Abstract
The order Hymenoptera holds great significance for humans, particularly in tropical and subtropical regions, due to its role as a pollinator of wild and cultivated flowering plants, parasites of destructive insects and honey producers. Despite this importance, limited attention has been given to the genetic diversity and molecular identification of Hymenopteran insects in most protected areas. This study provides insights into the first DNA barcode of Hymenopteran insects collected from Hazarganji Chiltan National Park (HCNP) and contributes to the global reference library of DNA barcodes. A total of 784 insect specimens were collected using Malaise traps, out of which 538 (68.62%) specimens were morphologically identified as Hymenopteran insects. The highest abundance of species of Hymenoptera (133/538, 24.72%) was observed during August and least in November (16/538, 2.97%). Genomic DNA extraction was performed individually from 90/538 (16.73%) morphologically identified specimens using the standard phenol-chloroform method, which were subjected separately to the PCR for their molecular confirmation via the amplification of cytochrome c oxidase subunit 1 (cox1) gene. The BLAST analyses of obtained sequences showed 91.64% to 100% identities with related sequences and clustered phylogenetically with their corresponding sequences that were reported from Australia, Bulgaria, Canada, Finland, Germany, India, Israel, and Pakistan. Additionally, total of 13 barcode index numbers (BINs) were assigned by Barcode of Life Data Systems (BOLD), out of which 12 were un-unique and one was unique (BOLD: AEU1239) which was assigned for Anthidium punctatum. This indicates the potential geographical variation of Hymenopteran population in HCNP. Further comprehensive studies are needed to molecularly confirm the existing insect species in HCNP and evaluate their impacts on the environment, both as beneficial (for example, pollination, honey producers and natural enemies) and detrimental (for example, venomous stings, crop damage, and pathogens transmission).
Introduction
In phylum Arthropoda, the dominant members belong to class Insecta, which comprises numerous insect species from orders such as Coleoptera, Diptera, Hymenoptera and Lepidoptera, all of which hold medical and agricultural interests [1]. Globally, only 20% of insect species have been fully described and named, leaving the majority of species unidentified [2]. These families constitute a major component in terrestrial metazoan biodiversity playing a crucial role in preserving ecological services [3], climate change monitoring [4] and providing beneficial ecosystem services to humans [5, 6]. However, Pakistan’s insect diversity has received minimal taxonomic attention due to a lack of taxonomic experts and subsequent lack of insect species descriptions [7, 8]. Additionally, the existing lack of evidence regarding population declines in the face of serious environmental issues such as overgrazing, deforestation, soil erosion and waterlogging [9].
Hymenoptera is the second most diverse order of Insects with over 150,000 described species of ants, wasps, bees and many others [10, 11]. The members of this order carry out the process of pollination and thus play a very significant role in maintaining the structure and function of the forest ecosystem [12–14]. They also exert significant influence over the characteristics of modern terrestrial environments [14, 15]. For example, Hymenoptera species display a broad spectrum of social behavior ranging from solitary lifestyles of parasitic wasps to complex nest networks of super-colonial wood ants and bumble bee family systems [15]. In addition, certain phytophagous hymenopterans can be beneficial to their host plants [12, 16]. These features make Hymenoptera an ideal order for understanding of evolutionary dynamics and cohesion of complex social groups of taxa.
Malaise traps have been widely used to assess the abundance and composition of various insect taxa specifically Hymenoptera and Diptera [17–19]. This type of traps consists large netting tent often made out of a fine mesh material, which are advantageous due to low maintenance requirements. Further, these traps provide a comprehensive snapshot of the local insect community [20–22] and several studies have highlighted their effectiveness in capturing Hymenopteran insects [23, 24].
Traditional morphological based approach to insect taxonomy has long been established as an effective means of specie description and identification [17]. However, these methods are challenged [18, 19, 25] by a variety of juvenile life stages of insects, individuals with fluctuating phenotypes and cryptic species that complicate distinct identification [18, 26–30]. To address this issue, the use of molecular techniques specifically DNA Barcoding have been employed to enable large-scale assessments of biodiversity [29–31]. The mitochondrial cytochrome c oxidase subunit 1 (cox1) gene is most commonly used as a genetic marker due to the presence of conserved region [26, 32] allowing for discrimination between various groups or biotypes or ‘cryptic species’ in a single species [33, 34]. The use of Barcode Index Numbers (BINs) in these studies boundaries by comprehensively presenting species diversity [35, 36]. In the order Hymenoptera, DNA barcoding evaluations have reported minor variation at the species level and demonstrated its ability to accurately identify different species [27, 37, 38].
Several studies on morpho-molecular characterization of faunal Hymenopteran fauna in Pakistan have been documented [39–45]. However, the exact distribution and dispersion of Hymenopteran insects in the Balochistan province remains poorly undocumented due to lack of comprehensive research on the subject. Morphology-based studies on Hymenopteran species in Balochistan have identified specific species such as Polistes gallicus (Linneus, 1767) and Vespula germanica (Fabricius, 1793) in district Quetta [46]. In district Killa Saifullah of Balochistan, two studies reported nine species including Polistes flavus (Cresson, 1868), Polistes greeniptica (Fabricius, 1804), Polistes wattii (Cameron 1900), Polistes olivaceous (DeGeer, 1773), Polistes indicus (Stolfa, 1934), Polistes stigma (Fabricius, 1793), Ropalidia brevata [47] and Vespa orientalis (Linnaeus, 1771) [48]. This study serves to bridge a current knowledge gap by performing the first-ever molecular identification of Hymenopteran insects that have been reported from the protected area (HCNP) of Balochistan. This process has the potential to assist taxonomists to accurately classifying these Hymenopteran insects.
Methods and materials
Ethical statement
The Advanced Studies and Research Board committee at the University of Balochistan, Quetta has approved this study under registration number UoB/Reg/GSO/1197.
Study location and Malaise traps setting
The present study was carried out in Hazarganji-Chiltan National Park (HCNP) (30°13’21.8"N 66°44’13.5"E), located 20 kilometers Southwest of Quetta city in the Balochistan province, Pakistan. This national park is renowned for its diverse fauna and flora that has been officially designated as the 25th national park in Pakistan. It falls under the International Union for Conservation of Nature (IUCN) Category-V classified as a protected landscape [49], and covers an area of approximately 1315.22 km2. The study area is located at an elevation of about 5500 feet and experiences an annual total precipitation of around 240 mm during the winter season. The average temperature in summer season (June–August) can reach up to 40°C, while the winter season (November–March) sees rainfall and snowfall with temperature dropping as low as -12°C [50].
Malaise traps made up of mash material were used. These traps were provided by the International Barcode of Life (IBOL) and designed especially for the capturing of insects. The geographic coordinates of each collection site were recorded using a global positioning system and data obtained were processed in Microsoft Excel 2013 (Microsoft 365®) to create a study map using ArcGIS v 10.3.1 (Fig 1). Five significant locations within the study area were selected for the placement of Malaise traps to collect the insect specimens. These locations include the sub-campus of Balochistan University of Information Technology and Management Sciences (BUITM) (30°05’14.6"N 66°56’01.7"E), Hazarganji Nullah (30°02’10.9"N 66°52’02.9"E), Kangari (30°03’08.9"N 66°55’02.9"E), Wadd (30°01’01.6"N 66°49’32.4"E) and Garak (30°07’37.36"N 66°43’33.52"E).
Fig 1. Map shows the sampling sites of the study area.
