Abstract
Human immunodeficiency virus type 1 Rev export depends upon the presence of the nuclear export signal (NES), a leucine-rich stretch of hydrophobic amino acids. Recently, the nuclear NES-binding receptor has been identified as CRM1 or exportin 1. Rev export has been shown to be CRM1 dependent. The function of the atypical NES-containing Rev-like proteins of equine infectious anemia virus (EIAV) and feline immunodeficiency virus (FIV) is inhibited by leptomycin B, a drug that specifically blocks NES-CRM1 interactions. These data suggest that the function of atypical NES-containing proteins is CRM1 dependent. In contrast to the inhibition of EIAV Rev and FIV Rev, the cytoplasmic accumulation of hepatitis B virus (HBV) posttranscriptional regulatory element (PRE)-containing and Mason-Pfizer monkey virus (MPMV) constitutive transport element (CTE)-containing RNAs is not inhibited by leptomycin B treatment. We conclude that the HBV PRE, like the MPMV CTE, functions independently of an NES receptor-exportin 1 interaction.
Human immunodeficiency virus type 1 (HIV-1) Rev and human T-cell leukemia virus type 1 Rex bind their respective RNA response elements, the Rev response element (RRE) and the Rex response element, and subsequently export singly spliced and genomic viral RNA to the cytoplasm. Rev and Rex export depends upon the presence of one class of nuclear export signals (NES) that contain hydrophobic amino acids, often leucines, and a core tetramer (reviewed in reference 16). Rev export is abolished if the NES is deleted or modified, but is restored if sequences encoding functional NES from other retroviruses or cellular proteins are provided (10–12, 17, 23, 38). The equine infectious anemia virus Rev-like protein (ERev) and the feline immunodeficiency virus (FIV) Rev-like protein (FIV Rev) possess atypical NES that differ significantly from those in Rev and Rex in the organization of the hydrophobic residues and lack a core tetramer (Fig. 1) (12, 23). However, ERev and FIV Rev NES are functionally interchangeable with the Rev NES (12, 23). Furthermore, microinjection studies have shown that the Rev NES can compete with the ERev NES (26). Hence, Rev, ERev, and FIV Rev are believed to travel the same NES-dependent export pathway.
FIG. 1.
Sequence alignment of typical and atypical NES. The core tetramer of typical NES is in boldface and bracketed. Leucines and chemically related amino acids involved in NES function are underlined.
Screens for low-molecular-weight inhibitors of Rev nuclear export identified leptomycin B (LMB) as an antibiotic which blocked this process (41). LMB played an important role in identifying the nuclear NES-binding receptor. LMB resistance in Schizosaccharomyces pombe was mapped to mutations within the protein CRM1 (chromosomal region maintenance) (14, 27). CRM1 has recently been identified as the nuclear NES-binding receptor (8, 9) or exportin 1 (7, 13, 28, 33). LMB has been shown to block Rev export (41) by binding exportin 1, thereby preventing the NES-exportin 1-RanGTP complex from assembling (7). LMB specifically blocks NES-dependent RNA export but does not block the mRNA or tRNA export pathways (7). The NES-dependent export of other proteins is also inhibited by LMB (13, 28, 35). Hence, the export of NES-containing proteins, such as Rev, is considered to be exportin 1 dependent and subsequently LMB sensitive.
The cytoplasmic accumulation of hepatitis B virus (HBV) surface RNA is facilitated by a cis-acting RNA element termed the posttranscriptional regulatory element (PRE) (19, 20). The PRE has been postulated to be an export element. Unlike the HIV-1 RRE, the HBV PRE functions independently of any virally encoded proteins. The PRE has been shown to induce the cytoplasmic accumulation of HIV-1-related RNAs from reporter vector pDM138, similar to Rev (3, 21). Using the same HIV-1-derived vector system, Roth and Dobbelstein recently reported that PRE-mediated activation of pDM138 can be inhibited by NES-containing proteins and concluded that the HBV PRE utilizes the Rev export pathway (31). These data suggest that cellular NES-containing proteins mediate HBV PRE function, exporting PRE-containing RNA through the exportin 1-dependent pathway.
