Abstract
Background:
Global plastic use has consistently increased over the past century with several different types of plastics now being produced. Much of these plastics end up in oceans or landfills leading to a substantial accumulation of plastics in the environment. Plastic debris slowly degrades into microplastics (MPs) that can ultimately be inhaled or ingested by both animals and humans. A growing body of evidence indicates that MPs can cross the gut barrier and enter into the lymphatic and systemic circulation leading to accumulation in tissues such as the lungs, liver, kidney, and brain. The impacts of mixed MPs exposure on tissue function through metabolism remains largely unexplored.
Objectives:
This study aims to investigate the impacts of polymer microspheres on tissue metabolism in mice by assessing the microspheres ability to translocate across the gut barrier and enter into systemic circulation. Specifically, we wanted to examine microsphere accumulation in different organ systems, identify concentration-dependent metabolic changes, and evaluate the effects of mixed microsphere exposures on health outcomes.
Methods:
To investigate the impact of ingested microspheres on target metabolic pathways, mice were exposed to either polystyrene () microspheres or a mixture of polymer microspheres consisting of polystyrene (), polyethylene (), and the biodegradability and biocompatible plastic, poly-(lactic-co-glycolic acid) (). Exposures were performed twice a week for 4 weeks at a concentration of either 0, 2, or via oral gastric gavage. Tissues were collected to examine microsphere ingress and changes in metabolites.
Results:
In mice that ingested microspheres, we detected polystyrene microspheres in distant tissues including the brain, liver, and kidney. Additionally, we report on the metabolic differences that occurred in the colon, liver, and brain, which showed differential responses that were dependent on concentration and type of microsphere exposure.
Discussion:
This study uses a mouse model to provide critical insight into the potential health implications of the pervasive issue of plastic pollution. These findings demonstrate that orally consumed polystyrene or mixed polymer microspheres can accumulate in tissues such as the brain, liver, and kidney. Furthermore, this study highlights concentration-dependent and polymer type-specific metabolic changes in the colon, liver, and brain after plastic microsphere exposure. These results underline the mobility within and between biological tissues of MPs after exposure and emphasize the importance of understanding their metabolic impact. https://doi.org/10.1289/EHP13435
Introduction
Over the past 50 years, global plastic production has grown exponentially. To date, metric tons of plastic are produced globally every year.1 Much of this plastic ends up in landfills or oceans where it may take several hundred years to degrade depending on composition and environmental factors.2 Exposure to light, heat, moisture, and microbes degrades plastic debris into microplastics (MPs), defined as plastic particles smaller than .3 MPs have become ubiquitous throughout our environment, and exposure to humans and animals is thought to occur through ingestion4–9 or inhalation.10,11 Multiple studies have reported MP detection in food,12 salt water,13 fresh water,14–16 farming soils,17 and crops used for both animal and human consumption.18 A 2019 review of over 50 existing studies on MPs suggests that consumption of common foods and beverages results in humans ingesting of plastic per week.19 It is currently estimated that by 2050, metric tons of plastic wastes will be released into the environment by bioturbation, atmospheric deposition, sewage irrigation, and landfills1,14,17,20 as well as exposure from indoor activities that include the use of laser printers, photocopiers, and three-dimensional (3D) printing as thoroughly reviewed in the following articles.21–25 With an estimated 3.2 (and growing) metric tons of MPs being released into the environment via commercial and household activities every year, MPs exposure is now unavoidable.26
In 2019, the World Health Organization (WHO) released a statement that, based on the limited available evidence, exposure to MPs poses a low concern for human health.27 However, subsequent reports of cellular and biochemical toxicity of MPs have made it clear that deleterious interactions between MPs and biological systems are concentration-dependent, meaning that the impacts of MPs exposures may increase with time.28 Ingestion is believed to be the most common route of MPs exposure.4–9,29–32 MP exposure in rats,33 mice,34,35 and zebrafish36,37 models has been shown to lead to gut microbiota dysbiosis. Gut dysbiosis is linked to numerous inflammatory and metabolic diseases,38–43 and studies in zebrafish exposed to MPs have been shown to induce intestinal injury and inflammation.36,44–48 In contrast to these reports, other studies have concluded MPs caused no intestinal histological damage in the colon of mice.49,50 In humans, recent evidence show MPs are abundant in colons collected after a colectomy51 and increased in the stool of individuals with inflammatory bowel disease52; however, the effects of MPs on gastrointestinal (GI) health is still being deciphered. Preliminary in vitro work has assessed the toxicity and uptake of micro- and nanoplastics in cell lines such as human colorectal adenocarcinoma cells (Caco-2)53 and human colon intestinal (HT29-MTX-E12)54 cells as well as a mixture of these cells.55,56 Additionally, studies in marine animals,46 rodent organoids,57 and immune and lung cell cultures58,59 have shown that MPs can alter cellular bioenergetics and induce oxidative stress and inflammation.
Exposing animals to MPs via the oral gastric route leads to the dissemination of MPs outside of the intestine, specifically the liver.60 Additionally, several studies in humans, mice, chickens, rats, and zebrafish have shown MP and nanoplastic (NP) ingestion results in their accumulation in tissues such as the placenta,61 liver,48,57 and kidney.62,63 The distribution into the liver caused metabolic changes in mice30,61 and fish.34,64,65 These studies highlight how MPs can cross the intestinal barrier and can cause extraintestinal manifestations (e.g., inflammation and oxidative stress). However, there is a lack of evidence regarding the effects of a combination of microplastics. The studies conducted so far have solely focused on investigating the impacts of polystyrene microplastics; they have not examined the metabolic changes that can take place in organs that have a direct interaction with the gut, such as the kidney and brain. As we begin to understand the environmentally relevant sizes, types, and concentrations of MPs humans are ingesting,9,66–68 researchers are able to further assess the effects of MPs using environmentally relevant concentrations, sizes, and types. The most common MPs include polyethylene (PE), polypropylene (PP), and polystyrene (PS).69 There are numerous other polymer types that have potential to become microplastics, such as polyester (PES), polyamide (PA), polyurethane (PU), polycarbonate (PC), polyvinyl chloride (PVC), and polyethylene terephthalate (PET) as well as synthetic textiles. Microbeads derived from PE, PP, and PS are commonly used in cosmetics and hygiene products.70 Other plastics such as poly(lactic-co-glycolic acid) (PLGA) is widely used for microencapsulation and prolonged delivery of materials. PLGA is biodegradable and recognized as being safe by several regulatory agencies in the US and Europe.71 To date, the systemic impacts of MP ingestion on metabolic pathways in organs have not been studied utilizing environmentally relevant concentrations and mixtures. Given PE and PS are two of the most common MPs that are purposely generated and utilized as microbeads and have leached into the environment,72 we set out to understand how PS microspheres () and mixed microspheres () exposure consisting of PS, PE, and PLGA at environmentally relevant concentrations cross the intestinal barrier and alter metabolism in the colon, liver, and brain. The addition of PLGA was used due to its utilization to encapsulate a broad range of therapeutic agents that delivered via the oral route. Specifically, we evaluated the systemic distribution and metabolic impacts of polystyrene and mixed polymer microsphere ingestion in mice after oral gavage. While not reflective of the myriad environmental MPs, plastic microspheres are a research model that controls for size, shape, and composition, while eliminating the contributions of endotoxin, pyrogens, metals, or other contaminants that may have interactive and confounding effects. Microscopic visualization and Raman spectroscopy were used to evaluate microsphere accumulation in the colon and translocation to the liver and brain, assess composition postexposure, and identify any physical or chemical changes associated with biological degradation. Targeted and untargeted metabolic profiling was then used to identify functional responses within the colon, liver, and brain. Moreover, we investigated sex-specific effects on the metabolome, conducting analyses with a total of males and females per exposure group. This study is one of the first to evaluate impacts of not only polystyrene microspheres but also mixed polymer microspheres at an equivalent concentration to what humans are estimated to consume per week.73
Methodology
Animal Model
Male and female C57BL/6 mice (8–12 wk of age at the beginning of the study) were obtained from Taconic Biosciences (Rensselaer, New York). Animals were housed in an Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC)-approved facility at the University of New Mexico Health Sciences Center. Animals were maintained at constant temperature (20–24°C), relative humidity (30%–60%), and on a 12-h light/dark cycle throughout the study. Animals were provided with normal chow and water ad libitum. All experiments were approved by the Institutional Animal Care and Use Committee of the University of New Mexico Health Sciences Center, in accordance with the National Institutes of Health guidelines for use of live animals. The University of New Mexico Health Sciences Center is accredited by the American Association for Accreditation of Laboratory Animal Care.
Exposure, Tissue Digestion, and Microplastic Isolation
Mice were exposed twice a week to polystyrene or mixed polymer microspheres via oral gastric gavage over a 4-wk period at (), (), and () of microspheres from Degradex (Phosphorex). Exposures were based on an estimated average of between 0.1 and of microplastics ingested by humans globally per week through all exposure pathways.73 Two different microsphere exposures were used for each concentration group; there was a control group (vehicle only) for each type of exposure, for a total of six groups. The two different microsphere exposures were a) polystyrene (PS) microspheres (Degradex; catalog number 127) and b) a mixed plastics treatment in a 1:1:1 ratio consisting of polystyrene (PS) (Degradex; catalog number 127), polyethylene (PE) (Cospheric; catalog number CPMS-0.96 ), and poly (lactic-co-glycolic acid) (PLGA) (Degradex; catalog number LG5000) microspheres. Prior to gavage, all microspheres were stored based on the manufacturer’s recommendations in a concentration suspended in deionized water with a small amount of surfactant and sodium azide as an antimicrobial agent at until exposure. Prior to administration, microspheres were washed using vehicle (deionized water). Microspheres were removed in dose concentrations for each gavage of or per week. These microspheres were centrifuged down at for 10 min to remove excess microsphere solution. Then same volume of fresh vehicle was added. Microspheres were vortexed and then respun in centrifuge at for 10 min, and washing cycle was repeated for a total of 3 rounds. Each week, the mice were exposed twice weekly at per exposure for a total of or per exposure for a total of . After 4 weeks, the mice were euthanized using isoflurane and exsanguinated, then systemically perfused for one minute and thirty seconds with ice cold saline to ensure removal of blood from major organs. Serum, brain, liver, kidney, and colon were isolated, snap frozen, and stored at . Samples are stored in glass vials to prevent plastic contamination from the postmortem procedures. All mice in each exposure group were housed in polycarbonate cages.
Digestion of the prefrontal cortex of the brain, left lobe of liver, and sagittal cross-section of kidney tissues was performed using the sample volume of 10% potassium hydroxide (KOH) prepared using deionized water. Samples were incubated at with agitation for 72 h. Samples were ultracentrifuged (Thermo Scientific Sorvall WX+) at for 4 h to isolate MPs into a pellet. The supernatant was removed, and the pellet was washed with 100% ethanol (EtOH) and centrifuged at for 10 min, followed by removal of excess EtOH. The process was repeated three times. After washing, the samples were resuspended in EtOH and stored at in glass tubes for processing.
To prevent adulteration from airborne and procedural contaminants, we used the following preventative measures: a) Latex gloves were worn during all experiments along with 100% cotton lab coats, b) all reagents were prepared using ultrapure water (GE LifeSciences; catalog number SH30529.02), c) all samples were stored in glass vials (DWK Life Sciences), d) all surfaces were covered in absorbent bench coat and cleaned with 70% ETOH prior to experiments, e) use of plastic containing equipment was kept to a bare minimum, f) and all tools were cleansed and autoclaved prior to use.
Visualization and Spectroscopic Characterization of Microplastics
Light microscopy.
Samples of isolated plastics were stored in 100% EtOH in glass tubes for a minimum of 24 h at before imaging. Slides were prepared by adding of sample, and MPs were identified and imaged via polarized light microscopy using an Olympus BX51 microscope. An ultraviolet light source was used to verify that the remaining solids were plastic in nature based on autofluorescence.
Raman spectroscopy.
Raman Spectroscopy analysis was processed on a WITec Alpha 300R Confocal Raman microscope with a laser on isolated microspheres from brain to confirm that microspheres of interest were polystyrene. Additionally, pristine polymeric microspheres that underwent 10% potassium hydroxide (KOH) digestion protocol were analyzed to determine if alteration occurred based on biological or chemical degradation. A series of peaks specific to the materials were generated. Substance-specific peaks from an in-house Infrared and Raman Characteristic Group Frequencies library [polystyrene (PS) microspheres (Degradex; catalog number 127); polyethylene (PE) (Cospheric; catalog number CPMS-0.96 ); and poly (lactic-co-glycolic acid) (PLGA) (Degradex; catalog number LG5000) microspheres] were compared to the generated data to identify the materials.
