Abstract
Federated multipartner machine learning has been touted as an appealing and efficient method to increase the effective training data volume and thereby the predictivity of models, particularly when the generation of training data is resource-intensive. In the landmark MELLODDY project, indeed, each of ten pharmaceutical companies realized aggregated improvements on its own classification or regression models through federated learning. To this end, they leveraged a novel implementation extending multitask learning across partners, on a platform audited for privacy and security. The experiments involved an unprecedented cross-pharma data set of 2.6+ billion confidential experimental activity data points, documenting 21+ million physical small molecules and 40+ thousand assays in on-target and secondary pharmacodynamics and pharmacokinetics. Appropriate complementary metrics were developed to evaluate the predictive performance in the federated setting. In addition to predictive performance increases in labeled space, the results point toward an extended applicability domain in federated learning. Increases in collective training data volume, including by means of auxiliary data resulting from single concentration high-throughput and imaging assays, continued to boost predictive performance, albeit with a saturating return. Markedly higher improvements were observed for the pharmacokinetics and safety panel assay-based task subsets.
Introduction
Already for six decades,1,2 the pharmaceutical field has been training Quantitative Structure Activity Relationship (QSAR) models, that relate the chemical structure of compounds or a descriptor representative of structure to their categorical (active or not) or quantitative (how active) readout in pharmacological assays.3 Classical QSAR modeling is a textbook example of supervised learning in that models are trained on given structure–activity example pairs and evaluated on distinct, unseen pairs. For well over five decades, QSAR models were nearly exclusively trained on a single task or target. While both descriptor and fitting approaches have gradually improved over time, the best option for single-task model improvement has remained generating more training pairs, requiring the experimental testing of more compounds in the corresponding assay.
Multitask modeling was first tentatively introduced to QSAR modeling about 25 years ago,4 but rose to prominence about a decade ago.5,6 The aim of multitask modeling is model improvement by information transfer across tasks, which is often embodied as a joint representation. Conceptually, multitask learning explores the same low-level relationships as single-task learning. But in contrast to single-task learning, multitask learning can prioritize those relationships that support multiple tasks, which tend to generalize better for individual tasks. Because compound coverage typically differs between assays, multitask models are usually also exposed to more compounds. As a result, when carefully applied, multitask models tend to match or outperform single-task models.7−12 Also, net benefits were shown to increase further as additional data and tasks were added, albeit less than linear.13
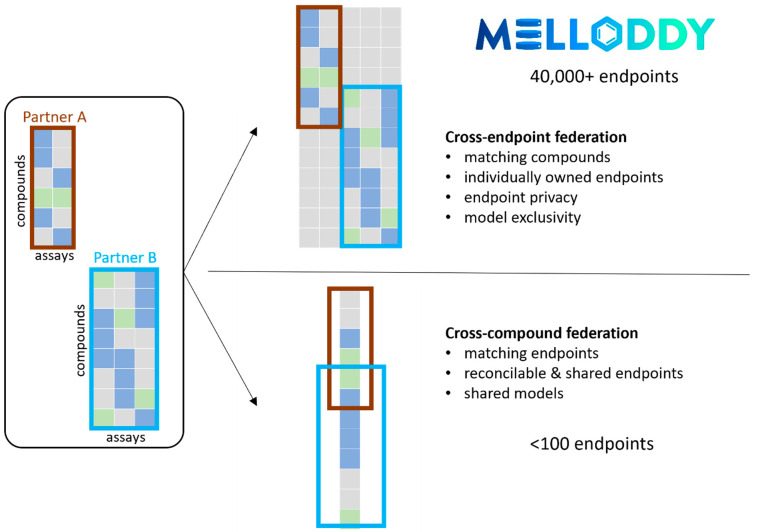
Federated learning enables machine learning across distributed data sets.14,15 Federated learning goes beyond applying conventional machine learning to federated data, i.e., distributed data that is somehow made accessible as a consolidated data set. In federated learning, the training process itself is distributed, often accommodating additional constraints, for example, a requirement to protect the confidentiality of data sets. Distributed QSAR data sets can be compound-wise or end-point-wise partitioned or show a mixed partition pattern. In the QSAR case, compound-wise partitioned data sets would document different sets of compounds with activity labels for the same assays and end-point-wise partitioned data sets would document a compound space with activity labels for different sets of end points or tasks (Figure 1). Existing federated learning solutions for QSAR modeling (and corresponding ones in other applications fields) have largely focused on the cross-compound federation of compound-wise partitioned data sets.16 Cross-compound federation can pose challenges. The identification of end points from different sources that are similar enough to be matched is nontrivial and requires end point disclosure; end-point-specific standardization may rely on data samples. Some potential participants owning bigger data volumes may be discouraged from maximal data commitment by substantially asymmetric data contributions yet equal entitlement to the resulting common task models from the usage rights perspective.
Figure 1.
Conceptual representation of the federated setup with two partners of different size, illustrating cross-end point and cross-compound federation. In practice, the number of end points amenable to cross-compound federation is far lower than to cross-end point federation, due to challenges in reconciliation across partners. Identical structures at different partners get identically represented, allowing implicit mapping through the machine learning algorithm without exchanging any sensitive information. Permission has been obtained to use the MELLODDY logo.
Here, we report the implementation and industrial application of a new and alternative approach to federated learning: one that in essence extends multitask learning across multiple parties, while protecting the confidentiality of the underlying data. Conceptually, this approach proposes cross-end point federation across end-point-wise partitioned data sets. Beyond generic standardized formatting, there is no attempt to end point or task matching, so the approach does not require the disclosure of end points. It also imparts more symmetry to usage rights: the party contributing data for some task becomes exclusively entitled to the model components specific to that task, encouraging maximal commitment of confidential data sets. Like in other multitask settings, a joint representation acts as the conduit of information. This joint representation is shared among the data participants but not with the operator of the system.
While, conceptually, cross-end point federation thus avoids some of the challenges of more established cross-compound federation, it may well pose questions of its own. For instance, the information transfer in multitask learning requires some level of commonality across assays and compounds; without it, no predictive benefits can be expected from a joint representation.17 In our privacy context, data composition cannot be shared, encumbering any direct attempt to optimize data composition across partners for information transfer during the modeling. Also, each task is not only defined but ultimately also evaluated by the data points of its (single) contributor, raising questions about whether that contributor-biased evaluation base can adequately assess the benefits of potential information inflow from other contributors. And to what extent would these and other constraints erode potential gains? The three-year MELLODDY project set out to study these questions in the context of a first-in-kind experiment in federated and privacy-preserving machine learning on sensitive industrial data at the relevant data warehouse scale.
Methods
Data and Data Preparation
Combining pharmacological and toxicological assay data of 10 pharma partners (i.e., Amgen, Astellas, AstraZeneca, Bayer, Boehringer Ingelheim, GSK, Janssen, Merck KGaA, Novartis, and Servier) in the multipartner learning, the data volume amounted to 2.6+ billion confidential experimental activity data points, documenting 21+ million chemical compounds and 40+ thousand assays. This corresponded to the vast majority of the partners’ data warehouses. The data set included both alive assays (where data are currently being generated) and historical assays (which have been discontinued). This distinction has been made because alive assays continue to be considered relevant, meet contemporary procedural and quality requirements, and are more likely to be longer running assays that have seen varied types of chemistry. In addition, data from publicly available sources18 were included.
Pharmacological and toxicological assay data can roughly be divided into three types: on-target activity, off-target activity, and ADME (Absorption, Distribution, Metabolism, and Excretion; describing the effect of the body on a drug). The categorization in this work follows these lines. Project specific assays typically covering on-target activity are designated by “Other” and include also phenotypic toxicity. The “ADME” category, in addition to ADME assays, includes physical chemistry assays given their importance to ADME properties. The “Panel” category includes assays used across discovery projects, typically for undesired off-target effects, such as from a generic safety panel.19
On- and off-target assay data will typically result from multiple measurements over a range of different concentrations, resulting in a single number summarizing the overall response. When that number falls outside of the concentration range, it cannot be quantified exactly, and is reported as censored data points (qualified by “>” or “<”). A significant fraction of the data is composed of censored data (i.e., 20% to 80% depending on the partner).
Before measuring over a range of concentrations, a promising activity has typically first been observed in single-concentration high-throughput screening (HTS).20 Imaging data typically result from plate-based imaging screens, where images are acquired by an automated microscope and then processed by image analysis software to generate dense fingerprints of cellular profiles.
