ABSTRACT
Maternal secretor status is one of the determinants of human milk oligosaccharides (HMOs) composition, which, in turn, influences the gut microbiota composition of infants. To understand if this change in gut microbiota impacts immune cell composition, intestinal morphology, and gene expression, 21-day-old germ-free C57BL/6 mice were transplanted with fecal microbiota from infants whose mothers were either secretors (SMM) or non-secretors (NSM) or from infants consuming dairy-based formula (MFM). For each group, one set of mice was supplemented with HMOs. HMO supplementation did not significantly impact the microbiota diversity; however, SMM mice had a higher abundance of genus Bacteroides, Bifidobacterium, and Blautia, whereas, in the NSM group, there was a higher abundance of Akkermansia, Enterocloster, and Klebsiella. In MFM, gut microbiota was represented mainly by Parabacteroides, Ruminococcaceae_unclassified, and Clostrodium_sensu_stricto. In mesenteric lymph node, Foxp3+ T cells and innate lymphoid cells type 2 were increased in MFM mice supplemented with HMOs, while in the spleen, they were increased in SMM + HMOs mice. Similarly, serum immunoglobulin A was also elevated in MFM + HMOs group. Distinct global gene expression of the gut was observed in each microbiota group, which was enhanced with HMOs supplementation. Overall, our data show that distinct infant gut microbiota due to maternal secretor status or consumption of dairy-based formula and HMO supplementation impacts immune cell composition, antibody response, and intestinal gene expression in a mouse model.
IMPORTANCE
Early life factors like neonatal diet modulate gut microbiota, which is important for the optimal gut and immune function. One such factor, human milk oligosaccharides (HMOs), the composition of which is determined by maternal secretor status, has a profound effect on infant gut microbiota. However, how the infant gut microbiota composition determined by maternal secretor status or consumption of infant formula devoid of HMOs impacts infant intestinal ammorphology, gene expression, and immune signature is not well explored. This study provides insights into the differential establishment of infant microbiota derived from infants fed by secretor or non-secretor mothers milk or those consuming infant formula and demonstrates that the secretor status of mothers promotes Bifidobacteria and Bacteroides sps. establishment. This study also shows that supplementation of pooled HMOs in mice changed immune cell composition in the spleen and mesenteric lymph nodes and immunoglobulins in circulation. Hence, this study highlights that maternal secretor status has a role in infant gut microbiota composition, and this, in turn, can impact host gut and immune system.
KEYWORDS: human milk oligosaccharides, HMO, immunity, gastrointestinal tract, neonatal, microbiome, secretor, non-secretor, formula
INTRODUCTION
Human milk (HM) is considered as personalized nutrition, made by the mother to fit the infant’s nutritional needs. HM contains nutrients such as vitamins, fatty acids, proteins, growth factors, immunoglobulins (Ig) such as IgG and IgE, extracellular vesicles, and its own microbiota. Human milk feeding is associated with several health benefits, providing optimal nutrients for healthy development and growth and acting as a source of antibodies to protect infants from early-life infections and allergies (1). In addition, HM promotes infants’ early gut microbiome seeding and programs the composition of microbiota. It also improves epithelial barrier function by increasing the expression of tight junction proteins and promotes innate immune mediators in epithelial cells (2). Multitudes of maternal, genetic, and environmental factors determine the composition of human milk. Human milk oligosaccharides (HMOs), the third most abundant component of human milk besides lactose and lipids, are crucial for optimal infant development (3–5). Infants cannot metabolize HMOs though their direct impact on the host immune system and gene expression has been previously shown in a mouse model (6, 7). HMOs work as a prebiotic factor because the distal intestinal microbiota uses HMOs as substrates shaping the gut microbiota composition. HMOs can also prevent the ability of pathobionts to bind to the host cells, thus protecting infants from enteric infections (4, 8).
Several factors influence HMOs composition (9–14). One such factor is the expression of the secretor gene (FUT2). Mothers with the functional FUT2 gene allele have active fucosyltransferase-2 (FUT2) enzyme. Active FUT2 enzyme adds fucose to the lactose backbone of HMO by α1-2-linkages and produces 2′-fucosyllactose (2′FL) in human milk (11, 15). Different percentages of the secretor population have been reported in studies depending on the geographical location. In an Australian cohort, 64.7% of mothers were secretors (16), in a Chinese cohort, 77% of mothers were secretors, whereas in a US cohort, 71.9% were reported to be secretors (17, 18). The mother’s secretor status is known to impact the development of child’s gut microbiota composition (16, 19, 20). Secretor individuals and infants consuming milk from secretor mothers have a higher abundance of Bifidobacterium species (16, 21). In addition, dietary 2-FL supplementation in a conventional mouse model enhanced the influenza antigen-specific immunoglobulins and the proliferation of antigen-specific T-cells (22). Importantly, infants consuming human milk containing a higher level of 2′-FL had a lower incidence of diarrheal diseases, including Campylobacter-related diarrhea (23). Similarly, supplementation of 2′-FL with dairy-based infant formula decreased the severity of experimentally induced necrotizing enterocolitis in mice (24).
However, whether microbiota programmed by maternal secretor milk mediates offspring gastrointestinal tract morphology, the immune response and intestinal gene expression is not well explored. To address this knowledge gap and understand the role of microbiota in infants consuming milk from secretor or non-secretor mothers, we transplanted the microbiota from these infants to germ-free mice. Germ-free mice have immature gastrointestinal and immune system development and function for their age compared to the conventional mice (25) making young germ-free mice a potential model to study infant-gut microbiota interaction. Microbiota from infants consuming dairy-based infant formula devoid of any HMOs were also transplanted to the germ-free mice as a control. To mimic HMOs content of human milk, one set of mice was supplemented with pooled HMOs and investigated its influence on the microbiota composition. Growth, gastrointestinal tract morphology, gene expression, and immune response were measured.
RESULTS
Maternal secretor status shapes infant microbiota differently from non-secretor or infant formula feeding
To determine whether the microbiota of infants differ across groups, 16S rRNA gene amplicons sequencing of inoculum that was used for fecal transplantation was carried out. A distinct microbiota abundance among the infants fed by secretor or non-secretor mothers or those consuming infant formula was observed (Fig. S1). While the microbiota of infants fed by secretor mothers (SMM) had a higher abundance of Alistipes, Anaerostipes, Bacteroides, Bifidobacterium, Blautia, Faecalibacillus, Enterococcus, Flavonifractor, Lactococcus, Mediterraneibacter, Parabacteroides, Phocaeicola, and Pseudoalteromonas, those fed by non-secretor mothers had a higher abundance of Blautia, Clostridium sensu stricto, Enterococcus, Eubacterium, Faecalibacterium, Haemophilus, Hungatella, Klebsiella, Lactobacillus, Odoribacter, Ruminococcus, and Veillonella. Similarly, feces of infants consuming milk formula had a higher abundance of Anaerotruncus, Clostridioides, Coprococcus, Hungatella, Lachnospira, Lacticaseibacillus, Parasutterella, Phascolarctobacterium, Phocaeicola, Ruthenibacterium, and Streptococcus (Fig. S1).
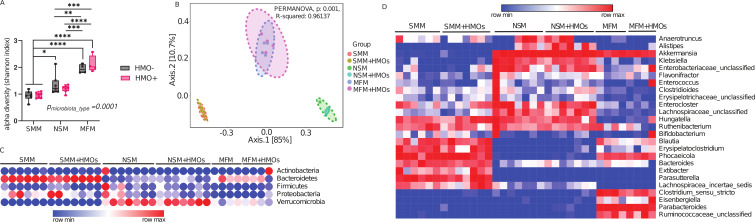
To determine whether the microbiota of infants consuming milk from secretor mothers (SMM), non-secretor mothers (NSM), or those consuming dairy-based infant formula (MFM) colonize the mouse gut, colon contents of the mice were subjected to 16S amplicon sequencing. A set of mice was supplemented with pooled human milk oligosaccharides (HMOs) resulting in a total of seven experimental groups: Germ-Free control (GF), SMM, SMM + HMOs, NSM, NSM + HMOs, MFM, and MFM + HMOs. Alpha diversity of MFM groups was significantly higher compared to SMM or NSM groups as shown by higher Shannon index values in the colon lumen (Fig. 1A). Interestingly, HMO supplementation did not change the alpha diversity in any of the groups studied. Similarly, beta diversity measured by Bray-Curtis dissimilarity significantly differed between SMM, NSM, and MFM groups although no impact of HMOs was seen (Fig. 1B). Microbiota from SMM and SMM + HMOs groups were dominated by phylum Bacteroidota (formerly, Bacteroidetes) (~89% abundance), whereas Verrucomicrobiota (formerly, Verrucomicrobia) (~33% abundance) phylum predominately belonged to NSM and NSM + HMOs groups (Fig. 1C). Several genera differed among the treatment groups (Fig. 1D). SMM and SMM + HMOs groups were predominantly represented by Bacteroides, Blautia, Erysipelatoclostrodium, Phocaeicola, Parasutterella, Bifidobacterium, and Extibacter (Fig. 1D; Fig S2 and 3), whereas NSM and NSM + HMOs groups had a higher abundance of Alistipes, Anaerotruncus, Akkermansia, Enterocloster, Lachnospiraceae_unclassified, and Klebsiella. Similarly, abundant taxa in MFM and MFM + HMOs groups were Clostridium_sensu_stricto, Parabacteroides, and Ruminococcacea_unclassified (Fig. 1D; Fig. S2 and 3). Genus Alistipes was abundant in NSM + HMOs in comparison to NSM (0.06% vs 0.18%) (Fig. S2 and 3). Furthermore, HMOs supplementation in MFM reduced the abundance of genus Clostridium_sensu_stricto (1.298% vs 0.75%) (Fig. S2). These data suggest that HMOs have minimal impact on the microbiota community structure and composition among the different groups after 14 days of oral administration in germ-free mice colonized on days 1 and 7. Comparison of the source of inoculum to the mouse colon microbiota suggests that the majority of the source microbiota successfully colonized in the mice (Fig. S1 to 3).
Fig 1.
Maternal secretor status shapes infant microbiota differently from non-secretor or infant formula feeding. (A) Alpha diversity as represented by Shannon index, (B) beta diversity shown as Bray-Curtis Dissimilarity, and (C) Phylum and (D) Genus level microbiota composition of mice colonized with microbiota from infants whose mothers were secretor or non-secretor or those consuming dairy-based formula. Group differences, the effect of microbiota, HMOs, and interaction were determined using the two-way ANOVA with Tukey’s multiple comparison tests in GraphPad Prism Version 10.0. 2 (www.graphpad.com), and adjusted P < 0.05 was considered significant in 1A. Group differences in Beta diversity were determined using a permutational multivariate analysis of variance (PERMANOVA) test. SMM = mice transplanted with fecal material from infants consuming milk from secretor mothers, NSM = mice transplanted with fecal material from infants consuming milk from non-secretor mothers, MFM = mice transplanted with fecal material from infants consuming dairy-based milk formula, HMO − = without human milk oligosaccharides supplementation, HMO+ = with human milk oligosaccharides.
Microbiota of infants fed by secretor or non-secretor mothers or those consuming infant formula alter gastrointestinal tract morphology
To determine how microbiota from infants consuming milk from mothers of different secretor status or those consuming infant formula alter growth and gastrointestinal tract (GIT) morphology, body weight gain, organ weight, and gastrointestinal tract morphology were evaluated. There was no difference in body weight gain and organ weights including liver, kidney, and cecum on day 14 of the colonization (Table S1). Similarly, no difference was observed in large intestine and small intestine length (Table S1). Histomorphometry analysis showed that the SMM + HMOs and NSM + HMOs group had higher ileum villi height when compared to the MFM group (Table 1). Interestingly, HMOs supplementation in the NSM group increased the ileum villi height, suggesting either direct or indirect impact of HMOs. Both MFM and MFM + HMOs groups showed significantly higher cecum gland length when compared to the NSM group (Table 1). These findings suggest that consuming milk from mothers with different secretor status or infant formula may have an impact on the microbiome that likely drives changes in gastrointestinal tract morphology.
TABLE 1.
Ileum villi height (µm) and cecum gland length (µm) of Germ-Free mice at day 35 of age colonized with microbiota from infants consuming milk from secretor or non-secretor mothers or those consuming dairy-based formulaa
| Location | SMM | SMM + HMOs | NSM | NSM + HMOs | MFM | MFM + HMOs |
|---|---|---|---|---|---|---|
| Ileum | ||||||
| Mean | 147.95abc | 165.55bd | 142.96cd | 190.53b | 120.89ac | 145.8abc |
| SEM | 8.66 | 9.27 | 7.71 | 10.67 | 7.07 | 8.53 |
| Cecum | ||||||
| Mean | 145.6ab | 137.4ab | 118.94a | 132.7ab | 163.96b | 168.07b |
| SEM | 8.52 | 7.94 | 6.08 | 6.9 | 9.6 | 9.84 |
Multiple comparison-adjusted pairwise contrasts were performed. P-value < 0.05 was considered a significant difference among the groups. Means in the same row without a common letter differ significantly.
