Abstract
The molecular mechanism of a reaction in solution is reflected in its transition-state ensemble and transition paths. We use a Bayesian formula relating the equilibrium and transition-path ensembles to identify transition states, rank reaction coordinates, and estimate rate coefficients. We also introduce a variational procedure to optimize reaction coordinates. The theory is illustrated with applications to protein folding and the dipole reorientation of an ordered water chain inside a carbon nanotube. To describe the folding of a simple model of a three-helix bundle protein, we variationally optimize the weights of a projection onto the matrix of native and nonnative amino acid contacts. The resulting one-dimensional reaction coordinate captures the folding transition state, with formation and packing of helix 2 and 3 constituting the bottleneck for folding.
Keywords: carbon nanotubes, chemical kinetics, protein folding, transition-state theory, Grotthuss mechanism
Identifying the molecular mechanism of a reaction in solution, such as protein folding or enzyme-catalyzed chemistry, poses serious challenges because of the large number of coupled degrees of freedom (1–5). The identification and characterization of populated intermediate states along reaction paths is only a first step. Understanding the mechanism at a molecular level requires in addition the characterization of the transitions between those populated intermediates. The goal then is (i) to identify what is common to the transitions in a rare molecular reaction (or significant subsets thereof), and (ii) to find coordinates that not only measure the progress of the reaction but also are useful to characterize the reaction dynamics. The former leads to the concept of a transition state, the latter to that of a reaction coordinate.
Transition states can be thought of as configurations “intermediate” between reactants and products. In one widely used definition (6–11), the ensemble of transition states comprises those configurations that have an equal probability of reaching reactant and product regions. The chance of proceeding to reactants or products first can be quantified by the splitting (or commitment) probability introduced by Onsager for ion-pair recombination (12). The splitting probability is defined as the fraction of trajectories reaching the reactant region first when initiated from a given configuration with random Maxwell–Boltzmann velocities, and possibly averaged over noise for stochastic dynamics. We note that one of the difficulties arising from the above definition of transition states is that splitting probabilities cannot be measured experimentally, not even in a single-molecule measurement with atomic resolution. The reason is that multiple initializations with precise atomic positions, including those of solvent molecules, are required. Here, we will show how this difficulty can be circumvented by calculating average splitting probabilities (13) from transition-path and equilibrium ensembles that can also be measured experimentally.
From a good reaction coordinate, one may expect a dynamically meaningful measure of the progress of a reaction. Formally, the projection operator formalism (1, 2) allows us to obtain the dynamics along a chosen coordinate, but for poor choices the projected dynamics will be highly non-Markovian with long-time memory effects. In contrast, for a well chosen coordinate, the dynamics will be essentially Markovian after a brief initial period accounting for “molecular collisions” (14). Qualitatively, this Markovian character implies that if all one knows is the value of the reaction coordinate for a configuration intermediate between reactants and products, one can predict the likely fate of a trajectory initiated from that configuration. Now the “likely fate” is the splitting probability! Thus, a good reaction coordinate should parameterize the splitting probability, such that the splitting probability of a configuration is a function of the corresponding reaction coordinate alone (15, 16). Berezhkovskii and Szabo (17) indeed found the optimal one-dimensional reaction coordinate to be normal to the surface of equal splitting probabilities [i.e., the separatrix (6, 18, 19)].
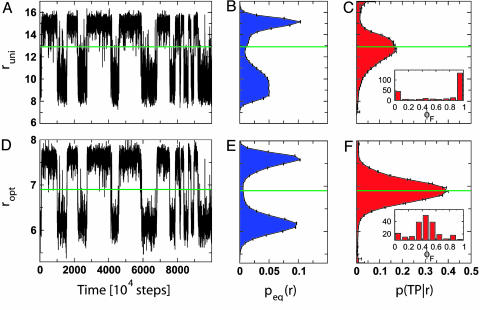
How does one identify transition states and good reaction coordinates for a rare molecular reaction in condensed phase? Answering this question requires access to an ensemble of reactive trajectories that can be obtained most efficiently from transition-path sampling (8–11, 13, 20, 21) or, if the transitions are sufficiently frequent, from long equilibrium trajectories. We note that partial information about reactive trajectories can also be obtained from single-molecule measurements. As illustrated in Fig. 1, a poor coordinate can often be distinguished from a good one almost immediately. Projected onto the poor coordinate, it may be possible to assign the state (i.e., reactant or product) in the context of the time-series history. However, equilibrium excursions from either state overlap in the projection. So if all one knows is the value of the reaction coordinate (say, r = r‡) without the preceding history, one cannot assign the state with confidence. In contrast, the good coordinate separates the states, and equilibrium excursions from either state do not overlap. Configurations with a reaction-coordinate value r = r‡ between reactant and product regions occur essentially only during transition paths, such that r ≈ r‡ should capture the configurations of the transition state. One of the objectives of this paper is to quantify such qualitative observations.
Fig. 1.
