Abstract
Frailty is classically associated with advanced age but is also an important predictor of clinical outcomes in comparatively young adults with cirrhosis. We examined the association of biological aging with frailty and post-transplant outcomes in a pilot of adults with cirrhosis undergoing liver transplantation (LT). Frailty was measured via the Liver Frailty Index (LFI). The primary epigenetic clock DNA methylation (DNAm) PhenoAge was calculated from banked peripheral blood mononuclear cells; we secondarily explored two first-generation clocks (Hannum; Horvath) and two additional second-generation clocks (GrimAge; GrimAge2). Twelve adults were included: seven frail (LFI ≥ 4.4, mean age 55 years) and five robust (LFI < 3.2, mean age 55 years). Mean PhenoAge age acceleration (AgeAccel) was + 2.5 years (P = 0.23) for frail versus robust subjects. Mean PhenoAge AgeAccel was + 2.7 years (P = 0.19) for subjects who were readmitted or died within 30 days of discharge post-LT versus those without this outcome. When compared with first-generation clocks, the second-generation clocks demonstrated greater average AgeAccel for subjects with frailty or poor post-LT outcomes. Measuring biological age using DNAm-derived epigenetic clocks is feasible in adults undergoing LT. While frail and robust subjects had the same average chronological age, average biological age as measured by second-generation epigenetic clocks tended to be accelerated among those who were frail or experienced a poor post-LT outcome. These results suggest that frailty in these relatively young subjects with cirrhosis may involve similar aging mechanisms as frailty classically observed in chronologically older adults and warrant validation in a larger cohort.
Supplementary Information
The online version contains supplementary material available at 10.1007/s11357-024-01076-5.
Keywords: Biological age, Epigenetic clock, Cirrhosis, Frailty
Introduction
Frailty is an important predictor of mortality and clinical outcomes in adults with cirrhosis even though these patients are relatively young while frailty is classically associated with advanced age [1–5]. Advanced age typically refers to chronological age, yet age-related vulnerabilities are likely driven by biological mechanisms that may be more or less advanced in individuals relative to their chronological age. Biological mechanisms of aging may similarly underlie frailty [6–8].
One emerging method for estimating biological age is by quantifying changes in DNA methylation (DNAm) using epigenetic clocks [9]. Multiple epigenetic clocks currently exist that are distinct in their derivations, tissue specificity, and associations with clinical outcomes [9–11]. These DNAm-based clocks can be divided into “first-generation” (e.g., Horvath [12]; Hannum [13]) and “second-generation” (e.g., PhenoAge [14]; GrimAge [15], and GrimAge2 [16]) clocks. The primary goal of the first-generation epigenetic clocks is to predict chronological age, whereas the primary goal of the second-generation epigenetic clocks is to predict aging-related phenotypes and outcomes, such as morbidity and mortality [16]. For example, DNAm PhenoAge (hereafter, PhenoAge) is trained on clinical predictors of age-associated mortality from National Health and Nutrition Examination Survey (NHANES) data and is associated with several medical conditions common in older populations [14], while GrimAge is based on DNAm surrogates of 12 plasma proteins and smoking pack-years [15]. The difference between an individual’s predicted age (via epigenetics) and chronological age adjusted by chronological age, called age acceleration (AgeAccel), predicts many diseases [17–19]. Thus, age is considered to be undesirably “accelerated” when biological age is greater than chronological age.
Cirrhosis is a unique context in which to study frailty because frailty strongly predicts clinical outcomes, even though this patient population is relatively chronologically young, and frailty may be reversible after transplant [20–22]. Prior studies have shown strong associations between a cirrhosis-specific Liver Frailty Index (LFI) score and worse clinical outcomes for frail patients [20, 23, 24]. LFI incorporates classical aging-related frailty measures including grip strength, chair stands, and balance. Frailty in this population is likely driven by both disease-related and aging-related mechanisms, with the former hypothesized to be more reversible than the latter after transplantation. It is unknown if biological mechanisms of aging play a role in mediating all frailty in these patients. In other words, it is unknown whether cirrhosis is driving accelerated, but potentially reversible, biological aging. Better understanding the mechanisms of frailty in this population could lead to new translational approaches to mitigate it and improve outcomes. There is already emerging utility for measuring biological age via epigenetic clocks in both transplant donors and recipients, which may improve the estimation of risk for post-transplant complications [25–28]. Less is known about the potential applications of measuring biological aging, such as via epigenetic clocks, in adults with cirrhosis undergoing liver transplant.
