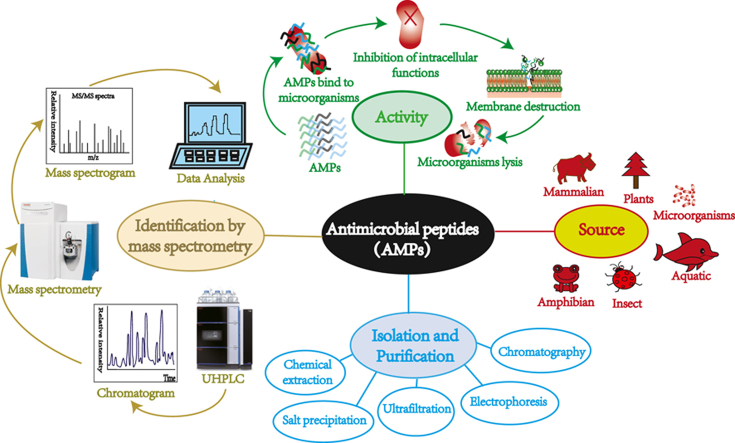
Abstract
Antimicrobial peptides (AMPs) constitute a group of small molecular peptides that exhibit a wide range of antimicrobial activity. These peptides are abundantly present in the innate immune system of various organisms. Given the rise of multidrug-resistant bacteria, microbiological studies have identified AMPs as potential natural antibiotics. In the context of antimicrobial resistance across various human pathogens, AMPs hold considerable promise for clinical applications. However, numerous challenges exist in the detection of AMPs, particularly by immunological and molecular biological methods, especially when studying of newly discovered AMPs in proteomics. This review outlines the current status of AMPs research and the strategies employed in their development, considering resent discoveries and methodologies. Subsequently, we focus on the advanced techniques of mass spectrometry for the quantification of AMPs in diverse samples, and analyzes their application, advantages, and limitations. Additionally, we propose suggestions for the future development of tandem mass spectrometry for the detection of AMPs.
Keywords: Mass spectrometry, LC-MS/MS, Antimicrobial peptides, Applications, Extraction
Graphical abstract
Highlights
-
•
MDR is increasingly common in pathogenic bacteria. The development and application of new antimicrobial drugs is imminent.
-
•
The diversity and potency of AMPs make them attractive targets for development as antimicrobial drugs.
-
•
Mass spectrometry has become a powerful tool in chemical and biological analysis, and has been applied in the study of AMPs.
-
•
Our study focuses on the application of MS/MS detecting AMPs in various samples and analyzes its advantages and limitations.
1. Introduction
Antimicrobial resistance poses a substantial threat to global health and development, with the prevalence of multidrug resistance becoming increasingly common among pathogenic bacteria. Drug-resistant pathogens are a serious concern in both hospital and non-hospital settings, presenting substantial challenges to the healthcare system, encompassing issues related to the identification, management, and control of infections caused by resistant organisms [1,2]. Therefore, the development and application of new antimicrobial drugs are imminent.
Antimicrobial peptides (AMPs) are short, structurally diverse peptides with a broad spectrum of antibacterial, antiviral, and antifungal activity [[3], [4], [5]]. Genetically encoded in nature, these peptides typically comprise 10–100 amino acids and are synthesized by diverse organisms as a defense mechanism against microbial invasion [[6], [7], [8]]. They have been reported to be inhibitors of central dogma processes, and blocking DNA, RNA, and protein synthesis [9]. AMPs play a crucial role in the immune defense of multicellular organisms and are currently under investigation for their potential as anti-infective drugs. Their unique ability to target multiple biological targets simultaneously gives AMPs a distinct advantage over traditional antibiotics [10]. An exemplification of this phenomenon is LL-37, which exhibits effects on bacterial cell membranes, in addition to its direct microbicidal, immunomodulatory, and antibiofilm properties [11,12]. The diversity and potency of AMPs make them highly appealing for the development of antimicrobial drugs [13], Currently, many AMPs are being evaluated in clinical trials [14,15]. AMPs have pharmacodynamic properties that effectively reduce the evolution of resistance in target microorganisms. Additionally, they may exhibit synergistic effects with other antimicrobial agents and traditional antibiotics. Overall, AMPs play a critical role in maintaining the health and well-being of organisms by protecting them from infection and regulating various biological processes. Accumulating evidence indicates that AMPs can be used in the treatment of microbial infections in humans [16].
However, for the clinical application of AMPs, comprehending their inherent biological properties to minimize the potential for unintended harm and to address the current challenge of antibiotic resistance faced by traditional antibiotics is critical [17,18]. Currently, AMPs are primarily detected by using molecular biological methods, immunological methods, and mass spectrometry (MS). Immunodetection assays, including ELISA, immunohistochemistry, and western blotting [19,20], have associated limitations. These include cross-reactivity between human heterophile antibodies and the antibodies contained in reagents as well as possible autoantibodies that can lead to error in the results due to defaults or excess values [21]. Additionally, matrix differences between patient sera (hemolysis, icterus, and lipemia) and calibrators present challenges. AMPs can also be detected using molecular research platforms, including polymerase chain reaction (PCR), microarray analysis, and next-generation sequencing (NGS) [[22], [23], [24]]. Potential disadvantages of molecular biological detection methods, such as PCR and RT-PCR, include limited sensitivity for detecting low levels of AMPs in complex samples. This may result in false-negative results or underestimation of the true AMP levels. The utilization of molecular techniques introduces the potential for false-positive findings due to contamination. To mitigate the risk of contamination, adequate controls and rigorous quality control measures must be employed. Molecular methods, such as NGS or microarray analysis, can be costly and time-consuming. Consequently, their utility for routine diagnostics or screenings may be limited.
In recent years, MS has emerged as a robust and indispensable tool in the field of chemical and biological analysis, and has been applied in the study of AMPs. MS overcomes these interferences of the detection of immunological methods by not relying on antibodies. It detects protein directly, providing information on the expression, post-translational modifications, and degradation products of the AMPs with higher sensitivity and specificity. The detection of AMPs by tandem mass spectroscopy (MS/MS) represents a real technological revolution in clinical diagnosis and treatment. Numerous studies exist, but most research on AMPs using MS/MS has primarily focused on two aspects: (1) extracting and characterizing of biologically active peptides for the discovery of new AMPs and (2) exploring possible functional and physiological properties. Therefore, these two aspects are outlined in this review.
The detection of AMP is an important and current research focus. To our we knowledge, systematic reviews focusing on the application of MS for AMP detection are currently lacking. Therefore, we aim to comprehensively review and summarize MS techniques in AMP research and application, particularly in the discovery and characterization of novel AMPs from natural sources. Additionally, studies related to AMP discovery, quantitative analysis, characterization, activity, and therapies are emphasized. Finally, this study presents a thorouth analysis of the advantages, limitations, and future perspectives of the methods employed. The discussion serves as a valuable reference for future research endeavors.
2. Sources of AMPs
AMPs can be classified based on various criteria, such as their structure, origin, and mechanism of action. Some of the commonly recognized types of AMPs, including plant AMPs, insect AMPs, and synthetic and engineered peptides. Note that this is not an exhaustive list, and new types of AMPs are continuously being discovered and characterized all the time. According to their source, AMPs can be divided into the following.
2.1. Bacteriophage/viral AMPs
Bacteriophages, also known as phages, are viral agents that specifically target and infect bacteria. Within the realm of phage research, numerous proteins have been isolated, including endolysins, depolymerases, and holins, have been isolated and identified as potential alternative antimicrobial agents [[25], [26], [27]]. Phage-encoded endolysins are bacteriolytic proteins that are synthesized during the final stages of the phage lytic cycle. These proteins play a crucial role in breaking down various components of the bacterial cell wall, thereby facilitating the release of phage progeny from the host cells [28]. Two distinct categories of phage amplification systems, namely phage-encoded lysis factors and phage tail complexes, are recognized [[29], [30], [31]].
2.2. Bacterial
In gram-positive bacteria, reports have indicated the occurrence of ribosomal and nonribosomal synthesis of AMPs [32]. Bacteriocins, synthesized by ribosomes in bacteria, are considered as bacterial AMPs. These peptides primarily exhibit activity primarily against bacteria closely related to the producer bacteria, necessitating defense mechanisms to counteract potential harmful effects [33]. Bacteriocins produced by gram-positive bacteria are classified into four categories: antibiotics, non-antibiotics, large bacteriocins, and bacteriocins with distinctive structures [34,35]. Numerous bacteriocins can be isolated from bacteria, with the majority originating gram-negative bacilli within the Enterobacteriaceae bacteria family. These AMPs have a narrow spectrum of antibacterial activity against gram-negative bacteria and are categorized as categories colicins, colicin-like, microcins, and phage tail-like bacteriocins [35].
2.3. Fungal AMPs
Fungal AMPs can be categorized into two primary groups: peptaibols and fungal defensins [36]. Peptaibols, originating mainly comes from the soil fungus Trichoderma [37], are short peptides, typically consisting of 5–21 amino acids. These peptides are characterized by a high proportion of non-proteinogenic amino acids, displaying antifungal activity [38,39]. Defensins, characterized by their short length and high cysteine content, are peptides that are ubiquitously found in microorganisms, plants, and animals [40,41]. Like other AMPs, defensins have demonstrated effectiveness as alternatives to current antifungal therapies, showing potential as novel therapeutic agents or drug leads [42,43].
2.4. Plant AMPs
Cysteine-rich AMPs, widely distributed in plants, which form part of the plant defense systems [44]. Seeds and fruits are particularly valuable sources of diverse AMPs in plants. Cysteine-rich AMPs exhibit persistent broad-spectrum antimicrobial activity, enabling plants to effectively defend against a wide array of pathogens [44,45]. Currently, plant AMPs are primarily classified according to their structure into α-hairpin proteins, defensins, heme-like peptides, desmin-type peptides, lipid transfer proteins, thionin, snake proteins and unclassified cysteine-rich AMP [46,47].
2.5. Animal AMPs
Various animals, including amphibians, fish, reptiles, birds, mammals, produce AMPs. Invertebrates, lacking adaptive immunity, rely on AMPs as a a vital component of their immune defense. In the case of the invertebrates under investigation, these peptides play a significant role in the immune response [48,49]. These invertebrate AMPs include defensins and cecropin found in insects, defensins found in mollusks and nematodes, macrodefensins found in horseshoe crabs, and invertebrate β-defensins found in crustaceans [[50], [51], [52]]. Vertebrate antimicrobial peptides (AMPs) exhibit a range of sizes, spanning from 15 to 200 residues, and fulfill crucial roles in the immediate defense response against pathogens [53]. These AMPs are found in fish, amphibians, reptiles, birds, and mammals. Fish, in particular, serve as a valuable source of AMPs, expressing various major classes such as cathelicidins, defensins, histone-derived peptides, hepcidins, and a fish-specific class of the cecropin family known as piscidins [54]. Reptiles and birds predominantly express cathelicidin and defensin family members as their AMPs [55,56].
The cathelicidin and defensin families constitute the primary groups of AMPs found in mammals. While additional mammalian AMPs, including platelet antimicrobial proteins, hepcidin, and dermocidin, exist outside cathelicidin or defensin families [57], LL-37 is the sole member of the cathelicidin family of AMPs in humans and it currently represents the most well-studied cathelicidin [58,59].
3. Application of MS in AMPs study
3.1. AMP isolation
Several pretreatment methods, such as liquid−liquid extraction, protein precipitation, and SPE, are employed for the analysis of biologically small molecular substances using MS/MS detection. Our previous study demonstrated that the magnetic bead method not only automates the sample pretreatment but also greatly reduces the cost compared with that of the SPE method [60]. However, the methods used for protein and peptide extraction are relatively complex. Obtaining peptides, derived from sources such as skin and limb secretions and bacteria, is a relatively complex process. Thus, the extraction of peptides suitable for research, along with considerations of quantity and purity of peptides, constitutes the current focus and challenge in AMPs research. The extraction and purification of AMPs present challenges and yield low quantities, thereby limiting their application in industry and scientific research [61]. Typically, AMPs possessing protease inhibitor activity are isolated through a series of purification steps, including salt extraction, ultrafiltration, and C18 reverse-phase chromatography.
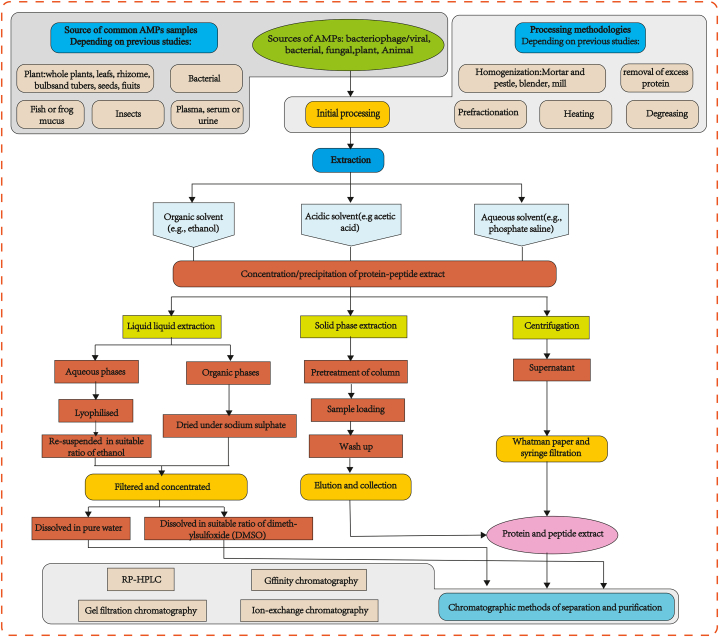
Nevertheless, the majority of existing methods are designed to target specific structural types of AMPs, restricting their ability to capture the full molecular diversity of AMPs within a given sample matrix. Barashkova, A S et al. [62] studied a general extraction method for the isolation of plant AMPs. The isolation of AMPs involves three main stages: 1) homogenization of material, 2) extraction, and 3) saturation purification of the extract. The extracts are typically fractionated using a series of liquid chromatography (LC) methods. The commonly used sample pretreatment and AMP extraction methods are presented in Fig. 1.
Fig. 1.
Typical procedures for isolation and purification of antimicrobial peptides [47,[62], [63], [64], [65]]
3.1.1. Sample preparation
AMPs can be derived from various parts of the plants such as roots, tubers, flowers, fruits, seeds, or the whole plant. Additionally, AMPs can be obtained from soil actinomycetes, molds, insects, and marine vertebrates. We extensively reviewed the preparation of AMPs samples from various sources. Several methods have been used to isolate AMPs, no uniform protocol capable of isolating all structural families of AMPs exists.
During the homogenization stage, some of the samples withstand mechanical damage. The disintegration method is selected based on the physical properties of the sample material. For instance, plant seeds and the dried material are ground in a coffee grinder, whereas frozen portions are crushed in a mortar and pestle in liquid nitrogen. Animal mucus secretions are ground in a blender with an extraction buffer [66]. The extraction process may include additional steps, such as heating [67], removal of excess protein, prefractionation [68], or removal of tannins [69], or degreasing [70], if necessary for large protein peptides or small molecules with higher content. Protein and peptides can be extracted using simple solvent extraction techniques [71]. The conventional methods for peptide extraction are detailed further, starting with simple solvent extraction, including aqueous, organic, and acidic extraction. Subsequently, multiple rounds of centrifugation and subsequent retrieval were performed using methodologies such as ultrafiltration, precipitation, or chromatography.
3.1.2. Selection of solvent
Currently, there are three distinct categories of extractants are employed for the extraction of AMP. The first category comprises water and water-based solutions, including salts and buffers. The second category encompasses organic-based solutions, specifically water solutions of ethanol, and the third category is acidic extractions (Fig. 1).
3.1.2.1. Aqueous extractions
Buffer was the first extractant used to isolate AMPs. The solvents most frequently employed in these investigations included saline, water, ammonium bicarbonate, and Tris-buffered saline. A maximum diversity of proteins and peptides are isolated during the extraction process. However, using of buffer as an extraction agent results in the extraction of various peptides, such as carbohydrates and secondary metabolites dissolved in water, reducing the total yield of the target compound.
