Abstract
Purpose
Glioma is the most common primary tumor in the brain, accounting for 81% of intracranial malignancies. Nowadays, cancer immunotherapy has become a novel and revolutionary treatment for patients with advanced, highly aggressive tumors. However, to date, there are no effective biomarkers to reflect the response of glioma patients to immunotherapy. In this study, we aimed to assess the clinical predictive value of ITGB2 in patients with glioma.
Methods
The correlation between ITGB2 expression levels and glioma progression was explored and validated using data from CGGA, TCGA, GEO datasets, and patient samples from our hospital. Univariate and multivariate cox regression models were developed to determine the predictive role of ITGB2 on the prognosis of patients with glioma. The relationship between ITGB2 and immune activation was then analyzed. Finally, we predicted the immunotherapy response in both high and low ITGB2 expression subgroups.
Results
ITGB2 was significantly elevated in gliomas with higher malignancy and predicted poor prognosis. In multivariate analysis, the hazard ratio for ITGB2 expression (low versus high) was 0.71 with 95% CI (0.59–0.85) (P < 0.001). Furthermore, we found that ITGB2 stratified glioma patients into high and low ITGB2 expression subgroups, exhibiting different clinical outcomes and immune activation status. At last, we demonstrated that glioma patients with high ITGB2 expression levels had better immunotherapy response.
Conclusions
This study demonstrated ITGB2 as a novel predictor for clinical prognosis and response to immunotherapy in gliomas. Assessing expression levels of ITGB2 is a promising method to discover patients that may benefit from immunotherapy.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00262-021-03022-2.
Keywords: Gliomas, Immunotherapy, Immune activation, T cell, ITGB2, Biomarker
Introduction
Glioma is the most common primary tumor in the brain, accounting for 81% of intracranial malignancies [1, 2]. At tissue level, gliomas originate from glial cells, and the main types include astrocytoma, oligodendroglioma, ependymoma, neuronal and mixed neuro-glioblastoma (e.g., ganglion glioma) [3]. Glioma is extremely harmful to human body; also, the median survival time of newly diagnosed glioma is only 12–18 months [4–6]. Although there are many treatment options for gliomas, including surgical treatment, chemotherapy, radiotherapy, and immunotherapy, the survival rate of glioma remains very low. The possible reason is that the heterogeneity of tumors and the complexity of epigenetics make it difficult to determine the therapeutic target of glioma, and the existence of the physiological blood–brain barrier limits the effect of drugs. Besides, the infiltrative nature of the tumor makes surgical treatment largely ineffective. Therefore, an in-depth understanding of the biological behaviors of tumor occurrence and progression will help provide more innovative and effective methods for clinical diagnosis and treatment in patients with glioma.
Previous studies confirmed that CTLA4 (cytotoxic T lymphocyte-associated protein 4), PDCD1 (PD1) (programmed cell death protein 1/programmed cell death 1), CD274 (PDL1) (cluster of differentiation 274/programmed death ligand 1) as immune checkpoints can prevent the immune system from killing cancer cells by inhibiting the auto-immunity [7, 8]. At the same time, the current main method of cancer treatment is based on the cancer immunotherapy with immune checkpoint blocker (ICB). Therefore, the use of anti-CTLA4, anti-PDCD1 (PD1) and anti-CD274 (PDL1) drugs will be promising means for the diagnosis and treatment of diseases, which has become the most effective method for advanced highly invasive anti-tumor patients to achieve therapeutic effect by regulating the state of T cell. However, in fact, only a few patients can benefit from immunotherapy currently. In addition, little is known about the immune checkpoints found to be synergistic with PD1 in glioma tissues, so it is crucial to find effective biomarkers associated with it.
ITGB2 (CD18) is one subunit of the β2 integrins, which are heterodimeric surface receptors expressed by leukocytes specially. It can connect the cytoskeleton and participate in intracellular signaling [9]. Also, combining 4 kinds of α subunits which are αL (CD11a, ITGA), αM (CD11b, ITGAM), αX (CD11c, ITGAX) and αD (CD11d, ITGAD) can become LAF-1 (leukocyte factor 1,CD11a/CD18) [10], MAC-1 (macrophage-1 antigen, CD11b/CD18) [11], CR4 (complement receptor 4,CD11c/CD18) [12] and CD11d/CD18 which bears structural similarity with MAC-1 [13]. In tumors, β2 integrin is involved in cell adhesion, stromal remodeling, and signal transduction among tumor cells and between tumor cells and tumor microenvironment to induce tumors to infiltration, angiogenesis and tumor-specific immune responses [14, 15]. Likewise, ITGB2 is closely related to the development of tumors. ITGB2 is involved in the development, metastasis and invasion of various tumors, such as liver cancer, colon cancer breast cancer and leukemia [16–18]. In addition, previous studies have shown that ITGB2 is involved in the development of glioma, but the role of ITGB2 in glioma progression and the potential molecular mechanism were poorly understood [19–23]. The aim of this study was to demonstrate that ITGB2 is a promising predictive target for glioma prognosis and immunotherapeutic response.
Methods and materials
Data source and expression analysis
Pan-cancer dataset in The Cancer Genome Atlas (TCGA) which consists of 33 kinds of cancer and adjacent tissue samples or GTEx expression matrices was analyzed with an online tool, UCSCXenaShiny [24] (https://hiplot.com.cn/advance/ucsc-xena-shiny). In this study, we analyzed both GBM and LGG. Gliovis [25] (http://gliovis.bioinfo.cnio.es/) was used to get all the expression matrices of gliomas, including 6 datasets containing 2336 samples: 642 grade II patients, 780 grade III patients and 914 grade IV patients (Table 1). Single cell RNA sequencing data are obtained from TISCH [26] with GEO ID: GSE131928 [27].
Table 1.
Datasets used in this study
Tumor samples collection
Human glioma tissues were considered exempt by the Human Investigation Ethical Committee of Shanghai General Hospital affiliated to Shanghai Jiao Tong University. Human tumor samples were consecutively recruited between January 2019 and January 2020 from the Department of Neurosurgery in Shanghai General Hospital. A total of 20 patients with glioma (LGG, n = 9; HGG, n = 11) underwent surgery for the first time and had not previously received radiotherapy or chemotherapy were collected.
Immunohistochemical (IHC) analysis
Patient tumor samples were fixed in 4% paraformaldehyde for 24 h and then embedded in paraffin wax. Paraffin blocks were cut into 5-µm sections and sealed with 5% BSA overnight at 4 °C, followed by staining with ITGB2 (Abcam, ab131044, USA). After washing with PBS, the sections were incubated with biotinylated anti-rabbit IgG (Vector Laboratories, CA, USA). The ABC method (Vector Laboratories) was utilized. Sections were observed using an AX-80 microscope (Olympus, Tokyo, Japan). Images were processed with Image J software, and relative expression was calculated.
Quantitative real-time PCR
Total RNA was extracted from human tumor tissues using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) and reverse transcripted using FastQuant RT kit (Tiangen, Shanghai, China). Real-time PCR was carried out using SuperReal SYBR Green kit (Tiangen, Shanghai, China) and Lightcycler 96 (Roche, Penzberg, Germany). The following commercially available Taqman gene expression assays were used: ITGB2: Hs00164957_m1. The PCR primers used were as follows:
ITGB2 forward: CCTGCAGATTGTTCCGGAGT; reverse: TGGGGCCACCTTTACTGAG;
PDCD1 forward: CAGTTCCAAACCCTGGTGGT; reverse: GGCTCCTATTGTCCCTCGTG.
CTLA4 forward: ACGGGACTCTACATCTGCAAGG; reverse: GGAGGAAGTCAGAATCTGGGCA.
TIM3 (HAVCR2) forward: GACTCTAGCAGACAGTGGGATC;
reverse: GGTGGTAAGCATCCTTGGAAAGG.
IDO1 forward: GCCTGATCTCATAGAGTCTGGC; reverse: TGCATCCCAGAACTAGACGTGC.
TIGIT forward: TGGTGGTCATCTGCACAGCAGT; reverse: TTTCTCCTGAGGTCACCTTCCAC.
The 2−ΔΔCT method was used to calculate the relative amplification of the promoter sequence of each gene.
Immune cells and bioinformatic analysis
The single sample gene set enrichment analysis (ssGSEA) was used to define an enrichment score to represent the degree of absolute enrichment of a gene set in each sample within a given dataset with R package “GSVA” [28]. Normalized enrichment scores could be calculated for each immune category. 28 types of immune cell’s gene set signatures were obtained from a previous study [29] (Additional file: Sheet 1).
To explore the association between ITGB2 expression levels and immune status, 25 immune-related gene sets from previous studies included innate and adaptive responses [30] (Additional file: Sheet 2). Gene set variation analysis (GSVA) was performed to obtain the immune profile of glioma samples with R package “GSVA”. GEPIA2021 [31] was utilized to investigate the expression levels of ITGB2 in different cell populations.
According to the median expression value of ITGB2, the CGGA dataset was divided into the group with high expression of ITGB2 (top 50%) and the group with low expression of ITGB2 (bottom 50%). Differentially expressed genes (DEGs) were analyzed by R package “limma”. Its biological significance is defined as |logFC| ≥ 1.5 and adj. p value < 0.05. The Gene Ontology Database (GO) that combines the international standard Gene function classification system and provides a dynamically changing standard vocabulary to comprehensively describe the attributes of genes and gene products in an organism and the Kyoto Encyclopedia of Genes and Genomes (KEGG) database for Gene function analysis and linkage of genomic information and functional information were used to annotate the Gene set with the R package “clusterProfiler” [32]. Functional enrichment was further studied by gene set enrichment analysis (GSEA) using R package “Pi” [33].