Here, we aim to investigate whether accelerated biological age, specifically PhenoAge AgeAccel, is associated with frailty and other clinical outcomes in a feasibility pilot of adults with cirrhosis undergoing liver transplantation. We then explored the consistency of our findings across four commonly used epigenetic clocks spanning both first- and second-generation methodologies.
Methods
Participants and study design
This pilot study was conducted as part of the Functional Assessment in Liver Transplantation (FrAILT) Study, a prospective study that enrolls ambulatory adults with cirrhosis awaiting liver transplantation. FrAILT Study participants who underwent liver transplantation at the University of California, San Francisco, from 2016 to 2019 and completed biospecimen collection at the time of liver transplantation (e.g., banked blood and liver tissue) were considered for this pilot. As part of the FrAILT Study protocol, all participants undergo in-person testing of physical frailty in the pre-transplant ambulatory setting using the Liver Frailty Index, calculated from grip strength, timed chair stands, and balance [23]. Participants who were classified as either frail (Liver Frailty Index ≥ 4.4) or robust (Liver Frailty Index < 3.2) were then selected in rank order to maximize differences between groups; subjects classified as pre-frail were excluded.
This study was performed in accordance with the ethical standards of the responsible committee on human experimentation, specifically approved by the University of California, San Francisco Institutional Review Board (#11–07513), and with the Declaration of Helsinki.
Additional data collection
Participant characteristics were abstracted from the electronic medical record, which included age, sex, race, ethnicity, body mass index (BMI), indication for liver transplant (e.g., cirrhosis due to alcohol use, hepatitis C virus, nonalcoholic steatohepatitis, cholestatic disease, or other), laboratory values to calculate the Model for End-stage Liver Disease Sodium (MELDNa) score, and medical co-morbidities (diabetes, hypertension).
DNA methylation analysis and epigenetic clock generation
Blood was collected at the start of liver transplantation, prior to administration of intravenous corticosteroids. DNA was extracted from peripheral blood mononuclear cells (PBMCs) through the centralized UCSF Institute for Human Genetics (IHG) Genomic Core Facility. Extracted DNA was then analyzed by the University of Minnesota Genomics Center, where the genome-wide DNA methylation (DNAm) status of 850,000 CpG dinucleotide sites was measured in triplicate using Illumina MethylationEPIC arrays [29]. The Illumina EPIC methylation array data were read and processed using the R package minfi (version 1.440) [30]. Detection P-values comparing the total signal for each probe to the background signal level were calculated to evaluate the quality of the samples. No samples with poor quality (mean detection P > 0.01) were identified. Quantile normalization (preprocessQuantile function in minfi) was used to normalize the data [31]. Our pre-planned DNAm clock, PhenoAge, was calculated using the MethylClock R package (version 1.4.0), which was also used to calculate the Horvath and Hannum clocks. GrimAge and GrimAge2 were calculated using R scripts generated by the authors of the original studies.
