Abstract
In this study, seven new pentacyclic triterpene glycosides, named dendrocinaosides A–G (1–7), and six known ones (8–13) were isolated from the whole plants of Dendrobium officinale. Their structures were determined by analyses of HR-ESI-MS, 1D and 2D NMR spectra. Compounds 1–4, 8, and 9 potentially inhibited α-glucosidase and α-amylase activities with the IC50 values ranging from 31.3 ± 2.2 to 42.4 ± 2.5 μM for anti α-glucosidase and from 36.5 ± 1.8 to 56.4 ± 2.0 μM for anti α-amylase activities, respectively, which were lower than that of the positive control, acarbose, showing IC50 values of 47.1 ± 1.4 μM for anti α-glucosidase and 145.7 ± 2.2 μM for anti α-amylase.
Seven new oleanane saponins were discovered from Dendrobium officinale. These saponins containing 29-noroleana-12,20(30)-dien-28-oic acid framework, caffeoyl, and coumaroyl moieties potentially inhibited α-glucosidase and α-amylase activities.
Introduction
The plant Dendrobium officinale Kimura & Migo (Orchidaceae family, Vietnamese name Lan Thạch Hộc) is an epiphytic herb that grows on large tree branches or on moist cliffs. The flat stem has longitudinal grooves divided into many segments, the stem is narrow, the tip is thicker, and the colour is light yellow. Short leaves with sheaths. The flowers are pink or white and pink, growing in short clusters between fallen leaves. The fruit is long and diamond shaped. This plant is distributed in the countries with tropical and subtropical climates such as China, Myanmar, Laos, and Vietnam. This plant has been used in traditional Chinese medicine for thousands of years used to protect the liver, treat cancer, gastritis, enhance immunity, stimulate digestion, reduce blood sugar and blood fat.1,2 In Vietnam, D. officinale has been found in the mountains and forests of Vinh Phu, Quang Tri, Quang Nam, Da Nang, Gia Lai, Lam Dong provinces. The previous studies reported terpenoids,3 bibenzyl derivatives,1–7 polysaccharides,8–15 are the main components of this plants. Some of which exhibited antitumor,7,14,15 anti-inflammatory activities8–13 and alleviates type 2 diabetes mellitus.9 Therefore, the whole plants of D. officinale were selected for further study. This paper reported the isolation, structural identification of thirteen triterpene glycosides (1–13), including seven undescribed compounds (1–7) and their anti α-glucosidase and α-amylase activities in vitro.
Experimental
General experimental procedures
The optical rotations were measured on a Jasco P2000 polarimeter. IR spectra were recorded on a PerkinElmer SpectrumTwo FTIR spectrometer. The NMR spectra were measured on a Bruker Avance 600 MHz FT-NMR spectrometer. The HR-ESI-MS were measured on a SCIEX X500 QTOF or Agilent 6530 QTOF system. The semi-preparative HPLC was performed on an Agilent 1260 infinity II system including binary pump, autosampler, DAD detector, and YMC J'sphere ODS-H80 (20 × 250 mm, 4 μm) HPLC column. Mobile phases were an isocratic system of methanol/water or acetonitrile/water at flow rate of 3 mL min−1. Thin layer chromatography was performed on pre-coated plates with silica gel or reversed phase C18 (RP18). Column chromatography was performed using silica gel (40–63 μm) or RP18 (150 μm) as adsorbents.
Plant material
Plant samples, Dendrobium officinale Kimura & Migo, were collected in Tam Dao, Vinh Phuc province, Vietnam during April 2022 and identified by Dr Nguyen The Cuong, Institute of Ecology and Biological Resources, VAST. Voucher specimen (NCCT-P160) was kept at the Institute of Marine Biochemistry, VAST.
Extraction and isolation
Dried powdered D. officinale whole plants (8.1 kg) was ultrasonically extracted with MeOH at 40 °C for three times (each, 20 L, 2 h) to obtain MeOH residue (300 g). This was suspended with 5.0 L of water and then successively partitioned with n-hexane, EtOAc to give n-hexane (DO1, 100 g), EtOAc (DO2, 28 g), and water-soluble extracts (DO3, 160 g). These fractions were screened for their anti α-glucosidase and α-amylase activities in vitro at the concentration of 200 μg mL−1. Fraction DO3 showed significant anti α-glucosidase (71.2%) and anti α-amylase (68.0%) activity. Therefore, this fraction was selected for further study. The DO3 fraction was run on a Diaion (HP-20) column using MeOH/H2O (1/4, 1/1, 3/4, and 1/0, each 3 L) as eluent to give four fractions, DO3A (45.6 g), DO3B (32 g), DO3C (27.0 g), and DO3D (18.5 g), respectively. Fraction DO3B (32 g) was isolated on an RP18 column eluting with MeOH/H2O (3/1) to get five fractions, DO3B1–DO3B5. Fraction DO3B2 (5.7 g) was chromatographed on a silica gel column eluting with CH2Cl2/MeOH (3/1) to get three fractions, DO3B2A-DO3B2C. Fraction DO3B2B (212 mg) was isolated on HPLC using CH3CN 35% in water to get two compounds 10 (22.5 mg, tR = 35.19 min) and 11 (20.3 mg, tR = 32.17 min). Fraction DO3B2C (183 mg) was purified on the HPLC using CH3CN 25% to get compound 12 (15.5 mg, tR = 59.31 min). Fraction DO3B4 (6.1 g) was chromatographed on a silica gel column eluting with CH2Cl2/MeOH (4/1) to get four fractions, DO3B4A–DO3B4D. Fraction DO3B4B (248 mg) was purified on the HPLC using CH3CN 35% to get two compounds, 8 (51.0 mg, tR = 34.00 min) and 9 (23.4 mg, tR = 43.46 min). Fraction DO3B4C (176 mg) was purified on the HPLC using CH3CN 31% to get two compounds, 5 (16.5 mg, tR = 37.70 min) and 6 (15.2 mg, tR = 25.53 min). Fraction DO3C (27.1 g) isolated on an RP18 column eluting with MeOH/H2O (4/1) to get four fractions, DO3C1–DO3C4. Fraction DO3C2 (2.7 g) was chromatographed on a silica gel column eluting with CH2Cl2/MeOH (5/1) to get six fractions, DO3C2A–DO3C2F. Fraction DO3C2B (178 mg) was purified on the HPLC using CH3OH 70% in H2O to get compound 1 (15.8 mg, tR = 45.56 min). Fraction DO3C2C (157 mg) was purified on the HPLC using CH3OH 70% in H2O to get compound 2 (7.0 mg, tR = 35.10 min). Fraction DO3C3 (1.9 g) was chromatographed on a silica gel column eluting with CH2Cl2/MeOH (6/1) to get three fractions, DO3C3A–DO3C3C. Fraction DOC3B (217 mg) was chromatographed on the HPLC using CH3CN 25% to get two compounds, 3 (18.2 mg, tR = 23.81 min) and 4 (17.0 mg, tR = 40.29 min). Fraction DO3D (18.5 g) isolated on an RP18 column eluting with MeOH/H2O (5/1) to get four fractions, DO3D1–DO3D4. Fraction DO3D2 (2.9 g) was chromatographed on a silica gel column eluting with CH2Cl2/MeOH (7/1) and then purified on the HPLC using CH3CN 25% to get compound 7 (19.0 mg, tR = 43.37 min). Fraction DO3D3 (1.8 g) was chromatographed on a silica gel column eluting with CH2Cl2/MeOH (8/1) and then purified on the HPLC using CH3OH 75% in H2O to get compound 13 (20.1 mg, tR = 38.5 min).
Dendrocinaoside A (1)
Colourless powder; mp 216–218 °C. [α]25D: −31.6 (c 0.1, MeOH); UV λmax: 220, 245, 331 nm. IR (KBr) ν cm−1: 3404, 2939, 1755, 1649, 1464, 1388, 1175, 1064; HR-ESI-MS m/z: 1227.5586 [M + Na]+, (calcd for [C61H88O24Na]+, 1227.5558, Δ = +2.3 ppm); m/z: 1205.5728 [M + H]+, (calcd for [C61H89O24]+, 1205.5739, Δ = −0.9 ppm). 1H- and 13C-NMR data were shown in the Table 1.