Quantifying the relative abundance of TIICs and predicting the response of the ITGB2 subgroup to immunotherapy
The cancer immunity cycle reflects the anticancer immune response and comprises seven steps: release of cancer cell antigens (Step 1), cancer antigen presentation (Step 2), priming and activation (Step 3), trafficking of immune cells to tumors (Step 4), infiltration of immune cells into tumors (Step 5), recognition of cancer cells by T cells (Step 6), and killing of cancer cells (Step 7). The activities of these steps determine the fate of the tumor cells. Xu et al. [34] evaluated the activities of these steps using a single sample gene set enrichment analysis (ssGSEA) based on the gene expression of individual samples (Additional file: Sheet 3). We also collected other therapeutic signatures, including oncogenic pathways that could shape a non-inflamed TME, targeted therapy-associated gene signatures (Additional file: Sheet 4), and gene signatures predicting radiotherapy responses [35].
Immune Cells Abundance Identifier (ImmuCellAI) (http://bioinfo.life.hust.edu.cn/ImmuCellAI#!/analysis) is a novel algorithm for estimating immune cell abundance based on RNA-seq or microarray data, which focuses on subsets of T cells that are associated with tumor progression and initiation [36]. It uses genome set markers to estimate the abundance of 24 immune cells from transcriptome data. Also, ImmuCellAI can be used to predict the response of ICB therapy with the ICB response prediction being checked. Genome set markers for the T cell subsets used in this study are listed in the supplementary material (Additional file: Sheet 5), including 18 T cell subtypes and 6 other immune cell types.
Patient’s response to a drug was a complex phenomenon, determined by a combination of genetic factors and the environment, so drug responses could be predicted by genomic characteristics. Glioma samples were scored using the GSVA method to predict the likely response of glioma to anti-PD1 drugs, and T-cell inflammation (TIS) signals from previous studies [30] were used and listed in Additional file: Sheet 6.
The subclass mapping (SubMap) method was also used to evaluate the correspondence of the two subgroups and the patients with different immunotherapy responses [37]. The P value is used to evaluate the similarity, and the lower the P value, the higher the similarity. In this study, TIS, SubMap, and ImmunCellAI were used to predict the response of glioma patients to immunotherapy.
Statistical analysis
All statistical analyses were performed using R software 4.0.1. Wilcoxon rank sum test and Fisher’s exact test were used to assess association between different factors. Correlation was assessed by Pearson correlation test. The association between clinical factors and overall survival was assessed using the Cox regression model in the survival analysis. Kaplan–Meyer survival curves were plotted and compared between subgroups using log-rank test with R packages “survival” and “survminer”. R package “meta” was used for meta-analysis. The ROC curve, sensitivity, specificity and the area under the curve (AUC) were generated using the R package “pROC”. The P value < 0.05 was considered significant for all statistical analysis.
Results
Pan-cancer analysis of ITGB2 expression
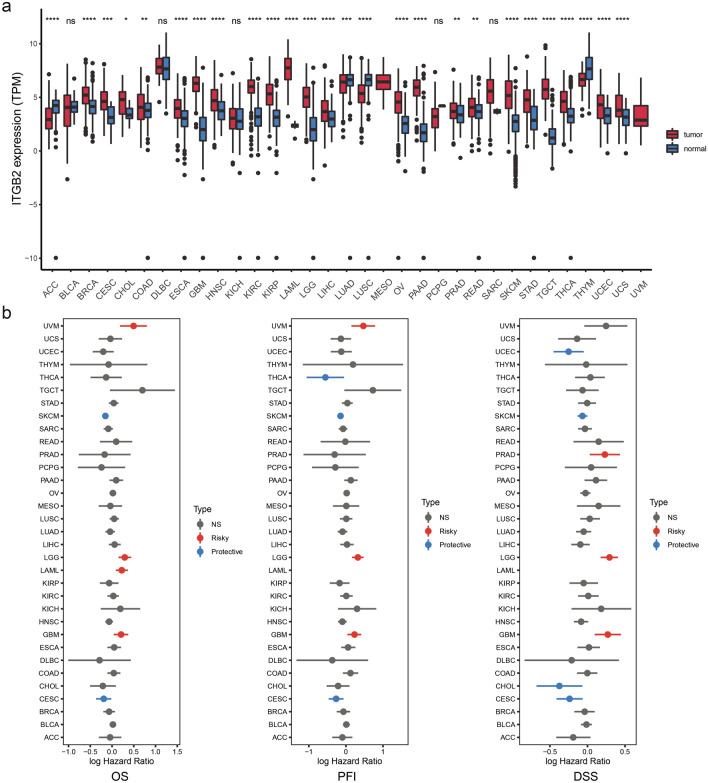
To investigate the expression of ITGB2 in tumors and normal tissues, we utilized an online tool, UCSCxenaShiny [24]. Pan-cancer analysis of ITGB2 expression revealed significant differences in the expression of ITGB2 levels between various tumors and adjacent tissues (or GTEX) (Fig. 1a). The expression of ITGB2 in breast invasive carcinoma (BRCA), esophageal carcinoma (ESCA), glioblastoma multiforme (GBM), head and neck squamous cell carcinoma (HNSC), kidney renal clear cell carcinoma (KIRC), kidney renal papillary cell carcinoma (KIRP), acute myeloid leukemia (LAML), brain lower grade glioma (LGG), liver hepatocellular carcinoma (LIHC), ovarian serous cystadenocarcinoma (OV), pancreatic adenocarcinoma (PAAD), skin cutaneous melanoma and stomach adenocarcinoma (SKCM), stomach adenocarcinoma (STAD), testicular germ cell tumors (TGCT), thyroid carcinoma (THCA), uterine corpus endometrial carcinoma (UCEC), and uterine carcinosarcoma (UCS) was higher than that in normal tissues (P < 0.05). Besides, ITGB2 expression in adrenocortical carcinoma (ACC), lung squamous cell carcinoma (LUSC) and thymoma (THYM) were lower than those in normal tissues (P < 0.05).
Fig. 1.
Pan-cancer analysis of ITGB2 expression. a UCSCXenaShiny was used to visualize ITGB2 mRNA expression in the cancer genome atlas (TCGA) pan-cancer datasets. *, P < 0.05; * *, P < 0.01; * * *, P < 0.001; * * * *, P < 0.0001, ns = no significance (Wilcoxon test). b Risk plot of correlation between ITGB2 with OS, PFI, DSS (red represents HR > 1 (risky) and P value < 0.05; blue represents HR < 1 (protective) and P value < 0.05; gray represents no significance with P value > 0.05)
Since the expression of ITGB2 varied significantly between a variety of tumors and normal tissues, we further investigated the relationship between ITGB2 expression levels and clinical outcomes. Patients in a cohort of 33 tumor types were then divided into high-expression and low-expression groups based on median of ITGB2 gene expression. Survival analysis revealed significant differences between several cancer types (Fig. 1b). Specifically, patients with high levels of ITGB2 expression showed shorter overall survival (OS), progression-free interval (PFI), and disease-specific survival (DSS) than patients with low levels of ITGB2 expression (Fig. 1b) in the LGG and GBM cohorts. In the following study, we focused on exploring the role of ITGB2 in gliomas.
High expression of ITGB2 infers a poor prognosis for glioma
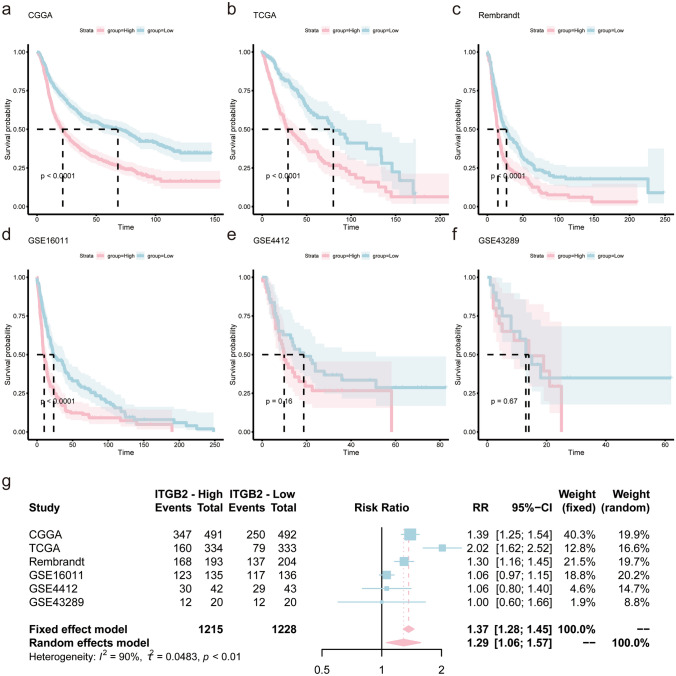
The above analysis showed that ITGB2 expression was significantly associated with the prognosis of glioma patients. To verify the reliability of this result, we further explored the prognostic value of ITGB2 in six independent datasets (Table 1). According to the median expression of ITGB2, patients in these datasets were divided into high expression subgroup and low expression subgroup. The log-rank test analysis (Kaplan–Meier curves) revealed that patients in CGGA(P < 0.0001), TCGA (P < 0.0001), Rembrandt (P < 0.0001) and GSE16011(P < 0.0001) with high expression of ITGB2 showed significantly poorer prognosis than low-expression subgroup (Fig. 2a, b, c, and d), while patients from the GSE43289 and GSE4412 datasets showed a similar trend but with no statistical significance (Fig. 2e, f).
