Abstract
Functionally relevant hepadnavirus-cell surface interactions were investigated with the duck hepatitis B virus (DHBV) animal model by using an in vitro infection competition assay. Recombinant DHBV pre-S polypeptides, produced in Escherichia coli, were shown to inhibit DHBV infection in a dose-dependent manner, indicating that monomeric pre-S chains were capable of interfering with virus-receptor interaction. Particle-associated pre-S was, however, 30-fold more active, suggesting that cooperative interactions enhance particle binding. An 85-amino-acid pre-S sequence, spanning about half of the DHBV pre-S chain, was characterized by deletion analysis as essential for maximal inhibition. Pre-S polypeptides from heron hepatitis B virus (HHBV) competed DHBV infection equally well despite a 50% difference in amino acid sequence and a much-reduced infectivity of HHBV for duck hepatocytes. These observations are taken to indicate (i) that the functionality of the DHBV pre-S subdomain, which interacts with the cellular receptor, is determined predominantly by a defined three-dimensional structure rather than by primary sequence elements; (ii) that cellular uptake of hepadnaviruses is a multistep process involving more than a single cellular receptor component; and (iii) that gp180, a cellular receptor candidate unable to discriminate between DHBV and HHBV, is a common component of the cellular receptor complex for avian hepadnaviruses.
Identification of cellular receptors and characterization of their interaction with the virus particle on the molecular level constitute a major focus of basic and medical virology, providing new perspectives for targeted antiviral prophylaxis and chemotherapy for a variety of viral pathogens, such as influenza viruses, picornaviruses, herpesviruses, adenoviruses, and human immunodeficiency virus (27, 41). Despite major efforts, a comparable state of knowledge has not been reached for the hepatitis B viruses (HBVs), a family of small, enveloped DNA viruses, with a major human pathogen as the prototype, causing acute and chronic liver infections in mammals and birds (25). Lack of knowledge on the mechanism of human HBV (HBV) entry reflects major experimental difficulties caused by the narrow host range and tissue tropism of the hepadnaviruses, which restrict studies of infection to primary cultures of human hepatocytes. Cell lines capable of supporting virus production from transfected viral DNA genomes are available but are refractory to HBV infection, as they lack the cellular receptor(s) and possibly also other properties essential for the initiation of infection (13). Thus, there is presently only circumstantial evidence supporting the assumption that HBV infection is initiated by the interaction with a virus-specific hepatocellular receptor. Numerous proteins have been proposed as receptor candidates from binding studies with viral envelope proteins or envelope-derived peptides (for a recent summary and review, see reference 2). However, as primary human hepatocytes are not readily obtainable in a reproducibly infectable state (8, 22), all of these initial results remain to be complemented by direct evidence from infection experiments.
Substantially more progress has been made with the duck hepatitis B virus (DHBV) animal model, which, by contrast, allows systematic infection experiments to be performed with cultured primary duck hepatocytes (PDHs) obtained from domestic animals (38). In particular, more is known about the role played by avian hepadnavirus viral surface proteins in cellular attachment and entry than is known for the case of HBV surface proteins. In DHBV, there are two nonglycosylated envelope proteins embedded in the lipid envelope: the major S protein, a transmembrane protein of 167 amino acids (aa) providing 80% of the surface protein content, and the larger L protein (large surface protein or pre-S/S polypeptide), in which the S protein is N-terminally extended by the hydrophilic pre-S domain of 161 amino acids (31). For comparison, HBV encodes three glycosylated envelope proteins, originally designated surface antigens or S antigens. The two envelope proteins of DHBV are present also in nucleocapsid-free virus-like particles (subviral particles [SVPs]) in ratios comparable to that in the virion. As in all hepadnavirus infections, DHBV SVPs are secreted from infected cells together with the mature virions in about a 1,000-fold excess. SVPs thus provide a valuable, readily available virus substitute for binding and infection studies aimed to elucidate basic features of hepadnavirus uptake. By using this tool, initial evidence for the presence of a titratable cellular DHBV receptor was obtained in infection competition and binding competition experiments with PDHs. Furthermore, by using recombinant SVPs produced in yeast and containing only a single envelope protein, the pre-S domain of the L protein was identified as being essential for receptor interaction. From these data it was concluded that DHBV infection was initiated by the specific attachment of the virus particle via surface-exposed pre-S domains to receptor molecules on the hepatocyte surface (17).
Several other observations indicating that the surface-exposed hydrophilic pre-S domain was a major factor in determining the narrow host range of the hepadnaviruses also emphasize the importance of the pre-S domain for processes occurring early in DHBV infection. A role of pre-S in host restriction was initially predicted from the selectively high sequence divergence between the pre-S gene products of avi- and ortho-hepadnaviruses (34). Evidence supporting this hypothesis was recently obtained in infection experiments with pseudotyped heron hepatitis B virus (HHBV), which demonstrated a 100-fold increase in the permissiveness of duck hepatocytes after complementation of the HHBV particle with chimeric L proteins containing DHBV pre-S sequences (4, 14).
Whereas these observations indicate that the DHBV pre-S domain contains the viral ligand, to date the interacting cellular partner on the duck hepatocyte surface has remained elusive. The only candidate protein of some promise has been gp180 (18), also designated p170 (37), a membrane-associated glycoprotein of approximately 180 kDa identified by its high affinity for DHBV L protein or recombinant pre-S fusion proteins. However, gp180 did not correlate, in other relevant properties, with the specifications expected for a DHBV receptor matching the tissue tropism or narrow host range of avian hepadnaviruses: (i) gp180 was found abundantly in duck tissues not supporting DHBV infection (18, 37); (ii) gp180 was shown to bind nondiscriminatingly to a pre-S polypeptide of HHBV (15), a virus unable to infect Pekin ducks (33); and (iii) gp180 has not yet been shown to confer susceptibility to DHBV infection upon transfection of LMH cells, a chicken hepatoma cell line that is refractory to infection but capable of virus production upon transfection of genomic DHBV. Thus, the molecules and mechanisms involved in the initiation of DHBV infection are understood only in part, and the cellular receptor unit(s) remains unknown.
In this report we address and resolve some of these pertinent issues. By characterizing the DHBV pre-S domain in detail in infection competition experiments, a pre-S subdomain was shown to be essential but not sufficient for infection. Together with data from an accompanying study (1), this study provides evidence that hepadnavirus uptake proceeds in at least two steps: initially, DHBV binds to a primary receptor molecule, which we propose to be gp180, as a component of a receptor complex of low species specificity and shared by all avian hepadnaviruses; subsequently, yet-unidentified interactions with a secondary receptor, which is likely to be the major determinant(s) of host range, lead to release of the nucleocapsid and productive infection.
MATERIALS AND METHODS
Plasmid constructs.
