Abstract
Background
Human cytomegalovirus (HCMV) is a common herpesvirus that can cause a range of symptoms, from mild conditions such as fevers to severe illnesses like pneumonia. Early and accurate diagnosis of HCMV infection is crucial, particularly for vulnerable populations with limited medical care. However, current diagnostic methods are often expensive, time-consuming, and require skilled technicians.
Materials and methods
We developed an HCMV-RPA-CRISPR diagnosis platform for the rapid and cost-effective detection of HCMV infection. This method utilizes recombinase polymerase amplification (RPA) to amplify the HCMV target gene isothermally without the need for thermal cycling equipment. The platform integrates the CRISPR/Cas12a system, significantly enhancing specificity and sensitivity. A total of 13 clinical blood samples were tested to evaluate the platform's effectiveness and accuracy. Additionally, a lateral flow assay (LFA) and fluorescence detection were incorporated for straightforward and rapid visual interpretation of the results.
Results
The assay effectively detected concentrations as low as a single copy of the positive control plasmid per microliter in under 1 h, without requiring specialized equipment or training. In clinical sample evaluations, both the fluorescence readout and LFA exhibited 100% sensitivity and specificity, identifying four HCMV-positive and nine HCMV-negative samples.
Conclusion
The HCMV-RPA-CRISPR diagnosis platform is comparably effective to qPCR for HCMV diagnosis. Its applicability in common clinical laboratories, clinics, and point-of-care settings, particularly in resource-limited environments, makes it a valuable tool for widespread HCMV screening and diagnosis.
Keywords: Human cytomegalovirus, HCMV, Point-of-care diagnostics, Recombinase polymerase amplification, CRISPR diagnosis
1. Introduction
Human cytomegalovirus (HCMV) is a double-stranded DNA virus with a high global incidence. Primary infections are often contracted in childhood, typically presenting as mild acute viral syndromes or remaining asymptomatic. However, HCMV can enter a latent state post-primary infection, persisting in the host for life and potentially reactivating under conditions of immunosuppression [[1], [2], [3]]. Primary HCMV infection or reactivation in immunocompromised individuals can lead to serious, life-threatening diseases. The risk of severe illness varies, with the highest mortality observed in hematopoietic stem cell or solid-organ transplant recipients [4].
The often-asymptomatic nature of HCMV and its potential for reactivation highlight the importance of rapid, point-of-care diagnostics. These are essential for early intervention, limiting transmission in healthcare settings, and managing risks during pregnancy, particularly in areas with limited resources. Timely and accessible diagnosis is key to cost-effective HCMV management, preventing substantial healthcare costs due to delayed or missed diagnoses.
Current diagnostic methods include virus culture, antibody or antigenemia assay, and quantitative polymerase chain reaction (qPCR) [[5], [6], [7], [8]]. Virus culture is technically challenging and time-consuming [9,10]. Antibody assays, which detect post-infection IgM and IgG antibodies [11], can differentiate between primary infection and reactivation but may not detect very early infection [12]. The antigenemia assay detects HCMV tegument protein pp65 using monoclonal antibodies, but it has several limitations, including being a manual test, requiring blood samples to be processed within 6–8 h of collection, and demanding high technical expertise for interpretation [13]. The nucleic acid-based qPCR technique is considered the gold standard for diagnosing various diseases, including HCMV, due to its high sensitivity and specificity. Its exponential DNA amplification allows for early detection compared to other diagnostic methods [14]. These methods, however, face challenges including long turnaround times and technical complexities.
The emergence of Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) technology, known for its precise DNA editing capabilities, offers an innovative alternative. CRISPR-based diagnostics, such as Specific High-Sensitivity Enzymatic Reporter UnLOCKing (SHERLOCK) and DNA Endonuclease-Targeted CRISPR Trans Reporter (DETECTR), represent a breakthrough in diagnostic technology. These methods harness the specific gene-editing properties of CRISPR to detect specific genetic sequences [15,16]. CRISPR-based diagnostics offer numerous advantages over traditional methods. These diagnostic methods can be conducted at a standard temperature of 37 °C, providing rapid and reliable results. This is particularly advantageous in resource-limited settings with inadequate diagnostic equipment compared to traditional PCR-based kits [15,[17], [18], [19]]. Furthermore, they hold the potential to detect multiple pathogens within a single sample, facilitating the diagnosis of infectious diseases in convenient multiplex assays [17,20,21]. Several CRISPR-based diagnostic tests have already been developed to detect contagious diseases, including coronavirus disease 2019, tuberculosis, and Zika. These tests have demonstrated accuracy comparable to conventional nucleic acid-based diagnostics, enabling faster detection [[22], [23], [24], [25]].
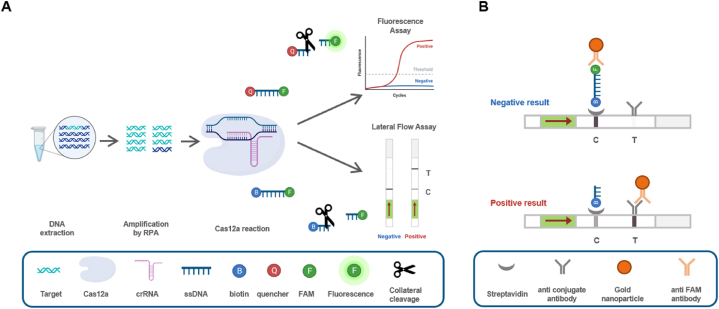
This study presents a rapid and sensitive diagnostic method for HCMV detection. The diagnostic approach combines pre-amplification of the HCMV gene using recombinase polymerase amplification (RPA), sequence-specific recognition and trans cleavage by CRISPR-Cas12a/CRISPR RNA (crRNA), and fluorescence detection or lateral flow assay (LFA) readout (Fig. 1). The aim of this research is to establish a novel and simplified diagnostic approach that utilizes CRISPR-based methodologies for the early detection of HCMV.