X-ray photoelectron spectroscopy (XPS) analysis.
A Kratos Ultra DLD spectrometer with a monochromatic Al source operating at 150W (1,486.6 eV) was used to perform XPS measurements. The spectrometer was operating at a pressure of Torr. Low energy electrons were used to accomplish a charge compensation, and all spectra charges were referenced and adjusted by the C1s region to 285 eV. All high-resolution C1s and survey spectra were acquired at pass energies of 160 and 20 eV and processed using CasaXPS software (version 2.3.25 PR1.0).
Metabolic Analysis of Colon, Liver, and Brain
Reagents used in this study were all liquid chromatography–mass spectrometry (LC-MS) grade, and all standard components for measuring metabolites were purchased from both Sigma-Aldrich and Fisher Scientific. Acetonitrile, ammonium acetate, acetic acid, and methanol (MeOH) were purchased from Fisher Scientific. Ammonium hydroxide was purchased from Sigma Aldrich. This study used deionized water that was produced by an in-house water purification system from EMD Millipore (Billerica, MA). Phosphate buffered saline (PBS) was purchased through GE Health care Life Sciences.
Tissue Preparation
To prepare each tissue sample for analysis, of the tissue of interest was homogenized in an Eppendorf tube using a Bullet Blender homogenizer (Next Advance). Each sample was homogenized in MeOH:PBS (a 4:1 vol/vol dilution). After completing the initial homogenization, an additional of MeOH:PBS was added, and samples were vortexed for 10 s. Thereafter, samples were stored at for 30 min, transferred to an ice bath, and sonicated for 30 min. Centrifugation at was then performed at for 10 min, and supernatant was transferred to a new Eppendorf tube. Drying of all samples was performed using a CentriVap Concentrator (Labconco) under vacuum, and all residue obtained was reconstituted in 40% PBS and 60% acetonitrile prior to MS analysis. A portion of all study samples were pooled together to obtain for quality control (QC) samples.
Targeted Metabolomics
The Agilent 1290 UPLC-6495 QQQ-MS system was utilized to perform liquid chromatography–tandem mass spectrometry (LC-MS/MS) experiments targeting metabolites in the tryptophan metabolic pathway.74 There were 28 metabolites targeted in this evaluation, including 3-hydroxy anthranilic acid, 3-hydroxykynurenine, 3-indolepropionic acid, 5-hydroxytryptophan, ADP ribose, anthranilic acid, HIAA, indole, indole-3-acetic acid, indole-3-lactic acid, indole-3-pyruvic acid, kynurenic acid, L-kynurenine, melatonin, NAD, NADH, N′-formylkynurenine, nicotinamide, nicotinamide mononucleotide, nicotinamide riboside, nicotinic acid, nicotinic acid adenine dinucleotide, nicotinic acid mononucleotide, quinolinic acid, serotonin, tryptamine, tryptophan, and xanthurenic acid. Each sample was injected using a volume of and analyzed in positive ionization mode. Chromatographic separations were conducted through a Waters XBridge BEH Amide column in hydrophilic interaction chromatography (HILIC) mode. The flow rate was set to , and the autosampler temperature and column compartment were maintained at and , respectively. The mobile phase consisted of two solvents: the first solvent containing ammonium acetate and ammonium hydroxide in 95% and 5% acetonitrile, and the second solvent containing ammonium acetate and ammonium hydroxide in 95% acetonitrile and 5% . An isocratic elution of 90% of the second solvent was performed for 1 min, followed by a decrease to 40% at timepoint minutes for 4 min, then gradually increasing back to 90% at timepoint minutes. The mass spectrometer was equipped with an electrospray ionization (ESI) source to acquire targeted data in multiple-reaction monitoring (MRM) mode. The LC-MS system employed the Agilent MassHunter workstation software, while the Agilent MassHunter Quantitative Data Analysis Software (version B.07.00) was used to integrate all extracted MRM peaks during analysis. Raw data is found in Excel Tables S3, S4, S7, S8, S11, and S12.
Untargeted LC-MS Metabolomics
The untargeted LC-MS metabolomics analysis was conducted using a Thermo Scientific Vanquish ultra-high-performance liquid chromatography (UHPLC) system with a Thermo Scientific Orbitrap Exploris 240 MS (Waltham, MA). Duplicate samples were analyzed in both negative and positive ionization modes. A Waters XBridge BEH Amide column (, particle size; Waters Corporation) was used for chromatographic separation in hydrophilic interaction liquid chromatography (HILIC) mode, with a flow rate of , and autosampler temperature was kept at with the column compartment set to . The mobile phase A contained 0.1% formic acid in water and the mobile phase B contained 0.1% formic acid in acetonitrile. Other LC conditions including gradients, autosampler, and column temperature were the same as those in targeted metabolomics described above. The mass spectrometer collected untargeted data at 70 to 800 m/z using an electrospray ionization (ESI) source. The spray voltage for the ion source is 3,500 V for positive ionization mode and 3,300 V for negative ionization mode. The Orbitrap resolution for MS1 full scan mode is 120,000. The top 20 scans were selected to trigger in MS2 mode, with resolutions of 60,000 for full scan and 30,000 for data-dependent MS2 (ddMS2), respectively. Additionally, the Higher-energy collisional dissociation (HCD) collision energy mode for MS2 is stepped with a normalized collision energy setting of 30%, 60%, and 150%. MS spectra peaks were identified using aqueous metabolites of in-house chemical standards (Sigma-Aldrich) (Excel Table S13), and compared several commercial MS databases embedded (mzCloud, Predict Composition, Chemspider, and Metabolika) in Thermo Scientific Compound Discoverer software 3.3. Limits of for mass accuracy and 100,000 for absolute intensity threshold were set for MS data extraction. Data annotation was based on isotopic pattern, retention time, exact mass, and MS/MS fragmentation patterns. Thermo Scientific Compound Discoverer 3.3 software was used for data processing of aqueous metabolomics data, and peak picking, alignment, and normalization was used for untargeted data. Quality control (QC) pools were established based on the coefficient of variation (CV) and signals showing up in of all samples to ensure high-quality data for analysis.75 In metabolomics, missing values that exist in more than 20% of samples may be removed from the data, which is called the “80% rule.” Finally, within six groups, the compounds have been identified as follows: brain (1,262 identified, 11,122 unidentified), colon (2,363 identified, 17,312 unidentified), liver (3,042 identified, 19,967 unidentified), Mixed polymer exposed Brain (MB) (2,674 identified, 21,016 unidentified), Mixed polymer exposed Colon (MC) (2,485 identified, 16,047 unidentified), and Mixed polymer exposed Liver (ML) (2,082 identified, 13,983 unidentified). Raw data is found in Excel Tables S1, S2, S5, S6, S9, and S10.
Statistical Analysis
All samples were analyzed and compared against untreated control mice exposed to vehicle using a false discovery rate, where appropriate. Pathway analysis and volcano plots were developed using MetaboAnalyst software 5.0 (www.metaboanalyst.ca) and validated with Reactome (https://reactome.org/)76,77 (Excel Table S14). Samples were normalized by sum against QC pools. Log transformation was performed, and all data was mean centered. -Value threshold was set to 0.05 with equal variance and 2.0-fold change threshold.
Results
Visualization of Systemic Microplastic Translocation
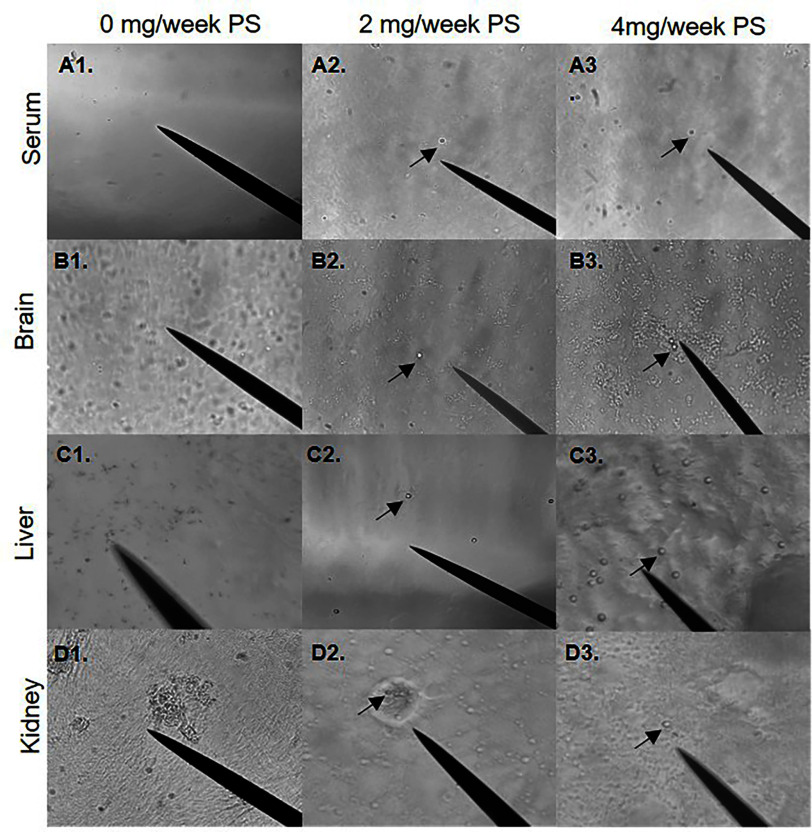
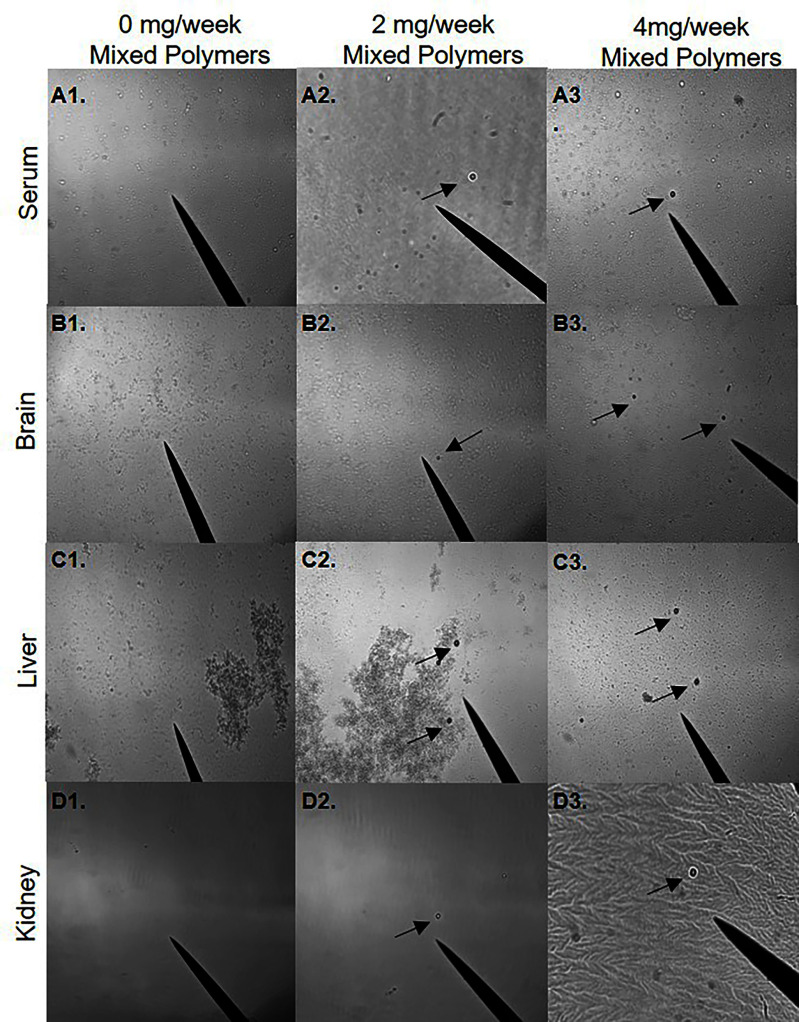
To determine whether orally administered microspheres could be translocated from the digestive system, polarized light microscopy was performed on serum, brain, liver, and kidney samples isolated from mice exposed to 0, 2, and polystyrene and mixed plastic [polystyrene, poly(lactic-co-glycolic acid), and polyethylene] microspheres for 4 weeks. The presence of polystyrene and mixed polymer microspheres was observed in the serum and in all three isolated tissues (Figure 1 and 2). Although not fully quantifiable with this visualization method, polystyrene and mixed polymer microspheres were readily more apparent in liver samples (Figure 1C) compared with brain and serum, with far fewer polystyrene and mixed polymer microspheres observed in the kidneys. These observations suggest that ingested microspheres may be able to translocate across the gut epithelium into the systemic circulation and accumulate differentially in the assessed organs.
