Abstract
The pSub201-pAAV/Ad plasmid cotransfection system was developed to eliminate homologous recombination which leads to generation of the wild-type (wt) adeno-associated virus type 2 (AAV) during recombinant vector production. The extent of contamination with wt AAV has been documented to range between 0.01 and 10%. However, the precise mechanism of generation of the contaminating wt AAV remains unclear. To characterize the wt AAV genomes, recombinant viral stocks were used to infect human 293 cells in the presence of adenovirus. Southern blot analyses of viral replicative DNA intermediates revealed that the contaminating AAV genomes were not authentic wt but rather wt AAV-like sequences derived from recombination between (i) AAV inverted terminal repeats (ITRs) in the recombinant plasmid and (ii) AAV sequences in the helper plasmid. Replicative AAV DNA fragments, isolated following amplification through four successive rounds of amplification in adenovirus-infected 293 cells, were molecularly cloned and subjected to nucleotide sequencing to identify the recombinant junctions. Following sequence analyses of 31 different ends of AAV-like genomes derived from two different recombinant vector stocks, we observed that all recombination events involved 10 nucleotides in the AAV D sequence distal to viral hairpin structures. We have recently documented that the first 10 nucleotides in the D sequence proximal to the AAV hairpin structures are essential for successful replication and encapsidation of the viral genome (X.-S. Wang et al., J. Virol. 71:3077–3082, 1997), and it was noteworthy that in each recombinant junction sequenced, the same 10 nucleotides were retained. We also observed that adenovirus ITRs in the helper plasmid were involved in illegitimate recombination with AAV ITRs, deletions of which significantly reduced the extent of wt AAV-like particles. Furthermore, the combined use of recombinant AAV plasmids lacking the distal 10 nucleotides in the D sequence and helper plasmids lacking the adenovirus ITRs led to complete elimination of replication-competent wt AAV-like particles in recombinant vector stocks. These strategies should be useful in producing clinical-grade AAV vectors suitable for human gene therapy.
Adeno-associated virus type 2 (AAV) is a nonpathogenic human parvovirus which contains a single-stranded, linear DNA genome of 4,680 nucleotides (45). AAV is dependent on coinfection with a helper virus, such as adenovirus or herpesvirus, for its optimal replication (3, 4), but in the absence of a helper virus, the wild-type (wt) AAV genome integrates into the host chromosome in a site-specific manner and establishes a latent infection (18–20, 39). Three elements of the AAV genome are required for the viral replicative life cycle. The first is a pair of inverted terminal repeats (ITRs) which fold into hairpin structures, and the 3′ end serves as a primer for AAV DNA replication (12, 23, 35, 38, 43). ITRs are also required for AAV genome encapsidation and integration (10, 38, 47). The second is the rep gene which codes for four viral replication (Rep) proteins (4, 5, 26). Rep proteins are also required for viral gene expression, DNA encapsidation, and site-specific integration (2, 5–8, 14–16, 21, 24, 25, 35, 40). And the third is the cap gene which encodes the viral capsid (Cap) proteins required for viral assembly (35, 45). ITRs are the sole cis-acting sequences required for viral DNA replication, encapsidation, and integration (37). Based on the fact that AAV is a nonpathogenic human parvovirus that can infect both dividing and nondividing cells (8, 30), and that it can stably integrate into the host chromosome, recombinant AAV vectors have been developed as a potentially useful alternative to the more commonly used retrovirus and adenovirus vectors for human gene therapy (4, 9, 26, 37, 41, 44, 44a).
The production of recombinant AAV utilizes a vector containing a transgene cassette flanked by the viral ITRs. Recombinant vectors are generated by cotransfecting the recombinant AAV plasmid and a helper plasmid into adenovirus-infected cells (36, 37). The helper plasmid contains the AAV rep and cap genes which provide Rep and Cap proteins in trans, respectively, required for efficient rescue of the recombinant AAV genome from the recombinant plasmid, followed by replication and encapsidation into progeny virions. Rescue, replication, and packaging of the AAV genome do not occur from the helper plasmid which lacks the AAV ITR. However, several reports have documented that wt AAV particles are also generated during recombinant AAV vector production (11, 17, 21, 26, 33, 42), but the underlying mechanism of generation of the wt AAV remains unknown. Whether the contaminating AAV is truly authentic wt AAV or whether these particles originate following recombination between the recombinant AAV and the helper plasmids also remains unclear. In pursuit of answers to these questions, we have carried out systematic analyses of the wt AAV genomes molecularly cloned from two different recombinant AAV vector stocks. Our data indicate that (i) the contaminating AAV is not authentic wt AAV but rather wt AAV-like, (ii) wt AAV-like particles are generated by nonhomologous recombination between the recombinant AAV and the helper plasmids, (iii) adenovirus ITRs in the helper plasmid contribute to generation of these particles, (iv) the recombinant sites in the AAV ITRs are crowded around the distal 10 nucleotides in the D sequence, and (v) removal of the adenovirus ITRs from the helper plasmid and deletion of the distal 10 nucleotides in the D sequence from the recombinant plasmid lead to elimination of replication-competent wt AAV-like particles.
MATERIALS AND METHODS
Cells, viruses, and plasmids.
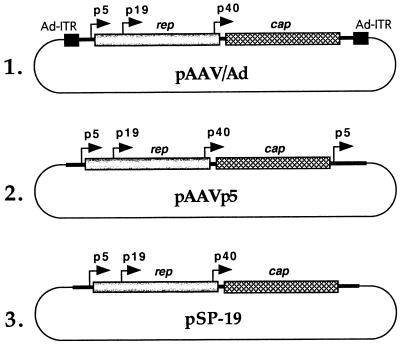
The human embryonic kidney cell line 293 was obtained from the American Type Culture Collection (Rockville, Md.), and the human nasopharyngeal carcinoma cell line KB was obtained from Asok C. Antony, Indiana University School of Medicine, Indianapolis. Cells were maintained as monolayer cultures in Iscove’s modified Dulbecco’s medium supplemented with 10% fetal bovine serum, penicillin, and streptomycin as previously described (27). The human adenovirus type 2 (Ad2) stock was obtained from Kenneth H. Fife, Indiana University School of Medicine, and propagated as previously described (28). The AAV helper plasmid containing the AAV coding sequences flanked by the Ad5 ITRs, pAAV/Ad (37), was supplied by Richard J. Samulski, University of North Carolina, Chapel Hill. An AAV helper plasmid lacking the Ad5 ITRs, pSP-19, has been described previously (48), and an additional AAV helper plasmid, pAAVp5, was constructed such that the AAVp5 promoter sequences were inserted downstream of the polyadenylation signal of AAV as follows. A 693-bp BalI-SacI DNA fragment containing the AAV p5 promoter sequences was isolated from plasmid pSub201 (36), digested with BbvI, and treated with the Klenow fragment of Escherichia coli DNA polymerase I. A 203-bp BalI-BbvI fragment was recovered, digested with XbaI, and ligated at the 3′ end of the AAV genome in plasmid pSP-19 partially digested with XbaI and completely digested with SpeI. Each of these three helper plasmids is shown schematically in Fig. 1. Recombinant AAV plasmids pCMVp-lacZ, containing the human cytomegalovirus (CMV) immediate-early promoter-driven β-galactosidase gene (31, 32), and pWP-8A, containing the genes for resistance to tetracycline and the herpesvirus thymidine kinase promoter-driven gene for resistance to neomycin (29), have also been described previously. The construction of plasmid pD-10, which contains deletions in the distal 10 nucleotides in the D sequence within the AAV ITRs, was described recently (49). The CMV promoter-driven lacZ gene sequences were also inserted in the pD-10 vector to generate a recombinant plasmid, pBK-2. Standard cloning techniques were used for constructing all recombinant plasmids (34).
