Abstract
Objective
Asthma, a chronic inflammatory disease in which type 2 T helper cells (Th2) play a causative role in the development of T2 asthma. N6-methyladenosine (m6A) modification, an mRNA modification, and methyltransferase-like 3 (METTL3) is involved in the development of T2 asthma by inhibiting Th2 cell differentiation. Sex determining region Y-box protein 5 (SOX5) is involved in regulating T cell differentiation, but its role in T2 asthma was unclear. The objective of this study was to explore the role of METTL3 and SOX5 in T2 asthma and whether there is an interaction between the two.
Materials and methods
Adults diagnosed with T2 asthma (n = 14) underwent clinical information collection and pulmonary function tests. In vivo and in vitro T2 asthma models were established using female C57BL/6 mice and human bronchial epithelial cells (HBE). The expressions of METTL3 and SOX5 were detected by Western blot and qRT-PCR and Western blot. Th2 cell differentiation was determined by flow cytometry and IL-4 level was detected by ELISA. m6A methylation level was determined by m6A quantitative assay. The relationship between METTL3 expression and clinical parameters was determined by Spearman rank correlation analysis. The function of METTL3 and SOX5 genes in asthma was investigated in vitro and in vivo. The RNA immunoprecipitation assay detected the specific interaction between METTL3 and SOX5.
Results
Patients with T2 asthma displayed lower METTL3 levels compared to healthy controls. Within this group, a negative correlation was observed between METTL3 and Th2 cells, while a positive correlation was noted between METTL3 and clinical parameters as well as Th1 cells. In both in vitro and in vivo models representing T2 asthma, METTL3 levels decreased significantly, while SOX5 levels showed the opposite trend. Overexpression of METTL3 gene in HBE cells significantly inhibited Th2 cell differentiation and increased m6A methylation activity. From a mechanism perspective, low METTL3 negatively regulates SOX5 expression through m6A modification dependence, while high SOX5 expression is positively associated with T2 asthma severity. Cell transfection experiments confirmed that METTL3 regulates Th2 cell differentiation and IL-4 release through SOX5.
Conclusions
Overall, our results indicate that METTL3 alleviates Th2 cell differentiation in T2 asthma by modulating the m6A methylation activity of SOX5 in bronchial epithelial cells. This mechanism could potentially serve as a target for the prevention and management of T2 asthma.
Keywords: T2 asthma, m6A, METTL3, SOX5, HBE cells, Th2
1. Introduction
Asthma is a heterogeneous chronic respiratory condition, with allergic asthma being the prevalent phenotype. The pathogenesis of this phenotype is driven by the immune mechanism orchestrated by T helper type 2 (Th2) cells, also known as type 2 (T2) asthma [1]. Th2 cells promote the infiltration of eosinophilic inflammatory cells into the airways by releasing interleukin-4 (IL-4), IL-5, IL-13, and other cytokines, contributing to the development of asthma [2]. While inhaled corticosteroids can effectively reduce the development of T2 asthma, the challenges in preventing and treating asthma persist. Further exploration of the pathogenesis of T2 asthma is essential to improve treatment strategies.
Multiple studies have indicated that human bronchial epithelial (HBE) cells have the ability to function as antigen presenting cells (APCs). When stimulated, these APCs present antigens to T cells, which facilitates the antigen presentation process and subsequent differentiation of T cells [3,4]. It has been reported that the use of HBE cells in the construction of asthma models can cause the increase of thymic stromal lymphopoietin, which is related to eosinophilia [5]. However, further investigation is needed to better understand the potential role of HBE cells as APCs in T2 asthma.
In gene expression, N6-methyladenosine (m6A) is an important mRNA modification. According to research, m6A modification is predominantly facilitated by m6A “writers”, “erasers” and “readers”. Methyltransferase-like 3 (METTL3), a vital component of the m6A methyltransferase complex and categorized as a “writer,” is crucial for regulating the epigenome [6]. METTL3's involvement in immune response regulation across various diseases is noteworthy. For instance, the targeted depletion of METTL3 in dendritic cells results in compromised cell phenotype and function, leading to an attenuated capacity to react to lipopolysaccharides (LPS). Consequently, this diminished response translates into a decreased capability to elicit T cell responses in vivo [7]. The deletion of METTL3 in macrophages restricts the expression of inflammatory factors upon LPS stimulation, diminishing effector responses. This scenario results in accelerated tumor growth and heightened vulnerability to bacterial infections [8]. Previous studies have shown that METTL3 reduces and negatively regulates Th2 cell differentiation in allergic asthma [9]. This has important implications for exploring potential targets for controlling T2 asthma.
Sex determining region Y-box protein 5 (SOX5) belongs to the SOX transcription factor family and participates in various physiological and pathological processes, including embryonic development, cell proliferation, apoptosis, and cellular senescence [10]. SOX5 expression has been found to elevate in rheumatoid arthritis, where it enhances the migration, invasion, and inflammatory response of fibroblast-like synovial cells, exacerbating the condition [11]. Ma et al. discovered that SOX5 plays a role in T cell differentiation, and decreasing SOX5 expression can impair the differentiation process of T cells [12]. Both SOX2 and SOX5 are from the SOX transcription factor family, and it has been reported that METTL3 regulates SOX2 expression through m6A methylation [13]. Currently, the functions of SOX5 in T2 asthma remain uncertain, as does whether it is a target of METTL3.
This study reveals a reduction in METTL3 expression in T2 asthma, showing a direct association with the severity of T2 asthma, and identifies SOX5 as a downstream target of METTL3. Moreover, upregulation of METTL3 inhibits Th2 cell differentiation in T2 asthma by modulating the m6A methylation activity of SOX5. In summary, our research indicates that targeting SOX5 through METTL3 holds promise as a potential therapeutic strategy for T2 asthma.
2. Materials and methods
2.1. Subjects
The Ethics Review Committee of the Second Xiangya Hospital of Central South University approved this study (Ethical Code: LYF2023066), and all participants provided signed informed consent. The study recruited participants on a voluntary basis who met specific eligibility criteria. The inclusion criteria stipulated that all participants were non-smokers or former smokers with a smoking history of fewer than 10 pack-years. Asthma control was evaluated using an asthma control test (ACT) with a scoring range of 0–25. A score above 20 denoted effective asthma management, while a score below 20 indicated suboptimal asthma control [14]. Flow cytometry was utilized to determine the proportion of Th2 cells in peripheral blood samples to identify T2 asthma. The research involved 14 patients with T2 asthma and 13 healthy controls (HCs) who were recruited from the Second Xiangya Hospital of Central South University. The inclusion criteria for patients with T2 asthma were as follows: (a) diagnosis of asthma based on GINA guidelines [15]; (b) no systemic corticosteroid therapy in the past three months; (c) FEV1%predicted above 60%; (d) intermittent presence of asthma symptoms, with ACT greater than 20; (e) aged 18 years or older; (f) treatment regimens at steps 4 or 5. Patients with conditions such as aspergillosis, cardiac asthma, or acute attacks, autoimmune disorders, hematological diseases, infection-related diseases, or women who were pregnant or nursing were excluded from the study. HCs did not have a background of long-term respiratory illness and were non-allergic. Fig. 1 provides a comprehensive flow diagram illustrating the process of recruiting voluntary patients for the study.
