Abstract
Primary coenzyme Q10 (CoQ10) deficiency is a group of inborn errors of metabolism caused by defects in CoQ10 biosynthesis. Biallelic pathogenic variants in COQ7, encoding mitochondrial 5-demethoxyubiquinone hydroxylase, have been reported in nine patients from seven families. We identified five new patients with COQ7-related primary CoQ10 deficiency, performed clinical assessment of the patients, and studied the functional effects of current and previously reported COQ7 variants and potential treatment options. The main clinical features included a neonatal-onset presentation with severe neuromuscular, cardiorespiratory and renal involvement and a late-onset disease presenting with progressive neuropathy, lower extremity weakness, abnormal gait, and variable developmental delay. Baker's yeast orthologue of COQ7, CATS, is required for growth on oxidative carbon sources and cat5Δ strain demonstrates oxidative growth defect. Expression of wild-type CAT5 could completely rescue the defect; however, yeast CAT5 harboring equivalent human pathogenic variants could not. Interestingly, cat5Δ yeast harboring p.Arg57Gln (equivalent to human p.Arg54Gln), p.Arg112Trp (equivalent to p.Arg107Trp), p.Ile69Asn (equivalent to p.Ile66Asn) and combination of p.Lys108Met and p.Leu116Pro (equivalent to the complex allele p.[Thr103Met;Leu111Pro]) partially rescued the growth defects, indicating these variants are hypomorphic alleles. Supplementation with 2,4 dihydroxybenzoic acid (2,4-diHB) rescued the growth defect of both the leaky and severe mutants. Overexpression of COQ8 and 2,4-diHB supplementation synergistically restored oxidative growth and respiratory defect. Overall, we define two distinct disease presentations of COQ7-related disorder with emerging genotype-phenotype correlation and validate the use of the yeast model for functional studies of COQ7 variants.
Introduction
Coenzyme Q (CoQ), also known as ubiquinone, is a lipid molecule present in all cell membranes. It has a benzoquinone ring and lipophilic polyisoprenoid tail[1]. The length of the isoprenoid tail is species dependent, but in humans, it is 10 and thus designated as CoQ10[2]. The majority of CoQ10 is made de novo in the cell, with the isoprenoid side chain synthesized via the mevalonate pathway and the benzoquinone ring synthesis involves a multi-protein complex encoded by nuclear COQ genes[1, 2]. CoQ10 acts as an electron shuttle between complexes I and II to complex III in the mitochondrial respiratory chain[2]. Ubiquinone has antioxidant properties and is essential cofactor for uncoupling proteins and several mitochondrial dehydrogenases involved in pyrimidine biosynthesis and fatty acid β-oxidation[2].
Primary CoQ10 deficiency (MIM 616733) is caused by variants in genes involved in the biosynthesis of CoQ10 and associated with variable phenotypes ranging from lethal progressive encephalomyopathy to isolated single organ involvement[3]. Genes known to cause CoQ10 deficiency include COQ2 (MIM 609825)[4, 5], COQ4 (MIM 612898)[6], COQ5 (MIM 616359)[7], COQ6 (MIM 614647)[8], COQ7 (MIM 601683)[9], COQ8A (MIM 606980)[10], COQ9 (MIM 612837)[11], ADCK3 (MIM 606980)[12], PDSS1 (MIM 607429)[13], PDSS2 (MIM 610564)[14] and HPDL (MIM 619026)[15].
COQ7 (MIM 601683) encodes the mitochondrial 5-demethoxyubiquinone (DMQ) hydroxylase (EC: 1.14.99.60), which converts 2-hexaprenyl-3-methyl-6-methoxy-1,4-benzenediol (DMQH2) to 2-hexaprenyl-3-methyl-6-methoxy-1,4,5-benzenetriol (DMeQH2); the latter is further converted to reduced form of CoQ10 by ubiquinone biosynthesis O-methyltransferase encoded by COQ3[16]. DMQ hydroxylase interacts with ubiquinone biosynthesis protein COQ9, encoded by COQ9, a lipid-binding protein[17].
Homozygous missense variants in COQ7 were first reported by Freyer et al[9] in a patient born to consanguineous parents with oligohydramnios, pulmonary hypoplasia, renal disease, hypotonia, sensorineural hearing loss, visual dysfunction, polyneuropathy, pre and post-natal growth restriction, feeding difficulties, developmental delay, developmental regression and elevation of plasma and cerebrospinal fluid (CSF) lactate[9]. Subsequently, five additional patients reported with progressive lower extremities spasticity, abnormal gait, muscle atrophy, and hearing impairment[18-22] (Table 1). More recently, a homozygous variant affecting the initiation codon of COQ7 was identified in 3 siblings with adolescent-onset progressive distal motor neuropathy[23]. Two additional patients, compound heterozygous for the variants p.(Ile66Asn) and p.(Tyr149Cys), with axonal neuropathy and neurodegenerative disorder were reported as part of a next-generation sequencing study in a cohort of 117 patients with probable or possible mitochondrial cause[24].
Table 1.
Clinical and biochemical characteristics of known patients with COQ7-associated primary CoQ10 deficiency
| Freyer et al, 2015 | Wang et al, 2017 | Kwong et al, 2019 | Hashemi et al, 2020 | Wang et al, 2022 |
Jacquier et al, 2022 (II-2) |
Jacquier et al, 2022 (II-1) |
Jacquier et al, 2022 (II-3) |
Sadr, et al, 2023 | Patient 1 |
Patient 2 |
Patient 3 |
Patient 4 |
Patient 5 |
|
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Variant | c.422T>A p.(Val141Glu) | c.332T>C (p.Leu111Pro) c.308C>T (p.Thr103Met) | c.599_600 delinsTAATGCATC (p.Lys200Ilefs*56) c.319C>Tp. (Arg107Trp) | c.332T>C (p.Leu111Pro) c.308C>T (p.Thr103Met) | c.161G>A (p.Arg54Gln) | c.3G>T (p.Met1?) | c.3G>T (p.Met1?) | c.3G>T (p.Met1?) | c.332T>C (p.Leu111Pro) c.308C>T (p.Thr103Met) | c.161G>A (p.Arg54Gln) c.446A>G (p.Tyr149Cys) | c.446A>G (p.Tyr149Cys) c.3G>T (p.Met1?) | c.161G>A (p.Arg54Gln) | c.161G>A (p.Arg54Gln) | c.3G>T (p.Met1?) |
| Zygosity | Homozygous | Homozygous (both) | Compound heterozygous | Homozygous (both) | Homozygous | Homozygous | Homozygous | Homozygous | Homozygous | Compound heterozygous | Compound heterozygous | Homozygous | Homozygous | Homozygous |
| Gender | Male | Female | Male | Female | Male | Male | Female | Male | Male | Male | Female | Female | Female | Female |
| Consanguinity | + | + | − | + | + | − | − | − | + | − | − | + | − | + |
| Gestational age when born | Full term | 37 weeks | 33 weeks | 38 weeks | 39 weeks | N/A | N/A | N/A | N/A | Full term | N/A | 32 weeks | 39 weeks | 38 weeks |
| Prenatal complications | Oligohydramnios, fetal lung hypoplasia, IUGR | Gestational diabetes | IUGR, cardiomegaly, TR, oligohydramnios | − | − | N/A | N/A | N/A | N/A | Oligohydramnios | N/A | Preterm labor. | − | − |
| SGA | + | − | + | − | − | N/A | N/A | N/A | N/A | − | N/A | − | − | N/A |
| Age at onset of symptoms | Birth | 14 months | Birth | 2 years | 15 months | 12 years | 9 years | 10 years | 13 months | Birth | 12 years | 3 years | 3 years | 5 years |
| Age at last examination/evaluation | 9 years | 6 years | 12 months | 9 years | 4.5 years | 36 years | 25 years | 42 years | 7 years | 15 years | 45 years | 9 years | 9 years | 33 years |
| Developmentaldelay | Moderate | − | Profound | − | + | − | − | -- | − | Severe | − | Mild | − | Mild |
| Intellectual disabilities | N/A | − | N/A | − | N/A | N/A | N/A | N/A | − | + | − | + | −. | + |
| Learning difficulties | + | + (Inattention) | N/A | + (Inattention) | N/A | N/A | N/A | N/A | − | + | + | + | − | + |
| Abnormal tone (hypo/hypertonia/spasticity) | Hypotonia, contracture of LE | Spastic paraparesis, mild truncal hypotonia | Generalized hypotonia | Spasticity | Hypotonia, spasticity, contracture LE | LE stiffness | N/A | N/A | Spasticity | Hypotonia, contracture LE | − | LE Spasticity | Spastic paraparesis | LE spasticity |
| LE Weakness | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| LE Spasticity | + | + | + | + | + | + | + | N/A | + | + | − | + | + | + |
| Muscularatrophy | + | + | + | − | N/A | + | N/A | + | − | − | − | + | − | + |
| Abnormal gait | + | + | N/A | + | + | + | + | + | + | + | + | + | + | + |
| Speech delay | − | N/A | N/A | − | + | N/A | N/A | N/A | − | + | − | + | − | + |
| Ophthalmologic | Visual dysfunction | − | Ptosis, bilateral visual loss | − | N/A | Myopia | N/A | N/A | − | Mild myopia | − | − | − | Microsaccadic eye movements during ocular pursuit |
| Audiologic | Mixed HL | Low frequency SNHL | Profound hearing impairment | Mild SNHL | N/A | − | N/A | N/A | Mild | SNHL | − | − | − | − |
| Cardiovascular | Systemic hypertension and LVH | N/A | Severe hypertrophic cardiomyopathy, TR and pericardial effusion | − | N/A | − | N/A | N/A | − | Mild dilation of ascending aorta | − | − | ASD | Arrhythmia |
| Respiratory | Lung hypoplasia with persistent pulmonary Hypertension of newborn | − | Central hypoventilation | − | N/A | N/A | N/A | N/A | − | Lung hypoplasia | − | Respiratory insufficiency inneonatal period. | − | − |
| Renal | Renal dysfunction with small dysplastic kidneys with impaired cortical differentiation | − | Multiple renal cysts and diffusely increased echogenicity with accentuation of cortico-medullary differentiation. | − | N/A | N/A | N/A | N/A | − | − | − | − | − | − |