Fig. 1.
Schematic illustration of HCMV diagnosis using the RPA-CRISPR Cas12a assay platform. (A) The viral DNA is extracted and amplified by RPA. The CRISPR-Cas12a system can recognize the target DNA and cleave it, as well as simultaneously cleave the ssDNA through collateral-cleavage activity. Both fluorescence readout and lateral flow assay strips visualize the cleaved ssDNA. (B) In the absence of target DNAs, the uncleaved ssDNA is captured in the C line and the gold nanoparticle is captured in the C line, indicating a negative result. In the presence of target DNAs, the labeled ssDNA undergoes cleavage through collateral cleavage by activated Cas12a, and the gold nanoparticle is captured in the T line, indicating a positive result. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
2. Materials and methods
2.1. Standard sample preparation
The sensitivity and effectiveness of the HCMV-RPA-CRISPR detection method were evaluated using cloned HCMV UL75 plasmid as the standard. The plasmid DNA was extracted using the Solg™ Plasmid Mini-prep Kit (SPM01-C200, SolGent, Korea), following the manufacturer's instructions.
2.2. RPA primer design and RPA reaction
The HCMV genome contains numerous polymorphisms, which have made it challenging to detect certain clinical species using previously reported primers designed to target polymorphic regions [26,27]. To overcome this challenge, the HCMV essential envelope glycoprotein H (gH, UL75) gene was identified as one of the most conserved genes in the HCMV genome [27].
We manually designed RPA primers specific to UL75, following the instructions provided by TwistDX (UK). The HCMV glycoprotein H (gH) genomic sequences of the Toledo strain (NCBI accession number, GU937742) were used as templates for primer design. The primers were synthesized by Bioneer (Korea) and listed in Table 1.
Table 1.
List of RPA primers, crRNA, and ssDNA used in this study.
| Name | Sequence (5′-3′) |
|---|---|
| UL75 RPA F1 | ATACTATGTATTCCATATGCCTCGATGTCTTT |
| UL75 RPA R1 | GTAGGTGTTAAGTCTCTGTTGGTATCTTTCTA |
| UL75 RPA F2 | CTACAATTACACAAAACGCACCTGGCCTCTTTT |
| UL75 RPA R2 | GATGAGCTAGCAACTGCGTAAAGTGTGATA |
| UL75 crRNA1 | UAAUUUCUACUAAGUGUAGAUGGUCAGAUCUACCUGGUUCA |
| UL75 crRNA2 | UAAUUUCUACUAAGUGUAGAUGCGUCUCUCCGUCGUAUGUA |
| Biotin ssDNA | FAM-TTATT-Biotin |
| FAM ssDNA | FAM-TTATT-BHQ1 |
The RPA reactions were performed following the instructions of the TwistAmp Basic Kit (TwistDX, UK). Briefly, the RPA TwistAmp freeze-dried enzyme pellet was rehydrated with 45.5 μl of a master-mix composed of 29.5 μl of rehydration buffer, 2.1 μl of each reverse/forward primer set (10 μM), and 11.8 μl of nuclease-free water. For each reaction tube, 2 μl of extracted DNA and 2.5 μl of Mg-acetate were added. The tube was mixed by inversion and briefly centrifuged. The reactions were performed at 37 °C using the ProFlex PCR System (Thermo Fisher, USA). The efficiency and specificity of the RPA primers were confirmed by gel electrophoresis and visualized using the Azure c300 imaging system (Azure Biosystems, USA).
2.3. CRISPR-Cas12a reaction
Two crRNAs were designed using the CRISPOR web tool (http://crispor.org) [28] to target the RPA amplicons. For LFA readout, single-stranded DNA (ssDNA) was labeled with the fluorophores 6-carboxyfluorescein (FAM) and biotin, as indicated in Table 1. For fluorescence detection, the ssDNA was labeled with FAM and Black Hole Quencher1 (BHQ1). The crRNAs and the ssDNAs were synthesized by Bioneer (Korea).
To detect Cas12a-mediated trans-cleavage activity using the LFA, 20 μL of the Cas12a reaction mixture was prepared with EnGen® Lba Cas12a (1 μM, M0653T, NEB, USA), NEBuffer r2.1 (B6002S, NEB, USA), crRNA (1 μM), RPA products, labeled ssDNA (10 μM), and distilled water. The mixture was incubated at 37 °C for 30 min using the ProFlex PCR System. After incubation, 20 μL of the reaction mixture was mixed with 100 μL of assay buffer of the HybriDetect – Universal Lateral Flow Assay Kit (MGHD1, Milenia Biotec, Germany) in a 1.5 mL tube.
To quantify the fluorescence, 100 μL of Cas12a reaction mixture was prepared and performed in a 96-well black polystyrene microplate (SPL, Korea). The fluorescence was detected and analyzed with the SpectraMax iD3 microplate reader (Molecular Devices, USA) at excitation and emission wavelengths of 470 nm and 520 nm, respectively. Emission was read in relative fluorescence units (RFU) to compare samples within each experiment.