Fig. 2.
High expression of ITGB2 infers a poor prognosis for glioma. Kaplan–Meier plots of ITGB2 in six glioma datasets, 95% CI (confidence interval) were also showed. Patients were divided into high and low expressed group by the medium expression level. a CGGA, b TCGA, c Rembrandt, d GSE4412, e GSE43289, and f GSE16011. g Forest plot of the RRs for patients with high ITGB2 expression compared to patients with low ITGB2 expression
The sample sizes of the six cohorts varied widely, with three having more than 500 samples and two having less than 200 samples. In order to improve the stability of the results, meta-analysis was performed and the results confirmed that patients with high expression of ITGB2 had shorter overall survival than patients with low expression (RR = 1.37, 95% CI 1.28–1.45, Fig. 2g).
To better understand the role of expression of ITGB2 in patients with glioma, we analyzed the CGGA dataset with clinical data of 1013 glioma patients. According to the ITGB2 levels, patients were divided into high expression group (n = 506) and low expression group (n = 507). Statistical analysis of the clinical data revealed that higher ITGB2 expression was more likely to be associated with older age (P < 0.001), poorer prognosis (P < 0.001), higher grade (P < 0.001), GBM type (P < 0.001), mesenchymal subtype (P < 0.001), IDH wild type (P < 0.001), and different therapeutic options (radiotherapy, P = 0.050; chemotherapy, P = 0.040), while there was no significant differences in gender and recurrence (Table 2).
Table 2.
Clinical characteristics of 1013 glioma patients in the CGGA dataset according to ITGB2 expression levels
| Variable | N | Overall, N = 10131 |
High, N = 5061 |
Low, N = 5071 |
p value2 |
|---|---|---|---|---|---|
| Age | 1012 | 43 (12) | 45 (13) | 42 (11) | < 0.001 |
| Gender | 1013 | 0.2 | |||
| Female | 421 (42%) | 200 (40%) | 221 (44%) | ||
| Male | 592 (58%) | 306 (60%) | 286 (56%) | ||
| Survival | 979 | 39 (35) | 31 (29) | 47 (39) | < 0.001 |
| Status | 985 | < 0.001 | |||
| Alive | 388 (39%) | 144 (29%) | 244 (49%) | ||
| Dead | 597 (61%) | 347 (71%) | 250 (51%) | ||
| Grade | 1013 | < 0.001 | |||
| II | 291 (29%) | 104 (21%) | 187 (37%) | ||
| III | 334 (33%) | 157 (31%) | 177 (35%) | ||
| IV | 388 (38%) | 245 (48%) | 143 (28%) | ||
| Histology | 1013 | ||||
| Anaplastic astrocytoma | 214 (21%) | 119 (24%) | 95 (19%) | ||
| Anaplastic oligoastrocytoma | 21 (2.1%) | 8 (1.6%) | 13 (2.6%) | ||
| Anaplastic oligodendroglioma | 94 (9.3%) | 29 (5.7%) | 65 (13%) | ||
| Astrocytoma | 175 (17%) | 85 (17%) | 90 (18%) | ||
| GBM | 388 (38%) | 245 (48%) | 143 (28%) | ||
| Oligoastrocytoma | 9 (0.9%) | 1 (0.2%) | 8 (1.6%) | ||
| Oligodendroglioma | 112 (11%) | 19 (3.8%) | 93 (18%) | ||
| Subtype | 432 | < 0.001 | |||
| Classical | 160 (37%) | 95 (35%) | 65 (41%) | ||
| Mesenchymal | 115 (27%) | 98 (36%) | 17 (11%) | ||
| Proneural | 157 (36%) | 80 (29%) | 77 (48%) | ||
| IDH_status | 961 | < 0.001 | |||
| Mutant | 529 (55%) | 209 (43%) | 320 (67%) | ||
| Wildtype | 432 (45%) | 273 (57%) | 159 (33%) | ||
| Recurrence | 1013 | 0.057 | |||
| Primary | 651 (64%) | 307 (61%) | 344 (68%) | ||
| Recurrent | 332 (33%) | 182 (36%) | 150 (30%) | ||
| Secondary | 30 (3.0%) | 17 (3.4%) | 13 (2.6%) | ||
| Radio_status | 927 | 0.050 | |||
| No | 162 (17%) | 68 (15%) | 94 (20%) | ||
| Yes | 765 (83%) | 386 (85%) | 379 (80%) | ||
| Chemo_status | 906 | 0.040 | |||
| No | 273 (30%) | 122 (27%) | 151 (33%) | ||
| Yes | 633 (70%) | 330 (73%) | 303 (67%) |
1Mean (SD); n (%)
2Welch two-sample t test; Pearson’s Chi-squared test
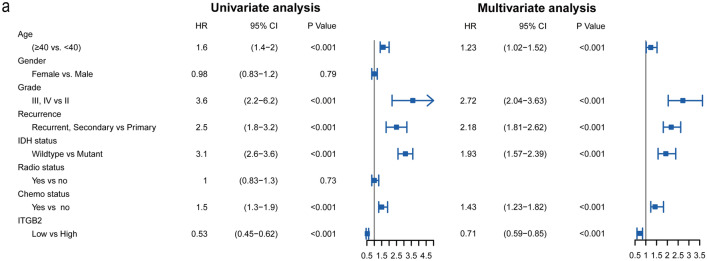
By utilizing the Cox regression model, we computed both univariate and multivariate hazard ratios for different variables of 1013 glioma patients. Univariate cox regression analysis (Fig. 3a) showed that ITGB2 expression level was an independent variable (low versus high, HR = 0.53, 95% CI (0.45–0.62)) to predict the outcome of glioma patients. Multiple cox regression analysis also revealed that ITGB2 expression level was an independent determinant (low versus high, HR = 0.71, 95% CI (0.59–0.85)) of the prognosis of patients with glioma after adjusting for age, grade, recurrence, IDH status and chemotherapy status.
Fig. 3.
Univariate and multivariate analysis for overall survival of glioma patients
These results above confirmed that the expression level of ITGB2 was significantly related to the OS of glioma patients. ITGB2 expression value was a stable factor for predicting the prognosis of glioma patients.
The expression level of ITGB2 increased with the progression of gliomas
After illustrating the prognostic value of ITGB2, we next investigated the correlation between ITGB2 expression level and tumor progression of glioma.
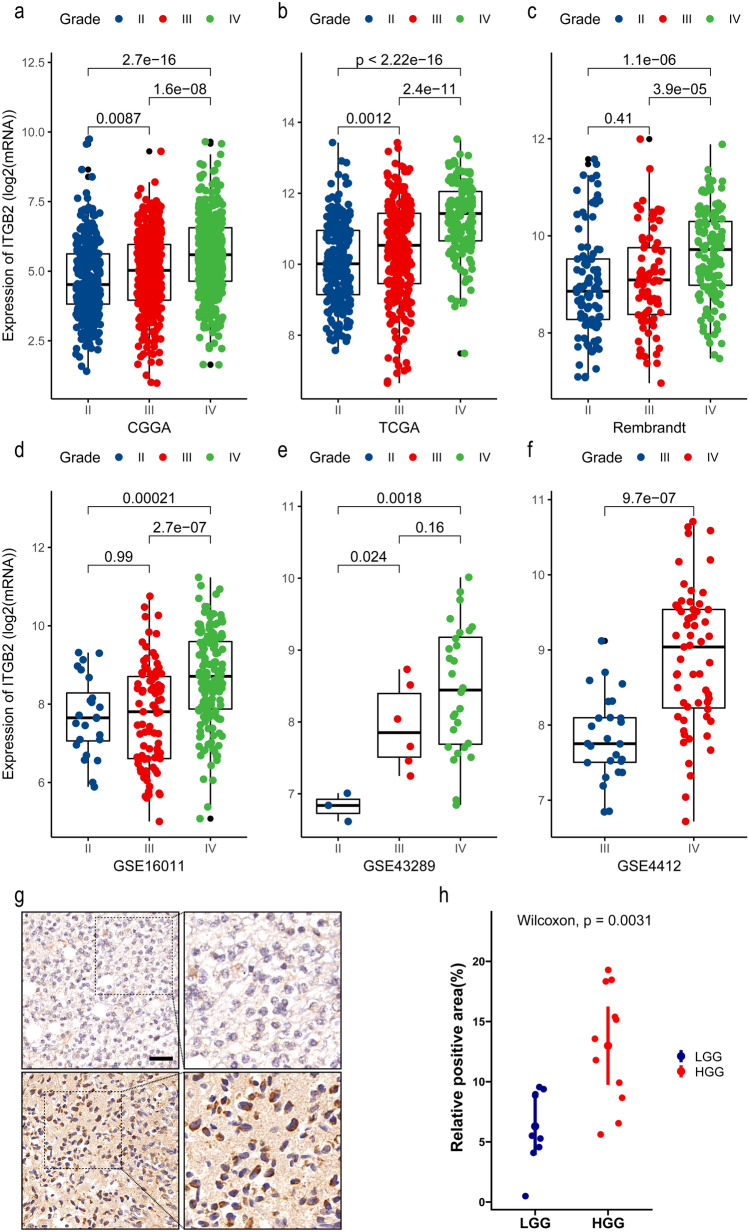
We observed that the expression level of ITGB2 increased in gliomas with high malignancy. In CGGA dataset, the ITGB2 expression was significantly higher in WHO grade III (n = 334) and grade IV (n = 388) than grade II (n = 291) (IV versus III: P < 0.001; IV versus II: P < 0.001; III versus III: P = 0.087, Fig. 4a). In the TCGA-GBMLGG dataset, a significant increase in ITGB2 expression was also noted in WHO grade IV (n = 150), and grade III (n = 244) than grade II (n = 226) (IV versus III: P < 0.001; IV versus II: P < 0.001; III versus III: P = 0.0012, Fig. 4b). Furthermore, a remarkable upward trend was also found in the Rembrandt dataset with 98 grade II, 85 grade III, and 130 grade IV patients (IV versus III: P < 0.001; IV versus II: P < 0.001; III versus II: P = 0.41, Fig. 4c). In addition, based on analysis of GEO datasets, we also found that the GSE16011 dataset with grade II (n = 24), grade III (n = 85), and grade IV (n = 159) glioma patients (IV versus III: P < 0.001; IV versus II: P = 0.00021; III versus III: P = 0.99, Fig. 4d), GSE43289 dataset with 3 grade II, 6 grade III, and 28 grade IV patients (IV versus III: P = 0.16; IV versus II: P = 0.0018; III versus II: P = 0.024, Fig. 4e), and GSE4412 dataset (26 grade III and 59 grade IV patients, P < 0.0001, Fig. 4f) all showed higher expression of ITGB2 in high grade gliomas.