Fig. 1.
Flow diagram of the study.
2.2. Data collection
Following the acquisition of written informed consent, the study documented participants' gender, age, and smoking status. Height and weight were measured to determine the body mass index (BMI) of each participant. Various laboratory tests were conducted, including measuring immunoglobulin E (IgE) levels, blood eosinophils, and blood neutrophils. Additionally, ACT, fractionated exhaled nitric oxide (FeNO) and pulmonary function test (PFT) outcomes were documented, encompassing measurements of FEV1, forced vital capacity (FVC), and FEV1%predicted.
2.3. Collecting blood samples and human CD4+ T cells, reagents, and culture condition
Using Ficoll-Paque, peripheral blood mononuclear cells (PBMCs) were isolated through density centrifugation [16,17]. In short, whole blood is collected and gently overlaid onto the liquid surface of the lymphocyte separation solution (TBD Science, Tianjin, China). Subsequently, centrifuge 600g for 21 min. The PBMCs precipitate was collected in the intermediate layer between the plasma and the separation solution, the cells were carefully cleaned twice with 1X phosphate buffered saline (PBS), and centrifuged for further use. To isolate human CD4+ T cells from PBMCs, magnetic bead separation (130–045–101, Miltenyi Biotec, Germany) was employed. The human CD4+ T cells that were isolated were subsequently cultured in complete RPMI 1640 culture medium (Gibco), enriched with 10% fetal bovine serum (Gibco), at a concentration of 1 × 106 cells/ml. To activate human CD4+ T cells, leukocyte activator (550583, BD Biosciences) was introduced, and the cells were cultured for 6 h. Following the incubation period, the cells were harvested for flow cytometry analysis. Additionally, serum and a fraction of PBMCs were obtained from all study participants and preserved at −80 °C for subsequent analysis.
2.4. Animal model
Since female mice are easily induced into allergic asthma [18], the Animal Center of the Second Xiangya Hospital of Central South University supplied a total of 12 C57BL/6 female mice, aged 7–8 weeks, for the study. These mice were carefully maintained in an environment that was free from specific pathogens, ensuring their overall health and well-being. The experimental procedures conducted in this study were approved by the Animal Care and Use Committee of the Second Xiangya Hospital of Central South University. A group of twelve female mice was randomly divided into either the normal control group or the T2 asthma group. Sensitization of the T2 asthma group was achieved by administering intraperitoneal injections of 100 μg ovalbumin (1 μg/μl, OVA, Sigma Aldrich) and 2 mg aluminum hydroxide (Sigma Aldrich) dissolved in 200 μl saline on day 0 and day 7. Subsequently, atomization with a 6% OVA solution was administered for 30 min on days 14–20 [19,20]. The normal control group received saline injections and atomization under the same conditions as the T2 asthma group, with the injection site, dosage, timing, and atomization duration all being consistent. On day 21, all mice were euthanized for further analysis.
2.5. Evaluation of airway hyperresponsiveness (AHR)
On day 21, methacholine (Mch)-induced airway resistance was assessed using direct plethysmography equipment (Buxco Electronics, USA) [21]. Firstly, the baseline lung resistance (RL) of each mouse was measured for a duration of 1 min. Subsequently, the mice's airways were exposed to 10 μl of aerosolized saline, followed by 10 μl of Mch at incremental doses. Following each dose, the RL measurements were taken once more.
2.6. Bronchoalveolar lavage fluid
Bronchoalveolar lavage fluid (BALF) was obtained by injecting 1 ml sterile saline into the lung through the trachea and then extracting and collecting. The process was repeated 3 times. After centrifugation at 1800 rpm at 4 °C for 5 min, particles containing inflammatory cells were re-suspended in PBS. The cells were fixed and underwent Wright-Giemsa staining. Following staining, a total of 200 cells were counted and differentially identified using a white light microscope counting chamber to quantify various types of inflammatory cells present in the BALF samples.
2.7. Analysis of histopathology
The lung tissue was preserved in 4% formalin and subsequently embedded in paraffin wax. The sections were cut to a thickness of 5 μm. Subsequently, these sections were stained with hematoxylin and eosin (H&E) and subjected to immunohistochemical analysis using specific antibodies: METTL3 antibody (Proteintech, China), SOX5 antibody (Proteintech, China), and eosinophilic antibody (anti-ECP, Proteintech, China). Following this, specific sections stained for each group were gathered and assessed to quantify the expression levels of eosinophil proteins, METTL3, and SOX5.
2.8. Cell asthma model and transfection
For the induction of T2 asthma, HBE cells were treated with 100 μg/ml HDM (10 μg/μl, Greer Laboratories, USA) for 24 h, while for establishing the normal control, an equal volume of 1X PBS was administered [22]. We utilized lentiviral short hairpin (sh) RNA targeting human METTL3 and SOX5 (sh-METTL3, sh-SOX5) to silence the expression of METTL3 and SOX5 in HBE cells, while a negative control (sh-NC) was obtained from GeneChem (Shanghai, China). The METTL3 shRNA sequence was 5’-GAAGACAAATCAACTGCAACG-3’ and the SOX5 shRNA sequence was 5’-GCACTTCAAATGACAACTTAA-3’. The overexpression plasmids for Human METTL3 and SOX5 (OE-METTL3, OE-SOX5) and the negative control (OE-NC) were acquired from HonorGene (Hunan, China). Transfection of shRNA and plasmid into HBE cells was performed using Lipofectamine 3000 (Invitrogen, USA) following the manufacturer's instructions. After 48 h, the cells were exposed to 100 μg/ml HDM for 24 h.
2.9. Isolation of mouse CD4+ T cells and cocultivation of human CD4+ T cells with HBE cells
The process of magnetic bead separation (130-117-043, Miltenyi Biotec, Germany) was employed to isolate CD4+ T cells from mouse spleen. Similarly, human CD4+ T cells (TCs) were also isolated using magnetic beads. These isolated cells were then cocultured with an established cell asthma model and transfected HBE cells (HBEs) at a 10:1 ratio (TCs: HBEs) for 24h. During cocultivation, the cells were cultured in complete RPMI 1640 medium supplemented with soluble anti-CD28 (1 μg/ml, eBioscience) and soluble anti-CD3e (0.5 μg/ml, eBioscience). Twenty-four hours later, the floating cells were gathered for flow cytometry analysis to examine T cell subsets. A portion of the cocultured human CD4+ T cells and supernatant were preserved for supplementary examinations. Moreover, total protein and RNA were extracted from HBE cells for additional analysis.