The simian type D retroviruses, Mason-Pfizer monkey virus (MPMV) and simian retrovirus types 1 and 2 contain cis-acting RNA export elements termed constitutive transport elements (CTEs) that induce nuclear export of viral unspliced mRNA (2, 5, 36, 42). The CTEs from all three simian viruses are very similar in structure and function (4, 36). The CTE can rescue Rev-deficient HIV-1 replication (36, 42). Additionally, CTE-containing intron lariats are exported when CTE is placed within the introns of pre-mRNAs injected into Xenopus oocyte nuclei (29, 32). CTE-containing RNA export is not inhibited by NES-bovine serum albumin conjugates but can instead compete for the mRNA export pathway (29, 32). In this way, the CTE has been demonstrated to be an export element that proceeds through an mRNA pathway and not the Rev pathway. Cellular factors bind the CTE and facilitate export in the absence of any virally encoded proteins.
We hypothesized that although ERev and FIV Rev contain atypical NES, their function requires CRM1 and would be inhibited by LMB. Additionally, if PRE function is mediated by CRM1, then the cytoplasmic accumulation of PRE-containing RNA should also be inhibited by LMB. Conversely, LMB should not affect CTE-mediated export, since the CTE does not export RNA through the CRM1-dependent Rev pathway in oocytes. We therefore used the MPMV CTE as a negative control. We tested the effect LMB had on ERev and FIV Rev function, as well as LMB effects on the cytoplasmic accumulation of PRE and CTE-containing RNA.
We employed a well-characterized chloramphenicol acetyltransferase (CAT) reporter system used to study the RNA export elements of complex retroviruses (17, 18, 23). The system utilizes pDM138, a plasmid reporter derived from the second intron of HIV-1 in which the HIV env gene is replaced by the CAT gene (18). When pDM138 is transiently transfected, the CAT gene is translated only if unspliced RNA is exported to the cytoplasm. Export of unspliced RRE-containing RNA depends on the presence of Rev. This laboratory and others have demonstrated that the PRE and CTE can induce the cytoplasmic accumulation of unspliced pDM138 reporter RNAs (3, 37).
To test whether LMB can inhibit FIV Rev NES function, 293 cells were transiently transfected with an RRE-containing plasmid (pDM128) (18), pDM128 and pRSVRev (18), or pDM128 and Δ3FIV (23). Δ3FIV is a fusion protein composed of NES-deleted Rev (Δ3) fused to FIV Rev and has been shown to export RRE-containing RNA (23). In a similar experiment, 293 cells were transiently transfected with pDM138ERRE-1, an ERev response element derivative (1), with or without an ERev expression vector (24, 34). β-Galactosidase expression vector pCH110 (Pharmacia) was included for transfection normalization purposes. Calcium phosphate transfections were done as previously described (3) except that the precipitations were divided in half and added to cells that either would or would not be treated with LMB. This was done to achieve similar transfection efficiencies for both conditions. After 18 h the medium on the cells was replaced with fresh medium containing 5 nM LMB or with medium alone. The cells were harvested 24 h later in reporter lysis buffer (Promega), cellular debris was pelleted, lysates were normalized for transfection efficiency with β-galactosidase, and normalized amounts of lysate were used for CAT assays as previously described (18). CAT assay results were quantitated on a Molecular Dynamics phosphorImager by using ImageQuaNT software. All experimental points were assayed in triplicate, and experiments were repeated at least twice. Representative experimental results are presented.
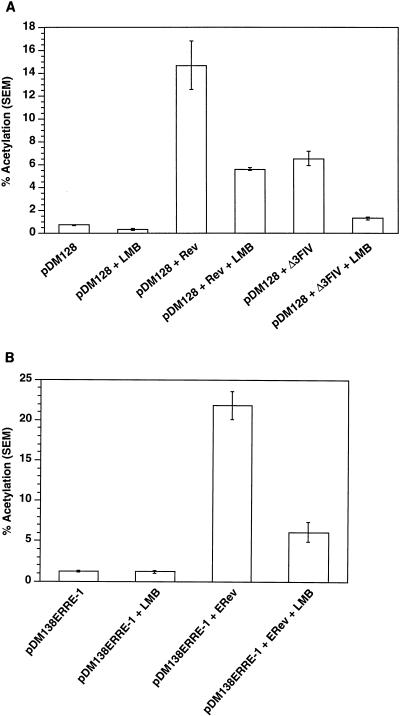
Figure 2A shows that LMB inhibited Rev-mediated CAT activity by 50% and Δ3FIV-mediated CAT activity by 80%. In addition, ERev-mediated CAT activity was inhibited by 72% in the presence of LMB (Fig. 2B). These results suggest that the function of atypical NES, like that of the typical Rev NES, is sensitive to LMB and therefore exportin 1 dependent.
