Abstract
Oxidation processes in mitochondria and different environmental insults contribute to unwarranted accumulation of reactive oxygen species (ROS). These, in turn, rapidly damage intracellular lipids, proteins, and DNA, ultimately causing aging and several human diseases. Cells have developed different and very effective systems to control ROS levels. Among these, removal of excessive amounts is guaranteed by upregulated expression of various antioxidant enzymes, through activation of the NF-E2-Related Factor 2 (NRF2) protein. Here, we show that Mitogen Activated Protein Kinase 15 (MAPK15) controls the transactivating potential of NRF2 and, in turn, the expression of its downstream target genes. Specifically, upon oxidative stress, MAPK15 is necessary to increase NRF2 expression and nuclear translocation, by inducing its activating phosphorylation, ultimately supporting transactivation of cytoprotective antioxidant genes.
Lungs are continuously exposed to oxidative damages induced by environmental insults such as air pollutants and cigarette smoke. Interestingly, we demonstrate that MAPK15 is very effective in supporting NRF2-dependent antioxidant transcriptional response to cigarette smoke of epithelial lung cells. Oxidative damage induced by cigarette smoke indeed represents a leading cause of disability and death worldwide by contributing to the pathogenesis of different chronic respiratory diseases and lung cancer. Therefore, the development of novel therapeutic strategies able to modulate cellular responses to oxidative stress would be highly beneficial. Our data contribute to the necessary understanding of the molecular mechanisms behind such responses and identify new potentially actionable targets.
Keywords: Cell signaling, Chronic respiratory diseases, Cigarette smoke, Gene expression, MAP kinases, Lung cancer
Highlights
-
•
MAPK15 controls NRF2 protein levels and nuclear localization.
-
•
MAPK15 induces NRF2 transcriptional activity, increasing the expression of its target genes.
-
•
MAPK15 directly phosphorylates the NRF2 protein.
-
•
MAPK15 activates PKC-dependent NRF2 Ser40 phosphorylation.
-
•
MAPK15 is necessary to induce the NRF2-dependent cytoprotective transcriptional program provoked by cigarette smoke.
Abbreviations
- ARE
antioxidant response elements
- COPD
chronic obstructive pulmonary disease
- CSE
condensed smoke extracts
- CHX
Cycloheximide
- DMF
dimethyl fumarate
- EPHX1
Epoxide hydrolase 1
- ERK
Extracellular signal-Regulated Kinase
- FACS
Fluorescence-Activated Cell Sorting
- FCCP
carbonyl cyanide 4-(trifluoromethoxy) phenylhydrazone
- FBS
fetal bovine serum
- GCLC
glutamate-cysteine ligase catalytic subunit
- GCLM
glutamate-cysteine ligase modifier subunit
- GeoMFI
Geometric Mean Fluorescent Intensity
- H2O2
hydrogen peroxide
- H2A.X
H2A histone family member X
- hAEC
human epithelial lung cells
- HO-1
Heme oxygenase 1
- JNK
c-Jun N-terminal kinases
- KD
kinase-dead
- MAPK15
Mitogen Activated Protein Kinase 15
- MGST1
Microsomal glutathione S-transferase 1
- MT2
Metallothionein-2
- NQO1
NAD(P)H dehydrogenase [quinone] 1
- NRF2
NF-E2-Related Factor 2
- PCA
Principal Component Analysis
- ROS
Reactive Oxygen Species
- RT-qPCR
quantitative reverse transcription polymerase chain reaction
- siMAPK15
MAPK15 specific siRNA
- SFN
sulforaphane
- siSCR
scrambled siRNA
- SOD3
Extracellular superoxide dismutase [Cu–Zn]
- SDC
Sodium Deoxycholate
- TPA
12-O-Tetradecanoylphorbol-13-acetate
- UGT1A6
UDP-glucuronosyltransferase 1-6
- ULK1
Unc-51 Like Autophagy Activating Kinase 1
- WT
wild-type
1. Introduction
NF-E2-Related Factor 2 (NRF2; encoded by NFE2L2) is a key transcription factor acting on Antioxidant Response Elements (ARE) contained in the promoters of numerous cytoprotective genes participating to the maintenance of the correct oxidative balance inside the cells, ultimately controlling their basal expression and strongly upregulating them upon oxidative damage [1]. Therefore, NRF2 is rightfully considered a master regulator of cellular responses against both endogenous and exogenous sources of oxidative stress [1]. In fact, even in the absence of external noxious stimuli, there are numerous intracellular sources of ROS. Among these, mitochondria are usually considered the major contributors because of the free radicals constantly generated during the oxidation processes necessary to supply energy to the cells [2]. Consequently, if not properly eliminated, continuous accumulation of ROS rapidly induces oxidation of intracellular lipids, proteins, and DNA, which has been implicated in many human disorders, from cardiovascular, respiratory, and metabolic diseases to cancer [3]. Not surprisingly, the incidence of all these conditions progressively increases during the aging process and an excessive intracellular amount of ROS has been frequently recognized as an important cause of cellular senescence [3]. Accordingly, NRF2 has been involved in regulating cell senescence phenotypes in vitro and aging features in vivo [4].
Because of their function, lungs and their epithelial cells are highly exposed to ROS mediated damage [5]. Indeed, in addition to their endogenous production, which is facilitated by high oxygen concentrations in inhaled air, they also suffer from being at the immediate interface with the environment, with continuous exposure to external insults such as air pollutants and cigarette smoke [6]. Accordingly, lungs have developed very effective systems of antioxidants located both inside and outside the cells, to cope with detrimental effects due to continuous oxidative stress. When external insults overcome the antioxidant capacity of the organ, either due to their high doses or prolonged duration, several lung diseases, such as asthma, chronic obstructive pulmonary disease (COPD), fibrosis, and cancer may result [7]. Interestingly, NRF2 has been identified as a key factor able to protect lungs from these pathological conditions and considered as a potential therapeutic target for its antioxidant effects [8]. There is, therefore, huge interest in understanding molecular mechanisms controlling its activity, to develop innovative pharmacological approaches against different diseases having oxidative stress and inflammation as underlying pathological features [1,9].
Mitogen Activated Protein Kinase (MAPK15), also known as Extracellular signal Regulated Kinase 7/8 (ERK7; ERK8) is an atypical member of the MAP kinase family whose activation has been mostly linked to induction of its expression and increased autophosphorylation events, while no direct upstream activating kinase has been identified so far [10,11]. Accumulating evidence now clearly point to a key role for MAPK15 in the responses to different types of stressful stimuli, overall contributing to molecular mechanisms maintaining cellular homeostasis. Indeed, MAPK15 has been involved in cellular responses to different physical (ionizing radiations) and chemical (chemotherapeutics, hydrogen peroxide) insults inducing DNA damage [12,13] and directly controls genomic stability by affecting PCNA stability and Telomerase activity [14,15]. In this context, it is also particularly important the role of MAPK15 in controlling the autophagic process, through Unc-51 Like Autophagy Activating Kinase 1 (ULK1)-dependent mechanisms [16,17], which allows cells to cope with different harmful stimuli [18,19]. Similarly, by regulating mitophagy, MAPK15 contributes to maintaining a “healthy” mitochondrial compartment, preventing excessive accumulation of ROS from these old and damaged organelles, which ultimately lead to chronic DNA damage and cellular senescence [20]. Consequently, modulation of MAPK15 activity may allow to interfere with oxidative stress-based pathogenetic mechanisms of different human diseases as we and others have already demonstrated in chronic obstructive pulmonary disease (COPD) and cancer, in vitro and in in vivo models [18,19,21]. In this context, while several reports have addressed the role of different MAP kinases in the control of NRF2 functions [22], no data are yet available regarding a specific role for the MAPK15 member of this family. Here, we demonstrate that MAPK15, by regulating the function of the NRF2 protein in lung cells, has yet another way to help cells to manage oxidative stress and may therefore open the opportunity for pharmacological or genetic interventions to prevent or treat very frequent and invalidating chronic respiratory diseases and well as deadly lung tumors.
2. Material and methods
2.1. Expression vectors
pCEFL-HA-MAPK15 and all its mutants were already described [16,23]. pGL4.37 [luc2P/ARE/Hygro] (Promega, 9PIE364), pLenti-NRF2, Myc-FLAG-tagged (Origene, RC204140L1) were purchased from indicated distributors. Vectors used for stable silencing of mouse cells (shSCR, scrambled control; shMAPK15, targeting mouse MAPK15) were already described [24]. The S40A point mutation in pLenti-NRF2_Myc-FLAG was introduced using the QuickChange XL Site-Directed Mutagenesis kit (Agilent, Santa Clara, CA), according to the manufacturer's instructions, with the following oligonucleotides: 5’-GTCGAGAAGTATTTGACTTCGCTCAGCGACGGAAAGAGTAT-3’ and 5’-ATACTCTTTCCGTCGCTGAGCGAAGTCAAATACTTCTCGAC-3'. To obtain the bacterially expressed NRF2 protein, we used pPROEX-HTc-Flag3-Nrf2, a gift from Yue Xiong (Addgene plasmid # 21553). Plasmids for the different dominant negative MAP kinases (pCDNA III MEK1 AA; pCEFL MEK5 AA; pCEFL GST MKK6 KR; pCDNA III SEK1 KR have been already described [25].
2.2. Statistical analysis
Fluorescence intensity was analyzed using the Quantitation Module of Volocity software (PerkinElmer Life Science, I40250). Significance (p-value) was assessed by pairwise Student's t-test, using GraphPad Prism8 software. Asterisks were attributed as follows: *p < 0.05, **p < 0.01, ***p < 0.001.
2.3. Cell culture and transfections
NCI–H358 cells were maintained in RPMI 1640 Medium supplemented with 10% fetal bovine serum (FBS), 2 mM l-glutamine and 100 units/ml penicillin-streptomycin. HeLa and 293T cells were maintained in DMEM supplemented with 10% FBS, 2 mM l-glutamine and 100 units/ml penicillin-streptomycin. Primary human airway epithelial cells (hAEC) were maintained in hAEC medium (Epithelix). For overexpression experiments, 3 × 105 cells were seeded in 6-well cell culture plates and transfected with 1 μg of each expression vector using Lipofectamine LTX (Life Technologies), according to manufacturer's instructions. NIH3T3 (CRL-1658) cells were maintained in DMEM supplemented with 10% calf serum, 2 mM l-glutamine and 100 units/ml penicillin-streptomycin. All cells were maintained at 37 °C in an atmosphere of 5% CO2/air.
2.4. Western blots
Total lysates were obtained by resuspending cellular pellets in RIPA buffer (50 mM TRIS-HCl pH = 8.0, 150 mM NaCl, 0.5% sodium deoxycholate, 0.1% SDS, 1% NP-40) with the addition of protease inhibitors (cOmplete Protease Inhibitor cocktail, EDTA-free; Roche Diagnostics) and phosphatase inhibitors (2 mM NaF, 2 mM Na3VO4; Sigma Aldrich). Total proteins were quantified by Bradford assay and the same quantity of lysates was used for Western blot analysis. Laemmli Loading Buffer 5X (250 mM Tris-HCl pH 6.8, 10% SDS, 50% glycerol, bromophenol blue) was added to protein samples, which were then heated for 5 min at 95 °C. Lysates were loaded on SDS-PAGE poly-acrylamide gel, transferred to Immobilon-P PVDF membrane (Merck Millipore), probed with appropriate antibodies, and revealed by enhanced chemiluminescence detection (ECL Plus; GE Healthcare). Densitometric analysis of western blots was performed with NIH Image J (National Institutes of Health).
2.5. Immunofluorescence
One × 104 cells were seeded in 12-well cell culture plates and transfected with each siRNA as described. When needed, samples were also transfected after 24 h, using Lipofectamine LTX (Life Technologies). Next, cells were washed with PBS, fixed with ice-cold methanol for 10 min, permeabilized with 0.2% Triton X-100 solution for 20 min and then blocked for 20 min with 2.5% BSA in PBS. Cells were then incubated with appropriate primary antibodies for 30 min, washed three times with PBS, and then incubated with appropriate Alexa Fluor 488-conjugated (Invitrogen, A21202) or Alexa Fluor 555-conjugated (Invitrogen, A31570) secondary antibodies. Nuclei were stained with 1.5 μM 4′,6-diamidino-2-phenylindole (DAPI) in PBS for 5 min. Coverslips were mounted in fluorescence mounting medium (Dako, S3023). Samples were visualized on a TSC SP5 confocal microscope (Leica, 5100000750) installed on an inverted LEICA DMI 6000CS (10741320) microscope using an oil immersion PlanApo 40 × 1.25 NA. Images were acquired using the LAS AF acquisition software (Leica).