Although aqueous extraction is the most commonly used extraction method, but the extract exhibits the lowest antimicrobial capacity. This is evident in studies comparing other extraction methods. In some studies, the aqueous extract did not show any antimicrobial activity [[72], [73], [74]]. Al-Rashed et al. (2018) [72] demonstrated that the aqueous extract did not display antimicrobial activity against the tested bacteria. Based on the well diffusion method, the acidic crude extract exhibited different antimicrobial activities on agar plates. Phosphate buffer is the most common extractant for isolating AMP [[75], [76], [77]]. Ben Brahim, R et al. (2022) [78] revealed that antimicrobial activity was not observed in extracts obtained using sulfuric acid, dichloromethane, and phosphate buffer extraction. However, some strains were growth inhibited by acetate and sodium acetate extracts. Other studies have reported a growth inhibitory effect on gram-negative and gram-positive bacteria with aqueous extracts [[79], [80], [81], [82], [83], [84]]. Gikas, E et al. (2023) [85] demonstrated the significant antimicrobial activity of urinary aqueous extracts against Escherichia coli and their relatively safety against mammalian cells. In a study by Ogbole, OO et al. (2021) [86], peptide fractions isolated using aqueous extracts from Euphorbia hirta leaf showed high antimicrobial activity against Coxsackievirus A13, Coxsackievirus A20, and Enterovirus C99.
Therefore, when detecting the biological activity of AMPs, special attention should be paid to the selection of extraction methods based on the characteristics of AMPs.
3.1.2.2. Organic extractions
Among organic extractions, ethanol is the most common solvent, followed by dichloromethane. Organically extracted AMPs have shown high antimicrobial activity [87,88]. This effectiveness can be attributed to two main factors: (1) the presence of hydrophobic groups in antimicrobial molecules, contributing to their affinity for membranes and ability to disrupt them [89]; (2) the use of organic solvents, which reduce interactions between hydrophobic groups in these extracts. Therefore, these extracts contain a high proportion of hydrophobic molecules, thereby hindering the aggregation of such hydrophobic molecules and facilitating their separation [90].
Studies have shown that both gram-positive and gram-negative bacteria were inhibited [[91], [92], [93]]. However, in a study by Křížkovská, B et al. (2023) [94], the majority of ethanol extracts from European herbs showed low or no toxicity against all strains. Subsequently, clinical multidrug-resistant (MDR) strains were tested to determine if ethanol extract could reverse drug-resistance phenotypes. The synergistic effects of breakpoint concentrations of antibiotics and nontoxins were observed. Neither the antibiotic nor the extract was active against the MDR strain when cultured alone. However, a combination of antibiotics and extracts reduced the viability of the MDR strain.
3.1.2.3. Acidic extractions
The most common solvent used for acid extraction was acetic acid, followed by trifluoroacetic acid. AMPs are also known as cationic host defense peptides [95,96]. Therefore, the first step in the extraction process was the extraction of proteins and peptides with basic properties during the aqueous extraction of the acid, thus simplifying further separation. The first thionins were isolated using sulfuric acid solutions in the 1970s [97]. Subsequently, the first AMP isolation and purification scheme was proposed. This scheme was modified and applied to the isolation of synthetic anti-LPS peptide Pep19–2.5 [98]. After the addition of 270 μL hydrochloric acid (10 M), the sample was hydrolyzed for 24 h at 120 °C. Subsequently, the solvent was evaporated in a vacuum evaporator for 4 h at 40 °C, and the pellet was resuspended in water to enrich to the applied concentration.
Overall, acidic extracts exhibit better antimicrobial activity than other extraction methods [72,78,99,100]. Ben Brahim, R. (2022) [78] tested the antimicrobial activity of Anthyllis sericea and Astragalus armatus obtained from different extractions against six bacterial strains. Acetate and sodium acetate extracts were active against the studied strains. However, the extraction products based on sulfuric acid, phosphate buffer, and dichloromethane did not show any activity against the different tested strains. Al-Rashed et al. (2018) [72] found that aqueous extracts of fish epidermal mucus did not show any antibacterial activity against the studied bacteria. However, acidic mucus extracts showed strong and variable inhibitory effects on the growth of the tested bacteria. Mucus samples inhibited the growth of Staphylococcus aureus, Escherichia coli, Salmonella, Bacillus subtilis, and Aeromonas hydrophila, but the sensitivity was not high.
3.1.3. Concentration and purification
After extraction, the next step involves saturation, purification, and concentration, which are crucial in the purification stage of downstream processing [101]. The typical purification methods include chemical extraction, salt precipitation, membrane separation and chromatography [65].
When adopting salt buffer, water, or acid solution as the extraction agent, most protein peptide saturation is achieved by ammonium sulfate solution [[102], [103], [104]]. When seeds with a high total protein content were used, this was done in one or two stages [103,105]. Dialysis is required to remove excess salt, which can be held against water, buffer solutions used during extraction, or solutions needed for further analysis [106]. Dialysis, as a protein separation and purification method, possesses limited purification capabilities. Therefore, a variety of separation methods should be used to effectively separate and purify proteins while preserving their biological properties. In recent years, desalination (low-pressure chromatography) has emerged as an alternative to dialysis. This technique involves the use of hydrophobic adsorbents, such as phenyl-agarose, reverse phase C8, and C18 [[107], [108], [109]]. RP-HPLC has become a primary purification method for peptides and other small compounds due to its good selectivity and high separation performance. Depending on the separation efficiency of each method, a combination of methods enables the purification of peptides with different purities or molecular weight ranges.
The combination of the aforementioned purification methods often enhances peptide purity. Currently, commonly used purification methods include a combination of gel filtration, ultrafiltration, ion exchange chromatography, and RP-HPLC [[110], [111], [112], [113]] (Fig. 1). In addition to the well-established peptide chemical extraction methods, there are emerging protein and peptide extraction methods such as microwave-assisted extraction, pulsed electric field, and ultrasound-assisted tooth extraction [114]. However, these methods are rarely used in currently studies. Therefore, summarizing all the accumulated experience seems meaningful. The existence of isolation protocols that allow extraction of a wide variety of peptides and comparison of results provides more possibilities for AMP studies.
3.2. AMPs detected through MS
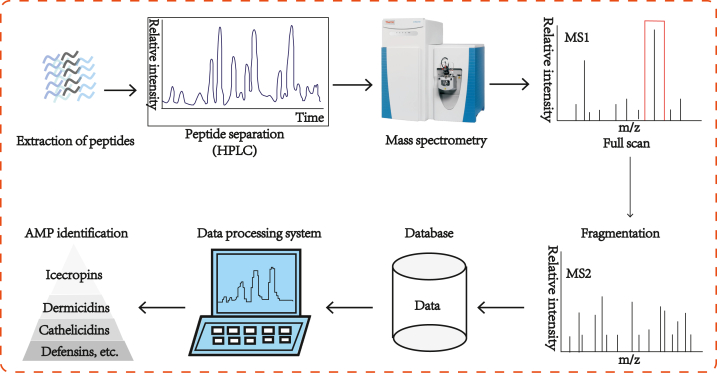
MS technology can identify and analyze AMPs with high sensitivity, high selectivity and high resolution, offering crucial technical assistance for their research and application. The detection of AMPs typically encompasses sample preparation, liquid chromatography separation, tandem mass spectrometry analysis, and data interpretation, as illustrated in Fig. 2.
Fig. 2.
Workflow diagram for the detection of antimicrobial peptides (AMPs) through mass spectrometry (MS).
3.2.1. MS detection
The peptide extracts were First separated and purified through chromatography. Methanol and acetonitrile are the most commonly used organic phases for HPLC [[115], [116], [117], [118], [119]]. Depending on the types of AMPs detected, an appropriate amount of formic acid, at a low proportion, can be added to either the aqueous phase or organic phase. The flow rate of liquid phase can then be adjusted appropriately for better substance retention. The most commonly used ion source is ESI [[120], [121], [122]]. After separation through LC, the peptide samples underwent ionization and filtration through the ion transport channel and were subsequently detected by the mass detector. The mass-to-charge ratio of the ionized peptides was detected using the mass spectrometer to obtain the primary mass spectrum. It was further detected through secondary MS, which is similar to the primary MS. The secondary MS material is selected using the quadrupole mass analyzer and transported to the collision cell for secondary fragmentation, producing a large number of secondary fragments. The secondary MS detects the fragmented peptide ions to obtain the secondary mass spectrum, whereas the primary MS detects the complete peptide ions.
Previous studies indicate that over 3100 natural AMPs have been described from groups representing life on Earth [4]. However, the research on the application of AMPs through MS primarily focuses on the discovery of new AMPs and the elucidation of their structures, often using high-resolution MS. Additionally, LC−MS/MS has been used to quantitatively detect AMPs. Iram, D et al. (2022) [123] reported the characterization of AMPs from Lactobacillus rhamnosus (C25) fermented goat milk, using high-resolution MS. Nanoflow LC−MS/MS analysis was performed on a Captive spray-Maxis-HD impact II QTOF (Bruker, Germany). Peptides were separated using a C-18, 3 μm nano-trap column (Bruker Magic C18AQ, 0.1 × 150 mm, 3 μm partials size, and 200 A° pore size). A gradient method was used, starting with 2% B at a flow rate of 5 μL/min for 2 min. The gradient increased was increased to 20% B at 46 min, followed by 45% B at 61 min. The gradient was then increased to 95% B in 2 min, which was maintained for 5 min, followed by 2% B at 12 min, resulting in a total run time of 84 min. Analytes were detected using LC−MS in positive ion mode.
Currently, the MS technology used in AMPs research primarily high-resolution MS, which is mainly used to discover new AMPs and study the amino acid composition of newly discovered AMPs. The quantitative detection of AMPs is mainly performed using LC−MS/MS. In this review, studies on the determination of AMPs through LC−MS over the past five years are summarized in Table 1.
Table 1.
General characteristics of AMPs detected by mass spectrometry in the most recent application.
| Type of Matrix | Pretreatment methods | Mobile phase | Column | Source | Mass Analyzer | Mass Spectrometry Technique |
Country | Year | References |
|---|---|---|---|---|---|---|---|---|---|
| Insects hemolymph | SDS-PAGE, LP-SEC, Acetone Protein precipitation, trypsin digestion | – | a glass column (150 mm*20 mm) | ESI+ | Aligent TOF/Q-TOF | FSCAN | India | 2022 | [124] |
| Insect hemolymph | Organic solvent extraction, followed by size exclusion RPLC and SDS-PAGE | 50% acetonitrile containing 0.05% TFA | ZORBAX‐XDB‐C18 analytical (4.6 × 250 mm) column (Agilent) | MALDI | MALDI‐TOF | – | India | 2021 | [125] |
| Food | Protein precipitation, Protease digestion, Ultrafiltration, HPLC, gel filtration chromatography | A, 100% acetonitrile; B, was water with 0.1% (m/v) trifluoroacetic acid (TFA) |
A C18 semi-prep column (10 × 250 mm, YMC, Japan) | ESI+ | QExactive (Thermo Fisher Scientific, USA) | FSCAN | China | 2019 | [126] |
| Human plasma or urine | SPE (Oasis WCX cartridge) | A, 0.2% FA in water; B, 0.2% FA in acetonitrile | Acquity UPLCR HSS T3 (2.1 mm × 100 mm, 1.8 μm) | ESI | AB 5500 triple quad MS | MRM | China | 2020 | [127] |
| Mouse plasma | Carboxylic acid magnetic bead affinity capture | A, 0.1% DFA (Difluoroacetic acid) in water;B, 0.1% DFA in acetonitrile | Waters 2.1 × 100 mm, 3.5 μm XSelect Peptide CSH C18 column | – | TSQ Quantiva triple Quad MS (Thermo Scientific) | SRM | USA | 2020 | [128] |
| DBS | Liquid liquid extraction, filtration | A, 0.2% FA in acetonitrile; B, 0.2% FA in water | XB-C18 (100 mm × 2.1 mm I.D., 2.6 μm) column Phonomenx Kinetex | ESI+ | 4500 Triple Quad MS | MRM | China | 2022 | [129] |
| Lactobacillus | Precipitation at 80% saturation of ammonium sulfate, separated with ultrafiltration, RPC |
A, 1% MeOH, 0.1% FA in pure water; B, 99.9% AcN, 0.1% FA in pure water | XP C18 trap column (3 μm, 120A 350 μm 0.5 mm) and a 3C18-CL-120 separation column (3 μm, 120A, 75 μm 150 mm) | Nano spray ESI+ | Triple TOF 5600+ MS | MRM | Russia | 2020 | [111] |
| Mouse serum and epithelial lining fluid | Protein precipitation | A, 0.1% formic acid in water; mobile phase B, 100% acetonitrile | Acquity UPLC HSS C18 column (100 mm × 2.1 mm internal diameter, 1.7 mm) | ESI+ | API 5500-Qtrap Triple Quad MS | MRM | USA | 2013 | [130] |
| Cation-adjusted Mueller-Hinton broth (CAMHB), human and rat plasma | Protein precipitation (vortexing, sonicating, and re-vortexing, followed by centrifugation) | A, water with 0.5% FA and 0.01% TFA; B, 50/50 ACN/methanol with 0.5% FA and 0.01% TFA | Waters XTerra MS™ column (C18 3.5 μm, 2.1 mm × 100 mm); Waters XSelect CSH™ column (C18 3.5 μm, 2.1 mm × 100 mm) | ESI+ | API 3000 Triple Quad MS | MRM | USA | 2017 | [131] |
| Goat milk | Protein precipitation | A, 0.1% (v/v) formic acid; B, 0.1% formic acid and 2% water in Optima acetonitrile | C-18, 3 μm nano-trap column (Bruker magic C18AQ, 0.1 × 150 mm, 3 μm partials size, and 200 A° pore size) | Captive spray + | Impact II QTOF | FSCAN | India | 2022 | [123] |
| Fish mucus | Acid solvent extraction, RP-HPLC | A, 0.1% formic acid B, acetonitrile |