FIG. 2.
Effect of LMB on the function of Rev and other Rev-like proteins in the CAT assay. (A) pDM128 (RRE-containing RNA) was transfected with pRSVRev or Δ3FIV or with neither in the absence or presence of LMB. LMB inhibits Rev- and Δ3FIV-mediated CAT activities of RRE-containing RNA by 50 and 80%, respectively. (B) pDM138ERRE-1-containing RNA either was or was not transfected with pRS-ERev in the presence and absence of LMB. LMB inhibits ERev-mediated CAT activity by 72%.
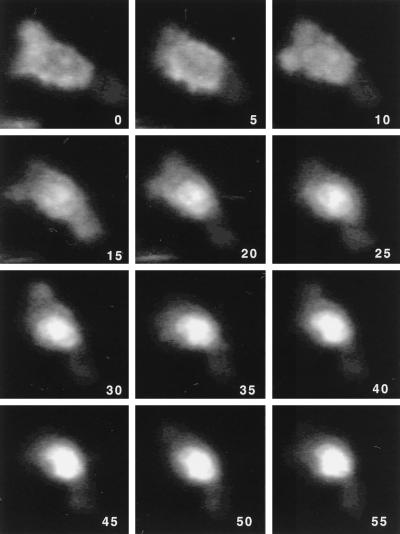
Treatment of Rev-expressing cells with actinomycin D has been shown to alter the subcellular localization of Rev from the nucleus to the cytoplasm (25, 41). Under these conditions, Rev shuttling is disrupted, leaving the export of Rev continuous while import is blocked. Conversely, blocking the export of a shuttling protein whose steady-state localization is cytoplasmic would shift the localization of the protein into the nucleus. We tested this hypothesis with LMB and an ERev/enhanced green fluorescence protein (eGFP) fusion protein (eGFP/ERev), which is cytoplasmic at high levels of expression but which functions like wild-type ERev in the CAT assay and presumably shuttles (15). 3T3 cells were plated onto coverslips in six-well plates and transiently transfected with 0.5 μg of eGFP/ERev DNA by using the polycation Superfect (Qiagen) according to the manufacturer’s protocol. Eighteen hours posttransfection, the medium was replaced with medium containing 5 nM LMB. The subcellular localization of the eGFP/ERev protein was then monitored under a fluorescence microscope (Nikon Diaphot 300) for several hours. As seen in Fig. 3, at equilibrium (time zero) the eGFP/ERev fusion protein is distributed throughout the cell with a significant amount in the cytoplasm. However, as time progresses during LMB treatment, the presence of the fusion protein in the cytoplasm diminishes, while a commensurate increase in nuclear localization is observed. After nearly 1 h of LMB treatment, the eGFP/ERev fusion protein is almost exclusively nuclear. This observation directly demonstrates that ERev export is perturbed by LMB treatment. Similar results have been reported for MAPKK, cyclin B1, actin, and c-abl (13, 35, 39, 40). The observed shift in subcellular localization of the eGFP/ERev fusion is due to import of the protein into the nucleus and subsequent inhibition of its export by LMB. The LMB-dependent disruption of eGFP/ERev shuttling correlates with the LMB sensitivity of ERev in the CAT assay (Fig. 2B) and further supports the conclusion that atypical NES-containing proteins function via a CRM1-dependent pathway.
FIG. 3.
LMB inhibition of eGFP/ERev shuttling visualized by fluorescence microscopy. An eGFP/ERev fusion protein construct was transfected into 3T3 cells. Eighteen hours posttransfection the medium was replaced with medium containing 5 nM LMB and the cells were placed under the fluorescence microscope. eGFP fluorescence was recorded over several hours. The time, in minutes post-LMB addition, is shown in the lower right corner of each image. At 30 min a noticeable difference in subcellular eGFP/ERev localization is apparent. By 55 min the localization of the eGFP/ERev protein is almost completely nuclear.