| Gastrointestinal | Feeding difficulties, FTT | − | Feeding difficulties, FTT | − | N/A | N/A | N/A | N/A | N/A | Feeding difficulties, FTT | − | − | − | − |
| Brain MRI | Normal | Normal | Subdural hematoma, basal ganglial and thalami hypodensities with abnormal lactate peak | Normal | Increased T2A and Flair signal in the supratentorial bilateral periventricular white matt er. | Normal | N/A | N/A | Normal | Left posterior temporal infarction. Elevated lactate peak. | Normal | Enlargement of lateral ventricles, mainly of post erior horns, with thinner of white matter adjacent to ventricles | Thinning of the corpus callosum | N/A |
| EMG | Peripheral sensor imotor polyneuropathy of axonal and demyelinating type | N/A | N/A | Normal | N/A | Pure motor axonal neuropathy affecting predominantly distal lower extremities. . | Pure motor axonal neuropathy affecting predominantly distal lower extremities. . | N/A | N/A | N/A | Motor neuropathy, primarily axonal process | Diffuse involvement of a second motor neuron or axonal motor neuropathy | Motor axonal neuropathy | Motor axonal impairment affecting the distal museulature of all extremities |
| Respiratory chain enzymes | Complex I + III and IV deficiency | N/A | Complex II + III deficiency with normal isolated activity of complex II and complex III | N/A | N/A | N/A | N/A | N/A | N/A | Normal | N/A | Decreased Complex II + III and IV activities | Normal | N/A |
| Biochemical findings | ||||||||||||||
| Blood (amino acids, carnitine/acylcarnitine profile, ie) | N/A | Normal | Elevated alanine | Normal | N/A | N/A | N/A | N/A | N/A | Normal | Increased long-chain acylcarnitine species (C12-C18) | Normal | Normal | N/A |
| Hyperlactatemia (mean value and range) | + | − | + | − | N/A | N/A | N/A | N/A | N/A | + | − | − | − | N/A |
| Urine (amino acids, organic acids, ie) | Moderately increased excretion of fumarate and malate | Normal | Increased lactate, pyruvate, 3-hydroxybutyrate, dicarboxylic aciduria and TCA cycle intermediates. | N/A | Normal | N/A | N/A | N/A | N/A | Increased quinolinic and oxalic acid in infancy. Repeat testing at 7 yo was normal | Increased 4-hydroxy phenylacetate | Normal | Normal | N/A |
| CSF (amino acids, lactate,ie) | Elevated CSF lactate and slightly increased albumin | Elevated CSF lactate, slight decrease in total protein | N/A | N/A | N/A | N/A | N/A | N/A | N/A | Elevated CSF lactate | N/A | N/A | Normal | N/A |
| Treatment | CoQ10 | CoQ10 11.4 mg/kg twice daily | CoQ10 20 mg/kg/d | CoQ10 | None | N/A | N/A | N/A | CoQ10 11.4 mg/kg/d | Ubiquinol 450 mg BID | CoQ10 800 mg/day | CoQ10 300 mg/d | None | N/A |
Abbreviation: ASD- atrial septal defect; CoQ10- Coenzyme Q10; CSF- cerebrospinal fluid; EMG- electromyography; FTT- failure to thrive; HL- hearing loss; IUGR- intrauterine growth retardation; LE- lower extremities; LVH- left ventricular hypertrophy; N/A- not available; SGA- small for gestational age; SNHL- sensorineural hearing loss; TCA- tricarboxylic acids; TR- tricuspid regurgitation. Shaded columns- patients with neonatal-onset presentation of COQ7-associated primary CoQ10 deficiency
Baker’s yeast (Saccharomyces cerevisiae) is a widely used model organism to study genetic disorders. Given its ability to grow in both oxidative and non-oxidative conditions, yeast has been extensively used for functional studies of mitochondrial disorders[25]. CoQ6, yeast’s homologue of CoQ10, has six isoprenoid tail[26]. A knockout of yeast CAT5, the homologue of human COQ7, lead to defective growth on non-fermentable carbon sources, including ethanol and glycerol[27].
Here, we report a new cohort of five patients with primary CoQ10 deficiency caused by biallelic variants in COQ7. We used a yeast model to evaluate functional effects of novel and previously reported COQ7 variants and study treatment options. Based on bioinformatic analyses and functional studies, the variants are disease causing. We establish the utility of the yeast model in CoQ10 biosynthesis defects, expand clinical and genotypic spectrum of COQ7-related primary CoQ10 deficiency, and examine potential treatment modalities.
Materials and Methods
We recruited 5 patients with predicted deleterious variants in COQ7 through a collaborative effort among multiple institutions to characterize the clinical and molecular spectrum associated with COQ7-related primary CoQ10 deficiency. The use of GeneMatcher facilitated the connection of clinical collaborators[28]. No interventions performed on patients and no biological specimens collected from participants for this research study. The guardians of patients signed consent forms for publication approved by Washington University IRB (Media Authorization for the Use and Disclosure of Protected Health Information) (Patient 1), HSHS hospital system (Authorization for Disclosure of Health Information) (Patient 2), SARAH Network Rehabilitation Hospitals (Patient 3), Mendelics Analise Genomica S.A. ("Mendelics") (Patient 4), Hospital das Clinicas da Faculdade de Medicina da Universidade de Sao Paulo (HCFMUSP) (Patient 5).
Next generation sequencing
Clinical trio exome sequencing (ES) for Patients 1 and 2 was performed by GeneDx (Gaithersburg, MD) and for Patients 3-5 by Mendelics (Sao Paulo, Brazil). Sanger sequencing was used to confirm the variants identified in the patients. Variant annotation and interpretation were performed according to companies’ pipeline. Variant classification is performed according to the American College of Medical Genetics recommendations[29].
Yeast strains and growth conditions
The yeast strain used in this study was BY4741 cat5::kanMX4 (MATa; his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 cat5::kanMX4) transformed with the empty vector pFL38 or the vector carrying the wild-type allele (wt), the humanized allele or the mutant alleles of CAT5. CAT5 was amplified and cloned under its natural promoter in the centromeric vector pFL38. CAT5 was mutagenized by a PCR Quick Change technique to obtain humanized and mutant alleles, and cloned into the pFL38 vector. The plasmid (empty, carrying the wild-type, humanized or mutant allele) was introduced in the BY4741 Δcat5 strain using the LiAc-ssDNA-PEG method, after growth in YPAD medium. For all the experiments, except for transformation, cells were grown in liquid SC medium (0.69% YNB without amino acids and 0.5% ammonium sulfate (Formedium™, UK), added of 1 g/L dropout mix without uracil) in constant shaking at 28 °C or in solid SC medium using 20 g/L agar for solidification (Formedium™, UK). Media were supplemented with various carbon sources (Carlo Erba Reagents, Italy) as indicated in the results and figures. For growth analyses, the strains were serially diluted, spotted, and grown at 28 °C and 37 °C on SC medium agar plates supplemented with a fermentable carbon source, 2% glucose, or an oxidative carbon source, 2% acetate.
Oxygen consumption assay
Mitochondrial respiratory activity in yeast was evaluated by measuring oxygen consumption, using a Clark-type oxygen electrode (Oxygraph System Hansatech Instruments England) at 30 °C with 1 ml of air-saturated respiration buffer (0.1 M phthalate–KOH pH 5.0, 0.5% glucose) from yeast cell suspensions cultured for 18 h at 28 °C in liquid SC medium supplemented with 0.6% glucose until exhaustion.
Protein quantification
Yeast cells were grown as for oxygen consumption assay and protein extraction was performed with the trichloroacetic acid method. Proteins were resuspended in Laemmli sample buffer pH 6.8, separated on 4–20% precast gels (Bio-Rad, USA) and Western blotted on nitrocellulose filters. Proteins were detected with primary antibodies for Cat5 (1:2000, kindly provided by C. Clarke from University of California) and Por1 (1:10000; Abcam ab110326), followed by fluorescent secondary antibodies (anti-rabbit StarBright™ Blue 520, anti-mouse StarBright™ Blue 700 1:5000; Bio-Rad, USA), signals were detected using Chemidoc MP Imaging System and quantified with Image Lab software (Bio-Rad, USA).
COQ8 overexpression and 2,4-diHB supplementation
To overexpress COQ8, COQ8 was amplified and cloned under its natural promoter in the vector pFL36. The BY4741 cat5Δ strains expressing cat5 variants on the pFL38 vector, were transformed with the empty vector pFL36 or with pFL36-COQ8 using the LiAc-ssDNA-PEG method, after growth in YPAD medium.
Growth analyses were performed in solid SC medium (without uracil and leucine) supplemented with glucose or acetate as carbon sources (Carlo Erba Reagents, Italy), and with or without the addition of 1mM or 2mM 2,4-dihydroxybenzoic acid (2,4-diHB) and incubated at 28°C. Oxygen consumption assay was performed as described above; when necessary 2mM 2,4-diHB was added in liquid SC medium supplemented with 0.6% glucose. 2,4-diHB was prepared at 1M in 100% ethanol and was conserved at −20°C.