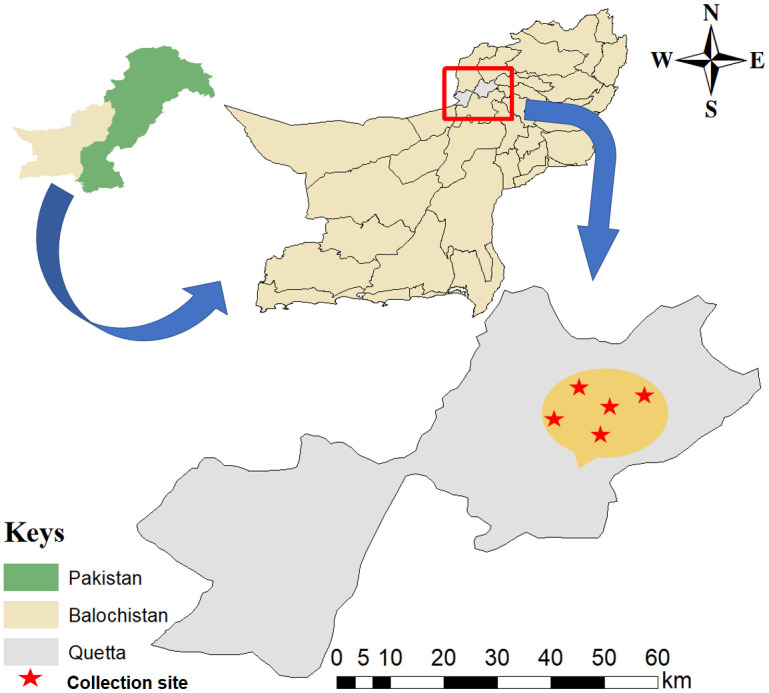
This map was created using of software ArcGIS v 10.3.1 (https://www.esri.com/en-us/arcgis/products/arcgis-desktop/resources).
Insect collection and preservation
Insects were collected using Malaise traps from April 2019 to November 2019 (a total of eight months of sampling). Specimens were collected in 500 mL plastic Nalgene® bottles containing 400 ml ethanol (95%) attached to each Malaise trap and then transferred into Whirl-Pak bag® containing 95% ethanol [51]. Specimen collection was performed on weekends (i.e., Saturday and Sunday). The dates of collection were marked on bags and these specimens were brought to the Entomology Laboratory at the University of Balochistan, Quetta for further molecular analyses.
Morphological identification
The collected specimens underwent a cleaning process using 70% ethanol and rinsed with distilled water to remove any external impurities. A stereomicroscope (Olympus SZ61, Japan) was used to identify the specimens by observing diagnostic features such as color pattern, wing venation, body shape, antennae and head. These characters were examined using established reference materials including standard published keys [52, 53] catalogs and electronic keys [54–56]. In the present study, we follow the family and subfamily classifications [18, 57–59] with additional resolution from the published articles [60, 61]. Identification was also performed by comparison with the help of other available Hymenopteran specimens already identified in the collections of Hymenopteran insects housed at National Insect Museum, National Agricultural Research Centre, Islamabad Pakistan (https://www.parc.gov.pk/).
Molecular analyses
Genomic DNA extraction was performed from each morphologically identified specimen using the standard phenol-chloroform method [62, 63]. Each specimen was individually homogenized in 200 μl of phosphate-buffered saline (pH = 7.4, 137 mM NaCl, 2.7 mM KCl, 8 mM Na2HPO4 and 2 mM KH2PO4). Homogenization was carried out by cutting one or two legs of each insect using sterile scissors and then grinding with a sterile pestle in a 1.5 mL Eppendorf tube. The resulting homogenate was used for DNA extraction. The quality and quantity of the extracted DNA were measured using NanoDrop (NanoQ, Optizen, Daejeon, South Korea) and then stored at -20°C for further analyses. The extracted genomic DNA from each morphologically identified specimen was used for conventional PCR (GE-96G, BIOER, Hangzhou, China) to amplify the universal genetic marker, partial fragments of cox1 gene (HC02198: 5’-TAA ACT TCA GGG TGA CCA AAA AAT CA-3’ and LCO1490: 5’-GGT CAA CAA ATC ATA AAG ATA TTG G-3’) [64]. The PCR cycling conditions were as follows: initial denaturation at 98 °C for 30s, followed by 40 cycles of denaturation at 98 °C for 10s, annealing at 63 °C for 20s, elongation at 72 °C for 25s and a final extension at 72 °C for 5 minutes. Each PCR reaction mixture was prepared in 20 μL, consisting of 1 μL of each primer (at a concentration of 10 pmol/μL), 4 μL PCR water, 2 μL (100 ng/μL) genomic DNA and 12 μL DreamTaq MasterMix (2X) (Thermo Fisher Scientific, USA). The PCR amplified products were run on a 2% agarose gel prepared in Tris borate EDTA (TBE) containing 2 μl ethidium bromide at a concentration of 0.2–0.5 μg/mL for staining purpose. The resulting bands were observed using Gel Documentation System (BioDoc-It™ Imaging Systems, UVP, LLC).
Phylogenetic analyses
The obtained amplified products were purified using GeneClean II Kit (Qbiogene, Illkirch, France) following the manufacturer’s protocol. The cox1 partial fragments were subjected to bidirectional sequencing through a commercial Korean company (Macrogen, Inc., Seoul, South Korea). The resulting bidirectional sequences were processed and refined by eliminating the poor reading and contaminated regions using SeqMan (V.5 DNASTAR, USA). The final trimmed and consensus sequences were further analyzed using Basic Local Alignment Search Tool (BLAST) [65] at National Center for Biotechnology Information (NCBI) (https://www.ncbi.nlm.nih.gov/). The sequences along with BINs and other related taxonomic information were recorded on BOLD (https://www.boldsystems.org/) and also deposited in the GenBank (NCBI). Then these sequences with high identity were downloaded in FASTA format from NCBI and were aligned using ClustalW and multiple alignments [66] that were further analyzed using BioEdit alignment editor tool (V.7.0.5, Raleigh, NC, USA) [67]. The Neighbor-Joining method employing the Kimura 2-parameter model was applied and 1000 bootstrap replicates were generated using Molecular Evolutionary Genetic Analysis (MEGA-X) software to construct the phylogenetic tree [68].
Results
Morphological identification
A total of 784 insects were captured using Malaise traps in the study area. The highest number of insects was collected at the BUITM sub-campus in Hazar Ganji (177/784, 22.58%), followed by Kangari (162/784, 20.66%), Hazarganji Nullah (154/784, 19.64%), Garak, (149/784, 19.01%) and Wadd (142/784, 18.11%). A total of 538/784 (68.62%) collected specimens were identified as Hymenopteran insects (S1 Fig). The most commonly identified Hymenopteran insects were Bethylidae sp. (59/538, 9.11%), followed by Tachysphex incertus (Radoszkowski, 1877) (48/538, 8.92%), Cerceris rybyensis (Linnaeus, 1771) (36/538, 8.55%), Tachytes freygessneri (43/538, 7.99%), Formicidae sp. and Lasioglossum sp. (42/538, 7.81%), Hymenoptera sp. (41/538, 7.62%), Anthidium punctatum, Camponotus compressus and Tachysphex sp. (39/538, 7.25%), Megachile leachella (38/538, 7.06%), Sphecidae sp. (37/538, 6.88%) and Evaniidae sp. (35/538, 6.51%) as presented in Table 1.