2.6. Analysis of gene expression
Total RNA was purified using QIAzol Lysis Reagent (Qiagen). Reverse transcription was performed with the QuantiTect Reverse Transcription Kit (Qiagen). The following primer pairs were used: m_HPRT_Forw 5’-GGCCCTCTGTGTGCTCAAG-3’; m_HPRT_Rev 5’-CTGATAAAATCTACAGTCATAGGAATGGA-3’; h_MAPK15_Forw 5’-TGGCCAGCGTACAACAGGT-3’; h_MAPK15_Rev 5’-CAGTCCCGTAGGCTTGGGAGTA-3’; m_MAPK15_Forw 5’- GCCCGGACGCAATCGCTCAA-3’; m_MAPK15_Rev 5’- GGGCTACGCGGAGGTTTGGG-3’; h_MAPK1_Forw 5’-GCCCATCTTTCCAGGGAAGCATTA-3’; h_MAPK1_Rev AGAGCTTTGGAGTCAGCATTTGGG-3’; h_HO-1_Forw 5’-CAACATCCAGCTCTTGAGG-3’; h_HO-1_Rev 5’-GGCAGAATCTTGCACTTTG-3’; h_NQO1_Forw 5’-GAAGAGCACTGATCGTACTGGC-3’; h_NQO1_Rev 5’-GGGTCCTTCAGTTTACCTGTGAT-3’; h_MGST1_Forw 5’-TCGTGACAAAGCAAATTGTCTGG-3’; h_MGST1_Rev 5’-CCATTACCTGGGTGAGGTCAA-3’; h_NRF2_Forw 5’-CAGCGACGGAAAGAGTATGA-3’; h_NRF2_Rev 5’-TGGGCAACCTGGGAGTAG-3’; h_GCLM_Forw 5’-GGAACCTGCTGAACTGGGG-3’; h_GCLM_Rev 5’-CCCTGACCAAATCTGGGTTGA-3’; h_GCLC_Forw 5’-GTTCTTGAAACTCTGCAAGAGAAG-3’; h_GCLC_Rev 5’-CCTTCAATCATGTAACTCC-3’; h_SLC7A11_Forw 5’-TGCCCAGATATGCATCGTCC-3’; h_SLC7A11_Rev 5’-GGGCAGATTGCCAAGATCTCA-3’; h_AKR1B10_Forw 5’-CAGCAACAGAGAGCAGGACG-3’; h_AKR1B10_Rev 5’-TGCTGACGATGAACAGGTCC-3’.
2.7. Knock-down of endogenous MAPK15
MAPK15-specific siRNA (5’-TTGCTTGGAGGCTACTCCCAA-3’) and control non-silencing siRNA (Scramble, 5’-AATTCTCCGAACGTGTCACGT-3’) were obtained from Qiagen. All siRNAs were transfected at a final concentration of 100 nM using Hiperfect (Qiagen) for HeLa cells or RNAiMAX (Life Technologies) for hAEC, NCI–H358, and 293T cells (1 × 105 cells seeded in 6-well cell culture plates). Samples were collected 72 h after transfection. When needed, co-transfection with pLenti-NRF2, Myc-FLAG-tagged or pGL4.37 [luc2P/ARE/Hygro] were performed 24 or 48 h after siRNA transfection, respectively.
2.8. ROS quantification
To quantify ROS production, we incubated, 0.5 μM or 1 μM of CM-H2DCFDA (Invitrogen, C6827) for 30 min in medium without serum according to the manufacturer's protocol. Samples were acquired on a FACSCanto II flow cytometer (BD Biosciences). Data were analyzed with FlowJo software. All analyses were performed in triplicate.
2.9. Reagents and antibodies
4′,6–Diamidino-2-fenilindolo diidrocloruro (DAPI) (VWR) were used at final concentration of and 1.5 μM; Carbonylcyanide-4-(trifluoromethoxy)-phenylhydrazone (FCCP) (Enzo Life Sciences) was used at final concentration of 30 μM CM-H2DCFDA (Invitrogen, C6827) Cycloheximide (CHX) (Merk, 01810); Sorafenib (MedChemExpress 284461-73-0); TMCB (Santa Cruz, sc-361383); sulforaphane (SFN) (Santa Cruz, sc-361383); dimethyl fumarate (DMF) (Santa Cruz, sc-239774); Go 6983 (Med Chem Express HY-13689); ProBlue Safe Stain (Coomassie) (Giotto Biotech, G00PB002); TPA, 12-O-Tetradecanoylphorbol-13-acetate (Med Chem Express Cat. No.: HY-18739). The following primary antibodies were used for western blots and confocal microscopy experiments: anti-MAPK15 (Invitrogen PA5-75930), anti-HA (Covance, MMS-101R) anti-MAPK1 (Santa Cruz, sc-154), anti-NRF2 (Cell Signaling, 12721), anti-phospho-NRF2_Ser40 (Abcam, ab180844), anti-IKBα (Santa Cruz, sc-371), anti-Flag (Sigma, F1804) anti-γH2A.X (Cell Signaling, 9718), anti-53BP1 (Novus, NB100-304), anti-NQO1 (Santa Cruz, sc-207495A), anti–HO–1 (Santa Cruz, sc-136960 for mouse samples and Genetex GTX101147 for human samples), anti-phospho-NRF2_Ser40 (Invitrogen, PA567520), anti-SLC7A11 (Cell Signaling, D2M7A); Anti-Phospho-PKC (pan) (βII Ser660) (Cell Signaling, 9371); anti-PKC (A-3) (Santa Cruz, sc-17769); anti-Myc (Cell Signaling, 2276); anti-Phospho-Ser/Thr-Pro (Upstate, 05–368). The following secondary antibodies were used for Western blot experiments: anti-mouse (Jackson ImmunoResearch, 115-036-003) and anti-rabbit (Jackson ImmunoResearch, 111-036-003) HRP-conjugated IgGs.
2.10. Luciferase assays
Cells were transfected with 5 ng of the ARE-luciferase reporter vector and 1 μg of different expression vectors. Twenty-four hours after transfection, cells were treated with FCCP or condensed smoke extracts (CSE) for the indicated time and concentration, then lysed in Passive Lysis Buffer (Promega) and luciferase activity in the cellular lysates was assessed on a Glomax 20/20 luminometer (Promega) using the Luciferase Assay System (Promega). Results were normalized for transfection efficiency by using a GFP coding vector (1 μg). All luciferase results represent the average ± S.D. of three independent experiments. All samples were read in triplicate.
2.11. Samples preparation, LC-MS/MS and bioinformatics analysis
A solution of 2% sodium deoxycholate (SDC)/100 mM ammonium bicarbonate was used to lyse the samples. The cell lysates were alkylated in the dark with 10 mM Iodoacetamide at room temperature for 30 min after being reduced with 5 mM tris(2-carboxyethyl)phosphine (TCEP) at 60 °C for 30 min. Bicinchoninic acid (BCA) assay was used for protein quantification. 60 μg of proteins, for each sample, were processed adding trypsin using an enzyme-to-protein ratio of 1:40 and incubated at 37 °C overnight. After digestion, all reaction mixtures were acidified with 1% formic acid to inhibit any residual enzyme activity and precipitate the SDC [26,27]. Digested samples were desalted with OASIS cartridges (Waters), brought to dryness, and reconstituted in 0.1% formic acid in water/acetonitrile (97/3, v/v). QExactive Plus Orbitrap mass spectrometer (Thermo Fischer Scientific) was used to conduct LC-MS/MS analysis. The Acquity UPLC™ peptide CSH C18 column, 1 mm × 100 mm, 1,7 μm, 130 (Waters) was used for the peptide separation at 50 °C with a flow rate of 100 μl/min. The mobile phases A and B used for the analysis were 0.1% formic acid in water and 0.1% formic acid in acetonitrile respectively. These experiments were performed using a data dependent analysis (DDA) setting to select the “top twelve” most-abundant ions for MS/MS analysis. Protein identification was performed using Proteome Discover 2.1 (Thermo Scientific) and Sequest algorithm by using Mus musculus database from UniprotKB (organism ID 10090). Moreover, the default peak-picking settings were used to process the raw MS files in MaxQuant (version 1.6.1.0) [28] and its integrated search engine Andromeda [29]. Protein identification and label free quantification (LFQ) by MaxQuant/Andromeda was based on at least one unique peptide with a minimum length of seven amino acids and a false discovery rate of 0.01 applied to both peptide and protein level. By default, the alignment time window size was 20 min, and the match time window size was 0.7 min. The cutoff for peptide and protein identifications was set at False Discovery Rate (FDR) of 0.01. The runs were automatically aligned by MaxQuant. The peak intensities across the whole series of measurements were compared using the free software Perseus (version 1.6.1.1) to analyze MaxQuant result files and provide quantitative information for all the peptides in the sample. Proteins identified from the reverse sequence database or based on a single modified peptide, as well as contaminant proteins identified from the contaminant sequence database, were filtered out. The LFQ intensities of proteins from the MaxQuant analysis were imported and transformed to logarithmic scale with base two. The protein quantification and calculation of statistical significance was carried out using two-way Student-t test and error correction (p value < 0.05) with the method of Benjamini–Hochberg. For further visualization, Principal Component Analysis (PCA) were performed to investigate underlying differences between samples and replicates in quantitative proteomics results. Moreover, volcano plots were used to show a summary distribution of differentially expressed proteins between samples. The volcano plot is an easy-to-interpret scatter plot that arranges values along dimensions of statistical (log10 p-value) significance. The proteins located on the upper left region and the upper right region are differentially expressed.
2.12. Condensed smoke extract preparation
CSE was obtained from Marlboro Red cigarettes (Phillip Morris, 13 mg/cig tar and 0.9 mg/cig nicotine). CSE was produced by blowing the smoke from 10 cigarettes into 100 ml of PBS at a rate of 1 cigarette/min. This was considered as 100% CSE. The 100% CSE was passed through the 0.22 μm filter, aliquoted, and stored at −80 °C.
2.13. Subcellular fractionation
Cytoplasmic and nuclear fractions were obtained by NE-PER Kit (Thermo Scientific, 78833), according to the manufacturer's instructions.
2.14. Kinase assay
Purified recombinant protein of full length MAPK15 was purchased from Origene (Cat. N. TP761629). The BL21 strain of Escherichia coli (E. coli) was transformed with the pPROEX-HTc vector encoding for the full-length form of the NRF2 protein. Bacterially expressed His-fusion proteins were purified as previously described [16].
MAPK15 purified protein (50 ng/sample) was incubated 30 min at 30 °C in kinase buffer [25 mM HEPES (pH = 7.6), 0.1 mM Na3VO4, 20 mM β-glycerophosphate, 2 mM DTT, 20 mM MgCl2, 25 μM ATP] with 2.5 μg/sample of the NRF2 protein. Reaction was stopped adding Laemmli buffer and resolved by SDS-PAGE followed by WB analysis or followed by peptide mass fingerprint (PMF) and phosphorylation detection. Kinase activity was estimated by densitometric analysis of western blots, performed with NIH Image J software (National Institutes of Health), or by mass spectrometry, as described below.
2.15. Peptide mass fingerprint (PMF) and phosphorylation investigation
Purified proteins from the kinase reaction samples were subjected to overnight in-solution digestion by trypsin (Promega) at 37 °C, after reduction with dithiothreitol (DTT, final concentration 10 mM, Merck Group) and alkylation with iodoacetamide (IAA, final concentration 25 mM, Merck Group) [30]. The resulting peptides were brought to dryness and reconstituted in 0.1% formic acid in water. LC-MS/MS analyses were performed using Q-Exactive HF-X Orbitrap mass spectrometer (Thermo Scientific). The peptide separation was carried out at 35 °C using a PepMap RSLC C18 column, 75 μm × 15 cm, 2 μm, 100 Å (Thermo Fisher) at a flow rate of 300 nl/min. The mobile phases A and B used for the analysis were 0.1% formic acid in water and 0.1% formic acid in 80% acetonitrile, respectively. The gradient started with 5% of B and then it was increased up to 90% in 60 min. The data were elaborated using BioPharma Finder 2.0 (Application Note 21682, Thermo Fisher) comparing the list of peaks obtained “in silico” simulating protein digestion of the expected aminoacidic sequence (retrieved from UniProtKB, accession code: Q16236) and the experimental data. Phosphorylation was set up as post-translational modification (PTMs) in addition to carbamidomethylation and methionine/tryptophan-oxidation.
