Abstract
Baculovirus can transiently transduce primary human and rat hepatocytes, as well as a subset of stable cell lines. To prolong transgene expression, we have developed new hybrid vectors which associate key elements from adeno-associated virus (AAV) with the elevated transducing capacity of baculovirus. The hybrid vectors contain a transgene cassette composed of the β-galactosidase (β-Gal) reporter gene and the hygromycin resistance (Hygr) gene flanked by the AAV inverted terminal repeats (ITRs), which are necessary for AAV replication and integration in the host genome. Constructs were derived both with and without the AAV rep gene under the p5 and p19 promoters cloned in different positions with respect to the baculovirus polyheidrin promoter. A high-titer preparation of baculovirus-AAV (Bac-AAV) chimeric virus containing the ITR–Hygr–β-Gal sequence was obtained with insect cells only when the rep gene was placed in an antisense orientation to the polyheidrin promoter. Infection of 293 cells with Bac-AAV virus expressing the rep gene results in a 10- to 50-fold increase in the number of Hygr stable cell clones. Additionally, rep expression determined the localization of the transgene cassette in the aavs1 site in approximately 41% of cases as detected by both Southern blotting and fluorescent in situ hybridization analysis. Moreover, site-specific integration of the ITR-flanked DNA was also detected by PCR amplification of the ITR-aavs1 junction in transduced human fibroblasts. These data indicate that Bac-AAV hybrid vectors can allow permanent, nontoxic gene delivery of DNA constructs for ex vivo treatment of primary human cells.
Gene therapy is a rapidly emerging field that aims to treat a variety of genetic or acquired diseases through the transfer of functional genetic material into cells both in vitro and in vivo (1, 30, 34). Critical to the success of gene therapy is the development of safe and efficient gene transfer vehicles. To date, various strategies have been developed for the transfer of therapeutic genes which include viral and nonviral vectors. All of these gene delivery systems, however, suffer from limitations in their applicability and efficacy (49). Among the viral vectors utilized for gene transfer protocols, adenovirus (Ad) vectors deliver genes to a wide variety of cell types and tissues independently of their proliferative state (9). However, the major disadvantage of this type of vector is the instability of the genes transferred into the target cell and the substantial pathology that develops at the site of gene transfer. These problems can be explained, at least in part, by a lack of integration of the recombinant Ad genome and by the development of a strong cellular immune response to the genetically corrected cells that express low levels of viral proteins (13, 53). Despite some improvements in reducing the immunogenicity of Ad vectors (3, 50, 55), their use for long-term expression of the therapeutic gene has yet to be fully assessed.
Retroviruses, the viral vectors currently most widely used, offer the desirable feature of being able to insert a gene of interest into the host genome, thus contributing to the stability of the transduced gene (12). However, retroviruses have a limited host range, and successful infection occurs only in mitotic cells, with the exception of the human immunodeficiency virus (31). Additionally, retroviruses integrate randomly into the host cell chromosome, thus raising some concern about the potential activation of transcriptionally silent oncogenes and the possible inactivation of tumor suppressor genes mediated by insertional mutagenesis (43).
The adeno-associated virus (AAV) is also used for gene delivery protocols. The lack of obvious pathogenic effects associated with AAV infection and the stability of the viral particle have elicited great interest in the use of AAV as a vector in the field of gene therapy (10). Additionally, AAV is the only known eukaryotic virus that preferentially integrates its DNA into a defined region of the host cell genome located on chromosome 19q13.3 (16, 28, 40). Although the mechanism of site-specific integration is still unclear, two viral elements, namely, the inverted terminal repeats (ITRs) and the Rep polypeptides, are required for targeting and inserting the viral DNA into the integration locus aavs1 (42). The two 145-nucleotide ITRs located at both ends of the viral genome are the minimal cis-acting sequences required in the integration process. The palindromic portions of the ITRs are capable of forming hairpin structures and serve as a self-priming origin for DNA replication, as well as a site for binding of AAV Rep proteins (20, 44, 48). The rep gene is transcribed from two promoters, p5 and p19. Transcription from the p5 promoter generates spliced and unspliced mRNAs which encode the Rep68 and Rep78 proteins, respectively. Two smaller proteins, Rep40 and Rep52, are encoded by spliced and unspliced mRNAs promoted by the p19 promoter, respectively (45). These proteins mediate replication and integration of the AAV genome, as well as packaging into viral particles (7). In particular, Rep68 and Rep78 interact with the terminal sequences only if the secondary structure of the termini is T shaped, a conformation that is believed to exist in virion DNA (4, 20). Additionally, these two large Rep polypeptides contain ATP-dependent helicase and strand-specific endonuclease activities which are thought to be important both for viral DNA replication and integration (21, 22). Furthermore, recent evidence has indicated that site-specific integration of an ITR-flanked DNA segment requires expression of the Rep proteins (5, 37) and that with only the expression of Rep78 or Rep68, highly efficient integration into the aavs1 site is obtained (46).
The major limitation of AAV-based vectors resides in the size constraint imposed by the inability of the mature AAV particles to package DNA fragments larger than 5 kb. Therefore, recombinant AAV vectors are generated by substituting the endogenous genes with exogenous DNA, resulting in the production of vectors that can no longer integrate in a site-specific manner. In fact, although these viruses can persist both in vitro and in vivo for prolonged periods of time, they do so either as episomes or by integrating randomly into the host chromosome (11, 14, 15, 24–27, 33, 38, 52).
Recently, the use of the Autographa californica multiple nuclear polyhedrosis virus as a vector for the delivery of genes into mammalian cells has been reported (8, 19). Although baculoviruses are normally utilized for the overexpression of recombinant proteins in insect cells (35) or as biopesticides (12), it was shown that A. californica multiple nuclear polyhydrosis virus can infect human hepatocytes, leading to an efficient transient expression of reporter genes under the control of an appropriate mammalian promoter (8, 19). In view of the ease of preparation and large cloning capacity of baculovirus (36), its use as vector for gene transfer holds great potential. However, both the inability of baculovirus to transduce cells in vivo (41) and the transient nature of the transferred DNA pose some limitations for its use. To determine whether the persistence of the baculovirus-transduced DNA can be prolonged, we introduced the AAV rep gene and a transgene cassette flanked by the AAV terminal repeats into a baculovirus backbone. We here show that a stable baculovirus-AAV (Bac-AAV) hybrid vector can be produced in insect cells at a high titer provided that the rep gene is cloned into an antisense orientation with respect to the baculovirus polyhedrin promoter. Additionally, we have determined that Bac-AAV infection of 293 cells results in highly efficient integration of the transgene cassette in the aavs1 site. Lastly, we observed that Bac-AAV can efficiently transduce human diploid fibroblasts, which results in the site-specific integration of ITR-flanked DNA. These findings extend the use of baculovirus as a vector for gene therapy.
MATERIALS AND METHODS
Vector constructs.
Recombinant baculoviruses were constructed by using the transfer vector pFastBac1 (pFB1) (Gibco-BRL). A β-galactosidase (β-Gal)-expressing cassette was derived from plasmid pCMV-β (Clontech). The EcoRI-HindIII fragment containing the cytomegalovirus (CMV) promoter, the β-Gal gene, and the simian virus 40 (SV40) polyadenylation signal was inserted in the modified sites of pFB1, generating plasmid pFB1Bac/CMV-β. To construct the chimeric Bac-AAV vectors, a p5-Rep-poly(A) cassette (ClaI and SpeI) was subcloned into the NspV and SpeI sites of pFB1, generating pFB1Rep-A.
A transgene cassette was assembled in plasmid pLitmus-28 (New England Biolabs) by three cloning steps, generating pLit/AAV-Hyg-β-Gal, which contains the following sequences: (i) the AAV ITRs derived from plasmid pSub201 (39) as a PvuII fragment and subcloned in the blunted XbaI site of pLitmus-28; (ii) the hygromycin resistance (Hygr) gene excised from plasmid pCEP-4 (Invitrogen) by digestion with NruI and NotI and inserted between the ITRs in the filled-in XbaI site; and (iii) the β-Gal cassette derived from plasmid CMV-β, which was introduced between the ITRs by blunt-end cloning into the filled-in NotI and HindIII sites.