C4 column (Vydac, 300A, 5 μm, 4.6 × 250 mm) | ESI+ | LC-MS/MS-QTOF | FSCAN | Tunisia | 2023 | [132] |
| Sea bass mucus | SPE | A, formic 5 acid in water (pH 2.55; B, and acetonitril | analytical C4 column (Vydac, 300A, 5 μm- 4.6 × 250 mm) | ESI+ | Agilent MSD 1946B Simple Quad MS | FSCAN | Tunisia | 2013 | [120] |
| Bacterial isolates | Trypsin digestion and Centrifugal filtration | A, 0.1% FA, 5% ACN, and 95% H2O; B, 0.1% FA, 95% ACN, and 5% H2O | C18, Agilent catalog no. G4240-62010 (160 nl, 150 mm) | ESI+ | LC-MS/MS Orbitrap Fusion/spectrometer Thermo Fisher | MRM | USA | 2019 | [122] |
| polymyxin B products | Centrifugal protein precipitation | A, water with 0.5% FA and 0.01% TFA; B, 50/50 ACN/methanol with 0.5% FA and 0.01% TFA | C18, 3.5 μm, 2.1 mm × 100 mm | ESI+ | API 3000 Triple Quad MS | MRM | USA | 2018 | [133] |
| Human plasma and epithelial lining fluid | Protein centrifugation precipitation | A, 0.1% (v/v) FA in water; B, 0.1% (v/v) FA in acetonitrile | C18 column (50 × 2.1 mm, 3 μm) | ESI+ | 4500 Triple Quad MS | MRM | China | 2023 | [134] |
| Ligilactobacillus salivarius cell-free supernatants | Protein centrifugation precipitation and RP-FPLC | A, 0,1% AF in H2O; B, 0.1% AF in ACN | C18 Picofrit column (Easy Spray Column, PepMap RSLC C18n) | ESI | Q-Exactive HF high-resolution MS | FSCAN | Spain | 2023 | [135] |
| The strain Bacillus amyloliquefaciens Ba168 | Protein centrifugation precipitation, trypsin digested, desalted on C18 containers , centrifuged, and concentrated |
A, 0.1% formic acid; B, 84% acetonitrile and 0.1% formic acid | C18 reversed-phase analytical column (10 cm length, 75 μm inner diameter, 3 μm resin) | ESI+ | Q Exactive MS | – | China | 2022 | [136] |
| Moss Physcomitrella | Homogenized, gel filtration, Protein precipitation and SPE on reverse-phase DSC18 cartridges | A, 0.1% (v/v) formic acid; B, 80% acetonitrile, 0.1% for mic acid | C18 trap column (3 μm, 120 Å, 350 m × 0.5 mm) and C18 column with a diameter of 75 μm (3 μm, Acclaim) | NanoSpray III ESI+ | QTRAP 4500 triple quadrupole MS | MRM | Russia | 2019 | [137] |
| Saccharomyces cerevisiae autolysate | Homogenized, protein precipitation, filtered, SDS-PAGE, FPLC Gel Filtration | A, water; B, acetonitrile | C18-AQ 3 μm column (15 cm length, 75 μm inner diameter) | ESI+ | Quadrupole-Orbitrap Q-Exactive Plus MS | FSCAN | Brazil | 2022 | [138] |
| Polymyxin B products | Centrifugation precipitation, SPE | acetonitrile:water:formic acid (30:70:0.1%; v/v/v) | C18 (1.8 μm, 2.1 mm i.d. x 50 mm) | ESI+ | Alliance e2695 Quattro triple-quadrupole MS | MRM | Kuwait | 2020 | [117] |
| Rubing cheese | Homogenization, Centrifugation precipitation, gastric trypsin digestion, Ultrafiltration | A, 0.1% formic acid in water; B, 0.1% formic acid acetonitrile solution | RP C18 (50 μm × 15 cm) | – | Q Exactive™ Plus-Orbitrap mass spectrometer | FSCAN | China | 2022 | [118] |
| Egg white filtrate | Homogenization, ultrafiltration, nanofiltration and centrifugation | A, 2% v/v acetonitrile, 0.08% v/v formic acid and 0.01% v/v TFA in deionized wate; B, 95% v/v acetonitrile, 0.08% v/v formic acid, and 0.01% v/v TFA in deionized water | concentrated on a C18 column (5 μm 100 Å pore, 300 μm i.d., 5 mm length; peptide separation on a C18 column (3 μm, 100 Å pore, 75 μm i.d., 150 mm length) |
Nano spray ESI+ | Q Exactive MS | – | U.K | 2021 | [115] |
| Seahorse peptide extracts | Homogenization, sonication pulses, centrifugation, filtration, reduction and alkylation reaction | 5%–38% ACN in 0.1% formic acid | ReproSil-Pur C18-AQ 3 μm resin (Dr. Maisch GmbH HPLC) | – | Fusion Lumos Orbitrap MS | FSCAN | Brazil | 2023 | [139] |
| Lactiplantibacillus plantarum B21 | concentration, n-butanol extraction and cation exchange chromatography | A, 0.1% (v/v) formic acid; B, 3% (v/v) ACN | C18 analytical column (length 15 cm, 5 μm) | ESI+ | LTQ™ Orbitrap Elite ETD | FSCAN | Australia | 2020 | [116] |
| Crude cheese whey | Protease digestion. filtration and centrifugation | A, 10 mM HCl; B, 10 mM HCl containing 80 mL 100 mL-1 acetonitrile | Inertsil ODS-3 (4.6 mm × 250 mm) | ESI+ | LCQ or LCMS 8040 | – | Japan | 2019 | [140] |
| Rhipicephalus microplus saliva | SDS-PAGE, Protease digestion and ultrafiltration centrifugation | A, 0.1% formic acid; B, 0.1% formic acid, 84% acetonitrile | C18 column (75 μm × 100 mm, 3 μm) | Nanocaptive spray ESI+ | Q Exactive MS | FSCAN | China | 2019 | [141] |
| Paenibacillus peoriae IBSD35 culture broth | 70% ammonium sulfate enriched, distilled water and desalted, centrifugation, and column chromatography, RP-HPLC, digests | A, 5% acetonitrile, 0.1% formic acid; B, 95% acetonitrile, 0.1% formic acid | 15 cm PicoFrit column (360 μm outer diameter, 75 μm inner diameter, 10 μm tip) filled with 1.9 μm of C18-resin | Nanocaptive spray ESI+ | Q Exactive MS | FSCAN | India | 2021 | [142] |
| B. velezensis 9D-6, liquid LB culture metabolites | liquid/liquid extraction, C18 RP-HPLC | A, 0.1% formic acid; B, acetonitrile | C18 RHLC on a 1260 Infinity Series | – | Q-Exactive Quadrupole-Orbitrap MS | FSCAN | Canada | 2019 | [143] |
| Chicken Muscle and Egg | Homogenization, centrifugation, filtration | A, 1% formic acid in water; B, 1% formic acid in acetonitrile | C18 (100 × 2.1 mm, 1.9 μm) | ESI+ | TSQ Quantis triple quadrupole MS | SRM | India | 2021 | [144] |
| Egg yolk hydrolysate | liquid/liquid extraction, Protease digestion | A, 0.05% (v/v) trifluoroacetic acid B, 0.05% trifluoroacetic acid in acetonitrile | C18 column, C18AQ-15 mm, 30 × 250 mm | CaptiveSpray+ | micro-TOF-Q II MS | SRM | Thailand | 2023 | [145] |
| Metabolites in the culture supernatants of Entomopathogenic bacterial symbiont | Centrifugation and filtration | A, 0.1% trifluoroacetic acid in water; B, acetonitrile | C18 analytical column,5 μm, 4.6 mm by 150 mm | ESI+ | Agilent 6120 quadrupole MS | – | France | 2022 | [146] |
| Peptides R9, penetratin | Homogenization, SPE | A, H2O/0.1% FA; B, ACN/0.1% FA | HSS T3 (2.1 × 50 mm, 1.8 μm) column | ESI+ | AB/Sciex 5500 or 6500 triple quadrupole system | MRM | USA | 2019 | [119] |
| Buthus occitanus crude venom | centrifugation precipitation, filtration, reduction and carboxamido methylated | A, [0.1% (v/v) formic acid; B, acetonitrile (ACN) and 0.1% (v/v) FA | C4 PepMap 300 particles with 5 μm size and 300 A pores) | Spray+ | Orbitrap Fusion MS | FSCAN | Morocco | 2021 | [147] |
| Skin secretion of the fungoid frog | RP-HPLC | A, water with 30% acetonitrile and 0.1% TFA | C-18 analytical column (Vydac 238 TP, 150 × 4.6 mm) | Nanocaptive spray ESI | SYNAPT G2 High Definition MS | – | India | 2018 | [148] |
| Fungus culture medium | Protein extraction, precipitation, chromatographic column separation | A, 0.1% formic acid in water; B, 0.1% formic acid in acetonitrile | C-18 analytical column (75 μm internal diameter and 70 cm in length) | Nanocaptive spray ESI | Q-Exactive Plus MS | FSCAN | USA | 2021 | [112] |
| Chinese yogurt | Ammonium sulfate precipitation or Organic solvent extraction, RP-HPLC | A, (2% acetonitrile, and 0.1% Formic acid; B, 80% acetonitrile, and 0.1% Formic acid | C18 reverse-phase column (Column, 10 cm long, 75 μm inner diameter, 3 μm resin) | ESI | Q Exactive MS | – | China | 2023 | [149] |
| Tree frog skin secretion | desalted and concentrated | A, 0.1% FA; B1, 80% ACN in 0.1% FA | C-18 column (15 cm × 50 μm Acclaim) | – | Q Exactive MS | FSCAN | Brazil | 2021 | [150] |
| Pichia pastoris | precipitated with ammonium sulfate, desalted by gel filtration, cation exchange, RP-FPLC | – | Ziptips C18 (Millipore) | ESI+ | triple quadrupole/ion trap MS 5500 QTRAP | MRM | Spain | 2019 | [151] |
| skin mucus of greater amberjack | Protein precipitation, 2-D Clean-Up Kit, SDS-PAGE | A, 0.1% formic acid; B, 80% acetonitrile, 0.1% formic acid | 300 μm × 5 mm Acclaim Pepmap precolumn (Thermo Scientific) | Nanocaptive spray ESI+ | Orbitrap Fusion MS | – | Spain | 2021 | [152] |
| Human Gut Bacterium Blautia obeum A2-162 | Immunoprecipitation, bound peptides eluted | A, water/0.1% formic acid; B, acetonitrile | C18 LC Column (C18, 2 μm, 500 × 0.75 mm) | Nanocaptive spray ESI+ | Orbitrap Fusion MS | – | UK | 2018 | [153] |
| Leptodactylus latrans Amphibian Skin Secretions | Lyophilized and resuspended | A, 0.1% formic acid in water; B, 0.1% formic acid in acetonitrile (ACN) | C18 column (75 μm × 10 cm, 1.7 μm BEH column) | Nanocaptive spray ESI+ | LTQ OrbitrapVelos MS | FSCAN | Argentina | 2018 | [154] |
| Animal tissues | Centrifugation, sonication, HLB SPE, HPLC-UV | A, ACN; B, 50% ACN in water; C, 50% ACN in water containing 0.02% FA | column (50 mm × 2.1 mm i.d., 2.6 μm) | ESI+ | Triple Quad 5500 triple-quadrupole MS | MRM | China | 2022 | [155] |
| Crude bacteriocins from both cell culture supernatant | Ammonium sulfate precipitation | – | C18 analytical column (180 mm length × 76 μm i.d., 3 μm) | – | Q Exactive MS | (DDA mode) | China | 2018 | [156] |
| Peptide Production from Donkey Milk by Selected Strains | centrifugation, filtration, SPE | A, 0.1% (v/v) formic acid in water; B, 0.1% (v/v) formic acid in acetonitrile | C18 column (0.5 × 100 mm, 2.7 μm) | DuoSpray Ion Source | 5600+ TripleTOF MS | (DDA) | Italy | 2021 | [157] |
| Moringa oleifera seeds | freeze-dried, Centrifugation, filtration | A, 2 % acetonitrile (containing 0.1 % formic acid) in water; B, 80 % acetonitrile (containing 0.1 % formic acid) in water | C18 column (75 μm × 250 mm | ESI+ | Q-Exactive MS | – | China | 2023 | [158] |
| plasma samples | HLB SPE | A, 0.1% formic acid in distilled water (v/v); B, 0.1% formic acid in acetonitrile (v/v) | XBridgeTM HILIC 3.5 μm 3 × 150 mm column | ESI+ | LCMS-8040 triple quadrupole MS | MRM | Thailand | 2020 | [159] |
| Ascidian tunic | Homogenized, centrifuged and digested with enzymes | A, 0.1% FA; B, 80% ACN | EASY-Spray column (ES800, PepMap RSLC, C18, 3 μm) | ESI+ | Q-Exactive Hybrid Quadrupole-Orbitrap MS | FSCAN | Portugal | 2020 | [160] |
| Fungal rice cultures | Sonication, centrifugation and filtration | A, 5 mM ammonium formate and 0.1% (v/v) formic acid water; B, 5 mM ammonium formate and 0.1% (v/v) formic acid in methanol | BEH C18 column (100 × 2.1 mm i.d., 1.7 μm particle size) | ESI | TQD triple quadrupole MS | – | Poland | 2020 | [161] |
| Fungal rice cultures | Centrifugation and filtration | A, 5 mM ammonium formate; B, (5 mM of ammonium formate in MeOH/water, 95:5, v/v) | C18 column (75 × 2.1 mm; 2.6 μm particles) | – | Q-Exactive Fourier-transform high-resolution | FSCAN | Poland | 2020 | [161] |
| Chicken Plasma | Lyophilized, centrifugation, gel filtration and reverse-phase chromatography | A, 0.1% formic acid in water; B, 0.1% formic acid in 80% acetonitrile | C18 column (2 μm, 100 Å, nanoViper, 75 μm i.d. × 15 cm) | Nanocaptive spray ESI+ | Hybrid quadrupole Q-Tof MS | – | Thailand | 2022 | [162] |
| Plasma | Ultrafiltration, centrifugation | – | – | ESI | Agilent Technologies 6490 Triple Quad | – | Belgium | 2019 | [163] |
| Canine Saliva | Centrifugation, digested | A, formic acid (0.1% v/v); B, acetonitrile with formic acid (0.1% v/v) | 150 mm Acclaim PepMap100 C18 column | – | LTQ-Orbitrap electron-transfer dissociation (ETD) | FSCAN | UK | 2022 | [164] |
| Serum samples | Protein precipitation | A, water with 0.1 % (v/v) formic acid; B, acetonitrile | column (ShimPack GIST-HP 150 mm × 3.0 mm) | ESI+ | AB Sciex Qtrap 3200 | MRM | Russia | 2021 | [165] |
| Octopus Skin Mucus |
Protein centrifugation precipitation, SDS-PAGE, trypsin digested | A, 0.1% FA; B, 98% acetonitrile (98% ACN) with 0.1% FA | column (EASY-Spray column, 50 cm × 75 μm ID, PepMap C18, 2 μm particles) | ESI+ | LTQ-Orbitrap-Elite MS | – | Spain | 2023 | [166] |
| Speckled anemone | Homogenization, centrifugation, sonication and filtration | A, 0.1% FA; B, 80% acetonitrile, 0.1% FA | Thermo RSLC pepmap100, 75 μm id, 100 Å pore size, 50 cm reversed-phase nano column | – | Q-Exactive™ Orbitrap MS | FSCAN | Australia | 2020 | [167] |
| Octopus ink sac | Homogenization, centrifugation, SDS-PAGE, trypsin digestion, chromatographic separation | A, 0.1% FA; B, 98% acetonitrile (98% ACN) with 0.1% FA | column (EASY-Spray column, 50 cm × 75 μm ID, PepMap C18, 2 μm particles) | ESI+ | LTQ-Orbitrap-Elite MS | – | Spain | 2023 | [168] |
| Burkholderia cepacia Complex | Centrifugation, filtration and digested | A, 0.1% FA, 2% ACN, 98% H2O; B, 0.1% FA, 100% ACN | HALO 2 Peptide ES-C18 2.1 × 100 mm 2 μm | ESI | Agilent ChipCube 6495 Triple Quadrupole MS | MRM | USA | 2020 | [169] |
| Samples from in vitro and in vivo | HLB-SPE | A, acetonitrile containing 0.1% formic acid; B, water with 0.1% formic acid | 2.5 μm, 2.1 mm × 50 mm XBridge™ Ethylene Bridged Hybrid (BEH) column | ESI+ | AgilentVarian 320 triple quadruple MS | MRM | Belgium | 2020 | [170] |
| Mice and human plasma | WCX- SPE | A, water containing 0.05 % formic acid; B, ACN containing 0.05 % formic acid | CSH C18 column, (75 × 3.0 mm, 2.5 μ) | ESI+ | API 5500 Q trap MS | MRM | India | 2019 | [171] |
| Animal feed | HLB-SPE | A, 0.1% formic acid in water; B, 0.1% formic acid in acetonitrile | BEH C18 column (2.1 × 50 mm, 1.7 μm) | ESI | TSQ triple quadrupole MS and Q-ToF MS | MRM | France | 2021 | [172] |
| Trichoderma | cold maceration and liquid extraction | A, 0.1% solution of acetic acid; B, methanol | C18 column (Phenomenex, 250 × 4.6 mm, 5 μm) | ESI+ | microtof II, Bruker Daltonics | – | Brazil | 2023 | [173] |
| Strains culture medium | Organic extraction, HPLC | A, 0.1% FA in ACN solution; B, 0.1% FA in water solution | Phenomenex Kinetex Biphenyl column (50 mm × 2.1 mm i.d., 2.6 μm) |
ESI+ | Triple Quad 5500 MS | MRM | China | 2022 | [155] |
Note: "-" indicates not described.