1H-NMR and 13C-NMR spectral data for 1–4.
| No. | 1 | 2 | 3 | 4 | ||||
|---|---|---|---|---|---|---|---|---|
| δ C | δ H (mult., J in Hz) | δ C | δ H (mult., J in Hz) | δ C | δ H (mult., J in Hz) | δ C | δ H (mult., J in Hz) | |
| 1 | 39.8 | 0.97 (m)/1.63 (m) | 39.7 | 0.97 (m)/1.63 (m) | 39.7 | 0.97 (m)/1.62 (m) | 39.7 | 0.96 (m)/1.62 (m) |
| 2 | 27.0 | 1.70(m)/1.86 (m) | 27.0 | 1.70(m)/1.85 (m) | 26.9 | 1.70(m)/185 (m) | 27.0 | 1.70(m)/1.85 (m) |
| 3 | 90.8 | 3.12 (dd, 12.0, 4.2) | 90.8 | 3.13 (dd, 12.0, 4.2) | 90.8 | 3.12 (dd, 12.0, 4.8) | 90.8 | 3.12 (dd, 12.0, 4.2) |
| 4 | 40.8 | — | 40.7 | — | 40.7 | — | 40.7 | — |
| 5 | 56.9 | 0.72 (br d, 11.4) | 57.5 | 0.76 (br d, 11.4) | 57.0 | 0.75 (br d, 11.4) | 57.0 | 0.75 (br d, 12.0) |
| 6 | 19.3 | 1.37 (m)/(1.49) | 19.3 | 1.37 (m)/1.50 (m) | 19.3 | 1.37 (m)/1.50 (m) | 19.3 | 1.35 (m)/1.48 (m) |
| 7 | 33.9 | 1.29 (m)/1.48 (m) | 33.9 | 1.30 (m)/1.48 (m) | 33.9 | 1.30 (m)/1.48 (m) | 33.9 | 1.29 (m)/1.46 (m) |
| 8 | 40.1 | — | 40.1 | — | 40.1 | — | 40.1 | — |
| 9 | 49.0 | 1.58 (m) | 49.0 | 1.58 (m) | 49.0 | 1.60 (m) | 49.0 | 1.58 (m) |
| 10 | 37.9 | — | 37.9 | — | 37.9 | — | 37.9 | — |
| 11 | 24.6 | 1.90–1.92 (m) | 24.6 | 1.90–1.92 (m) | 24.7 | 1.90–1.92 (m) | 24.6 | 1.90–1.92 (m) |
| 12 | 124.3 | 5.34 (t, 3.0) | 124.2 | 5.31 (t, 3.0) | 124.2 | 5.28 (t, 3.6) | 123.9 | 5.28 (t, 3.6) |
| 13 | 144.2 | — | 144.0 | — | 144.8 | — | 144.8 | — |
| 14 | 42.9 | — | 42.9 | — | 42.8 | — | 42.9 | — |
| 15 | 28.9 | 1.13 (m)/1.80 (m) | 28.9 | 1.10 (m)/1.78 (m) | 29.0 | 1.10 (m)/1.78 (m) | 28.9 | 1.08 (m)/1.77 (m) |
| 16 | 24.2 | 1.85 (m)/2.17a | 24.1 | 1.77(m)/2.12 (td, 13.2, 3.6) | 24.0 | 1.77(m)/2.07 (td, 13.2, 2.4) | 24.0 | 1.72 (m)/2.05 (td, 13.2, 3.0) |
| 17 | 48.5 | — | 47.9 | — | 48.4 | — | 48.3 | — |
| 18 | 48.4 | 2.75 (dd, 13.8, 4.8) | 44.7 | 2.81 (dd, 13.8, 4.2) | 41.4 | 2.90 (dd, 13.2, 4.2) | 41.8 | 2.90 (dd, 13.2, 3.6) |
| 19 | 42.6 | 2.10 (dd, 13.8, 4.6) 2.57 (t, 13.8) | 47.6 | 1.47a 2.00 (t, 13.8) | 41.4 | 1.11a 1.83 (t, 13.8) | 41.4 | 1.12a 1.83 (t, 13.4) |
| 20 | 149.6 | — | 71.3 | — | 36.8 | — | 36.8 | — |
| 21 | 30.9 | 2.15a 2.24 (td, 13.2, 3.6) | 35.6 | 1.51 (m)/1.65 (m) | 29.5 | 1.18 (m)/1.52 (m) | 29.3 | 1.18 (m)/1.50 (m) |
| 22 | 38.5 | 1.55 (m)/1.91 (m) | 39.8 | 1.67 (m)/1.75 (m) | 32.6 | 1.65 (m)/1.74 (m) | 32.6 | 1.65 (m)/1.74 (m) |
| 23 | 28.5 | 0.95 (s) | 28.5 | 0.93 (s) | 28.5 | 0.93 (s) | 28.5 | 0.93 (s) |
| 24 | 16.9 | 0.73 (s) | 16.9 | 0.73 (s) | 16.9 | 0.72 (s) | 16.9 | 0.72 (s) |
| 25 | 16.1 | 0.95 (s) | 16.0 | 0.95 (s) | 16.0 | 0.93 (s) | 16.0 | 0.94 (s) |
| 26 | 17.8 | 0.80 (s) | 18.0 | 0.80 (s) | 17.8 | 0.79 (s) | 17.8 | 0.79 (s) |
| 27 | 26.3 | 1.18 (s) | 26.2 | 1.18 (s) | 26.3 | 1.17 (s) | 26.3 | 1.17 (s) |
| 28 | 177.3 | — | 177.6 | — | 178.0 | — | 178.0 | — |
| 29 | — | — | — | — | 74.2 | 3.21 (s) | 74.3 | 3.21 (s) |
| 30 | 107.5 | 4.64 (br s) 4.65 (br s) | 25.0 | 1.25 (s) | 19.7 | 0.95 (s) | 19.7 | 0.95 (s) |
Overlapped signal.
| 3-O-Arabinose | 3-O-Arabinose | 3-O-Arabinose | 3-O-Arabinose | |||||
|---|---|---|---|---|---|---|---|---|
| 1′ | 105.2 | 4.49 (d, 7.8) | 105.2 | 4.49 (d, 7.2) | 105.2 | 5.49 (d, 7.2) | 105.2 | 5.48 (d, 7.8) |
| 2′ | 74.2 | 5.15 (dd, 9.0, 7.8) | 74.2 | 5.15 (dd, 9.0, 7.2) | 74.2 | 5.15 (dd, 9.0, 7.2) | 74.2 | 5.15 (dd, 9.0, 7.2) |
| 3′ | 72.8 | 3.76 (dd, 9.0, 3.0) | 72.8 | 3.76 (br d, 9.0) | 72.8 | 3.76 (br d, 9.0) | 72.8 | 3.76 (br d, 9.0) |
| 4′ | 70.2 | 3.88 (br s) | 70.2 | 3.88 (br s) | 70.2 | 3.88 (br s) | 70.2 | 3.87 (br s) |
| 5′ | 67.1 | 3.62 (d, 12.0) 3.92 (dd, 12.0, 2.4) | 67.1 | 3.61 (br s, 12.0) 3.92 (br d, 12.0) | 67.1 | 3.61 (br s, 12.0) 3.92 (br d, 12.0) | 67.1 | 3.61 (br s, 11.4) 3.92 (br d, 11.4) |
| 28-O-Glucose | 28-O-Glucose | 28-O-Glucose | 28-O-Glucose | |||||
|---|---|---|---|---|---|---|---|---|
| 1′′ | 95.9 | 5.32 (d, 7.8) | 95.8 | 5.34 (d, 7.8) | 95.8 | 5.35 (d, 7.8) | 95.8 | 5.35 (d, 7.8) |
| 2′′ | 73.8 | 3.33a | 73.8 | 3.33a | 73.8 | 3.34a | 73.9 | 3.34a |
| 3′′ | 78.2 | 3.40a | 78.2 | 3.40a | 78.2 | 3.40a | 78.2 | 3.40a |
| 4′′ | 71.0 | 3.40a | 71.0 | 3.40a | 71.0 | 3.40a | 71.0 | 3.42a |
