Abstract
The phylogenetically conserved catalytic core domain of human immunodeficiency virus type 1 (HIV-1) integrase contains elements necessary for specific recognition of viral and target DNA features. In order to identify specific amino acids that determine substrate specificity, we mutagenized phylogenetically conserved residues that were located in close proximity to the active-site residues in the crystal structure of the isolated catalytic core domain of HIV-1 integrase. Residues composing the phylogenetically conserved DD(35)E active-site motif were also mutagenized. Purified mutant proteins were evaluated for their ability to recognize the phylogenetically conserved CA/TG base pairs near the viral DNA ends and the unpaired dinucleotide at the 5′ end of the viral DNA, using disintegration substrates. Our findings suggest that specificity for the conserved A/T base pair depends on the active-site residue E152. The phenotype of IN(Q148L) suggested that Q148 may be involved in interactions with the 5′ dinucleotide of the viral DNA end. The activities of some of the proteins with mutations in residues in close proximity to the active-site aspartic and glutamic acids were salt sensitive, suggesting that these mutations disrupted interactions with DNA.
Integrase is the retroviral protein responsible for inserting a double-stranded DNA copy of the viral genome into the host chromosome. To accomplish this essential step in viral replication, integrase catalyzes two chemical reactions. In the first chemical step, a dinucleotide is cleaved from the 3′ ends of the newly synthesized viral DNA. The newly exposed hydroxyls on the 3′ ends are used in the second chemical step to attack phosphodiester bonds on opposite strands of the target DNA, joining the 3′ viral DNA ends to the target DNA. The gapped structure that results is repaired by an essentially uncharacterized process. The virally encoded integrase protein and phylogenetically conserved sequences present at the ends of the double-stranded DNA genome are required for integration.
In vitro assays have been developed to allow detailed analysis of integrase-catalyzed reactions. A synthetic oligonucleotide model of the viral DNA end can serve as a substrate for 3′-end processing and integration (16). In vitro, integrase can also catalyze the reverse of integration, or disintegration (15). The substrate for this reaction resembles an intermediate in the integration reaction: a single viral DNA end joined to target DNA. Integrase can catalyze a cleavage-ligation reaction on this substrate in which the 3′ hydroxyl on target DNA 5′ to the site of joining is used as a nucleophile to attack the junction between the viral DNA and target DNA 3′ to the site of joining. The products are a continuous target DNA strand and a free viral DNA end. Disintegration substrates have proven to be extremely useful for determining the catalytic requirements of integrase and for analyzing mutant integrase proteins that have defects in requirements for the forward reaction (11, 27, 37, 43, 44, 69). End processing, integration, and disintegration all require a divalent metal ion but no exogenous energy source (5, 15).
Mutational analysis has led to the definition of three functional domains of integrases. The N terminus of integrase contains a phylogenetically conserved zinc finger motif and binds zinc in vitro (8, 11, 72). Zinc binding promotes tetramerization of human immunodeficiency virus type 1 (HIV-1) integrase and enhances end-processing and integration activities (72). The enzymatic properties of proteins with mutations in the conserved histidine or cysteine pairs, or with a deletion of this domain, suggest that this domain may be involved in integrase-integrase and/or integrase-substrate interactions (26, 35, 37, 69, 72). The C terminus of integrase possesses sequence- and metal ion-independent DNA binding activity (28, 55, 70). Biochemical methods have demonstrated that this isolated domain exists as a dimer in solution (1, 46), and in the case of HIV-1 integrase, the solution structure has been solved as a dimer (24, 46). The core domains of all retroviral integrases contain a phylogenetically conserved DD(35)E motif. Mutations in any of the three acidic residues composing this motif have parallel detrimental effects on 3′-end-processing, integration, and disintegration activities, arguing for the existence of a single active site that is directly involved in catalysis of all three reactions (20, 27, 39, 44, 67). The proximity of these residues in the crystal structures of the avian sarcoma virus (ASV) and HIV-1 integrase core domains supports the hypothesis that they form a catalytic center (6, 7, 22, 23). This isolated domain was crystallized as a dimer with an extensive hydrophobic interface in the case of both HIV-1 and ASV integrase. Biochemical methods have demonstrated that this domain exists as a dimer in solution (34, 36). Upon soaking the ASV integrase core domain crystal in Mg2+ (or Mn2+), an Mg2+ (or Mn2+) ion was bound by the two aspartic acid residues, suggesting that these residues coordinate a divalent metal ion required for catalysis (7). The core domain is the most conserved of the three functional domains of integrase, having some amino acid homology, but much more striking structural homology, with the RNase H domain of HIV-1 reverse transcriptase, Escherichia coli RNase H, Mu transposase, and the Holliday junction resolvase RuvC (for a review, see reference 57), all of which are proteins that catalyze substitution reactions on phosphodiester bonds.
We were interested in identifying amino acids that interact with DNA substrates and control the specificity of the chemical reactions catalyzed by integrase. Specificity for certain substrate features certainly exists. (i) In vivo, mutations in viral DNA end sequences, especially the phylogenetically invariant CA/TG dinucleotide at the viral DNA end, result in a replication-incompetent virus (58). Furthermore, mutations in these base pairs in model viral DNA substrates result in up to a 100-fold reduction in end processing (9, 16, 40, 43, 61, 63, 64, 66). (ii) Viral DNA end substrates that lack the terminal two nucleotides on the 5′ end are not joined processively after 3′ end processing in vitro, indicating that this dinucleotide is critical for stable enzyme-viral DNA end interactions (10, 25). (iii) Integrase prefers to integrate into bent regions on target DNA (3, 49, 51–53). The core domain influences the target site preference of the integration reaction (38, 50, 65). The determinants of integrase’s substrate specificity have proven elusive. The finding that the isolated HIV-1 integrase core domain could recognize key features of both viral and target DNAs led us to focus our attention on this domain (31). The three-dimensional information from the crystal structure of the HIV-1 integrase core domain (22, 23) in conjunction with homology alignments of related integrases allowed us to identify conserved residues near the DD(35)E motif that could potentially be involved in substrate interactions. Using site-directed mutagenesis, we evaluated 13 residues for participation in specificity: 10 near the DD(35)E motif, as well as the 3 acidic residues themselves. More than one substitution was made at most positions. The specificities of these mutant enzymes for viral and target DNAs were evaluated by using in vitro assays.
MATERIALS AND METHODS
Construction of mutant integrase genes.
Mutant integrase genes were constructed in a synthetic integrase gene (GenBank accession no. AF029884) in a pUC19 vector. The nearest two unique restriction sites flanking the site to be mutagenized were used to make a replacement with duplex oligonucleotides containing the appropriate ends and the mutation. After transformation into DH5α electrocompetent cells, plasmid DNA was prepared from single colonies. The region containing the replacement and its junctions was sequenced, and a larger portion of the gene containing the mutation was subcloned into a vector containing the entire synthetic gene under the control of the T7 promoter. This parent gene contained an F185K mutation and an N-terminal hexahistidine tag to facilitate purification of the mutant proteins. All of the mutant proteins had an N-terminal hexahistidine tag. The only protein that lacked the F185K mutation was IN(D116N). After sequencing to ensure that the appropriate mutation had been transferred to the expression construct, the construct was transformed into the BL21 expression strain.
Integrase expression.
Single colonies were tested for expression of integrase by growing them to late log phase in Luria broth (LB) and then inducing integrase expression by the addition of 0.25 mM isopropyl-β-d-thiogalactopyranoside (IPTG). A sample of the culture before induction and after 3 h of induction at 37°C was boiled in sodium dodecyl sulfate protein gel loading buffer for 3 min and electrophoresed on a sodium dodecyl sulfate–10% polyacrylamide gel. Western analysis was performed on nitrocellulose electroblots of these gels by using polyclonal serum from an HIV-1-infected patient as the primary antibody and either horseradish peroxidase- or alkaline phosphatase-conjugated goat anti-human antibody (Bio-Rad) as the secondary antibody. Cultures expressing integrase were frozen at −80°C in 50% glycerol.
Large-scale growth for purification.
Integrase-expressing strains were streaked from the glycerol stock onto LB plates containing 50 μg of ampicillin per ml. An individual colony was used to inoculate 500 ml of LB containing 50 μg of ampicillin per ml. After overnight growth at 37°C, 250 ml of this culture was added to 2 liters of LB containing 50 μg of ampicillin per ml. The culture was grown at 30°C for 3 h, and then IPTG was added to a final concentration of 0.4 mM. After 3 h of induction, the culture was centrifuged for 20 min at 5,000 × g at 4°C to pellet the bacteria. Cell pellets were frozen at −20°C.
Integrase purification.
