Abstract
During the analysis of Ig superfamily members within the available rainbow trout (Oncorhynchus mykiss) EST gene index, we identified a unique Ig heavy-chain (IgH) isotype. cDNAs encoding this isotype are composed of a typical IgH leader sequence and a VDJ rearranged segment followed by four Ig superfamily C-1 domains represented as either membrane-bound or secretory versions. Because teleost fish were previously thought to encode and express only two IgH isotypes (IgM and IgD) for their humoral immune repertoire, we isolated all three cDNA isotypes from a single homozygous trout (OSU-142) to confirm that all three are indeed independent isotypes. Bioinformatic and phylogenetic analysis indicates that this previously undescribed divergent isotype is restricted to bony fish, thus we have named this isotype “IgT” (τ) for teleost fish. Genomic sequence analysis of an OSU-142 bacterial artificial chromosome (BAC) clone positive for all three IgH isotypes revealed that IgT utilizes the standard rainbow trout VH families, but surprisingly, the IgT isotype possesses its own exclusive set of DH and JH elements for the generation of diversity. The IgT D and J segments and τ constant (C) region genes are located upstream of the D and J elements for IgM, representing a genomic IgH architecture that has not been observed in any other vertebrate class. All three isotypes are primarily expressed in the spleen and pronephros (bone marrow equivalent), and ontogenically, expression of IgT is present 4 d before hatching in developing embryos.
Keywords: development, immunoglobulin
A key hallmark of the vertebrate adaptive immune system is the generation of antigen-specific antibodies from B cells through the process of V(D)J recombination. This process is restricted solely to gnathostomes (jawed vertebrates); no evidence for either antibody gene fragments or the VDJ-recombinase machinery has been identified in agnathan fish (lampreys and hagfish) (1, 2). The specific effector function (complement fixation, recognition by phagocytic cells, and secretion in mucosal tissues) depends on the nature of the isotype constant (C) region. In mammals, there are five Ig isotypes that possess distinct effector functions for secretory immunity. IgM is the only antibody isotype found universally in gnathostomes, and until 1997, teleosts (bony fish) were thought to possess only IgM; however, research on catfish (3) and, later, on Atlantic salmon (4) provided evidence for the existence of IgD in teleosts. The genomic location of the teleost delta gene (δ) immediately downstream of μ, coupled with modest sequence identity to mammalian δ and coexpression of IgM and IgD in catfish B cell lines, solidified the relationship of teleost IgD to that of mammals. Expression of teleost IgD, like that of mammals, is achieved by alternative splicing from the rearranged VDJ gene to the Cδ genes, with the exception that the teleost IgD message retains the first exon of Cμ1, thus forming a chimeric antibody isotype. After the discovery of Ig heavy-chain (IgH) genes in elasmobranchs [IgW and nurse shark new antigen receptor (NAR)] and, later, the teleost δ gene, researchers have continually speculated on the primordial origins of antibody isotypes (5–7). In this article, we describe the isolation and characterization of a teleost IgH (IgT) isotype from rainbow trout that shares some properties with other vertebrate IgH isotypes, but the gene itself occupies a position within the trout IgH locus that has not been described in any gnathostome.
Materials and Methods
Fish. Rainbow trout [Oncorhynchus mykiss (Onmy), Clear Springs Foods, Buhl, ID] were maintained at 12°C by using standard biofiltration systems. OSU-142 and Hot Creek homozygous trout (8) were provided by Gary Thorgaard (Washington State University).
Primers. For the PCR primers used in this study, see Table 1, which is published as supporting information on the PNAS web site.
cDNA and Bacterial Artificial Chromosome (BAC) Library Screening. An OSU-142 splenic ZAP Express (Stratagene cDNA library was screened (1.2 × 106 plaque-forming units) sequentially with specific probes as listed in Table 1. Positive clones were first analyzed by PCR with Cμ,Cδ, and Cτ intron-spanning primer sets on which positive clones were fully sequenced. The OSU-142 4.5X genomic BAC library (9) was screened by using [32P]dCTP-labeled single-exon probes for Cμ4, Cδ7, and Cτ4 under stringent conditions. BAC-clone minipreps were used as templates for PCR using the single-exon primer sets with OSU-142 genomic DNA (100 ng) serving as a positive control to assess the gene content.
BAC Sequencing and Annotation. OnmyBAC-IgH.1 was processed by using the Qiagen Large Construct kit for the construction of a BAC DNA shotgun library. BAC DNA was sheared into 1- to 3-kbp fragments, subcloned into pBSK+, sequenced to nine times coverage, and assembled by using the phred-phrapconsed software package (10, 11). Only Phred values of >20 were used for the assembly. The BAC clone was annotated by using GenScan (http://genes.mit.edu/GENSCAN.html) in combination with manual sequence analysis (macvector, Accelrys, San Diego).
Physical Mapping of the Trout IgH Regions. In situ chromosomal hybridization and karyotyping procedures have been described in ref. 9.
Expression of IgH Isotypes. RNA isolation, RT-PCR, and Northern blotting protocols have been described in ref. 12. Probes were identical to those used for the cDNA library screening procedure. Blots were washed at 65°C and exposed for 14 h (for IgM and EfTU-1) and for 7 d (for IgD and IgT). Total RNA (1 μg) was reverse transcribed (Promega) into first-strand cDNA by using random hexamers. One-tenth of the cDNA products were used for a qualitative assessment of expression by PCR using the following conditions for 30 cycles: 94°C for 15 sec, 58°C for 30 sec, and 72°C for 30 sec followed by 72°C for 10 min. The products were sequenced for authenticity.