The first-generation clocks Horvath [12] and Hannum [13] were optimized to reflect age-related DNAm changes and thus predict chronological age. Better prediction of chronological age may worsen the prediction of age-related health outcomes that are mediated by biological aging [32]. The second-generation clocks PhenoAge [14] and GrimAge [15, 16] were optimized to predict clinical outcomes related to aging and mortality, rather than chronological age per se. The development of DNAm PhenoAge occurred in two steps using data from the National Health and Nutrition Examination Survey (NHANES) [14]. First, a composite biomarker index (PhenoAge) was trained to predict aging-related mortality. Second, the epigenetic clock (DNAm PhenoAge) was trained to predict the PhenoAge composite index. DNAm PhenoAge was validated for predicting mortality and various aging conditions in four large longitudinal studies [14]. An alternative second-generation epigenetic clock, GrimAge, incorporated DNAm markers of key plasma proteins that increase with age, and a methylation marker of smoking pack-years [15]. The seven included DNAm-based surrogates for plasma proteins were adrenomedullin levels (DNAm ADM), beta-2 microglobulin (DNAm B2M), cystatin C (DNAm cystatin C), growth differentiation factor 15 (DNAm GDF-15), leptin (DNAm leptin), plasminogen activation inhibitor 1 (DNAm PAI-1), and tissue inhibitor metalloproteinase 1 (DNAm TIMP-1) [15]. An update to GrimAge (“GrimAge2”) included two additional DNAm-based estimators of (log transformed) plasma proteins: high-sensitivity C-reactive protein (DNAm logCRP) and hemoglobin A1C (DNAm logA1C) [16]. The DNAm estimation of each plasma protein included in GrimAge2 was analyzed for each study subject.
Age acceleration (AgeAccel) was calculated for each sample as the residual of the regression of chronological age and predicted age from the epigenetic clock. This provides a normalized measure indicating whether an individual appears to be biologically older (age acceleration > 0) or younger (age acceleration < 0) compared to their age peers. This age acceleration metric has been shown to minimize spurious associations [19].
Statistical analysis
The percentage of variation in each epigenetic clock that was accounted for by chronological age was calculated using a coefficient of determination (R2) derived from a simple linear regression.
Group differences between frail and robust participants were calculated using parametric statistics. All analyses were two-tailed, and a P-value was set at 0.05. Adjustments for potential confounders were not made due to the small sample of participants. Statistical analyses and figure generation were performed using Prism v9.4.1.