FIG. 1.
Schematic representation of the recombinant AAV helper plasmids. The three viral promoters are marked by arrows, and the viral rep and cap genes are represented by shaded and cross-hatched boxes, respectively. The Ad ITRs are denoted by closed boxes, and the plasmid vector backbones are indicated by thin lines.
Packaging of recombinant AAV.
DNA transfections were performed by the calcium phosphate coprecipitation method essentially as previously described (34). Briefly, 15 μg of recombinant AAV plasmid (pCMVp-lacZ, pWP-8A, or pBK-2) and 15 μg of the AAV helper plasmid (pAAVp5, pAAV/Ad, or pSP-19) were used per 15-cm-diameter dish of 70% confluent 293 cells. Eight hours posttransfection, the medium was replaced with fresh medium containing 20 PFU of Ad2. The cultures were incubated at 37°C in a CO2 incubator for 65 to 72 h, and cells were harvested. The cell pellets were subjected to three cycles of freezing and thawing. CsCl was added to a final density of 1.4 g/cm3 and centrifuged in an SW50.1 swinging-bucket rotor at 35,000 rpm for 48 h at 20°C. Fractions with refractive indices of 1.371 to 1.374 were pooled and dialyzed in 1× phosphate-buffered saline, followed by exhaustive digestion with DNase I. Clarified supernatants were heated at 56°C for 30 min to inactivate Ad2. Equivalent amounts were analyzed on quantitative DNA slot blots, using 32P-labeled DNA probes specific for wt AAV, lacZ, or neo sequences as previously described (22).
AAV DNA replication and amplification assays.
Approximately 70% confluent 293 cells in 10-cm-diameter dishes were coinfected with recombinant AAV (multiplicity of infection of 10) and Ad2 (10 PFU). Seventy-two hours postinfection, low-Mr DNA samples were isolated by the procedure described by Hirt (13). Equivalent amounts of low-Mr DNA, with or without prior digestion with restriction endonucleases, were analyzed by Southern blotting using a 32P-labeled DNA probe specific for AAV coding sequences. To amplify the replication-competent wt AAV in the recombinant vector stocks, 293 cells were coinfected with recombinant AAV (multiplicity of infection of 10) and Ad2 (10 PFU) as described above. Seventy-two hours postinfection, cells were harvested and subjected to three cycles of freezing and thawing. Clarified culture supernatants were used to infect 293 cells with 10 PFU of Ad2 again. This process was repeated for a total of four rounds.
Molecular cloning and sequencing of the wt AAV-like genomes.
After four rounds of amplification, the supernatants were used to infect 293 cells with 10 PFU of Ad2. Low-Mr DNA samples were isolated 72 h postinfection, digested with restriction endonuclease BalI, and cloned into the EcoRV site of plasmid pBluescript II SK(+). Bacterial colonies containing plasmids with AAV sequences were selected by in situ colony hybridization using 32P-labeled AAV DNA as a probe. Nucleotide sequence analyses of each of the inserted full-length AAV genomes were carried out with T3 and T7 primers as previously described (50).
RESULTS
Contamination of the recombinant AAV vector stocks with wild-type AAV-like particles.
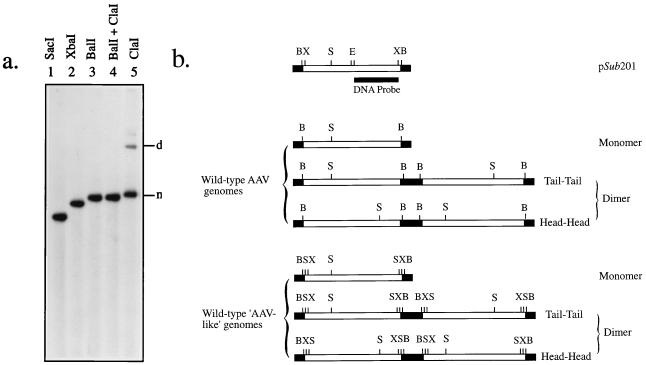
Several groups have reported that recombinant AAV stocks contain various levels of contaminating wild-type AAV particles (11, 17, 21, 26, 33, 42). There are two possible sources of this contamination. First, the helper adenovirus stocks may contain low levels of wt AAV, which is unlikely to be the case; second, the contaminating AAV particles are not truly authentic AAV but are generated by recombination events involving the recombinant AAV and the helper plasmids. Indeed, wt AAV genomes present in the recombinant vector stocks were not exactly like the wt AAV genome because of differences in the patterns of hybridization following digestion with various restriction endonucleases and Southern blot analyses. For example, when recombinant vCMVp- lacZ vector stocks, produced following cotransfection with plasmids pCMVp-lacZ and pAAVp5 and purified on CsCl gradients, were used to infect Ad2-infected 293 cells and low- Mr DNA were analyzed by using the AAV right-end EcoRI-XbaI DNA fragment as a probe, the results shown in Fig. 2a were obtained. It is clear that the replicated AAV genomes could be digested with restriction enzymes SacI, XbaI, and BalI but not with ClaI. Based on these results, the following conclusions can be drawn. First, the recombinant AAV vector preparations are free of the authentic wt AAV contamination since the wt AAV genome contains only one SacI site and no XbaI sites; second, the contaminating AAV genomes are derived from the helper plasmid since there are three SacI and two XbaI sites in the helper plasmid (Fig. 2b), and digestion with SacI or XbaI generated the DNA fragments of the expected sizes. Thus, these contaminating AAV particles are wt AAV-like particles.