Plasmid pLit/AAV-Hyg-β-Gal was digested with SpeI and AvrII, and the 6.5-kb fragment containing the transgene cassette was subcloned into the modified pFB1 vector (digested with XbaI and AvrII) or pFB1RepA to generate Bac-ITR and Bac/ITR-RepA, respectively. Similarly, to generate plasmid Bac-RepS, the p5-Rep fragment was inserted in the SfuI site of pFB1. Bac-ITR/RepS was derived from Bac-Rep-S by introducing the transgene cassette as a AvrII-SpeI fragment into the SpeI site.
Recombinant baculoviruses were produced according to the manufacturer’s instructions (Gibco-BRL). Viruses were propagated in Sf9 insect cells according to standard methods (36). Budded virus from insect cell culture medium was filtered on a 0.22-μm (pore size) filter and concentrated by ultrafiltration in a 45 Ti rotor (30,000 rpm, 75 min). The viral pellet was resuspended in phosphate-buffered saline (PBS), and the titers of the virus were determined by plaque assay on Sf9 insect cells.
Cell culture.
293, Huh-7, and MRC-5 cells were maintained in Dulbecco modified Eagle medium supplemented with 10% fetal calf serum, 2 mM glutamine, 100 U of penicillin per ml, and 100 μg of streptomycin per ml. Cells were grown in 10-cm-diameter dishes (Falcon) at 37°C in 5% CO2. Stock cells were routinely passaged every 3 days by treatment with trypsin (0.05%) and EDTA (0.53 mM) and then replated at cell densities appropriate for exponential growth.
To isolate stable cell clones, the infected cells from a single 6-cm-diameter plate were plated at a density of 2 × 104 cells per plate in 10 plates containing selection medium (Dulbecco modified Eagle medium, 10% fetal calf serum, and 300 μg of hygromycin per ml). Hygromycin-resistant (Hygr) clones were isolated after 10 days of selection and then expanded and processed for genomic DNA extraction, Southern blotting, and fluorescence in situ hybridization (FISH) analysis. Assays for β-Gal activity were carried out using the β-Gal enzyme assay system (Promega) as described by the manufacturer. β-Gal was detected histochemically in cells fixed with 0.5% glutaraldehyde solution in PBS and by incubation for 4 h with staining solution (4 mM K4[Fe(CN)6], 4 mM K3[Fe(CN)6], 40 mM MgCl2, 0.4-mg/ml X-Gal [5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside] in PBS).
Western blot analysis.
The expression of AAV Rep proteins was evaluated by Western blotting with a polyclonal antibody which recognizes all four Rep isoforms (47). A total of 2 × 106 Sf9 cells were infected at a multiplicity of infection (MOI) of 10, and 72 h later cells were collected, washed in PBS, and lysed by freezing and thawing three times in lysis buffer [10 mM HEPES buffer [pH 8.0], 0.6 M NaCl]. Portions (200 μg) of proteins of each sample were separated by sodium dodecyl sulfate (SDS)–12.5% polyacrylamide gel electrophoresis and then transferred onto a nitrocellulose membrane. The nitrocellulose membrane was immersed in 1% skimmed milk in Tris-buffered saline (20 mM Tris-HCl, 500 mM NaCl [pH 7.5]) (blocking buffer) for 20 min at room temperature. The anti-Rep polyclonal antiserum diluted in blocking buffer was applied to the nitrocellulose membrane and incubated for 1 h at room temperature. The membrane was then washed repeatedly with blocking buffer. The bands were visualized with ECL reagents according to the manufacturer’s instructions (Amersham).
Southern blot analysis.
Baculovirus genomic DNA was prepared from 1-ml portions of the third viral passage according to standard protocols (35). One microgram of viral DNA was digested with EcoRV, which releases two fragments of 4 and 10 kb. After electrophoresis in 1% agarose gel, the digested fragments were transferred onto a nylon membrane (Hybond N+), processed according to the manufacturer’s instructions, and hybridized overnight at 65°C in Church buffer (7% SDS, 0.25 M NaPi [pH 7.2], 1 mM EDTA [pH 8.0], bovine serum albumin [0.1 g/ml]) with random primed 32P-labeled probes. The following probes were used: for the gentamicin gene a 1.3-kb EcoRV-BamHI fragment derived from the plasmid pFastBac1 and for the transgene cassette a 6.5-kb AvrII-SpeI fragment derived from plasmid pLit/AAV-Hyg-β-Gal.
To determine site-specific integration of the ITR-DNA fragment, genomic DNA was prepared as previously described (37) and processed as described above. Filters were first hybridized with a probe specific for the transgene cassette; the hybridized probe was then removed by boiling the filters in 0.2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and 1% SDS for 10 min, and the same filters were then hybridized to a probe covering nucleotides 1 to 3,525 of aavs1.
FISH.
A 6.5-kb DNA fragment corresponding to the transgene cassette and an 80-kb aavs1 DNA fragment isolated by screening a genomic DNA library were labeled by using the Nick Translation Kit (Boehringer Mannheim) according to the manufacturer’s instructions and used as probes in chromosome analysis.
The chromosome spreads from selected clones were prepared according to typical cytogenetic techniques (29). Cytogenetic preparations were pretreated with pepsin solution and dehydrated by washing with cold 70, 90, and 100% ethanol. The preparations were then denatured with a 50% formamide solution. For each sample, 200 ng of probe, 2 μg of human Cot-1 DNA, and 9 μg of sonicated salmon sperm DNA were precipitated and resuspended in hybridization buffer (50% formamide, 2× SSC, 1% bovine serum albumin, and 10% dextran sulfate). Probes were denatured for 8 min at 80°C and subsequently incubated for 10 min at 37°C to allow preannealing of repeated sequences. Finally, the hybridization solution was placed on the samples, covered with coverslips, and incubated overnight at 37°C in a moist chamber. The samples were then washed three times in 50% formamide and three times in 2× SSC at 42°C. Visualization of the biotin-labeled probe was carried out by repeated incubations with Cy3-avidin (Amersham), biotinylated anti-avidin D (Vector Laboratories), and again with Cy3-avidin. The digoxigenin-labeled probe was detected by using mouse anti-digoxigenin antibody, digoxigenin-labeled anti-mouse antibody, and fluorescein isothiocyanate (FITC)-labeled anti-digoxigenin antibody (Boehringer Mannheim). Alternatively, FITC-avidin (Vector Laboratories) and rhodamine-labeled antidigoxigenin (Boehringer Mannheim) were used. After immunodetection, slides were counterstained with 200 ng of 4′,6-diamidino-2-phenylindole (DAPI). UV excitation was used to locate metaphases, and photographic images were taken with a charge-coupled-device (CCD) camera (Photometrics) with green (FITC) or blue-violet (Cy3 or rhodamine) illumination. Images were processed with Adobe Photoshop on an Apple Quadra computer.
PCR amplification of the ITR-aavs1 junction.
Integration of ITR-flanked DNA in the aavs1 site was determined by PCR by using nested primer pairs that flank the AAV-chromosome junction as previously described (17, 37): primers 16s (AAV), 5′-GTAGCATGGCGGGTTAATCA, and 15a (aavs1), 5′-GCGCGCATAAGCCAGTAGAGC, were used in the first round of PCR amplification with 0.5 mg of genomic DNA as the substrate. After an initial incubation for 4 min at 94°C, the reaction mixture was subjected to 30 cycles of PCR amplification for 1 min at 94°C, 1 min at 55°C, and 2 min at 72°C. One percent of the amplification product was diluted into a new reaction mixture containing a set of nested primers with the following sequences: 17s (AAV), 5′-TTAACTACAAGGAACCCCTA, and Cr2 (aavs1), 5′-ACAATGGCCAGGGCCAGGCAG. The PCR parameters were the same as for the first amplification. For molecular cloning of the amplified junction fragments, the product of the second round of amplification was purified on a 1% agarose gel and subcloned by blunt end ligation into plasmid pZERO-2.1 (Invitrogen). Sequencing was performed by standard chain termination protocols.
RESULTS
Construction of hybrid Bac-AAV vectors.