Microbiota of infants consuming milk from secretor or non-secretor mothers or infant formula impact immune cells composition
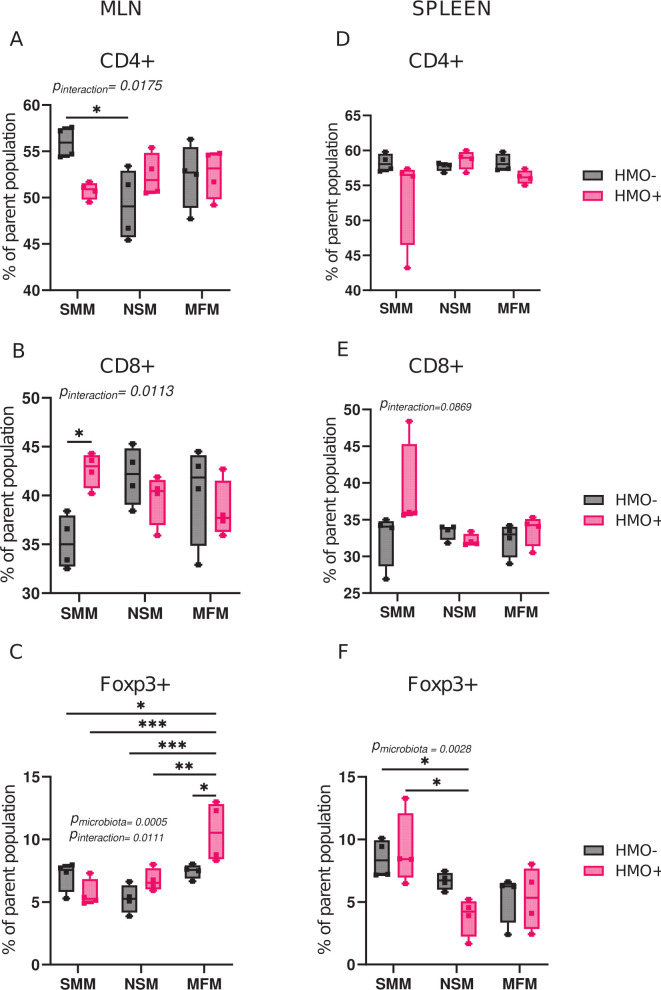
To elucidate the effect of microbiota of infants fed by secretor or non-secretor mothers or those consuming infant formula on immune cell composition, the innate and adaptive immune cell composition of mesenteric lymph nodes (MLN) and spleen of colonized germ-free mice were determined. In MLN, the percentage of CD4+ T cells was higher in the SMM group than in the NSM group, whereas no significant differences were observed among other groups (Fig. 2A). HMO supplementation increased the CD8+ T cells percentage in MLN (Fig. 2B) of SMM in comparison to SMM alone. Interestingly, the percentage of Foxp3+ T regulatory cells in MLN increased after HMO supplementation only in the MFM group (Fig. 2C). In the spleen, microbiota had no significant effect on CD4+ T cells and CD8+ T cells population among the groups (Fig. 2D and E), while the SMM and SMM + HMOs groups showed higher Foxp3+ T cells relative to NSM + HMO groups.
Fig 2.
Microbiota of infants consuming milk from secretor or non-secretor mothers or infant formula impact T cells. Percentage of cytotoxic CD8+ T-lymphocytes (Live CD90.2 + CD8b+), CD4+ T-helper cells (Live CD90.2 + CD4+), and Foxp3 T-regulatory cells (Live CD90.2 + CD4+Foxp3+) in mesenteric lymph nodes (A–C) and spleen (D–F) in mice transplanted with microbiota from infants consuming milk from secretor or non-secretor mothers or those consuming milk formula and those supplemented with HMOs. Cell populations are shown as the percentage of the parent population (Fig. S1). Group differences, the effect of microbiota, HMOs, and their interactions were determined using two-way ANOVA with Tukey’s multiple comparison test in GraphPad Prism Version 10.0.2 (www.graphpad.com). P-adjusted <0.05 was considered significant. Group differences are indicated by “*” system, ***P < 0.001, **P < 0.01, *P < 0.05. The P-values for the effects of microbiota or HMO factor or their interactions are included when significant. SMM = mice transplanted with fecal material from infants consuming milk from secretor mothers, NSM = mice transplanted with fecal material from infants consuming milk from non-secretor mothers, MFM = mice transplanted with fecal material from infants consuming dairy-based formula, HMO− = without human milk oligosaccharides supplementation, HMO+ = with human milk oligosaccharides.
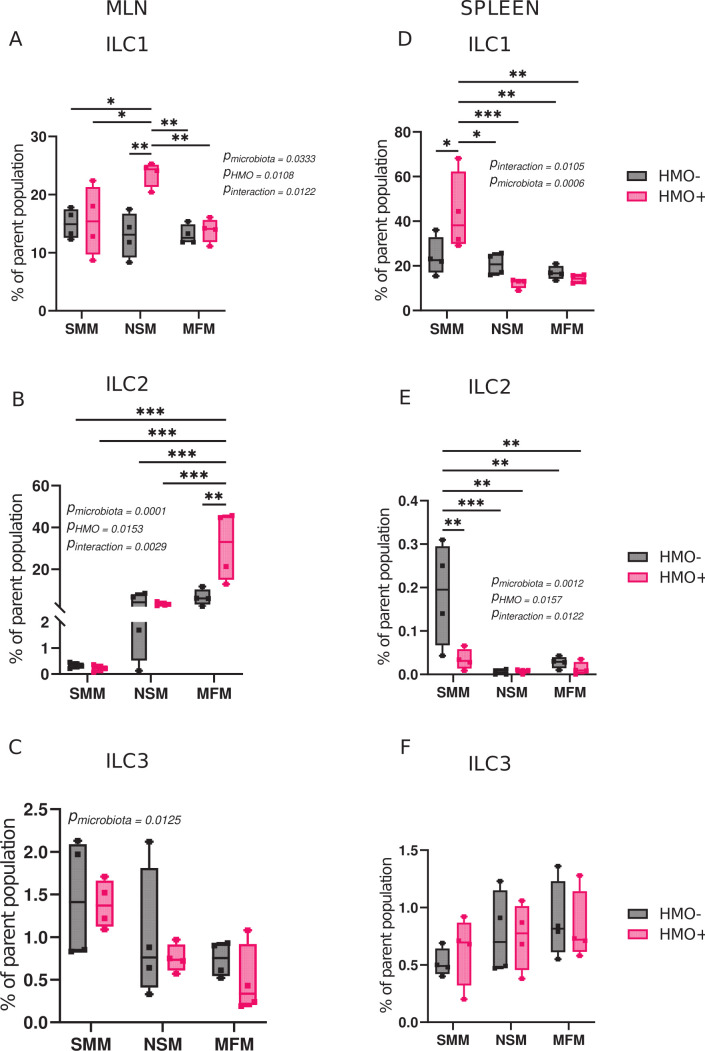
Likewise, HMO supplementation in the NSM group showed a higher percentage of innate lymphoid cells 1 (ILC1) population in the MLN (Fig. 3A). Innate lymphoid cells 2 (ILC2) which are involved in type 2 immune response were higher in HMO-supplemented MFM groups in the MLN (Fig. 3B). Variation among microbiota treatment groups was large for innate lymphoid cells 3 (ILC3), and no two-treatment means were significantly different from one another (Fig. 3C). In spleen, the SMM group supplemented with HMOs showed higher ILC1 in comparison to all the other groups (Fig. 3D). In addition, SMM group had a higher percentage of ILC2 than other groups (Fig. 3E). Notably, ILC2 percentage in the spleen was less than 0.5% of the parent population. There were no differences observed in B cells and the innate immune cell composition among the groups in MLN and spleen (Fig. S4).
Fig 3.
Microbiota of infants consuming milk from secretor or non-secretor mothers or infant formula impact innate lymphoid cells. Percentage of Innate Lymphoid Cell (ILC), ILC1 (Live CD90.2 + CD8b-TCRβ-GATA3-RORγt-Tbet+), ILC2 (Live CD90.2 + CD8b-TCRβ-GATA3+), and ILC3 (Live CD90.2 + CD8b-TCRβ-RORγt+) in mesenteric lymph nodes (A–C) and spleen (D–F) in mice transplanted with microbiota from infants consuming milk from secretor or non-secretor mothers or those consuming milk formula and those supplemented with HMOs. Cell populations are shown as the percentage of the parent population (Fig. S1). Group differences, the effect of microbiota, HMOs, and their interactions were determined using two-way ANOVA with Tukey’s multiple comparison test in GraphPad Prism Version 10.0.2 (www.graphpad.com). P-adjusted <0.05 was considered significant. Group differences are indicated by “*” system, ***P < 0.001, **P < 0.01, *P < 0.05. The P-values for the effects of microbiota or HMO factor or their interactions are included when significant. SMM = mice transplanted with fecal material from infants consuming milk from secretor mothers, NSM = mice transplanted with fecal material from infants consuming milk from non-secretor mothers, MFM = mice transplanted with fecal material from infants consuming dairy-based formula, HMO− = without human milk oligosaccharides supplementation, HMO+ = with human milk oligosaccharides.
Microbiota of infants fed by secretor or non-secretor mothers or those consuming infant formula alter serum immunoglobulins
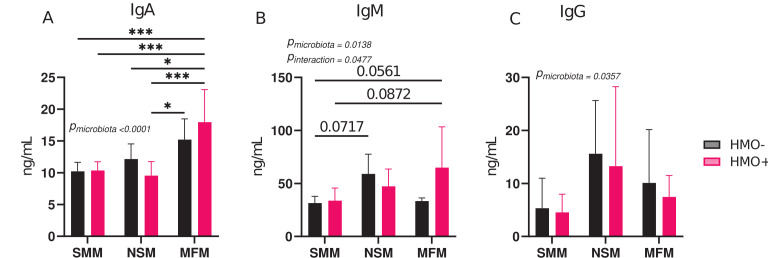
To determine whether the different gut microbiota from infants impact serum immunoglobulins of transplanted mice, immunoglobulins A (IgA), G (IgG), and M (IgM) were quantified using enzyme-linked immunosorbent assay (ELISA). Mice colonized with MFM and supplemented with HMOs had significantly higher IgA levels than other groups (Fig. 4A). Similarly, supplementation with HMOs in MFM groups tended to increase serum IgM when compared to SMM and SMM + HMOs groups (Fig. 4B). There was weak evidence that NSM has a higher level of serum IgM than the SMM group (P = 0.07) (Fig. 4B). No significant differences were observed among the groups for serum IgG levels (Fig. 4C).
Fig 4.
Microbiota of infants fed by secretor or non-secretor mothers or those consuming infant formula alter serum immunoglobulins. (A) Serum Immunoglobulin A (IgA), (B) Immunoglobulin M (IgM), and (C) Immunoglobulin G (IgG) in mice transplanted with microbiota from infants consuming milk from secretor or non-secretor mothers or those consuming milk formula and those supplemented with HMOs. Group differences, the effect of microbiota, HMOs, and their interactions were determined using two-way ANOVA with Tukey’s multiple comparison test in GraphPad Prism Version 10.0.2 (www.graphpad.com). P-adjusted <0.05 was considered significant. Group differences are indicated by “*” system, ***P < 0.001, **P < 0.01, *P < 0.05. The P-values for the effects of microbiota or HMO factor or their interactions are included when significant. SMM = mice transplanted with fecal material from infants consuming milk from secretor mothers, NSM = mice transplanted with fecal material from infants consuming milk from non-secretor mothers, MFM = mice transplanted with fecal material from infants consuming dairy-based formula, HMO− = without human milk oligosaccharides supplementation, HMO+ = with human milk oligosaccharides.