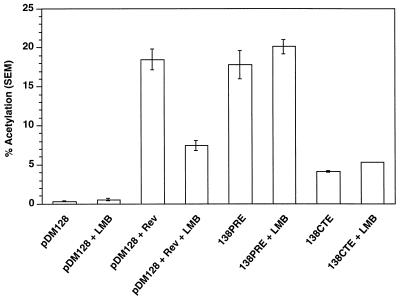
To test if the cytoplasmic accumulation of PRE- and CTE-containing RNAs is LMB sensitive, 293 cells were transiently transfected with pDM128 and pRSVRev, 138PRE (PRE-containing RNA [3]), and 138CTE (MPMV CTE-containing RNA [37]). Transfection, LMB treatment, normalization, and CAT assays were identical to those of the previous experiment. Figure 4 shows that while Rev-mediated CAT activity is inhibited 60% by LMB, the cytoplasmic accumulation of PRE- and CTE-containing RNA is not affected. These data suggest that PRE and CTE function does not require CRM1. Furthermore, the data imply that PRE- and CTE-binding proteins do not contain NES. However, at this point we can only conclude that PRE and CTE function is not mediated by the concerted efforts of NES-containing proteins and CRM1. Since CRM1 is reportedly not involved in the export of mRNA (7) and since the CTE has been shown to export RNA via the mRNA pathway (32), the CRM1 independence of CTE-induced RNA export is expected. Nevertheless, these data are added evidence that CTE-containing RNA does not travel through an NES-saturable pathway.
FIG. 4.
Effect of LMB on the function of the PRE and CTE in the CAT assay. pDM128 (RRE-containing RNA) either was or was not transfected with pRSVRev in the presence and absence of LMB; 138PRE (PRE-containing RNA) and 138CTE (CTE-containing RNA) were transfected in the presence and absence of LMB. LMB inhibits Rev export of RRE-containing RNA by 60%. LMB does not affect the cytoplasmic accumulation of PRE- or CTE-containing RNAs.
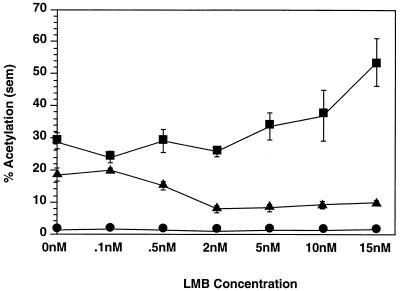
Wolff et al. have shown that LMB inhibits Rev-dependent but not Rev-independent gene expression (41). We also observed that LMB inhibits Rev-dependent gene expression but not PRE-mediated gene expression (Fig. 4). To rule out the possibility that the difference in LMB sensitivity between RRE-containing RNA and PRE-containing RNA was dependent on the concentration of LMB we used (5 nM), we performed transient transfections and CAT assays on pDM138 (empty vector), pDM128 and pRSVRev, and 138PRE in which the LMB concentration was titrated over a broad range. The dose response of RNA expression to LMB is depicted in Fig. 5. As can be seen in Fig. 5, while there is maximal inhibition of Rev-dependent gene expression at 2 nM concentrations of LMB, there is no decrease in PRE-mediated gene expression. In fact, PRE-mediated gene expression increased at higher concentrations of LMB. There was no effect on pDM138 CAT expression over the range of LMB concentration. This difference in dose responsiveness to LMB between Rev-dependent gene expression and PRE-mediated gene expression further supports the conclusion that PRE-containing RNA is not dependent upon an NES-exportin 1 interaction in transit to the cytoplasm. Additionally, the increase in expression of PRE-containing RNA at higher concentrations of LMB suggests that the PRE does not associate with any exportin 1-related proteins less sensitive to LMB.
FIG. 5.
Dose response curve of LMB effects on Rev-dependent and PRE-mediated gene expression in the CAT assay. pDM128 (RRE-containing RNA) and pRSVRev (▴), 138PRE (PRE-containing RNA) (■), and pDM138 (empty vector) (•) were transfected in the presence of increasing concentrations of LMB. Rev-dependent CAT expression decreased until it was maximally inhibited at 2 nM LMB. PRE-mediated CAT expression was not affected between 0 and 2 nM LMB and increased between 2 and 15 nM LMB. pDM138 CAT expression was not affected by LMB.