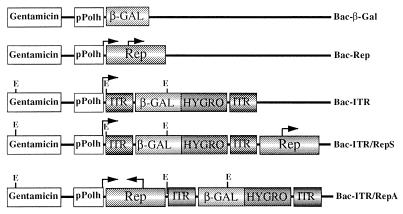
To take advantage of the ease of manipulation of baculovirus and of the unique ability of AAV virus to preferentially integrate its viral DNA into a defined region of human chromosome 19, we constructed a series of baculovirus vectors carrying AAV components known to be required for excision and integration of the AAV genome in the aavs1 site in human cells. Figure 1 shows the vectors used in this study. To determine the transduction efficiency of the recombinant baculoviruses and to establish stable cell clones from the infected cells, the β-Gal and Hygr genes were inserted between the AAV ITRs and cloned downstream of the baculovirus polyheidrin promoter (pPolh) (Bac-ITR). Additionally, the AAV Rep gene under the control of its own promoters p5 and p19 was cloned outside of the ITRs either in the sense orientation (Bac-ITR/RepS) or in the antisense orientation (Bac-ITR/RepA) with respect to the pPolh promoter. Lastly, baculoviruses carrying the CMV–β-Gal expression cassette (Bac–β-Gal) or the rep gene (Bac-Rep) were also constructed as controls for transduction efficiency and Rep expression. Viruses were produced at high titers (ranging from 1 × 109 to 9 × 109 PFU/ml) and used to infect mammalian cells.
FIG. 1.
Schematic representation of the recombinant baculoviruses used in this study. Baculovirus transfer plasmids were derived from pFastBac1 as described in Materials and Methods. The E. coli β-Gal gene, the Hygr gene (HYGRO), the rep gene (Rep), and the AAV ITRs are indicated. The expression of the β-Gal and Hygr genes is driven by the CMV and TK promoters, respectively. The p5 and p19 promoters regulate expression of the rep gene. Transcription initiation sites of the baculovirus polyhedrin promoter (pPolh) and of the p5 and p19 promoters are indicated by an arrow. Relevant EcoRV restriction sites are indicated with an E.
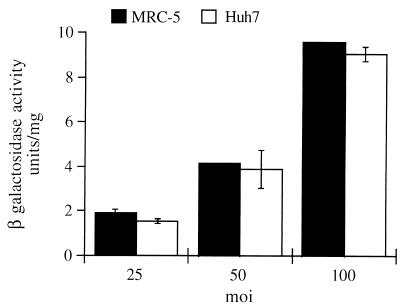
In an initial functional analysis of the transducing capacity of the recombinant baculovirus vectors, we observed that the transient β-Gal gene expression in infected strain 293 cells generally increased as a function of the MOI, a finding in agreement with published data (references 8 and 18 and data not shown). To determine whether the transducing capacity of baculovirus could be extended to other cell types, baculovirus transduction efficiency in human diploid fibroblast MRC-5 was compared to that observed in the human hepatoma cell line Huh-7. The hepatoma cell line has been shown to be extremely susceptible to baculovirus transduction (19). Figure 2 shows β-Gal gene expression detected in MRC-5 and Huh-7 cells infected with Bac–β-Gal at different MOIs (25, 50, and 100) at 48 h postinfection. These two cell lines showed comparable levels of β-Gal, thus indicating that these cells are equally susceptible to baculovirus infection; similar results were obtained upon infection of primary rat fibroblasts (data not shown). These results indicate that baculovirus transducing capacity can also be extended to primary cultures that are not necessarily of hepatic origin.
FIG. 2.
Baculovirus-mediated expression of β-Gal in MRC-5 and Huh-7 cells. Cells (105) were plated and infected with Bac–β-Gal at the MOI indicated. At 48 h postinfection total cell extracts were prepared, normalized for protein content, and assayed for β-Gal activity. Each column reflects the average of results of two independent assays, with the error bars representing the standard deviations.
Stability of recombinant Bac-AAV genomes.
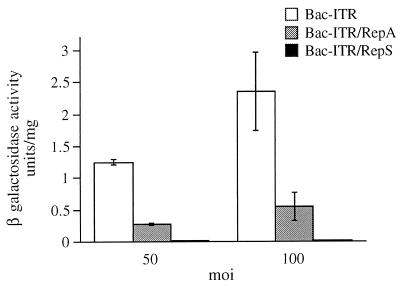
When the transduction efficiencies of Bac-ITR, Bac-ITR/RepA, and Bac-ITR/RepS were compared by infecting 293 cells at MOIs of 50 and 100, we found that the Bac-ITR virus efficiency was significantly higher than that of Bac-ITR/RepA. However, both viruses displayed a similar increase in β-Gal gene expression as a function of MOI. In contrast, very little, if any, β-Gal gene expression could be detected upon infection of 293 cells with Bac-ITR/RepS (Fig. 3).
FIG. 3.
Baculovirus-mediated expression of β-Gal activity in strain 293 cells infected with different constructs (Bac-ITR, Bac-ITR/RepS, and Bac-ITR/RepA) at MOIs of 50 and 100. Infection of 293 cells, preparation of cell extracts, and the β-Gal assay were carried out as indicated in Materials and Methods.
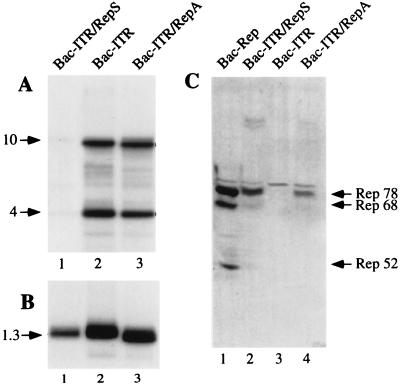
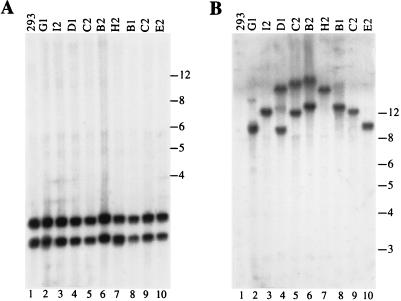
To determine whether the Bac-ITR/RepS genome had undergone partial rearrangement during virus amplification, thus explaining its low transducing efficiency, we examined the integrity of Bac-AAV genomes by Southern blot analysis. Genomic DNA was digested with EcoRV and resolved on an agarose gel. EcoRV was chosen because it cleaves once within the ITR-flanked transgene sequence and once within the gentamicin gene, releasing three fragments of 10, 4, and 1.3 kb. Figure 4 shows the viral DNA genomic blots hybridized with a probe specific for the transgene cassette (Fig. 4A) and for the gentamicin gene (Fig. 4B). The gentamicin gene is present in all virus genomes and therefore was used as an internal control. Quantification of transgene probe hybridization, normalized for that of gentamicin, indicated that the hybridization signal of Bac-ITR/RepA was 11.4-fold higher than that of Bac-ITR/RepS (Fig. 4A, compare lane 3 to lane 1), whereas no significant difference was observed between Bac-ITR/RepA and Bac-ITR (Fig. 4A, compare lane 3 to lane 2). Thus, these data suggest that the low transduction efficiency of Bac-ITR/RepS may be ascribed to the loss or rearrangement of the ITR-flanked transgene cassette during the process of baculovirus amplification.
FIG. 4.
Stability of recombinant Bac-AAV viruses. (A) Southern blot analysis of baculovirus genomes. One microgram of viral DNA prepared from the third viral passage was digested with EcoRV, fractionated on 1% agarose gel, transferred to a nylon membrane, and then hybridized with a probe specific for the transgene sequence. The expected bands of 4 and 10 kb are indicated by an arrow. (B) The same filter was stripped and hybridized with a probe specific for the gentamicin gene, which recognizes a 1.3-kb band. The intensity of the bands (indicated by arrows) in both panels A and B were quantified with a phosphorimager according to the manufacturer’s instructions. (C) Western blot detection of Rep isoforms expressed in Sf9 cells infected with different baculoviruses at an MOI of 10. At 2 days postinfection the total cell extracts were analyzed for the presence of Rep proteins. Rep78, Rep68, and Rep52 are indicated by an arrow.
