Abstract
Fusobacterium nucleatum (Fn), a bacterium present in the human oral cavity and rarely found in the lower gastrointestinal tract of healthy individuals1, is enriched in human colorectal cancer (CRC) tumours2–5. High intratumoural Fn loads are associated with recurrence, metastases and poorer patient prognosis5–8. Here, to delineate Fn genetic factors facilitating tumour colonization, we generated closed genomes for 135 Fn strains; 80 oral strains from individuals without cancer and 55 unique cancer strains cultured from tumours from 51 patients with CRC. Pangenomic analyses identified 483 CRC-enriched genetic factors. Tumour-isolated strains predominantly belong to Fn subspecies animalis (Fna). However, genomic analyses reveal that Fna, considered a single subspecies, is instead composed of two distinct clades (Fna C1 and Fna C2). Of these, only Fna C2 dominates the CRC tumour niche. Inter-Fna analyses identified 195 Fna C2-associated genetic factors consistent with increased metabolic potential and colonization of the gastrointestinal tract. In support of this, Fna C2-treated mice had an increased number of intestinal adenomas and altered metabolites. Microbiome analysis of human tumour tissue from 116 patients with CRC demonstrated Fna C2 enrichment. Comparison of 62 paired specimens showed that only Fna C2 is tumour enriched compared to normal adjacent tissue. This was further supported by metagenomic analysis of stool samples from 627 patients with CRC and 619 healthy individuals. Collectively, our results identify the Fna clade bifurcation, show that specifically Fna C2 drives the reported Fn enrichment in human CRC and reveal the genetic underpinnings of pathoadaptation of Fna C2 to the CRC niche.
Subject terms: Bacterial genomics, Colorectal cancer, Phylogeny, Pathogens
A study reveals that Fusobacterium nucleatum subspecies animalis is bifurcated into two distinct clades, and shows that only one of these dominates the colorectal cancer niche, probably through increased colonization of the human gastrointestinal tract.
Main
Fn, a member of the oral microbiota, has gained attention as an emerging cancer-associated bacterium. Worldwide, unbiased genomic analyses have revealed an enrichment of Fn in human CRC relative to non-cancerous colorectal tissues9. Previous work by our group and others demonstrated that patients with CRC tumours harbouring high levels of Fn have poorer survival5, that Fusobacterium colonizes regions of patient tumours with immune and epithelial cell functions supportive of cancer progression10, that Fusobacterium persists in metastatic disease6 and that microbiome modulation targeting Fn could change the course of this disease6,11. Moreover, exogenous Fn infection in animal and cellular models has supported a cancer-promoting role for this bacterium6,9–14. However, considerable strain-to-strain variation in Fn genotypic and phenotypic features has been described12–16. Such heterogeneity has raised challenges with reproducing Fn cancer-inducing phenotypes in animal and cellular models16,17 and it has been proposed that only a select group of Fn strains may possess carcinogenic capabilities17.
Here, leveraging a comprehensive collection of human CRC Fn strains and carrying out extensive comparative genomics, we reveal that a select clade within Fn subspecies predominates the CRC niche. In vitro and in vivo functional studies demonstrate that this clade is highly virulent in the context of CRC, and should be a primary focus in subsequent mechanistic studies on Fn pathogenicity in CRC and for the development of targeted inhibitors.
Niche-enriched Fn genes and subspecies
Classical microbiology culture approaches have re-emerged as valuable tools to functionally assess members of tissue-associated microbiomes. Here we carried out Fusobacterium targeted culture on 130 human CRC tumours from which 65 Fusobacterium CRC-associated strains were obtained from 59 unique patients. Given that Fn is predominantly an oral pathobiont, we included 81 Fusobacterium strains isolated from the oral cavity of individuals without cancer, as a control group. These oral strains were obtained from the American Type Culture Collection (ATCC) and the Korean Collection for Oral Microbiology (KCOM) repositories. Using PacBio long-read single-molecule real-time18 sequencing, we generated complete and closed genomes, with corresponding epigenetic methylomes, for 146 unique Fusobacterium strains (Fig. 1a and Supplementary Tables 1 and 2), 92% of which belonged to the species Fn (n = 135; n = 55 CRC associated and n = 80 oral associated; Fig. 1b). As Fn is the most frequently detected species in CRC tumours2,3, we therefore focused our analysis on a comparison of these 55 CRC-associated and 80 oral-associated Fn genomes.
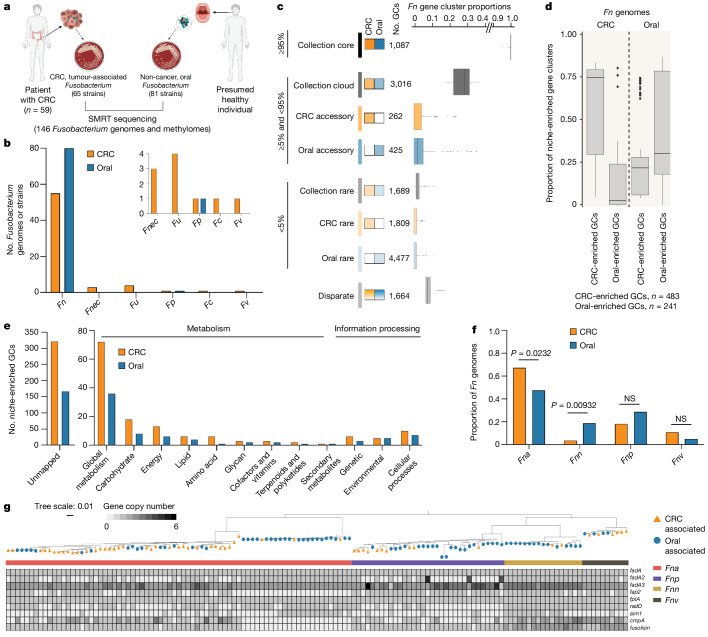
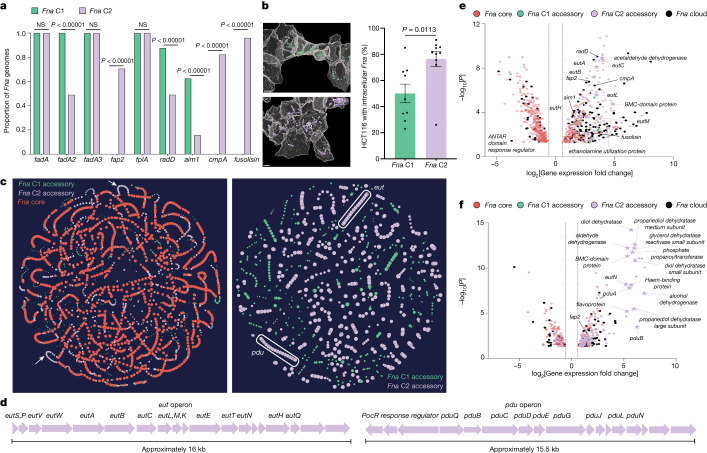
Fig. 1. Fn niche features.
a, A schematic of Fusobacterium strain collection (n = 146) and the sequencing strategy for unique strains. SMRT, single-molecule real-time sequencing. b, A column graph depicting the proportion of Fusobacterium genomes, subset by species, within the CRC (orange) and oral (blue) niches. The inset shows all non-Fn species of Fusobacterium (Fnec, F. necrophorum; Fu, F. ulcerans; Fp, F. pseudoperiodonticum; Fc, F. canifelinum; Fv, F. varium). c, The composition of the Fn pangenome subset by niche. Anvi’o21 gene cluster (GC) prevalence was used to define core (≥95%), accessory (≥5% and <95%) and rare (<5%) features conserved in both CRC-associated and oral-associated strains (collection core, ≥95% in all strains within the collection; collection cloud, ≥5% and <95% in all strains within the collection; collection rare, <5% in all strains within the collection). Disparate features are those that do not fall into any of the other noted bins. d, The proportion of niche-enriched gene clusters across CRC-associated and oral-associated Fn genomes. The plot box shows the 25th percentile, median and 75th percentile. The plot whiskers indicate the minima and maxima. e, KofamKOALA KEGG orthologue analysis27 of niche-enriched gene clusters. f, A column graph depicting the proportion of Fn genomes, grouped by subspecies, within the CRC and oral niche. Statistical analysis was carried out using a two-sample z-test, two-tailed. NS, not significant. g, Gene presence–absence heat map of canonical Fn virulence factors (fadA (refs. 38,39,42), fap2 (ref. 34), fplA (ref. 33), radD (refs. 36,62), aim1 (ref. 35), cmpA (ref. 37) and fusolisin32) across Fn subspecies, in which each column represents an individual genome (Fna n = 75, Fnn n = 17, Fnp n = 33, Fnv n = 10). The heat map is organized using an rpoB gene-based phylogenetic tree. For each genome, the tree end points indicate the niche origin (CRC (orange); oral (blue)) and the bar colour indicates the Fn subspecies (Fna (red); Fnn (gold); Fnp (purple); Fnv (brown)). The graphics in a were created using BioRender.com.
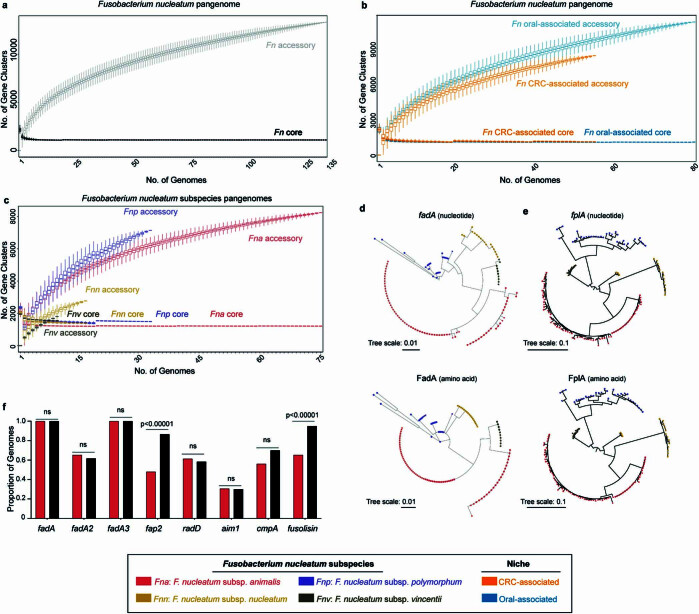
Given that Fn strains in human CRC tumours are predicted to originate from the human oral cavity19,20, but are rare members of the lower gastrointestinal (GI) tract microbiota of individuals without cancer1, we reasoned that CRC-associated Fn strains harbour an additional genetic repertoire to facilitate their colonization in human CRC tumours. To test this, we examined our 135 Fn genomes using the analysis and visualization platform for ‘omics data (Anvi’o) workflow for microbial pangenomics21. Pangenomic analysis identifies all genes present in a species (‘pangenome’) and discerns between gene content conserved among most members (‘core genome’) and gene content shared among subsets of members (‘accessory genomes’)22,23. We observed that accessory genome size increases as the number of sampled Fn genomes increases, supportive of previous proposals that Fn has an open pangenome24 (Extended Data Fig. 1a). To account for uneven sampling25 between CRC (n = 55) and oral (n = 80) genomes, we subset our analysis on the basis of niche and found that CRC-associated Fn strains have a smaller accessory genome compared to that of oral-associated strains (Fig. 1c, Extended Data Fig. 1b and Supplementary Table 1). Functional enrichment analysis26 identified 483 and 241 gene clusters significantly enriched (q < 0.05) in CRC and oral strains, respectively (Fig. 1d and Supplementary Table 3). Kyoto Encyclopedia of Genes and Genomes (KEGG) orthologue analysis27 of the 724 niche-enriched gene clusters revealed that mapped gene clusters (31.2%) were predominantly involved in putative metabolic functions and pathways (Fig. 1e and Supplementary Table 3).
Extended Data Fig. 1. Fn genetic characterization by niche and subspecies.
a, Size of the Fn pangenome split by the core genome (≥95%) (black) and accessory (<95%) genome (grey). n = 10,000 random subsamplings of 135 Fn genomes. Data is plotted as median ± s.d. b, Size of the Fn pangenome split by CRC-associated (orange) and oral-associated (blue) niche origin, with respective core and accessory genomes labeled. n = 10,000 random subsamplings of 55 Fn CRC-associated and 80 Fn oral-associated genomes. Data is plotted as median ± s.d. c, Size of the Fn pangenome split by Fn subspecies, Fna (red), Fnn (gold), Fnp (purple), Fnv (brown), with respective core and accessory genomes labeled. n = 10,000 random subsamplings of 75 Fna, 17 Fnn, 33 Fnp, and 10 Fnv genomes. Data is plotted as median ± s.d. d-e, Maximum-likelihood dendrograms of d, fadA and e, fplA nucleotide and amino acid sequences. For each genome, tree end points indicate Fn subspecies; Fna (red), Fnn (gold), Fnp (purple), Fnv (brown). f, Column graph depicts the proportion of Fn genomes containing canonical Fn virulence factors, subset by Fna (red), non-Fna (black) subspecies. Statistical analysis performed using two sample Z test, two-tailed. NS, not significant.
Previously published studies using orthogonal approaches have observed a differential distribution of Fn subspecies in tumour tissue from patients with CRC and mucosal biopsy specimens from patients with inflammatory bowel disease28,29 (IBD). As there is sufficient genetic heterogeneity between the four Fn subspecies that reclassification into separate species has been proposed30, we increased the resolution of our analyses to the subspecies level (Fna, Fn subspecies nucleatum (Fnn), Fn subspecies polymorphum (Fnp) and Fn subspecies vincentii (Fnv); Methods). It is notable that studies seeking to delineate the contributions of Fn in CRC predominately use the model strains Fnn ATCC 25586 (ref. 7), an oral isolate, Fnn ATCC 23726 (refs. 20,31), a urogenital isolate, and Fna 7_1 (ref. 29), an isolate from a patient with IBD. Here, analysis of the proportion of Fn subspecies by niche found that of the four Fn subspecies, only Fna is significantly associated with the CRC niche (two-sample z-test, two-tailed, P = 0.0232; Fig. 1f), validating previous studies28, whereas Fnn is significantly enriched in the oral niche (two-sample z-test, two-tailed, P = 0.00932; Fig. 1f). Thus, we reasoned that the repertoire of genetic factors associated with colonization or virulence in the CRC niche would not be fully represented in Fnn model strains and indeed show that Fnn ATCC 25586 contains only 17.60% of CRC-enriched gene clusters (Supplementary Table 3). This supports the use of Fna strains such as Fna 7_1, which has been shown to induce colonic tumours in mouse models3, for mechanistic studies. Comparison of Fn subspecies pangenomes showed that Fna has the smallest core genome compared to those of other subspecies, suggestive of further unresolved Fna genetic heterogeneity (Extended Data Fig. 1c).