Bacterial pellets were thawed on ice. Pellets were resuspended in 50 ml of lysis buffer (20 mM Tris [pH 8.0], 0.1 mM EDTA, 2 mM β-mercaptoethanol, 0.5 M NaCl, 5 mM imidazole, and 0.2 mg of lysozyme per ml). The resuspended pellets were sonicated on ice for 25 s five times at intervals of 2 min and then incubated on ice for 30 min. This lysate was centrifuged in a Sorvall centrifuge for 45 min at 39,000 × g in an SS-34 Sorvall rotor at 4°C. The pellet was Dounce homogenized 30 times in TNM (20 mM Tris [pH 8.0], 1 M NaCl, 2 mM β-mercaptoethanol) containing 5 mM imidazole. The resuspended pellet was then stirred at 4°C for 1 h. The mixture was centrifuged in a Beckman ultracentrifuge for 1 h at 85,500 × g in a Beckman SW-28 swinging-bucket rotor at 4°C. The supernatant was gravity loaded onto a 1-ml Ni2+-nitrilotriacetic acid agarose (Ni-NTA resin; Qiagen) column preequilibrated with TNM containing 5 mM imidazole. The column was then washed with 20 column volumes of TNM containing 5 mM imidazole, followed by 20 column volumes of TNM containing 40 mM imidazole. Integrase was eluted with a step gradient. The Bradford protein assay (Bio-Rad) was used to determine that integrase eluted in the fractions containing 100 and 200 mM imidazole. Peak fractions were pooled, diluted 1:10 with TNM buffer, and gravity loaded onto a second 0.5-ml Ni-NTA column. Washing and elution were carried out as described for the 1-ml column. Peak fractions were pooled, 3-[(3-cholamidopropyl)-dimethyl-ammonio]-1-propanesulfonate (CHAPS) was added to a final concentration of 10 mM, and the fractions were dialyzed against buffer containing 20 mM HEPES (pH 7.5), 10 mM dithiothreitol (DTT), 300 mM NaCl, 10 mM CHAPS, and 10% glycerol. The protein concentration was determined by the Bradford method with purified integrase as a standard. The concentration of the integrase used as a standard had been determined by amino acid analysis. Aliquots were frozen in liquid nitrogen and stored at −80°C. The concentrations of integrase cited throughout this report are concentrations of integrase promoters.
Construction of substrates.
All oligonucleotides were purchased from Operon Technologies, Inc. (Emeryville, Calif.). Oligonucleotides were purified by electrophoresis on a 15 or 20% denaturing polyacrylamide gel prior to use. T4 polynucleotide kinase (New England Biolabs) and [γ-32P]ATP (Amersham; 3,000 Ci/mmol) were used for all 5′ end labelling. Ymer disintegration substrates were made from four oligonucleotides annealed together to form the structure of one viral DNA end joined to target DNA; a Ymer substrate contained 19 bp of viral DNA joined to 30 bp of target DNA. Dumbbell disintegration substrates consisted of a single oligonucleotide folded upon itself to form the structure of one viral DNA end joined to target DNA in which the viral DNA portion was 5 bp in length and the target DNA portion was 10 bp in length. Disintegration substrates were 5′ end labelled and annealed as described by Chow et al. (15). For mutant Ymer disintegration substrates, oligonucleotides with the indicated sequence were used in the construction. Viral DNA end substrates, both blunt ended and prerecessed, were 5′ end labelled and annealed as described by Ellison and Brown (25).
Activity assays.
Activity assays were performed under a variety of conditions. For Table 1 and Fig. 2, 3′ end processing was measured by using a blunt-ended U5 viral DNA end substrate. The reaction mixtures included 300 nM integrase and 50 nM viral DNA end substrate incubated in a solution containing 7.5 mM MnCl2, 12 mM DTT, 50 μg of bovine serum albumin (BSA) per ml, 2 mM CHAPS, 2% glycerol, 22 mM HEPES (pH 7.5), and either 25 or 90 mM NaCl, as indicated. All end-processed and integrated products were included in the quantitation of 3′-end-processing activity. The amount of product for each reaction analyzed was divided by the amount of product for a parallel reaction containing IN(F185K) and is therefore expressed as a percentage in Table 1. The 3′-end-processing reaction mixtures were incubated for 30 min, the integration reaction mixtures were incubated for 10 min, and the disintegration reaction mixtures were incubated for 4 min, all at 37°C. All incubation times were determined to be in the linear range for IN(F185K) in kinetic experiments performed under similar conditions. A prerecessed viral DNA substrate was used in the integration reactions, and a Ymer disintegration substrate was used in the disintegration reactions. The disintegration reaction conditions were identical to those described above except that 150 mM integrase and 50 mM Ymer substrate were used and the final NaCl concentration was 50 mM. Reactions with a Ymer substrate without the 5′ dinucleotide were done in parallel (data not shown). Reactions were stopped by the addition of an equal volume of formamide loading buffer (95% formamide, 50 mM EDTA, 0.1% bromophenol blue, and 0.1% xylene cyanol). Samples were heated to 90°C for 2 min before electrophoresis on a 15% denaturing polyacrylamide gel. Substrate and product were quantitated with a Molecular Dynamics Phosphorimager.
TABLE 1.
Diminished activities and enhanced salt sensitivities of selected mutants
| IN mutanta | Activity [% of IN(F185K) activity]
|
||||
|---|---|---|---|---|---|
| 3′ end processing
|
Integration
|
Disintegration | |||
| 25 mM NaCl | 90 mM NaCl | 25 mM NaCl | 90 mM NaCl | ||
| Q62A | 4.5 | 2.2b | 7.6 | 2.7b | 7.6 |
| Q62N | 19 | 17 | 11 | 7.7 | 6.3 |
| T66A | 22 | 13 | 53 | 28 | 49 |
| H67S | 140 | 118 | 133 | 105 | 52 |
| E92A | 25c | 21 | 24c | 14 | 49 |
| E92N | 24c | 21 | 26c | 15 | 61 |
| N117S | 30 | 9.1b | 35 | 8.2b | 39 |
| N117Q | 40 | 31 | 59 | 48 | 73 |
| N120S | 165 | 116 | 148 | 79b | 120 |
| N120Q | 103 | 50b | 78 | 28b | 102 |
| Q148L | NDd | 17 | ND | 47 | 17 |
| N155E | 3.8e | 3.1e | 7.9 | 6.4 | 5.6 |
| N155K | 2.2e | 3.1e | 8.1 | 6.8 | 2.8 |
| N155L | 18 | 10 | 16 | 7.0b | 12 |
| K156E | 11 | 3.0b | 9.0 | 1.3b | 7.9 |
| K159N | 21 | 8.7b | 23 | 6.9b | 17 |
| K159S | 26 | 9.5b | 36 | 9.1b | 22 |
| F185K | 100 | 100 | 100 | 100 | 100 |
All mutant proteins also contained the IN(F185K) mutation.
Salt sensitive.
Reaction mixtures contained 45 mM (not 25 mM) NaCl.
ND, not determined.
IN(N155E) and IN(N155K) preparations had nuclease contamination that prevented accurate quantitation of the 3′-end-processed products; only the amounts of integrated products are given in this case, as a percentage of the integrated products in a matched reaction with IN(F185K). In all other cases, all end-processing and integration products are included in the calculation of end-processing activity. Integration could be detected with IN(N155E) and IN(N155K) by using a blunt-ended viral end substrate, indicating that these mutant proteins could, in fact, carry out end processing.
FIG. 2.
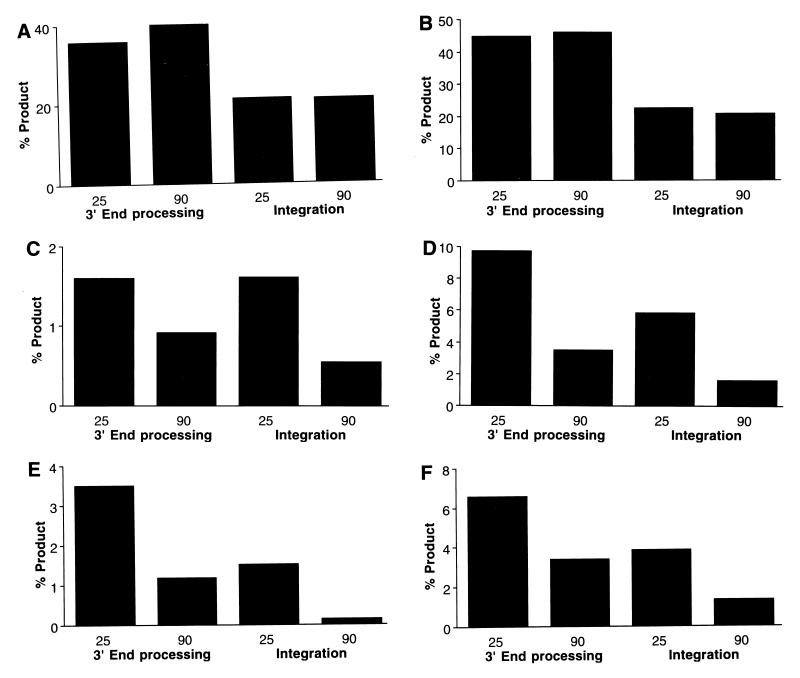
Salt sensitivities of mutant integrase proteins. The end-processing and integration activities of mutant integrase proteins were measured in buffers containing 25 or 90 mM NaCl. The y axis represents the total amount of substrate that had been converted to product. IN(F185K) and IN(H67S) (A and B, respectively) had similar activities at both tested NaCl concentrations. However, the activities of a subset of the mutant integrase proteins were lower at the higher NaCl concentration: IN(Q62A) (C), IN(N117S) (D), IN(K156E) (E), and IN(K159N) (F). Other mutants with similar salt sensitivities are noted in the text and in Table 1.