Abbreviations: SELDI-TOF, Surface-enhanced laser desorption/ionization time-of-flight; IDM, Interaction discovery mapping; RP‐HPLC, Reverse‐phase high‐pressure liquid chromatography; DBS, Dried blood spots; SPE, Solid phase extraction; MALDI, Matrix-assisted laser desorption/ionization; FSCAN, Full scan; MRM, Multiple reaction monitoring; SRM, Selective reaction monitoring; DDA, Data-dependent acquisition; LTQ, Linear trap quadrupole; LP-SEC: Low-pressure size exclusion chromatography; SDS-PAGE: Sodium dodecyl sulfate−polyacrylamide gel electrophoresis; IEC: Ion exchange chromatography; FPLC: Fast-performance liquid chromatography.
3.2.2. Data analysis
Three commonly used methods exist for the analysis of MS data. The first is the database search [[151], [156], [168]], which involves comparing theoretical enzyme digestion with theoretical fragments to identify peptides that match the MS spectrum. The identification information obtained through this method is derived from the sequences generated by known coding regions in the database. Cochet, MF et al. (2021) [115] used the database to match the mass spectra for data analysis, identifying peptides from the MS/MS spectra using the X!Tandem pipeline software [Plateforme d’Analyze Proteomique de Paris Sud-Ouest (PAPPSO) at INRAE, Jouy-en-Josas, Fragnce; http://pappso.inra.fr]. The search was conducted against a database consisting of reviewed proteins of Gallus gallus, which included 2262 proteins downloaded from the common Repository of Adventitious Protein (http://thegpm.org/crap). The database search parameters were defined as follows: non-specific enzyme cleavage was employed, allowing a mass error of 0.05 Da was allowed for fragment ions and 10 ppm for parent ions. Putative modifications considered in the search included methionine oxidation and serine phosphorylation. Furthermore, a minimum score corresponding to an e-value below 0.05 was deemed necessary for the accurate identification of peptides.
The second method is de novo sequencing [174,154], which is a type of secondary spectrum that doed no rely on the database for directly deducing suitable peptides matching. It addresses the challenge of parsing peptides that are not present in the database, such as peptides produced by variable bases in non-coding regions. The de novo sequencing steps for AMPs analysis included mass spectrum data acquisition, peak identification, mass-to-charge ratio extraction, fragment spectrum extraction, fragment spectrum analysis, deduction of amino acid sequence, generation of candidate sequence, evaluation of candidate sequence, and verification of the sequence. De novo sequencing is a complex task that dependent on instrumental resolution, data quality, and analyst experience. Professional mass spectrophotologists usually possess extensive experience in this area to ensure that the inferred sequence is accurate and reliable. It can be combined with other experimental techniques and database information to improve the reliability of amino acid sequence prediction. Grady, EN et al. (2019) [143] reported the utilization of MS/MS spectra for de novo sequencing of peptide-containing compounds to validate potential compounds identified using Antibase.
The third method is Spectrum library search [139,142], which involves is the rapid detection and identification of known peptides in the sample, followed by obtaining by pre-defined peptide screens through spectrum search. Feng, LL et al. (2019) [141] reported that LC–MS/MS data obtained in its raw form underwent a search using the Mascot engine v.2.2 (Matrix Science, London, UK) against established protein libraries that have been previously published. Proteins were obtained from the Uniprot database (Uniprot_Ixodidae_80119_20180302.fasta; 80119 protein sequences were downloaded on March 02, 2018). The Mascot search parameters utilized were as follows: trypsin was used as the enzyme, with a maximum of two allowed missed cleavages; peptide mass tolerance was set at 20 ppm; and fragment mass tolerance was set at 0.1 Da. Fixed modifications included carbamidomethyl (C), iTRAQ8plex (N-term), and iTRAQ8plex (K). Peptides were considered identified when the false discovery rate (FDR) was below 0.
With the rapid development of instrumental technology and the combined application of multi-omics techniques, the scale and complexity of data are increasing. Faced with of the challenge of processing and analyzing high-throughput protein and peptide MS data, many comprehensive data analysis software platforms are available to assist us in the efficient and time-saving analysis and processing of such data. PEAKS Studio software (Waterloo, ON, Canada) is commonly used for data acquisition and analysis [[145], [175], [176], [177], [178]]. Additionally, software such as Proteome Discoverer [179,160], Mascot software [180,157], and ProteinLynx Global SERVER [[148], [181], [182]] are widely used in protein polypeptide analysis.
3.3. Application of MS in the functional research of AMPs
The role of mass spectrometers in the field of AMP research is primarily focused on the following aspects.
3.3.1. Discovery and characterization of new AMPs from natural sources
Over the past few decades, a variety of AMPs have been found, with peptides exhibiting similar properties were extracted from diverse organisms, including mammals, insects, birds, fish, and plants. Due to their low concentration in bodily fluids, a comprehensive purification approach is necessary for obtaining pure peptides. The limited quantity of peptides obtained necessitates the use of highly sensitive analytical techniques like MS for sequencing purposes, making MS one of the main methods for their structure identification [183,184]. Liu, J et al. (2017) [183] outlines the basic steps of an accurate mass determination method in the positive ion mode of offline low-energy CID ESI Qq-TOF MS/MS. This method is commonly used for de novo sequencing of peptides to elucidate the structure of AMPs. Rodriguez, A et al. (2023) [185] performed peptide sequencing analysis with NanoLC-ESI−MS/MS for peptides from the marine shellfish. Punginelli, D et al. (2023) [186] demonstrated that nine new peptides were further identified through high-resolution MS analysis and database searches. Two synthetic peptides, one from a green leaf and one from a rhizome of Oceania, were identified and showed anti-biofilm activities against Staphylococcus aureus, Escherichia coli and Pseudomonas aeruginosa.
Research in this area uses MS-based screening approaches to identify new AMPs from natural sources, such as plants, animals, and microbes. This represents the most common approach currently employed in studies applying MS to detect AMPs.
3.3.2. Quantitative analysis of AMPs
Research in this area is dedicated to developing methods for quantifying AMPs accurately in complex samples, including biological fluids and tissues. The goal is to identify potential biomarkers for infectious or inflammatory diseases by measuring AMP levels under different conditions. Wang, Y et al. (2023) [187] have effectively developed a method using UPLC−MS/MS for the precise measurement of colistin levels in both plasma and kidney tissue samples. This method has been successfully used to analyze the pharmacokinetics of colistin sulfate in rats, and the relationship between the renal accumulation of colistin and the administration time has been explored.
However, several challenges persist. First, the variability of sample preparation and the cumbersome nature of sample pretreatment before MS quantitative detection are notable. To achieve a specific detection concentration and purity of the target substance, pretreatment conditions should be explored before applying each quantification method [188,189]. Second, the detection range of MS is limit despite its high sensitivity and specificity [[171], [190], [191]]. After establishing the methodology, the performance of the linear detection range was verified. He, J et al. (2013) [130] established an MS method for the quantification of polymyxin B in mouse serum, demonstrating a linear concentration range of 0.0065–3.2 mg/L. Third, the complexity of the sample matrix is a critical consideration. A diverse range of biological samples, such as blood, urine, cerebrospinal fluid, skin mucus, and tissue, are used for the extraction of AMPs. Numerous substances present in these biological samples can exert matrix effects on the target substances. These matrix effects pose challenges to the direct analysis of MS, particularly when conducting prompt quantitative investigations [192]. In a study conducted by Song et al. (2019) [193], various cyclic polypeptide antibiotics exhibited significant matrix effects in different feeds, resulting in signal inhibition up to 50%. Matrix effect studies constitute one of the crucial factors in the development of various MS methodologies [194,129].
3.3.3. Localization of AMPs in tissues and cells
Mass spectrometry imaging (MSI) is an emerging technology with substantial potential in biomedicine, including advancements in biomarker discovery, metabolomics studies, drug applications, and clinical diagnostics [195,196]. Considerable achievements have been realized in using MSI to visualize the distribution of AMPs in tissues and cells.
Lu, J et al. (2023) [197] performed MSI [MALDI-7090 tandem time-of-flight (TOF)-MS] analysis of rat spinal cord 1 h after polymyxin B was administered via intraventricular injection at a clinically relevant dose of 0.5 mg/kg. MSI of brain slices revealed a distinct distribution pattern of polymyxin B, with most of it clustered in the lateral ventricle on the injection side and subsequently into the third and fourth ventricles. No polymyxin B was detected in the left ventricle and brain tissue. This mechanistic understanding, combined with pharmacokinetic/pharmacodynamic drug delivery strategies, will provide valuable insights for the development of secure and efficient intraventricular/intrathecal polymyxin regimens for treating central nervous system (CNS) infections caused by MDR gram-negative bacteria. Nilsson, A et al. (2015) [198] used these techniques to measure the distribution of polymyxin drugs and their metabolites. MSI provides a powerful alternative for tissue homogenization analysis and labeling or antibody imaging, enabling simultaneous detection of the spatial distribution of drugs and drug metabolites. Gonzalez, DJ et al. (2012) [199] identified novel phenol-soluble regulatory protein derivatives in methicillin-resistant Staphylococcus aureus using MSI. This work suggests that MSI can be used as a useful tool to extend our understanding of an important family of small peptide virulence factors.
3.3.4. Development of AMP-based therapies
This line of research focuses on using MS to optimize the pharmacokinetics and efficacy of AMP-based therapies. The rapid detection of AMPs in body fluids through LC−MS/MS can be used for the monitoring of therapeutic drugs. The application of AMPs in therapeutic monitoring primarily involves studying antibacterial activity against different pathogens in vitro.
Wang, X et al. (2023) [158] successfully isolated a novel AMP, named MCNDCGA peptide (also referred to as MOp3), from the seeds of Moringa oleifera. This peptide exhibited a significant inhibitory effect against Staphylococcus aureus, with a minimum inhibitory concentration (MIC) of 2 mg/mL. Parra, ALC et al. (2022) [200] investigated AMPs from ornamental tobacco floral nectar, selecting six peptides for additional characterization, synthesis, and assessment of their antimicrobial efficacy against plant pathogenic fungi and bacteria. The results demonstrated that all six peptides exhibited certain antibacterial activity. Cuesta, SA et al. (2021) [201] reported two novel crizole septin-16 and septin-17 peptides. These peptides, initially discovered through molecular cloning and MS/MS, were synthesized using solid-phase peptide synthesis. Their MICs and hemolytic activity were subsequently assessed through experimental testing. The results showed that crizocseptin exhibited antibacterial activity against Escherichia coli, Staphylococcus aureus, and Candida albicans, with low hemolytic effects. Polymyxin is predominantly used in the treatment of MDR infections in clinical practice, with numerous studies focusing on the detection of polymyxin through LC−MS/MS [111,[127], [131], [134]].
The objective is to explore the structure and activity of the new AMPs and to identify potential therapeutic applications.
Overall, research on AMPs through MS involves a range of techniques, including LC, MS/MS, MALDI, imaging, and other advanced MS-based approaches. The specific type of research will depend on the specific questions being addressed and the available resources. In this review, we provide a comprehensive summary of the studies conducted on the detection of AMPs using MS, encompassing different types and species (Table 1).
4. Limitations and future perspectives
4.1. Challenges in the application of MS for the detection of AMP
Currently, the application of LC−MS/MS in AMPs research primarily centers on the discovery of new AMPs, quantification, and antibacterial activity detection. The mass of peptides required to assess AMP mechanisms, structures, and functions may vary from a few micrograms to many milligrams. Therefore, a key challenge in AMP research lies in optimizing extraction or production methods to effectively fulfill the requirements for characterization and functional exploration. simultaneously, the selection of extraction methods and solvents during sample pretreatment and extraction greatly influences the amount and activity of AMPs, posing another research challenge. Researchers must tailor pre-treatment and extraction conditions according to the properties of the respective AMP and subsequently choose the appropriate method.
The application of MS technology to investigate the localization of AMPs in tissues or cells represents a new research hotspot. MSI technology facilitates the localization and semi-quantitatively analysis of AMP expression and its interaction with other biomolecules. While the development of MSI technology brings new prospects, it also introduces challenges for researchers. On the one hand, a higher level of professional and technical expertise is required among scientific research staff. On the other hand, research on MS equipment and auxiliary tools demands elevated standards, entailing substantial investments in equipment and space.
4.2. Prospects of MS applied to AMP detection
Recent studies have demonstrated the utilization of "applied proteomics" approaches to investigate the diverse biological activities of AMPs. Digital mining using proteomics technology combined with bioinformatics analysis has revealed the medicinal value of various AMPs. To enhance the practical application of AMPs, a comprehensively analysis of their societal cost-effectiveness is essential. Studies have identified several AMPs with cytolytic activities. The discovery and development of AMPs possessing biological functions and devoid of toxic side effects in the human body for clinical application necessitate extensive clinical experiments for validation. Future research endeavors can focus on innovating novel AMPs, considering genetic engineering techniques focus on synthesis of high-value AMPs for biomedical applications. Furthermore, given recent advancements in precision genome editing technology, the synthesize of AMPs with no cytotoxicity becomes feasible, offering a potential avenue for treating infectious diseases and addressing the global drug resistance problem. With the in-depth study of AMPs, the effect of AMPs on the human immune system has emerged as a key research focus. In the future, the integration of MS/MS technology with other immunodetection technologies, such as immunoassay, can enhance the dimension and depth of AMPs research.
5. Conclusion
MS holds immense potential in the study of protein peptides due to its high sensitivity and specificity, particularly with the recent advancements in high-resolution MS technology. It not only facilitates peptide isolation but also serves the purpose of species identification. The capability to analyze multiple samples simultaneously contributes time and cost savings, overcoming many limitations associated with alternative technologies.
The review aimed to comprehend the methodological studies on the detection of AMP using MS/MS technology, encompassing the entire operation cycle from sample separation to MS identification. Despite sincere efforts to summarize comprehensive information and laboratory processes experience, this review is not exhaustive. Given the complexity and diversity of AMP sources, which present additional challenges to the technical application of MS/MS, a truly comprehensive review is nearly impossible. However, the content is anticipated to be sufficient to provide insights for the future stud on AMPs.
Funding statement
This work was supported by the Key Research, Development, and Promotion Projects of Henan Province (222102310171, 222102310328 and 222102310067), and Medical Science and Technology Project of Henan Province (LHGJ20210636 and LHGJ20200607).
Ethics declarations
Review and approval by an ethics committee was not needed because this review article was prepared using already available references and does not involve new human and animal experiments.
Data availability statement
Data sharing is not applicable to this article as no new data were created or analyzed in this review article.