As Fna is enriched in the CRC tumour niche, we tested whether Fn virulence factors previously described as important for host colonization are more prevalent in Fna than in other Fn subspecies. Fn type Va autotransporter virulence factors include fusolisin32, a serine protease that damages host tissue and inactivates immune effectors, FplA (ref. 33), a phospholipase autotransporter that binds to host phosphoinosite-signalling lipids, and the Fap2 (ref. 34), Aim1 (ref. 35), RadD (ref. 36) and CmpA (ref. 37) adhesins that mediate Fn interactions with either host cells or other bacterial species. As the role of fplA, aim1, radD and cmpA in CRC remains unclear, to ensure a comprehensive analysis, we queried the presence of these virulence factors across our Fn genome collection. An additional adhesin, FadA (refs. 38,39), mediates Fn attachment to and invasion of host epithelial cells40,41, with two additional FadA homologues recently identified42. Previously analysed fadA distribution in a limited number of Fusobacterium genomes has suggested fadA absence from passively invading species and increased incidence in highly invasive species13, although this distribution does not always coincide with in vivo invasion assays13,38,42,43. We found that although fplA and fadA are well conserved in Fn, their nucleotide and amino acid sequences segregate by Fn subspecies, perhaps indicative of variable interactions with host ligands (Extended Data Fig. 1d,e). Our results show that none of these canonical virulence factors is significantly associated with Fna compared to other Fn subspecies, suggesting that additional unknown genetic factors facilitate the enrichment of Fna in CRC (Extended Data Fig. 1f). However, we observed that a subset of Fna strains lacked fap2, cmpA and fusolisin and, by rpoB gene analysis, these Fna strains seemed to form a distinct Fna clade (Fig. 1g).
An Fna clade enriched in the CRC niche
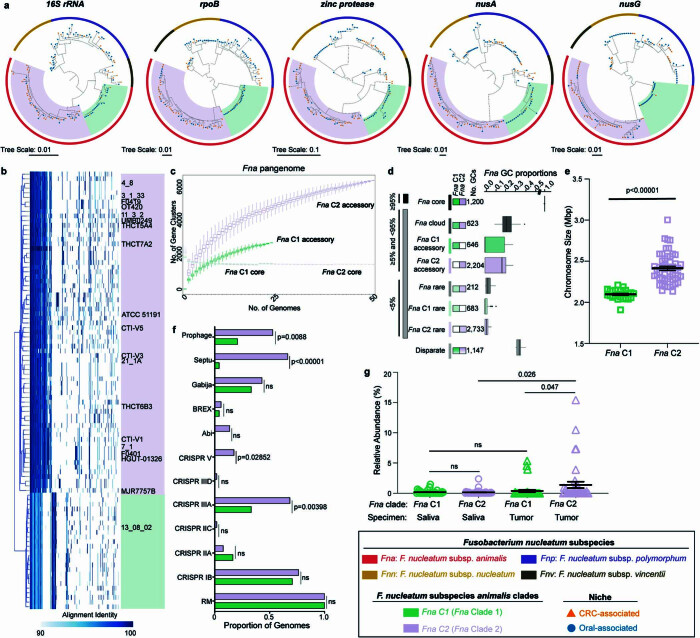
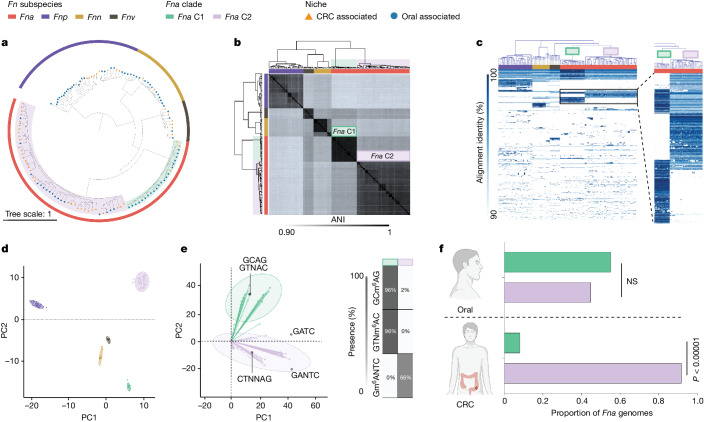
To further examine the observation that Fna strains form two distinct clades, we compared phylogenetic trees of housekeeping genes previously used for Fn subspecies typing28,44. Analysis of these single-marker genes supported the observation that two distinct Fna clades exist, which we call Fna clade 1 (Fna C1) and Fna clade 2 (Fna C2; Extended Data Fig. 2a). Beyond these single-marker genes, genome-wide differences between Fna clades are supported by a kSNP45 reference-free whole-genome phylogeny (Fig. 2a). To quantify the relatedness of Fna C1 to Fna C2, we compared the average nucleotide identity (ANI), a well-established index that measures the percentage of similarity between genomes, with an established 95% species threshold46. Between Fna clades the ANI ranged from 91.61% to 93.11%, comparable to the ANI between other Fn subspecies (Fig. 2b and Supplementary Tables 4 and 5). Further, we visualized the patterns of protein-coding genes present across Fna genomes using the Genes in Genomes-Map (GiG-map) tool and found that Fna C1 and Fna C2 have distinct protein-coding gene content (Fig. 2c). This was further supported by principal component analysis (PCA) of Anvi’o gene cluster presence–absence (Fig. 2d). Notably, the frequently used Fna 7_1 strain groups with Fna C2 (Extended Data Fig. 2b).
Extended Data Fig. 2. Phylogenetics and genomics of Fna clades.
a, Maximum-likelihood phylogenetic trees of Fn single marker genes, 16 S rRNA, rpoB, zinc protease, nusA and nusG. For each genome (n = 135), tree end points indicate niche origin, CRC (orange) or oral (blue), bar color indicates Fn subspecies (Fna (red), Fnn (gold), Fnp (purple), Fnv (brown)), and background color indicates Fna clades (Fna C1 (green) and Fna C2 (lavender)). b, Genes-in-genomes map (GiG-map) visualization of protein coding gene content across Fna genomes. Boxes highlight Fna clades, Fna C1 (green) and Fna C2 (lavender). Previously published NCBI genomes are labeled by strain name. c, Size of the Fna pangenome split by Fna clade, Fna C1 (green) and Fna C2 (lavender), with respective core and accessory genomes labeled. n = 10,000 random subsamplings of 24 Fna C1 and 51 Fna C2 genomes. Data is plotted as median ± s.d. d, Composition of Fna pangenome subset by clade. Anvi’o gene cluster (GC) prevalence was used to define core (≥95%), accessory (≥5% and <95%), and rare (<5%) features conserved in both Fna C1 and Fna C2 strains (“Fna core” (≥95% in all Fna strains), “Fna cloud” ( ≥ 5% and <95% in all Fna strains), “Fna rare” (<5% in all strains) or unique for strains from each clade. Disparate features are those that do not fall into any of the other noted bins. Plot box shows 25th percentile, median, and 75th percentile. Plot whiskers indicate minima and maxima. e, Column graph indicates chromosome sizes in Fna C1 (n = 24) and Fna C2 (n = 51). Data is plotted as mean ± s.e.m. Statistical analysis performed using Welch’s T-test, two-tailed. f, Column graph depicts the proportion of Fna genomes containing innate bacterial genetic defense systems, subset by Fna clades, Fna C1 (green) and Fna C2 (lavender). Statistical analysis performed using two sample Z test, two-tailed. NS, not significant. g, Graph shows the percent relative abundance of Fna C1 (green) and Fna C2 (lavender) in paired saliva (circle) or tumor biopsy (triangle) samples from 39 patients with colorectal adenocarcinomas47. Data is plotted as mean ± s.e.m. Statistical analysis performed using Welch’s T-test, paired. NS, not significant.
Fig. 2. Genetic and epigenetic characteristics of Fna clades.
a, A kSNP45 maximum-likelihood whole-genome phylogenetic tree. For each Fn genome (n = 135), the tree end points indicate the niche origin (CRC (orange); oral (blue)) and the bar colour indicates the Fn subspecies (Fna (red); Fnn (gold); Fnp (purple); Fnv (brown)). Within Fna, the background colour indicates the Fna clade (Fna C1 (green); Fna C2 (lavender)). b, A clustered dendrogram of the ANI matrix. The bar colour indicates the Fn subspecies (Fna (red); Fnn (gold); Fnp (purple); Fnv (brown)). The Fna clades are highlighted with green and lavender boxes. ANI values are reported in Supplementary Tables 4 and 5. c, A GiG-map visualization of the protein-coding gene content across Fn genomes. The top bar colour indicates the Fn subspecies (Fna (red); Fnn (gold); Fnp (purple); Fnv (brown)) and the box colour indicates the Fna clade (Fna C1 (green); Fna C2 (lavender)). The inset on the right highlights groups of protein-coding genes that are distinct between Fna C1 and Fna C2. An interactive GiG-map dataset is available at https://fredhutch.github.io/fusopangea/. d, PCA of Anvi’o gene clusters by presence and absence in each genome. The colours indicate the Fn subspecies and Fna clades (Fnn (gold); Fnp (purple); Fnv (brown); Fna C1 (green); Fna C2 (lavender)). The ellipses are drawn to 95% confidence. e, Left: PCA of Fna genome-wide methyl-modified nucleotide sequences. The ellipses are drawn to 95% confidence. The overlay of the PCA biplot shows the top five nucleotide motifs that are driving the Fna clade bifurcation. Right: a table indicating the distribution of each motif across the Fna clades. The colour indicates the Fna clade (Fna C1 (green); Fna C2 (lavender)). f, A column graph depicting the proportion of Fna CRC-associated and Fna oral-associated genomes, subset by Fna clade (Fna C1 (green); Fna C2 (lavender)). The statistical analysis was carried out using a two-sample z-test, two-tailed. NS, not significant. The graphics in f were created using BioRender.com.
Therefore, we reassessed the genetic, epigenetic and ecological properties of Fna as two genetically distinct clades. Comparison of the Fna clade pangenomes showed that Fna C1 and Fna C2 had similar core genome sizes, although Fna C2 had a larger accessory genome (Extended Data Fig. 2c,d), suggesting that Fna C2 strains harbour additional genetic factors that may be beneficial during colonization of CRC tumours. Consistent with this, comparisons of individual genome size and content indicated that Fna C2 strains have significantly larger chromosome sizes (Welch’s t-test, two-tailed, P < 0.00001; Extended Data Fig. 2e), more extrachromosomal plasmids (Supplementary Tables 1 and 6) and a greater number of innate genetic defences and mobile genetic elements (Extended Data Fig. 2f) compared to Fna C1 strains. PCA analysis of Fna methylomes indicated that Fna C1 and Fna C2 are also epigenetically distinct (Fig. 2e). The methyl-modified DNA motifs most influencing this epigenetic bifurcation are GTNm6AC (100% Fna C1, 0% Fna C2), GCm6AG (100% Fna C1, 0% Fna C2) and Gm6ANTC (0% Fna C1, 63% Fna C2; Fig. 2e and Supplementary Table 7). Although both Fna clades are present in the oral cavity with non-significant differences, only Fna C2 is significantly associated with the CRC niche (two-sample z-test, two-tailed, P < 0.00001; Fig. 2f). We further tested this observation on publicly available 16S rRNA gene sequencing data from paired tumour tissue and saliva samples from patients with CRC47. To gain resolution to the Fna clade level, we identified Fna clade-specific amplicon sequence variants48 (Methods and Supplementary Table 8). Supportive of our observations, our data show that Fna C2 is significantly enriched in tumour samples compared to Fna C1 (t-test, paired, P = 0.047; Extended Data Fig. 2g). However, there was no statistically significant difference between Fna clades in paired oral samples, indicating that although both Fna clades are present in the oral cavity of patients with CRC, only Fna C2 is enriched in the tumour niche (Extended Data Fig. 2g).
Human lower GI tract Fna C2 enrichment
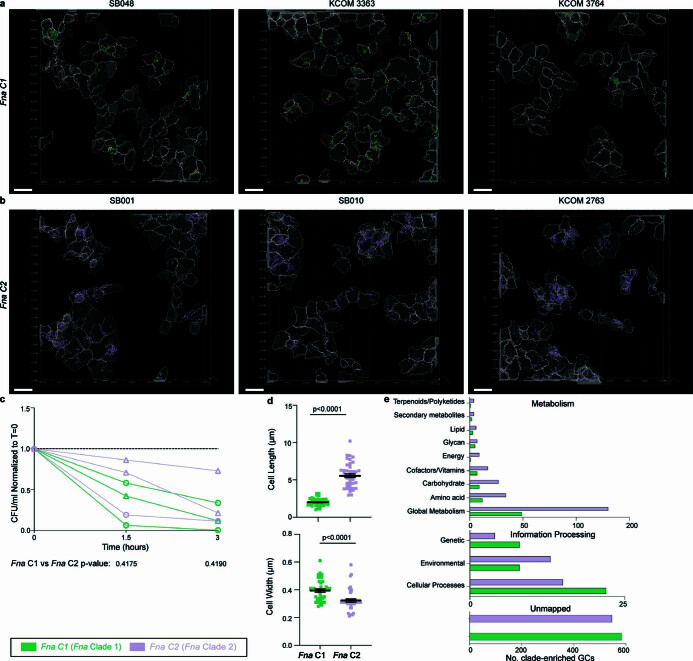
Pangenome analysis revealed that Fna is composed of two distinct clades but only Fna C2 is enriched in the CRC niche. Low levels of Fna C1 in this niche could be due to poor tumour colonization potential, an inherent lack of virulence or tumour-supportive factors that are possessed by Fna C2, or Fna C1 being unable to evade immune clearance. To interrogate these possibilities and reveal Fna clade-specific genetic factors, we applied a comprehensive inter-Fna clade comparative analysis across all 75 Fna genomes (24 Fna C1, 51 Fna C2). Canonical Fn virulence factors including the adhesins encoded by radD, aim1 and fadA2 were significantly enriched in Fna C1 compared to Fna C2 (two-sample z-test, two-tailed, P < 0.00001; Figs. 1g and 3a) suggesting that their role may be particularly important in the oral cavity. Conversely, as noted, fap2, cmpA and fusolisin are absent from Fna C1 and significantly associated with Fna C2 (two-sample z-test, two-tailed, P < 0.00001; Figs. 1g and 3a). Given the reported epithelial31 and immune cell49 interactions of Fap2 in CRC, its association with Fna C2 supports its increased adherence and invasion potential in this niche. Co-culture of Fna strains from each clade with a human colon cancer cell line (HCT116) demonstrate that Fna C2 has a significantly higher level of cancer epithelial cell invasion compared to Fna C1 strains (Fig. 3b and Extended Data Fig. 3a,b; Welch’s t-test, two-tailed, P = 0.0113), indicative of differential invasion potential and/or aerotolerance of individual strains (Extended Data Fig. 3c). Further, the Fna clades are morphologically distinct, with Fna C2 cells being significantly longer (Fna C1: 2.01 μm average, Fna C2: 5.26 μm average) and thinner (Fna C1: 0.39 μm average, Fna C2: 0.33 μm average) than Fna C1 cells (Extended Data Fig. 3d; Welch’s t-test, two-tailed, P < 0.00001 length, P < 0.00001 width). As bacterial morphology can affect colonization of host niches and susceptibility to host defences50, physical differences between Fna clade cells are noteworthy.