3. Results
3.1. A proteomic approach identifies MAPK15 as an upstream regulator of the expression of NRF2 target genes
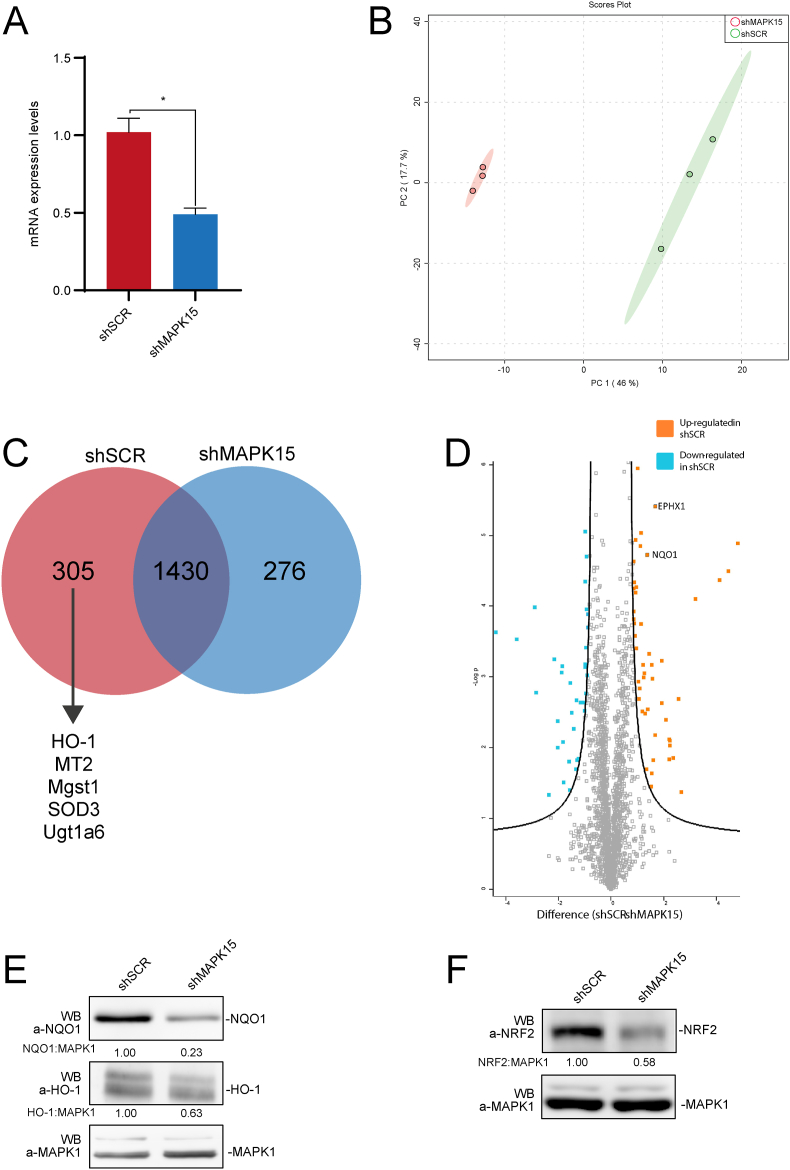
A shotgun approach was performed to investigate the proteomic profile of model murine NIH3T3 cells interfered for MAPK15 expression by a classical shRNA approach (shMAPK15) and compared with control/scrambled shRNA (shSCR) transfected cells (Fig. 1A). Only high confidence proteins were considered (total of 1735 and 1706 proteins, respectively), deriving from the triplicates of each condition. Specifically, 2011 quantifiable unique proteins with association to a known Mus musculus gene were found. Next, proteomics datasets were further categorized and examined with various statistical tools to provide a high-level understanding of the similarities and differences between shSCR and shMAPK15 samples. Principal Component Analysis (PCA) was next employed, to explore the sample separation and cluster. The first two main PCA components explained 63.1% of the variation (Fig. 1B) confirming a sizeable percentage. All triplicates of shSCR and shMAPK15 samples included in the analysis appeared to cluster in two distinct groups, reinforcing the evidence of a clear different proteomic datasets between shSCR and shMAPK15 (Fig. 1B).
Fig. 1.
Proteomic analysis. (A) NIH3T3 cells, stably transduced with SCR- or MAPK15-specific shRNA, were collected and subjected to quantitative reverse transcription polymerase chain reaction (RT-qPCR), to monitor mRNA expression of MAPK15. (B). Principal Component Analysis (PCA)-biplot showing a PCA of shSCR and shMAPK15 samples, analyzed by mass spectrometry. The percentage of the variance contributed by each principal component is indicated in the axis: 46.0% for the first component and 17.1% for the second component. (C) Venn diagram showing samples overlap, i.e., proteins uniquely identified in shSCR (305), in shMAPK15 (276) and the common subset of 1430 proteins. (D) Volcano plot to investigate the upregulated (in orange) and downregulated (in blue) among the common subsets of proteins, in comparison between control (scrambled, shRNA) and shMAPK15 cells. (E-F) NIH3T3 cells, stably transduced with shSCR- or shMAPK15-specific shRNA, were lysed, and then subjected to WB analysis, as indicated. One experiment, representative of 3 independent experiments is shown. Densitometric analysis of bands is also shown. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
A numerical comparison between the shSCR and shMAPK15 subsets was assessed and shown in a Venn diagram (Fig. 1C). We found that 305 proteins were unique in shSCR, 276 were unique in shMAPK15, whereas 1.430 were expressed in both samples (and further analyzed in Fig. 1D). The “overlap index”, based on Jaccard similarity coefficient, was 0.71. For shSCR, 17.58% of the total proteome was unique, while 16.18% was unique for shMAPK15. Among the 305 unique proteins absent in shMAPK15 treated samples, we noticed several reported NRF2 target genes, e.g., Heme Oxygenase 1 (HO-1), Metallothionein-2 (MT2), Microsomal glutathione S-transferase 1 (MGST1), Extracellular superoxide dismutase [Cu–Zn] (SOD3) and UDP-glucuronosyltransferase 1–6 (UGT1A6). The common subset was also further investigated with a volcano plot analysis (Fig. 1D), showing a label-free quantification (LFQ) intensity distribution of differentially expressed proteins between shSCR and shMAPK15 samples. The proteins located on the upper left region and the upper right region (S0 = 1, FDR<0.05) are differentially expressed. Among them, NAD(P)H dehydrogenase [quinone] 1 (NQO1) and Epoxide hydrolase 1 (EPHX1), two additional NRF2 target genes [31], also resulted downregulated in shMAPK15 in comparison with shSCR samples. Ultimately, to confirm the reliability of our proteomic approach, we demonstrated, by Western blot (WB), the downregulation of representative NRF2 target genes identified as positive hits by our analysis, namely NQO1 and HO-1 (Fig. 1E), overall suggesting a specific role for MAPK15 in controlling NRF2 activity and functions. In line with this hypothesis, although NRF2 was not detected during the proteomic analysis, possibly because its level fell below the detection limit of the approach, also this protein was strongly downregulated in MAPK15 interfered cells, when analyzed by WB (Fig. 1F).
3.2. Endogenous MAPK15 controls NRF2 protein levels and its transcriptional activity
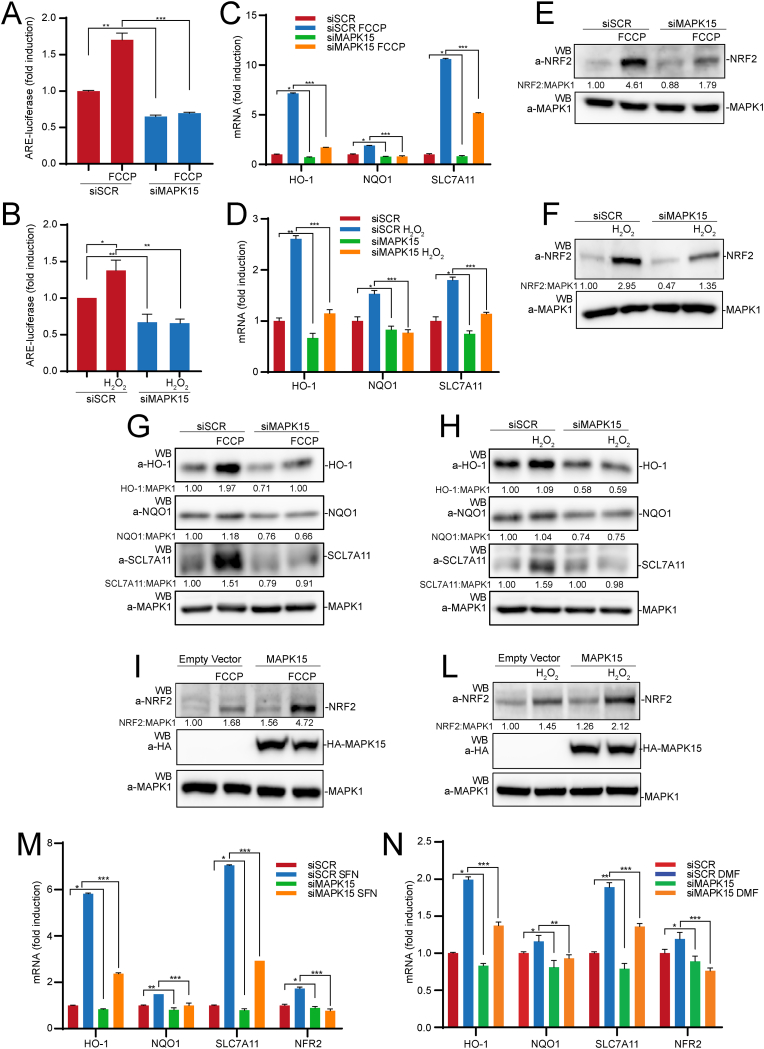
Based on these proteomic data, we next investigated a direct role for MAPK15 in regulating the expression of antioxidant genes by affecting NRF2 activity. Indeed, downregulation of MAPK15 endogenous expression by specific siRNA [20] (Fig. S1A) readily inhibited NRF2 transcriptional activity in HeLa cells, when measured by using an ARE-luciferase reporter gene, either in unstimulated conditions, and upon treatment with 2-[2-[4-(trifluoromethoxy)phenyl]hydrazinylidene]-propanedinitrile (FCCP) (Fig. 2A), which is able to strongly increase intracellular ROS levels in these cells [20], and hydrogen peroxide (H2O2) (Fig. 2B). Accordingly, MAPK15 downregulation also inhibited mRNA expression of representative NRF2-regulated genes, namely HO-1, NQO-1 and SLC7A11 both in basal conditions and upon stimulation with FCCP (Fig. 2C) and H2O2 (Fig. 2D). Of notice, we also observed that MAPK15 downregulation in 293T (Fig. S2A) and HeLa (Fig. S2B) cells reduced NRF2 gene transcription, a result that was nonetheless not unexpected based on the evidence of positive feedback loops affecting this gene, thanks to NRF2 ability to bind its own promoter [32]. In line with these data, reduced MAPK15 expression also affected NRF2 protein level in basal conditions, but especially prevented its upregulation upon treatment with stimuli able to increase intracellular ROS, namely FCCP (Fig. 2E) and H2O2 (Fig. 2F).
Fig. 2.