2.10. Flow cytometry
Both human and mouse CD4+ T cells were treated with a leukocyte activation cocktail and incubated at 37 °C with 5% CO2 for 6 h before being harvested for flow cytometry analysis. After the 6-h incubation, the cells were stained with a cell viability marker (Fixable Viability Stain 510 antibody, BD Pharmingen). Subsequently, the cells were treated with surface markers BB515-anti-human-CD4 (BD Pharmingen) and FITC-anti-mouse-CD4 (eBioscience) antibodies for staining. After staining, the cells were fixed and permeabilized with the Cytofix/Cytoperm Soln Kit (BD Pharmingen). Following the cells underwent intracellular staining with the following antibodies: BV421-anti-human-IL-4, PE-Cy [7]-anti-human-IFN-γ, PE-anti-mouse-IL-4 (all from BD Pharmingen), and PerCP/Cyanine 5.5-anti-mouse-IFN-γ (Biolegend) in permeabilization buffer. Isotype controls were employed in the control group for comparison purposes. Flow cytometry analysis was performed using FACS CantoⅡ (Becton Dickinson), and the data were analyzed using FlowJo version X software.
2.11. Western blot
The lung and HBE cells underwent crushing and lysis. The resulting lysed proteins were subjected to electrophoresis and transferred to a PVDF membrane. Following this, the membranes were incubated overnight at 4 °C with appropriately diluted METTL3, β-actin (Proteintech, Wuhan, China), and SOX5 antibodies. Subsequently, images were captured using a chemiluminescent gel imaging system, and the band intensity was quantified using Image J software (National Institutes of Health). The relative expression levels of METTL3 and SOX5 were determined by calculating the ratio of the gray value of the target band to the gray value of β-actin.
2.12. Quantitative real-time PCR
Total RNA was extracted from PBMCs, lung tissue, spleen tissue, human CD4+ T cells, and HBE cells using TRIzol reagent (Invitrogen). Subsequently, the first strand cDNA was synthesized with the PrimeScript RT reverse transcriptase (Takara, Japan). Real-time fluorescence quantitative PCR (qRT-PCR) was carried out using the SYBR Premix Ex Taq (Takara, Japan), with mouse β-actin and human GAPDH serving as internal reference genes. The expression levels of the target genes were determined using the 2−ΔΔCt method based on the cycle threshold (Ct) values obtained from the samples. Primers for the target genes were synthesized by Sangon Biotechnology (Shanghai, China) and the primer sequences can be found in Table 2.
Table 2.
Primer sequences of the study
| Gene symbol | Forward | Reverse |
|---|---|---|
| human METTL3 | 5′-CCAGCACAGCTTCAGCAGTTCC-3′ | 5′-GCGTGGAGATGGCAAGACAGATG-3′ |
| human SOX5 | 5′-TGCAGCAACACCAGGCTTAG-3′ | 5′-TCAGAGCTGGCATGTGAGGA-3′ |
| human T-bet | 5′-GCAACGCTTCCAACACGCATATC-3′ | 5′-GAGTAATCTCGGCATTCTGGTAGGC-3′ |
| human GATA3 | 5′-CATCACCACCTACCCGCCCTAC-3′ | 5′-GTTCACACACTCCCTGCCTTCTG-3′ |
| human GAPDH | 5′-CAGGAGGCATTGCTGATGAT-3′ | 5′-GAAGGCTGGGGCTCATTT-3′ |
| mouse METTL3 | 5′-CGCTGCCTCCGATGTTGATCTG-3′ | 5′-CTGACTGACCTTCTTGCTCTGCTG-3′ |
| mouse SOX5 | 5′-AGCGACCAGCCTCTCCGTATG-3′ | 5′-GCCTCTCACTCTCCTCCTCTTCC-3′ |
| mouse T-bet | 5′-ATCACTAAGCAAGGACGGCGAATG-3′ | 5′-ACCAAGACCACATCCACAAACATCC-3′ |
| mouse GATA3 | 5′-TCTGGAGGAGGAACGCTAATGGG-3′ | 5′-CGGGTCTGGATGCCTTCTTTCTTC-3′ |
| mouse β-actin | 5′-GTGCTATGTTGCTCTAGACTTCG-3′ | 5′-ATGCCACAGGATTCCATACC-3′ |
2.13. Enzyme-linked immunosorbent assay
We used enzyme-linked immunosorbent assay (ELISA) kits to measure the levels of IFN-γ and IL-4 in mouse serum, patient serum, and cell supernatant. Specifically, the mouse IFN-γ (CSB-E04578 m, Cusabio, China), mouse IL-4 (CSB-E04634 m, Cusabio, China), human IFN-γ (KE00146, Proteintech, China), and human IL-4 (KE00232, Proteintech, China) were utilized for this purpose.
2.14. RNA immunoprecipitation assay
The RNA immunoprecipitation (RIP) assay was performed using the Imprint RIP Kit from Sigma Aldrich, following the manufacturer's instructions. In summary, 5 μg of anti-METTL3 and anti-rabbit IgG (Proteintech, China) were incubated with 50 μL magnetic beads, which were then added to the cell lysate and incubated overnight at 4 °C. Subsequently, the RNA-protein IP complexes were subjected to 6 washes and treated with proteinase K digestion buffer to eliminate the proteins. The RNA was extracted using the phenol-chloroform RNA extraction method, purified for qRT-PCR analysis, and normalized to the input.
2.15. RNA m6A quantification
The m6A level in total RNA extracted from HBE cells was assessed using the Epiquik m6A RNA methylation quantification kit (Epigentek, USA) following the manufacturer's protocol. In brief, 200 ng of RNA, along with an m6A standard, was immobilized in assay wells, and incubated with the capture antibody solution and detection antibody solution as instructed. The m6A level was quantified colorimetrically by measuring the absorbance of each well at a wavelength of 450 nm and subsequently calculated based on the standard curve.
2.16. Statistics analysis
Statistical analysis was carried out using SPSS 26.0 software (IBM Corp.), and the graphs were generated using GraphPad Prism 9.0.0 software (GraphPad Software Inc). Each experiment was repeated at least three times. Continuous variables were expressed as mean ± standard deviation (M ± SD) or median (interquartile range [IQR]), while categorical variables were presented as the number (percentage). Group differences were assessed using Student's t-test, the Mann-Whitney U test, and the chi-square test for categorical variables. Spearman's rank correlation test was used to evaluate variable correlations. A statistically significant difference was defined as a P-value <0.05.
3. Results
3.1. Characteristics of the population
Table 1 displayed the demographic profiles, lung function indices, and biochemical indices of the 27 participants, comprising of 14 patients with T2 asthma and 13 HCs. No significant variations in smoking history, age, gender, or blood neutrophil counts were observed between HCs and patients with T2 asthma. Nevertheless, distinct variances were noted between the two groups regarding BMI, IgE, FeNO, blood eosinophils, and lung function indices. In comparison to the T2 asthma group, those classified as HCs exhibited a higher percentage of Th1 cells and elevated levels of IFN-γ. Conversely, they displayed a decreased percentage of Th2 cells and lower levels of IL-4. It is interesting to note that T2 asthma group exhibited higher percentages of Th2 cells and IL-4 levels, while simultaneously displaying lower percentages of Th1 cells and IFN-γ levels compared to HCs. Table 1 provided comprehensive details regarding the characteristics of the participants.
Table 1.