The stable-complex assay was performed as follows. First, 50 nM blunt-ended U5 viral DNA end substrate was incubated with 300 nM integrase in 10 μl for 5 min at 37°C in a solution containing 26 mM HEPES (pH 7.5), 90 mM NaCl, 7.5 mM MnCl2, 3 mM DTT, 3 mM CHAPS, 3% glycerol, and 50 μg of BSA per ml. A 3-μl aliquot was removed from the reaction mixture and added to an equal volume of formamide loading buffer. Three microliters of either TEN (10 mM Tris [pH 8.0], 1 mM EDTA, 50 mM NaCl) or TEN containing 15 pmol of a nonviral duplex competitor oligonucleotide (A227/A228) (the sequence is reported in reference 19) per μl was then added to the remaining mix, and the incubation was continued for 25 min at 37°C. To assess only the integration activity under these conditions, reactions with the prerecessed U5 viral DNA end substrate were performed in parallel without the addition of target DNA and stopped with formamide loading buffer after incubation at 37°C for 10 min. Disintegration assays were also performed in parallel with 150 nM integrase, 50 nM Ymer disintegration substrate, and 50 mM NaCl. These reaction mixtures were incubated for 5 min at 37°C before the reactions were stopped with an equal volume of formamide loading buffer.
IN(Q148L) and IN(F185K) were assayed for turnover by using dumbbell disintegration substrates with or without the 5′ dinucleotide (14, 15). Duplicate reactions were performed with the following conditions: 1.5 μM dumbbell disintegration substrate, 150 nM integrase, 60 mM NaCl, 7.5 mM MnCl2, 12 mM DTT, 50 μg of BSA per ml, 2 mM CHAPS, 2% glycerol, and 22 mM HEPES, pH 7.5. Aliquots were removed from the reaction mixtures at 5, 10, 15, 20, 30, 45, 60, 90, and 120 min, and reactions were stopped by the addition of an equal volume of formamide loading buffer. Samples were heated to 90°C for 2 min before electrophoresis on a 20% denaturing polyacrylamide gel for 1.5 h at 65 W. Results for duplicate samples were quantitated with a Phosphorimager and averaged.
For the mutants with mutations in the aspartic and glutamic acid residues of the DD(35)E motif, the reactions listed in Table 2 were performed at pH 7.5 under the following conditions: 60 mM NaCl, 7.5 mM MnCl2, 12 mM DTT, 50 μg of BSA per ml, 2 mM CHAPS, 2% glycerol, and 22 mM HEPES, pH 7.5. Integrase (150 nM) and Ymer disintegration substrate (50 nM) were incubated together for 60 min in the case of the mutants assayed at pH 7.5. A 5-min time point from the reaction with IN(F185K) was used to predict the amount of conversion in a linear reaction at 60 min. The activities of the mutants given were calculated by dividing the amount of product in the reaction by the amount of product in a parallel reaction with IN(F185K). Reactions listed in the lower part of Table 2 were performed under the following conditions: 75 nM integrase, 50 nM Ymer disintegration substrate, 90 mM NaCl, 7.5 mM MnCl2, 3 mM DTT, 3 mM CHAPS, 3% glycerol, 6 mM HEPES (pH 7.5), and 30 mM Tris (pH 8.5). The reference activity of IN(F185K) under these conditions was measured in the same manner as described above. The mutants assayed at pH 8.5 were allowed to incubate for 30 min at 37°C. Samples were analyzed as described for the other mutants.
TABLE 2.
Activities of D64, D116, and E152 mutantsa
| IN mutant | Disintegration [% of IN(F185K) activity] |
|---|---|
| D64R | 0 |
| D116E | 2 |
| D116N | 0.001 |
| D116R | 0.025 |
| E152D | 0.175 |
| E152H | 0.018 |
| D64RD116R | 0 |
| F185K | 100 |
| D64C | 1.7 |
| D116C | 18 |
| E152C | 0.001 |
| F185K | 100 |
All reactions were measured in the linear phase of their kinetics. The reactions in the top part of the table were performed with 150 nM integrase and 100 nM Ymer substrate at pH 7.5. The reactions in the bottom part of the table were performed with 70 nM integrase and 100 nM Ymer substrate at pH 8.5.
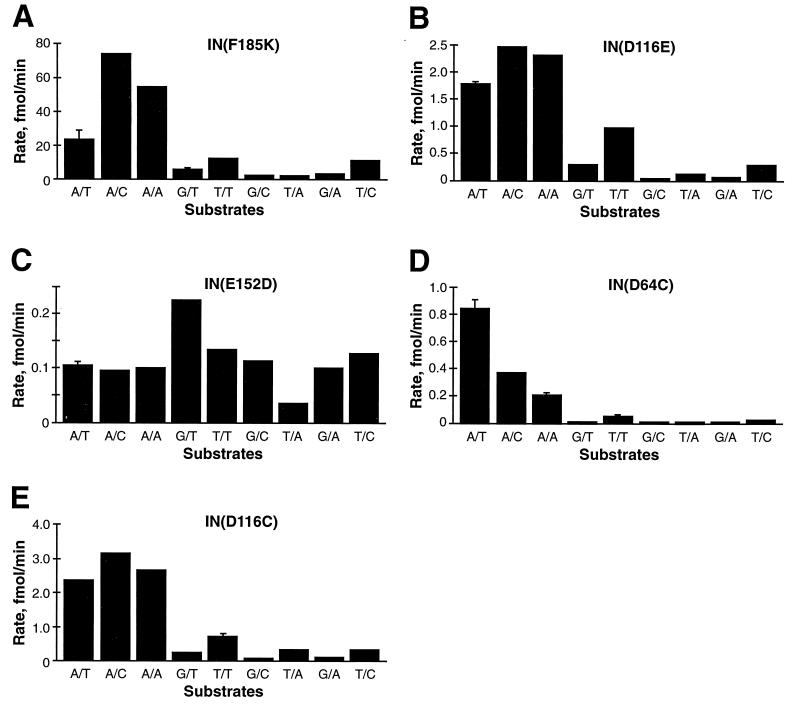
Rates for IN(E152D), IN(D116E), and IN(F185K) were determined at pH 7.5 with the following conditions: 150 nM integrase, 100 nM Ymer disintegration substrate, 60 mM NaCl, 5 mM MnCl2, 12 mM DTT, 50 μg of BSA per ml, 2 mM CHAPS, 2% glycerol, and 22 mM HEPES, pH 7.5. Aliquots were removed from the reaction mixture at timed intervals and added to an equal volume of formamide loading buffer, and the amounts of substrate and product were quantitated with a Phosphorimager. Rates were calculated by plotting product yield on the y axis versus time on the x axis and obtaining the slope of the line by a least-squares fit. Five time points were used for each rate determination. Time intervals were chosen such that the amount of substrate converted to product was ≤20%. For each protein the rate was measured in triplicate for at least one substrate in order to determine a standard deviation. Although the standard deviation within a single experiment was relatively small, the absolute rates measured in different experiments varied by as much as 2.5-fold. However, the specificity (i.e., relative rates measured for different substrates) was always fundamentally similar. The rates given in Fig. 4 were the result of parallel assays performed in a single experiment for each protein.
FIG. 4.
Sensitivities of mutant integrase proteins to substitutions for the conserved subterminal A/T base pair of the viral DNA end. The rate of disintegration was calculated by plotting the amount of substrate converted to product at each time point and determining the slope of the line by the least-squares method. The y axis represents the calculated rates. In a few cases where a standard deviation was calculated by determining rates for a substrate in triplicate, this is indicated by an error bar. The base pair substituted at the position of the phylogenetically conserved subterminal A/T base pair is indicated below each bar. Rates given for IN(D64C) and IN(D116C) were determined at pH 8.5; rates given for IN(F185K), IN(D116E), and E(152D) were determined at pH 7.5. Rates were also determined for IN(F185K) at pH 8.5 and were approximately twofold lower for each substrate (data not shown).