| 5′′ | 78.1 | 3.52 (m) | 78.1 | 3.52 (m) | 78.2 | 3.52 (m) | 78.1 | 3.52 (m) |
| 6′′ | 69.4 | 3.78a 4.08 (d, 12.0, 1.8) | 69.5 | 3.80a 4.09 (d, 12.0, 1.8) | 69.4 | 3.82a 4.09 (d, 12.0, 1.8) | 69.4 | 3.82a 4.09 (br d, 12.0) |
| 6′′-O-Glucose | 6′′-O-Glucose | 6′′-O-Glucose | 6′′-O-Glucose | |||||
|---|---|---|---|---|---|---|---|---|
| 1′′′ | 104.2 | 4.41 (d, 7.8) | 104.3 | 4.41 (d, 7.8) | 104.2 | 4.42 (d, 7.8) | 104.2 | 4.43 (d, 7.8) |
| 2′′′ | 75.3 | 3.24 (dd, 9.0, 7.8) | 75.3 | 3.24a | 75.3 | 3.24 (dd, 9.0, 7.8) | 75.3 | 3.25 (dd, 9.0, 7.8) |
| 3′′′ | 76.7 | 3.48 (t, 9.0) | 76.7 | 3.48 (t, 9.0) | 76.7 | 3.48 (t, 9.0) | 76.7 | 3.48 (t, 9.0) |
| 4′′′ | 79.6 | 3.56 (t, 9.0) | 79.7 | 3.56a | 79.7 | 3.55 (t, 9.0 | 79.6 | 3.55 (t, 9.0 |
| 5′′′ | 76.8 | 3.30 (m) | 76.9 | 3.31 (m) | 76.9 | 3.31 (m) | 76.9 | 3.31 (m) |
| 6′′′ | 61.9 | 3.67 (dd, 11.4, 4.2) 3.84 (dd, 11.4, 1.8) | 61.9 | 3.67a 3.82a | 61.9 | 3.67a 3.82a | 61.9 | 3.67 (dd, 11.4, 5.4) 3.83a |
| 4′′′-O-Rhamnose | 4′′′-O-Rhamnose | 4′′′-O-Rhamnose | 4′′′-O-Rhamnose | |||||
|---|---|---|---|---|---|---|---|---|
| 1′′′′ | 102.9 | 4.86 (d, 1.2) | 103.0 | 4.86 (d, 1.2) | 103.0 | 4.85 (d, 1.2) | 103.0 | 4.86 (d, 1.2) |
| 2′′′′ | 72.4 | 3.86 (dd, 3.0, 1.2) | 72.4 | 3.85a | 72.4 | 3.85a | 72.4 | 3.85a |
| 3′′′′ | 72.2 | 3.66 (dd, 9.0, 3.0) | 72.2 | 3.65a | 72.2 | 3.65a | 72.2 | 3.64a |
| 4′′′′ | 73.8 | 3.43 (t, 9.0) | 73.8 | 3.43a | 73.8 | 3.4a | 73.8 | 3.43a |
| 5′′′′ | 70.7 | 3.98 (m) | 70.7 | 3.98a | 70.7 | 3.97a | 70.7 | 3.98a |
| 6′′′′ | 17.8 | 1.29 (d, 6.6) | 17.8 | 1.29 (d, 6.6) | 17.8 | 1.28 (d, 6.6) | 17.8 | 1.29 (d, 6.6) |
| Caffeoyl | Caffeoyl | Coumaroyl | Caffeoyl | |||||
|---|---|---|---|---|---|---|---|---|
| 1′′′′′ | 127.8 | — | 127.8 | — | 127.0 | — | 127.8 | — |
| 2′′′′′ | 115.1 | 7.05 (d, 1.8) | 115.0 | 7.05 (d, 1.8) | 131.1 | 7.46 (d, 8.0) | 115.1 | 7.05 (d, 1.8) |
| 3′′′′′ | 146.9 | — | 146.9 | — | 117.0 | 6.83 (d, 8.0) | 146.8 | — |
| 4′′′′′ | 149.4 | — | 149.6 | — | 161.7 | — | 149.6 | — |
| 5′′′′′ | 116.7 | 6.80 (d, 7.8) | 116.6 | 6.80 (d, 8.0) | 117.0 | 6.83 (d, 8.0) | 116.6 | 6.80 (d, 8.0) |
| 6′′′′′ | 122.9 | 6.95 (dd, 7.8, 1.8) | 122.9 | 6.95 (dd, 8.0, 1.8) | 131.1 | 7.46 (d, 8.0) | 122.9 | 6.95 (dd, 8.0, 1.8) |
| 7′′′′′ | 147.1 | 7.58 (d, 15.6) | 147.1 | 7.58 (d, 16.2) | 146.8 | 7.65 (d, 15.6) | 147.1 | 7.58 (d, 16.2) |
| 8′′′′′ | 115.4 | 6.29 (d, 15.6) | 115.5 | 6.29 (d, 16.2) | 115.3 | 6.35 (d, 15.6) | 115.5 | 6.29 (d, 16.2) |
| 9′′′′′ | 168.6 | — | 168.6 | — | 168.6 | — | 168.6 | - |
Dendrocinaoside B (2)
Colorless powder; mp 207–208 °C. [α]25D: −43.5 (c 0.1, MeOH); UV λmax: 220, 245, 335 nm. IR (KBr) ν cm−1: 3422, 2942, 1732, 1639, 1460, 1390, 1228, 1063; HR-ESI-MS m/z: 1223.5854 [M + H]+, (calcd for [C61H91O25]+, 1223.5844, Δ = +0.8 ppm); 1H- and 13C-NMR data were shown in the Table 1.
Dendrocinaoside C (3)
Colorless powder; mp 212–214 °C. [α]25D: −28.0 (c 0.1, MeOH); UV λmax: 230, 315 nm. IR (KBr) ν cm−1: 3414, 2939, 1730, 1642, 1548, 1467, 1391, 1376, 1254, 1170, 1074; HR-ESI-MS m/z: 1221.6071 [M + H]+, (calcd for [C62H93O24]+, 1221.6051, Δ = +1.6 ppm); 1H- and 13C-NMR data were shown in the Table 1.
Dendrocinaoside D (4)
Colorless powder; mp 221–223 °C. [α]25D: −33.8 (c 0.1, MeOH); UV λmax: 219, 245, 300, 330 nm. IR (KBr) ν cm−1: 3444, 2939, 1731, 1751, 1698, 1634, 1456, 1259, 1170, 1075; HR-ESI-MS m/z 1235.5839 [M–H]−, (calcd for [C62H91O25]−, 1235.5855, Δ = −1.3 ppm), m/z 1271.5568 [M + 35Cl]−, (calcd for [C62H92O2535Cl]−, 1271.5521, Δ = −4.2 ppm), m/z 1273.5563 [M + 37Cl]−, (calcd for [C62H92O2537Cl]−, 1273.5592, Δ = −2.3 ppm); 1H- and 13C-NMR data were shown in the Table 1.
Dendrocinaoside E (5)
Colorless powder; mp 198–199 °C. [α]25D: −45.2 (c 0.1, MeOH); UV (MeOH) λmax (nm): terminal absorption at 190–240 nm; IR (KBr) ν cm−1: 3441, 2943, 1715, 1645, 1069; HR-ESI-MS m/z 1075.5679 [M + H]+, (calcd for [C53H87O22]+, 1075.5684, Δ = −0.5 ppm), m/z 1097.5476 [M + Na]+, (calcd for [C53H86O22Na]+, 1097.5503, Δ = −2.5 ppm); 1H- and 13C-NMR data were shown in the Table 2.
1H-NMR and 13C-NMR spectral data for 5–7.