Table 1. Collection sites, number of insects collected and their morpho-molecular characterization.
| Collection sites | No. of insects collected | Morphologically Identified species | Number | Molecular Characterization | Sequences |
|---|---|---|---|---|---|
| BUITM sub-campus | 177 | Tachysphex incertus | 10 | 2 | 2 |
| Tachytes freygessneri | 9 | 2 | 2 | ||
| Anthidium punctatum | 8 | 2 | 2 | ||
| Megachile leachella | 9 | 1 | 1 | ||
| Cerceris rybyensis | 9 | 2 | 2 | ||
| Camponotus compressus | 8 | 1 | 1 | ||
| Lasioglossum sp. | 9 | 1 | 1 | ||
| Tachysphex sp. | 10 | 2 | 2 | ||
| Hymenoptera sp. | 8 | 1 | 1 | ||
| Evaniidae sp. | 7 | 2 | 2 | ||
| Formicidae sp. | 10 | 1 | 1 | ||
| Sphecidae sp. | 8 | 1 | 1 | ||
| Bethylidae sp. | 12 | 2 | 2 | ||
| Hazarganji Nullah | 154 | Tachysphex incertus | 11 | 1 | 1 |
| Tachytes freygessneri | 7 | 1 | 1 | ||
| Anthidium punctatum | 9 | 2 | 2 | ||
| Megachile leachella | 8 | 1 | 1 | ||
| Cerceris rybyensis | 10 | 2 | 2 | ||
| Camponotus compressus | 7 | 1 | 1 | ||
| Lasioglossum sp. | 8 | 1 | 1 | ||
| Tachysphex sp. | 5 | 1 | 1 | ||
| Hymenoptera sp. | 8 | 2 | 2 | ||
| Evaniidae sp. | 7 | 2 | 2 | ||
| Formicidae sp. | 7 | 1 | 1 | ||
| Sphecidae sp. | 6 | 1 | 1 | ||
| Bethylidae sp. | 9 | 1 | 1 | ||
| Kangari | 162 | Tachysphex incertus | 8 | 2 | 2 |
| Tachytes freygessneri | 10 | 1 | 1 | ||
| Anthidium punctatum | 6 | 2 | 2 | ||
| Megachile leachella | 7 | 1 | 1 | ||
| Cerceris rybyensis | 8 | 2 | 2 | ||
| Camponotus compressus | 6 | 1 | 1 | ||
| Lasioglossum sp. | 9 | 1 | 1 | ||
| Tachysphex sp. | 8 | 1 | 1 | ||
| Hymenoptera sp. | 9 | 1 | 1 | ||
| Evaniidae sp. | 7 | 2 | 2 | ||
| Formicidae sp. | 9 | 1 | 1 | ||
| Sphecidae sp. | 10 | 1 | 1 | ||
| Bethylidae sp. | 11 | 2 | 2 | ||
| Wadd | 142 | Tachysphex incertus | 10 | 2 | 2 |
| Tachytes freygessneri | 9 | 1 | 1 | ||
| Anthidium punctatum | 7 | 1 | 1 | ||
| Megachile leachella | 8 | 2 | 2 | ||
| Cerceris rybyensis | 9 | 1 | 1 | ||
| Camponotus compressus | 10 | 1 | 1 | ||
| Lasioglossum sp. | 7 | 1 | 1 | ||
| Tachysphex sp. | 10 | 2 | 2 | ||
| Hymenoptera sp. | 6 | 1 | 1 | ||
| Evaniidae sp. | 8 | 1 | 1 | ||
| Formicidae sp. | 7 | 1 | 1 | ||
| Sphecidae sp. | 5 | 2 | 2 | ||
| Bethylidae sp. | 8 | 2 | 2 | ||
| Garak | 149 | Tachysphex incertus | 9 | 2 | 2 |
| Tachytes freygessneri | 8 | 1 | 1 | ||
| Anthidium punctatum | 9 | 1 | 1 | ||
| Megachile leachella | 6 | 2 | 2 | ||
| Cerceris rybyensis | 10 | 2 | 2 | ||
| Camponotus compressus | 8 | 1 | 1 | ||
| Lasioglossum sp. | 9 | 1 | 1 | ||
| Tachysphex sp. | 6 | 1 | 1 | ||
| Hymenoptera sp. | 10 | 1 | 1 | ||
| Evaniidae sp. | 6 | 2 | 2 | ||
| Formicidae sp. | 9 | 1 | 1 | ||
| Sphecidae sp. | 8 | 1 | 1 | ||
| Bethylidae sp. | 9 | 1 | 1 | ||
| Total collection | 784 | Tachysphex incertus | 48 | 9 | 9 |
| Tachytes freygessneri | 43 | 6 | 6 | ||
| Anthidium punctatum | 39 | 8 | 8 | ||
| Megachile leachella | 38 | 7 | 7 | ||
| Cerceris rybyensis | 36 | 9 | 9 | ||
| Camponotus compressus | 39 | 5 | 5 | ||
| Lasioglossum sp. | 42 | 5 | 5 | ||
| Tachysphex sp. | 39 | 7 | 7 | ||
| Hymenoptera sp. | 41 | 6 | 6 | ||
| Evaniidae sp. | 35 | 9 | 9 | ||
| Formicidae sp. | 42 | 5 | 5 | ||
| Sphecidae sp. | 37 | 6 | 6 | ||
| Bethylidae sp. | 59 | 8 | 8 | ||
| Total | 538 | 90 | 90 | ||
Seasonal distribution
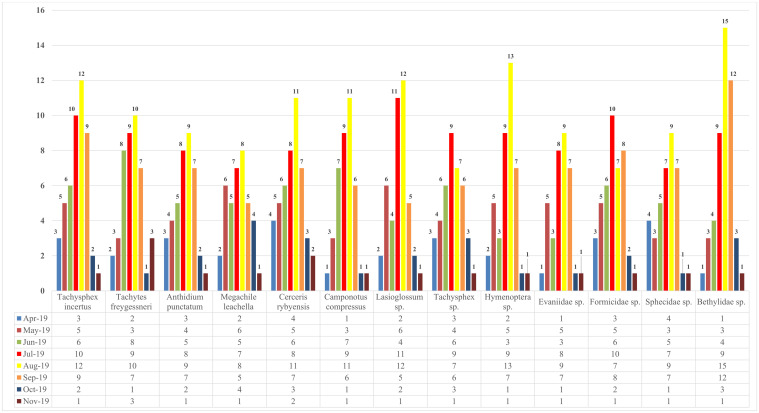
Out of 784 captured Hymenopteran insects, only 538 (68.62%) morphologically identified insects were reported on monthly basis. The highest number (133/538, 24.72%) of insects were reported in August, followed by the second highest count in July (114/538, 21.19%), September 93/538 (17.29%), June 68/538 (12.64%), May 57/538 (10.59%), April 31/538 (5.76%), October 26/538 (4.83%), while November had the least number of insects (16/538, 2.97%). This data indicates that the distribution of Hymenopteran insects follows a Gaussian distribution pattern (Fig 2).
Fig 2. Month-wise (April 2019 to November 2019) distribution of Hymenopteran insects.
Molecular confirmation
A total of 90 (16.73%) out of 538 (one or two specimens of morphologically identified Hymenopteran insects from each study location) were used for molecular confirmation. The remainder of the insect species in the Malaise traps belonged to other insect orders (unpublished data, manuscript under preparation). Of these, six insect species were identified at specie level, namely Tachysphex incertus (Radoszkowski, 1877) (n = 9), Cerceris rybyensis (Linnaeus, 1771) (n = 9), Anthidium punctatum (Latreille, 1809) (n = 8), Megachile leachella (Curtis 1828) (n = 7), Tachytes freygessneri (Kohl, 1881) (n = 6), Camponotus compressus (Fabricius 1787) (n = 5). Two insects were identified at genera level including Tachysphex sp. (n = 7), and Lasioglossum sp. (n = 5). Four different Hymenopteran insects were identified at family level including Evaniidae sp. (n = 9), Bethylidae sp. (n = 8), Sphecidae sp. (n = 6) and Formicidae sp. (n = 5), while one group of insects was identified at the level of order as Hymenoptera sp. (n = 6) as shown in Table 1.