Rates for IN(D64C), IN(D116C), and IN(F185K) were determined at pH 8.5 under the following conditions: 75 nM integrase, 50 nM Ymer disintegration substrate, 98 mM NaCl, 5 mM MnCl2, 3 mM DTT, 3 mM CHAPS, 3% glycerol, 6 mM HEPES (pH 7.5), and 30 mM Tris (pH 8.5). Rates were calculated as described for reactions performed at pH 7.5.
RESULTS
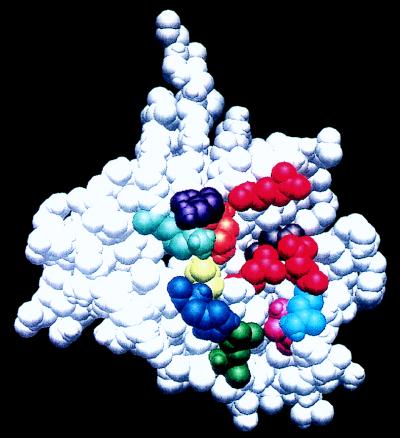
The premise of these experiments was that phylogenetically conserved residues near the active site of HIV-1 integrase were likely candidates to participate in recognizing critical substrate features. We chose to mutate the amino acids shown mapped onto the structure of the HIV-1 integrase core domain in Fig. 1. The three acidic residues of the phylogenetically conserved DD(35)E motif are depicted in red. Nine of the 10 residues mutagenized are shown; the 10th, Q148, is not resolved in this structure but is predicted to be near the DD(35)E residues. The selection of residues potentially involved in specificity was based on the existing crystal structure for HIV-1 integrase (22, 23); any conformational change that occurs upon DNA or divalent metal ion binding might cause us to miss residues whose position relative to the DD(35)E motif is affected by such a change. Moreover, potential interactions with other integrase promoters in a multimeric complex could not be predicted in this way.
FIG. 1.
Amino acids targeted for mutagenesis. The three-dimensional structure of the HIV-1 integrase catalytic core domain is shown (22, 23). The amino acids chosen for mutagenesis are in color: Q62 is grey, T66 is yellow, H67 is dark blue, E92 is green, N117 is light blue, N120 is magenta, N155 is orange, K156 is purple, and K159 is light green. The residues that define the DD(35)E active-site motif (D64, D116, and E152) are red. A Q148 mutant was also analyzed in this work; this residue has not been resolved in the crystal structure. This picture was generated by using the program GRASP.
All mutations were made in the context of full-length integrase with the F185K mutation and an N-terminal hexahistidine tag. Mutant enzymes were overexpressed in E. coli and purified by Ni2+ affinity chromatography. Mutant proteins were assayed for 3′-end-processing, integration, and disintegration activities under a variety of conditions. Parameters varied included protein concentration, substrate concentration, and salt concentration.
3′-end-processing, integration, and disintegration activities of mutants.
Table 1 summarizes the activities of the mutant integrase proteins relative to those of the F185K parent protein, which is considered to be wild type in vitro (35). The only tested position at which substitution did not appear to have an effect on catalysis of 3′ end processing, integration, or disintegration under any conditions was H67. Serine was the only residue substituted for histidine at this position. This residue is therefore probably not involved in enzyme-substrate interactions (or protein-protein interactions) that are critical for catalysis. Substitutions at position E92 resulted in mutant proteins that displayed parallel reductions in end-processing, integration, and disintegration activities. The identity of the substitution (A or N) did not have a large effect on the phenotype of the mutant.
Several of the mutations increased the sensitivity of integrase to the ionic strength of the reaction buffer. IN(F185K) had virtually identical activities in solutions containing 25 and 90 mM NaCl (Fig. 2A). However, the integration and end-processing activities of IN(Q62A), IN(N117S), IN(K156E), IN(K159N), and IN(K159S) were reduced by at least 50% when the salt concentration was raised from 25 to 90 mM NaCl (Table 1). Figure 2 depicts the end-processing and integration activities of wild-type and some mutant integrases in reaction mixtures containing 25 or 90 mM NaCl. IN(H67S) (Fig. 2B) was unaffected by the change in ionic strength. The activity profiles for four of the salt-sensitive mutants, i.e., IN(Q62A) (Fig. 2C), IN(N117S) (Fig. 2D), IN(K156E) (Fig. 2E), and IN(K159N) (Fig. 2F), are shown. The integration activities of IN(N120Q), IN(N120S), and IN(N155L) were more affected by the increase in ionic strength than were the 3′-end-processing activities. The enhanced salt sensitivity of several of these mutants was dependent on the identity of the replaced amino acid. Substitutions at positions Q62 (N), N117 (Q), and N155 (E or K) resulted in mutant proteins that did not display salt-sensitive activity.
IN(Q148L) displayed an unusual phenotype; the 3′-end-processing activity of this mutant protein [17% of that of IN(F185K)] was more impaired than its integration activity [47% of that of IN(F185K)] as measured on a prerecessed viral DNA end substrate. Furthermore, the yield of integration products following 3′ end processing by IN(Q148L) was disproportionately low.
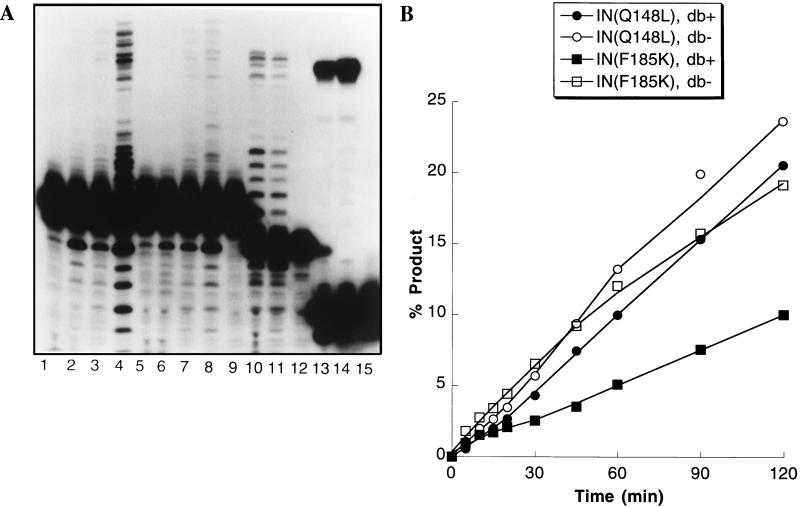
The 3′-end-processing and integration reactions normally proceed without an intervening release of the bound viral DNA substrate. Thus, the stable complex between integrase and the viral DNA can be maintained in the presence of a molar excess of competitor DNA, allowing the 3′-end-processing and integration reactions to proceed processively. In contrast, IN(Q148L) was unable to interact stably with a viral DNA end when an 80-fold molar excess of nonviral competitor DNA was added after preincubation [Fig. 3A; compare the difference in end-processed product generated by IN(Q147L) in the reactions analyzed in lanes 2 and 6 to the difference in end-processed product generated by IN(F185K) in the reactions analyzed in lanes 4 and 8].
FIG. 3.
(A) IN(Q148L) does not catalyze end processing and integration processively. Mixtures for the reactions analyzed in lanes 1 to 9 each contained a 50 nM concentration of the blunt-ended viral DNA substrate. Samples analyzed in lanes 1 and 5 were taken at 5 min from reaction mixtures containing 300 nM IN(Q148L). Samples analyzed in lanes 3 and 7 were taken at 5 min from reaction mixtures containing 300 nM IN(F185K). After 5 min, either 3 μl of TEN or 3 μl of TEN containing 15 pmol of A227/A228 per μl was added to 7 μl of the reaction mixture, and the incubation was continued for 25 min at 37°C. Samples analyzed in lanes 2 and 6 contain reactions with IN(Q148L) in which 3 μl of TEN or TEN containing 15 pmol of competitor DNA per μl was added, respectively. Samples analyzed in lanes 4 and 8 were taken from reactions with IN(F185K) to which TEN or TEN containing 15 pmol of competitor DNA per μl was added, respectively. Integrase was omitted from the reactions analyzed in lanes 9, 12, and 15. Mixtures for the reactions analyzed in lanes 10 to 12 contained a 50 nM concentration of the prerecessed viral DNA end substrate and were incubated at 37°C for 10 min. Mixtures for the reactions analyzed in lanes 10 and 11 contained 300 nM IN(Q148L) and 300 nM IN(F185K), respectively. Mixtures for the reactions analyzed in lanes 13 to 15 contained 50 nM Ymer disintegration substrate and were incubated at 37°C for 5 min. Mixtures for the reactions analyzed in lanes 13 and 14 contained 150 nM IN(Q148L) and 150 nM IN(F185K), respectively. Integration of 3′-end-processed viral DNA ends by IN(Q148L) is poor in the presence of a molar excess of competitor DNA (compare lanes 2 and 6), whereas the integration of 3′-end-processed viral DNA ends by IN(F185K) proceeds normally under these conditions (compare lanes 4 and 8). (B) Turnover of IN(Q148L) is insensitive to the presence of the 5′ dinucleotide of the viral DNA end. Parallel reactions were performed in duplicate with 1.5 μM substrate and 150 nM integrase. Results from duplicate experiments were averaged. A dumbbell disintegration substrate either with (db+) or without (db−) the 5′ overhang was used. IN(Q148L) and IN(F185K) had approximately the same rates of turnover in reactions with the (db−) substrate. However, while IN(F185K) showed a lower rate of turnover in reactions with the db+ substrate, turnover of IN(Q148L) was unaffected by inclusion of the 5′ overhang.