| No. | 5 | 6 | 7 | |||
|---|---|---|---|---|---|---|
| δ C | δ H (mult., J in Hz) | δ C | δ H (mult., J in Hz) | δ C | δ H (mult., J in Hz) | |
| 1 | 39.8 | 0.99 (m)/1.63 (m) | 39.8 | 0.98 (m)/1.64 (m) | 39.78 | 0.98 (m)/1.64 (m) |
| 2 | 27.0 | 1.72 (m)/1.87 (m) | 27.0 | 1.72 (m)/1.86 (m) | 27.0 | 1.70 (m)/1.87 (m) |
| 3 | 90.7 | 3.15 (dd, 11.4, 4.2) | 90.7 | 3.16 (dd, 11.4, 4.2) | 90.7 | 3.15 (dd, 11.4, 4.2) |
| 4 | 40.7 | — | 40.8 | — | 40.8 | — |
| 5 | 57.1 | 0.80 (br d, 11.4) | 57.1 | 0.80 (br d, 11.4) | 57.1 | 0.80 (br d, 11.4) |
| 6 | 19.4 | 1.43 (m)/1.56 (m) | 19.4 | 1.43 (m)/1.56 (m) | 19.4 | 1.42 (m)/1.55 (m) |
| 7 | 33.9 | 1.33 (m)/1.52 (m) | 33.9 | 1.33 (m)/1.52 (m) | 34.0 | 1.33 (m)/1.50 (m) |
| 8 | 40.1 | — | 40.2 | — | 40.2 | — |
| 9 | 49.0 | 1.60 (m) | 49.0 | 1.60 (m) | 49.0 | 1.59 (m) |
| 10 | 37.9 | — | 38.0 | — | 37.9 | — |
| 11 | 24.6 | 1.92–1.94 (m) | 24.6 | 1.90–1.92 (m) | 24.6 | 1.90–1.93 (m) |
| 12 | 124.0 | 5.31 (t, 3.6) | 123.8 | 5.34 (t, 3.0) | 123.9 | 5.29 (t, 3.0) |
| 13 | 144.6 | — | 144.6 | — | 144.8 | — |
| 14 | 42.9 | — | 42.9 | — | 42.9 | — |
| 15 | 28.9 | 1.10 (m)/1.78 (m) | 28.9 | 1.10 (m)/1.80 (m) | 28.9 | 1.10 (m)/1.80 (m) |
| 16 | 24.3 | 1.76 (m)/2.10 (td, 13.2, 3.0) | 23.8 | 1.74 (m)/2.04 (td, 13.2, 3.0) | 24.1 | 1.73 (m)/2.07 (td, 13.8, 2.4) |
| 17 | 49.0 | — | 47.7 | — | 48.4 | — |
| 18 | 41.9 | 2.85 (dd, 13.2, 4.2) | 41.5 | 3.09 (dd, 13.8, 4.2) | 41.8 | 2.91 (dd, 13.8, 4.2) |
| 19 | 42.5 | 1.40a/1.62a | 46.1 | 1.47a 1.84 (t, 13.8) | 41.4 | 1.11a 1.83 (t, 13.8) |
| 20 | 36.1 | — | 69.8 | — | 36.8 | — |
| 21 | 29.9 | 1.31 (m)/1.52 (m) | 34.1 | 1.50 (m)/1.56 (m) | 29.3 | 1.19 (m)/1.50 (m) |
| 22 | 32.8 | 1.58 (m)/1.72 (m) | 32.5 | 1.57 (m)/1.94 (m) | 32.5 | 1.66 (m)/1.74 (m) |
| 23 | 28.6 | 1.06 (s) | 28.6 | 1.06 (s) | 28.6 | 1.06 (s) |
| 24 | 17.0 | 0.87 (s) | 17.0 | 0.88 (s) | 17.0 | 0.87 (s) |
| 25 | 16.1 | 0.98 (s) | 16.2 | 0.98 (s) | 16.1 | 0.98 (s) |
| 26 | 17.9 | 0.82 (s) | 18.0 | 0.84 (s) | 17.9 | 0.83 (s) |
| 27 | 26.3 | 1.19 (s) | 26.3 | 1.17 (s) | 26.3 | 1.19 (s) |
| 28 | 178.1 | — | 178.0 | — | 178.0 | — |
| 29 | 28.0 | 0.92 (s) | 31.5 | 1.20 (s) | 74.3 | 3.21 (s) |
| 30 | 66.3 | 3.45a/3.54a | — | — | 19.6 | 0.95 (s) |
Overlapped signal.
| 3-O-Arabinose | 3-O-Arabinose | 3-O-Arabinose | ||||
|---|---|---|---|---|---|---|
| 1′ | 107.1 | 4.30 (d, 6.6) | 107.1 | 4.30 (d, 6.6) | 107.1 | 4.30 (d, 6.6) |
| 2′ | 72.8 | 3.59 (dd, 9.0, 6.6) | 72.8 | 3.59 (dd, 9.0, 6.6) | 72.8 | 3.58 (dd, 9.0, 6.6) |
| 3′ | 74.3 | 3.52a | 74.3 | 3.52a | 74.3 | 3.53a |
| 4′ | 69.5 | 3.82 (br s) | 69.5 | 3.82 (br s) | 69.5 | 3.82 (br s) |
| 5′ | 66.3 | 3.53a 3.84a | 66.3 | 3.53a 3.84a | 66.3 | 3.53a 85 (dd, 9.6, 2.4) |
| 28-O-Glucose | 28-O-Glucose | 28-O-Glucose | ||||
|---|---|---|---|---|---|---|
| 1′′ | 95.8 | 5.35 (d, 7.8) | 96.1 | 5.32 (d, 7.8) | 95.8 | 5.38 (d, 7.8) |
| 2′′ | 73.9 | 3.35a | 73.8 | 3.36a | 73.8 | 3.35a |
| 3′′ | 78.2 | 3.40a | 78.2 | 3.43a | 78.2 | 3.43a |
| 4′′ | 71.1 | 3.42a | 71.1 | 3.43a | 71.0 | 3.44a |
| 5′′ | 78.0 | 3.52 (m) | 78.0 | 3.52 (m) | 77.8 | 3.54 (m) |
| 6′′ | 69.6 | 3.78 (dd, 12.0, 5.4) 4.10 (dd, 12.0, 1.8) | 69.8 | 3.78 (dd, 12.0, 5.4) 4.10 (dd, 12.0, 1.2) | 69.6 | 3.78 (dd, 11.4, 5.4) 4.10 (dd, 11.4, 1.8) |
| 6′′-O-Glucose | 6′′-O-Glucose | 6′′-O-Glucose | ||||
|---|---|---|---|---|---|---|
| 1′′′ | 104.4 | 4.41 (d, 7.8) | 104.6 | 4.39 (d, 7.8) | 104.7 | 4.37 (d, 7.8) |
| 2′′′ | 75.3 | 3.25 (dd, 9.0, 7.8) | 75.3 | 3.25 (dd, 9.0, 7.8) | 75.2 | 3.22 (dd, 9.0, 7.8) |
| 3′′′ | 76.7 | 3.48 (t, 9.0) | 76.8 | 3.48 (t, 9.0) | 78.0 | 3.37 (t, 9.0) |
| 4′′′ | 79.6 | 3.54a | 79.8 | 3.54a | 71.6 | 3.31a |
| 5′′′ | 76.9 | 3.31 (m) | 76.9 | 3.33 (m) | 78.0 | 3.27 (m) |
| 6′′′ | 62.0 | 3.68 (dd, 11.4, 5.4) 3.82a | 62.0 | 3.68 (dd, 11.4, 5.4) 3.82a | 62.0 | 3.69 (dd, 12.0, 6.6) 3.87 (dd, 12.0, 1.8) |
Dendrocinaoside F (6)
Colorless powder; mp 192–194 °C. [α]25D: −31.0 (c 0.1, MeOH); UV (MeOH) λmax (nm): terminal absorption at 190–240 nm; IR (KBr) ν cm−1:3412, 2937, 1737, 1645, 1457, 1389, 1066; HR-ESI-MS m/z 1061.5553 [M + H]+, (calcd for [C52H85O22]+, 1061.5527, Δ = +2.5 ppm), m/z 1083.5373 [M + Na]+, (calcd for [C52H84O22Na]+, 1083.5347, Δ = +2.4 ppm); 1H- and 13C-NMR data were shown in the Table 2.
Dendrocinaoside G (7)
Colorless powder; mp 194–196 °C. [α]25D: −46.8 (c 0.1, MeOH); UV (MeOH) λmax (nm): terminal absorption at 190–250 nm; IR (KBr) ν cm−1: 3415, 2942, 1715, 1612, 1456, 1418, 1078; HR-ESI-MS m/z 929.5128 [M + H]+, (calcd for [C47H77O18]+, 929.5105, Δ = +2.5 ppm), m/z 951.4913 [M + Na]+, (calcd for [C47H76O18Na]+, 951.4913, Δ = −1.7 ppm); 1H- and 13C-NMR data were shown in the Table 2.
Acid hydrolysis of compounds 1–7
Acid hydrolysis of compounds 1–7 were the same as described in previous reports16,17 and referred to ESI.†
α-Glucosidase and α-amylase inhibitory assay
The α-glucosidase and α-amylase inhibitory assay protocols are the same as described in previous reports18,19 and referred to ESI.†
Results and discussion
The whole part methanol extract of D. officinale was separated into n-hexane, EtOAc, and water-soluble fractions. The water portion was isolated on a Diaion HP20 column chromatography, then on silica gel or ODS reversed phase, and further purified by semipreparative HPLC to afford thirteen pentacyclic triterpene glycosides (1–13).