Fig. 3. Inter Fna-clade comparative analyses.
a, A gene presence versus absence column graph depicting the proportion of Fna genomes containing canonical Fn virulence factors, subset by clade (Fna C1 (green); Fna C2 (lavender)). The statistical analysis was carried out using a two-sample z-test, two-tailed. b, Left: computational confocal analysis of colon cancer epithelial cells (HCT116; grey) co-incubated with representative Fna C1 (green) or Fna C2 (lavender) strains. Scale bars, 4 μm. Right: a bar plot demonstrating the percentage of HCT116 cells with intracellular Fna; n = 3 biological replicates with 3 analysed z-stacks. Data are plotted as mean ± s.e.m. The statistical analysis was carried out using Welch’s t-test, two-tailed. c, A PPanGGOLiN51 map of the Fna pangenome. Each node represents a gene group, syntenic nodes represent neighbouring genes, the size indicates relative presence across Fna genomes, and the colour depicts pangenome partition (Fna core (red); Fna C1 accessory genome (green); Fna C2 accessory genome (lavender)). The white arrows (left panel) and oblongs (right panel) indicate Fna C2-associated putative eut and pdu operons. d, Schematics of these Fna C2 operons. An interactive PPanGGOLiN map is available at https://fredhutch.github.io/fusopangea/. e,f, Differentially expressed genes (log2[fold change] ≥ 0.58 and ≤−0.58 with −log10[P value] ≥ 1.30) in a representative Fna C2 strain, SB010, exposed to EA (e) or 1,2-PD (f) compared to unexposed SB010 control. To highlight SB010-unique content, genes also differentially expressed under the same exposure conditions in a representative Fna C1 strain, KCOM 3764, have been removed (Extended Data Fig. 4b,c). The vertical dotted lines indicate the threshold of significant gene expression, defined as log2[fold change] ≥ 0.58 and ≤−0.58. The statistical analysis was carried out using glmQLFTest, two-sided. The data point colours indicate Fna core (red), Fna C1 accessory (green), Fna C2 accessory (lavender) or Fna cloud (black; present in ≥5% and <95% in all Fna strains) genes. The stars indicate eut and pdu operon genes.
Extended Data Fig. 3. Morphological and genomic differences between Fna clades.
Representative Fna C1 and Fna C2 strains co-cultured with human colon cancer cells (HCT116). a-b, Computational analysis of confocal imaging. Independent masks for cancer epithelial cells (grey), and intracellular bacterial cells (Fna C1 green; Fna C2 lavender) were generated. Masks were used to calculate the percent of HCT116 cells with intracellular Fna (Fig. 3b) (see Methods). Scale bar is 20 μm. c, Bacterial aerotolerance was assessed through serial dilution plating at start, mid-point, and endpoint of co-culture. Graph shows resulting bacterial colony forming units per millilter, standardized to start point for each strain. Dashed line indicates normalization equal to one. Statistical analysis performed using a Welch’s T-test, two-tailed. d, Bar plots indicate Fna cell length and cell width as measured from confocal microscopy images, subset by Fna clades, Fna C1 (green) or Fna C2 (lavender); n = 45 cells per Fna clade. Data is plotted as mean ± s.e.m. Statistical analysis performed using Welch’s T-test, two-tailed. e, KofamKOALA KEGG27 ortholog mapping of Fna clade-enriched gene clusters.
We reasoned that delineation of Fna clade-unique genome content could reveal hitherto unknown genetic factors enabling Fna C2 transit to and survival within the human colonic niche. Predominant genetic differences between Fna clades are consistent with Fna C2 having increased nutrient scavenging mechanisms and enhanced metabolic potential (Extended Data Fig. 3e and Supplementary Table 9). As functionally related bacterial genes often form co-regulated units (operons), we implemented the Partitioned PanGenome Graph of Linked Neighbors (PPanGGOLiN)51 tool to assess whether Fna clade-unique genetic factors formed putative operons (Fig. 3c). Consistent with Anvi’o analysis, the PPanGGOLiN analysis showed that Fna C2 syntenic blocks were predominantly associated with metabolic mechanisms (Supplementary Table 10). Thus, the pathoadaptation of Fna C2 to the CRC niche is multifactorial, and in addition to canonical Fn virulence factors is potentially facilitated by enhanced metabolic capabilities.
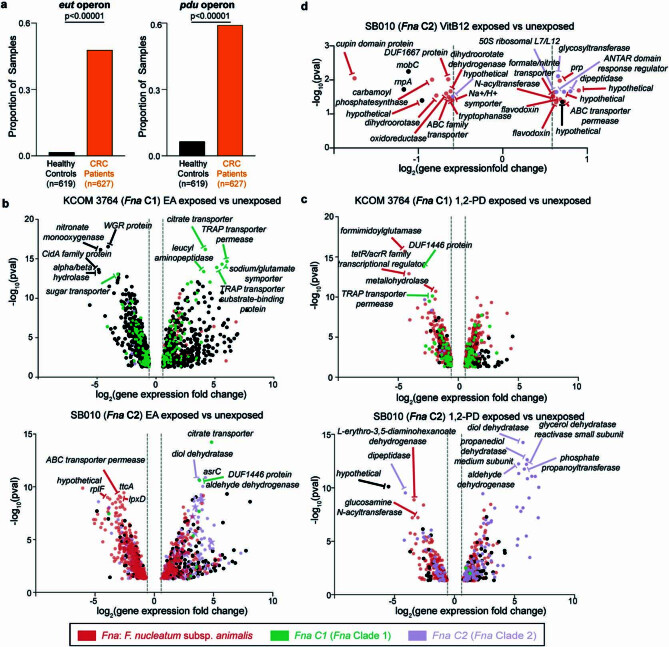
To validate our inter-Fna clade pangenomic approach, we focused on two Fna C2-associated putative operons consistent with ethanolamine (EA) metabolism (eut) and 1,2-propanediol (1,2-PD) metabolism (pdu; Fig. 3d). These operons contribute about 20% of Fna C2-unique gene content (Supplementary Table 10). Enteric pathogens not only gain a competitive advantage through direct metabolism of EA and 1,2-PD, but also exploit their intestinal specificity. Sensing through eut and pdu activates global regulators of virulence and induces transcriptional profiles consistent with GI niche adaptation52. Analysis of stool metagenomic datasets from publicly available cohorts of patients with CRC (n = 627) and healthy individuals (n = 619) indicates that the eut and pdu operons are significantly enriched in patients with CRC (two-sample z-test, two-tailed, eut P < 0.00001, pdu P < 0.00001; Extended Data Fig. 4a).
Extended Data Fig. 4. Fna clade transcriptomic responses to intestinal metabolites.
a, Bar plots demonstrate the proportion of stool metagenomic samples from patients with CRC or healthy controls in which putative eut and pdu operons were detected. Statistical analysis performed using two sample Z test, two-tailed. Cohort sample sizes are indicated at the bottom of each panel. b-c, Differentially expressed genes (with log2-transformed fold change ≥ 0.58 and ≤ −0.58 with -log10(p-value) ≥ 1.30) of a representative Fna C1 strain (KCOM 3764) and a representative Fna C2 strain (SB010) under (b) ethanolamine (EA) or (c) 1,2-propanediol (1,2-PD) exposure as compared to their respective unexposed controls. Top five significant (-log10(p-value) ≥ 1.30) upregulated and downregulated genes are labeled. d, Differentally expressed genes (with log2-transformed fold change ≥ 0.58 and ≤ −0.58 with -log10(p-value) ≥ 1.30) in SB010 under Vitamin B12 exposure alone as compared to unexposed control. All differentially expressed genes labeled. For b-d, vertical dotted lines indicate the threshold of significant gene expression, defined as log2-transformed fold change ≥ 0.58 and ≤ −0.58. Statistical analysis performed using glmQLFTest, 2-sided. Data point colors indicate whether a gene is categorized as part of the Fna core genome (red), Fna C1-associated accessory genome (green), or Fna C2-associated accessory genome (lavender) by PPanGGOLiN.
Motivated by the conservation of eut and pdu in Fna C2, and their absence in Fna C1, we assessed global transcriptomic responses of Fna cells following exposure to these intestinal-associated metabolites. RNA sequencing of representative Fna C1 and Fna C2 strains after exposure to EA or 1,2-PD indicated that both Fna clades have significant transcriptomic changes (Extended Data Fig. 4b–d and Supplementary Tables 11–15). As Fna C1 is deficient in both eut and pdu, we reasoned that significant transcriptomic changes following exposure to EA or 1,2-PD in Fna C1 would be independent of these operons. Thus, through a subtractive approach, we focused on differentially expressed genes (t-test, two-tailed, P < 0.05) in Fna C2 cells that were not differentially expressed (t-test, two-tailed, P > 0.05) in Fna C1 cells. Our results demonstrate that in Fna C2, eut and pdu genes are transcriptionally upregulated in response to EA and 1,2-PD, respectively (Fig. 3e,f).
Furthermore, Fna C2 cells exposed to EA or 1,2-PD significantly upregulated 13.02% of Fna C2-associated genes, including canonical Fn virulence factors. Although present in both clades, radD and aim1 are upregulated in the presence of EA in Fna C2 but not Fna C1 cells (Fig. 3e). Virulence factors uniquely present in Fna C2 are additionally upregulated when Fna C2 cells are exposed to EA (cmpA, fusolisin and fap2) or 1,2-PD (fap2; Fig. 3e,f). The upregulation of Fna C2-associated genes and virulence factors known to be important for interactions with human epithelial cells suggests that following their transit to the human GI tract, sensing of these molecules could induce Fna C2 transcriptional profiles consistent with extra-oral niche adaptation.
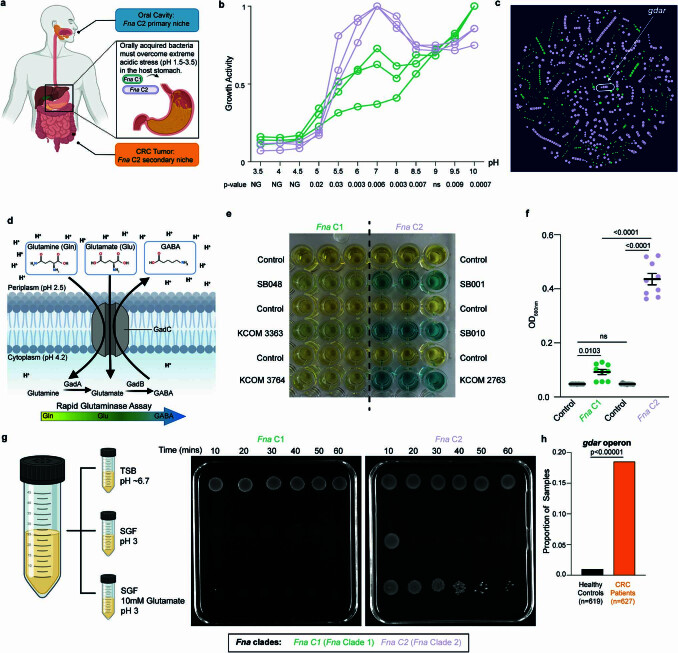
This also led us to reconsider how Fna C2 might be translocating to extra-oral tumour niches. Previous studies suggest that oral fusobacteria travel to CRC tumours through the bloodstream during transient bacteremia caused by activities such as daily hygiene practices or dental procedures20. Our identification of transcriptionally active eut and pdu operons suggests that direct descent through the GI tract, with subsequent infiltration of CRC tumours through the lumen, may be an additional pathway used by Fna C2. Yet, for GI transit to be a viable route of dissemination, Fna C2 would need to overcome the deleterious effects of extreme acid stress encountered in the stomach (pH 1.5–3.5; Extended Data Fig. 5a). Assessing the preferential growth pH, we observed that both Fna clades are sensitive to pH below 4.5 (Extended Data Fig. 5b). From pH 5.5 to 8.5, Fna C2 strains had a significantly higher level of growth activity compared to Fna C1, with maximum growth activity at pH 7 (Extended Data Fig. 5b). Under basic conditions (pH 9.5–10), Fna C1 strains had significantly higher growth activity compared to Fna C2, with maximum growth activity at pH 10 (Extended Data Fig. 5b). Pangenome analysis also revealed a putative glutamate-dependent acid resistance (GDAR) system conserved across all Fna C2, but absent in Fna C1 (Extended Data Fig. 5c and Supplementary Table 10). The GDAR system, found in pathogenic and commensal gut bacteria, is one of the most potent acid resistance mechanisms53, with glutamate being the only component necessary for the system to operate at pH ≤3 (Extended Data Fig. 5d). Using a colorimetric pH change assay, we tested the conversion of glutamine through glutamate into γ-aminobutyric acid in the presence of Fna C1 and Fna C2 and found that the level of this conversion is significantly higher in the presence of Fna C2 strains (Extended Data Fig. 5e,f).
Extended Data Fig. 5. Differences in pH preference and acid resistance across Fna clades.
a, Schematic of potential gastrointestinal route from the oral primary niche to the CRC tumor secondary niche. b, Plot indicates growth activity as measured in Biolog PM10 plates for representative Fna C1 (green) and Fna C2 (lavender) strains. Data is plotted as the normalized average across duplicates. Statistical analysis at each pH performed using Welch’s T-test, two-tailed. NG = no growth. c, PPanGGOLiN51 map of Fna pangenome. Each node represents a gene group, syntenic nodes represent neighboring genes, size indicates relative presence across Fna genomes, and color depicts elements in the Fna C1-associated accessory genome (green), and the Fna C2-associated accessory genome (lavender). White arrow indicates putative glutamate-dependent acid resistance (gdar) operon. d, Schematic depicts mechanism of GDAR acid resistance system. e, Qualitative and f, quantitative measurements of colorimetric assay measuring pH change indicative of conversion of glutamine to glutamate (yellow to green) and conversion of glutamate to γ-aminobutyric acid (GABA) (green to blue) in the presence of representative Fna C1 and Fna C2 strains. n = 3 technical triplicates of 3 biological replicates of 3 strains per Fna clade. Data is plotted as mean ± s.e.m and statistical analysis performed using an ANOVA. g, Schematic of experiment testing the effects of pH stress by exposure to simulated gastric fluid (SGF) at pH 3 or SGF supplemented with 10 mM glutamate at pH 3. Plates show resulting growth for a representative Fna C1 and a representative Fna C2 strain over the course of an hour exposure, as compared to tryptic soy broth (TSB) control at pH ~6.7. h, Bar plots demonstrate the proportion of stool metagenomic samples from patients with CRC (orange) or healthy controls (black) in which a putative gdar operon was detected. Statistical analysis performed using two sample Z test, two-tailed. Cohort sample sizes are indicated at the bottom of each panel. The graphics in a,d,g were created using BioRender.com.
To further mimic effects of pH stress during gastric transit, we exposed Fna clades to simulated gastric fluid at pH 3. Both were non-viable after 10 min of exposure to simulated gastric fluid. However, in the presence of supplemented glutamate, Fna C2 survived for an extended period (about 60 min), which was not observed for Fna C1 lacking GDAR (Extended Data Fig. 5g). Analysis of stool metagenomic datasets indicated that gdar operons are significantly enriched in patients with CRC compared to healthy individuals (two-sample z-test, two-tailed, P < 0.00001; Extended Data Fig. 5h). Thus, in addition to active eut and pdu systems, differences in pH preference and acid resistance mechanisms may contribute to the ability of Fna C2 to access the GI and tumour niches.