Phylogenetic analysis outputs
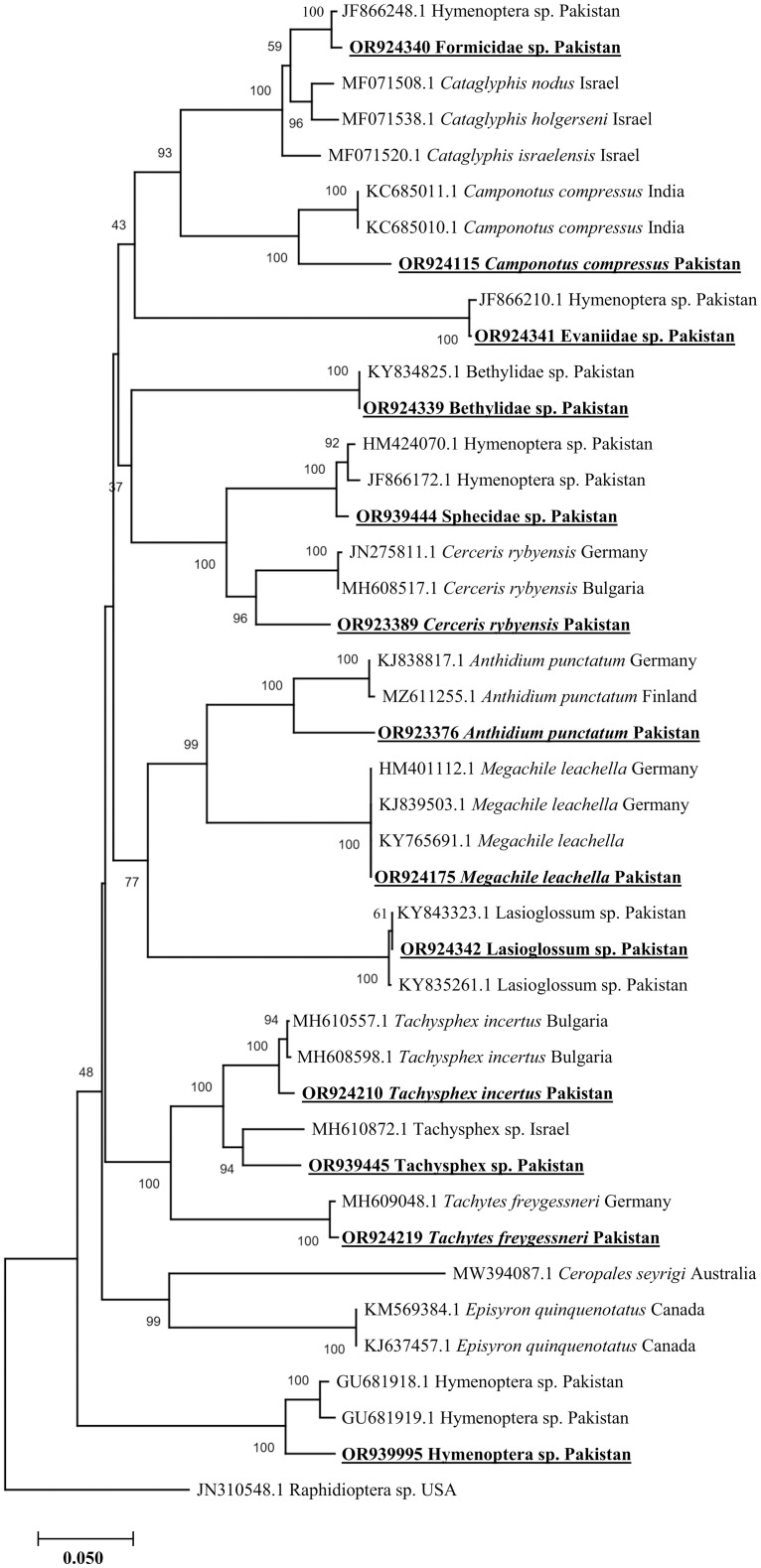
A phylogenetic tree was constructed using the Neighbor-Joining (NJ) method for 13 sequences (Fig 3). The obtained identical sequences were treated as consensus sequences for each Hymenopteran insect. The BLAST analysis revealed sequence similarities ranging from 91.64% to 100% which provided strong support for the reliability of phylogenetic tree fidelity. For example, Tachysphex incertus (658 bp) exhibited a 98.48% identity with Tachysphex incertus, Tachytes freygessneri (658 bp) showed 99.54% similarity with Tachytes freygessneri, Anthidium punctatum (614 bp) had 92.83%-93.16% identity with Anthidium punctatum, Megachile leachella (539 bp) showed 100% with Megachile leachella, Cerceris rybyensis (658 bp) showed 91.64% identity with Cerceris rybyensis, Camponotus compressus (617 bp) showed 93.37% with Camponotus compressus, Lasioglossum sp. (637 bp) showed 99.84% to 100% with Lasioglossum sp., Tachysphex sp. (658 bp) showed 93.92% with Tachysphex sp., Hymenoptera sp. (603 bp) showed 94.53% to 95.02% with Hymenoptera sp., Evaniidae sp. (655 bp) showed 99.48% with Hymenoptera sp., Formicidae sp. (580 bp) showed 99.14% with Hymenoptera sp., Sphecidae sp. (638 bp) showed 98.43% to 98.59% with Hymenoptera sp. and Bethylidae sp. (557 bp) had 100% with Bethylidae sp. These clusters were samples from various nations across the world including Australia, Bulgaria, Canada, Finland, Germany, India, Israel and Pakistan (Fig 3).
Fig 3. For order Hymenoptera, the phylogenetic tree based on cox1 partial fragments sequences was constructed via the Neighbor-Joining method with the Kimura 2-parameter model.
The bootstrap values (1000 replications) are shown at each node. Raphidioptera sp. (JN310548.1) from USA was selected as an outgroup. The obtained sequences of this study are marked in bold and also underlined.
BOLD and GenBank databases
All the details regarding specific identified species along with their specific specimen numbers, images, their BINs, and accession numbers are available in BOLD and GenBank databases as presented in Table 2. The results of the BOLD database analysis showed that 13 BINs were assigned for the obtained consensus sequences with six of these assigned to the species level, two to the genera level, four to the family level, and one to the order level. Of the 12 BINs that had been previously assigned (un-unique), one unique BIN (BOLD: AEU1239) was assigned to Anthidium punctatum in the study region, indicating the potential presence of a new species in HCNP, Balochistan.
Table 2. Identification of Hymenoptera insect species through DNA barcoding method and their close matching results are presented.
| Specimen code | Species | Own BINS | Nearest BINS | Accession number (NCBI) |
|---|---|---|---|---|
| AQBL-2 | Tachytes freygessneri (Kohl, 1881) | BOLD:ACH384 | BOLD:ADM7708 | OR924219 |
| AQBL-7 | Anthidium punctatum (Latreille, 1809) | BOLD:AEU1239 | BOLD:AEH2201 | OR923376 |
| AQBL-8 | Cerceris rybyensis (Linnaeus,1771) | BOLD:AEJ5559 | BOLD:ACY9461 | OR923389 |
| AQBL-9 | Tachysphex incertus (Radoszkowski, 1877) | BOLD:AAV7059 | BOLD:ACX5234 | OR924210 |
| AQBL-10 | Tachysphex sp. * | BOLD:AET4720 | BOLD:ACZ0155 | OR939445 |
| AQBL-16 | Evaniidae sp. ⁑ | BOLD:AAQ0495 | BOLD:ABW3800 | OR924341 |
| AQBL-20 | Sphecidae sp. ⁑ | BOLD:AAG8316 | BOLD:ADU2012 | OR939444 |
| AQBL-21 | Bethylidae sp. ⁑ | BOLD:ACD9427 | BOLD:AEA2483 | OR924339 |
| AQBL-27 | Hymenoptera sp. ⁑* | BOLD:AES2580 | BOLD:AAG8309 | OR939995 |
| AQBL-28 | Lasioglossum sp. * | BOLD:ACA2731 | BOLD:ADJ7874 | OR924342 |
| AQBL-38 | Megachile leachella (Curtis, 1828) | BOLD:AAD2767 | BOLD:AAN4635 | OR924175 |
| AQBL-45 | Formicidae sp. ⁑ | BOLD:AAQ0513 | BOLD:AEK5824 | OR924340 |
| AQBL-41 | Camponotus compressus (Fabricius 1787) | BOLD:ADZ9693 | BOLD:AEE5073 | OR924115 |
Symbol used in the above table: (*) denotes the representation of genera, (⁑) signifies the representation of families, while (⁑*) represents the order Hymenoptera.