Clinical characteristics of participants.
| Items | Total | HCs | T2 asthma | P value |
|---|---|---|---|---|
| Subjects, n (%) | 27 | 13 (48.1) | 14 (51.9) | |
| Age(y), M ± SD | 42.74 ± 7.31 | 44.31 ± 6.87 | 41.28 ± 7.66 | 0.290 |
| Sex M/F, n/n (%/%) | 11/16 (40.7/59.3) | 4/9 (30.8/69.2) | 7/7 (50.0/50.0) | 0.440 |
| BMI (kg/m2), M ± SD | 23.88 ± 2.23 | 22.41 ± 1.45 | 25.24 ± 1.96 | <0.001 |
| Smoking history, n (%) | 0.901 | |||
| Never-smoker | 19 (70.4) | 9 (69.2) | 10 (71.4) | |
| Ex-smoker | 8 (29.6) | 4 (30.8) | 4 (28.6) | |
| ACT, M ± SD | 22.64 ± 1.50 | |||
| Lung function indexes, median (IQR) | ||||
| FEV1 (L) | 2.4 (1.8–2.6) | 2.5 (2.3–2.9) | 1.9 (1.7–2.4) | 0.002 |
| FEV1/FVC (%) | 78.1 (74.6–82.3) | 81.4 (80.5–87.1) | 74.7 (72.5–77.5) | <0.001 |
| FEV1%predicted (%) | 95.2 (80.4–102.3) | 102.4 (96.1–109.6) | 81.6 (71.5–95.4) | <0.001 |
| Biochemical indexes, median (IQR)) | ||||
| IgE (mg/l) | 318.8 (229.4–612.6) | 249.6 (174.0–464.1) | 588.8 (252.6–945.7) | 0.014 |
| IL-4 (pg/ml) | 17.59 ± 7.05 | 10.69 ± 2.0 | 23.98 ± 2.02 | <0.001 |
| IFN-γ (pg/ml) | 15.29 ± 5.92 | 21.11 ± 2.06 | 9.89 ± 0.88 | <0.001 |
| FeNO (ppb) | 22.96 ± 9.69 | 15.38 ± 5.68 | 30.00 ± 6.86 | <0.001 |
| Blood eosinophils ( × 109) | 0.18 ± 0.08 | 0.13 ± 0.07 | 0.22 ± 0.08 | 0.005 |
| Blood neutrophils ( × 109) | 3.87 ± 1.18 | 3.78 ± 1.21 | 3.94 ± 1.19 | 0.730 |
| Th2 (%), M ± SD | 3.39 ± 1.53 | 1.84 ± 0.13 | 4.83 ± 0.07 | <0.001 |
| Th1 (%), M ± SD | 3.34 ± 1.53 | 4.89 ± 0.08 | 1.90 ± 0.13 | <0.001 |
Note: Comparisons were determined using Student's t-test, the Mann-Whitney U test and the chi-square test between the two groups. P < 0.05 was considered statistically significant.
Abbreviations: M, male; F, female; BMI, body mass index; ACT, asthma control test; FEV1, forced expiratory volume in 1 s; FVC, forced vital capacity; IgE, immune globulin E; FeNO, fractionated exhaled nitric oxide; Th2, T-helper cell type 2; Th1, T-helper cell type 1; M ± SD, mean ± standard deviation; IQR, interquartile range.
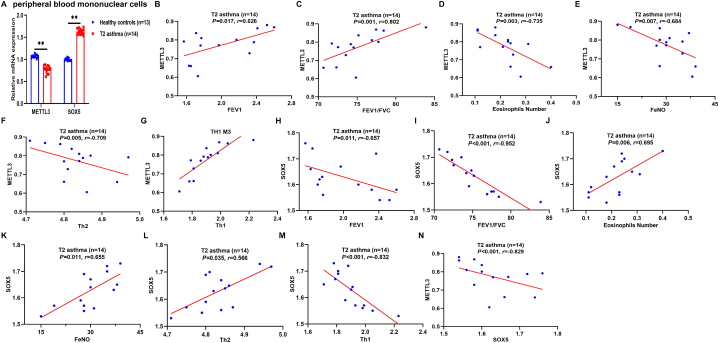
3.2. Low METTL3 expression in peripheral blood of T2 asthma patients is correlated with disease severity
METTL3 has been reported to reduce and negatively regulate Th2 cell differentiation in T2 asthma, while SOX5 is involved in T cell differentiation but has not been reported in T2 asthma [9,12]. To this end, the expression of METTL3 and SOX5 in two groups of HCs and T2 asthma patients was detected by qRT-PCR. Compared to the HCs group, reduced METTL3 mRNA levels and increased SOX5 mRNA levels were observed in the T2 asthma group (Fig. 2A). Based on these findings, we postulated a potential association between METTL3 and SOX5 and the severity of T2 asthma. Subsequently, the Spearman rank correlation test was employed to examine the relationship between the expression of METTL3 and SOX5 mRNA and the severity of T2 asthma. Specifically, METTL3 expression exhibited a positive correlation with FEV1 (P = 0.017, r = 0.626; Fig. 2B), FEV1/FVC (P = 0.001, r = 0.802; Fig. 2C), and Th1 (P < 0.001, r = 0.873; Fig. 2G), while displaying negative correlations with blood eosinophils (P = 0.003, r = -0.735; Fig. 2D), FeNO (P = 0.007, r = -0.684; Fig. 2E), and Th2 (P = 0.005, r = -0.709; Fig. 2F), validating the existence of established eosinophilic airway inflammation in individuals with T2 asthma. On the other hand, SOX5 expression was negatively correlated with FEV1 (P = 0.011, r = -0.657; Fig. 2H), FEV1/FVC (P < 0.001, r = -0.952; Fig. 2I), Th1 (P < 0.001, r = -0.832; Fig. 2M), and METTL3 (P < 0.001, r = -0.829; Fig. 2N), but positively correlated with blood eosinophils (P = 0.006, r = 0.695; Fig. 2J), FeNO (P = 0.011, r = 0.655; Fig. 2K), and Th2 (P = 0.035, r = 0.566; Fig. 2L). These findings suggest that low METTL3 expression and high SOX5 expression are associated with T2 asthma severity and potentially contribute to the onset and progression of T2 asthma.
Fig. 2.
Low METTL3 expression in peripheral blood of patients with T2 asthma is correlated with disease severity. (A) The expression of METTL3 and SOX5 mRNA in peripheral blood of the two groups was detected by qRT-PCR. (B–G) The correlation between METTL3 levels and FEV1 (B), FEV1/FVC (C), eosinophils number (D), FeNO (E), Th2 cells (F), and Th1 cells (G). (H–N) The correlation between SOX5 levels and FEV1 (H), FEV1/FVC (I), eosinophils number (J), FeNO (K), Th2 cells (L), Th1 cells (M), and METTL3 (N). Correlations were determined by Spearman's rank correlation test. P < 0.05 was considered statistically significant. FEV1, forced expiratory volume in 1 s; FVC, forced vital capacity; METTL3, methyltransferase-like 3; SOX5, sex determining region Y-box protein 5; Th2, T-helper cell type 2; Th1, T-helper cell type 1.
