Abstract
Strain GDVII and other members of the GDVII subgroup of Theiler’s murine encephalomyelitis virus (TMEV) are highly virulent and cause acute polioencephalomyelitis in mice. Neither viral persistence nor demyelination is demonstrated in the few surviving mice. On the other hand, strain DA and other members of the TO subgroup of TMEV are less virulent and establish a persistent infection in the spinal cord, which results in a demyelinating disease. We previously reported that GDVII does not actively replicate in a murine macrophage-like cell line, J774-1, whereas DA strain productively infects these cells (M. Obuchi, Y. Ohara, T. Takegami, T. Murayama, H. Takada, and H. Iizuka, J. Virol. 71:729–733, 1997). In the present study, we used recombinant viruses between these strains of the two subgroups to demonstrate that the DA L coding region of DA strain is important for virus growth in J774-1 cells. Additional experiments with a mutant virus indicate that L* protein, which is synthesized out of frame with the polyprotein from an additional alternative initiation codon in the L coding region of TO subgroup strains, is a key determinant responsible for the cell-type-specific restriction of virus growth. L* protein may play a critical role in the DA-induced restricted demyelinating infection by allowing growth in macrophages, a major site for virus persistence.
Theiler’s murine encephalomyelitis virus (TMEV) includes a number of murine picornavirus strains that are divided into two subgroups on the basis of their different biological activities. Strain GDVII and other members of the GDVII subgroup produce acute fatal polioencephalomyelitis in mice. In the few surviving mice, no virus persistence is observed (10). In contrast, strain DA and other members of the TO subgroup persistently infect the spinal cord, resulting in demyelination. The white matter disease caused by TO subgroup strains is an excellent experimental model of the human demyelinating disease multiple sclerosis (15). The precise mechanisms of viral persistence and demyelination of TO subgroup strains remain poorly understood.
Macrophages may be a critical cell in the TMEV-induced demyelinating disease. Studies suggest that the virus persists primarily in macrophages (1, 5, 9, 12) and that virus persistence is required for TMEV-induced demyelination (2, 11). Macrophages persistently infected with TO subgroup strains may secrete cytokines or other factors which induce myelin breakdown, resulting in demyelination. In a previous study (14), we compared infection of a murine macrophage-like cell line, J774-1, with two TMEV subgroup strains. Strain GDVII did not actively replicate in J774-1 cells, whereas strain DA productively infected the cells. In the present study, we compared the infection of intratypic recombinant and mutant viruses in J774-1 cells to identify the molecular determinants responsible for TMEV growth in J774-1 cells. These findings may be important in our understanding of virus replication and multiplication in macrophages and the pathogenesis of TMEV-induced demyelinating disease.
MATERIALS AND METHODS
Cells.
J774-1, an H-2d macrophage-like cell line derived from a tumor of a female BALB/c mouse, and BHK-21 cells, a baby hamster kidney-derived fibroblast cell line permissive for TMEV infection, were maintained as previously described (14).
Recombinant and mutant viruses.
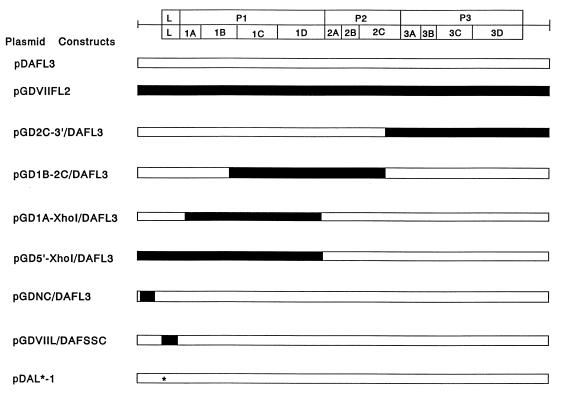
A series of chimeric and mutant cDNAs are shown in Fig. 1. Chimeric cDNA clones constructed by replacing part of pDAFL3 with segments from pGDVIIFL2 as previously described (6, 18, 20) are named by first listing 5′ and 3′ junctions of the GDVII segment, then adding a slash, and finally adding DAFL3. pDAFL3 and pGDVIIFL2 are wild-type full-length infectious cDNAs from strains DA and GDVII, respectively (6, 18). pGD2C-3′/DAFL3 contains a segment of pGDVIIFL2 from a StuI site at nucleotide (nt) 4843 in 2C to the 3′ terminus. pGD1B-2C/DAFL3 contains a pGDVIIFL2 segment from an NcoI site at nt 1964 in 1B to the same StuI site. pGD1A-XhoI/DAFL3 contains a pGDVIIFL2 segment from a SauI site at nt 1332 in 1A to a silent XhoI site, which was engineered to leave a perfect P1/P2 junction following XhoI digestion (20). pGD5′-XhoI/DAFL3 contains a pGDVIIFL2 segment from the 5′ terminus to the same XhoI site. pGDNC/DAFL3 contains a pGDVIIFL2 segment from an EcoRI site at nt 280 to a KpnI site at nt 936. pDAFSSC was generated by introducing six silent restriction enzyme cleavage sites into the pDAFL3 leader (L) coding region (8). In pGDVIIL/DAFSSC, the DA L coding region was substituted by GDVII L. pDAL*-1 has a mutation of the AUG at nt 1079 to ACG, which is an alternative, out-of-frame, initiation codon used for the synthesis of L* protein (7).