Microbiota of infants consuming milk from secretor or non-secretor mothers or those consuming infant formula impacts gene expression in the distal small intestine
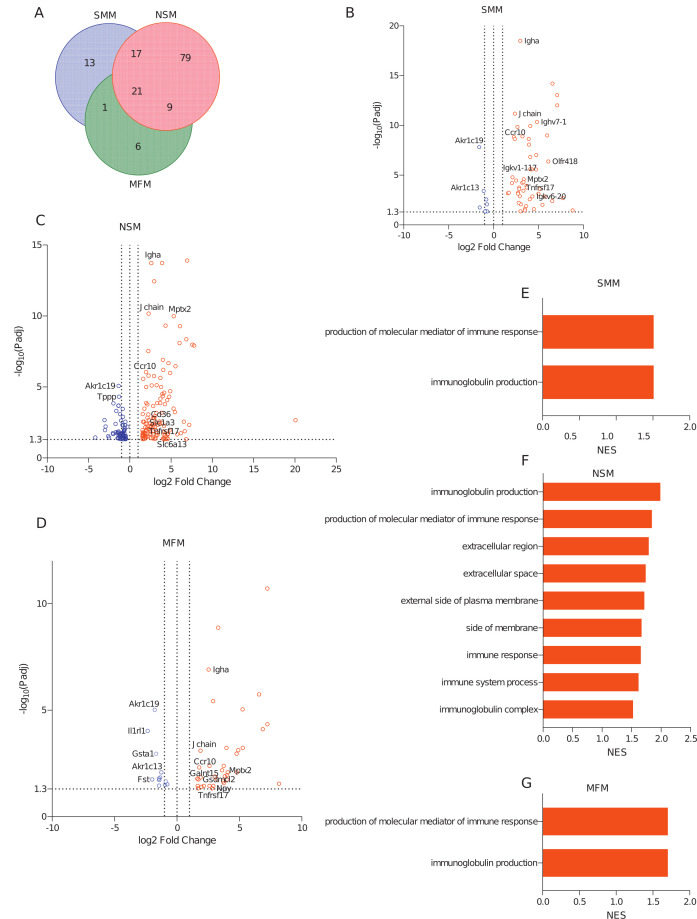
To evaluate the host response at the transcriptome level in the intestine, mRNA sequencing from the distal small intestine (ileum) was performed. Differentially expressed genes (DEGs) between the treatment groups and the germ-free (GF) controls are shown in Fig. 5 (Fig. 5B through D; Table S2). In the ileum, 52, 126, and 37 genes were significantly upregulated compared to germ-free controls (Fig. 5A through D; Table S2, sheets “Up in SMM vs GF,” “Up in NSM vs GF,” and “Up in MFM vs GF”), while 8, 63, and 12 were downregulated (Fig. S5A; Table S2, sheets “Down in SMM vs GF,” “Down in NSM vs GF,” and “Down in MFM vs GF,” Fig. 5B through 5D) in SMM, NSM, and MFM groups when compared to germ-free controls, respectively (log2 fold change >1.5, adjusted P-value < 0.05, pairwise comparison). Among these differentially expressed genes, 21 genes were significantly upregulated (Fig. 5A) and 5 genes were significantly downregulated (adjusted P-value < 0.05) (Fig. S5A) in all three groups compared to GF controls (Table S4, sheets “Upregulated Genes in HMO−,” “Downregulated Genes in HMO−”). Interestingly, although SMM and NSM had several common upregulated genes (17 genes), only one upregulated gene was shared between SMM and MFM groups (Fig. 5A; Table S4, sheet “Upregulated Genes in HMO−”). The genes that were upregulated in all three groups were mainly related to immunoglobulin and immune responses, including immunoglobulin alpha (IgA), tumor necrosis factor receptor superfamily member 17 (Tnfrsf17), Chemokine receptor 10 (CCR10), and mucosal pentraxin 2 (Mptx2) (Table S4, sheet “Upregulated Genes in HMO−”). The unique genes (13 genes) that were upregulated in SMM were predominantly related to immunoglobulin structures (Table S4, sheet “Upregulated Genes in HMO−”), whereas, in NSM (79 unique genes), genes were related to solute carriers (Slc1a3, Slc6a13), a transcriptional factor (Irf4), antimicrobial peptides (Defa36, Defa28, Defa35, Defa31, Defa33), and fatty acid uptake (CD36) (Table S4, sheet “Upregulated Genes in HMO−”). In the MFM group (six unique genes), a neuropeptide-related gene, Npy, and a pore-forming protein-related gene, Gsdmcl2 were significantly upregulated (Fig. 5D, Table S4, sheet “Upregulated Genes in HMO−”). Among the downregulated genes, 50 unique genes were suppressed in NSM ileum, while only 2 unique genes were downregulated in the MFM group (Fig S5A; Table S4, sheet “Downregulated Genes in HMO−”, Fig. 5C). The unique genes in NSM include claudin 23 (Cldn23), interferon-gamma receptor (Ifngr1), a transmembrane protein (Tmem42), and a solute carrier family 6 gene (Slc6A20b). One of the suppressed genes in the MFM group was solute carrier family 23 (Slc23a2) (Table S4, sheet “Downregulated Genes in HMO−”, Fig. 5C).
Fig 5.
Microbiota of infants consuming milk from secretor or non-secretor mothers or those consuming infant formula impacts gene expression in the distal small intestine. DEGs and activated pathways in the distal small intestine (ileum) of mice transplanted with microbiota from infants consuming milk from secretor or non-secretor mothers or those consuming milk. Each treatment is compared to the germ-free controls. (A) Venn-diagram showing shared and unique DEGs among three groups when compared to germ-free controls, (B–D) Volcano plots showing DEGs in SMM (B), NSM (C), and MFM (D) groups compared to GF controls. Significantly upregulated genes are represented by red circles (log2 fold change >1.5, adjusted P-value < 0.05, pairwise comparison). (E–G) Barplots showing the significantly activated pathways in SMM (E), NSM (F), and MFM (G) groups compared to GF controls. Pathways are ranked based on normalized enrichment score (NES) and significant pathways are shown (pairwise comparison, P-adjusted <0.05). SMM = mice transplanted with fecal material from infants consuming milk from secretor mothers, N = 6; NSM = mice transplanted with fecal material from infants consuming milk from non-secretor mothers, N = 7; MFM = mice transplanted with fecal material from infants consuming dairy-based formula, N = 4.
To understand the pathways altered due to microbiota, all genes obtained from RNA-sequencing analysis were subjected to Gene Set Enrichment Analysis (GSEA). GSEA revealed that biological processes (BP) related to immunoglobulin production and production of molecular mediators of immune responses were activated in all three groups (Fig. 5E–5G). In contrast, biological processes like immunoglobulin production, production of molecular mediator of immune response, immune response, immune system process, and cellular components like extracellular regions and immunoglobulin complexes were activated only in the NSM group (Fig. 5F).
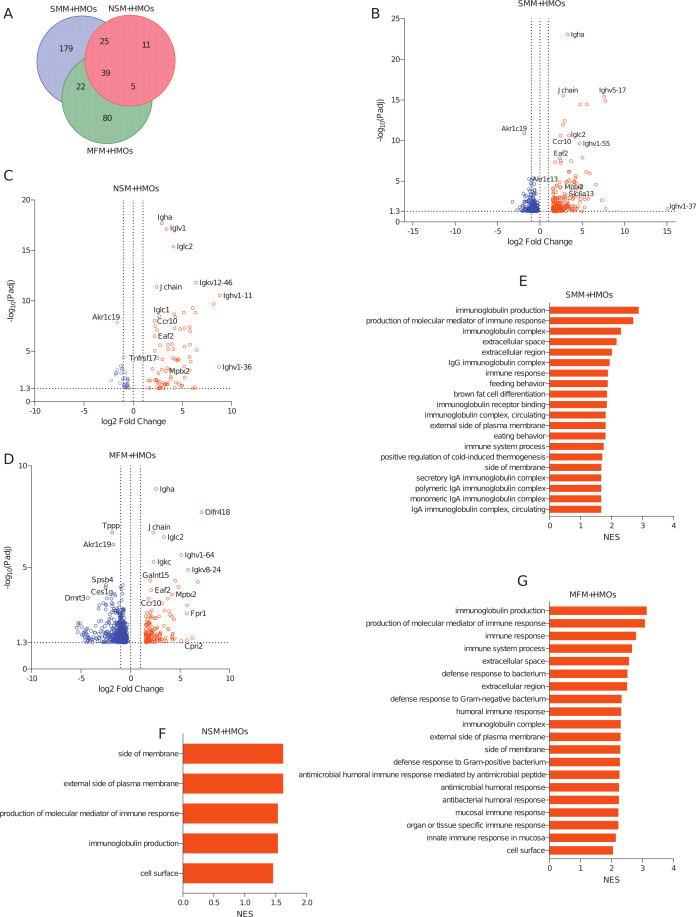
In addition, when mice were supplemented with HMOs, 265 genes in SMM + HMOs, 80 genes in NSM + HMOs, and 146 genes in MFM + HMOs groups were upregulated when compared to GF controls (Fig. 6A through D; Table S4, sheet “Upregulated Genes in HMO+”, log2 fold change >1.5, adjusted P-value < 0.05, pairwise comparison). Likewise, 424, 31, and 621 genes were downregulated in SMM + HMOs, NSM + HMOs, and MFM + HMOs groups when compared to the GF controls, respectively (Fig. S5E; Table S4, sheet “Downregulated Genes in HMO+”, Fig. 6B–6D,). Among these genes, 39 were significantly upregulated and 27 were downregulated in all three groups (Fig. 6A; Fig. S5E; Table S4, sheets “Upregulated Genes in HMO+”, “Downregulated Genes in HMO+”). The 39 genes upregulated in all three treatment groups were predominantly related to immunoglobulins and immune responses (Table S4, sheet “Upregulated Genes in HMO+”). Besides the shared genes, several unique genes were changed due to HMO, specifically in SMM + HMOs and MFM + HMOs groups (179 and 80 genes, respectively, Fig. 6A; Table S4, sheet “Upregulated Genes in HMO+”). Among the GSEA pathways that were activated, five pathways, such as production of molecular mediator of immune response, immunoglobulin production, pathways related to cell surface, side of membrane, and external side of plasma membrane, were activated in all three groups (Fig. 6E through 6G; Table S6, sheet “Pathway UP in Ileum in HMO+”).
Fig 6.
HMOs supplementation in mice colonized with microbiota of infants consuming milk from secretor or non-secretor mothers or those consuming infant formula impacts gene expression in the distal small intestine. DEGs and activated pathways in the distal small intestine (ileum) of mice transplanted with microbiota from infants consuming milk from secretor or non-secretor mothers or those consuming milk formula and supplemented with HMOs. Each treatment is compared to the germ-free controls. (A) Venn-diagram showing shared and unique DEGs among the groups when compared to germ-free controls, (B) volcano plots showing DEGs in SMM + HMOs, (C) NSM + HMOs, and (D) MFM + HMOs groups compared to GF controls. Significantly upregulated genes are represented by red circles (log2 fold change >1.5, adjusted P-value < 0.05, pairwise comparison). (E–G) Barplots showing the significantly activated pathways in SMM + HMOs (E), NSM + HMOs (F), and MFM + HMOs (G) groups compared to GF controls. Pathways are ranked based on normalized enrichment score (NES) and significant pathways are shown (pairwise comparison, P-adjusted <0.05).SMM + HMOs = mice transplanted with fecal material from infants consuming milk from secretor mothers and supplemented with HMOs, N = 7; NSM + HMOs = mice transplanted with fecal material from infants consuming milk from non-secretor mothers and supplemented with HMOs, N = 7; MFM + HMOs = mice transplanted with fecal material from infants consuming dairy-based formula, N = 4.
While in the absence of HMOs fewer pathways were upregulated in all three groups compared to GF controls, (Fig. 5E–5G), HMOs supplementation induced activation of several pathways compared to the control, specifically in SMM + HMOs and MFM + HMOs groups, with 20 pathways being shared between these two groups (Table S6, sheet “Pathway UP in Ileum in HMO+”). Among the unique pathways activated in SMM + HMOs, they were related to NK T cell activation, B and T cell activation and proliferation, G protein-coupled receptor activity, and peroxisome proliferator-activated receptor signaling pathways (Table S6, sheet “Pathway UP in Ilum in HMO+”). Interestingly, only in the MFM + HMOs group, pathways related to antimicrobial response, defense response to bacterium, defense response to Gram-negative bacterium and Gram-positive bacterium, and antimicrobial humoral response were upregulated (Fig. 6G; Table S6, sheet “Pathway UP in Ilum in HMO+”).
Microbiota of infants consuming milk from secretor or non-secretor mothers or those consuming infant formula impacts gene expression in the large intestine
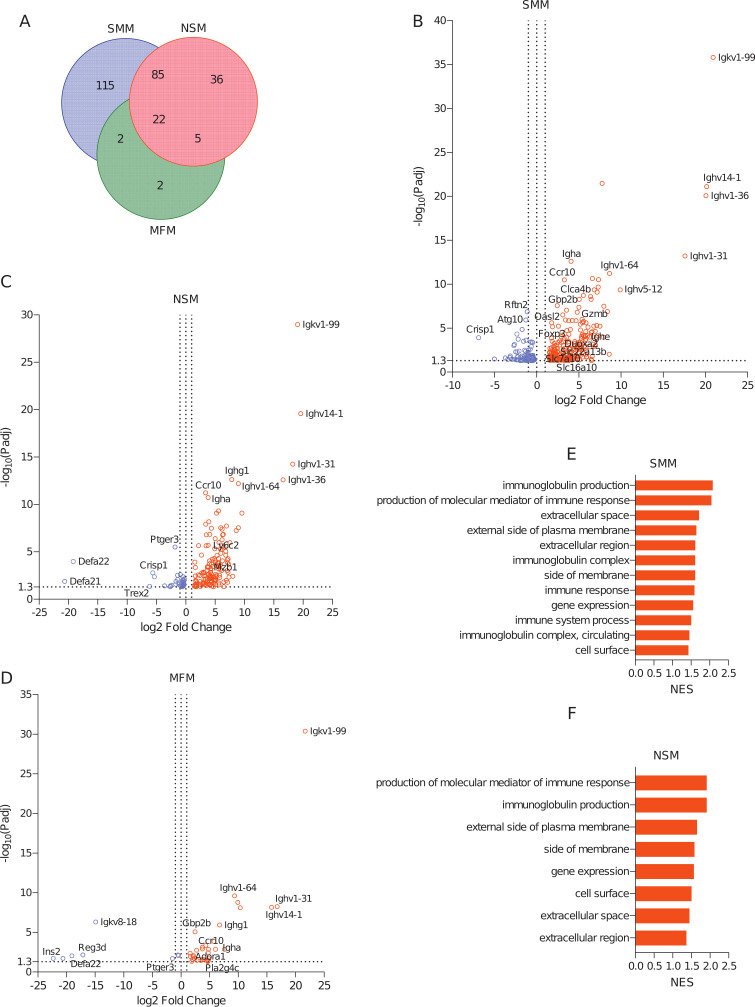
In the colon, 224, 148, and 32 genes were upregulated (Fig. 7A; Table S3, sheets “Up in SMM vs GF,” “Up in NSM vs GF,” and “Up in MFM vs GF”) and 100, 47, and 8 genes were downregulated (Fig. S6A; Table S3, sheets “Down in SMM vs GF,” “Down in NSM vs GF,” and “Down in MFM vs GF”) in SMM, NSM, and MFM groups, respectively, compared to GF controls (log2 fold change >1.5, adjusted P-value < 0.05, pairwise comparison). Among the upregulated genes, 22 genes were common among all three groups and were predominately related to immunoglobulins and immune response (Table S5, sheet “Upregulated Genes in HMO−”). Specifically, an immunoglobulin A related gene, Igha, and chemokine receptor 10, Ccr10, were upregulated. Similar to small intestinal (ileum) gene expression, the number of upregulated genes common between SMM and NSM (85 DEGs) was higher than between MFM and SMM (2 DEGs) or NSM (5 DEGs) (Table S5, sheet “Upregulated Genes in HMO−”). The unique upregulated genes in SMM (115 DEGs) were represented by genes related to solute carrier family (Slc22a13b, Slc16a6, Slc16a10, Slc7a10), phospholipase (Pla2g5, Pla2g12b, Pla2g2a), granzyme (Gzmb), apolipoprotein (Apol9a, Apol9b), regulatory T cells (Foxp3), transcription factor (Irf4), and glutathione transferase (Gsta1, Gstm3) (Fig. 7B; Table S5, sheet “Upregulated Genes in HMO−”). In NSM, marginal zone B-related gene, Mzb1, and lymphocyte antigen, Ly6c2, were upregulated, while in MFM, two unique genes, Adora1 and Pla2g4c, were upregulated (Fig. 7C and D, Table S5, sheet “Upregulated Genes in HMO−”). Also, the majority of downregulated genes were from the colon of SMM and NSM groups, with 13 downregulated DEGs shared between these groups (Fig. S6A; Table S5, sheet “Downregulated Genes in HMO−”). In SMM, 86 unique genes were downregulated, whereas in NSM and MFM, 31 and 5 individual genes were downregulated compared to the GF controls, respectively (Fig. S6A; Table S5, sheet “Downregulated Genes in HMO−”). GSEA analysis showed biological processes related to immunoglobulin production, and production of molecular mediators of immune response were activated in SMM and NSM groups (Fig. 7E and F; Table S6, sheet “Pathway UP in colon in HMO−”), whereas immune response and immune system pathways were activated only in SMM (Fig. 7E; Table S6, sheet “Pathway UP in colon in HMO−”). However, no such pathways were activated in the MFM group (Table S6, sheet “Pathway UP in colon in HMO−”).