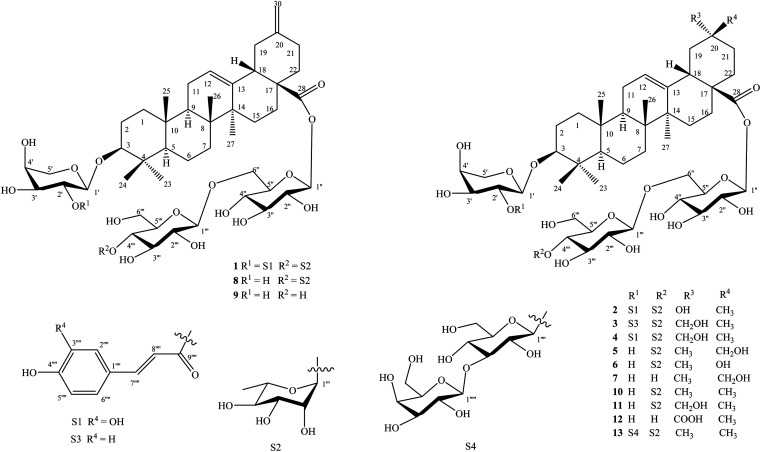
The known saponins were identified to be yemuoside YM14 (8),20 yemuoside YM11 (9),20 3-O-α-l-arabinopyranosyl-oleanolic acid 28-O-α-l-rhamnopyranosyl-(1→4)-β-d-glucopyranosyl-(1→6)-β-d-glucopyranosyl ester (10),21 3-O-α-l-arabinopyranosyl-29-hydroxyoleanolic acid 28-O-α-l-rhamnopyranosyl-(1→4)-β-d-glucopyranosyl-(1→6)-β-d-glucopyranosyl ester (11),22 3-O-α-l-arabinopyranosyl-serratagenic acid 28-O-β-d-glucopyranosyl-(1→6)-β-d-glucopyranosyl ester (12),23 3-O-β-d-galactopyranosyl-(1→3)-β-d-glucopyranosyl-(1→2)-α-l-arabinopyranosyl-oleanolic acid 28-O-α-l-rhamnopyranosyl-(1→4)-β-d-glucopyranosyl-(1→6)-β-d-glucopyranosyl ester (13)24 by comparisons of their physical and NMR data with those reported in the literature. New saponins 1–7 differed from above known ones by either containing addional coumaroyl, caffeoyl, and sugar moiety or loss of methyl group at C-20 in the oleanane-type backbone.
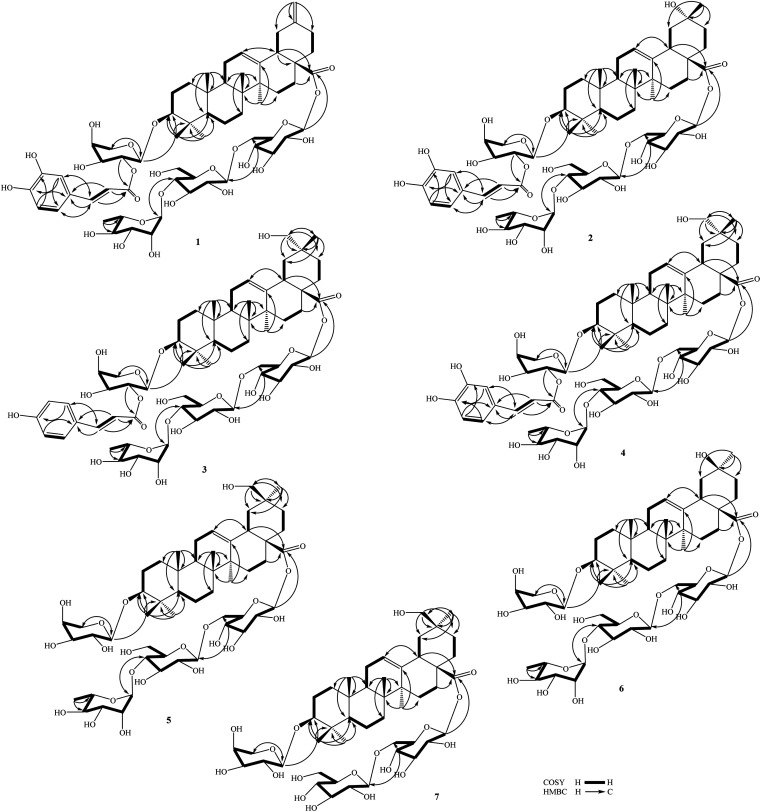
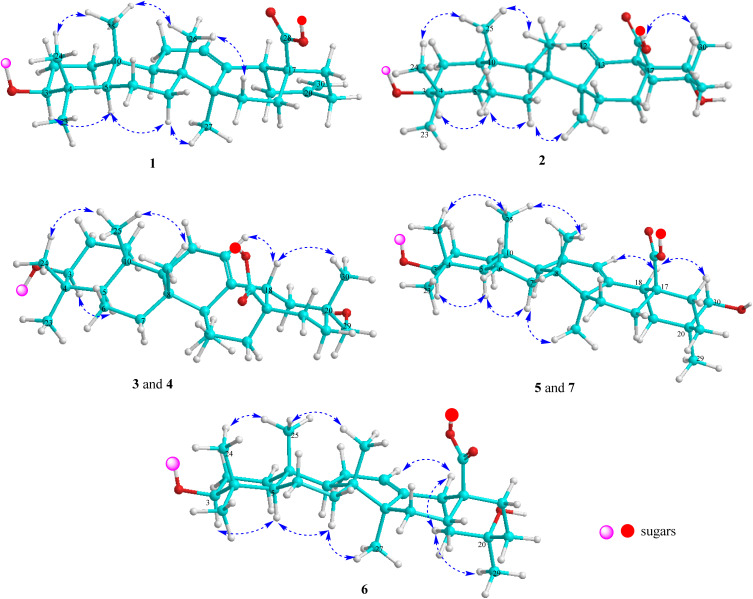
Compound 1 (Fig. 1) had a molecular formula, C61H88O24, as determined by the sodium adductive ion [M + Na]+ at m/z 1227.5586 (calcd for [C61H88O24Na]+, 1227.5558) and proton adductive ion [M + H]+ at m/z 1205.5728 (calcd for [C61H89O24]+, 1205.5739) in the HR-ESI-MS. The IR spectrum of 1 suggested the presence of hydroxy (3404 cm−1), carbonyl (1755 cm−1), aromatic ring and olefinic double bond (1649 cm−1), and C–O–C (1064 cm−1) functional groups. The 1H-, 13C-NMR, and HSQC spectra of 1 showed signals corresponding to five quaternary methyl groups [δC/δH: 28.5 (C-23)/0.95 (H3-23), 16.9 (C-24)/0.73 (H3-24), 16.1 (C-25)/0.95 (H3-25), 17.8 (C-26)/0.80 (H3-26), 26.3 (C-27)/1.18 (H3-27)], two double bonds [δC 124.3 (CH, C-12)/δH 5.34 (t, J = 3.0 Hz) and δC 144.2 (C, C-13)] and [δC 149.6 (C, C-20) and δC 107.5 (CH2, C-30)/δH 4.64 and 4.65 (each 1H, br s)], one trans-caffeoyl group [δH 7.05 (d, 1.8 Hz), 6.80 (d, 7.8 Hz), 6.95 (dd, 7.8, 1.8 Hz), 7.58 (d, 15.6 Hz), and 6.29 (d, 15.6 Hz)] and δC 168.6 (C O), four sugar moieties [δC 105.2/δH 4.49 (d, 7.8 Hz, H-1′), δC 95.9/δH 5.32 (d, 7.8 Hz, H-1′′), δC 104.2/δH 4.41 (d, 7.8 Hz, H-1′′′), δC 102.9/δH 4.86 (d, 1.2 Hz, H-1′′′′)], and a carboxylate group [δC 177.3, C-28]. The NMR data of 1 were similar to the corresponding data of compound 8 (yemuoside YM14)20 except the additional signals due to the caffeoyl group (Table 1). The aglycone of 1 was indicated to be 29-noroleana-12,20(30)-dien-28-oic acid and the sugar moieties, 28-O-α-rhamnopyranosyl-(1→4)-β-glucopyranosyl-(1→6)-β-d-glucopyranosyl ester and 3-O-α-arabinopyranosyl, were indicated by HSQC, COSY, and HMBC correlations as shown in Fig. 2, in comparison with the corresponding NMR data of compound 8 (yemuoside YM14).20 The caffeoyl moiety linked to C-2′ of the arabinose sugar as confirmed by 1H–1H COSY cross peak of H-1′ (δH 4.49)/H-2′ (δH 5.15) and HMBC correlation from H-2′ (δH 5.15) to C-9′′′′′ (δC 168.6). The HMBC correlations from H3-27 (δH 1.18) to C-13 (δC 144.2) and from H2-30 (δH 4.64 and 4.65) to C-19 (δC 42.6)/C-20 (δC 149.6)/C-21 (δC 30.9) indicated the location of double bonds at C-12/C-13 and C-20/C-30. The small J value (1.2 Hz) of an rhamnose anomeric proton at δH 4.86 suggested α-form of this glycosidic linkage, and the large J values (7.8 Hz) of the glucose anomeric protons at δH 5.32 and 4.41 suggested β-form of these glycosidic linkages, while the large J values (7.8 Hz) of the arabinose anomeric proton at δH 4.49 indicated α-form of this glycosidic linkage.20 The trans-configuration of the caffeoyl moiety was evidenced by large J value of the olefinic protons (15.6 Hz). In addition, proton H-3 (δH 3.12) showed NOESY correlation with H-5 (δH 0.77) suggesting alpha/axial orientation of H-3 (Fig. 3). Acid hydrolysis of 1 yielded d-glucose, l-arabinose, and l-rhamnose which were identified by comparison with authentic samples (d-glucose, l-arabinose, and l-rhamnose, Sigma-Aldrich) via thin-layer chromatography, and from the positive sign of the optical rotations. Consequently, the structure of 1 was established to be 3-O-{(2-trans-caffeoyl)-α-l-arabinopyranosyl}-29-noroleana-12,20(30)-dien-28-oic acid 28-O-α-l-rhamnopyranosyl-(1→4)-β-d-glucopyranosyl-(1→6)-β-d-glucopyranosyl ester and named as dendrocinaoside A.