FIG. 1.
A series of chimeric and mutant cDNAs generated to delineate sequences responsible for virus growth in J774-1 cells. The position of the TMEV coding area is shown at the top. The GDVII genome and segments from it are shown as solid bars, and the DA genome is shown as open bars. The asterisk indicates a point mutation of the L coding region of pDAL*-1, changing the L* initiation AUG to ACG (but not changing the coding region for the polyprotein).
Kinetics of virus growth.
The kinetics of the growth of the recombinant and mutant viruses in J774-1 cells were examined as previously described (14). A 35-mm-diameter plastic culture dish containing 3 × 106 cells was infected at a multiplicity of infection of 10 PFU per cell. The culture supernatants and cell lysates were harvested at the indicated times and subjected to titer determination by a standard plaque assay on BHK-21 cells.
Pulse-labeling experiment of viral and cellular proteins.
Radiolabeling of viral and cellular proteins was performed as previously described (14). Briefly, 3 × 106 cells in a 35-mm-diameter plastic culture dish were infected with each virus at a multiplicity of infection of 10 PFU per cell and incubated in methionine-free medium. At the indicated times postinoculation (p.i.), the cells were pulse-labeled with 0.1 mCi of l-[35S]methionine (1,000 Ci/mmol; Amersham) per ml in methionine-free medium for 3 h. The cells were then scraped off the dish and dissolved in sample buffer (0.01 M Tris-HCl [pH 6.8], 1 mM EDTA, 2.5% sodium dodecyl sulfate, 5% 2-mercaptoethanol, 10% glycerol, 0.001% bromophenol blue) after being washed with phosphate-buffered saline. The cell lysates were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis in a 12% polyacrylamide gel.
RNase protection assay.
To prepare the antisense probe for mapping the 5′ end of the viral RNA, pDAFL3 was cleaved at the HincII site in the viral genome (nt 7783). The linearized plasmid was transcribed in an in vitro transcription system with T3 RNA polymerase (Stratagene, La Jolla, Calif.) in the presence of [α-32P]UTP as described by the manufacturer. The radiolabeled 362-nt RNA probe contained 310 nt of sequence complementary to the 3′ region of the viral genome and 52 nt of vector sequence (16–18).
The RNase protection assay was performed with the RPA II RNase protection assay kit (Ambion Inc., Austin, Tex.). Total RNA was extracted and purified from 7 × 106 virus-infected J774-1 cells by the guanidine thiocyanate method (4). A 10-μg portion of RNA and 100,000 cpm of 32P-labeled probe were hybridized for 16 h at 42°C and treated with RNase solution as specified by the manufacturer. The RNase-resistant fragments were detected by electrophoresis on a 5% polyacrylamide gel containing 8 M urea. The gels were dried and exposed to a type BAS-III imaging plate (Fuji Photo Film Co., Ltd., Kanagawa, Japan) and analyzed with a Storm 860 bioimaging analyzer (Molecular Dynamics Japan, Tokyo, Japan).
RESULTS
Growth of recombinant viruses in J774-1 cells.