Examples of “poor” and “good” reaction coordinates for the folding of a 47-residue Gō-like protein model. The poor coordinate (runi) is the projection of the contact map onto a matrix with uniform weights, whereas the good coordinate (ropt) projects the contact map onto a matrix with optimized weights. (A and D) Time series of each reaction coordinate for the same simulation segment. (B and E) The equilibrium probability of the reaction coordinate peq(r). (C and F) The probability of being on a transition path given the value of r, p(TP|r). The green horizontal lines indicate “transition states” r = r‡. Only 1/10th of the trajectories were used in the optimization to avoid “over-fitting,” but the complete trajectories were used to test r with p(TP|r). Insets in C and F show the distributions of splitting probabilities for configurations at the maximum r = r‡ of p(TP|r).
In the following, we will first introduce a probabilistic relation between the equilibrium and transition-path ensembles. This Bayesian expression allows us to define and identify a transition-state ensemble. Moreover, it immediately leads to a quantitative measure for the quality of a reaction coordinate that will allow us to search systematically for optimal reaction coordinates. The Bayesian relation between the equilibrium and transition-path ensembles also leads to an expression for the rate coefficient and to a simple transition-path sampling algorithm. To illustrate the variational method of identifying reaction coordinates, we will study the folding of a simple protein model. Transition-path sampling and rate calculations will be illustrated for the slow dipolar reorientation of a hydrogen-bonded water chain inside a carbon nanotube.
Theory
Bayesian Relation Between Equilibrium and Transition-Path Ensembles. In the following, we consider a molecular system with deterministic Newtonian or stochastic Langevin dynamics in phase or configuration space. Following ref. 13, we define transition paths as those trajectory segments that exit from the reactant region A and reach the product region B without crossing back into A, and vice versa. Because of the requirement of no recrossing into A and B, the definitions of reactant and product regions can be rather stringent and include only configurations in the densely populated regions. (Incidentally, dividing up long equilibrium trajectories into segments separated by transition paths that connect the “bottoms” of the free energy wells also provides a convenient way of assigning intermediate configurations to reactant and product states.)
We can now construct probability distributions in phase space peq(x) and p(x|TP) for the equilibrium ensemble and the transition paths, respectively. x is a point in the full phase space in which the dynamics is Markovian. These probability distributions are related to each other through a Bayesian expression for conditional probabilities,
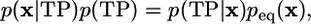 |
[1] |
where we have introduced two probabilities (with values between 0 and 1): p(TP|x) is the probability for being on a transition path (TP), given that the system is in x; and p(TP) is the fraction of time spent in transition paths, averaged over long equilibrium trajectories. Transition states can now be identified as those points with the highest probability p(TP|x) that trajectories passing through them are reactive (13), i.e., form transition paths between reactants and products.
Relation to Splitting Probabilities. As shown in ref. 13, the conditional probability p(TP|x) of being in a transition path is directly related to the splitting probabilities φA(x) of reaching the reactant region A first and φB(x) of reaching the product region B first on trajectories initiated from x: p(TP|x) = φA(x) φB(x) + φA(x)φB(x), where x = (–p, q) is a point in phase space with the same position q as x = (p, q), but reversed momenta p. In particular, for diffusive dynamics (i.e., Langevin dynamics in the overdamped limit), we have p(TP|q) = 2φA(q)φB(q) with φB(q) = 1 – φA(q). p(TP|q) reaches its maximum of ½ exactly on the stochastic separatrix (6, 18, 19) where φA(q) = φB(q) = 1/2. For diffusive dynamics, the definition of transition states as points with the highest probability p(TP|q) that trajectories passing through them are transition paths is thus equivalent to that using a “separatrix” or commitment probabilities (6–11).
Test for Reaction Coordinates. The Bayesian relation, Eq. 1, can be generalized for projected dynamics. For a reaction coordinate r = r(x), we have (13)
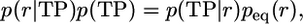 |
[2] |
where p(TP|r) is the conditional average of p(TP|x) with an equilibrium weight:
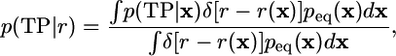 |
[3] |
with δ(r) Dirac's delta function.
For a good reaction coordinate r = r(x), p(TP|r) should have a single sharp and high peak, collapsing the transition states with a high value of p(TP|x) into a single value of r. With Eq. 2, different reaction coordinates r(x) and r′(x) can be compared quantitatively even without knowing the normalizing factor p(TP) in Eq. 2, because p(TP) is identical for all projections. Furthermore, the same criterion can be used in a variational search for optimal reaction coordinates, as shown below. Fig. 1 C and F shows normalized p(TP|r) for projections onto a poor and good coordinate: in the latter case, the maximum of p(TP|r) is considerably higher and approaches the diffusive limit of 0.5. In practice, the equilibrium probabilities peq(r) can be obtained from umbrella sampling (in any suitable coordinate) and the transition path probabilities p(r|TP) from transition-path sampling (8–11, 13, 20, 21) or, if feasible, both can be obtained from long equilibrium simulations with multiple transitions.