Fig 7.
Microbiota of infants consuming milk from secretor or non-secretor mothers or those consuming infant formula impacts gene expression in the large intestine. DEGs and activated pathways in the large intestine (colon) of mice transplanted with microbiota from infants consuming milk from secretor or non-secretor mothers or those consuming milk. Each treatment is compared to the germ-free controls. (A) Venn-diagram showing shared and unique DEGs among three groups when compared to germ-free controls, (B–D) Volcano plots showing DEGs in SMM (B), NSM (C), and MFM (D) groups, compared to GF controls. Significantly upregulated genes are represented by red circles (log2 fold change >1.5, adjusted P-value < 0.05, pairwise comparison). (E–G) Barplots showing the significantly activated pathways in SMM (E), NSM (F), and MFM (G) groups compared to GF controls. Pathways are ranked based on NES and significant pathways are shown (pairwise comparison, P-adjusted < 0.05). SMM = mice transplanted with fecal material from infants consuming milk from secretor mothers, N = 6; NSM = mice transplanted with fecal material from infants consuming milk from non-secretor mothers, N = 7; MFM = mice transplanted with fecal material from infants consuming dairy-based formula, N = 4.
Among the suppressed pathways, in SMM, genes were overrepresented by pathways related to nuclear components and protein modifications (Fig. S6B; Table S6, sheet “Pathway DOWN in Colon in HMO−”). Similarly, in NSM colon, pathways related to molecular function, cellular processes, and intracellular components were suppressed (Fig. S6C; Table S6, sheet “Pathway DOWN in Colon in HMO−”). In the MFM group, majority of the pathways related to cellular responses were suppressed (Fig. S6D; Table S6, sheet “Pathway DOWN in Colon in HMO−”).
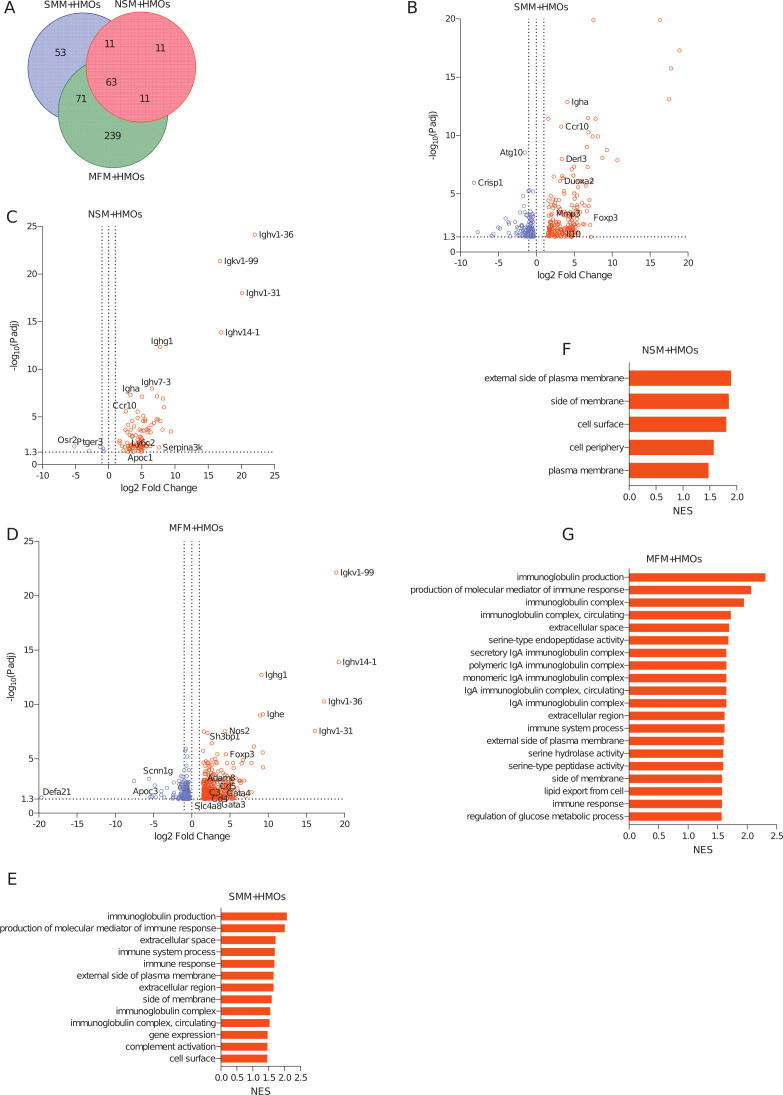
When mice were supplemented with HMOs, 198 genes from SMM + HMOs, 96 genes from NSM + HMOs, and 384 genes from MFM + HMOs groups were upregulated in the colon when compared to the GF controls (Fig. 8A through D; Table S3, sheets “Up in SMM + HMOs vs GF,” “Up in NSM + HMOs vs GF,” and “Up in MFM + HMOs vs GF,” log2 fold change > 1.5, adjusted P-value < 0.05, pairwise comparison). Likewise, 131, 5, and 324 genes were downregulated in SMM + HMOs, NSM + HMOs, and MFM + HMOs groups compared to the germ-free controls, respectively (Fig. 8B through D; Fig. S6E; Table S3, sheets “Down in SMM + HMOs vs GF,” Down in NSM + HMOs vs GF,” and “Down in MFM + HMOs vs GF”). Among these differentially expressed genes, 63 were upregulated in all three groups (Fig. 8A; Table S5, sheet “Upregulated Genes in HMO+”). These DEGs were mainly related to immunoglobulins and immune responses, including immunoglobulin E (Ighe) and immunoglobulin A (Igha) (Table S5, sheet, “Upregulated Genes in HMO+”). Likewise, there were 71 significantly upregulated genes common between SMM + HMOs and MFM + HMOs groups (Table S5, sheet, “Upregulated Genes in HMO+”). Though genes encoding immunoglobulins (Igha, Ighe) were upregulated in all three HMOs supplemented groups, the Foxp3 gene was upregulated only in the SMM + HMOs and MFM + HMOs groups (Fig. 8B and D; Table S5, sheet “Upregulated Genes in HMO+”). The unique genes upregulated in SMM + HMOs include genes involved in the regulation of inflammation, like Mfsd2a and IL10 (Fig. 8B; Table S5, sheet “Upregulated Genes in HMO+”). Similarly, in the NSM + HMOs group, genes encoding apolipoprotein, Apof, Apoc1, and Apoa2 were upregulated (Table S5, sheet, “Upregulated Genes in HMO+”). Two hundred thirty-nine unique genes were differentially higher in the MFM + HMOs group. These genes were related to lymphocyte activation (Cd3e, Cd4, Cd8a, Cd28, Cd300e), chemokine receptors (Ccr2, Ccr5, Cxcr2), transcriptions factors (Gata3, Gata4), solute carrier family (Slc14a1, Slc4a8, Slc7a10, Slco5a1), and tumor necrosis factor superfamily (Tnfrsf13b, Tnfrsf9, Tnfsf8) (Table S5, sheet “Upregulated Genes in HMO+”). GSEA analysis also revealed that majority of changes in the colon were in the MFM + HMOs group, followed by SMM + HMOs (Fig. 8E and G; Table S6, sheet “Pathway UP in Colon in HMO+”). Though some pathways related to cellular components were activated in all three groups, the pathways activated in SMM + HMOs were dominated by genes related to immune system, immune response, immunoglobulin production, and complement activation (Fig. 8E; Table S6, sheet “Pathway UP in Colon in HMO+”). In MFM + HMOs, cellular components and biological functions related to IgA were upregulated (Fig. 8G; Table S6, sheet “Pathway UP in Colon in HMO+”). However, in NSM + HMOs group, only five of the cellular components-related pathways were upregulated compared to the GF controls (Fig. 8F; Table S6, sheet “Pathway UP in Colon in HMO+”). Among the suppressed, pathways related to metabolic processes and cellular components were suppressed in the SMM + HMOs group, whereas in NSM + HMOs groups, the pathways were represented by cellular processes, cellular response, and signaling (Fig. S6F and G; Table S6, sheet “Pathway DOWN in Colon in HMO+”). Similarly, in MFM + HMOs group, pathways representing cellular response, defense response to bacteria, and cell cycles were downregulated (Fig. S6H; Table S6, sheet “Pathway DOWN in Colon in HMO+”).
Fig 8.
HMOs supplementation in mice colonized with microbiota of infants consuming milk from secretor or non-secretor mothers or those consuming infant formula impacts gene expression in the distal small intestine. DEGs and activated pathways in the large intestine (colon) of mice transplanted with microbiota from infants consuming milk from secretor or non-secretor mothers or those consuming milk formula and supplemented with HMOs. Each treatment is compared to the germ-free controls. (A) Venn-diagram showing shared and unique DEGs among the groups when compared to germ-free controls, (B) Volcano plots showing DEGs in SMM + HMOs, (C) NSM + HMOs, and (D) MFM + HMOs groups compared to GF controls. Significantly upregulated genes are represented by red circles (log2 fold change > 1.5, adjusted P-value < 0.05, pairwise comparison). (E–G) Barplots showing the significantly activated pathways in SMM + HMOs (E), NSM + HMOs (F), and MFM + HMOs (G) groups compared to GF controls. Pathways are ranked based on normalized enrichment score and significant pathways are shown (pairwise comparison, P-adjusted < 0.05).SMM + HMOs = mice transplanted with fecal material from infants consuming milk from secretor mothers and supplemented with HMOs, N = 7; NSM + HMOs = mice transplanted with fecal material from infants consuming milk from non-secretor mothers and supplemented with HMOs, N = 7; MFM + HMOs = mice transplanted with fecal material from infants consuming dairy-based formula, N = 4.
DISCUSSION
Maternal, dietary, and environmental factors determine early seeding of gut microbiota in neonates. The establishment of gut microbiota is essential for infant growth and development, which is influenced, in part, by dietary intake in infancy. HMOs are composed of diverse and complex carbohydrates (8), whose representation is determined by several factors, including mothers’ secretor status (26). Previous literature has suggested that maternal secretor status plays a role in infant gut microbiota composition (16). Likewise, infants consuming infant formula devoid of HMOs contain distinct microbiota compared to those consuming human milk (27–29). Indeed, the present study showed characteristic microbial community structure in the feces of infants fed by secretor or non-secretor mothers or those consuming infant formula (Fig. S1). Bifidobacterium and Bacteroides were the two abundant bacterial genera in the feces of infants fed by secretor mothers. Several previous studies have also shown a higher abundance of Bifidobacterium species in breastfed children of secretor mothers (16, 20, 30, 31). Bifidobacterium has also been shown to be associated with secretor genotype in adults (21). In contrast to our findings, a study in a Chinese cohort showed enrichment of the genus Bifidobacterium in infants fed by non-secretor mothers though a positive correlation between maternal secretor status and specific Bifidobacterium OTUs was observed (19). In a Danish cohort, no significant differences were found in the relative abundance of Bifidobacterium-related amplicon sequence variants in the feces of infants fed by secretor or non-secretor mothers, possibly due to the older age of the infants (5 months) in the study (20, 32, 33). Rural vs urban status have revealed distinct microbiota composition in infants (34–37). In our cohort samples, we observe more Bacteroides than Bifidobacteria unlike in rural population. Our data and the published literature suggest that maternal secretor status and likely the housing environment and urbanization of the population are likely factors in infant microbiota composition.