Sequencing and Comparative Bioinformatics. DNA sequencing of cloned cDNAs was performed by using cycle-sequencing chemistry (Applied Biosystems). Amino acid sequences were broken down into individual Ig superfamily C domains by using blastp (Swiss Institute of Bioinformatics, Basel) conserved-domain analysis and manual inspection. Individual Ig C domains were aligned by using clustalw (European Bioinformatics Institute, Cambridge, U.K.) with the open-gap and gap-extension penalties set at 10 and 0.5, respectively. Individual domains from nurse shark new antigen receptor (NAR) and sandbar shark IgW (5, 7) were included only if they displayed >20% identity to individual Cτ domains. From this alignment, unrooted phylogenetic trees were constructed by using the neighbor-joining method with mega 2.1 (13) (random tie breaking) and Poisson distance correction (gaps ignored). Trees were bootstrapped 1,000 times, and only those branch sites with bootstrap values of >50% are shown. Transmembrane regions were identified by using tmpred (www.chnet.org), and N-linked glycosylation sites were predicted by using the netnglyc 1.0 server (www.cbs.dtu.dk/services/NetNGlyc). Similarity searches were mainly performed by using blast programs at www.ncbi.nlm.nih.gov, coupled with sequence extraction(s) from ensembl (www.ensembl.org) and blat (http://genome.ucsc.edu/cgi-bin/hgBlat) searches with the available zebrafish scaffolds. The following supercontigs and BAC clones were used for the zebrafish IgH locus: ctg14038, ctg1404, ctg14057, BX649502, and BX510335.
Results and Discussion
IgH Genes in Rainbow Trout. During tblastn (Netherlands Bioinformatics Centre, Nijmegen, The Netherlands) analysis of the available rainbow trout EST gene index at the National Center for Biotechnology Information and The Institute for Genomic Research (www.tigr.org/tdb/tgi/) by using the Ig superfamily C domain from trout TAPBP as the query (9), a sequence was discovered that displayed high identity to IgH C domains but was divergent from all known IgH genes in teleost fish. PCR primers were then developed to amplify a homologous probe from splenic cDNA for screening a splenic cDNA library derived from a single homozygous trout, OSU-142 (8). Because these cDNAs represent an isotype that had yet to be described in fish, we also cloned IgM and IgD from the same library to confirm that the newly identified IgH isotype did indeed represent a third expressed IgH isotype in trout. From the screening process, a single expressed IgM gene was found, but duplicate forms of IgD and the unique IgH isotype were identified. We have named the unique teleost IgH isotype “IgT” (τ) (for teleost) because bioinformatic analysis indicates that this isotype is restricted to teleost fish.
OSU-142 IgM and IgD Sequences. Our group and others (14, 15) have previously reported cDNAs encoding secreted and membrane-bound forms of rainbow trout IgM. We used a combination of cDNA probes corresponding to the Cμ1 and -2 domains to screen a homozygous OSU-142 directional splenic cDNA library. Atlantic salmon encode two μ genes per haplotype, whereas gel filtration analysis suggested that rainbow trout express only one μ gene, along with allotypic variants (16). To investigate this issue, 35 full-length and partial IgM-positive clones were identified and sequenced from the 5′ end of the cDNAs. Four independent clones were completely sequenced (GenBank accession nos. AY870256–AY870259). The clones coded for the secreted version of IgM, with each containing different VDJ rearrangements and displaying 100% nucleotide identity for all four μ C domains, supporting the finding that trout express a single Cμ gene per haplotype.
IgD cDNAs and genomic clones have been identified in Atlantic salmon (4). For the isolation of rainbow trout IgD, trout ESTs were identified that displayed >96% amino acid identity to an Atlantic salmon δ7 query by using tblastn. This match was used to design homologous PCR primers to amplify a δ7 rainbow trout cDNA probe. cDNA library screening of the homozygous spleen library resulted in 12 full-length and partial clones corresponding to IgD. Similar to IgD from all teleost fish, the trout IgD clones were chimeric IgHs in that the first C domain is encoded by Cμ1 as a result of alternative splicing. It is thought that this organization allows association of Ig light chain mediated by Cμ1 because the δ1 sequence lacks residues for light-chain binding (3). The full-length trout δ clones revealed a single Ig C domain organization for the OSU-142 IgD clones (μ1-δ1-δ2a-δ3a-δ4a-δ2b-δ7). Analysis of Atlantic salmon has shown intraspecies cis-duplication of δ2–4 (4), thus the “a” and “b” exon nomenclature for the trout δ gene. None of the IgD clones contained the δ5 and δ6 that are typical of teleost IgD, but duplicated forms of IgD were identified during the screen that displayed 94% amino acid identity across the Cδ domains, including the presence of nine conserved N-glycosylation sites (see Fig. 5, which is published as supporting information on the PNAS web site). In channel catfish, the IgH locus encodes two distinct δ genes that represent both the membrane-bound and secreted forms of IgD (17). cDNAs representing secreted versions of trout IgD were not found.