For respiratory complexes subunits quantification, yeast cells were grown as for oxygen consumption assay and protein extraction was performed with the trichloroacetic acid method. Proteins were resuspended in Laemmli sample buffer pH 6.8, separated on 12% running gels and Western blotted on nitrocellulose filters. Proteins were detected with primary antibodies for SdhA (1:1000, ABclonal A2594), Core2 (1:500, kindly provided by B.L. Trumpower), Atp4 (1:10000, kindly provided by J. Velours), Cox2 (1:2500, Abcam Ab110271) and Pgk1 (1:20000, Abcam Ab113687), followed by fluorescent secondary antibodies (anti-rabbit StarBright™ Blue 520, anti-mouse StarBright™ Blue 700 1:7500; Bio-Rad, USA), signals were detected using Chemidoc MP Imaging System and quantified with Image Lab software (Bio-Rad, USA).
Homology modelling
Homology modelling of human 5-demethoxyubiquinone hydroxylase in the 33-187 amino acid range (UniProt sequence code Q99807) was based on the crystal structure of a putative bacterioferritin-related protein (Protein Data Bank, PDB, code 2VZB). The protein sequence alignment (with annotated protein secondary structure elements, and the amino acids coordinating two iron ions) is shown in Supplementary File 1. The protein model was built with SCWRL4[30]. The two iron atoms were bound to the model in positions corresponding to those observed in the putative bacterioferritin-related protein.
Results
Patients description
Patient 1
Patient 1 is a 19-year-old male of Northern European ancestry. He was born full term via vaginal delivery to a 35-year-old mother after a pregnancy complicated by oligohydramnios requiring amnio infusion. His birth weight was 3070g (28th centile), length was 52.5cm (92nd centile), and occipito-frontal circumference (OFC) was 33cm (12th centile). Apgar scores were 6, and 9 at 1 and 5 minutes, respectively. He had poor tone, and decreased air entry bilaterally. He was intubated and admitted to the NICU, requiring conventional mechanical ventilation for 4 days. He had multiple pneumothoraces, pulmonary hypoplasia based on chest X-rays, and feeding difficulties. He received total parenteral nutrition on day of life (DOL) 2-9. He required placement of nasogastric (NG) tube due to poor suck. He had posturing at 3 weeks of life, which prompted brain imaging studies that showed hemorrhagic left middle cerebral artery infarction. Brain imaging performed at 1 month of age revealed well-defined area of cortical and subcortical left temporal infarction and early tissue loss. EEG at the time was normal. He was discharged from the NICU at 8 weeks of age.
At 4 months of age, he was admitted for failure to thrive (FTT), and noted to have hypotonia, opisthotonus, flexion contractures of the lower extremities, and myotonic jerks of his upper extremities. MRI at that time, revealed delayed myelination, increased signal in the pontine tegmentum, and minimal left temporal encephalomalacia, and brain magnetic resonance spectroscopy (MRS) showed lactate peaks in the pons and deep white matter. He had elevated lactate in blood at 8.2 mmol/L (Nl: 0.5-2.2), and CSF at 5.04 mmol/L, (Nl: 0.8-2.4), elevated pyruvate at 0.16mM, (Nl: 0.03-0.12) with elevated lactate:pyruvate ratio at 32, (10-20). Due to continued poor feeding, he required G-button feeding between 4 to 9 months of age. Additional details of his workup are provided in Supplementary Material.
The patient has had global developmental delay since birth, with no head control until age 9 months, and was not able to sit unsupported until age 13 months. He was started on levocarnitine and a “mitochondrial cocktail” including vitamin E, ascorbic acid, pyridoxine hydrochloride, thiamine hydrochloride, and coenzyme Q. At that point, he was given the diagnosis of Leigh disease and enrolled into a dichloroacetic acid treatment trial, and continued this medication for ~7 years. He continued receiving the vitamin cocktail (CoQ10 dose was 5-10 mg/kg/day) until his COQ7-related disorder was established. His CoQ10 was then switched to ubiquinol 450 mg twice daily (~20 mg/kg/day).
He had bilateral inguinal hernia repair and orchiopexy at the age of two. He was diagnosed with moderate sensorineural hearing loss (SNHL) at the age of seven. He was diagnosed with ADHD at the age of 8 and displayed autistic traits, marked echolalia, and gesture imitations of others. Ophthalmologic examination showed mild myopic refractive error but no retinal abnormalities. At the age of 12, he had neurodevelopmental testing that showed intelligence quotient (IQ) of 44. He presented to our institution at age 14 years.
The proband’s family history is notable for two male siblings who died on day of life 1. They both had oligohydramnios and lung hypoplasia. They had no genetic testing and the underlying diagnosis was unknown. The proband has a 21-year-old healthy brother. The proband’s mother had one first trimester miscarriage. Parents are non-consanguineous and both are alive and healthy.
On physical examination at age 19 years, patient’s height was 160.4cm (16th centile), weight 44.8kg (19th centile), OFC was 53.8cm (40th centile). The proband exhibited echolalia, anticipatory anxiety, and dysarthria, and spoke in short sentences. He could follow simple commands but needed prompting for more complex commands. He displayed axial and appendicular hypotonia. The proximal and distal muscle strength (Medical Research Council Grade) in upper extremities was 5/5, bilateral tibialis anterior was 3/5, and bilateral hamstring and quadriceps was 4/5. He had significantly reduced ankle range of motion and mild bilateral plantarflexion contractures. There was mild appendicular ataxia bilaterally and his gait was widely based.
Patient 2
Patient 2 is a 45-year-old female who began experiencing muscle weakness symptoms around 12 years of age, such as tripping while climbing stairs. Since then, she had slowly but inexorably progressive weakness of her lower limbs. At the age of 16, she was clinically diagnosed with Charcot-Marie-Tooth (CMT). She began to notice weakness in her upper limbs in late twenties. Around the same time, she also experienced a mild decrease in temperature and pain sensation in a glove-stock distribution, as well as bilateral foot drop. At the age of 41, she was dependent on scooter for transportation. At the time, a spinal MRI revealed degenerative changes of the cervical and thoracic spines and brain MRI revealed nonspecific T2 hyperintensity in the left subcortical frontal white matter. An EMG was performed, which showed abnormal motor nerve conduction velocities (motor neuropathy) consistent with a primary axonal process. Creatine kinase level was normal. She had a history of learning disabilities and dropped out of school in the 11th grade. She had no history of developmental delay, hearing or vision loss, renal or cardiac abnormalities, or spasticity. Of note, her sister, who has similar symptoms but is more severely affected, was wheelchair-bound by the age of 30. Further details regarding her sister are unavailable.
Upon her latest evaluation at the age of 44, she was able to ambulate around her home with a walker, but is not able to walk independently. Physical examination revealed mild interosseous muscle wasting on both hands, with limited active extension of her fingers but not passive extension, as well as lower extremity weakness and muscle atrophy.
The patient started taking over the counter CoQ10 (800 mg per day) after her diagnosis was confirmed, but stopped after 3 months due to lack of improvement of her symptoms.
Patient 3
Patient 3 is a 9-year-old Brazilian female who is the second child of consanguineous parents (first cousins). She met all of her early developmental milestones and could walk independently by 14 months. At the age of 3 years, she was noted to have tiptoe gait, which progressed to feet deformity and lower limb spasticity requiring Achilles tendon lengthening at the age of 4 8/12. At the age of five, she developed pes cavus deformity with hammertoes and ankle flexion deformity. When she was six years old, she began exhibiting upper and lower limb weakness, as well as spastic gait. Her physical examination at the time revealed tendon hyperreflexia, extensor plantar response, limb muscular atrophy, and finger fasciculation. At the age of 8 years, she had nerve conduction studies, which revealed decreased amplitude in right deep peroneal and right tibial nerves, with normal conduction velocity in both nerves; these findings were consistent with axonal neuropathy. EMG showed abnormal morphology of motor unit potentials (large amplitude, long duration and increased firing frequency) and reduced recruitment pattern. Her brain MRI revealed enlargement of lateral ventricles, mainly of posterior horns, with thinner of white matter adjacent to ventricles. She was also diagnosed with intellectual disabilities and was unable to read despite receiving educational support. She has never had any seizures, dystonia, ataxia or behavioral problems. Her growth parameters were appropriate for her age.
The patient was prescribed CoQ10 (300 mg per day) after her diagnosis with COQ7-related disorder was established. She continued receiving CoQ10 despite the lack of improvement in her neurological manifestations.
Patient 4
Patient 4 is a 9-year-old Brazilian female who was born at term via uncomplicated Cesarian section following an unremarkable antenatal history. By 15 months, she had met all of her early developmental milestones and could walk independently. She also had normal for her age language development and cognitive performance at school. When she was three years old, she began exhibiting bilateral equine posturing of her feet and pes cavus deformity with her right lower extremity being more severely affected. Her medical history was otherwise unremarkable except for an atrial septum defect that required surgical repair. Family history was notable for pes cavus deformity in her paternal grandmother. Her neurological examination at age 9 years showed tiptoe gait, pes cavus, hammertoes and foot eversion. Passive range of motion of her Achilles heels was restricted to 90 degrees bilaterally, but neither spasticity nor clonus were present. There was bilateral lower extremity weakness with predominant distal musculature involvement, graded as 3/5 in foot dorsiflexion and 4/5 in plantar flexion and knee flexion. Nerve conduction studies were normal at the age of 7 years old, but an EMG was not performed.
Patient 5
Patient 5 is a 33-year-old Brazilian female who was born to consanguineous parents (first degree cousins), with an unremarkable gestational and perinatal history. She met all of her developmental milestones until the age of five, when she began displaying inward rotation of her right foot. She had a learning disability and was diagnosed with depression when she was 15 years old. At the age of 20 years, she developed paresthesia and feet atrophy and started exhibiting frequent falls. When she was 25, she was diagnosed with cardiac arrhythmia and started on metoprolol and enalapril. At the age of 33, her physical examination revealed distal atrophy, bilateral hammer toes, curved feet, lower limb spasticity, distal weakness, Achilles and brachioradialis hyperreflexia, and an absent plantar reflex. Dysmetria, an ataxic gait, dysarthria, and microsaccadic eye movements during ocular pursuit were also present. EMG showed normal motor nerve conduction velocities and latencies, with low muscle action potential amplitudes and signs of chronic reinnervation and fibrillations, suggesting a motor axonal impairment affecting the distal musculature of all extremities.