Discussion
The current study aimed to expand scientific understating of Hymenoptera diversity in Balochistan given their lack of documentation. Previous faunistic studies relied on morphological analysis of species with medical and economic importance [46, 69–73]. These morphological studies are essential for determining species abundance in a given geographical area and monitoring the long-term shifts in population trends and diversity [73]. The results of these investigations indicates the biodiversity of insects and their importance in Pakistan’s protected areas such as HCNP [74]. Though these areas have received minimal research attention notably on Hymenopteran. In order to gain reliable data on Hymenopteran diversity, molecular technique was used in this study for their molecular confirmation.
In the present study, different Hymenopteran insects were molecularly identified, and corresponding BINs were generated, in which one was unique that was indicating the lack of representation in barcode database. In the IBOL database, all species codes and their morphologically identified images were displayed. DNA barcoding serves as an invaluable tool for large-scale, high-throughput taxonomic classifications offering potential implications for biodiversity investigations in protected regions. Despite this potential capacity, the diversity of Hymenopteran insects in Balochistan has not been adequately explored. For instance, Ashfaq et al. [75] conducted extensive research on identification of dominant insect orders including Coleoptera, Hemiptera, Hymenoptera and Lepidoptera that were captured by using Malaise traps in Pakistan. Studies have demonstrated that the HCNP, Hingol National Park (HNP) and other protected areas in Balochistan serve as biodiversity hotspots for various insects including bees, wasps, ants and sawflies [76]. Limited information regarding different Hymenopteran insects pose significant economic and health risks to local population of insects in Balochistan [77]. There have been noteworthy contributions of using DNA barcoding to identify various Hymenoptera bee species [78]. Insects in the Himalayas [79], conserved moderate-climate regions in Canada [80], South African savannah termite species [81], insect species in the Amazon jungle [82], New York city community garden bees [83] and insects from Thailand’s national parks [84] have been identified via DNA barcoding.
Our findings are consistent with previous investigations, which have used a combination of basic morphological and DNA barcode approaches [27, 85, 86]. Notably, one study relied mainly on the morphological features and reported almost 70% identification rate of various insects; this figure is closely matched by the 68.62% rate obtained in our findings [86]. The application of molecular identification has been widely accepted as a successful species identification method, solving issues which arise due to morphological similarities and concealed variations within cryptic species [87]. Several studies have used cox1 gene for the molecular identification of insect species [8, 38, 78]. By conducting phylogenetic analysis of sequences obtained in this study, we determined an evolutionary linkage between various obtained insects and their corresponding species, each showing the percent similarity ranging from 91.64% to 100%. Furthermore, genomes data from Canada, Germany, United States of America, United Kingdom, Pakistan and India have produced similar phylogenetic patterns. The cladding of the obtained sequences of different insects in phylogenetic tree is representing that the obtained insects are similar as in the aforementioned various countries [8, 78, 86, 88–90].
In this study, collected data regarding seasonal variations were analyzed that have impacts on the distribution of Hymenopteran insects. The average temperature from May to September was high and creating an ideal environment for insects to proliferate. However, a decrease in average temperature from September to April resulted in decline of their population. The average population of Hymenopteran insects showed a pattern of Gaussian distribution with highest number recorded in August (24.72%) and the least in November (2.97%). The Gaussian distribution has been described in other studies that were presenting the various Hymenopteran characteristics such as time spent on host feeding, host acceptance, host suitability, time spent walking, and body size [91, 92]. The seasonal variations of Hymenopteran insects can be better understood by studying the influence of environmental factors such as humidity, temperature, precipitation, and food availability [93, 94]. The obtained results can help to develop the mitigation strategies to overcome the substantial economic and health concerns of the local insect population in Pakistan [77]. Other influencing factors such as shortage of food, competition for resources, temperature, snow-fall and habitat destruction can also contribute to the survivorship of insects during the colder months [95, 96].
This study is limited by a few factors such as it’s a short-term focus on seasonal collections which may not fully account the yearly fluctuation of Hymenopteran insects’ population, thereby potentially obscuring meaningful distribution trends and species with distinct life cycles or behaviors. Moreover, the availability of DNA barcodes in majority of specimens was likely hindered by probable contamination of samples, resulting the degradation of DNA. In order to gain a more substantial insights into the ecology, behavior and evolution of the Hymenopteran insects in other protected areas, future studies should build upon the foundations established by this study. This study does not present any evidence of cryptic species, though it does emphasize the difficulty of species identification due to the presence of cryptic species and morphological convergence [97].
Conclusion
Fron the current study, we conclude that the HCNP has a rich of Hymenopteran insects’ fauna. DNA barcoding confirmed a total of six different species at their specie level, two at genus level, four at family level, and one at order level. The BOLD database has identified 13 BINs for Hymenopteran insects, of which one unique BIN was obtained for Anthidium punctatum. Furthermore, temperature variability throughout the year was found to exhibits a Gaussian distribution pattern. To further validate the molecular identities of insect species, it is recommended to perform further comprehensive studies in order to know about the diversification of insect species in other protected areas of Pakistan.
Supporting information
Figure presents a taxonomically arranged list of morphologically identified Hymenopterans insects: (A) Anthidium punctatum (B) Bethylidae sp. (C) Camponotus compressus (D) Ceropales seyrigi (D) Evaniidae sp. (F) Formicidae sp. (G) Lasioglossum sp. (H) Megachile leachella (I) Hymenoptera sp. (J) Sphecidae sp. (K) Tachysphex incertus (L) Tachysphex sp. The datasets presented in the study have been deposited to the NCBI GenBank repository (ncbi.nlm.nih.gov), and the obtained accession numbers are OR923389 (Cerceris rybyensis), OR924175 (Megachile leachella), OR923376 (Anthidium punctatum), OR924115 (Camponotus compressus), OR924210 (Tachysphex incertus), OR924219 (Tachytes freygessneri), OR939445 (Tachysphex sp.), OR924342 (Lasioglossum sp.), OR924339 (Bethylidae sp.), OR924340 (Formicidae sp.), OR924341 (Evaniidae sp.), OR939444 (Sphecidae sp.), and OR939995 (Hymenoptera sp.).
(TIF)
Acknowledgments
Authors are thankful to the workers at HCNP who assist us in collecting of Hymenopteran fauna. We are thankful to Prof. Dr. Muhammad Ashfaq Ahmed (University of Balochistan) for the editing the revised manuscript.
Data Availability
All relevant data are within the manuscript and its Supporting information files.
Funding Statement
The author(s) received no specific funding for this work.
References
- 1.Kobayashi S, Maldonado JE, Gaete A, Araya I, Aguado-Norese C, Cumplido N, et al. DNA sequencing in the classroom: complete genome sequence of two earwig (Dermaptera; Insecta) species. Biol Res. 2023;56(1):6. doi: 10.1186/s40659-023-00414-9 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scheffers BR, Joppa LN, Pimm SL, Laurance WF. What we know and don’t know about Earth’s missing biodiversity. Trends Ecol Evol. 2012;27(9):501–10. doi: 10.1016/j.tree.2012.05.008 . [DOI] [PubMed] [Google Scholar]
- 3.Adams WM, Aveling R, Brockington D, Dickson B, Elliott J, Hutton J, et al. Biodiversity conservation and the eradication of poverty. Science. 2004;306(5699):1146–9. doi: 10.1126/science.1097920 [DOI] [PubMed] [Google Scholar]
- 4.McMahon SM, Harrison SP, Armbruster WS, Bartlein PJ, Beale CM, Edwards ME, et al. Improving assessment and modelling of climate change impacts on global terrestrial biodiversity. Trends Ecol Evol. 2011;26(5):249–59. doi: 10.1016/j.tree.2011.02.012 . [DOI] [PubMed] [Google Scholar]
- 5.Stork NE. How many species of insects and other terrestrial arthropods are there on Earth? Annu Rev Entomol. 2018;63:31–45. doi: 10.1146/annurev-ento-020117-043348 . [DOI] [PubMed] [Google Scholar]
- 6.Noriega JA, Hortal J, Azcárate FM, Berg MP, Bonada N, Briones MJ, et al. Research trends in ecosystem services provided by insects. Basic Appl Ecol. 2018;26:8–23. doi: 10.1016/j.baae.2017.09.006 [DOI] [Google Scholar]
- 7.Ashfaq M, Akhtar S, Rafi MA, Mansoor S, Hebert PD. Mapping global biodiversity connections with DNA barcodes: Lepidoptera of Pakistan. PLoS One. 2017;12(3):e0174749. doi: 10.1371/journal.pone.0174749 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ashfaq M, Sabir JS, El-Ansary HO, Perez K, Levesque-Beaudin V, Khan AM, et al. Insect diversity in the Saharo-Arabian region: Revealing a little-studied fauna by DNA barcoding. PLoS One. 2018;13(7):e0199965. doi: 10.1371/journal.pone.0199965 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baig MB, Al-Subaiee FS. Biodiversity in Pakistan: key issues. Biodiversity. 2009;10(4):20–9. [Google Scholar]
- 10.Aguiar AP, Jennings JT. Order Hymenoptera, family Stephanidae. Arthropod fauna of the United Arab Emirates. 2010;3:299–305. [Google Scholar]
- 11.Boyle N, Skvarla M, McNeil D, Reagle N. Introduction to Insects. 2021 September:1–26.