The three recombinant viral genomes produced in Escherichia coli did not show any detectable rearrangement (data not shown); therefore, we hypothesized that loss of the β-Gal and Hygr expression cassette occurred during Bac-ITR/RepS amplification in insect cells. Specifically, in view of the role of the Rep polypeptides in promoting the selective excision and amplification of the ITR-flanked DNA (44, 48), we speculated that loss of the transgene cassette could be associated with the expression of Rep isoforms in Sf9 cells. To verify this hypothesis, insect cells were infected with the Bac-AAV vectors and the Rep expression pattern was assessed by Western blotting. As shown in Fig. 4C, Rep78, Rep68, and Rep52 could be clearly detected in Bac-Rep infected cells (Fig. 4C, lane 1), whereas no Rep polypeptides could be detected in Bac-ITR cell lysates (Fig. 4C, lane 2). Interestingly, the expression level of Rep polypeptides differed between Bac-ITR/RepS and Bac-ITR/RepA cell lysates (Fig. 4C, compare lanes 2 to lanes 4). rep gene expression was lower in cells infected with the Bac-AAV carrying the rep gene in the antisense orientation with respect to pPolh than in the corresponding virus carrying the rep gene in a sense orientation. Thus, in agreement with the Southern blot analysis of genomic DNA, these results suggest that the ITR-flanked cassette is destabilized in Bac-AAV virus genome when Rep proteins are expressed above a certain threshold.
Bac-AAV infection of 293 cells.
Recent studies have shown that transfection of strain 293 cells with a plasmid carrying the rep gene and a green fluorescence protein expression cassette inserted between the AAV terminal repeats results in a highly efficient integration of the ITR-flanked DNA (5, 37, 46). On the basis of this observation we wanted to determine whether the delivery of AAV components mediated by baculovirus can establish more stable cell clones. To this end, 293 cells were infected with Bac–β-Gal, Bac-ITR/RepA, and Bac-ITR vectors, and the transduction efficiencies of these viruses and the stable integration of the ITR-flanked DNA were compared by measuring β-Gal expression and the production of Hygr clones.
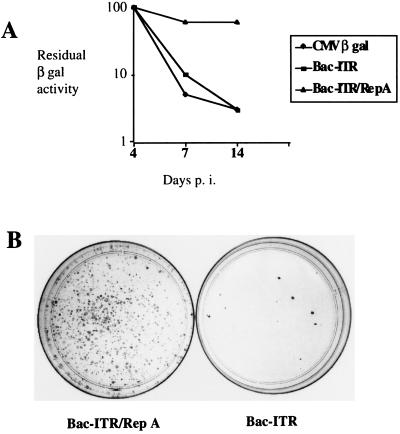
Figure 5A shows the reporter activity measured in infected cells cultured in the absence of selection as a function of time (up to 14 days postinfection). Although the transduction efficiencies determined at day 1 postinfection were similar with all three vectors (data not shown), the residual β-Gal activity at day 14 postinfection was much higher in the Bac-ITR/RepA-infected cells than in those infected with either Bac-ITR, which does not contain the rep gene, or Bac–β-Gal, which contains only the β-Gal expression cassette.
FIG. 5.
(A) Time course detection of β-Gal activity in strain 293 cells infected with different baculovirus vectors (CMV–β-Gal, Bac-ITR, and Bac-ITR/RepA) at an MOI of 100. Reporter activities were measured at days 4, 7, and 14 postinfection on a subset of the cell passages. β-Gal activities detected at days 7 and 14 are expressed as percentages of the activity detected at day 4, which is arbitrarily considered to be 100%. (B) Hygr clones derived from Bac-ITR/RepA- or Bac-ITR-infected 293 cells. From the experiment described in panel A, cell aliquots were collected at day 4 postinfection and plated in hygromycin-selective medium. At 14 days postinfection the clones were fixed and stained with 10% Giemsa and counted under a microscope. Only clones containing more than 50 cells were scored.
To assess the influence of the AAV components on the frequency of integration events, a sample of cells infected with either Bac-ITR or Bac-ITR/RepA was collected at day 4 postinfection and plated in hygromycin selection medium. As shown in Fig. 5B, infection of strain 293 cells with Bac-ITR/RepA resulted in a significantly higher number of stable cell clones than did infection of cells with Bac-ITR. The increase in the number of Hygr clones ranged in several experiments from 10- to 50-fold (data not shown).
Five hundred Hygr clones were obtained by plating 20,000 cells infected with Bac-ITR/RepA. Based on the cloning efficiency of the 293 cells (50%) and on the percentage of the cells transduced by Bac-ITR/RepA (50%), we estimate that approximately 10% of the transduced cells carrying the Hygr gene integrated into the host genome. In addition, the majority of the Hygr clones (>90%) expressed β-Gal genes (as determined by histochemical staining of the infected cells), suggesting that the entire ITR-flanked transgene cassette had been inserted into the host chromosome. These results indicate that under both selective and nonselective conditions, baculovirus delivery of the rep gene allows a very efficient transduction of ITR-flanked cassette, probably by mediating its insertion into the human genome.
Site-specific integration of ITR-flanked DNA.
It has been shown that the rep gene mediates site-specific integration of ITR-flanked DNA fragment into the aavs1 site (5, 37, 46). To verify the efficiency of site-specific integration mediated by the Bac-AAV vectors, Hygr clones from strain 293 cells infected with Bac-ITR/RepA and Bac-ITR were isolated and expanded and total genomic DNA was then subjected to Southern blot analysis. The rearrangement of the aavs1 site resulting from the integration of the ITR-flanked transgene cassette was assessed by using aavs1- and Hyg–β-Gal-specific probes. Site-specific integration of the transduced transgene cassette was scored when the aavs1-specific probe recognized additional bands that were absent in mock-infected cells and when the same additional bands were also detected by the transgene probe.
The analysis of some of the strain 293 clones derived from infection with Bac-ITR is shown in Fig. 6. The genomic DNA extracted from nine independent clones was digested with ApaI and analyzed by Southern blotting. ApaI was chosen because it cleaves once in the aavs1 sequence but does not cleave within the transgene. Thus, the insertion of the transduced DNA into the aavs1 site should be easily detectable by the altered pattern of one of the aavs1-derived restriction fragments. The aavs1-specific probe recognizes two bands of approximately 2.5 and 2.8 kb (Fig. 6A, lanes 2 to 10). Additionally, the hybridization pattern of the infected clone DNA is identical to that of the mock-infected cell DNA (Fig. 6A, lane 1). Single or double bands ranging in size from 8 to 20 kb were detected in selected DNA clones when the same genomic blot was hybridized with a probe specific for the transgene cassette. Thus, in all of the clones analyzed no obvious rearrangement of the aavs1 site could be ascribed to the infection of Bac-ITR, indicating that the integration of the transgene occurred at sites other than aavs1.
FIG. 6.
Southern blot analysis of strain 293 Hygr clones derived from Bac-ITR infection. Ten micrograms of genomic DNA from each clone was digested with ApaI, fractionated on 1% agarose gel, and transferred to a nylon membrane. (A) Hybridization to an aavs1 probe. The two detected bands of roughly 2.5 and 2.8 kb correspond to the aavs1 preintegration site. (B) Same membrane after rehybridization to a transgene-specific probe. The positions of molecular size standards (in kilobases) are indicated.
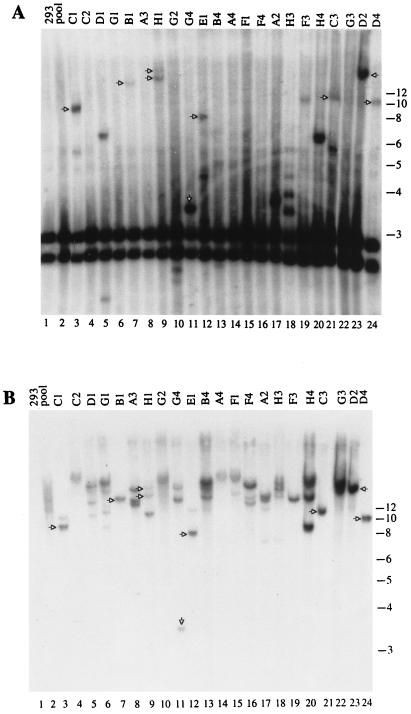
Figure 7 shows the hybridization pattern obtained with the clones derived from the Bac-ITR/RepA-infected cells. In 13 of 22 clones (59%) (Fig. 7A, clones C1, D1, B1, H1, G4, E1, A2, H3, F3, H4, C3, D2, and D4), rearrangement of the aavs1 region was apparent. The same genomic blot was then hybridized with a transgene probe to ascertain the presence of the transgene sequences within the rearranged aavs1 bands. In 8 of 13 clones (61%) (Fig. 7B, clones C1, B1, H1, G4, E1, C3, D2, and D4), bands were detected which hybridized to the transgene probe matching those obtained with aavs1 probe (shown with an arrow), indicating that the transgene was indeed inserted in the aavs1 site. Thus, site-specific integration occurred in 8 of 22 clones (36%) analyzed. Additional transgene bands detected only with the transgene probe were present in half the clones analyzed. These bands might be due to rearrangements in the aavs1 region flanking the transgene sequences (40) or to multiple insertions at sites other than aavs1.