When 21-day-old germ-free mice were transplanted with the infant fecal microbiota, several bacterial taxa successfully engrafted into the mice. Mice gavaged with microbiota from infants consuming infant formula devoid of HMOs (MFM) had higher gut microbial alpha diversity when compared to mice colonized with microbiota from infants consuming milk from secretor (SMM) or non-secretor mothers (NSM) (Fig. 1A). These data are in agreement with previous findings of higher microbial diversity in infants consuming infant formula compared to infants consuming human milk (38). Similarly, higher alpha diversity was observed in piglets fed dairy-based infant formula than those provided human milk (39, 40). Bacteroides was one of the abundant genera in the mice colonized with microbiota from infants fed by secretor mothers (SMM). A higher abundance of Bacteroides spp. and related proteins was previously shown in the cecal lumen of human milk-fed piglets relative to those consuming milk formula (27). In addition, several studies conducted worldwide suggest that distinct microbiota are present in human milk-fed infants with differential abundance of Bacteroides and/or Bifidobacterium, further supporting elevated Bacteroides abundances in our findings (41). In the majority of these studies, secretor status and milk HMOs composition were not identified, which is a knowledge gap filled in our study. In addition, a potential probiotic bacterium, Blautia, also successfully colonized in the SMM group (Fig. 1D; Fig. S2) (42). Interestingly, 2′-FL HMO can increase the abundance of Blautia. Though Blautia itself was not found to utilize 2′-FL, it can utilize fucose released from 2′-FL used by Bifidobacterium bifidum as shown via co-culture experiments (43). The higher abundances of both Bifidobacterium and Blautia in the SMM group, which were transplanted with microbiota from infants fed by secretor mothers (i.e., milk containing abundant 2′-FL), suggests potential cross-feeding between these two microbial species observed in in vitro may also be relevant in vivo. One of the predominant bacterial genera in NSM and NSM + HMOs groups was a mucin-degrading bacterium, Akkermansia, which also utilizes HMOs and allows other gut microbiota to cross-feed on the byproducts of the substrate, though its HMOs utilization capacity is reported to be strain-dependent (44, 45). However, why this bacterium is abundant only in NSM and NSM + HMOs groups and not in SMM/SMM + HMOs groups needs further exploration. It is possible that elevated abundance of Bifidobacteria that use short-chain HMOs might outcompete Akkermansia. Clostridium_sensu_stricto, a bacterium which had the highest abundance in MFM mice, has been positively associated with antibiotic exposure during infancy (46). Interestingly, HMOs supplementation in these mice significantly reduced the abundance of this microbe, suggesting a potential beneficial effect of HMOs in formula-fed infants. Parabacteroides was another abundant microbe in the MFM and MFM + HMOs groups whose higher abundance has also been previously reported in milk formula-fed piglets (28).
Microbiome composition has a role in intestinal morphology (47, 48). In our experimental settings, HMOs supplementation in mice colonized with NSM microbiota increased the ileum villus height compared to NSM and MFM groups though no differences were observed between SMM and SMM + HMOs and MFM and MFM + HMOs groups. Previous studies have reported increased intestinal villus height in suckling rats and newborn piglets receiving 2′-FL HMOs (49, 50). Though pooled HMOs were used in our study, the increase in ileum villus height in HMOs groups compared to the NSM only groups suggests that this effect may arise through the impact of 2′-FL HMOs, as the NSM group was colonized with microbiota from infants who were consuming milk devoid of 2′-FL. Similarly, HMOs supplementation increased ileum villus height in SMM + HMOs and NSM + HMOs groups compared to the MFM group though this effect was not seen when the MFM group was fed with HMOs. In contrast to these findings, in our previous study, villus height was decreased in germ-free mice supplemented with HMOs for 14 days (7). Since there was an absolute absence of microbiota in that study, the increase in villus height observed in the current study implicates microbiota and HMOs interaction effects.
Early-life microbiota establishment is essential for the optimal development of the infant immune system, whereas inadequate microbiota colonization during early life can have long-lasting effects on infants. In germ-free mice, the immune system is not well developed, signifying the importance of microbiota in immune system development (51, 52). HMOs can also modulate both innate and adaptive immune cell composition either directly or via modulating microbiota or microbial metabolites (5, 7, 53). In our study, HMOs supplementation elevated CD8+ T cells in the SMM group and Foxp3+ T regulatory cells (Tregs) in the MFM group in the MLN. In a previous study, where rats were given 2′-FL reported an increased percentage of CD8+ T cells in MLN (49). Though no difference in CD8+ T cells between MFM and MFM + HMOs groups was observed in our study, another study demonstrated the elevated percentage of CD8+ T cells in MLN of HMOs-fed pigs compared to formula-fed pigs (54). This effect of HMOs, only observed with specific microbiota, indicates an interaction between the HMOs and microbiota type. The localized increase in Foxp3+ Tregs in the MLN in the MFM + HMOs group could be attributed to the first exposure of the MFM microbiota to HMOs, as MFM microbiota were derived from infants fed milk formula devoid of any HMOs. In support of this observation, there was a significant upregulation of Foxp3 gene in the colon of MFM mice supplemented with HMOs. However, such changes were not observed in the MFM only group. In addition, in the spleen, the SMM and SMM + HMOs groups had the highest percentage of Foxp3+ Tregs, as well as upregulation of Foxp3 gene in the colon of both groups, which may be attributed to the elevated abundances of Bacteroides and Bifidobacterium in these groups. Interestingly, upregulation of Foxp3 gene in large intestine has been shown in HMO supplemented germ-free mice, suggesting HMO could have direct effect on Foxp3 gene expression (7). Bacteroides fragilis was shown to promote the development of Foxp3+ T cells in the mono-colonized mouse gut (55). Similarly, polysaccharide A derived from B. fragilis can induce the production of Foxp3+ T cells from CD4 T cells (55, 56). Bifidobacterium is also known to enhance Foxp3 cell numbers in peripheral blood and intestine (57, 58). These findings suggest that higher abundance of beneficial microbiota like Bacteroides and Bifidobacterium in the infants who were consuming HMO-containing milk may increase intestinal Foxp3 gene expression as well as splenic Foxp3+ T regulatory cells.
Innate lymphoid cells (ILCs) are a heterogeneous group of lymphoid-like cells lacking specific antigen receptors (59). These cells are primarily located at mucosal barriers, lymphoid organs, and various other tissues. Our study showed that the effect of HMOs on ILC composition was tissue and microbiota-specific (Fig. 3). In MLN, HMOs increased the abundance of ILC1 in the NSM group and ILC2 in the MFM group, while in the spleen, HMOs elevated ILC1 and decreased ILC2 in the SMM group. An in vitro study reported that HMOs such as 2′-FL can induce Th1 type IFN-gamma and IL-10 secretion and reduce Th-2 type IL-13 in peripheral blood mononuclear cells (60). These findings suggest that though HMOs can impact the innate lymphoid cells composition in lymphoid tissues, their impact can be different depending on the microbiota type (i.e., secretor vs non-secretor) and previous exposure of microbiota to the HMOs.
In the ileum, the main pathways upregulated in all groups were related to immunoglobulin production and production of molecular mediators of immune response, regardless of the microbiota type. This robust response to fecal microbiota transplantation in germ-free mice could be due to the young age of mice and their initial lack of microbes as their immune system is not well-developed. Germ-free mice lack immunoglobulin A-secreting intestinal B cells, and colonization of these mice with microbiota induces IgA production (61, 62). In our study, a gene encoding IghA was one of the common upregulated genes in all microbiota-transplanted mice. However, we also observed unique gene expression profiles in each group, suggesting the possibility of distinct microbiota-dependent effect on the ileum transcriptome. Interestingly, 17 upregulated genes were common between SMM and NSM, and only one gene was common between SMM and MFM. The larger number of genes shared could be attributed to colonization of mice with microbiota from the breast-fed infants. HMOs supplementation increased overall global ileal gene expression in mice relative to groups without HMOs. As the addition of HMOs did not induce significant differences in microbiota composition, these changes could be attributed to either the direct effect of HMOs or the function of microbiota (i.e., production of metabolites), which was not evaluated in our study due to sample limitations. Several unique pathways were also activated in the HMOs-supplemented SMM and MFM groups (Table S7). Specifically, in the MFM + HMOs group, pathways related to antibacterial response were upregulated, including defense response to gram-positive/gram-negative bacteria, response to lipopolysaccharides, and antibacterial humoral responses. In the same group, innate lymphoid cells (ILC2) and Foxp3+ T cells were also elevated in mesenteric lymph nodes, as well as IgA in the serum. All these findings support the notion that HMOs could have a protective effect on formula-fed children. In contrast, breastfed infants have been already exposed to the HMOs; thus, major changes in immune cell composition or antibody response were not observed.
In the colon, Igha was upregulated in all SMM, NSM, and MFM groups similar to the ileum transcriptome, whereas a gene encoding immunoglobulin E (IgE), Ighe, was upregulated only in SMM and NSM groups. Since the SMM and NSM groups were colonized with microbiota from breast-fed infants, the upregulation of Ighe gene could be attributed to the microbiome associated with human milk exposure rather than the mother’s secretor status. A gene related to Foxp3 was upregulated only in SMM. Gut-resident Foxp3 regulatory cells contribute to tissue homeostasis and induce tolerance to gut microbiota during the postnatal period (63–65). Certain commensal species of Bacteroides can elicit the production of Foxp3+ Tregs and have been shown to be associated with high numbers of Tregs (55, 56). Supporting these reports, higher abundances of Bacteroides spp. were seen in the colon contents of SMM mice. Though HMOs supplementation did not change Foxp3 gene expression in SMM and NSM groups, it elevated the expression of Foxp3 transcripts in MFM groups. It is likely that SMM and NSM groups had been exposed to human milk, and the functional outputs of these microbiota were programmed. In addition, in MLN of MFM, an elevated Foxp3+ Treg percentage due to HMOs supplementation fits with our previous observation (7). This suggests that even without robust change in gut microbiota composition, HMOs can induce Foxp3 gene expression in mice. HMOs also promoted the expression of a gene related to anti-inflammatory marker, Il10, in SMM group. A previous study had showed that 2′-FL can enhance IL-10 and Th1-type IFN-gamma production and reduce Th2-type IL-13 in a cell-culture settings (60, 66). In our study, HMOs supplementation in SMM group increased splenic ILC1s, reduced ILC2s, and upregulated large intestine IL10 expression. It is also noteworthy that, although fewer pathways were significantly activated in SMM/NSM groups in the large intestine compared to the control, no such pathways were activated in MFM group. However, HMOs induced a pronounced impact on MFM group with several unique pathways activated in this group. Specifically, multiple pathways related to IgA immunoglobulin were activated in HMOs supplemented MFM groups. Interestingly, unlike in the ileum, pathways related to antimicrobial responses were not activated in MFM groups. This suggests that HMOs can change the gene expression in ileum and colon differently in MFM group. Similarly, to ileal gene expression, very few common genes were upregulated in SMM and MFM, but HMOs supplementation increased the common upregulated genes in the SMM and MFM groups. These findings suggest that the impact of HMOs is more pronounced in mice colonized microbiota which were naïve to HMOs, leading to robust modulation in gene expression.
Conclusion
Utilizing a germ-free mouse model, the impact of microbiota derived from infants fed by secretor or non-secretor mothers or those consuming infant formula on intestinal morphology, innate and adaptive immune system, and intestinal gene expression was evaluated. This study validated previous findings that formula-fed infants have higher gut microbiota diversity. We also demonstrated a minimum effect of the supplementation with pool of human milk oligosaccharides for 14 days in the overall gut microbiota composition in mice. However, distinct gut microbiota abundances were observed between the treatment groups, irrespective of the HMOs supplementation. This study also showed distinct adaptive immune responses and innate lymphoid cell composition due to different microbiota and supplementation of HMOs. At the transcriptome level, microbiota-specific changes were seen in both the ileum and colon. Several features related to immune response and immunoglobulin production were common among the three groups. Unlike gut microbiota composition, robust changes were seen in gene expression levels due to HMOs supplementation. Specifically, the changes were mainly seen in the mice transplanted with microbiota derived from infants consuming infant formula devoid of HMOs. Overall, we report the impact of mothers’ milk secretor status or infant formula consumption on infant microbiome and show its impact on immune system and intestinal gene expression using a mouse model.
Limitations
First, the secretor status of the infants who consumed the human milk or infant formula was not determined. Second, wild-type mice’s intestines are rich in fucosylated glycans, and transplanted microbiota could be changed due to these glycans. Third, in this study, day 21 old germ-free mice were transplanted with the human infant fecal inoculum. Though at this age, the immune system and intestinal tract is immature compared to the conventional mice of similar age (67, 68), using younger mice pups could be more translational to young infants. Fourth, pooled HMOs were used in the study. Thus, future studies will be interesting to investigate the effect of individual HMOs like 2'FL, which is the major HMO determining the maternal secretor status and leveraging FUT2 knockout mice.