2.4. Sensitivity and specificity of HCMV-RPA-CRISPR
The sensitivity of the HCMV-RPA-CRISPR was determined by using a 1:10 serially diluted UL75 plasmid DNA (ranging from 10−1 to 105 copies/μL). Comparative analysis included testing against HCMV, herpes simplex virus (HSV-1 and HSV-2), and varicella-zoster virus (VZV). These viruses were propagated in specific cell lines: HFF (human foreskin fibroblast; ATCC, SCRC-1041) for HCMV, Vero (African green monkey kidney cells; ATCC, CCL-81) for HSV-1 and HSV-2, and MeWo (human melanoma; ATCC, HTB-65) cells for VZV. The culture medium used was Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% Cosmic Calf Serum (CCS) and 1% penicillin/streptomycin for HFF and Vero cells, while MeWo cells were cultured with 2% CCS. After infection with the respective viruses, the cells were incubated for several days. Harvesting was performed when cytopathic effects (CPE) were observed in over 80% of the cells. Viral genomic DNAs were then extracted from either the media of virus-infected cell cultures (for HSV-1, HSV-2, and HCMV) or directly from the virus-infected cells (for VZV) using the Solg™ Genomic DNA Prep Kit (SGD41-C100, SolGent, Korea), following the manufacturer's instructions.
2.5. Quantitative PCR
Quantitative PCR (qPCR) was performed to compare and validate the specificity of the HCMV-RPA-CRISPR assay. The extracted DNAs from 13 clinical samples were analyzed using the TOPreal™ SYBR Green qPCR High-ROX PreMIX (RT501 M, enzynomics, Korea) in the Mic qPCR cycler (Bio Molecular Systems, Australia) with the HCMV gH specific primer pair reported previously [29]. A cycle threshold of less than 33 was considered positive based on in-hospital screening results.
2.6. Clinical sample preparation
For clinical sample preparation, peripheral blood was collected from suspected HCMV patients after obtaining written informed consent. Plasma samples were collected by centrifuging the blood samples at 15,000 rcf for 5 min at room temperature to separate the plasma. Total genomic DNA of the plasma samples was extracted using the Solg™ Genomic DNA Prep Kit (SGD41-C100, SolGent, Korea), following the manufacturer's instructions. This procedure was approved by the Institutional Review Board (IRB) of Jeju National University Hospital (IRB no. 2023-05-027).
2.7. Statistics and reproducibility
Statistical significances were analyzed using Prism 8 (GraphPad Software, version 8.0.1). The data involving endpoint fluorescence and threshold time to positive were all displayed with error bars representing mean ± standard deviation (SD) from three or more replicates. The two-tailed unpaired t-test was applied to investigate the differences between groups. Unless otherwise specified, each image for visual detection shown in the corresponding figure is representative of at least two independent experiments.
3. Results
3.1. Establishment of the HCMV-RPA-CRISPR diagnosis platform
An HCMV diagnosis platform was established using the RPA-CRISPR-Cas12a system (Fig. 1A). DNA extracted from the patient's blood was subjected to an RPA reaction using target-specific RPA primers for 20 min at 37 °C to amplify the target gene sequence. The RPA product was then combined with a CRISPR-Cas12a mixture containing crRNA and ssDNA, and the reaction was carried out for 30 min at 37 °C. For the fluorescence assay, BHQ1-and FAM-labeled ssDNA were added. If the result was positive, the collateral cleavage activity would cleave the ssDNA, resulting in a fluorescent signal. For the LFA, biotin- and FAM-labeled ssDNA were added. The LFA is based on the cleavage of a double-labeled reporter (FAM–biotin); the presence of many reporters causes anti-FAM antibody–gold nanoparticle conjugates (Au-NP) to gather at the C (control) line on the strip. In our experiments, when the activated CRISPR/Cas12a cleaved the ssDNA FAM–biotin reporter, the band intensity markedly increased on the T (test) line and decreased on the C line (Fig. 1B) [30]. The final product was mixed with the LFA buffer, the strip was immersed, and the results were visible within 3 min. A strong signal on the C (control) line near the buffer indicated a negative diagnosis, while a strong signal on the T (test) line indicated a positive diagnosis.
3.2. Optimal conditions for HCMV-RPA-CRISPR
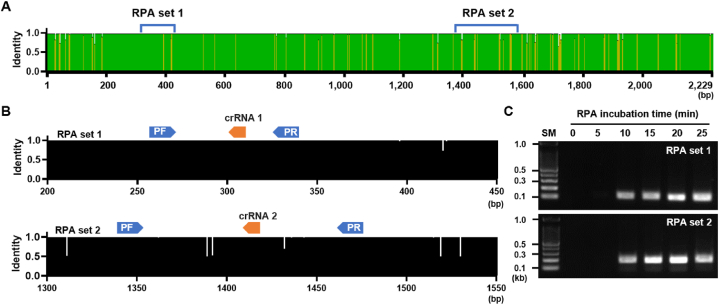
To design RPA primers for the UL75 gene, we obtained and analyzed sequence information from NCBI for areas with low genetic variation. Two sets of RPA primers were designed (Fig. 2A), along with two corresponding crRNAs, to enable CRISPR-Cas12a recognition of the sequence amplified by RPA (Fig. 2B).