Fig. 4.
The expression level of ITGB2 increased with the progression of gliomas. The X-axis represents the WHO grade, and the Y-axis represents ITGB2 expression values (log2). Based on Wilcoxon test. a CGGA, b TCGA, c Rembrandt, d GSE16011, e GSE43289, and f GSE4412. g Representations (left) and quantification of immunohistochemistry (right) positive areas of ITGB2 in low-grade glioma (LGG) and high-grade glioma (HGG)
To further validate above results, IHC for ITGB2 was performed to assess ITGB2 expression in patient-derived glioma tissue samples. Consistent with previous results, in comparison with low-grade gliomas (LGG), a significant increase in ITGB2 was observed in high-grade gliomas (HGG) (Fig. 4g and h).
In conclusion, the expression of ITGB2 increased with the development of gliomas, suggesting that ITGB2 may be involved in the malignant progression of gliomas.
ITGB2 is associated with immune activation and immune infiltration in glioma
It is known that the prognosis of glioma is related to the infiltration and activation of immune cells [38]. ITGB2, which is one subunit of the β2 integrins, plays an essential role in the immune system and affects the abundance and activation of immune cells (Supplementary Fig. 1a). It has been confirmed by research that CD4+ T cells stimulated by dendritic cells will decrease its proliferation and Th1 polarization when lacking LFA-1. And it is suggested that the interaction between LFA-1 and ICAM-1 reduces the threshold of T cell stimulation [39].
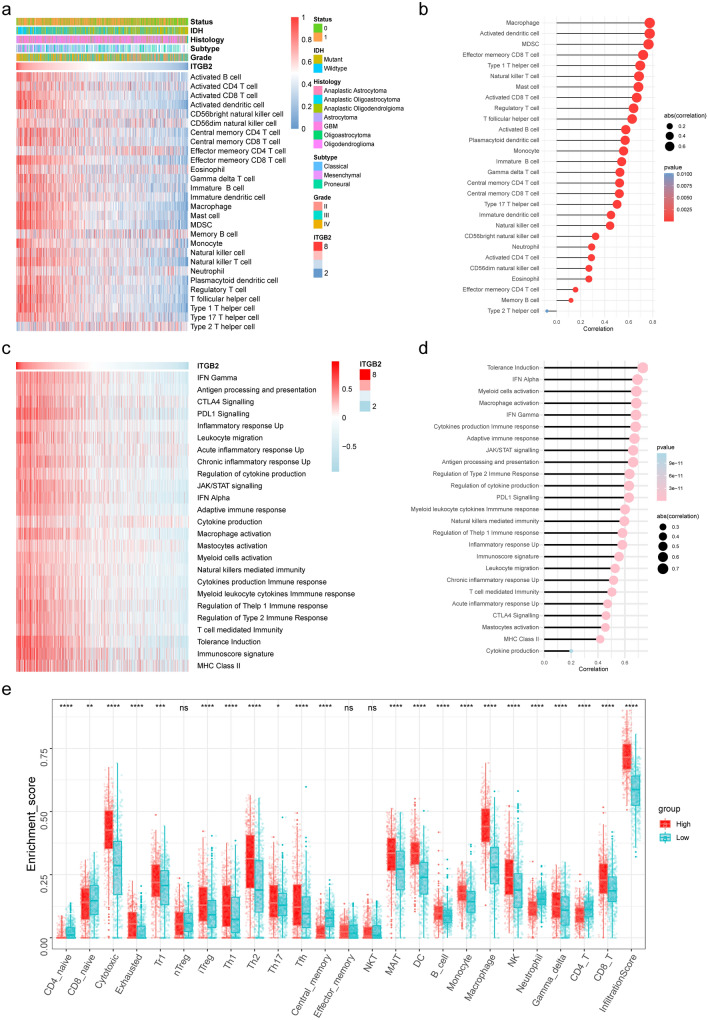
To further validate these conclusions, we evaluated the correlation between ITGB2 expression and the level of immune cells infiltration. The relative quantity of the 28 immune cells from the CGGA dataset was systematically estimated using the ssGSEA algorithm (Fig. 5a). The correlation of ITGB2 expression with infiltrating levels of immune cells was evaluated by Pearson method, which revealed close relationships between ITGB2 with T cells, macrophages, and B cells (Fig. 5b). Previous study reported that ITGB2 is expressed on immune cells [40]. Similarly, we demonstrated that ITGB2 was predominantly located on monocytes/macrophages and CD8+ exhausted T cells in glioma (based on single cell RNA sequencing data obtained from TISCH [26] with GEO ID: GSE131928 [27]) (Supplementary Fig. 2a, b, c), which further implied ITGB’s role in remodeling the immune microenvironment. Furthermore, we utilized GEPIA2021 [31] (http://gepia2021.cancer-pku.cn/) to investigate the expression of ITGB2 in macrophages of different states, and the results demonstrated that ITGB2’s expression was significantly higher in M2 macrophages compared to M0 and M1 macrophages (Supplementary Fig. 2d, e), indicating that ITGB2 is involved in macrophage polarization, which was an essential step for the remodeling of tumor immune microenvironment (TIME). These above findings suggest that ITGB2 activation in combination with the polarization of M2 macrophages and infiltration of exhausted T cells led to remodeling of TIME and marked tumor progression.
Fig. 5.
ITGB2 is associated with immune infiltration and immune activation in gliomas. a Heatmap showing ITGB2-associated relative abundance of 28 immune cells in gliomas, annotations show corresponding clinical features of each sample. b The correlation between the ssGSEA scores of 28 immune cells and the expression of ITGB2 in gliomas. c Heatmap showing ITGB2-associated GSVA scores of 25 innate and adaptive immunity-related gene sets. d The correlation between the GSVA scores of 25 innate and adaptive immunity-related gene sets and the expression of ITGB2 in gliomas. e The fraction of TILCs in ITGB2 high and low subgroups. Within each group, the scattered dots represent TILCs expression values. The thick line represents the median value. The bottom and top of the boxes are the 25th and 75th percentiles, interquartile range. The statistical difference of two subgroups was compared through the Kruskal–Wallis test
Also, we analyzed the KEGG pathways and GO terms with the DEGs (differentially expressed genes). According to the KEGG analysis results, complement and coagulation cascades, pertussis, focal adhesion and ECM−receptor interaction were remarkably enriched (Supplementary Fig. 3a). Among the biological process terms of GO, most of DEGs were enriched in neutrophil degranulation, neutrophil activation involved in immune response, and collagen-containing extracellular matrix (Supplementary Fig. 3b, c).
Gene set enrichment analysis (GSEA) was also used to explore the mechanisms of ITGB2 in gliomas. The CGGA expression data were analyzed with “MsigdbC2REACTOME” (REACTOME gene set, based on R package “Pi”). The enrichment results are shown in Additional file 1: Sheet 6. Results showed that various immune activation and tumor progression-associated genes were enriched, especially in cytokine signaling in immune system, cell cycle and PD-1 signaling (Supplementary Fig. 3d), reflecting relatively enhanced tumor progression and activated inflammation.