Figure 1.
Visualization of systemic polystyrene microsphere translocation. Visualization of polystyrene microspheres resuspended from isolated pellet in 100% EtOH. The black arrow indicates polystyrene microspheres. (A1–A3) Five-micrometer polystyrene microspheres in serum (20×). (B1–B3) Five-micrometer polystyrene microspheres in brain (20×). (C1–C3) Five-micrometer polystyrene microspheres in liver (40×). (D1–D3) Five-micrometer polystyrene microspheres in kidney (40×). Mice were exposed twice a week for 4 wk to a low dose of or a high dose of with polystyrene microspheres via oral gavage. Images are representative of . Note: EtOH, ethanol.
Figure 2.
Visualization of systemic mixed polymer microsphere translocation. Visualization of mixed polymer microspheres resuspended from isolated pellet in 100% EtOH. The black arrow indicates microspheres. (A1–A3) Five-micrometer mixed polymers [polystyrene (PS), polyethylene (PE), and poly-(lactic-co-glycolic acid) (PLGA)] microspheres in serum (20×). (B1–B3) Five-micrometer mixed polymers (PS, PE, PLGA) in brain (20×). (C1–C3) Five-micrometer mixed polymers (PS, PE, PLGA) in liver (20×). (D1–D3) Five-micrometer mixed polymers (PS, PE, PLGA) in kidney (40×). Mice were exposed twice a week for 4 wk to a low dose of or a high dose of with of mixed polymers via oral gavage. Images are representative of . Note: EtOH, ethanol.
Using Raman spectroscopy and XPS analysis, we wanted to validate that the brain microspheres isolated and viewed under polarized light microscopy were truly polystyrene microspheres. The Raman spectra of the original polystyrene microspheres were consistent with those particulates found in brain isolates (Figure S1A), and further matched a library polystyrene standard spectra, validating that the systemically translocated and recovered microspheres were polystyrene (Figure 1B2,B3). The microsphere sample recovered from brain isolate, however, did show peak shift differences compared to fresh polystyrene, potentially indicating modification of the surface chemistry or accumulation of other biochemicals (i.e., a corona effect). This observation led us to question whether the alteration was due to the KOH digestion or biological degradation. To address this, we compared the recovered polystyrene microspheres to naïve polystyrene microspheres digested with KOH under the same conditions used in our isolation protocol (Figure S1B). The Raman spectra for the naive polystyrene microspheres subjected to our isolation protocol was similar to that of the KOH-digested microspheres, suggesting that the KOH digestion did not alter the structure of the polystyrene microspheres.
Given that the liver appeared to have the highest concentrations of microspheres, we wanted to investigate the chemical and surface composition of polystyrene microspheres recovered from the livers of exposed mice compared to naïve polystyrene microspheres using XPS survey scan mode (Figure S1C–E). XPS analysis revealed greater levels of surface potassium and nitrogen in the recovered microspheres, consistent with a biochemical adherence or interaction. XPS analysis also identified fluorine on the surface of the microspheres isolated from liver tissue. We assume that this fluorine peak resulted from the surface adsorption of isoflurane, which was used as a general anesthetic prior to euthanasia in our studies. This fluorine peak may serve as a useful indicator that plastics were obtained from an anesthetized subject, as opposed to derived from ex vivo processing or storage of tissue samples in polystyrene containers.
Untargeted Metabolic and Pathway Analysis in the Colon
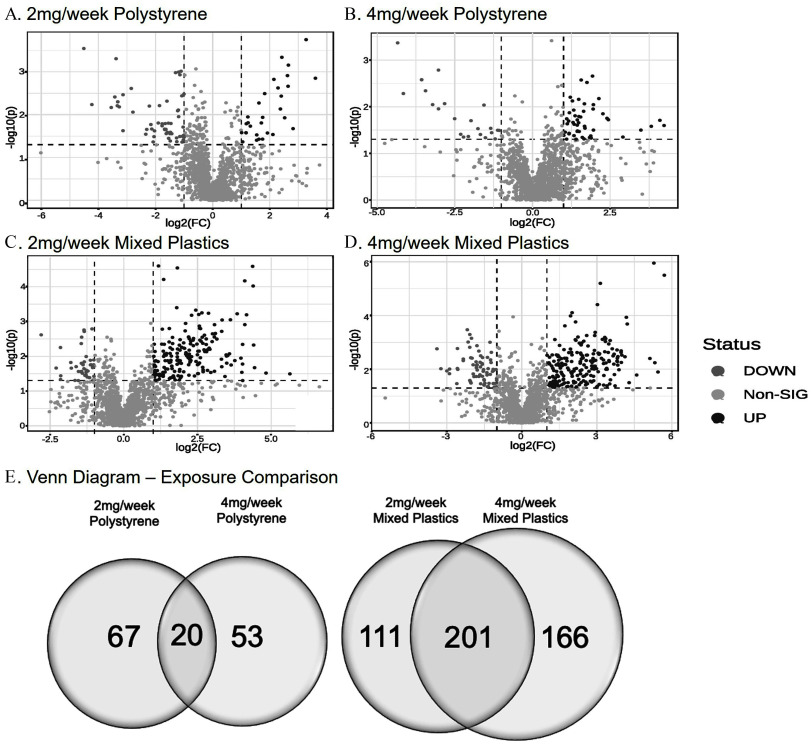
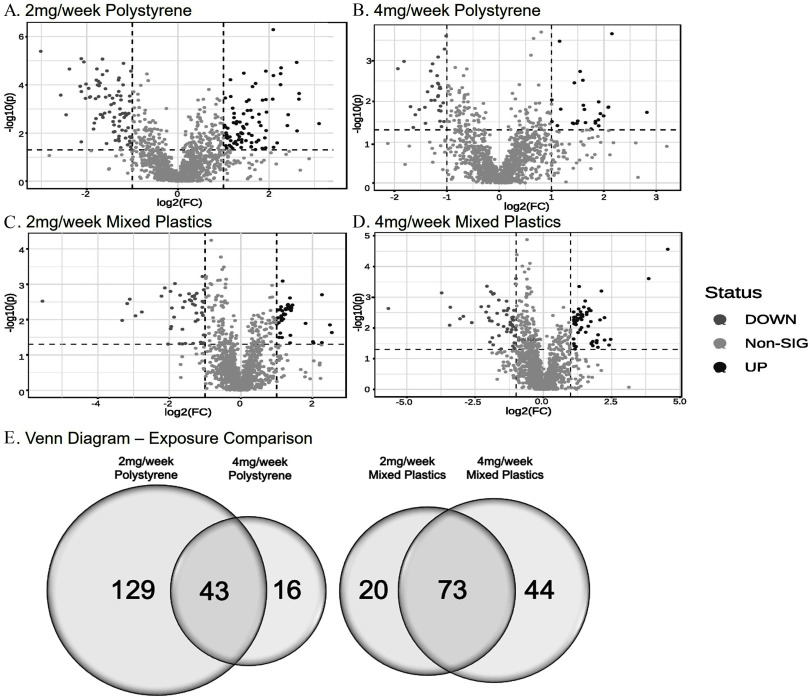
Untargeted metabolomics was performed on colonic tissue metabolites in response to environmentally relevant oral polystyrene and mixed polymer microsphere exposure. Volcano plots for each exposure group showed both significantly higher and lower metabolite levels when compared to control mice (Figure 3A–D). Following the 4-wk exposures, 140 metabolites in the polystyrene-exposed group and 478 metabolites in the mixed plastics-exposed group were significantly different from control (; Figure 3E). We observed that 67 metabolites were uniquely different in the polystyrene group (Table S1; Excel Table S1) and 53 metabolites were uniquely different in the polystyrene group (Table S2; Excel Table S2), with 20 (14.3%) significantly different metabolites occurring in both concentration groups (Figure 3E). The mixed plastics exposure groups showed a much higher metabolic response, with 111 uniquely different metabolites in the exposure group (Table S3; Excel Table S3) and 166 uniquely different metabolites in the group (Table S4; Excel Table S4). The mixed plastic groups shared 201 (42.0%) metabolites that differed from controls (Figure 3E).
Figure 3.
Untargeted metabolomics of colon. Untargeted metabolomic analysis in colon tissue of mice exposed to (A) polystyrene, (B) polystyrene, (C) mixed polymer, or (D) mixed polymer. Data plotted as log(2) fold change (). (E) Venn diagram representing the significantly different metabolites following microsphere exposures ( as compared to control). Mice were exposed twice a week for weeks with polystyrene microspheres or mixed polymers [polystyrene, polyethylene, and poly-(lactic-co-glycolic acid)] at (low dose) or (high dose); per group. Source data can be found in Excel Tables S1 and S2.
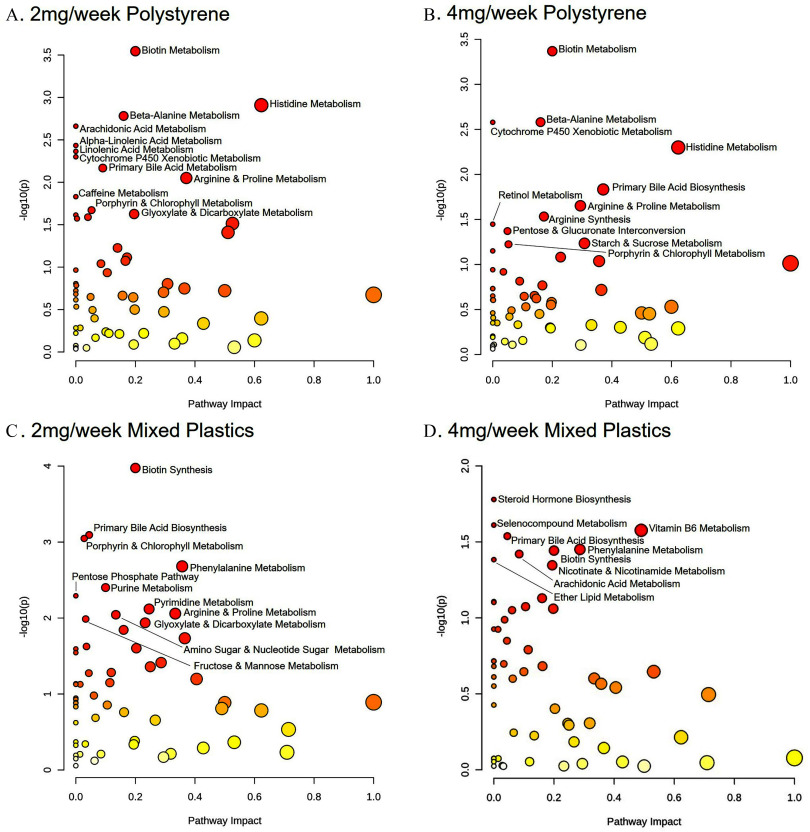
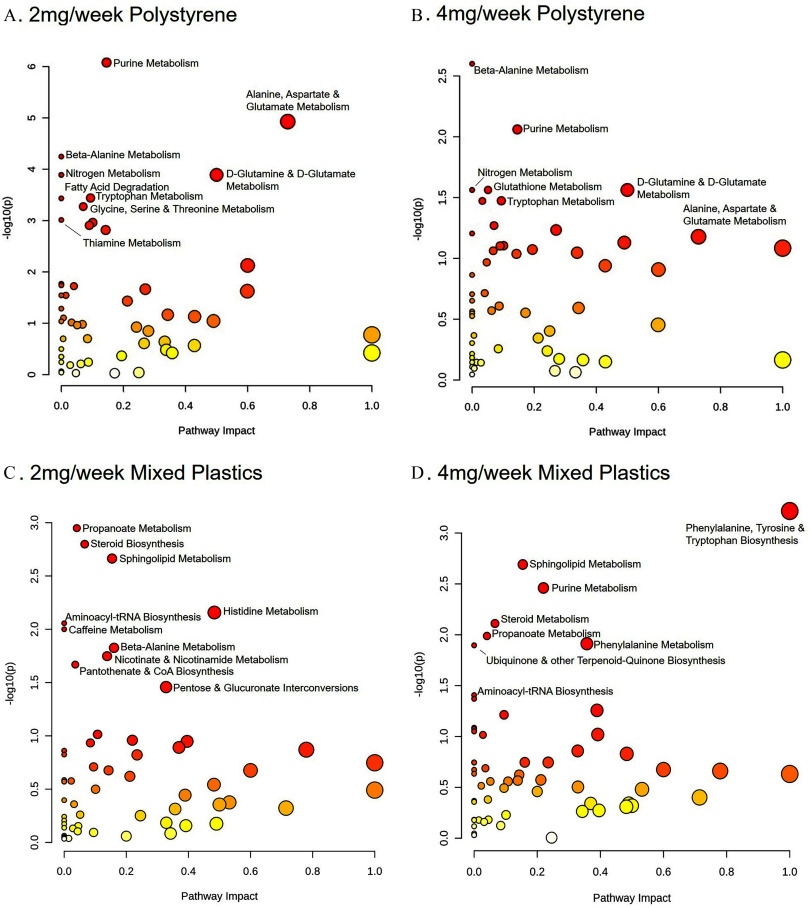
To further understand these changes in metabolites, metabolic pathway analysis was performed. Pathway analysis of metabolite levels in colons from mice exposed to and polystyrene compared to controls revealed significant differences from controls () in shared pathways contributing to a) biotin metabolism, b) histidine metabolism, c) metabolism, d) arginine and proline metabolism, e) cytochrome P450 xenobiotic metabolism, and f) porphyrin and chlorophyll metabolism (Figure 4A,B). Interestingly, both mixed plastics exposed groups exhibited some overlap in metabolic pathway when compared to the polystyrene only groups including a) porphyrin and chlorophyll metabolism, b) primary bile acid biosynthesis, c) arginine and proline metabolism, d) arachidonic acid metabolism, and e) the pentose phosphate pathway (Figure 4A–D). Whereas, the and mixed plastics groups only shared differences in pathways linked to the a) primary bile acid biosynthesis and b) the biotin synthesis pathway.