MATERIALS AND METHODS
Microbiota inoculum
Human milk from mothers and fecal material from 3- to 3.5-month-old exclusively breastfed infants who were enrolled in the ongoing Happy Tummies Study (clinical trial# NCT03493711) at Arkansas Children’s Nutrition Center were collected. Similarly, fecal material from infants who were exclusively consuming dairy-based infant formula since birth was also collected. Parents were advised to collect fecal material in a sterile vial and asked to store at −20°C until transported to the research center. At the research center, samples were transferred immediately to −80°C. The Institutional Review Board of University of Arkansas of Medical Sciences approved the human fecal samples for studies. Human milk secretor status of the mothers was determined in Dr. Lars Bode’s Laboratory at the University of California-San Diego (UCSD) based on the presence of 2′-FL (69). Fecal samples from infants consuming secretor (N = 5) or non-secretor milk (N = 5) or infant formula (N = 5) were pooled, aliquoted into 100 mg, and transferred into a Coy Anaerobic Chamber (Coy Lab, Grass Lake, MI). One hundred milligram of fecal material was mixed in 1 mL reduced 1× phosphate-buffered saline containing 0.10% resazurin (Sigma, Catalog # R7017-5G) and 0.05% L-cysteine (MP Bio, Catalog # 02101444-CF). The solution was vortexed vigorously for 5 min and allowed to settle down for 5 min. The supernatant was transferred into a sterile Eppendorf tube and exited the anaerobic chamber for transplantation into mice.
Animal studies
All animal experiments were conducted in accordance with the Institutional Animal Care and Use Committee at the University of Arkansas for Medical Sciences. At 21–23 days of age, C57BL/6 germ-free mice were distributed across groups (N = 8–13 per group) based on body weights. HMOs were isolated and purified from the donor human milk by LB’s Laboratory at University of California (UC)-San Diego (69). A set of mice per group (N = 4–7 per group) received 100 µL of microbiota inoculum (total amount 10 mg/mouse) and another set of animals received both microbiota inoculum (same amount) and 15 mg of HMOs per mouse (N = 4–7 per group). HMOs were gavaged orally for 14 days, whereas fecal material was gavaged twice, on days 1 and 7. Germ-free control mice (N = 6 per group) received the vehicle. Body weights were recorded daily throughout the experiment. At day 35 of age, mice were anesthetized with isoflurane. Blood was collected in a tube via retroorbital bleeding. After breath cessation, mice were subjected to cervical dislocation. Small intestine (SI), large intestine (LI) lengths, and organ weights were measured. Tissue samples were processed fresh or fixed in formalin or flash-frozen in liquid nitrogen and stored at −80°C. Blood was spun down at 3,000 rpm for 10 min in 4°C, and clear serum was collected, aliquoted, and stored immediately at −80°C. Samples were processed for 16S sequencing, histomorphometry, ELISA, flow cytometry, and transcriptome analyses.
Histomorphometry analyses
The distal ileum (3 cm), proximal colon (2 cm), and cecal sections were fixed in formalin. Fixed tissues were cut, paraffin-embedded, and processed for staining with hematoxylin and eosin (H and E). Histomorphometry analyses of SI crypt depth and villi height and the LI and cecum gland depth were carried out to evaluate the GIT morphology. All measurements were made using Aperio Image software by a board-certified pathologist, Dr. Tanya LeRoith. Crypts immediately adjacent or as near as possible to measured villi were selected and the crypt depth was measured using the pen tool. Crypts had to be intact and entirely within the plane of the section.
Flow cytometry
Mesenteric lymph nodes and spleen were collected from each mouse in a tube containing cold Roswell Park Memorial Institute media (RPMI) (Invitrogen, Catalog # 11875093) containing 20% FBS media and immediately stored on ice. Using a 3-mL syringe plunger, spleen and MLN tissues were squished through a 70-µm strainer placed on top of a sterile 50-mL conical tube. Ten milliliters of cold RPMI + 20% FBS media was added into each conical tube and spun down at 3,000 rpm for 5 min at 4°C. Supernatant was aspirated without disturbing the cell pellet; 5 mL cold RPMI + 20% FBS media was added and vortexed to mix. The MLN cells were then poured onto a 40-µm strainer placed on top of a sterile 50 mL conical tube and spun down at 3,000 rpm for 5 min at 4°C. Supernatant was aspirated and cell pellet was resuspended in 1 mL cold flow cytometry staining buffer (Thermofisher, Catalog #00-4222-26).
Spleen cells were resuspended in 2 mL lysis buffer kept at room temperature, vortexed to mix, and incubated for 10 min at room temperature. After incubation, 3 mL cold RPMI + 20% FBS media was added, vortexed to mix, and spun down at 3,000 rpm for 5 min at 4°C. Supernatant was removed, and 5 mL cold RPMI + 20% FBS was added and vortexed to mix. Single-cell suspension was obtained in 2 mL cold flow cytometry staining buffer as described above for MLN.
Single-cell suspensions were counted using Vi-Cell Blu Cell counter (Beckman Coulter, Catalog # C19196) and 2 × 106 splenocytes and 8 × 105 MLN cells were stained with live/dead stain followed by the Fc receptor block (Innovex, NB309) and antibodies for various immune cell populations. Antibody information is provided in Table S7. Stained cells were analyzed using BD LSRFortessa (UAMS flow cytometry core facility). Gating for T cells, innate lymphoid cells, B cells, and myeloid cells are shown in Fig. S7.
ELISA
Serum Immunoglobulin A, IgG, and IgM were measured using ELISA using commercial mouse kits (Invitrogen, catalog # 88-50450, 88-50400, and 88-50470) as per the manufacturer instructions. The plates were read using an Omega Polar Star at 450 nm wavelength.
16S rRNA Sequencing and microbial community composition analysis
16S rRNA gene amplicon sequencing (V4-V5 region) and analysis was performed as described previously (70). Briefly, extracted DNA was amplified using KAPA HiFi hot start ready mix (catalog number: KK2602, Kapa Biosystems, Wilmington, MA) according to the manufacturer’s instructions and using the universal bacterial primers 515FB (GTGYCAGCMGCCGCGGTAA) and 926R (CCGYCAATTYMTTTRAGTTT) (71). PCR parameters were as follows: initial denaturation of 98°C for 3 min, followed by 20 cycles of 10 s denaturation at 98°C, 30 s annealing at 50°C, and 30 s extension at 72°C. A 10-min final extension at 72°C was performed, and products were held thereafter at 4°C. PCRs were cleaned up to remove unincorporated dNTPs and primers using the Axygen AxyPrep PCR Clean-up Kit (Axygen Scientific, Corning Life Sciences, Tewksbury, MA); amplicons were barcoded using the TruSeq dual-index approach (using primers TS-501-516 and TS-701-724; Integrated DNA Technologies, Coralville, IA) and extracted again using the AxyPrep PCR Clean-up Kit. DNA concentrations were quantitated using the Qubit dsDNA HS Assay Kit (Invitrogen, Carlsbad, CA) and pooled equally. Pool quality was measured using an Agilent Bioanalyzer (Agilent, Santa Clara, CA), and sequencing was performed using an Illumina MiSeq 2 × 250 paired end run at the Purdue Genome Facility.
Amplicon sequences were analyzed using mothur v. 1.44.3 (72) following the MiSeq SOP (https://mothur.org/wiki/miseq_sop/) with the previously described modifications (73). Briefly, contigs were constructed by merging read pairs and reads with errors in barcodes or primer sequences, ambiguous bases, homopolymers longer than 9 bases, and lengths longer than 411 bp were removed. Screened reads were aligned to the mothur-formatted SILVA bacterial reference database v.132, preclustered allowing up to 3 differences, and chimeras were identified and removed using the mothur-formatted Ribosomal Database Project classifier v.18, to which species epithets had been added (74). Sequence taxonomy classifications were accepted over a bootstrap value of 95 and reads classified as eukaryota, mitochondria, chloroplasts or of unknown phylogeny were removed. Samples were clustered using the OptiClust method (75) and OTU Tables were rarefied to 4,450 reads per sample for ecological analyses. Alpha-diversity (Shannon Index), Beta diversity (Bray-Curtis dissimilarity), taxa abundance from the OTUs Tables were determined using MicrobiomeAnalyst, a web-based platform for microbiome analysis (76).
Transcriptome analyses
Total RNA from colon and ileum was extracted using QIAzol (QIAgen, Catalog #79306) and 1-bromo-3-chloropropane (Millipore Sigma, Catalog #B9673). RNA was purified using RNeasy mini kit following the manufacturer’s instructions. The RNA concentration was measured by QUBIT fluorometer (Invitrogen, Catalog # 33226) and messenger RNA was purified from total RNA using poly-T oligo-attached magnetic beads. After fragmentation, the first strand cDNA was synthesized using random hexamer primers followed by the second-strand cDNA synthesis. The library was ready after end repair, A-tailing, adapter ligation, size selection, amplification, and purification. The library was checked with QUBIT fluorometer and real-time PCR for quantification and bionalyzer for size distribution detection. Quantified libraries were pooled and sequenced on the Illumina platform at Novogene Corporation Inc. (Sacramento, CA).
Bulk RNA-Seq analysis
Sequenced FASTQ files were subject to quality control and adapter sequence trimming with FastQC (v0.11.9) (77) and TrimGalore (v0.6.5) (78). Trimmed reads were then aligned with STAR (79) (v2.7.9a) against the mouse GRCm39_107 reference genome. Aligned reads were processed with featureCounts (80) (subread v2.0.1) to produce a gene-count matrix for all samples. All downstream processing and analysis of the data were performed using R (v4.3.0, R Core Team, 2022) (81). Differential expression analysis was performed using DESeq2 (82) (v1.40.0). Genes were termed as differentially expressed at an adjusted P-value < 0.05 and | log2FoldChange | > 1.5. Genes were mapped from ENSEMBL IDs to the appropriate gene symbols and ENTREZ IDs using the bitr function from clusterProfiler (83) (4.9.0) and subjected to Gene Set Enrichment Analysis (84). Several gene sets were used as reference databases for the pathway analysis, including Gene Ontology, C2, C5, and C7 gene sets from MSigDB (85), extracted using the msigdbr (v7.5.1, Dolgalev 2022) package, and tested with the gseGO and GSEA functions from clusterProfiler. Pathways were considered significantly activated or suppressed with adjusted P-value < 0.05. P-values were adjusted using Benjamini-Hochberg correction for both the differential expression and pathway analyses.
Statistical analyses
Differences in Shannon index among the groups were determined using a two-way ANOVA with Tukey’s multiple comparison test. Beta diversity was calculated using the Bray-Curtis dissimilarity statistics and visualized using principal coordinate analysis, and the differences among the groups were tested using a permutational multivariate analysis of variance (PERMANOVA) test. Histomorphometry parameters among groups within each tissue type (log-transformed) were modeled with a linear mixed-effects model accounting for repeated measurements within each individual and compared using multiple-pairwise comparison with Sidak’s adjustment. Group differences, the effect of microbiota, HMOs and interaction on body weight gain, organ weights, and immune cells composition were determined using a two-way ANOVA with Tukey’s multiple comparison tests in GraphPad Prism Version 9.3.1 (www.graphpad.com). Adjusted P < 0.05 was considered significant. Mixed-effects models were fit using the packages lme4 (v1.1-31 (86) and emmeans (v1.8.3, Lenth 2022) in R (v4.2.1, R Core Team 2022) (81, 86, 87).
ACKNOWLEDGMENTS
Germ-free mouse breeder pairs were obtained from National Gnotobiotic Rodent Resource Center, University of North Caroline School of Medicine and breeding was carried out by the Dr. Yeruva team members.
This work was supported by USDA-ARS [6026-51000-012-000D]. Transcriptome data analyses were supported, in part, using the University of Pittsburgh HTC cluster, which is supported by NIH award number S10OD028483.
M.G. and L.Y. designed and conceived the overall study. L.Y. supervised the research and received funding. B.T.S. and D.R. analyzed the transcriptome data. R.F. processed the tissues for histomorphometry. L.B. provided the HMOs for the study. S.R.L. and T.Y. collected and analyzed the microbiota data. T.L. evaluated the histology slides. QR analyzed the histology data. S.M.C. and L.C. assisted with germ-free mouse facility. A.A. assisted with Happy Tummies study. All authors approved the final manuscript.
Contributor Information
Laxmi Yeruva, Email: laxmi.yeruva@usda.gov.
Joshua E. Elias, Chan Zuckerberg Biohub, Stanford, California, USA
FUNDING
DATA AVAILABILITY
Sequencing data are deposited under BioProject accession number PRJNA1081588 (16S rRNA) and GEO accession number GSE260491 (RNA-sequencing).
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/msystems.00294-24.
Figures S1 to S7.
Changes in body weight, organ weight, and intestinal length.
Differentially expressed genes in the distal small intestine (ileum).
Differentially expressed genes in the large intestine (colon).
Unique or common differentially expressed genes in the distal small intestine (ileum).
Unique or common differentially expressed genes in the large intestine (colon).
Unique or common pathways in the distal small intestine (ileum) or large intestine (colon).