All data preparation steps that are independent of a partner’s specific data warehouse setup were performed by each partner independently according to a common protocol, including compound standardization and featurization to ECFP6 chemical fingerprints,22 folded to 32k bits, using the MELLODDY-TUNER21 package. This ensured that identical structures get identically represented by all partners, allowing implicit mapping through the machine learning algorithm without explicitly mapping the structures up front. This approach does not require the exchange of descriptors or assay data across partners or the platform operator. To further increase security, the fingerprints were shuffled prior to training employing a key secret to the platform operator. Thus, shuffling is also required during inference with the trained models.
Modeling
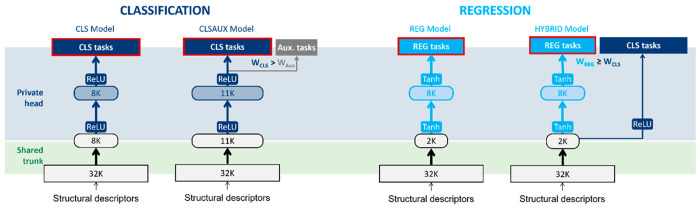
Two main modeling modalities can be distinguished: regression, where a continuous value (assay measurement) was predicted directly, and binary classification, where a label (active/inactive) was predicted relative to a threshold on the assay measurements. Hybrid applies to the case where both classification and regression tasks were trained simultaneously with a single network, with the classification nodes outputting a probability for each task and the regression nodes outputting a continuous value. To this end, the nonlinear activation function was adjusted for classification (ReLU (and softmax)) and regression (Tanh). As shown in Figure 2, adding a layer dedicated to regression was found to be beneficial.
Figure 2.
Overview of the different training modalities with layer sizes commonly optimal for partners for the federated setting (see SI for extensive optimal hyperparameters).
Following the assumption (validated below) that more data integration leads toward more comprehensive and superior predictivity, partners maximized their data contributions to maximize the likelihood of cross-company learning synergies. Doing so implied the addition of data points which may not be routinely considered for modeling, such as low-volume assay data, censored data, multiple thresholds, or data originating from HTS or imaging experiments. As such, it was at the partners’ discretion whether to subject any assay to the data processing pipeline, and cross-partner overlap was not required. The assay data would then form the basis for the tasks to be modeled provided the data volume and quality criteria were met (Table 1). For classification, highly imbalanced tasks were allowed if deemed relevant, while for regression, an additional requirement on the spread of values was imposed after harmonization to standard units to maximize conformity between partners.
Table 1. Minimal Task Data Volume Quora for Training and Evaluation by Assay Type.
| model entry | training quorum | evaluation quorum | |
|---|---|---|---|
| classification | 25 actives and 25 inactives per task | 10 actives and 10 inactives per fold | |
| classification: AUX-HTS | 10 actives and 10000 measurements per task | not applicable | |
| classification: AUX-PL | 10 actives and 10000 measurements per task | not applicable | |
| regression | passed classification training quorum; min standard deviation per task = 10–12 | 50 data points out of which 25 uncensored per task | 50 data points out of which 25 uncensored per fold |
| regression: pIC50-type assays | as above | as above | as above and standard deviation >0.5 per fold |
A large portion of the data participated in the training of the model but was disregarded for performance evaluation. For classification, this included so-called auxiliary tasks based on HTS and image-based data. For regression it was observed that regression-focused hybrid models including auxiliary classification tasks, were superior to classification-focused or balanced hybrid models (Figure S5). It is hypothesized that, since regression tasks were subject to stricter data volume and quality quora (Table 1), new and useful information was brought in from adding classification tasks to regression tasks but not vice versa.
Furthermore, the automatic multithresholding approach resulted in multiple tasks per assay, of which only one was considered for performance evaluation. Likewise, tasks not meeting the data volume quora and all censored data contributed to the model training but were equally disregarded for performance evaluation. Both types of tasks were not considered auxiliary tasks. Hence the term auxiliary data, as it is used here, is narrower than the data used for training but not for evaluation. Instead, it is meant to designate the HTS and image-based data for classification and the classification tasks for regression in a hybrid setting.
Given the challenges posed by the high-volume, high-dimensional, and sparse nature of the input and target matrices, SparseChem23 was well suited for modeling through feedforward neural networks. In the federated setting, the SparseChem models were conceptually split into a private head for every partner containing the output layers for the partner’s distinct tasks, and a shared trunk part common to all partners (Figure 2).24,25 On the platform, the weights of the common trunk could be trained in a federated way by applying secure aggregation26 of the individual gradients from each minibatch of the contributing partners.
Evaluation
Metrics in Labeled Space
Machine learning models are typically evaluated by comparing their predictions with the ground truth on a labeled data set not used for training. For classification models the area under the receiver operator characteristic curve (AUC-ROC) or precision recall curve (AUC-PR) is typically used.27 AUC-ROC values range between 0 and 1 for systematically wrong and perfect predictors, respectively, with 0.5 being the value of a random predictor. AUC-ROC is symmetric, meaning that it is identical for the prediction of active compounds (typically the minority class) and inactive compounds. AUC-PR is considered more informative when the prediction of the minority class (the “actives”) in a highly unbalanced data set is of interest.28 AUC-PR has, however, the disadvantage that it depends on the class ratio, and therefore AUC-PR values cannot easily be compared across data sets or tasks in a multitask setting.
For regression models, the Pearson correlation between ground truth and predicted values, the root-mean-square error of the prediction (RMSE), and the coefficient of determination (R2) are common performance metrics.29,30R2 describes the fraction of variance around the mean of experimental observations that is explained by the model; R2 = 1 – (SSres/SStot) with SS being the residual and total sum of squares, respectively. It has 1.0 as upper bound for a perfect model, and no lower bound. Negative values are observed if the unexplained variance around model predictions (SSres/n) exceeds the variance around the mean (SStot/n) of n experimental observations.
The above metrics have each their individual scale, and the performance of a random predictor is not always well-defined. This can be addressed by calculating for each task the performance difference on a relative scale describing to what extent the task’s performance gap between the baseline model and a perfect model is closed by the model of interest to arrive at the Relative Improvement of Proximity to Perfection (RIPtoP). The performance of the baseline model, perfect model, and model of interest is estimated by metricbaseline, metricperfect, and metricMoI, respectively, with a perfect model being only theoretically attainable for realistic and sizable tasks:
| 1 |
This benefits from the fact that the performance of a perfect model is well-defined in all metrics (1 for AUC-ROC, AUC-PR, R2 and conformal efficiency, and 0 for RMSE). In our case the baseline model was a model trained with data of a single partner only, whereas the model of interest was the federated, multipartner model. The performance metrics depend on the data set used to calculate them.31 In drug discovery, the ability of models to extrapolate outside of the space of their training data to novel chemical compound classes is desirable, and to evaluate this, the train–validation–test fold split needs to be designed accordingly. One way to achieve this is to assign complete chemical classes to a fold. In federated machine learning, this must be done consistently across all data owners in a privacy-preserving way, guaranteeing that the same scaffold at different partners will be assigned to the same fold. For this purpose, a deterministic fold splitting procedure using rule-based scaffold assignment32 has been developed that can be executed independently by the partners with MELLODDY-TUNER.21 This led to more conservative performance assessments compared with a random split while retaining an almost even distribution of compounds across all folds in a (partial) 5-fold cross-validation. After a 5-fold scaffold-based compound split, models are initially trained on three folds, using a fourth fold to select HPs followed by retraining on those four seen folds, and assessed on the last, unseen fold.
Applicability Domain Metrics
Performance metrics such as AUC-ROC, AUC-PR, and R2 require a labeled data set for evaluation. In the context of federated learning, this implies that each partner can apply such metrics to only one’s own tasks and compounds. Given the limited overlap of chemical libraries between partners, one might expect federated learning to inform the model, particularly in those regions of chemical space from other partners, where no labels are available. Using labeled metrics, the best estimate of the predictive performance in such unlabeled spaces is to assume that the values for the labeled data set will be representative for the unlabeled data, regardless of its characteristics. This highlights the need for complementary metrics not requiring labeled data such as uncertainty metrics. In this context, the applicability domain (AD) indicates the chemical space where a model is estimated to make predictions of sufficient reliability.33−35
For classification, the conformal prediction (CP) framework36 is used for this reliability estimation; conformal efficiency (CE), the fraction of predictions estimated to be confident, is a proxy for the size of the AD relative to the chemical space considered.37
For regression, the confidence metric of the spread in predicted values from an ensemble model38 was explored but dropped after only a very limited relationship with performance metrics could be established.