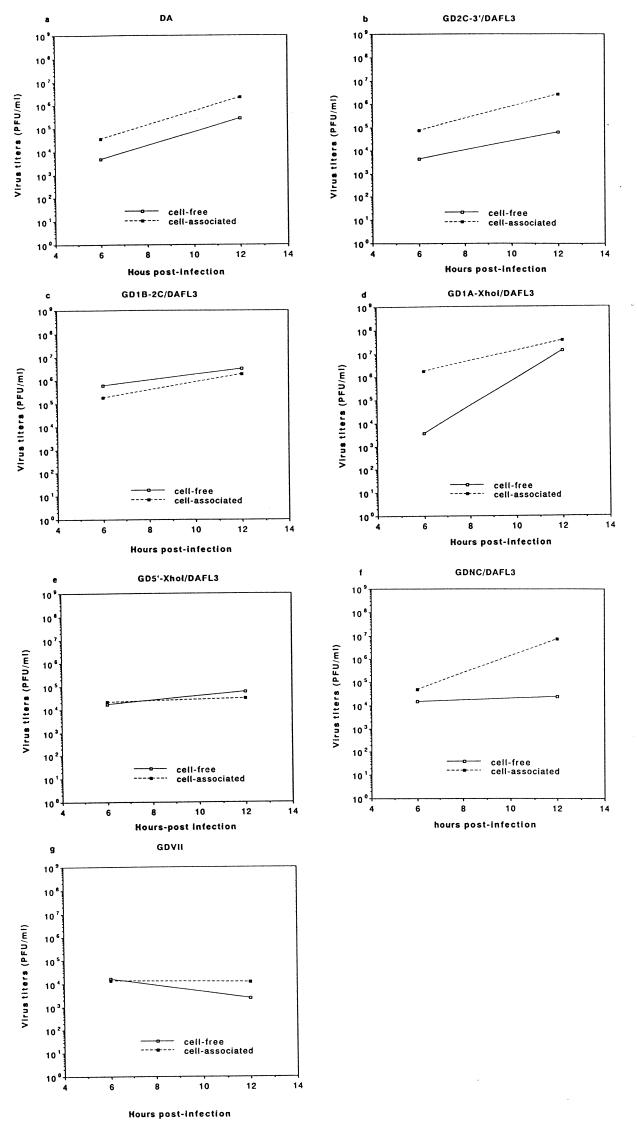
Since our previous report (14) demonstrated that infectivity data collected at 6 and 12 h p.i. reliably indicated whether a virus grew in J774-1 cells, virus titers at these times were first examined. Figure 2 shows the infectivity titers following infection of J774-1 cells with five recombinant viruses and the parental DA and GDVII viruses. As expected, the 12-h-p.i. levels of cell-free and cell-associated DA virus were more than 2 log units higher than the 6-h-p.i. levels. On the other hand, the 12-h-p.i. levels of cell-free and cell-associated GDVII viruses were lower than the 6-h-p.i. levels. For infection with GD2C-3′/DAFL3, GD1B-2C/DAFL3, and GD1A-XhoI/DAFL3 recombinant viruses, the levels of cell-free and cell-associated viruses were approximately 4 × 103 to 2 × 106 PFU/ml at 6 h p.i. and increased to 6 × 104 to 4 × 107 PFU/ml at 12 h p.i. The 12-h-p.i. levels were 1 to 3 log units higher than the 6-h-p.i. levels. These results indicated growth of those recombinant viruses in J774-1 cells. In contrast, the levels of cell-free and cell-associated GD5′-XhoI/DAFL3 viruses were approximately 2 × 104 PFU/ml at 6 h p.i. and remained at a similar level at 12 h p.i., demonstrating that they did not grow in J774-1 cells. The above results suggested that a determinant(s) for virus growth in J774-1 cells was present in the 5′ untranslated region or the L coding region. To further narrow the critical region, we infected J774-1 cells with GDNC/DAFL3 virus, which contains most of the GDVII 5′ untranslated region. The level of cell-associated GDNC/DAFL3 virus increased from 5 × 104 PFU/ml at 6 h p.i. to approximately 7 × 106 PFU/ml at 12 h p.i. The level of cell-free GDNC/DAFL3 virus at 6 h p.i. (1.5 × 104 PFU/ml) increased only slightly at 12 h p.i. (2.4 × 104 PFU/ml); however, further studies indicated that the level of cell-free virus increased to 3 × 106 PFU/ml at 24 h p.i. (data not shown). In summary, virus growth occurred after infection with GDNC/DAFL3, GD2C-3′/DAFL3, GD1B-2C/DAFL3, and GD1A-XhoI/DAFL3 but not GD5′-XhoI/DAFL3, suggesting that the DA L coding region is important for virus growth in J774-1 cells. To further clarify the determinants critical for restriction of virus growth, experiments were performed with GDVIIL/DAFSSC and DAL*-1 mutant viruses.