Fna C2 affects intestinal tumorigenesis
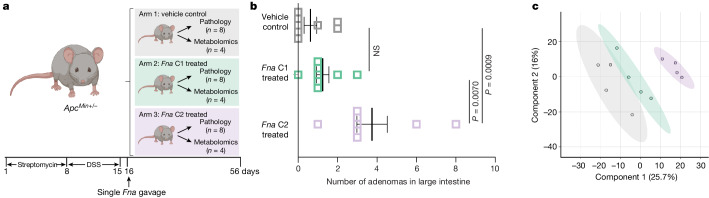
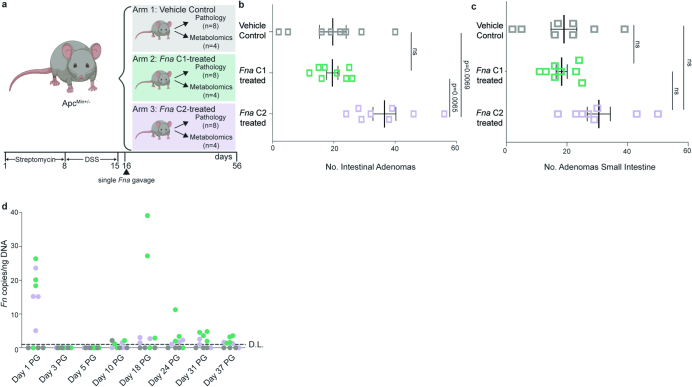
As Fna C2-enriched gene clusters were predominantly associated with enhanced metabolic potential (Extended Data Fig. 3e and Supplementary Table 10), we sought to determine whether Fna treatment of the dextran sodium sulfate-induced colitis ApcMin+/− mouse model54 of CRC affected intestinal tumorigenesis and metabolic pathways in vivo (Fig. 4a). To capture a higher proportion of Fna clade-specific accessory genes (Extended Data Fig. 2d), a mix of three representative strains for each clade was used. Following the administration of a single oral gavage of Fna C1 mix, Fna C2 mix or vehicle control, we observed a significant increase in the number of intestinal adenomas in Fna C2-treated mice compared to both Fna C1 and vehicle control independently (Extended Data Fig. 6a,b; analysis of variance (ANOVA), P = 0.0065 and P = 0.0069, respectively), specifically in the large intestine (Fig. 4b; ANOVA, P = 0.0070 and P = 0.0009, respectively; Extended Data Fig. 6c). There was no significant difference in adenoma burden between Fna C1 treatment and vehicle control mice. Low-level Fn was inconsistently detected during the course of the study (Extended Data Fig. 6d). We carried out liquid chromatography–mass spectrometry global metabolomics on intestinal tissue from each treatment arm for comparative metabolite analysis (Supplementary Table 16). Partial least squares discriminant analysis of measured intestinal metabolites demonstrated that Fna C2-treated mice formed a distinct cluster away from other treatment arms, suggesting a differential metabolic profile. However, intestinal metabolites from Fna C1-treated and vehicle control mice had more similar metabolite profiles, clustering together (Fig. 4c).
Fig. 4. Fna C2 impact on intestinal tumorigenesis and metabolism.
a, A schematic of the study with 6–8-week-old ApcMin+/− mice receiving streptomycin and dextran sodium sulfate (DSS) treatment to alter the native microbiome and induce colitis, respectively. Mice were orally gavaged with vehicle control (arm 1) or a mix of three representative Fna C1 (arm 2) and Fna C2 (arm 3) strains. A strain mix was used to capture a higher proportion of Fna clade-specific accessory genes (Extended Data Fig. 2d). The mice were monitored until the end-point at 6 weeks post-gavage when they reached 15–17 weeks of age. b, A plot indicating the number of adenomas in the large intestine by treatment arm (vehicle control (grey); Fna C1 treated (green); Fna C2 treated (lavender); n = 8 mice per arm). The data are plotted as mean ± s.e.m. The statistical analysis was carried out using one-way ANOVA. c, Partial least squares discriminant analysis of detected intestinal metabolites (n = 1,296). The colours represent the treatment arm (vehicle control (grey); Fna C1 treated (green); Fna C2 treated (lavender)). The graphics in a were created using BioRender.com.
Extended Data Fig. 6. Intestinal adenoma burden and fecal Fusobacterium load in Fna treated mice.
a, Schematic of study with ApcMin+/− mice orally gavaged with vehicle control (Arm 1) or representative Fna C1 (Arm 2) and Fna C2 (Arm 3) strains post streptomycin and dextran sodium sulfate (DSS) to clear the native microbiome and induce colitis, respectively. b-c, Plots indicate the number of adenomas in b, the small and large intestines combined and c, the small intestine by treatment arm, vehicle control (grey), Fna C1-treated (green), or Fna C2-treated (lavender). Data is plotted as mean ± s.e.m; n = 8 mice per treatment arm. Statistical analysis performed using one-way ANOVA. d, Fusobacterium-targeted qPCR was performed on fecal pellets from one-day post-gavage (PG) to study endpoint. Fn presence in each sample is plotted as Fn copies per ng of fecal DNA with a detection limit (D.L.) of 1 Fn copy. Each sample was run in triplicate, and samples with ≥2 duplicates with detectable signal are included. The graphics in a were created using BioRender.com.
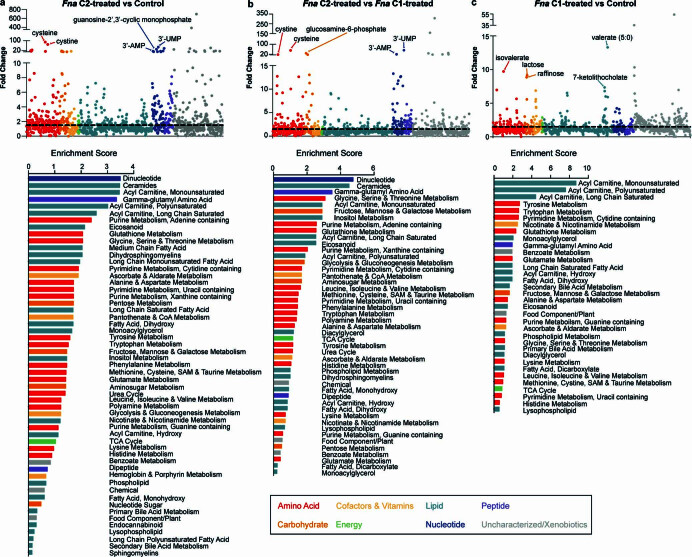
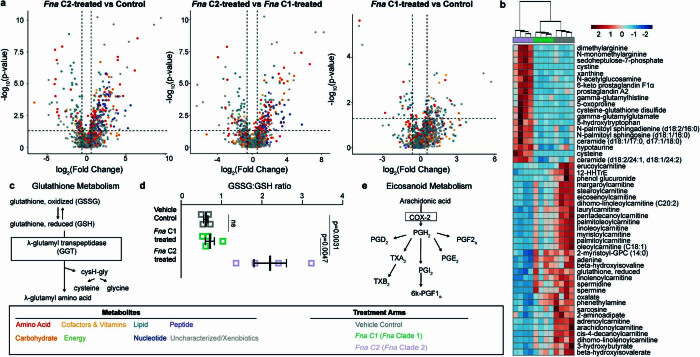
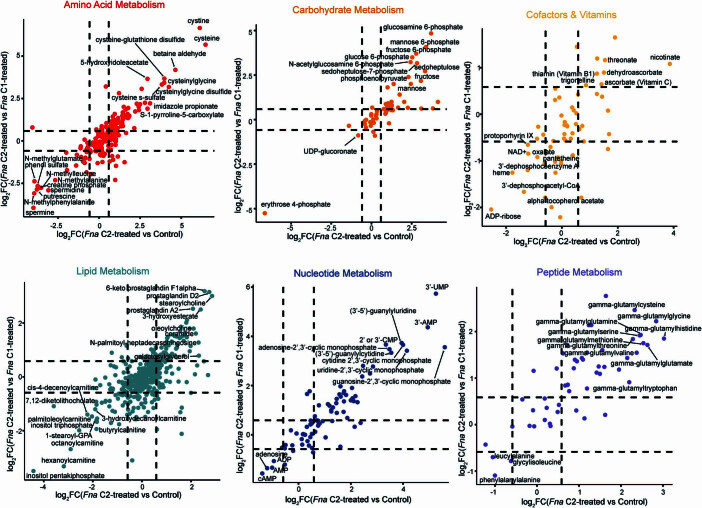
Of 1,296 metabolites measured (Extended Data Fig. 7a–c and Supplementary Table 16), comparative analysis demonstrated a significant enrichment in glutathione metabolism and γ-glutamyl amino acid pathways in Fna C2-treated mice, compared to both Fna C1-treated and vehicle control mice (Extended Data Figs. 7a–c, 8a and 9 and Supplementary Table 17). Specifically, we observed a significant increase in the levels of precursors to γ-glutamyl-cysteinyl-glycine (GSH) synthesis, including cysteine and γ-glutamylcysteine, decreased levels of glutathione in its reduced form (GSH) and significantly higher levels of the GSH degradation product 5-oxoproline (Extended Data Figs. 8b and 9 and Supplementary Table 16). Consistent with γ-glutamyl amino acid generation from reduced glutathione in the presence of γ-glutamyl transpeptidase, increased levels of γ-glutamyl amino acids and cysteine, glycine and cysteinyl-glycine were also observed (Extended Data Fig. 8b,c and Supplementary Table 16). GSH deficiency or an elevated ratio of oxidized (GSSG) to reduced (GSH) forms of glutathione increases the vulnerability of mammalian cells to oxidative stress, inflammation and tumour progression55. The GSSG/GSH ratio of Fna C2-treated mice significantly increased by 3.5- and 3.0-fold compared to the control and Fna C1-treated groups respectively, suggesting increased oxidative stress (Extended Data Fig. 8d; one-way ANOVA, P = 0.0031 and P = 0.0047, respectively). Studies have demonstrated that metabolism of GSH by γ-glutamyl transpeptidase can exert pro-oxidant effects. In cancer cells, this is a source of endogenous reactive oxygen species that can facilitate persistent oxidative stress and contribute to genomic instability56,57. Consistent with this, significantly increased levels of other markers of oxidative stress including cystine and cysteine-glutathione disulfide and significantly decreased levels of polyamines capable of scavenging reactive oxygen species, including putrescine, spermidine and spermine, were observed in Fna C2-treated mice (Extended Data Figs. 8b and 9). Notably, recent work from our group demonstrated that Fusobacterium was predominantly associated with epithelial cells harbouring severe chromosomal abnormalities10, one of the most common forms of genomic instability in cancer.
Extended Data Fig. 7. Intestinal metabolite changes in Fna treated mice.
Scatter plots shows fold change for 1,296 detected metabolites from a, Fna C2 treated mice compared to control, b, Fna C2 treated mice compared to Fna C1 treated mice, and c, Fna C1 treated mice compared to control colored by metabolic categorization. Top five characterized metabolites are labeled. Plots below indicate the pathway enrichment score all enriched pathways. Bars are colored by metabolic categorization.
Extended Data Fig. 8. Altered metabolites in Fna treated mice.
a, Plots show the log2-transformed fold change in metabolite level between treatment arms by the -log10(p-value). Points indicate individual metabolites colored by their metabolic categorization. Dotted lines indicate the threshold of significant gene expression, defined as log2-transformed fold change ≥ 0.58 and ≤ −0.58 (vertical lines) and a -log10(p-value) ≥ 1.30 (horizontal line). Statistical analysis performed on natural log-transformed values using one-sided T-test. b, Clustered heatmap of top fifty metabolites across study arms. Dendrogram groups individual samples by similarity of metabolite profile. Bar color for each sample indicates treatment arm, vehicle control (grey), Fna C1-treated (green), or Fna C2-treated (lavender). c, Schematic of glutathione metabolic pathway. d, Plot demonstrates the ratio between oxidized (GSSG) and reduced (GSH) glutathione levels for each treatment arm, vehicle control (grey), Fna C1-treated (green), or Fna C2-treated (lavender); n = 4 mice per arm. Data is plotted as mean ± s.e.m. Statistical analysis performed using one-way ANOVA. e, Schematic of eicosanoid metabolic pathway.
Extended Data Fig. 9. Significantly altered intestinal metabolites in Fna C2-treated mice.
Plots of individual, characterized metabolites, with metabolic categories having > 10 significantly altered metabolites shown. Each plot shows the log2-transformed log change between Fna C2-treated mice compared to Fna C1-treated mice versus the log2-transformed log change between Fna C2-treated mice compared to control mice. Dashed lines indicate the threshold of significantly altered metabolites, defined as log2-transformed fold change (FC) ≥ 0.58 and ≤ −0.58. Upper right quadrant indicates metabolites that are significantly elevated in Fna C2-treated mice as compared to both Fna C1-treated and control mice. Lower left quadrant indicates metabolites that are significantly lower in Fna C2-treated mice as compared to both Fna C1-treated and control mice. Top metabolites in each of these quadrants are labeled.
In addition to their role in combating oxidative stress, polyamines are able to suppress inflammation through inhibition of macrophage cytokine synthesis58. Consistent with increased inflammation, significantly higher levels of N-monomethylarginine and dimethylarginine were observed in Fna C2-treated mice compared to other treatment arms (Extended Data Fig. 8b). Both of these metabolites inhibit the synthesis of the anti-inflammatory agent nitric oxide. Furthermore, we observed significantly higher levels of pro-inflammatory prostaglandins and ceramides, including prostaglandin A2, N-palmitoyl-sphingosine and N-palmitoyl-sphingadienine (Extended Data Fig. 8b). Ceramides can also be metabolized by cancer cells to reduce tumour cell apoptosis and proliferation59. Other metabolites that promote cancer cell proliferation and metastasis across a range of cancers include eicosanoids, which are similarly significantly increased in Fna C2-treated mice compared to Fna C1-treated or vehicle control mice (Extended Data Figs. 7a–c and 8e and Supplementary Table 17). This included increased levels of 6-keto prostaglandin F1-α (Extended Data Fig. 8b) through COX2 (also known as PTGS2) metabolism of arachidonic acid. Notably, COX2 was previously reported to be one of the most upregulated genes in Fn-associated human colorectal tumours3. Overall, our results demonstrate the ability of Fna C2, but not Fna C1, to metabolically affect the intestinal milieu towards pro-oncogenic conditions.