Results
A total of 12 patients with cirrhosis who underwent liver transplantation were included in this pilot study: seven frail and five robust (Table 1). For both groups, the average age was 55 years and approximately 40% of the subjects were women. Compared with robust participants, the frail participants were more likely to have a history of diabetes, hypertension, or non-alcoholic steatohepatitis, and higher average MELD-Na scores at the time of liver transplantation.
Table 1.
Baseline characteristics of FrAILT pilot study participants by frailty status
| Characteristic | Frail (N = 7) | Robust (N = 5) |
|---|---|---|
| Age (years)* | 55 ± 8 | 55 ± 8 |
| Female sex, n (%) | 3 (43%) | 2 (40%) |
| Race, n (%) | ||
| White | 7 (100%) | 4 (80%) |
| Asian | 0 (0%) | 1 (20%) |
| Hispanic or Latino ethnicity, n (%) | 1 (14%) | 1 (20%) |
| Medical history, n (%) | ||
| Diabetes | 3 (43%) | 0 (0%) |
| Hepatocellular carcinoma | 1 (14%) | 2 (40%) |
| Hypertension | 5 (71%) | 0 (0%) |
| Etiology of liver disease, n (%) | ||
| Alcohol-associated cirrhosis | 2 (29%) | 1 (20%) |
| Non-alcoholic steatohepatitis | 4 (57%) | 1 (20%) |
| Chronic hepatitis C | 1 (14%) | 1 (20%) |
| Cholestatic disease | 0 (0%) | 1 (20%) |
| Other | 0 (0%) | 1 (20%) |
| Body mass index (kg/m2)* | 32.4 ± 8.3 | 26.5 ± 7.6 |
| MELD-Na at liver transplant* | 24 ± 11 | 16 ± 9 |
| Liver Frailty Index* | 4.9 ± 0.4 | 2.7 ± 0.2 |
*Average ± standard deviation
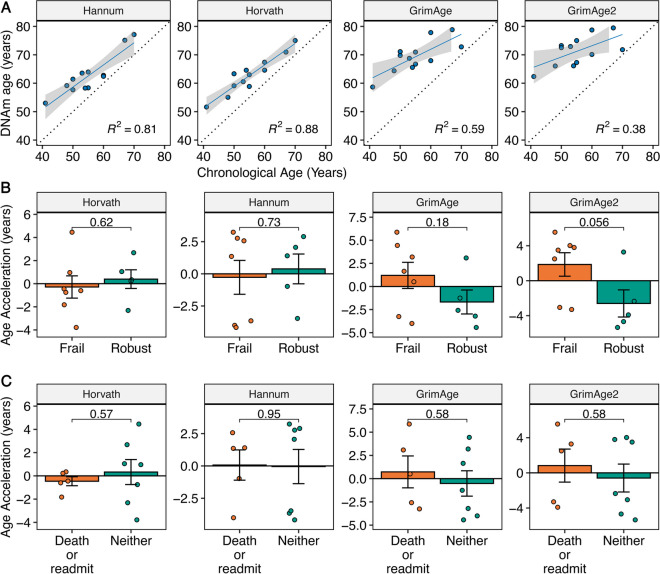
First, we investigated the correlation between the biological age estimated by PhenoAge and the corresponding chronological age (Fig. 1A). Sixty-six percent of the variation in biological age estimated by PhenoAge was explained by chronological age (R2 = 0.66, Fig. 1A). We explored the correlation between biological age, estimated by the other four clocks, and corresponding chronological age, which demonstrated a greater correlation for the first-generation clocks compared with the second-generation clocks (Fig. 2A).
Fig. 1.
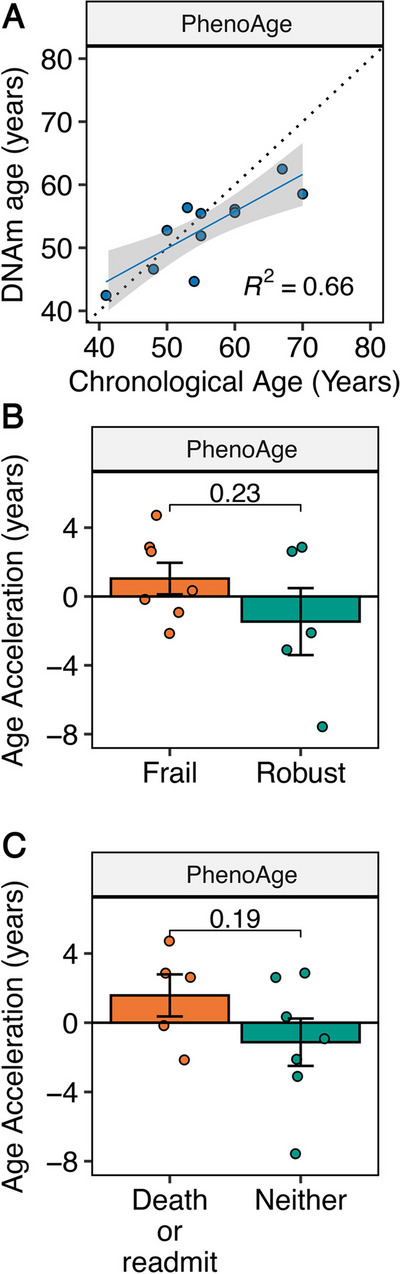
DNA methylation (DNAm) PhenoAge data for pilot study of FrAILT participants. A Linear regression of DNAm PhenoAge versus chronological age. B DNAm PhenoAge age acceleration in years by LFI frailty status (frail, LFI ≥ 4.4; robust LFI < 3.2). P-value by unpaired T test. C DNAm age acceleration in years by clinical outcome (death or readmission within 30 days of discharge versus neither). P-value by unpaired T test
Fig. 2.