To confirm the CAT assay results, we directly analyzed RNA by Northern blot analysis. For the Northern blotting, 293 cells were transiently transfected with 138PRE, pDM138, and pDM128 plus pRSVRev. Two DNA-CaPO4 precipitates were prepared for each reporter and used to transfect two plates (10 cm in diameter) each, which then either were or were not treated with LMB as in previous experiments. This was done to achieve similar transfection efficiencies for both conditions. The cells were harvested and lysed with 0.5% Nonidet P-40 (NP-40), and cytoplasmic RNA was isolated with RNA STAT (Tel-Test). Nuclei were washed several times in 0.5% NP-40–20% glycerol, and nuclear RNA was isolated with RNA STAT. Then, 5 μg of nuclear RNA and 10 μg of cytoplasmic RNA from each transfection were run on a 1% agarose–formaldehyde gel and transferred to a nylon membrane (Stratagene). The Northern blot was prehybridized in QuikHyb (Stratagene) for 1 h at 65°C and then hybridized with 32P-labeled CAT and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) probes for 2 h at 65°C. The blot was washed in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) at room temperature and then in 0.1× SSC at 65°C and exposed to X-ray film, and an autoradiograph was obtained.
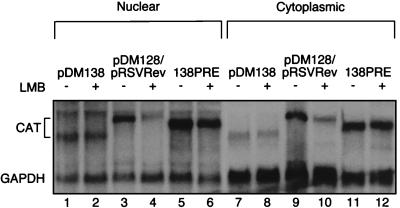
The Northern blot analysis shown in Fig. 6 confirms the CAT assay results in Fig. 4 and 5. The cytoplasmic accumulation of RRE-containing RNA is inhibited in the presence of LMB (compare lanes 9 and 10), while the cytoplasmic accumulation of PRE-containing RNA is not (compare lanes 11 and 12). LMB has no effect on the nuclear accumulation of pDM138 or PRE-containing RNA. Hence, LMB does not appear to affect the levels of RNA substrate available for export. Interestingly, in the presence of LMB, the amount of RRE-containing RNA in the nucleus is diminished (compare lanes 3 and 4). This suggests that the disruption of CRM1 function causes unspliced RRE-containing RNA to be unstable in the nucleus, even in the presence of Rev. In this situation, the unspliced RNA is presumably being spliced or degraded. This observation is consistent with the report that Rev increases the stability of nuclear HIV RNA (22). The ability of Rev to stabilize nuclear RRE-containing RNAs led Malim and Cullen to suggest a model in which RNA export and stability may be closely linked. Consistent with this model, our observation of an LMB-dependent decrease in pDM138RRE nuclear RNA in the presence of Rev suggests that Rev binding alone is not sufficient for the stabilization of nuclear HIV RNA. Alternatively, LMB may lower the amount of RRE-containing RNA in the nucleus available for export, which would reduce the amount observed in the cytoplasm. We do not favor this interpretation, however, since LMB has no such effect on pDM138 or PRE-containing RNA. LMB has no effect on pDM138 CAT or GAPDH expression in either compartment. This RNA analysis is further evidence that LMB affects exportin 1-dependent and, therefore, Rev-dependent gene expression but not PRE-mediated or cellular mRNA (GAPDH) gene expression. On the basis of these results we conclude that the HBV PRE and Rev do not travel through the same export pathway, contrary to the conclusion reached by Roth and Dobbelstein (31).
FIG. 6.
Northern blot analysis of nuclear and cytoplasmic RRE- and PRE-containing RNA in the presence or absence of LMB. Nuclear (lanes 1 to 6) and cytoplasmic (lanes 7 to 12) RNA of pDM138 (empty vector), pDM128 (RRE-containing RNA) and pRSVRev, and 138PRE (PRE-containing RNA) was transfected in the presence or absence of LMB. LMB inhibited Rev-induced cytoplasmic accumulation of RRE-containing RNA (compare lanes 9 and 10). Cytoplasmic accumulation of PRE-containing RNA was not affected by LMB (compare lanes 11 and 12). LMB had no effect on pDM138 CAT expression.
We have demonstrated here that like that of the typical NES of Rev, the function of atypical NES from the Rev-like proteins of other complex retroviruses is susceptible to inhibition by LMB and is therefore CRM1 dependent. Alternatively, proteins that contain atypical NES may bind a different NES receptor(s) related to exportin 1, which may also be sensitive to LMB, and export through other pathways. A recent report suggests that the export of NES-containing proteins may be through the mRNA pathway as well as the Rev pathway (6, 30). This suggests that exportin 1 might be a factor common to both pathways or that other exportin 1-like proteins function in pathways other than that used by Rev. However, our observation that LMB has no effect on the expression of cellular mRNAs for GAPDH (Fig. 6) and histone 2B (data not shown) argues against this interpretation.