FIG. 7.
Southern blot analysis of strain 293 Hygr clones derived from Bac-ITR/RepA infection. Conditions of infection and DNA analysis were as described in Fig. 6. (A) Hybridization to aavs1 probe. (B) Same membrane hybridized to transgene probe. The upshifted bands that are annealed to both probes are considered to be indicative of site-specific integration and are indicated with arrows. The positions of the molecular size standards (in kilobases) are indicated.
To confirm the site-specific integration of the transduced ITR-DNA cassette and to differentiate between single and multiple integration events into different sites, FISH analysis of metaphase spreads was performed on infected strain 293 cells by using the aavs1- and transgene-specific probes. Metaphases were scored as positive only if both probes were colocalized on both sister chromatids of a given chromosome.
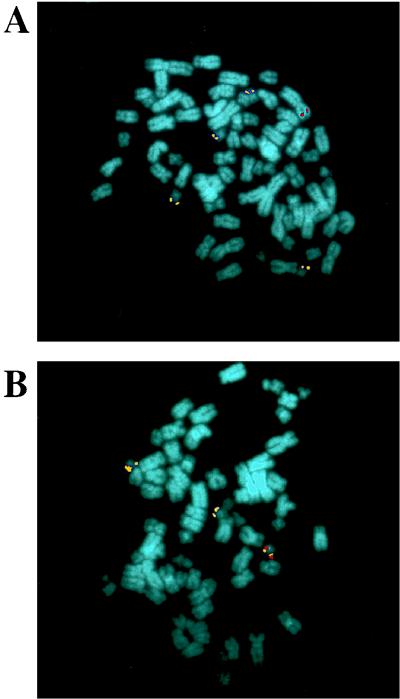
At 10 days after infection with Bac-ITR/RepA, 400 Hygr clones were pooled and analyzed by FISH. The transgene probe colocalized with the aavs1 probe in 10 of the 24 (41%) metaphases analyzed (Fig. 8B), whereas in the remaining metaphases the transgene probe was located on different chromosomes that were not detected by the aavs1 probe (Fig. 8A). The aavs1 probe annealed to three or four chromosomes 19 in most of metaphases analyzed, in agreement with the polyploid nature of this cell line. No hybridization of the transgene probe was detected on mock-infected cells (data not shown). Although FISH analysis is not strictly quantitative, the site-specific integration frequency of 41% established by FISH analysis of pooled cell clones is in good agreement with the integration frequency of 36% derived from Southern blot analysis of single clones. The colocalization of the transgene and aavs1 probes was also observed upon FISH analysis of metaphases derived from the individual clones that had been previously characterized by Southern blotting (data not shown).
FIG. 8.
In situ hybridization of metaphase chromosomes from a pool of 400 strain 293 Hygr clones derived from Bac-ITR/RepA infection. Chromosome preparations were hybridized with a Hyg–β-Gal-specific (in red) or aavs1-specific (in yellow) probe as described in Materials and Methods. (A) The transgene is integrated into a chromosome other than chromosome 19. (B) Colocalization of the transgene and of the aavs1 probe.
Analysis of ITR-aavs1 junctions.
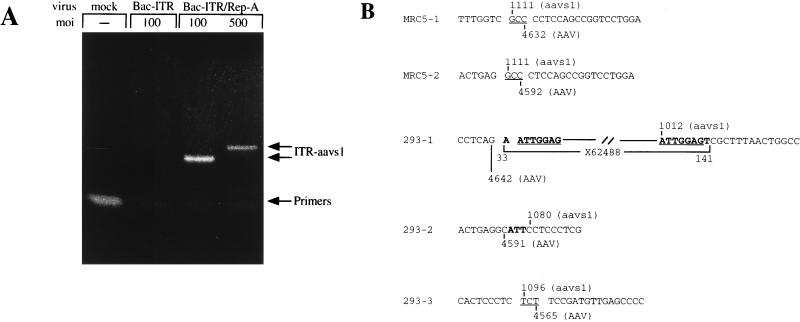
To precisely identify the integration site of the transgene cassette, 293 and MRC-5 cells were infected with Bac-ITR/RepA and the genomic DNA was extracted and subjected to amplification of the ITR-aavs1 junction. For this purpose, two sets of nested primers specific for the AAV terminal repeat and chromosome 19 were used (17, 37). As a control, genomic DNA from cells infected with Bac-ITR was also analyzed. As shown in Fig. 9A, specific DNA bands were amplified from MRC-5 cells infected with Bac-ITR/RepA at MOIs of 100 and 500. The size difference of the DNA bands probably reflects the different junction species amplified from the population of transduced cells. In contrast, no specific product was detected with mock- or Bac-ITR-infected cells. A similar protocol was utilized to amplify the ITR-aavs1 junction from Bac-ITR/RepA-infected 293 cells. The amplified DNA bands were cloned and sequenced, and the sequence data is shown in Fig. 9B. In MRC-5 cells, the insertion of the ITR-flanked transgene cassette was mapped at nucleotide 1111 of aavs1, where a short homology between the AAV ITR and aavs1 can be identified in the GCC triplet. A deletion of 43 and 83 bases within the ITR sequence was also identified. Similarly, the ITR-aavs1 junctions amplified from pooled Hygr 293 clones indicated that the insertion of the transgene cassette had occurred at nucleotides 1012, 1080, and 1096 of aavs1. Interestingly, in clone 293-2 an insertion of three nucleotides was detected at the junction between the ITR and aavs1. A larger insert of 101 nucleotides was identified in clone 293-1. This insert is partially homologous to sequence X62488 deposited in the GenBank database by Samulsky and coworkers and is derived from amplification of the ITR-aavs1 junction from AAV-infected cells (40). Taken together, these results indicate that, analogous to what has been observed following AAV infection and in transfection of AAV-derived plasmids (5, 37, 42, 46), baculovirus-mediated transduction of human cells results in the integration of the ITR-flanked DNA cassette in a specific region of the aavs1 site.
FIG. 9.
(A) PCR amplification of the ITR-aavs1 junction in baculovirus-infected strain 293 cells. Cells were infected at the indicated MOI and were collected 4 days postinfection; total DNA was prepared and subjected to nested PCR amplification as described in Materials and Methods. Bands corresponding to primers and amplified junctions are indicated. (B) ITR-aavs1 junction sequences from Bac-ITR/RepA-infected 293 and MRC-5 cells. Common bases at the ITR-aavs1 junction are underlined. Sequences not belonging to ITR or aavs1 are in boldface. X62488 was identified in GenBank with a Blast search. Sequences are depicted in ITR, aavs1 order.
DISCUSSION
In this study, we described the construction of a novel hybrid virus which combines the remarkable transducing efficiency of baculovirus with the unique property of site-specific integration characteristic of AAV. The Bac-AAV vector constructed displays the following properties: genome stability upon virus amplification, highly efficient transduction of primary cells, and targeted integration of the ITR-flanked DNA. Thus, this hybrid virus is ideal for the delivery of large DNA segments to both culture and primary cell lines where the gene of interest flanked by the AAV terminal repeats can be maintained and expressed for an extended period of time.