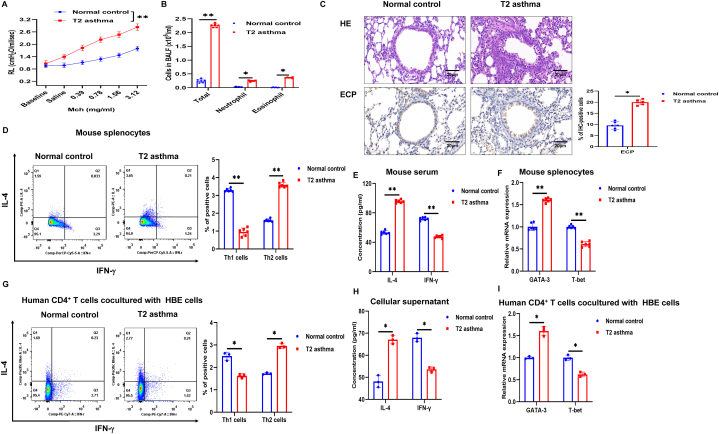
3.3. T2 asthma mediated by Th2 cells
To gain further insights into the function of METTL3 in T2 asthma, experimental models of T2 asthma were developed in mice and HBE cells. On day 21, methacholine-induced airway resistance and BALF cells were examined to evaluate the development of T2 asthma. The results showed that after methacholine stimulation, the RL of T2 asthma mice significantly increased compared to normal control mice (Fig. 3A). In the BALF of the two groups, total cell counts, eosinophil counts and neutrophil counts were elevated in T2 asthma mice compared to the normal control, with eosinophil counts showing a primary increase in T2 asthma (Fig. 3B). Histological examination of the lungs revealed a notable increase in peribronchial inflammatory cell infiltration in mice with T2 asthma compared to normal control mice (Fig. 3C). Immunohistochemical analysis of eosinophil cationic protein (ECP) indicated a substantial increase in eosinophil infiltration within the lungs of the T2 asthma group compared to the normal control group (Fig. 3C). T-bet is a critical transcription factor for Th1 cell differentiation, while GATA3 is essential for Th2 cell differentiation. In the T2 asthma group, there was a significant increase in Th2 cells, serum IL-4 levels, and GATA3 mRNA expression in mouse splenocytes compared to the normal control group (Fig. 3D–F).
Fig. 3.
T2 asthma mediated by Th2 cells. (A) Lung resistance in the normal control and T2 asthma groups. (B) Two groups of BALF cells: total, neutrophil, and eosinophil cells. (C) Lung tissues of the two groups were stained with H&E and immunohistochemical staining performed with [eosinophil antibody (anti-ECP)]. Scale bar, 20 μm. (D) Th1 and Th2 cells were detected by flow cytometry in mouse splenocytes. (E) Mouse serum levels of IL-4 and IFN-γ were detected by ELISA. (F) The expression of GATA3 and T-bet mRNA in mouse splenocytes were detected by qRT-PCR. (G) Th1 and Th2 cells were detected by flow cytometry in human CD4+ T cells. (H) Cellular supernatant levels of IL-4 and IFN-γ were detected by ELISA. (I) The expression of GATA3 and T-bet mRNA in human CD4+ T cells were detected by qRT-PCR. *P < 0.05. **P < 0.01. IHC, immunohistochemistry.
In the current research, it has been discovered that HBE cells possess the function of APCs, which play a role in influencing T cell differentiation [3,4]. This function is crucial in the sensitization and pathogenesis of asthma. To induce T2 asthma, HBE cells were treated with HDM or PBS for 24 h, followed by co-culturing with human CD4+ T cells. The study findings indicated a rise in Th2 cells in the T2 group when compared to the normal control group, whereas Th1 cells were predominantly elevated in the normal control group (Fig. 3G). Furthermore, the levels of IL-4 in the cellular supernatant and the expression of GATA3 mRNA in human CD4+ T cells from the T2 asthma group were markedly higher compared to those in the normal control group (Fig. 3H and I). These findings suggest that HBE cells can act as APCs, and that exposure to HDM can trigger the emergence of Th2 cell-mediated T2 asthma.
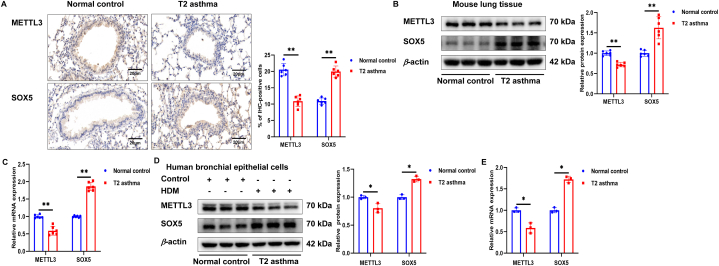
3.4. Expression of METTL3 and SOX5 in T2 asthma
The previous results (Fig. 2A) indicated that patients with T2 asthma exhibited decreased METTL3 and increased SOX5 levels. In our in vivo study, histological examination of lung tissue from the T2 asthma group revealed reduced METTL3 staining intensity and heightened SOX5 staining intensity compared to the normal controls (Fig. 4A). Additionally, the levels of METTL3 and SOX5 proteins and mRNA in lung tissue and human bronchial epithelial (HBE) cells from the T2 asthma group aligned with the findings from the lung tissue staining (Fig. 4B–E). These findings suggest that METTL3 and SOX5 may play pivotal roles in the pathogenesis of T2 asthma.
Fig. 4.
Expression of METTL3 and SOX5 in T2 asthma. (A) Lung tissues of each group were stained for anti-METTL3 and anti-SOX5. Scale bar, 20 μm. (B and C) The expression of METTL3 and SOX5 protein and mRNA in lung tissues of each group was detected by western blot and qRT-PCR. (D and E) The expression of METTL3 and SOX5 protein and mRNA in HBE cells of each group was detected by western blot and qRT-PCR. *P < 0.05. **P < 0.01.
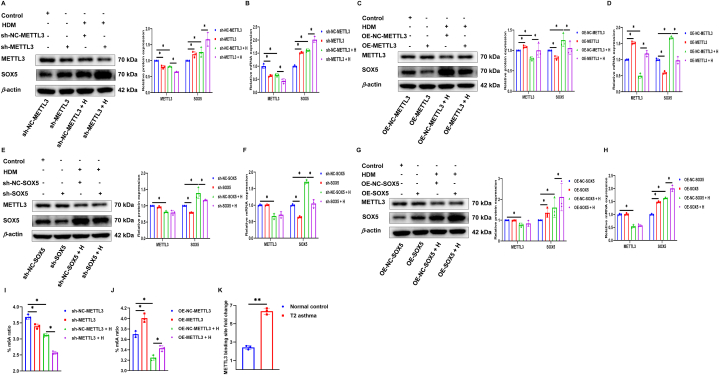
3.5. METTL3 is needed to maintain SOX5 silencing
Subsequently, to delve deeper into the molecular mechanism linking METTL3 to T2 asthma, we investigated whether there was an association between METTL3 and SOX5 in HBE cells with or without exposure to HDM. Initially, we successfully demonstrated gene silencing or overexpression of METTL3 in HBE cells in the presence or absence of HDM. The outcomes revealed that the levels of SOX5 mRNA and protein increased or decreased with METTL3 gene silencing or overexpression compared to the control group (Fig. 5A–D). Additionally, we successfully conducted silencing or overexpression of SOX5 in HBE cells exposed to and unexposed to HDM. Furthermore, we assessed the expression of METTL3 and found that there were no notable alterations in the mRNA and protein levels of METTL3 when the SOX5 gene was silenced or overexpressed (Fig. 5E–H).