Loss of MAPK15 expression decreases NRF2 activity and protein level. (A) HeLa cells were transfected with scrambled siRNA (siSCR) or siRNA against MAPK15 (siMAPK15). After 48 h they were also transfected with the ARE luciferase reporter vector. Twenty-four hours after transfection, cells were treated with vehicle or 30 μM FCCP, for 6 h. Then, samples were lysed, and luciferase activity was measured in cell extracts. Data are represented as fold induction of the normalized luciferase activity compared to control cells transfected with GFP. All luciferase results represent the average ± S.D. of three independent experiments. All samples were read in triplicate. (B) Same as in (A) but treating cells with vehicle or 300 μM H2O2 for 10 min and harvested after 1 h. (C) HeLa cells were transfected with siSCR or siMAPK15 and after 72 h samples were treated with vehicle or FCCP 30 μM for 6 h, then collected and subjected to RT-qPCR, to monitor mRNA expression of HO-1, NQO1 and SLC7A11. (D) Same as in (C) but treating cells with 300 μM H2O2 for 2 h (E) HeLa cells were transfected with siSCR or siMAPK15 and after 72 h, samples were treated with 30 μM FCCP for 6 h. Then samples were harvested and subjected to SDS-PAGE followed by WB. One experiment, representative of 3 independent experiments is shown. Densitometric analysis of bands is shown. (F) Same as in (E) but treating cells with 300 μM H2O2 for 30 min. (G) HeLa cells were transfected with siSCR or siMAPK15 and after 72 h, samples were treated with 30 μM FCCP for 6 h. Then samples were harvested and subjected to SDS-PAGE followed by WB for HO-1, NQO1 and SLC7A11. One experiment, representative of 3 independent experiments is shown. Densitometric analysis of bands is shown. (H) Same as in (G) but treating cells with 300 μM H2O2 for 2 h. (I) HeLa cells were transfected with an empty vector or MAPK15_WT. After 24 h, cells were treated with FCCP 30 μM for 6 h. Then samples were harvested and subjected to SDS-PAGE followed by WB. One experiment, representative of 3 independent experiments is shown. Densitometric analysis of bands is shown. (L) Same as in (I) but treating cells with 300 μM H2O2 for 30 min. (M) HeLa cells were transfected with siSCR or siMAPK15 and after 72 h were treated with 10 μM of Sulforaphane (SFN) or vehicle for 6 h. Then samples were collected and subjected to RT-qPCR, to monitor mRNA expression of NQO1, HO-1, SLC7A11 and NRF2. (N) Same as in (M) but treating cells with 5 μM of dimethyl fumarate (DMF) for 3 h.
In line with the effects on mRNA expression of NRF2 target genes, MAPK15 interference also prevented upregulation of the corresponding proteins, namely HO-1, NQO1 and SCL7A11, induced by FCCP (Fig. 2G) and H2O2 (Fig. 2H). As an alternative and complementary approach, we next activated MAPK15 downstream signaling pathways by overexpressing its cDNA [17], and demonstrated both increased NRF2 expression in unstimulated conditions and strong cooperation with FCCP (Fig. 2I) and H2O2 (Fig. 2L). Ultimately, to confirm the ability of MAPK15 to control NRF2-dependent antioxidant responses also in conditions not directly inducing cellular ROS, we used available pharmacological NRF2 inducers, namely sulforaphane (SFN) (Fig. 2M) and dimethyl fumarate (DMF) (Fig. 2N) to induce mRNA expression of NRF2 target genes (HO-1, NQO1, SCL7A11 and NRF2 itself) and successfully inhibited this effect by downregulating MAPK15 expression.
3.3. MAPK15 directly phosphorylates the NRF2 protein
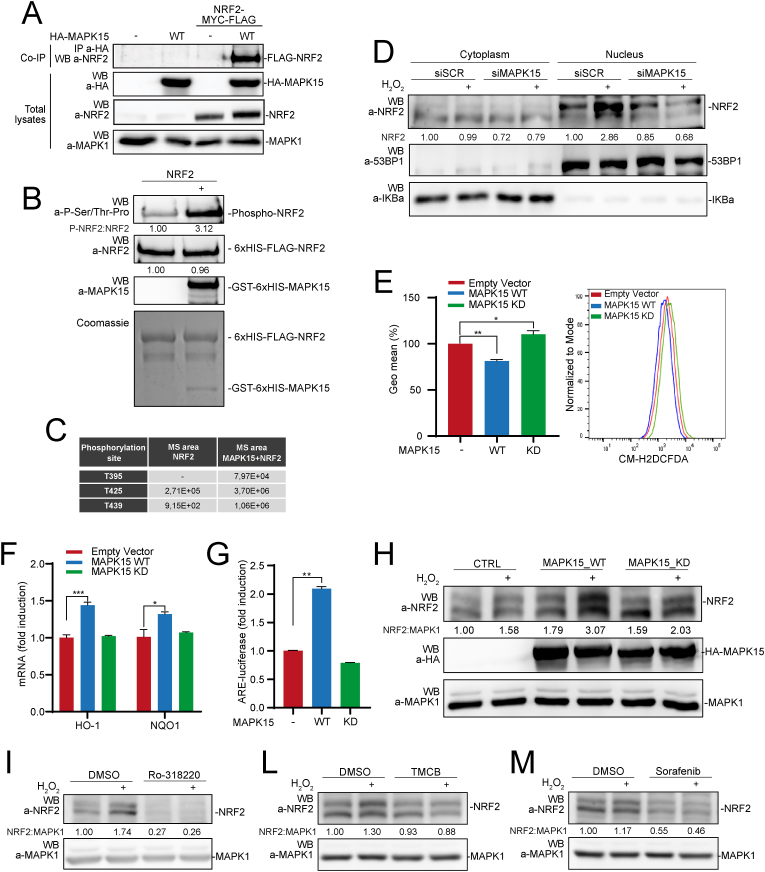
Cellular mechanisms impinging on NRF2 activation are rather complex and often based on protein-protein interactions allowing multiple post-translational modifications, among which phosphorylation by several different kinases [22], ultimately affecting NRF2 protein levels, nuclear localization, and transcriptional activity [33]. Indeed, MAPK15 coimmunoprecipitated with NRF2, in vivo (Fig. 3A), and directly phosphorylated this protein, as demonstrated by an in vitro kinase assay using recombinant MAPK15 and NRF2 proteins (Fig. 3B). Noteworthy, in this assay, we scored phosphorylation by using antibodies recognizing proline-directed serine and threonine phosphorylated residues, which are the expected “targets” for MAP kinases such as MAPK15 [34], further confirming the specificity of the experimental approach. Accordingly, analysis of in vitro kinase reactions by mass spectrometry demonstrated increased phosphorylation of three specific NRF2 threonine residues followed by prolines (Thr395, Thr425 and Thr439) (Fig. 3C and Fig. S3).
Fig. 3.
MAPK15 directly phosphorylates NRF2 and regulates its nuclear translocation. (A) 293T cells were transiently transfected with NRF2-MYC-FLAG plus empty vector or HA-MAPK15_WT. After 24 h, lysates were immunoprecipitated with anti-HA antibodies and subjected to SDS-PAGE followed by WB, to detect coimmunoprecipitated NRF2 protein. Total lysates were also analyzed for expression of indicated proteins. One experiment, representative of 3 independent experiments, is shown. (B) NRF2 protein purified from bacteria was subjected to kinase assay in presence or absence of recombinant, bacterially purified MAPK15. Samples from kinase reactions were resolved by SDS-PAGE, and NRF2 phosphorylation was demonstrated by WB using specific anti-phospho-Ser/Thr-Pro antibodies. Coomassie staining and WB analysis (with indicated antibodies) of kinase reactions were used as loading controls. (C) Kinase reactions prepared as in (B) were resolved by Mass Spectrometry analysis, to determine the identity and amounts of phosphorylated residues. (D) 293T cells were transfected with siSCR or siMAPK15. After 72 h, they were treated with 300 μM H2O2 for 1 h and then subjected to nucleo-cytoplasmic fractionation. Amounts of NRF2 in the different fractions were analyzed by WB. Normalization of cytosolic and nuclear lysates was performed by WB, using anti-IKBα and anti-53BP1 antibodies, respectively. One experiment, representative of 3 independent experiments, is shown. Densitometric analysis of bands is shown. (E) Representative Fluorescence-Activated Cell Sorting (FACS) histograms or corresponding Geometric Mean Fluorescent Intensity (GeoMFI) bars of CM-H2DCFDA (1 μM) fluorescence 24 h after 293T cells transfection using empty vector, MAPK15_WT or MAPK15_KD plasmids. Bars represent the average ± S.D. of 3 independent experiments (n = 3). (F) RT-qPCR was used to monitor mRNA expression of NQO1 and HO-1 in 293T cells transiently overexpressing empty vector, MAPK15_WT or MAPK15_KD. (G) 293T cells were co-transfected with ARE luciferase reporter vector plus empty vector or MAPK15_WT or MAPK15_KD. Twenty-four hours after transfections, samples were lysed and the luciferase activity was measured in cell extracts. Data are represented as fold induction of the normalized luciferase activity with respect to control cells transfected with GFP. All luciferase results represent the average ± S.D. of three independent experiments. All samples were read in triplicate. (H) 293T cells were transiently co-transfected with NRF2-MYC-FLAG plus empty vector or MAPK15_WT or MAPK15_KD. After 24 h, samples were treated with 300 μM H2O2, for 1 h. Lysates were analyzed by WB with indicated antibodies. One experiment, representative of 3 independent experiments, is shown. Densitometric analysis of bands is indicated. (I) 293T cells were treated with vehicle or 2 μM Ro-318220, for 6 h. During the last hour of Ro-318220 treatment, samples were treated with 300 μM H2O2, for 1 h. Then samples were collected and subjected to SDS-PAGE followed by WB. One experiment, representative of 3 independent experiments, is shown. Densitometric analysis of bands is shown. (L) Same as in (I), but cells were treated with vehicle or TMCB 10 μM, for 24 h. (M) Same as in (I), but cells were treated with vehicle or Sorafenib 10 μM, for 6 h.
As Thr395 and Thr439 have been already described as target of CDK5 phosphorylation and demonstrated to control NRF2 translocation to the nucleus [35], we next hypothesized that the same residues may be used by MAPK15 to control this process. Indeed, we observed reduced nuclear accumulation of NRF2 after oxidative stress induced by H2O2 (Fig. 3D) and FCCP (Fig. S4), in MAPK15 downregulated cells, ultimately demonstrating a specific role for this MAP kinase in nuclear translocation of the NRF2 protein, which is a prerequisite for its transcriptional activity. To further support a role for MAPK15 kinase activity in controlling NRF2 functions by directly phosphorylating NRF2, we next demonstrated that a MAPK15 kinase-dead mutant (MAPK15_KD) [17], was unable to reduce cellular ROS levels (Fig. 3E), to upregulate the expression of the NRF2-regulated genes HO-1 and NQO1 (Fig. 3F), and to stimulate the transcription of the ARE-luciferase reporter gene (Fig. 3G), when compared to its wild-type (WT) counterpart in transfected 293T cells. Moreover, such mutant was also unable to cooperate with oxidative stress (H2O2) in increasing NRF2 protein levels, as compared to the overexpressed WT protein (Fig. 3H).
To confirm these data also with a different approach, we next set up to investigate the effect of endogenous MAPK15 chemical inhibition on the stabilization of NRF2 induced by oxidative stress. Indeed, when we treated 293T cells with three different and unrelated MAPK15 pharmacological inhibitors, Ro-318220 (Fig. 3I) [16], TMCB (Fig. 3L) [36] and Sorafenib (Fig. 3M) (https://www.kinase-screen.mrc.ac.uk/inhibitor-and-results/16), we observed a significant reduction of NRF2 protein levels both in basal conditions and upon cellular exposure to oxidative stress by H2O2 stimulation (Fig. 3I-M), overall supporting the ability of MAPK15 to directly phosphorylate specific NRF2 residues involved in protein nuclear translocation and transactivating potential.