The terminal dinucleotide at the 5′ ends of viral DNA, which is left unpaired after end processing, has been shown to be critical for the stability of the integrase-viral DNA complex (10, 25). Thus, under multiple-turnover conditions, wild-type integrase has a higher rate of turnover on disintegration substrates that lack the 5′ dinucleotide, presumably because the absence of stabilizing interactions with this dinucleotide facilitates dissociation from the viral DNA product (31). This dinucleotide had little effect on the rate of disintegration by IN(Q148L) under multiple-turnover conditions, while turnover of IN(F185K) was significantly reduced by this dinucleotide, as expected (Fig. 3B). IN(Q148L) did not display any reduction in reintegration of the viral DNA product in disintegration reactions.
The specificities of the mutants cited in Table 1 for the phylogenetically conserved subterminal CA/TG dinucleotide pairs of the viral DNA end did not differ substantially from that of IN(F185K).
Disintegration activities and substrate specificities of mutant integrases with alterations in the putative active-site residues D64, D116, and E152.
We evaluated the effects of several substitutions in each of the three acidic active-site residues: D64, D116, and E152. Mutations in these residues have previously been reported to have parallel detrimental effects on 3′ end processing, integration, and disintegration, reducing catalytic activity by several orders of magnitude (20, 27, 44, 67). This DD(35)E motif is the most phylogenetically conserved feature in retroviral integrases and bacterial transposases; the subterminal CA/TG base pairs in the viral DNA are also universally conserved in retroviruses and are a common feature of bacterial transposons. It therefore seemed possible that the conserved aspartic acid and glutamic acid residues themselves could be responsible in some part for specificity for the CA/TG sequence. In order to measure the substrate specificities of proteins with mutations in the aspartic acid or glutamic acid active-site residues, substitutions that would generate proteins with measurable catalytic activity had to be found. Since the disintegration assay is the most sensitive to very low levels of integrase activity, we used disintegration substrates with mutations in the CA/TG base pairs to assay these mutant proteins for their substrate specificity. Since integrase shows similar specificities for these base pairs in the integration and disintegration reactions (14, 69) and since the orientations of viral DNA and target DNA with respect to the active site are similar for integration and disintegration (32), the interactions mediating specificity for the CA/TG sequence should be similar for both reactions.
The disintegration activities of the D64, D116, and E152 mutants were compared to that of IN(F185K). The results are summarized in Table 2. All of these mutants had some ability to complement, in vitro, a mutant integrase protein lacking the N-terminal 50 amino acids, suggesting that the mutants were folded properly. The mutants listed in the top part of Table 2 were assayed for disintegration activity at pH 7.5. IN(D64C) and IN(D116C) displayed a pH optimum different from that of wild-type integrase, with more disintegration activity at pH 8.5, and thus were assayed at this pH. This shift was presumably due to the higher pKa of cysteine (pKa = 8.5) than of aspartic acid or glutamic acid (pKa = 4.4). IN(F185K) displayed a slight reduction in activity at pH 8.5. IN(D64C), IN(D116C), IN(D116E), and IN(E152D) had sufficient disintegration activity to be analyzed for their specificities for the conserved A/T and C/G base pairs of the viral DNA end.
The results of the specificity analysis with disintegration substrates with mutations at the conserved A/T base pair are presented in Fig. 4. At both the A/T and C/G base pairs (data not shown), eight different base pair substitutions were tested. The substitutions can be categorized into three types: a mutant base paired with a complementary mutant base, a mutant base mispaired with a wild-type base, or a mutant base mispaired with a mutant base. The use of substrates with these three categories of substitutions allowed the effects of structure (matched or mismatched) to be distinguished from the effects of specific base substitution on the measured rate (61). In all cases, with the exception of substrates in which the wild-type A was mispaired with a mutant base, the rate of disintegration catalyzed by IN(F185K) was lower for mutant substrates than for the wild-type substrate (Fig. 4A). The specificity profile of IN(F185K) at pH 7.5 is shown; the specificity profile of IN(F185K) was virtually identical at pH 8.5, but the absolute rates were roughly twofold lower (data not shown). Although the overall rates were lower, the specificity profiles displayed by IN(D116C) and IN(D116E) on disintegration substrates with substitutions in either the A/T or C/G base pair were very similar to that displayed by IN(F185K) (compare Fig. 4A, B, and E). This result implies that D116 does not play a role in the recognition of either of these base pairs.
In contrast to the specificity displayed by IN(F185K), IN(E152D) displayed a marked indifference to mutations in the A/T base pair (compare Fig. 4C and A). IN(E152D) appeared to have the lost the ability to distinguish the wild-type A/T base pair from other paired or mispaired bases at this position in a disintegration substrate. Thus, E152 appears to play a role in the specific recognition of the A/T base pair by the wild-type integrase protein. The rates at which IN(E152D) could catalyze disintegration of substrates with mutations in the C/G base pair were sufficiently low that they could not be accurately measured. However, extended incubations suggested that IN(E152D) retained specificity analogous to that displayed by IN(F185K) for the C/G base pair (data not shown).
The rates determined for IN(D64C) on wild-type and mutant disintegration substrates revealed that the activity of this mutant protein was very sensitive to any change in the conserved A/T (Fig. 4D) or C/G (data not shown) base pair. Interestingly, this mutant catalyzed disintegration of substrates in which the A/T base pair had been changed to A/C or A/A at a rate slightly lower than the rate at which it catalyzed disintegration of the wild-type substrate. This contrasts with the results seen with IN(F185K), which disintegrated these mutant substrates at a rate slightly higher than that observed with a wild-type substrate (compare Fig. 4A and D). In this context, IN(D64C) appeared to have an increased specificity for the wild-type T base. Due to its sensitivity to mutations in the disintegration substrate and its low basal activity, the activity of IN(D64C) on some mutant substrates (A/T changed to G/T, G/C, T/A, or G/A) could be only roughly estimated.
To date, no binding or cross-linking assay has been described in which the effects of mutations in the DNA substrate correlate well with their effects on catalytic activity. Engelman et al. (28) found that IN(D64N), IN(D116N), and IN(E152Q) could all cross-link to a model U5 viral DNA end, indicating that these residues in the wild-type protein did not contribute to the nonspecific DNA binding measured by the cross-linking assay. Using a similar cross-linking assay, Drelich et al. (21) observed a seemingly paradoxical result: IN(D116N) and IN(D116A) did not cross-link to a model U5 viral DNA end, but IN(D64A) and IN(E152A) did, suggesting that D64 and E152 were not involved in interacting with the viral DNA but that D116 was. Several of the active-site mutants listed in Table 2 were analyzed for binding of Ymer disintegration and viral DNA end substrates by using a nitrocellulose filter binding assay. Our results indicated that the active-site mutants tested [including IN(D116N)] all retained DNA binding activity. The level of binding activity measured in this assay had no consistent correlation with the catalytic activity of the protein (data not shown).
DISCUSSION
The catalytic core domain of HIV-1 integrase is responsible for recognition of the subterminal phylogenetically conserved CA/TG dinucleotide pair near the viral DNA end, for the 5′-terminal dinucleotide, and for target DNA adjacent to the site of joining (31). We used model substrates to analyze the substrate specificities of integrase mutants that contained changes in residues near the critical acidic residues of the phylogenetically conserved DD(35)E motif or in these acidic residues themselves. We identified specific residues involved in recognition of the subterminal A/T base pair and the 5′ dinucleotide at the viral DNA ends.