Fig. 5.
METTL3 is needed to maintain SOX5 silencing. (A–D) Western blot and qRT-PCR were used to detect the expression of METTL3 and SOX5 protein and mRNA in METTL3 gene silencing or overexpression with or without HDM exposure. (E–H) Western blot and qRT-PCR were used to detect the expression of METTL3 and SOX5 protein and mRNA in SOX5 gene silencing or overexpression with or without HDM exposure. (I and J) The levels of m6A in METTL3 gene silencing or overexpression with or without HDM exposure was detected by m6A RNA measurement quantitative assay. (K) The interaction between METTL3 and SOX5 mRNA was analyzed by RIP-qRT-PCR assay with or without HDM exposure. *P < 0.05. **P < 0.01.
As a writer of m6A, we further investigated whether METTL3 has a regulatory effect on m6A modification in T2 asthma. In fact, in RNA methylation quantification, as expected, silencing or overexpression of METTL3 reduced or increased the overall modification of m6A in HBE cells (Fig. 5I and J). To demonstrate the direct interaction between METTL3 and SOX5, and to determine the m6A-dependent mechanism regulated by SOX5, the interaction between METTL3 and SOX5 mRNA was examined using RIP in HBE cells with or without HDM exposure. RIP experiments showed a significant increase in METTL3 binding to SOX5 after exposure to HDM, indicating a direct interaction between METTL3 and SOX5 (Fig. 5K). These results suggest that METTL3 and SOX5 are involved in the pathogenesis of T2 asthma through m6A-dependent modification, and that METTL3 negatively regulates SOX5 expression through m6A methylation modification.
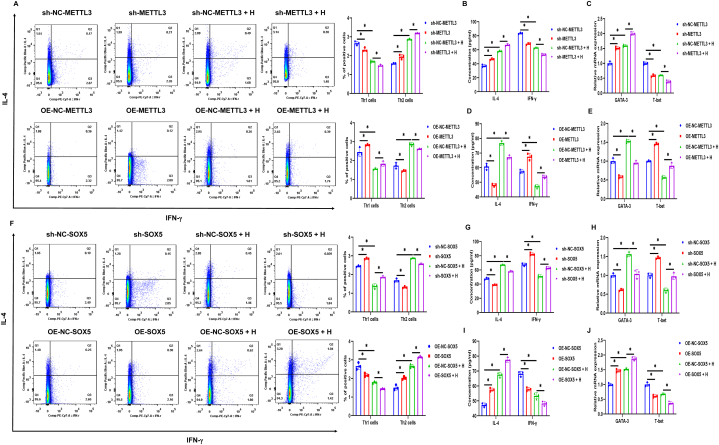
3.6. Differentiation of Th2 cells under METTL3 and SOX5 overexpression or silencing
T2 asthma is primarily driven by Th2 cells, with existing knowledge indicating that METTL3 plays a negative regulatory role in Th2 cell differentiation. Nevertheless, the specific role of SOX5 in the pathogenesis of T2 asthma remains elusive. To address this, we conducted METTL3 and SOX5 gene transfection experiments in HBE cells and co-cultured these cells with human CD4+ T cells. Interestingly, it was observed that the expression levels of Th2 cells, IL-4, and GATA3 increased or decreased when METTL3 was silenced or overexpressed. In contrast, a consistent trend was observed in Th1 cells, IFN-γ, and T-bet (Fig. 6A–E). Meanwhile, when SOX5 was overexpressed or silenced, as expected, the expressions of Th2 cells, IL-4, and GATA3 tended to increase or decrease, while those of Th1 cells, IFN-γ, and T-bet tended to decrease or increase (Fig. 6F–J). These findings suggest that METTL3 inhibits Th2 cell differentiation by negatively regulating SOX5 expression, a mechanism that may contribute to the exploration of therapeutic targets for T2 asthma.
Fig. 6.
Differentiation of Th2 cells under METTL3 and SOX5 overexpression or silencing. (A–E) After the METTL3 gene was transfected in HBE cells with or without HDM exposure and cocultured with human CD4+ T cells for 24 h, the expression of Th2 and Th1 cells (A), the levels of IL-4, IFN-γ in the cell supernatant (B and D), and the expression of GATA3 and T-bet mRNA in human CD4+ T cells (C and E). (F–J) After the transfection of the SOX5 gene in HBE cells with or without HDM exposure and cocultured with human CD4+ T cells for 24 h, the expression of Th2 and Th1 cells (F), the levels of IL-4, IFN-γ (G and I), and the expression of GATA3 and T-bet mRNA (H and J). *P < 0.05.
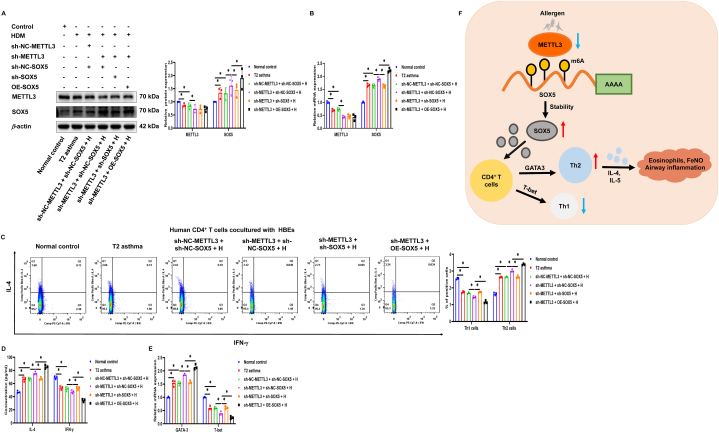
3.7. Silencing SOX5 attenuates low METTL3-induced Th2 cell differentiation
METTL3 has the ability to downregulate the expression of SOX5 via m6A methylation modification, and both are involved in the modulation of Th2 cell differentiation. To explore whether METTL3's influence on Th2 cell differentiation is dependent on SOX5, we performed co-transfection studies involving METTL3 and SOX5 in HBE cells exposed to HDM, followed by co-culturing with human CD4+ T cells for additional evaluation. When METTL3 was silenced, there was a notable increase in the expression of SOX5, as indicated by the results (Fig. 7A and B). Furthermore, interestingly, the enhancing effect of low METTL3 on Th2 cell differentiation was significantly reversed by SOX5 silencing, suggesting that up-regulation of SOX5 is the reason why low METTL3 expression induces preferential Th2 cell differentiation in T2 asthma. Concurrently, in the context of METTL3 silencing, upregulation of SOX5 was observed to enhance the expression of Th2 cells, GATA3, and IL-4, while decreasing the expression of IFN-γ, Th1 cells, and T-bet. These data suggest that METTL3 inhibits Th2 cell differentiation in T2 asthma by inhibiting SOX5 expression (Fig. 7C–E).