Figure 4.
Colonic metabolome pathway analysis. Metabolomic pathway analysis of differences in the colon following oral microsphere exposure in mice exposed to (A) polystyrene, (B) polystyrene, (C) mixed polymers, or (D) mixed polymers. Mice were exposed twice a week for weeks with polystyrene microspheres or mixed polymers [polystyrene, polyethylene, and poly-(lactic-co-glycolic acid)] at (low dose) or (high dose); per group. Source data can be found in Excel Tables S1 and S2.
Nicotinate and Nicotinamide Metabolic Pathway in the Colon
We also performed targeted metabolic analysis of 28 metabolites associated with nicotinate and nicotinamide metabolic pathway (Figure S2A–D; Table S13). Analysis of colon tissue isolated from mice exposed to polystyrene only revealed one metabolite that was more highly secreted, 5-hydroxytryptophan (), in the exposure group and () in the exposure group (Table S13). Melatonin () was lower in the concentration group (Figure S2A,B; Table S13A). The mixed plastics group shared a higher level of kynurenic acid ( in , in ), xanthurenic acid ( in , in ), melatonin ( in , in ), and N′-formylkynurenine ( in , in ) metabolites (Figure S2C,D; Table S14). A significantly lower level of the nicotinamide () and ADP ribose () metabolites was noted in the mixed plastic group (Table S14A) while the mixed plastics group exhibited a significantly lower level of NAD (), indole (), nicotinic acid (), and nicotinic acid adenine dinucleotide () (Table S14B).
Oral Gastric Exposure of Plastic Microspheres Effects on Liver Metabolome
Untargeted metabolomics was performed on liver tissues from all microsphere-exposed animals compared to nonexposed mice. Volcano plots for each exposure and concentration group showed patterns of metabolite differences (from control) within each group (Figure 5). We observed that 188 metabolites in the polystyrene-exposed group and 137 metabolites in the mixed plastics-exposed group were significantly different in the plastic microsphere exposure group compared to control (). We observed that 129 (68.6%) metabolites were uniquely different in the polystyrene group (Table S5; Excel Table S5) and 16 (8.5%) metabolites were uniquely different in the polystyrene group (Table S6; Excel Table S6), with 43 (22.9%) significantly metabolites that were different from control in common between the two different concentrations of PS groups (Figure 5E). In contrast, the mixed plastics exposure groups showed a much higher metabolic response, with 20 (14.6%) uniquely different metabolites in the exposure group (Table S7; Excel Table S7) and 44 (32.1%) uniquely different metabolites in the group (Table S8; Excel Table S8), and 73 (53.3%) metabolites that were different from control shared among both mixed polymer concentrations.
Figure 5.
Untargeted metabolomics of liver. Untargeted metabolomic analysis in the liver of mice exposed to (A) polystyrene, (B) polystyrene, (C) mixed plastics, or (D) mixed plastics. Data plotted as log(2) fold difference (). (E) Venn diagram representing the significantly different metabolites following microplastic exposures ( as compared to control). Mice were exposed twice a week for weeks with polystyrene microspheres or mixed polymers [polystyrene, polyethylene, and poly-(lactic-co-glycolic acid)] at (low dose) or (high dose); per group. Source data can be found in Excel Tables S5 and S6.
To identify the key metabolic pathways altered in the liver following oral microsphere exposure, metabolic pathway analysis was again performed. When comparing the impact on liver metabolic pathways between the and polystyrene-exposed groups, we observed significant differences () in both PS groups in the following pathways: a) alanine, aspartate, and glutamate metabolism; b) beta-alanine metabolism; c) D-glutamine and D-glutamate metabolism; d) nitrogen metabolism; e) purine metabolism; and f) tryptophan metabolism (Figure 6A,B). The beta-alanine and purine metabolic pathways were also altered in the and mixed plastic groups, respectively (Figure 6C,D). The and mixed plastic groups showed overlap in the a) amino acetyl-tRNA biosynthesis pathway, b) propanoate metabolism, and c) sphingolipid metabolism pathway (Figure 6C,D). The and mixed plastic groups also showed a difference in steroid biosynthesis and steroid metabolism, respectively. Interestingly, among all microsphere-treated groups, there were several pathways associated with amino acids biosynthesis or metabolism that was differentially regulated when compared to untreated mice.
Figure 6.
Hepatic metabolome pathway analysis. Metabolomic pathway analysis of alterations in the liver following oral MP exposure in mice exposed to (A) polystyrene, (B) polystyrene, (C) mixed polymer, or (D) mixed polymer. Mice were exposed twice a week for weeks with polystyrene microspheres or mixed polymers [polystyrene, polyethylene, and poly-(lactic-co-glycolic acid)] at (low dose) or (high dose); per group. Source data can be found in Excel Tables S5, S6, and S14.
Livers were next examined for metabolites associated with the nicotinate and nicotinamide metabolic pathway via targeted metabolomics. Liver samples from animals exposed to only polystyrene microspheres showed a shared higher expression of nicotinic acid mononucleotide (NMN) and lower expression of 5-hydroxy indoleacetic acid (HIAA) and L-kynurenine in the exposure and exposure groups (Figure S3A,B). The mixed plastic-exposed animals showed metabolite differences in both exposure groups; however, the metabolites being significantly higher between these groups were not comparable. In the mixed plastic exposure group, melatonin was higher and 5-hydroxytryptophan and HIAA were lower (Figure S3C). In contrast, the mixed plastics exposure group showed a significantly higher expression of metabolites nicotinamide riboside, 3-hydroxykynureneine, nicotinic acid adenine dinucleotide, and NAD and a significantly lower expression of N′-formylkynurenine, tryptophan, and indole-3-lactic-acid (Figure S3D).
Oral Gastric Exposure of PS-Microspheres and Mixed Plastics Effects on Brain Metabolome
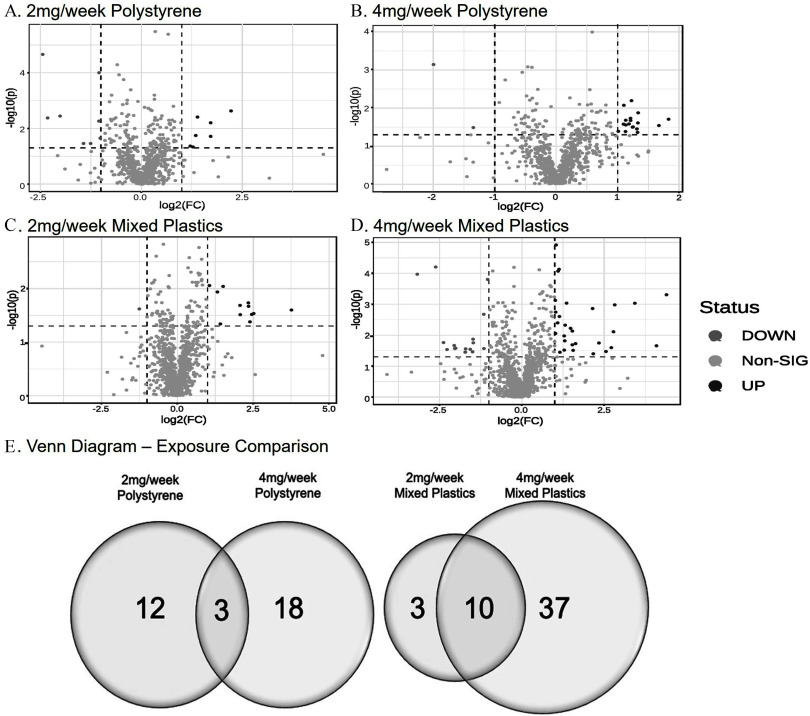
To compare PS vs. mixed plastic exposure, the prefrontal cortex of the brain was collected to perform untargeted metabolomics. Volcano plots for each exposure group showed patterns of metabolite changes within each group (Figure 7A–D). We observed that 33 metabolites in the polystyrene-exposed group and 50 metabolites in the mixed plastic-exposed group were significantly different due to plastic microsphere exposure (). We also observed that 12 (36.4%) metabolites were differently altered in the polystyrene group (Table S9; Excel Table S9) and 18 (54.5%) metabolites were uniquely different in the polystyrene group (Table S10; Excel Table S10), with 3 (9.1%) metabolites significantly different from control in both concentration groups (Figure 7E). In contrast, the mixed plastics exposure groups showed a much higher metabolic response, with 3 (6%) uniquely different metabolites in the group (Table S11; Excel Table S11) and 37 (74%) uniquely different metabolites in the group (Table S12; Excel Table S12), and 10 (20%) metabolites different from control shared between both of the mixed plastic groups.
Figure 7.
Untargeted metabolomics of brain. Untargeted metabolomic analysis of brain isolates from mice exposed to (A) polystyrene, (B) polystyrene, (C) mixed polymer, or (D) mixed plastics. Data plotted as log(2) fold change (). (E) Venn diagram representing the significantly different metabolites following microplastic exposures ( as compared to control). Mice were exposed twice a week for weeks with polystyrene microspheres or mixed polymers [polystyrene, polyethylene, and poly-(lactic-co-glycolic acid)] at (low dose) or (high dose); per group. Source data can be found in Excel Tables S9 and S10.