We have demonstrated that the cytoplasmic accumulation of HBV PRE-containing and MPMV CTE-containing RNA is not inhibited by LMB treatment and therefore is independent of an NES-CRM1 interaction. The fact that PRE and CTE function is not affected by LMB suggests that the PRE may utilize the mRNA pathway as does the CTE. However, we have observed an important functional difference between the two cis-acting elements. The CTE has been reported to rescue Rev-deficient HIV-1 replication (36, 42). While we also find that the CTE is able to rescue Rev-deficient HIV-1 replication, in our hands the PRE cannot rescue Rev-deficient HIV-1 replication in an analogous manner (data not shown). This implies that there is a fundamental difference between the CTE and PRE. Further experiments are necessary to better delineate the function of the PRE and, if it is an export element, to determine which pathway it utilizes.
Acknowledgments
This work was supported by National Institutes of Health grant AI35477 and Arthur and Larry Kramer (T.J.H.); a Ford Foundation Postdoctoral Fellowship (G.C.O.); National Cancer Institute training grant T32 CA64041 (J.E.D.); a National Science Foundation Graduate Research Fellowship (M.E.H.); and the James B. Pendleton Trust Microscopy Facility.
We thank Barbara Wolff for LMB and Allison Bocksruker for help in preparing the manuscript.
REFERENCES
- 1.Belshan M, Harris M E, Shoemaker A E, Hope T J, Carpenter S. Biological characterization of Rev variation in equine infectious anemia virus. J Virol. 1998;72:4421–4426. doi: 10.1128/jvi.72.5.4421-4426.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bray M, Prasad S, Dubay J W, Hunter E, Jeang K-T, Rekosh D, Hammarskjöd M-L. A small element from mason-pfizer monkey virus genome makes human immunodeficiency virus type 1 expression and replication Rev-independent. Proc Natl Acad Sci USA. 1994;91:1256–1260. doi: 10.1073/pnas.91.4.1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Donello J, Smith G J, Beeche A A, Lucero G, Hope T J. The hepatitis B virus posttranscriptional regulatory element is composed of two subelements. J Virol. 1996;70:4345–4351. doi: 10.1128/jvi.70.7.4345-4351.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ernst R K, Bray M, Rekosh D, Hammarskjöld M-L. Secondary structure and mutational analysis of the Mason-Pfizer monkey virus RNA constitutive transport element. RNA. 1997;3:210–222. [PMC free article] [PubMed] [Google Scholar]
- 5.Ernst R K, Bray M, Rekosh D, Hammarskjöld M-L. A structured retroviral RNA element that mediates nucleocytoplasmic export of intron-containing RNA. Mol Cell Biol. 1997;17:135–144. doi: 10.1128/mcb.17.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fischer U, Huber J, Boelens W C, Mattaj I W, Luhrmann R. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell. 1995;82:475–483. doi: 10.1016/0092-8674(95)90436-0. [DOI] [PubMed] [Google Scholar]
- 7.Fornerod M, Ohno M, Yoshida M, Mattaj I W. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. . (Comments.) [DOI] [PubMed] [Google Scholar]
- 8.Fornerod M, van Baal S, Valentine V, Shapiro D N, Grosveld G. Chromosomal localization of genes encoding CAN/Nup214-interacting proteins—human CRM1 localizes to 2p16, whereas Nup88 localizes to 17p13 and is physically linked to SF2p32. Genomics. 1997;42:538–540. doi: 10.1006/geno.1997.4767. [DOI] [PubMed] [Google Scholar]
- 9.Fornerod M, van Deursen J, van Baal S, Reynolds A, Davis D, Murti K G, Fransen J, Grosveld G. The human homologue of yeast CRM1 is in a dynamic subcomplex with CAN/Nup214 and a novel nuclear pore component Nup88. EMBO J. 1997;16:807–816. doi: 10.1093/emboj/16.4.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fridell R A, Benson R E, Hua J, Bogerd H P, Cullen B R. A nuclear role for the Fragile X mental retardation protein. EMBO J. 1996;15:5408–5414. [PMC free article] [PubMed] [Google Scholar]