Plasmids for recombinant expression of pre-S polypeptides in Escherichia coli were constructed by inserting the respective coding sequences into a slightly modified version of the expression vector pQE 8 (Qiagen), as shown in Fig. 1A. For this purpose, pre-S-encoding gene fragments from DHBV subtype 3, 16, or 26 (24, 34, 35), HHBV subtype 4 (33), and HBV subtype ayw (7) were amplified by PCR from template plasmids with Pfu polymerase (Stratagene) and oligonucleotide primers introducing a BamHI site before and a KpnI site after the coding sequences (Fig. 1A). DHBV DNA templates (subtype 26) encoding L proteins with internal deletion mutations (6) were kindly provided by Hans Will (Hamburg, Germany).
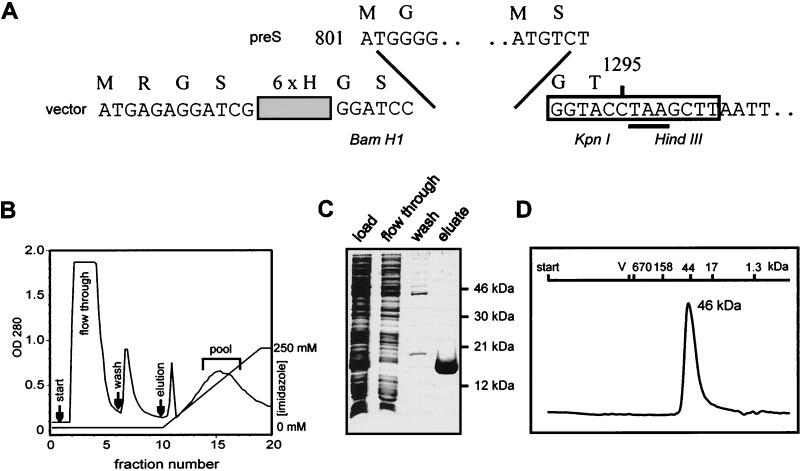
FIG. 1.
Expression of pre-S-coding sequences in E. coli and purification of recombinant proteins. (A) Pre-S-coding sequences were inserted into the prokaryotic expression vector pQE 8 (Qiagen) modified by a KpnI/HindIII linker (boxed sequence) as described in Materials and Methods. The construct encoding full-length pre-S and the first 4 aa from the S region is shown. Numbers 801 and 1295 indicate the first and the last nucleotides of the subcloned fragment, which is followed by a stop codon (underlined). The six amino-terminal histidine residues and six additional vector-encoded amino acids are shown in one-letter code above the nucleotide sequence. (B) Affinity chromatography with an Ni2+-nitrilotriacetic acid column. Hexahistidine pre-S fusion proteins were purified from total cellular lysates of recombinant E. coli as described in Materials and Methods and shown here for the DHBV pre-S fragment from aa 1 to 119. Elution was performed with an imidazole gradient from 0 to 250 mM as indicated. Pooled fractions are marked by a bracket. OD280, optical density at 280 nm. (C) Silver-stained SDS gel of fractions eluted from the affinity column shown in panel B. Molecular masses of marker proteins are indicated on the right. (D) Elution profile of renatured DHBV pre-S on a Superdex 200 column (Pharmacia). V0 and the elution volumes of thyroglobulin (670 kDa), gamma globulin (158 kDa), ovalbumin (44 kDa), myoglobin (17 kDa), and vitamin B12 (1.3 kDa), used for calibration, are indicated at the top.
Expression, purification, and exclusion chromatography of recombinant pre-S polypeptides from E. coli.
E. coli M15pREP4 cells (Qiagen) were transformed with the respective expression plasmids and grown in 1 liter of TB medium (with 100 μg of ampicillin per ml and 25 μg of kanamycin per ml) to an optical density of 0.8 to 1.0 (λ = 600 nm). Gene expression was induced with IPTG (isopropyl-β-d-thiogalactopyranoside) at a concentration of 1 mM. At 3 h after induction, cells were harvested by centrifugation at 4,000 × g for 20 min. The bacterial pellet was washed with phosphate-buffered saline (PBS) and either stored at −20°C or immediately lysed by incubation with 25 ml of solubilization buffer (6 M guanidine hydrochloride, 100 mM NaPi, 10 mM Tris, pH 8.0) by using a Dounce homogenizer and a Bronson Sonifier. The lysate was clarified by centrifugation at 100,000 × g for 15 min, and the supernatant was applied to an Ni2+-nitrilotriacetic acid agarose (Qiagen) column (bed volume, 8 ml) connected to a fast protein liquid chromatography system (Pharmacia). After equilibration with 7 M urea–100 mM NaPi–10 mM Tris (pH 8.0), unspecifically bound proteins were eluted at pH 6.3. Elution of histidine-tagged pre-S polypeptides was achieved with a linear imidazole gradient from 0 to 250 mM imidazole in 10 to 15 bed volumes. Fractions of 3 ml were collected and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Fractions containing purified pre-S polypeptides were pooled and either immediately dialyzed as described below or stored frozen at −20°C.
Pre-S polypeptides to be used for infection competition experiments were dialyzed stepwise against the following buffers containing decreasing concentrations of salt and urea to completely remove any traces of denaturing agents: (i) 4 M urea–50 mM NaPi, pH 6.0 to 6.3; (ii) 2 M urea–20 mM NaPi, pH 6.3; and (iii) 20 mM NaPi, pH 6.3 (three times with 5 liters). Protein concentrations were calculated from the absorbance at 280 nm based on the respective theoretical extinction coefficients. The integrity of all proteins was controlled by SDS-PAGE after dialysis.
Gel filtration of DHBV pre-S was performed on a calibrated Superdex 200 column (1.6 by 60 cm; Pharmacia) connected to a fast protein liquid chromatography system (Pharmacia) and equilibrated in 5% sucrose–150 mM NaCl–25 mM NaPi, pH 7.0 (temperature, 4°C; flow rate, 2.2 ml/min; sample volume, 0.5 ml). The column was calibrated with thyroglobulin (670 kDa), gamma globulin (158 kDa), ovalbumin (44 kDa), myoglobin (17 kDa), and vitamin B12 (1.3 kDa).
gp180 binding competition assay.
A DHBV pre-S–Sepharose and liver cell extract was prepared, and the binding reaction and Western blot analysis were carried out as described elsewhere (1). As a competitor, DHBV pre-S polypeptide or DHBV-positive duck serum was added to the binding reaction mixture. The amount of DHBV L protein was estimated from the DHBV DNA content of the serum, assuming 1,000 SVPs per DNA-containing virion and 20 molecules of L proteins per SVP (17).
Chemically synthesized DHBV pre-S polypeptides.