Fig. 7.
Silencing SOX5 attenuates low METTL3-induced Th2 cell differentiation. (A and B) Western blot and qRT-PCR were used to detect the expression of METTL3 and SOX5 protein and mRNA in METTL3 gene silencing and SOX5 gene silencing or overexpression with or without HDM exposure. (C–E) After the METTL3 gene and SOX5 gene were transfected in HBE cells with or without HDM exposure and cocultured with human CD4+ T cells for 24 h, the expression of Th2 and Th1 cells (C), the levels of IL-4, IFN-γ (D), and the expression of GATA3 and T-bet mRNA (E). (F) The schematic outlines the mechanistic basis of this study. *P < 0.05.
4. Discussion
The m6A modification, a prevalent mRNA alteration, involves in the onset and progression of diverse diseases. This study presents evidence of the impact of m6A modification on the advancement of T2 asthma using HBE cells. In short, we demonstrated that METTL3 is lowly expressed in patients with T2 asthma and is negatively correlated with disease severity, and that low-expressed METTL3 correlates with poorer lung function. METTL3 functional transfection experiments showed that METTL3 negatively regulates Th2 cell differentiation in T2 asthma, and low expression of METTL3 could enhance Th2 cell differentiation. Mechanistically, we verified that METTL3 promotes the degradation of SOX5 mRNA in a process dependent on m6A, consequently suppressing the differentiation of Th2 cells. Silencing SOX5 resulted in a notable suppression of Th2 cell differentiation, while heightened SOX5 expression in peripheral blood exhibited a positive association with airway inflammation in individuals with T2 asthma. In conclusion, this research underscores the significance of METTL3 in the immunopathogenesis of T2 asthma, the key aspect of which is the inhibition of Th2 cell differentiation through m6A modifiation-dependent down-regulation of SOX5 (Fig. 7F).
METTL3, a crucial component of the m6A methyltransferase complex, serves as an active contributor to m6A methylation modifications and participates in immune responses across various diseases, such as breast cancer, infection, and kidney injury [[23], [24], [25], [26]]. Within this investigation, a marked decrease in METTL3 expression was observed in the T2 asthma cohort as compared to the normal control group at the clinical, cellular, and animal levels. At the same time, low METTL3 was associated with severity in patients with T2 asthma and promoted airway inflammation. This is consistent with previous reports [9]. Low METTL3 promotes airway inflammation and aggravates allergic asthma by promoting the activation of M2-type macrophages [9]. This indicates that m6A modification may have a crucial involvement in the progression of T2 asthma. T2 asthma is a common clinical phenotype mediated primarily by Th2 cells. In this study, we found that METTL3 transfection in antigen-presenting HBE cells significantly enhanced Th2 cell differentiation by silencing METTL3, further highlighting the critical role of METTL3 in T2 asthma. Mast cells play a vital role in type 2 asthma, and studies have demonstrated that mast cells deficient in METTL3 exhibit notably heightened reactions to acute stimuli [27].
SOX5, belonging to the SOX transcription factor family, participates in various physiological and pathological processes including cell growth, tumor proliferation and invasion [28]. Liu et al. discovered a notable upregulation of SOX5 in dilated cardiomyopathy, contributing to disease progression and the elevation of inflammatory factor levels [29]. The present study found that when SOX5 is silenced, it can significantly inhibit the Th2 cell response. It has been found that SOX5 is involved in the occurrence of respiratory conditions, including the overlap syndrome of chronic obstructive pulmonary disease and asthma [30,31]. A study has shown that Th17 cell differentiation is inhibited by reducing SOX5 in CD4 T cell selenophenin I deficiency [12]. In this study, SOX5 expression was increased when METTL3 was silenced in HBE cells. Mechanistically, METTL3 negatively regulates SOX5 expression in an m6A activity-dependent manner. Further co-transfection experiments showed that SOX5 silencing significantly reversed the enhancement effect of low METTL3 on Th2 cell differentiation. It should be noted that the expression of SOX5 was elevated in the peripheral blood of individuals with T2 asthma and exhibited a positive correlation with disease severity. These findings help explore potential therapeutic targets for T2 asthma.
In summary, we demonstrate the pivotal role of m6A modification in the development of T2 asthma. The combined network of the METTL3/SOX5/Th2 axis highlights the mechanism of m6A activity-dependent epigenetic regulation. The findings from this study contribute to the identification of potential targets for the prevention and treatment of T2 asthma.
Data availability statement
The data used to support the findings of this study are included within the article.
Funding statement
This study was supported by the National Natural Science Foundation of China (No. 82100039, No. 82160009, No. 82170039, No. 82260008), the Guangxi Natural Science Foundation (No. 2022GXNSFAA035452) and the 2022 Basic Research Plan of Guizhou Province (Natural Science Project) (Qian Ke He foundation-ZK [2022] General 425).
CRediT authorship contribution statement
Zhifeng Chen: Writing – original draft, Methodology, Formal analysis, Data curation, Conceptualization. Yulin Shang: Methodology. Xiufeng Zhang: Data curation. Wentao Duan: Methodology. Jianmin Li: Data curation. Liming Zhu: Formal analysis. Libing Ma: Data curation. Xudong Xiang: Funding acquisition, Formal analysis. Jingsi Jia: Methodology, Conceptualization. Xiaoying Ji: Funding acquisition, Formal analysis, Conceptualization. Subo Gong: Funding acquisition, Data curation, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e28884.
Contributor Information
Jingsi Jia, Email: jingsijia@csu.edu.cn.
Xiaoying Ji, Email: 183572401@qq.com.