CRediT authorship contribution statement
Panpan Fang: Writing – original draft, Project administration, Funding acquisition, Data curation, Conceptualization. Songlin Yu: Writing – review & editing, Software, Project administration, Methodology, Formal analysis. Xiaoli Ma: Writing – original draft, Visualization, Validation, Resources, Investigation, Data curation. Lian Hou: Supervision, Resources, Methodology. Tiewei Li: Validation, Resources, Investigation, Data curation. Kaijie Gao: Investigation, Funding acquisition, Data curation. Yingyuan Wang: Resources, Project administration, Methodology. Qianqian Sun: Visualization, Validation, Software. Lujun Shang: Resources, Investigation. Qianqian Liu: Visualization, Methodology. Manjie Nie: Supervision, Investigation. Junmei Yang: Writing – review & editing, Project administration, Funding acquisition, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e28484.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Golli A.L., Cristea O.M., Zlatian O., Glodeanu A.D., Balasoiu A.T., Ionescu M., Popa S. Prevalence of multidrug-resistant pathogens causing Bloodstream infections in an intensive care unit. Infect. Drug Resist. 2022;15:5981–5992. doi: 10.2147/IDR.S383285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ren Y., Chakraborty T., Doijad S., Falgenhauer L., Falgenhauer J., Goesmann A., Schwengers O., Heider D. Multi-label classification for multi-drug resistance prediction of Escherichia coli. Comput. Struct. Biotechnol. J. 2022;20:1264–1270. doi: 10.1016/j.csbj.2022.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hoelscher M.P., Forner J., Calderone S., Krämer C., Taylor Z., Loiacono F.V., Agrawal S., Karcher D., et al. Expression strategies for the efficient synthesis of antimicrobial peptides in plastids. Nat. Commun. 2022;13(1):5856. doi: 10.1038/s41467-022-33516-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lazzaro B.P., Zasloff M., Rolff J. Antimicrobial peptides: application informed by evolution. Science. 2020;368(6490) doi: 10.1126/science.aau5480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spohn R., Daruka L., Lázár V., Martins A., Vidovics F., Grézal G., Méhi O., Kintses B., et al. Integrated evolutionary analysis reveals antimicrobial peptides with limited resistance. Nat. Commun. 2019;10(1):4538. doi: 10.1038/s41467-019-12364-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brogden K.A., Ackermann M., McCray P.B., Jr., Tack B.F. Antimicrobial peptides in animals and their role in host defences. Int. J. Antimicrob. Agents. 2003;22(5):465–478. doi: 10.1016/s0924-8579(03)00180-8. [DOI] [PubMed] [Google Scholar]
- 7.Jhong J.H., Chi Y.H., Li W.C., Lin T.H., Huang K.Y., Lee T.Y. dbAMP: an integrated resource for exploring antimicrobial peptides with functional activities and physicochemical properties on transcriptome and proteome data. Nucleic Acids Res. 2019;47(D1):D285–d297. doi: 10.1093/nar/gky1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maróti G., Kereszt A., Kondorosi E., Mergaert P. Natural roles of antimicrobial peptides in microbes, plants and animals. Res. Microbiol. 2011;162(4):363–374. doi: 10.1016/j.resmic.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 9.Patrzykat A., Friedrich C.L., Zhang L., Mendoza V., Hancock R.E. Sublethal concentrations of pleurocidin-derived antimicrobial peptides inhibit macromolecular synthesis in Escherichia coli. Antimicrob. Agents Chemother. 2002;46(3):605–614. doi: 10.1128/AAC.46.03.605-614.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eckert R. Road to clinical efficacy: challenges and novel strategies for antimicrobial peptide development. Future Microbiol. 2011;6(6):635–651. doi: 10.2217/fmb.11.27. [DOI] [PubMed] [Google Scholar]
- 11.Bucki R., Leszczyńska K., Namiot A., Sokołowski W. Cathelicidin LL-37: a multitask antimicrobial peptide. Arch. Immunol. Ther. Exp. 2010;58(1):15–25. doi: 10.1007/s00005-009-0057-2. [DOI] [PubMed] [Google Scholar]
- 12.Zasloff M. Antimicrobial peptides of multicellular organisms: my perspective. Adv. Exp. Med. Biol. 2019;1117:3–6. doi: 10.1007/978-981-13-3588-4_1. [DOI] [PubMed] [Google Scholar]
- 13.Czaplewski L., Bax R., Clokie M., Dawson M., Fairhead H., Fischetti V.A., Foster S., Gilmore B.F., et al. Alternatives to antibiotics-a pipeline portfolio review. Lancet Infect. Dis. 2016;16(2):239–251. doi: 10.1016/S1473-3099(15)00466-1. [DOI] [PubMed] [Google Scholar]
- 14.Koo H.B., Seo J. Antimicrobial peptides under clinical investigation. Peptide Science. 2019;111 [Google Scholar]
- 15.Magana M., Pushpanathan M., Santos A.L., Leanse L., Fernandez M., Ioannidis A., Giulianotti M.A., Apidianakis Y., et al. The value of antimicrobial peptides in the age of resistance. Lancet Infect. Dis. 2020;20(9):e216–e230. doi: 10.1016/S1473-3099(20)30327-3. [DOI] [PubMed] [Google Scholar]
- 16.Fry D.E. Antimicrobial peptides. Surg. Infect. 2018;19(8):804–811. doi: 10.1089/sur.2018.194. [DOI] [PubMed] [Google Scholar]
- 17.Petrosillo N., Ioannidou E., Falagas M.E. Colistin monotherapy vs. combination therapy: evidence from microbiological, animal and clinical studies. Clin. Microbiol. Infect. 2008;14(9):816–827. doi: 10.1111/j.1469-0691.2008.02061.x. [DOI] [PubMed] [Google Scholar]
- 18.Vriens K., Cools T.L., Harvey P.J., Craik D.J., Spincemaille P., Cassiman D., Braem A., Vleugels J., et al. Synergistic activity of the plant defensin HsAFP1 and caspofungin against Candida albicans biofilms and planktonic cultures. PLoS One. 2015;10(8) doi: 10.1371/journal.pone.0132701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang P., Wang X., Yang X., Liu Z., Wu M., Li G. Budesonide suppresses pulmonary antibacterial host defense by down-regulating cathelicidin-related antimicrobial peptide in allergic inflammation mice and in lung epithelial cells. BMC Immunol. 2013;14:7. doi: 10.1186/1471-2172-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wehkamp U., Jost M., Wehkamp K., Harder J. Dysregulated expression of antimicrobial peptides in skin lesions of patients with cutaneous T-cell lymphoma. Acta Derm. Venereol. 2020;100(1) doi: 10.2340/00015555-3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang R., Ma X., Zou Y., Qiu L., Wang D., Tang Y., Cao Y., Yu S., et al. Total serum vitamin B12 (cobalamin) LC-MS/MS assay as an arbiter of clinically discordant immunoassay results. Clin. Chem. Lab. Med. 2023;61(1):86–92. doi: 10.1515/cclm-2022-0523. [DOI] [PubMed] [Google Scholar]
- 22.Huang H., Lee W.Y., Zou H., Li H., Zhang S., Li H., Lin J. Antimicrobial peptides in Dendrobium officinale: genomic parameters, peptide structures, and gene expression patterns. Plant Genome. 2023 doi: 10.1002/tpg2.20348. [DOI] [PubMed] [Google Scholar]
- 23.Jaleel L.K., Umran M.A., Kaddo K.B.J., Ad'hiah A.H. Evaluation of human β-defensins in the cerebrospinal fluid of suspected meningitis. Biomed Rep. 2023;18(1):10. doi: 10.3892/br.2022.1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shang C., Ye T., Zhou Q., Chen P., Li X., Li W., Chen S., Hu Z., et al. Genome-wide identification and bioinformatics analyses of host defense peptides snakin/GASA in mangrove plants. Genes. 2023;14(4) doi: 10.3390/genes14040923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chandran C., Tham H.Y., Abdul Rahim R., Lim S.H.E., Yusoff K., Song A.A. Lactococcus lactis secreting phage lysins as a potential antimicrobial against multi-drug resistant Staphylococcus aureus. PeerJ. 2022;10 doi: 10.7717/peerj.12648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Danis-Wlodarczyk K.M., Wozniak D.J., Abedon S.T. Treating bacterial infections with bacteriophage-based enzybiotics: in vitro. In Vivo and Clinical Application, Antibiotics (Basel) 2021;10(12) doi: 10.3390/antibiotics10121497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Łusiak-Szelachowska M., Weber-Dąbrowska B., Górski A. Bacteriophages and lysins in biofilm control. Virol. Sin. 2020;35(2):125–133. doi: 10.1007/s12250-019-00192-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mirski T., Lidia M., Nakonieczna A., Gryko R. Bacteriophages, phage endolysins and antimicrobial peptides - the possibilities for their common use to combat infections and in the design of new drugs. Ann. Agric. Environ. Med. 2019;26(2):203–209. doi: 10.26444/aaem/105390. [DOI] [PubMed] [Google Scholar]
- 29.Parisien A., Allain B., Zhang J., Mandeville R., Lan C.Q. Novel alternatives to antibiotics: bacteriophages, bacterial cell wall hydrolases, and antimicrobial peptides. J. Appl. Microbiol. 2008;104(1):1–13. doi: 10.1111/j.1365-2672.2007.03498.x. [DOI] [PubMed] [Google Scholar]
- 30.Scholl D. Phage tail-like bacteriocins. Annu Rev Virol. 2017;4(1):453–467. doi: 10.1146/annurev-virology-101416-041632. [DOI] [PubMed] [Google Scholar]
- 31.Schroven K., Aertsen A., Lavigne R. Bacteriophages as drivers of bacterial virulence and their potential for biotechnological exploitation. FEMS Microbiol. Rev. 2021;45(1) doi: 10.1093/femsre/fuaa041. [DOI] [PubMed] [Google Scholar]
- 32.Tajbakhsh M., Karimi A., Fallah F., Akhavan M.M. Overview of ribosomal and non-ribosomal antimicrobial peptides produced by Gram positive bacteria. Cell. Mol. Biol. 2017;63(10):20–32. doi: 10.14715/cmb/2017.63.10.4. [DOI] [PubMed] [Google Scholar]
- 33.Draper L.A., Ross R.P., Hill C., Cotter P.D. Lantibiotic immunity. Curr. Protein Pept. Sci. 2008;9(1):39–49. doi: 10.2174/138920308783565750. [DOI] [PubMed] [Google Scholar]
- 34.Acedo J.Z., Chiorean S., Vederas J.C., van Belkum M.J. The expanding structural variety among bacteriocins from Gram-positive bacteria. FEMS Microbiol. Rev. 2018;42(6):805–828. doi: 10.1093/femsre/fuy033. [DOI] [PubMed] [Google Scholar]
- 35.Simons A., Alhanout K., Duval R.E. Bacteriocins, antimicrobial peptides from bacterial origin: overview of their biology and their impact against multidrug-resistant bacteria. Microorganisms. 2020;8(5) doi: 10.3390/microorganisms8050639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kyle K.E., Puckett S.P., Caraballo-Rodríguez A.M., Rivera-Chávez J., Samples R.M., Earp C.E., Raja H.A., Pearce C.J., et al. Trachymyrmex septentrionalis ants promote fungus garden hygiene using Trichoderma-derived metabolite cues. Proc. Natl. Acad. Sci. U.S.A. 2023;120(25) doi: 10.1073/pnas.2219373120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bissett J., Gams W., Jaklitsch W., Samuels G.J. Accepted Trichoderma names in the year 2015, IMA fungus. 2015;6(2):263–295. doi: 10.5598/imafungus.2015.06.02.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ayers S., Ehrmann B.M., Adcock A.F., Kroll D.J., Carcache de Blanco E.J., Shen Q., Swanson S.M., Falkinham J.O., 3rd, et al. Peptaibols from two unidentified fungi of the order Hypocreales with cytotoxic, antibiotic, and anthelmintic activities. J. Pept. Sci. 2012;18(8):500–510. doi: 10.1002/psc.2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lam Y.T.H., Ricardo M.G., Rennert R., Frolov A., Porzel A., Brandt W., Stark P., Westermann B., et al. Rare glutamic acid methyl ester peptaibols from sepedonium ampullosporum damon KSH 534 exhibit promising antifungal and anticancer activity. Int. J. Mol. Sci. 2021;22(23) doi: 10.3390/ijms222312718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gao X., Ding J., Liao C., Xu J., Liu X., Lu W. Defensins: the natural peptide antibiotic. Adv. Drug Deliv. Rev. 2021;179 doi: 10.1016/j.addr.2021.114008. [DOI] [PubMed] [Google Scholar]
- 41.Mygind P.H., Fischer R.L., Schnorr K.M., Hansen M.T., Sönksen C.P., Ludvigsen S., Raventós D., Buskov S., et al. Plectasin is a peptide antibiotic with therapeutic potential from a saprophytic fungus. Nature. 2005;437(7061):975–980. doi: 10.1038/nature04051. [DOI] [PubMed] [Google Scholar]
- 42.Bogdanov I.V., Fateeva S.I., Voropaev A.D., Ovchinnikova T.V., Finkina E.I. Immunomodulatory effects of the pea defensin Psd1 in the caco-2/immune cells Co-culture upon Candida albicans infection. Int. J. Mol. Sci. 2023;24(9) doi: 10.3390/ijms24097712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Silva P.M., Gonçalves S., Santos N.C. Defensins: antifungal lessons from eukaryotes. Front. Microbiol. 2014;5:97. doi: 10.3389/fmicb.2014.00097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ma H., Feng Y., Cao Q., Jia J., Ali M., Shah D., Meyers B.C., He H., et al. Evolution of antimicrobial cysteine-rich peptides in plants. Plant Cell Rep. 2023;42(9):1517–1527. doi: 10.1007/s00299-023-03044-3. [DOI] [PubMed] [Google Scholar]
- 45.Srivastava S., Dashora K., Ameta K.L., Singh N.P., El-Enshasy H.A., Pagano M.C., Hesham A.E., Sharma G.D., et al. Cysteine-rich antimicrobial peptides from plants: the future of antimicrobial therapy. Phytother Res. 2021;35(1):256–277. doi: 10.1002/ptr.6823. [DOI] [PubMed] [Google Scholar]
- 46.Tam J.P., Wang S., Wong K.H., Tan W.L. Antimicrobial peptides from plants. Pharmaceuticals. 2015;8(4):711–757. doi: 10.3390/ph8040711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tang S.S., Prodhan Z.H., Biswas S.K., Le C.F., Sekaran S.D. Antimicrobial peptides from different plant sources: isolation, characterisation, and purification. Phytochemistry. 2018;154:94–105. doi: 10.1016/j.phytochem.2018.07.002. [DOI] [PubMed] [Google Scholar]
- 48.Tincu J.A., Taylor S.W. Antimicrobial peptides from marine invertebrates. Antimicrob. Agents Chemother. 2004;48(10):3645–3654. doi: 10.1128/AAC.48.10.3645-3654.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu R., Patocka J., Nepovimova E., Oleksak P., Valis M., Wu W., Kuca K. Marine invertebrate peptides: antimicrobial peptides. Front. Microbiol. 2021;12 doi: 10.3389/fmicb.2021.785085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bazaliński D., Przybek-Mita J., Lisowicz K., Skórka M., Więch P. Defensins of Lucilia sericata larvae and their influence on wound repair processes in practical assessment-A study of three cases. Int. J. Environ. Res. Publ. Health. 2023;20(7) doi: 10.3390/ijerph20075357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bulet P., Stöcklin R., Menin L. Anti-microbial peptides: from invertebrates to vertebrates. Immunol. Rev. 2004;198:169–184. doi: 10.1111/j.0105-2896.2004.0124.x. [DOI] [PubMed] [Google Scholar]
- 52.Zhu S., Gao B. Evolutionary origin of β-defensins. Dev. Comp. Immunol. 2013;39(1–2):79–84. doi: 10.1016/j.dci.2012.02.011. [DOI] [PubMed] [Google Scholar]
- 53.Avila E.E. Functions of antimicrobial peptides in vertebrates. Curr. Protein Pept. Sci. 2017;18(11):1098–1119. doi: 10.2174/1389203717666160813162629. [DOI] [PubMed] [Google Scholar]
- 54.Masso-Silva J.A., Diamond G. Antimicrobial peptides from fish. Pharmaceuticals. 2014;7(3):265–310. doi: 10.3390/ph7030265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sugiarto H., Yu P.L. Avian antimicrobial peptides: the defense role of beta-defensins. Biochem. Biophys. Res. Commun. 2004;323(3):721–727. doi: 10.1016/j.bbrc.2004.08.162. [DOI] [PubMed] [Google Scholar]