Results
In the following, the main results of the MELLODDY project are presented by comparing the performance of multitask single-partner models trained locally to multitask multipartner models trained in the distributed federated setup, using a variety of visualizations that highlight different aspects of the performance differences. All results are presented on a relative scale to facilitate comparison across different partners and metrics employing RIPtoP as described above.
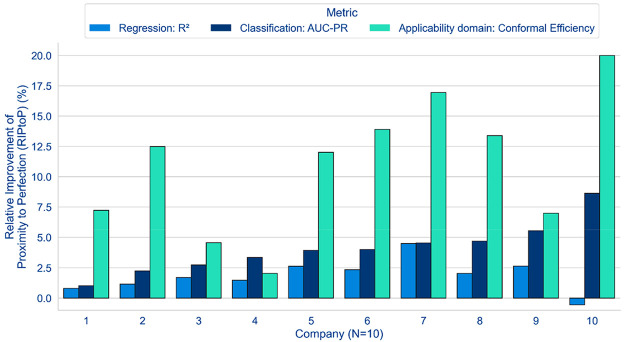
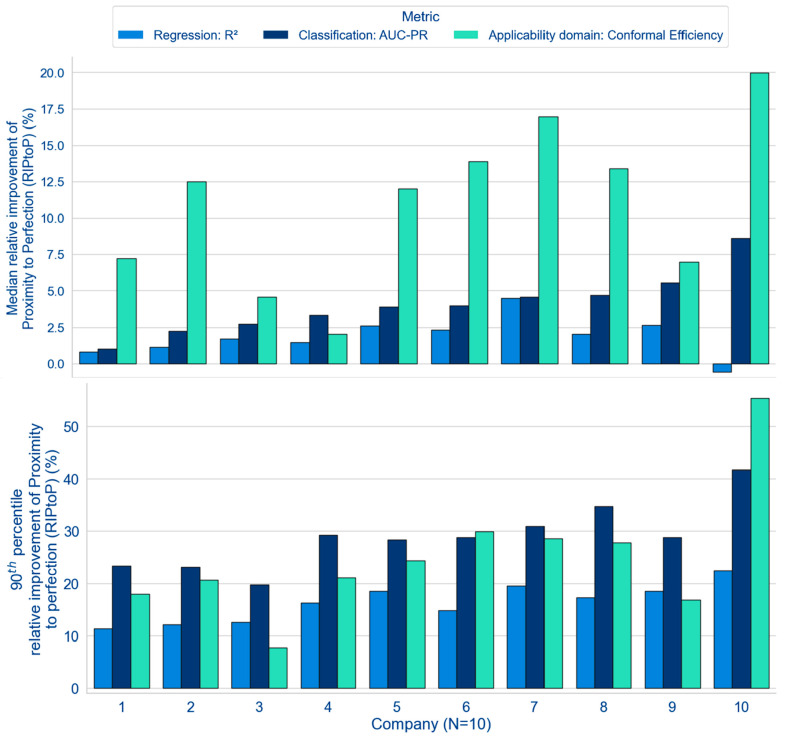
Figure 3 presents an overview of the results for the MELLODDY project performance differences across-companies between optimal multi- and single-partner models (i.e., with or without auxiliary data). Results based on the median task performance clearly demonstrate the benefit for the federated run over the single-partner run in almost all cases, as highlighted by the positive median delta values (y-axis) for the classification metric (AUC-PR) and regression (R2) and for the applicability domain (conformal efficiency; Figure 3, top). Figure S13 highlights that the evidence of federated superiority was robust, regardless of the alternative to the RIPtoP that was selected.
Figure 3.
Performance deltas (between multi- and single-partner runs) based on median (top) and 90th percentile (bottom) across companies for their respective optimal model (either with or without auxiliary data).
The next sections will present a detailed breakdown of the federated performance gains, starting with an analysis of the classification models in terms of predictive performance and applicability domain metrics, followed by a comparison to the regression model performance.
Classification
The overview at the top of Figure 3 outlines a clear trend toward a positive effect of federated learning on the models’ classification metric. Eight partners reported a median RIPtoP(AUC-PR) of more than 2.5%, with one partner over 7.5%, while all partners reported some positive influence of the federated learning. All partners observed at least 20% RIPtoP(AUC-PR) for the best 10% of their models, with one partner reporting over 40% (Figure 3, bottom). The performance of the classification models is further explored in Figure 4.
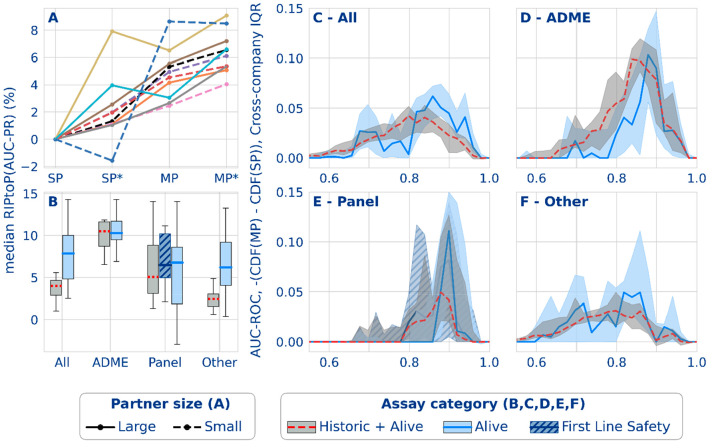
Figure 4.
Classification performance results from the federated run. (A) Effect of multipartner (MP) and auxiliary data (*) on the median AUC-PR task performance for 5 smaller (dashed lines) and 5 larger (solid lines) partners. (B) Distribution of median AUC-PR task performance (RIPtoP(AUC-PR)) over partners. (C–F) Difference between the empirical cumulative distribution functions (CDFs) from single- and multipartner models for different assay types based on AUC-ROC. The difference between the cumulative proportion of tasks in the multi- versus single-partner models (y-axis) is shown for the binned performance (x-axis). The line plots indicate the median probability difference for that bin over all partners. The interquartile ranges are indicated by the shaded envelope. Mind that AUC-ROC is shown here due to its stable baseline of 0.5 for a random classifier.
Figure 4A analyses the influence of collective training data volume on the single- and multipartner models (and how this influences the choice of the optimal model shown in Figure 3). The figure outlines a positive effect for increases in collective training data volume from the single-partner (SP), single-partner with auxiliary data (SP*), multipartner (MP) and multipartner with auxiliary data (MP*) models. Each model performance is normalized to the single-partner baseline and shows a general concordance between partners for the benefit of the inclusion of auxiliary data and also for other partners’ data through federated learning. 9/10 partners had both SP* and MP* preference. A further breakdown of the effect of auxiliary data on performance is included in Figure S8, suggesting that auxiliary data does not consistently additionally enable a model to leverage the federated training (MP*-SP* performance is not consistently greater than MP-SP performance).
Taken together, this indicates that increases in collective training data volume, including by means of auxiliary data resulting from single concentration high-throughput and imaging assays, boosted predictive performances. However, the improvements resulting from the multipartner federation and the introduction of auxiliary data were not linearly additive but rather indicate saturation effects.
Furthermore, Figure 4A partitions the group of partners by the data volume. The effect of a task’s data volume on its change in performance upon going from single-task learning to multitask learning has been studied in the past,17,39 revealing that correlated labels across tasks in similar molecular structures must be present for a performance gain, while the absence of similar molecular structures led to absence of effect. Seeing MP federated learning as a further extension of SP multitask learning, likewise correlated tasks of a minimal size to support the correlation might have the best chances to gain due to federated learning. However, Figure 4A shows no consistent relationship between the performance and the partner size. Assuming the larger partners have larger shares of larger tasks, this could indicate that there is no strong size effect on the performance gains of the individual tasks. Exploratory task-level analyses confirmed this, with an upward trend toward the largest tasks for RIPtoP(AUC-PR), although that effect was caused by a relatively small number of tasks (Figures S22 and S23).