3.4. MAPK15 induces PKC-dependent NRF2 serine 40 phosphorylation and protein stabilization
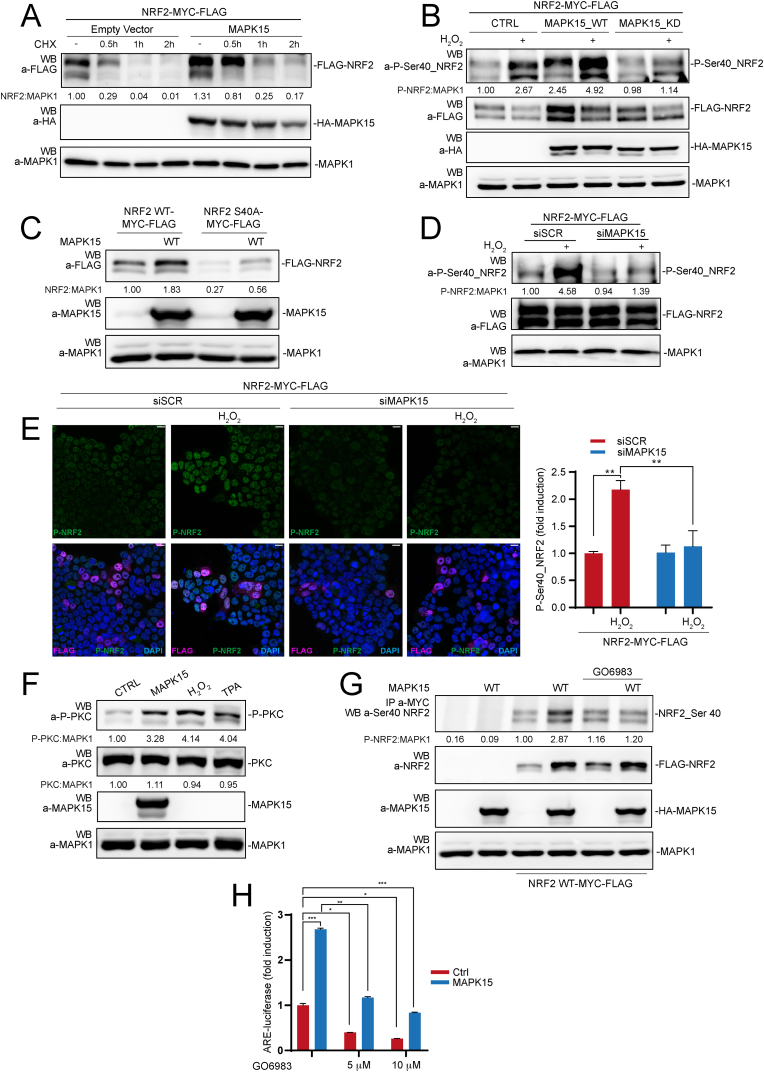
Under normal conditions, NRF2 is a very short-lived protein but its stability can be rapidly increased by oxidative or electrophilic stress, by inhibiting its interaction with the Kelch-like ECH-associated protein-1 (KEAP1), which serves as the primary ubiquitin E3 ligase adaptor mediating ubiquitination-dependent proteasomal degradation of NRF2 [9]. As NRF2 protein levels were clearly affected by modulating MAPK15 expression (see Fig. 2 above), we decided to next investigate molecular mechanisms responsible for this regulation. Accordingly, we first confirmed that MAPK15 increased the stability of a NRF2 transfected protein (to avoid the interfering bias of transcriptional regulation of the endogenous gene) (Fig. 4A). Next, as phosphorylation of NRF2 at Ser40 by protein kinase C (PKC) has been demonstrated to prevent its proteasomal degradation [37,38], we took advantage of an anti-phospho-Ser40-NRF2 antibody to ascertain potential effects of MAPK15 on the phosphorylation of this specific residue. Indeed, we demonstrated that overexpression of the wild-type MAPK15 cDNA (MAPK15_WT) strongly induced Ser40_NRF2 phosphorylation, both in unstimulated 293T cells and in H2O2 stimulated cells, while MAPK15_KD was unable to induce Ser40_NRF2 phosphorylation in basal conditions and even prevented this post-translational modification upon H2O2 stimulation (Fig. 4B). Accordingly, mutation of Ser40 in NRF2 to alanine (NRF2-S40A) strongly reduced MAPK15-dependent NRF2 stabilization (Fig. 4C), overall supporting the important, although not exclusive role of this residue in the control of NRF2 protein levels downstream of MAPK15. Correspondingly, MAPK15 downregulation by specific siRNA (Fig. S1B) inhibited oxidative stress-dependent phosphorylation of NRF2 on Ser40, when measured by both WB (Fig. 4D) and confocal microscopy (Fig. 4E) analysis.
Fig. 4.
MAPK15 induces PKC-dependent NRF2 Serine 40 phosphorylation and protein stabilization. (A) 293T cells were transiently co-transfected with NRF2-MYC-FLAG plus empty vector or MAPK15_WT. After 24 h, they were treated with 50 μM Cycloheximide (CHX), for indicated times. Lysates were analyzed by WB for the indicated proteins. One experiment, representative of 3 independent experiments, is shown. Densitometric analysis of bands is indicated. (B) 293T cells were transiently co-overexpressed with NRF2-MYC-FLAG plus empty vector or MAPK15_WT or MAPK15_KD. After 24 h, samples were treated with 300 μM H2O2, for 1 h, and the lysates were subjected to SDS-PAGE followed by WB and analyzed for indicated proteins. One experiment, representative of 3 independent experiments, is shown. Densitometric analysis of bands is indicated. (C) 293T cells were transiently co-transfected with NRF2-MYC-FLAG or NRF2-MYC-FLAG S40A, plus empty vector or MAPK15_WT. After 24 h. Lysates were analyzed by WB for the indicated proteins. One experiment, representative of 3 independent experiments, is shown. Densitometric analysis of bands is indicated. (D) 293T cells were transfected with siSCR or siMAPK15. Then, after 24 h, they were transfected with NRF2-MYC-FLAG. After additional 48 h, cells were treated with 300 μM H2O2 for 1 h and lysates were analyzed by WB for indicated proteins. One experiment, representative of 3 independent experiments, is shown. Densitometric analysis of bands is indicated. (E) 293T cells were transfected with siSCR or siMAPK15 and, after 24 h, transfected with empty vector or NRF2-MYC-FLAG. After a total of 72h, samples were treated with 300 μM H2O2, for 1 h, then fixed and subjected to immunofluorescence analysis. Scale bars correspond to 10 μm. The accompanied graph shows intensitometric analysis of phospho-NRF2 S40A fluorescence from five representative microscopy fields signal per cell ± SD. (F) 293T cells were transiently transfected with empty vector or MAPK15_WT or treated with 300 μM H2O2 or 200 nM TPA for 1 h, as positive controls. Lysates were analyzed by WB for the indicated proteins. One experiment, representative of 3 independent experiments, is shown. Densitometric analysis of bands is indicated. (G) 293T cells were transiently transfected with NRF2-MYC-FLAG or NRF2-MYC-FLAG_S40A plus empty vector or HA-MAPK15_WT. After 24 h, samples were treated, as indicated, with 10 μM GO6983 for 2 h. Lysates were next immunoprecipitated with anti-MYC antibodies and subjected to SDS-PAGE followed by WB to detect phospho-NRF2-S40 protein. Total lysates were also analyzed for expression of indicated proteins. One experiment, representative of 3 independent experiments, is shown. (H) 293T cells were transfected with ARE luciferase reporter vector plus empty vector or MAPK15_WT. Twenty-four hours after transfection, samples were treated with 5 μM or 10 μM GO6983, for 2 h. Then, samples were lysed, and luciferase activity was measured in cell extracts. Data are represented as fold induction of the normalized luciferase activity compared to control cells transfected with GFP. All luciferase results represent the average ± S.D. of three independent experiments. All samples were read in triplicate.
As serine 40 in NRF2 is not followed by a proline, it does not represent the typical recognition sequence for proline-directed Ser/Thr kinases such as MAP kinase family members [34]. Consequently, we explored the possibility that MAPK15 may induce phosphorylation of Ser40 on NRF2 by activating its already known kinase, i.e., PKC [37,38]. Indeed, MAPK15 activated PKC at levels comparable to the 12-O-Tetradecanoylphorbol-13-acetate (TPA) and H2O2 positive controls, when scored by anti phospho-PKC WB (Fig. 4F) [39,40] and a specific PKC inhibitor, GO6983 [40], strongly reduced MAPK15-dependent Ser40_NRF2 phosphorylation (Fig. 4G). Accordingly, GO6983 also readily inhibited MAPK15-dependent NRF2 transcriptional activity, when measured by using the ARE-luciferase reporter gene (Fig. 4H). Overall, these data therefore demonstrate that MAPK15 is able to activate PKC and, through this kinase, induce phosphorylation of the key Ser40 residue in NRF2, ultimately opposing to its proteasomal degradation.
3.5. NRF2 overactivation prevents increased oxidative stress and DNA damage induced by MAPK15 downregulation
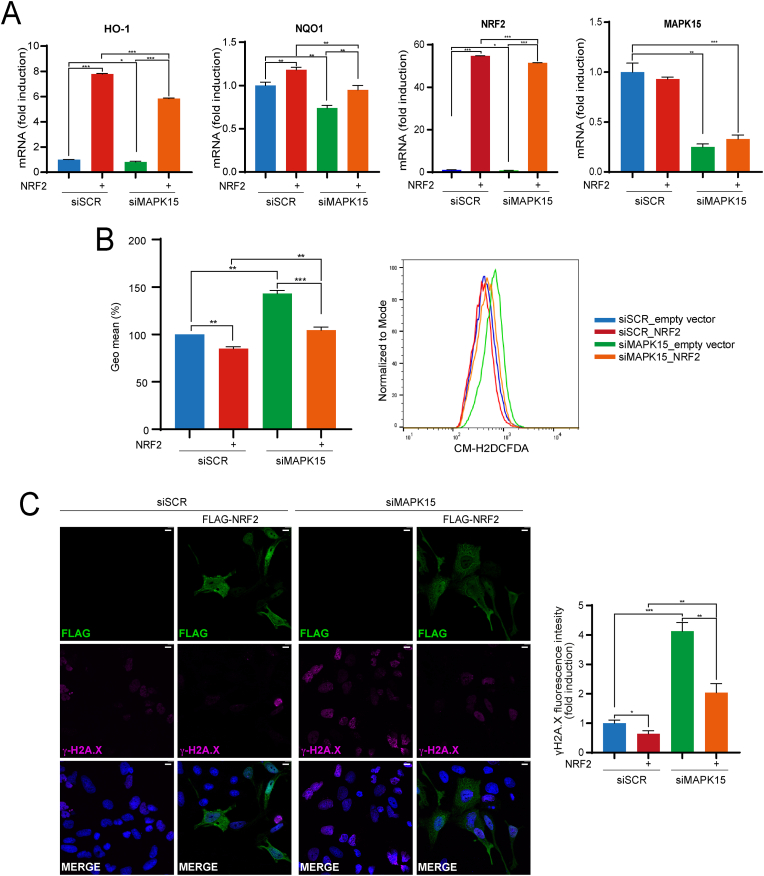
Reduced efficacy in NRF2-dependent cellular response to oxidative stress causes increased ROS which, in turn, can damage several cellular components, including nuclear DNA, possibly contributing to genomic instability and cancer [41]. Accordingly, in line with the suggested role of MAPK15 in supporting NRF2 activity, we and others have repeatedly demonstrated, in MAPK15-downregulated cells, very high levels of phosphorylated γH2A histone family member X (γH2A.X) foci, which specifically scores DNA damage [15,19,20]. As several mechanisms have been proposed to explain this observation [15,19,20], we next evaluated if also reduced NRF2 activity may contribute to increased ROS levels and DNA damage, in MAPK15-downregulated cells. To address this point, we decided to transfect NRF2 cDNA, to artificially increase its transcriptional and antioxidant activities and rescue ROS-increasing and DNA-damaging phenotypes dependent on MAPK15 downregulation [42]. Indeed, NRF2 overexpression significatively rescued the expression of HO-1 and NQO-1 genes (Fig. 5A) and, consequently, reduced ROS levels (Fig. 5B) and nuclear DNA damage (Fig. 5C) determined by MAPK15 RNA interference in HeLa cells.
Fig. 5.
Hyperactivation of the NRF2 pathway rescues MAPK15-dependent cellular phenotypes. (A) HeLa cells were transfected with siSCR or siMAPK15. After 24 h, samples were transfected with empty vector or NRF2-MYC-FLAG. After a total of 72h, samples were subjected to RT-qPCR to monitor mRNA expression of the indicated genes. (B) Representative FACS histograms and corresponding GeoMFI bars of CM-H2DCFDA fluorescence from HeLa cells transfected with siSCR or siMAPK15, after 24 h samples were transfected with empty vector or NRF2-MYC-FLAG. After a total of 72h, samples underwent FACS analysis for CM-H2DCFDA 1 μM fluorescence. Bars represent the average ± S. D of 3 independent experiments (n = 3). (C) HeLa cells were transfected with siSCR or siMAPK15, after 24 h samples were transfected with empty vector or NRF2-MYC-FLAG. After a total of 72 h, cells were fixed and subjected to immunofluorescence analysis. Scale bars correspond to 10 μm. The accompanied graph shows intensitometric analysis of DNA damage through nuclear γH2A.X fluorescence from five representative microscopy fields signal per cell ± SD.