- 56.van Hoek M.L. Antimicrobial peptides in reptiles. Pharmaceuticals. 2014;7(6):723–753. doi: 10.3390/ph7060723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ageitos J.M., Sánchez-Pérez A., Calo-Mata P., Villa T.G. Antimicrobial peptides (AMPs): ancient compounds that represent novel weapons in the fight against bacteria. Biochem. Pharmacol. 2017;133:117–138. doi: 10.1016/j.bcp.2016.09.018. [DOI] [PubMed] [Google Scholar]
- 58.Chernomordik F., Cercek B., Zhou J., Zhao X., Lio N.W.M., Chyu K.Y., Shah P.K., Dimayuga P.C. Impaired tolerance to the autoantigen LL-37 in acute coronary syndrome. Front. Immunol. 2023;14 doi: 10.3389/fimmu.2023.1113904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Memariani H., Memariani M. Antibiofilm properties of cathelicidin LL-37: an in-depth review. World J. Microbiol. Biotechnol. 2023;39(4):99. doi: 10.1007/s11274-023-03545-z. [DOI] [PubMed] [Google Scholar]
- 60.Yu S., Zhou W., Yu J., Li M., Zhang S., Jiang X., Wang H., Ma X., et al. An automated magnetic bead extraction method for measuring plasma metanephrines and 3-methoxytyramine using liquid chromatography tandem mass spectrometry. Anal. Bioanal. Chem. 2022;414(11):3541–3549. doi: 10.1007/s00216-022-03984-x. [DOI] [PubMed] [Google Scholar]
- 61.Sampaio de Oliveira K.B., Leite M.L., Rodrigues G.R., Duque H.M., da Costa R.A., Cunha V.A., de Loiola Costa L.S., da Cunha N.B., et al. Strategies for recombinant production of antimicrobial peptides with pharmacological potential. Expet Rev. Clin. Pharmacol. 2020;13(4):367–390. doi: 10.1080/17512433.2020.1764347. [DOI] [PubMed] [Google Scholar]
- 62.Barashkova A.S., Rogozhin E.A. Isolation of antimicrobial peptides from different plant sources: does a general extraction method exist? Plant Methods. 2020;16:143. doi: 10.1186/s13007-020-00687-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Díaz-Puertas R., Adamek M., Mallavia R., Falco A. Fish skin mucus extracts: an underexplored source of antimicrobial agents. Mar. Drugs. 2023;21(6) doi: 10.3390/md21060350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lee Y., Bilung L.M., Sulaiman B., Chong Y.L. The antibacterial activity of fish skin mucus with various extraction solvents and their in-vitro evaluation methods. Int. Aquat. Res. 2020;12(1):1–21. [Google Scholar]
- 65.Wen Q., Zhang L., Zhao F., Chen Y., Su Y., Zhang X., Chen P., Zheng T. Production technology and functionality of bioactive peptides. Curr. Pharmaceut. Des. 2023;29(9):652–674. doi: 10.2174/1381612829666230201121353. [DOI] [PubMed] [Google Scholar]
- 66.Hayashida P.Y., da Silva Junior P.I. Insights into antimicrobial peptides from limacus flavus mucus. Curr. Microbiol. 2021;78(8):2970–2979. doi: 10.1007/s00284-021-02552-3. [DOI] [PubMed] [Google Scholar]
- 67.Dos Santos L.A., Taveira G.B., Ribeiro S.F.F., Pereira L.D.S., Carvalho A.O., Rodrigues R., Oliveira A.E.A., Machado O.L.T., et al. Purification and characterization of peptides from Capsicum annuum fruits which are α-amylase inhibitors and exhibit high antimicrobial activity against fungi of agronomic importance. Protein Expr. Purif. 2017;132:97–107. doi: 10.1016/j.pep.2017.01.013. [DOI] [PubMed] [Google Scholar]
- 68.Monaci L., Quintieri L., Caputo L., Visconti A., Baruzzi F. Rapid profiling of antimicrobial compounds characterising B. subtilis TR50 cell-free filtrate by high-performance liquid chromatography coupled to high-resolution Orbitrap™ mass spectrometry. Rapid Commun. Mass Spectrom. 2016;30(1):45–53. doi: 10.1002/rcm.7408. [DOI] [PubMed] [Google Scholar]
- 69.Claeson P., Göransson U., Johansson S., Luijendijk T., Bohlin L. Fractionation protocol for the isolation of polypeptides from plant biomass. J. Nat. Prod. 1998;61(1):77–81. doi: 10.1021/np970342r. [DOI] [PubMed] [Google Scholar]
- 70.Jiao M., Chen L., He Y., Wu L., Mei H. Identification of proteins in housefly (Musca domestica) larvae powder by LC-MS/MS and their potential medical relevance. RSC Adv. 2019;9(52):30545–30555. doi: 10.1039/c9ra05854k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sable R., Parajuli P., Jois S. Peptides, peptidomimetics, and polypeptides from marine sources: a wealth of natural sources for pharmaceutical applications. Mar. Drugs. 2017;15(4) doi: 10.3390/md15040124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Al-Rasheed A., Handool K.O., Garba B., Noordin M.M., Bejo S.K., Kamal F.M., Daud H.H.M. Crude extracts of epidermal mucus and epidermis of climbing perch Anabas testudineus and its antibacterial and hemolytic activities. The Egyptian Journal of Aquatic Research. 2018;44(2):125–129. [Google Scholar]
- 73.García M., Molina J.P., Sainz J., Santamaría A., Medina S., Aguinaga J. Antibacterial activity evaluation of the Nile tilapia Oreochromis niloticus (Linnaeus, 1758) skin mucus, against Vibrio bacteria affecting the white shrimp Penaeus vannamei. Latin American Journal of Aquatic Research. 2019;47:580–585. [Google Scholar]
- 74.Subhashini S. Screening of antibacterial and cytotoxic activity of extracts from epidermis and epidermal mucus of barbonymus schwanenfeldii (tinfoil barb FISH) Int. J. Renew. Energy Technol. 2013;2:492–497. [Google Scholar]
- 75.Al Akeel R., Al-Sheikh Y., Mateen A., Syed R., Janardhan K., Gupta V.C. Evaluation of antibacterial activity of crude protein extracts from seeds of six different medical plants against standard bacterial strains. Saudi J. Biol. Sci. 2014;21(2):147–151. doi: 10.1016/j.sjbs.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Al Akeel R., Mateen A., Syed R., Alqahtani M.S., Alqahtani A.S. Alanine rich peptide from Populus trichocarpa inhibit growth of Staphylococcus aureus via targetting its extracellular domain of Sensor Histidine Kinase YycGex protein. Microb. Pathog. 2018;121:115–122. doi: 10.1016/j.micpath.2018.05.010. [DOI] [PubMed] [Google Scholar]
- 77.Al Akeel R., Mateen A., Syed R., Alyousef A.A., Shaik M.R. Screening, purification and characterization of anionic antimicrobial proteins from foeniculum vulgare. Molecules. 2017;22(4) doi: 10.3390/molecules22040602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ben Brahim R., Ellouzi H., Fouzai K., Asses N., Neffati M., Sabatier J.M., Bulet P., Regaya I. Optimized chemical extraction methods of antimicrobial peptides from roots and leaves of extremophilic plants: Anthyllis sericea and Astragalus armatus collected from the Tunisian desert. Antibiotics (Basel) 2022;11(10) doi: 10.3390/antibiotics11101302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Caruso G., Maricchiolo G., Genovese L., Pasquale F., Caruso R., Denaro M.G., Delia S., Laganà P. Comparative study of antibacterial and haemolytic activities in sea bass, European eel and blackspot seabream. Open Mar. Biol. J. 2014;8:10–16. [Google Scholar]
- 80.Guardiola F.A., Cuesta A., Abellán E., Meseguer J., Esteban M.A. Comparative analysis of the humoral immunity of skin mucus from several marine teleost fish. Fish Shellfish Immunol. 2014;40(1):24–31. doi: 10.1016/j.fsi.2014.06.018. [DOI] [PubMed] [Google Scholar]
- 81.Haniffa M.A., Viswanathan S., Jancy D., Poomari K., Manik S.A. An, Antibacterial studies of fish mucus from two marketed air-breathing fishes - Channa striatus and Heteropneustes fossilis. International Research Journal of Microbiology. 2014;5:22–27. [Google Scholar]
- 82.Kumari S., Tyor A.K., Bhatnagar A. Evaluation of the antibacterial activity of skin mucus of three carp species. Int. Aquat. Res. 2019;11(3):225–239. [Google Scholar]
- 83.Sewify G.H., Hamada H.M., Alhadrami H.A. In vitro evaluation of antimicrobial activity of alimentary canal extracts from the red palm weevil, Rhynchophorus ferrugineus olivier larvae. BioMed Res. Int. 2017;2017 doi: 10.1155/2017/8564601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tyor A., Kumari S. Biochemical characterization and antibacterial properties of fish skin mucus of fresh water fish. Hypophthalmichthys nobilis. 2016;8:132–136. [Google Scholar]
- 85.Kumar R., Kaushik J.K., Mohanty A.K., Kumar S. Identification of bioactive components behind the antimicrobial activity of cow urine by peptide and metabolite profiling. Anim Biosci. 2023;36(7):1130–1142. doi: 10.5713/ab.22.0249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ogbole O.O., Akinleye T.E., Nkumah A.O., Awogun A.O., Attah A.F., Adewumi M.O., Adeniji A.J. In vitro antiviral activity of peptide-rich extracts from seven Nigerian plants against three non-polio enterovirus species C serotypes. Virol. J. 2021;18(1):161. doi: 10.1186/s12985-021-01628-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Alves M.J., Ferreira I.C., Dias J., Teixeira V., Martins A., Pintado M. A review on antimicrobial activity of mushroom (Basidiomycetes) extracts and isolated compounds. Planta Med. 2012;78(16):1707–1718. doi: 10.1055/s-0032-1315370. [DOI] [PubMed] [Google Scholar]
- 88.Subramanian S., Ross N.W., MacKinnon S.L. Comparison of antimicrobial activity in the epidermal mucus extracts of fish. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2008;150(1):85–92. doi: 10.1016/j.cbpb.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 89.Yount N.Y., Bayer A.S., Xiong Y.Q., Yeaman M.R. Advances in antimicrobial peptide immunobiology. Biopolymers. 2006;84(5):435–458. doi: 10.1002/bip.20543. [DOI] [PubMed] [Google Scholar]
- 90.Afkarian M., Bhasin M., Dillon S.T., Guerrero M.C., Nelson R.G., Knowler W.C., Thadhani R., Libermann T.A. Optimizing a proteomics platform for urine biomarker discovery. Mol. Cell. Proteomics. 2010;9(10):2195–2204. doi: 10.1074/mcp.M110.000992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cunsolo V., Schicchi R., Chiaramonte M., Inguglia L., Arizza V., Cusimano M.G., Schillaci D., Di Francesco A., et al. Identification of new antimicrobial peptides from mediterranean medical plant charybdis pancration. Steinh.) Speta, Antibiotics (Basel). 2020;9(11) doi: 10.3390/antibiotics9110747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Goraya J., Wang Y., Li Z., O'Flaherty M., Knoop F.C., Platz J.E., Conlon J.M. Peptides with antimicrobial activity from four different families isolated from the skins of the North American frogs Rana luteiventris, Rana berlandieri and Rana pipiens. Eur. J. Biochem. 2000;267(3):894–900. doi: 10.1046/j.1432-1327.2000.01074.x. [DOI] [PubMed] [Google Scholar]
- 93.Isidorov V., Zalewski A., Zambrowski G., Swiecicka I. Chemical composition and antimicrobial properties of honey bee venom. Molecules. 2023;28(10) doi: 10.3390/molecules28104135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Křížkovská B., Hoang L., Brdová D., Klementová K., Szemerédi N., Loučková A., Kronusová O., Spengler G., et al. Modulation of the bacterial virulence and resistance by well-known European medicinal herbs. J. Ethnopharmacol. 2023;312 doi: 10.1016/j.jep.2023.116484. [DOI] [PubMed] [Google Scholar]
- 95.Campos M.L., de Souza C.M., de Oliveira K.B.S., Dias S.C., Franco O.L. The role of antimicrobial peptides in plant immunity. J. Exp. Bot. 2018;69(21):4997–5011. doi: 10.1093/jxb/ery294. [DOI] [PubMed] [Google Scholar]
- 96.Mookherjee N., Anderson M.A., Haagsman H.P., Davidson D.J. Antimicrobial host defence peptides: functions and clinical potential. Nat. Rev. Drug Discov. 2020;19(5):311–332. doi: 10.1038/s41573-019-0058-8. [DOI] [PubMed] [Google Scholar]
- 97.Okada T., Yoshizumi H., Terashima Y. A lethal toxic substance for brewing yeast in wheat and barley: Part I. Assay of toxicity on various grains, and sensitivity of various yeast strainsPart II. Isolation and some properties of toxic principle. Agric. Biol. Chem. 1970;34(7):1084–1094. [Google Scholar]
- 98.Wohlfart S., Kilian M., Storck P., Gutsmann T., Brandenburg K., Mier W. Mass spectrometric quantification of the antimicrobial peptide pep19-2.5 with stable isotope labeling and acidic hydrolysis. Pharmaceutics. 2021;13(9) doi: 10.3390/pharmaceutics13091342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Guzmán F., Wong G., Román T., Cárdenas C., Alvárez C., Schmitt P., Albericio F., Rojas V. Identification of antimicrobial peptides from the microalgae tetraselmis suecica (kylin) butcher and bactericidal activity improvement. Mar. Drugs. 2019;17(8) doi: 10.3390/md17080453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Patel M., Ashraf M.S., Siddiqui A.J., Ashraf S.A., Sachidanandan M., Snoussi M., Adnan M., Hadi S. Profiling and role of bioactive molecules from Puntius sophore (Freshwater/Brackish fish) skin mucus with its potent antibacterial, antiadhesion, and antibiofilm activities. Biomolecules. 2020;10(6):920. doi: 10.3390/biom10060920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tafreshi S.H., Mirdamadi S., Khatami S. Comparison of different nisin separation and concentration methods: industrial and cost-effective perspectives. Probiotics Antimicrob Proteins. 2020;12(3):1226–1234. doi: 10.1007/s12602-019-09607-9. [DOI] [PubMed] [Google Scholar]
- 102.El-Araby M.M., El-Shatoury E.H., Soliman M.M., Shaaban H.F. Characterization and antimicrobial activity of lectins purified from three Egyptian leguminous seeds. Amb. Express. 2020;10(1):90. doi: 10.1186/s13568-020-01024-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Vieira Bard G.C., Nascimento V.V., Oliveira A.E., Rodrigues R., Da Cunha M., Dias G.B., Vasconcelos I.M., Carvalho A.O., et al. Vicilin-like peptides from Capsicum baccatum L. seeds are α-amylase inhibitors and exhibit antifungal activity against important yeasts in medical mycology. Biopolymers. 2014;102(4):335–343. doi: 10.1002/bip.22504. [DOI] [PubMed] [Google Scholar]
- 104.Wang S., Rao P., Ye X. Isolation and biochemical characterization of a novel leguminous defense peptide with antifungal and antiproliferative potency. Appl. Microbiol. Biotechnol. 2009;82(1):79–86. doi: 10.1007/s00253-008-1729-2. [DOI] [PubMed] [Google Scholar]
- 105.Ribeiro S.F., Carvalho A.O., Da Cunha M., Rodrigues R., Cruz L.P., Melo V.M., Vasconcelos I.M., Melo E.J., et al. Isolation and characterization of novel peptides from chilli pepper seeds: antimicrobial activities against pathogenic yeasts. Toxicon. 2007;50(5):600–611. doi: 10.1016/j.toxicon.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 106.Dehghanifar S., Keyhanfar M., Emtiazi G. Production and partial purification of thermostable bacteriocins from Bacillus pumilus ZED17 and DFAR8 strains with antifungal activity. Mol. Biol. Res. Commun. 2019;8(1):41–49. doi: 10.22099/mbrc.2019.31563.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.da Silva M.S., Gomes V.M., Taveira G.B., de Azevedo Dos Santos L., Maracahipes Á C., Rodrigues R., de Oliveira Carvalho A., Fernandes K.V.S., et al. Bifunctional inhibitors from capsicum chinense seeds with antimicrobial activity and specific mechanism of action against phytopathogenic fungi. Protein Pept. Lett. 2021;28(2):149–163. doi: 10.2174/0929866527666200617124221. [DOI] [PubMed] [Google Scholar]