Fna C2 enrichment in human CRC cohorts
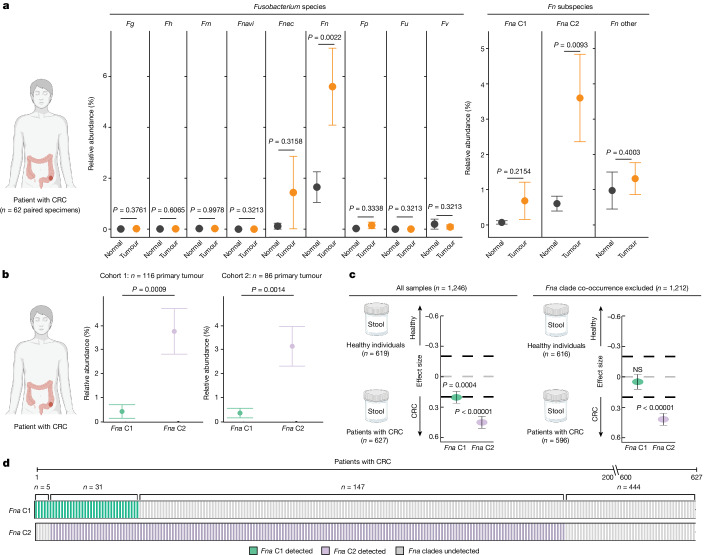
As Fna C2 strains are both significantly enriched in the CRC niche (Fig. 2f) and increase intestinal tumorigenesis in our mouse model compared to Fna C1 (Fig. 4b and Extended Data Fig. 6a–c), we next sought to determine the prevalence and abundance of these Fna clades in human tissue and stool specimens through culture-independent approaches. We carried out bacterial 16S rRNA gene sequencing on resected tumour tissue from 116 patients with treatment-naive CRC (CRC cohort 1) and on adjacent normal tissue from 62 of these patients (Supplementary Table 18). Comparing the percentage relative abundance of different Fusobacterium species between paired tumour and adjacent normal tissue (n = 62 patients), we observed that Fn was the only Fusobacterium species significantly enriched in tumour tissue compared to adjacent normal (Fig. 5a and Supplementary Table 19), supportive of previous reports2,60 (t-test, paired, P = 0.0022). However, using the Fna clade-specific amplicon sequence variants to resolve Fn to a higher taxonomic resolution that includes Fna C1, Fna C2 and non-Fna subspecies of Fn, we demonstrate that only Fna C2 is significantly enriched in tumour compared to paired normal tissue (Fig. 5a, and Supplementary Tables 8 and 19; t-test, paired, P = 0.0093). As neither Fna C1 nor non-Fna subspecies of Fn are significantly enriched, this suggests that it is specifically Fna C2 that is driving the previously reported enrichment of Fn in human CRC tumours. Furthermore, across two independent patient cohorts (CRC cohort 1 n = 116 and CRC cohort 2 n = 86), we demonstrate that within CRC tumour tissue, Fna C2 is significantly enriched compared to Fna C1 (Fig. 5b and Supplementary Tables 20 and 21; t-test, paired, cohort 1 P = 0.0009, cohort 2 P = 0.0014), supporting our observations at the Fna strain level (Fig. 2f).
Fig. 5. Fn in human tissue microbiome and stool metagenomic specimens.
a, Plots showing the relative abundance for Fusobacterium species (Fg, F. gonidiaformans; Fh, F. hwasookii; Fm, F. mortiferum; Fnavi, F. naviforme; left plot), and Fn subspecies and Fna clades (right plot) using microbial 16S rRNA gene sequencing of paired tumour (orange) and normal adjacent (black) tissue (n = 62 patients with CRC). Amplicon sequence variants were used to obtain Fna clade resolution (Extended Data Fig. 10 and Supplementary Table 8). The data are plotted as mean ± s.e.m. The statistical analysis was carried out using one-sided t-test, paired. b, Plots showing the relative abundance for Fna C1 (green) and Fna C2 (lavender) within patient primary colorectal tumour tissue from two independent cohorts (cohort 1 (n = 116) this study; cohort 2 (n = 86) BioProject PRJNA362951). The data are plotted as mean ± s.e.m. The statistical analysis was carried out using one-sided t-test, paired. c, Fna C1 and Fna C2 detection in stool metagenomic data from patients with CRC and healthy individuals. The left plot shows the pooled effect sizes for Fna C1 (green) and Fna C2 (lavender) calculated using a meta-analysis of standardized mean differences and a random-effects model on MetaPhlAn4 (ref. 63) species-level genome bin abundances on all CRC samples (n = 627) and samples from healthy individual (n = 619). The right plot shows the effect sizes for Fna C1 and Fna C2 calculated using the same approach, but here samples in which Fna C1 co-occurred with Fna C2 were excluded. The data are plotted as mean ± s.e.m. The statistical significance was assessed by a Wald test, two-sided. All P values are corrected using the Benjamini–Yakuteli method. d, Fna C1 and Fna C2 presence in stool metagenomes of patients with CRC. The bars indicate individual stool samples from patients with CRC (n = 627) and are coloured by Fna C1 and Fna C2 detection (Fna C1 detected (green); Fna C2 detected (lavender); Fna undetected (grey)). The lower brackets indicate the number of stool samples in which Fna C1 occurred independently (n = 5), Fna C2 occurred independently (n = 147), Fna clades co-occurred (n = 31) or Fna clades were not detected (n = 444). The graphics in a–c were created using BioRender.com.
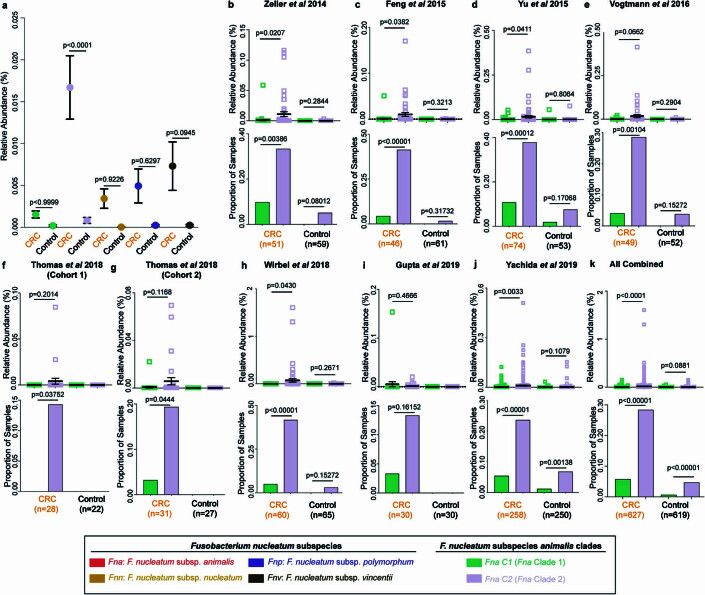
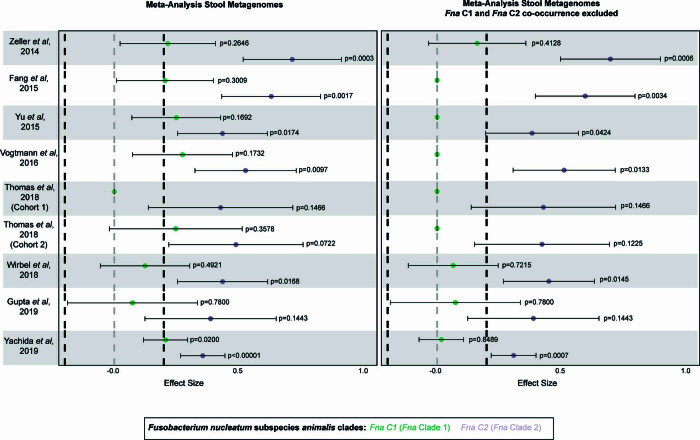
We next sought to determine whether the prevalence of Fna clades differed between patients with CRC and healthy individuals. To do so, we analysed stool metagenomic datasets from publicly available cohorts of patients with CRC (n = 627) and healthy individuals (n = 619; Extended Data Fig. 10 and Supplementary Table 22). Fna was detected in 29.2% of stool samples from patients with CRC and 4.8% of stool samples from healthy individuals (Supplementary Table 23). Meta-analysis of standardized mean differences by a random-effects model for Fna C1 and Fna C2 demonstrated that both Fna clades have a significant pooled effect size associated with CRC (Fna C1 effect size = 0.21, 95% confidence interval (0.09, 0.32), P = 4.45 × 10−4; Fna C2 effect size = 0.45, 95% confidence interval (0.34, 0.56), P = 5.55 × 10−15; Fig. 5c, Extended Data Fig. 11 and Supplementary Table 24). However, the effect size for Fna C2 was larger than that for Fna C1. Notably, in the absence of Fna C2 co-occurrence, Fna C1 was not significantly associated with CRC (Fig. 5c, Extended Data Fig. 11 and Supplementary Table 25). Although synergistic interactions between CRC-enriched microbes have previously been reported61, it is not clear whether Fna C1 co-occurrence with Fna C2 results in a compounding pathogenic effect. Similar to our observation in CRC tumour tissue (Supplementary Tables 20 and 21), our data show that Fna C2 is more prevalent and abundant in the stool of patients with CRC than Fna C1 (Fig. 5d and Extended Data Fig. 10) and is furthermore the only Fn subgroup significantly enriched in the stool of patients with CRC compared to healthy individuals (Extended Data Fig. 10a). These culture-independent human specimen analyses support our strain-level genomic discovery that Fna C2 is the dominant CRC-associated Fna clade (Fig. 2). This further highlights the significance of our in vitro (Fig. 3) and in vivo (Fig. 4) findings demonstrating the increased virulence and tumorigenic potential of Fna C2 compared to Fna C1.
Extended Data Fig. 10. Detection of Fn subspecies and Fna clades in human stool metagenomes.
Detection of a, Fn subspecies (Fnn, Fnv, Fnp) and a-k, Fna clades (Fna C1 and Fna C2) in stool metagenomic data from previously published independent cohorts from patients with CRC and healthy controls. a, Plot shows the percent relative abundance of each Fn subspecies and Fna clade in stool samples from patients with CRC (n = 627) or healthy controls (n = 619). Data is plotted as mean ± s.e.m and statistical analysis performed using a one-way ANOVA. b-k, Samples are plotted both by (b-k) individual cohorts and (k) pooled results. In each panel, top plot shows the percent relative abundances of Fna C1 (green) and Fna C2 (lavender) in each sample. Data is plotted as mean ± s.e.m and statistical analysis performed using Welch’s T-test, paired. Bottom plot demonstrates the proportion of stool samples from patients with CRC and the proportion of stool samples from healthy controls in which Fna C1 and Fna C2 were detected. Data is plotted as mean + s.d. and statistical analysis performed using two sample Z test, two-tailed. Cohort sample sizes are indicated at the bottom of each panel.
Extended Data Fig. 11. Meta-analysis of Fna in human stool metagenomes.
Fna C1 and Fna C2 detection in stool metagenomic data from previously published independent cohorts from patients with CRC and healthy controls plotted by individual cohorts. Left plot shows the effect sizes for Fna C1 and Fna C2 calculated across all samples (CRC n = 627, healthy control n = 619) using a meta-analysis of standardized mean differences and a random effects model on MetaPhlAn463 species-level genome bins (SGB) abundances. Right plot shows the effect sizes for Fna C1 and Fna C2 calculated using the same approach but with samples where Fna C1 co-occurred with Fna C2 excluded (CRC n = 596, healthy control n = 616). Data is plotted as mean ± s.e.m. Statistical significance assessed by Wald test, two-sided. All p-values are corrected via the Benjamini-Yakuteli method.
Discussion
Advances in next-generation sequencing have revealed the presence of bacterial communities within human tumour tissues. A key challenge for cancer microbiome research is to move beyond the characterization of microbial composition in tumours towards functional studies that determine whether, and how, these microbes are contributing to disease. In CRC, Fn gained early and continued attention owing to the fact that this bacterium was rarely detected in the lower GI tract of healthy individuals1, yet enriched within the CRC tumour microbiome2,60. Fn species are normal members of the human oral microbiota, and strains from the oral cavity are thought to seed CRC tumours19,20. However, the noted genetic and phenotypic heterogeneity12–15 of Fn led to an open question of whether Fn strains that colonize and dominate human tumours harbour distinct genetic attributes that contribute to CRC initiation or progression. Through large-scale culturing, sequencing and comparative genomic analyses of human CRC and non-cancer oral Fn strains, we revealed the distinct CRC-enriched genetic factors of Fn. Further, we identified that these CRC-enriched factors were predominantly present within a specific clade of Fna. This was mirrored by our discovery that Fna is bifurcated into two distinct clades: Fna C1, which is largely restricted to the oral cavity, and Fna C2, which dominates the human CRC tumour niche. Notably, only Fna C2 induced tumours and altered intestinal metabolism towards increased oxidative stress within a CRC animal model. Further, comparative genomic analysis between Fna clades revealed the genetic elements that cumulatively engender the pathoadaptation of Fna C2 to the CRC niche. Given the power of using Fna C1 as a comparative group for Fna C2, we created an interactive website to enable the exploration of Fna pangenomic datasets designated The Fusobacterium Pangenome Atlas at https://fredhutch.github.io/fusopangea/. Collectively, this work demonstrates that Fna C2 is a highly virulent subgroup of Fn that should be the primary focus for mechanistic studies and therapeutic drug design in CRC.
Methods
Fusobacterium strain isolation from tumour tissue from patients with CRC
Fusobacterium strains were isolated from CRC tumour tissue specimens from patients from North America and Europe as previously described6. Briefly, tissue sections were minced with a scalpel, and spread plated on selective fastidious anaerobe agar (FAA) plates (Oxoid, Thermo Fisher Scientific) supplemented with 7% or 10% defibrinated horse blood (DHB; Lampire Biological Laboratories, Fisher Scientific) with josamycin, vancomycin and norfloxacin at 3, 4 and 1 μg ml−1, respectively (Sigma Aldrich). Plates were incubated at 37 °C in anaerobic conditions (AnaeroGen Gas Generating Systems, Oxoid, Thermo Fisher Scientific) and inspected for growth every 2 days. Colonies were picked and streak purified, and colony PCR was carried out on selected bacterial colonies as previously described6 with 16S rRNA gene universal primers (342F and 1492R). Colony PCR products were sent for Sanger sequencing, and BLASTn analysis of trace sequences was used to confirm bacterial species identity. Cultures were suspended in tryptic soy broth (TSB) and 40% glycerol and stored at −80 °C.
Fusobacterium strain isolation from Korean Collection for Oral Microbiology and ATCC ampoules
Fusobacterium strains from the Korean Collection for Oral Microbiology (KCOM) collection were isolated from the oral cavity as previously described44. Strains from the ATCC and KCOM repositories were grown from ampoules on Schaedler agar plates supplemented with vitamin K1 and 5% defibrinated sheep blood (Becton Dickinson) and FAA plates (Oxoid, Thermo Fisher Scientific) supplemented with 7% DHB (Lampire Biological Laboratories, Fisher Scientific). Plates were incubated at 37 °C in a Bactron600 anaerobic chamber (Sheldon Manufacturing) for 5–7 days. Cultures were suspended in Schaedler broth with vitamin K1 and 30% glycerol and stored at −80 °C.
High molecular weight genomic DNA extraction
Fusobacterium strains were cultured under anaerobic conditions at 37 °C (AnaeroGen Gas Generating Systems, Oxoid, Thermo Fisher Scientific) for 48–72 h on FAA plates (Oxoid, Thermo Fisher Scientific) supplemented with 10% DHB (Lampire Biological Laboratories, Fisher Scientific) and plates for CRC-associated strains were further supplemented with josamycin, vancomycin and norfloxacin at 3, 4 and 1 μg ml−1, respectively (Sigma Aldrich). High molecular weight genomic DNA was extracted using the MasterPure Gram Positive DNA Purification Kit (Epicentre, Lucigen). Cells from two plates were resuspended in 1.5 ml 1× PBS and collected by centrifugation. Pellets were processed according to the manufacturer’s instructions, modified by doubling all reagent volumes and removing vortexing steps to prevent DNA shearing. High molecular weight genomic DNA was quantified using a Qubit fluorometer (Thermo Fisher Scientific).