3.6. MAPK15 is a key player in the response to oxidative stress induced by cigarette smoke in normal and tumor lung cells
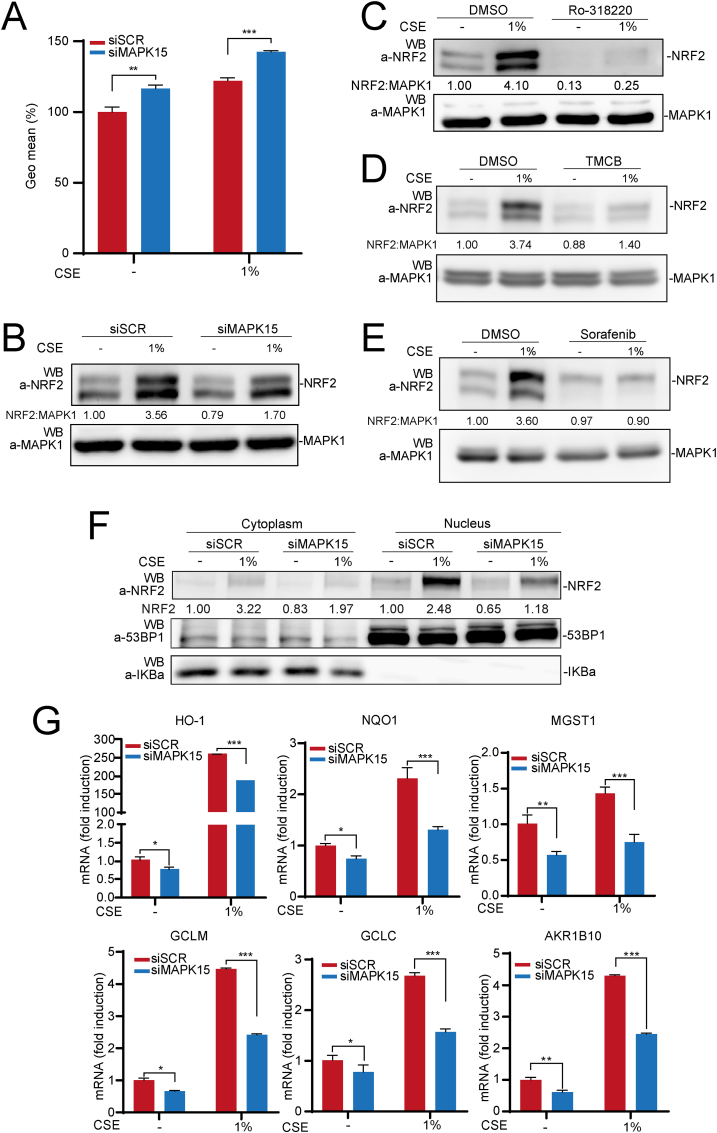
Chronic respiratory lung diseases and tumors affecting this organ collectively represent leading causes of disability and death worldwide and chronic use of tobacco has been clearly recognized as a primary trigger for a large proportion of these cases [43,44]. Among many reported pathogenetic mechanisms exerted by cigarette smoke to affect lung tissue, oxidative stress surely has a key role [45], for example by inducing accumulation of mutagenic lesions which may be responsible for both cancer initiation and progression [46]. We, therefore, next investigated the possibility of an involvement of MAPK15 in controlling NRF2-dependent antioxidant responses elicited by condensed smoke extracts (CSE), in normal and cancer lung cells. Indeed, CSE strongly increased ROS levels in normal human epithelial lung cells (hAEC), and this effect was further amplified by MAPK15 downregulation (Fig. 6A), which was particularly efficient in these cells (Fig. S1C). Next, CSE readily elicited an antioxidant response by increasing NRF2 protein levels, an effect that was reduced by ∼50% in MAPK15 interfered cells (Fig. 6B). Importantly, also a pharmacological approach, by using the Ro-318220, TMCB and Sorafenib MAPK15 inhibitors, strongly reduced NRF2 protein levels both in unstimulated conditions and upon CSE treatment of hAEC cells (Fig. 6C–, D and E, respectively) and of 293T cells (Fig. S5), supporting the possibility of developing new drugs modulating cellular responses to oxidative stress by acting on MAPK15. Remarkably, we also noticed that, upon CSE treatment, NRF2 mostly accumulated in the nucleus, while MAPK15 downregulation efficaciously prevented this outcome (Fig. 6F), supporting the key role of this kinase in controlling the effective transactivating potential of NRF2 also in normal lung cells. Indeed, MAPK15 downregulation resulted particularly effective also in reducing the expression of several NRF2 target genes induced by CSE in these cells (Fig. 6G), overall suggesting a role for MAPK15 in preventing and/or limiting excessive oxidative stress and its long-term pathogenetic effects in the lungs. Interestingly, among such genes, we also found AKR1B10, which is expressed at detectable levels only in hAEC cells among all those we analyzed in our study (Fig. S6A), demonstrating reduced mRNA (Fig. 6G) and protein (Fig. S6B) levels upon MAPK15 downregulation, both in basal and in CSE-induced cells.
Fig. 6.
MAPK15 controls cytoprotective responses of normal human epithelial lung cells to cigarette smoke. (A) GeoMFI bars of CM-H2DCFDA fluorescence from hAEC cells transfected with siSCR or siMAPK15. After 72 h of transfection, samples were treated for 4 h with 1% CSE. Bars represents the results represent the average ± S. D of 3 independent experiments (n = 3). (B) hAEC were transfected with siSCR or siMAPK15. After 72 h, cells were treated for 4 h with 1% CSE. Next, lysates were analyzed by WB for indicated proteins. One experiment, representative of 3 independent experiments is shown. Densitometric analysis of bands is indicated. (C) hAEC cells were treated with vehicle or 2 μM Ro-318220 for 6 h. During the last 4 h of Ro-318220 treatment, samples were also treated with 1% CSE. Then, samples were collected and analyzed by WB. One experiment, representative of 3 independent experiments is shown. Densitometric analysis of bands is shown. (D) hAEC cells were treated with vehicle or TMCB 10 μM, for 24 h. During the last hours of TMCB treatment, samples were treated with 1% CSE 4 h. Then samples were collected and subjected to SDS-PAGE followed by WB. One experiment, representative of 3 independent experiments, is shown. Densitometric analysis of bands is shown. (E) hAEC cells were treated with vehicle or Sorafenib 10 μM, for 24 h. During the last hours of TMCB treatment, samples were treated with 1% CSE 4 h. Then samples were collected and subjected to SDS-PAGE followed by WB. One experiment, representative of 3 independent experiments, is shown. Densitometric analysis of bands is shown. (F) hAEC were transfected with siSCR or siMAPK15. After 72 h, cells were first treated for 4 h with 1% CSE, and then subjected to fractionation using the NE-PER fractionation kit and analyzed by WB. Cytosolic NRF2 was normalized with IKBα, while nuclear NRF2 was normalized with 53BP1. One experiment, representative of 3 independent experiments, is shown. Densitometric analysis of bands is shown. (G) hAEC cells were transfected with siSCR or siMAPK15, for 72 h. Next, samples were treated with 1% CSE and subjected to RT-qPCR to monitor mRNA expression of the indicated genes.
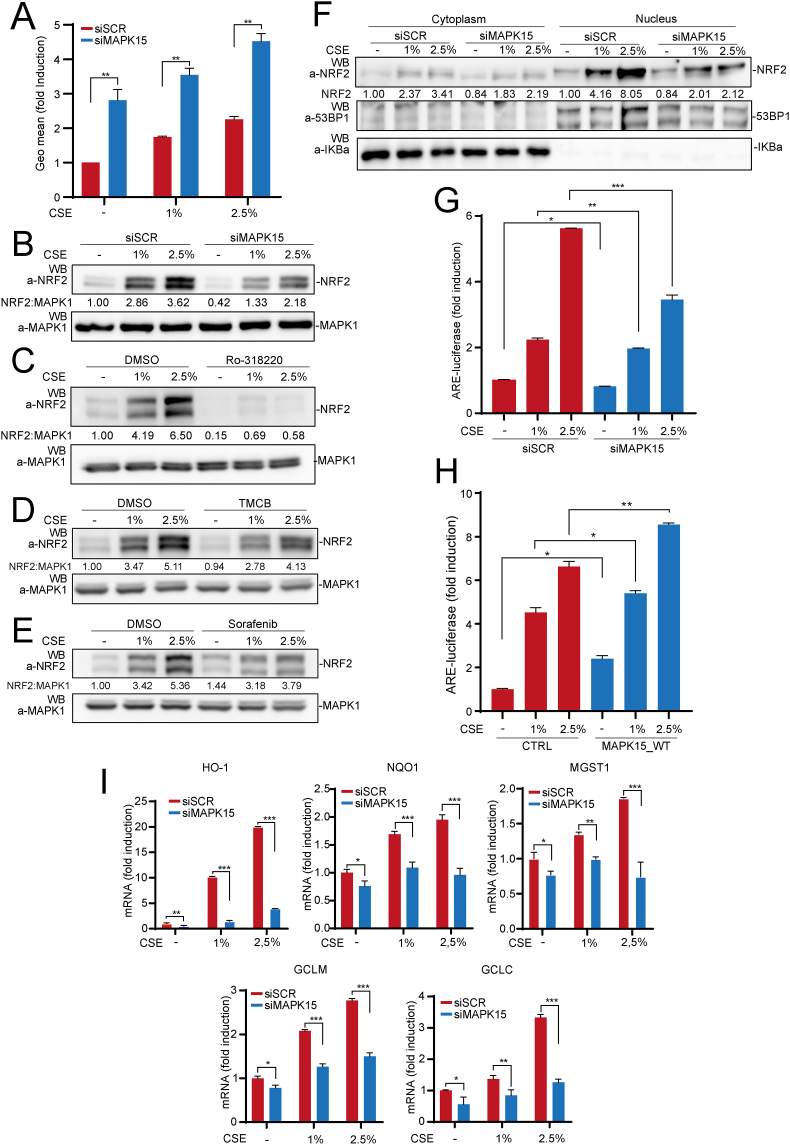
Analogously, upon treatment with CSE, MAPK15 was able to control NRF2-dependent antioxidant responses also in a lung cancer cell model. For this, we choose NCI–H358 cells, which express high levels of the MAPK15 protein, can be readily interfered by specific siRNA for MAPK15 (Fig. S1D) and show rapid dose-dependent increase in ROS levels, upon CSE stimulation (Fig. 7A, red bars). Conversely, we currently excluded from our analysis cells such as A549 and NCI–H460, since they constitutively express high levels of activated NRF2 protein independently of oxidative stimuli because of specific tumor driver DNA mutations/alterations [47]. In NCI–H358 cells, downregulation of MAPK15 strongly cooperated with CSE treatment to increase ROS levels (Fig. 7A) and reduce NRF2 protein levels (Fig. 7B), the latter effect being very efficiently phenocopied by using three different MAPK15 pharmacological inhibitors, Ro-318220 (Fig. 7C), TMCB (Fig. 7D) and Sorafenib (Fig. 7E). Importantly, the inhibitory effect of MAPK15 downregulation was particularly marked on the active, nuclear fraction of NRF2, when this was induced by CSE treatment (Fig. 7F). Indeed, we also directly confirmed inhibition of its transactivating function on the ARE-luciferase reporter gene, when stimulated with CSE (Fig. 7G). Accordingly, overexpression of the wild-type MAPK15 protein was able to cooperate with CSE to enhance its effects on NRF2 transactivation of the ARE reporter plasmid (Fig. 7H). Correspondingly, MAPK15 downregulation reduced the expression of several NRF2 target genes, in unstimulated NCI–H358 cells, while efficaciously preventing their increase triggered by oxidative stress induced by CSE (Fig. 7I), overall demonstrating the ability of lung cancer cells to take advantage of MAPK15 to stimulate NRF2 dependent antioxidant responses.
Fig. 7.