- 108.Figueredo S., Mastroianni J.R., Tai K.P., Ouellette A.J. Expression and purification of recombinant alpha-defensins and alpha-defensin precursors in Escherichia coli. Methods Mol. Biol. 2010;618:47–60. doi: 10.1007/978-1-60761-594-1_4. [DOI] [PubMed] [Google Scholar]
- 109.Kini S.G., Wong K.H., Tan W.L., Xiao T., Tam J.P. Morintides: cargo-free chitin-binding peptides from Moringa oleifera. BMC Plant Biol. 2017;17(1):68. doi: 10.1186/s12870-017-1014-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Calderón-de la Sancha F.J., Carrasco-Navarro U., Santander G., Barrios-González J., Mejía A. Novel antimicrobial activity of protein produced by Streptomyces lividans TK24 against the phytopathogen Clavibacter michiganensis. Arch. Microbiol. 2022;204(11):687. doi: 10.1007/s00203-022-03290-1. [DOI] [PubMed] [Google Scholar]
- 111.Pavlova A.S., Ozhegov G.D., Arapidi G.P., Butenko I.O., Fomin E.S., Alemasov N.A., Afonnikov D.A., Yarullina D.R., et al. Identification of antimicrobial peptides from novel Lactobacillus fermentum strain. Protein J. 2020;39(1):73–84. doi: 10.1007/s10930-019-09879-8. [DOI] [PubMed] [Google Scholar]
- 112.Swift C.L., Louie K.B., Bowen B.P., Olson H.M., Purvine S.O., Salamov A., Mondo S.J., Solomon K.V., et al. Anaerobic gut fungi are an untapped reservoir of natural products. Proc. Natl. Acad. Sci. U. S. A. 2021;118(18) doi: 10.1073/pnas.2019855118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Yu J., Hu Y., Xue M., Dun Y., Li S., Peng N., Liang Y., Zhao S. Purification and identification of antioxidant peptides from enzymatic hydrolysate of spirulina platensis. J. Microbiol. Biotechnol. 2016;26(7):1216–1223. doi: 10.4014/jmb.1601.01033. [DOI] [PubMed] [Google Scholar]
- 114.Ayswaria R., Vijayan J., Nathan V.K. Antimicrobial peptides derived from microalgae for combating antibiotic resistance: current status and prospects. Cell Biochem. Funct. 2023;41(2):142–151. doi: 10.1002/cbf.3779. [DOI] [PubMed] [Google Scholar]
- 115.Cochet M.F., Baron F., Bonnassie S., Jan S., Leconte N., Jardin J., Briard-Bion V., Gautier M., et al. Identification of new antimicrobial peptides that contribute to the bactericidal activity of egg white against Salmonella enterica serovar enteritidis at 45 °C. J. Agric. Food Chem. 2021;69(7):2118–2128. doi: 10.1021/acs.jafc.0c06677. [DOI] [PubMed] [Google Scholar]
- 116.Golneshin A., Gor M.C., Williamson N., Vezina B., Van T.T.H., May B.K., Smith A.T. Discovery and characterisation of circular bacteriocin plantacyclin B21AG from Lactiplantibacillus plantarum B21. Heliyon. 2020;6(8) doi: 10.1016/j.heliyon.2020.e04715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Matar K.M., Al-Refai B. Quantification of colistin in plasma by liquid chromatography-tandem mass spectrometry: application to a pharmacokinetic study. Sci. Rep. 2020;10(1):8198. doi: 10.1038/s41598-020-65041-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wei G., Wang D., Wang T., Yang C., Shi Y., Huang A. Insights into in vitro digestion properties and peptide profiling of Chinese rubing PDO cheese prepared using different acidification technology. Food Res. Int. 2022;158 doi: 10.1016/j.foodres.2022.111564. [DOI] [PubMed] [Google Scholar]
- 119.Wen J., Wang W., Lee K.J., Choi B.K., Harradine P., Salituro G.M., Hittle L. Quantitation of super basic peptides in biological matrices by a generic perfluoropentanoic acid-based liquid chromatography-mass spectrometry method. J. Am. Soc. Mass Spectrom. 2019;30(9):1779–1789. doi: 10.1007/s13361-019-02257-9. [DOI] [PubMed] [Google Scholar]
- 120.Fekih-Zaghbib S., Fildier A., Barrek S., Bouhaouala-Zahar B. A complementary LC-ESI-MS and MALDI-TOF approach for screening antibacterial proteomic signature of farmed European sea bass mucus. Fish Shellfish Immunol. 2013;35(2):207–212. doi: 10.1016/j.fsi.2013.04.017. [DOI] [PubMed] [Google Scholar]
- 121.Fekih-Zaghbib S., Ksouri A., Bouhaouala-Zahar B. Differences in fish mucus proteomes identify potential antimicrobial peptide biomarkers. Dev. Comp. Immunol. 2023;145 doi: 10.1016/j.dci.2023.104730. [DOI] [PubMed] [Google Scholar]
- 122.Wang H., Strich J.R., Drake S.K., Chen Y., Youn J.H., Rosenberg A.Z., Gucek M., Dekker J.P., et al. Rapid identification of New Delhi metallo-β-lactamase (NDM) using tryptic peptides and LC-MS/MS. Antimicrob. Agents Chemother. 2019;63(9) doi: 10.1128/AAC.00461-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Iram D., Kindarle U.A., Sansi M.S., Meena S., Puniya A.K., Vij S. Peptidomics-based identification of an antimicrobial peptide derived from goat milk fermented by Lactobacillus rhamnosus (C25) J. Food Biochem. 2022;46(12) doi: 10.1111/jfbc.14450. [DOI] [PubMed] [Google Scholar]
- 124.Jain V., Mishra P.K., Mishra M., Prakash V. Constitutive expression and discovery of antimicrobial peptides in Zygogramma bicolorata (Coleoptera: chrysomelidae) Proteins. 2022;90(2):465–475. doi: 10.1002/prot.26239. [DOI] [PubMed] [Google Scholar]
- 125.Chowdhury T., Mandal S.M., Dutta S., Ghosh A.K. Identification of a novel proline-rich antimicrobial protein from the hemolymph of Antheraea mylitta. Arch. Insect Biochem. Physiol. 2021;106(3) doi: 10.1002/arch.21771. [DOI] [PubMed] [Google Scholar]
- 126.Gao X., Chen Y., Chen Z., Xue Z., Jia Y., Guo Q., Ma Q., Zhang M., et al. Identification and antimicrobial activity evaluation of three peptides from laba garlic and the related mechanism. Food Funct. 2019;10(8):4486–4496. doi: 10.1039/c9fo00236g. [DOI] [PubMed] [Google Scholar]
- 127.Liu X., Yu Z., Wang Y., Wu H., Bian X., Li X., Fan Y., Guo B., et al. Therapeutic drug monitoring of polymyxin B by LC-MS/MS in plasma and urine. Bioanalysis. 2020;12(12):845–855. doi: 10.4155/bio-2020-0051. [DOI] [PubMed] [Google Scholar]
- 128.O'Neill M.J., Chan K., Jaynes J.M., Knotts Z., Xu X., Abisoye-Ogunniyan A., Guerin T., Schlomer J., et al. LC-MS/MS assay coupled with carboxylic acid magnetic bead affinity capture to quantitatively measure cationic host defense peptides (HDPs) in complex matrices with application to preclinical pharmacokinetic studies. J. Pharm. Biomed. Anal. 2020;181 doi: 10.1016/j.jpba.2020.113093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Huang X., Liu X., Wang Y., Zhang J. Determination of polymyxin B in dried blood spots using LC-MS/MS for therapeutic drug monitoring. J. Chromatogr., B: Anal. Technol. Biomed. Life Sci. 2022;1192 doi: 10.1016/j.jchromb.2022.123131. [DOI] [PubMed] [Google Scholar]
- 130.He J., Gao S., Hu M., Chow D.S., Tam V.H. A validated ultra-performance liquid chromatography-tandem mass spectrometry method for the quantification of polymyxin B in mouse serum and epithelial lining fluid: application to pharmacokinetic studies. J. Antimicrob. Chemother. 2013;68(5):1104–1110. doi: 10.1093/jac/dks536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Covelli J., Ruszaj D., Straubinger R., Li J., Rao G.G. The development and validation of a simple liquid chromatography-tandem mass spectrometry method for polymyxin B1 and B2 quantification in different matrices. J. Chromatogr., B: Anal. Technol. Biomed. Life Sci. 2017;1065–1066:112–118. doi: 10.1016/j.jchromb.2017.09.031. [DOI] [PubMed] [Google Scholar]
- 132.Fekih-Zaghbib S., Ksouri A., Bouhaouala-Zahar B. Differences in fish mucus proteomes identify potential antimicrobial peptide biomarkers. Dev. Comp. Immunol. 2023 doi: 10.1016/j.dci.2023.104730. [DOI] [PubMed] [Google Scholar]
- 133.Diep J.K., Covelli J., Sharma R., Ruszaj D.M., Kaye K.S., Li J., Straubinger R.M., Rao G.G. Comparison of the composition and in vitro activity of polymyxin B products. Int. J. Antimicrob. Agents. 2018;52(3):365–371. doi: 10.1016/j.ijantimicag.2018.05.014. [DOI] [PubMed] [Google Scholar]
- 134.Zhang B., Li X., Chen Y., Chen B., Cheng Y., Lin H., Que W., Liu M., et al. Determination of polymyxin B in human plasma and epithelial lining fluid using LC-MS/MS and its clinical application in therapeutic drug monitoring. J. Pharm. Biomed. Anal. 2023;227 doi: 10.1016/j.jpba.2023.115291. [DOI] [PubMed] [Google Scholar]
- 135.Sevillano E., Peña N., Lafuente I., Cintas L.M., Muñoz-Atienza E., Hernández P.E., Borrero J. Nisin S, a novel nisin variant produced by ligilactobacillus salivarius P1CEA3. Int. J. Mol. Sci. 2023;24(7) doi: 10.3390/ijms24076813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Lu M., Chen Y., Li L., Ma Y., Tong Z., Guo D., Sun P., An D. Analysis and evaluation of the flagellin activity of Bacillus amyloliquefaciens Ba168 antimicrobial proteins against Penicillium expansum. Molecules. 2022;27(13) doi: 10.3390/molecules27134259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Fesenko I., Azarkina R., Kirov I., Kniazev A., Filippova A., Grafskaia E., Lazarev V., Zgoda V., et al. Phytohormone treatment induces generation of cryptic peptides with antimicrobial activity in the Moss Physcomitrella patens. BMC Plant Biol. 2019;19(1):9. doi: 10.1186/s12870-018-1611-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Santos M., Freitas C.S., Verissimo da Costa G.C., Pereira P.R., Paschoalin V.M.F. Identification of antibacterial peptide candidates encrypted in stress-related and metabolic Saccharomyces cerevisiae proteins. Pharmaceuticals. 2022;15(2) doi: 10.3390/ph15020163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Correa C.N., Fiametti L.O., de Barros G.M., de Castro L.M. Revealing natural intracellular peptides in gills of seahorse Hippocampus reidi. Biomolecules. 2023;13(3) doi: 10.3390/biom13030433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Elbarbary H.A., Ejima A., Sato K. Generation of antibacterial peptides from crude cheese whey using pepsin and rennet enzymes at various pH conditions. J. Sci. Food Agric. 2019;99(2):555–563. doi: 10.1002/jsfa.9214. [DOI] [PubMed] [Google Scholar]
- 141.Feng L.L., Liu L., Cheng T.Y. Proteomic analysis of saliva from partially and fully engorged adult female Rhipicephalus microplus (Acari: ixodidae) Exp. Appl. Acarol. 2019;78(3):443–460. doi: 10.1007/s10493-019-00390-4. [DOI] [PubMed] [Google Scholar]
- 142.Ngashangva N., Mukherjee P., Sharma K.C., Kalita M.C., Indira S. Analysis of antimicrobial peptide metabolome of bacterial endophyte isolated from traditionally used medicinal plant Millettia pachycarpa benth. Front. Microbiol. 2021;12 doi: 10.3389/fmicb.2021.656896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Grady E.N., MacDonald J., Ho M.T., Weselowski B., McDowell T., Solomon O., Renaud J., Yuan Z.C. Characterization and complete genome analysis of the surfactin-producing, plant-protecting bacterium Bacillus velezensis 9D-6. BMC Microbiol. 2019;19(1):5. doi: 10.1186/s12866-018-1380-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Kumar H., Kumar D., Nepovimova E., Oulkar D., Kumar A., Azad R.M.R., Budakoti S.K., Upadhyay N.K., et al. Determination of colistin B in chicken muscle and egg using ultra-high-performance liquid chromatography-tandem mass spectrometry. Int. J. Environ. Res. Publ. Health. 2021;18(5) doi: 10.3390/ijerph18052651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Pimchan T., Tian F., Thumanu K., Rodtong S., Yongsawatdigul J. Isolation, identification, and mode of action of antibacterial peptides derived from egg yolk hydrolysate. Poultry Sci. 2023;102(7) doi: 10.1016/j.psj.2023.102695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Lanois-Nouri A., Pantel L., Fu J., Houard J., Ogier J.C., Polikanov Y.S., Racine E., Wang H., et al. The odilorhabdin antibiotic biosynthetic cluster and acetyltransferase self-resistance locus are niche and species specific. mBio. 2022;13(1) doi: 10.1128/mbio.02826-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Daoudi K., Malosse C., Lafnoune A., Darkaoui B., Chakir S., Sabatier J.M., Chamot-Rooke J., Cadi R., et al. Mass spectrometry-based top-down and bottom-up approaches for proteomic analysis of the Moroccan Buthus occitanus scorpion venom. FEBS Open Bio. 2021;11(7):1867–1892. doi: 10.1002/2211-5463.13143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Vineeth Kumar T., Asha R., Shyla G., George S. Identification and characterization of novel host defense peptides from the skin secretion of the fungoid frog, Hydrophylax bahuvistara (Anura: ranidae) Chem. Biol. Drug Des. 2018;92(2):1409–1418. doi: 10.1111/cbdd.12937. [DOI] [PubMed] [Google Scholar]
- 149.Ismael M., Wang T., Yue F., Cui Y., Yantin Q., Qayyum N., Lü X. A comparison of mining methods to extract novel bacteriocins from Lactiplantibacillus plantarum NWAFU-BIO-BS29. Anal. Biochem. 2023;661 doi: 10.1016/j.ab.2022.114938. [DOI] [PubMed] [Google Scholar]
- 150.Mariano D.O., Sciani J.M., Antoniazzi M.M., Jared C., Conceição K., Pimenta D.C. Quantity - but not diversity - of secreted peptides and proteins increases with age in the tree frog Pithecopus nordestinus. J. Venom. Anim. Toxins Incl. Trop. Dis. 2021;27 doi: 10.1590/1678-9199-JVATITD-2020-0105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Arbulu S., Jiménez J.J., Gútiez L., Feito J., Cintas L.M., Herranz C., Hernández P.E. Cloning and expression of synthetic genes encoding native, hybrid- and bacteriocin-derived chimeras from mature class IIa bacteriocins, by Pichia pastoris (syn. Komagataella spp.) Food Res. Int. 2019;121:888–899. doi: 10.1016/j.foodres.2019.01.015. [DOI] [PubMed] [Google Scholar]
- 152.Fernández-Montero Á., Torrecillas S., Montero D., Acosta F., Prieto-Álamo M.J., Abril N., Jurado J. Proteomic profile and protease activity in the skin mucus of greater amberjack (Seriola dumerili) infected with the ectoparasite Neobenedenia girellae - an immunological approach. Fish Shellfish Immunol. 2021;110:100–115. doi: 10.1016/j.fsi.2021.01.001. [DOI] [PubMed] [Google Scholar]
- 153.Gherghisan-Filip C., Saalbach G., Hatziioanou D., Narbad A., Mayer M.J. Processing and structure of the lantibiotic peptide nso from the human gut bacterium blautia obeum A2-162 analysed by mass spectrometry. Sci. Rep. 2018;8(1) doi: 10.1038/s41598-018-28248-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Siano A., Humpola M.V., de Oliveira E., Albericio F., Simonetta A.C., Lajmanovich R., Tonarelli G.G. Leptodactylus latrans Amphibian skin secretions as a novel source for the isolation of antibacterial peptides. Molecules. 2018;23(11) doi: 10.3390/molecules23112943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Song X., Turiel E., Yang J., Martín-Esteban A., He L. Determination of polypeptide antibiotics in animal tissues using liquid chromatography tandem mass spectrometry based on in-line molecularly imprinted solid-phase extraction. J. Chromatogr. A. 2022;1673 doi: 10.1016/j.chroma.2022.463192. [DOI] [PubMed] [Google Scholar]