PacBio single-molecule real-time sequencing and genome assembly
Single-molecule real-time sequencing18 was carried out on a PacBio Sequel instrument (Pacific Biosciences) or a PacBio Sequel II instrument (Pacific Biosciences) at the University of Minnesota Genomics Center. Sequencing reads were processed using Microbial Assembly pipeline within Pacific Biosciences’ SMRTAnalysis pipeline v.9.0.0.92188. Additional assembly was carried out using Flye assembler v.2.8 as needed (https://github.com/fenderglass/Flye).
Fusobacterium species typing
Fusobacterium genomes were subtyped to the species level and Fn genomes were further subtyped to the subspecies level on the basis of a cumulative score of individual marker genes. Marker genes previously used for Fusobacterium typing were used: the 16S rRNA gene, rpoB and a zinc metalloprotease gene30. From each complete, closed genome, its species or subspecies classification was first analysed by all three marker genes individually. Each marker gene was isolated and analysed using BLASTn, with the top hit by percentage identity noted. For each possible species or subspecies, a confidence score was calculated as the number of concordant subspecies results divided by the number of marker genes present. For each genome, its final classification was determined by the highest confidence score. Results for this analysis are noted in Supplementary Table 1. Phylogenetic classifications were further tested using GTDB-Tk (ref. 64; https://github.com/Ecogenomics/GTDBTk) as listed in Supplementary Table 2.
Pangenomic analyses
Pangenome analysis was carried out using the Anvi’o workflow21, the PPanGGOLiN tool51 and the GiG-map tool (https://github.com/FredHutch/gig-map) to characterize the Fn pangenome across 135 Fn genomes, and to characterize the Fna pangenome across 51 Fna genomes. For Fn genomes, Anvi’o thresholds were set to a minbit of 0.9 and an MCL of 2, and PPanGGOLiN thresholds were set to 90% identity and 90% coverage. For Fna genomes, Anvi’o thresholds were set to a minbit of 0.9 and an MCL of 7, and PPanGGOLiN thresholds were set to 90% identity and 90% coverage. For both genome sets, GiG-map was run with default settings. PPanGGOLiN’s alignment feature was used to map resulting Anvi’o gene clusters to their corresponding PPanGGOLiN nodes. To assess the size of the pangenome as the number of sampled genomes increases, the Fn and Fna Anvi’o-derived pangenomes were independently sampled for combinations up to 10,000 or otherwise randomly subsampled 10,000 times from 1 to 135 genomes and 1 to 75 genomes, respectively. This approach was subset by niche and clade as appropriate.
Genomic dendrograms
Individual gene and protein sequences were aligned through MEGA X (ref. 65) using the MUSCLE clustering algorithm from which a maximum-likelihood dendrogram was generated. kSNP3 (ref. 45) with a k-mer size of 13, resulting in a fraction of core k-mers of 0.217, was used to generate a maximum-likelihood phylogeny of the 135 Fn genomes in our collection. Final images were generated using the interactive tree of life tool, v.5 (ref. 66).
Identification of Fn canonical virulence factors
To query the presence of canonical Fn virulence genes in our collection of Fn genomes, we used the Operon Contextualization Across Prokaryotes to Uncover Synteny tool (https://github.com/FredHutch/octapus) with a minimum percentage identity threshold of 60%.
Identification of Fn genetic defence systems and prophage
The presence of innate bacterial defence systems was queried using the Prokaryotic Antiviral Defense Locator67 and intact prophage presence was analysed using the Phage Search Tool Enhanced Release68,69 tools.
PCA
PCA of Fn Anvi’o-derived gene content was carried out on a gene cluster presence–absence matrix using the R prcomp function in the stats package, v.3.6.2. PCA of Fna methylated nucleotide motifs was carried out on a methylated motif presence–absence matrix (Supplementary Table 7) using the PCA function in the R factoextra package, v.1.0.7.
Fn and HCT116 co-culture assays
The human colon cancer epithelial cell line HCT116 was purchased from ATCC. The cell line was not authenticated. Mycoplasma testing was carried out using the MycoProbe Mycoplasma Detection Kit (R&D Systems). HCT116 cells were cultured in McCoys 5A with l-glutamine (Corning) supplemented with 10% (v/v) fetal bovine serum (Sigma) and incubated at 37 °C in 5% CO2. HCT116 cells were seeded at 1.25 × 106 cells per well into 6-well plates with a glass coverslip at the bottom of each well (Nunclon Delta Surface, Thermo Scientific) and allowed to adhere for 16 h. Resuspended cultures of Fna C1 (SB048, KCOM 3363 and KCOM 3764) and Fna C2 (SB001, SB010 and KCOM 2763) strains were prepared in McCoys. Bacterial membranes were stained with 5 µg ml−1 FM 4-64FX (Molecular Probes). Each bacterial strain was co-incubated with HCT116 cells in wells at a multiplicity of infection of 100:1. These bacterial–eukaryotic co-cultures were incubated for 3 h at 37 °C in 5% CO2. Bacterial viability was assessed at time (T) = 0, T = 1.5 and T = 3 h by preparing serial dilutions for each strain and plating 50 µl of each dilution on FAA plates (Oxoid, Thermo Fisher Scientific) supplemented with 10% DHB (Hemostat, Fisher Scientific). Plates were incubated at 37 °C in a Bactron600 anaerobic chamber (Sheldon Manufacturing) for 2 days until colonies were counted. After incubation, wells were washed four times with PBS with gentle swirling to remove unattached bacterial and HCT116 cells. Cells were fixed in 4% paraformaldehyde in PBS for 30 min at room temperature. Following fixation, cells were washed three times in PBS and then permeabilized with 0.2% (v/v) Triton X-100 in PBS for 4 min at room temperature. Cells were washed three times in PBS and then stained for 20 min at room temperature with two drops per millilitre of NucBlue Fixed Cell Stain ReadyProbes (Invitrogen) and ActinGreen 488 ReadyProbes (Invitrogen) to stain DNA and actin, respectively. A dissecting microscope was used to visually confirm that cells remained on the coverslips after processing. Samples were viewed with a Leica SP8 confocal laser scanning microscope (Leica) for image acquisition. Three z-stacks of each co-culture were taken using a 63× oil lens and the following parameters: 1,024 × 1,024 resolution, pixel size 100.21 nm, speed 600, zoom factor 1.9 and z-step 0.3 mm.
Computational analysis to determine intracellular Fn
Confocal z-stacks from bacterial–eukaryotic co-cultures of HCT116 cells co-incubated with Fna C1 (SB048, KCOM 3363 and KCOM 3764) or Fna C2 (SB001, SB010 and KCOM 2763) strains were imported into Imaris. All measurements were carried out on three different z-stacks per biological replicate, with three biological replicates. In Imaris, the bacterial surface volumes were created using the fluorescence of the FM 4-64FX membrane stain (surface detail 0.223 mm, background subtraction using diameter of largest sphere of 0.5 mm). The eukaryotic cell detection tool was used to define and ID cells using the nuclear stain and the actin stain. The nuclei were split by seed points. The detected eukaryotic cells were exported to create a cell surface mask. To define intracellular bacterial cells, the bacterial surface was classified by the shortest distance to the eukaryotic cell surface (min to −0.0000001 distance to eukaryotic cell membrane). This new classification was exported as a new ‘intracellular bacterial cell’ surface. To assess the number of eukaryotic cells with intracellular bacteria, the number of objects defined by the eukaryotic cell surface mask with internal objects defined by the ‘intracellular bacterial cell’ surface mask was counted. Statistical comparison of the percentage of HCT116 cells with intracellular Fna bacterial cells by Fna clade was carried out by applying a Welch’s t-test using GraphPad Prism v.7.0 software (GraphPad Software).
Cell length and width measurements
Fna C1 and Fna C2 strain cell dimensions were measured using Fiji with the Bioformats Plugin (required to import Leica.lif files). First, the scale of the image was set by going to Analyze, then Set Scale, and then Set 1 mm to equal 9.979 pixels (pixel size 100.21 nm). Measurements were then captured using the freehand straight-line tool from the brightest point on each cell membrane stain. Statistical comparison of cell lengths and cell width by Fna clade was carried out by applying a Welch’s t-test using GraphPad Prism v.7.0 software (GraphPad Software).
RNA sequencing
Fn strains SB010 and KCOM 3764 were grown on FAA plates (Oxoid, Thermo Fisher Scientific) supplemented with 10% DHB (Fisher Scientific). Plates were incubated at 37 °C in a Bactron600 anaerobic chamber (Sheldon Manufacturing) for 2 days. Subsequent lawns were prepared on FAA + 10% DHB plates and incubated at 37 °C in a Bactron600 anaerobic chamber for 2 days. Cells were resuspended in TSB (Becton Dickinson) and standardized to an optical density at 600 nm (OD600nm) of 0.5. The culture was split into triplicates for each condition and incubated under anaerobic conditions at 37 °C for 4 h. The conditions were as follows: TSB broth alone, TSB supplemented with 50 mM 1,2-PD (Fisher Scientific) and 20 nM vitamin B12 (Fisher Scientific) or TSB supplemented with 15 mM EA (Fisher Scientific) and 20 nM vitamin B12, for 4 h at 37 °C under anaerobic conditions. SB010 was further incubated in TSB supplemented with 20 nM vitamin B12 under the same conditions. Cells were pelleted at 8,000 r.p.m. for 5 min and washed once in 1× PBS and pelleted again under the same conditions. Cells were then washed once in RNAlater (Thermo Fisher) and pelleted again, and all supernatant was removed before storage at −80 °C. RNA was extracted using the RNeasy Extraction Kit (Qiagen) for Illumina Stranded RNA library preparation with RiboZero Plus rRNA depletion. RNA library was sequenced to a minimum read count of 12 million paired-end reads.
Mouse model experiments
Multiple intestinal neoplasia (ApcMin+/−) mice were purchased (Jackson Laboratory, strain No. 002020). Female mice aged 6–8 weeks old were used for two experimental trials with three treatment arms each. Mice were randomly assigned to treatment arms. Mice were treated with streptomycin (2 mg ml−1; Sigma Aldrich) in drinking water for 7 days and then treated with 1.5% dextran sodium sulfate (MP Biomedical) in drinking water for 7 days to induce colitis and facilitate colonic tumours. Mice were then supplied with normal water for 24 h before receiving an oral gavage of Fna strains. Treatment arm 1 mice each received a 200 µl volume of PBS vehicle control, arm 2 mice each received 1 × 109 Fna clade 1 (Fna C1) cells in a 200 µl volume, and arm 3 mice each received 1 × 109 Fna clade 2 (Fna C2) cells in a 200 µl volume. The Fna C1 slurry was an equal mix of strains KCOM 3363, KCOM 3764 and SB048, and the Fna C2 slurry was an equal mix of strains SB001, SB010 and KCOM 2763. Strain mixes instead of single-strain representatives were chosen to capture a greater proportion of Fna clade-specific genes. Fna strains were grown on FAA plates (Oxoid, Thermo Fisher Scientific) supplemented with 10% DHB (Fisher Scientific). Plates were incubated at 37 °C in a Bactron600 anaerobic chamber (Sheldon Manufacturing) for 2–3 days. Subsequent lawns were prepared on FAA + 10% DHB plates and incubated at 37 °C in a Bactron600 anaerobic chamber for 2 days. For each Fna strain, cells were resuspended in PBS. Strain mixes were prepared by volume on the basis of OD600nm standardized by each strain’s colony-forming units per millilitre at OD600nm = 1 (Fna C1: KCOM 3363 6.71 × 107, KCOM 3764 7.27 × 107, SB048 1.97 × 108; Fna C2: SB001 7.61 × 107, SB010 5.00 × 108, KCOM 2763 1.82 × 108) for an equal mix of cells from each Fna C1 and each Fna C2 strain. Mice were monitored until the end-point (6 weeks post-gavage) when the mice were 15–17 weeks old. The Fred Hutchinson Cancer Center Animal Care and Use Committee approved all experimental protocols (IACUC PROTO202100004). All animal work complied with relevant ethical guidelines. Mice were housed on a 12-h light/12-h dark cycle with controlled temperature (65–75 °F (about 18–23 °C)) and humidity (40–60%). Maximal tumour size depended on the number of palpable tumours (1 tumour, maximum 2 cm diameter; 2 tumours, maximum 1.5 cm diameter; ≥3 tumours, maximum under veterinary discretion) and these limits were not exceeded. Intestinal sections from all mice (n = 8 per arm) were blindly assessed by pathology for intestinal adenoma load. To assess differences in intestinal adenoma load by treatment arm, P values were calculated by applying a one-way ANOVA using GraphPad Prism v.7.0 software (GraphPad Software).
Intestinal metabolomics analysis
Metabolomic profiling was conducted using ultrahigh-performance liquid chromatography–tandem mass spectrometry by the metabolomics provider Metabolon on intestinal tissue sections from mice from the second mouse study (n = 4). The global discovery panel used by Metabolon includes 5,400+ metabolites in 70 major pathways, including metabolites of both eukaryotic and bacterial origin. Metabolic pathway enrichment analysis was carried out by Metabolon. Further analysis, including partial least squares discriminant analysis on detected metabolites and heat map clustering were carried out on sample-normalized data using MetaboAnalyst70, v.5.
Mouse faecal DNA extraction and quantitative PCR
DNA was extracted from mouse faecal samples using the Zymo Quick-DNA Microprep Kit (Zymo Research) according to the manufacturer’s instructions. A custom TaqMan primer and probe set was used to amplify Fusobacterium genus DNA (Integrated DNA Technologies) as previously described71. The cycle threshold (Ct) values for the Fusobacterium genus were normalized to the input amount of mouse faecal genomic DNA in each reaction and were assayed in at least duplicate in 20-µl reactions containing 1× final concentration TaqMan Universal PCR Master Mix (Applied Biosystems) and the Fusobacterium TaqMan primer and probe, in a 96-well optical PCR plate. A positive control and non-template control were included in each quantitative PCR run. Fusobacterium copy numbers were estimated following the generation of a standard curve with pure Fna C1 and Fna C2 DNA input. Amplification and detection of DNA was carried out with the QuantStudio 3 Real-Time PCR System (Applied Biosystems) using the following reaction conditions: 10 min at 95 °C and 40 cycles of 15 s at 95 °C and 1 min at 60 °C. Ct was calculated using the automated settings (Applied Biosystems). The primer and probe sequences for the TaqMan assay are as follows: Fusobacterium genus forward primer, 5′-AAGCGCGTCTAGGTGGTTATGT-3′; Fusobacterium genus reverse primer, 5′-TGTAGTTCCGCTTACCTCTCCAG-3′; Fusobacterium genus FAM probe, 5′-CAACGCAATACAGAGTTGAGCCCTGCATT-3′.