FIG. 2.
Growth of the parental and recombinant viruses in J774-1 cells. The culture supernatants (solid lines) and cell lysates (broken lines) of infected J774-1 cells were harvested at 6 and 12 h p.i. and subjected to titer determination by a standard plaque assay on BHK-21 cells. Data are expressed as the mean of three independent experiments. (a and g) parental DA and GDVII viruses, respectively; (b to f) recombinants: GD2C-3′/DAFL3 (b), GD1B-2C/DAFL3 (c), GD1A-XhoI/DAFL3 (d), GD5′-XhoI/DAFL3 (e), and GDNC/DAFL3 (f). Details of the construction of these viruses are provided in Materials and Methods and shown in Fig. 1.
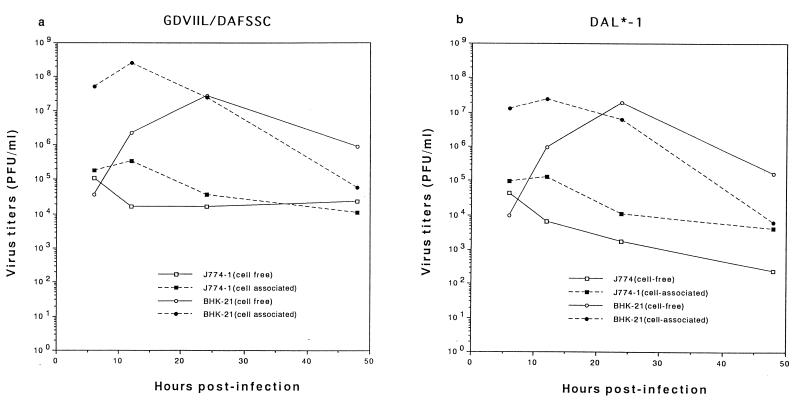
Growth of GDVIIL/DAFSSC and DAL*-1 mutant viruses in BHK-21 and J774-1 cells.
Since our previous study (14) showed that the infection of TMEV in BHK-21 cells is different from that in J774-1 cells, the growth of GDVIIL/DAFSSC and DAL*-1 mutant viruses was examined and compared following infection of BHK-21 and J774-1 cells at several time points (Fig. 3). In BHK-21 cells, infection with GDVIIL/DAFSSC and DAL*-1 mutant viruses led to levels of cell-free and cell-associated viruses which peaked at 12 and 24 h p.i., respectively, and ranged from 107 to 109 PFU/ml. In contrast, the kinetics of growth of these two viruses in J774-1 cells were remarkably different. The 12-h-p.i. titers of both cell-associated GDVIIL/DAFSSC and DAL*-1 viruses were minimally higher than the 6-h-p.i. titers, gradually decreasing to approximately 104 PFU/ml at 48 h p.i. This tendency was even more prominent for cell-free viruses, where there was no apparent increase of cell-free virus titers but instead steadily decreasing titers. The results indicated that both GDVIIL/DAFSSC and DAL*-1 viruses were able to grow in BHK-21 cells but did not grow in J774-1 cells.
FIG. 3.
Kinetics of GDVIIL/DAFSSC and DAL*-1 virus growth in BHK-21 and J774-1 cells. The culture supernatants (solid lines) and cell lysates (broken lines) of infected BHK-21 or J774-1 cells were harvested at the indicated times and subjected to titer determination by a standard plaque assay on BHK-21 cells. Data are expressed as the mean of three independent experiments. (a) GDVIIL/DAFSSC; (b) DAL*-1. Details of the construction of these viruses are provided in Materials and Methods and shown in Fig. 1.
Viral and cellular protein synthesis of GDVIIL/DAFSSC and DAL*-1 mutant virus following infection of J774-1 cells.