FIG. 2.
(a) Southern blot analyses for identification of replication-competent wt AAV-like genomes. Following four rounds of amplification, low-Mr DNA samples were digested with the indicated restriction endonucleases and analyzed by Southern blotting using the right half of the AAV DNA (EcoRI-XbaI DNA fragment) probe. m and d denote the monomeric and dimeric replicative forms of the AAV genome. (b) Schematic representation of the AAV DNA replicative intermediates. The expected approximate sizes of DNA fragments generated by enzymes SacI (S), XbaI (X), BalI (B) from the wt and the wt AAV-like DNA are indicated. The EcoRI (E)-XbaI (X) DNA probe specific for the right-half of the AAV genome from plasmid pSub201 is depicted as a thick line, and AAV ITRs are shown as closed boxes.
Strategies for molecular cloning and sequencing of the wt AAV-like genomes.
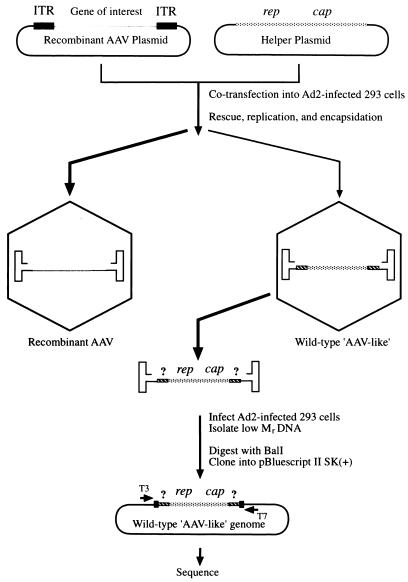
To investigate the mechanism of generation of these wt AAV-like particles, it became necessary to characterize these genomes at the nucleotide sequence level. Figure 3 illustrates how these wt AAV-like particles might be generated during recombinant AAV vector production as well as the strategy to characterize these wt AAV-like genomes. Briefly, following cotransfection of the recombinant AAV plasmid containing a gene of interest and the AAV helper plasmid containing the viral rep and cap genes in Ad2-infected 293 cells, most virions contain the recombinant AAV genome; however, a small population of virions contain the wt AAV-like genome which is comprised of AAV ITRs (derived from the recombinant AAV plasmid) and the viral rep and cap genes (derived from the helper plasmid), all of which are required for AAV replication and encapsidation. The viral genomes were amplified following four rounds of amplification in Ad2-infected 293 cells. Low-Mr DNA samples were digested with BalI restriction endonuclease, which cleaves within the AAV hairpin structure at nucleotide 121 in plasmid pSub201 (36), and molecularly cloned and sequenced as described in Materials and Methods. BalI digestion of these replication-competent AAV-like genomes would be expected to yield one fragment containing the AAV D sequence and the putative recombination junction sites since our previous studies have documented that the first 10 nucleotides in the D sequence are required for high-efficiency replication and encapsidation of the AAV genome (47–49). Indeed, a single DNA fragment was detected on Southern blots following digestion with BalI (Fig. 2a). A total of 24 recombinant plasmids were sequenced, 22 of which contained intact AAV genomes. These plasmids could be divided into six groups, A, B, C, D, E, and F, the left and the right junction sequences from which are presented in Fig. 4. The left junction in plasmids from group A contains the first 19 nucleotides of the D sequence and the left end of the AAV genome. Based on the sequence, it is clear that this genome is not the authentic wt AAV since it lacks the portion between the D sequence and the AAV p5 promoter. The right junction in plasmids from group A contains the same 19 nucleotides of the D sequence, 30 additional nucleotides that match the left end of plasmids from group A, and the right end of the helper plasmid. Thus, these wt AAV-like particles contain all of the elements required for DNA replication, such as the AAV ITR, as well as the rep and the cap genes. Furthermore, these data suggest that the wt AAV-like particles are generated by nonhomologous recombination between the recombinant AAV plasmid and the helper plasmid. The 30 nucleotides at the right end of plasmid A are derived from the left end of the helper plasmid, which suggests that the recombination event first occurred at the left end of the genome. The sequence at the right end of plasmids from group A arose most probably from repair and/or recombination between the left and the right ends of the recombinant AAV genome and not from that between the recombinant AAV and the helper plasmid DNA. In plasmids from group B, both ends contain the entire D sequence, but the recombination junctions between the AAV ITR derived from the recombinant plasmid and the AAV genome derived from the helper plasmid are completely different. Similarly, in plasmids from group C, the left and right ends contain 17 and 19 nucleotides in the D sequence, respectively, but the recombination junctions between the AAV ITR and the AAV genome are totally different. These results suggest that recombination events involving the left and right ends are independent of each other. In plasmids from group D, the left end is the same as the left end in plasmids from group A, but the right end is different from the right end in plasmids from group A. In the plasmid from group E, the left end is the same as the left end in plasmids from group A, but the right end is the same as the right end in plasmids from group C. In the plasmid from group F, the nucleotide sequence of the left end is the same as that of the left end in plasmids from group C, and the right end is the same as the right end in plasmids from group B. The frequencies of these recombination events are presented in Table 1. It appears that the Ra ITR is repaired from the La ITR in approximately 9% of the clones. The plasmid in group E is derived from recombination between plasmids in groups A and C, and the plasmid in group F is derived from recombination between plasmids in groups C and B, which together constitute approximately 9% of the clones. However, in approximately 82% of the clones examined, the recombination event involving each ITR occurs independently.
FIG. 3.
Experimental strategy for cloning the wt AAV-like genomes from recombinant vector stocks. These particles generated during the recombinant vector production are amplified through four successive rounds of infection of adenovirus-infected 293 cells. Low-Mr DNA is digested with restriction endonuclease BalI, and DNA fragments are cloned in a pBluescript SK(+) plasmid vector. AAV sequence-positive clones are subjected to nucleotide sequencing using the T3 and T7 primers.
FIG. 4.
Nucleotide sequences of the left and right junctions between AAV ITRs derived from the recombinant AAV-lacZ vector and the AAV sequence derived from the helper plasmid (pAAVp5). The D sequence, downstream from the BalI site (CCAA), is shown in outline shadow font, and the AAV sequences from the helper plasmid are shown in bold italic font. The sequence shown in outline italic font in the right junction in plasmids in group A (Form Ra) represents a duplication of the same sequence from the left junction (Form La). The underlined nucleotide pairs indicate the recombination junctions.