Correlation between ITGB2 and immune phenotypes and TIICs of gliomas
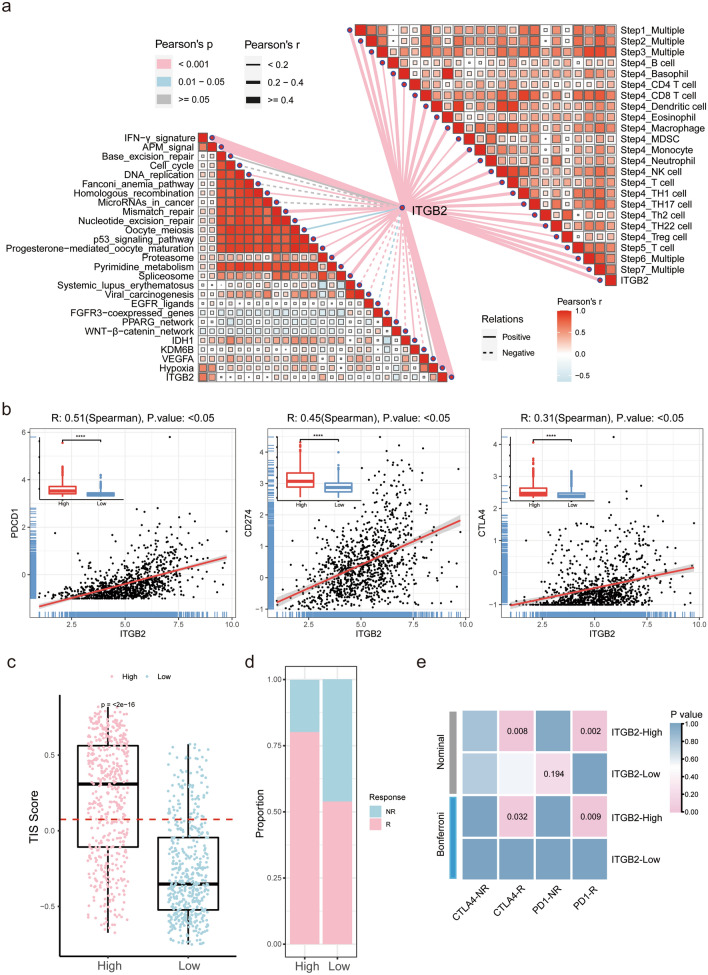
To further explore the presence of malignant gliomas with active immune phenotype, an artificially managed set of genes associated with adaptive and innate immune responses was used to quantify the immunophenotype [30]. As shown in heatmap, with the ITGB2 expression increased, the immune phenotype trended to be “hot”. The quantification of the result showed a high correlation between the expression of ITGB2 with PDL1 signaling (r = 0.62, P < 0.05), CTLA4 signaling (r = 0.45, P < 0.05), and T cell-mediated immunity (r = 0.50, P < 0.05) via Spearman’s test (Fig. 5c, d), which further confirmed the findings in GSEA results.
Considering the correlation between ITGB2 and T cells, the relative abundances of 24 types of tumor infiltrating immune cells (TIICs) of gliomas were quantified with ImmuCellAI [36]. It is worth noting that the proportion of TIICs was significantly different between ITGB2 high and low subgroups (Fig. 5e).
The activities of the cancer immunity cycle are a direct comprehensive performance of the functions of the chemokine system and other immunomodulators. In the high-ITGB2 subgroup, activities of the majority of the steps in the cycle were found to be up-regulated, including the release of cancer cell antigens (Step 1), priming and activation (Step 3), and trafficking of immune cells to tumors (Step 4) (CD8 T cell recruiting, macrophage recruiting, Th1 cell recruiting, NK cell recruiting, DC recruiting, and TH17 recruiting). Subsequently, the increased activities of these steps may increase the infiltration levels of effector TIICs in the glioma immune microenvironment (Fig. 6a, top-right).
Fig. 6.
Correlation between ITGB2 and immune phenotypes and TIICs of gliomas and ITGB2’s potential in predicting immunotherapy responses of gliomas. a (Top right) Correlations between ITGB2 and the steps of the cancer immunity cycle. (Bottom left) Correlations between ITGB2 and the enrichment scores of immunotherapy-predicted pathways. b The correlation between the expression of ITGB2 and PDCD1 (PD-1), CD274 (PDL-1) and CTLA4 in CGGA cohort. c T-cell inflammatory signature (TIS) scores across ITGB2 subgroups. A point presents a single glioma sample. Red line indicates the median value. d Rates of the different anti-PD1 and anti-CTLA4 responses of patients from the CGGA cohort predicted by the ImmunCellAI in the high or low ITGB2 subgroups. e SubMap analysis revealed that ITGB2-high subgroup could be more sensitive to immunotherapy (Bonferroni corrected P value < 0.05)
Subgroups divided by ITGB2 expression predict potential immunotherapy responses of gliomas
We analyzed the correlations between ITGB2 and the ICB response-related signatures [35]. ITGB2 positively correlated with the enrichment scores for immunotherapy-related positive signatures, such as IFN γ signature (Fig. 6a, bottom-left). The above findings suggested that ITGB2 was closely associated with T cells, which play an important role in immunosurveillance evasion in malignant gliomas [41].
Interestingly, TIICs levels were up-regulated in the high-ITGB2 group, while immune activation and tumor progression were both enriched in ITGB2 high group. This may be due to the high expression of PD1 (PDCD1)/PDL1 (CD274) and CTLA4 in the high-ITGB2 group. We used the linear regression model and found ITGB2 showed significant correlations with PDCD1(r = 0.51, P < 0.05), CD274(r = 0.45, P < 0.05) and CTLA4(r = 0.31, P < 0.05) (Fig. 6b); the same conclusions were also drawn in analysis of TCGA GBMLGG dataset (Supplementary Fig. 4a). ITGB2-high subgroup was characterized by immune activation and also accompanied with immune suppression, this feature explained why immune activation enriched in ITGB2-high subgroup, but did not hinder tumor progression.
To verify transcriptome results from public datasets, 20 patients from Shanghai general hospital were included in our study and quantitative real-time PCR was utilized to investigate the correlation between expression levels of ITGB2 and immune checkpoints, and the results showed that ITGB2 was positively correlated with PDCD1 (r = 0.58, p = 0.0072), CTLA4 (r = 0.51, p = 0.0219), TIM3 (HAVCR2) (r = 0.58, p = 0.0079), TIGIT (r = 0.59, p = 0.0065) and IDO1 (r = 0.52, p = 0.0179) (Supplementary Fig. 4b). Meanwhile, we used CGGA and TCGA datasets to verify the correlation between ITGB2 and these immune checkpoints, and the results were consistent with the conclusions obtained from the above experiments (CTLA4 (r = 0.24, CGGA; r = 0.24, TCGA), TIM3(HAVCR2) (r = 0.77, CGGA; r = 0.95, TCGA), TIGIT (r = 0.24, CGGA; r = 0.22, TCGA) and IDO1 (r = 0.39, CGGA; r = 0.52, TCGA)) (Supplementary Fig. 4c, d); ITGB2 was positively correlated with the expression of PDCD1, CTLA4, TIM3, TIGIT and IDO1, which indicated a hypothetic treatment as immune checkpoint.
To further validate this hypothesis, we utilized T cell inflammatory signature (TIS) scores in high and low ITGB2 subgroups. Patients with high ITGB2 expression get higher scores in the TIS signature (Fig. 6c) (P < 0.001), reporting to be correlated with response to anti-PDL1 checkpoint inhibitor pembrolizumab, which supports the hypothesis. Meanwhile, ImmunCellAI also suggested that high levels of ITGB2 tended to more likely respond to immunotherapy (80.0%, 405/506, CGGA) than low ITGB2 subgroup (54.0%, 274/507, CGGA) (Fig. 6d), with high predictive efficacy of ITGB2 for immunotherapy response in glioma patients (AUC: CGGA 69.05% (67.83–72.94%)) (Supplementary Fig. 5a).
We also utilized the SubMap algorithm to compare the similarity of the expression profiles between the two ITGB2 subgroups of glioma patients and 47 previous melanoma patients with detailed immunotherapeutic information, and revealed that patients in ITGB2-high subgroup were more responsive to anti-PD1 and anti-CTLA4 treatment (Bonferroni corrected P value = 0.032 and 0.009, respectively) (Fig. 6e), which was consistent with the previous conclusions.
To sum up, ITGB2 may be a good index for quantifying the tumor immune microenvironment and prediction for immunotherapy responses of gliomas.
Discussion
Due to the high heterogeneity of glioma, the individual case is highly variable [42]. Therefore, the treatment of glioma needs comprehensive consideration based on the individual prognostic factors, clinical symptoms, side effects and tumor progression [43]. Genetic examination can be used to guide radiotherapy and chemotherapy. For an instance, people with mutations in isocitrate dehydrogenase gene 1 (IDH1) and 2 (IDH2) have a more favorable prognosis and clinical response after radiotherapy and chemotherapy [44, 45]. Besides, patients with methylated MGMT status have predictive value on the benefit of chemotherapy [46–48] and the ones with 1p19q co-deletion considered not to administrated with radiotherapy [49, 50], while there are still lack of biomarkers to guide adjuvant immunotherapy. Thus, we aim to explore the underlying mechanism of ITGB2 in the progression of glioma, and its potential immune activation and sensitivity to immunotherapy responses in patients with glioma.
ITGB2, also known as CD18, is the key subunit of β2 integrin, which actively participates in the immune response of the body. At present, β2 integrin, considered as a target therapy for autoimmune diseases, has attached much attention. It is well known that T cells, which express LFA-1, are closely related to tumor progression clinical prognosis, and response to immunotherapy in both of human and mice [41]. However, there are few researches clarifying the relationship between ITGB2 and tumor immunotherapeutic target. In our present study, we found that ITGB2 is highly expressed in a variety of cancers, especially in glioma. We observed that patients with high levels of ITGB2 showed shorter OS, PFI, and DSS than the one with low ITGB2 expression. In order to further explore the correlation between ITGB2 and glioma, the data from CGGA were divided into high- and low-ITGB2 subgroups according to the median value of ITGB2 expression. The results showed that high ITGB2 expression was more likely to be associated with age, poor prognosis, high grade, GBM type, mesenchymal subtype, IDH wild type, and different therapeutic options. Therefore, we speculated that ITGB2 can be served as a predictor for clinical prognosis of glioma patients.