Biolog PM10 phenotype microarray plates
Biolog PM10 plates and corresponding IF-0a and IF-10b solutions were pre-reduced under anaerobic conditions at 4 °C overnight (AnaeroGen Gas Generating Systems, Oxoid, Thermo Fisher Scientific). Fna strains were grown on FAA plates (Oxoid, Thermo Fisher Scientific) supplemented with 10% DHB (Fisher Scientific). Plates were incubated at 37 °C in a Concept1000 anaerobic chamber (BakerRuskinn) for 24 h. Under these same anaerobic conditions, Fna cells were resuspended in 2 ml of pre-reduced IF-0a and normalized across all samples to OD600nm = 0.179 as recommended by Biolog. The final suspension was prepared by combining 0.75 ml of normalized bacterial suspension with 11.25 ml of mix B (100 ml pre-reduced IF-10b with 1.2 ml dye mix D, and 11.18 ml pre-reduced sterile water) to a final volume of 12 ml. For each PM10 plate well, 100 μl of final suspension was added. The PM10 plate was then equilibrated to aerobic conditions at room temperature for 10 min, and then incubated under anaerobic, hydrogen-free conditions for 24 h at 37 °C (AnaeroGen Gas Generating Systems, Oxoid, Thermo Fisher Scientific). Plates were imaged and absorbance at 590 nm was quantified using a plate reader (Biotek).
Glutaminase assay
Fna strains were grown on FAA plates (Oxoid, Thermo Fisher Scientific) supplemented with 10% DHB (Fisher Scientific) in a Concept1000 anaerobic chamber (BakerRuskinn) at 37 °C for 2 days. Sterile cotton swabs were used to resuspend cells in TSB (Becton Dickinson) supplemented with 2.5% yeast extract (Becton Dickinson) and 0.4 mg ml−1 l-cysteine (Alfa Aesar). Fna strains were grown in liquid culture in a Concept1000 anaerobic chamber (BakerRuskinn) at 37 °C for about 20 h. For each strain, 0.75 ml of culture standardized to OD600nm = 1 was spun down at 7830 r.p.m. The cell pellet was resuspended in 1 ml of Gls solution. Gls solution contains 0.2 g l-glutamine (Sigma Aldrich), 0.01 g bromocresol green (Sigma Aldrich), 18 g sodium chloride (Sigma Aldrich), 0.6 ml Triton X-100 (Sigma Aldrich) and 200 ml deionized water. Gls solution is filter sterilized post pH adjustment to 3.1. For each strain, a 300 μl volume was aliquoted into a conical-bottom 96-well plate in triplicate and incubated anaerobically at 37 °C for 2 h. The plate was spun down for 1 min at 3,000 r.p.m. The supernatant was transferred to a flat-bottom 96-well plate and absorbance at 600 nm was quantified using a plate reader (Biotek).
Acid resistance in simulated gastric fluid
Fna strains were grown on FAA plates (Oxoid, Thermo Fisher Scientific) supplemented with 10% DHB (Fisher Scientific) in a Concept1000 anaerobic chamber (BakerRuskinn) at 37 °C for 1–2 days. The cells were resuspended in 50 ml TSB (Becton Dickinson) supplemented with 2.5% yeast extract (Becton Dickinson) and 0.4 mg ml−1 l-cysteine (Alfa Aesar). The cells were grown in liquid culture in a Concept1000 anaerobic chamber (BakerRuskinn) at 37 °C for 25 h. All strains were standardized to an OD600nm = 1 in 5 ml of supplemented TSB, simulated gastric fluid (Biochemazone) at pH 3 or simulated gastric fluid supplemented with 10 mM glutamate (Sigma Aldrich) at pH 3. Every 10 min, 10 μl of each suspension was spotted on FAA + 10% DHB plates. Plates were incubated anaerobically in a Concept1000 anaerobic chamber (BakerRuskinn) at 37 °C for 3 days.
Patient specimens
All patient tumour tissue included in the analysis was diagnosed colorectal adenocarcinoma. For patient cohort 1, patients signed an informed consent for the collection and analysis of their tumour specimens. The use of patient specimens for this work was approved by the Fred Hutchinson Cancer Center Institutional Review Board under protocol numbers RG 1006552, 1005305, 1006664 and 1006974. Patient age, sex and ethnicity were not selection criteria for specimen acquisition. For microbial culturing efforts, primary CRC tumours that were treatment naive were prioritized. For patient cohort 2, samples from BioProject PRJNA362951 were used.
Bacterial 16S rRNA gene sequencing
DNA was extracted from patient tissue as described previously6 and processed with the ZymoBIOMICS Service - Targeted Metagenomic Sequencing (Zymo Research). Bacterial V3–V4 16S ribosomal RNA gene-targeted sequencing was carried out. The V3–V4 targeting primers have been custom-designed by Zymo Research to provide the best coverage of the 16S gene while maintaining high sensitivity. They are based on the general bacterial 16S rRNA gene primers 341F (CCTACGGGNGGCWGCAG) and 805R (GACTACHVGGGTATCTAATCC), which amplify the V3–V4 region of the 16S rRNA gene. The amplification was carried out at a higher annealing temperature to ensure only bacterial sequences were amplified. An extraction control was included and showed no amplification during the library preparation (run to 42 cycles). The sequencing library was prepared using the AccuBIOME Amplicon Sequencing Kit (Zymo Research), in which PCR reactions were carried out in real-time PCR machines to prevent PCR chimera formation. The amplicon libraries were cleaned up with Zymo Research’s Select-a-Size DNA Clean & Concentrator (>200-base-pair fragments were kept), quantified with TapeStation, normalized and pooled together. The final library was quantified with quantitative PCR and sequenced on an Illumina MiSeq with a v3 reagent kit (600 cycles). The sequencing was carried out with >10% PhiX mix and in paired-end mode. Raw sequence reads were trimmed with Trimmomatic-0.33 (ref. 72). Fna clade-specific amplicon sequence variants were designed by the provider CosmosID. We provided 16S rRNA gene sequences for all Fna C1 and Fna C2 strains. As the 16S sequence of Fna C1 branched closely with Fnv (Extended Data Fig. 2a), we additionally provided the 16S rRNA gene sequences for all Fnv strains, to ensure the specificity of an Fna C1 amplicon sequence variant that would not detect Fnv. A custom SILVA database was generated using these 16S rRNA gene sequences and SILVA 138.1 SSU Ref. NR99 version, and the DADA2 version of the species training set. First, all sequences in the SILVA database that matched with supplied sequences were removed from SILVA. Next, the custom sequences were added into the SILVA database file, in which the species names were appended on the basis of supplied metadata info (Fna C1, Fna C2 or Fnv). Analysis on this database was then run through the nf-core AmpliSeq pipeline, with the parameters --FW_primer CCTACGGGRSGCAGCA, --RV_primer GACTACHVGGGTATCT, --trunc_qmin 20, --trunc_rmin 0.2, --max_ee 6, --min-frequency 1, --picrust, and -- dada_ref_tax_custom.
Meta-analysis of Fna clades in relation to CRC using publicly available shotgun metagenomic samples
To study the association between each Fna clade and CRC, we profiled shotgun stool metagenomic samples from 9 publicly available cohorts (Supplementary Table 22), for a total of 627 patients with CRC and 619 healthy individuals using MetaPhlAn4 (ref. 63; https://github.com/biobakery/biobakery/wiki/metaphlan4) against an Fna clade-specific database generated from our Fna genomes, which are available at the National Centre for Biotechnology Information (NCBI) under the BioProject accession number PRJNA549513. A distinct species-level genome bin (SGB)73 could be identified for each Fn subspecies and Fna clade (Fna C1: SGB6013, Fna C2: SGB6007, Fnn: SGB6011, Fnp: SGB6001, Fnv: SGB6014). Each SGB was associated with the sample condition fitting an ordinary least squares model of the shape: arcsin-squared-root-transformed SGB abundance ~ study condition + C(sex) + age + BMI + sequencing depth of sample. For each model, an adjusted standardized mean difference between the two study conditions was extracted as previously described74: standardized mean difference = (t × (n1 + n2))/(sqrt(n1 + n2) × sqrt(n1 + n2 − 2)), in which t defines the t-score of the corresponding variable, n1 is the number of samples in the zero class, n2 is the number of samples in the one class, and n1 + n2 − 2 are the degrees of freedom for the model. Corresponding standard errors were computed as: s.e. = sqrt(((n1 + n2 − 1)/(n1 + n2 −3)) × (4/(n1 + n2)) × (1 + (((standardized mean difference)2)/8))). Statistical significance was assessed by the two-tailed Wald test. Effect sizes were pooled and analysed using random-effect meta-analysis75 using the Paule–Mandel heterogeneity estimator76. The statistical significance of the meta-analysis was computed as the z-score of the null hypothesis that the average effect is zero75. All P values were corrected using the Benjamini–Yakuteli method.
Mapping of putative eut, pdu and gdar operons in publicly available metagenomic samples
To assess the presence of putative eut, pdu and gdar system operons in patients with CRC compared to healthy individuals, we profiled shotgun stool metagenomic samples from 9 publicly available cohorts (Supplementary Table 22), for a total of 627 patients with CRC and 619 healthy individuals. Metagenomic samples were mapped against the Fna SB010 eut, pdu and gdar operons using Bowtie2 (version 2.4.5, --sensitive parameter)77. Breadth and depth of coverage of each gene in the operons was assessed using the breadth_depth.py script of the CMSeq tool (parameters --minqual 30 --mincov 1)78. Detected genes had a breadth of coverage threshold above 50%. For eut and pdu results, putative operons had a threshold of presence of 90% of eut and pdu genes relative to the Fna SB010 operon structures. For gdar results, putative operons had a threshold of presence of 100% of gdar genes relative to the Fna SB010 operon structure.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Online content
Any methods, additional references, Nature Portfolio reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at 10.1038/s41586-024-07182-w.
Supplementary information
Legends for Supplementary Tables 1–25.
Supplementary Tables 1–25.
Source data
Acknowledgements
This research was supported by the Experimental Histopathology Shared Resource of the Fred Hutch/University of Washington Cancer Consortium (P30 CA015704), the Comparative Medicine Shared Resource of the Fred Hutch/University of Washington Cancer Consortium (P30 CA015704) and the Cellular Imaging Shared Resource of the Fred Hutch/University of Washington Cancer Consortium (P30 CA015704). Scientific Computing Infrastructure at the Fred Hutchinson Cancer Center was funded by the Office of Research Infrastructure Programs grant S10OD028685. Research reported in this publication was supported by the National Institute of Dental and Craniofacial Research of the National Institutes of Health under award numbers R01 DE027850 and R21 DE033533 (both to C.D.J.), the National Cancer Institute under award number R00 CA229984-03 (to S.B.), start-up funds provided by the Fred Hutchinson Cancer Center (to S.B. and C.D.J.), support from the W.M. Keck Research Foundation (to S.B. and C.D.J.), the Washington Research Foundation Postdoctoral Fellowship (to M.Z.-R.) and the Bio & Medical Technology Development Program of the National Research Foundation funded by the Korean government (2013M3A9B8013860 and 2017M3A9B8065844; to J.-K.K.). We thank A. Baryiames and C. Becker for microbiology and cell culture support; M. Stepanovica and A. McGlinchey for analysis support; D. Raftery for discussions; C. Watson and A. Koehne for pathology review; E. Cromwell, S. Masunaga, A. J. Santo, J. Rivera and U. Demirkol for assistance with animal studies; and H. M. Johnston for guidance. The graphics in Figs. 1a, 2f, 4a and 5a–c and Extended Data Figs. 5a,d,g and 6a were created using BioRender.com.
Extended data figures and tables
Author contributions
M.Z.-R., S.B. and C.D.J. designed the study and wrote the paper. S.B., E.S., K.D.L. and A.G.K. processed patient tissue specimens. M.Z.-R., K.D.L., S.C.Y., S.-N.P., Y.K.L., J.-K.K. and S.B. carried out microbial isolation and culture. M.Z.-R., D.S.J., S.L.C. and C.D.J. carried out DNA isolation, genome sequencing and genome assembly. M.Z.-R., S.S.M., H.B., H.W. and D.S.J. carried out pangenomic analyses on genomes and methylomes. A.B.M. and P.M. analysed stool metagenomes and carried out meta-analysis. M.Z.-R. carried out RNA sequencing experiments and analysis. K.D.L. carried out co-culture assays and subsequent microscopy-based cell invasion analysis. M.Z.-R., K.D.L., Y.W. and A.G.K. carried out mouse trials and downstream processing of mouse samples. M.Z.-R. analysed metabolite data. E.F.M. carried out the glutaminase assays and phenotyping using Biolog PM10 plates. M.Z.-R., A.D.W. and S.B. carried out statistical analyses. M.Z.-R., F.E.D., N.S., S.B. and C.D.J. obtained funding and supervised computational and wet lab experiments. All authors read and provided edits to the paper and contributed to the final version.
Peer review
Peer review information
Nature thanks Cynthia Sears and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Data availability
All genomes from this study are available at the NCBI under the BioProject accession number PRJNA549513 and all methylomes are available in the Restriction Enzyme Database (REBASE). Raw sequencing data from RNA-sequencing experiments are available in the NCBI Sequence Read Archive repository under the BioProject accession number PRJNA937266. Raw sequencing data from 16S rRNA sequencing experiments are available in the NCBI Sequence Read Archive repository under the BioProject accession number PRJNA1064180. Source data are provided with this paper.
Competing interests
S.B. has consulted for GlaxoSmithKline and BiomX. C.D.J. has consulted for Series Therapeutics and Azitra. S.B. is an inventor on US patent application no. PCT/US2018/042966, submitted by the Broad Institute and Dana-Farber Cancer Institute, which covers the targeting of Fusobacterium for the treatment of CRC. S.B., C.D.J. and M.Z.-R. are inventors on US patent application no. F053-0188USP1/22-158-US-PSP, submitted by the Fred Hutchinson Cancer Center, which covers the modulation of cancer-associated microbes. K.D.L. is employed by NanoString Technologies at present. The remaining authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors jointly supervised this work: Susan Bullman, Christopher D. Johnston
Contributor Information
Susan Bullman, Email: sbullman@fredhutch.org.
Christopher D. Johnston, Email: johnston@fredhutch.org
Extended data
is available for this paper at 10.1038/s41586-024-07182-w.
Supplementary information
The online version contains supplementary material available at 10.1038/s41586-024-07182-w.