Characterization of IgT. Screening of the homozygous cDNA library yielded several full-length and partial clones for rainbow trout IgT. Thirteen clones, representing full-length and truncated versions of IgT, were completely sequenced. Three full-length cDNA clones were composed of a leader peptide, a rearranged VDJ segment, and four Ig superfamily C domains ending in either a C-terminal transmembrane domain or a secretory tail (Fig. 1A). Five different trout VH families of the 11 documented VH families (18) for rainbow trout were represented among the IgT cDNA clones. Clone 8bb (GenBank accession no. AY870265; 2,326 bp) contains an ORF of 600 amino acids, whereas clone 2bb (GenBank accession no. AY870264; 2,019 bp) encodes for 551 amino acids. A duplicated version of IgT was found during the library screen that represented half of all clones identified in which the IgT duplicates share 96% amino acid identity across the Cτ domains (Fig. 1 A). Analysis of clone 8bb revealed that it terminated in an acid-rich domain followed by a transmembrane region and a positively charged, short cytoplasmic tail. The membrane-bound (8bb) and secreted form (2bb) of IgT have apparent molecular masses of 63 and 58 kDa, respectively, excluding the leader and not taking into consideration the presence of two N-linked glycosylation sites within the third and fourth C domains. Strikingly, initial blastx/p analysis of Cτ1–4 showed the highest identity to teleost fish and mammalian IgM sequences. Genomic analysis of the available fugu scaffolds (www.ensembl.org) revealed a τ-like gene located on scaffold 3494, ≈8.3 kbp upstream of the fugu μ gene (19). This region encodes an exon resembling Cτ1 (33% identity to Cτ1), an exon similar to Cτ4, and finally, an exon coding for a TM1 domain. Using these particular exons, blastx analysis revealed membrane-bound and secreted versions of a fugu IgH isotype (AB201354 and AB201355) in GenBank that, in accordance with scaffold 3494, encodes an isotype composed of two C domains. Analysis of the recent tetraodon assembly, however, including GenScan analysis of chromosome 3 (position 8.15 Mbp) that houses the μ and δ genes, yielded inconclusive results for the presence of a τ-like gene. Recently, however, a likely zebrafish IgT orthologue (IgZ, AY643750), which displays 59% amino acid similarity to IgT (Fig. 1), has been deposited in GenBank. The group working on IgZ also deposited a trout EST sequence (GenBank accession no. AY773715, 80% amino acid identity) that represents a distant allele to our IgT clones. AY773715 is from an EST library (20) derived from a Kamloop (BC, Canada) strain of rainbow trout, representing an unrelated strain, because the OSU-142 homozygous rainbow trout were developed from a strain from Lake Shasta, CA.
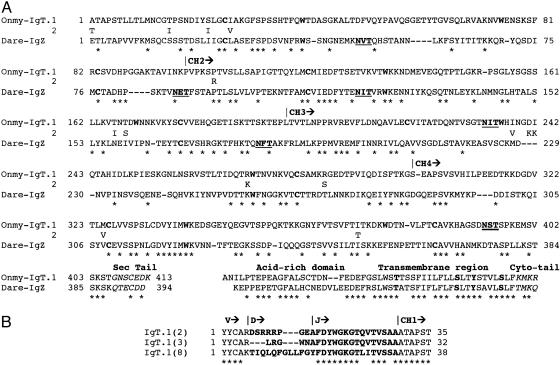
Fig. 1.
Comparative analysis of IgT in teleost fish. (A) Amino acid alignment of the rainbow trout (Onmy) IgT and zebrafish (Dare) IgZ C domains. C regions (CH) are based on cDNA and germ-line sequences for rainbow trout. Cysteine and tryptophan residues typical of the Ig fold are in boldface type. Potential N-linked glycosylation sites are underlined, and residues differing between the IgT duplicates are shown immediately under the IgT.1 sequence. * denotes identity, and – indicates gaps. S, T, and Y residues found in the transmembrane region typical of associating with B cell coreceptors CD79A/B are in boldface type. (B) CDR3 junctions for three IgT VDJ rearrangements.
Cysteine and tryptophan residues required for the Ig fold (21) are found in all four rainbow trout Cτ domains, but the Cζ3 domain differs in that the first cysteine is found in the position normally occupied by tryptophan. The additional cysteine within the CH1 domain of IgT and IgZ (Cys-13 and Cys-14, respectively) implies that these molecules can associate with light chain. However, the highest level of amino acid identity (80%) was found among the τ and ζ transmembrane regions. The transmembrane region contains residues typical of B cell receptors, including hydrophobic and hydrophilic residues consistent with the CART domain and Thr, Ser, and Tyr residues known to be essential for association with the B cell coreceptors CD79A/B (22). The short secretory tails of both IgT and IgZ contain a single Cys residue and several acidic amino acids. Pathogen neutralization provided by mucosal immunity is an essential first line of defense for vertebrates. This neutralization is carried out by the polymeric IgA isotype, and comparative analysis has determined that the PX3NXS/TL/VX4E/DX4CY motif is typically required for multimeric polymerization, where the N-linked glycosylation site (underlined) and penultimate cysteine are critical for association of the J chain (23, 24). IgT contains an N-linked glycosylation site near the end of CH4 and a Cys residue in the secretory tail, but overall, this region bears little similarity to the motif required for J-chain association. In addition, IgT lacks the obvious hinge features typical of IgA, but interestingly, there are five Pro residues near the CH1 and -2 junction, suggesting that this region may be flexible. The third isotype of Xenopus (25, 26), IgX, is considered to be an IgA analog because IgX plasma cells represent ≈50% of total Ig-producing lymphocytes in the gut, and IgX forms polymers as large as IgM but, similar to IgT, the secretory domain of IgX shows little resemblance to the J-chain motif, aside from ending with Cys-Tyr. Only through biochemical analysis will the function and polymerization potential of IgT be determined.