TABLE 1.
Summary of recombination junctions in the wt AAV-like genomesa
| Group | Left and right junction sequences | No. of clones/22 plasmids | Frequency (%) |
|---|---|---|---|
| A | La + Ra | 4 | 18.2 |
| B | Lb + Rb | 6 | 27.3 |
| C | Lc + Rc | 8 | 36.4 |
| D | La + Rd | 2 | 9.1 |
| E | La + Rc | 1 | 4.5 |
| F | Lc + Rb | 1 | 4.5 |
The nucleotide sequences obtained from 22 different plasmids containing the wt AAV-like genome were categorized into six distinct groups (A to F), and the left (L) and right (R) recombination junction sequences were grouped into four forms (a to d).
The recombination junctions are illustrated in Fig. 5, which indicates that most of the recombination events are clustered in the distal 10 nucleotides in the D sequence. Interestingly, however, there was no clear pattern of recombination sites in the AAV helper plasmid DNA (data not shown). When the left junction (form La) in plasmids from group A is compared with the right junction (form Rc) in plasmids from group C (Fig. 4), the recombination sites in the ITRs are the same but the rest of the sequences are different. Taken together, these data indicate that the genesis of wt AAV-like genomes is a consequence of multiple, independent, nonhomologous yet nonrandom recombination events involving the recombinant AAV and the helper plasmids as well as recombinant AAV-like genomes.
FIG. 5.
Summary of nucleotide sequences of the left and the right junctions in AAV ITRs from the recombinant AAV-lacZ vector. The sequence at the top represents the AAV D sequence (outline shadow font) downstream from the BalI site. The three left and four right end sequences in the wt AAV-like genomes are shown in the middle. The sequence at the bottom indicates the recombination sites in AAV ITRs. The underlined nucleotides form the junctions, and each asterisk represents the frequency of the recombination events.
Role of adenovirus ITRs in recombination.
To corroborate that the distal 10 nucleotides in the D sequence were indeed the hot spots for recombination, the wt AAV-like genomes amplified from a different recombinant AAV vector (vTc.Neo) generated by cotransfection of the recombinant AAV plasmid, pWP-8A, and a different AAV helper plasmid, pAAV/Ad, were analyzed as described above except that each of the ends was analyzed independently. The nucleotide sequences of six left ends and three right ends of these genomes are presented in Fig. 6. These data further provide strong evidence that most of the recombination events involve the distal 10 nucleotides in the D sequence and that the recombination is nonhomologous. Interestingly, however, upon closer examination, it became evident that each of the junction sequences also contained the Ad5 ITR sequences. The nucleotides in the Ad5 ITR sequence involved in recombination events are highlighted in Fig. 7. When a helper plasmid that lacked the Ad5 ITRs was used to generate recombinant AAV vectors, nearly a fivefold reduction in the recombination frequency leading to generation of wt AAV-like particles was observed. These data are summarized in Fig. 8. Based on sequence analyses of 7 independent clones that were obtained with a helper plasmid that contained Ad5 ITRs, there were five sites of recombination that led to generation of biologically active wt AAV-like particles during packaging of recombinant AAV, compared with 22 additional clones generated with a helper plasmid that lacked the Ad5 ITRs in which there were three sites of recombination. Similar results were obtained when the extent of generation of wt AAV-like particles was determined by quantitative DNA slot blot analyses (Table 2). Taken together, these results demonstrate that the Ad5 ITRs play a role in illegitimate recombination during the generation of the wt AAV-like particles.
FIG. 6.
Nucleotide sequences of the left and right junctions between AAV ITRs from the recombinant vector pWP-8A and the AAV sequence from the pAAV/Ad helper plasmid. The D sequence is shown in outline shadow font, and the helper plasmid sequences are shown in bold italic font. The underlined nucleotides indicate the recombination junctions, and the asterisks represent the recombination frequency.
FIG. 7.
Nucleotide sequences of the junction fragments involving the adenovirus ITRs. The adenovirus ITR sequence is shown in bold italic font at the top. The sequence in the middle corresponds to the D sequence downstream from the BalI site in the recombinant AAV plasmid pWP-8A. The recombination sites in the Ad5 ITR sequences are indicated by the underlined nucleotides. The asterisks represent the recombination frequency.
FIG. 8.
Nucleotide sequence analyses of recombinant junctions in the left ITR of the wt AAV-like genomes. The AAV D sequence, starting with the BalI site (nucleotide 122) is shown in outline font, and the rest of the AAV DNA sequence is shown in bold font. +Ad5-ITR denotes the helper plasmid that contains the Ad5 ITRs (pAAV/Ad), and −Ad5-ITR denotes the helper plasmid that lacks the Ad5-ITRs (pSP-19) which were used as helper plasmids to generate the recombinant AAV vector stocks. The underlined nucleotides represent the recombination sites, and the numbers indicate the observed frequency of recombination events in 7 clones for the former and in 22 clones for the latter that were analyzed.
TABLE 2.
Extent of contamination with the wt AAV-like physical particles in vector stocks generated from various combinations of recombinant and helper plasmids
| Recombinant plasmida | Helper plasmidb | Physical titer of contaminating wt AAV-like particles (%)c |
|---|---|---|
| pCMVp-lacZ | pAAV/Ad | 2.0 × 109 (4.0) |
| pCMVp-lacZ | pSP-19 | 4.0 × 108 (0.8) |
| pBK-2 | pAAV/Ad | 5.0 × 108 (1.0) |
| pBK-2 | pSP-19 | 7.0 × 107 (0.1) |
Plasmids pCMVp-lacZ and pBK-2 contain the same CMV promoter-driven lacZ gene in recombinant AAV vector backbone containing 20 and 10 nucleotides, respectively, in the D sequences in the viral ITRs.
The AAV coding sequences are flanked by adenovirus ITRs in plasmid pAAV/Ad, but deleted in plasmid pSP-19.
Contaminating wt AAV-like genomes in 5 × 1010 particles of each of the recombinant vCMVp-lacZ vector stocks per ml were detected and quantitated on DNA slot blots as previously described (22).
Strategies for elimination of the wt AAV-like particles.