- 11.Fridell R A, Bogerd H P, Cullen B R. Nuclear export of late HIV-1 mRNAs occurs via a cellular protein export pathway. Proc Natl Acad Sci USA. 1996;93:4421–4424. doi: 10.1073/pnas.93.9.4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fridell R A, Partin K M, Carpenter S, Cullen B R. Identification of the activation domain of equine infectious anemia virus Rev. J Virol. 1993;67:7317–7323. doi: 10.1128/jvi.67.12.7317-7323.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fukuda M, Eisuke N, et al. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature. 1997;390:308. doi: 10.1038/36894. [DOI] [PubMed] [Google Scholar]
- 14.Hamamoto T, Gunji S, Tsuji H, Beppu T. Leptomycins A and B, new antifungal antibiotics. I. Taxonomy of the producing strains and their fermentation, purification, and characterization. J Antibiot. 1983;36:639–645. doi: 10.7164/antibiotics.36.639. [DOI] [PubMed] [Google Scholar]
- 15.Harris M E, Gontarek R R, Derse D, Hope T J. Differential requirements for alternative splicing and nuclear export functions of equine infectious anemia virus Rev protein. Mol Cell Biol. 1998;18:3889–3899. doi: 10.1128/mcb.18.7.3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hope T J. Viral RNA export. Chem Biol. 1997;4:335–344. doi: 10.1016/s1074-5521(97)90124-1. [DOI] [PubMed] [Google Scholar]
- 17.Hope T J, Bond B L, McDonald D, Klein N P, Parslow T G. Effector domains of human immunodeficiency virus type 1 Rev and human T-cell leukemia virus type 1 Rex are functionally interchangeable and share an essential peptide motif. J Virol. 1991;65:6001–6007. doi: 10.1128/jvi.65.11.6001-6007.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hope T J, Huang X, McDonald D, Parslow T. Steroid-receptor fusion of the human immunodeficiency virus type 1 Rev transactivator: mapping cryptic functions of the arginine-rich motif. Proc Natl Acad Sci USA. 1990;87:7787–7791. doi: 10.1073/pnas.87.19.7787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang J, Liang T J. A novel hepatitis B virus (HBV) genetic element with Rev response element-like properties that is essential for expression of HBV gene products. Mol Cell Biol. 1993;13:7476–7486. doi: 10.1128/mcb.13.12.7476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang Z-M, Yen T S B. Hepatitis B virus RNA element that facilitates accumulation of surface gene transcripts in the cytoplasm. J Virol. 1994;68:3193–3199. doi: 10.1128/jvi.68.5.3193-3199.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang Z M, Zang W Q, Yen T S. Cellular proteins that bind to the hepatitis B virus posttranscriptional regulatory element. Virology. 1996;217:573–581. doi: 10.1006/viro.1996.0152. [DOI] [PubMed] [Google Scholar]
- 22.Malim M H, Cullen B R. Rev and the fate of pre-mRNA in the nucleus: implications for the regulation of RNA processing in eukaryotes. Mol Cell Biol. 1993;13:6180–6189. doi: 10.1128/mcb.13.10.6180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mancuso V A, Hope T J, Zhu L, Derse D, Phillips T, Parslow T G. Posttranscriptional effector domains in the Rev proteins of feline immunodeficiency virus and equine infectious anemia virus. J Virol. 1994;68:1998–2001. doi: 10.1128/jvi.68.3.1998-2001.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martarano L, Stephens R, Rice N, Derse D. Equine infectious anemia virus trans-regulatory protein Rev controls viral mRNA stability, accumulation, and alternative splicing. J Virol. 1994;68:3102–3111. doi: 10.1128/jvi.68.5.3102-3111.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meyer B E, Malim M H. The HIV-1 Rev trans-activator shuttles between the nucleus and the cytoplasm. Genes Dev. 1994;8:1538–1547. doi: 10.1101/gad.8.13.1538. [DOI] [PubMed] [Google Scholar]
- 26.Meyer B E, Meinkoth J L, Malim M H. Nuclear transport of human immunodeficiency virus type 1, visna virus, and equine infectious anemia virus Rev proteins: identification of a family of transferable nuclear export signals. J Virol. 1996;70:2350–2359. doi: 10.1128/jvi.70.4.2350-2359.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nishi K, Yoshida M, Fujiwara D, Nishikawa M, Horinouchi S, Beppu T. Leptomycin B targets a regulatory cascade of CRM1, a fission yeast nuclear protein, involved in control of higher order chromosome structure and gene expression. J Biol Chem. 1994;269:6320–6324. [PubMed] [Google Scholar]