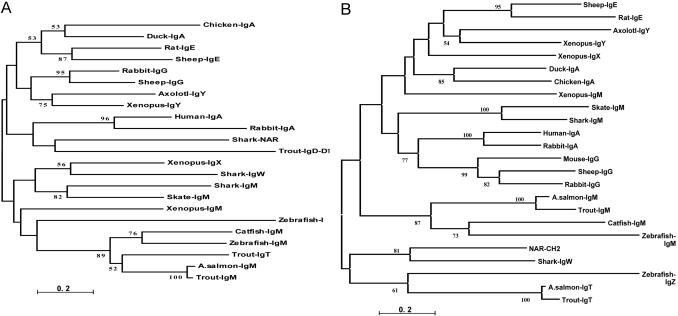
Phylogenetic Assessment of the τ Gene. Our initial analysis of GenBank by using IgT (tblastn) revealed the presence of likely orthologs in zebrafish and Atlantic salmon. blastp domain-by-domain comparison of the C regions displayed intriguing features of the τ lineage. Significant matches (amino acid identity) were as follows: CH1 salmonid μ (52%), CH2 zebrafish ζ (34%), and surprisingly, vertebrate Ig light chains (33%), CH3 shark and NAR CH2 and -4 (28%), zebrafish ζ (27%), CH4 Xenopus μ and pig γ1 (32%), and zebrafish ζ (36%). Because antibody isotypes contain various numbers of Ig C domains, ranging from two to seven (not including intraspecies-duplicated domains, as found in teleost δ), we performed a systematic phylogenetic analysis of individual Ig C domains to reveal potential relationships to the τ lineage (Fig. 2). δ lineages were not compared because they showed low amino acid identity to τ. As shown in Fig. 2 A, the phylogenetic analysis of CH1, a domain required for light-chain association, indicates that rainbow trout τ CH1 forms a tight clade with the teleost μ lineage, as supported by high bootstrap values. In support of this finding, trout μ and τ CH1 domains share 51% amino acid identity, implying a close relationship with the salmonid μ lineage. The zebrafish ζ CH1 domain lies within the greater μ clade, but its relationship is difficult to resolve because bootstrap values for this branch were <50%. CH2 analysis (see Fig. 6, which is published as supporting information on the PNAS web site) confirms a relationship between the τ and μ lineages, because both τ and ζ cluster with elasmobranch μ CH2. τ and ζ display 34% identity and an average identity of 25% to both shark and skate μ CH2. For both the CH3 and CH4 analyses, a partial Atlantic salmon IgT EST was included to provide resolving power for the phylogenetic analyses because these domains showed the highest degree of divergence through blastp analysis. Interestingly, the τ and ζ CH3 genes formed a distinct clade adjacent to NAR CH2 and shark IgW CH3 domains (Fig. 2B), whereas the trout τ CH3 sequence exhibits ≈26% identity and ≈40% amino acid similarity with these two elasmobranch genes. Within the τ/ζ clade, trout and salmon τ displayed 85% identity, whereas τ and ζ CH3 were only 27% identical. Finally, the phylogenetic relationship of vertebrate IgH CH4 domains indicates that the τ/ζ clade is unique and that the nearest adjacent clades, representing teleost IgM and amphibian IgY (IgG analogs) (27), are separated by considerable genetic distance (see Fig. 7, which is published as supporting information on the PNAS web site). Taken together, these results imply that the τ lineage displays some homology to the teleost μ lineage but most likely represents a distinct IgH lineage in teleosts.
Fig. 2.
Phylogenetic relationships for individual vertebrate IgH C domains; individual Ig C domains were aligned by using clustalw, and trees were generated by using the neighbor-joining method, with genetic distances obtained by Poisson correction (scale bar below the tree). (A)CH1 domain. (B)CH3 domain. GenBank accession nos. follow. IgA: duck, U27222; chicken, S40610; human, BAC85198; and rabbit, X82116. IgE: rat, AAA41365; sheep, M84356; and horse, U15150. IgG: rabbit, K00752; sheep, X69797; and mouse, J00453. IgY: axolotl, X69492; and Xenopus, X15114. NAR and IgW: nurse shark NAR, U18701 and sandbar shark IgW, U40560. IgX: Xenopus, X13779. IgM: skate, M35185; shark, Y00840; Xenopus, J03631; Atlantic salmon, S48652; catfish, M27230; trout, AY870256; and zebrafish, CAI11475. IgT/Z: rainbow trout, AY870265; Atlantic salmon, TC29571; and zebrafish, AY643750.