- 12.Nyman T, Farrell BD, Zinovjev AG, Vikberg V. Larval habits, host-plant associations, and speciation in nematine sawflies (Hymenoptera: Tenthredinidae). Evolution. 2006;60(8):1622–37. doi: 10.1111/j.0014-3820.2006.tb00507.x . [DOI] [PubMed] [Google Scholar]
- 13.Grimaldi D, Engel MS. Evolution of the Insects. New York, NY, USA: Cambridge University Press; 2005. [Google Scholar]
- 14.Archibald S, Rasnitsyn AP, Brothers DJ, Mathewes RW. Modernisation of the Hymenoptera: ants, bees, wasps, and sawflies of the early Eocene Okanagan Highlands of western North America. The Canadian Entomologist. 2018;150(2):205–57. doi: 10.4039/tce.2017.59 [DOI] [Google Scholar]
- 15.Resh VH, Cardé RT. Encyclopedia of insects: Academic press; 2009. [Google Scholar]
- 16.Schultner E, Saramäki J, Helanterä H. Genetic structure of native ant supercolonies varies in space and time. Mol Ecol. 2016;25(24):6196–213. doi: 10.1111/mec.13912 . [DOI] [PubMed] [Google Scholar]
- 17.Ulyshen MD, Hanula JL, Horn S. Using Malaise traps to sample ground beetles (Coleoptera: Carabidae). The Canadian Entomologist. 2005;137(2):251–6. doi: 10.4039/N04-035 [DOI] [Google Scholar]
- 18.van Achterberg K. Can Townes type Malaise traps be improved? Some recent developments. Entomologische berichten. 2009;69(4):129–35. [Google Scholar]
- 19.Aguiar AP, Santos BF. Discovery of potent, unsuspected sampling disparities for Malaise and Möricke traps, as shown for Neotropical Cryptini (Hymenoptera, Ichneumonidae). J. Insect Conserv. 2010;14:199–206. doi: 10.1007/s10841-009-9246-x [DOI] [Google Scholar]
- 20.Vårdal H, Taeger A. The life of René Malaise: from the wild east to a sunken island. Zootaxa. 2011;3127:38–52. doi: 10.11646/zootaxa.3127.1.2 [DOI] [Google Scholar]
- 21.Lin X, Stur E, Ekrem T. Exploring genetic divergence in a species-rich insect genus using 2790 DNA barcodes. PloS one. 2015;10(9):e0138993. doi: 10.1371/journal.pone.0138993 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaczmarek M, Entling MH, Hoffmann C. Using Malaise Traps and Metabarcoding for Biodiversity Assessment in Vineyards: Effects of Weather and Trapping Effort. Insects. 2022;13(6):507. doi: 10.3390/insects13060507 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karlsson D, Hartop E, Forshage M, Jaschhof M, Ronquist F. The Swedish Malaise trap project: a 15 year retrospective on a countrywide insect inventory. Biodivers Data J. 2020;8. doi: 10.3897/BDJ.8.e47255 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Skvarla MJ, Larson JL, Fisher JR, Dowling AP. A review of terrestrial and canopy Malaise traps. Ann Entomol Soc Am. 2021;114(1):27–47. doi: 10.1093/aesa/saaa044 [DOI] [Google Scholar]
- 25.Volpato A, Ahmed KS, Williams CD, Day MF, O’Hanlon A, Ruas S, et al. Using Malaise traps to assess aculeate Hymenoptera associated with farmland linear habitats across a range of farming intensities. Insect Conserv Divers. 2020;13(3):229–38. doi: 10.1111/icad.12383 [DOI] [Google Scholar]
- 26.Midgley JM, Villet MH. Metrological framework for selecting morphological characters to identify species and estimate developmental maturity of forensically significant insect specimens. Forensic Sci Res. 2021;6(1):75–83. doi: 10.1080/20961790.2020.1794347 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hebert PD, Cywinska A, Ball SL, DeWaard JR. Biological identifications through DNA barcodes. Proceedings of the Royal Society of London Series B: Proc Biol Sci. 2003;270(1512):313–21. doi: 10.1098/rspb.2002.2218 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barrett RD, Hebert PD. Identifying spiders through DNA barcodes. Can J. Zool. 2005;83(3):481–91. doi: 10.1139/z05-024 [DOI] [Google Scholar]
- 29.Ball SL, Armstrong KF. DNA barcodes for insect pest identification: a test case with tussock moths (Lepidoptera: Lymantriidae). Can J Forest Res. 2006;36(2):337–50. doi: 10.1139/x05-276 [DOI] [Google Scholar]
- 30.Jinbo U, Kato T, Ito M. Current progress in DNA barcoding and future implications for entomology. Entomol Sci. 2011;14(2):107–24. doi: 10.1111/j.1479-8298.2011.00449.x [DOI] [Google Scholar]
- 31.Navajas M, Fenton B. The application of molecular markers in the study of diversity in acarology: a review. Exp Appl Acarol. 2000;24:751–74. doi: 10.1023/a:1006497906793 . [DOI] [PubMed] [Google Scholar]
- 32.Hebert PD, Ratnasingham S, De Waard JR. Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proc Biol Sci. 2003. Aug 7;270 Suppl 1(Suppl 1):S96–9. doi: 10.1098/rsbl.2003.0025 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brunner P, Fleming C, Frey J. A molecular identification key for economically important thrips species (Thysanoptera: Thripidae) using direct sequencing and a PCR-RFLP-based approach. Agricultural and Forest Entomology. 2002;4(2):127–36. doi: 10.1046/j.1461-9563.2002.00132.x [DOI] [Google Scholar]
- 34.Rugman-Jones PF, Hoddle MS, Stouthamer R. Nuclear-mitochondrial barcoding exposes the global pest western flower thrips (Thysanoptera: Thripidae) as two sympatric cryptic species in its native California. J Econ Entomol. 2010;103(3):877–86. doi: 10.1603/ec09300 . [DOI] [PubMed] [Google Scholar]