Exome sequencing identified variants in COQ7
ES of Patient 1 identified two COQ7 variants: a maternally-inherited variant, c.446A>G;(p.Tyr149Cys) and a paternally-inherited pathogenic variant, c.161G>A; (p.Arg54Gln). Both p.Tyr149Cys and p.Arg54Gln variants have been reported in gnomAD with very low frequency (0.00001416 and 0.00001193, respectively) (Last accessed on Nov 13, 2022). In silico analysis using various bioinformatic tools (Revel[31], Varity[32], SIFT[33], MT[34]) predicts deleterious effects on protein structure and/or function. The results of in silico analysis and classification based on ACMG criteria of all variants in this study[29] are presented in Table 2. All disease-associated variants, including those reported in this study, are shown in Figure 1A.
Table 2.
In silico prediction and ACMG classification of the missense variant
| c.446A>G;(p.Tyr149Cys) | c.161G>A;(p.Arg54Gln) | c.3G>T (p.Met1?). | |
|---|---|---|---|
| Revel | Deleterious (moderate) (0.83) | Deleterious (supporting) (0.7) | Uncertain (0.35) |
| Varity | Deleterious (0.96) | Deleterious (0.9) | Deleterious (0.96) |
| SIFT | Deleterious (supporting) (0) | Uncertain (0.001) | Deleterious (supporting) (0) |
| MT | Deleterious (1) | Deleterious (1) | Deleterious (1) |
| CADD score | 27.2 | 27.0 | 24.9 |
| gnomAD allele frequency | 0.00001416 | 0.00001193 | 0 |
| ACMG Classification | PS3, PM2, PP3, PP4 | PS3, PM2, PP3, PP4 | PVS1, PM2, PP4, PP5 |
| Interpretation | Likely Pathogenic | Likely pathogenic | Likely Pathogenic |
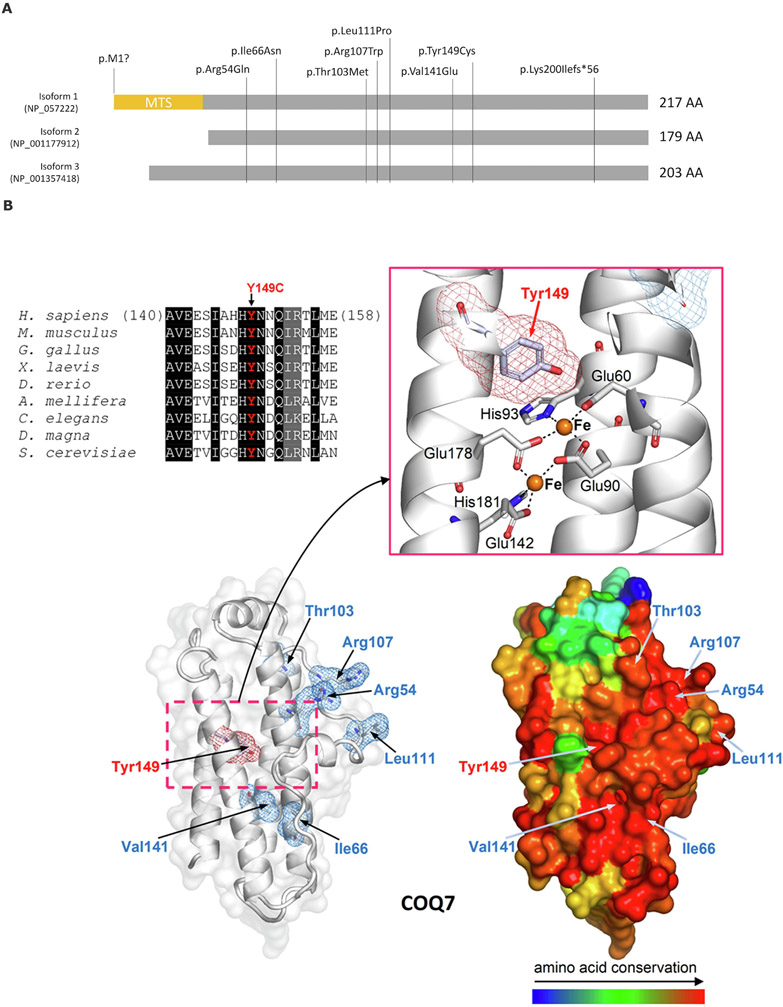
Figure 1. COQ7 protein structure and known disease-associated variants.
(A) Three main isoforms of COQ7 proteins and all known disease-associated variants reported. MTS-mitochondrial targeting sequence (adapted from Jacquier, et al[23]). (B) Multiple sequence alignment highlights the conservation of Tyr149. Columns with identical and similar residues are shadowed, respectively, in black and gray. The homology model of 5-demethoxyubiquinone hydroxylase (based on PDB structure 2VZB; see Materials and Methods for details) is shown with the site of Tyr149Cys replacement (Tyr149 is highlighted by red meshes) and of the previously reported variants p.Arg54Gln, p.Arg107Trp, p.Leu111Pro, and p.Val141Glu (highlighted by blue meshes). On the left, the COQ7 model is presented showing the protein structure as white ribbons and transparent protein surface (the zoomed view details Tyr149, the two active iron ions, represented by orange spheres, and the metal-coordinating residues). On the right, the same model is represented as protein surface colored according to amino acid conservation (as inferred from the multiple sequence alignment of homologous proteins from the following organisms/UniProt entries: H. sapiens, Q99807; M. musculus, P97478; G. gallus, A0A1D5NTT1; X. laevis, Q640V8; D. rerio, F1QW05; A. mellifera, Q6J4P7; C. elegans, P48376; D. magna, A0A0P6DRG9; S. cerevisiae, P41735).
ES of Patient 2 also identified two COQ7 variants: the above mentioned missense variant, c.446A>G;(p.Tyr149Cys) and a missense variant that is predicted to affect the start codon, c.3G>T (p.Met1?). The two variants were confirmed to present in trans. The variant p.Met1? has not been reported in gnomAD (Last accessed on Nov 13, 2022).
ES of Patients 3 and 4 revealed that they are homozygous for the above mentioned pathogenic variants, c.161G>A (p.Arg54Gln) and Patient 5 was homozygous for the c.3G>T (p.Met1?) variant.
Homology modelling
Homology modelling showed that p.Tyr149Cys variant affects a highly conserved residue in the core of a four helix bundle of the 5-demethoxyubiquinone hydroxylase, specifically close to the site hosting the catalytical di-iron center (Fig. 1B). Thus, p.Tyr149Cys, by substituting cysteine for tyrosine, most likely impairs the function of the bi-metallic center, whose redox activity is extremely sensitive to the type and conformation of surrounding residues, or may even cause domain unfolding.
Arg54 is found in a large conserved region on the protein surface (Figure 1), suggesting that it may be involved in protein-protein interactions. However, replacing Arg54 with glutamine, which is also hydrophilic like arginine, partially preserves the chemical-physical properties of the affected site, resulting in limited function damage and explaining the variant's mild pathogenic effect.
The previously reported variant, p.Ile66Asn, introduces a hydrophilic asparagine that disrupts a hydrophobic interaction between two helices in the four helical bundle, resulting in a domain fold alteration. p.Val141Glu is also expected to be detrimental to the domain fold because the affected valine stabilizes the four helical bundle through interactions with other hydrophobic residues that glutamic acid, an anionic residue, cannot achieve. p.Leu111Pro, and p.Arg107Trp cluster on the protein surface nearby p.Arg54Gln, but differ from the latter in that they involve non-conserved amino acid substitutions, potentially causing more disruptive effects. The conservation of the affected region, as hypothesized above, suggests that it may function as an interface for intermolecular interactions. Finally, p.Thr103Met replaces a solvent-exposed threonine, a polar residue, with the hydrophobic methionine, possibly causing conformational reorganization of the protein affected region.
Yeast complementation study
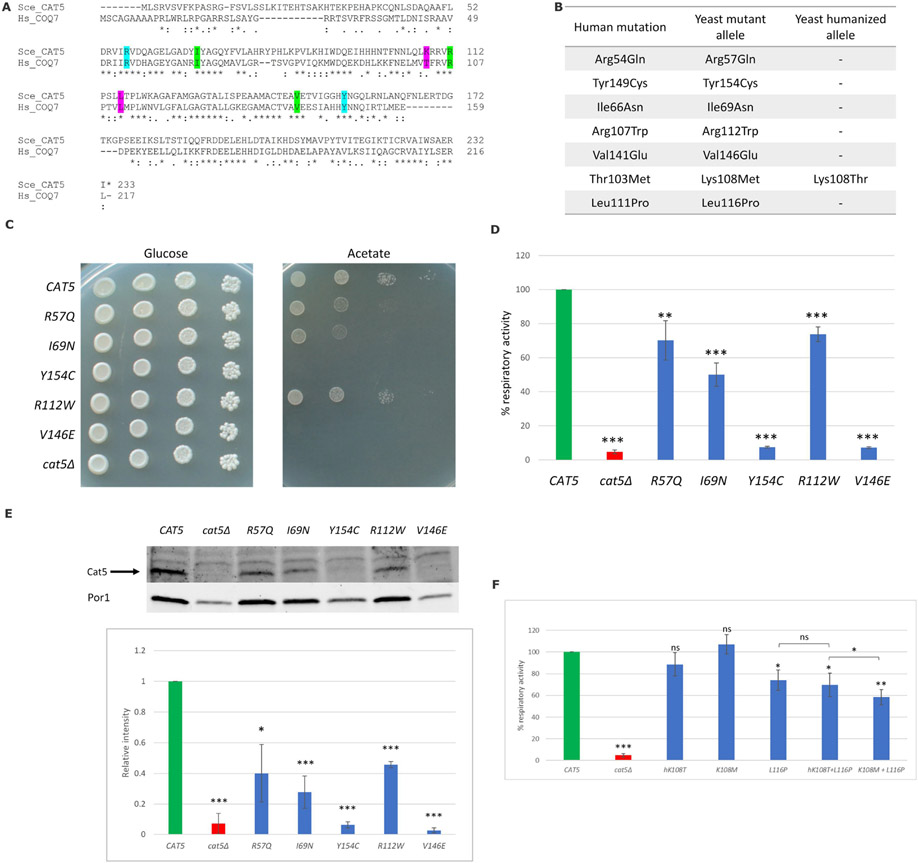
To study the functional effect of the COQ7 variants in the patients of this cohort and previously reported variants, we used the yeast S. cerevisiae as a model taking advantage of the presence of the ortholog gene CAT5. As shown by protein alignment (Fig 2A), the human residues p.Arg54, p.Ile66, p.Arg107, p.Leu111, p.Val141 and p.Tyr149, equivalent respectively to yeast p.Arg57, p.Ile69, p.Arg112, p.Leu116, p.Val146 and p.Tyr154, are conserved from yeast to humans, while the human residue p.Thr103, equivalent to yeast p.Lys108, is not conserved but lies in a conserved stretch. Therefore, for the latter we have created both the so-called humanized version and the putative pathological allele to compare their effect as previously described[35].