- 156.Yi L., Luo L., Lü X. Efficient exploitation of multiple novel bacteriocins by combination of complete genome and peptidome. Front. Microbiol. 2018;9:1567. doi: 10.3389/fmicb.2018.01567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Cirrincione S., Luganini A., Lamberti C., Manfredi M., Cavallarin L., Giuffrida M.G., Pessione E. Donkey milk fermentation by lactococcus lactis subsp. cremoris and Lactobacillus rhamnosus affects the antiviral and antibacterial milk properties. Molecules. 2021;26(16) doi: 10.3390/molecules26165100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Wang X., He L., Huang Z., Zhao Q., Fan J., Tian Y., Huang A. Isolation, identification and characterization of a novel antimicrobial peptide from Moringa oleifera seeds based on affinity adsorption. Food Chem. 2023;398 doi: 10.1016/j.foodchem.2022.133923. [DOI] [PubMed] [Google Scholar]
- 159.Wacharachaisurapol N., Phasomsap C., Sukkummee W., Phaisal W., Chanakul A., Wittayalertpanya S., Chariyavilaskul P., Puthanakit T. Greater optimisation of pharmacokinetic/pharmacodynamic parameters through a loading dose of intravenous colistin in paediatric patients. Int. J. Antimicrob. Agents. 2020;55(6) doi: 10.1016/j.ijantimicag.2020.105940. [DOI] [PubMed] [Google Scholar]
- 160.Matos A., Domínguez-Pérez D., Almeida D., Agüero-Chapin G., Campos A., Osório H., Vasconcelos V., Antunes A. Shotgun proteomics of ascidians tunic gives new insights on host-microbe interactions by revealing diverse antimicrobial peptides. Mar. Drugs. 2020;18(7) doi: 10.3390/md18070362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Urbaniak M., Waśkiewicz A., Trzebny A., Koczyk G., Stępień Ł. Cyclodepsipeptide biosynthesis in hypocreales fungi and sequence divergence of the non-ribosomal peptide synthase genes. Pathogens. 2020;9(7) doi: 10.3390/pathogens9070552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Tian F., Rodtong S., Thumanu K., Hua Y., Roytrakul S., Yongsawatdigul J. Molecular insights into the mode of action of antibacterial peptides derived from chicken plasma hydrolysates. Foods. 2022;11(22) doi: 10.3390/foods11223564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Fage D., Deprez G., Wolff F., Hites M., Jacobs F., Van Bambeke F., Cotton F. Investigation of unbound colistin A and B in clinical samples using a mass spectrometry method. Int. J. Antimicrob. Agents. 2019;53(3):330–336. doi: 10.1016/j.ijantimicag.2018.10.017. [DOI] [PubMed] [Google Scholar]
- 164.M M.G., Pasha S., Inui T., Chapple I., Harris S., Holcombe L. A mass spectrometric approach to the proteomic profiling of the Canis lupus familiaris acquired enamel pellicle on hydroxyapatite discs. J. Vet. Dent. 2022;39(3):241–249. doi: 10.1177/08987564221097188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Burkin M.A., Galvidis I.A., Surovoy Y.A., Plyushchenko I.V., Rodin I.A., Tsarenko S.V. Development of ELISA formats for polymyxin B monitoring in serum of critically ill patients. J. Pharm. Biomed. Anal. 2021;204 doi: 10.1016/j.jpba.2021.114275. [DOI] [PubMed] [Google Scholar]
- 166.Pérez-Polo S., Imran M.A.S., Dios S., Pérez J., Barros L., Carrera M., Gestal C. Identifying natural bioactive peptides from the common Octopus (Octopus vulgaris cuvier, 1797) skin mucus by-products using proteogenomic analysis. Int. J. Mol. Sci. 2023;24(8) doi: 10.3390/ijms24087145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Mitchell M.L., Tonkin-Hill G.Q., Morales R.A.V., Purcell A.W., Papenfuss A.T., Norton R.S. Tentacle transcriptomes of the speckled anemone (actiniaria: actiniidae: oulactis sp.): venom-related components and their domain structure. Mar. Biotechnol. 2020;22(2):207–219. doi: 10.1007/s10126-020-09945-8. [DOI] [PubMed] [Google Scholar]
- 168.Imran M.A.S., Carrera M., Pérez-Polo S., Pérez J., Barros L., Dios S., Gestal C. Insights into common Octopus (Octopus vulgaris) ink proteome and bioactive peptides using proteomic approaches. Mar. Drugs. 2023;21(4) doi: 10.3390/md21040206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Wang H., Cissé O.H., Bolig T., Drake S.K., Chen Y., Strich J.R., Youn J.H., Okoro U., et al. A phylogeny-informed proteomics approach for species identification within the burkholderia cepacia complex. J. Clin. Microbiol. 2020;58(11) doi: 10.1128/JCM.01741-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Qi B., Gijsen M., Van Brantegem P., De Vocht T., Deferm N., Abza G.B., Nauwelaerts N., Wauters J., et al. Quantitative determination of colistin A/B and colistin methanesulfonate in biological samples using hydrophilic interaction chromatography tandem mass spectrometry. Drug Test. Anal. 2020;12(8):1183–1195. doi: 10.1002/dta.2812. [DOI] [PubMed] [Google Scholar]
- 171.Puttrevu S.K., Laxman T.S., Tripathi A.K., Yadav A.K., Verma S.K., Mishra A., Pradhan R., Verma N.K., et al. Liquid chromatography-tandem mass spectrometry based method development and validation of S016-1271 (LR8P), a novel cationic antimicrobial peptide for its application to pharmacokinetic studies. J. Pharm. Biomed. Anal. 2019;169:116–126. doi: 10.1016/j.jpba.2019.01.046. [DOI] [PubMed] [Google Scholar]
- 172.Gaugain M., Raynaud A., Bourcier S., Verdon E., Hurtaud-Pessel D. Development of a liquid chromatography-tandem mass spectrometry method to determine colistin, bacitracin and virginiamycin M1 at cross-contamination levels in animal feed. Food Addit. Contam. Part A Chem Anal Control Expo Risk Assess. 2021;38(9):1481–1494. doi: 10.1080/19440049.2021.1922760. [DOI] [PubMed] [Google Scholar]
- 173.Castro G.S., Sousa T.F., da Silva G.F., Pedroso R.C.N., Menezes K.S., Soares M.A., Dias G.M., Santos A.O., et al. Characterization of peptaibols produced by a marine strain of the fungus Trichoderma endophyticum via mass spectrometry, genome mining and phylogeny-based prediction. Metabolites. 2023;13(2) doi: 10.3390/metabo13020221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Ng C.C.A., Zhou Y., Yao Z.P. Algorithms for de-novo sequencing of peptides by tandem mass spectrometry: a review. Anal. Chim. Acta. 2023;1268 doi: 10.1016/j.aca.2023.341330. [DOI] [PubMed] [Google Scholar]
- 175.Li W., Petruzziello F., Zhao N., Zhao H., Ye X., Zhang X., Rainer G. Separation and identification of mouse brain tissue microproteins using top-down method with high resolution nanocapillary liquid chromatography mass spectrometry. Proteomics. 2017;17(12) doi: 10.1002/pmic.201600419. [DOI] [PubMed] [Google Scholar]
- 176.Ngoh Y.Y., Gan C.Y. Identification of Pinto bean peptides with inhibitory effects on α-amylase and angiotensin converting enzyme (ACE) activities using an integrated bioinformatics-assisted approach. Food Chem. 2018;267:124–131. doi: 10.1016/j.foodchem.2017.04.166. [DOI] [PubMed] [Google Scholar]
- 177.Nyimanu D., Kay R.G., Kuc R.E., Brown A.J.H., Gribble F.M., Maguire J.J., Davenport A.P. In vitro metabolism of synthetic Elabela/Toddler (ELA-32) peptide in human plasma and kidney homogenates analyzed with mass spectrometry and validation of endogenous peptide quantification in tissues by ELISA. Peptides. 2021;145 doi: 10.1016/j.peptides.2021.170642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Wei X., Liu P.N., Mooney B.P., Nguyen T.T., Greenlief C.M. A comprehensive study of gradient conditions for deep proteome discovery in a complex protein matrix. Int. J. Mol. Sci. 2022;23(19) doi: 10.3390/ijms231911714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Gangadharappa B.S., Rajashekarappa S., Sathe G. Proteomic profiling of Serratia marcescens by high-resolution mass spectrometry. Bioimpacts. 2020;10(2):123–135. doi: 10.34172/bi.2020.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Bouyssié D., Gonzalez de Peredo A., Mouton E., Albigot R., Roussel L., Ortega N., Cayrol C., Burlet-Schiltz O., et al. Mascot file parsing and quantification (MFPaQ), a new software to parse, validate, and quantify proteomics data generated by ICAT and SILAC mass spectrometric analyses: application to the proteomics study of membrane proteins from primary human endothelial cells. Mol. Cell. Proteomics. 2007;6(9):1621–1637. doi: 10.1074/mcp.T600069-MCP200. [DOI] [PubMed] [Google Scholar]
- 181.Jan K., Ahmed I., Dar N.A., Farah M.A., Khan F.R., Shah B.A., Fazio F. LC-MS/MS based characterisation and differential expression of proteins in Himalayan snow trout, Schizothorax labiatus using LFQ technique. Sci. Rep. 2023;13(1) doi: 10.1038/s41598-023-35646-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Stelzhammer V., Amess B., Martins-de-Souza D., Levin Y., Ozanne S.E., Martin-Gronert M.S., Urday S., Bahn S., et al. Analysis of the rat hypothalamus proteome by data-independent label-free LC-MS/MS. Proteomics. 2012;12(22):3386–3392. doi: 10.1002/pmic.201100642. [DOI] [PubMed] [Google Scholar]
- 183.Liu J., Jiang J. Mass spectrometry approaches for determining the structure of antimicrobial peptides. Methods Mol. Biol. 2017;1548:89–99. doi: 10.1007/978-1-4939-6737-7_7. [DOI] [PubMed] [Google Scholar]
- 184.Stegemann C., Hoffmann R. Sequence analysis of antimicrobial peptides by tandem mass spectrometry. Methods Mol. Biol. 2008;494:31–46. doi: 10.1007/978-1-59745-419-3_3. [DOI] [PubMed] [Google Scholar]
- 185.Rodriguez A., Martell-Huguet E.M., González-García M., Alpízar-Pedraza D., Alba A., Vazquez A.A., Grieshober M., Spellerberg B., et al. Identification and characterization of three new antimicrobial peptides from the marine mollusk nerita versicolor (gmelin, 1791) Int. J. Mol. Sci. 2023;24(4) doi: 10.3390/ijms24043852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Punginelli D., Catania V., Abruscato G., Luparello C., Vazzana M., Mauro M., Cunsolo V., Saletti R., et al. New bioactive peptides from the mediterranean seagrass posidonia oceanica (L.) delile and their impact on antimicrobial activity and apoptosis of human cancer cells. Int. J. Mol. Sci. 2023;24(6) doi: 10.3390/ijms24065650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Wang Y., Yu X., Chen C., Zhang X., Ye Z., Yang J., Chen Y., Xiang Z., et al. Development of UPLC-MS/MS method for the determination of colistin in plasma and kidney and its application in pharmacokinetics. J. Pharm. Biomed. Anal. 2023;233 doi: 10.1016/j.jpba.2023.115440. [DOI] [PubMed] [Google Scholar]
- 188.Li J., Shimko K.M., He C., Patterson B., Bade R., Shiels R., Mueller J.F., Thomas K.V., et al. Direct injection liquid chromatography-tandem mass spectrometry as a sensitive and high-throughput method for the quantitative surveillance of antimicrobials in wastewater. Sci. Total Environ. 2023;900 doi: 10.1016/j.scitotenv.2023.165825. [DOI] [PubMed] [Google Scholar]
- 189.Tong L.Y., Xu B.Z., Nie X.M., Wang X.J., Ma J.H., Guo W., Li G.R., Gong Y.K., et al. [Determination of 22 mycotoxins in milk by liquid chromatography-quadrupole/orbitrap mass spectrometry] Se Pu. 2023;41(11):986–994. doi: 10.3724/SP.J.1123.2023.07010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Fu Q., Li X., Zheng K., Ke Y., Wang Y., Wang L., Yu F., Xia X. Determination of colistin in animal tissues, egg, milk, and feed by ultra-high performance liquid chromatography-tandem mass spectrometry. Food Chem. 2018;248:166–172. doi: 10.1016/j.foodchem.2017.12.029. [DOI] [PubMed] [Google Scholar]
- 191.Gardner M.S., Rowland M.D., Siu A.Y., Bundy J.L., Wagener D.K., Stephenson J.L. Comprehensive defensin assay for saliva. Anal. Chem. 2009;81(2):557–566. doi: 10.1021/ac801609r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Boison J.O., Lee S., Matus J. A multi-residue method for the determination of seven polypeptide drug residues in chicken muscle tissues by LC-MS/MS. Anal. Bioanal. Chem. 2015;407(14):4065–4078. doi: 10.1007/s00216-015-8644-z. [DOI] [PubMed] [Google Scholar]
- 193.Song X., Huang Q., Zhang Y., Zhang M., Xie J., He L. Rapid multiresidue analysis of authorized/banned cyclopolypeptide antibiotics in feed by liquid chromatography-tandem mass spectrometry based on dispersive solid-phase extraction. J. Pharm. Biomed. Anal. 2019;170:234–242. doi: 10.1016/j.jpba.2019.03.050. [DOI] [PubMed] [Google Scholar]
- 194.Berska J., Bugajska J., Sztefko K. A liquid chromatography-tandem mass spectrometry method for simultaneously determining meropenem and linezolid in blood and cerebrospinal fluid. Ann Lab Med. 2024;44(2):174–178. doi: 10.3343/alm.2023.0250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Abdelmoula W.M., Lopez B.G., Randall E.C., Kapur T., Sarkaria J.N., White F.M., Agar J.N., Wells W.M., et al. Peak learning of mass spectrometry imaging data using artificial neural networks. Nat. Commun. 2021;12(1):5544. doi: 10.1038/s41467-021-25744-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Neagu A.N. Proteome imaging: from classic to modern mass spectrometry-based molecular histology. Adv. Exp. Med. Biol. 2019;1140:55–98. doi: 10.1007/978-3-030-15950-4_4. [DOI] [PubMed] [Google Scholar]
- 197.Lu J., Zhu Y., Parkington H.C., Hussein M., Zhao J., Bergen P., Rudd D., Deane M.A., et al. Transcriptomic mapping of neurotoxicity pathways in the rat brain in response to intraventricular polymyxin B. Mol. Neurobiol. 2023;60(3):1317–1330. doi: 10.1007/s12035-022-03140-7. [DOI] [PubMed] [Google Scholar]
- 198.Nilsson A., Goodwin R.J., Swales J.G., Gallagher R., Shankaran H., Sathe A., Pradeepan S., Xue A., et al. Investigating nephrotoxicity of polymyxin derivatives by mapping renal distribution using mass spectrometry imaging. Chem. Res. Toxicol. 2015;28(9):1823–1830. doi: 10.1021/acs.chemrestox.5b00262. [DOI] [PubMed] [Google Scholar]
- 199.Gonzalez D.J., Okumura C.Y., Hollands A., Kersten R., Akong-Moore K., Pence M.A., Malone C.L., Derieux J., et al. Novel phenol-soluble modulin derivatives in community-associated methicillin-resistant Staphylococcus aureus identified through imaging mass spectrometry. J. Biol. Chem. 2012;287(17):13889–13898. doi: 10.1074/jbc.M112.349860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Parra A.L.C., Freitas C.D.T., Souza P.F.N., von Aderkas P., Borchers C.H., Beattie G.A., Silva F.D.A., Thornburg R.W. Ornamental tobacco floral nectar is a rich source of antimicrobial peptides. Plant Sci. 2022;324 doi: 10.1016/j.plantsci.2022.111427. [DOI] [PubMed] [Google Scholar]
- 201.Cuesta S.A., Reinoso C., Morales F., Pilaquinga F., Morán-Marcillo G., Proaño-Bolaños C., Blasco-Zúñiga A., Rivera M., et al. Novel antimicrobial cruzioseptin peptides extracted from the splendid leaf frog, Cruziohyla calcarifer. Amino Acids. 2021;53(6):853–868. doi: 10.1007/s00726-021-02986-w. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this review article.