- 35.Ortiz AS, Rubio RM, Guerrero JJ, Garre MJ, Serrano J, Hebert PD, et al. Close congruence between Barcode Index Numbers (bins) and species boundaries in the Erebidae (Lepidoptera: Noctuoidea) of the Iberian Peninsula. Biodivers Data J. 2017;(5). doi: 10.3897/BDJ.5.e19840 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huemer P, Wieser C, Stark W, Hebert PD, Wiesmair B. DNA barcode library of megadiverse Austrian Noctuoidea (Lepidoptera)–a nearly perfect match of Linnean taxonomy. Biodivers Data J. 2019;7. doi: 10.3897/BDJ.7.e37734 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilson JJ, Sing KW, Floyd RM, Hebert PD. DNA barcodes and insect biodiversity. Insect biodiversity: science and society. 2017:575–92. doi: 10.7717/peerj.13267 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.deWaard JR, Levesque-Beaudin V, deWaard SL, Ivanova NV, McKeown JT, Miskie R, et al. Expedited assessment of terrestrial arthropod diversity by coupling Malaise traps with DNA barcoding. Genome. 2019;62(3):85–95. doi: 10.1139/gen-2018-0093 . [DOI] [PubMed] [Google Scholar]
- 39.Inayatullah M. To the knowledge of Vespidae (Hymenoptera) of Pakistan. Zootaxa. 2012;3318:26–50. [Google Scholar]
- 40.Siddiqui JA, Bodlah I, Carpenter JM, Naeem M, Ahmad M, Bodlah MA. Vespidae (Hymenoptera) of the Pothwar region of Punjab, Pakistan. Zootaxa. 2015;3914(5):501–24. [DOI] [PubMed] [Google Scholar]
- 41.Faiz A, Rafi MA, Zia A, Shah A, Shah SW, Khan RU, et al. Wasp fauna of (Eumeninae, Vespinae and Polistinae) in forests of Gilgit-Baltistan (Pakistan). Pure and Applied Biology. 2016;5(4):1. doi: http%3A//doi.org/10.19045/bspab.2016.50073 [Google Scholar]
- 42.Rafi MA, Carpenter JM, Qasim M, Shehzad A, Zia A, Khan MR, et al. The vespid fauna of Pakistan. Zootaxa. 2017;4362(1):1–28. doi: 10.11646/zootaxa.4362.1.1 [DOI] [PubMed] [Google Scholar]
- 43.Rasool M, Zahid M, Shah M. Solitary Wasps (Hymenoptera: Vespidae: Eumeninae) of Swat Pakistan, with two species newly reported from the country and three unidentified species. J Entomol Zool Stud. 2017;5(2):648–53. [Google Scholar]
- 44.Khan AK, Khan MR, Rafi MA, Qasim M. Wasp fauna of (Eumeninae, Vespinae and Polistinae) of district Poonch, Azad Jammu and Kashmir (Pakistan). J Entomol Zool Stud. 2017;5(6):1587–90. [Google Scholar]
- 45.Arsalan M, Abbas A, Gul SU, Rehman HU, Jawad SM, Shah W, et al. Biodiversity of wasps species collected from district Karak, KP, Pakistan. J Entomol Zool Stud. 2018;6(2):21–3. [Google Scholar]
- 46.Durrani S, Khan IA, Rafi MA, Makai G, Qasim M, Mengal F, et al. Wasps (Hymenoptera: Vespidae and Bradynobaenidae) from Quetta, Balochistan (Pakistan). University of Sindh Journal of Animal Sciences. 2018;2(3):35–41. [Google Scholar]
- 47.Das BP, Gupta VK. The social wasps of India and the adjacent countries. The social wasps of India and the adjacent countries. 1989. [Google Scholar]
- 48.Naz F, Kakar A, Nasim M, Mohammad W, Khan A, Naseeb N. Faunal diversity of paper wasp species and oriental hornet from tehsil Killa Saifullah of Zhob division, Pakistan. Pure and Applied Biology (PAB). 2020;9(1):1148–62. [Google Scholar]
- 49.Afsar S, Bano S. Mapping the Endangered/Key Species of Hazarganji-Chiltan National Park through geo-spatial technology. Inter J Biol Biotech. 2013;10:229–35. [Google Scholar]
- 50.Haneef M, Kakar A, Naseem M, Kurd A, Rafiq N, Kakar B, et al. Incidence of ectoparasite in chiltan wild goat (Artiodactyla: Caprinae) native of Hazarganji chiltan national park (HCNP), Balochistan, Pakistan. Pure and Applied Biology (PAB). 2019;8(1):389–96. [Google Scholar]
- 51.Bartholomew CS, Prowell D. Pan compared to malaise trapping for bees (Hymenoptera: Apoidea) in a longleaf pine savanna. Journal of the Kansas Entomological Society. 2005;78(4):390–2. [Google Scholar]
- 52.Mason WR, Huber JT. Order Hymenoptera. Hymenoptera of the world: an identification guide to families Minister of Supply and Services, Canada. 1993:4–12. [Google Scholar]
- 53.Marsh PM. Hymenoptera of the world: an identification guide to families. American Entomologist. 1994;40(2):115–6. doi: 10.1093/ae/40.2.115 [DOI] [Google Scholar]
- 54.Engel MS, Rasmussen C, Ayala R, de Oliveira FF. Stingless bee classification and biology (Hymenoptera, Apidae): a review, with an updated key to genera and subgenera. ZooKeys. 2023;1172:239. doi: 10.3897/zookeys.1172.104944 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Özbek H, Dathe HH. The bees of the genus Hylaeus Fabricius, 1793 of Turkey, with keys to the subgenera and species (Hymenoptera: Anthophila, Colletidae). Beiträge zur Entomologie = Contributions to Entomology. 2020;70(2):273–346. [Google Scholar]
- 56.Oh S-H, An S-L, Lee J-W. Review of Korean Latibulus (Hymenoptera: Ichneumonidae: Cryptinae) and a key to the world species. The Canadian Entomol. 2012;144(4):509–25. doi: 10.4039/tce.2012.54 [DOI] [Google Scholar]
- 57.Rasheed MT, Bodlah I, e Fareen AG, Wachkoo AA, Huang X, Akbar SA. A checklist of ants (Hymenoptera: Formicidae) in Pakistan. Sociobiology. 2019;66(3):426–39. doi: http%3A//doi.org/10.13102/sociobiology.v66i3.4330 [Google Scholar]
- 58.Augul RS, Abdul-Rassoul MS, Kaddou IK. Identification key to species of Sphecini (Hymenoptera: Sphecidae: Sphecinae) in Iraq. J Biodivers Environ Sci. 2015;6(1):111–21. [Google Scholar]
- 59.Moghaddam MG, Turrisi GF. Taxonomic and faunistic study of Aulacidae (Hymenoptera, Evanioidea) from Iran, with illustrated key to species. Zoosyst Evol. 2018;94(1):95–108. doi: 10.3897/zse.94.22501 [DOI] [Google Scholar]
- 60.Mohyuddin G, Bashir A, Mahmood A, Sharif T, Waheed I, Ahmed S. Taxonomic studies of family (Formicidae: Hymenoptera) six genera from district Faisalabad Punjab Pakistan. J Entomol Zool Stud. 2020;8(1):1384–9. [Google Scholar]
- 61.Lanes GO, Kawada R, Azevedo CO, Brothers DJ. Revisited morphology applied for systematics of flat wasps (Hymenoptera, Bethylidae). Zootaxa. 2020;4752(1):1–127. doi: 10.11646/zootaxa.4752.1.1 . [DOI] [PubMed] [Google Scholar]
- 62.Sanbrook J, Fritsch E, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed1989. 31 p.