It is known that the prognosis of glioma is related to the infiltration and activation of immune cells [38]. The following bioinformatics analysis showed there was close relationship between ITGB2 and immune progress, which indicated ITGB2’s role in glioma immune microenvironment. In this study, we observed that ITGB2-high group is characterized by immune activation and also accompanied with immune suppression; this feature explains why immune activation enriched in ITGB2-high subgroup, but did not hinder tumor progression. Previous study reported that ITGB2 is expressed on immune cells [40]. Similarly, we demonstrated that ITGB2 was predominantly located on monocytes/macrophages and CD8+ exhausted T cells in glioma, which further implied ITGB’s role in remodeling the immune microenvironment. Moreover, the expression of ITGB2 was significantly higher in M2 macrophages compared to M0 and M1 macrophages. The above findings suggest a critical role for ITGB2 in the remodeling of the immune component of TIME of glioma.
Generally, tumor cells can form a pro-tumor progression microenvironment together with stromal cells, immune cells, vascular endothelial cells, and their secretory factors and extracellular matrix components [51]. These immune cells and associated stromal components, which can be recruited and activated by tumor cells, constructed an anti-tumor inflammatory microenvironment during the early stage of tumor formation, thereby impeding tumor development [52]. With tumor progression, immune cells that infiltrated in the tumor microenvironment, not only play an anti-tumor role, but also promote immune evasion of tumors and tumor growth [53, 54]. In recent decades, tumor immunotherapy tended to be a research focus and has achieved remarkable results in tumor therapy [55, 56]. As is shown in our study, we found that DEGs between ITGB2 subgroups were enriched in immune response and inflammatory response. According to the results of GSEA, we found that a variety of pathways related with immune activation and tumor progression were enriched, especially cytokine signal and PD1 signal transduction. We clarified that the immune microenvironment in glioma with high ITGB2 levels tended to be “hot”. Furthermore, we found that the ITGB2-high subgroup expressed higher levels of PD1, PDL1 and CTLA4 compared with ITGB2-low subgroup, indicating that ITGB2-high subgroup is more likely to be in a state of immune suppression, which inhibits the function of immune cells. Based on these findings, we investigated the relationship between ITGB2 and immune checkpoints and sensitivity of different subgroups of ITGB2 to immunotherapy response. The analysis showed that strong correlations were shown between ITGB2 and immune checkpoints, suggesting that ITGB2 could be assumed as a new immune checkpoint for immunotherapy of glioma. We speculated that ITGB2 could serve as a therapeutic target for glioma and broaden its immunological application.
Immune checkpoint blockade (ICB) uses immune checkpoint inhibitors to block inhibitory signaling and directly stimulate the activation of cytotoxic T lymphocytes to achieve anti-tumor effects [57, 58], with the promoting function in killing ability of T cells against cancer cells. Although the immune system can recognize malignant tumor cells, due to the up-regulation of suppressive immune checkpoints in the tumor microenvironment, the inactivation of anti-tumor T cells leads to the ineffective immune response to cancer. Because both of CTLA4 and PD1 are commonly expressed on the surface of T cells, ICB can reduce the size of tumors and anti-CTLA4 monoclonal antibody and PD1/PDL1 can enhance their immune response to cancer by blocking drugs CTLA4 and PD1/PDL1 signaling pathways [59, 60]. In our study, the relative abundances of 24 types of immune cells in the TIME of gliomas were quantified with ImmuCellAI, and we found that the relative abundance of many kinds of T cells changed significantly in the high expression group of ITGB2. Additionally, we found that patients in high ITGB2 subgroups get higher TIS scores, reporting to be correlated with response to anti-PDL1 checkpoint inhibitor pembrolizumab. In order to further demonstrate the predictive value of ITGB2 for immunotherapy response, we used ImmuCellAI and SubMap algorithm to predict the possibility of immunotherapy response in patients with glioma, all the results confirmed the predictive value of ITGB2 in predicting the immunotherapy response of glioma patients. Notably, it is the first time to illustrate the ITGB2 as a novel diagnostic and therapeutic target for gliomas.
Despite these findings, there are existing limitations. The data of samples were obtained from CGGA, TCGA, and GEO database, and the particular information about the extent of surgical resection was not provided, which is a critical factor for overall survival. Therefore, further analysis with more detailed clinical information should be presented in the following studies. Besides, we lack sufficient clinical data to validate the predictive value of ITGB2 for glioma immunotherapy response, which require further effort in our future studies. At last, we did not systematically investigate the detailed mechanisms involved in the regulation of TIME by ITGB2, which we will focus on studying in future researches.
In conclusion, our study illustrated that ITGB2 can be a novel effective indicator for predicting the clinical stage, prognosis and immune response of patients with glioma. It is reasonable to suggest that the ITGB2 can be a practical target for immunotherapy in patients with glioma. These results are of great clinical significance, which will be conducive to the precise treatment of patients with glioma.
Supplementary Information
Below is the link to the electronic supplementary material.
ITGB2 expression and correlation with immune signatures in pan-cancer. (a) ITGB2 has a significant relationship with tumor microenvironment in various tumors. Supplementary Figure 2. ITGB2 involved in remodeling tumor immune microenvironment.(a) U-MAP plot demonstrating clustering obtained from single cell RNA sequencing (GSE131928). Clusters annotations: CD8Tex, Mono/Macro, AC-like Malignant, MES-like Malignant, OPC-like Malignant, NPC-like Malignant, Oligodendrocyte. (b-c) ITGB2 specifically expressed in Mono/Macro and CD8Tex. (d-e) GEPIA2021 demonstrated that ITGB2's expression was significantly higher in M2 macrophages compared to M0 and M1 macrophages. Supplementary Figure 3. Gene function enrichment analysis between ITGB2 high and low subgroups. (a) KEGG results for differential expression genes between ITGB2 high and low subgroups. The X-axis represents gene counts and the Y-axis represents different enriched pathways (log2 fold change> 1.5). (b) GO (Gene Ontology) results for differential expression genes (Cut-off criteria for DEGs significance was adj. p value< 0.05 and the absolute value of the log2 fold change> 1.5). The X-axis represents gene ratio and the Y-axis represents different enriched pathways (BP: biological progress; CC: cellular component; MF: molecular function). (c) The plot of top three GO pathways for differential expression genes. (d) Rank-based gene set enrichment analysis shows significantly activated immune related pathways in ITGB2 high subgroup compared with ITGB2 low subgroup (LFC, log fold change). Supplementary Figure 4. Validation of correlation between ITGB2 and immune checkpoints in CGGA and TCGA cohorts. (a) The correlation between PDCD1, CD274, CTLA4 and ITGB2 in TCGA dataset. (b) Correlation between mRNA expression of ITGB2 and PDCD1, CTLA4, TIM3 (HAVCR2), IDO1 and TIGIT in tumor tissues obtained from glioma patients (n = 20). (c) The correlation between TIGIT, IDO1, TIM3(HAVCR2) and ITGB2 in CGGA dataset. (d) The correlation between TIGIT, IDO1, TIM3(HAVCR2) and ITGB2 in TCGA dataset. Supplementary Figure 5. Association between ITGB2 expression and immunotherapeutic response. (a) ROC curves for ITGB2 in predicting the immunotherapy response of glioma patients. (PDF 10377 KB)
Marker genes of 28 immune cells. Supplementary Sheet 2. Marker genes of 25 immune-related pathways. Supplementary Sheet 3. Marker genes of the steps of the cancer immunity cycle. Supplementary Sheet 4. Marker genes of immunotherapy-predicted pathways. Supplementary Sheet 5. Marker genes of TIICs (Based on ImmuCellAI). Supplementary Sheet 6. Marker genes of TIS scores. Supplementary Sheet 7. GSEA result of ITGB2 high vs low subgroup. (XLSX 66 KB)
Acknowledgements
The authors would like to thank Zhenyu Song from Fudan University for his generous and outstanding help in this research design. We thank openbiox community and Hiplot team (https://hiplot.com.cn) for providing technical assistance and valuable tools for data analysis and visualization.
Abbreviations
- ACC
Adrenocortical carcinoma
- BLCA
Bladder urothelial carcinoma
- BP
Biological process
- BRCA
Breast invasive carcinoma
- CC
Cellular component
- CESC
Cervical squamous cell carcinoma and endocervical adenocarcinoma
- CGGA
Chinese glioma genome atlas
- CHOL
Cholangiocarcinoma
- COAD
Colon adenocarcinoma
- CTLA4
Cytotoxic T-lymphocyte-associated protein 4
- DEG
Differential expressed gene
- DLBC
Lymphoid neoplasm diffuse large B-cell lymphoma
- DSS
Disease-specific survival
- ESCA
Esophageal carcinoma
- GBM
Glioblastoma multiforme
- GEO
Gene expression omnibus
- GO
Gene ontology
- GSEA
Gene set enrichment analysis
- HNSC
Head and neck squamous cell carcinoma
- ImmuCellAI
Immune cells abundance identifier
- ICB
Immune checkpoint blockades
- IDO1
Indoleamine 2,3-dioxygenase
- IHC
Immunohistochemistry
- ITGB2
Integrin beta-2
- KEGG
Kyoto encyclopedia of genes and genomes
- KICH
Kidney chromophobe
- KIRC
Kidney renal clear cell carcinoma
- KIRP
Kidney renal papillary cell carcinoma
- LCML
Chronic myelogenous leukemia
- LGG
Brain lower-grade glioma
- LIHC
Liver hepatocellular carcinoma
- LUAD
Lung adenocarcinoma
- LUSC
Lung squamous cell carcinoma
- MESO
Mesothelioma
- MF
Molecular function
- OS
Overall survival
- OV
Ovarian serous cystadenocarcinoma
- PAAD
Pancreatic adenocarcinoma
- PCPG
Pheochromocytoma and paraganglioma
- PD-1
Programmed cell death 1
- PDL-1 (CD274)
Programmed cell death 1 ligand 1
- PFI
Progression-free interval
- PRAD
Prostate adenocarcinoma
- READ
Rectum adenocarcinoma
- SARC
Sarcoma
- SKCM
Skin cutaneous melanoma
- SubMap
Subclass mapping
- STAD
Stomach adenocarcinoma
- TCGA
The cancer genome atlas
- TGCT
Testicular germ cell tumors
- THCA
Thyroid carcinoma
- THYM
Thymoma
- TIICs
Tumor-infiltrating immune cells
- TIGIT
T cell immunoreceptor with Ig and ITIM domains
- TIM3 (HAVCR2)
(T-cell immunoglobulin and mucin domain-containing protein 3/hepatitis A virus cellular receptor 2)
- TIS
T-cell inflammation
- UCEC
Uterine corpus endometrial carcinoma
- UCS
Uterine carcinosarcoma
- UVM
Uveal melanoma
Author contributions
HX, AZ and XH collected clinical data. HX, WW performed data analysis. HX, AZ and ML designed and wrote the manuscript. WW and ML contributed to discussion.