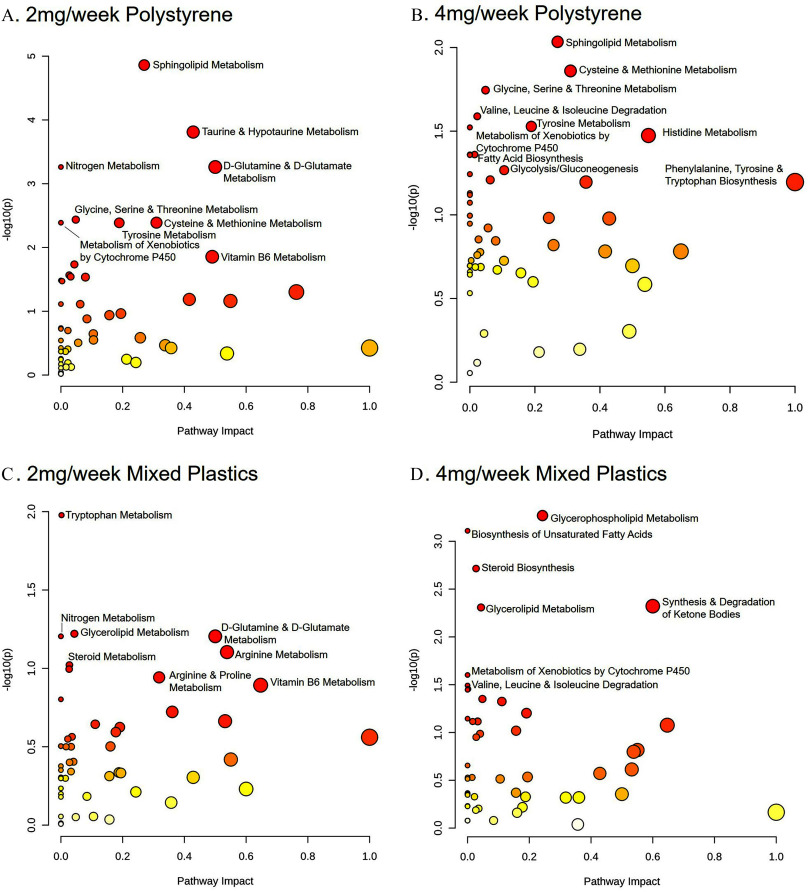
To investigate the key metabolic pathways altered by plastic microsphere exposure in the brain, metabolic pathway analysis was again performed. When comparing the brain isolates from and polystyrene-only exposed samples, we observed significant modulation () of the following pathways: a) cysteine and methionine metabolism; b) glycine, serine, and threonine metabolism; c) sphingolipid metabolism; d) tyrosine metabolism; and e) the xenobiotic metabolism regulated by cytochrome P450 (Figure 8A,B). Only the xenobiotic metabolism by cytochrome P450 pathway was shared by the mixed plastic exposure group. Whereas both mixed plastic exposure groups shared a significant modulation in a) glycerolipid metabolism and b) the steroid biosynthesis pathway (Figure 8C,D). Both the exposure to PS or mixed polymers displayed alterations in pathways associated with a) D-glutamine and D-glutamate metabolism and b) nitrogen metabolism. The exposure to PS or mixed polymers showed significantly alterations in pathways associated with the valine, leucine, and isoleucine degradation.
Figure 8.
Brain metabolome pathway analysis. Metabolomic pathway analysis of alterations in the brain following oral MP exposure in mice exposed to (A) polystyrene, (B) polystyrene, (C) mixed polymer, or (D) mixed polymer. Mice were exposed twice a week for weeks with polystyrene microspheres or mixed polymers [polystyrene, polyethylene, and poly-(lactic-co-glycolic acid)] at (low dose) or (high dose); per group. Source data can be found in Excel Tables S9 and S10.
When performing targeted metabolic analysis in the brain for metabolites associated with the nicotinate and nicotinamide metabolic pathway, we found animals exposed to polystyrene only showed higher expression of quinolinic acid and lower of nicotinic acid and tryptamine in the exposure while the concentration group did not show any significant differences in expression of metabolites (Figure S4A,B). However, we did see trends of metabolites such as melatonin and nicotinic acid being lower and quinolinic acid and ADP ribose being higher, but these were not always significant. The mixed polymer-exposed group showed multiple metabolite differences in both concentration groups, with NADH being higher and L-kynurenine, HIAA, and 3-hydroxykynurenine being significantly lower in both the and exposure groups (Figure S4C,D).
Discussion
There is no doubt that all living organisms are being exposed to microplastics. While the physiological effects of MP pollution on marine organisms are well-documented,78–81 the impacts on terrestrial organisms including humans are only beginning to be elucidated. The most common route of exposure appears to be through our diet.9,80–83 Inhalation intake can also contribute to gut exposure through contaminated mucus ingestion.84–87 To support this, there have been numerous reports showing in mammals and other species that MPs can accumulate in the gastrointestinal (GI) tract due to their detection in the stool and tissue.51,52,66,88 Moreover,recent studies showing MP can accumulate in human blood and lungs10,89,90 suggest that MP can pass the various barriers of the body including the GI tract. Furthermore, this systemic MP accumulation could drastically be increased in individuals with underlying conditions especially those that show signs of increased intestinal permeability such as inflammatory bowel disease (IBD), celiac disease, obesity, and metabolic dysfunction-associated steatotic liver disease (MASLD).91–96 A few studies have performed both targeted and untargeted metabolomics in the serum,33,34,60 liver,97 and stool66 of micro- and nanoplastic-exposed mice; however, these mice were exposed to a single-type of MP (or NP). Given humans are exposed to a plethora of plastics, we set out to identify, quantify, and compare the colon metabolome of mice exposed to both concentrations of PS or mixed plastics after a 4-wk exposure.
Similar studies have only exposed mice to a single type of MP and then performed metabolomics on either the serum, liver, or stool.34,60,66,68,97,98 However, humans are being exposed to a plethora of plastics and the assessment of mixed plastic exposure in animal models is critical to understand the true effects of plastic pollution and health outcomes. Our primary focus was on particle uptake, translocation, and impact on biological microenvironments. However, the concern for polymer clearance is an essential aspect to understanding the overall fate and impact of ingested polymers. Clearance mechanisms play a critical role in determining the concentration of these particles within biological tissues. GI issues that need to be considered on this topic include but are not limited to GI motility (contraction and transit), intestinal epithelial absorption, and stool consistency. A limitation of our research is that we did not investigate estimating clearance rates. We do recognize the significance of microplastics being cleared, as microplastics have been found in stool samples from both health and disease-state individuals showing they can be cleared after ingestion.52,66,99 Nevertheless, our findings provide further support that plastic microspheres can become embedded in other internal organs after ingestion. After oral gastric plastic microsphere exposure in a healthy mouse, we found that microspheres could be detected in distant organs (i.e., brain, liver, and kidney). We hypothesize that the microspheres pass the intestinal epithelial barrier and gut vascular barrier and translocate via the systemic circulation to these organs. Moreover, we show that a four-week PS alone or mixed plastic exposure can impact various metabolic pathways in the colon, liver, and brain of mice when compared to unexposed mice. The colon, liver, and brain showed the most common dysregulated pathways, which showed a link to amino acids. Additionally, we observed differences in metabolic pathways related to purines, pyrimidines, and glutamate, which are products of amino acid metabolism, in our mice exposed to microplastics compared to controls. Amino acids are fundamental for human health as they influence numerous physiological processes, and disruptions in amino acid metabolism have been linked to numerous inflammatory and metabolic diseases.100–104
Interestingly, our metabolomics data not only showed differences in the colon, liver, and brain metabolome when comparing our mixed plastic exposure to PS alone groups, but also showed a difference when examining the two concentrations for each group. This became more apparent when we performed targeted metabolomics on the metabolites associated with the nicotinate and nicotinamide metabolic pathway. The greatest impact on the host metabolome was in the following order: colon, liver, and then brain. One reason for these fewer metabolic changes in the brain as compared to the colon and liver may be due to the overall greater apparent accumulation of microspheres in these tissues and the crosstalk between the gut and liver. Other reasons for more extensive metabolic alterations occurring in the colon and liver may be due to the role that these tissues play in overall break down, digestion, detoxification, and synthesis of consumed products.
In this study, the prefrontal cortex was the only region of the brain evaluated. Thus, we cannot rule out that other portions of the brain may accumulate plastic microspheres and this could cause further changes in the brain metabolome. Additionally, we could not precisely pinpoint the microspheres location due to tissue homogenization. This is a limitation because we acknowledge that the microspheres may be trapped within the vasculature of the brain and have not crossed the blood–brain barrier. Nevertheless, there are still altered metabolic pathways in the brain after polystyrene and mixed polymer microsphere exposure. One alteration of interest was the modulation of the xenobiotic pathway by cytochrome P450 seen in the brain of animals exposed to of PS and mixed plastics. Although metabolism is primarily carried out in the liver, xenobiotic metabolism by cytochrome P450 alteration in the brain as a result of plastic microsphere exposure could point to potential neurotoxic effects. This data supports the various studies showing that plastic microspheres can be neurotoxic in mice105–109 and a more recent paper showing nanoplastics also may affect mouse brain function.106 A very recent publication has shown microplastics can be detected in the brains of mice after 3 weeks of exposure that ultimately affected behavior.110 These reports suggest that ingestion of MP/NP over time could cause adverse neurodevelopmental outcomes or trigger the development of neurodegenerative diseases.111 While this finding requires further investigation, the effects of MPs on our central nervous system may be an interesting avenue for future research.
The results of this study may serve as a model to explore chronic exposure outcomes associated with mixed plastic exposure. Thus, leading to novel techniques to identify their potential risks on human health and future quantitative method development to establish MP-associated metabolite presence. Taken together, our data highlight potential risks that the different types, mixtures, and concentrations of plastic microsphere exposure can impact health outcomes.
Limitation of Study
Currently, there is substantial research on possible connections of microplastic exposure and poor health outcomes in wildlife, but there has been limited investigation into long-term human health outcomes.112 When ingested, MPs have the potential to expose organisms to higher concentrations of monomers, polymers, or chemicals associated with the manufacturing process that could potentiate their toxicity.113 It is believed the micro- and nanoplastics that organisms are ingesting contain chemicals that can further exacerbate plastic-associated toxicity, and this has been reviewed elsewhere.114,115 The microplastics utilized in this study were commercially bought and do not contain chemicals such as phthalates, bisphenol A (BPA), or polyfluorinated alkyl substances (PFAs).116–118 These chemical additives could have an extra layer of problems such as a variety of health effects not limited to altered immune and thyroid function, kidney disease, liver disease, lipid and insulin dysregulation, cancers, and altered reproductive and development outcomes, but we believe our study shows that plastic microsphere exposure can have far-reaching effects after ingestion. Although there is still ongoing research to identify and understand the widespread human health risk of MPs, the current study helped to identify potential organ-specific metabolic pathway alterations that are associated with different types, mixtures, and concentrations of plastic microspheres. Further investigation will need to be performed to identify if these metabolic alterations may play a role in inflammation, immune regulation, metabolism, multiorgan dysfunction, and even potentially exacerbate conditions such as IBD, MASLD, and obesity.119 Lastly, future studies should focus on novel techniques to adequately identify and quantify microplastics in tissues as well as specific plasticizers. This could help elucidate the functional impacts of altered metabolites due to systemic uptake and distribution of MPs. In pursuit of advancing our study’s future direction, we are currently immersed in the development of novel methodology for the quantitation of microplastics within tissue samples. To achieve this, we are employing pyrolysis-gas chromatography–mass spectrometry (Py-GC/MS). This innovative approach promises to provide us with a more comprehensive understanding of the presence and concentration of microplastics in biological tissues, thereby significantly contributing to our knowledge of the environmental and health implications associated with these ubiquitous contaminants. We believe Py-GC/MS may help in further identification by quantifying total mass of plastics present. Nevertheless, it is limited in being able to identify particle size, shape, and chemical composition and can be destructive to the sample which will not allow for any other analysis after completion of pyrolysis protocols. The current techniques (e.g., Raman and microscopic visualization) used in this study also have their own limitations. For Raman spectroscopy, while valuable in identifying polymer types, it does have limitations in providing detailed information about size, shape, and surface properties of the particles. Additionally, there is a specific sample size that limits the detection of nanoparticles. Similarly, microscopic visualization is great to visualize the particles but are unable to distinguish between plastic and nonplastic materials. Therefore, new techniques will need to be developed to further characterize and identify microplastics in biological samples.
Supplementary Material
Acknowledgments
M.M.G. and A.S.R. performed all analysis, tissue collection, and isolation with the help from S.D.M., J.L.M.H., C.F., E.E.H., D.P.S., R.T., J.G.E., A.B., R.P.H., S.L., G.H., K.J.K., J.Y.C., R.R.G., and J.G.I. H.G. and Y.J. contributed to metabolomics and analysis. Exposures were performed by M.M.G., A.S.R., S.L., and G.H. M.M.G., M.J.C., and E.F.C. participated in writing the manuscript. M.M.G., J.G.I., M.J.C., and E.F.C. designed the study, analyzed data, and wrote the paper. All authors approved the final version of the manuscript.
We would like to acknowledge Jesse Benson Hesch for her valuable contribution to the editing of this manuscript. Her expertise and meticulous attention to detail significantly enhanced the quality of our work and preparation of this manuscript.