The strategy implemented in the construction of the hybrid Bac-AAV vector is based on the observation that baculovirus readily infects hepatocytes without any apparent toxic effect on the transduced cells (8, 19). Interestingly, baculovirus can infect not only hepatic cells, but also other cell types such as the human fibroblast cell strain MRC-5 (Fig. 2), human embryonic kidney cells strain 293 (Fig. 3), HeLa cells (54), and primary rat fibroblasts (data not shown). The efficiency of baculovirus transduction of these cell types is quite high and reaches levels comparable to those observed with hepatoma cell line Huh-7 (data not shown). Thus, although the mechanism of viral uptake is not clear, these results indicate that the ability of baculovirus to express a reporter gene is quite general and can be observed in a variety of cell types.
Rep polypeptides are expressed in Sf9 cells (Fig. 4), indicating that the p5 and p19 promoters that govern transcription of the rep gene are also functional in insect cells. Rep proteins inhibit the replication of a number of viruses, including SV40 (6), human immunodeficiency type 1 (2), bovine papillomavirus type 1 (18), and Ad (51). Apparently, rep expression does not interfere with baculovirus replication since hybrid Bac-AAV viruses carrying the rep gene grow to titers comparable to those of other recombinant baculoviruses (data not shown). Interestingly, hybrid vectors carrying both the rep gene and the ITR-flanked transgene cassette can be amplified only where the rep gene is positioned in an antisense orientation with respect to the pPolh promoter (Fig. 1 and 4). The reduction in Rep protein expression in cells infected with Bac-ITR/RepA may be explained, at least in part, by the transcription of antisense Rep RNA mediated by the pPolh promoter, which is known to be very active in these cells (36). The mechanism by which the expression of Rep polypeptides may compromize the stability of the Bac-AAV genome is not clear, but it can be assumed that the excision of the ITR-flanked transgene cassette is mediated by the expression of Rep polypeptides upon baculovirus amplification in Sf9 cells. Although AAV replication requires Ad helper functions (7), it has been demonstrated by transfection studies of AAV plasmids that overexpressed Rep proteins can excise the ITR-flanked DNA, converting it into monomer-sized replicative intermediate molecules even in the absence of Ad functions (32). Thus, although we have not assessed the functionality of Rep polypeptides in insect cells, a similar event may have occurred in Sf9 cells infected with the Bac-ITR/RepS, leading to disruption of the Bac-AAV genome structure. The stability of the Bac-ITR/RepA may in turn be explained by the reduced amount of Rep polypeptides which, in the context of the Sf9 cells, may not have been sufficient to mediate the excision of the ITR-flanked cassette.
The correlation between the level of rep gene expression and the loss of the ITR-flanked DNA has important implications for the construction of mammalian hybrid vectors carrying both the rep gene and the ITR-flanked DNA. If these recombinant vectors are amplified in mammalian cells, the efficiency of rep-mediated excision of the ITR cassette may jeopardize the integrity of the genome structure of the recombinant virus. Thus, the construction of chimeric vectors carrying these genetic elements may require a tight control of rep gene expression. Interestingly, the construction of a hybrid herpesvirus-AAV vector was recently reported (23). This vector prolongs expression of the β-Gal reporter gene, albeit only for a few days. The overall genomic structure of the hybrid virus was not described, and thus it is entirely possible that, analogous to our observations with Bac-ITR/RepS, the expression of the Rep polypeptides may have caused partial loss of the ITR-flanked cassette.
The prolonged β-Gal expression in the presence of the rep gene (Fig. 5) reflects an increase in integration levels stimulated by the Rep polypeptides, demonstrating the validity of the Bac-AAV vector in ensuring prolonged persistence of the transgene cassette. This conclusion is also supported by the observation that the number of Hygr strain 293 clones obtained with Bac-ITR/RepA infection is 10- to 50-fold higher than with Bac-ITR alone (Fig. 5). A similar observation based on transfection of 293 cells with plasmids carrying the rep gene has been recently reported (5, 46). We do not know if the higher transduction efficiency based on the expression of rep is restricted to strain 293 cells and may be attributed to the E1A-mediated induction of the p5 promoter or if the enhanced integration is observable in other cell types. To this end, we are currently examining the integration efficiency of Bac-AAV in other cell types.
The fate of the ITR-flanked transgene in selected clones was assessed by Southern blot analysis, FISH, and PCR amplification of the ITR-aavs1 junction. In agreement with several reports (5, 37, 37a, 46), targeting of the ITR-flanked DNA to aavs1 is dependent on the expression of the rep gene since no integration of the ITR-transgene in chromosome 19 was detected upon infection of the 293 cells with Bac-ITR (Fig. 6). In contrast, Bac-ITR/RepA infection of 293 cells results in a 36 to 41% frequency of site-specific integration (Fig. 7 and 8). The frequency of integration observed upon baculovirus infection of 293 cells is comparable to that detected in these cells upon infection with recombinant AAV virus or with the transfection of AAV-derived plasmids (5, 37, 42, 46). However, in a study of bronchial epithelial cells, in 94% of the cells containing integrated wild-type AAV, the virus genome was mapped on chromosome 19 by FISH (25). Additionally, approximately 70% of latently infected cell lines that were analyzed in another survey were found to contain AAV inserted into aavs1 (42). Thus, the integration efficiency detected in other cell types with wild-type AAV appears to be significantly greater than that observed in 293 cells upon either plasmid transfection or Bac-AAV infection. The biological significance of this difference in integration efficiency is not clear; however, it is tempting to speculate that it may reflect differences in the concentration of specific host factors that may contribute to targeting to the aavs1 site of the AAV-derived DNA.
Southern blot analysis of the selected strain 293 clones revealed the presence of several bands corresponding to rearranged aavs1 sequences that can be accounted for by the insertion of the transgene cassette (Fig. 7). Also, several bands hybridized only by the aavs1 probe indicate that rearrangement of the aavs1 as a consequence of rep expression may have taken place, much as has been observed upon transfection of AAV-derived plasmids and subsequent selection of neomycin-resistant 293 clones (5). Interestingly, in half of the clones analyzed the probe specific for the β-Gal–Hygr genes detected an additional band which varied in size. An additional band was also detected in clones infected with Bac-ITR. Although this band appears less intense, it probably represents a second integration event. In line with this hypothesis, FISH analysis revealed the presence of second integration events in a minority of the metaphases analyzed (Fig. 8).
The 6.5-kb transgene cassette inserted between the ITRs contains the Hygr gene and the β-Gal gene that were coselected in the presence of hygromycin in almost all of the clones analyzed (data not shown). Although this cassette is 2 kb longer than the wild-type virus genome, site-specific integration of the ITR-flanked DNA was observed when the rep gene was carried on the same virus (Fig. 7 and 8). We have not examined the fine structure of the ITR-flanked DNA inserted into the aavs1 site. However, the size of the bands hybridized by both the transgene and aavs1 probes is consistent in most clones with insertion of the full-length ITR-flanked transgene cassette. This possibility is also supported by the observation that both reporter genes are expressed in all clones and, most importantly, in those clones where only one upshifted band is detected. It is, however, possible that fine rearrangements at the integration sites have taken place, disrupting the structure of the ITR-flanked DNA without interfering with the expression of both genes.
The observation that rep-mediated site-specific integration in 293 cells is accompanied by rearrangements of the integration site and, possibly, of the inserted transgene cassette is not unprecedented (5). Thus, it is not surprising that rearrangement of the aavs1 region is detected upon infection of 293 cells with Bac-ITR/RepA and subsequent selection of Hygr clones. Although the biological reasons for such rearrangements are not clear, it is important to point out that such genome rearrangements have been mostly documented with established cell lines. It would be of interest to determine whether the same rearrangements are detected in primary cultures. Nonetheless, this result indicates that integration is not limited by the size of the DNA flanked by the AAV terminal repeats and can occur with DNA segments larger than the wild-type AAV genome. Important large genes (such as the dystrophin gene) can, at least in theory, be transduced and integrated by using this system.
rep-mediated integration also occurs in the human fibroblast cell strain MRC-5 after Bac-ITR/RepA infection. This conclusion is based on the PCR amplification of the ITR-aavs1 junction that localizes the insertion of the ITR-flanked DNA cassette in the same region of aavs1 as described previously for AAV (40) (Fig. 9). Thus, although no further work has been carried out to characterize the structure of the inserted DNA or to determine the efficiency of site-specific integration, this result extends the use of the hybrid Bac-AAV virus to human diploid cells. The ITR-aavs1 junctions of the infected MRC-5 cells show similarities to those generated by wild-type AAV. In fact, as previously reported (40), several nucleotides are found in common between the ITR and aavs1 at the viral-cellular junction (Fig. 9). In contrast, the insertion of a short stretch of nucleotides in between the ITR-cellular junction was detected in two junctions derived from strain 293 infected cells. The mechanism by which these insertions have been generated is not clear, but it is unlikely that they are artifacts of PCR amplification since the longer insertion shows 90-nucleotide sequence identity to a similar sequence reported by Samulsky and coworkers (40). We have observed similar insertions in Huh-7 cells infected with wild-type AAV (37). Although the biological significance of these insertions is not clear, it may be related to differences between the diploid cell strain MRC-5 and the polyploid cell lines Huh-7 and 293.