Subo Gong, Email: gsb510@csu.edu.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Al-Shaikhly T., Murphy R.C., Parker A., et al. Location of eosinophils in the airway wall is critical for specific features of airway hyperresponsiveness and T2 inflammation in asthma. Eur. Respir. J. 2022;60(2) doi: 10.1183/13993003.01865-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maison N., Omony J., Illi S., et al. T2-high asthma phenotypes across lifespan. Eur. Respir. J. 2022;60(3) doi: 10.1183/13993003.02288-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qu X., Li M., Liu H.J., et al. Role of bombesin receptor activated protein in the antigen presentation by human bronchial epithelial cells. J. Cell. Biochem. 2013;114(1):238–244. doi: 10.1002/jcb.24366. [DOI] [PubMed] [Google Scholar]
- 4.Lee H.S., Park D.E., Lee J.W., et al. IL-23 secreted by bronchial epithelial cells contributes to allergic sensitization in asthma model: role of IL-23 secreted by bronchial epithelial cells. Am. J. Physiol. Lung Cell Mol. Physiol. 2017;312(1):L13–l21. doi: 10.1152/ajplung.00114.2016. [DOI] [PubMed] [Google Scholar]
- 5.Yang X., Su B., Liu J., et al. A CpG-Oligodeoxynucleotide suppresses Th2/Th17 inflammation by inhibiting IL-33/ST2 signaling in mice from a model of adoptive dendritic cell transfer of smoke-induced asthma. Int. J. Mol. Sci. 2023;24(4) doi: 10.3390/ijms24043130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen T., Hao Y.J., Zhang Y., et al. m(6)A RNA methylation is regulated by microRNAs and promotes reprogramming to pluripotency. Cell Stem Cell. 2015;16(3):289–301. doi: 10.1016/j.stem.2015.01.016. [DOI] [PubMed] [Google Scholar]
- 7.Wang H., Hu X., Huang M., et al. Mettl3-mediated mRNA m(6)A methylation promotes dendritic cell activation. Nat. Commun. 2019;10(1):1898. doi: 10.1038/s41467-019-09903-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tong J., Wang X., Liu Y., et al. Pooled CRISPR screening identifies m(6)A as a positive regulator of macrophage activation. Sci. Adv. 2021;7(18) doi: 10.1126/sciadv.abd4742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han X., Liu L., Huang S., et al. RNA m(6)A methylation modulates airway inflammation in allergic asthma via PTX3-dependent macrophage homeostasis. Nat. Commun. 2023;14(1):7328. doi: 10.1038/s41467-023-43219-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.She Z.Y., Yang W.X. SOX family transcription factors involved in diverse cellular events during development. Eur. J. Cell Biol. 2015;94(12):547–563. doi: 10.1016/j.ejcb.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 11.Shi Y., Wu Q., Xuan W., et al. Transcription factor SOX5 promotes the migration and invasion of fibroblast-like synoviocytes in part by regulating MMP-9 expression in collagen-induced arthritis. Front. Immunol. 2018;9:749. doi: 10.3389/fimmu.2018.00749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma C., Hoffmann F.W., Nunes L.G., et al. Selenoprotein I deficiency in T cells promotes differentiation into tolerant phenotypes while decreasing Th17 pathology. J. Leukoc. Biol. 2022;112(6):1387–1397. doi: 10.1002/JLB.1A0122-080R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li T., Hu P.S., Zuo Z., et al. METTL3 facilitates tumor progression via an m(6)A-IGF2BP2-dependent mechanism in colorectal carcinoma. Mol. Cancer. 2019;18(1):112. doi: 10.1186/s12943-019-1038-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nathan R.A., Sorkness C.A., Kosinski M., et al. Development of the asthma control test: a survey for assessing asthma control. J. Allergy Clin. Immunol. 2004;113(1):59–65. doi: 10.1016/j.jaci.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 15.Levy M.L., Bateman E.D., Allan K., et al. Global access and patient safety in the transition to environmentally friendly respiratory inhalers: the Global Initiative for Asthma perspective. Lancet (London, England) 2023;402(10406):1012–1016. doi: 10.1016/S0140-6736(23)01358-2. [DOI] [PubMed] [Google Scholar]
- 16.Tang F.S., Hansbro P.M., Burgess J.K., Ammit A.J., Baines K.J., Oliver B.G. A novel immunomodulatory function of neutrophils on rhinovirus-activated monocytes in vitro. Thorax. 2016;71(11):1039–1049. doi: 10.1136/thoraxjnl-2015-207781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bruscia E.M., Zhang P.X., Satoh A., et al. Abnormal trafficking and degradation of TLR4 underlie the elevated inflammatory response in cystic fibrosis. J. Immunol. 2011;186(12):6990–6998. doi: 10.4049/jimmunol.1100396. Baltimore, Md : 1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang H.Y., Mitzner W. Sex differences in mouse models of asthma. Can. J. Physiol. Pharmacol. 2007;85(12):1226–1235. doi: 10.1139/Y07-116. [DOI] [PubMed] [Google Scholar]
- 19.Hoffman S.M., Tully J.E., Nolin J.D., et al. Endoplasmic reticulum stress mediates house dust mite-induced airway epithelial apoptosis and fibrosis. Respir. Res. 2013;14(1):141. doi: 10.1186/1465-9921-14-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ni Y., Hao J., Hou X., et al. Dephosphorylated polymerase I and transcript release factor prevents allergic asthma exacerbations by limiting IL-33 release. Front. Immunol. 2018;9:1422. doi: 10.3389/fimmu.2018.01422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Locke N.R., Royce S.G., Wainewright J.S., Samuel C.S., Tang M.L. Comparison of airway remodeling in acute, subacute, and chronic models of allergic airways disease. Am. J. Respir. Cell Mol. Biol. 2007;36(5):625–632. doi: 10.1165/rcmb.2006-0083OC. [DOI] [PubMed] [Google Scholar]
- 22.Liu D., He L., Ding N., et al. Bronchial epithelial cells of young and old mice directly regulate the differentiation of Th2 and Th17. Biosci. Rep. 2019;39(2) doi: 10.1042/BSR20181948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang J.N., Wang F., Ke J., et al. Inhibition of METTL3 attenuates renal injury and inflammation by alleviating TAB3 m6A modifications via IGF2BP2-dependent mechanisms. Sci. Transl. Med. 2022;14(640) doi: 10.1126/scitranslmed.abk2709. [DOI] [PubMed] [Google Scholar]
- 24.Wan W., Ao X., Chen Q., et al. METTL3/IGF2BP3 axis inhibits tumor immune surveillance by upregulating N(6)-methyladenosine modification of PD-L1 mRNA in breast cancer. Mol. Cancer. 2022;21(1):60. doi: 10.1186/s12943-021-01447-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Winkler R., Gillis E., Lasman L., et al. m(6)A modification controls the innate immune response to infection by targeting type I interferons. Nat. Immunol. 2019;20(2):173–182. doi: 10.1038/s41590-018-0275-z. [DOI] [PubMed] [Google Scholar]
- 26.Mao Y., Jiang F., Xu X.J., et al. Inhibition of IGF2BP1 attenuates renal injury and inflammation by alleviating m6A modifications and E2F1/MIF pathway. Int. J. Biol. Sci. 2023;19(2):593–609. doi: 10.7150/ijbs.78348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leoni C., Bataclan M., Ito-Kureha T., Heissmeyer V., Monticelli S. The mRNA methyltransferase Mettl3 modulates cytokine mRNA stability and limits functional responses in mast cells. Nat. Commun. 2023;14(1):3862. doi: 10.1038/s41467-023-39614-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li G., Wang K., Wang J., Qin S., Sun X., Ren H. miR-497-5p inhibits tumor cell growth and invasion by targeting SOX5 in non-small-cell lung cancer. J. Cell. Biochem. 2019;120(6):10587–10595. doi: 10.1002/jcb.28345. [DOI] [PubMed] [Google Scholar]
- 29.Liu Y., Jiang B., Cao Y., et al. High expression levels and localization of Sox5 in dilated cardiomyopathy. Mol. Med. Rep. 2020;22(2):948–956. doi: 10.3892/mmr.2020.11180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hardin M., Cho M., McDonald M.L., et al. The clinical and genetic features of COPD-asthma overlap syndrome. Eur. Respir. J. 2014;44(2):341–350. doi: 10.1183/09031936.00216013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zou H., Wang S., Wang S., et al. SOX5 interacts with YAP1 to drive malignant potential of non-small cell lung cancer cells. Am. J. Cancer Res. 2018;8(5):866–878. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data used to support the findings of this study are included within the article.