Abstract
Background:
Chronic lung allograft dysfunction in lung transplant recipients (LTxRs) has two phenotypes: obstructive bronchiolitis obliterans syndrome (BOS) and restrictive allograft syndrome (RAS). Our goal was to define distinct immunologic markers of exosomes from LTxRs with BOS or RAS.
Methods:
Plasma was collected from LTxRs with BOS (n=18), RAS (n=13), and from stable LTxRs (n=5). Antibodies to lung self-antigens (SAgs) were determined by ELISA. Exosomes were isolated by ultracentrifugation. Donor specific antibodies to HLA were quantified using Luminex. Exosomes were characterized for lung SAgs, transcription factors, 20S proteasome, HLA class I and II, and polymeric immunoglobulin receptor protein using western blot. Exosome miRNA was analyzed using NanoString. The exosome-induced immune response was determined in mice.
Results:
LTxRs with RAS, but not BOS, had donor specific antibodies at diagnosis. CIITA, NFkB, polymeric immunoglobulin receptor protein, 20S proteasome, HLA-DQ, and HLA-DR were significantly higher in RAS exosomes than in BOS exosomes. RAS plasma had high levels of proinflammatory cytokines and distinct exosomal miRNA. Immunization of C57BL/6 mice with RAS exosomes showed severe inflammation and peribronchial fibrosis, whereas BOS exosomes induced patchy inflammation and fibrosis.
Conclusion:
LTxRs with BOS or RAS had exosomes with distinct molecular and immunologic profiles. RAS samples had a higher concentration of proinflammatory factors, HLA class II, lung SAgs, and antibodies to HLA class II molecules, indicating severe allograft injury. Mice immunized with RAS exosomes developed lesions in airways, pleura, interlobular septum, and alveoli, whereas BOS exosomes induced mild to patchy inflammation with lung fibrosis.
Keywords: Lung transplantation, restrictive allograft syndrome, bronchiolitis obliterans syndrome, exosomes
Introduction
Lung transplantation (LTx) is a life-saving intervention for patients with end-stage lung disease. However, long-term survival is limited by chronic lung allograft dysfunction (CLAD).1 CLAD is an umbrella term to describe the clinical manifestations of a range of pathologic processes in the airway and parenchyma of the lung allograft that leads to deterioration in lung function after LTx.2 CLAD is sub-classified into two distinct clinical entities: restrictive allograft syndrome (RAS) and bronchiolitis obliterans syndrome (BOS), the most common form of CLAD.3, 4 RAS is characterized by pleuroparenchymal fibrosis and a more aggressive clinical course.5, 6
Exosomes are small membrane-bound endosomal vesicles, measuring <200 nm in diameter, released into circulation from cells.7, 8 Exosomes are involved in intercellular communication, with critical roles for several physiological processes including stress and inflammation.9 We have shown that lung transplant recipients (LTxRs) with BOS have increased concentrations of exosomes containing the lung self-antigens (SAgs) Kα1 tubulin (Kα1T) and collagen V (Col-V).10, 11 The goal of this study was to identify distinct molecular and immunologic features involved in inflammation, induction of major histocompatibility complex (MHC) molecules, and antibody synthesis in exosomes isolated from LTxRs with RAS (RAS exosomes) or BOS (BOS exosomes). Toward this, we measured the proinflammatory transcription factor nuclear factor-κB (NFkB), the MHC regulating trans-coactivator CIITA, human leukocyte antigen (HLA) class I, HLA-DR, HLA-DQ, 20S proteasome, and polymeric immunoglobulin receptor protein (PIGR) as well as the ability of RAS and BOS exosomes to induce immune responses in mice.