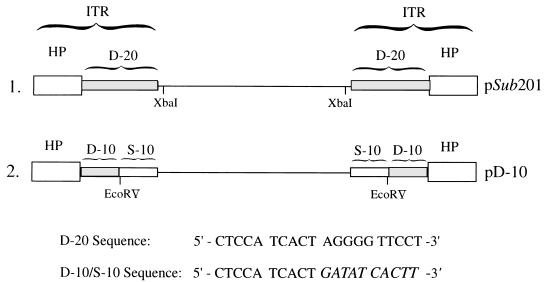
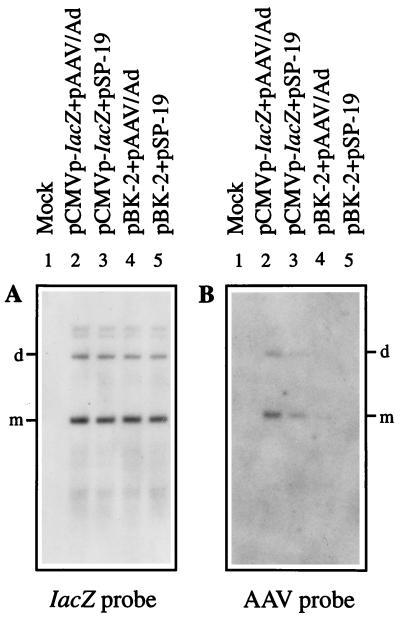
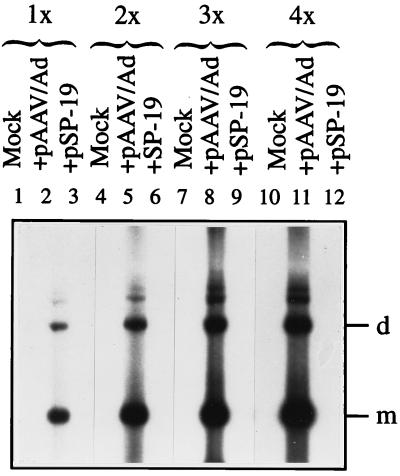
From the foregoing, it stood to reason that removal of the distal 10 nucleotides in the D sequence from the recombinant AAV vector and removal of the Ad5 ITRs from the AAV helper plasmid might prove beneficial in substantially reducing, if not completely eliminating, the generation of the wt AAV-like particles during recombinant AAV vector production. This possibility was experimentally tested as follows. The potential hot spots of recombination were deleted in a recombinant AAV plasmid, pD-10 (Fig. 9), the construction of which has recently been reported (49). This vector was used to generate a recombinant AAV plasmid, pBK-2, containing the CMV promoter-driven lacZ gene. Four sets of recombinant vCMVp-lacZ vector stocks were generated either with pCMVp-lacZ (pD-20) or with pBK-2 (pD-10), with pAAV/Ad and pSP-19 (prep/cap) as helper plasmids. Quantitative DNA slot blot analysis of each of the stocks revealed that contamination with the wt AAV-like genomes was highest in vectors generated from pCMVp-lacZ plus pAAV/Ad and lowest in vectors generated from pBK-2 plus pSP-19. These results, summarized in Table 2, suggest that both adenovirus ITRs and the distal 10 nucleotides in the AAV-ITRs promote the generation of the wt AAV-like particles. Equivalent amounts of each of the vector stocks were also used to infect Ad2-infected 293 cells under identical conditions. Low-Mr DNA isolated 72 h postinfection were analyzed by Southern blotting using DNA probes specific for lacZ or AAV sequences, respectively. These results are shown in Fig. 10. It is evident that the lacZ probe detected the characteristic monomeric and dimeric replicative forms of the recombinant AAV genomes, the hybridization intensities of which were roughly the same in all vector preparations (A). Interestingly, however, when a replicate Southern blot was probed with the AAV probe, no AAV DNA replicative intermediates could be detected in the recombinant AAV vector stocks prepared with plasmids pBK-2 and pSP-19. The extent of generation of replication-competent wt AAV-like particles was most pronounced in recombinant vector stocks generated with pCMVp-lacZ plus pAAV/Ad, followed by that with pCMVp-lacZ plus pSP-19 and pBK-2 plus pAAV/Ad (Fig. 10B). Even when the vector stocks generated with plasmids pBK-2 and pSP-19 were amplified through four successive rounds of amplification in Ad2-infected 293 cells, the AAV probe failed to detect any replication-competent wt AAV-like particles, whereas abundant hybridization signals were detected in vector stocks generated with plasmids pCMVp-lacZ and pAAV/Ad even after one round of amplification (Fig. 11). Although a low level of wt AAV-like genomes was generated even in the absence of adenovirus ITRs and the distal 10 nucleotides in the AAV D sequence (Table 2), these genomes were replication incompetent (Fig. 10). Thus, these results corroborate our contention that removal of the distal 10 nucleotides in the D sequence from the recombinant AAV vector and removal of the adenovirus ITRs from the AAV helper plasmid are sufficient to eliminate generation of replication-competent wt AAV-like particles during recombinant AAV vector production.
FIG. 9.
Schematic structures of pSub201 and pD-10 recombinant AAV vectors. The D sequence is shown as a shaded box in plasmid pSub201. In plasmid pD-10, the distal 10 nucleotides in the D sequence have been replaced by a substitute (S) sequence described previously (47, 48). The relevant restriction endonuclease sites (XbaI in pSub201 and EcoRV in pD-10) for cloning a gene of interest are also indicated. HP, hairpin.
FIG. 10.
Southern blot analyses of replication of the recombinant AAV-lacZ and the wt AAV-like genomes generated from recombinant plasmid pCMVp-lacZ (in pSub201) or pBK-2 (CMVp-lacZ in pD-10) with a helper plasmid containing (pAAV/Ad) or lacking (pSP-19) Ad5-ITRs, respectively. Equivalent amounts of low-Mr DNA isolated at 72 h posttransfection from adenovirus-coinfected 293 cells were analyzed by Southern blotting using lacZ-specific (A) and AAV-specific (B) DNA probes. Autoradiography was performed for 48 h (A) and 4 days (B). m and d denote the monomeric and dimeric viral replicative DNA intermediates, respectively.
FIG. 11.
Southern blot analyses of replication of wt AAV-like genomes present in recombinant vCMVp-lacZ vector stocks generated from the recombinant AAV plasmid containing the entire D sequence (pCMVp-lacZ) and the helper plasmid containing Ad5 ITRs (pAAV/Ad) (lanes 2, 5, 8, and 11) or from the recombinant AAV plasmid containing deletions in the distal 10 nucleotides in the D sequence (pBK-2) and the helper plasmid, pSP-19, lacking Ad5 ITRs (pSP-19) (lanes 3, 6, 9, and 12). Equivalent amounts of low-Mr DNA isolated at 72 h postinfection from adenovirus-coinfected 293 cells were obtained following the indicated rounds of amplification (1× to 4×) and analyzed by Southern blotting using an AAV-specific DNA probe. In lanes 1, 4, 7, and 10, low-Mr DNA from mock-infected 293 cells were analyzed. m and d denote the monomeric and dimeric replicative forms of the AAV genome, respectively.