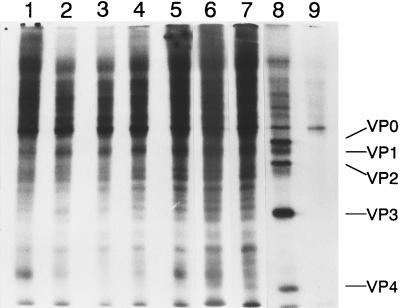
To further characterize GDVIIL/DAFSSC and DAL*-1 viruses, viral and cellular protein synthesis was analyzed in J774-1 cells by a pulse-label experiment. As previously published (14), newly synthesized virus-specific proteins with some evidence of the shutoff of host cellular protein were clearly demonstrated 12 h after DA infection of J774-1 cells (Fig. 4, lane 8) and there was a remarkable inhibition of both cellular and viral protein synthesis 12 h after GDVII infection of J774-1 cells (lane 9). For GDVIIL/DAFSSC and DAL*-1 virus infection of J774-1 cells, there was only a minimum amount of viral protein synthesis throughout the 12-h period of observation. In addition, no prominent inhibition of cellular protein synthesis was observed (lanes 2 to 7).
FIG. 4.
Protein synthesis in TMEV-infected J774-1 cells. J774-1 cells infected with GDVIIL/DAFSSC virus (lane 2 to 4) or DAL*-1 virus (lanes 5 to 7) at various times p.i. were labeled for 3 h with l-[35S]methionine as described in Materials and Methods. Lanes: 2 and 5, labeled at 3 h p.i.; 3 and 6, labeled at 6 h p.i.; 4 and 7, labeled at 9 h p.i.; 8 and 9, DA- and GDVII-infected J774-1 cells, respectively, labeled at 9 h p.i.; 1, mock-infected J774-1 cells.
Viral RNA synthesis of DAL*-1 mutant virus following infection of J774-1 cells.
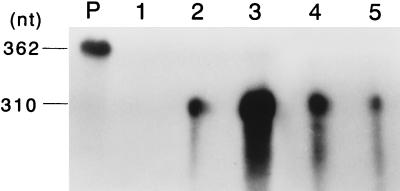
To examine whether the limited viral growth and viral protein synthesis occur at the level of viral RNA replication, viral RNA synthesis of DAL*-1 virus was analyzed by an RNase protection assay and compared with that of the parental DA strain. A representative gel pattern is shown in Fig. 5. In J774-1 cells infected with the parental DA strain, viral genomic RNA clearly increased from 3 to 9 h p.i. On the other hand, in J774-1 cells infected with DAL*-1 virus, a decrease of viral genomic RNA was observed from 3 to 9 h. Similar patterns were also found in several independent experiments (data not shown). The data suggested that the restriction of replication of DAL*-1 virus in infection of J774-1 cells occurred at the step of viral RNA synthesis, as previously reported for the parental GDVII strain (14).
FIG. 5.
Genomic RNA of the parental DA and mutant DAL*-1 viruses following infection of J774-1 cells. Total RNA was extracted from J774-1 cells 3 h (lanes 2 and 4) and 9 h (lanes 3 and 5) after infection of DA (lanes 2 and 3) or DAL*-1 (lanes 4 and 5) virus. A 10-μg portion of RNA was hybridized with [α-32P]UTP-labeled riboprobe and treated with RNase solution. The RNase-resistant fragments were denatured, electrophoresed on a 5% polyacrylamide–8 M urea gel, and analyzed with a bioimaging analyzer. Lanes P and 1 contained [α-32P]UTP-labeled probe and mock-infected J774-1 cells, respectively.
DISCUSSION
A number of investigators have attempted to delineate determinants for the TMEV-induced disease phenotypes. Recent studies have implicated the L coding region, which is located at the start of the open reading frame that is used to synthesize the polyprotein and has an unknown function (19). A previously published study showed that the L coding region affects virus expression in certain cell types (8) and indicated the probability that L protein plays a role in DA-induced demyelinating disease (8). Of interest is the recent recognition that the L coding region of TO subgroup strains (DA, BeAn, TO4/WW, and Yale) has an additional alternative, out-of-frame open reading frame starting at the AUG at nt 1079 (7, 13) which synthesizes a 17-kDa protein called L* in vivo and in vitro (3, 7). In contrast, an ACG rather than an AUG is present in a genomic region of the virulent GDVII subgroup strains that corresponds in location to the L* initiation codon of DA (7, 13); therefore, GDVII subgroup strains fail to synthesize L* protein (7). The importance of L* protein to the late white matter disease was demonstrated by noting that DAL*-1 virus, which has a mutation of the DAL* AUG to ACG (but with no change in the polyprotein coding region), has markedly decreased demyelinating activity (3).