DNA methylation (DNAm) data for first-generation (Hannum; Horvath) and additional second-generation (GrimAge; GrimAge2) epigenetic clocks for the pilot study of FrAILT participants. A Linear regression of DNAm age versus chronological age for each epigenetic clock. B DNAm age acceleration in years by LFI frailty status (frail, LFI ≥ 4.4; robust LFI < 3.2) for each epigenetic clock. P-value by unpaired T test. C DNAm age acceleration in years by clinical outcome (death or readmission within 30 days of discharge versus neither) for each epigenetic clock. P-value by unpaired T test
We next explored the average differences in AgeAccel by either pre-transplant frailty status or post-transplant clinical outcome. Our pre-planned analysis focused on the second-generation clock PhenoAge, which we hypothesized was more likely to show an association with frailty due to its design. The mean PhenoAge AgeAccel was + 2.5 years (P = 0.23) for frail versus robust subjects (Fig. 1B). When comparing subjects by frailty status, differences in AgeAccel for frail versus robust subjects were clustered by clock type (Fig. 2B). The mean AgeAccel for the first-generation clocks was − 0.7 years (P > 0.5) when comparing frail versus robust subjects (Fig. 2B). However, the other second-generation clocks showed mean AgeAccel of + 2.9 years (GrimAge; P = 0.18) and + 4.5 years (GrimAge2; P = 0.056) (Fig. 2B). We next explored post-transplant clinical outcomes by creating a composite outcome encompassing death and/or readmission within 30 days of hospital discharge following a liver transplant. There were five subjects (three frail) who were readmitted to the hospital or died within 30 days of hospital discharge, and seven subjects without this outcome. The mean PhenoAge AgeAccel was + 2.7 years (P = 0.19) for those with the composite outcome versus those without (Fig. 1C). As with frailty status, differences in AgeAccel by clinical outcome were clustered by clock type. When comparing subjects by clinical outcome, the first-generation clocks demonstrated mean AgeAccel differences of less than 1 year (P > 0.5) for those who experienced this poor clinical outcome versus those without this outcome (Fig. 2C). In contrast, the second-generation clocks tended, on average, to demonstrate accelerated aging for the subjects with poor clinical outcomes (Fig. 2C).
The subcomponents of GrimAge2, the DNAm predictors of individual plasma proteins, show varied associations with specific clinical conditions such as steatohepatitis and adiposity [16]. Therefore, we explored which subcomponents of GrimAge2 were most associated with frailty in this cohort (Supplementary Fig. 1). We found that frail subjects demonstrated increased mean DNAm logCRP compared with robust subjects (P = 0.032) (Supplementary Fig. 1A).
Discussion
In this pilot of 12 adults with cirrhosis undergoing liver transplantation, we established that measuring biological age using epigenetic clocks derived from DNA methylation data is feasible. As expected, we found greater correlations between chronological age and the biological ages estimated by the first-generation epigenetic clocks than the biological ages estimated by the second-generation epigenetic clocks, as the latter were developed to predict biological aging-related mortality rather than chronological age per se. The relatively lower variation in chronological age explained by the second-generation clocks in this cohort was expected, given that the cohort is relatively young with a narrow range of ages—yet a wide variation in frailty status. In aggregate, both frail and robust subjects had the same average chronological age, but we observed that second-generation estimates of biological age tended, on average, to be accelerated among those who were physically frail and decelerated among those who were physically robust. As hypothesized, we also observed a trend for AgeAccel, when estimated using second-generation clocks, to be greater in subjects who experienced a poor clinical outcome (death or hospital readmission). While neither of these comparisons met the threshold for statistical significance (P < 0.05), the consistent direction of these differences across the three second-generation clocks provides provocative evidence to justify further investigation in an adequately powered cohort. Lastly, we found that among the subcomponents of GrimAge2, DNAm estimations of CRP showed the strongest difference between frail and non-frail subjects.
To our knowledge, this is the first study of the relationship between biological age and frailty in a cirrhosis/liver transplant cohort. Cirrhosis and liver transplant is a uniquely powerful model for studying the relationship between disease, aging, and frailty. Most patients are relatively young, but recent research has revealed that they become frail in ways highly reminiscent of classical aging-related frailty, a frailty index is strongly predictive of outcomes, and, most interestingly, frailty is to some extent reversible after transplant. It is not yet clear if frailty driven primarily by a single disease process (cirrhosis) is the same process as the classical multisystem aging/multimorbidity-driven frailty in old age. A cirrhosis/liver transplant cohort provides an opportunity to separate frailty from advanced chronological age, and indeed, we found a pilot signal that biological aging is accelerated in these young but frail patients. Importantly, this signal was only present for second-generation clock estimates, which likely reflects how these clocks are optimized to predict aging phenotypes, as opposed to the first-generation clocks that are optimized to predict chronological age. These pilot data suggest that the pathophysiology of frailty in cirrhosis may indeed involve similar mechanisms of aging as frailty in older adults, either in addition to or downstream of cirrhosis-specific pathophysiology. A prior study found similar biological age acceleration in chronic kidney disease which was ameliorated by renal transplantation [33], supporting the hypothesis that biological aging may be downstream of multiple disease-specific pathophysiologies and therefore a plausible target for both risk prediction and therapeutics across multiple conditions. It is possible that optimal prediction of clinical outcomes in such multifactorial conditions can be made with the same epigenetic clock designed to predict crosscutting aging phenotypes.