The DD(35)E active-site motif is shared by proteins in a superfamily of polynucleotidyl transferases. The two conserved aspartic acids have been shown to coordinate a divalent metal ion in the crystal structure of the core domain of ASV integrase. Substituting cysteine for one of the aspartic acids in the active site of the related TnsA (59) or MuA (2) transposase changes the metal ion preference from Mg2+ to Mn2+, strongly suggesting that the aspartic acid residue is part of an essential metal ion binding site in both enzymes. Substituting cysteine for D64 or D116 of HIV-1 integrase (the residues analogous to the aspartic acids coordinating a metal ion in the active site of ASV integrase) resulted in mutant proteins with an altered pH optimum and in 50- or 5-fold-less catalytic activity than in the wild-type protein, respectively. Thus, the ability of each of these side chains alone to provide only a single coordination site for a metal ion is sufficient to coordinate a divalent metal ion in the active site of integrase. In the case of ASV integrase, only one of the two carboxyl oxygens of each of the active-site aspartic acids coordinates the divalent metal ion (7). Although these substitutions of cysteine for aspartate might have been expected to alter the metal ion preference of integrase, sulfur interacts preferentially with Mn2+ over Mg2+, and wild-type HIV-1 integrase already has an essentially absolute preference for Mn2+ as a cofactor for disintegration in vitro.
In contrast to the aspartic acid residues, the glutamic acid residue of the DD(35)E motif of HIV-1 integrase was relatively intolerant of substitutions. Substitution of cysteine at position E152 resulted in a protein with 105-fold-lower disintegration activity than IN(F185K) under substrate-excess conditions, indicating that this unnatural side chain was not compatible with catalysis. The most conservative substitution, that of aspartic acid, for E152 was the only substitution that resulted in a mutant protein with enough activity to allow catalytic rates to be measured for disintegration substrates with mutations in the subterminal viral DNA A/T base pair. In contrast to IN(F185K) and other active-site mutants, IN(E152D) displayed a marked indifference to mutations in the phylogenetically conserved viral A/T base pair. IN(E152D) did not display a gross DNA binding defect (data not shown), arguing that this altered substrate specificity was achieved downstream of DNA binding, perhaps in a subsequent step of docking into the active site. This altered specificity, along with the intolerance to substitution, suggests that in addition to, or instead of, playing a direct role in the chemistry of the reaction, E152 may play a role in the recognition of the A/T base pair that is essential for efficient catalysis. E152 is located in a loop that demonstrates flexibility in the crystal structures of many related proteins, including ASV and HIV-1 integrases and MuA. The finding that a mutation in E152 affects recognition of the A/T base pair suggests that E152 may normally interact with this base pair; the salient effect of this interaction may be to stabilize this peptide in an active conformation. The simplest model for an E152-A/T interaction that stabilizes the geometry of the active site would be a base-specific interaction. However, more indirect interactions, for example, base-specific interactions between other residues and the A/T base pair, which act to specifically position E152 in an active orientation, would also account for these results.
Substitution of cysteine at D64 resulted in a hyperspecific phenotype; that is, the activity of this mutant protein on substrates mutated at the conserved A/T or C/G base pair compared to its activity on a wild-type substrate was much lower than would be predicted based on the specificity profile of IN(F185K). One possible explanation for this phenotype is that the mutation, either directly or indirectly, caused a loss of nonspecific interactions with terminal nucleotides of the viral DNA, making the mutant protein more reliant on remaining base-specific interactions to position the phylogenetically conserved viral DNA CA/TG base pairs. If these specific interactions were then removed by mutating the substrate, the activity of IN(D64C) would be disproportionately compromised compared to the activity of IN(F185K).
When Q148 was replaced with a leucine, the result was a mutant protein with reduced processivity for end processing and integration and an apparent loss of interactions with the unpaired 5′-terminal dinucleotide of the viral DNA strand. IN(Q148L) did not display altered specificity for the phylogenetically conserved viral DNA CA/TG base pairs (data not shown), implying that Q148 is not involved in specific recognition of this viral sequence. In a previous mutational analysis of HIV-1 integrase, IN(Q148L) was found to generate more cyclic dinucleotide product in the 3′-end-processing reaction than wild-type integrase when Mn2+ was used as a cofactor (68). This residue is located in the same flexible loop as E152, which has proven to be difficult to resolve in the HIV-1 integrase core crystal structure and in the crystal structures of related enzymes. This loop may be flexible for an as-yet-unappreciated mechanistic reason that relates to interactions with DNA and possibly to distortion of the substrate. Substrate distortion has been shown to be important for many polynucleotidyl transferase reactions (56, 60–62). Perhaps Q148 promotes unpairing at the viral terminus by interacting with the 5′-terminal two nucleotides of the viral DNA on the unprocessed strand. IN(Q148L) may therefore produce a preponderance of cyclic dinucleotide product because an Mn2+-dependent mechanism that promotes unpairing of the terminal base pairs in a manner that favors cyclization is the only pathway available to the mutant for fraying. Whereas numerous substitutions for E152 have been shown to abolish HIV-1 replication (41, 45, 71), the role of Q148 in HIV-1 growth has yet to be determined.
The salt sensitivity observed in a subset of the mutant proteins may be explained if the wild-type residues at these positions are normally involved in nonspecific interactions with the DNA substrates. Depending on the substitution, a particular mutant amino acid side chain might maintain or disrupt a stabilizing interaction. Disruption of an interaction with the DNA substrate (or creation of a new, unfavorable interaction) would make the mutant more dependent on remaining substrate interactions. If favorable electrostatic interactions were then disrupted by raising the salt concentration, the mutant protein might display disproportionately reduced activity compared to wild-type protein. Although we cannot exclude the possibility that disruptions of protein-protein interactions might explain the salt sensitivity of some of the mutants, the location of the altered amino acids near the active site places them in position to interact with DNA. Furthermore, these residues are located on the outside faces of the core dimer (as opposed to the adjoining faces with the large hydrophobic interface), arguing against their involvement in dimerization of the core domain.
Analysis of critical specific enzyme-substrate interactions may provide information that facilitates the development of enzyme inhibitors that may be useful in the treatment of HIV infection. To date, screens for potential antiviral drugs directed at integrase have sought inhibitors of its catalytic activity (12, 13, 17, 18, 29, 30, 33, 42, 47, 48, 54). An alternative approach to targeting antiviral agents at integrase would be deregulation of substrate specificity. Treatment of retrovirally infected cells with a compound that caused integrase to lose specificity could result in inappropriate suicidal cleavage of viral replication intermediates, preventing successful integration and viral replication (4). The identification of specific residues involved in determining the substrate specificity of HIV-1 integrase helps provide a rational foundation for such a strategy.
ACKNOWLEDGMENTS
This work was supported by a grant from the NIH and by the Howard Hughes Medical Institute. P.O.B. is an associate investigator of the Howard Hughes Medical Institute.
We thank T. Heuer for helpful discussions and for generating the picture of HIV-1 integrase in GRASP (Fig. 1) and K. Reich, D. Herschlag, and P. O’Brien for helpful discussions.
REFERENCES
- 1.Andrake M, Skalka A M. Multimerization determinants reside in both the catalytic core and C terminus of avian sarcoma virus integrase. J Biol Chem. 1995;270:29299–29306. doi: 10.1074/jbc.270.49.29299. [DOI] [PubMed] [Google Scholar]
- 2.Baker, T. Personal communication.