Figure 4B shows the median task performance distribution aggregated across companies and split by assay type, including assays that are alive (see SI for exact assay type definitions). Results showed a higher RIPtoP(AUC-PR) for both panel and ADME tasks than for the remaining ones (other), suggesting that the occurrence of similar panel and ADME assays at a number of pharma companies, causing task correlations across partners, was beneficial for the predictive performance.17 In the category “All”, the higher improvements for the alive assays compared to the general picture can be ascribed to the fact that the alive assays had a higher proportion of panel and ADME assays.
We further postulate a different compound exposure for panel and ADME assays versus that for other assays. Other assays are often project specific on-target assays exposed to compounds from the chemical series that a given medicinal chemistry project is exploring. Panel and ADME assays on the other hand observe broad chemistry from across multiple projects and often competitor compounds resynthesized for characterization. This potential commonality in chemistry with other partners could contribute to the observed federated benefit.
Figure 4C presents the delta between the cumulative distribution functions (CDFs) of the task performances of the SP and MP models. As the value of a CDF at a certain performance threshold will correspond to the fraction of tasks that have a lower or equal performance, the difference of the CDFs shows a clear benefit for the MP model, as indicated by the positive values, corresponding to a shift toward a larger proportion of tasks being assigned to a higher performance. Overall, the benefits in federated learning applied to tasks over the full AUC-ROC range, as outlined by the wide spread of the lines above zero across the bins. They were most apparent for tasks in useful, intermediate performance regions.
These findings are important demonstrations of the usefulness of the federated learning approach since it is most impactful to improve tasks in this intermediate performance range: moderately improving tasks with a low baseline performance still makes the model unusable, while improving tasks with a baseline close to perfection might not have significant impact on the prediction results.
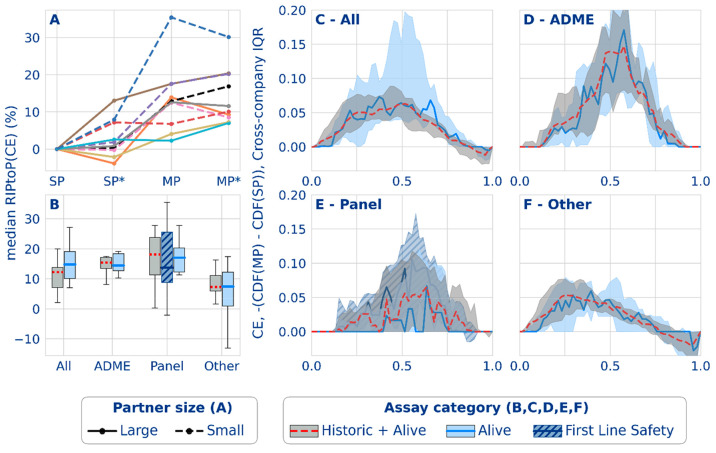
The median task performances in Figure 3 show a clear trend toward a positive effect of federated learning on the models’ applicability domain. As outlined previously,37 conformal efficiency is related to a model’s predictivity and can be considered a proxy for the size of the applicability domain of that model. Six partners reported a median RIPtoP(CE) of more than 10%, with 1 at ∼20%, and 4 partners reported between 0% and 10%. One partner reported >50% improvement, and eight partners between 10% and 30% for the best 10% of their models (Figure 3, bottom). In Figure 5A, contrary to the RIPtoP(AUC-PR), no clearly positive effect can be observed upon inclusion of auxiliary data: MP* was not consistently higher than MP for all partners, and neither was SP* compared to SP. This suggests that the main boost to the applicability domain was caused by the inclusion of the others’ data in the training and that addition of auxiliary data was less important. Neither smaller nor larger partners show a consistently higher improvement, suggesting that the volume of a partner’s training data was not a determining factor, and larger partners too have the potential to extend the applicability domain. Regarding assay types, Figure 5B shows similar patterns to the results on the RIPtoP(AUC-PR). Panel and ADME tasks showed a relatively high RIPtoP(CE), as do the alive assays compared to the remaining ones (other).
Figure 5.
Classification applicability domain results from the federated run. (A) Effect of multipartner (MP) and auxiliary data (*) on the median task performance for 5 smaller (dashed lines) and 5 larger (solid lines) partners. (B) Distribution of median task performance (RIPtoP(CE)) over partners. (C–F) Difference between the empirical cumulative distribution functions (CDFs) from single- and multipartner models for different assay types based on CE. The difference between the cumulative proportion of tasks in the multi- versus single-partner models (y-axis) is shown for the binned performance (x-axis). The line plots indicate the median probability difference for a bin over partners. The interquartile ranges are indicated with the shaded envelope.
Figure 5C illustrates that the benefits apply to tasks over the full range of conformal efficiency. The peaks in the delta CDF for panel and ADME assays occurred at higher values (0.6) than the peaks for the other assays (0.4), mirroring the equivalent plot on RIPtoP(AUC-PR) and reflecting the better overall performance of the panel and ADME assays. Interestingly, for the other assays, a consistent dip across partners toward negative delta CDF values at 0.85 and higher conformal efficiencies could be observed, which was not present for ADME and panel assays. The effect can be explained by the observation that, on one hand, SP models have top efficiencies much closer to perfection, and typically, such tasks predict almost all compounds confidently negative (inactive). Considering the historical hit rates of these tasks, this is considered an instance of overconfidence. MP models on the other hand have more information to support positive predictions, resulting in more predictions with both class labels but also confident (single class label) positive predictions. Since the predictions with both class labels will not contribute to the efficiency, this results in lower values in the MP case, reflected in negative delta CDF values. Finally, Figure S12 shows detailed results confirming previous findings.37 Examples include higher gains in unlabeled space compared to those in labeled space.
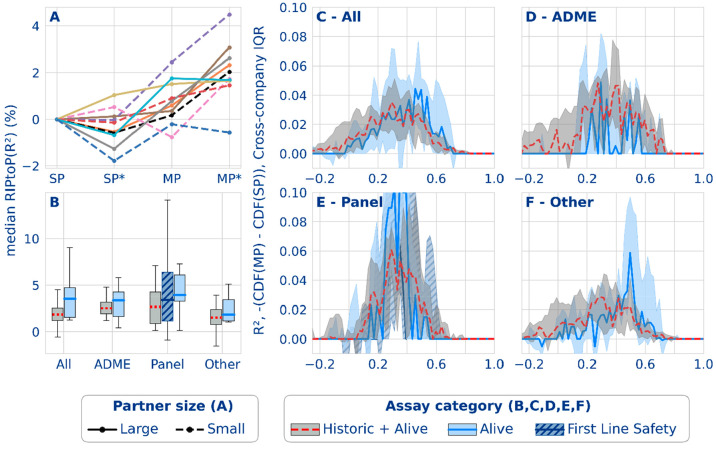
Regression
The general trends observed for the regression models were mostly in line with the classification results; however, subtle differences existed. For all but one partner, the multipartner models exceeded single-partner performance measured by median RIPtoP on the regression metric, R2 (Figure 3, top), albeit with smaller magnitude compared to the classification metric AUC-PR. This finding is repeated by the 90th percentile RIPtoPs. Turning to alternative performance metrics, MP gains on the correlation coefficient are positive for all partners (Figure S20). The magnitude of the relative performance gains was directly related to the location of the baseline performance on the metric scale. While average SP baseline AUC-PR values typically ranged between 0.6 and 0.8 on a scale of 0.0 to 1.0, average R2 values were at lower values (around 0.3 to 0.4) on a scale of −∞ to 1.0 (SI Figures S9–11). As demonstrated in Figure S19, relative performance improvements closer to the end of the scale (observed in typical classification model performance measured in AUC-PR) were emphasized by the RIPtoP metric, rightfully accounting for the increasing difficulty to improve an already good baseline. Upon switching RIPtoP to relative improvement or absolute delta, the ordering of the classification and regression federated gains was comparable or flipped, suggesting that both classification and regression models benefitted equally from federated learning (Figure S18). An additional aspect that is shared between both model types is that they both benefitted from extended data volume in the form of auxiliary data.
Figure 6A shows that the hybrid approach mainly benefitted the federated setup. Only for three partners it improved the SP model, while for 8/10 partners hybrid was the optimal model on the MP side. Auxiliary data improved classification models in 9/10 cases for both SP and MP models. Aggregated over all partners, the median RIPtoP(R2) performance increases by 1.8% on the MP level when replacing a plain regression model by the hybrid approach.