We also found a statistically significant association between frailty and the GrimAge2 DNAm estimation of CRP, but not its other nine DNAm-based components. The relationship between elevated CRP and frailty is established in other cohorts using circulating blood-based measures [34], and some studies [35] suggest DNAm-based CRP measures may outperform circulating CRP as a marker of chronic inflammation. The association between frailty and elevated DNAm-based CRP identified in this cohort supports the hypothesis that the frailty experienced by these relatively young patients with cirrhosis may be similar to prototypical age-related frailty.
A larger study might be able to distinguish to what degree (or in which aspects) frailty and the biological aging that underlies it can be reversed after the resolution of the precipitating disease (i.e., after liver transplant). Reversing (accelerated) biological aging in the context of cirrhosis may provide a proof of concept for reversing biological aging in other contexts. This pathophysiological question could have important implications for the clinical management of cirrhosis, by providing better prognostication of patients whose frailty will best recover after transplant, or who are at the highest risk for unfavorable outcomes, as well as potentially suggesting pre- or peri-transplant therapies targeting biological mechanisms of aging.
Several limitations are noted. This pilot cohort is small and incorporates a single blood specimen collection timepoint due to our initial goal of establishing feasibility and testing this concept in this population. Thus, there may be residual confounding by factors that are different between groups that we are unable to control for due to the small number of participants. We selected participants from the non-contiguous frail and robust categories to focus on between-group differences and so are unable to analyze the frailty score as a continuous variable. For the purposes of this study, we limited our investigation to five commonly used epigenetic clocks spanning two main methodologies, though future studies may benefit from the incorporation of additional or customized epigenetic clocks to estimate age in different physiological systems.
Conclusions
Measuring biological age via epigenetic clock analysis is feasible in adults with cirrhosis undergoing liver transplant. Second-generation clock measures of AgeAccel may be associated with both frailty and clinical outcomes in these relatively young patients. DNA methylation-based measures of CRP are also associated with frailty in these subjects. A larger study is needed to determine if biological age provides predictive power for frailty-related outcomes independent of clinical frailty measures.
Supplementary Information
Below is the link to the electronic supplementary material.
Funding
This work was supported by a UCSF Bakar Aging Research Institute (BARI) Investigator Award and Buck Institute for Research on Aging intramural funds. Original sample collection was supported in part by R21AG067554.
Dr. LaHue is supported by R03AG074035, Larry L. Hillblom Foundation (A137420), UCSF Claude D. Pepper Older Americans Independence Center P30 AG044281, and the UCSF Bakar Aging Research Institute.
Dr. Furman is supported by Buck Bioinformatics Core, P01AG066591, U54AG075932, P01AI153559.
Dr. Lai is supported by R01AG059183, K24AG080021, P30DK026743.
Dr. Newman is supported by NIH R01AG068025 and Buck Institute Institutional Funds.
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Jennifer C. Lai and John C. Newman contributed equally to this work.