Figure 2. Functional studies of COQ7 variants using a yeast model system.
(A) Alignment (Clustal Omega) of the human COQ7 and the yeast Cat5 proteins. The residues affected in our cohort’s patients are highlighted in cyan, residues affected in previously reported patients are highlighted in green, and two additional residues identified in compound homozygous state in patients are highlighted in pink. Amino acids are indicated as conserved (*), with strongly similar properties (:) or with weakly similar properties (.); (B) Amino acid changes found in the patients and the corresponding humanized and mutant yeast alleles; (C) The cat5Δ strain harboring wild-type, mutant alleles or the empty vector were serially diluted and spotted on SC (without uracil) agar plates supplemented with the fermentable carbon source 2% glucose or the non-fermentable carbon source 2% acetate and incubated at 28°C; (D) Cells were grown at 28°C in SC medium without uracil supplemented with 0.6% glucose. Values are represented as the mean of at least three values ±SD. The green bar indicates the wild-type strain; the blue bars indicate strains carrying the alleged pathological mutations; the red bar indicates the null mutant strain. Statistical analysis was performed using a one-tail paired Student’s t-test: ** p < 0.01, *** p < 0.001; (E) Representative Western blot on total protein extract using an anti-Cat5 polyclonal antibody and anti-Por1 as loading control; signals were first normalized to Porin and then to the wild-type to which the value 1.0 was assigned. Densitometric analysis is reported in the histogram below the blot and was performed on at least three independent blots using Image Lab Software (Bio-Rad). Statistical analyses were performed using a one-tail paired Student’s t-test: * p < 0.05, *** p < 0.001; (F) Cells were grown at 28°C in SC medium without uracil supplemented with 0.6% glucose. Values are represented as the mean of at least three values ±SD. The green bar indicates the wild-type strain; the blue bars indicate strains carrying the humanized allele or the alleged pathological mutations; the red bar indicates the null mutant strain. Statistical analyses were performed using a one-tail paired Student’s t-test: ns: not significant, * p < 0.05, ** p < 0.01, *** p < 0.001. All comparisons are referred to the wild-type unless indicated with brackets.
To study the effect of the presumed pathological amino acid substitutions p.Arg54Gln and p.Tyr149Cys identified in our cohort, the corresponding yeast CAT5 codons were directly mutated, producing the mutant alleles cat5R57Q and cat5Y154C (Fig. 2B). The previously reported human pathogenic variants, p.Ile66Asn, p.Arg107Trp and p.Val141Glu, were also investigated using strains carrying the corresponding yeast mutant alleles (cat5I69N, cat5R112W and cat5V146E) (Fig. 2B).
To evaluate the effect of these variants on mitochondrial functions, we determined the strains’ oxidative growth ability using a spot assay analysis. CAT5-5-knock out yeast (cat5Δ) transformed with the empty vector was unable to grow on oxidative carbon sources (e.g., acetate); however, re-expression of the wild-type CAT5 allele rescued the growth defect (Fig. 2C). The expression of the cat5R57Q, cat5I69N and cat5R112W alleles resulted in a mild but significant growth defect on acetate. The expression of the cat5Y154C and cat5V146E led to a complete inability to grow on media containing oxidative carbon sources, similar to cat5Δ (Fig. 2C). These results show that the variants equivalent to those identified in this cohort's patients, cat5R57Q and cat5Y154C, are deleterious in yeast, confirming the pathogenicity of the human variants p.Arg54Gln and p.Tyr149Cys, respectively.
To better characterize these CAT5 variants, the respiratory activity was assessed by measuring oxygen consumption rate. When compared to the wild-type strain, all variants showed significant reduction of the respiratory rate, but to varying degrees. The strain cat5Y154C had a nearly complete reduction of oxygen consumption rate (7.4% residual activity), comparable to the cat5Δ null mutant, suggesting an almost complete loss of function of the protein. Whereas the strain cat5R57Q showed a mild reduction in oxygen consumption rate (70.2% residual activity), a partial respiratory activity was retained, indicating that this variant does not result in a complete loss of function of the Cat5 protein. The strain cat5I69N resulted in a severe reduction in oxygen consumption (50.1% residual activity), while the strain cat5R112W resulted in a mild reduction in oxygen consumption (73.7% residual activity), similar to the cat5R57Q strain. The strain cat5V146E, on the other hand, resulted in a nearly complete loss in oxygen consumption rate (7.2% residual activity), similar to the strain cat5Y154C and the cat5Δ null mutant (Fig. 2D).
To assess the effect of the variants on protein stability, we measured the steady-state level of the Cat5 proteins using Western blot analyses. As with the cat5Δ null mutant, almost no protein was detected in the strains cat5Y154C and cat5v146E; whereas cat5R57Q, cat5I69N and cat5R112W strains showed significant reduction in the protein amount (Fig. 2E), indicating that all mutations alter protein stability in yeast but to different extents. An alternative, but less likely, interpretation could be a decreased binding affinity of the antibody due to altered protein structure.
Human p.Thr103Met and p.Leu111Pro were also studied in yeast, either alone or in cis on the same allele. In addition to strains carrying the p.Leu116Pro (cat5L116P) or p.Lys108Met (cat5K108M) substitutions and its humanized strain cat5hK108T, the strain cat5K108M+L116P and its control cat5hK108T+L116P were created to simulate the homozygous patients for this complex allele. We performed growth test analyses that showed no differences in growth at both permissive (28 °C) and stress (37 °C) temperatures (data not shown). There were no significant differences in oxygen consumption between the wild-type strain, the humanized strain cat5hK108T (88.7% vs. wt), and the mutant strain cat5K108M (107.1% vs. wt), whereas the strain cat5L116P showed a significant reduction in oxygen consumption rate (74.2% vs. wt). The strain cat5hK108T+L116P had a defect similar to cat5L116P (69.7% vs. wt), whereas the strain cat5K108M+L116P had a worsening of the defect (58.4% vs. wt) (Fig. 2F). These results suggest that the substitution p.Leu116Pro, equivalent to human p.Leu111Pro, is slightly deleterious, whereas p.Lys108Met, equivalent to the human p.Thr103Met, causes a defect only when it is in cis as a complex allele with p.Leu116Pro, resulting in aggravation of the mutant phenotype. Western blot analyses did not show significant differences in any of these alleles (data not shown).
COQ8 overexpression and 2,4-diHB supplementation
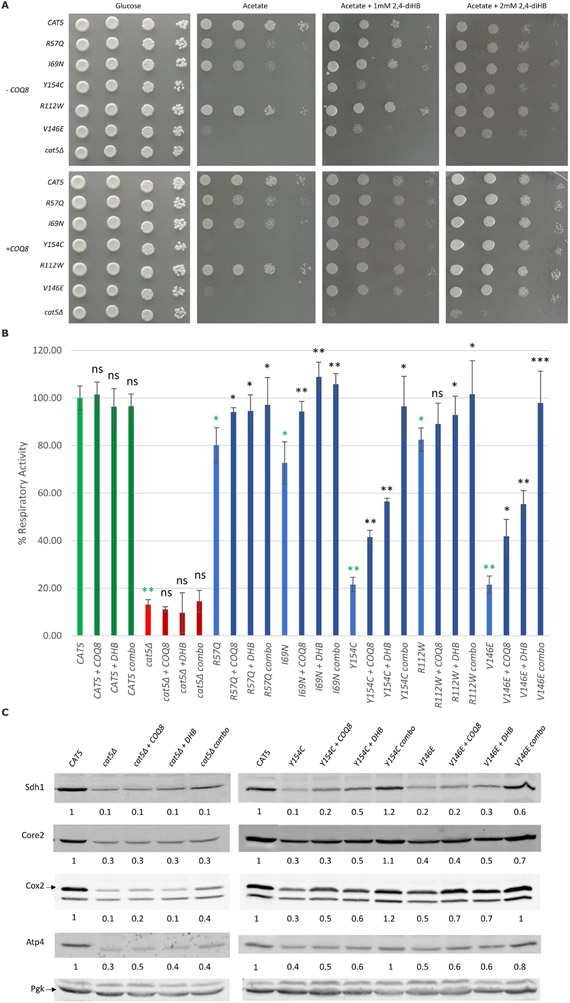
We evaluated the effect of COQ8 overexpression, which promotes the stabilization of the CoQ complex[36] and/or 2,4-diHB supplementation, a structural analog of 4-HB (4-hydroxybenzoic acid), the native precursor of the CoQ benzoquinone ring, on the mutants that presented an oxidative growth defect (cat5R57Q, cat5169N, cat5R112W, cat5Y154C, and cat5V146E); the wild-type strain and the null mutant strain were used as controls.