The results presented in this work indicate that transgene excision from a double-stranded DNA baculovirus backbone vector and subsequent integration into the human genome occur with significant frequency. Thus, this study can be considered an important step towards improving the use of baculovirus as a vector for gene transfer since it demonstrates the feasibility of using hybrid Bac-AAV virus for in vitro gene transfer in human cells. Baculovirus infection in vivo, like retrovirus infection, is hampered by serum components, namely, by complement factors (41). Therefore, the use of this system may be limited to ex vivo gene delivery. However, the data presented here indicate that baculovirus infection and permanent transduction of the target cells is not limited to hepatic cells. The demonstration that hybrid viral vectors carrying specific genetic elements of the AAV virus can direct integration of a selected DNA sequence is an important starting point for the construction of chimeric viral vectors suitable for in vivo gene delivery.
ACKNOWLEDGMENTS
We thank A. Sgura and D. Cimini for in situ hybridization analysis of infected cells and E. Fattori, A. Nicosia, and C. Toniatti for critical review and helpful suggestions. We also thank J. Clench for editorial assistance, M. Emili for graphics, and P. Neuner for oligonucleotide synthesis.
REFERENCES
- 1.Anderson W F. Human gene therapy. Science. 1992;256:808–813. doi: 10.1126/science.1589762. [DOI] [PubMed] [Google Scholar]
- 2.Antoni B A, Rabson A B, Miller I L, Tremple J P, Chejanovsky N, Carter B J. Adeno-associated virus Rep protein inhibits human immunodeficiency virus production in human cells. J Virol. 1991;59:284–291. doi: 10.1128/jvi.65.1.396-404.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Armentano D, Sookdeo C C, Hehir K M, Gregory R J, George J A S T, Prince G A, Wadsworth S C, Smith A E. Characterization of an adenovirus gene transfer vector containing an E4 deletion. Hum Gene Ther. 1995;6:1343–1353. doi: 10.1089/hum.1995.6.10-1343. [DOI] [PubMed] [Google Scholar]
- 4.Ashktorab H, Srivastava A. Identification of nuclear proteins that specifically interact with adeno-associated virus type 2 inverted terminal repeat hairpin DNA. J Virol. 1989;63:3034–3039. doi: 10.1128/jvi.63.7.3034-3039.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balagué C, Kalla M, Zhang W W. Adeno-associated virus Rep78 protein and terminal repeats enhance integration of DNA sequences into the cellular genome. J Virol. 1997;71:3299–3306. doi: 10.1128/jvi.71.4.3299-3306.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bantel-Schaal U, zur Hausen H. Adeno-associated viruses inhibit SV40 DNA amplification and herpes simplex virus replication in SV40-transformed hamster cells. Virology. 1988;164:64–74. doi: 10.1016/0042-6822(88)90620-4. [DOI] [PubMed] [Google Scholar]
- 7.Berns K I. Parvoviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Vol. 2. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2173–2197. [Google Scholar]
- 8.Boyce F M, Bucher N L R. Baculovirus mediated gene transfer into mammalian cells. Proc Natl Acad Sci USA. 1996;93:2348–2352. doi: 10.1073/pnas.93.6.2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bramson J L, Graham F L, Gauldie J. The use of adenoviral vectors for gene therapy and gene transfer in vivo. Curr Biol. 1995;6:590–595. doi: 10.1016/0958-1669(95)80097-2. [DOI] [PubMed] [Google Scholar]
- 10.Carter B J. The promise of adeno-associated virus vectors. Nat Biotechnol. 1996;14:1725–1727. [Google Scholar]
- 11.Clark K R, Sferra T J, Johnson P R. Recombinant adeno-associated viral vectors mediate long-term transgene expression in muscle. Hum Gene Ther. 1997;8:659–669. doi: 10.1089/hum.1997.8.6-659. [DOI] [PubMed] [Google Scholar]
- 12.Cory J S, Hirst M L, Hails R S, Goulson D, Green B M, Carty T M, Possee R D, Cayley P J, Bishop D H L. Field trial of a genetically improved baculovirus insecticide. Nature. 1994;370:138–140. [Google Scholar]
- 13.Dai Y, Schwarz E M, Gu D, Zhang W, Sarvetnick N, Verma I M. Cellular and humoral immune responses to adenoviral vectors containing factor IX gene: tolerization of factor IX and vector antigens allows for long-term expression. Proc Natl Acad Sci USA. 1995;92:1401–1405. doi: 10.1073/pnas.92.5.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fisher K J, Jooss K, Alston J, Yang Y, Haecker S E, High K, Pathak R, Raper S E, Wilson J M. Recombinant adeno-associated virus for muscle directed gene therapy. Nat Med. 1997;3:306–312. doi: 10.1038/nm0397-306. [DOI] [PubMed] [Google Scholar]
- 15.Flotte T R, Afione S A, Zeitlin P L. Adeno-associated virus vector gene expression occurs in non-dividing cells in the absence of vector DNA integration. J Respir Cell Mol Biol. 1994;11:517–521. doi: 10.1165/ajrcmb.11.5.7946381. [DOI] [PubMed] [Google Scholar]
- 16.Giraud C, Winocour E, Berns K I. Site-specific integration by adeno-associated virus is directed by a cellular sequence. Proc Natl Acad Sci USA. 1994;91:10039–10043. doi: 10.1073/pnas.91.21.10039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goodman S, Xiao X, Donahue R E, Moulton A, Miller J, Walsh C, Young N S, Samulski R J, Nienhuis A W. Recombinant adeno-associated virus-mediated gene transfer into hematopoietic progenitor cells. Blood. 1994;84:1492–1500. [PubMed] [Google Scholar]
- 18.Hermonat P L. Inhibition of bovine papilloma virus plasmid DNA replication by adeno-associated virus. Virology. 1992;189:329–333. doi: 10.1016/0042-6822(92)90710-7. [DOI] [PubMed] [Google Scholar]
- 19.Hofmann C, Sandig V, Jennings G, Rudolph M, Schlag P, Strauss M. Efficient gene transfer into human hepatocytes by baculovirus vectors. Proc Natl Acad Sci USA. 1995;92:10099–10103. doi: 10.1073/pnas.92.22.10099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Im D S, Mucyczka N. Factors that bind to adeno-associated virus terminal repeats. J Virol. 1989;63:3095–3104. doi: 10.1128/jvi.63.7.3095-3104.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Im D S, Mucyczka N. The AAV origin binding protein Rep68 is an ATP-dependent site-specific endonuclease with DNA helicase activity. Cell. 1990;61:447–457. doi: 10.1016/0092-8674(90)90526-k. [DOI] [PubMed] [Google Scholar]
- 22.Im D S, Mucyczka N. Partial purification of adeno-associated virus Rep78, Rep68, Rep52, and Rep40 and their biochemical characterization. J Virol. 1992;66:1119–1128. doi: 10.1128/jvi.66.2.1119-1128.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnston K M, Jacoby D, Pechan P A, Fraefel C, Borghesani P, Schuback D, Dunn R J, Smith F I, Breakfield X O. HSV/AAV hybrid amplicon vectors extend transgene expression in human glioma cells. Hum Gene Ther. 1997;8:359–370. doi: 10.1089/hum.1997.8.3-359. [DOI] [PubMed] [Google Scholar]
- 24.Kapitt M G, Leone P, Samulski R J, Xiao X, Pfaff D W, O’Malley K L, During M J. Long-term gene expression and phenotypic correction using adeno-associated virus vectors in the mammalian brain. Nat Genet. 1994;8:148–154. doi: 10.1038/ng1094-148. [DOI] [PubMed] [Google Scholar]