Features of the IgT VDJ Region. From analysis of full-length IgT clones, it was clear that all three rainbow trout IgH isotypes use the standard 11 VH families. Our initial analysis showed that two different VH genes (from VH8 and -11) (18) were used by both IgM and IgT clones within the cDNA library that was derived from a single homozygous trout. The other full-length and partial clones for IgT represented three additional VH families from trout, making a total of five VH families represented within this initial survey of IgT VH diversity. VDJ recombination generates the third complementarity-determining region (CDR3) that is known to have an important function in antigen recognition. Although the trout IgH isotypes use the same VH families, IgT uses different D and J segments to generate the IgT CDR3. Trout IgM CDR3s are compact, with an average size of 4–5 aa generated by both VDJ recombination and P and N nucleotide addition (28), whereas our initial inspection of three IgT clones found that they display a CDR3 range of 5–10 aa (Fig. 1B), based on three clones derived from the IgH.1 locus. In addition, comparative modeling suggests an extended CDR3 loop for IgT clone 8 that is reminiscent of the large CDR3 loops associated with NAR (29) (data not shown). Retention and usage of a broad size range for the IgT CDR3 is likely beneficial for the secretory immune response because a diverse range of epitopes could be recognized.
The IgH.1 Locus in Rainbow Trout. The murine IgH locus encodes five different Ig isotypes with the following genomic organization: Vn-Dn-J4-(S)Cμ-Cδ-(S)Cγ3-(S)Cγ1-(S)Cγ2b-(S)Cγ2a-(S)Cε-(S)Cα. The Cμ and Cδ genes are joined to recombined VDJ genes by RNA processing, whereas genes located further downstream are joined to the functional VDJ segment through a process known as class-switch recombination, involving intrachromosomal deletional rearrangement focused on regions of repetitive-switch DNA found upstream of each CH-chain region (30). The genomic organization of IgH genes in elasmobranchs is quite different from the mammalian organization because repeated blocks of joined and nonjoined VDJ and CH occur in what is termed the multicluster arrangement (VDJ-C)n. IgH genes in teleosts possess a gene organization similar to that of higher vertebrates (translocon configuration), whereas the Ig light-chain genes are of the multicluster type; thus, bony fish posses a chimeric organization for the H- and L-chain genes (2).
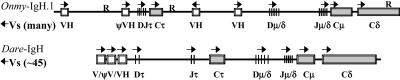
To investigate the IgH genomic architecture in trout, an OSU-142 BAC library was screened by using a mixture of μ-, δ-, and τ-exon-specific probes. Thirteen IgH+ BAC clones were identified through PCR analysis and were found to be comprised of one μ single-positive, six μ/δ double-positives, three μ/τ double-positives, two τ single-positives, and a single triple-positive clone. This latter clone, OnmyBAC-IgH.1, was completely sequenced, resulting in two major contigs of 107 and 14 kbp. The physical map of OnmyBAC-IgH.1 (Fig. 3 Upper), starting at the 5′ end, is composed of a proximal VH gene, a retroposon element, and a VH pseudogene followed by three IgH D and two J elements and four exons encoding the IgT C domains. These particular D and J elements are the ones used to build up a complete IgT gene and possess standard recombination-signal sequences (RSSs). As mentioned previously, CDR3 regions are larger for IgT than for IgM. One factor to account for this difference is that the first Dτ element is 37 bp long (open in all three reading frames), of which 27 bp are used within IgT clone 8 (Fig. 1B). DH elements are typically 12–16 nucleotides long. The second and third Dτ genes are typical in size (DH2, 13 bp and DH3, 15 bp). Over 30% of the IgT cDNA clones were represented as sterile transcripts (JCτ1Cτ2Cτ3Cτ4sec), such as IgT clone 15 (GenBank accession no. AY870266). Clone 15 initiates between Dτ3 and Jτ1, and the ORF begins with a methionine codon within the RSS of τJH1. Sterile transcripts were also found for trout IgD (GenBank accession no. AY870262) and have also been noted for trout IgL (31). Unlike teleost IgM, in which the membrane-bound form is generated by splicing the TM segment to the end of μ exon 3, the membrane version of IgT is generated by splicing to a cryptic splice site near the very end of τ exon 4, thus retaining all four Ig C domains for both the secreted and membrane-bound versions of IgT, similar to that of IgM from amphibians, birds, and mammals (2). Between the τ TM exons and the two additional VH genes, genscan analysis detected an intact transposable element of the piggyBac class (Fig. 3 Upper). Coupled with other retroposon-like short interspersed nuclear elements (SINEs) and repetitive elements, these elements may have contributed to the architecture of the rainbow trout IgH locus. The position of the second and third VH genes implies that they are used by IgM, and given the reverse orientation of the second VH gene, it is likely used by means of an inversion mechanism. Finally, the last portion of the trout IgH.1 locus encodes six DH and five JH elements (all with standard RSSs) that are used by the μ and δ genes.
Fig. 3.
Genomic organization of the rainbow trout and zebrafish IgH loci. Two major sequence contigs (107 and 14 kbp) resulted for the trout IgH.1 locus, which have been merged into a single contig within the δ gene. The three trout VH genes are members of the VH2, -8, and -5 families, respectively (18). The zebrafish locus (≈75 kbp) is based on ensembl supercontigs ctg14038, ctg1404, and ctg14057 and BAC sequences BX649502 and BX510335. ψ, pseudogenes; R, repetitive regions; →, transcriptional orientation.