Methods
Study population
We retrospectively analyzed 18 LTxRs with BOS, 13 with RAS and 5 time-matched stable LTxRs (controls). All patients underwent LTx at St. Joseph’s Hospital and Medical Center, Phoenix, Arizona, had plasma available, and consented to participate. The Institutional Review Board approved this study (IRB# PHXB16-0027-10-18). Patient demographics, transplant, and laboratory data were collected via chart abstraction (Table S1). CLAD (BOS or RAS) was diagnosed by spirometry and radiographic findings according to the International Society for Heart and Lung Transplantation guidelines.2, 6
Exosome isolation and validation
Exosomes were isolated from plasma at the time of RAS or BOS diagnosis by the ultracentrifugation method previously described,12 followed by 0.22-micron filtration. Exosome purity and size were determined by NanoSight (Malveran NS300).13
Characterization of exosomes using western blot
Total exosome protein (15μg) was resolved in polyacrylamide gel electrophoresis and transferred into a polyvinylidene difluoride membrane. The membrane was blocked with 5% BSA in 1x phosphate buffered saline and was probed with exosome- marker CD9 (312102, BioLegend), collagen V (Abcam), and Ka1 tubulin (sc-12462-R, Santa Cruz Biotechnology). 20S proteasome subunit a3 (sc-58414, Santa Cruz Biotechnology), HLA-class I (Abcam), HLA-DQ (Abcam), HLA-DR (Abcam), NFkB (Cell Signaling Technologies), CIITA (Abcam), and PIGR (R&D Systems) were measured using specific antibodies (Abs). Binding was determined using secondary Abs conjugated with horseradish peroxidase. The blots were washed with PBS Tween (Thermo Fisher Scientific), developed using chemiluminescent horseradish peroxidase substrate (WBKLS0500, Millipore Sigma), and exposed using the Odyssey CLx Imaging System (LICOR Biosciences). The band intensity of target protein was quantified using ImageJ software and normalized with CD9.
microRNA (miRNA) analysis of isolated exosomes
The NanoString nCounter Human miRNA Expression Assay Kit (NanoString) was used to profile 800 human miRNAs. miRNAs were analyzed with NanoString assays and quantified after background correction and median normalization.14 See Supplement for experimental details and analysis.
Determination of Abs to lung SAgs
The presence of Abs to lung SAgs (Col-V, Kα1T) was detected using ELISA as previously described.15
Determination of donor specific antibodies (DSA) to HLA
HLA antibodies were analyzed with Luminex single antigen beads (R&D Systems) at the time of RAS or BOS diagnosis and also at 6-, 12-, and 24-months post-transplant.
Measurement of cytokines
The Magnetic Luminex assay kit (Invitrogen) was used according to the manufacturer’s protocol. Briefly, 50μl of human serum (1:50 dilution) was added to the multiplex beads coated in the wells and incubated for 2 hours at room temperature. Cytokines with known concentrations were used as standards. The cytokines bound in the beads were detected using biotinylated anti-human multicytokine reporter and streptavidin-PE detection. The plate was washed and read using Bio-Plex Luminex (Bio-Rad Laboratories) and cytokine concentrations were measured using a standard curve (mean fluorescence intensity).
Immunization of mice with exosomes and development of Abs to lung SAgs (Kα1T, Col-V)
Three groups of C57BL/6 mice were immunized with: (i) saline control (n=3), (ii) BOS exosomes (n=3), and (iii) RAS exosomes (n=3). Details of immunizations and detection of Abs to Kα1T and Col-V were previously described.16, 17
Enzyme-linked immunospot (EliSPOT)
Cellular immune responses and the number of cells secreting cytokines specific to Kα1T and Col-V were enumerated using EliSPOT (R&D Systems). Splenocytes from mice immunized with exosomes or control were isolated by Ficoll-Hypaque gradient centrifugation and analyzed by EliSPOT .18
Histology and immunohistochemistry
Mice were euthanized and lungs were harvested, fixed in neutralized 10% formalin, and embedded in paraffin blocks. For trichrome staining, 4- to 5-mm thick sections were cut and mounted on slides. Slides were scanned and analyzed using Aperio Image (Leica Biosystems).16
Statistical analysis
Data was analyzed using Prism 8.0 software (GraphPad, Inc.). The Ab levels for lung SAgs, optical density of exosomes containing lung SAgs, and cytokine levels in plasma from LTxRs in stable condition (controls) and LTxRs with BOS or RAS were compared using unpaired student’s t-test. Data was expressed as mean and standard error. P-values <0.05 were considered statistically significant. Statistical significance of the change in exosome protein abundance between each groups (stable LTxRs vs RAS, stable LTxRs vs BOS and BOS vs RAS for all 9 exosome proteins) was determined using student’s t-test.