Figure 6.
Regression performance results from the federated run. (A) Effect of multipartner (MP) and auxiliary data (*) on the median task performance for 5 smaller (dashed lines) and 5 larger (solid lines) partners. (B) Distribution of median task performance RIPtoP(R2)) over partners. (C–F) Difference between the empirical cumulative distribution functions (CDFs) from single- and multipartner models for different assay types based on R2. The difference between the cumulative proportion of tasks in the multi- versus single-partner models (y-axis) is shown for the binned performance (x-axis). The line plots indicate the median probability difference for a bin over partners. The interquartile ranges are indicated with the shaded envelope.
Trends in performance gains across assay categories were reproduced in regression with the exception of the ADME tasks that showed the largest improvements in classification but dropped to being comparable to the panel tasks in regression (Figure 6B). A potential reason for the lower multipartner benefit on ADME data in regression could be the increased heterogeneity of ADME end points compared to the standardized dose–response data for non-ADME assays (panel and other). Despite unit standardization of comparable assays across partners, scale normalization (see SI), and a strong overlap across partners, diversity of the data units and scales might still pose a challenge for training federated regression models as opposed to classification models, which are less sensitive since they are trained on binary binned data.
As observed for the classification models, also in regression, the benefit of the federation was most apparent for tasks in useful performance regions (see Figure 6C–F). These tasks had already an acceptable baseline performance which was boosted further by federated learning. As described above, the regression performance scale was shifted to smaller values for the regression metric (R2) compared to that for the classification metric (AUC-PR).
Discussion
The topic of federated learning is one that has attracted much visionary discussion and theoretical exploration but much less effort in concrete application on actual data sets at relevant scale. Insofar that federated learning has already been applied to drug discovery, it has been mostly in the context of cross-compound federation of compound-wise separated data sets documenting activities in a limited and predefined set of generic assays.16,40−42 In that setting, participants must disclose the assays to federate over, which rules out the involvement of more sensitive assays. Second, in principle, participants (and often the operator) get access to the same resulting joint model. This constant return may discourage some potential participants to prepare, involve, and hence risk more than the minimally required data volume. Finally, more than one collaborative effort has been marred by underestimating the challenge of reconciling readouts of somewhat to very different assays even if those were designed to document the same mechanism or target. Beyond material and equipment differences, various extents of divergence of protocols, modality (e.g., biochemical binding assays versus cellular functional assays), or activity direction (agonistic versus antagonistic activities on the same target) can be at play.19
Here we propose an alternative approach: cross-end point federation of end-point-wise separated data sets, one that was inspired by the predictive benefits of multitask learning extended across partners by federation17,43 and designed to incentivize maximal data involvement by all partners. Generally, a task is defined by the labels provided by a given task owner, who also becomes the exclusive owner of the head model for that task. This obviates the general need for task disclosure or reconciliation. The prospect of receiving more and better private head models encourages participants to involve tasks for many assays and maximal data volumes per task. Our approach also enabled increased privacy comfort. In contrast to other state-of-the-art solutions,44,45 the private underlying data and resulting head models never leave the respective owner-controlled architectures, in any form. Information is exchanged as trunk model updates, and secure aggregation26 protects the participants’ privacy, i.e., it prevents the attribution of inferred information to the contributing participant(s). In combination with industry standard security protocols, this incentivization scheme and increased privacy comfort proved a success: all ten pharmaceutical participants involved the vast majority of their SAR data warehouses. To the best of our knowledge, this is the first federation experiment at actual SAR warehouse scale. The collective data volume of 2.6+ billion confidential experimental activity data points, documenting 21+ million physical small molecules and 40+ thousand assays exceeds that of individual participants by almost an order of magnitude on average; it is several orders of magnitude bigger than any other federated or collaborative efforts in drug discovery known to us to date.
Cross-end point federation enabled the improvement of models for tens of thousands of assays, compared to the dozens or at best hundreds of commonly used assays that are typically considered to be practically compatible with cross-compound federation (under the somewhat tenuous assumption of near-perfect reconciliation of readouts across partners). Indeed, for all ten partners, the majority of classification or regression tasks benefitted, and for the majority of those partners, they benefitted with a RIPtoP of more than 4% in AUC-PR and 2% in R2. This indicates that, in practice, the information transfer occurred generally and broadly across a vast spectrum of assays, many of which would not be amenable to cross-compound federation. Notably, a core cross-end point federation scheme can in principle be extended to enable cross-compound federation by mapping common assays to a shared head model.25 To secure the benefits of cross-end point federation, such cross-compound extension may then best be reserved to a limited set of amenable assays, such as some safety panel assays that happen to be outsourced by multiple pharma partners to common contract research organizations.19 Small off-line exercises proved promising (Figure S6). However, this promise did not yet materialize in preliminary experiments at scale that extended cross-end point federation across the full data set with cross-compound federation of a few dozen commonly outsourced safety panel assays. The results suggested that in contrast to the seemingly robust cross-end point modality, a cross-compound extension modality may be more dependent on flawless data preparation and vulnerable to the imbalance between fused and nonfused tasks. While promising, this avenue requires follow-up studies beyond the scope of the original project.
On the magnitude of the observed predictive improvement, there are a few considerations. First, it is important to note that we compare multipartner and single-partner models that are built using the very same multitask modeling approach. Hence, the baseline is a model that is already a multitask model empowered at the scale of a SAR data warehouse. This is important because single partner studies have shown that with a growing number of covered tasks13 or data points46 the predictive performance of multitask models increases consistently but sublinearly, i.e., it gradually slows down. Our results show that predictive performance keeps benefitting without plateauing from adding auxiliary or partner tasks or both beyond the single SAR data warehouse scale, but it does so at a modest pace.
Second, performance improvement clearly depends on the metric used. Here we introduced RIPtoP normalization to mitigate the challenge of differences in distribution of baseline AUC-PR and R2 values among the pharma partners. Importantly, metrics like AUC-PR are ultimately evaluated using data points from the same unique data owner who provided the task-defining data points, while any benefits of federation are driven by data points from other data owners. Any owner-specific data biases may therefore favor the baseline and disfavor the federated performance and hence underestimate performance improvement from federation. We have elsewhere shown that conformal efficiency37 may mitigate such metric biases: a proxy for the size of the applicability domain of a model, i.e., the set of compounds for which a model is estimated to return predictions meeting a predefined confirmation rate, it correlates with AUC-ROC but is less dependent on the choice of evaluation set. Interestingly, this metric shows more pronounced predictive performance gains for classification than for AUC-PR, which suggests that federated models may generalize better. Prospective experimental validation of this hypothesis will require comparative analysis of model robustness over time (see SI for details) and hence remains of out scope of this work.
Lastly, any benefit assessment should also evaluate cost. The MELLODDY experiment has demonstrated that cross-end point federation boosts predictive model performance more often than not and for a notable portion of assays prominently so. On the other hand, while a lot of technological progress has been realized during the project, federated learning to date comes at non-negligible cost and nonzero risk, and it requires building a transparent and reciprocally beneficial case. Opportunity cost is one aspect to work into the equation; what is the cost for physical compound availability and testing that would lead to a similar and similarly robust increase in predictive performance of assays of interest, which importantly requires defining those assays of interests? Another aspect is model update planning. Here a minimally required data volume growth may come to mind, which would depend on several factors, but considering the sublinear performance gains, the lower bound could be set to a 2-fold increase of data of comparable quality and overlap. This can be realized by adding partners or further unlocking alternative data sources, such as images, omics readouts, or target structures.