The null mutant cat5Δ was unable to grow in the presence of 2,4-diHB without COQ8 overexpression; however, in presence of COQ8 overexpression, cat5Δ showed partial growth in a 2,4-diHB dose-dependent manner. The leaky mutant strains (cat5R57Q, cat5I69N and cat5R112W) overexpressing COQ8 showed improvement of oxidative growth on acetate, whereas the severe mutants (cat5Y154C and cat5V146E) had almost no effect, suggesting that the stabilization of the complex has a beneficial effect depending on the residual activity of Cat5 mutant protein (Fig. 3A). We saw a significant improvement in oxidative growth on acetate, but to different extents, for all mutants when we added 1mM 2,4-diHB to the medium; for severe mutants, the beneficial effect of 2,4-diHB was strongly increased when COQ8 was overexpressed (Fig. 3A). We observed a dose-dependent response by increasing the concentration of 2,4-diHB; specifically, with 2mM 2,4-diHB supplementation, severe mutants cat5Y154C and cat5V146E showed a significant reduction of the growth defect even without COQ8 overexpression. Respiratory activity was assessed to further characterize the beneficial effect of COQ8 overexpression and/or 2 mM 2,4-diHB supplementation. No significant difference was identified between the wild-type strain, the wild-type strain overexpressing COQ8 (+ COQ8), the wild-type strain after 2,4-diHB supplementation (+ DHB), and the wild type with both the treatments (combo). Similarly, no significant differences were observed when the null mutant strain received same treatments (non-treated, + COQ8, + DHB, combo). On the other hand, all mutant strains showed significant improvement using the two treatments individually (+ COQ8, + DHB) or together (combo). The only exception is the cat5R112W strain overexpressing COQ8, probably due to the milder defect of this strain, making it more difficult to appreciate small variations. Notably, the cat5Y154C, and cat5V146E strains, which had significantly decreased oxygen consumption, demonstrated a more pronounced beneficial effect, leading to a complete restoration of respiratory activity when both treatments were combined (combo, Fig. 3B).
Figure 3. 2,4-Dihydrobenzoic acid supplementation and overexpression of COQ8 synergistically rescued oxidative growth defect of mutant cat5Δ strain.
(A) Oxidative growth: the cat5Δ strain harboring wild-type and mutant alleles or the empty vector with or without COQ8 overexpression (± COQ8) were serially diluted and spotted on SC agar plates supplemented with the fermentable carbon source 2% glucose or the non-fermentable carbon source 2% acetate, with or without the addition of 2,4-dihydroxybenzoic acid (2,4-diHB) and incubated at 28°C (B) Respiratory activity: Non-treated strains, strains with COQ8 overexpression (+ COQ8), with 2 mM 2,4-diHB supplementation (+ DHB) or with both treatments (combo) were grown at 28°C in SC medium without uracil and leucine supplemented with 0.6% glucose. Values are represented as the mean of at least three values ±SD. The green bars indicate the wild-type strains; the blue bars indicate strains carrying different COQ7 variants; the red bars indicate the null mutant strains. Light colors for the non-treated strains, and dark colors for the treated strains. Statistical analysis was performed using a one-tail paired Student’s t-test comparing the non-treated mutant strains with the non-treated wild type strain (green asterisks) or comparing the treated strains to the relative non-treated strain (black asterisks/ns). ns: not significant, * p<0.05, ** p<0.01, *** p<0.001. (C) Representative Western blots for the evaluation of respiratory complexes subunits levels on the wild-type strain, cat5Δ strain and strains carrying Y154C and V146E variants with COQ8 overexpression (+ COQ8), 2,4-diHB supplementation (+ DHB) and with both the treatments (combo). Antibodies used: Sdh1 for complex II; Core2 for complex III; Cox2 for complex IV; Atp4 for complex V; Pgk as loading control. The quantification was performed on two independent blots using Image Lab Software (Bio-Rad).
A reduction of the levels of some mitochondrial proteins was previously reported for the null mutant cat5Δ [37]. Therefore, we measured by Western blot the levels of the respiratory complexes subunits Sdh1 (complex II), Core2 (complex III), Cox2 (complex IV) and Atp4 (complex V) to better understand the mechanism underlying the beneficial effect of the treatments (+ COQ8, + DHB, combo).
A significant reduction in all proteins studied was detected for the severe mutants (carrying Y154C and V146E variants) and for the null mutant strain (cat5Δ), while no significant difference was visible for the leaky mutants (carrying R57Q, I69N, and R112W variants, data not shown). COQ8 overexpression and/or 2,4-diHB supplementation were tested on cat5Δ, cat5Y154C and cat5V146E to evaluate the potential beneficial effect. No significant improvement was detectable for the cat5Δ strain with the different treatments (+ COQ8, + DHB, combo); while an appreciable improvement in protein quantity was detected for the cat5Y154C and cat5V146E strains, especially when both treatments were combined (Fig. 3C). These results suggest that the improvement of oxidative growth and respiratory activity through COQ8 overexpression and 2,4-diHB supplementation could be partially due to increased levels of the respiratory complexes.
Discussion
We report the largest cohort of patients with COQ7-associated primary CoQ10 deficiency and present functional studies with diagnostic and therapeutic implications. According to the clinical data from our cohort and previously reported patients, there are two main clinical phenotypes of COQ7-associated primary CoQ10 deficiency: a neonatal-onset presentation with severe neuromuscular, cardiorespiratory and renal involvement and a late-onset disease presenting with lower extremity weakness, abnormal gait, progressive neuropathy and variable developmental delay (Table 1). Patients with neonatal-onset disease, including Patient 1 in our study and two previously reported patients[9, 20], had prenatal complications such as oligohydramnios (3/3) and IUGR (2/3). Hypotonia, spasticity, moderate-to-profound developmental delay, contractures and hearing impairment (sensorineural or mixed) are also common among neonatal-onset patients. All patients with neonatal-onset disease had respiratory involvement: two patients had lung hypoplasia and one had central hypoventilation. Two of the three patients had cardiac and renal involvement. One patient had cardiomyopathy, which led to heart failure triggered by sepsis and early death.
The disease course was more variable in patients with later-disease (11/14) [18, 19, 21, 23]. The age of onset ranged from 14 months to 12 years. Lower extremity weakness and abnormal gait were the presenting symptoms of all reported patients. Three of the patients (3/7) in this group had hearing impairment, which was less severe than the patients in the neonatal onset group (denominator is different from 11 based on availability of data in previous reports). Mild developmental delay (3/8), speech delay (3/11) and learning disabilities (5/7) are also present among patients with the late-onset disease. In this cohort, patient 5 was found to have an adult-onset cardiac arrhythmia. There are no reports of renal or respiratory involvements in late-onset disease.
The most common variant in known patients is p.Arg54Gln, which was found in homozygous state in 3 patients and heterozygous in one patient, accounting for 7 out of 26 alleles (27%) (Siblings with same genotype were counted once). The variant p.Met1? was found in 2 families in a homozygous state and in 1 heterozygote patient and accounts for 19% (5/26) of alleles, while the complex allele p.[Thr103Met;Leu111Pro] was found in 3 homozygotes, accounting for ~23% (6/26) of alleles. The p.Tyr149Cys variant accounts for ~11% (3/26) of all alleles. All previously reported patients harbor missense variants in at least one allele. A single frameshift variant, p.Lys200Ilefs*56, was identified in a previously reported patient who was heterozygote for p.Arg107Trp; this patient exhibited severe phenotype and early death[20]. These findings indicate that COQ7 is essential for mitochondrial function and that its complete loss is fatal in human.
Despite the small number of patients with COQ7-associated primary CoQ10 deficiency, there is some genotype-phenotype correlation. The 3 patients who are homozygous for p.Arg54Gln (2 in this cohort and 1 reported by Wang et al, 2022[19]) had a milder phenotype with primarily muscular involvement. These patients had no hearing loss, visual impairment, cardiomyopathy or renal dysfunction. Functional studies in fibroblasts of the previously reported patient showed approximately 45% reduction of CoQ10 level and drastic decrease in COQ7 expression[19]. Protein homology revealed that the replacement of Arg54 by a glutamine partially preserves the chemical-physical properties. Residual activity of p.Arg54Gln is also observed in CAT5-knockout yeast strain harboring p.Arg57Gln. These findings indicate that p.Arg54Gln is likely a hypomorphic allele. The CAT5-knock out yeast strain harboring p.Tyr154Cys, equivalent to p.Tyr149Cys, has complete oxidative growth defect, indicating a severe impact on enzyme function. Furthermore, Western blot shows a drastic reduction of the yeast protein steady-state level. Protein modelling also indicates major impact of the variant on protein function. The p.Tyr149Cys variant has never been reported in a homozygous state suggesting a severe phenotype that can lead to lethality. A recent study described 3 siblings homozygous for p.Met1? and who exhibited adolescent-onset distal motor neuropathy[23]. Patient 2 and Patient 5 also harbor this variant that affects the initiation codon of isoform 1, a main isoform in motor neuron and fibroblast (Figure 1A)[23]. The second initiation codon is predicted to cause a frameshift and nonfunctional protein, but this is unlikely because of the milder phenotypes, while the third initiation codon is in-frame and predicted to cause a truncated protein lacking 38 amino acid residues from N-terminus involving a mitochondrial transport signaling (MTS). The loss of MTS was shown to cause protein mis-localization in other mitochondrial proteins[38], but localization of COQ7 was not affected[23]. Instead, the protein level of COQ7 was severely reduced in patients’ fibroblasts, likely from translational defect [23]. Investigation in neuroblastoma cell line showed that COQ7 isoform 1 was severely affected, while the two other isoforms which use different initiation codons could partially compensate for COQ7 isoform 1 function[23]. These results, in combination with later-onset disease and milder presentation of the patients harboring p.Met1?, indicate that p.Met1? is also a hypomorphic allele.