Estimating Reaction Rates. p(TP) is the fraction of time spent in transition paths, averaged over long equilibrium trajectories. Dividing by the average duration of a transition path, 〈tTP〉, one obtains the number of crossings between reactant and product regions per unit time (13). This relation can be used to estimate rate coefficients for the two-state model 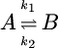 ,
,
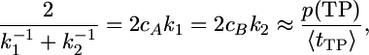 |
[4] |
where cA and cB are the equilibrium mole fractions of reactants and products, respectively.
Transition-Path Sampling by Shooting. Transition-path sampling (8–11, 20, 21) provides a powerful method to create and examine reactive trajectories alone. To avoid storing of intermediate paths, one can perform transition-path sampling by shooting from a single arbitrary dividing surface (13). The computational efficiency is determined by p(TP|r) and will therefore be lower for a poor reaction coordinate. Initial configurations q1, q2,..., qn with reaction coordinates r‡ ≈ r1 ≈ r2 ≈... ≈ rn, can be obtained, e.g., by saving structures along an umbrella sampling run biased toward r‡. From each of those structures, one (or several) trajectory pairs would be initiated with Maxwell–Boltzmann momenta pi and –pi. If a trajectory pair starting in qi ends in opposite regions A and B, the combined trajectory [with the momenta of one segment reversed (11)] forms a transition path and enters the transition-path ensemble with a relative weight of (13)
 |
[5] |
where the sum is over the points of intersection of the trajectory with the dividing surface ri = r(qi) from which the trajectory was started, and vk = dr/dt at time t = tk is the velocity normal to the dividing surface at the kth intersection. In combination with Eq. 4, we obtain an estimate of the reaction rate coefficients (13),
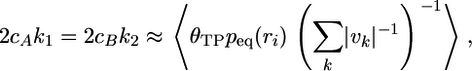 |
[6] |
with θTP = 1 if the trajectory pair forms a transition path, and 0 otherwise. The average 〈...〉 in Eq. 6 is over combined forward and backward paths initiated from an equilibrium ensemble of phase points (pi, qi).
Results
Folding of a Small “Two-State” Protein. Protein folding is an intrinsically high-dimensional problem, yet many proteins are experimentally found to populate essentially only two states at equilibrium (22). Transitions between those states (folded and unfolded) are highly cooperative (23). It should thus be possible to find low-dimensional coordinates that accurately describe the process. In the following, we will identify and characterize such coordinates for a simple protein model.
As an example, we have used a small (47 residue) three-helix bundle protein that folds fast in experiments (24). A Gō-like model of this protein was built from the experimental structure (25) by using a standard procedure (26). The principal feature of this model is that favorable interactions occur only between residues in contact in the native state. Simulations were run by using the charmm code (27).
The protein model exhibits essentially two-state transitions, as monitored by a standard Gō-model reaction coordinate, the fraction of native contacts Q (28): Langevin dynamics trajectories at the folding temperature (Tf) hop frequently between an “unfolded” state having Q ≈ 0.4 and a folded state with Q ≈ 0.9 (the folding and unfolding times τf and τu are ≈4 × 106 time steps, respectively, at Tf). Therefore, in this case all analysis can be performed on long equilibrium simulations, without the need for transition-path sampling.
Although Q turns out to be quite a good reaction coordinate (29), having a maximal p(TP|Q) of 0.40, we can attempt to construct an equally good (or better) contact-based coordinate with limited a priori information (as would be the case for simulations with a more “realistic” transferable potential). Because folding trajectories are highly heterogeneous in Cartesian space, we have chosen to follow the simulations in contact space. Each configuration is described by a “contact matrix” q. A reaction coordinate rw can be defined by projecting the contact matrix onto an arbitrary weight matrix w,  . The goal then is to find a set of 1,081 = 47 × 46/2 weights wij (with wij = wji and wii = 0) that correspond to a good reaction coordinate. In the contact matrix, qij is 1 when the distance between i and j is <12.0 Å and 0 otherwise. This definition does not discriminate between native and nonnative contacts, and thus differs somewhat from that used for the fraction of native contacts, Q, where the cutoff distance for residue pairs is proportional to their distance in the native structure (26). Therefore, Q cannot strictly be recovered with the basis set used here.
. The goal then is to find a set of 1,081 = 47 × 46/2 weights wij (with wij = wji and wii = 0) that correspond to a good reaction coordinate. In the contact matrix, qij is 1 when the distance between i and j is <12.0 Å and 0 otherwise. This definition does not discriminate between native and nonnative contacts, and thus differs somewhat from that used for the fraction of native contacts, Q, where the cutoff distance for residue pairs is proportional to their distance in the native structure (26). Therefore, Q cannot strictly be recovered with the basis set used here.