The MELLODDY project provides a very concrete example of a realistic and economically relevant application of privacy-preserving cross-end point federation at data warehouse scale, to the best of our knowledge, the first example in the field of drug discovery. It has focused on assessing the predictive benefits from cross-partner over single-partner learning. To this end, the project settled early on a robust workhorse predictive technology that had been battle-tested in the drug discovery field, namely, feedforward neural networks processing ECFP-encoded fingerprints. Given their direct compatibility with the technology, Gobbi 2D pharmacophore fingerprints,47 atom pairs,48 and topological torsion49 fingerprints were explored in off-line simulated partner exercises but showed no clear advantage over ECFP fingerprints. MELLODDY has not explored alternative state-of-the-art SAR modeling approaches like graph-convolutional neural networks50,51 or transformers.52 In the absence of a compelling rationale that these methods would favor the federation case, the extensive methodological refactoring required for their inclusion in a privacy-preserving cross-end point federation scheme fell out of scope of the current project. Going forward, the models can be used as-is by individual partners to support drug discovery with direct predictions against included tasks or alternatively be leveraged using more flexible downstream machine learning methods. In the latter case, the model predictions could be added as side information or the shared trunk of the federated models, which uniquely embeds information from multiple companies, could be added to a pool of advanced compound descriptors like CDDD.53
Conclusions
The MELLODDY project is the first realization of cross-end point federated learning in drug discovery across 10 pharma partners, at an unprecedented data warehouse scale. The approach extends the benefits of multitask learning, known from single-partner applications, to the multipartner setting, without compromising the confidentiality of the underlying data.
For all the partners, the majority of classification or regression tasks benefitted, and for the majority of those partners, they benefitted with a Relative Improvement of Proximity to Perfection (RIPtoP) of more than 4% in AUC-PR and 2% in R2 or of more than 12.5% in AUC-PR and 4.8% in R2 for at least one partner. Due to partner-specific biases, these conventional metrics may underestimate the predictive benefit from cross-end point federation as suggested by a median RIPtoP in conformal efficiency of at least 12% for the majority of partners and exceeding 20% for one partner. The best overall predictive performance was obtained after adding auxiliary data in the form of HTS or image-based data.
Models for ADME and panel assays showed more pronounced predictive performance improvements compared with more partner-specific assays, probably driven by the occurrence of similar assays at multiple partners.
As an outlook, we see the onset of MELLODDY making a mark on pharmaceutical companies, whether through best-practice data processing with MELLODDY-TUNER or computationally efficient large-scale modeling with SparseChem. The models as they are can be employed to guide decision making or optimize resources, or they can serve as generators for input features or side information. We believe that the operational and scientific achievements of the MELLODDY project have shown the potential of federated learning in real life. The current and other scientific publications and open-source software libraries that this project establishes in its wake may inspire future collaborative modeling efforts in drug discovery and beyond.
Acknowledgments
At all partner sites, we are grateful to our many colleagues who advanced this project through scientific discussions, IT, and administrative support: Anne Bonin, Florian Boulnois, Marc Daxer, Fang Du, Pierre Farmer, Oleksandr Fedorenko, Oliver Fortmeier, Grégori Gerebtzoff, Peter Grandsard, Anke Hackl, André Hildebrandt, Holger Hoefling, Dieter Kopecky, Stefan Korte, Jimmy Kromann, Daniel Kuhn, Peter Kutchukian, Paula Marin Zapata, Risto Milani, Floriane Montanari, Frank Morawietz, Britta Nisius, Aileen Novero, Carl Petersson, Jordon Rahaman, Dak Rojnuckarin, and Nikolaus Stiefl. Specials thanks go out to our project managers Tinne Boeckx and Evelyn Verstraete. Luc Geeraert is thanked for excellent scientific writing contributions. The sponsored provisioning of computing infrastructure by AWS is gratefully acknowledged. This research received funding from the Flemish Government (AI Research Program). Y.M., A.Ar., J.S., M.O. and R.L. are affiliated to Leuven.AI - KU Leuven institute for AI, B-3000, Leuven, Belgium. This project has received funding from the Innovative Medicines Initiative 2 Joint Undertaking under grant agreement No 831472. This Joint Undertaking receives support from the European Union’s Horizon 2020 research and innovation programme and EFPIA.
Data Availability Statement
The massive underlying data sets amount to the bulk of the private QSAR warehouses of the partners, which guarantees industrial and economic relevance. However, it absolutely precludes the publication of the underlying massive and private data sets or the models that were derived.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jcim.3c00799.
Author Contributions
Conceptualization: M.O., J.S., A.Ar., R.L., D.A.C., C.L., D.G., N.D.H., H.F., E.B., B.B., O.E., A.H.G., Y.M., M.N.G., A.S., H.C. Data curation: W.H., L.M., T.M., N.S., L.F., A.Z., A.P., L.H., R.N., P.S., A.Ar., N.D., R.J., A.Af., S.V., S.H., M.H., N.H., M.K., P.E., H.F., S.P., A.G., A.H.G., A.S., H.C. Formal analysis: W.H., L.M., T.M., N.S., L.F., A.Z., A.P., L.H., M.O., R.N., P.S., J.S., A.Ar., N.D., A.Af., R.L., S.H., M.K., H.F., S.P., A.G., Y.M., H.C. Funding acquisition: D.A.C., D.G., H.F., E.B., B.B., F.R., A.H.G., Y.M., M.N.G., A.S., H.C. Investigation: W.H., L.M., T.M., N.S., L.F., A.Z., A.P., L.H., M.O., R.N., J.S., A.Ar., N.D., R.J., A.Af., R.L., S.V., S.H., M.H., B.P., M.T., N.R., M.K., C.L., P.A., D.M., H.F., S.P., Y.S., A.G., O.E., Y.M., M.N.G. Methodology: W.H., L.M., T.M., N.S., L.F., A.Z., A.P., L.H., M.O., N.F., J.S., A.Ar., R.J., R.L., B.P., M.K., P.A., H.F., S.P., E.B., F.R., Y.M., M.N.G., A.S., H.C. Project administration: T.M., N.D., F.Z., M.K., D.G., H.F., E.B., B.B., A.G., A.H.G., Y.M., M.N.G., A.S., H.C. Resources: W.H., L.M., N.S., L.F., N.H., D.A.C., M.K., N.D.H., H.F., E.B., B.B., A.H.G., M.N.G., H.C. Software: W.H., L.M., T.M., N.S., L.F., A.Z., A.P., L.H., M.O., P.S., J.S., A.Ar., N.D., R.J., R.L., M.T., N.H., A.D., H.F., E.B., M.N.G., A.S. Supervision: N.F., J.S., A.Ar., N.D., R.L., A.D., N.R., D.A.C., C.L., D.G., D.M., N.D.H., H.F., S.P., G.A., B.B., Y.S., F.R., O.E., A.H.G., Y.M., M.N.G., A.S., H.C. Validation: W.H., L.M., T.M., N.S., L.F., A.Z., A.P., L.H., M.O., P.S., J.S., A.Ar., R.L., M.K., H.F., A.G., H.C. Visualization: W.H., L.M., T.M., N.S., L.F., A.Z., H.F., S.P., H.C. Writing – original draft: W.H., L.M., T.M., N.S., L.F., A.Z., A.P., L.H., R.N., N.F., N.D., B.P., S.P., Y.M., A.S., H.C. Writing – review & editing: W.H., L.M., T.M., N.S., L.F., A.Z., A.P., L.H., M.O., R.N., A.Ar., B.P., N.H., A.D., M.K., P.A., H.F., S.P., G.A., B.B., A.G., F.R., Y.M., A.S., H.C.
The authors declare the following competing financial interest(s): M.N.G. and M.T. are employed and own stocks in the company Owkin commercializing the underlying Federated Learning Platform based on the open source Substra software. The remaining authors have no conflicts of interest to declare.
An SI file was added October 20, 2023.