MAPK15 controls cytoprotective responses of NCI–H358 lung cancer cells to cigarette smoke. (A) GeoMFI bars of CM-H2DCFDA fluorescence from NCI–H358 cells transfected with siSCR or siMAPK15. After 72 h of transfection, samples were treated for 4 h with 1% or 2.5% CSE. Bars represent the average ± S. D of 3 independent experiments (n = 3). (B) NCI–H358 were transfected with siSCR or siMAPK15 and, after 72 h, were treated with 1% or 2.5% CSE for 4 h. Next, lysates were analyzed by WB for indicated proteins. One experiment, representative of 3 independent experiments is shown. Densitometric analysis of bands is indicated. (C) NCI–H358 cells were treated with vehicle or 2 μM Ro-318220 for 6 h. During the last 4 h of Ro-318220 treatment, samples were also treated with 1% or 2.5% CSE. Then samples were collected and analyzed by WB. One experiment, representative of 3 independent experiments, is shown. Densitometric analysis of bands is shown. (D) Same as in (C), but cells were treated with vehicle or TMCB 10 μM, for 24 h. (E) Same as in (C), but cells were treated with vehicle or 10 μM Sorafenib, for 24 h. (F) NCI–H358 were transfected with siSCR or siMAPK15. After 72 h, cells were first treated for 4 h with 1% or 2.5% CSE, and then subjected to fractionation using the NE-PER fractionation kit. Lysates were analyzed by WB. Cytosolic NRF2 were normalized with IKBα, while nuclear NRF2 was normalized with 53BP1. One experiment, representative of 3 independent experiments is shown. Densitometric analysis of bands is shown. (G) NCI–H358 cells transfected with siSCR or siMAPK15. After 48 h, cells were also transfected with the ARE luciferase reporter vector. Twenty-four hours after transfection, cells were treated with 1% or 2.5% CSE for 4 h. Then, samples were lysed, and the luciferase activity was measured in cell extracts. Data are represented as fold induction of the normalized luciferase activity compared to control cells transfected with GFP. All luciferase results represent the average ± S.D. of three independent experiments. All samples were measured in triplicate. (H) NCI–H358 cells were transfected with ARE luciferase reporter vector and empty vector or MAPK15_WT. Twenty-four hours after transfection, samples were lysed, and the luciferase activity was measured in cell extracts. Data are represented as fold induction of the normalized luciferase activity with respect to control cells transfected with GFP. All luciferase results represent the average ± S.D. of 3 independent experiments. All samples were read in triplicate. (I) NCI–H358 cells were transfected with siSCR or siMAPK15 for 72h. Next, samples were treated for 4 h with 1% or 2.5% CSE and subjected to RT-qPCR to monitor mRNA expression of the indicated genes.
4. Discussion
An imbalance between production and disposal of ROS, resulting in excessive formation of damaging oxidative species, has been postulated to play a role in several human diseases. Reducing oxidative stress, possibly by targeting NRF2 activity, therefore holds many promises with the aim of preventing or treating them [1,9]. Nonetheless, the negligible effect of small scavenging molecules in clinical trials and the fear for a cancer promoting potential of constitutive NRF2 activation strongly support research in this field to develop effective and safe antioxidant therapies [1]. In this context, our current demonstration that MAPK15 finely regulates the activation of NRF2 in normal and cancer cells in basal conditions and upon pro-oxidative stimuli may have important new implications in the way we plan to approach prevention and treatment of oxidative stress in human diseases. Indeed, MAPK15 can control multiple steps involved in preventing intracellular damages due to excessive ROS accumulation, making this kinase an almost ideal target for preventing physio-pathological consequences of oxidative stress, i.e., inflammation, aging and cancer. To date, MAPK15 has been indeed demonstrated to control mitochondrial fitness through the mitophagic process [20], to enhance NRF2-dependent responses, removing ROS once they are formed (this report) and, to directly participate to DNA damage response by affecting PCNA stability [15]. Based on these observations and on previous reports involving this kinase in COPD [21], we turned our attention to a potential role in controlling NRF2 function in the lung cells, which strongly depend on this protein to respond to oxidative stress, particularly when caused by cigarette smoke [48]. Indeed, we show that MAPK15 is effective in regulating NRF2 activation in response to cigarette smoke, both in normal and in cancer pulmonary cells, suggesting potential medical benefit from modulation of MAPK15 activity for a wide range of lung diseases. Still, while inhibition of MAPK15 activity would be desirable to reduce NRF2-dependent cancer cell progression and drug resistance, several chronic respiratory diseases would possibly benefit from MAPK15 activation, to improve NRF2-based antioxidant defenses of normal cells [1]. Therefore, each approach would come with a downside: MAPK15 inhibition in lung cancer patients could induce or accelerate aging-related disorders in normal tissues, while MAPK15 stimulation in patients with COPD or other chronic respiratory diseases could possibly favor tumor progression from mutated cells. Moreover, it should be considered that, while pharmacological inhibitors against MAPK15 are already available or could be easily optimized [49], no drug able to directly activate MAPK15 is currently known, making a genetic approach more feasible to stimulate this kinase and support antioxidant defenses. Nonetheless, while we have here clearly demonstrated the potential for MAPK15 inhibitors in repressing NRF2 basal and stress-induced activity, we have also shown that overexpression of the MAP kinase is sufficient to stimulate NRF2 activity and cytoprotective transcriptional response in lung cells, allowing optimism regarding the possibility of increasing protective cellular responses to pro-oxidative environmental insults such as cigarette smoke.
The activity of all known typical members of the MAP kinases family (ERKs, JNKs, p38s and ERK5) has been already associated to the regulation of NRF2 activity, although the specific roles and molecular mechanisms have still to be completely clarified, based on seemingly contradictory results observed using different stimuli and cell lines, as recently reviewed by Liu and coll [22]. Indeed, for example, p38s have been reported to both positively and negatively regulate NRF2 antioxidant activity, while ERKs and JNKs are more often suggested to positively regulate NRF2 activity [22]. On the other hand, when we comparatively inhibited most MAP kinase pathways, in our cellular settings, by a kinase dead mutant approach, we observed very strong reduction of CSE-induced NRF2 activity only upon inhibition of MAPK15, p38 and ERK5 signaling pathways, while inhibiting ERKs and JNKs had more limited or no effect, respectively (Fig. S7). While these data can be explained by context-dependent effects exerted by the different typical MAP kinases, due to the use of cells of different origin (mostly cancer cells) and of a variety of different stimuli, our current results regarding MAPK15 strongly support the ability of this kinase to stimulate NRF2 activity in multiple different cell types, both normal and tumoral. Accordingly, we also tested several different stimuli by using both ROS producing agents (H2O2, FCCP, CSE) and direct pharmacological NRF2 inducers (sulforaphane and dimethyl fumarate), and always obtained consistent results for MAPK15, overall suggesting a general role for this kinase in controlling NRF2-dependent antioxidant responses in different tissues and in response to multiple stimuli. Actually, MAPK15 functions have been mostly related to the ability of cells of responding to different cellular stresses, often represented by ROS-inducing stimuli [12,13,20], thereby suggesting a strong specialization of this kinase in the management of oxidative stress: consequently, the ability of MAPK15 to directly regulate NRF2 activity is surely advantageous to reach the final aim of counteracting the deleterious effects of multiple and very different physical and chemical oxidative stimuli. Importantly, while we have now demonstrated the ability of MAPK15 to directly phosphorylate NRF2 and increase its protein levels and nuclear translocation, the cascade of molecular events determining MAPK15-dependent activation of NRF2 functions will surely warrant further investigation. Particularly, accurate mapping of MAPK15-dependent phosphorylation sites in NRF2 will be necessary to improve our understanding of the molecular details regulating these mechanisms. In this regard, our mass spectrometry analysis already suggested three proline-directed threonine phosphorylation sites in NRF2, two of which, Thr395 and Thr439, have been already described as target of CDK5 [35], and demonstrated to control NRF2 translocation to the nucleus, suggesting that the same residues may be used by MAPK15 to regulate the same process. Accordingly, we show that MAPK15 indeed controls oxidative stress-induced nuclear translocation. At the same time, we here demonstrate that MAPK15 is also able to activate PKC and, through this kinase, induce phosphorylation of NRF2 in Serine 40 which, intriguingly, prevents its KEAP1-mediated proteasomal degradation but has no effect on its nuclear translocation and binding to an ARE sequence [22,37]. This, therefore, allows us to speculate that MAPK15 is able to induce NRF2 stabilization and its nuclear translocation by two independent mechanisms impinging, the former on PKC-dependent NRF2 phosphorylation on Ser40, and the latter on direct proline-directed phosphorylation of important NRF2 threonine residues (i.e., T395 and T439), overall inducing a strong NRF2-dependent antioxidant cellular response (Fig. S8). As we also observed that the overexpression of the MAPK15_KD mutant can increase NRF2 protein level but is completely unable to induce its activity, our hypothesis is that kinase independent MAPK15 mechanisms impinging on PKC activation may be involved in the control of NRF2 stability. Overall, this suggests that MAPK15 can independently control some of the existing multiple layers of NRF2 regulation [9], allowing finer and sharper modulation of the extent and accuracy of NRF2 responses to multiple stimuli or even determine a specific transcriptional program in different cell types. Ultimately, the identification of NRF2 as a novel MAPK15 effector will positively contribute to the understanding of the biology of this MAP kinase, to clarify a still superficial knowledge of its functions which anyhow point to an important place in the molecular mechanisms used by cells to manage oxidative stress and prevent the onset and progression of serious human diseases.
5. Concluding remarks
In conclusion, our study provides valuable information to comprehend the molecular mechanisms underlying key cytoprotective responses against ROS and suggests novel targeted approaches for managing pathologies in which oxidative stress acts as a primary cause of the disorder or as a secondary contributor to disease progression.
Funding
This work was supported by Next Generation EU [DM 1557 October 11, 2022], in the context of the National Recovery and Resilience Plan, Investment PE8 – Project Age-It: “Ageing Well in an Ageing Society”. The views and opinions expressed are only those of the authors and do not necessarily reflect those of the European Union or the European Commission. Neither the European Union nor the European Commission can be held responsible for them. This work was also supported by specific funding from Regione Toscana/Istituto per lo Studio, la Prevenzione e la Rete Oncologica (ISPRO).
Data availability
Data will be made available on request.
CRediT authorship contribution statement
Lorenzo Franci: Data curation, Formal analysis, Investigation, Writing – original draft. Giulia Vallini: Data curation, Investigation. Franca Maria Bertolino: Investigation. Vittoria Cicaloni: Data curation, Formal analysis. Giovanni Inzalaco: Investigation, Visualization. Mattia Cicogni: Investigation. Laura Tinti: Investigation. Laura Calabrese: Investigation. Virginia Barone: Investigation. Laura Salvini: Data curation, Supervision. Pietro Rubegni: Supervision. Federico Galvagni: Investigation, Supervision, Writing – review & editing. Mario Chiariello: Conceptualization, Funding acquisition, Writing – original draft, Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We thank Angela Strambi and David Colecchia for their critical reading of the manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2024.103131.
Contributor Information
Lorenzo Franci, Email: lorenzofranci4@gmail.com.
Giulia Vallini, Email: giuliavallini19@gmail.com.
Franca Maria Bertolino, Email: francamariabertolino@gmail.com.
Vittoria Cicaloni, Email: v.cicaloni@toscanalifesciences.org.
Giovanni Inzalaco, Email: giov.inzalaco@gmail.com.
Mattia Cicogni, Email: m.cicogni@toscanalifesciences.org.
Laura Tinti, Email: l.tinti@toscanalifesciences.org.
Laura Calabrese, Email: laura.calabrese@unisi.it.
Virginia Barone, Email: virginia.barone@unisi.it.
Laura Salvini, Email: l.salvini@toscanalifesciences.org.
Pietro Rubegni, Email: rubegni@unisi.it.
Federico Galvagni, Email: federico.galvagni@unisi.it.