We started our search from the least biased initial guess of equal weights wuni. The projection of a trajectory segment at Tf onto wuni is shown in Fig. 1 A. Because there are generally more contacts formed in the folded state than the unfolded, especially with a Gō-like potential, the corresponding reaction coordinate runi turns out to be a reasonably good order parameter (i.e., it separates folded and unfolded states). However, it is relatively poor at identifying reactive states, having a maximal value of p(TP|runi) of only 0.17 (Fig.1C; transition paths spanned Q = 0.4–0.9 without recrossings). In addition, there is a significant transition-path probability p(TP|runi) for some large values of runi, which is caused by contacts formed only on a few transition paths that are not representative of most transitions.
To improve on this initial guess, we use p(TP|r) as a target function for a variational optimization procedure. Specifically, we optimize the maximum of a Gaussian fit to p(TP|r), to ensure that all reactive configurations are condensed into a single peak in p(TP|r). This procedure also suppresses subsidiary peaks from atypical contacts corresponding to configurations q where p(TP|q) is large, but both p(q|TP) and peq(q) are vanishingly small. We use a Monte Carlo optimization procedure in which we modify only relative weights by randomly changing two elements, wij and wkl, in such a way as to preserve the magnitude  of w. Monte Carlo moves include swapping, sign reversal, and reassigning fractions of the total weight. Reprojections on a trial coordinate can then be evaluated efficiently, because only the changes arising from the two altered elements need to be calculated. Starting from the initial uniform matrix of weights wuni, we applied this procedure recursively, accepting only moves that increased the maximum of p(TP|r), to generate an optimal matrix wopt. As shown in Fig. 1F, wopt gives a sharply peaked distribution of p(TP|ropt) with a maximum of 0.39.
of w. Monte Carlo moves include swapping, sign reversal, and reassigning fractions of the total weight. Reprojections on a trial coordinate can then be evaluated efficiently, because only the changes arising from the two altered elements need to be calculated. Starting from the initial uniform matrix of weights wuni, we applied this procedure recursively, accepting only moves that increased the maximum of p(TP|r), to generate an optimal matrix wopt. As shown in Fig. 1F, wopt gives a sharply peaked distribution of p(TP|ropt) with a maximum of 0.39.
To test whether individual configurations at the maximum of p(TP|ropt) were indeed part of the transition-state ensemble, we calculated the distribution of the folding probability φF [or pfold (7)]. Starting from 240 configurations close to the maximum of p(TP|ropt), we initiated 100 trajectories each with random Maxwell–Boltzmann velocities at Tf. The fraction of runs that fold first provides an estimate of the φF value of the starting configuration. The distribution of φF for the 240 configurations is sharply peaked close to 0.5 (Fig. 1F Inset); further, the mean value of 2φF(1 – φF) is 0.38, within the statistical error of the maximum in the distribution of p(TP|ropt), as expected for diffusive dynamics. In contrast, the distribution of φF for structures having maximal values of p(TP|runi) is peaked at 0 and 1. Unlike the optimized coordinate ropt, the uniform projection runi captures few transition-state configurations (Fig. 1C Inset), as is expected from the substantially smaller maximum of p(TP|runi). Even though ropt is a good reaction coordinate, it is not unique. We find that near-optimal weight matrices generated by Metropolis Monte Carlo search in weight space (at nonzero effective temperature) are degenerate, reflecting strong correlations among contacts (i.e., presence of one contact implying likely presence of another). Indeed, reaction coordinates of similar quality could be found even if only 100 of 1,081 contacts were included in the optimization (where the search randomly varied both the set of 100 contacts and their weights; Fig. 5, which is published as supporting information on the PNAS web site).
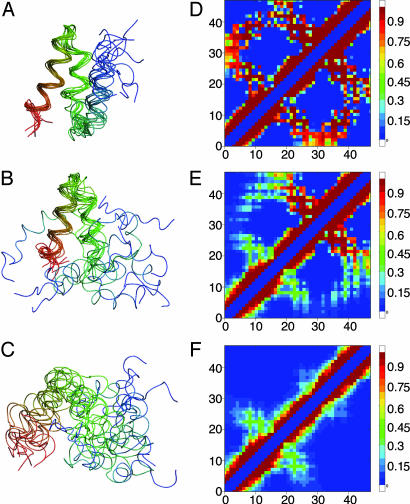
The optimized reaction coordinate ropt can be used to gain insight into the folding mechanism. By selecting snapshots from the trajectories corresponding to certain ropt projections, we can identify critical events on the folding paths. Fig. 2 shows 10 structures each for three values of ropt that are not significantly populated at equilibrium. On the unfolded side of the peak in p(TP|ropt) (ropt = 6.5; Fig. 2 A), a largely unstructured ensemble is obtained, although there is some local helical structure. The remarkable feature of the transition state (ropt = 6.9; Fig. 2B) is that the structure of helices 2 and 3 is almost completely native-like, whereas helix 1 is partly formed, but not docked against helices 2 and 3. On the folded side of the barrier (ropt = 7.4; Fig. 2C), all three helices are native-like, with slight disorder in helix 1. On the basis of this analysis, we identify the formation and packing of helices 2 and 3 as the bottleneck of folding.