References
- 1.Segata N, et al. Composition of the adult digestive tract bacterial microbiome based on seven mouth surfaces, tonsils, throat and stool samples. Genome Biol. 2012;13:R42. doi: 10.1186/gb-2012-13-6-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kostic AD, et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 2012;22:292–298. doi: 10.1101/gr.126573.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kostic AD, et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe. 2013;14:207–215. doi: 10.1016/j.chom.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Flanagan L, et al. Fusobacterium nucleatum associates with stages of colorectal neoplasia development, colorectal cancer and disease outcome. Eur. J. Clin. Microbiol. Infect. Dis. 2014;33:1381–1390. doi: 10.1007/s10096-014-2081-3. [DOI] [PubMed] [Google Scholar]
- 5.Mima K, et al. Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis. Gut. 2016;65:1973–1980. doi: 10.1136/gutjnl-2015-310101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bullman S, et al. Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science. 2017;358:1443–1448. doi: 10.1126/science.aal5240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu T, et al. Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell. 2017;170:548–563. doi: 10.1016/j.cell.2017.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Serna G, et al. Fusobacterium nucleatum persistence and risk of recurrence after preoperative treatment in locally advanced rectal cancer. Ann. Oncol. 2020;31:1366–1375. doi: 10.1016/j.annonc.2020.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.LaCourse, K. D., Johnston, C. D. & Bullman, S. The relationship between gastrointestinal cancers and the microbiota. Lancet Gastroenterol. Hepatol. 6, 498–509 (2021). [DOI] [PMC free article] [PubMed]
- 10.Galeano Niño JL, et al. Effect of the intratumoral microbiota on spatial and cellular heterogeneity in cancer. Nature. 2022;611:810–817. doi: 10.1038/s41586-022-05435-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.LaCourse KD, et al. The cancer chemotherapeutic 5-fluorouracil is a potent Fusobacterium nucleatum inhibitor and its activity is modified by intratumoral microbiota. Cell Rep. 2022;41:111625. doi: 10.1016/j.celrep.2022.111625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Allen-Vercoe E, Strauss J, Chadee K. Fusobacterium nucleatum: an emerging gut pathogen? Gut Microbes. 2011;2:294–298. doi: 10.4161/gmic.2.5.18603. [DOI] [PubMed] [Google Scholar]
- 13.Manson McGuire A, et al. Evolution of invasion in a diverse set of Fusobacterium species. mBio. 2014;5:e01864-14. doi: 10.1128/mBio.01864-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holt RA, Cochrane K. Tumor potentiating mechanisms of Fusobacterium nucleatum, a multifaceted microbe. Gastroenterology. 2017;152:694–696. doi: 10.1053/j.gastro.2017.01.024. [DOI] [PubMed] [Google Scholar]
- 15.Ponath F, Zhu Y, Cosi V, Vogel J. Expanding the genetic toolkit helps dissect a global stress response in the early-branching species Fusobacterium nucleatum. Proc. Natl Acad. Sci. USA. 2022;119:e2201460119. doi: 10.1073/pnas.2201460119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Queen J, et al. Comparative analysis of colon cancer-derived Fusobacterium nucleatum subspecies: inflammation and colon tumorigenesis in murine models. mBio. 2022;13(1):e02991-21. doi: 10.1128/mbio.02991-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tomkovich S, et al. Locoregional effects of microbiota in a preclinical model of colon carcinogenesis. Cancer Res. 2017;77:2620–2632. doi: 10.1158/0008-5472.CAN-16-3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eid J, et al. Real-time DNA sequencing from single polymerase molecules. Science. 2009;323:133–138. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- 19.Komiya Y, et al. Patients with colorectal cancer have identical strains of Fusobacterium nucleatum in their colorectal cancer and oral cavity. Gut. 2019;68:1335–1337. doi: 10.1136/gutjnl-2018-316661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abed J, et al. Colon cancer-associated Fusobacterium nucleatum may originate from the oral cavity and reach colon tumors via the circulatory system. Front. Cell. Infect. Microbiol. 2020;10:400. doi: 10.3389/fcimb.2020.00400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eren AM, et al. Anvi’o: an advanced analysis and visualization platform for ‘omics data. PeerJ. 2015;3:e1319. doi: 10.7717/peerj.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tettelin, H. et al. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome”. Proc. Natl Acad. Sci. USA102 13950–13955 (2005). [DOI] [PMC free article] [PubMed]
- 23.Tettelin, H. & Medini, D. (eds) The Pangenome: Diversity, Dynamics and Evolution of Genomes (Springer, 2020). [PubMed]
- 24.Ang MY, et al. Comparative genome analysis of Fusobacterium nucleatum. Genome Biol. Evol. 2016;8:2928–2938. doi: 10.1093/gbe/evw199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Horesh G, et al. Different evolutionary trends form the twilight zone of the bacterial pan-genome. Microb. Genomics. 2021;7:000670. doi: 10.1099/mgen.0.000670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shaiber A, et al. Functional and genetic markers of niche partitioning among enigmatic members of the human oral microbiome. Genome Biol. 2020;21:292. doi: 10.1186/s13059-020-02195-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aramaki T, et al. KofamKOALA: KEGG ortholog assignment based on profile HMM and adaptive score threshold. Bioinformatics. 2020;36:2251–2252. doi: 10.1093/bioinformatics/btz859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Borozan I, et al. Molecular and pathology features of colorectal tumors and patient outcomes are associated with Fusobacterium nucleatum and its subspecies animalis. Cancer Epidemiol. Biomarkers Prev. 2022;31:210–220. doi: 10.1158/1055-9965.EPI-21-0463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Strauss J, et al. Invasive potential of gut mucosa-derived Fusobacterium nucleatum positively correlates with IBD status of the host. Inflamm. Bowel Dis. 2011;17:1971–1978. doi: 10.1002/ibd.21606. [DOI] [PubMed] [Google Scholar]
- 30.Kook J-K, et al. Genome-based reclassification of Fusobacterium nucleatum subspecies at the species level. Curr. Microbiol. 2017;74:1137–1147. doi: 10.1007/s00284-017-1296-9. [DOI] [PubMed] [Google Scholar]
- 31.Abed J, et al. Fap2 mediates Fusobacterium nucleatum colorectal adenocarcinoma enrichment by binding to tumor-expressed Gal-GalNAc. Cell Host Microbe. 2016;20:215–225. doi: 10.1016/j.chom.2016.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bachrach G, Rosen G, Bellalou M, Naor R, Sela MN. Identification of a Fusobacterium nucleatum 65 kDa serine protease. Oral Microbiol. Immunol. 2004;19:155–159. doi: 10.1111/j.0902-0055.2004.00132.x. [DOI] [PubMed] [Google Scholar]
- 33.Casasanta MA, et al. A chemical and biological toolbox for Type Vd secretion: characterization of the phospholipase A1 autotransporter FplA from Fusobacterium nucleatum. J. Biol. Chem. 2017;292:20240–20254. doi: 10.1074/jbc.M117.819144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Coppenhagen-Glazer S, et al. Fap2 of Fusobacterium nucleatum is a galactose-inhibitable adhesin involved in coaggregation, cell adhesion, and preterm birth. Infect. Immun. 2015;83:1104–1113. doi: 10.1128/IAI.02838-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaplan CW, et al. Fusobacterium nucleatum apoptosis-inducing outer membrane protein. J. Dent. Res. 2005;84:700–704. doi: 10.1177/154405910508400803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaplan CW, Lux R, Haake SK, Shi W. The Fusobacterium nucleatum outer membrane protein RadD is an arginine-inhibitable adhesin required for inter-species adherence and the structured architecture of multispecies biofilm. Mol. Microbiol. 2009;71:35–47. doi: 10.1111/j.1365-2958.2008.06503.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lima BP, Shi W, Lux R. Identification and characterization of a novel Fusobacterium nucleatum adhesin involved in physical interaction and biofilm formation with Streptococcus gordonii. MicrobiologyOpen. 2017;6:e00444. doi: 10.1002/mbo3.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Han YW, et al. Identification and characterization of a novel adhesin unique to oral fusobacteria. J. Bacteriol. 2005;187:5330–5340. doi: 10.1128/JB.187.15.5330-5340.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu M, et al. FadA from Fusobacterium nucleatum utilizes both secreted and nonsecreted forms for functional oligomerization for attachment and invasion of host cells. J. Biol. Chem. 2007;282:25000–25009. doi: 10.1074/jbc.M611567200. [DOI] [PubMed] [Google Scholar]
- 40.Fardini Y, et al. Fusobacterium nucleatum adhesin FadA binds vascular endothelial cadherin and alters endothelial integrity: VE-cadherin is a novel receptor for F. nucleatum. Mol. Microbiol. 2011;82:1468–1480. doi: 10.1111/j.1365-2958.2011.07905.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rubinstein MR, et al. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/β-catenin signaling via its FadA adhesin. Cell Host Microbe. 2013;14:195–206. doi: 10.1016/j.chom.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Umaña A, et al. Utilizing whole Fusobacterium genomes to identify, correct, and characterize potential virulence protein families. J. Bacteriol. 2019;201:e00273-19. doi: 10.1128/JB.00273-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gursoy UK, Pöllänen M, Könönen E, Uitto V-J. Biofilm formation enhances the oxygen tolerance and invasiveness of Fusobacterium nucleatum in an oral mucosa culture model. J. Periodontol. 2010;81:1084–1091. doi: 10.1902/jop.2010.090664. [DOI] [PubMed] [Google Scholar]
- 44.Kim H-S, et al. Application of rpoB and zinc protease gene for use in molecular discrimination of Fusobacterium nucleatum subspecies. J. Clin. Microbiol. 2010;48:545–553. doi: 10.1128/JCM.01631-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gardner SN, Slezak T, Hall BG. kSNP3.0: SNP detection and phylogenetic analysis of genomes without genome alignment or reference genome. Bioinformatics. 2015;31:2877–2878. doi: 10.1093/bioinformatics/btv271. [DOI] [PubMed] [Google Scholar]
- 46.Richter M, Rosselló-Móra R. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl Acad. Sci. USA. 2009;106:19126–19131. doi: 10.1073/pnas.0906412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Russo E, et al. From adenoma to CRC stages: the oral-gut microbiome axis as a source of potential microbial and metabolic biomarkers of malignancy. Neoplasia. 2023;40:100901. doi: 10.1016/j.neo.2023.100901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Callahan BJ, McMurdie PJ, Holmes SP. Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME J. 2017;11:2639–2643. doi: 10.1038/ismej.2017.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gur C, et al. Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity. 2015;42:344–355. doi: 10.1016/j.immuni.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang DC, Blair KM, Salama NR. Staying in shape: the impact of cell shape on bacterial survival in diverse environments. Microbiol. Mol. Biol. Rev. 2016;80:187–203. doi: 10.1128/MMBR.00031-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gautreau G, et al. PPanGGOLiN: depicting microbial diversity via a partitioned pangenome graph. PLoS Comput. Biol. 2020;16:e1007732. doi: 10.1371/journal.pcbi.1007732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pacheco AR, Sperandio V. Enteric pathogens exploit the microbiota-generated nutritional environment of the gut. Microbiol. Spectr. 2015;3:3.3.09. doi: 10.1128/microbiolspec.MBP-0001-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Biase DD, Pennacchietti E. Glutamate decarboxylase‐dependent acid resistance in orally acquired bacteria: function, distribution and biomedical implications of the gadBC operon. Mol. Microbiol. 2012;86:770–786. doi: 10.1111/mmi.12020. [DOI] [PubMed] [Google Scholar]
- 54.Tanaka T, et al. Dextran sodium sulfate strongly promotes colorectal carcinogenesis in ApcMin/+ mice: inflammatory stimuli by dextran sodium sulfate results in development of multiple colonic neoplasms. Int. J. Cancer. 2006;118:25–34. doi: 10.1002/ijc.21282. [DOI] [PubMed] [Google Scholar]
- 55.Kennedy L, Sandhu JK, Harper M-E, Cuperlovic-Culf M. Role of glutathione in cancer: from mechanisms to therapies. Biomolecules. 2020;10:1429. doi: 10.3390/biom10101429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pompella A, De Tata V, Paolicchi A, Zunino F. Expression of γ-glutamyltransferase in cancer cells and its significance in drug resistance. Biochem. Pharmacol. 2006;71:231–238. doi: 10.1016/j.bcp.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 57.Hanigan, M. H. Gamma-glutamyl transpeptidase. Adv. Cancer Res.122, 103–141 (2014). [DOI] [PMC free article] [PubMed]
- 58.Rao JN, Xiao L, Wang J-Y. Polyamines in gut epithelial renewal and barrier function. Physiology. 2020;35:328–337. doi: 10.1152/physiol.00011.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Morad SAF, Cabot MC. Ceramide-orchestrated signalling in cancer cells. Nat. Rev. Cancer. 2013;13:51–65. doi: 10.1038/nrc3398. [DOI] [PubMed] [Google Scholar]
- 60.Castellarin M, et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012;22:299–306. doi: 10.1101/gr.126516.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dejea CM, et al. Patients with familial adenomatous polyposis harbor colonic biofilms containing tumorigenic bacteria. Science. 2018;359:592–597. doi: 10.1126/science.aah3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Engevik MA, et al. Fusobacterium nucleatum adheres to Clostridioides difficile via the RadD adhesin to enhance biofilm formation in intestinal mucus. Gastroenterology. 2021;160:1301–1314. doi: 10.1053/j.gastro.2020.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Blanco-Miguez, A. et al. Extending and improving metagenomic taxonomic profiling with uncharacterized species with MetaPhlAn 4. Nat. Biotechnol. 41, 1633–1644 (2023). [DOI] [PMC free article] [PubMed]
- 64.Chaumeil P-A, Mussig AJ, Hugenholtz P, Parks DH. GTDB-Tk: a toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics. 2020;36:1925–1927. doi: 10.1093/bioinformatics/btz848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Letunic I, Bork P. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021;49:W293–W296. doi: 10.1093/nar/gkab301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Payne LJ, et al. Identification and classification of antiviral defence systems in bacteria and archaea with PADLOC reveals new system types. Nucleic Acids Res. 2021;49:10868–10878. doi: 10.1093/nar/gkab883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhou Y, Liang Y, Lynch KH, Dennis JJ, Wishart DS. PHAST: a fast phage search tool. Nucleic Acids Res. 2011;39:W347–W352. doi: 10.1093/nar/gkr485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Arndt D, et al. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016;44:W16–W21. doi: 10.1093/nar/gkw387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xia J, Psychogios N, Young N, Wishart DS. MetaboAnalyst: a web server for metabolomic data analysis and interpretation. Nucleic Acids Res. 2009;37:W652–W660. doi: 10.1093/nar/gkp356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Martin FE, Nadkarni MA, Jacques NA, Hunter N. Quantitative microbiological study of human carious dentine by culture and real-time PCR: association of anaerobes with histopathological changes in chronic pulpitis. J. Clin. Microbiol. 2002;40:1698–1704. doi: 10.1128/JCM.40.5.1698-1704.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pasolli E, et al. Extensive unexplored human microbiome diversity revealed by over 150,000 genomes from metagenomes spanning age, geography, and lifestyle. Cell. 2019;176:649–662. doi: 10.1016/j.cell.2019.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nakagawa S, Cuthill IC. Effect size, confidence interval and statistical significance: a practical guide for biologists. Biol. Rev. 2007;82:591–605. doi: 10.1111/j.1469-185X.2007.00027.x. [DOI] [PubMed] [Google Scholar]
- 75.Borenstein, M., Hedges, L. V., Higgins, J. P. T & Rothstein H. R. Introduction to Meta-Analysis (Wiley, 2021).
- 76.Veroniki AA, et al. Methods to estimate the between‐study variance and its uncertainty in meta‐analysis. Res. Synth. Methods. 2016;7:55–79. doi: 10.1002/jrsm.1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zolfo M, et al. Detecting contamination in viromes using ViromeQC. Nat. Biotechnol. 2019;37:1403–1408. doi: 10.1038/s41587-019-0334-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Legends for Supplementary Tables 1–25.
Supplementary Tables 1–25.
Data Availability Statement
All genomes from this study are available at the NCBI under the BioProject accession number PRJNA549513 and all methylomes are available in the Restriction Enzyme Database (REBASE). Raw sequencing data from RNA-sequencing experiments are available in the NCBI Sequence Read Archive repository under the BioProject accession number PRJNA937266. Raw sequencing data from 16S rRNA sequencing experiments are available in the NCBI Sequence Read Archive repository under the BioProject accession number PRJNA1064180. Source data are provided with this paper.