Funding was supported in part by the National Center for Research Resources and the National Center for Advancing Translational Sciences of the National Institutes of Health (NIH) through NIH grant 1R01 ES032037-01A1 (E.F.C.), in part by UL1TR001449 (E.F.C.), in part by NIH grant P20GM121176 (E.F.C.; J.G.I.) and P20GM130422 (M.J.C.), and in part by NIH grant K12GM088021 (M.M.G.; ASERT-IRACDA), in part by NIMHD grant P50MD015706 (J.G.I.), in part by pilot funding from P20GM130422 (E.E.H.), and in part by P30CA118100.
Studies were conducted with full approval by the Institutional Animal Care and Use Committees of the University of New Mexico.
Conclusions and opinions are those of the individual authors and do not necessarily reflect the policies or views of EHP Publishing or the National Institute of Environmental Health Sciences.
References
- 1.Geyer R, Jambeck JR, Law KL. 2017. Production, use, and fate of all plastics ever made. Sci Adv 3(7):e1700782, PMID: , 10.1126/sciadv.1700782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bajt O. 2021. From plastics to microplastics and organisms. FEBS Open Bio 11(4):954–966, PMID: , 10.1002/2211-5463.13120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson PJ, Warrack S, Langen V, Challis JK, Hanson ML, Rennie MD. 2017. Microplastic contamination in Lake Winnipeg, Canada. Environ Pollut 225:223–231, PMID: , 10.1016/j.envpol.2017.02.072. [DOI] [PubMed] [Google Scholar]
- 4.Pham DT, Kim J, Lee S-H, Kim J, Kim D, Hong S, et al. 2023. Analysis of microplastics in various foods and assessment of aggregate human exposure via food consumption in Korea. Environ Pollut 322:121153, PMID: , 10.1016/j.envpol.2023.121153. [DOI] [PubMed] [Google Scholar]
- 5.Irnidayanti Y, Soegianto A, Brabo AH, Abdilla FM, Indriyasari KN, Rahmatin NM, et al. 2023. Microplastics in green mussels (Perna viridis) from Jakarta Bay, Indonesia, and the associated hazards to human health posed by their consumption. Environ Monit Assess 195(7):884, PMID: , 10.1007/s10661-023-11535-9. [DOI] [PubMed] [Google Scholar]
- 6.Daniel DB, Ashraf PM, Thomas SN. 2020. Microplastics in the edible and inedible tissues of pelagic fishes sold for human consumption in Kerala, India. Environ Pollut 266(pt 2):115365, PMID: , 10.1016/j.envpol.2020.115365. [DOI] [PubMed] [Google Scholar]
- 7.Daniel DB, Ashraf PM, Thomas SN, Thomson KT. 2021. Microplastics in the edible tissues of shellfishes sold for human consumption. Chemosphere 264(pt 2):128554, PMID: , 10.1016/j.chemosphere.2020.128554. [DOI] [PubMed] [Google Scholar]
- 8.Mohsen M, Lin C, Abdalla M, Liu S, Yang H. 2023. Microplastics in canned, salt-dried, and instant sea cucumbers sold for human consumption. Mar Pollut Bull 192:115040, PMID: , 10.1016/j.marpolbul.2023.115040. [DOI] [PubMed] [Google Scholar]
- 9.Cox KD, Covernton GA, Davies HL, Dower JF, Juanes F, Dudas SE. 2019. Human consumption of microplastics. Environ Sci Technol 53(12):7068–7074, PMID: , 10.1021/acs.est.9b01517. [DOI] [PubMed] [Google Scholar]
- 10.Amato-Lourenco LF, Oliveira RC, Junior GR, Galvao LDS, Ando RA, Mauad T. 2021. Microplastics inhalation: evidence in human lung tissue. European Respiratory Journal 58(suppl 65):PA1792, 10.1183/13993003.congress-2021.PA1792. [DOI] [Google Scholar]
- 11.Vianello A, Jensen RL, Liu L, Vollertsen J. 2019. Simulating human exposure to indoor airborne microplastics using a breathing thermal manikin. Sci Rep 9(1):8670, PMID: , 10.1038/s41598-019-45054-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Udovicki B, Andjelkovic M, Cirkovic-Velickovic T, Rajkovic A. 2022. Microplastics in food: scoping review on health effects, occurrence, and human exposure. Int J Food Contamination 9(1):7, 10.1186/s40550-022-00093-6. [DOI] [Google Scholar]
- 13.Vivekanand AC, Mohapatra S, Tyagi VK. 2021. Microplastics in aquatic environment: challenges and perspectives. Chemosphere 282:131151, PMID: , 10.1016/j.chemosphere.2021.131151. [DOI] [PubMed] [Google Scholar]
- 14.Ren Z, Gui X, Xu X, Zhao L, Qiu H, Cao X. 2021. Microplastics in the soil-groundwater environment: aging, migration, and co-transport of contaminants – a critical review. J Hazard Mater 419:126455, PMID: , 10.1016/j.jhazmat.2021.126455. [DOI] [PubMed] [Google Scholar]
- 15.Schymanski D, Oßmann BE, Benismail N, Boukerma K, Dallmann G, von der Esch E, et al. 2021. Analysis of microplastics in drinking water and other clean water samples with micro-Raman and micro-infrared spectroscopy: minimum requirements and best practice guidelines. Anal Bioanal Chem 413(24):5969–5994, PMID: , 10.1007/s00216-021-03498-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schymanski D, Goldbeck C, Humpf HU, Furst P. 2018. Analysis of microplastics in water by micro-Raman spectroscopy: release of plastic particles from different packaging into mineral water. Water Res 129:154–162, PMID: , 10.1016/j.watres.2017.11.011. [DOI] [PubMed] [Google Scholar]
- 17.Nizzetto L, Futter M, Langaas S. 2016. Are agricultural soils dumps for microplastics of urban origin? Environ Sci Technol 50(20):10777–10779, PMID: , 10.1021/acs.est.6b04140. [DOI] [PubMed] [Google Scholar]
- 18.Priya AK, Jalil AA, Dutta K, Rajendran S, Vasseghian Y, Qin J, et al. 2022. Microplastics in the environment: recent developments in characteristic, occurrence, identification and ecological risk. Chemosphere 298:134161, PMID: , 10.1016/j.chemosphere.2022.134161. [DOI] [PubMed] [Google Scholar]
- 19.De Wit W, Biguad N. 2019. No Plastic in Nature: Assessing Plastic Ingestion From Nature to People. Geneva, Switzerland: WWF International. [Google Scholar]
- 20.Rillig MC, Ingraffia R, de Souza Machado AA. 2017. Microplastic incorporation into soil in agroecosystems. Front Plant Sci 8:1805, PMID: , 10.3389/fpls.2017.01805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Watson-Wright C, Singh D, Demokritou P. 2017. Toxicological implications of released particulate matter during thermal decomposition of nano-enabled thermoplastics. NanoImpact 5:29–40, PMID: , 10.1016/j.impact.2016.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Singh D, Schifman LA, Watson-Wright C, Sotiriou GA, Oyanedel-Craver V, Wohlleben W, et al. 2017. Nanofiller presence enhances polycyclic aromatic hydrocarbon (PAH) profile on nanoparticles released during thermal decomposition of nano-enabled thermoplastics: potential environmental health implications. Environ Sci Technol 51(9):5222–5232, PMID: , 10.1021/acs.est.6b06448. [DOI] [PubMed] [Google Scholar]
- 23.Pirela SV, Martin J, Bello D, Demokritou P. 2017. Nanoparticle exposures from nano-enabled toner-based printing equipment and human health: state of science and future research needs. Crit Rev Toxicol 47(8):683–709, PMID: , 10.1080/10408444.2017.1318354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sotiriou GA, Singh D, Zhang F, Chalbot M-CG, Spielman-Sun E, Hoering L, et al. 2016. Thermal decomposition of nano-enabled thermoplastics: possible environmental health and safety implications. J Hazard Mater 305:87–95, PMID: , 10.1016/j.jhazmat.2015.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Singh D, Marrocco A, Wohlleben W, Park H-R, Diwadkar AR, Himes BE, et al. 2022. Release of particulate matter from nano-enabled building materials (NEBMs) across their lifecycle: potential occupational health and safety implications. J Hazard Mater 422:126771, PMID: , 10.1016/j.jhazmat.2021.126771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boucher J, Friot D. 2017. Primary Microplastics in the Oceans: A Global Evaluation of Sources. Gland, Switzerland: International Union for Conservation of Nature. [Google Scholar]
- 27.WHO (World Health Organization). 2019. WHO Calls for More Research into Microplastics and Crackdown on Plastic Pollution. Geneve, Switzerland: WHO. [Google Scholar]
- 28.Hirt N, Body-Malapel M. 2020. Immunotoxicity and intestinal effects of nano- and microplastics: a review of the literature. Part Fibre Toxicol 17(1):57, PMID: , 10.1186/s12989-020-00387-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Montero V, Chinchilla Y, Gómez L, Flores A, Medaglia A, Guillén R, et al. 2023. Human health risk assessment for consumption of microplastics and plasticizing substances through marine species. Environ Res 237(pt 1):116843, PMID: , 10.1016/j.envres.2023.116843. [DOI] [PubMed] [Google Scholar]
- 30.Shruti VC, Perez-Guevara F, Kutralam-Muniasamy G. 2020. Metro station free drinking water fountain- a potential “microplastics hotspot” for human consumption. Environ Pollut 261:114227, PMID: , 10.1016/j.envpol.2020.114227. [DOI] [PubMed] [Google Scholar]
- 31.Van Cauwenberghe L, Janssen CR. 2014. Microplastics in bivalves cultured for human consumption. Environ Pollut 193:65–70, PMID: , 10.1016/j.envpol.2014.06.010. [DOI] [PubMed] [Google Scholar]
- 32.Ziino G, Nalbone L, Giarratana F, Romano B, Cincotta F, Panebianco A. 2021. Microplastics in vacuum packages of frozen and glazed icefish (Neosalanx spp.): a freshwater fish intended for human consumption. Ital J Food Saf 10(4):9974, PMID: , 10.4081/ijfs.2021.9974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao N, Zhao M, Jin H. 2023. Microplastic-induced gut microbiota and serum metabolic disruption in Sprague-Dawley rats. Environ Pollut 320:121071, PMID: , 10.1016/j.envpol.2023.121071. [DOI] [PubMed] [Google Scholar]
- 34.Jin Y, Lu L, Tu W, Luo T, Fu Z. 2019. Impacts of polystyrene microplastic on the gut barrier, microbiota and metabolism of mice. Sci Total Environ 649:308–317, PMID: , 10.1016/j.scitotenv.2018.08.353. [DOI] [PubMed] [Google Scholar]
- 35.Lu L, Wan Z, Luo T, Fu Z, Jin Y. 2018. Polystyrene microplastics induce gut microbiota dysbiosis and hepatic lipid metabolism disorder in mice. Sci Total Environ 631–632:449–458, PMID: , 10.1016/j.scitotenv.2018.03.051. [DOI] [PubMed] [Google Scholar]
- 36.Jin Y, Xia J, Pan Z, Yang J, Wang W, Fu Z. 2018. Polystyrene microplastics induce microbiota dysbiosis and inflammation in the gut of adult zebrafish. Environ Pollut 235:322–329, PMID: , 10.1016/j.envpol.2017.12.088. [DOI] [PubMed] [Google Scholar]
- 37.Zhao Y, Qin Z, Huang Z, Bao Z, Luo T, Jin Y. 2021. Effects of polyethylene microplastics on the microbiome and metabolism in larval zebrafish. Environ Pollut 282:117039, PMID: , 10.1016/j.envpol.2021.117039. [DOI] [PubMed] [Google Scholar]
- 38.Sata Y, Marques FZ, Kaye DM. 2020. The emerging role of gut dysbiosis in cardio-metabolic risk factors for heart failure. Curr Hypertens Rep 22(5):38, PMID: , 10.1007/s11906-020-01046-0. [DOI] [PubMed] [Google Scholar]