Polypeptides synthesized were as follows: DPS 1 (aa 60 to 139), QNQGAWPAGAGRRVGLSNPTPQEI PQPQWTPEEDQKAREAFRRYQEERPPETTTIPPSSPPQWKLQPGDP LLGNQSLLE; DPS 3 (aa 80 to 139), PQEIPQPQWTPEEDQKAREAFRRYQEERPPETTTIPPSSPPQWKLQPGDDPLLGNQSLLE; DPS 4 (aa 98 to 139), EAFRRYQEERPPETTTIPPSSPPQWKLQPGDDPLLGNQSLLF; DPS 6 (aa 115 to 139), PPSSPPQWKLQPGDDPLLGNQSLLE; DPS 7 (aa 82 to 121), KEIPQPQWTPEEDQKAREAFKQANEERPPETTTIPPSSPPQ; DPS 9 (aa 82 to 121), KEIPQPQYAEDDDQKAREAFRRYQEERPPETTTIPPSSPPQ; DPS 12 (aa 82 to 121), KEIPQPQWTPEEDQKAREAFRRYQEERPPETTTIPPSSPPQ; and DPS 13 (four repeats of DPS 12 plus VP1), (KEIPQPQWTPEEDQK AREAFRRYQEERPPETTTIPPSSPPQK)4K2LRGDLQVLAQKVARTLCA. Polypeptide FMDV VP 1 (aa 144 to 159) (LRGDLQVLAQKVARTL) was included as a control. Alterations from the DHBV subtype 16 amino acid sequence are indicated in boldface. For competition experiments the polypeptides were resuspended in PBS and used at concentrations ranging from 16 to 160 μM.
Protein analysis.
SDS-PAGE and Tricine-SDS-PAGE were performed with 15% polyacrylamide-bisacrylamide according to the methods of Laemmli (20) and Schägger and von Jagow (30). Prior to loading, proteins were dissolved in SDS sample buffer and boiled for 5 min. After electrophoresis, gels were either directly stained with Coomassie brilliant blue R250 or fixed in 30% ethanol–10% acetic acid and stained with silver as described previously (12).
Preparation of SVPs from DHBV and HHBV.
SVPs from DHBV were isolated from sera of infected ducklings by a method initially developed for HBV (11). Virus was inactivated by UV irradiation. The concentration of viral envelope proteins in SVPs was determined by SDS-PAGE followed by Coomassie blue staining, relative to carboanhydrase and recombinant DHBV pre-S polypeptides as standards. Viral particles from HHBV were obtained from medium supernatants of LMH cells transfected with an overlength genome of HHBV subtype 4 as described previously (33).
Preparation of PDHs and infection competition assay.
PDHs were prepared and cultivated as previously described (29). For infection competition assays, 8 × 105 cells, cultivated for 3 to 8 days in 12-well plates, were infected with 4 × 107 DNA-containing DHBV particles (determined by DNA dot blotting) in the presence or absence of inactivated SVPs or recombinant pre-S polypeptides. For the latter, stock solutions in 150 μl of 20 mM NaPi (pH 6.2) were supplemented with 350 μl of maintenance medium containing infectious DHBV serum and incubated for 12 h at 37°C. Cells were washed twice with PBS and cultured for an additional 9 days. Culture supernatants were collected between days 5 and 9. Viral particles were pelleted by centrifugation at 100,000 × g for 1 h, resuspended in 100 μl of PBS, and quantified by DNA dot blotting. The supernatants of the 100,000 × g spin were used for determination of duck hepatitis B virus e antigen (DHBeAg) by immuno-dot blotting (29). All assays were analyzed with a Molecular Dynamics PhosphorImager.
RESULTS
Recombinant DHBV pre-S polypeptides compete DHBV infection.
Inhibition of DHBV infection by SVPs has been shown to depend on the presence of the pre-S domain of the large envelope protein (17). To test whether the isolated pre-S polypeptide, corresponding to residues 1 to 161 of the L protein, was sufficient for causing this effect, pre-S polypeptides from DHBV and mutants thereof, as well as pre-S polypeptides from other hepadnavirus species, were synthesized in E. coli. DNA fragments containing the corresponding reading frames were amplified by PCR and subcloned into a bacterial expression vector such that they were preceded amino terminally by a 12-aa leader sequence including a hexahistidine tag (Fig. 1A). With this system, recombinant proteins were obtained in high yield (approximately 20 mg/liter of E. coli culture), but in most cases they were found to be localized in inclusion bodies. Purification of the His-tagged polypeptides by affinity chromatography was therefore performed routinely under denaturing conditions (see Materials and Methods). This single purification step (exemplified for the DHBV pre-S polypeptide from aa 1 to 119 in Fig. 1B and C) resulted in essentially pure polypeptides as judged by SDS-PAGE and silver staining. Denaturing agents were removed by stepwise dialysis of the pooled peak fractions. During this procedure, only minor precipitation occurred, an observation indicating that no significant protein aggregation had taken place during removal of the denaturation agent. This conclusion was confirmed by data from two-dimensional (2D) nuclear magnetic resonance analysis (40) and from surface plasmon resonance spectroscopy (BIAcore), which also indicated that DHBV pre-S molecules were structurally homogeneous and not aggregated (39). That the full-length recombinant DHBV pre-S polypeptide represented a homogeneous population of properly refolded molecules was also ascertained by its migration as a single peak in gel filtration on a Superdex 200 column (Fig. 1D). The apparent molecular mass observed (46 kDa) significantly exceeds the calculated mass for the 177-aa polypeptide (19,822 Da), which was confirmed by electrospray mass spectrometry, suggesting that the DHBV pre-S polypeptide might form a dimer under the conditions chosen. However, no change in the elution position in calibrated gel filtration was observed in the presence of denaturing agents such as 7 M urea or 6 M guanidinium hydrochloride (data not shown), indicating that the DHBV pre-S polypeptide most likely exists as a monomer whose increased apparent molecular mass is due to a nonglobular structure.
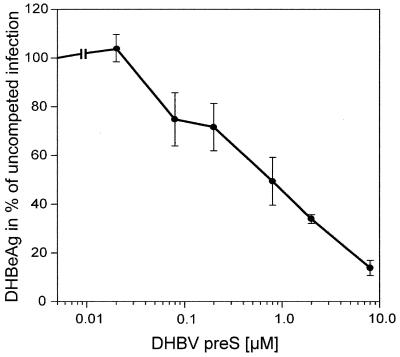
To examine whether the recombinant DHBV pre-S polypeptides were biologically active, they were tested in the infection competition assay previously used in experiments with the L protein as part of the DHBV SVP (17). PDHs were infected with DHBV in the presence of increasing amounts of pre-S polypeptide, and infection inhibition was assayed by determining the relative amounts of DHBeAg secreted into the culture medium (29). The results obtained (Fig. 2) demonstrated that the E. coli-derived DHBV full-length pre-S polypeptide (subtype 16) affected DHBV infection in a concentration-dependent manner, with 50% inhibition being achieved at concentrations of 0.4 to 0.8 μM under the conditions chosen. Pre-S polypeptides from DHBV subtypes 3 and 26, which differ in 7 and 15 amino acids, respectively, had the same specific competition activity as the pre-S protein of DHBV subtype 16. Subtype 16 was used in all experiments unless specified otherwise.
FIG. 2.
Competition of DHBV infection by recombinant DHBV pre-S polypeptide. PDHs (8 × 105) were infected with 4 × 107 DHBV particles in the presence of increasing concentrations of E. coli-derived DHBV pre-S protein (subtype 16). Virus replication between days 5 and 9 postinfection was determined by immuno-dot blot analysis of DHBeAg secreted into the culture medium and is presented as a percentage of the value for an uncompeted control infection (for details, see Materials and Methods). Each point represents the average of three independent experiments; bars indicate the standard deviations.
