.F., Tenstad O., Fronton L., Otteneder M.B., Wiig H. Interstitial IgG antibody pharmacokinetics assessed by combined in vivo- and physiologically-based pharmacokinetic modelling approaches. J. Physiol. 2017;595:7311–7330. doi: 10.1113/JP274819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eppihimer M.J., Russell J., Langley R., Vallien G., Anderson D.C., Granger D.N. Differential expression of platelet-endothelial cell adhesion molecule-1 (PECAM-1) in murine tissues. Microcirculation. 1998;5:179–188. [PubMed] [Google Scholar]
- 25.Ayyar V.S., Jusko W.J. Transitioning from Basic toward Systems Pharmacodynamic Models: Lessons from Corticosteroids. Pharmacol. Rev. 2020;72:414–438. doi: 10.1124/pr.119.018101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun Y.N., DuBois D.C., Almon R.R., Pyszczynski N.A., Jusko W.J. Dose-dependence and repeated-dose studies for receptor/gene-mediated pharmacodynamics of methylprednisolone on glucocorticoid receptor down-regulation and tyrosine aminotransferase induction in rat liver. J. Pharmacokinet. Biopharm. 1998;26:619–648. doi: 10.1023/a:1020746822634. [DOI] [PubMed] [Google Scholar]
- 27.Sayers E.J., Peel S.E., Schantz A., England R.M., Beano M., Bates S.M., Desai A.S., Puri S., Ashford M.B., Jones A.T. Endocytic Profiling of Cancer Cell Models Reveals Critical Factors Influencing LNP-Mediated mRNA Delivery and Protein Expression. Mol. Ther. 2019;27:1950–1962. doi: 10.1016/j.ymthe.2019.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sahay G., Querbes W., Alabi C., Eltoukhy A., Sarkar S., Zurenko C., Karagiannis E., Love K., Chen D., Zoncu R., et al. Efficiency of siRNA delivery by lipid nanoparticles is limited by endocytic recycling. Nat. Biotechnol. 2013;31:653–658. doi: 10.1038/nbt.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sago C.D., Krupczak B.R., Lokugamage M.P., Gan Z., Dahlman J.E. Cell Subtypes Within the Liver Microenvironment Differentially Interact with Lipid Nanoparticles. Cell. Mol. Bioeng. 2019;12:389–397. doi: 10.1007/s12195-019-00573-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Di J., Du Z., Wu K., Jin S., Wang X., Li T., Xu Y. Biodistribution and Non-linear Gene Expression of mRNA LNPs Affected by Delivery Route and Particle Size. Pharm. Res. (N. Y.) 2022;39:105–114. doi: 10.1007/s11095-022-03166-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pardi N., Muramatsu H., Weissman D., Karikó K. In vitro transcription of long RNA containing modified nucleosides. Methods Mol. Biol. 2013;969:29–42. doi: 10.1007/978-1-62703-260-5_2. [DOI] [PubMed] [Google Scholar]
- 32.Baiersdörfer M., Boros G., Muramatsu H., Mahiny A., Vlatkovic I., Sahin U., Karikó K. A Facile Method for the Removal of dsRNA Contaminant from In Vitro-Transcribed mRNA. Mol. Ther. Nucleic Acids. 2019;15:26–35. doi: 10.1016/j.omtn.2019.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jayaraman M., Ansell S.M., Mui B.L., Tam Y.K., Chen J., Du X., Butler D., Eltepu L., Matsuda S., Narayanannair J.K., et al. Maximizing the potency of siRNA lipid nanoparticles for hepatic gene silencing in vivo. Angew. Chem. Int. Ed. Engl. 2012;51:8529–8533. doi: 10.1002/anie.201203263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Howard M.D., Hood E.D., Greineder C.F., Alferiev I.S., Chorny M., Muzykantov V. Targeting to endothelial cells augments the protective effect of novel dual bioactive antioxidant/anti-inflammatory nanoparticles. Mol. Pharm. 2014;11:2262–2270. doi: 10.1021/mp400677y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kagan L., Gershkovich P., Wasan K.M., Mager D.E. Dual physiologically based pharmacokinetic model of liposomal and nonliposomal amphotericin B disposition. Pharm. Res. (N. Y.) 2014;31:35–45. doi: 10.1007/s11095-013-1127-z. [DOI] [PubMed] [Google Scholar]
- 36.Anderson B.R., Muramatsu H., Jha B.K., Silverman R.H., Weissman D., Karikó K. Nucleoside modifications in RNA limit activation of 2'-5'-oligoadenylate synthetase and increase resistance to cleavage by RNase L. Nucleic Acids Res. 2011;39:9329–9338. doi: 10.1093/nar/gkr586. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting the findings described in this manuscript are available from the corresponding author upon reasonable request.