Fig. 2.
Structures representing different values of the folding coordinate ropt. Residues are colored on a blue–green–red color scale from the N to C terminus and aligned with the native structure by using residues 17–47 (helices 2 and 3) only. (A) Folded side of the barrier (ropt = 7.4). (B) Transition state at the maximum of p(TP|ropt) (ropt = 6.9). Note that helix 1 is at least partially formed, but not properly docked against the scaffold of helices 2 and 3. (C) Unfolded side of the barrier (ropt = 6.5). (D–F) Fraction of contacts 〈qij〉 present in the folded state (ropt > 7.2) (D), transition state (6.89 < ropt < 6.91) (E), and unfolded state (ropt < 6.5) (F). Note that helical contacts are overemphasized by the 12-Å contact cutoff.
Analysis of the contact maps for the folded and unfolded protein, and for the transition state identified above (Fig. 2 D–F) provides further information on the mechanism. The only significant tertiary contacts present in the denatured state are those between helices 1 and 2, which are present ≈30% of the time. In the transition state, the additional contacts present are principally between helices 2 and 3 (80% formed), as suggested by Fig. 2B, and to a lesser extent between helices 1 and 3. The relative importance of H2–H3 and H1–H3 interhelical contacts is also reflected in the native-like appearance of the average weight matrix obtained by Metropolis Monte Carlo sampling, and of restricted weight matrices with only 100 contacts (Fig. 6, which is published as supporting information on the PNAS web site). Of the helix–helix interaction energies in the native state, those between helices 2 and 3 (H2–H3) are strongest (–7.6 kcal·mol–1), with weaker H1–H2 and H1–H3 interactions (–5.6 and –3.0 kcal·mol–1, respectively). In summary, the strongest tertiary interaction (H2–H3) is absent from the unfolded state (which may not at first be expected for a Gō-like model), and its formation is the “decisive” step during folding.
Collective Dipole Flip of Ordered Water Chain in Nanotube. In the previous example, we extracted transition paths from long equilibrium simulations. Next, we will perform transition-path sampling to collect otherwise rare reactive trajectories.
Molecular dynamics (MD) simulations of a short carbon nanotube segment dissolved in water showed that water filled the tube, forming hydrogen-bonded wires with collective dipole orientations either up or down along the tube axis (30, 31). The characteristic time for reorientations of the dipole chain was estimated to be in the range of 2–3 ns, three orders of magnitude slower than that of an individual water molecule in the bulk fluid. Dipole reorientation is an essential step in the Grotthuss mechanism (32) of proton transfer along one-dimensionally ordered water chains (33, 34). In a molecular model of the biological proton pump cytochrome c oxidase, water-chain reorientations induced by the changes in the electric fields in the protein active site provide the coupling of redox chemistry to vectorial proton translocation across the membrane (35).
In the following, we explore the dipole flip of ordered water chains, first to illustrate the transition-path sampling algorithm based on Eq. 5 and to test the accuracy of the rate estimates, Eqs. 4 and 6, and then to gain insight into the mechanism by which such reorientations occur. As reaction coordinate, we chose the total dipole moment Mz of water molecules inside the pore projected onto its axis. A continuous and differentiable 3D sigmoidal-type weight was used in summing the contributions of water dipole moments to Mz for the tumbling nanotube. To obtain an accurate equilibrium distribution of Mz, we performed 1-ns umbrella sampling runs with a harmonic bias for 12 overlapping windows. The simulation setup was as in ref. 30, with amber code and parameters for the carbons (36), the three-site water model TIP3P (37), a temperature of 300 K, and a pressure of 1 bar (1 bar = 100 kPa). The simulation system contained one (6,6)-type nanotube of ≈13.4-Å length and ≈8.1-Å diameter together with ≈1,000 water molecules in a cubic box under periodic boundary conditions. Transition-path sampling was performed by running trajectories at constant energy and volume “forward” and “backward” in time (i.e., with initial momenta ±p) from starting configurations in the region near the barrier, Mz = 0. The transition-path simulations were terminated when |Mz| exceeded 9 debye (1 debye + 3.3356 × 10–30 m·C; TIP3P dipole moment is 2.35 debye). The resulting 1,174 transition paths with a combined length of ≈15 ns were weighted according to Eq. 5. Initial Maxwell–Boltzmann velocities for the rigid water molecules were created by using a rigid-body representation. With the leap-frog-type integrator using velocities at half steps, care was taken that the forward and backward paths were continuous and reversible at the starting configuration.