- 3.Bor Y C, Bushman F D, Orgel L E. In vitro integration of human immunodeficiency virus type 1 cDNA into targets containing protein-induced bends. Proc Natl Acad Sci USA. 1995;92:10334–10338. doi: 10.1073/pnas.92.22.10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown P O. Integration. In: Coffin J, Hughes S, Varmus H, editors. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1997. [PubMed] [Google Scholar]
- 5.Brown P O, Bowerman B, Varmus H E, Bishop J M. Correct integration of retroviral DNA in vitro. Cell. 1987;49:347–356. doi: 10.1016/0092-8674(87)90287-x. [DOI] [PubMed] [Google Scholar]
- 6.Bujacz G, Jaskolski M, Alexandratos J, Wlodawer A, Merkel G, Katz R A, Skalka A M. High-resolution structure of the catalytic domain of avian sarcoma virus integrase. J Mol Biol. 1995;253:333–346. doi: 10.1006/jmbi.1995.0556. [DOI] [PubMed] [Google Scholar]
- 7.Bujacz G, Jaskolski M, Alexandratos J, Wlodawer A, Merkel G, Katz R A, Skalka A M. The catalytic domain of avian sarcoma virus integrase: conformation of the active site residues in the presence of divalent cations. Structure. 1996;4:89–96. doi: 10.1016/s0969-2126(96)00012-3. [DOI] [PubMed] [Google Scholar]
- 8.Burke C J, Sanyal G, Bruner M W, Ryan J A, LaFemina R L, Robbins H L, Zeft A S, Middaugh C R, Cordingley M G. Structural implications of spectroscopic characterization of a putative zinc finger peptide from HIV-1 integrase. J Biol Chem. 1992;267:9639–9644. [PubMed] [Google Scholar]
- 9.Bushman F D, Craigie R. Activities of human immunodeficiency virus (HIV) integration protein in vitro: specific cleavage and integration of HIV DNA. Proc Natl Acad Sci USA. 1991;88:1339–1343. doi: 10.1073/pnas.88.4.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bushman F D, Craigie R. Integration of human immunodeficiency virus DNA: adduct interference analysis of required DNA sites. Proc Natl Acad Sci USA. 1992;89:3458–3462. doi: 10.1073/pnas.89.8.3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bushman F D, Engelman A, Palmer I, Wingfield R C. Domains of the integrase protein of human immunodeficiency virus type 1 responsible for polynucleotidyl transfer and zinc binding. Proc Natl Acad Sci USA. 1993;90:3428–3432. doi: 10.1073/pnas.90.8.3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carteau S, Mouscadet J F, Goulaouic H, Subra F, Auclair C. Inhibitory effect of the polyanionic drug suramin on the in vitro HIV DNA integration reaction. Arch Biochem Biophys. 1993;305:606–610. doi: 10.1006/abbi.1993.1468. [DOI] [PubMed] [Google Scholar]
- 13.Carteau S, Mouscadet J F, Goulaouic H, Subra F, Auclair C. Inhibition of the in vitro integration of Moloney murine leukemia virus DNA by the DNA minor groove binder netropsin. Biochem Pharmacol. 1994;47:1821–1826. doi: 10.1016/0006-2952(94)90311-5. [DOI] [PubMed] [Google Scholar]
- 14.Chow S A, Brown P O. Substrate features important for recognition and catalysis by human immunodeficiency virus type 1 integrase identified by using novel DNA substrates. J Virol. 1994;68:3896–3907. doi: 10.1128/jvi.68.6.3896-3907.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chow S A, Vincent K A, Ellison V, Brown P O. Reversal of integration and DNA splicing mediated by integrase of human immunodeficiency virus. Science. 1992;255:723–726. doi: 10.1126/science.1738845. [DOI] [PubMed] [Google Scholar]
- 16.Craigie R, Fujiwara T, Bushman F. The IN protein of Moloney murine leukemia virus processes the viral DNA ends and accomplishes their integration in vitro. Cell. 1990;62:829–837. doi: 10.1016/0092-8674(90)90126-y. [DOI] [PubMed] [Google Scholar]
- 17.Cushman M, Sherman P. Inhibition of HIV-1 integration protein by aurintricarboxylic acid monomers, monomer analogs, and polymer fractions. Biochem Biophys Res Commun. 1992;185:85–90. doi: 10.1016/s0006-291x(05)80958-1. [DOI] [PubMed] [Google Scholar]
- 18.Cushman M, Golebiewski W M, Pommier Y, Mazumder A, Reymen D, De Clercq E, Graham L, Rice W G. Cosalane analogues with enhanced potencies as inhibitors of HIV-1 protease and integrase. J Med Chem. 1995;38:443–452. doi: 10.1021/jm00003a007. [DOI] [PubMed] [Google Scholar]
- 19.Dotan I, Scottoline B P, Heuer T S, Brown P O. Characterization of recombinant murine leukemia virus integrase. J Virol. 1995;69:456–468. doi: 10.1128/jvi.69.1.456-468.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Drelich M, Wilhelm R, Mous J. Identification of amino acid residues critical for endonuclease and integration activities of HIV-1 integrase protein in vitro. Virology. 1992;188:459–468. doi: 10.1016/0042-6822(92)90499-f. [DOI] [PubMed] [Google Scholar]
- 21.Drelich M, Haenggi M, Mous J. Conserved residues Pro-109 and Asp-116 are required for interactions of the human immunodeficiency virus type 1 integrase protein with its viral DNA substrate. J Virol. 1993;67:5041–5044. doi: 10.1128/jvi.67.8.5041-5044.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dyda, F. Personal communication.
- 23.Dyda F, Hickman A B, Jenkins T M, Engelman A, Craigie R, Davies D R. Crystal structure of the catalytic domain of HIV-1 integrase: similarity to other polynucleotidyl transferases. Science. 1994;266:1981–1986. doi: 10.1126/science.7801124. [DOI] [PubMed] [Google Scholar]
- 24.Eijkelenboom A P A M, Puras-Lutzke R A, Boelems R, Plasterk R H A, Kaptein R, Hard K. The DNA-binding domain of HIV-1 integrase has an SH3-like fold. Nat Struct Biol. 1995;2:807–810. doi: 10.1038/nsb0995-807. [DOI] [PubMed] [Google Scholar]
- 25.Ellison V, Brown P O. A stable complex between integrase and viral DNA ends mediates human immunodeficiency integration in vitro. Proc Natl Acad Sci USA. 1994;91:7316–7320. doi: 10.1073/pnas.91.15.7316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ellison V, Gerton J, Vincent K, Brown P O. An essential interaction between distinct domains of HIV-1 integrase mediates assembly of the active multimer. J Biol Chem. 1995;270:3320–3326. doi: 10.1074/jbc.270.7.3320. [DOI] [PubMed] [Google Scholar]
- 27.Engelman A, Craigie R. Identification of conserved amino acid residues critical for human immunodeficiency virus type 1 integrase function in vitro. J Virol. 1992;66:6361–6369. doi: 10.1128/jvi.66.11.6361-6369.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Engelman A, Hickman A, Craigie R. The core and carboxyl-terminal domains of the integrase protein of human immunodeficiency virus type 1 each contribute to nonspecific DNA binding. J Virol. 1994;68:5911–5917. doi: 10.1128/jvi.68.9.5911-5917.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fesen M R, Kohn K W, Leteurtre F, Pommier Y. Inhibitors of human immunodeficiency virus integrase. Proc Natl Acad Sci USA. 1993;90:2399–2403. doi: 10.1073/pnas.90.6.2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fesen M R, Pommier Y, Leteurtre F, Hiroguchi S, Yung J, Kohn K W. Inhibition of HIV-1 integrase by flavones, caffeic acid phenethyl ester (CAPE) and related compounds. Biochem Pharmacol. 1994;48:595–608. doi: 10.1016/0006-2952(94)90291-7. [DOI] [PubMed] [Google Scholar]
- 31.Gerton J, Brown P O. The core domain of HIV-1 integrase recognizes key features of its DNA substrates. J Biol Chem. 1997;272:25809–25815. doi: 10.1074/jbc.272.41.25809. [DOI] [PubMed] [Google Scholar]
- 32.Gerton, J., D. Herschlag, and P. O. Brown. 1997. Unpublished data.