Fig. 2.
Design and screening of RPA primers. (A) Map of nucleotide variation in the UL75 gDNA region of the 103 HCMV strains and location of the target region for RPA. (B) Conservation of target region for RPA and crRNAs among all 103 HCMV strains. (C) Gel electrophoresis image of RPA product amplified with two different primer sets. RPA reactions were performed for the indicated time. The original agarose gel image is provided in the supplementary data.
Efficient RPA primers were selected using the UL75 plasmid as a template. RPA analysis was conducted at 37 °C for various reaction times. RPA sets 1 and 2 successfully amplified DNA of the expected size (RPA set 1: 118 bp, RPA set 2: 187 bp) after 10 min, with no non-specific bands (Fig. 2C). These results indicated that both RPA sets specifically and rapidly amplified the target sequence. Based on the gel electrophoresis results, the 20-min reaction sample exhibited the strongest band intensity. Therefore, we determined that a 20-min reaction time would be optimal for further experiments.
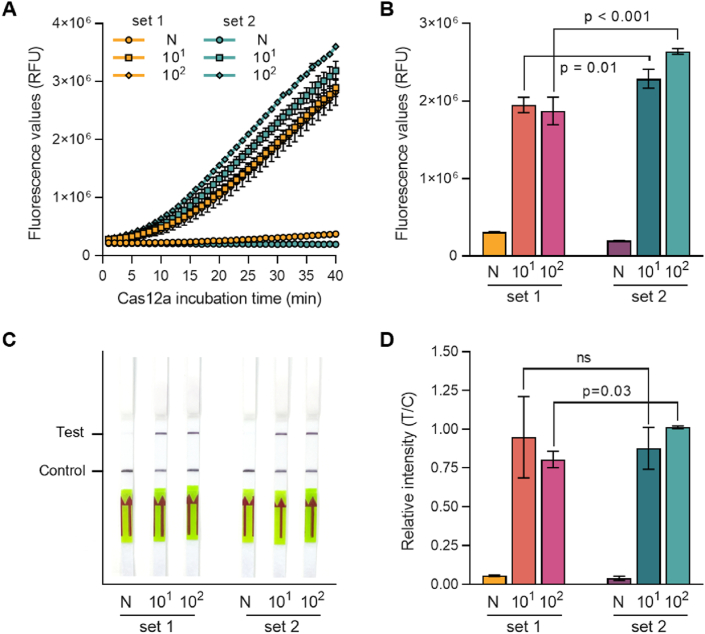
To determine the optimal CRISPR crRNA for target detection, two RPA-crRNA sets (crRNA 1 for RPA set 1 and crRNA 2 for RPA set 2) were tested using a reference plasmid of UL75 at concentrations of 1×101 and 1×102 copies/μL. The sensitivity of Cas12a-based detection of RPA-amplified HCMV UL75 sequences was evaluated using real-time fluorescence readouts. The fluorescence of the reaction products was measured using a plate reader every minute. The RPA-CRISPR-based detection system successfully detected HCMV, while no significant signal was observed in the negative control (N) (Fig. 3A). The fluorescence of the reaction using RPA set 2 and crRNA 2 showed higher signal intensity than that using RPA set 1 and crRNA 1 (Fig. 3A). After 30 min of reaction, the fluorescence of set 2 increased in both samples with a template concentration of 1×102 and 1×103 copies/μL (Fig. 3B). The RPA-CRISPR reaction was performed under these reaction conditions. The diagnosis results were confirmed using the LFA. As a result, the T-line band was noticeably enhanced in all samples except for the negative control (Fig. 3C and D). These experimental results established optimal assay conditions: an RPA reaction at 37 °C for 20 min using RPA primer set 2 and a CRISPR reaction at 37 °C for 30 min using crRNA 2.
Fig. 3.
Evaluation of RPA-crRNA. Two different RPA-crRNA sets were evaluated using fluorescence readout and LFA with UL75 plasmid DNA (101 and 102 copies/μl). (A) Fluorescence signals were detected during the CRISPR-Cas12a reaction by a plate reader every minute. (B) Fluorescence values after 30 min of the CRISPR-Cas12a reactions in panel A. Error bars indicate SD (n = 3). (C) Diagnosis results were read out using LFA strips after 30 min of Cas12a reaction. The photo was taken 3 min after the strips were dipped. (D) Relative band intensity of LFA strips in panel C. Error bars indicate SD (n = 3). Comparisons between groups were performed using the two-tailed unpaired Student's t-test. N: negative control, no template sample.