A Benjamini-Hochberg test for multiple correction test was performed on our group of interest for all exosome proteins i.e. BOS vs RAS LTxR (Table S2).
Antibody levels and cytokine production in splenocytes were compared using student’s t-test in different groups of mice.
Results
Lung transplant recipients
Patient demographics including age, sex, ethnicity, and underlying diagnosis were not significantly different between LTxRs diagnosed with RAS or BOS (Table S1). All patients had sufficiently long survival to have a certain CLAD phenotype (BOS/RAS) and an impact on the analysis (i.e., 1 year). Samples selected for BOS and RAS patients are at the time of diagnosis. All BOS samples selected for the current study fall within 1-3 years of transplant after the diagnosis of BOS (>1 year of transplant). All the RAS samples selected for study fall within one year of transplant at the time of diagnosis of RAS 6-12 months of transplant. Time matched samples were used as stable. In both cohorts (BOS/RAS), patients with a persistent decrease in FEV1 of 10-20% in the absence of other causes as per ISHLT guidelines.2 At the time of diagnosis pulmonary function testing FEV1 (liters) was 1.36±0.65 for BOS cohort, 1.16±0.56 for RAS cohort Table S1.
Characterization of exosomes
The mean size of the paricles used in our study was <200 nm, in agreement with the exosome size described by the International Society for Extracellular Vesicles.19 A representative NanoSight image is provided in Figure S1.
Development of Abs to Col-V and Kα1T
Antibodies to Col-V were detected in 4 of 18 LTxRs with BOS (mean concentration, 126±13.74 ng/ml). Antibodies to Kα1T were detected in 2 of 18 LTxRs with BOS (both also had Abs to Col-V; mean concentration, 130.0±13.52 ng/ml). Antibodies to Col-V were detected in 2/13 RAS LTxR (143.0 ±1.41 ng/ml) and Kα1T was detected in 1/13 LTxRs with RAS (166.0ng/ml). None of the LTxRs in stable condition had significant levels of Abs to lung SAgs.
LTxRs with RAS developed DSA to mismatched donor HLA at time of diagnosis
LTxRs with RAS demonstrated persistent Abs to HLA class II (DQ, DR and DP). 12/13 LTxRs with RAS developed DSA within 6 months post-transplant and had detectable DSA to HLA class II at diagnosis. Eight of the LTxRs with RAS developed persistent Abs to HLA class II (DQ, DR and DP). 3/13 LTxRs with RAS also developed Abs to HLA class I at 12 months post-transplant. In contrast, none of the 18 LTxRs with BOS had DSA to HLA class I. However, 2 LTXRs with BOS developed DSA specific to HLA-DQ and HLA-DR at diagnosis (12 months post-transplant). Serial analysis of DSA in LTxRs with RAS or BOS is presented in Table 1.
Table 1.