Yeast studies facilitated the validation of pathogenicity of all the studied variants, allowing us to broaden the genotypic spectrum of this disorder and characterize the functional impact of the variants on mitochondrial function. Although a heterologous complementation approach could allow to study any mutation, the homologous complementation approach showed to be useful to study several variants due to the high similarity of protein sequences.
The variants studied can be divided into two groups: those that cause a leaky defect (yeast p.Arg57Gln, p.Ile69Asn, p.Arg112Trp and p.Leu116Pro) and those that cause a severe defect (yeast p.Tyr154Cys and p.Val146Glu). Accordingly, human p.Arg54Gln, p.Ile66Asn, p.Arg107Trp and p.Leu111Pro variants are hypomorphic alleles, whereas human p.Tyr149Cys and p.Val141Glu are amorphic alleles. The latter two were observed in patients with neonatal onset disease, indicating that the yeast study correlates well with the clinical presentation. While p.Arg107Trp was found in a patient with neonatal-onset disease who also has a frameshift variant, p.Lys200Ilefs*56, which was predicted to cause complete loss of the protein [20]. The variant p.Lys108Met, which is equivalent to human p.Thr103Met, did not cause an oxidative defect, but did worsen the phenotype of the strain carrying p.Leu116Pro (equivalent to human p.Leu111Pro)[21].
The importance of early diagnosis of CoQ10 deficiencies is highlighted by its response to treatment with CoQ10 supplementation[39, 40]. Supplementation can improve myopathy and renal involvement; however, it does not halt the progression of neurologic phenotypes[41]. In our cohort, Patient 1-3 received CoQ10 supplementation but no significant neurologic improvement has been observed. Patients with primary CoQ10 deficiency respond differently to CoQ10 supplementation depending on the underlying genetic defect, organ damage at the time of treatment initiation, CoQ10 pharmaceutical formulations, dose, and its dosing interval [3, 42] Since oral CoQ10 supplementation is less effective once severe organ damage has already occurred, the lack of response to treatment in three of our patients may indicate prior or even prenatal neuronal damage that could not be repaired. A synthetic substrate of CoQ10 biosynthesis, 2,4-diHB, was shown to bypass COQ7 hydroxylase and restore respiratory enzyme activities in fibroblasts of patients with severe COQ7 defects[21, 23]. However, it also acts as competitive inhibitor of COQ7 hydroxylase, and causes the reduction of CoQ10 biosynthesis in fibroblasts of patients with residual COQ7 hydroxylase activity[21]. Our data show that overexpression of COQ8 could restore the growth of COQ7 leaky mutants but not cat5Δ strain, indicating that the effect of COQ8 to restore oxidative growth depends on the presence of Coq7. Coq8 functions to stabilize multiple enzymes in CoQ biosynthesis, including Coq7[37]. Supplementation of 2mM 2,4-diHB partially rescues the growth defect and the reduction of respiratory activity of both the leaky and severe mutations, possibly due to an increase in the levels of respiratory complexes. Overexpression of COQ8 and 2 mM 2,4-diHB supplementation fully restore oxidative growth and respiratory activity for all the mutants. These results suggest additional therapeutic approaches should be studied in patients with COQ7-related disorder.
In this work, we studied and characterized a large cohort of patients with COQ7-related primary CoQ10 deficiency. We defined two disease presentations: early-onset and late onset, each with a unique natural history and prognosis. We established Baker’s yeast as a model organism to study the functional effects of COQ7 variants. We utilized this model to study COQ7 variants and potential treatment options. We demonstrated that a higher residual activity of COQ7 correlates with milder clinical severity, supporting an emerging genotype to phenotype relationship. We also used homology modelling to study the effect of the variants on protein structure and predict their functional impact. Despite expanding the genotypic and phenotypic spectra of this ultra-rare disorder, larger cohorts of patients are required to better define the genotype to phenotype correlation, the natural history of the disease, biomarkers of the disorders, and whether specific treatments can improve clinical findings.
Supplementary Material
Acknowledgements
We thank the patients and their families for their cooperation with this article. We thank Dr. CF Clarke for providing the anti-Cat5/Coq7 antibody.
Details of funding
The work has benefitted from the equipment and the framework of the COMP-HUB Initiative, funded by the “Departments of Excellence” program of the Italian Ministry for Education, University and Research (MIUR, 2018-2022). We are grateful for the support of the Consorzio Interuniversitario per le Biotecnologie (CIB).
Footnotes
Conflict of Interest
The authors declare no conflict of interest
Details of ethics approval:
No interventions performed on patients and no biological specimens collected from participants. The guardian (s) of patients signed a consent form for publication approved by Washington University IRB (Media Authorization for the Use and Disclosure of Protected Health Information) or other local academic institutions.
A patient consent statement
Consent was obtained from the patient’s family for publication of this report.
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Tran UC, Clarke CF, Endogenous Synthesis of Coenzyme Q in Eukaryotes Mitochondrion 7 (2007) S62–S71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Stefely JA, Pagliarini DJ, Biochemistry of Mitochondrial Coenzyme Q Biosynthesis Trends Biochem Sci 42 (2017) 824–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Desbats MA, Lunardi G, Doimo M, Trevisson E, Salviati L, Genetic bases and clinical manifestations of coenzyme Q10 (CoQ10) deficiency Journal of Inherited Metabolic Disease 38 (2015) 145–156. [DOI] [PubMed] [Google Scholar]
- [4].Quinzii C, Naini A, Salviati L, Trevisson E, Navas P, Dimauro S, Hirano M, A mutation in para-hydroxybenzoate-polyprenyl transferase (COQ2) causes primary coenzyme Q10 deficiency Am J Hum Genet 78 (2006) 345–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Diomedi-Camassei F, Di Giandomenico S, Santorelli FM, Caridi G, Piemonte F, Montini G, Ghiggeri GM, Murer L, Barisoni L, Pastore A, Muda AO, Valente ML, Bertini E, Emma F, COQ2 nephropathy: a newly described inherited mitochondriopathy with primary renal involvement J Am Soc Nephrol 18 (2007) 2773–2780. [DOI] [PubMed] [Google Scholar]
- [6].Brea-Calvo G, Haack TB, Karall D, Ohtake A, Invernizzi F, Carrozzo R, Kremer L, Dusi S, Fauth C, Scholl-Burgi S, Graf E, Ahting U, Resta N, Laforgia N, Verrigni D, Okazaki Y, Kohda M, Martinelli D, Freisinger P, Strom TM, Meitinger T, Lamperti C, Lacson A, Navas P, Mayr JA, Bertini E, Murayama K, Zeviani M, Prokisch H, Ghezzi D, COQ4 mutations cause a broad spectrum of mitochondrial disorders associated with CoQ10 deficiency Am J Hum Genet 96 (2015) 309–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Malicdan MCV, Vilboux T, Ben-Zeev B, Guo J, Eliyahu A, Pode-Shakked B, Dori A, Kakani S, Chandrasekharappa SC, Ferreira CR, Shelestovich N, Marek-Yagel D, Pri-Chen H, Blatt I, Niederhuber JE, He L, Toro C, Taylor RW, Deeken J, Yardeni T, Wallace DC, Gahl WA, Anikster Y, A novel inborn error of the coenzyme Q10 biosynthesis pathway: cerebellar ataxia and static encephalomyopathy due to COQ5 C-methyltransferase deficiency Hum Mutat 39 (2018) 69–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Heeringa SF, Chernin G, Chaki M, Zhou W, Sloan AJ, Ji Z, Xie LX, Salviati L, Hurd TW, Vega-Warner V, Killen PD, Raphael Y, Ashraf S, Ovunc B, Schoeb DS, McLaughlin HM, Airik R, Vlangos CN, Gbadegesin R, Hinkes B, Saisawat P, Trevisson E, Doimo M, Casarin A, Pertegato V, Giorgi G, Prokisch H, Rotig A, Nurnberg G, Becker C, Wang S, Ozaltin F, Topaloglu R, Bakkaloglu A, Bakkaloglu SA, Muller D, Beissert A, Mir S, Berdeli A, Varpizen S, Zenker M, Matejas V, Santos-Ocana C, Navas P, Kusakabe T, Kispert A, Akman S, Soliman NA, Krick S, Mundel P, Reiser J, Nurnberg P, Clarke CF, Wiggins RC, Faul C, Hildebrandt F, COQ6 mutations in human patients produce nephrotic syndrome with sensorineural deafness J Clin Invest 121 (2011) 2013–2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Freyer C, Stranneheim H, Naess K, Mourier A, Felser A, Maffezzini C, Lesko N, Bruhn H, Engvall M, Wibom R, Barbaro M, Hinze Y, Magnusson M, Andeer R, Zetterstrom RH, von Dobeln U, Wredenberg A, Wedell A, Rescue of primary ubiquinone deficiency due to a novel COQ7 defect using 2,4-dihydroxybensoic acid J Med Genet 52 (2015) 779–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Zhang L, Ashizawa T, Peng D, Primary coenzyme Q10 deficiency due to COQ8A gene mutations Mol Genet Genomic Med 8 (2020) e1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Danhauser K, Herebian D, Haack TB, Rodenburg RJ, Strom TM, Meitinger T, Klee D, Mayatepek E, Prokisch H, Distelmaier F, Fatal neonatal encephalopathy and lactic acidosis caused by a homozygous loss-of-function variant in COQ9 Eur J Hum Genet 24 (2016) 450–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Gerards M, van den Bosch B, Calis C, Schoonderwoerd K, van Engelen K, Tijssen M, de Coo R, van der Kooi A, Smeets H, Nonsense mutations in CABC1/ADCK3 cause progressive cerebellar ataxia and atrophy Mitochondrion 10 (2010) 510–515. [DOI] [PubMed] [Google Scholar]