3.3. Sensitivity and specificity of CRISPR-Cas12a-LFA detection for HCMV
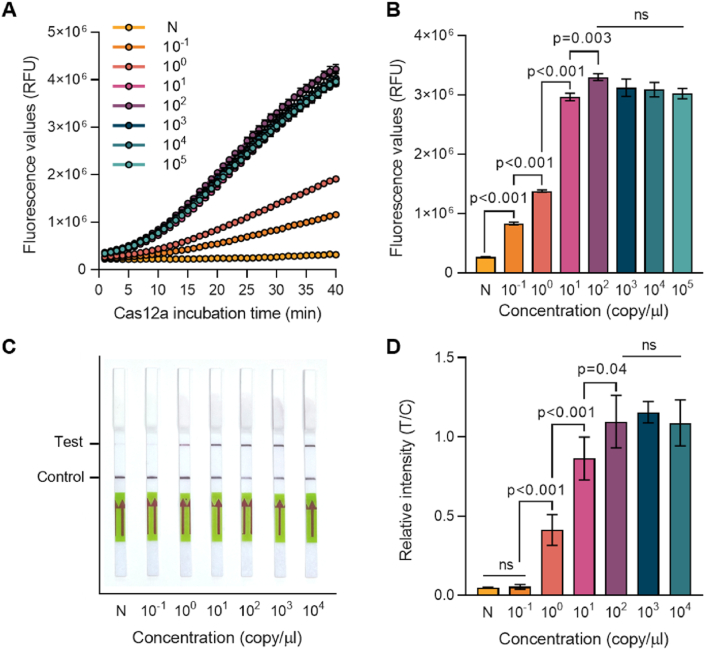
To determine the limit of detection (LoD), RPA-CRISPR reactions were performed using a serially diluted UL75 plasmid. Fluorescence generated during the 40-min CRISPR reaction confirmed the results. Positive samples showed an increased fluorescence signal, while negative samples did not (Fig. 4A). Samples with DNA template concentrations of 1×101 copies/μL or higher showed a rapid increase in fluorescence after 10 min. Saturation was observed at 1×101 copies/μL when comparing the fluorescence signals at 30 min of reaction (Fig. 4B). Even samples with lower quantities of template, specifically 1×10−1 and 1×10° copies/μl, showed fluorescence signals, indicating that detection was possible down to a final concentration of 1×10−1 copies/μL. The same template confirmed the LFA reaction results. The CRISPR-Cas12a reaction was performed for 30 min using ssDNA labeled with FAM and biotin, ensuring that detection was possible up to 1×10° copies/μL (Fig. 4C and D).
Fig. 4.
LoD determination of RPA-CRISPR-Cas12a HCMV diagnosis platform. RPA-CRISPR-Cas12a reactions were performed using serially diluted UL75 plasmid. (A) Fluorescence signals were detected during the CRISPR-Cas12a reaction by a plate reader every minute. (B) Fluorescence values after 30 min of the CRISPR-Cas12a reactions in panel A. Error bars indicate SD (n = 3). (C) Diagnosis results were read out using LFA strips after 30 min of Cas12a reaction. The photo was taken 3 min after the strips were dipped. (D) Relative band intensity of LFA strips in panel C. Error bars indicate SD (n = 3). Comparisons between groups were performed using the two-tailed unpaired Student's t-test. N: negative control, no template sample.
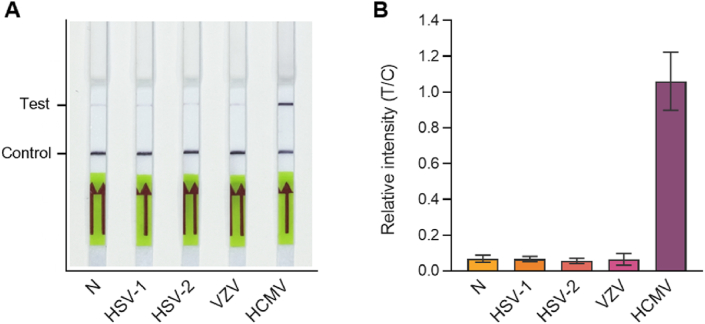
The HCMV RPA-CRISPR platform was highly specific to HCMV and could differentiate it from other herpesviruses. Viral DNA from HSV-1, HSV-2, VZV, and HCMV was used for the diagnosis assay. The assay results demonstrated that the HCMV-RPA-CRISPR assay platform could specifically identify only HCMV (<b>Fig. 5A and B</b>).
Fig. 5.
Specificity of RPA-CRISPR-Cas12a HCMV diagnosis platform. RPA-CRISPR-Cas12a reactions were performed using viral DNA of HSV-1, HSV-2, VZV, and HCMV (A). Diagnosis results were read out using LFA after 30 min of CRISPR-Cas12a reaction. (B) Relative band intensity of the LFA strips in panel A. Error bars represent the results of two independent experiments. N: negative control, no template sample.
3.4. Validation with clinical samples
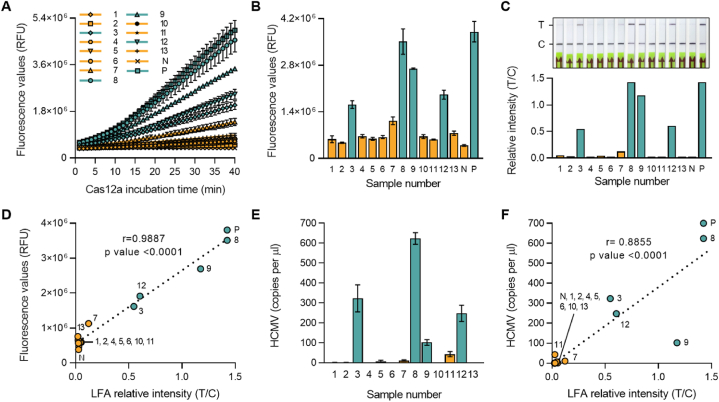
To evaluate the effectiveness of the HCMV-RPA-CRISPR assay platform in clinical samples, 13 blood samples were tested, including a UL75 plasmid DNA as a positive control (P) as well as a no-template negative control (N). The clinical samples were initially tested at Jeju National University Hospital and were diagnosed using conventional qPCR with four positives and nine negatives among the 13 samples. Using the HCMV-RPA-CRISPR assay with Fluorescence and LFA readout, all four HCMV positive samples were identified as positive, and all nine negative samples were identified as negative. These results demonstrated that the HCMV-RPA-CRISPR method was consistent with qPCR-based clinical diagnosis (Fig. 6A–C). A linear regression analysis was conducted to examine the relationship between fluorescence and LFA readout. The results indicate a strong positive correlation in both readout methods (Fig. 6D–F, Table 2). As shown in Fig. 6, the HCMV-RPA-CRISPR diagnosis method distinguished four positive and nine negative samples with 100% sensitivity and specificity. Furthermore, statistical analysis using a two-tailed unpaired Student's t-test revealed that HCMV-RPA-CRISPR could reliably distinguish positive from negative samples (P value < 0.0001).