- 63.Sambrook J, Russell DW. Purification of nucleic acids by extraction with phenol: chloroform. Cold Spring Harbor Protocols. 2006;2006(1):pdb. prot4455. doi: 10.1101/pdb.prot4455 [DOI] [PubMed] [Google Scholar]
- 64.Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol. 1994;3(5):294–9. [PubMed] [Google Scholar]
- 65.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–10. doi: 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- 66.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22(22):4673–80. doi: 10.1093/nar/22.22.4673 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hall T, Biosciences I, Carlsbad C. BioEdit: an important software for molecular biology. GERF Bull Biosci. 2011;2(1):60–1. [Google Scholar]
- 68.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Bio evol. 2018;35(6):1547. doi: 10.1093/molbev/msy096 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ahmed HA, Noor S, Sani IA, Kanwal S, Khudaidad S, Khawar M. Ant fauna (Hymenoptera: Formicidae) of Quetta, Balochistan, Pakistan. Serranga. 2013;18(2):47–59. [Google Scholar]
- 70.Rafi MA, Durrani S, Khan IA, Parveen G, Qasim M, Ullah Q, et al. Faunistic study of bees (Hymenoptera: Apoidea) form Quetta, Pakistan. University of Sindh J Anim Sci (USJAS). 2019;3(2). [Google Scholar]
- 71.Ayub M, Hazara A, Kazmi S. Life history of honey bee, Apis mellifera, L.(Hymenoptera: Apidae) in Quetta and Dhadar [Pakistan]. Balochistan J Agri Sci (Pakistan). 2001;2(2):48–51. [Google Scholar]
- 72.Hina A, Sabina N, Imran S, Safoora K, Shereen K, Masooma K. Ants fauna (Hymenoptera: Formicidae) of Quetta, Balochistan, Pakistan. Serangga. 2013;18:47–59. [Google Scholar]
- 73.Cameron P. On a new genus and some new species of aculeate Hymenoptera collected by Lieut.-Col. CG Nurse in Baluchistan. J Bombay Natur Hist Soci. 1907;18(1):130. [Google Scholar]
- 74.Noor S, Ahmed HA, Mengal F, Durrani S, Rasheed S, Abang F, et al. An annotated list of the butterfly fauna of Quetta, Pakistan. J Entomol Zool Stud. 2018;6:771–7. [Google Scholar]
- 75.Ashfaq M, Khan AM, Rasool A, Akhtar S, Nazir N, Ahmed N, et al. A DNA barcode survey of insect biodiversity in Pakistan. PeerJ. 2022;10:e13267. doi: 10.7717/peerj.13267 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Aslam S, Siddiqui S, Ullah U, Manzoor U, Lateef T, Samreen N, et al. Vertebrate wildlife of Pakistan: A review. Canadian J Pure Appl Sci. 2022;16(2):5483–95. [Google Scholar]
- 77.Sarwar M. Insect vectors involving in mechanical transmission of human pathogens for serious diseases. Inter J Bioinfor Biomed Engin. 2015;1(3):300–6. [Google Scholar]
- 78.Quicke DL, Alex Smith M, Janzen DH, Hallwachs W, Fernandez-Triana J, Laurenne NM, et al. Utility of the DNA barcoding gene fragment for parasitic wasp phylogeny (Hymenoptera: Ichneumonoidea): data release and new measure of taxonomic congruence. Mol Ecol Resour. 2012;12(4):676–85. doi: 10.1111/j.1755-0998.2012.03143.x . [DOI] [PubMed] [Google Scholar]
- 79.Srinivas G, Jayappa A, Patel A. Diversity of flower visitors in brinjal (Solanum melongena L.). Advances. 2016;5(3):834–7. [Google Scholar]
- 80.Ratnasingham S, Hebert PD. BOLD: The Barcode of Life Data System (http://www.barcodinglife.org). Mol Ecol Notes. 2007;7(3):355–64. doi: 10.1111/j.1471-8286.2007.01678.x . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Davies AB, Eggleton P, van Rensburg BJ, Parr CL. Seasonal activity patterns of African savanna termites vary across a rainfall gradient. Insectes Sociaux. 2015;62:157–65. [Google Scholar]
- 82.Lewinsohn TM, Freitas AVL, Prado PI. Conservation of terrestrial invertebrates and their habitats in Brazil. Conserv Biol. 2005;19(3):640–5. [Google Scholar]
- 83.Matteson KC, Ascher JS, Langellotto GA. Bee richness and abundance in new York city urban gardens. Annals Entomol Soci America. 2008;101(1):140–50. doi: 10.1603/0013-8746(2008)101[140:BRAAIN]2.0.CO;2 [DOI] [Google Scholar]
- 84.Sharkey M, Stoelb S. Revision of Agathacrista new genus (Hymenoptera, Braconidae, Agathidinae, Agathidini). J Hymeno Res. 2013;33:99–112. doi: 10.3897/jhr.33.4373 [DOI] [Google Scholar]
- 85.Ivanova NV, Dewaard JR, Hebert PD. An inexpensive, automation-friendly protocol for recovering high-quality DNA. Mol Ecol Notes. 2006;6(4):998–1002. doi: 10.1111/j.1471-8286.2006.01428.x [DOI] [Google Scholar]
- 86.Schmidt S, Schmid-Egger C, Morinière J, Haszprunar G, Hebert PD. DNA barcoding largely supports 250 years of classical taxonomy: identifications for Central European bees (Hymenoptera, Apoidea partim). Mol Ecol Resour. 2015;15(4):985–1000. doi: 10.1111/1755-0998.12363 . [DOI] [PubMed] [Google Scholar]
- 87.Xiao J-H, Wang N-X, Li Y-W, Murphy RW, Wan D-G, Niu L-M, et al. Molecular approaches to identify cryptic species and polymorphic species within a complex community of fig wasps. PLoS One. 2010;5(11):e15067. doi: 10.1371/journal.pone.0015067 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Koch JB, Lozier J, Strange JP, Ikerd H, Griswold T, Cordes N, et al. USBombus, a database of contemporary survey data for North American Bumble Bees (Hymenoptera, Apidae, Bombus) distributed in the United States. Biodiver Data J. 2015;(3). doi: 10.3897/BDJ.3.e6833 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sheffield CS, Heron J, Gibbs J, Onuferko TM, Oram R, Best L, et al. Contribution of DNA barcoding to the study of the bees (Hymenoptera: Apoidea) of Canada: progress to date. The Canadian Entomol. 2017;149(6):736–54. doi: 10.4039/tce.2017.49 [DOI] [Google Scholar]
- 90.Harshana A, Dey D. Taxonomic studies on the ant genus Lepisiota Santschi, 1926 (Hymenoptera: Formicidae: Formicinae) in India, with description of four new species. Oriental Insects. 2023;57(3):785–818. doi: 10.1080/00305316.2022.2125096 [DOI] [Google Scholar]
- 91.Le Goff GJ, Berthe J, Tougeron K, Dochy B, Lebbe O, Renoz F, et al. Effect of the instar of the pear psyllid Cacopsylla pyri (Hemiptera: Psyllidae) on the behaviour and fitness of the parasitoid Trechnites insidiosus (Hymenoptera: Encyrtidae). European J Entomol. 2021;118. doi: 10.14411/eje.2021.028 [DOI] [Google Scholar]
- 92.Zhou JC, Liu QQ, Wang QR, Ning SF, Che WN, Dong H. Optimal clutch size for quality control of bisexual and Wolbachia-infected thelytokous lines of Trichogramma dendrolimi Matsumura (Hymenoptera: Trichogrammatidae) mass reared on eggs of a substitutive host, Antheraea pernyi Guérin-Méneville (Lepidoptera: Saturniidae). Pest Manag Sci. 2020;76(8):2635–44. [DOI] [PubMed] [Google Scholar]
- 93.Yadav N, Singh P. Seasonal abundance of insect pests on mung bean and its correlation with abiotic factors. J Entomol Res. 2013;37(4):297–9. [Google Scholar]
- 94.Schwarz B, Vázquez DP, CaraDonna PJ, Knight TM, Benadi G, Dormann CF, et al. Temporal scale-dependence of plant–pollinator networks. Oikos. 2020;129(9):1289–302. doi: 10.1111/oik.07303 [DOI] [Google Scholar]
- 95.Marchioro CA, Foerster LA. Biotic factors are more important than abiotic factors in regulating the abundance of Plutella xylostella L., in Southern Brazil. Revista Brasileira de Entomologia. 2016;60:328–33. doi: 10.1016/j.rbe.2016.06.004 [DOI] [Google Scholar]
- 96.Dar SA, Ansari MJ, Al Naggar Y, Hassan S, Nighat S, Zehra SB, et al. Causes and reasons of insect decline and the way forward. 2021. doi: 10.5772/intechopen.98786 [DOI] [Google Scholar]
- 97.Adams BJ, Li E, Bahlai CA, Meineke EK, McGlynn TP, Brown BV. Local-and landscape-scale variables shape insect diversity in an urban biodiversity hot spot. Ecol Applica. 2020;30(4):e02089. doi: 10.1002/eap.2089 . [DOI] [PMC free article] [PubMed] [Google Scholar]