DISCUSSION
To obtain recombinant AAV vectors that are free of any contaminating wt AAV, we must first understand the underlying molecular mechanism of generation of the wt AAV-like particles during recombinant AAV vector production. To this end, we considered the following three approaches. The first approach was to purify these wt AAV-like particles, release the single-stranded virion DNA, anneal the single strands, and molecularly clone the duplex DNA into plasmid vectors for sequence analyses. This was not pursued given the extremely low titers of wt AAV-like particles coupled with the potential difficulties in cloning these intact genomes (35, 38). Some of these difficulties included the complexity of AAV ITR structures, incomplete reannealing, and the variability in junction sequences in the wt AAV-like single-stranded genomes. The second approach was to resort to amplification of the wt AAV-like genomes by PCR. This was also considered less desirable since amplification of the replication-defective genomes could not be circumvented, and both ends of the wt AAV-like genomes could not be simultaneously analyzed (1). We devised a third approach in which the replication-competent replicative DNA intermediates of wt AAV-like genomes were cloned into a plasmid and subjected to nucleotide sequence analyses. The use of restriction enzyme BalI ensured that both monomeric and dimeric replicative forms would be represented and that the cloning step would be more efficient since the bulk of the AAV ITRs would be excluded, leading to simultaneous analyses of both ends of the viral genomes. However, a number of inherent experimental restrictions could not be overcome. For example, in order to be cloned and analyzed, AAV DNA intermediates must be digestable with BalI. In order to be amplified, recombinants must be efficiently rescued and packaged, eliminating ITR and proximal D-sequence mutations. Furthermore, since all recombinants are unlikely to replicate at the same rate, there might be preferential selection of those species that replicate more efficiently. And finally, since rep and cap gene functions can be provided in trans, mixtures of recombinants could be generated in which otherwise defective mutants could trans-complement another.
Previous studies have documented that significant sequence homology between the recombinant AAV plasmid and the helper plasmid leads to replication-competent wt AAV (26). To eliminate this problem, Samulski et al. (37) subsequently developed the pSub201-pAAV/Ad packaging system in which all homologous sequences between the two plasmids were deleted. However, despite this advance, we (21, 33) and others (11, 17, 26, 42) have consistently detected the presence of wt AAV sequences, ranging between 0.01 and 10%, in recombinant AAV vector stocks. Extensive sequence analyses of the recombination junctions in the present study revealed that the wt AAV-like particles are generated by nonhomologous recombination.
The presence of Ad5 ITRs in the helper plasmid seemed to promote recombination between the recombinant AAV and the helper plasmids. There are two stretches of homology between Ad5 ITR and AAV ITR sequences (AGGGG and GGGGT), but only the latter was involved in only one of the clones examined. Additional stretches of homology between the Ad5 ITR and the noncoding sequences in the recombinant AAV plasmid, pWP-8A, also exist (TCATC, TTAT, TAATGA, AGTTT, and GGGAA [Fig. 7]); however, most of the recombination sites share either no homology or only one or two nucleotides between these sequences. These data suggest that for the most part, four to six nucleotides are not sufficient to mediate homologous recombination between the two plasmid sequences. Thus, we conclude that the observed recombination events are illegitimate. Although remarkably similar observations have recently been made by Allen et al. (1), these authors examined only six clones (four from the left end and two from the right end) generated by PCR amplification, which would not discriminate between replication-competent and replication-defective wt AAV-like genomes. Furthermore, the two ends of the viral genomes were analyzed independently. In contrast, we examined recombination junctions at both ends simultaneously in a substantially larger population (22 clones) of replication-competent wt AAV-like genomes in our studies. Our data also indicate that most of the recombination events involve the distal 10 nucleotides in the D sequence. It is likely that recombination events occur in the proximal 10 nucleotides in the D sequence as well, but since the first 10 nucleotides of the D sequence are indispensable for replication and encapsidation of the AAV genome (47–49, 51), we surmise that such recombination events lead to production of replication-defective wt AAV-like particles.
A number of approaches have been used to obtain wt AAV-free stocks of recombinant AAV vectors (1, 9, 37, 46). Our studies offer a relatively simple strategy to achieve the same objective. Although we did not detect significant differences in the recombinant viral titers when helper plasmids either contain the Ad5 ITRs or lack them (Table 2), we suggest that it might be prudent to avoid the use of a helper plasmid that contains adenovirus ITRs to minimize generation of wt AAV-like particles. Furthermore, since removal of the distal 10 nucleotides in the D sequence from the next generation of pD- 10- based recombinant AAV vectors leads to complete elimination of replication-competent wt AAV-like particles, when a helper plasmid lacking adenovirus ITRs is used, such a combined approach should be useful in generating clinical-grade vectors for human gene therapy.
ACKNOWLEDGMENTS
We thank Richard J. Samulski for providing plasmid pAAV/Ad.
This research was supported in part by Public Health Service grants (HL-48342, HL-53586, HL-58881, and DK-49218; Centers for Excellence in Molecular Hematology) from the National Institutes of Health and a grant from the Phi Beta Psi Sorority. A.S. was supported by an Established Investigator Award from the American Heart Association.