Fig. 1. Structures of compounds 1–13 isolated from the stems of D. officinale.
Fig. 2. Key HMBC and COSY correlations of compounds 1–7.
Fig. 3. Important NOESY correlations of compounds 1–7.
Compound 2 had a molecular formula, C61H90O25, as determined by the proton adductive ion at m/z 1223.5854, (calcd for [C61H91O25]+, 1223.5844), indicating 17 degrees of unsaturation. The IR pectrum of 2 suggested the presence of hydroxy (3422 cm−1), carbonyl (1732 cm−1), aromatic ring and olefinic double bond (1639 cm−1), and C–O–C (1063 cm−1) functional groups. The NMR data of 2 were very similar to those of 1 except for the absence of C-20/C-30 double bond signals and the additional signals of one quaternary methyl group [δC 25.0 (C-30)/δH 1.25 (3H, s, H3-30) and one oxygenated quaternary carbon at δC 71.3 (C-20). The HMBC correlations from H-30 to C-19 (δC 47.6, CH2)/C-20 (δC 71.3, C)/C-21 (δC 35.5, CH2) were observed. These data suggested that the Δ20(30) double bond was replaced by one hydroxy group at C-20 of the 29-noroleanolic acid aglycone. In addition, the caffeoyl, the sugar moieties 28-O-α-rhamnopyranosyl-(1→4)-β-glucopyranosyl-(1→6)-β-d-glucopyranosyl ester and 3-O-α-arabinopyranosyl were identified by comparison the NMR data with those of 1 (Table 1) and further confirmed by HSQC, COSY, and HMBC correlations (Fig. 2). The HMBC correlations from H-2′ to C-9′′′′′ (C O), from H-1′ ara to C-3, from H-1′′′′ rha to C-4′′′, from H-1′′′ to C-6′′, and from H-1′′ to C-28 were observed. The large 3J2,3 value (12.0 Hz) of H-3 together with the NOE cross peak of H-3/H-5 indicated alpha/axial orientation of H-3. In addition, The large 3J18,19 value (13.8 Hz) and H3-30 (δH 1.25) showed NOE cross peak with H-18 (δH 2.81) confirmed beta/axial orientations of H-18 and the methyl group at C-20 (or 20α-hydroxy group) (Fig. 3). The low J value for rhamnose H-1′′′′ proton (1.2 Hz) indicated an α-form, and high J values for the two glucose anomeric protons H-1′′ and H-1′′′ (7.8 Hz) suggested β-forms for the glycosidic linkages. In addition, the high J value for arabinose H-1′ proton (7.2 Hz) suggested α-form of this glycosidic linkage. Acid hydrolysis of 2 yielded d-glucose, l-arabinose, and l-rhamnose which were identified by comparison with authentic samples (d-glucose, l-arabinose, and l-rhamnose, Sigma-Aldrich) via thin-layer chromatography, and from the positive sign of the optical rotations. Thus, compound 2 was determined to be 3-O-[(2-trans-caffeoyl)-α-l-arabinopyranosyl]-20α-hydroxy-29-noroleanolic acid 28-O-α-l-rhamnopyranosyl-(1→4)-β-d-glucopyranosyl-(1→6)-β-d-glucopyranosyl ester and named as dendrocinaoside B.
The molecular formula of 3 was determined to be C62H92O24 from the pseudo molecular ion peak [M + H]+ at m/z 1221.6071 (calcd for [C62H93O24]+, 1221.6051), indicating 17 degrees of unsaturation. The IR spectrum of 3 suggested the presence of hydroxy, carboxylate, aromatic ring, double bond, and C–O–C functionalities. The 13C NMR of 3 displayed 62 carbons, comprising 30 from an oleanolic acid aglycone, 9 from a coumaroyl moiety, and 23 from four sugar moieties.16 The NMR data for 3 closely resembled those of compound 2 (Table 1), with the exception of additional signals for one oxygenated methylene group at C-20 [δC 74.2/δH 3.21 (2H, s)] and a caffeoyl group was replaced by a coumaroyl group (Table 1). The sugar moieties, 3-O-α-l-arabinopyranosyl and 28-O-α-l-rhamnopyranosyl-(1→4)-β-d-glucopyranosyl-(1→6)-β-d-glucopyranosyl ester were identified. The observed HMBC correlations from H-7′′′′′ and H-8′′′′′ to C-9′′′′′, from H-2′ to C-9′′′′′, and from H-1′ to C-3 indicated the coumaroyl group linked to C-2′ of the arabinose sugar, and this sugar linked to C-3 of the aglycone by an ether linkage. Protons H-1′′′′ (δH 4.85), H-1′′′ (δH 4.42), and H-1′′ (δH 5.35) showed HMBC correlations with C-4′′′ (δC 79.6), C-6′′ (δC 69.4), and C-28 (δC 178.0), respectively, confirming rhamnopyranosyl-(1→4)-glucopyranosyl-(1→6)-glucopyranosyl moiety, which linked to C-28 by an ester linkage. The hydroxy group was at C-29 as determined by HMBC correlations from H-30 (δH 0.95) to C-19 (δC 41.4)/C-20 (δC 36.8)/C-21 (δC 29.5)/C-29 (δC 74.2), from H-29 (δH 3.21) to C-19/C-20/C-21/C-30 (δC 19.7), and from NOESY cross peak between H-18 (δH 2.90) and H-30 (δH 0.95) (Fig. 2 and 3). All the 3J1,2 values of the anomeric protons were remarkably like those of 1 and 2 (Table 1) suggesting the same form of glycosidic linkages. Acid hydrolysis of 3 yielded d-glucose, l-arabinose, and l-rhamnose, which were identified by comparison with authentic samples (d-glucose, l-arabinose, and l-rhamnose, Sigma-Aldrich) via thin-layer chromatography, and from the positive sign of the optical rotations. Thus, compound 3 was determined to be 3-O-[(2-trans-coumaroyl)-α-l-arabinopyranosyl]-29-hydroxyoleanolic acid 28-O-α-l-rhamnopyranosyl-(1→4)-β-d-glucopyranosyl-(1→6)-β-d-glucopyranosyl ester and named as dendrocinaoside C.
The molecular formula of 4 was determined to be C62H92O25 from the pseudo molecular ion peak [M–H]− at m/z 1235.5839 (calcd for [C62H91O25]−, 1235.5855), indicating 17 degrees of unsaturation. The IR spectrum of 4 suggested the presence of hydroxy, carboxylate, aromatic ring, double bond, and C–O–C functionalities. The 1H, 13C NMR, HSQC, and HMBC spectra of 4 were almost similar to those of 3 except for the coumaroyl signals were replaced by caffeoyl signals with the presence of additional signals for an aromatic ABX coupled proton system [δH 7.05 (d, 1.8 Hz), 6.80 (d, 8.0 Hz), and 6.95 (dd, 8.0, 1.8 Hz)] (Table 1). The hydroxy group was at C-29 as determined by HMBC correlations from H3-30 to C-19/C-20/C-21/C-29 and from H2-29 to C-19/C-20/C-21/C-30 along with NOESY cross peak from H-18 to H3-30. H-3 proton was alpha/axial orientation as evident by the high 3J2,3 value (12.0 Hz) along with NOESY cross peak between H-3 and H-5. The trans-configuration of the coumaroyl group was indicated by high J value (16.2 Hz) of the double bond. All the 3J1,2 values of the anomeric protons were terribly like those of 1–3 (Table 1) suggesting the same form of glycosidic linkages. Acid hydrolysis of 4 yielded d-glucose, l-arabinose, and l-rhamnose, which were identified by comparison with authentic samples (d-glucose, l-arabinose, and l-rhamnose, Sigma-Aldrich) via thin-layer chromatography, and from the positive sign of the optical rotations. Thus, compound 4 was determined to be 3-O-[(2-trans-caffeoyl)-α-l-arabinopyranosyl]-29-hydroxyoleanolic acid 28-O-α-l-rhamnopyranosyl-(1→4)-β-d-glucopyranosyl-(1→6)-β-d-glucopyranosyl ester and named as dendrocinaoside D.