- 33.Hazuda D, Felock P, Hastings J, Pramanik B, Wolfe A, Goodarzi G, Vora A, Brackmann K, Grandgenett D. Equivalent inhibition of half-site and full-site retroviral strand transfer reactions by structurally diverse compounds. J Virol. 1997;71:807–811. doi: 10.1128/jvi.71.1.807-811.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hickman A B, Palmer I, Engelman A, Craigie R, Wingfield P. Biophysical and enzymatic properties of the catalytic domain of HIV-1 integrase. J Biol Chem. 1994;269:29279–29287. [PubMed] [Google Scholar]
- 35.Jenkins T M, Engelman A, Ghirlando R, Craigie R. A soluble active mutant of HIV-1 integrase: involvement of both the core and carboxy-terminal domains in multimerization. J Biol Chem. 1996;271:7712–7718. doi: 10.1074/jbc.271.13.7712. [DOI] [PubMed] [Google Scholar]
- 36.Jenkins T M, Hickman A B, Dyda F, Ghirlando R, Davies D R, Craigie R. Catalytic domain of human immunodeficiency virus type 1 integrase: identification of a soluble mutant by systematic replacement of hydrophobic residues. Proc Natl Acad Sci USA. 1995;92:6057–6061. doi: 10.1073/pnas.92.13.6057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jonsson C B, Roth M J. Role of the His-Cys finger of Moloney murine leukemia virus integrase protein in integration and disintegration. J Virol. 1993;67:5562–5571. doi: 10.1128/jvi.67.9.5562-5571.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Katzman M, Sudol M. Mapping domains of retroviral integrase responsible for viral DNA specificity and target site selection by analysis of chimeras between human immunodeficiency virus type 1 and visna virus integrases. J Virol. 1995;69:5687–5696. doi: 10.1128/jvi.69.9.5687-5696.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kulkosky J, Jones K S, Katz R A, Mack A M S. Residues critical for retroviral integrative recombination in a region that is highly conserved among retroviral/retrotransposon integrases and bacterial insertion sequence transposases. Mol Cell Biol. 1992;12:2331–2338. doi: 10.1128/mcb.12.5.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.LaFemina R L, Callahan P L, Cordingly M G. Substrate specificity of recombinant human immunodeficiency virus integrase protein. J Virol. 1991;65:5624–5630. doi: 10.1128/jvi.65.10.5624-5630.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.LaFemina R L, Schneider C L, Robbins H L, Callahan P L, LeGrow K, Roth E, Schleif W A, Emini E A. Requirement of active human immunodeficiency virus type 1 integrase enzyme for productive infection of human T-lymphoid cells. J Virol. 1992;66:7414–7419. doi: 10.1128/jvi.66.12.7414-7419.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.LaFemina R L, Graham P L, LeGrow K, Hastings J C, Wolfe A, Young S D, Emini E A, Hazuda D J. Inhibition of human immunodeficiency virus integrase by bis-catechols. Antimicrob Agents Chemother. 1995;39:320–324. doi: 10.1128/aac.39.2.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leavitt A D, Rose R B, Varmus H E. Both substrate and target oligonucleotide sequences affect in vitro integration mediated by human immunodeficiency virus type 1 integrase protein produced in Saccharomyces cerevisiae. J Virol. 1992;66:2359–2368. doi: 10.1128/jvi.66.4.2359-2368.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leavitt A D, Shiue L, Varmus H E. Site-directed mutagenesis of HIV-1 integrase demonstrates differential effects on integrase functions in vitro. J Biol Chem. 1993;268:2113–2119. [PubMed] [Google Scholar]
- 45.Leavitt A D, Robles G, Alesandro N, Varmus H E. Human immunodeficiency virus type 1 integrase mutants retain in vitro integrase activity yet fail to integrate viral DNA efficiently during infection. J Virol. 1996;70:721–728. doi: 10.1128/jvi.70.2.721-728.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lodi P J, Ernst J A, Kuszewski J, Hickman A B, Engelman A, Craigie R, Clore G M, Gronenborn A M. Solution structure of the DNA binding domain of HIV-1 integrase. Biochemistry. 1995;34:9826–9833. doi: 10.1021/bi00031a002. [DOI] [PubMed] [Google Scholar]
- 47.Mazumder A, Gupta M, Perrin D M, Sigman D S, Rabinovitz M, Pommier Y. Inhibition of human immunodeficiency virus type 1 integrase by a hydrophobic cation: the phenanthroline-cuprous complex. AIDS Res Hum Retroviruses. 1995;11:115–125. doi: 10.1089/aid.1995.11.115. [DOI] [PubMed] [Google Scholar]
- 48.Mouscadet J F, Carteau S, Goulaouic H, Subra F, Auclair C. Triplex-mediated inhibition of HIV DNA integration in vitro. J Biol Chem. 1994;269:21635–21638. [PubMed] [Google Scholar]
- 49.Muller H P, Varmus H E. DNA bending creates favored sites for retroviral integration: an explanation for preferred insertion sites in nucleosomes. EMBO J. 1994;13:4707–4714. doi: 10.1002/j.1460-2075.1994.tb06794.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pahl A, Flugel R M. Characterization of the human spuma retrovirus integrase by site-directed mutagenesis, by complementation analysis, and by swapping the zinc finger domain of HIV-1. J Biol Chem. 1995;270:2957–2966. doi: 10.1074/jbc.270.7.2957. [DOI] [PubMed] [Google Scholar]
- 51.Pruss D, Bushman F D, Wolffe A P. Human immunodeficiency virus integrase directs integration to sites of severe DNA distortion within the nucleosome core. Proc Natl Acad Sci USA. 1994;91:5913–5917. doi: 10.1073/pnas.91.13.5913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pruss D, Reeves R, Bushman F D, Wolffe A P. The influence of DNA and nucleosome structure on integration events directed by HIV integrase. J Biol Chem. 1994;269:25031–25041. [PubMed] [Google Scholar]
- 53.Pryciak P M, Varmus H E. Nucleosomes, DNA-binding proteins, and DNA sequence modulate retroviral integration target site selection. Cell. 1992;69:769–780. doi: 10.1016/0092-8674(92)90289-o. [DOI] [PubMed] [Google Scholar]
- 54.Puras-Lutzke R A, Eppens N A, Weber P A, Houghten R A, Plasterk R H. Identification of a hexapeptide inhibitor of the human immunodeficiency virus integrase protein by using a combinatorial chemical library. Proc Natl Acad Sci USA. 1995;92:11456–11460. doi: 10.1073/pnas.92.25.11456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Puras-Lutzke R A, Vink C, Plasterk R H A. Characterization of the minimal DNA-binding domain of the HIV integrase protein. Nucleic Acids Res. 1994;22:4125–4131. doi: 10.1093/nar/22.20.4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ramsden D A, McBlane J F, van Gent D C, Gellert M. Distinct DNA sequence and structural requirements for the two steps of V(D)J recombination signal cleavage. EMBO J. 1996;15:3197–3206. [PMC free article] [PubMed] [Google Scholar]
- 57.Rice P, Craigie R, Davies D R. Retroviral integrases and their cousins. Curr Opin Struct Biol. 1996;6:76–83. doi: 10.1016/s0959-440x(96)80098-4. [DOI] [PubMed] [Google Scholar]
- 58.Roth M J, Schwartzberg P L, Goff S P. Structure of the termini of DNA intermediates in the integration of retroviral DNA: dependence on IN function and terminal DNA sequence. Cell. 1989;58:47–54. doi: 10.1016/0092-8674(89)90401-7. [DOI] [PubMed] [Google Scholar]
- 59.Sarnovsky R L, May E W, Craig N L. The Tn7 transposase is a heterotrimeric complex in which DNA breakage and joining activities are distributed between different gene products. EMBO J. 1996;15:6348–6361. [PMC free article] [PubMed] [Google Scholar]
- 60.Savilahti H, Rice P A, Mizuuchi K. The phage Mu transpososome core: DNA requirements for assembly and function. EMBO J. 1995;14:4893–4903. doi: 10.1002/j.1460-2075.1995.tb00170.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Scottoline B P, Chow S, Ellison V, Brown P O. Disruption of the terminal base pairs of retroviral DNA during integration. Genes Dev. 1997;11:371–382. doi: 10.1101/gad.11.3.371. [DOI] [PubMed] [Google Scholar]
- 62.Scottoline, B. P., T. Heuer, and P. O. Brown. 1997. Unpublished data.
- 63.Sherman P A, Fyfe J A. Human immunodeficiency virus integration protein expressed in E. coli possesses selective DNA cleaving activity. Proc Natl Acad Sci USA. 1990;87:5119–5123. doi: 10.1073/pnas.87.13.5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sherman P A, Dickson M L, Fyfe J A. Human immunodeficiency virus type 1 integration protein: DNA sequence requirements for cleaving and joining reactions. J Virol. 1992;66:3593–3601. doi: 10.1128/jvi.66.6.3593-3601.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shibagaki Y, Chow S A. Central domain of retroviral integrase is responsible for target site selection. J Biol Chem. 1997;272:8361–8369. doi: 10.1074/jbc.272.13.8361. [DOI] [PubMed] [Google Scholar]
- 66.van den Ent F, Vink C, Plasterk R H A. DNA substrate requirements for different activities of the human immunodeficiency virus type 1 integrase protein. J Virol. 1994;68:7825–7832. doi: 10.1128/jvi.68.12.7825-7832.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.van Gent D C, Groeneger A A, Plasterk R H. Mutational analysis of the integrase protein of human immunodeficiency virus type 2. Proc Natl Acad Sci USA. 1992;89:9598–9602. doi: 10.1073/pnas.89.20.9598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.van Gent D C, Groeneger A A M O, Plasterk R H A. Identification of amino acids in HIV-1 integrase involved in site-specific hydrolysis and alcoholysis of viral DNA termini. Nucleic Acids Res. 1993;21:3373–3377. doi: 10.1093/nar/21.15.3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vincent K A, Ellison V, Chow S A, Brown P O. Characterization of human immunodeficiency virus type 1 integrase expressed in Escherichia coli and analysis of variants with amino-terminal mutations. J Virol. 1993;67:425–437. doi: 10.1128/jvi.67.1.425-437.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vink C, Oude Groeneger A M M, Plasterk R H A. Identification of the catalytic and DNA-binding region of the human immunodeficiency virus type 1 integrase protein. Nucleic Acids Res. 1993;21:1419–1425. doi: 10.1093/nar/21.6.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wiskerchen M, Muesing M A. Human immunodeficiency virus type 1 integrase: effects of mutations on viral ability to integrate, direct viral gene expression from unintegrated viral DNA templates, and sustain viral propagation in primary cells. J Virol. 1995;69:376–386. doi: 10.1128/jvi.69.1.376-386.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zheng R, Jenkins T M, Craigie R. Zinc folds the N-terminal domain of HIV-1 integrase, promotes multimerization, and enhances catalytic activity. Proc Natl Acad Sci USA. 1996;93:13659–13664. doi: 10.1073/pnas.93.24.13659. [DOI] [PMC free article] [PubMed] [Google Scholar]