We were unable to fully join OnmyBAC-IgH.1 into a single contig because of the presence of a repetitive element (≈900 bp) that has a core motif of TATAACAGTAGCGAGGC27 and is located between the δ2B (terminus of the major contig) and δ3B exons. The second contig encodes the δ3B, δ4B, δ2C (not present in Atlantic salmon), and δ7 C domains as well as the two transmembrane domains for IgD, but remote blastn/x analysis of all shotgun clones (>2,600 clones) was negative for exons resembling teleost δ5 or δ6. Given the position of the unique repetitive element and the presence of several repetitive elements (SINEs and Tc1 elements) between δ2C and δ7, δ5 and δ6 were likely deleted from this locus. Overall, the trout and zebrafish IgH loci (Fig. 3 Upper) are quite similar in size, content, and organization, thus providing support for the supposition that τ and ζ are likely orthologous genes.
The AID gene is essential for class-switch recombination and somatic hypermutation of Ig genes (30). Because an AID homolog has been reported in catfish (32), the question was raised whether class-switch recombination of the trout Ig isotypes was possible, but bioinformatic and manual sequence analysis did not reveal any obvious repetitive regions similar to those for amphibian, avian, or mammalian switch regions (27, 33). Our analysis of cDNA clones indicated that at least some of the trout IgH locus has been duplicated, given the presence of duplicated versions of the τ and δ genes from a single homozygous trout. To formally test this hypothesis, we physically mapped four of the IgH-positive BAC clones, including OnmyBAC-IgH.1, by using in situ hybridization on trout chromosomal spreads. All BAC clones hybridized with the short and long arm of chromosomes 1 and 12, respectively (see Fig. 8, which is published as supporting information on the PNAS web site), thus supporting the hypothesis that trout possess two IgH loci.
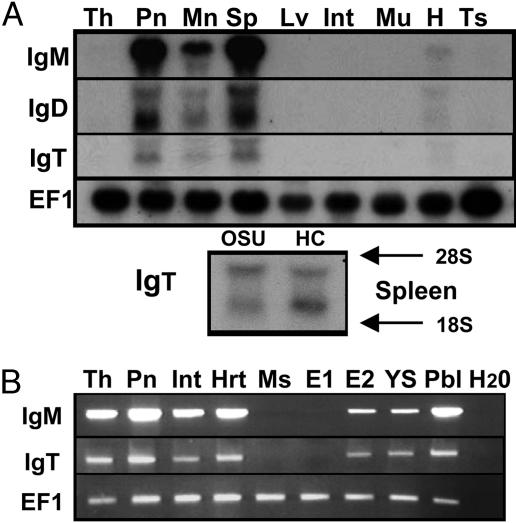
Expression of IgH Isotypes in Naïve Tissues. The tissue distribution for all three IgH isotypes was examined by Northern blot and RT-PCR analysis. In trout, the thymus and pronephros are the primary lymphoid tissues because they express key markers involved in VDJ recombination, namely Rag and TdT (34). The trout spleen is the major secondary lymphoid-organ tissue involved in antigen processing and is a major source of B cells and MHC class II expression (35). Northern blot analysis (Fig. 4A) demonstrates that rainbow trout μ, δ, and τ genes are primarily expressed within the spleen, pronephros, and mesonephros, with weak expression noted in the thymus and heart after prolonged exposures. Overall, IgM is expressed at the highest levels, followed by IgD and IgT. A single highly expressed transcript is present for IgM, whereas two mRNA transcripts are observed for trout IgD. The shorter transcript for IgD (3 kb) is expressed at higher levels, compared with the 3.8-kb transcript, and corresponds with the size of trout full-length IgD clones 1 and 17. Two transcripts are also present for IgT, corresponding to the sizes of the membrane-bound (2.1 kb) and secreted (2.4 kb) versions of IgT. A similar pattern of expression for IgT was observed for the splenic samples derived from two different clonal lines of trout.
Fig. 4.
Tissue-specific expression of IgM, IgD, and IgT in naïve rainbow trout. (A) Northern blot analysis (12 μg) by using RNA from 1-yr-old rainbow trout (Clear Springs strain). Th, thymus; Pn, pronephros; Mn, mesonephros; Sp, spleen; Lv, liver; Int, intestine; Mu, muscle; H, heart; Ts, testis. OSU-142 (OSU) and Hot Creek (HC) spleen samples. (B) RT-PCR analysis of IgM and IgT in select tissues. Hrt, heart; Ms, mesonephros; E1, day-12 embryo; E2, day-22 embryo; YS, yolk sac fry; Pbl, peripheral blood lymphocytes.
For a more sensitive analysis, we then examined the expression of IgM and IgT by RT-PCR (Fig. 4B). Ontogenically, Rag1 and -2 expression commence at ≈10 d postfertilization in trout with the first sIgM+ (surface IgM) B cells occurring in the pronephros at 4 d posthatch (34). Typically, the embryonic period from fertilization to hatching in trout is 25–26 d at 12°C. A major event during B cell development is the rearrangement and expression of IgH during the transition from the progenitor to the pre-B cell stage before IgL chain rearrangement and expression. In the present study, we demonstrate that neither IgM nor IgT C regions are expressed within day-12 embryos, but both are expressed, albeit at different levels, within embryos 4 d before hatching and within yolk-sac fry (3 d posthatch), before the presence of sIgM+ B cells in the pronephros. Thus, both isotypes are expressed early and throughout trout development. Both genes are also expressed in the thymus, intestine, and heart, but muscle is negative. The expression of both genes in the intestine is relatively weak, but our analysis was carried out with naïve trout, and expression is likely different during an immune response. Finally, Ficoll-purified peripheral blood lymphocytes highly express IgM message and modest levels of IgT mRNA, thus the expression in the heart is most likely due to circulating B cells. Salmonid blood is a rich source of leukocytes (>1–2 × 107 per milliliter), and FACS analysis demonstrates that 25–35% of the lymphoid gate is composed of sIgM+ B cells (data not shown). Thus, given the amount of total lymphocytes in blood and the expression of IgT, IgT+ B cells likely represent a small but significant pool of B cells.