- 28.Ossareh N B, Bachelerie F, Dargemont C. Evidence for a role of CRM1 in signal-mediated nuclear protein export. Science. 1997;278:141–144. doi: 10.1126/science.278.5335.141. [DOI] [PubMed] [Google Scholar]
- 29.Pasquinelli A, Ernst R K, Lund E, Grimm C, Zapp M L, Rekosh D, Hammarskjöld M-L, Dahlberg J E. The constitutive transport element (CTE) of Mason-Pfizer monkey virus (MPMV) accesses a cellular mRNA export pathway. EMBO J. 1997;16:7500–7510. doi: 10.1093/emboj/16.24.7500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pasquinelli A E, Powers M A, Lund E, Forbes D, Dahlberg J E. Inhibition of mRNA export in vertebrate cells by nuclear export signal conjugates. Proc Natl Acad Sci USA. 1997;94:14394–14399. doi: 10.1073/pnas.94.26.14394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roth J, Dobbelstein M. Export of hepatitis B virus RNA on a Rev-like pathway: inhibition by the regenerating live inhibitory factor IkBa. J Virol. 1997;71:8933–8939. doi: 10.1128/jvi.71.11.8933-8939.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saavedra C, Felber B, Izaurralde E. The simian retrovirus-1 CTE unlike the HIV-1 RRE, utilises factors required for the export of cellular mRNAs. Curr Biol. 1997;7:619–628. doi: 10.1016/s0960-9822(06)00288-0. [DOI] [PubMed] [Google Scholar]
- 33.Stade K, Ford C S, Guthrie C, Weis K. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- 34.Stephens R M, Derse D, Rice N R. Cloning and characterization of cDNAs encoding equine infectious anemia virus Tat and putative Rev proteins. J Virol. 1990;64:3716–3725. doi: 10.1128/jvi.64.8.3716-3725.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taagepera S, McDonald D, Loeb J E, Whitaker L L, McElroy A, Wang J Y J, Hope T J. Nuclear-cytoplasmic shuttling of c-abl tyrosine kinase. Proc Natl Acad Sci USA, 1998;95:7457–7462. doi: 10.1073/pnas.95.13.7457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tabernero C, Zolotukhin A S, Valentin A, Pavlakis G N, Felber B K. The posttranscriptional control element of the simian retrovirus type 1 forms an extensive RNA secondary structure necessary for its function. J Virol. 1996;70:5998–6011. doi: 10.1128/jvi.70.9.5998-6011.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tang H, Gaietta G M, Fischer W H, Ellisman M H, Wong-Staal F. A cellular cofactor for the constitutive transport element of type D retrovirus. Science. 1997;276:1412–1415. doi: 10.1126/science.276.5317.1412. [DOI] [PubMed] [Google Scholar]
- 38.Tiley L S, Malim M H, Cullen B R. Conserved functional organization of the human immunodeficiency virus type 1 and visna virus Rev proteins. J Virol. 1991;65:3877–3881. doi: 10.1128/jvi.65.7.3877-3881.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Toyoshima F, Moriguchi T, Wada A, Fukuda M, Nishida E. Nuclear export of cyclin B1 and its possible role in the DNA damage-induced G2 checkpoint. EMBO J. 1998;17:2728–2735. doi: 10.1093/emboj/17.10.2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wada A, Fukuda M, Mishima M, Nishida E. Nuclear export of actin: a novel mechanism regulating the subcellular localization of a major cytoskeletal protein. EMBO J. 1998;17:1635–1641. doi: 10.1093/emboj/17.6.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wolff B, Sanglier J J, Wang Y. Leptomycin B is an inhibitor of nuclear export: inhibition of nucleo-cytoplasmic translocation of the human immunodeficiency virus type 1 (HIV-1) Rev protein and Rev-dependent mRNA. Chem Biol. 1997;4:139–147. doi: 10.1016/s1074-5521(97)90257-x. [DOI] [PubMed] [Google Scholar]
- 42.Zolotukhin A S, Valentin A, Pavlakis G N, Felber B K. Continuous propagation of RRE(−) and Rev(−)RRE(−) human immunodeficiency virus type 1 molecular clones containing a cis-acting element of simian retrovirus type 1 in human peripheral blood lymphocytes. J Virol. 1994;68:7944–7952. doi: 10.1128/jvi.68.12.7944-7952.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]