Development of DSA to HLA class I and II in lung transplant recipients with RAS
| RAS LTxRs (n=13) |
Time post-transplant | |||||
|---|---|---|---|---|---|---|
| 6 months | 12 months | 24 months | ||||
| HLA Class I |
HLA Class II |
HLA Class I | HLA Class II |
HLA Class I | HLA Class II |
|
| LTxR 1 | NO DSA | DQ4, DPA1, DR4 | NO DSA | DQ4, DPA1, DR4 | NO DSA | DQ4, DPA1, DR4 |
| LTxR 2 | NO DSA | NO DSA | A02, A23 | NO DSA | NO DSA | NO DSA |
| LTxR 3 | NO DSA | DR52 | NO DSA | NO DSA | NO DSA | DR51, DR52, DQA1, DQ6 |
| LTxR 4 | NO DSA | DPA1, DRB1, DR4, DR16, DR10 | NO DSA | DPA1, DRB1 | NO DSA | NO DSA |
| LTxR 5 | NO DSA | DR12, DR4, DQ7 | NO DSA | DRB1, DR4, DR12, DQ7, DR52, DR58 | NO DSA | DR4, DQ7, DRB1 |
| LTxR 6 | NO DSA | DR53, DQA05 | A2, A24, A34, A66, A69, Cw9, Cw10 | DR04, DQA05, DR53, DP11 | NO DSA | DR04, DQA05, DR53, DP11 |
| LTxR 7 | NO DSA | DQA03 | NO DSA | DQA03 | NO DSA | DQA03 |
| LTxR 8 | NO DSA | DPA1, DR51 | NO DSA | DPA1, DR51, | NO DSA | DPA1, DR51 |
| LTxR 9 | Cw15, | DQ7, DRB3 | A68 | DQ7, DR52 | Deceased | Deceased |
| LTxR 10 | NO DSA | DQA 03 | Deceased | Deceased | ||
| LTxR 11 | NO DSA | DPA1 | NO DSA | NO DSA | Deceased | Deceased |
| LTxR 12 | NO DSA | DR4, DPA1 DQ4 | NO DSA | NO DSA | Deceased | Deceased |
| LTxR 13 | NO DSA | DR52, DRB2, DR1, DR17, DQ5 | NO DSA | DR52, DR1, DR17 | Deceased | Deceased |
| BOS LTxRs (n=12) | ||||||
| LTxR 1 | NO DSA | DQA3 | NO DSA | NO DSA | NO DSA | NO DSA |
| LTxR 2-7 | NO DSA | NO DSA | NO DSA | NO DSA | NO DSA | NO DSA |
| LTxR 8 | Cw15 | DQ7, DQA03, DQA05 | NO DSA | DQ7, DQA03, DQA05 | NO DSA | DQ7, DQA1, DR4 |
| LTxR 9 | NO DSA | NO DSA | NO DSA | NO DSA | Cw15 | DR4, DQ7, DRB3 |
| LTxR 10 | NO DSA | NO DSA | NO DSA | NO DSA | NO DSA | DPB1, DRB3, DQ7, DQ6 |
| LTxR 11 | A1, | DQA1, DR17 | NO DSA | DQA1, DQA2, D52, D17 | A1, Cw7 | DQA1, DQA2, D52, D17 |
| LTxR 12 | Cw5, A1, A11 | DQ6, DQA1, DQA2 | NO DSA | NO DSA | NO DSA | NO DSA |
| LTxR 13-18 | NO DSA | NO DSA | NO DSA | NO DSA | NO DSA | NO DSA |
All LTxRs are followed up to two years for development of DSA. All DSA development was measured using Luminex platform. Development of DSA to class I and II is less frequent in BOS as compared to RAS LTxRs.
BOS, bronchiolitis obliterans syndrome; DSA, donor specific antibodies; HLA, human leukocyte antigen; LTxR, lung transplant recipient; RAS, restrictive allograft syndrome.
Greater quantities of HLA-DR, HLA-DQ, NFkB, 20S proteasome, PIGR, and CIITA in RAS exosomes and increased levels of lung SAg (Kα1T) in BOS exosomes
The concentrations of HLA-DR and HLA-DQ were significantly higher in the exosomes isolated from LTxRs with RAS compared to BOS (p=.0002 and p<0.0001, respectively). RAS exosomes also contained higher levels of NFkB (p<0.0003), 20S proteasome (p<0.0001), PIGR (p<0.0001), and CIITA (p=0.1434) than BOS exosomes (Figure 1A, B; Table 2). HLA class I, detected using monoclonal Abs to framework determinant (W6/32), was higher on RAS exosomes, however, not statistically significant (Figure 2A, B; Table 2). The concentration of Kα1T was higher in in exosomes from LTxRs with BOS (Kα1T, p=0.0201) but Col-V was higher in RAS but not statistically significant (p=0.0537) (Figure 3A, B; Table 2).