In summary, we have provided information about a unique IgH isotype that we have named IgT, for teleost fish. Phylogenetic analysis suggests a possible relationship of the τ C-region gene with the fish μ lineage, but it is clear that τ represents a divergent IgH isotype whose role in the trout immune response awaits further investigation. A second major feature of the analysis was the discovery that the τ locus possesses its own set of D and J elements, and the position of these elements, along with the τ gene upstream of μ, provides an IgH genomic architecture that has not been seen in any other vertebrate class. In addition, by analyzing a single homozygous trout, we conclusively showed that the IgD and IgT genes are duplicated in rainbow trout, a feature that is likely because of the tetraploid ancestry of all salmonid fish. Finally, we hypothesize that, given the architecture of the trout and zebrafish IgH loci and the absence of obvious switch regions in trout, progenitor B cells, through epigenic factors, likely become committed to either IgT or IgM lineages during B cell development, in a process similar to that of T cell lineage commitment.
Supplementary Material
Acknowledgments
We thank Nil Ratan Saha and Louis Du Pasquier for their comments on this manuscript and Li Li and Sara Ho (Lark Technologies, Houston) for their assistance with the BAC assembly. This work was supported in part by U.S. Department of Agriculture National Research Initiative Competitive Grants Program (NRICGP) Grant 2004-05635 and National Science Foundation Grant MCB-0453924 (to J.D.H.).
Author contributions: J.D.H. designed research; J.D.H., E.D.L., and R.B.P. performed research; J.D.H. analyzed data; and J.D.H. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: BAC, bacterial artificial chromosome; IgH, immunoglobulin heavy chain; Onmy, Oncorhynchus mykiss; NAR, new antigen receptor.
Data deposition: cDNA and BAC sequences reported in this paper have been deposited in the GenBank database (accession nos. AY870256–AY870268 and AY872256–AY872257).
Note. While this paper was under review at PNAS, Danilova et al. (36) reported their analysis of an isotype, IgZ, in zebrafish. Similar to the findings presented here, zebrafish IgZ and IgM do not share D and J elements, implying that IgT and IgZ are distinct B cell receptors compared to IgM. However, in adult zebrafish, IgZ is limited to the primary lymphoid tissue, whereas IgT is expressed in a variety of trout tissues, suggesting functional differences between the IgT and IgZ isotypes. Finally, phylogenetic analysis indicates an orthologous relationship for IgZ and IgT, but whether they are truly functional orthologues awaits further experimentation.
References
- 1.Eason, D. D., Cannon, J. P., Haire, R. N., Rast, J. P., Ostrov, D. A. & Litman, G. W. (2004) Semin. Immunol. 16, 215–226. [DOI] [PubMed] [Google Scholar]
- 2.Flajnik, M. F. (2002) Nat. Rev. Immunol. 2, 688–698. [DOI] [PubMed] [Google Scholar]
- 3.Wilson, M., Bengten, E., Miller, N. W., Clem, L. W., Du Pasquier, L. & Warr, G. W. (1997) Proc. Natl. Acad. Sci. USA 94, 4593–4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hordvik, I., Thevarajan, J., Samdal, I., Bastani, N. & Krossoy, B. (1999) Scand. J. Immunol. 50, 202–210. [DOI] [PubMed] [Google Scholar]
- 5.Bernstein, R. M., Schluter, S. F., Shen, S. & Marchalonis, J. J. (1996) Proc. Natl. Acad. Sci. USA 93, 3289–3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Greenberg, A. S., Hughes, A. L., Guo, J., Avila, D., McKinney, E. C. & Flajnik, M. F. (1996) Eur. J. Immunol. 26, 1123–1129. [DOI] [PubMed] [Google Scholar]
- 7.Greenberg, A. S., Avila, D., Hughes, M., Hughes, A., McKinney, E. C. & Flajnik, M. F. (1995) Nature 374, 168–173. [DOI] [PubMed] [Google Scholar]
- 8.Young, W. P., Wheeler, P. A., Fields, R. D. & Thorgaard, G. H. (1996) J. Hered. 87, 77–80. [DOI] [PubMed] [Google Scholar]