Funding
This research was funded by Natural Science Foundation of Shanghai (18ZR1430400).
Data availability
Publicly available datasets were analyzed in this study. These data can be found here: http://gliovis.bioinfo.cnio.es/ and http://tisch.comp-genomics.org/home/. The supplementary material for this article can be found online. All processed data and R codes used in this study can be obtained from the corresponding author on reasonable request.
Declarations
Conflict of interests
The authors declare that they have no competing interests.
Ethical approval
The study was approved by the Human Investigation Ethical Committee of Shanghai General Hospital, and the written informed consent was obtained from all patients.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Houshi Xu, Anke Zhang and Xiaying Han have contributed equally.
Contributor Information
Wei Wang, Email: wangweiocean@163.com.
Meiqing Lou, Email: Meiqing_Lou2020@163.com.
References
- 1.Ostrom QT, et al. Epidemiology of intracranial gliomas. Prog Neurol Surg. 2018;30:1–11. doi: 10.1159/000464374. [DOI] [PubMed] [Google Scholar]
- 2.Ostrom QT, et al. The epidemiology of glioma in adults: a “state of the science” review. Neuro Oncol. 2014;16(7):896–913. doi: 10.1093/neuonc/nou087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Louis DN, et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114(2):97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bush NAO, Chang SM, Berger MS. Current and future strategies for treatment of glioma. Neurosurg Rev. 2017;40(1):1–14. doi: 10.1007/s10143-016-0709-8. [DOI] [PubMed] [Google Scholar]
- 5.Zeng T, Cui D, Gao L. Glioma: an overview of current classifications, characteristics, molecular biology and target therapies. Front Biosci (Landmark edition) 2015;20:1104–1115. doi: 10.2741/4362. [DOI] [PubMed] [Google Scholar]
- 6.Cui H, et al. NF-YC in glioma cell proliferation and tumor growth and its role as an independent predictor of patient survival. Neurosci Lett. 2016;631:40–49. doi: 10.1016/j.neulet.2016.08.003. [DOI] [PubMed] [Google Scholar]
- 7.Latchman Y, et al. PD-L2 is a second ligand for PD-1 and inhibits T cell activation. Nat Immunol. 2001;2(3):261–268. doi: 10.1038/85330. [DOI] [PubMed] [Google Scholar]
- 8.Parry RV, et al. CTLA-4 and PD-1 receptors inhibit T-cell activation by distinct mechanisms. Mol Cell Biol. 2005;25(21):9543–9553. doi: 10.1128/MCB.25.21.9543-9553.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Adorno-Cruz V, Liu H. Regulation and functions of integrin α2 in cell adhesion and disease. Gen Dis. 2019;6(1):16–24. doi: 10.1016/j.gendis.2018.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jung S, Yuki K. Differential effects of volatile anesthetics on leukocyte integrin macrophage-1 antigen. J Immunotoxicol. 2016;13(2):148–156. doi: 10.3109/1547691X.2015.1019596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Varga G, et al. Active MAC-1 (CD11b/CD18) on DCs inhibits full T-cell activation. Blood. 2007;109(2):661–669. doi: 10.1182/blood-2005-12-023044. [DOI] [PubMed] [Google Scholar]
- 12.Lukácsi S, et al. The role of CR3 (CD11b/CD18) and CR4 (CD11c/CD18) in complement-mediated phagocytosis and podosome formation by human phagocytes. Immunol Lett. 2017;189:64–72. doi: 10.1016/j.imlet.2017.05.014. [DOI] [PubMed] [Google Scholar]
- 13.Yakubenko VP, Yadav SP, Ugarova TP. Integrin alphaDbeta2, an adhesion receptor up-regulated on macrophage foam cells, exhibits multiligand-binding properties. Blood. 2006;107(4):1643–1650. doi: 10.1182/blood-2005-06-2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bednarczyk M, et al. β2 integrins-multi-functional leukocyte receptors in health and disease. Int J Mol Sci. 2020;21(4):1402. doi: 10.3390/ijms21041402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cooper J, Giancotti FG. Integrin signaling in cancer: mechanotransduction, stemness, epithelial plasticity, and therapeutic resistance. Cancer Cell. 2019;35(3):347–367. doi: 10.1016/j.ccell.2019.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu M, et al. LncRNA ITGB2-AS1 could promote the migration and invasion of breast cancer cells through up-regulating ITGB2. Int J Mol Sci. 2018;19(7):1866. doi: 10.3390/ijms19071866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chang CM, et al. Innate immunity gene polymorphisms and the risk of colorectal neoplasia. Carcinogenesis. 2013;34(11):2512–2520. doi: 10.1093/carcin/bgt228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ma J, et al. Innate immune cell-derived microparticles facilitate hepatocarcinoma metastasis by transferring integrin α(M)β2 to tumor cells. J Immunol (Baltimore, Md.: 1950) 2013;191(6):3453–3461. doi: 10.4049/jimmunol.1300171. [DOI] [PubMed] [Google Scholar]
- 19.Almeida J, et al. Adipocyte proteome and secretome influence inflammatory and hormone pathways in glioma. Metab Brain Dis. 2019;34(1):141–152. doi: 10.1007/s11011-018-0327-y. [DOI] [PubMed] [Google Scholar]
- 20.Rajaraman P, et al. Common variation in genes related to innate immunity and risk of adult glioma. Cancer Epidemiol Biomarkers Prev Publ Am Assoc Cancer Res Cosponsored Am Soc Prev Oncol. 2009;18(5):1651–1658. doi: 10.1158/1055-9965.EPI-08-1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gladitz J, Klink B, Seifert M. Network-based analysis of oligodendrogliomas predicts novel cancer gene candidates within the region of the 1p/19q co-deletion. Acta Neuropathol Commun. 2018;6(1):49. doi: 10.1186/s40478-018-0544-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou P, et al. No association of VAMP8 gene polymorphisms with glioma in a Chinese han population. Int J Clin Exp Pathol. 2015;8(5):5681–5687. [PMC free article] [PubMed] [Google Scholar]
- 23.Chen Y, et al. VAMP8 facilitates cellular proliferation and temozolomide resistance in human glioma cells. Neuro Oncol. 2015;17(3):407–418. doi: 10.1093/neuonc/nou219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang S, Liu X. The UCSCXENATOOLS R package: a toolkit for accessing genomics data from UCSC xena platform, from cancer multi-omics to single-cell RNA-seq. J Open Sour Softw. 2019;4(40):1627. doi: 10.21105/joss.01627. [DOI] [Google Scholar]
- 25.Bowman RL, et al. GlioVis data portal for visualization and analysis of brain tumor expression datasets. Neuro Oncol. 2017;19(1):139–141. doi: 10.1093/neuonc/now247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun D, et al. TISCH: a comprehensive web resource enabling interactive single-cell transcriptome visualization of tumor microenvironment. Nucleic Acids Res. 2020;49(D1):D1420–D1430. doi: 10.1093/nar/gkaa1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neftel C, et al. An integrative model of cellular states, plasticity, and genetics for glioblastoma. Cell. 2019;178(4):835–849. doi: 10.1016/j.cell.2019.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jia Q, et al. Local mutational diversity drives intratumoral immune heterogeneity in non-small cell lung cancer. Nat Commun. 2018;9(1):5361. doi: 10.1038/s41467-018-07767-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.García-Mulero S, et al. Lung metastases share common immune features regardless of primary tumor origin. J Immunother Cancer. 2020;8(1):e000491. doi: 10.1136/jitc-2019-000491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li C, et al. GEPIA2021: integrating multiple deconvolution-based analysis into GEPIA. Nucleic Acids Res. 2021;49(W1):W242–W246. doi: 10.1093/nar/gkab418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu G, et al. Clusterprofiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fang H, et al. A genetics-led approach defines the drug target landscape of 30 immune-related traits. Nat Genet. 2019;51(7):1082–1091. doi: 10.1038/s41588-019-0456-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu L, et al. TIP: a web server for resolving tumor immunophenotype profiling. Can Res. 2018;78(23):6575–6580. doi: 10.1158/0008-5472.CAN-18-0689. [DOI] [PubMed] [Google Scholar]
- 35.Auslander N, et al. Robust prediction of response to immune checkpoint blockade therapy in metastatic melanoma. Nat Med. 2018;24(10):1545–1549. doi: 10.1038/s41591-018-0157-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miao YR, et al. ImmuCellAI: a unique method for comprehensive T-cell subsets abundance prediction and its application in cancer immunotherapy. Adv Sci (Weinheim, Baden-Wurttemberg, Germany) 2020;7(7):1902880. doi: 10.1002/advs.201902880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hoshida Y, et al. Subclass mapping: identifying common subtypes in independent disease data sets. PloS One. 2007;2(11):e1195. doi: 10.1371/journal.pone.0001195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grabowski MM, et al. Immune suppression in gliomas. J Neurooncol. 2021;151(1):3–12. doi: 10.1007/s11060-020-03483-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Varga G, et al. LFA-1 contributes to signal I of T-cell activation and to the production of T(h)1 cytokines. J Invest Dermatol. 2010;130(4):1005–1012. doi: 10.1038/jid.2009.398. [DOI] [PubMed] [Google Scholar]