REFERENCES
- 1.Allen J M, Debelak D J, Reynolds T C, Miller A D. Identification and elimination of replication-competent adeno-associated virus (AAV) that can arise by non-homologous recombination during AAV vector production. J Virol. 1997;71:6816–6822. doi: 10.1128/jvi.71.9.6816-6822.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ashktorab H, Srivastava A. Identification of nuclear proteins that specifically interact with adeno-associated virus type 2 inverted terminal repeat hairpin DNA. J Virol. 1989;63:3034–3039. doi: 10.1128/jvi.63.7.3034-3039.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berns K I, Bohenzky R A. Adeno-associated viruses: an update. Adv Virus Res. 1987;32:243–306. doi: 10.1016/s0065-3527(08)60479-0. [DOI] [PubMed] [Google Scholar]
- 4.Berns K I, Giraud C. Biology of adeno-associated virus. Curr Top Microbiol Immunol. 1996;218:1–23. doi: 10.1007/978-3-642-80207-2_1. [DOI] [PubMed] [Google Scholar]
- 5.Berns K I, Kotin R M, Labow M A. Regulation of adeno-associated virus DNA replication. Biochim Biophys Acta. 1988;951:425–429. doi: 10.1016/0167-4781(88)90116-9. [DOI] [PubMed] [Google Scholar]
- 6.Chejanovsky N, Carter B J. Mutagenesis of an AUG codon in the adeno-associated virus rep gene: effects on viral DNA replication. Virology. 1989;173:120–128. doi: 10.1016/0042-6822(89)90227-4. [DOI] [PubMed] [Google Scholar]
- 7.Chiorini J A, Wiener S M, Yang L, Smith R H, Safer B, Kilcoin N P, Liu Y, Urcelay E, Kotin R M. The roles of AAV Rep proteins in gene expression and targeted integration. Curr Top Microbiol Immunol. 1996;218:25–33. doi: 10.1007/978-3-642-80207-2_2. [DOI] [PubMed] [Google Scholar]
- 8.Flotte T R, Afione S A, Zeitlin P L. Adeno-associated virus vector gene expression occurs in nondividing cells in the absence of vector DNA integration. Am J Respir Cell Mol Biol. 1994;11:517–521. doi: 10.1165/ajrcmb.11.5.7946381. [DOI] [PubMed] [Google Scholar]
- 9.Flotte T R, Carter B J. Adeno-associated virus vectors for gene therapy. Gene Ther. 1995;2:357–362. [PubMed] [Google Scholar]
- 10.Giraud C, Winocour E, Berns K I. Recombinant junctions formed by site-specific integration of adeno-associated virus genome into an episome. J Virol. 1995;69:6917–6924. doi: 10.1128/jvi.69.11.6917-6924.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hargrove P W, Vanin E F, Kurtzman G J, Nienhuis A W. High-level globin expression mediated by a recombinant adeno-associated virus genome that contains the 3′ γ-globin gene regulatory element and integrates as tandem copies in erythroid cells. Blood. 1997;89:2167–2175. [PubMed] [Google Scholar]
- 12.Hauswirth W W, Berns K I. Origin and termination of adeno-associated virus DNA replication. Virology. 1977;78:488–499. doi: 10.1016/0042-6822(77)90125-8. [DOI] [PubMed] [Google Scholar]
- 13.Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967;26:365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- 14.Im D-S, Muzyczka N. Factors that bind to adeno-associated virus terminal repeats. J Virol. 1989;63:3095–3104. doi: 10.1128/jvi.63.7.3095-3104.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Im D-S, Muzyczka N. The AAV origin binding protein Rep68 is an ATP-dependent site-specific endonuclease with DNA helicase activity. Cell. 1990;61:447–457. doi: 10.1016/0092-8674(90)90526-k. [DOI] [PubMed] [Google Scholar]
- 16.Im D-S, Muzyczka N. Partial purification of adeno-associated virus Rep78, Rep68, Rep52, and Rep40 proteins and their biochemical characterization. J Virol. 1992;66:1119–1128. doi: 10.1128/jvi.66.2.1119-1128.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koeberl D D, Alexander I E, Halbert C L, Russell D W, Miller A D. Persistent expression of human clotting factor IX from mouse liver after intravenous injection of adeno-associated virus vectors. Proc Natl Acad Sci USA. 1997;94:1426–1431. doi: 10.1073/pnas.94.4.1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kotin R M, Berns K I. Organization of adeno-associated virus DNA in latently infected Detroit 6 cells. Virology. 1989;170:460–467. doi: 10.1016/0042-6822(89)90437-6. [DOI] [PubMed] [Google Scholar]
- 19.Kotin R M, Menninger J C, Ward D C, Berns K I. Mapping and direct visualization of a region-specific viral DNA integration site on chromosome 19q13-qter. Genomics. 1991;10:831–834. doi: 10.1016/0888-7543(91)90470-y. [DOI] [PubMed] [Google Scholar]
- 20.Kotin R M, Siniscalco M, Samulski R J, Zhu X, Hunter L, Laughlin C A, McLaughlin S, Muzyczka N, Rocchi M, Berns K I. Site-specific integration by adeno-associated virus. Proc Natl Acad Sci USA. 1990;87:2211–2215. doi: 10.1073/pnas.87.6.2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kube D M, Ponnazhagan S, Srivastava A. Encapsidation of adeno-associated virus type 2 Rep proteins in wild-type and recombinant progeny virions: Rep-mediated growth inhibition of primary human cells. J Virol. 1997;71:7361–7371. doi: 10.1128/jvi.71.10.7361-7371.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kube D M, Srivastava A. Quantitative DNA slot blot analysis: inhibition of DNA binding to membranes by magnesium ions. Nucleic Acids Res. 1997;25:3375–3376. doi: 10.1093/nar/25.16.3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lusby E, Fife K H, Berns K I. Nucleotide sequence of the inverted terminal repetition in adeno-associated virus DNA. J Virol. 1980;34:402–409. doi: 10.1128/jvi.34.2.402-409.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCarty D M, Pereira D J, Zolotukhin I, Zhou X, Ryan J H, Muzyczka N. Identification of linear DNA sequences that specifically bind the adeno-associated virus rep protein. J Virol. 1994;68:4988–4997. doi: 10.1128/jvi.68.8.4988-4997.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCarty D M, Ryan J H, Zolotukhin S, Zhou X, Muzyczka N. Interaction of the adeno-associated virus rep protein with a sequence within the palindrome of the viral terminal repeat. J Virol. 1994;68:4998–5006. doi: 10.1128/jvi.68.8.4998-5006.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muzyczka N. Use of adeno-associated virus as a general transduction vector for mammalian cells. Curr Top Microbiol Immunol. 1992;158:97–129. doi: 10.1007/978-3-642-75608-5_5. [DOI] [PubMed] [Google Scholar]
- 27.Nahreini P, Srivastava A. Rescue and replication of the adeno-associated virus 2 genome in mortal and immortal human cells. Intervirology. 1989;30:74–85. doi: 10.1159/000150078. [DOI] [PubMed] [Google Scholar]