To exclude the possibility that competition of infection was caused by an interaction of pre-S polypeptides with viral particles rather than with a cell surface receptor, we incubated iodinated DHBV pre-S polypeptide (125I-DpreS) with purified SVPs and analyzed the components on a calibrated Superose 12HR gel filtration column. As expected, SVPs eluted in the void volume, whereas 125I-DpreS eluted exclusively at 46 kDa, without aggregate formation, regardless of the presence or absence of SVPs (data not shown). Taken together, these results indicate that the DHBV pre-S polypeptide is sufficient for competition by interacting with a cell surface receptor and, moreover, that posttranslational modifications of pre-S known to occur in animal cells but not in E. coli (e.g., myristylation and phosphorylation [9, 23]) are not a prerequisite for this inhibition.
Terminal deletion mutants define an 85-aa pre-S polypeptide as being essential for efficient receptor interaction.
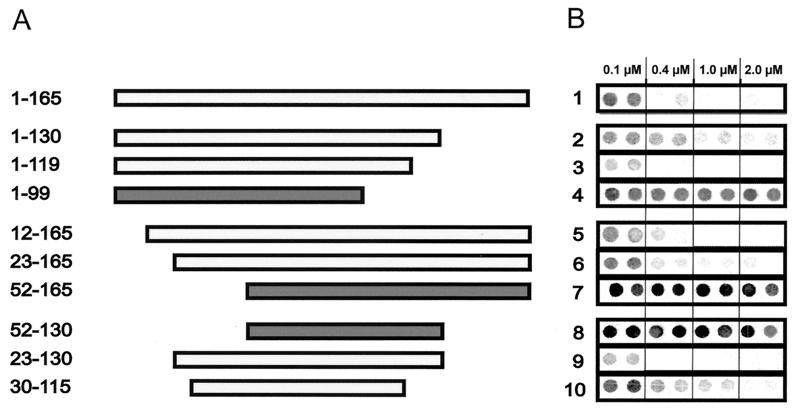
To delineate the region within the DHBV pre-S domain that is required for competition activity, we produced terminally truncated pre-S polypeptides from appropriately deleted pre-S reading frames. Nine of the mutants tested are schematically shown in Fig. 3A: three carboxy-terminally deleted mutants (DpreS1-130, DpreS1-119, and DpreS1-99), three amino-terminally deleted mutants (DpreS12-165, DpreS23-165, and DpreS52-165), and three combined mutants lacking both C-terminal and N-terminal parts of DHBV pre-S (DpreS52-130, DpreS23-130, and DpreS30-115). Mutant proteins were purified and characterized as described for Fig. 1B and C and then tested for their ability to compete with DHBV infection in comparison with full-length DpreS1-165 as a reference. The results of the DHBeAg immuno-dot blots from this analysis are shown in Fig. 3B, and the biological activities of the above-mentioned deletion mutants are shown in Fig. 3A. Removal of the first 12 or 23 aa (DpreS12-165 and DpreS23-165) (rows 5 and 6) as well as deletion of the last 31 or 42 aa of pre-S (DpreS1-130 and DpreS1-119) (rows 2 and 3) did not significantly alter infection competition, an observation indicating that the respective amino acids of both termini were dispensable for pre-S activity. This conclusion was confirmed by testing DpreS23-130, a polypeptide that combines the terminal deletions (row 9). This compound was again found to be as active as the preS1-165 reference protein (row 1). Further shortening at both termini to DpreS30-115 still resulted in a fully inhibition competent pre-S polypeptide (row 10), whereas pre-S chains with more extensive deletions, such as DpreS1-99, DpreS52-165, or DpreS52-130, were unable to compete DHBV infection (rows 4, 7, and 8).
FIG. 3.
Competition of DHBV infection by terminally deleted DHBV pre-S polypeptides. PDHs (8 × 105) were infected with 4 × 107 DNA-containing particles in the presence of increasing concentrations of either full-length or terminally deleted recombinant DHBV pre-S polypeptides. (A) Schematic drawing of the amino- and carboxy-terminal deletion mutants tested. Numbers on the left indicate the limits of the polypeptides, which are depicted as bars. Note that the entire pre-S sequence covers aa 1 to 161 (Fig. 1A). Bars in light gray correspond to polypeptides that behave in infection inhibition like full-length polypeptide (aa 1 to 165); bars in dark gray represent polypeptide mutants that lost activity. (B) DHBeAg dot blot analysis of culture medium from PDHs collected between days 5 and 9 postinfection in the presence of the pre-S polypeptides depicted in panel A. Polypeptide concentrations are indicated at the top. DHBeAg was quantified as described in Materials and Methods. Each dot corresponds to a single well of a 12-well culture plate. Two independent experiments for each concentration are shown. Apparent differences in competition activity relative to the reference in row 1 (visible in rows 2 and 10) were preparation dependent and could not be reproduced in independent experiments using another polypeptide preparation.
This clear-cut difference was observed with all of the terminal deletions shown in Fig. 3 even at polypeptide concentrations of up to 10 μM (not shown). In contrast, intermediate, although much reduced, specific activities were obtained with two additional deletion mutants, DpreS38-115 and DpreS43-115. These polypeptides displayed about a 15-fold-lower inhibition competence, as protein concentrations of 8 μM were required for 50% inhibition (not shown), indicating that the pre-S sequences between aa 30 and 51 contain elements that contribute to efficient pre-S–receptor interaction. Taken together, these data define an 85-aa domain in the center of the pre-S sequence (aa 30 to 115) as a minimal, fully active competitor polypeptide. This minimal competitor showed no detectable difference from the full-length pre-S protein in specific activity as ascertained in numerous infection competition experiments.
Short deletions within the receptor binding domain abolish pre-S competition.
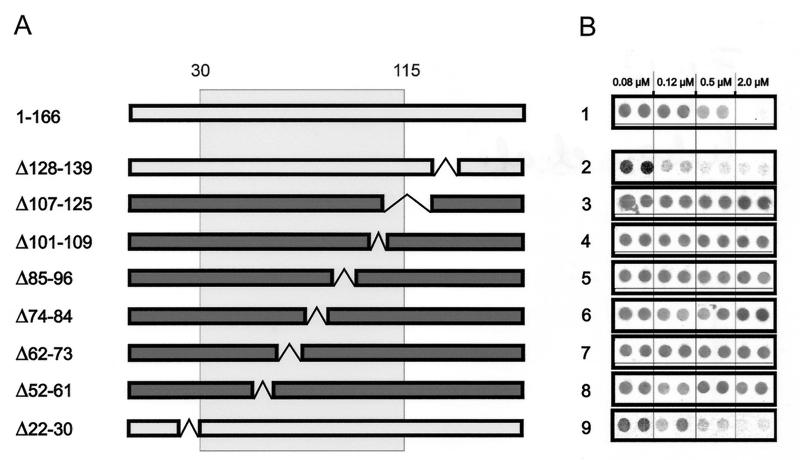
To identify sequence elements within the minimal competitor that are essential for pre-S–receptor interaction, the above-described competition analysis was extended by testing a series of internal pre-S deletion mutants initially constructed by Fernholz (6) and subcloned after PCR amplification into the pQE expression vector. Mutant proteins were purified and applied for infection competition assays as described in Materials and Methods. Stability of the polypeptides during infection competition was ascertained by SDS-PAGE and Western blotting of cell culture supernatants after infection.