Fig. 6.
HCMV diagnosis of patient blood samples using RPA-CRISPR/Cas12a-LFA and qPCR. (A) Fluorescence signals were detected during the CRISPR-Cas12a reaction by a plate reader every minute. (B) Fluorescence values after 30 min of Cas12a reaction in panel A. Error bars indicate SD (n = 3). Comparisons between groups were performed using the two-tailed unpaired Student's t-test. (C) Diagnosis results were read out using an LFA strip after 30 min of CRISPR-Cas12a reaction. The photo was taken 3 min after the strips were dipped (top). Relative band intensity of the LFA strip (bottom). (D) Linear regression analysis between fluorescence values after 30 min of Cas12a reaction and relative band intensities of LFA strips. (E) Predicted viral copy numbers were calculated by qPCR. Error bars represent the results of three independent technical replicates. (F) Linear regression analysis between calculated viral copy numbers and relative band intensities of LFA strips. Green bars and dots indicate positive diagnosed patient samples, and yellow bars and dots indicate negative patient samples. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Table 2.
Clinical validation of HCMV-RPA-CRISPR diagnosis. PPA: Positive Prediction Agreement, NPA: Negative Prediction Agreement.
| RPA-CRISPR-Cas12a | qPCR diagnosis |
PPA | NPA | ||
|---|---|---|---|---|---|
| Positive | Negative | ||||
| Fluorescence | Positive | 4 | 0 | 4 of 4 = 100% | 9 of 9 = 100% |
| Negative | 0 | 9 | |||
| LFA | Positive | 4 | 0 | 4 of 4 = 100% | 9 of 9 = 100% |
| Negative | 0 | 9 | |||
4. Discussion
Effective and timely diagnosis of HCMV infections is vital, particularly in vulnerable populations. This study introduces a novel diagnostic method utilizing CRISPR-Cas12a technology combined with RPA for rapid and affordable detection of HCMV. The advantages of this technique, such as its simplicity, speed, and cost-effectiveness, make it suitable for resource-limited settings lacking skilled technicians.
Among various disease diagnosis methods, the nucleic acid amplification test (NAAT) is highly effective due to its early detection and high sensitivity and specificity. However, conventional PCR-based strategies have limitations due to expensive equipment and complex laboratory procedures [31]. Our proposed CRISPR-Cas12a method offers all the advantages of traditional NAATs but does not require a thermal cycler, making it feasible even in small hospitals without specialized laboratories. Our newly developed the HCMV-RPA-CRISPR assay exhibits high sensitivity, capable of detecting a single copy of viral DNA (Fig. 4A and B). We compared the diagnostic accuracy of the HCMV-RPA-CRISPR assay with that of quantitative PCR. A linear regression analysis was used to examine the relationship between calculated viral copy numbers and relative band intensities of LFA strips. The result showed a strong correlation between the two diagnostic methods. Furthermore, our assay exhibited high specificity in HCMV detection without cross-reactivity with other herpes viruses (Fig. 5).
RPA and LFA readouts offer several advantages. RPA enhances sensitivity by amplifying the target nucleic acid sequence. Additionally, activated CRISPR-Cas12a cleaves thousands of ssDNA in the LFA, further amplifying the signal [16]. This user-friendly design allows non-specialized personnel to perform the assay with minimal training, expanding access to accurate HCMV diagnosis, particularly in regions with limited-skilled technicians. The simplified workflow reduces the turnaround time compared to traditional methods, and the affordability of the technique makes it suitable for resource-limited settings.
While preparing our manuscript, a study by Monk et al. was published, demonstrating the use of the CRISPR-Cas12a system combined with PCR and fluorescence detection for HCMV detection [32]. Although PCR is known for its specificity and high sensitivity, it requires costly equipment, limiting its application to well-equipped hospitals or laboratories. In contrast, our study introduces a distinct approach that utilizes RPA and CRISPR diagnostic systems with LFA detection, eliminating the need for expensive equipment and skilled technicians. This straightforward workflow and rapid turnaround time enhance practicality and efficiency, particularly in resource-limited settings.
In this study, plasma samples were analyzed using our newly developed HCMV-RPA-CRISPR assay. Evaluations using a wider variety of samples, such as urine, respiratory samples, and amniotic fluid, could expand the technique's scope. Investigating patient tracking for quantitative monitoring of viral load using a rapid and simple HCMV-RPA-CRISPR diagnosis platform might be beneficial, aiding in determining the optimal time to end antiviral therapy. For the presented results, the threshold was arbitrarily set based on confirmation from the hospital (Fig. 6). Standardization of the HCMV-RPA-CRISPR assay using international units, the World Health Organization (WHO) standard for HCMV, and increasing the number of patient samples will be implemented to establish precise thresholds by direct comparison with current qPCR analysis.