- 39.Aron-Wisnewsky J, Prifti E, Belda E, Ichou F, Kayser BD, Dao MC, et al. 2019. Major microbiota dysbiosis in severe obesity: fate after bariatric surgery. Gut 68(1):70–82, PMID: , 10.1136/gutjnl-2018-316103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boursier J, Mueller O, Barret M, Machado M, Fizanne L, Araujo-Perez F, et al. 2016. The severity of nonalcoholic fatty liver disease is associated with gut dysbiosis and shift in the metabolic function of the gut microbiota. Hepatology 63(3):764–775, PMID: , 10.1002/hep.28356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kaur N, Chen CC, Luther J, Kao JY. 2011. Intestinal dysbiosis in inflammatory bowel disease. Gut Microbes 2(4):211–216, PMID: , 10.4161/gmic.2.4.17863. [DOI] [PubMed] [Google Scholar]
- 42.Joossens M, Huys G, Cnockaert M, De Preter V, Verbeke K, Rutgeerts P, et al. 2011. Dysbiosis of the faecal microbiota in patients with Crohn’s disease and their unaffected relatives. Gut 60(5):631–637, PMID: , 10.1136/gut.2010.223263. [DOI] [PubMed] [Google Scholar]
- 43.Tamboli CP, Neut C, Desreumaux P, Colombel JF. 2004. Dysbiosis in inflammatory bowel disease. Gut 53(1):1–4, PMID: , 10.1136/gut.53.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Limonta G, Mancia A, Benkhalqui A, Bertolucci C, Abelli L, Fossi MC, et al. 2019. Microplastics induce transcriptional changes, immune response and behavioral alterations in adult zebrafish. Sci Rep 9(1):15775, PMID: , 10.1038/s41598-019-52292-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Qiao R, Deng Y, Zhang S, Wolosker MB, Zhu Q, Ren H, et al. 2019. Accumulation of different shapes of microplastics initiates intestinal injury and gut microbiota dysbiosis in the gut of zebrafish. Chemosphere 236:124334, PMID: , 10.1016/j.chemosphere.2019.07.065. [DOI] [PubMed] [Google Scholar]
- 46.Qiao R, Sheng C, Lu Y, Zhang Y, Ren H, Lemos B. 2019. Microplastics induce intestinal inflammation, oxidative stress, and disorders of metabolome and microbiome in zebrafish. Sci Total Environ 662:246–253, PMID: , 10.1016/j.scitotenv.2019.01.245. [DOI] [PubMed] [Google Scholar]
- 47.Wan Z, Wang C, Zhou J, Shen M, Wang X, Fu Z, et al. 2019. Effects of polystyrene microplastics on the composition of the microbiome and metabolism in larval zebrafish. Chemosphere 217:646–658, PMID: , 10.1016/j.chemosphere.2018.11.070. [DOI] [PubMed] [Google Scholar]
- 48.Lu Y, Zhang Y, Deng Y, Jiang W, Zhao Y, Geng J, et al. 2016. Uptake and accumulation of polystyrene microplastics in zebrafish (Danio rerio) and toxic effects in liver. Environ Sci Technol 50(7):4054–4060, PMID: , 10.1021/acs.est.6b00183. [DOI] [PubMed] [Google Scholar]
- 49.Stock V, Böhmert L, Lisicki E, Block R, Cara-Carmona J, Pack LK, et al. 2019. Uptake and effects of orally ingested polystyrene microplastic particles in vitro and in vivo. Arch Toxicol 93(7):1817–1833, PMID: , 10.1007/s00204-019-02478-7. [DOI] [PubMed] [Google Scholar]
- 50.Sun H, Chen N, Yang X, Xia Y, Wu D. 2021. Effects induced by polyethylene microplastics oral exposure on colon mucin release, inflammation, gut microflora composition and metabolism in mice. Ecotoxicol Environ Saf 220:112340, PMID: , 10.1016/j.ecoenv.2021.112340. [DOI] [PubMed] [Google Scholar]
- 51.Ibrahim YS, Tuan Anuar S, Azmi AA, Wan Mohd Khalik WMA, Lehata S, Hamzah SR, et al. 2021. Detection of microplastics in human colectomy specimens. JGH Open 5(1):116–121, PMID: , 10.1002/jgh3.12457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yan Z, Liu Y, Zhang T, Zhang F, Ren H, Zhang Y. 2022. Analysis of microplastics in human feces reveals a correlation between fecal microplastics and inflammatory bowel disease status. Environ Sci Technol 56(1):414–421, PMID: , 10.1021/acs.est.1c03924. [DOI] [PubMed] [Google Scholar]
- 53.Wu B, Wu X, Liu S, Wang Z, Chen L. 2019. Size-dependent effects of polystyrene microplastics on cytotoxicity and efflux pump inhibition in human caco-2 cells. Chemosphere 221:333–341, PMID: , 10.1016/j.chemosphere.2019.01.056. [DOI] [PubMed] [Google Scholar]
- 54.Herrala M, Huovinen M, Järvelä E, Hellman J, Tolonen P, Lahtela-Kakkonen M, et al. 2023. Micro-sized polyethylene particles affect cell viability and oxidative stress responses in human colorectal adenocarcinoma Caco-2 and HT-29 cells. Sci Total Environ 867:161512, PMID: , 10.1016/j.scitotenv.2023.161512. [DOI] [PubMed] [Google Scholar]
- 55.DeLoid GM, Cao X, Bitounis D, Singh D, Llopis PM, Buckley B, et al. 2021. Toxicity, uptake, and nuclear translocation of ingested micro-nanoplastics in an in vitro model of the small intestinal epithelium. Food Chem Toxicol 158:112609, PMID: , 10.1016/j.fct.2021.112609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.DeLoid GM, Cao X, Coreas R, Bitounis D, Singh D, Zhong W, et al. 2022. Incineration-generated polyethylene micro-nanoplastics increase triglyceride lipolysis and absorption in an in vitro small intestinal epithelium model. Environ Sci Technol 56(17):12288–12297, PMID: , 10.1021/acs.est.2c03195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cheng W, Li X, Zhou Y, Yu H, Xie Y, Guo H, et al. 2022. Polystyrene microplastics induce hepatotoxicity and disrupt lipid metabolism in the liver organoids. Sci Total Environ 806(pt 1):150328, PMID: , 10.1016/j.scitotenv.2021.150328. [DOI] [PubMed] [Google Scholar]
- 58.Merkley SD, Moss HC, Goodfellow SM, Ling CL, Meyer-Hagen JL, Weaver J, et al. 2022. Polystyrene microplastics induce an immunometabolic active state in macrophages. Cell Biol Toxicol 38(1):31–41, PMID: , 10.1007/s10565-021-09616-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.El Hayek E, Castillo E, In JG, Garcia M, Cerrato J, Brearley A, et al. 2023. Photoaging of polystyrene microspheres causes oxidative alterations to surface physicochemistry and enhances airway epithelial toxicity. Toxicol Sci 193(1):90–102, PMID: , 10.1093/toxsci/kfad023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Deng Y, Zhang Y, Lemos B, Ren H. 2017. Tissue accumulation of microplastics in mice and biomarker responses suggest widespread health risks of exposure. Sci Rep 7:46687, PMID: , 10.1038/srep46687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cary CM, DeLoid GM, Yang Z, Bitounis D, Polunas M, Goedken MJ, et al. 2023. Ingested polystyrene nanospheres translocate to placenta and fetal tissues in pregnant rats: potential health implications. Nanomaterials (Basel) 13(4):720, PMID: , 10.3390/nano13040720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xiong X, Gao L, Chen C, Zhu K, Luo P, Li L. 2023. The microplastics exposure induce the kidney injury in mice revealed by RNA-seq. Ecotoxicol Environ Saf 256:114821, PMID: , 10.1016/j.ecoenv.2023.114821. [DOI] [PubMed] [Google Scholar]
- 63.Meng X, Yin K, Zhang Y, Wang D, Lu H, Hou L, et al. 2022. Polystyrene microplastics induced oxidative stress, inflammation and necroptosis via NF-kappaB and RIP1/RIP3/MLKL pathway in chicken kidney. Toxicology 478:153296, PMID: , 10.1016/j.tox.2022.153296. [DOI] [PubMed] [Google Scholar]
- 64.Cheung LTO, Lui CY, Fok L. 2018. Microplastic contamination of wild and captive flathead grey mullet (Mugil cephalus). Int J Environ Res Public Health 15(4):597, PMID: , 10.3390/ijerph15040597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jing J, Zhang L, Han L, Wang J, Zhang W, Liu Z, et al. 2022. Polystyrene micro-/nanoplastics induced hematopoietic damages via the crosstalk of gut microbiota, metabolites, and cytokines. Environ Int 161:107131, PMID: , 10.1016/j.envint.2022.107131. [DOI] [PubMed] [Google Scholar]
- 66.Schwabl P, Köppel S, Königshofer P, Bucsics T, Trauner M, Reiberger T, et al. 2019. Detection of various microplastics in human stool: a prospective case series. Ann Intern Med 171(7):453–457, PMID: , 10.7326/M19-0618. [DOI] [PubMed] [Google Scholar]
- 67.Blackburn K, Green D. 2022. The potential effects of microplastics on human health: what is known and what is unknown. Ambio 51(3):518–530, PMID: , 10.1007/s13280-021-01589-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wright SL, Kelly FJ. 2017. Plastic and human health: a micro issue? Environ Sci Technol 51(12):6634–6647, PMID: , 10.1021/acs.est.7b00423. [DOI] [PubMed] [Google Scholar]
- 69.Ziani K, Ioniță-Mîndrican C-B, Mititelu M, Neacșu SM, Negrei C, Moroșan E, et al. 2023. Microplastics: a real global threat for environment and food safety: a state of the art review. Nutrients 15(3):617, PMID: , 10.3390/nu15030617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fendall LS, Sewell MA. 2009. Contributing to marine pollution by washing your face: microplastics in facial cleansers. Mar Pollut Bull 58(8):1225–1228, PMID: , 10.1016/j.marpolbul.2009.04.025. [DOI] [PubMed] [Google Scholar]
- 71.Han FY, Thurecht KJ, Whittaker AK, Smith MT. 2016. Bioerodable PLGA-based microparticles for producing sustained-release drug formulations and strategies for improving drug loading. Front Pharmacol 7:185, PMID: , 10.3389/fphar.2016.00185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rochman CM, Kross SM, Armstrong JB, Bogan MT, Darling ES, Green SJ, et al. 2015. Scientific evidence supports a ban on microbeads. Environ Sci Technol 49(18):10759–10761, PMID: , 10.1021/acs.est.5b03909. [DOI] [PubMed] [Google Scholar]
- 73.Senathirajah K, Attwood S, Bhagwat G, Carbery M, Wilson S, Palanisami T. 2021. Estimation of the mass of microplastics ingested - a pivotal first step towards human health risk assessment. J Hazard Mater 404(pt B):124004, PMID: , 10.1016/j.jhazmat.2020.124004. [DOI] [PubMed] [Google Scholar]
- 74.Scieszka D, Hunter R, Begay J, Bitsui M, Lin Y, Galewsky J, et al. 2022. Neuroinflammatory and neurometabolomic consequences from inhaled wildfire smoke-derived particulate matter in the Western United States. Toxicol Sci 186(1):149–162, PMID: , 10.1093/toxsci/kfab147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wei R, Wang J, Su M, Jia E, Chen S, Chen T, et al. 2018. Missing value imputation approach for mass spectrometry-based metabolomics data. Sci Rep 8(1):663, PMID: , 10.1038/s41598-017-19120-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Griss J, Viteri G, Sidiropoulos K, Nguyen V, Fabregat A, Hermjakob H. 2020. ReactomeGSA - efficient multi-omics comparative pathway analysis. Mol Cell Proteomics 19(12):2115–2125, PMID: , 10.1074/mcp.TIR120.002155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Milacic M, Beavers D, Conley P, Gong C, Gillespie M, Griss J, et al. 2023. The reactome pathway knowledgebase 2024. Nucleic Acids Res 52(D1):D672–D678, 10.1093/nar/gkad1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Barnes DK, Galgani F, Thompson RC, Barlaz M. 2009. Accumulation and fragmentation of plastic debris in global environments. Philos Trans R Soc Lond B Biol Sci 364(1526):1985–1998, PMID: , 10.1098/rstb.2008.0205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Auta HS, Emenike CU, Fauziah SH. 2017. Distribution and importance of microplastics in the marine environment: a review of the sources, fate, effects, and potential solutions. Environ Int 102:165–176, PMID: , 10.1016/j.envint.2017.02.013. [DOI] [PubMed] [Google Scholar]