Supplementary Material
References
- Hansch C.; Maloney P. P.; Fujita T.; Muir R. M. Correlation of Biological Activity of Phenoxyacetic Acids with Hammett Substituent Constants and Partition Coefficients. Nature 1962, 194, 178–180. 10.1038/194178b0. [DOI] [Google Scholar]
- Hansch C.; Fujita T. ρ-σ-π Analysis. A Method for the Correlation of Biological Activity and Chemical Structure. J. Am. Chem. Soc. 1964, 86, 1616–1626. 10.1021/ja01062a035. [DOI] [Google Scholar]
- Muratov E. N.; Bajorath J.; Sheridan R. P.; Tetko I. V.; Filimonov D.; Poroikov V.; Oprea T. I.; Baskin I. I.; Varnek A.; Roitberg A.; Isayev O.; Curtalolo S.; Fourches D.; Cohen Y.; Aspuru-Guzik A.; Winkler D. A.; Agrafiotis D.; Cherkasov A.; Tropsha A. QSAR without Borders. Chem. Soc. Rev. 2020, 49, 3525–3564. 10.1039/D0CS00098A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y.; Chen K. X.; Jiang H. L.; Ji R. Y. QSAR/QSTR of Fluoroquinolones: An Example of Simultaneous Analysis of Multiple Biological Activities Using Neural Network Method. Eur. J. Med. Chem. 1998, 33, 647–658. 10.1016/S0223-5234(98)80023-8. [DOI] [Google Scholar]
- González-Díaz H.; Prado-Prado F. J.; Santana L.; Uriarte E. Unify QSAR Approach to Antimicrobials. Part 1: Predicting Antifungal Activity against Different Species. Bioorg. Med. Chem. 2006, 14, 5973–5980. 10.1016/j.bmc.2006.05.018. [DOI] [PubMed] [Google Scholar]
- Unterthiner T.; Mayr A.; Klambauer G.; Steijaert M.; Wegner J.; Ceulemans H.; Hochreiter S. Deep Learning as an Opportunity in Virtual Screening. Adv. Neural Inf. Process. Syst. 2014, 27, 1–9. [Google Scholar]
- Göller A. H.; Kuhnke L.; Montanari F.; Bonin A.; Schneckener S.; ter Laak A.; Wichard J.; Lobell M.; Hillisch A. Bayer’s in Silico ADMET Platform: A Journey of Machine Learning over the Past Two Decades. Drug Discovery Today 2020, 25, 1702. 10.1016/j.drudis.2020.07.001. [DOI] [PubMed] [Google Scholar]
- Mayr A.; Klambauer G.; Unterthiner T.; Steijaert M.; Wegner J. K.; Ceulemans H.; Clevert D. A.; Hochreiter S. Large-Scale Comparison of Machine Learning Methods for Drug Target Prediction on ChEMBL. Chem. Sci. 2018, 9, 5441–5451. 10.1039/C8SC00148K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenselink E. B.; Ten Dijke N.; Bongers B.; Papadatos G.; Van Vlijmen H. W. T.; Kowalczyk W.; Ijzerman A. P.; Van Westen G. J. P. Beyond the Hype: Deep Neural Networks Outperform Established Methods Using a ChEMBL Bioactivity Benchmark Set. J. Cheminform. 2017, 9, 1–14. 10.1186/s13321-017-0232-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsundar B.; Liu B.; Wu Z.; Verras A.; Tudor M.; Sheridan R. P.; Pande V. Is Multitask Deep Learning Practical for Pharma?. J. Chem. Inf. Model. 2017, 57, 2068–2076. 10.1021/acs.jcim.7b00146. [DOI] [PubMed] [Google Scholar]
- Dahl G. E.; Jaitly N.; Salakhutdinov R.. Multi-Task Neural Networks for QSAR Predictions. arXiv 2014, 1406.1231, https://arxiv.org/abs/1406.1231.
- Wenzel J.; Matter H.; Schmidt F. Predictive Multitask Deep Neural Network Models for ADME-Tox Properties: Learning from Large Data Sets. J. Chem. Inf. Model. 2019, 59, 1253–1268. 10.1021/acs.jcim.8b00785. [DOI] [PubMed] [Google Scholar]
- Ramsundar B.; Kearnes S.; Riley P.; Webster D.; Konerding D.; Pande V.. Massively Multitask Networks for Drug Discovery. arXiv 2015, 1502.02072, https://arxiv.org/abs/1502.02072. [Google Scholar]
- McMahan H. B.; Moore E.; Ramage D.; Hampson S.; Agüera y Arcas B. Communication-Efficient Learning of Deep Networks from Decentralized Data. Proc. 20th Int. Conf. Artif. Intell. Stat. AISTATS 2017 2017, 54, 1273–1282. [Google Scholar]
- Yang Q.; Liu Y.; Chen T.; Tong Y. Federated Machine Learning: Concept and Applications. ACM Trans. Intell. Syst. Technol. 2019, 10, 1–19. 10.1145/3298981. [DOI] [Google Scholar]
- Hanser T.; Bastogne D.; Basu A.; Davies R.; Delaunois A.; Fowkes A.; Harding L.; Johnston C.; Korlowski; Kotsampasakou E.; Plante J.; Rosenbrier-Ribeiro L.; Rowell P.; Sabnis Y.; Sartini A.; Sibony A.; Werner S.; White A.; Yukawa T.. Using privacy-preserving federated learning to enable pre-competitive cross-industry knowledge sharing and improve QSAR models. 2022Society of Toxicology (SOT) Annual Meeting. https://www.lhasalimited.org/wp-content/uploads/2023/02/Using-privacy-preserving-federated-learning-to-enable-pre-competitive-cross-industry-knowledge-sharing-and-improve-QSAR-models.pdf.
- Xu Y.; Ma J.; Liaw A.; Sheridan R. P.; Svetnik V. Demystifying Multitask Deep Neural Networks for Quantitative Structure-Activity Relationships. J. Chem. Inf. Model. 2017, 57, 2490–2504. 10.1021/acs.jcim.7b00087. [DOI] [PubMed] [Google Scholar]
- Gaulton A.; Hersey A.; Nowotka M. L.; Bento A. P.; Chambers J.; Mendez D.; Mutowo P.; Atkinson F.; Bellis L. J.; Cibrian-Uhalte E.; Davies M.; Dedman N.; Karlsson A.; Magarinos M. P.; Overington J. P.; Papadatos G.; Smit I.; Leach A. R. The ChEMBL Database in 2017. Nucleic Acids Res. 2017, 45, D945–D954. 10.1093/nar/gkw1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowes J.; Brown A. J.; Hamon J.; Jarolimek W.; Sridhar A.; Waldron G.; Whitebread S. Reducing Safety-Related Drug Attrition: The Use of in Vitro Pharmacological Profiling. Nat. Rev. Drug Discovery 2012, 11, 909–922. 10.1038/nrd3845. [DOI] [PubMed] [Google Scholar]
- Wildey M. J.; Haunso A.; Tudor M.; Webb M.; Connick J. H.. High-Throughput Screening. Platform Technologies in Drug Discovery and Validation; Goodnow R. A., Jr., Ed.; Annual Reports in Medicinal Chemistry; Elsevier Inc., 2017; Vol. 50, pp 149–195, 10.1016/bs.armc.2017.08.004. [DOI] [Google Scholar]
- MELLODDY-TUNER. https://github.com/melloddy/MELLODDY-TUNER.
- Rogers D.; Hahn M. Extended-Connectivity Fingerprints. J. Chem. Inf. Model. 2010, 50, 742–754. 10.1021/ci100050t. [DOI] [PubMed] [Google Scholar]
- Arany A.; Simm J.; Oldenhof M.; Moreau Y.. SparseChem: Fast and Accurate Machine Learning Model for Small Molecules. arXiv 2022, 2203.04676, https://arxiv.org/abs/2203.04676.
- Galtier M.; Marini C.. Substra: A Framework for Privacy-Preserving, Traceable and Collaborative Machine Learning. arXiv 2019, 1910.11567, https://arxiv.org/abs/1910.11567.