Meanwhile, this platform has important limitations. DNA extraction from patient samples has been a major obstacle to point-of-care diagnostics because they require expensive equipment such as centrifuges. However, recent advancements have introduced simpler methods for nucleic acid extraction from patient samples, and we are currently working on incorporating these methods into our platform. Additionally, while RPA-CRISPR technology excels in detecting the presence of pathogens, it may not be as effective in quantifying the viral load, which is crucial for assessing the severity of an infection and monitoring treatment responses. Although we have demonstrated a semi-quantitative tendency in LFA, the platform currently requires further refinement. We are actively testing more samples to improve the platform's ability to quantify viral load, enhancing its utility in clinical settings. This development is essential for a more comprehensive understanding of infection dynamics and for tailoring patient-specific treatment strategies.
In conclusion, combining RPA and CRISPR-Cas12a technology presents a promising and innovative approach for the rapid and affordable diagnosis of HCMV infection. This user-friendly method can potentially revolutionize HCMV diagnostics, particularly in resource-limited settings. This technique's simplicity, speed, and cost-effectiveness can significantly improve HCMV management and healthcare outcomes for affected individuals.
Ethical approval statement
This study was reviewed and approved by the Institutional Review Board (IRB) of Jeju National University Hospital (IRB no. 2023-05-027). Written informed consent was obtained from all subjects and/or their legal guardians.
Funding
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Ministry of Science and ICT (2021R1A2C1010313) and the Ministry of Education (RS2023-00270936), Republic of Korea, and the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI) funded by the Ministry of Health & Welfare (HI22C1510), Republic of Korea.
Data availability statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
CRediT authorship contribution statement
Kihye Shin: Writing – review & editing, Writing – original draft, Validation, Methodology, Investigation, Conceptualization. Gil Myeong Seong: Writing – review & editing, Resources, Conceptualization. Jeong Rae Yoo: Writing – review & editing, Resources. Eui Tae Kim: Writing – review & editing, Supervision, Project administration, Funding acquisition, Data curation.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We thank members of the Kim lab and Professor Lisa N. Akhtar (Northwestern University Feinberg School of Medicine) for insightful discussion and input. We thank Professor Jin-Hyun Ahn (Sungkyunkwan University School of Medicine) for providing the standard plasmid encoding UL75.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e28726.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Griffiths P., Reeves M. Pathogenesis of human cytomegalovirus in the immunocompromised host. Nat. Rev. Microbiol. 2021;19(12):759–773. doi: 10.1038/s41579-021-00582-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gandhi M.K., Khanna R. Human cytomegalovirus: clinical aspects, immune regulation, and emerging treatments. Lancet Infect. Dis. Dec 2004;4(12):725–738. doi: 10.1016/s1473-3099(04)01202-2. (in eng) [DOI] [PubMed] [Google Scholar]
- 3.Zuhair M., et al. Estimation of the worldwide seroprevalence of cytomegalovirus: a systematic review and meta-analysis. Rev. Med. Virol. May 2019;29(3):e2034. doi: 10.1002/rmv.2034. (in eng) [DOI] [PubMed] [Google Scholar]
- 4.Ariza-Heredia E.J., Nesher L., Chemaly R.F. Cytomegalovirus diseases after hematopoietic stem cell transplantation: a mini-review. Cancer Lett. Jan 1 2014;342(1):1–8. doi: 10.1016/j.canlet.2013.09.004. (in eng) [DOI] [PubMed] [Google Scholar]
- 5.A Ross S., Novak Z., Pati S., B Boppana S. Overview of the diagnosis of cytomegalovirus infection. Infect. Disord. - Drug Targets. 2011;11(5):466–474. doi: 10.2174/187152611797636703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De la Hoz R.E., Stephens G., Sherlock C. Diagnosis and treatment approaches of CMV infections in adult patients. J. Clin. Virol. 2002;25:1–12. doi: 10.1016/s1386-6532(02)00091-4. [DOI] [PubMed] [Google Scholar]
- 7.Lazzarotto T., Guerra B., Lanari M., Gabrielli L., Landini M.P. New advances in the diagnosis of congenital cytomegalovirus infection. J. Clin. Virol. 2008;41(3):192–197. doi: 10.1016/j.jcv.2007.10.015. 2008/03/01/ [DOI] [PubMed] [Google Scholar]
- 8.Razonable R.R., et al. Clinical diagnostic testing for human cytomegalovirus infections. J. Infect. Dis. 2020;221(Supplement_1):S74–S85. doi: 10.1093/infdis/jiz601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cannon M.J., Schmid D.S., Hyde T.B. Review of cytomegalovirus seroprevalence and demographic characteristics associated with infection. Rev. Med. Virol. Jul 2010;20(4):202–213. doi: 10.1002/rmv.655. [DOI] [PubMed] [Google Scholar]