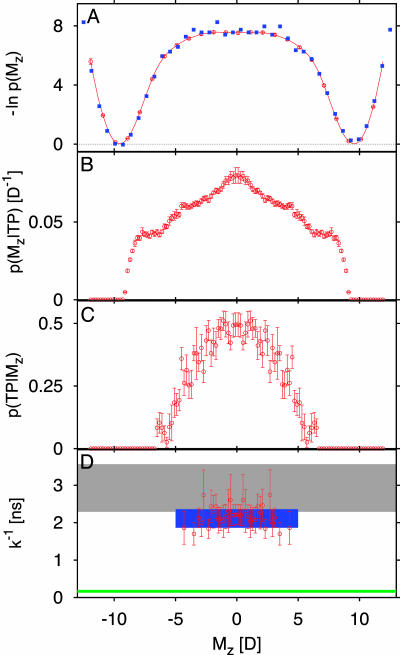
Fig. 3 summarizes the results for the thermodynamics and kinetics of the water-dipole flip. We find that Eqs. 4 and 6 give accurate rate coefficients when compared to long (66-ns) equilibrium simulations. In comparison with the slow dipolar reorientation with a rate coefficient of ≈1/(2 ns), transition paths are fast, with an average duration of only ≈2.0 ps. This time for flipping a chain of five or six water molecules is comparable to the characteristic time of ≈2 ps for reorientation of a single TIP3P water molecule in the bulk fluid. The distribution of transition-path durations tTP is well approximated by a gamma distribution of mean 2 ps and standard deviation 1.4 ps (not shown). On average, the transition paths cross the Mz = 0 dividing surface seven times, with a roughly exponential distribution of the (odd) number of crossings, suggesting “diffusive” dynamics on the broad barrier top (Fig. 3D). Accordingly, the relative weights of the paths created by straightforward shooting, Eq. 5, are distributed broadly, and only about 15% of the paths contribute to the top 95% of the relative weights.
Fig. 3.
Dipole flip of water chain inside carbon nanotube. (A) Equilibrium free energy surface as a function of the total dipole moment (D, debye unit) of the water chain from umbrella sampling (red line) and equilibrium MD (blue squares). (B) Probability density of the dipole moment in the transition-path ensemble. (C) Transition-path probability p(TP|Mz). (D) Reciprocal rate coefficient k–1 for dipole flip from ≈66-ns equilibrium MD simulations (gray shaded area indicates ± one estimated standard deviation), 〈tTP〉/p(TP) (blue rectangle indicates ± one estimated standard deviation; red symbols with error bars show estimates from individual histogram bins), and transition-state theory (green line) (3, 42).
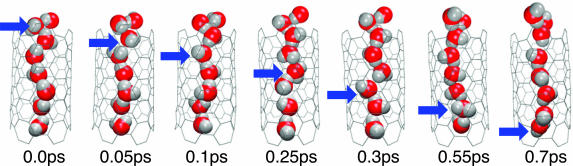
The molecular configurations along transition paths differ significantly from those seen in equilibrium trajectories. In particular, we find that the dipole moment distribution p(Mz|TP) in the transition-path ensemble peaks near Mz = 0. At equilibrium, Mz = 0 is at the center of an ≈7–8 kBT high free-energy barrier and thus rarely visited. The terrace-like steps in p(Mz|TP) indicate that during transition paths, dipole flips of individual water molecules have discrete character. Indeed, if we analyze individual transition paths, we find a step-like reorientation. As shown in Fig. 4, rather than collectively reorienting the dipoles of the water molecules, a hydrogen-bonding defect moves through the tube without significant translational motion of the water molecules, consistent with the static equilibrium analysis of a water chain in vacuum by Pomès and Roux (33). In the nanotube system, a D defect [formed by a water molecule accepting two hydrogen bonds and donating none (34)] is energetically preferred over an L defect (formed by a water molecule donating two hydrogen bonds and accepting none), as is expected from an earlier analysis of the preferred water-dipole orientation at the openings of the tube (38). Correspondingly, defects enter almost exclusively from the end at which pore water donates hydrogen bonds to the surrounding solvent.
Fig. 4.
Snapshots of water-chain reorientation inside carbon nanotube along the transition path with the highest relative weight, Eq. 5. Blue arrows indicate the progression of a hydrogen-bond defect along the water chain over a period of 0.7 ps.
The transition-path probability p(TP|Mz) reaches ≈0.5 near Mz = 0, the maximum expected for diffusive dynamics (Fig. 3), suggesting that the dipole moment Mz is a good reaction coordinate. To test whether Mz = 0 captures the transition state we have calculated the splitting probabilities for 45 equilibrium configurations with dipole moments near Mz = 0 by using 50 trajectories each with Maxwell–Boltzmann initial velocities. We find that the distribution of the resulting splitting probabilities is indeed narrowly peaked at 0.5, with a standard deviation of about 0.125, close to that expected from random binomial variations in 50 trials.