- 80.Setala O, Fleming-Lehtinen V, Lehtiniemi M. 2014. Ingestion and transfer of microplastics in the planktonic food web. Environ Pollut 185:77–83, PMID: , 10.1016/j.envpol.2013.10.013. [DOI] [PubMed] [Google Scholar]
- 81.Huerta Lwanga E, Mendoza Vega J, Ku Quej V, Chi JDLA, Sanchez Del Cid L, Chi C, et al. 2017. Field evidence for transfer of plastic debris along a terrestrial food chain. Sci Rep 7(1):14071, PMID: , 10.1038/s41598-017-14588-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kosuth M, Mason SA, Wattenberg EV. 2018. Anthropogenic contamination of tap water, beer, and sea salt. PLoS One 13(4):e0194970, PMID: , 10.1371/journal.pone.0194970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Liebezeit G, Liebezeit E. 2014. Synthetic particles as contaminants in German beers. Food Addit Contam Part A Chem Anal Control Expo Risk Assess 31(9):1574–1578, PMID: , 10.1080/19440049.2014.945099. [DOI] [PubMed] [Google Scholar]
- 84.Möller W, Häussinger K, Winkler-Heil R, Stahlhofen W, Meyer T, Hofmann W, et al. 2004. Mucociliary and long-term particle clearance in the airways of healthy nonsmoker subjects. J Appl Physiol (1985) 97(6):2200–2206, PMID: , 10.1152/japplphysiol.00970.2003. [DOI] [PubMed] [Google Scholar]
- 85.Lomer MC, Thompson RP, Powell JJ. 2002. Fine and ultrafine particles of the diet: influence on the mucosal immune response and association with Crohn’s disease. Proc Nutr Soc 61(1):123–130, PMID: , 10.1079/pns2001134. [DOI] [PubMed] [Google Scholar]
- 86.Beamish LA, Osornio-Vargas AR, Wine E. 2011. Air pollution: an environmental factor contributing to intestinal disease. J Crohns Colitis 5(4):279–286, PMID: , 10.1016/j.crohns.2011.02.017. [DOI] [PubMed] [Google Scholar]
- 87.Fournier SB, D’Errico JN, Adler DS, Kollontzi S, Goedken MJ, Fabris L, et al. 2020. Nanopolystyrene translocation and fetal deposition after acute lung exposure during late-stage pregnancy. Part Fibre Toxicol 17(1):55, PMID: , 10.1186/s12989-020-00385-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sandys O, Te Velde A. 2022. Raising the alarm: environmental factors in the onset and maintenance of chronic (low-grade) inflammation in the gastrointestinal tract. Dig Dis Sci 67(9):4355–4368, PMID: , 10.1007/s10620-021-07327-1. [DOI] [PubMed] [Google Scholar]
- 89.Leslie HA, van Velzen MJM, Brandsma SH, Vethaak AD, Garcia-Vallejo JJ, Lamoree MH. 2022. Discovery and quantification of plastic particle pollution in human blood. Environ Int 163:107199, PMID: , 10.1016/j.envint.2022.107199. [DOI] [PubMed] [Google Scholar]
- 90.Jenner LC, Rotchell JM, Bennett RT, Cowen M, Tentzeris V, Sadofsky LR. 2022. Detection of microplastics in human lung tissue using muFTIR spectroscopy. Sci Total Environ 831:154907, PMID: , 10.1016/j.scitotenv.2022.154907. [DOI] [PubMed] [Google Scholar]
- 91.Munkholm P, Langholz E, Hollander D, Thornberg K, Orholm M, Katz KD, et al. 1994. Intestinal permeability in patients with Crohn’s disease and ulcerative colitis and their first degree relatives. Gut 35(1):68–72, PMID: , 10.1136/gut.35.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Büning C, Geissler N, Prager M, Sturm A, Baumgart DC, Büttner J, et al. 2012. Increased small intestinal permeability in ulcerative colitis: rather genetic than environmental and a risk factor for extensive disease? Inflamm Bowel Dis 18(10):1932–1939, PMID: , 10.1002/ibd.22909. [DOI] [PubMed] [Google Scholar]
- 93.Turpin W, Lee S-H, Raygoza Garay JA, Madsen KL, Meddings JB, Bedrani L, et al. 2020. Increased intestinal permeability is associated with later development of Crohn’s disease. Gastroenterology 159(6):2092–2100, PMID: , 10.1053/j.gastro.2020.08.005. [DOI] [PubMed] [Google Scholar]
- 94.Teshima CW, Dieleman LA, Meddings JB. 2012. Abnormal intestinal permeability in Crohn’s disease pathogenesis. Ann NY Acad Sci 1258:159–165, PMID: , 10.1111/j.1749-6632.2012.06612.x. [DOI] [PubMed] [Google Scholar]
- 95.Visser J, Rozing J, Sapone A, Lammers K, Fasano A. 2009. Tight junctions, intestinal permeability, and autoimmunity: celiac disease and type 1 diabetes paradigms. Ann NY Acad Sci 1165:195–205, PMID: , 10.1111/j.1749-6632.2009.04037.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Portincasa P, Bonfrate L, Khalil M, Angelis MD, Calabrese FM, D’Amato M, et al. 2021. Intestinal barrier and permeability in health, obesity and NAFLD. Biomedicines 10(1):83, PMID: , 10.3390/biomedicines10010083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang Q, Wu Y, Zhang W, Shen T, Li H, Wu J, et al. 2022. Lipidomics and transcriptomics insight into impacts of microplastics exposure on hepatic lipid metabolism in mice. Chemosphere 308(pt 3):136591, PMID: , 10.1016/j.chemosphere.2022.136591. [DOI] [PubMed] [Google Scholar]
- 98.Shi C, Han X, Guo W, Wu Q, Yang X, Wang Y, et al. 2022. Disturbed gut-liver axis indicating oral exposure to polystyrene microplastic potentially increases the risk of insulin resistance. Environ Int 164:107273, PMID: , 10.1016/j.envint.2022.107273. [DOI] [PubMed] [Google Scholar]
- 99.Wu P, Lin S, Cao G, Wu J, Jin H, Wang C, et al. 2022. Absorption, distribution, metabolism, excretion and toxicity of microplastics in the human body and health implications. J Hazard Mater 437:129361, PMID: , 10.1016/j.jhazmat.2022.129361. [DOI] [PubMed] [Google Scholar]
- 100.Handzlik MK, Gengatharan JM, Frizzi KE, McGregor GH, Martino C, Rahman G, et al. 2023. Insulin-regulated serine and lipid metabolism drive peripheral neuropathy. Nature 614(7946):118–124, PMID: , 10.1038/s41586-022-05637-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Grohmann U, Mondanelli G, Belladonna ML, Orabona C, Pallotta MT, Iacono A, et al. 2017. Amino-acid sensing and degrading pathways in immune regulation. Cytokine Growth Factor Rev 35:37–45, PMID: , 10.1016/j.cytogfr.2017.05.004. [DOI] [PubMed] [Google Scholar]
- 102.Hasegawa T, Iino C, Endo T, Mikami K, Kimura M, Sawada N, et al. 2020. Changed amino acids in NAFLD and liver fibrosis: a large cross-sectional study without influence of insulin resistance. Nutrients 12(5):1450, PMID: , 10.3390/nu12051450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gaggini M, Carli F, Rosso C, Buzzigoli E, Marietti M, Della Latta V, et al. 2018. Altered amino acid concentrations in NAFLD: impact of obesity and insulin resistance. Hepatology 67(1):145–158, PMID: , 10.1002/hep.29465. [DOI] [PubMed] [Google Scholar]
- 104.Lamichhane S, Kemppainen E, Trošt K, Siljander H, Hyöty H, Ilonen J, et al. 2019. Circulating metabolites in progression to islet autoimmunity and type 1 diabetes. Diabetologia 62(12):2287–2297, PMID: , 10.1007/s00125-019-04980-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yang ZS, Bai YL, Jin CH, Na J, Zhang R, Gao Y, et al. 2022. Evidence on invasion of blood, adipose tissues, nervous system and reproductive system of mice after a single oral exposure: nanoplastics versus microplastics. Biomed Environ Sci 35(11):1025–1037, PMID: , 10.3967/bes2022.131. [DOI] [PubMed] [Google Scholar]
- 106.Yang Q, Dai H, Cheng Y, Wang B, Xu J, Zhang Y, et al. 2023. Oral feeding of nanoplastics affects brain function of mice by inducing macrophage IL-1 signal in the intestine. Cell Rep 42(4):112346, PMID: , 10.1016/j.celrep.2023.112346. [DOI] [PubMed] [Google Scholar]
- 107.Jin H, Yang C, Jiang C, Li L, Pan M, Li D, et al. 2022. Evaluation of neurotoxicity in BALB/c mice following chronic exposure to polystyrene microplastics. Environ Health Perspect 130(10):107002, PMID: , 10.1289/EHP10255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Liang B, Huang Y, Zhong Y, Li Z, Ye R, Wang B, et al. 2022. Brain single-nucleus transcriptomics highlights that polystyrene nanoplastics potentially induce Parkinson’s disease-like neurodegeneration by causing energy metabolism disorders in mice. J Hazard Mater 430:128459, PMID: , 10.1016/j.jhazmat.2022.128459. [DOI] [PubMed] [Google Scholar]
- 109.Yin K, Wang D, Zhao H, Wang Y, Zhang Y, Liu Y, et al. 2022. Polystyrene microplastics up-regulates liver glutamine and glutamate synthesis and promotes autophagy-dependent ferroptosis and apoptosis in the cerebellum through the liver-brain axis. Environ Pollut 307:119449, PMID: , 10.1016/j.envpol.2022.119449. [DOI] [PubMed] [Google Scholar]
- 110.Gaspar L, Bartman S, Coppotelli G, Ross JM. 2023. Acute exposure to microplastics induced changes in behavior and inflammation in young and old mice. Int J Mol Sci 24(15):12308, PMID: , 10.3390/ijms241512308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Silva-Adaya D, Garza-Lombó C, Gonsebatt ME. 2021. Xenobiotic transport and metabolism in the human brain. NeuroToxicology 86:125–138, PMID: , 10.1016/j.neuro.2021.08.004. [DOI] [PubMed] [Google Scholar]
- 112.Vethaak AD, Legler J. 2021. Microplastics and human health. Science 371(6530):672–674, PMID: , 10.1126/science.abe5041. [DOI] [PubMed] [Google Scholar]
- 113.Prata JC, da Costa JP, Lopes I, Duarte AC, Rocha-Santos T. 2020. Environmental exposure to microplastics: an overview on possible human health effects. Sci Total Environ 702:134455, PMID: , 10.1016/j.scitotenv.2019.134455. [DOI] [PubMed] [Google Scholar]
- 114.Chang W-H, Herianto S, Lee C-C, Hung H, Chen H-L. 2021. The effects of phthalate ester exposure on human health: a review. Sci Total Environ 786:147371, PMID: , 10.1016/j.scitotenv.2021.147371. [DOI] [PubMed] [Google Scholar]
- 115.Yong CQY, Valiyaveettil S, Tang BL. 2020. Toxicity of microplastics and nanoplastics in mammalian systems. Int J Environ Res Public Health 17(5):1509, PMID: , 10.3390/ijerph17051509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sharma P, Vishwakarma R, Varjani S, Gautam K, Gaur VK, Farooqui A, et al. 2022. Multi-omics approaches for remediation of bisphenol A: toxicity, risk analysis, road blocks and research perspectives. Environ Res 215(pt 2):114198, PMID: , 10.1016/j.envres.2022.114198. [DOI] [PubMed] [Google Scholar]
- 117.Barhoumi B, Sander SG, Tolosa I. 2022. A review on per- and polyfluorinated alkyl substances (PFASs) in microplastic and food-contact materials. Environ Res 206:112595, PMID: , 10.1016/j.envres.2021.112595. [DOI] [PubMed] [Google Scholar]
- 118.Arrigo F, Impellitteri F, Piccione G, Faggio C. 2023. Phthalates and their effects on human health: focus on erythrocytes and the reproductive system. Comp Biochem Physiol C Toxicol Pharmacol 270:109645, PMID: , 10.1016/j.cbpc.2023.109645. [DOI] [PubMed] [Google Scholar]
- 119.Horn DL, Bettcher LF, Navarro SL, Pascua V, Neto FC, Cuschieri J, et al. 2021. Persistent metabolomic alterations characterize chronic critical illness after severe trauma. J Trauma Acute Care Surg 90(1):35–45, PMID: , 10.1097/TA.0000000000002952. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.