As shown in Fig. 4 (rows 2 and 9), deletions outside the receptor binding domain, such as those in DpreSΔ128-139 and DpreSΔ22-30, did not affect pre-S competition, confirming that the terminal sequences of the pre-S chain do not substantially contribute to receptor binding as determined by our assay. In contrast and rather unexpectedly, deletions affecting various parts of the central pre-S domain all led to a complete loss of infection inhibition, as shown in Fig. 4 for mutants DpreSΔ107-125, DpreSΔ101-109, DpreSΔ85-96, DpreSΔ74-84, DpreSΔ62-73, and DpreSΔ52-61 (rows 3 to 8). Competition competence was abrogated even in the case of the 4-aa deletions in DpreSΔ62-65 and DpreSΔ67-70 (data not shown). This sensitivity to internal deletions covering essentially all of the binding domain suggests that a defined tertiary structure rather than a patchwork of sequential elements characterizes the interaction of the DHBV pre-S subdomain defined above with its cellular receptor on the molecular level.
FIG. 4.
Competition of DHBV infection of PDHs by internally deleted pre-S polypeptides. (A) Schematic drawing of the mutant polypeptides (DHBV pre-S subtype 26). Deletions are shown by gaps within the bars. Bars in light gray indicate protein mutants that behave like full-length pre-S; bars in dark gray represent protein mutants that lost their ability to compete infection. The minimal competitor region (aa 30 to 115) as determined by analysis of terminally deleted DHBV pre-S polypeptides is indicated as a box. Note that DHBV pre-S subtype 26 encodes an additional amino acid at position 146. The full-length protein therefore contains 166 aa. (B) Immuno-dot blot analysis of culture medium from PDHs in the presence of different internal pre-S mutants (rows 2 to 9) compared with full-length DHBV pre-S (row 1). DHBeAg was quantified as described in Materials and Methods. Two independent experiments for each concentration are shown.
A low interaction potential of a 40-aa core DHBV pre-S sequence increases upon oligomerization.
While clear results were obtained with the great majority of the terminal and internal deletions tested, some residual activity was observed, as discussed above, with constructs DpreS38-115 and DpreS43-115. This suggested that the N-terminal part of the minimal binding domain, although contributing to binding, was not strictly essential for pre-S–receptor interaction. This interpretation is supported by the results obtained in an earlier infection competition study using a series of chemically synthesized DHBV pre-S polypeptides (16). In these experiments, which are summarized in Table 1, an 80-mer encompassing aa 60 to 139 of DHBV pre-S, as well as a series of related peptides which had been terminally shortened in steps of 20 aa, was tested. Competition activity of DHBV pre-S polypeptides containing aa 60 to 139 (DPS 1), 80 to 139 (DPS 3), and 82 to 121 (DPS 12) was reproducibly detected only at 160 μM, the highest concentration employed, which is about 200-fold above the concentration required for half-maximal inhibition by recombinant DHBV pre-S polypeptides containing the minimal competitor sequence (pre-S aa 30 to 115).
TABLE 1.
Potential of chemically synthesized pre-S polypeptides to inhibit DHBV infection
| Peptide (aa)a | Activity (Ki, μM)b |
|---|---|
| DPS 1 (60–139) | + (ND) |
| DPS 3 (80–139) | + (ND) |
| DPS 4 (98–139) | − (ND) |
| DPS 6 (115–139) | − (ND) |
| DPS 7 (82–121) | − (ND) |
| DPS 9 (82–121) | − (ND) |
| DPS 12 (82–121) | + (100) |
| DPS 13 ([82–121]4VP1) | + (4) |
| FMDV VP1 (144–159) | − (ND) |
Amino acid sequences correspond to the DHBV-16 pre-S sequence. DPS 7 and DPS 9 contained amino acid exchanges at positions 101 to 104 and 88 to 92, respectively (for details, see Materials and Methods).
+ and −, greater and less than 40% inhibition of infection of PDHs detected at a concentration of 160 μM polypeptide, respectively. Ki, concentration of peptide monomer required for 50% inhibition of DHBV infection of PDH (at a multiplicity of infection of 150). ND, not determined.
The results of this analysis defined a core pre-S sequence between positions 80 and 121 which was equally active as the initial 80-mer but sensitive to removal of an additional 20 aa from either end as well as to mutational alterations changing blocks of 5 or 4 aa, respectively, within the core sequence. Interestingly, the competition activity of the core 40-mer was increased more than 40-fold upon tetramerization by linkage to a branched support during chemical synthesis. With this tetrameric polypeptide, half-maximal inhibition of DHBV infection was achieved at about 4 μM peptide monomer (Table 1). These data suggest that the low affinity detected with the monomeric core 40-mer may be enhanced by a simultaneous and cooperative interaction with several receptor molecules on the hepatocyte surface.
Pre-S domains in SVPs compete more effectively than monomeric pre-S polypeptides.
In the experiments described above, we analyzed recombinant DHBV pre-S chains, indicated to be in a monomeric, nonaggregated state (Fig. 1D), for their interaction with a cell surface receptor. Infection competition by synthetic pre-S peptides had suggested that ligand oligomerization enhanced competition competence. It was therefore of interest to determine the specific activity of DHBV pre-S chains presented as a part of the L protein on the surface of the DHBV particle. These experiments were performed similarly to the one shown in Fig. 2 for the recombinant pre-S polypeptide, using an SVP preparation that had been standardized for its pre-S content relative to the recombinant polypeptide. In three independent dilution series, 24, 56, and 36 nM particle-associated pre-S were found to be required for 50% inhibition of DHBV infection. Taking into consideration that about half of the pre-S domains are not exposed on the particle surface (10, 36), these data indicate that the particle-associated pre-S chains display an approximately 30-fold higher activity than the recombinant DHBV pre-S (0.6 μM for half-maximal inhibition [Fig. 2]). The alternative explanation that only a minor fraction of recombinant pre-S polypeptide was properly folded seems highly unlikely in view of the biophysical properties of the pre-S polypeptide, as already discussed in the context of Fig. 1D.
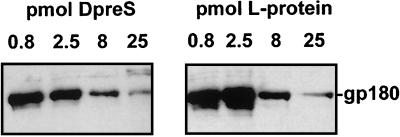
Further evidence for the assumption that our preparations of recombinant polypeptides were structurally homogenous, and also functionally equivalent to native DHBV pre-S chains, was obtained in a binding competition experiment comparing the affinities of pre-S chains to gp180, the duck glycoprotein for which we have good evidence that it is a cellular receptor for avian hepadnaviruses (see below and reference 1). In this experiment (Fig. 5), no significant differences in affinity to gp180 were detected between recombinant pre-S and pre-S domains in DHBV L protein derived from detergent-disrupted SVPs. We therefore conclude that the much-enhanced activity of the particle-associated pre-S chains probably reflects their particular spatial arrangement at the surface of the virus particle and possibly also the ability of the particle to interact with more than a single receptor molecule on the hepatocyte.