- 9.Phillips, R. B., Zimmerman, A., Noakes, M. A., Palti, Y., Morasch, M. R., Eiben, L., Ristow, S. S., Thorgaard, G. H. & Hansen, J. D. (2003) Immunogenetics 55, 561–569. [DOI] [PubMed] [Google Scholar]
- 10.Ewing, B. & Green, P. (1998) Genome Res. 8, 186–194. [PubMed] [Google Scholar]
- 11.Gordon, D., Abajian, C. & Green, P. (1998) Genome Res. 8, 195–202. [DOI] [PubMed] [Google Scholar]
- 12.Hansen, J. D., Strassburger, P., Thorgaard, G. H., Young, W. P. & Du Pasquier, L. (1999) J. Immunol. 163, 774–786. [PubMed] [Google Scholar]
- 13.Kumar, S., Tamura, K., Jakobsen, I. B. & Nei, M. (2001) Bioinformatics 17, 1244–1245. [DOI] [PubMed] [Google Scholar]
- 14.Lee, M. A., Bengten, E., Daggfeldt, A., Rytting, A. S. & Pilstrom, L. (1993) Mol. Immunol. 30, 641–648. [DOI] [PubMed] [Google Scholar]
- 15.Hansen, J., Leong, J. A. & Kaattari, S. (1994) Mol. Immunol. 31, 499–501. [DOI] [PubMed] [Google Scholar]
- 16.Hordvik, I., Berven, F. S., Solem, S. T., Hatten, F. & Endresen, C. (2002) Mol. Immunol. 39, 313–321. [DOI] [PubMed] [Google Scholar]
- 17.Bengten, E., Quiniou, S. M., Stuge, T. B., Katagiri, T., Miller, N. W., Clem, L. W., Warr, G. W. & Wilson, M. (2002) J. Immunol. 169, 2488–2497. [DOI] [PubMed] [Google Scholar]
- 18.Roman, T., Andersson, E., Bengten, E., Hansen, J., Kaattari, S., Pilstrom, L., Charlemagne, J. & Matsunaga, T. (1996) Immunogenetics 43, 325–326. [DOI] [PubMed] [Google Scholar]
- 19.Saha, N. R., Suetake, H., Kikuchi, K. & Suzuki, Y. (2004) Immunogenetics 56, 438–447. [DOI] [PubMed] [Google Scholar]
- 20.Rexroad, C. E., III, Lee, Y., Keele, J. W., Karamycheva, S., Brown, G., Koop, B., Gahr, S. A., Palti, Y. & Quackenbush, J. (2003) Cytogenet. Genome Res. 102, 347–354. [DOI] [PubMed] [Google Scholar]
- 21.Lesk, A. M. & Chothia, C. (1982) J. Mol. Biol. 160, 325–342. [DOI] [PubMed] [Google Scholar]
- 22.Campbell, K. S., Backstrom, B. T., Tiefenthaler, G. & Palmer, E. (1994) Semin. Immunol. 6, 393–410. [DOI] [PubMed] [Google Scholar]
- 23.Yoo, E. M., Coloma, M. J., Trinh, K. R., Nguyen, T. Q., Vuong, L. U., Morrison, S. L. & Chintalacharuvu, K. R. (1999) J. Biol. Chem. 274, 33771–33777. [DOI] [PubMed] [Google Scholar]
- 24.Wiersma, E. J., Chen, F., Bazin, R., Collins, C., Painter, R. H., Lemieux, R. & Shulman, M. J. (1997) J. Immunol. 158, 1719–1726. [PubMed] [Google Scholar]
- 25.Hsu, E., Flajnik, M. F. & Du Pasquier, L. (1985) J. Immunol. 135, 1998–2004. [PubMed] [Google Scholar]
- 26.Mussmann, R., Du Pasquier, L. & Hsu, E. (1996) Eur. J. Immunol. 26, 2823–2830. [DOI] [PubMed] [Google Scholar]
- 27.Du Pasquier, L., Robert, J., Courtet, M. & Mussmann, R. (2000) Immunol. Rev. 175, 201–213. [DOI] [PubMed] [Google Scholar]
- 28.Roman, T., De Guerra, A. & Charlemagne, J. (1995) Eur. J. Immunol. 25, 269–273. [DOI] [PubMed] [Google Scholar]
- 29.Roux, K. H., Greenberg, A. S., Greene, L., Strelets, L., Avila, D., McKinney, E. C. & Flajnik, M. F. (1998) Proc. Natl. Acad. Sci. USA 95, 11804–11809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Honjo, T., Muramatsu, M. & Fagarasan, S. (2004) Immunity 20, 659–668. [DOI] [PubMed] [Google Scholar]
- 31.Daggfeldt, A., Bengten, E. & Pilstrom, L. (1993) Immunogenetics 38, 199–209. [DOI] [PubMed] [Google Scholar]
- 32.Saunders, H. L. & Magor, B. G. (2004) Dev. Comp. Immunol. 28, 657–663. [DOI] [PubMed] [Google Scholar]
- 33.Lundqvist, M. L., Middleton, D. L., Hazard, S. & Warr, G. W. (2001) J. Biol. Chem. 276, 46729–46736. [DOI] [PubMed] [Google Scholar]
- 34.Hansen, J. D. & Zapata, A. G. (1998) Immunol. Rev. 166, 199–220. [DOI] [PubMed] [Google Scholar]
- 35.Ohta, Y., Landis, E., Boulay, T., Phillips, R. B., Collet, B., Secombes, C. J., Flajnik, M. F. & Hansen, J. D. (2004) J. Immunol. 173, 4553–4560. [DOI] [PubMed] [Google Scholar]
- 36.Danilova, N., Bussmann, J., Jekosch, K. & Steiner, L. A. (2005) Nat. Immunol. 6, 295–302. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.