References
- 1.Clegg A, Young J, Iliffe S, Rikkert MO, Rockwood K. Frailty in elderly people. The Lancet. 2013;381(9868):752–762. doi: 10.1016/S0140-6736(12)62167-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lai JC, Dodge JL, Sen S, Covinsky K, Feng S. Functional decline in patients with cirrhosis awaiting liver transplantation: results from the functional assessment in liver transplantation (FrAILT) study. Hepatology. 2016;63(2):574–580. doi: 10.1002/hep.28316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lai JC, Dodge JL, McCulloch CE, Covinsky KE, Singer JP. Frailty and the burden of concurrent and incident disability in patients with cirrhosis: a prospective cohort study. Hepatol Commun. 2020;4(1):126–133. doi: 10.1002/hep4.1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lai JC, Dodge JL, Kappus MR, et al. Changes in frailty are associated with waitlist mortality in patients with cirrhosis. J Hepatol. 2020;73(3):575–581. doi: 10.1016/j.jhep.2020.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lai JC, Shui AM, Duarte-Rojo A, et al. Frailty, mortality, and health care utilization after liver transplantation: from the multicenter functional assessment in liver transplantation (FrAILT) study. Hepatology. 2022;75(6):1471–1479. doi: 10.1002/hep.32268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guyonnet S, Rolland Y, Takeda C, et al. The INSPIRE bio-resource research platform for healthy aging and geroscience: focus on the human translational research cohort (the INSPIRE-T cohort) J Frailty Aging. 2021;10(2):110–120. doi: 10.14283/jfa.2020.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McCartney DL, Stevenson AJ, Walker RM, et al. Investigating the relationship between DNA methylation age acceleration and risk factors for Alzheimer’s disease. Alzheimers Dement (Amst) 2018;10:429–437. doi: 10.1016/j.dadm.2018.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.López-Otín C, Blasco MA, Partridge L, Serrano M, Kroemer G. Hallmarks of aging: an expanding universe. Cell. 2023;186(2):243–278. doi: 10.1016/j.cell.2022.11.001. [DOI] [PubMed] [Google Scholar]
- 9.Horvath S, Raj K. DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nat Rev Genet. 2018;19(6):371–384. doi: 10.1038/s41576-018-0004-3. [DOI] [PubMed] [Google Scholar]
- 10.Jansen R, Han LK, Verhoeven JE, et al. An integrative study of five biological clocks in somatic and mental health. Tyler JK, Hägg S, Hägg S, Belsky DW, van den Akker E, Le Couteur DG, eds. eLife. 2021;10:e59479. doi: 10.7554/eLife.59479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fransquet PD, Wrigglesworth J, Woods RL, Ernst ME, Ryan J. The epigenetic clock as a predictor of disease and mortality risk: a systematic review and meta-analysis. Clin Epigenetics. 2019;11(1):62. doi: 10.1186/s13148-019-0656-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013;14(10):R115. doi: 10.1186/gb-2013-14-10-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hannum G, Guinney J, Zhao L, et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell. 2013;49(2):359–367. doi: 10.1016/j.molcel.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levine ME, Lu AT, Quach A, et al. An epigenetic biomarker of aging for lifespan and healthspan. Aging (Albany NY). 2018;10(4):573–591. doi: 10.18632/aging.101414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu AT, Quach A, Wilson JG, et al. DNA methylation GrimAge strongly predicts lifespan and healthspan. Aging (Albany NY) 2019;11(2):303–327. doi: 10.18632/aging.101684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu AT, Binder AM, Zhang J, et al. DNA methylation GrimAge version 2. Aging. 2022;14(23):9484–9549. doi: 10.18632/aging.204434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma Q, Li BL, Yang L, et al. Association between phenotypic age and mortality in patients with multivessel coronary artery disease. Dis Markers. 2022;2022:4524032. doi: 10.1155/2022/4524032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Han WM, Apornpong T, Gatechompol S, et al. Association of phenotypic aging marker with comorbidities, frailty and inflammatory markers in people living with HIV. BMC Geriatr. 