FIG. 5.
Recombinant DHBV pre-S polypeptide binds gp180 with affinities comparable to those of particle-derived DHBV L protein. Western blots detecting gp180, bound to immobilized DHBV pre-S polypeptide in the presence of increasing concentrations of competing free DHBV pre-S polypeptide (DpreS) or DHBV L protein derived from serum of a DHBV-infected duckling (L-protein), are shown. Numbers above the lanes indicate the amounts of free competitor present during binding of gp180 to immobilized DHBV pre-S. The gp180-specific band is indicated at the right.
The pre-S polypeptide of HHBV inhibits DHBV infection.
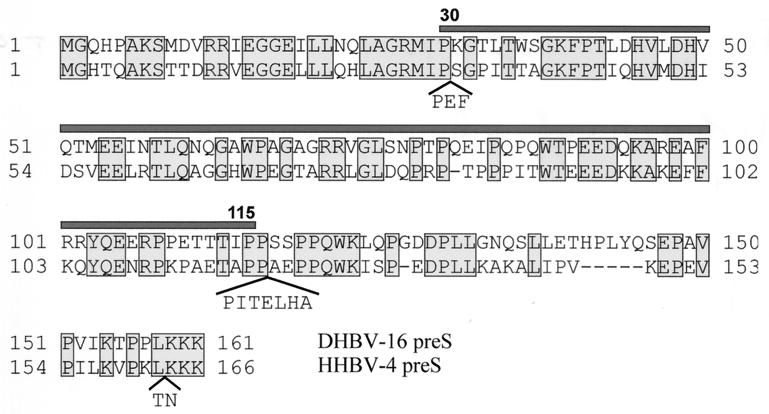
It has been speculated that the host discrimination between the avian hepadnaviruses DHBV and HHBV is the result of sequence diversion of the corresponding pre-S domains. Indeed, the DHBV and HHBV pre-S domains differ by 50% in amino acid sequence, an observation suggesting discrimination at the level of receptor interaction (see Fig. 7). To test whether such a difference in species specificity was indeed detectable, an E. coli-derived HHBV pre-S polypeptide, as well as HHBV SVPs produced from transfected LMH cells, were analyzed for their ability to interfere competitively with DHBV infection. Recombinant HBV pre-S polypeptide, prepared by the protocol used for the avian pre-S proteins, was included to represent the more distantly related mammalian hepadnaviruses, which show no significant homology to the corresponding DHBV pre-S sequence (34).
FIG. 7.
Comparison of the amino acid sequences of the pre-S domains of DHBV (subtype 16, upper row) and HHBV (subtype 4, lower row). Amino acid positions are indicated by numbers beside the rows. Conserved amino acids are boxed. Insertions within the HHBV pre-S sequence are shown in brackets. The minimal competitor region representing the receptor binding domain (aa 30 to 115) is indicated by the bar at the top of the DHBV pre-S sequence.
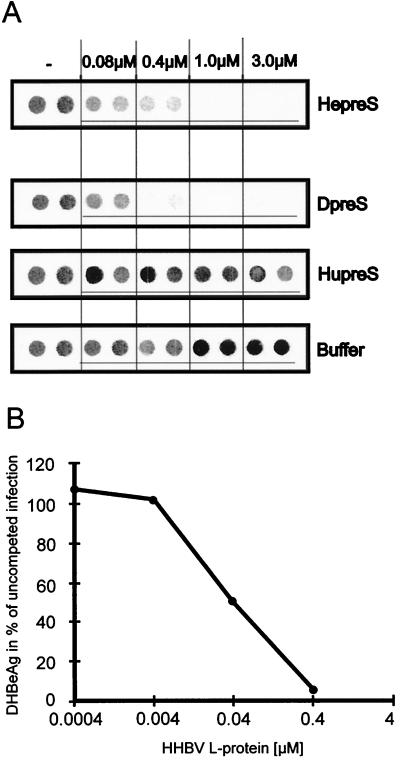
Figure 6A shows the results of DHBeAg immuno-dot blots from PDH cells infected with DHBV in the presence of competing HHBV pre-S, DHBV pre-S, or HBV pre-S. As expected, there was no competition by the HBV pre-S polypeptide. HHBV pre-S, however, inhibited DHBV infection effectively. Indeed, no difference between the HHBV and DHBV pre-S polypeptides was detected, within the limits of sensitivity of our assay. This functional equivalence between two polypeptides with markedly different primary sequences was confirmed by the results of infection inhibition assays with SVPs from HHBV. As shown in Fig. 6B, HHBV SVPs displayed inhibition comparable to that of DHBV SVPs, with 50% inhibition being achieved at a pre-S concentration of about 30 nM HHBV L-protein. We thus conclude that DHBV and HHBV interact with closely related primary receptor molecules on their respective host hepatocytes. Therefore, the marked difference in host specificity in productive infection by HHBV and DHBV, which results in a 100-fold-reduced infectivity of HHBV for PDHs (4), must be related to a step in viral uptake subsequent to the initial high-affinity binding of the virus particle to the cell surface.
FIG. 6.
Competition of DHBV infection with heterologous pre-S proteins. Infection competition was carried out as described in the legends to Fig. 2 and 3. (A) Immuno-dot blot analysis of culture medium from PDHs between days 5 and 9 postinfection with DHBV in the presence of HHBV pre-S polypeptide (HepreS), DHBV pre-S polypeptide (DpreS), HBV pre-S polypeptide (HupreS), or buffer. Polypeptide concentrations are indicated at the top. For each concentration, two independent measurements are shown. (B) Competition of DHBV infection by SVPs from HHBV. Amounts of secreted DHBeAg are presented as a percentage of the value for an uncompeted control infection.
DISCUSSION
By using recombinant DHBV pre-S polypeptides in an infection competition assay, we have characterized functionally important virus-cell surface interactions in hepadnavirus infection. Several observations were made which contribute substantially to our knowledge of the molecular mechanisms of events critical early in hepadnavirus infection.
An unusually extended domain of about 80 aa spanning about half of the DHBV pre-S chain was found to be required for efficient interaction with a cell surface receptor. This functionally defined receptor interaction domain was highly sensitive to internal deletions, suggesting that it requires integrity to form a particular 3D structure with key residues interacting with the receptor and is not represented by short primary sequence motifs as identified with some other viruses (41) and also initially proposed for HBV (26). The presence of a particular 3D structure in the pre-S domain is also indicated by aberrantly high apparent molecular weights of pre-S polypeptides in calibrated gel filtration, as exemplified in Fig. 1D. Since deletions outside the receptor interaction domain had no influence on infection competition (Fig. 4, rows 2 and 9), this domain appears to fold independently within the pre-S moiety. We are currently establishing the 3D structure of this domain (40), which may permit us to decipher common structural motifs that are important for receptor interaction. We also note that the domain defined as receptor interacting (aa 30 to 115) is contained within a region defined by linker scan mutations to be essential for DHBV infectivity but not for virus formation (aa 6 to 114) (21).