- Oldenhof M.; Ács G.; Pejó B.; Schuffenhauer A.; Holway N.; Sturm N.; Dieckmann A.; Fortmeier O.; Boniface E.; Mayer C.; Gohier A.; Schmidtke P.; Niwayama R.; Kopecky D.; Mervin L.; Rathi P. C.; Friedrich L.; Formanek A.; Antal P.; Rahaman J.; Zalewski A.; Heyndrickx W.; Oluoch E.; Stößel M.; Vančo M.; Endico D.; Gelus F.; de Boisfossé T.; Darbier A.; Nicollet A.; Blottière M.; Telenczuk M.; Nguyen V. T.; Martinez T.; Boillet C.; Moutet K.; Picosson A.; Gasser A.; Djafar I.; Simon A.; Arany A.; Simm J.; Moreau Y.; Engkvist O.; Ceulemans H.; Marini C.; Galtier M. Industry-Scale Orchestrated Federated Learning for Drug Discovery. Proc. AAAI Conf. Artif. Intell. 2023, 37, 15576–15584. 10.1609/aaai.v37i13.26847. [DOI] [Google Scholar]
- Ács G.; Castelluccia C. I Have a DREAM! (DiffeRentially PrivatE SmArt Metering). Lect. Notes Comput. Sci. (including Subser. Lect. Notes Artif. Intell. Lect. Notes Bioinformatics) 2011, 6958, 118–132. 10.1007/978-3-642-24178-9_9. [DOI] [Google Scholar]
- Davis J.; Goadrich M. The Relationship between Precision-Recall and ROC Curves. ICML 2006 - Proceedings of the 23rd International Conference on Machine Learning 2006, 233–240. 10.1145/1143844.1143874. [DOI] [Google Scholar]
- Saito T.; Rehmsmeier M. The Precision-Recall Plot Is More Informative than the ROC Plot When Evaluating Binary Classifiers on Imbalanced Datasets. PLoS One 2015, 10, e0118432. 10.1371/journal.pone.0118432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chicco D.; Warrens M. J.; Jurman G. The Coefficient of Determination R-Squared Is More Informative than SMAPE, MAE, MAPE, MSE and RMSE in Regression Analysis Evaluation. PeerJ. Comput. Sci. 2021, 7, e623. 10.7717/peerj-cs.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S. Correlation and Causation. J. Agric. Res. 1921, XX, 557–585. [Google Scholar]
- Sheridan R. P. Time-Split Cross-Validation as a Method for Estimating the Goodness of Prospective Prediction. J. Chem. Inf. Model. 2013, 53, 783–790. 10.1021/ci400084k. [DOI] [PubMed] [Google Scholar]
- Simm J.; Humbeck L.; Zalewski A.; Sturm N.; Heyndrickx W.; Moreau Y.; Beck B.; Schuffenhauer A. Splitting Chemical Structure Data Sets for Federated Privacy-Preserving Machine Learning. J. Cheminform. 2021, 13, 1–14. 10.1186/s13321-021-00576-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingspohn W.; Mathea M.; Ter Laak A.; Heinrich N.; Baumann K. Efficiency of Different Measures for Defining the Applicability Domain of Classification Models. J. Cheminform. 2017, 9, 1–17. 10.1186/s13321-017-0230-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragos H.; Gilles M.; Alexandre V. Predicting the Predictability: A Unified Approach to the Applicability Domain Problem of Qsar Models. J. Chem. Inf. Model. 2009, 49, 1762–1776. 10.1021/ci9000579. [DOI] [PubMed] [Google Scholar]
- Norinder U.; Carlsson L.; Boyer S.; Eklund M. Introducing Conformal Prediction in Predictive Modeling. A Transparent and Flexible Alternative to Applicability Domain Determination. J. Chem. Inf. Model. 2014, 54, 1596–1603. 10.1021/ci5001168. [DOI] [PubMed] [Google Scholar]
- Shafer G.; Gammerman A.; Vovk V.. Algorithmic Learning in a Random World; Springer, 1981; Vol. 3. [Google Scholar]
- Heyndrickx W.; Arany A.; Simm J.; Pentina A.; Sturm N.; Humbeck L.; Mervin L.; Zalewski A.; Oldenhof M.; Schmidtke P.; Friedrich L.; Loeb R.; Afanasyeva A.; Schuffenhauer A.; Moreau Y.; Ceulemans H. Conformal Efficiency as a Metric for Comparative Model Assessment Befitting Federated Learning. Artif. Intell. Life Sci. 2023, 3, 100070. 10.1016/j.ailsci.2023.100070. [DOI] [Google Scholar]
- Lakshminarayanan B.; Pritzel A.; Blundell C. Simple and Scalable Predictive Uncertainty Estimation Using Deep Ensembles. NIPS ’17: Proceedings of the 31st International Conference on Neural Information Processing Systems 2017, 6405–6416. 10.5555/3295222.3295387. [DOI] [Google Scholar]
- Ma J.; Sheridan R. P.; Liaw A.; Dahl G. E.; Svetnik V. Deep Neural Nets as a Method for Quantitative Structure-Activity Relationships. J. Chem. Inf. Model. 2015, 55, 263–274. 10.1021/ci500747n. [DOI] [PubMed] [Google Scholar]
- Verras A.; Waller C. L.; Gedeck P.; Green D. V. S.; Kogej T.; Raichurkar A.; Panda M.; Shelat A. A.; Clark J.; Guy R. K.; Papadatos G.; Burrows J. Shared Consensus Machine Learning Models for Predicting Blood Stage Malaria Inhibition. J. Chem. Inf. Model. 2017, 57, 445–453. 10.1021/acs.jcim.6b00572. [DOI] [PubMed] [Google Scholar]
- Bosc N.; Felix E.; Arcila R.; Mendez D.; Saunders M. R.; Green D. V. S.; Ochoada J.; Shelat A. A.; Martin E. J.; Iyer P.; Engkvist O.; Verras A.; Duffy J.; Burrows J.; Gardner J. M. F.; Leach A. R. MAIP: A Web Service for Predicting Blood-Stage Malaria Inhibitors. J. Cheminform. 2021, 13, 13. 10.1186/s13321-021-00487-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S.; Xue D.; Chuai G.; Yang Q.; Liu Q. FL-QSAR: A Federated Learning-Based QSAR Prototype for Collaborative Drug Discovery. Bioinformatics 2021, 36, 5492–5498. 10.1093/bioinformatics/btaa1006. [DOI] [PubMed] [Google Scholar]
- Smith V.; Chiang C.; Sanjabi M.; Talwalkar A.. Federated Multi-Task Learning. In Advances in Neural Information Processing Systems; MIT Press, 2017; Vol. 30. [Google Scholar]
- Ma R.; Li Y.; Li C.; Wan F.; Hu H.; Xu W.; Zeng J. Secure Multiparty Computation for Privacy-Preserving Drug Discovery. Bioinformatics 2020, 36, 2872–2880. 10.1093/bioinformatics/btaa038. [DOI] [PubMed] [Google Scholar]
- Martin E. J.; Zhu X. W. Collaborative Profile-QSAR: A Natural Platform for Building Collaborative Models among Competing Companies. J. Chem. Inf. Model. 2021, 61, 1603–1616. 10.1021/acs.jcim.0c01342. [DOI] [PubMed] [Google Scholar]
- de la Vega de León A.; Chen B.; Gillet V. J. Effect of Missing Data on Multitask Prediction Methods. J. Cheminform. 2018, 10, 1–12. 10.1186/s13321-018-0281-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobbi A.; Poppinger D. Genetic Optimization of Combinatorial Libraries. Biotechnol. Bioeng. 1998, 61, 47–54. . [DOI] [PubMed] [Google Scholar]
- Smith D. H.; Carhart R. E.; Venkataraghavan R. Atom Pairs as Molecular Features in Structure-Activity Studies: Definition and Applications. J. Chem. Inf. Comput. Sci. 1985, 25, 64–73. 10.1021/ci00046a002. [DOI] [Google Scholar]
- Nilakantan R.; Bauman N.; Venkataraghavan R.; Dixon J. S. Topological Torsion: A New Molecular Descriptor for SAR Applications. Comparison with Other Descriptors. J. Chem. Inf. Comput. Sci. 1987, 27, 82–85. 10.1021/ci00054a008. [DOI] [Google Scholar]
- Gilmer J.; Schoenholz S. S.; Riley P. F.; Vinyals O.; Dahl G. E. Neural Message Passing for Quantum Chemistry. 34th Int. Conf. Mach. Learn. ICML 2017 2017, 3, 2053–2070. [Google Scholar]
- Yang K.; Swanson K.; Jin W.; Coley C.; Eiden P.; Gao H.; Guzman-Perez A.; Hopper T.; Kelley B.; Mathea M.; Palmer A.; Settels V.; Jaakkola T.; Jensen K.; Barzilay R. Analyzing Learned Molecular Representations for Property Prediction. J. Chem. Inf. Model. 2019, 59, 3370–3388. 10.1021/acs.jcim.9b00237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaswani A.; Shazeer N.; Parmar N.; Uszkoreit J.; Jones L.; Gomez A. N.; Kaiser Ł.; Polosukhin I.. Attention Is All You Need. In Advances in Neural Information Processing Systems; MIT Press, 2017; Vol. 30. [Google Scholar]
- Winter R.; Montanari F.; Noé F.; Clevert D. A. Learning Continuous and Data-Driven Molecular Descriptors by Translating Equivalent Chemical Representations. Chem. Sci. 2019, 10, 1692–1701. 10.1039/C8SC04175J. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The massive underlying data sets amount to the bulk of the private QSAR warehouses of the partners, which guarantees industrial and economic relevance. However, it absolutely precludes the publication of the underlying massive and private data sets or the models that were derived.