- 40.Zhang X, et al. ITGB2-mediated metabolic switch in CAFs promotes OSCC proliferation by oxidation of NADH in mitochondrial oxidative phosphorylation system. Theranostics. 2020;10(26):12044–12059. doi: 10.7150/thno.47901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lanitis E, et al. Mechanisms regulating T-cell infiltration and activity in solid tumors. Ann Oncol Off J Eur Soc Med Oncol. 2017;28(Suppl_12):xii18–xii32. doi: 10.1093/annonc/mdx238. [DOI] [PubMed] [Google Scholar]
- 42.Baumert BG, et al. Temozolomide chemotherapy versus radiotherapy in high-risk low-grade glioma (EORTC 22033–26033): a randomised, open-label, phase 3 intergroup study. Lancet Oncol. 2016;17(11):1521–1532. doi: 10.1016/S1470-2045(16)30313-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Douw L, et al. Cognitive and radiological effects of radiotherapy in patients with low-grade glioma: long-term follow-up. Lancet Neurol. 2009;8(9):810–818. doi: 10.1016/S1474-4422(09)70204-2. [DOI] [PubMed] [Google Scholar]
- 44.Dang L, Jin S, Su SM. IDH mutations in glioma and acute myeloid leukemia. Trends Mol Med. 2010;16(9):387–397. doi: 10.1016/j.molmed.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 45.Laurence MG, et al. Oncogenic activities of IDH1/2 mutations: from epigenetics to cellular signaling. Trends Cell Biol. 2017;27(10):738–752. doi: 10.1016/j.tcb.2017.06.002. [DOI] [PubMed] [Google Scholar]
- 46.Bady P, et al. MGMT methylation analysis of glioblastoma on the Infinium methylation beadchip identifies two distinct CpG regions associated with gene silencing and outcome, yielding a prediction model for comparisons across datasets, tumor grades, and CIMP-status. Acta Neuropathol. 2012;124(4):547–560. doi: 10.1007/s00401-012-1016-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van den Bent MJ, et al. MGMT-STP27 methylation status as predictive marker for response to PCV in anaplastic oligodendrogliomas and oligoastrocytomas. A report from EORTC study 26951. Clin Cancer Res Off J Am Assoc Cancer Res. 2013;19(19):5513–5522. doi: 10.1158/1078-0432.CCR-13-1157. [DOI] [PubMed] [Google Scholar]
- 48.Wick W, et al. Prognostic or predictive value of MGMT promoter methylation in gliomas depends on IDH1 mutation. Neurology. 2013;81(17):1515–1522. doi: 10.1212/WNL.0b013e3182a95680. [DOI] [PubMed] [Google Scholar]
- 49.Staedtke V, aDzaye OD, Holdhoff M. Actionable molecular biomarkers in primary brain tumors. Trends Cancer. 2016;2(7):338–349. doi: 10.1016/j.trecan.2016.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Khan IN, et al. Current and emerging biomarkers in tumors of the central nervous system: possible diagnostic, prognostic and therapeutic applications. Semin Cancer Biol. 2018;52(1):85–102. doi: 10.1016/j.semcancer.2017.07.004. [DOI] [PubMed] [Google Scholar]
- 51.Xiao Y, et al. Multi-omics profiling reveals distinct microenvironment characterization and suggests immune escape mechanisms of triple-negative breast cancer. Clin Cancer Res Off J Am Assoc Cancer Res. 2019;25(16):5002–5014. doi: 10.1158/1078-0432.CCR-18-3524. [DOI] [PubMed] [Google Scholar]
- 52.Chen Y-P, et al. Genomic analysis of tumor microenvironment immune types across 14 solid cancer types: immunotherapeutic implications. Theranostics. 2017;7(14):3585–3594. doi: 10.7150/thno.21471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mellman I, Coukos G, Dranoff G. Cancer immunotherapy comes of age. Nature. 2011;480(7378):480–489. doi: 10.1038/nature10673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gil Del Alcazar CR, Alečković M, Polyak K. Immune escape during breast tumor progression. Cancer Immunol Res. 2020;8(4):422–427. doi: 10.1158/2326-6066.CIR-19-0786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Del Paggio JC. Immunotherapy: cancer immunotherapy and the value of cure. Nat Rev Clin Oncol. 2018;15(5):268–270. doi: 10.1038/nrclinonc.2018.27. [DOI] [PubMed] [Google Scholar]
- 56.Patel JM, et al. Peritumoral administration of DRibbles-pulsed antigen-presenting cells enhances the antitumor efficacy of anti-GITR and anti-PD-1 antibodies via an antigen presenting independent mechanism. J Immunother Cancer. 2019;7(1):311. doi: 10.1186/s40425-019-0786-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.López-Soto A, Gonzalez S, Folgueras AR. IFN signaling and ICB resistance: time is on tumor's side. Trends Cancer. 2017;3(3):161–163. doi: 10.1016/j.trecan.2017.01.004. [DOI] [PubMed] [Google Scholar]
- 58.Jiang P, et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat Med. 2018;24(10):1550–1558. doi: 10.1038/s41591-018-0136-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rotte A. Combination of CTLA-4 and PD-1 blockers for treatment of cancer. J Exp Clin Cancer Res CR. 2019;38(1):255. doi: 10.1186/s13046-019-1259-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wei SC, et al. Distinct cellular mechanisms underlie anti-CTLA-4 and anti-PD-1 checkpoint blockade. Cell. 2017;170(6):1120–1133.e17. doi: 10.1016/j.cell.2017.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
ITGB2 expression and correlation with immune signatures in pan-cancer. (a) ITGB2 has a significant relationship with tumor microenvironment in various tumors. Supplementary Figure 2. ITGB2 involved in remodeling tumor immune microenvironment.(a) U-MAP plot demonstrating clustering obtained from single cell RNA sequencing (GSE131928). Clusters annotations: CD8Tex, Mono/Macro, AC-like Malignant, MES-like Malignant, OPC-like Malignant, NPC-like Malignant, Oligodendrocyte. (b-c) ITGB2 specifically expressed in Mono/Macro and CD8Tex. (d-e) GEPIA2021 demonstrated that ITGB2's expression was significantly higher in M2 macrophages compared to M0 and M1 macrophages. Supplementary Figure 3. Gene function enrichment analysis between ITGB2 high and low subgroups. (a) KEGG results for differential expression genes between ITGB2 high and low subgroups. The X-axis represents gene counts and the Y-axis represents different enriched pathways (log2 fold change> 1.5). (b) GO (Gene Ontology) results for differential expression genes (Cut-off criteria for DEGs significance was adj. p value< 0.05 and the absolute value of the log2 fold change> 1.5). The X-axis represents gene ratio and the Y-axis represents different enriched pathways (BP: biological progress; CC: cellular component; MF: molecular function). (c) The plot of top three GO pathways for differential expression genes. (d) Rank-based gene set enrichment analysis shows significantly activated immune related pathways in ITGB2 high subgroup compared with ITGB2 low subgroup (LFC, log fold change). Supplementary Figure 4. Validation of correlation between ITGB2 and immune checkpoints in CGGA and TCGA cohorts. (a) The correlation between PDCD1, CD274, CTLA4 and ITGB2 in TCGA dataset. (b) Correlation between mRNA expression of ITGB2 and PDCD1, CTLA4, TIM3 (HAVCR2), IDO1 and TIGIT in tumor tissues obtained from glioma patients (n = 20). (c) The correlation between TIGIT, IDO1, TIM3(HAVCR2) and ITGB2 in CGGA dataset. (d) The correlation between TIGIT, IDO1, TIM3(HAVCR2) and ITGB2 in TCGA dataset. Supplementary Figure 5. Association between ITGB2 expression and immunotherapeutic response. (a) ROC curves for ITGB2 in predicting the immunotherapy response of glioma patients. (PDF 10377 KB)
Marker genes of 28 immune cells. Supplementary Sheet 2. Marker genes of 25 immune-related pathways. Supplementary Sheet 3. Marker genes of the steps of the cancer immunity cycle. Supplementary Sheet 4. Marker genes of immunotherapy-predicted pathways. Supplementary Sheet 5. Marker genes of TIICs (Based on ImmuCellAI). Supplementary Sheet 6. Marker genes of TIS scores. Supplementary Sheet 7. GSEA result of ITGB2 high vs low subgroup. (XLSX 66 KB)
Data Availability Statement
Publicly available datasets were analyzed in this study. These data can be found here: http://gliovis.bioinfo.cnio.es/ and http://tisch.comp-genomics.org/home/. The supplementary material for this article can be found online. All processed data and R codes used in this study can be obtained from the corresponding author on reasonable request.