Another interesting observation was that the inhibitory potential of pre-S domains on the SVP was about 30-fold enhanced relative to that of pre-S chains from E. coli. However, the recombinant pre-S polypeptide and monomeric DHBV L chains interacted equally well with gp180 (Fig. 5), the DHBV receptor protein discussed below. Therefore, the enhanced activity of SVPs in infection competition most likely reflects an ordered 2D arrangement of pre-S chains as part of the membrane-anchored L protein at the particle surface. Such a presentation may conceivably facilitate cooperative pre-S–receptor interactions, as were observed with oligomers of a core pre-S polypeptide (Table 1). A possibility that we cannot exclude on the basis of our present experiments is that pre-S binding is stabilized by virus-cell interactions involving the S moiety of L protein, the S protein subunits as such, or the phospholipids of the DHBV envelope.
Finally, and much to our surprise, infection competition activity was not affected by the 50% amino acid differences present in the pre-S domain of HHBV (Fig. 7). This observation further supports the model that structural elements play the dominant role in pre-S–receptor interaction. Moreover, with respect to host discrimination, it defies the assumption that virus docking is the primary discriminatory event leading to the different host ranges of DHBV and HHBV, although reduced virus binding to host cells with reduced susceptibility to DHBV infection suggests that this step may also play a role in host specificity (28). Interestingly, the amino acid sequence of HHBV pre-S, compared to that of DHBV pre-S, contains two insertions at the borders of the receptor binding domain (Fig. 7). This observation argues for the evolutionary conservation of structural rather than sequential elements, thereby providing a high degree of variability in surface epitopes, allowing evasion of recognition by host defense systems.
A candidate cellular receptor fitting the above definition of a primary receptor is glycoprotein gp180, a transmembrane protein that has been classified as a member of the carboxypeptidase D family (19, 32). Support for the notion that gp180 may be the molecule that we address by our functional assay comes from comparison of the pre-S amino acid sequences that are required for competition of either DHBV infection (this report) or physical gp180 binding: using the same set of internal deletion mutants, we observed that the functionally important part of pre-S (aa 30 to 115) is nearly superimposable with the domain required for pre-S binding (1). In a similar approach, and with results essentially consistent with our quantitative results, Ishikawa et al. (15) had earlier mapped the gp180 binding site to aa 43 to 108, in contrast to a mapping to aa 87 to 102 by Tong et al. (37). Moreover, our functional characterization of a cell surface receptor that does not discriminate between HHBV and DHBV has resolved the conflict arising from the puzzling observation, initially reported by Ishikawa et al. (15), that HHBV pre-S also binds duck gp180, which we have confirmed and extended by including heron gp180 in a similar analysis (1). Finally, this complementary report (1) clearly demonstrates by confocal microscopy that gp180 mediates cellular uptake of DHBV particles and furthermore that gp180 is a Golgi-resident protein which cycles to and from the plasma membrane. Considering this dynamic receptor presentation, it seems impossible to use data from infection competition at elevated temperatures (as performed here and in a previous study [17]) to reliably evaluate the number of virus binding sites at the cell surface.
Taking these data together, we propose that the cellular uptake of hepadnaviruses includes at least two steps involving distinct pre-S determinants which contribute differentially to host discrimination. The initial step apparently involves the interaction of a defined pre-S subdomain with gp180, a primary receptor with little species specificity, at least between the two distantly related avian viruses studied here. The initial binding event is then followed by a species-specific interaction(s) with presently unknown components probably involving the more amino-terminal part of pre-S (14). Discrimination of virus entry at the level of secondary coreceptors exists in the case of human immunodeficiency virus, where different strains dock to CD4 but use distinct chemokine receptors as obligatory cofactors in infection (3, 5). In analogy, gp180 appears to be only a first component of a complex virus entry machinery that comprises an additional (yet unknown) coreceptor(s) and cellular factors that complete virus infection.
ACKNOWLEDGMENTS
We thank Bärbel Glass for the preparation of PDHs; Christa Kuhn for DHBV SVPs and antibodies; Rainer Frank for advice, chemical synthesis of polypeptides, and mass spectrometric analysis; Hans Will for providing the DHBV subtype 26 deletion mutants; Elizabeth Grgacic and Eckard Wimmer for critical reading of the manuscript; and Karin Coutinho for expert editorial assistance.
K.M.B. thanks the Boehringer Ingelheim Fonds for a fellowship. This work was supported by a grant from the Deutsche Forschungsgemeinschaft (SFB 229) and by the Fonds der Chemischen Industrie.
REFERENCES
- 1.Breiner K M, Urban S, Schaller H. Carboxypeptidase D (gp180), a Golgi-resident protein, functions in the attachment and entry of avian hepatitis B viruses. J Virol. 1998;72:8094–8104. doi: 10.1128/jvi.72.10.8098-8104.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.DeMeyer S, Gong J Z, Suwandhi W, van Pelt J, Soumillon A, Yap S H. Organ and species specificity of hepatitis B virus (HBV) infection: a review of literature with a special reference to preferential attachment of HBV to human hepatocytes. J Viral Hepat. 1997;4:145–153. doi: 10.1046/j.1365-2893.1997.00126.x. [DOI] [PubMed] [Google Scholar]
- 3.Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Di M P, Marmon S, Sutton R E, Hill C M, Davis C B, Peiper S C, Schall T J, Littman D R, Landau N R. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 4.Fehler, F., S. Seitz, and H. Schaller. Unpublished data.