- 28.Nahreini P, Srivastava A. Rescue of the adeno-associated virus 2 genome correlates with alterations in DNA-modifying enzymes in human cells. Intervirology. 1992;33:109–115. doi: 10.1159/000150239. [DOI] [PubMed] [Google Scholar]
- 29.Nahreini P, Woody M J, Zhou S Z, Srivastava A. Versatile adeno-associated virus 2-based vectors for constructing recombinant virions. Gene. 1993;124:257–262. doi: 10.1016/0378-1119(93)90402-o. [DOI] [PubMed] [Google Scholar]
- 30.Podsakoff G, Wong K K, Jr, Chatterjee S. Stable and efficient gene transfer into nondividing cells by adeno-associated virus-based vectors. J Virol. 1994;68:5656–5666. doi: 10.1128/jvi.68.9.5656-5666.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ponnazhagan S, Mukherjee P, Yoder M C, Wang X-S, Zhou S Z, Kaplan J, Wadsworth S, Srivastava A. Adeno-associated virus 2-mediated gene transfer in vivo: organ-tropism and expression of transduced sequences in mice. Gene. 1997;190:203–210. doi: 10.1016/s0378-1119(96)00576-8. [DOI] [PubMed] [Google Scholar]
- 32.Ponnazhagan S, Wang X-S, Woody M J, Luo F, Kang L Y, Nallari M L, Munshi N C, Zhou S Z, Srivastava A. Differential expression in human cells from the p6 promoter of human parvovirus B19 following plasmid transfection and recombinant adeno-associated virus 2 (AAV) infection: human megakaryocytic leukemia cells are non-permissive for AAV infection. J Gen Virol. 1996;77:1111–1122. doi: 10.1099/0022-1317-77-6-1111. [DOI] [PubMed] [Google Scholar]
- 33.Ponnazhagan S, Yoder M C, Srivastava A. Adeno-associated virus type 2-mediated transduction of murine hematopoietic cells with long-term repopulating ability and sustained expression of a human globin gene in vivo. J Virol. 1997;71:3098–3104. doi: 10.1128/jvi.71.4.3098-3104.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. pp. 1.53–1.110. [Google Scholar]
- 35.Samulski R J, Berns K I, Tan M, Muzyczka N. Cloning of adeno-associated virus into pBR322: rescue of intact virus from the recombinant plasmid in human cells. Proc Natl Acad Sci USA. 1982;79:2077–2081. doi: 10.1073/pnas.79.6.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Samulski R J, Chang L-S, Shenk T. A recombinant plasmid from which an infectious adeno-associated virus genome can be excised in vitro and its use to study viral replication. J Virol. 1987;61:3096–3101. doi: 10.1128/jvi.61.10.3096-3101.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Samulski R J, Chang L-S, Shenk T. Helper-free stocks of recombinant adeno-associated viruses: normal integration does not require viral gene expression. J Virol. 1989;63:3822–3828. doi: 10.1128/jvi.63.9.3822-3828.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Samulski R J, Srivastava A, Berns K I, Muzyczka N. Rescue of adeno-associated virus from recombinant plasmids: gene correction within the terminal repeats of AAV. Cell. 1983;33:135–143. doi: 10.1016/0092-8674(83)90342-2. [DOI] [PubMed] [Google Scholar]
- 39.Samulski R J, Zhu X, Xiao X, Brook J, Houseman D E, Epstein N, Hunter L A. Targeted integration of adeno-associated virus (AAV) into human chromosome 19. EMBO J. 1991;10:3941–3950. doi: 10.1002/j.1460-2075.1991.tb04964.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Senapathy P, Tratschin J-D, Carter B J. Replication of adeno-associated virus DNA. Complementation of naturally occurring rep− mutants by a wild-type genome or an ori− mutant and correction of terminal palindrome deletions. J Mol Biol. 1984;179:1–20. doi: 10.1016/0022-2836(84)90303-6. [DOI] [PubMed] [Google Scholar]
- 41.Shaughnessy E, Lu D, Chatterjee S, Wong K K., Jr Parvoviral vectors for the gene therapy of cancer. Semin Oncol. 1996;23:159–171. [PubMed] [Google Scholar]
- 42.Snyder R O, Miao C H, Patijn G A, Spratt K, Danos O, Nagy D, Gown A M, Winther B, Meuse L, Cohen L K, Thompson A R, Kay M A. Persistent and therapeutic concentrations of human factor IX in mice after hepatic gene transfer of recombinant AAV vectors. Nat Genet. 1997;16:270–276. doi: 10.1038/ng0797-270. [DOI] [PubMed] [Google Scholar]
- 43.Srivastava A. Replication of the adeno-associated virus DNA termini in vitro. Intervirology. 1987;27:138–147. doi: 10.1159/000149732. [DOI] [PubMed] [Google Scholar]
- 44.Srivastava A. Parvovirus-based vectors for human gene therapy. Blood Cells. 1994;20:531–536. [PubMed] [Google Scholar]
- 44a.Srivastava A. Delivery system for gene therapy: adeno-associated virus 2. In: Quesenberry P J, Stein G S, Forget B, Weissman S, editors. Stem cell biology and gene therapy. New York, N.Y: Wiley-Liss, Inc.; 1998. pp. 257–288. [Google Scholar]
- 45.Srivastava A, Lusby E W, Berns K I. Nucleotide sequence and organization of the adeno-associated virus 2 genome. J Virol. 1983;45:555–564. doi: 10.1128/jvi.45.2.555-564.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vincent K A, Piraino S T, Wadsworth S C. Analysis of recombinant adeno-associated virus packaging and requirements for rep and cap gene products. J Virol. 1997;71:1897–1905. doi: 10.1128/jvi.71.3.1897-1905.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang X-S, Ponnazhagan S, Srivastava A. Rescue and replication signals of the adeno-associated virus 2 genome. J Mol Biol. 1995;250:573–580. doi: 10.1006/jmbi.1995.0398. [DOI] [PubMed] [Google Scholar]
- 48.Wang X-S, Ponnazhagan S, Srivastava A. Rescue and replication of adeno-associated virus type 2 as well as vector DNA sequences from recombinant plasmids containing deletions in the viral inverted terminal repeats: selective encapsidation of viral genomes in progeny virions. J Virol. 1996;70:1668–1677. doi: 10.1128/jvi.70.3.1668-1677.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang X-S, Qing K Y, Ponnazhagan S, Srivastava A. Adeno-associated virus type 2 DNA replication in vivo: mutation analyses of the D sequence in viral inverted terminal repeats. J Virol. 1997;71:3077–3082. doi: 10.1128/jvi.71.4.3077-3082.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang X-S, Srivastava A. A novel terminal resolution-like site in the adeno-associated virus type 2 genome. J Virol. 1997;71:1140–1146. doi: 10.1128/jvi.71.2.1140-1146.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang X-S, Srivastava A. Rescue and autonomous replication of adeno-associated virus type 2 genomes containing Rep-binding site mutations in the viral p5 promoter. J Virol. 1998;72:4811–4818. doi: 10.1128/jvi.72.6.4811-4818.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]