Figure 1:
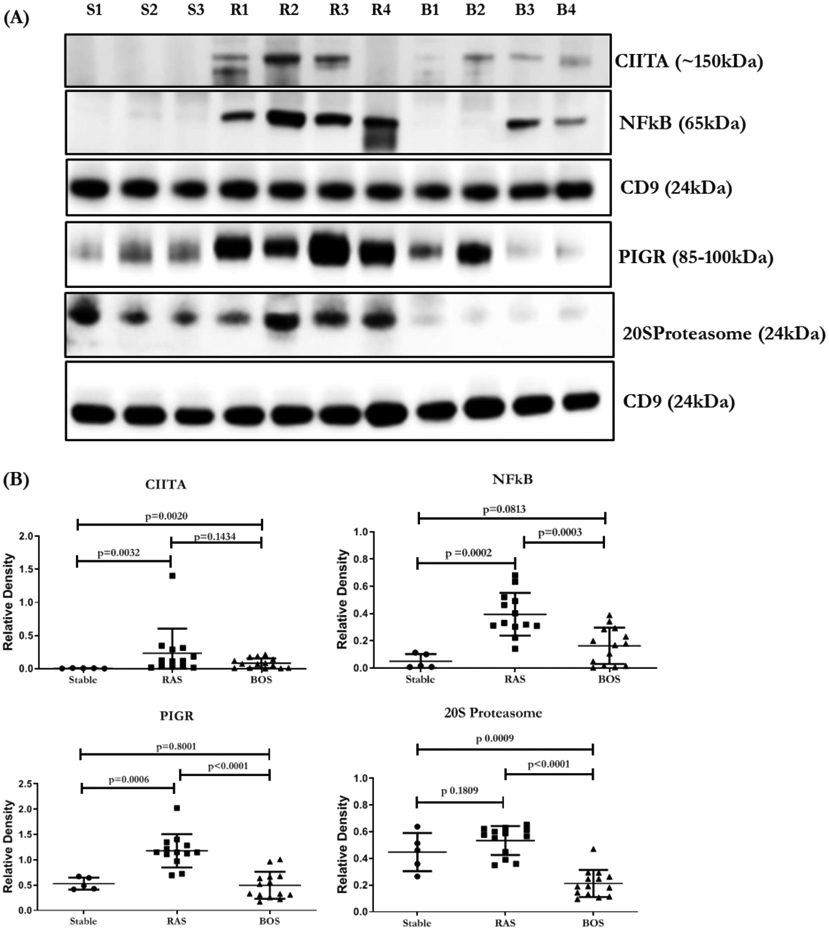
(A) Western blot of exosomal proteins, transcription factors CIITA, NFkB, 20S proteasome and PIGR, from LTxRs with stable LTxRs (S1-S3), or RAS (R1-R4) and BOS (B1-B4). (B) Densitometry and statistical analysis of western blots for stable n=5, RAS n=13 and BOS n=14. CD9 in the figure is used to normalize the blots for CIITA, NFkB, PIGR and 20S proteasome. All the graphs are represented as dot plots with Mean (Longest horizontal line) and SD (Vertical Bar Line). *p<0.05, **p<0.01 and ***p<0.001. BOS, bronchiolitis obliterans syndrome; RAS, restrictive allograft syndrome
Table 2.
Characterization of exosome contents from lung transplant recipients with BOS or RAS compared to stable recipients (controls without CLAD). Significantly different proteins (p<0.05) in BOS vs RAS after student’s t-test and multiple testing by Benjamini Hochberg correction.
| Exosomal proteins |
Stable (n=5) |
BOS (n=14) Fold change |
RAS (n=13) Fold change |
p values BOS vs RAS |
P-Adj (BH) |
|---|---|---|---|---|---|
| Collagen V | 1 | 2.44 | 3.84 | 0.0537 | 0.0690 |
| Kα1 tubulin | 1 | 1.85 | 1.22 | 0.0201 | 0.0301 |
| HLA-DQ | 1 | 1.43 | 3.35 | 0.0001 | 0.0003 |
| HLA-DR | 1 | 3.98 | 12.05 | 0.0002 | 0.00045 |
| HLA class I | 1 | 10.06 | 11.17 | 0.5127 | 0.5127 |
| NFkB | 1 | 3.33 | 8.08 | 0.0003 | 0.00054 |
| CIITA | 1 | 12.5 | 25.8 | 0.1434 | 0.16132 |
| PIGR | 1 | 0.93 | 2.22 | 0.0001 | 0.0003 |
| 20S proteasome | 1 | 0.47 | 1.19 | 0.0001 | 0.0003 |
All values are normalized to CD9 blots. Fold change values are compared to stable (controls without CLAD). p values and p-Adj values are represented after multiple correction test between BOS and RAS LTxR groups.