Antibodies used for immune cell for flow cytometry.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Atyeo C, Alter G. 2021. The multifaceted roles of breast milk antibodies. Cell 184:1486–1499. doi: 10.1016/j.cell.2021.02.031 [DOI] [PubMed] [Google Scholar]
- 2. Noel G, In JG, Lemme-Dumit JM, DeVine LR, Cole RN, Guerrerio AL, Campbell JD, Kovbasnjuk O, Pasetti MF. 2021. Human breast milk enhances intestinal mucosal barrier function and innate immunity in a healthy pediatric human enteroid model. Front Cell Dev Biol 9:685171. doi: 10.3389/fcell.2021.685171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Maessen SE, Derraik JGB, Binia A, Cutfield WS. 2020. Perspective: human milk oligosaccharides: fuel for childhood obesity prevention Adv Nutr 11:35–40. doi: 10.1093/advances/nmz093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bode L. 2012. Human milk oligosaccharides: every baby needs a sugar mama. Glycobiology 22:1147–1162. doi: 10.1093/glycob/cws074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Plaza-Díaz J, Fontana L, Gil A. 2018. Human milk oligosaccharides and immune system development. Nutrients 10:1038. doi: 10.3390/nu10081038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhang S, Li T, Xie J, Zhang D, Pi C, Zhou L, Yang W. 2021. Gold standard for nutrition: a review of human milk oligosaccharide and its effects on infant gut microbiota. Microb Cell Fact 20:108. doi: 10.1186/s12934-021-01599-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rosa F, Sharma AK, Gurung M, Casero D, Matazel K, Bode L, Simecka C, Elolimy AA, Tripp P, Randolph C, Hand TW, Williams KD, LeRoith T, Yeruva L. 2022. Human milk oligosaccharides impact cellular and inflammatory gene expression and immune response. Front Immunol 13:907529. doi: 10.3389/fimmu.2022.907529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Newburg DS, Ruiz-Palacios GM, Morrow AL. 2005. Human milk glycans protect infants against enteric pathogens. Annu Rev Nutr 25:37–58. doi: 10.1146/annurev.nutr.25.050304.092553 [DOI] [PubMed] [Google Scholar]
- 9. Menzel P, Vogel M, Austin S, Sprenger N, Grafe N, Hilbert C, Jurkutat A, Kiess W, Binia A. 2021. Concentrations of oligosaccharides in human milk and child growth. BMC Pediatr 21:481. doi: 10.1186/s12887-021-02953-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kunz C, Meyer C, Collado MC, Geiger L, García-Mantrana I, Bertua-Ríos B, Martínez-Costa C, Borsch C, Rudloff S. 2017. Influence of gestational age, secretor, and Lewis blood group status on the oligosaccharide content of human milk. J Pediatr Gastroenterol Nutr 64:789–798. doi: 10.1097/MPG.0000000000001402 [DOI] [PubMed] [Google Scholar]
- 11. Han SM, Derraik JGB, Binia A, Sprenger N, Vickers MH, Cutfield WS. 2021. Maternal and infant factors influencing human milk oligosaccharide composition: beyond maternal genetics. J Nutr 151:1383–1393. doi: 10.1093/jn/nxab028 [DOI] [PubMed] [Google Scholar]
- 12. Spevacek AR, Smilowitz JT, Chin EL, Underwood MA, German JB, Slupsky CM. 2015. Infant maturity at birth reveals minor differences in the maternal milk metabolome in the first month of lactation. J Nutr 145:1698–1708. doi: 10.3945/jn.115.210252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Thurl S, Munzert M, Boehm G, Matthews C, Stahl B. 2017. Systematic review of the concentrations of oligosaccharides in human milk. Nutr Rev 75:920–933. doi: 10.1093/nutrit/nux044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. McGuire MK, Meehan CL, McGuire MA, Williams JE, Foster J, Sellen DW, Kamau-Mbuthia EW, Kamundia EW, Mbugua S, Moore SE, Prentice AM, Kvist LJ, Otoo GE, Brooker SL, Price WJ, Shafii B, Placek C, Lackey KA, Robertson B, Manzano S, Ruíz L, Rodríguez JM, Pareja RG, Bode L. 2017. What's normal? Oligosaccharide concentrations and profiles in milk produced by healthy women vary geographically. Am J Clin Nutr 105:1086–1100. doi: 10.3945/ajcn.116.139980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Soyyılmaz B, Mikš MH, Röhrig CH, Matwiejuk M, Meszaros-Matwiejuk A, Vigsnæs LK. 2021. The mean of milk: a review of human milk oligosaccharide concentrations throughout lactation. Nutrients 13:2737. doi: 10.3390/nu13082737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Smith-Brown P, Morrison M, Krause L, Davies PSW. 2016. Mothers secretor status affects development of childrens microbiota composition and function: a pilot study. PLoS One 11:e0161211. doi: 10.1371/journal.pone.0161211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fan Y, Vinjamuri A, Tu D, Lebrilla CB, Donovan SM. 2023. Determinants of human milk oligosaccharides profiles of participants in the STRONG kids 2 cohort. Front Nutr 10:1105668. doi: 10.3389/fnut.2023.1105668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wu J, Wu S, Huo J, Ruan H, Xu X, Hao Z, Wei Y. 2020. Systematic characterization and longitudinal study reveal distinguishing features of human milk oligosaccharides in China. Curr Dev Nutr 4:zaa113. doi: 10.1093/cdn/nzaa113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liu F, Yan J, Wang X, Wang C, Chen L, Li Y, Chen J, Guo H. 2021. Maternal fucosyltransferase 2 status associates with the profiles of human milk oligosaccharides and the fecal microbiota composition of breastfed infants. J. Agric. Food Chem 69:3032–3043. doi: 10.1021/acs.jafc.0c04575 [DOI] [PubMed] [Google Scholar]
- 20. Lewis ZT, Totten SM, Smilowitz JT, Popovic M, Parker E, Lemay DG, Van Tassell ML, Miller MJ, Jin Y-S, German JB, Lebrilla CB, Mills DA. 2015. Maternal fucosyltransferase 2 status affects the gut bifidobacterial communities of breastfed infants. Microbiome 3:13. doi: 10.1186/s40168-015-0071-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wacklin P, Mäkivuokko H, Alakulppi N, Nikkilä J, Tenkanen H, Räbinä J, Partanen J, Aranko K, Mättö J. 2011. Secretor genotype (FUT2 gene) is strongly associated with the composition of Bifidobacteria in the human intestine. PLoS One 6:e20113. doi: 10.1371/journal.pone.0020113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Xiao L, Leusink-Muis T, Kettelarij N, van Ark I, Blijenberg B, Hesen NA, Stahl B, Overbeek SA, Garssen J, Folkerts G, van’t Land B. 2018. Human milk oligosaccharide 2'-Fucosyllactose improves innate and adaptive immunity in an influenza-specific murine vaccination model. Front Immunol 9:452. doi: 10.3389/fimmu.2018.00452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Morrow AL, Ruiz-Palacios GM, Altaye M, Jiang X, Guerrero ML, Meinzen-Derr JK, Farkas T, Chaturvedi P, Pickering LK, Newburg DS. 2004. Human milk oligosaccharides are associated with protection against diarrhea in breast-fed infants. J Pediatr 145:297–303. doi: 10.1016/j.jpeds.2004.04.054 [DOI] [PubMed] [Google Scholar]
- 24. Good M, Sodhi CP, Yamaguchi Y, Jia H, Lu P, Fulton WB, Martin LY, Prindle T, Nino DF, Zhou Q, Ma C, Ozolek JA, Buck RH, Goehring KC, Hackam DJ. 2016. The human milk oligosaccharide 2'-fucosyllactose attenuates the severity of experimental necrotising enterocolitis by enhancing mesenteric perfusion in the neonatal intestine. Br J Nutr 116:1175–1187. doi: 10.1017/S0007114516002944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lynch CM, Nagpal J, Luczynski P, Neufeld KM, Dinan TG, Clarke G, Cryan JF. 2024. Germ-free animals: a key tool in unraveling how the microbiota affects the brain and behavior, p 401–454. In Niall Hyland CS (ed), The Gut-Brain axis, 2nd ed. Academic Press. [Google Scholar]
- 26. Azad MB, Robertson B, Atakora F, Becker AB, Subbarao P, Moraes TJ, Mandhane PJ, Turvey SE, Lefebvre DL, Sears MR, Bode L. 2018. Human milk oligosaccharide concentrations are associated with multiple fixed and modifiable maternal characteristics, environmental factors, and feeding practices. J Nutr 148:1733–1742. doi: 10.1093/jn/nxy175 [DOI] [PubMed] [Google Scholar]
- 27. Rosa F, Zybailov BL, Glazko GV, Rahmatallah Y, Byrum S, Mackintosh SG, Bowlin AK, Yeruva L. 2021. Milk formula diet alters bacterial and host protein profile in comparison to human milk diet in neonatal piglet model. Nutrients 13:3718. doi: 10.3390/nu13113718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Saraf MK, Piccolo BD, Bowlin AK, Mercer KE, LeRoith T, Chintapalli SV, Shankar K, Badger TM, Yeruva L. 2017. Formula diet driven microbiota shifts tryptophan metabolism from serotonin to tryptamine in neonatal porcine colon. Microbiome 5:77. doi: 10.1186/s40168-017-0297-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Odiase E, Frank DN, Young BE, Robertson CE, Kofonow JM, Davis KN, Berman LM, Krebs NF, Tang M. 2023. The gut microbiota differ in exclusively breastfed and formula-Fed United States infants and are associated with growth status. J Nutr 153:2612–2621. doi: 10.1016/j.tjnut.2023.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Boudry G, Charton E, Le Huerou-Luron I, Ferret-Bernard S, Le Gall S, Even S, Blat S. 2021. The relationship between breast milk components and the infant gut microbiota. Front Nutr 8:629740. doi: 10.3389/fnut.2021.629740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bai Y, Tao J, Zhou J, Fan Q, Liu M, Hu Y, Xu Y, Zhang L, Yuan J, Li W, Ze X, Malard P, Guo Z, Yan J, Li M. 2018. Fucosylated human milk oligosaccharides and N-glycans in the milk of Chinese mothers regulate the gut microbiome of their breast-fed infants during different Lactation stages. mSystems 3:e00206-18. doi: 10.1128/mSystems.00206-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Laursen MF, Pekmez CT, Larsson MW, Lind MV, Yonemitsu C, Larnkjær A, Mølgaard C, Bode L, Dragsted LO, Michaelsen KF, Licht TR, Bahl MI. 2021. Maternal milk microbiota and oligosaccharides contribute to the infant gut microbiota assembly. ISME Commun 1:21. doi: 10.1038/s43705-021-00021-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chia LW, Mank M, Blijenberg B, Aalvink S, Bongers RS, Stahl B, Knol J, Belzer C. 2020. Bacteroides thetaiotaomicron fosters the growth of butyrate-producing Anaerostipes caccae in the presence of lactose and total human milk carbohydrates. Microorganisms 8:1513. doi: 10.3390/microorganisms8101513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, Collini S, Pieraccini G, Lionetti P. 2010. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci U S A 107:14691–14696. doi: 10.1073/pnas.1005963107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Seppo AE, Bu K, Jumabaeva M, Thakar J, Choudhury RA, Yonemitsu C, Bode L, Martina CA, Allen M, Tamburini S, Piras E, Wallach DS, Looney RJ, Clemente JC, Järvinen KM. 2021. Infant gut microbiome is enriched with Bifidobacterium longum ssp. infantis in old order mennonites with traditional farming lifestyle. Allergy 76:3489–3503. doi: 10.1111/all.14877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jackson CM, Mahmood MM, Järvinen KM. 2022. Farming lifestyle and human milk: modulation of the infant microbiome and protection against allergy. Acta Paediatr 111:54–58. doi: 10.1111/apa.16147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dhakal S, Wang L, Antony L, Rank J, Bernardo P, Ghimire S, Bondra K, Siems C, Lakshmanappa YS, Renu S, Hogshead B, Krakowka S, Kauffman M, Scaria J, LeJeune JT, Yu Z, Renukaradhya GJ. 2019. Amish (rural) vs. non-Amish (urban) infant fecal microbiotas are highly diverse and their transplantation lead to differences in mucosal immune maturation in a humanized germfree piglet model. Front Immunol 10:1509. doi: 10.3389/fimmu.2019.01509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ma J, Li Z, Zhang W, Zhang C, Zhang Y, Mei H, Zhuo N, Wang H, Wang L, Wu D. 2020. Comparison of gut microbiota in exclusively breast-fed and formula-fed babies: a study of 91 term infants. Sci Rep 10:15792. doi: 10.1038/s41598-020-72635-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Brink LR, Matazel K, Piccolo BD, Bowlin AK, Chintapalli SV, Shankar K, Yeruva L. 2019. Neonatal diet impacts bioregional microbiota composition in piglets fed human breast milk or infant formula. J Nutr 149:2236–2246. doi: 10.1093/jn/nxz170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Charton E, Bourgeois A, Bellanger A, Le-Gouar Y, Dahirel P, Romé V, Randuineau G, Cahu A, Moughan PJ, Montoya CA, Blat S, Dupont D, Deglaire A, Le Huërou-Luron I. 2022. Infant nutrition affects the microbiota-gut-brain axis: comparison of human milk vs. infant formula feeding in the piglet model. Front Nutr 9:976042. doi: 10.3389/fnut.2022.976042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Borewicz K, Suarez-Diez M, Hechler C, Beijers R, de Weerth C, Arts I, Penders J, Thijs C, Nauta A, Lindner C, Van Leusen E, Vaughan EE, Smidt H. 2019. The effect of prebiotic fortified infant formulas on microbiota composition and dynamics in early life. Sci Rep 9:2434. doi: 10.1038/s41598-018-38268-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Liu X, Mao B, Gu J, Wu J, Cui S, Wang G, Zhao J, Zhang H, Chen W. 2021. Blautia-a new functional genus with potential probiotic properties? Gut Microbes 13:1–21. doi: 10.1080/19490976.2021.1875796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Horigome A, Hashikura N, Yoshida K, Xiao JZ, Odamaki T. 2022. 