- 5.Feng Y, Broder C C, Kennedy P E, Berger E A. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 6.Fernholz D. Studien zur Synthese und Funktion der Hüllproteine bei Hepatitis B Viren. Ph.D. thesis. Munich, Germany: Ludwig-Maximilians-Universität; 1992. [Google Scholar]
- 7.Galibert F, Mandart E, Fitoussi F, Tiollais P, Charnay P. Nucleotide sequence of the hepatitis B virus genome (subtype ayw) cloned in E. coli. Nature. 1979;281:646–650. doi: 10.1038/281646a0. [DOI] [PubMed] [Google Scholar]
- 8.Galle P R, Hagelstein J, Kommerell B, Volkmann M, Schranz P, Zentgraf H. In vitro experimental infection of primary human hepatocytes with hepatitis B virus. Gastroenterology. 1994;106:664–673. doi: 10.1016/0016-5085(94)90700-5. [DOI] [PubMed] [Google Scholar]
- 9.Grgacic E V, Anderson D A. The large surface protein of duck hepatitis B virus is phosphorylated in the pre-S domain. J Virol. 1994;68:7344–7350. doi: 10.1128/jvi.68.11.7344-7350.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guo J T, Pugh J C. Topology of the large envelope protein of duck hepatitis B virus suggests a mechanism for membrane translocation during particle morphogenesis. J Virol. 1997;71:1107–1114. doi: 10.1128/jvi.71.2.1107-1114.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heermann K H, Goldmann U, Schwartz W, Seyffarth T, Baumgarten H, Gerlich W H. Large surface proteins of hepatitis B virus containing the pre-S sequence. J Virol. 1984;52:396–402. doi: 10.1128/jvi.52.2.396-402.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heukeshoven J, Dernick R. Improved silver staining procedure for fast staining in PhastSystem Development Unit. I. Staining of sodium dodecyl sulfate gels. Electrophoresis. 1988;9:28–32. doi: 10.1002/elps.1150090106. [DOI] [PubMed] [Google Scholar]
- 13.Hild M, Weber O, Schaller H. Glucagon treatment interferes with an early step of duck hepatitis B virus infection. J Virol. 1998;72:2600–2606. doi: 10.1128/jvi.72.4.2600-2606.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ishikawa T, Ganem D. The pre-S domain of the large viral envelope protein determines host range in avian hepatitis B viruses. Proc Natl Acad Sci USA. 1995;92:6259–6263. doi: 10.1073/pnas.92.14.6259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ishikawa T, Kuroki K, Lenhoff R, Summers J, Ganem D. Analysis of the binding of a host cell surface glycoprotein to the pre-S protein of duck hepatitis B virus. Virology. 1994;202:1061–1064. doi: 10.1006/viro.1994.1440. [DOI] [PubMed] [Google Scholar]
- 16.Klingmüller U. Interaktion des Enten Hepatitis B Virus (DHBV) mit primären Entenleberzellen: Charakterisierung des viralen Liganden und Nachweis des zellulären Rezeptors. Ph.D. thesis. Heidelberg, Germany: Ruprecht-Karls-Universität; 1992. [Google Scholar]
- 17.Klingmüller U, Schaller H. Hepadnavirus infection requires interaction between the viral pre-S domain and a specific hepatocellular receptor. J Virol. 1993;67:7414–7422. doi: 10.1128/jvi.67.12.7414-7422.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuroki K, Cheung R, Marion P L, Ganem D. A cell surface protein that binds avian hepatitis B virus particles. J Virol. 1994;68:2091–2096. doi: 10.1128/jvi.68.4.2091-2096.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuroki K, Eng F, Ishikawa T, Turck C, Harada F, Ganem D. gp180, a host cell glycoprotein that binds duck hepatitis B virus particles, is encoded by a member of the carboxypeptidase gene family. J Biol Chem. 1995;270:15022–15028. doi: 10.1074/jbc.270.25.15022. [DOI] [PubMed] [Google Scholar]
- 20.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 21.Lenhoff R J, Summers J. Coordinate regulation of replication and virus assembly by the large envelope protein of an avian hepadnavirus. J Virol. 1994;68:4565–4571. doi: 10.1128/jvi.68.7.4565-4571.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mabit H, Vons C, Dubanchet S, Capel F, Franco D, Petit M A. Primary cultured normal human hepatocytes for hepatitis B virus receptor studies. J Hepatol. 1996;24:403–412. doi: 10.1016/s0168-8278(96)80160-7. [DOI] [PubMed] [Google Scholar]
- 23.Macrae D R, Bruss V, Ganem D. Myristylation of a duck hepatitis B virus envelope protein is essential for infectivity but not for virus assembly. Virology. 1991;181:359–363. doi: 10.1016/0042-6822(91)90503-4. [DOI] [PubMed] [Google Scholar]
- 24.Mandart E, Kay A, Galibert F. Nucleotide sequence of a cloned duck hepatitis B virus genome: comparison with woodchuck and human hepatitis B virus sequences. J Virol. 1984;49:782–792. doi: 10.1128/jvi.49.3.782-792.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mason, W. S., and C. Seeger (ed.). 1991. Hepadnaviruses. Molecular biology and pathogenesis. Curr. Top. Microbiol. Immunol. 168.
- 26.Neurath A R, Kent S B, Strick N, Parker K. Identification and chemical synthesis of a host cell receptor binding site on hepatitis B virus. Cell. 1986;46:429–436. doi: 10.1016/0092-8674(86)90663-x. [DOI] [PubMed] [Google Scholar]
- 27.Norkin L C. Virus receptors: implications for pathogenesis and the design of antiviral agents. Clin Microbiol Rev. 1995;8:293–315. doi: 10.1128/cmr.8.2.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pugh J C, Di Q, Mason W S, Simmons H. Susceptibility to duck hepatitis B virus infection is associated with the presence of cell surface receptor sites that efficiently bind viral particles. J Virol. 1995;69:4814–4822. doi: 10.1128/jvi.69.8.4814-4822.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rigg R J, Schaller H. Duck hepatitis B virus infection of hepatocytes is not dependent on low pH. J Virol. 1992;66:2829–2836. doi: 10.1128/jvi.66.5.2829-2836.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schägger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 31.Schlicht H J, Kuhn C, Guhr B, Mattaliano R J, Schaller H. Biochemical and immunological characterization of the duck hepatitis B virus envelope proteins. J Virol. 1987;61:2280–2285. doi: 10.1128/jvi.61.7.2280-2285.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Song L, Fricker L D. Purification and characterization of carboxypeptidase D, a novel carboxypeptidase E-like enzyme, from bovine pituitary. J Biol Chem. 1995;270:25007–25013. doi: 10.1074/jbc.270.42.25007. [DOI] [PubMed] [Google Scholar]
- 33.Sprengel R, Kaleta E F, Will H. Isolation and characterization of a hepatitis B virus endemic in herons. J Virol. 1988;62:3832–3839. doi: 10.1128/jvi.62.10.3832-3839.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sprengel R, Kuhn C, Will H, Schaller H. Comparative sequence analysis of duck and human hepatitis B virus genomes. J Med Virol. 1985;15:323–333. doi: 10.1002/jmv.1890150402. [DOI] [PubMed] [Google Scholar]
- 35.Sprengel R, Schneider R, Marion P L, Fernholz D, Wildner G, Will H. Comparative sequence analysis of defective and infectious avian hepadnaviruses. Nucleic Acids Res. 1991;19:4289. doi: 10.1093/nar/19.15.4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Swameye I, Schaller H. Dual topology of the large envelope protein of duck hepatitis B virus: determinants preventing pre-S translocation and glycosylation. J Virol. 1997;71:9434–9441. doi: 10.1128/jvi.71.12.9434-9441.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tong S, Li J, Wands J R. Interaction between duck hepatitis B virus and a 170-kilodalton cellular protein is mediated through a neutralizing epitope of the pre-S region and occurs during viral infection. J Virol. 1995;69:7106–7112. doi: 10.1128/jvi.69.11.7106-7112.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tuttleman J S, Pugh J C, Summers J W. In vitro experimental infection of primary duck hepatocyte cultures with duck hepatitis B virus. J Virol. 1986;58:17–25. doi: 10.1128/jvi.58.1.17-25.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Urban, S., C. Kruse, and G. Multhaup. A soluble form of the avian hepatitis B virus receptor: biochemical characterization and functional analysis of the receptor ligand complex. Submitted for publication. [DOI] [PubMed]
- 40.Urban, S., U. Marx, and P. Rösch. Unpublished data.
- 41.Wimmer E, editor. Cellular receptors for animal viruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]