To explore whether Mz alone also captures the dynamics of the collective dipole flip, we have constructed a one-dimensional Langevin model for motion along Mz. From the equipartition theorems for the kinetic and potential energies, and the time integral of the Mz-correlation function of a Langevin oscillator, we obtain an effective mass of 2.46 × 10–6 ps2/Å3 and a friction coefficient of 216 ps–1 by using the simulation data for the harmonically biased simulation near the barrier top, Mz = 0. Langevin simulations on the full free energy surface (Fig. 3A) give a rate coefficient for dipole reorientation of ≈1/(2.1 ns), a transition-path duration of ≈1.6 ps, an average number of barrier crossings of 6.7 per transition path, and p(TP|Mz = 0) = 0.50, all in excellent agreement with the actual MD data [≈1/(2 ns), 2.0 ps, 7, and 0.5, respectively]. That Mz alone is a good reaction coordinate may appear somewhat surprising, considering that the dipole moment of the water chain is strongly coupled to the water solvent surrounding the tube through electrostatic interactions. Indeed, the rate of dipole reorientation for water wires across equivalent (6,6) nanotubes in 2D membranes is much lower because of the high cost of moving an effectively charged hydrogen bonding defect (34) through a low-dielectric environment (single transition during ≈20-ns equilibrium MD; unpublished results). The solvent-dependent rate without explicit solvent component in the reaction coordinate suggests fast solvent relaxation outside the tube. Consequently, solvent effects are well described by a potential of mean force and an effective friction for the Mz dynamics. Finally, we note that if the polarity of the pore is lowered (30), we observe an entirely different dipole-flip mechanism in which the tube first empties, and then water reenters the tube with the opposite dipole orientation.
Conclusions
We have shown that a Bayesian relation between the equilibrium ensemble and the transition-path ensemble can be used to locate transition states, optimize reaction coordinates, and estimate reaction rate coefficients. For a simple model of a three-helix bundle protein, we constructed a reaction coordinate by variationally optimizing the weights of a projection onto the amino acid contact matrix. Starting from a poor initial coordinate with uniform weights, we obtained a projection that accurately located the transition-state ensemble. By comparing protein configurations along the coordinate, we could identify the bottleneck of folding as the formation and packing of two helices, with the third helix relatively disordered at the transition state. We have also described a simple transition-path sampling algorithm and tested it for the dipole reorientation of an ordered water chain inside a carbon nanotube. The transition paths were used to estimate the rate of dipolar reorientations, giving a result in agreement with long equilibrium MD simulations.
In the protein-folding example considered here, a good basis set for the variational optimization of reaction coordinates could be guessed relatively easily. In general, that may not be the case. To expand the basis set, linear combinations of projections onto principal component axes may be useful for peptides and proteins (39, 40); nonlinear functions can be optimized in the same way if required. In addition, inclusion of solvent coordinates (9, 15, 41) may be necessary to construct a search space that covers degrees of freedom essential for the reaction kinetics and mechanism. Applications of our approach to more complex systems are expected to aid in the development of novel and unanticipated reaction coordinates.
Supplementary Material
Acknowledgments
We thank A. Szabo, W. A. Eaton, and A. Berezhkovskii for many helpful discussions.
Author contributions: R.B.B. and G.H. designed research; R.B.B. and G.H. performed research; R.B.B. and G.H. analyzed data; and R.B.B. and G.H. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviation: MD, molecular dynamics.
References
- 1.Zwanzig, R. (2001) Nonequilibrium Statistical Mechanics (Oxford Univ. Press, New York).
- 2.Berne, B. J. & Pecora, R. (1976) Dynamic Light Scattering (Wiley, New York).
- 3.Chandler, D. (1978) J. Chem. Phys. 68, 2959–2970. [Google Scholar]
- 4.Berne, B. J., Borkovec, M. & Straub, J. E. (1988) J. Phys. Chem. 92, 3711–3725. [Google Scholar]
- 5.Hänggi, P., Talkner, P. & Borkovec, M. (1990) Rev. Mod. Phys. 62, 251–341. [Google Scholar]
- 6.Klosek, M. M., Matkowsky, B. J. & Schuss, Z. (1991) Ber. Bunsen Ges. Phys. Chem. 95, 331–337. [Google Scholar]
- 7.Du, R., Pande, V. S., Grosberg, A. Y., Tanaka, T. & Shakhnovich, E. S. (1998) J. Chem. Phys. 108, 334–350. [Google Scholar]
- 8.Geissler, P. L., Dellago, C. & Chandler, D. (1999) J. Phys. Chem. B 103, 3706–3710. [Google Scholar]
- 9.Bolhuis, P. G., Dellago, C. & Chandler, D. (2000) Proc. Natl. Acad. Sci. USA 97, 5877–5882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bolhuis, P. G., Chandler, D., Dellago, C. & Geissler, P. L. (2002) Annu. Rev. Phys. Chem. 53, 291–318. [DOI] [PubMed] [Google Scholar]
- 11.Dellago, C., Bolhuis, P. G. & Geissler, P. L. (2002) Adv. Chem. Phys. 123, 1–78. [Google Scholar]
- 12.Onsager, L. (1938) Phys. Rev. 54, 554–557. [Google Scholar]
- 13.Hummer, G. (2004) J. Chem. Phys. 120, 516–523. [DOI] [PubMed] [Google Scholar]
- 14.Hummer, G. & Kevrekidis, I. G. (2003) J. Chem. Phys. 118, 10762–10773. [Google Scholar]
- 15.Ma, A. & Dinner, A. R. (February 11, 2005) J. Phys. Chem. B, http://dx.doi.org/10.1021/jp045546c.