- 10.Ramanan P., Razonable R.R. Cytomegalovirus infections in solid organ transplantation: a review. Infection & Chemotherapy. 2013;45(3):260. doi: 10.3947/ic.2013.45.3.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen J., Hu L., Wu M., Zhong T., Zhou Y.H., Hu Y. Kinetics of IgG antibody to cytomegalovirus (CMV) after birth and seroprevalence of anti-CMV IgG in Chinese children. Virol. J. 2012;9:304. doi: 10.1186/1743-422X-9-304. Dec 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Revello M.G., Gerna G. Diagnosis and management of human cytomegalovirus infection in the mother, fetus, and newborn infant. Clin. Microbiol. Rev. 2002;15(4):680–715. doi: 10.1128/CMR.15.4.680-715.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.The T.H., et al. Cytomegalovirus antigenemia. Rev. Infect. Dis. Sep-Oct 1990;12(Suppl 7):S734–S744. https://www.ncbi.nlm.nih.gov/pubmed/2173103 [Online]. Available: [PubMed] [Google Scholar]
- 14.Humar A., et al. Clinical utility of quantitative cytomegalovirus viral load determination for predicting cytomegalovirus disease in liver transplant recipients. Transplantation. Nov 15 1999;68(9):1305–1311. doi: 10.1097/00007890-199911150-00015. [DOI] [PubMed] [Google Scholar]
- 15.Kellner M.J., Koob J.G., Gootenberg J.S., Abudayyeh O.O., Zhang F. SHERLOCK: nucleic acid detection with CRISPR nucleases. Nat. Protoc. Oct 2019;14(10):2986–3012. doi: 10.1038/s41596-019-0210-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen J.S., et al. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science. Apr 27 2018;360(6387):436–439. doi: 10.1126/science.aar6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaminski M.M., Abudayyeh O.O., Gootenberg J.S., Zhang F., Collins J.J. CRISPR-based diagnostics. Nat. Biomed. Eng. 2021;5(7):643–656. doi: 10.1038/s41551-021-00760-7. [DOI] [PubMed] [Google Scholar]
- 18.Freije C.A., Sabeti P.C. Detect and destroy: CRISPR-based technologies for the response against viruses. Cell Host Microbe. 2021;29(5):689–703. doi: 10.1016/j.chom.2021.04.003. 2021/05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rajan A., ShrivastavaJanhawi S., Kumar A., Singh A.K., Arora P.K. CRISPR-Cas system: from diagnostic tool to potential antiviral treatment. Appl. Microbiol. Biotechnol. 2022;106(18):5863–5877. doi: 10.1007/s00253-022-12135-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jolany Vangah S., Katalani C., Booneh H.A., Hajizade A., Sijercic A., Ahmadian G. CRISPR-based diagnosis of infectious and noninfectious diseases. Biol. Proced. Online. 2020;22:22. doi: 10.1186/s12575-020-00135-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brogan D.J., Akbari O.S. CRISPR diagnostics: advances toward the point of care. Biochemistry. Mar 24 2022 doi: 10.1021/acs.biochem.2c00051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ding X., et al. Ultrasensitive and visual detection of SARS-CoV-2 using all-in-one dual CRISPR-Cas12a assay. Nat. Commun. 2020;11(1):4711. doi: 10.1038/s41467-020-18575-6. 2020/09/18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Broughton J.P., et al. CRISPR-Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. Jul 2020;38(7):870–874. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen Q., et al. CRISPR–Cas12‐based field‐deployable system for rapid detection of synthetic DNA sequence of the monkeypox virus genome. J. Med. Virol. 2023;95(1) doi: 10.1002/jmv.28385. [DOI] [PubMed] [Google Scholar]
- 25.Myhrvold C., et al. Field-deployable viral diagnostics using CRISPR-Cas13. Science. 2018;360(6387):444–448. doi: 10.1126/science.aas8836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sowmya P., Madhavan H.N., Therese K.L. Failure to genotype Human Cytomegalovirus by PCR-RFLP method due to sequence variation within the primer binding site. J Virol Methods. Jun 2006;134(1–2):250–251. doi: 10.1016/j.jviromet.2005.12.001. (in eng) [DOI] [PubMed] [Google Scholar]
- 27.Pignatelli S., Dal Monte P., Rossini G., Landini M.P. Genetic polymorphisms among human cytomegalovirus (HCMV) wild-type strains. Rev. Med. Virol. Nov-Dec 2004;14(6):383–410. doi: 10.1002/rmv.438. (in eng) [DOI] [PubMed] [Google Scholar]
- 28.Concordet J.-P., Haeussler M. CRISPOR: intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucleic Acids Res. 2018;46(W1):W242–W245. doi: 10.1093/nar/gky354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fukushima E., et al. Identification of a highly conserved region in the human cytomegalovirus glycoprotein H gene and design of molecular diagnostic methods targeting the region. J. Virol Methods. 2008;151(1):55–60. doi: 10.1016/j.jviromet.2008.03.022. 2008/07/01/ [DOI] [PubMed] [Google Scholar]
- 30.Zhang Y., Ying J.Y. Homogeneous immunochemical assay on the lateral flow strip for measurement of DNase I activity. Anal. Chem. Oct 20 2015;87(20):10193–10198. doi: 10.1021/acs.analchem.5b02658. [DOI] [PubMed] [Google Scholar]
- 31.Afzal A. Molecular diagnostic technologies for COVID-19: limitations and challenges. J. Adv. Res. Nov 2020;26:149–159. doi: 10.1016/j.jare.2020.08.002. (in eng) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Monk C.H., et al. Development of a CRISPR-Cas12a rapid diagnostic for human cytomegalovirus. Antivir. Res. 2023;215 doi: 10.1016/j.antiviral.2023.105624. 2023/07/01/ [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.