- 25.Kearns W G, Afione S A, Fulmer S B, Pang M G, Erikson D, Egan M, Landrum M J, Flotte T R, Cutting G R. Recombinant adeno-associated virus (AAV-CFTR) vectors do not integrate in a site-specific fashion in an immortalized epithelial cell line. Gene Ther. 1996;3:748–755. [PubMed] [Google Scholar]
- 26.Kessler P D, Podsakoff G M, Chen X, McQuiston S A, Colosi P C, Matelis L A, Kurtzman G J, Byrne B J. Gene delivery to skeletal muscle results in sustained expression and systemic delivery of a therapeutic protein. Proc Natl Acad Sci USA. 1996;93:14082–14087. doi: 10.1073/pnas.93.24.14082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koebler D D, Alexander I E, Halbert C L, Russell D W, Miller A D. Persistent expression of human clotting factor IX from mouse liver after intravenous injection of adeno-associated virus vectors. Proc Natl Acad Sci USA. 1997;94:1426–1431. doi: 10.1073/pnas.94.4.1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kotin R M, Siniscalco M, Samulski R J, Zhu X, Hunter L, Laughlin C A, McLaughlin S, Muzyczka N, Rocchi M, Berns K I. Site-specific integration by adeno-associated virus. Proc Natl Acad Sci USA. 1990;87:2211–2215. doi: 10.1073/pnas.87.6.2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lawrence J B, Villnave C A, Singer R H. Sensitive, high-resolution chromatin and chromosome mapping in situ: presence and orientation of two closely integrated copies of EBV in a lymphoma line. Cell. 1988;52:51–61. doi: 10.1016/0092-8674(88)90530-2. [DOI] [PubMed] [Google Scholar]
- 30.Ledley F D. Are contemporary methods for somatic gene therapy suitable for clinical applications? Cin Invest Med. 1993;16:78–88. [PubMed] [Google Scholar]
- 31.Lewis P, Hensel M, Emerman M. Human immunodeficiency virus infection of cells arrested in the cell cycle. EMBO J. 1992;11:3053–3058. doi: 10.1002/j.1460-2075.1992.tb05376.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li J, Samulski R J, Xiao X. Role of highly regulated Rep gene expression in adeno-associated virus vector production. J Virol. 1997;71:5236–5243. doi: 10.1128/jvi.71.7.5236-5243.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McLaughlin S K, Collis P, Hermonat P L, Muzyczka N. Adeno-associated virus general transduction vectors: analysis of proviral structure. J Virol. 1988;62:1963–1973. doi: 10.1128/jvi.62.6.1963-1973.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller A D. Human gene therapy comes of age. Nature. 1992;357:455–460. doi: 10.1038/357455a0. [DOI] [PubMed] [Google Scholar]
- 35.Miller L K. Baculoviruses: high-level expression in insect cells. Curr Opin Genet Dev. 1993;3:97–101. doi: 10.1016/s0959-437x(05)80348-x. [DOI] [PubMed] [Google Scholar]
- 36.O’Reilly D R, Miller L K, Luckow V A. Baculovirus expression vectors: a laboratory manual. W. H. New York, N.Y: Freeman & Co.; 1992. [Google Scholar]
- 37.Pieroni, L., and N. La Monica. Unpublished data.
- 37a.Ponnazhagan S, Erikson D, Kearns W G, Zhou S Z, Nahreini P, Wang X-S, Srivastava A. Lack of site-specific integration of the recombinant adeno-associated virus 2 genomes in human cells. Hum Gene Ther. 1997;8:275–284. doi: 10.1089/hum.1997.8.3-275. [DOI] [PubMed] [Google Scholar]
- 38.Russel D W, Miller A D, Alexander I E. Adeno-associated virus vectors preferentially transduce cells in S phase. Proc Natl Acad Sci USA. 1994;91:8915–8919. doi: 10.1073/pnas.91.19.8915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Samulski R J, Chang L-S, Shenk T. A recombinant plasmid from which an infectious adeno-associated virus genome can be excised in vitro and its use to study viral replication. J Virol. 1987;61:3096–3101. doi: 10.1128/jvi.61.10.3096-3101.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Samulski R J, Zhu X, Xiao X, Brook J D, Housman D E, Epstein N, Hunter L A. Targeted integration of adeno-associated virus (AAV) into human chromosome 19. EMBO J. 1991;10:3941–3950. doi: 10.1002/j.1460-2075.1991.tb04964.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sandig V, Hoffman C, Steinert S, Jennings G, Schlag P, Strauss M. Gene transfer into hepatocytes and human liver tissue by baculovirus vectors. Hum Gene Ther. 1996;7:1937–1945. doi: 10.1089/hum.1996.7.16-1937. [DOI] [PubMed] [Google Scholar]
- 42.Shelling A N, Smith M. Targeted integration of transfected and infected adeno-associated virus vectors containing the neomycin resistance gene. Gene Ther. 1994;1:165–169. [PubMed] [Google Scholar]
- 43.Smith A E. Viral vectors in gene therapy. Annu Rev Microbiol. 1995;49:807–838. doi: 10.1146/annurev.mi.49.100195.004111. [DOI] [PubMed] [Google Scholar]
- 44.Snyder R O, Samulski R J, Muzyczka N. In vitro resolution of covalently joined AAV chromosome ends. Cell. 1990;60:105–113. doi: 10.1016/0092-8674(90)90720-y. [DOI] [PubMed] [Google Scholar]
- 45.Srivastava A, Lubsy E W, Berns K I. Nucleotide sequence and organization of the adeno-associated virus 2 genome. J Virol. 1983;45:555–564. doi: 10.1128/jvi.45.2.555-564.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Surosky R T, Urabe M, Godwin S G, McQuiston S A, Kurtzman G J, Ozawa K, Natsoulis G. Adeno-associated virus Rep proteins target DNA sequences to a unique locus in human genome. J Virol. 1997;71:7951–7959. doi: 10.1128/jvi.71.10.7951-7959.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Toniatti, C. Unpublished data.
- 48.Urcelay E, Ward P, Wiener S M, Safer B, Kotin R. Asymmetric replication in vitro from a human sequence element is dependent on adeno-associated virus Rep proteins. J Virol. 1995;69:2038–2046. doi: 10.1128/jvi.69.4.2038-2046.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Verma I M, Somia N. Gene therapy—promises, problems and prospects. Nature. 1997;389:239–242. doi: 10.1038/38410. [DOI] [PubMed] [Google Scholar]
- 50.Wang Q, Jia X-C, Finer M H. A packaging cell line for propagation of recombinant adenovirus vectors containing two lethal gene-region deletions. Gene Ther. 1995;2:775–783. [PubMed] [Google Scholar]
- 51.Weitzman M D, Fisher K J, Wilson J M. Recruitment of wild-type and recombinant adeno-associated virus into adenovirus replication centers. J Virol. 1996;70:1845–1854. doi: 10.1128/jvi.70.3.1845-1854.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xiao X, Li J, Samulski R J. Efficient gene transfer into muscle tissue of immunocompetent mice by adeno-associated virus vector. J Virol. 1996;70:8098–8108. doi: 10.1128/jvi.70.11.8098-8108.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang Y, Li Q, Ertl H C, Wilson J M. Cellular and humoral immune responses to viral antigens crate barriers to lug-directed gene therapy with recombinant adenoviruses. J Virol. 1995;69:2004–2015. doi: 10.1128/jvi.69.4.2004-2015.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yap C-C, Ishii K, Aoki Y, Aizaki H, Tani H, Shimuzu H, Ueno Y, Miyamura T, Matsuura Y. A hybrid baculovirus-T7 RNA polymerase system for recovery of an infectious virus from cDNA. Virology. 1997;231:192–200. doi: 10.1006/viro.1997.8537. [DOI] [PubMed] [Google Scholar]
- 55.Yeh P, Dedieu J F, Orsini C, Vigne E, Denefle P, Perricaudet M. Efficient dual transcomplementation of adenovirus E1 and E4 regions from a 293-derived cell line expressing a minimal E4 functional unit. J Virol. 1996;70:559–565. doi: 10.1128/jvi.70.1.559-565.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]