2'-fucosyllactose increases the abundance of Blautia in the presence of extracellular fucosidase-possessing bacteria. Front Microbiol 13:913624. doi: 10.3389/fmicb.2022.913624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kostopoulos I, Elzinga J, Ottman N, Klievink JT, Blijenberg B, Aalvink S, Boeren S, Mank M, Knol J, de Vos WM, Belzer C. 2020. Akkermansia muciniphila uses human milk oligosaccharides to thrive in the early life conditions in vitro. Sci Rep 10:14330. doi: 10.1038/s41598-020-71113-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Luna E, Parkar SG, Kirmiz N, Hartel S, Hearn E, Hossine M, Kurdian A, Mendoza C, Orr K, Padilla L, Ramirez K, Salcedo P, Serrano E, Choudhury B, Paulchakrabarti M, Parker CT, Huynh S, Cooper K, Flores GE. 2022. Utilization efficiency of human milk oligosaccharides by human-associated Akkermansia is strain dependent. Appl Environ Microbiol 88. doi: 10.1128/AEM.01487-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Barnett DJM, Endika MF, Klostermann CE, Gu F, Thijs C, Nauta A, Schols HA, Smidt H, Arts ICW, Penders J. 2023. Human milk oligosaccharides, antimicrobial drugs, and the gut microbiota of term neonates: observations from the KOALA birth cohort study. Gut Microbes 15:2164152. doi: 10.1080/19490976.2022.2164152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lin L, Zhang J. 2017. Role of intestinal microbiota and metabolites on gut homeostasis and human diseases. BMC Immunol 18:2. doi: 10.1186/s12865-016-0187-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Vicentini FA, Keenan CM, Wallace LE, Woods C, Cavin JB, Flockton AR, Macklin WB, Belkind-Gerson J, Hirota SA, Sharkey KA. 2021. Intestinal microbiota shapes gut physiology and regulates enteric neurons and glia. Microbiome 9:210. doi: 10.1186/s40168-021-01165-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Azagra-Boronat I, Massot-Cladera M, Mayneris-Perxachs J, Knipping K, Van’t Land B, Tims S, Stahl B, Garssen J, Franch À, Castell M, Rodríguez-Lagunas MJ, Pérez-Cano FJ. 2019. Immunomodulatory and prebiotic effects of 2'-fucosyllactose in suckling rats. Front Immunol 10:1773. doi: 10.3389/fimmu.2019.01773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Cilieborg MS, Sangild PT, Jensen ML, Østergaard MV, Christensen L, Rasmussen SO, Mørbak AL, Jørgensen CB, Bering SB. 2017. Alpha1,2-fucosyllactose does not improve intestinal function or prevent Escherichia coli F18 diarrhea in newborn pigs. J Pediatr Gastroenterol Nutr 64:310–318. doi: 10.1097/MPG.0000000000001276 [DOI] [PubMed] [Google Scholar]
- 51. Olszak T, An D, Zeissig S, Vera MP, Richter J, Franke A, Glickman JN, Siebert R, Baron RM, Kasper DL, Blumberg RS. 2012. Microbial exposure during early life has persistent effects on natural killer T cell function. Science 336:489–493. doi: 10.1126/science.1219328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hapfelmeier S, Lawson MAE, Slack E, Kirundi JK, Stoel M, Heikenwalder M, Cahenzli J, Velykoredko Y, Balmer ML, Endt K, Geuking MB, Curtiss R, McCoy KD, Macpherson AJ. 2010. Reversible microbial colonization of germ-free mice reveals the dynamics of IgA immune responses. Science 328:1705–1709. doi: 10.1126/science.1188454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kong C, Elderman M, Cheng L, de Haan BJ, Nauta A, de Vos P. 2019. Modulation of intestinal epithelial glycocalyx development by human milk oligosaccharides and non-digestible carbohydrates. Mol Nutr Food Res 63:e1900303. doi: 10.1002/mnfr.201900303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Comstock SS, Li M, Wang M, Monaco MH, Kuhlenschmidt TB, Kuhlenschmidt MS, Donovan SM. 2017. Dietary human milk oligosaccharides but not prebiotic oligosaccharides increase circulating natural killer cell and mesenteric lymph node memory T cell populations in noninfected and rotavirus-infected neonatal piglets. J Nutr 147:1041–1047. doi: 10.3945/jn.116.243774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Round JL, Mazmanian SK. 2010. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci U S A 107:12204–12209. doi: 10.1073/pnas.0909122107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Telesford KM, Yan W, Ochoa-Reparaz J, Pant A, Kircher C, Christy MA, Begum-Haque S, Kasper DL, Kasper LH. 2015. A commensal symbiotic factor derived from Bacteroides fragilis promotes human CD39+Foxp3+ T cells and Treg function. Gut Microbes 6:234–242. doi: 10.1080/19490976.2015.1056973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Konieczna P, Groeger D, Ziegler M, Frei R, Ferstl R, Shanahan F, Quigley EMM, Kiely B, Akdis CA, O’Mahony L. 2012. Bifidobacterium infantis 35624 administration induces Foxp3 T regulatory cells in human peripheral blood: potential role for myeloid and plasmacytoid dendritic cells. Gut 61:354–366. doi: 10.1136/gutjnl-2011-300936 [DOI] [PubMed] [Google Scholar]
- 58. Verma R, Lee C, Jeun EJ, Yi J, Kim KS, Ghosh A, Byun S, Lee CG, Kang HJ, Kim GC, Jun CD, Jan G, Suh CH, Jung JY, Sprent J, Rudra D, Castro C, Molinaro A, Surh CD, Im SH. 2018. Cell surface polysaccharides of Bifidobacterium bifidum induce the generation of Foxp3+ regulatory T cells. Sci Immunol 3:eaat6975. doi: 10.1126/sciimmunol.aat6975 [DOI] [PubMed] [Google Scholar]
- 59. Kumar V. 2014. Innate lymphoid cells: new paradigm in immunology of inflammation. Immunol Lett 157:23–37. doi: 10.1016/j.imlet.2013.11.003 [DOI] [PubMed] [Google Scholar]
- 60. Ayechu-Muruzabal V, Overbeek SA, Kostadinova AI, Stahl B, Garssen J, Van’t Land B, Willemsen LEM. 2020. Exposure of intestinal epithelial cells to 2'-fucosyllactose and CpG enhances galectin release and instructs dendritic cells to drive Th1 and regulatory-type immune development. Biomolecules 10:784. doi: 10.3390/biom10050784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Tsuji M, Suzuki K, Kitamura H, Maruya M, Kinoshita K, Ivanov II, Itoh K, Littman DR, Fagarasan S. 2008. Requirement for lymphoid tissue-inducer cells in isolated follicle formation and T cell-independent immunoglobulin A generation in the gut. Immunity 29:261–271. doi: 10.1016/j.immuni.2008.05.014 [DOI] [PubMed] [Google Scholar]
- 62. Rousseaux A, Brosseau C, Le Gall S, Piloquet H, Barbarot S, Bodinier M. 2021. Human milk oligosaccharides: their effects on the host and their potential as therapeutic agents. Front Immunol 12:680911. doi: 10.3389/fimmu.2021.680911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Al Nabhani Z, Dulauroy S, Marques R, Cousu C, Al Bounny S, Déjardin F, Sparwasser T, Bérard M, Cerf-Bensussan N, Eberl G. 2019. A weaning reaction to microbiota is required for resistance to immunopathologies in the adult. Immunity 50:1276–1288. doi: 10.1016/j.immuni.2019.02.014 [DOI] [PubMed] [Google Scholar]
- 64. Knoop KA, Gustafsson JK, McDonald KG, Kulkarni DH, Coughlin PE, McCrate S, Kim D, Hsieh CS, Hogan SP, Elson CO, Tarr PI, Newberry RD. 2017. Microbial antigen encounter during a preweaning interval is critical for tolerance to gut bacteria. Sci Immunol 2:eaao1314. doi: 10.1126/sciimmunol.aao1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Cosovanu C, Neumann C. 2020. The many functions of Foxp3+ regulatory T cells in the intestine. Front Immunol 11:600973. doi: 10.3389/fimmu.2020.600973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Dinleyici M, Barbieur J, Dinleyici EC, Vandenplas Y. 2023. Functional effects of human milk oligosaccharides (HMOs). Gut Microbes 15:2186115. doi: 10.1080/19490976.2023.2186115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Abrams GD, Bauer H, Sprinz H. 1963. Influence of the normal flora on mucosal morphology and cellular renewal in the ileum. A comparison of germ-free and conventional mice. Lab Invest 12:355–364. [PubMed] [Google Scholar]
- 68. Hooper LV, Littman DR, Macpherson AJ. 2012. Interactions between the microbiota and the immune system. Science 336:1268–1273. doi: 10.1126/science.1223490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Jantscher-Krenn E, Zherebtsov M, Nissan C, Goth K, Guner YS, Naidu N, Choudhury B, Grishin AV, Ford HR, Bode L. 2012. The human milk oligosaccharide disialyllacto-N-tetraose prevents necrotising enterocolitis in neonatal rats. Gut 61:1417–1425. doi: 10.1136/gutjnl-2011-301404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Tuncil YE, Thakkar RD, Marcia ADR, Hamaker BR, Lindemann SR. 2018. Divergent short-chain fatty acid production and succession of colonic microbiota arise in fermentation of variously-sized wheat bran fractions. Sci Rep 8:16655. doi: 10.1038/s41598-018-34912-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Walters W, Hyde ER, Berg-Lyons D, Ackermann G, Humphrey G, Parada A, Gilbert JA, Jansson JK, Caporaso JG, Fuhrman JA, Apprill A, Knight R. 2016. Improved bacterial 16S rRNA gene (V4 and V4-5) and fungal internal transcribed spacer marker gene primers for microbial community surveys. mSystems 1:e00009-15. doi: 10.1128/mSystems.00009-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. doi: 10.1128/AEM.01541-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Yao T, Chen MH, Lindemann SR. 2020. Structurally complex carbohydrates maintain diversity in gut-derived microbial consortia under high dilution pressure. FEMS Microbiol Ecol 96:fiaa158. doi: 10.1093/femsec/fiaa158 [DOI] [PubMed] [Google Scholar]
- 74. Cole JK, Hutchison JR, Renslow RS, Kim Y-M, Chrisler WB, Engelmann HE, Dohnalkova AC, Hu D, Metz TO, Fredrickson JK, Lindemann SR. 2014. Phototrophic biofilm assembly in microbial-mat-derived unicyanobacterial consortia: model systems for the study of autotroph-heterotroph interactions. Front Microbiol 5:109. doi: 10.3389/fmicb.2014.00109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Westcott SL, Schloss PD. 2017. OptiClust, an improved method for assigning amplicon-based sequence data to operational taxonomic units. mSphere 2:e00073-17. doi: 10.1128/mSphereDirect.00073-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Lu Y, Zhou G, Ewald J, Pang Z, Shiri T, Xia J. 2023. MicrobiomeAnalyst 2.0: comprehensive statistical, functional and integrative analysis of microbiome data. Nucleic Acids Res 51:W310–W318. doi: 10.1093/nar/gkad407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data. Available from: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
- 78. Felix Krueger FJ, Ewels P, Afyounian E, Weinstein M, Schuster-Boeckler B, Hulselmans G. 2023. FelixKrueger/TrimGalore: v0.6.10 - DOI via zenodo (0.6.10). Zenodo. Available from: 10.5281/zenodo.7598955 [DOI] [Google Scholar]
- 79. Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29:15–21. doi: 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Liao Y, Smyth GK, Shi W. 2014. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30:923–930. doi: 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- 81. Team RC. 2022. R: a language and environment for statistical computing, on R foundation for statistical computing, Vienna, Austria. https://www.R-project.org/.
- 82. Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15. doi: 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Yu G, Wang LG, Han Y, He QY. 2012. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS: J Integr Biol 16:284–287. doi: 10.1089/omi.2011.0118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102:15545–15550. doi: 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P. 2015. The molecular signatures database hallmark gene set collection. Cell Syst 1:417–425. doi: 10.1016/j.cels.2015.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Bates D, Mächler M, Bolker BM, Walker SC. 2015. Fitting linear mixed-effects models using Lme4. J Stat Softw 67:1–48. doi: 10.18637/jss.v067.i01 [DOI] [Google Scholar]
- 87. Russell VL. 2022. emmeans: estimated marginal means, aka least-squares means. R package version 1.8.3. https://CRAN.R-project.org/package=emmeans.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figures S1 to S7.
Changes in body weight, organ weight, and intestinal length.
Differentially expressed genes in the distal small intestine (ileum).
Differentially expressed genes in the large intestine (colon).
Unique or common differentially expressed genes in the distal small intestine (ileum).
Unique or common differentially expressed genes in the large intestine (colon).
Unique or common pathways in the distal small intestine (ileum) or large intestine (colon).
Antibodies used for immune cell for flow cytometry.
Data Availability Statement
Sequencing data are deposited under BioProject accession number PRJNA1081588 (16S rRNA) and GEO accession number GSE260491 (RNA-sequencing).