We previously reported the rather surprising result that GDVII strain fails to replicate in a murine macrophage-like cell line, J774-1, while DA productively infects these cells (14). We were especially interested in this observation because macrophages are a major site of persistence of TO subgroup strains (1, 5, 9, 12) and because virus persistence is required for DA-induced demyelinating disease (2, 11).
To identify the determinants for the cell type restriction, we investigated the growth of a number of recombinant and mutant TMEV strains in J774-1 cells. GD2C-3′/DAFL3, GD1B-2C/DAFL3, and GD1A-XhoI/DAFL3 viruses grew similarly to the parental DA strain in J774-1 cells, suggesting that a segment of GDVII from the 1A coding region to the 3′ terminus does not affect virus growth in J774-1 cells. GDNC/DAFL3 also grew well; however, there was some delay in virus release. This delay may result from an influence of the GDVII 5′ noncoding region when present in the DA genome. The finding is being investigated further. On the other hand, GD5′-XhoI/DAFL3 did not grow, suggesting that a determinant in the DA L coding region is critical for virus growth in J774-1 cells. This suggestion was confirmed by noting that growth of GDVIIL/DAFSSC virus, which contains the L coding region of GDVII substituted for that of DA, is severely restricted in J774-1 cells. Growth in J774-1 cells was also restricted following infection with DAL*-1 virus. Since the only difference in the proteins produced following the parental DA and DAL*-1 virus infections is the synthesis of L* protein, the reason for virus growth in J774-1 cells following infection with the parental DA virus and recombinant viruses that contain the DA L region (which has the L* AUG) is presumably the synthesis of L* protein. In the case of DAL*-1 virus, GDVII virus, and recombinant viruses that do not contain the DA L region (which lacks the L* AUG) and do not synthesize L* protein, the growth in J774-1 cells was not observed. We considered further experiments testing the growth in macrophages of a GDVII mutant virus which contained the L* AUG. Initial studies, however, suggest that the presence of the L* AUG within the GDVII genome is not sufficient for the synthesis of L*, making this approach difficult to pursue. L* protein may play a critical role in the demyelinating disease because it permits growth in macrophages with subsequent persistence in these cells, leading to demyelination.
A previously published study (8) showed that GDVIIL/DAFSSC and DAL*-1 viruses grew well in BHK-21 cells but required a high MOI for growth in L-929 cells whereas wild-type DA virus grew well in both cell lines at high and low MOIs (8). We found that these viruses grew well in BHK-21 cells but failed to multiply efficiently in J774-1 cells even at an MOI of 10. These results suggest that the mechanism for restriction of virus growth in J774-1 cells is different from that in L-929 cells and/or that the L coding region contains another determinant(s) for cell-type-specific growth besides L*.
The present study demonstrated that recombinant or mutant TMEV that does not synthesize the L* protein has a restricted growth in J774-1 cells. On the other hand, DAL*-1 virus does not demonstrate a shutoff of host cell synthesis in the presence of a minimal production of viral proteins, as is the case for GDVII virus (14). These findings suggest that the effects of GDVII infection on the synthesis of cellular and viral proteins in J774-1 cells are not solely related to the presence or absence of L* protein.
ACKNOWLEDGMENTS
This work was supported by a Grant-in-Aid for Scientific Research from the Ministry of Education, Science, Sports and Culture; a grant for Project Research from Kanazawa Medical University (P96-1); a grant from the Japan Health Sciences Foundation; and a grant from the National Institutes of Health (to R.P.R.).