- 16.Rhee, Y. M. & Pande, V. S. (January 27, 2005) J. Phys. Chem. B, http://dx.doi.org/10.1021/jp045544s. [DOI] [PubMed]
- 17.Berezhkovskii, A. & Szabo, A. (2005) J. Chem. Phys. 122, 014503. [DOI] [PubMed] [Google Scholar]
- 18.Ryter, D. (1987) Physica A 142, 103–121. [Google Scholar]
- 19.Ryter, D. (1987) J. Stat. Phys. 49, 751–765. [Google Scholar]
- 20.Pratt, L. R. (1986) J. Chem. Phys. 85, 5045–5048. [Google Scholar]
- 21.Dellago, C., Bolhuis, P. G., Csajka, F. S. & Chandler, D. (1998) J. Chem. Phys. 108, 1964–1977. [Google Scholar]
- 22.Jackson, S. E. (1998) Folding Des. 3, R81–R91. [DOI] [PubMed] [Google Scholar]
- 23.Rhoades, E., Cohen, M., Schuler, B. & Haran, G. (2004) J. Am. Chem. Soc. 126, 14686–14687. [DOI] [PubMed] [Google Scholar]
- 24.Wang, T., Zhu, Y. & Gai, F. (2004) J. Phys. Chem. B 108, 3694–3697. [Google Scholar]
- 25.Johannson, M. U., de Chāteau, M., Wikström, M., Forsén, S., Drakenberg, T. & Björk, L. (1997) J. Mol. Biol. 266, 859–865. [DOI] [PubMed] [Google Scholar]
- 26.Karanicolas, J. & Brooks, C. L., III (2002) Protein Sci. 11, 2351–2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brooks, B. R., Bruccoleri, R. E., Olafson, B. D., States, D. J., Swaminathan, S. & Karplus, M. (1983) J. Comput. Chem. 4, 187–217. [Google Scholar]
- 28.Nymeyer, H., García, A. E. & Onuchic, J. N. (1998) Proc. Natl. Acad. Sci. USA 95, 5921–5928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Socci, N. D., Onuchic, J. N. & Wolynes, P. G. (1996) J. Chem. Phys. 104, 5860–5868. [Google Scholar]
- 30.Hummer, G., Rasaiah, J. C. & Noworyta, J. P. (2001) Nature 414, 188–190. [DOI] [PubMed] [Google Scholar]
- 31.Vaitheeswaran, S., Rasaiah, J. C. & Hummer, G. (2004) J. Chem. Phys. 121, 7955–7965. [DOI] [PubMed] [Google Scholar]
- 32.de Grotthuss, C. J. T. (1806) Ann. Chim. 58, 54–74. [Google Scholar]
- 33.Pomès, R. & Roux, B. (1998) Biophys. J. 75, 33–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dellago, C., Naor, M. M. & Hummer, G. (2003) Phys. Rev. Lett. 90, 105902. [DOI] [PubMed] [Google Scholar]
- 35.Wikström, M., Verkhovsky, M. I. & Hummer, G. (2003) Biochim. Biophys. Acta 1604, 61–65. [DOI] [PubMed] [Google Scholar]
- 36.Cornell, W. D., Cieplak, P., Bayley, C. I., Gould, I. R., Merz, K. M., Jr., Ferguson, D. M., Spellmeyer, D. C., Fox, T., Caldwell, J. W. & Kollman, P. A. (1995) J. Am. Chem. Soc. 117, 5179–5197. [Google Scholar]
- 37.Jorgensen, W. L., Chandrasekhar, J., Madura, J. D., Impey, R. W. & Klein, M. L. (1983) J. Chem. Phys. 79, 926–935. [Google Scholar]
- 38.Waghe, A., Rasaiah, J. C. & Hummer, G. (2002) J. Chem. Phys. 117, 10789–10795. [Google Scholar]
- 39.García, A. E. (1992) Phys. Rev. Lett. 68, 2696–2699. [DOI] [PubMed] [Google Scholar]
- 40.Hummer, G., García, A. E. & Garde, S. (2001) Proteins Struct. Funct. Genet. 42, 77–84. [PubMed] [Google Scholar]
- 41.McCormick, T. A. & Chandler, D. (2003) J. Phys. Chem. B 107, 2796–2801. [Google Scholar]
- 42.Montgomery, J. A., Jr., Chandler, D. & Berne, B. J. (1979) J. Chem. Phys. 70, 4056–4066. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.