- [13].Mollet J, Giurgea I, Schlemmer D, Dallner G, Chretien D, Delahodde A, Bacq D, de Lonlay P, Munnich A, Rotig A, Prenyldiphosphate synthase, subunit 1 (PDSS1) and OH-benzoate polyprenyltransferase (COQ2) mutations in ubiquinone deficiency and oxidative phosphorylation disorders J Clin Invest 117 (2007) 765–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Lopez LC, Schuelke M, Quinzii CM, Kanki T, Rodenburg RJ, Naini A, Dimauro S, Hirano M, Leigh syndrome with nephropathy and CoQ10 deficiency due to decaprenyl diphosphate synthase subunit 2 (PDSS2) mutations Am J Hum Genet 79 (2006) 1125–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Banh RS, Kim ES, Spillier Q, Biancur DE, Yamamoto K, Sohn ASW, Shi G, Jones DR, Kimmelman AC, Pacold ME, The polar oxy-metabolome reveals the 4-hydroxymandelate CoQ10 synthesis pathway Nature 597 (2021) 420–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Manicki M, Aydin H, Abriata LA, Overmyer KA, Guerra RM, Coon JJ, Dal Peraro M, Frost A, Pagliarini DJ, Structure and functionality of a multimeric human COQ7:COQ9 complex Mol Cell 82 (2022) 4307–4323 e4310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Lohman DC, Forouhar F, Beebe ET, Stefely MS, Minogue CE, Ulbrich A, Stefely JA, Sukumar S, Luna-Sanchez M, Jochem A, Lew S, Seetharaman J, Xiao R, Wang H, Westphall MS, Wrobel RL, Everett JK, Mitchell JC, Lopez LC, Coon JJ, Tong L, Pagliarini DJ, Mitochondrial COQ9 is a lipid-binding protein that associates with COQ7 to enable coenzyme Q biosynthesis Proc Natl Acad Sci U S A 111 (2014) E4697–4705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Hashemi SS, Zare-Abdollahi D, Bakhshandeh MK, Vafaee A, Abolhasani S, Inanloo Rahatloo K, DanaeeFard F, Farboodi N, Rohani M, Alavi A, Clinical spectrum in multiple families with primary COQ(10) deficiency Am J Med Genet A 185 (2021) 440–452. [DOI] [PubMed] [Google Scholar]
- [19].Wang Y, Gumus E, Hekimi S, A novel COQ7 mutation causing primarily neuromuscular pathology and its treatment options Mol Genet Metab Rep 31 (2022) 100877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Kwong AK, Chiu AT, Tsang MH, Lun KS, Rodenburg RJT, Smeitink J, Chung BH, Fung CW, A fatal case of COQ7-associated primary coenzyme Q(10) deficiency JIMD Rep 47 (2019) 23–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Wang Y, Smith C, Parboosingh JS, Khan A, Innes M, Hekimi S, Pathogenicity of two COQ7 mutations and responses to 2,4-dihydroxybenzoate bypass treatment J Cell Mol Med 21 (2017) 2329–2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Sadr Z, Zare-Abdollahi D, Rohani M, Alavi A, A founder mutation in COQ7, p.(Leu111Pro), causes pure hereditary spastic paraplegia (HSP) in the Iranian population Neurol Sci (2023). [DOI] [PubMed] [Google Scholar]
- [23].Jacquier A, Theuriet J, Fontaine F, Mosbach V, Lacoste N, Ribault S, Risson V, Carras J, Coudert L, Simonet T, Latour P, Stojkovic T, Piard J, Cosson A, Lesca G, Bouhour F, Allouche S, Puccio H, Pegat A, Schaeffer L, Homozygous COQ7 mutation: a new cause of potentially treatable distal hereditary motor neuropathy Brain (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Theunissen TEJ, Nguyen M, Kamps R, Hendrickx AT, Sallevelt S, Gottschalk RWH, Calis CM, Stassen APM, de Koning B, Mulder-Den Hartog ENM, Schoonderwoerd K, Fuchs SA, Hilhorst-Hofstee Y, de Visser M, Vanoevelen J, Szklarczyk R, Gerards M, de Coo IFM, Hellebrekers D, Smeets HJM, Whole Exome Sequencing Is the Preferred Strategy to Identify the Genetic Defect in Patients With a Probable or Possible Mitochondrial Cause Front Genet 9 (2018) 400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Ceccatelli Berti C, di Punzio G, Dallabona C, Baruffini E, Goffrini P, Lodi T, Donnini C, The Power of Yeast in Modelling Human Nuclear Mutations Associated with Mitochondrial Diseases Genes (Basel) 12 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Awad AM, Bradley MC, Fernandez-Del-Rio L, Nag A, Tsui HS, Clarke CF, Coenzyme Q(10) deficiencies: pathways in yeast and humans Essays Biochem 62 (2018) 361–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Tran UC, Marbois B, Gin P, Gulmezian M, Jonassen T, Clarke CF, Complementation of Saccharomyces cerevisiae coq7 mutants by mitochondrial targeting of the Escherichia coli UbiF polypeptide: two functions of yeast Coq7 polypeptide in coenzyme Q biosynthesis J Biol Chem 281 (2006) 16401–16409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Sobreira N, Schiettecatte F, Valle D, Hamosh A, GeneMatcher: a matching tool for connecting investigators with an interest in the same gene Hum Mutat 36 (2015) 928–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, Voelkerding K, Rehm HL, A.L.Q.A. Committee, Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology Genet Med 17 (2015) 405–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Krivov GG, Shapovalov MV, Dunbrack RL Jr., Improved prediction of protein side-chain conformations with SCWRL4 Proteins 77 (2009) 778–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Ioannidis NM, Rothstein JH, Pejaver V, Middha S, McDonnell SK, Baheti S, Musolf A, Li Q, Holzinger E, Karyadi D, Cannon-Albright LA, Teerlink CC, Stanford JL, Isaacs WB, Xu J, Cooney KA, Lange EM, Schleutker J, Carpten JD, Powell IJ, Cussenot O, Cancel-Tassin G, Giles GG, MacInnis RJ, Maier C, Hsieh CL, Wiklund F, Catalona WJ, Foulkes WD, Mandal D, Eeles RA, Kote-Jarai Z, Bustamante CD, Schaid DJ, Hastie T, Ostrander EA, Bailey-Wilson JE, Radivojac P, Thibodeau SN, Whittemore AS, Sieh W, REVEL: An Ensemble Method for Predicting the Pathogenicity of Rare Missense Variants Am J Hum Genet 99 (2016) 877–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Wu Y, Li R, Sun S, Weile J, Roth FP, Improved pathogenicity prediction for rare human missense variants Am J Hum Genet 108 (2021) 1891–1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Vaser R, Adusumalli S, Leng SN, Sikic M, Ng PC, SIFT missense predictions for genomes Nat Protoc 11 (2016) 1–9. [DOI] [PubMed] [Google Scholar]
- [34].Schwarz JM, Cooper DN, Schuelke M, Seelow D, MutationTaster2: mutation prediction for the deep-sequencing age Nat Methods 11 (2014) 361–362. [DOI] [PubMed] [Google Scholar]
- [35].Sommerville EW, Ng YS, Alston CL, Dallabona C, Gilberti M, He L, Knowles C, Chin SL, Schaefer AM, Falkous G, Murdoch D, Longman C, de Visser M, Bindoff LA, Rawles JM, Dean JCS, Petty RK, Farrugia ME, Haack TB, Prokisch H, McFarland R, Turnbull DM, Donnini C, Taylor RW, Gorman GS, Clinical Features, Molecular Heterogeneity, and Prognostic Implications in YARS2-Related Mitochondrial Myopathy JAMA Neurol 74 (2017) 686–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Xie LX, Ozeir M, Tang JY, Chen JY, Jaquinod SK, Fontecave M, Clarke CF, Pierrel F, Overexpression of the Coq8 kinase in Saccharomyces cerevisiae coq null mutants allows for accumulation of diagnostic intermediates of the coenzyme Q6 biosynthetic pathway J Biol Chem 287 (2012) 23571–23581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].He CH, Xie LX, Allan CM, Tran UC, Clarke CF, Coenzyme Q supplementation or overexpression of the yeast Coq8 putative kinase stabilizes multi-subunit Coq polypeptide complexes in yeast coq null mutants Biochim Biophys Acta 1841 (2014) 630–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Wongkittichote P, Tungpradabkul S, Wattanasirichaigoon D, Jensen LT, Prediction of the functional effect of novel SLC25A13 variants using a S. cerevisiae model of AGC2 deficiency J Inherit Metab Dis 36 (2013) 821–830. [DOI] [PubMed] [Google Scholar]
- [39].DiMauro S, Quinzii CM, Hirano M, Mutations in coenzyme Q10 biosynthetic genes Journal of Clinical Investigation 117 (2007) 587–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Desbats MA, Vetro A, Limongelli I, Primary coenzyme Q10 deficiency presenting as fatal neonatal multiorgan failure 23 (2015) 1254–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Aure K, Benoist JF, Ogier de Baulny H, Romero NB, Rigal O, Lombes A, Progression despite replacement of a myopathic form of coenzyme Q10 defect Neurology 63 (2004) 727–729. [DOI] [PubMed] [Google Scholar]
- [42].Emmanuele V, Lopez LC, Berardo A, Naini A, Tadesse S, Wen B, D'Agostino E, Solomon M, DiMauro S, Quinzii C, Hirano M, Heterogeneity of coenzyme Q10 deficiency: patient study and literature review Arch Neurol 69 (2012) 978–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.