2022;22(1):1010. doi: 10.1186/s12877-022-03720-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.El Khoury LY, Gorrie-Stone T, Smart M, et al. Systematic underestimation of the epigenetic clock and age acceleration in older subjects. Genome Biol. 2019;20(1):283. doi: 10.1186/s13059-019-1810-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lai JC, Covinsky KE, McCulloch CE, Feng S. The Liver Frailty Index improves mortality prediction of the subjective clinician assessment in patients with cirrhosis. Am J Gastroenterol. 2018;113(2):235–242. doi: 10.1038/ajg.2017.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lai JC, Segev DL, McCulloch CE, Covinsky KE, Dodge JL, Feng S. Physical frailty after liver transplantation. Am J Transplant. 2018;18(8):1986–1994. doi: 10.1111/ajt.14675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lai JC, Shui AM, Duarte-Rojo A, et al. Association of frailty with health-related quality of life in liver transplant recipients. JAMA Surg. 2023;158(2):130–138. doi: 10.1001/jamasurg.2022.6387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lai JC, Covinsky KE, Dodge JL, et al. Development of a novel Frailty Index to predict mortality in patients with end-stage liver disease. Hepatology. 2017;66(2):564–574. doi: 10.1002/hep.29219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kardashian A, Ge J, McCulloch CE, et al. Identifying an optimal Liver Frailty Index cutoff to predict waitlist mortality in liver transplant candidates. Hepatology. 2021;73(3):1132–1139. doi: 10.1002/hep.31406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peters FS, Manintveld OC, Betjes MGH, Baan CC, Boer K. Clinical potential of DNA methylation in organ transplantation. J Heart Lung Transplant. 2016;35(7):843–850. doi: 10.1016/j.healun.2016.02.007. [DOI] [PubMed] [Google Scholar]
- 26.Schaenman J, Zhou X, Guo R, et al. DNA methylation age is more closely associated with infection risk than chronological age in kidney transplant recipients. Transplant Direct. 2020;6(8):e576. doi: 10.1097/TXD.0000000000001020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vasco M, Benincasa G, Fiorito C, et al. Clinical epigenetics and acute/chronic rejection in solid organ transplantation: an update. Transplant Rev. 2021;35(2):100609. doi: 10.1016/j.trre.2021.100609. [DOI] [PubMed] [Google Scholar]
- 28.Alsaggaf R, Katta S, Wang T, et al. Epigenetic aging and hematopoietic cell transplantation in patients with severe aplastic anemia. Transplant Cell Therapy. 2021;27(4):313.e1–313.e8. doi: 10.1016/j.jtct.2021.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pidsley R, Zotenko E, Peters TJ, et al. Critical evaluation of the Illumina MethylationEPIC BeadChip microarray for whole-genome DNA methylation profiling. Genome Biol. 2016;17(1):208. doi: 10.1186/s13059-016-1066-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aryee MJ, Jaffe AE, Corrada-Bravo H, et al. Minfi: a flexible and comprehensive bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. 2014;30(10):1363–1369. doi: 10.1093/bioinformatics/btu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Touleimat N, Tost J. Complete pipeline for Infinium(®) Human Methylation 450K BeadChip data processing using subset quantile normalization for accurate DNA methylation estimation. Epigenomics. 2012;4(3):325–341. doi: 10.2217/epi.12.21. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Q, Vallerga CL, Walker RM, et al. Improved precision of epigenetic clock estimates across tissues and its implication for biological ageing. Genome Med. 2019;11(1):54. doi: 10.1186/s13073-019-0667-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neytchev O, Erlandsson H, Witasp A, et al. Epigenetic clocks indicate that kidney transplantation and not dialysis mitigate the effects of renal ageing. J Intern Med. 2024;295(1):79–90. 10.1111/joim.13724 [DOI] [PubMed]
- 34.Soysal P, Stubbs B, Lucato P, et al. Inflammation and frailty in the elderly: a systematic review and meta-analysis. Ageing Res Rev. 2016;31:1–8. doi: 10.1016/j.arr.2016.08.006. [DOI] [PubMed] [Google Scholar]
- 35.Verschoor CP, Vlasschaert C, Rauh MJ, Paré G. A DNA methylation based measure outperforms circulating CRP as a marker of chronic inflammation and partly reflects the monocytic response to long-term inflammatory exposure: a Canadian longitudinal study on aging analysis. Aging Cell. 2023;22(7):e13863. doi: 10.1111/acel.13863. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.