Mario Chiariello, Email: mario.chiariello@cnr.it.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Forman H.J., Zhang H. Targeting oxidative stress in disease: promise and limitations of antioxidant therapy. Nat. Rev. Drug Discov. 2021;20:689–709. doi: 10.1038/s41573-021-00233-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chance B., Sies H., Boveris A. Hydroperoxide metabolism in mammalian organs. Physiol. Rev. 1979;59:527–605. doi: 10.1152/physrev.1979.59.3.527. [DOI] [PubMed] [Google Scholar]
- 3.Davalli P., Mitic T., Caporali A., Lauriola A., D'Arca, Ros D., Senescence Cell. Novel molecular mechanisms in aging and age-related diseases. Oxid. Med. Cell. Longev. 2016;2016 doi: 10.1155/2016/3565127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yagishita Y., Chartoumpekis D.V., Kensler T.W., Wakabayashi N. NRF2 and the moirai: Life and death decisions on cell fates. Antioxidants Redox Signal. 2023;38:684–708. doi: 10.1089/ars.2022.0200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thimmulappa R.K., Chattopadhyay I., Rajasekaran S. Oxidative stress mechanisms in the pathogenesis of environmental lung diseases. Oxidative Stress in Lung Diseases. 2020;2:103–137. doi: 10.1007/978-981-32-9366-3_5. [DOI] [Google Scholar]
- 6.Chakraborti S., Parinandi N.L., Ghosh R., Ganguly N.K., Chakraborti T. vol. 488. 2019. (Oxidative Stress in Lung Diseases). [Google Scholar]
- 7.Rogers L.K., Cismowski M.J. Oxidative stress in the lung - the essential paradox. Curr Opin Toxicol. 2018;7:37–43. doi: 10.1016/j.cotox.2017.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mizumura K., Maruoka S., Shimizu T., Gon Y. Role of Nrf2 in the pathogenesis of respiratory diseases. Respir Investig. 2020;58:28–35. doi: 10.1016/j.resinv.2019.10.003. [DOI] [PubMed] [Google Scholar]
- 9.Cuadrado A., Rojo A.I., Wells G., Hayes J.D., Cousin S.P., Rumsey W.L., Attucks O.C., Franklin S., Levonen A.L., Kensler T.W., Dinkova-Kostova A.T. Therapeutic targeting of the NRF2 and KEAP1 partnership in chronic diseases. Nat. Rev. Drug Discov. 2019;18:295–317. doi: 10.1038/s41573-018-0008-x. [DOI] [PubMed] [Google Scholar]
- 10.Hadwiger J.A., Aranda R.G., Fatima S. Atypical MAP kinases - new insights and directions from amoeba. J. Cell Sci. 2023;136 doi: 10.1242/jcs.261447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lau A.T.Y., Xu Y.M. Regulation of human mitogen-activated protein kinase 15 (extracellular signal-regulated kinase 7/8) and its functions: a recent update. J. Cell. Physiol. 2018;234:75–88. doi: 10.1002/jcp.27053. [DOI] [PubMed] [Google Scholar]
- 12.Klevernic I.V., Martin N.M., Cohen P. Regulation of the activity and expression of ERK8 by DNA damage. FEBS Lett. 2009;583:680–684. doi: 10.1016/j.febslet.2009.01.011. [DOI] [PubMed] [Google Scholar]
- 13.Li Z., Li N., Shen L., Fu J. Quantitative proteomic analysis identifies MAPK15 as a potential regulator of radioresistance in nasopharyngeal carcinoma cells. Front. Oncol. 2018;8:548. doi: 10.3389/fonc.2018.00548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cerone M.A., Burgess D.J., Naceur-Lombardelli C., Lord C.J., Ashworth A. High-throughput RNAi screening reveals novel regulators of telomerase. Cancer Res. 2011;71:3328–3340. doi: 10.1158/0008-5472.CAN-10-2734. [DOI] [PubMed] [Google Scholar]
- 15.Groehler A.L., Lannigan D.A. A chromatin-bound kinase, ERK8, protects genomic integrity by inhibiting HDM2-mediated degradation of the DNA clamp PCNA. J. Cell Biol. 2010;190:575–586. doi: 10.1083/jcb.201002124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Colecchia D., Strambi A., Sanzone S., Iavarone C., Rossi M., Dall'Armi C., Piccioni F., Verrotti di Pianella A., Chiariello M. MAPK15/ERK8 stimulates autophagy by interacting with LC3 and GABARAP proteins. Autophagy. 2012;8:1724–1740. doi: 10.4161/auto.21857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Colecchia D., Dapporto F., Tronnolone S., Salvini L., Chiariello M. MAPK15 is part of the ULK complex and controls its activity to regulate early phases of the autophagic process. J. Biol. Chem. 2018;293:15962–15976. doi: 10.1074/jbc.RA118.002527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Colecchia D., Rossi M., Sasdelli F., Sanzone S., Strambi A., Chiariello M. MAPK15 mediates BCR-ABL1-induced autophagy and regulates oncogene-dependent cell proliferation and tumor formation. Autophagy. 2015;11:1790–1802. doi: 10.1080/15548627.2015.1084454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rossi M., Colecchia D., Ilardi G., Acunzo M., Nigita G., Sasdelli F., Celetti A., Strambi A., Staibano S., Croce C.M., Chiariello M. MAPK15 upregulation promotes cell proliferation and prevents DNA damage in male germ cell tumors. Oncotarget. 2016;7:20981–20998. doi: 10.18632/oncotarget.8044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Franci L., Tubita A., Bertolino F.M., Palma A., Cannino G., Settembre C., Rasola A., Rovida E., Chiariello M. MAPK15 protects from oxidative stress-dependent cellular senescence by inducing the mitophagic process. Aging Cell. 2022;21 doi: 10.1111/acel.13620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang M., Fang L., Zhou L., Molino A., Valentino M.R., Yang S., Zhang J., Li Y., Roth M. MAPK15-ULK1 signaling regulates mitophagy of airway epithelial cell in chronic obstructive pulmonary disease. Free Radic. Biol. Med. 2021;172:541–549. doi: 10.1016/j.freeradbiomed.2021.07.004. [DOI] [PubMed] [Google Scholar]
- 22.Liu T., Lv Y.F., Zhao J.L., You Q.D., Jiang Z.Y. Regulation of Nrf2 by phosphorylation: consequences for biological function and therapeutic implications. Free Radic. Biol. Med. 2021;168:129–141. doi: 10.1016/j.freeradbiomed.2021.03.034. [DOI] [PubMed] [Google Scholar]
- 23.Iavarone C., Acunzo M., Carlomagno F., Catania A., Melillo R.M., Carlomagno S.M., Santoro M., Chiariello M. Activation of the Erk8 mitogen-activated protein (MAP) kinase by RET/PTC3, a constitutively active form of the RET proto-oncogene. J. Biol. Chem. 2006;281:10567–10576. doi: 10.1074/jbc.M513397200. [DOI] [PubMed] [Google Scholar]
- 24.Pietrobono S., Franci L., Imperatore F., Zanini C., Stecca B., Chiariello M. MAPK15 controls hedgehog signaling in medulloblastoma cells by regulating primary ciliogenesis. Cancers. 2021;13:4903. doi: 10.3390/cancers13194903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chiariello M., Marinissen M.J., Gutkind J.S. Multiple mitogen-activated protein kinase signaling pathways connect the cot oncoprotein to the c-jun promoter and to cellular transformation. Mol. Cell Biol. 2000;20:1747–1758. doi: 10.1128/MCB.20.5.1747-1758.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin Y., Zhou J., Bi D., Chen P., Wang X., Liang S. Sodium-deoxycholate-assisted tryptic digestion and identification of proteolytically resistant proteins. Anal. Biochem. 2008;377:259–266. doi: 10.1016/j.ab.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 27.Zhou J., Zhou T., Cao R., Liu Z., Shen J., Chen P., Wang X., Liang S. Evaluation of the application of sodium deoxycholate to proteomic analysis of rat hippocampal plasma membrane. J. Proteome Res. 2006;5:2547–2553. doi: 10.1021/pr060112a. [DOI] [PubMed] [Google Scholar]
- 28.Cox J., Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 29.Cox J., Neuhauser N., Michalski A., Scheltema R.A., Olsen J.V., Mann M. Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 2011;10:1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- 30.Sethi M.K., Downs M., Zaia J. Serial in-solution digestion protocol for mass spectrometry-based glycomics and proteomics analysis. Mol Omics. 2020;16:364–376. doi: 10.1039/d0mo00019a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suzuki T., Takahashi J., Yamamoto M. Molecular basis of the KEAP1-NRF2 signaling pathway. Mol. Cell. 2023;46:133–141. doi: 10.14348/molcells.2023.0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kwak M.K., Itoh K., Yamamoto M., Kensler T.W. Enhanced expression of the transcription factor Nrf2 by cancer chemopreventive agents: role of antioxidant response element-like sequences in the nrf2 promoter. Mol. Cell Biol. 2002;22:2883–2892. doi: 10.1128/MCB.22.9.2883-2892.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ngo V., Duennwald M.L. Nrf2 and oxidative stress: a general overview of mechanisms and implications in human disease. Antioxidants. 2022;11:2345. doi: 10.3390/antiox11122345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Coulombe P., Meloche S. Atypical mitogen-activated protein kinases: structure, regulation and functions. Biochim. Biophys. Acta. 2007;1773:1376–1387. doi: 10.1016/j.bbamcr.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 35.Jimenez-Blasco D., Santofimia-Castaño P., Gonzalez A., Almeida A., Bolaños J.P. Astrocyte NMDA receptors' activity sustains neuronal survival through a Cdk5-Nrf2 pathway. Cell Death Differ. 2015;22:1877–1889. doi: 10.1038/cdd.2015.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pagano M.A., Bain J., Kazimierczuk Z., Sarno S., Ruzzene M., Di Maira G., Elliott M., Orzeszko A., Cozza G., Meggio F., Pinna L.A. The selectivity of inhibitors of protein kinase CK2: an update. Biochem. J. 2008;415:353–365. doi: 10.1042/BJ20080309. [DOI] [PubMed] [Google Scholar]
- 37.Bloom D.A., Jaiswal A.K. Phosphorylation of Nrf2 at Ser40 by protein kinase C in response to antioxidants leads to the release of Nrf2 from INrf2, but is not required for Nrf2 stabilization/accumulation in the nucleus and transcriptional activation of antioxidant response element-mediated NAD(P)H:quinone oxidoreductase-1 gene expression. J. Biol. Chem. 2003;278:44675–44682. doi: 10.1074/jbc.M307633200. [DOI] [PubMed] [Google Scholar]
- 38.Huang H.C., Nguyen T., Pickett C.B. Phosphorylation of Nrf2 at Ser-40 by protein kinase C regulates antioxidant response element-mediated transcription. J. Biol. Chem. 2002;277:42769–42774. doi: 10.1074/jbc.M206911200. [DOI] [PubMed] [Google Scholar]
- 39.Konishi H., Yamauchi E., Taniguchi H., Yamamoto T., Matsuzaki H., Takemura Y., Ohmae K., Kikkawa U., Nishizuka Y. Phosphorylation sites of protein kinase C delta in H2O2-treated cells and its activation by tyrosine kinase in vitro. Proc. Natl. Acad. Sci. U. S. A. 2001;98:6587–6592. doi: 10.1073/pnas.111158798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Radaszkiewicz K.A., Beckerová D., Woloszczuková L., Radaszkiewicz T.W., Lesáková P., Blanářová O.V., Kubala L., Humpolíček P., Pachernik J. 12-O-Tetradecanoylphorbol-13-acetate increases cardiomyogenesis through PKC/ERK signaling. Sci. Rep. 2020;10 doi: 10.1038/s41598-020-73074-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Di Micco R., Krizhanovsky V., Baker D., d'Adda di Fagagna F. Cellular senescence in ageing: from mechanisms to therapeutic opportunities. Nat. Rev. Mol. Cell Biol. 2021;22:75–95. doi: 10.1038/s41580-020-00314-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liang K.J., Woodard K.T., Weaver M.A., Gaylor J.P., Weiss E.R., Samulski R.J. AAV-Nrf2 promotes protection and Recovery in animal models of oxidative stress. Mol. Ther. 2017;25:765–779. doi: 10.1016/j.ymthe.2016.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Labaki W.W., Han M.K. Chronic respiratory diseases: a global view. Lancet Respir. Med. 2020;8:531–533. doi: 10.1016/S2213-2600(20)30157-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Proctor R.N. Tobacco and the global lung cancer epidemic. Nat. Rev. Cancer. 2001;1:82–86. doi: 10.1038/35094091. [DOI] [PubMed] [Google Scholar]
- 45.Arunachalam K., Anand K., Palanisamy S., Anathy V. Editorial: oxidative stress related to cellular metabolism in lung health and diseases. Front. Pharmacol. 2022;13 doi: 10.3389/fphar.2022.1015423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Albino A.P., Huang X., Jorgensen E.D., Gietl D., Traganos F., Darzynkiewicz Z. Induction of DNA double-strand breaks in A549 and normal human pulmonary epithelial cells by cigarette smoke is mediated by free radicals. Int. J. Oncol. 2006;28:1491–1505. doi: 10.3892/ijo.28.6.1491. [DOI] [PubMed] [Google Scholar]
- 47.Taguchi K., Yamamoto M. The KEAP1-NRF2 system in cancer. Front. Oncol. 2017;7:85. doi: 10.3389/fonc.2017.00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rangasamy T., Cho C.Y., Thimmulappa R.K., Zhen L., Srisuma S.S., Kensler T.W., Yamamoto M., Petrache I., Tuder R.M., Biswal S. Genetic ablation of Nrf2 enhances susceptibility to cigarette smoke-induced emphysema in mice. J. Clin. Invest. 2004;114:1248–1259. doi: 10.1172/JCI21146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Strambi A., Mori M., Rossi M., Colecchia D., Manetti F., Carlomagno F., Botta M., Chiariello M. Structure prediction and validation of the ERK8 kinase domain. PLoS One. 2013;8 doi: 10.1371/journal.pone.0052011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.