REFERENCES
- 1.Aubert C, Chamorro M, Brahic M. Identification of Theiler’s virus infected cells in the central nervous system of the mouse during demyelinating disease. Microb Pathog. 1987;3:319–326. doi: 10.1016/0882-4010(87)90002-7. [DOI] [PubMed] [Google Scholar]
- 2.Chamorro M, Aubert C, Brahic M. Demyelinating lesions due to Theiler’s virus are associated with ongoing central nervous system infection. J Virol. 1986;57:992–997. doi: 10.1128/jvi.57.3.992-997.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen H-H, Kong W-P, Zhang L, Ward P L, Roos R P. A picornaviral protein synthesized out of frame with the polyprotein plays a key role in a virus-induced immune-mediated demyelinating disease. Nat Med. 1995;1:927–931. doi: 10.1038/nm0995-927. [DOI] [PubMed] [Google Scholar]
- 4.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 5.Clatch R J, Miller S D, Metzner R, Dal Canto M C, Lipton H L. Monocytes/macrophages isolated from the mouse central nervous system contain infectious Theiler’s murine encephalomyelitis virus (TMEV) Virology. 1990;176:244–254. doi: 10.1016/0042-6822(90)90249-q. [DOI] [PubMed] [Google Scholar]
- 6.Fu J, Stein S, Rosenstein L, Bodwell T, Routbort M, Semler B L, Roos R P. Neurovirulence determinants of genetically engineered Theiler viruses. Proc Natl Acad Sci USA. 1990;87:4125–4129. doi: 10.1073/pnas.87.11.4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kong W-P, Roos R P. Alternative translation initiation site in the DA strain of Theiler’s murine encephalomyelitis virus. J Virol. 1991;65:3395–3399. doi: 10.1128/jvi.65.6.3395-3399.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kong W-P, Ghadge G D, Roos R P. Involvement of cardiovirus leader in host cell-restricted virus expression. Proc Natl Acad Sci USA. 1994;91:1796–1800. doi: 10.1073/pnas.91.5.1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Levy M, Aubert C, Brahic M. Theiler’s virus replication in brain macrophages cultured in vitro. J Virol. 1992;66:3188–3193. doi: 10.1128/jvi.66.5.3188-3193.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lipton H L. Persistent Theiler’s murine encephalomyelitis virus infection in mice depends on plaque size. J Gen Virol. 1980;46:169–177. doi: 10.1099/0022-1317-46-1-169. [DOI] [PubMed] [Google Scholar]
- 11.Lipton H L, Calenoff M, Bandyopadhyay P, Miller S D, Dal Canto M C, Gerety S, Jensen K. The 5′ noncoding sequences from a less virulent Theiler’s virus dramatically attenuate GDVII neurovirulence. J Virol. 1991;65:4370–4377. doi: 10.1128/jvi.65.8.4370-4377.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lipton H L, Twaddle G, Jelachich M L. The predominant virus antigen burden is present in macrophages in Theiler’s murine encephalomyelitis virus-induced demyelinating disease. J Virol. 1995;69:2525–2533. doi: 10.1128/jvi.69.4.2525-2533.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Michiels T, Jarousse N, Brahic M. Analysis of the leader and capsid coding regions of persistent and neurovirulent strains of Theiler’s virus. Virology. 1995;214:550–558. doi: 10.1006/viro.1995.0066. [DOI] [PubMed] [Google Scholar]
- 14.Obuchi M, Ohara Y, Takegami T, Murayama T, Takada H, Iizuka H. Theiler’s murine encephalomyelitis virus subgroup strain-specific infection in a murine macrophage-like cell line. J Virol. 1997;71:729–733. doi: 10.1128/jvi.71.1.729-733.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ohara Y, Roos R P. The antibody response in Theiler’s virus infection: new perspectives on multiple sclerosis. Prog Med Virol. 1987;34:156–179. [PubMed] [Google Scholar]
- 16.Ohara Y, Stein S, Fu J, Stillman L, Klaman L, Roos R P. Molecular cloning and sequence determination of DA strain of Theiler’s murine encephalomyelitis viruses. Virology. 1988;164:245–255. doi: 10.1016/0042-6822(88)90642-3. [DOI] [PubMed] [Google Scholar]
- 17.Pevear D C, Borkowski J, Calenoff M, Oh C K, Ostrowski B, Lipton H L. Insights into Theiler’s virus neurovirulence based on a genomic comparison of the neurovirulent GDVII and less virulent BeAn strains. Virology. 1988;165:1–12. doi: 10.1016/0042-6822(88)90652-6. [DOI] [PubMed] [Google Scholar]
- 18.Roos R P, Stein S, Ohara Y, Fu J, Semler B L. Infectious cDNA clones of the DA strain of Theiler’s murine encephalomyelitis virus. J Virol. 1989;63:5492–5496. doi: 10.1128/jvi.63.12.5492-5496.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rueckert R R. Picornaviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, Chanock R M, Melnick J L, Monath T P, Roizman B, Straus S E, editors. Fields virology. 3rd ed. Vol. 1. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 609–654. [Google Scholar]
- 20.Zhang L, Senkowski A, Shim B, Roos R P. Chimeric cDNA studies of Theiler’s murine encephalomyelitis virus neurovirulence. J Virol. 1993;67:4404–4408. doi: 10.1128/jvi.67.7.4404-4408.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]