Compound 5 (Fig. 1) has a molecular formula, C53H86O22, as determined by its HR-ESI-MS (found m/z 1075.5679 [M + H]+), (calcd for [C53H87O22]+, 1075.5684, Δ = −0.5 ppm), found m/z 1097.5476 [M + Na]+, (calcd for [C53H86O22Na]+, 1097.5503, Δ = −2.5 ppm), indicating 11 degrees of unsaturation. The IR spectrum of 5 supported the presence of hydroxy (3441 cm−1), carbonyl (1715 cm−1), aromatic ring (1645 cm−1), and C–O–C (1069 cm−1) functional groups. The NMR spectra of 5 were like those of 4 except for the absence of the caffeoyl group signals. The oxygenated methylene group was located at C-20 identified by HMBC correlation from its protons (δH 3.45) and the neighbour methyl protons (δH 0.92) to C-19/C-20/C-21. However, the carbon chemical shifts for the oxygenated methylene group (δC 66.3) and methyl group (δC 28.0) were quite different from those of 4 (δC 74.3 and 19.7, respectively) suggesting 30-hydroxy group in 5. This was further confirmed by NOESY cross peak between H-18 (δH 2.85) and H-30 (δH 3.45). Proton H-3 was axial orientation as indicated by NOESY cross peak between H-3 and H-5, along with the high 3J2,3 value (11.4 Hz) of H-3. In the HMBC spectrum, H-1′ correlated to C-3, H-1′′ correlated to C-28, H-1′′′ correlated to C-6′′, and H-1′′′′ correlated to C-4′′′ confirming 3-O-arabinopyranosyl and 28-O-rhamnopyranosyl-(1→4)-glucopyranosyl-(1→6)-glucopyranosyl moieties. All the 3J1,2 values of the anomeric protons were similar to those of 1–4 (Table 1) suggesting the same form of glycosidic linkages. Acid hydrolysis of 5 yielded d-glucose, l-arabinose, and l-rhamnose, which were identified by comparison with authentic samples (d-glucose, l-arabinose, and l-rhamnose, Sigma-Aldrich) via thin-layer chromatography, and from the positive sign of the optical rotations. Thus, compound 5 was determined to be 3-O-α-l-arabinopyranosyl-30-hydroxyoleanolic acid 28-O-α-l-rhamnopyranosyl-(1→4)-β-d-glucopyranosyl-(1→6)-β-d-glucopyranosyl ester and named as dendrocinaoside E.
The molecular formula of 6 was C52H84O22 determined from its HR-ESI-MS (found m/z 1061.5553 [M + H]+, (calcd for [C52H85O22]+, 1061.5527, Δ = +2.5 ppm), found m/z 1083.5373 [M + Na]+, (calcd for [C52H84O22Na]+, 1083.5347, Δ = +2.4 ppm)), indicating 11 degrees of unsaturation. The IR spectrum of 6 suggested the presence of hydroxy (3412 cm−1), carbonyl (1737 cm−1), aromatic ring (1645 cm−1), and C–O–C (1066 cm−1) functional groups. The NMR spectra of 6 were closely related to those of compound 2 except for the absence of the caffeoyl group signals (Tables 1 and 2). The arabinose (δC 107.1, 72.8, 74.3, 69.5, and 66.3), a rhamnose (δC 102.9, 72.4, 72.2, 73.8, 70.7, and 17.9), and two glucose sugars were identified. These sugar moieties and their glycosidic linkages were suggested to be like those of compounds 1–5 determined by HSQC, COSY, HMBC spectra (Fig. 2), along with 3J1,2 values of the anomeric protons and acid hydrolysis results. The hydroxy group was at C-20 confirmed by HMBC correlation from H-29 to C-19/C-20/C-21. The carbon chemical shift of the methyl group at C-20 (δC 31.5) in 6 was vast different from that in 2 (δC 25.0). This evidence suggested 20α-hydroxy in 6 instead of 20β-hydroxy in 2. This was further confirmed by NOESY cross peaks between Halpha/axial-19 (δH 1.84, dd, J = 13.8, 13.8 Hz) and H3-29 (δH 1.20). Thus, compound 6 was determined to be 3-O-α-l-arabinopyranosyl]-20β-hydroxy-29-noroleanolic acid 28-O-α-l-rhamnopyranosyl-(1→4)-β-d-glucopyranosyl-(1→6)-β-d-glucopyranosyl ester and named as dendrocinaoside F.
Compound 7 had a molecular formula of C47H76O18, as determined by its HR-ESI-MS (found m/z 929.5128 [M + H]+), (calcd for [C47H77O18]+, 929.5105, Δ = +2.5 ppm), found m/z 951.4913 [M + Na]+, (calcd for [C47H76O18Na]+, 951.4924, Δ = −1.7 ppm)), indicating 10 degrees of unsaturation. The IR spectrum suggested the presence of hydroxy, carboxylate, double bond, and ether functional groups. The NMR data of 7 were closely related to those of compound 5 except for the absence of rhamnose moieties. For the aglycone of 7, one Δ12-double bond [δC 123.9/δH 5.29 (t, 3.0 Hz) and δC 144.8], the oxygenated methylene group [δC 74.3/δH 3.21 (2H, s)], one methine carbinol group (δC 90.7/δH 3.15 (dd, 11.4, 4.2 Hz), and six quaternary methyl groups were identified (Table 2). In addition, three anomeric protons at δH 4.30, 5.38, and 4.37 showed HSQC corrections with carbons at δC 107.1, 95.8, and 104.7, respectively, suggesting one arabinose and two glucose sugars. In the HMBC spectrum, H-1′ correlated with C-3, H-1′′′ correlated with C-6′′, and H-1′′ correlated with C-28 confirming the arabinose linked to C-3, one glucose linked to C-6 of the remaining glucose, which linked to C-28 by an ester linkage. The high 3J1,2 values of the anomeric protons at δH 4.30 (J = 6.6 Hz), 5.38 (J = 7.8 Hz), and 4.37 (J = 7.8 Hz) suggesting α-form of the arabinosyl linkage and β-form of the glucosyl linkages. H-3 proton was axial orientation as supported by the high 3J2,3 value (11.4 Hz) of H-3 at δH 3.15, and by NOESY cross peak from H-3 to H-5. In addition, H-18 showed NOESY cross peak with H3-30 (δH 0.95) indicating this methyl group was beta/axial orientation (or 29-hydroxy group). Acid hydrolysis of 7 yielded d-glucose and l-arabinose, which were identified by comparison with authentic samples (d-glucose and l-arabinose, Sigma-Aldrich) via thin-layer chromatography, and from the positive sign of the optical rotations. Thus, compound 7 was determined to be 3-O-α-l-arabinopyranosyl-29-hydroxyoleanolic acid 28-O-β-d-glucopyranosyl-(1→6)-β-d-glucopyranosyl ester and named as dendrocinaoside G.
In the digestive system, α-amylase and α-glucosidase are two important enzymes that hydrolyse starch yielding oligosaccharide and finally glucose. Inhibition of α-amylase and/or α-glucosidase slow down the hydrolysis processes which are helpful to reduce levels of glucose in the blood. The α-amylase and/or α-glucosidase inhibitors therefore could be lead compounds for new drug developments in the treatment of type 2 diabetes. Therefore, compounds 1–13 were screened for both anti α-glucosidase and α-amylase activities at the concentration of 200 μM (Table S1†). Acarbose, an antidiabetic drug, was used as a positive control in both tests. Compounds 1–11 showed anti α-glucosidase activity and compounds 1–4, 6, 8, 9, 11–13 showed anti α-amylase activity with inhibitory percentages over 50%. Therefore, further evaluation of α-glucosidase and α-amylase inhibition of these compounds were assayed to determine their IC50 values. The results (Table 3) indicated that compounds 1–4, 8, and 9 potentially inhibited both anti α-glucosidase (IC50 values: 31.3 ± 2.2 to 42.4 ± 2.5 μM) and anti α-amylase (IC50 values: 36.5 ± 1.8 to 56.4 ± 2.0 μM) activities, which were lower than that of the positive control, acarbose, showing IC50 values of 47.1 ± 1.4 μM (anti α-glucosidase) and 145.7 ± 2.2 μM (anti α-amylase). These results suggested that 29-noroleana-12,20(30)-dien-28-oic acid framework and the presence of the caffeoyl or coumaroyl moieties may play significant role in anti α-glucosidase and α-amylase activities of the isolated saponins, and further study on anti-diabetes of this plant should be continued.
α-Glucosidase and α-amylase inhibitory effects of 1–13.
| Compounds | Inhibition (IC50, μM) | |
|---|---|---|
| α-Glucosidase | α-Amylase | |
| 1 | 31.3 ± 2.2 | 52.1 ± 2.3 |
| 2 | 37.7 ± 1.7 | 45.3 ± 1.9 |
| 3 | 33.1 ± 1.5 | 56.4 ± 2.0 |
| 4 | 33.8 ± 1.6 | 36.5 ± 1.8 |
| 5 | 105.1 ± 2.8 | >200 |
| 6 | 99.6 ± 3.4 | 135.2 ± 3.3 |
| 7 | 148.1 ± 2.5 | >200 |
| 8 | 40.5 ± 1.9 | 44.5 ± 2.3 |
| 9 | 42.4 ± 2.5 | 39.8 ± 2.5 |
| 10 | 107.7 ± 2.9 | >200 |
| 11 | 136.5 ± 3.7 | 148.6 ± 4.2 |
| 12 | >200 | 127.4 ± 3.1 |
| 13 | >200 | 166.5 ± 3.7 |
| Acarbose a | 47.1 ± 1.4 | 145.7 ± 2.2 |
Acarbose was used as a positive control.
Conclusions
The bio-guided fractionation study on the whole plants of Dendrobium officinale led to the isolation of thirteen saponins, including seven new compounds namely dendrocinaosides A–G. Their chemical structures were determined by spectroscopic methods, including NMR spectroscopy and HRESIMS analysis. Compounds 1–4, 8, and 9 potentially inhibited α-glucosidase (IC50 values from 31.3 ± 2.2 to 42.4 ± 2.5 μM) and α-amylase (IC50 values from 36.5 ± 1.8 to 56.4 ± 2.0 μM), which were lower than that of the positive control, acarbose.
Author contributions
PH Yen, PV Kiem, BH Tai, contributed to research idea and writing; DTT Hang, LDT Lam, DT Dung, DT Trang, DTH Yen, NH Hoang, PTT Huong, NV Dung, NA Bang, ND Duy contributed to isolation; PH Yen, PV Kiem, BH Tai contributed to structure elucidation and bioassay.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
This research is supported by Vietnam Ministry of Science and Technology under grant number ĐTĐLCN.65/22.
Electronic supplementary information (ESI) available: HR-ESI-MS, 1D, and 2D- NMR spectra of new compounds of new compounds would be found. See DOI: https://doi.org/10.1039/d4ra01483a
Notes and references
- Zhang P. Zhang X. Zhu X. Hua I. F. J. Agric. Food Chem. 2023;71:14870–14889. doi: 10.1021/acs.jafc.3c04154. [DOI] [PubMed] [Google Scholar]
- Chen W. Lu J. Zhang J. Wu J. Yu L. Qin L. Zhu B. Front. Pharmacol. 2021;12:726528. doi: 10.3389/fphar.2021.726528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W. M. Ye Q. H. Dai J. Q. Martin M. T. Zhu J. P. Planta Med. 2003;69:1136–1140. doi: 10.1055/s-2003-818005. [DOI] [PubMed] [Google Scholar]
- Yang L. Liu S. J. Luo H. R. Cui J. Zhou J. Wang X. J. Sheng J. Hu J. M. J. Asian Nat. Prod. Res. 2015;17:125–131. doi: 10.1080/10286020.2014.942294. [DOI] [PubMed] [Google Scholar]
- Liu Y. Li X. Sui S. Tang J. Chen D. Kang Y. Xie K. Liu J. Lan J. Wu L. Chen R. Peng Y. Dai J. Acta Pharm. Sin. B. 2023;13:1771–1785. doi: 10.1016/j.apsb.2022.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D. K. Shao H. Y. Yang L. Hu J. M. Nat. Prod. Bioprospect. 2022;12:1–11. doi: 10.1007/s13659-022-00323-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C. Zhang C. Zhuo Y. Gong B. Xu W. Zhang G. Int. J. Mol. Sci. 2023;24:15375. doi: 10.3390/ijms242015375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. Guo H. Dong Y. Yuan S. Wei X. Zhang Y. Dong L. Wang F. Bai T. Yang Y. Chin. J. Nat. Med. 2024;22:4–14. doi: 10.1016/S1875-5364(24)60558-3. [DOI] [PubMed] [Google Scholar]
- Liu H. Xing Y. Wang Y. Ren X. Zhang D. Dai J. Xiu Z. Yu S. Dong Y. Foods. 2023;12:2310. doi: 10.3390/foods12122310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J. Chen H. Nie Q. Huang X. Nie S. Int. J. Biol. Macromol. 2020;164:1939–1948. doi: 10.1016/j.ijbiomac.2020.08.007. [DOI] [PubMed] [Google Scholar]
- Tang Y. Zhang X. Lin Y. Sun J. Chen S. Wang W. Li J. Molecules. 2023;28:3071. doi: 10.3390/molecules28073071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W. Ruan W. Wang Z. Faseb. J. 2022;36:e22504. doi: 10.1096/fj.202200353RRR. [DOI] [PubMed] [Google Scholar]
- Wang K. Yang X. Wu Z. Wang H. Li Q. Mei H. You R. Zhang Y. Front. Pharmacol. 2020;11:240. doi: 10.3389/fphar.2020.00240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K. Zhou X. Wang J. Zhou Y. Qi W. Chen H. Nie S. Xie M. Carbohydr. Polym. 2021;264:118018. doi: 10.1016/j.carbpol.2021.118018. [DOI] [PubMed] [Google Scholar]
- Pang C. Zhang X. Huang M. Xie G. Liu S. Ye X. Zhang X. Transl. Cancer Res. 2020;9:2683–2691. doi: 10.21037/tcr.2020.02.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voutquenne-Nazabadioko L. Gevrenova R. Borie N. Harakat D. Sayagh C. Weng A. Thakur M. Zaharieva M. Henry M. Phytochemistry. 2013;90:114–127. doi: 10.1016/j.phytochem.2013.03.001. [DOI] [PubMed] [Google Scholar]
- Ha T. T. T. Dung N. T. Tai B. H. Kiem P. V. J. Nat. Med. 2023;77:238–249. doi: 10.1007/s11418-022-01663-w. [DOI] [PubMed] [Google Scholar]
- Trang D. T. Tai B. H. Hoang N. H. Cuc N. T. Bang N. A. Dung D. T. Yen D. T. H. Huong P. T. T. Dung N. V. Hang D. T. T. Yen P. H. Kiem P. V. Chem. Biodivers. 2024;21:e202400124. doi: 10.1002/cbdv.202400124. [DOI] [PubMed] [Google Scholar]
- Hanh T. T. H. Chau N. M. Tram L. H. Luyen N. T. Binh P. T. Minh C. V. Nam N. H. Dat N. T. Pharm. Biol. 2014;52:74–77. [Google Scholar]
- Wang H. B. Yu D. Q. Liang X. T. Watanabe N. Tamai M. Omura S. Planta Med. 1989;55:303–306. doi: 10.1055/s-2006-962013. [DOI] [PubMed] [Google Scholar]
- Shao C. J. Kasai R. Xu J. D. Tanaka O. Chem. Pharm. Bull. 1988;36:601–608. [Google Scholar]
- Shao C. J. Kasai R. Xu J. D. Tanaka O. Chem. Pharm. Bull. 1989;37:42–45. [Google Scholar]
- Wei X. Gao D. F. Abe Y. Tanaka Y. Zhu H. T. Wang D. Yang C. R. Zhang Y. J. Nat. Prod. Res. 2020;34:1373–1379. doi: 10.1080/14786419.2018.1512987. [DOI] [PubMed] [Google Scholar]
- Abdel-Gawad M. M. El-Sayed M. M. Abdel-Hameed E. S. Al-Azhar J. Pharm. Sci. 1997;20:52–65. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.