BOS, bronchiolitis obliterans syndrome; CLAD, chronic lung allograft dysfunction; RAS, restrictive allograft syndrome.
Figure 2:
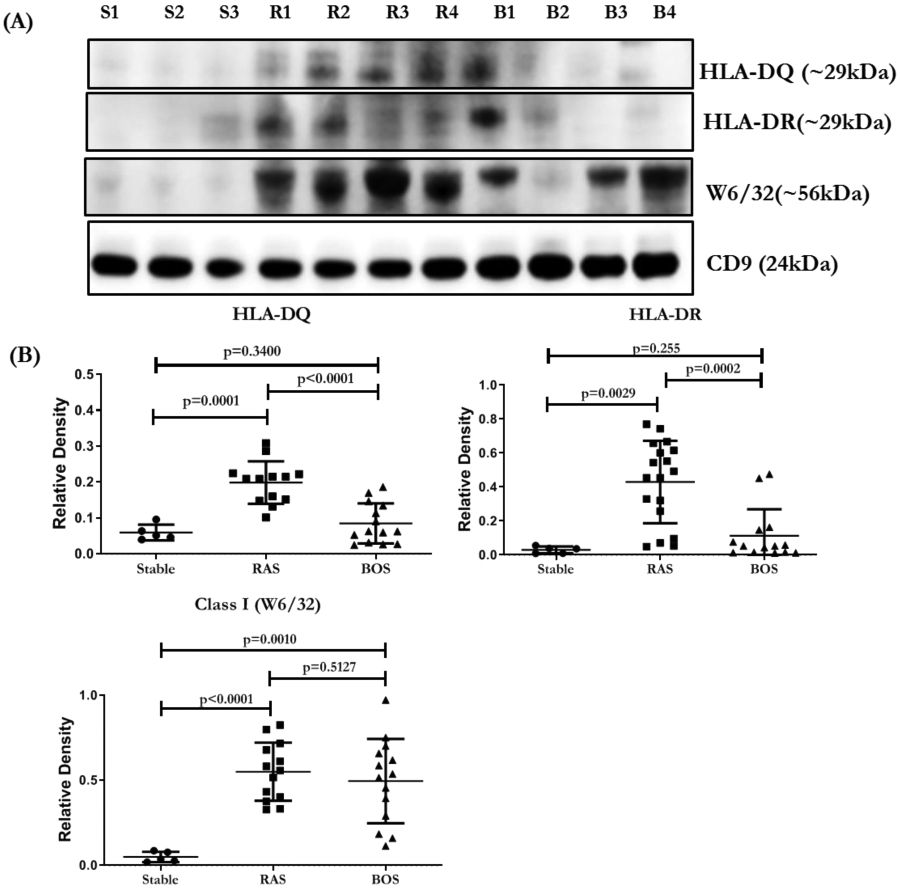
(A) Western blot of exosomal proteins HLA-DQ, HLA-DR and class I (W6/32) from LTxRs with stable LTxRs (S1-S3), or RAS (R1-R4) and BOS (B1-B4). (B) Densitometry and statistical analysis of western blots for stable n=5, RAS n=13 and BOS n=14. CD9 given in the figure is used to normalize the blots for HLA-DQ, HLA-DR and W6/32. All the graphs are represented as dot plots with Mean (Longest horizontal line) and SD (Vertical Bar Line). p<0.05, **p<0.01 and ***p<0.001. BOS, bronchiolitis obliterans syndrome; RAS, restrictive allograft syndrome.
Figure 3:
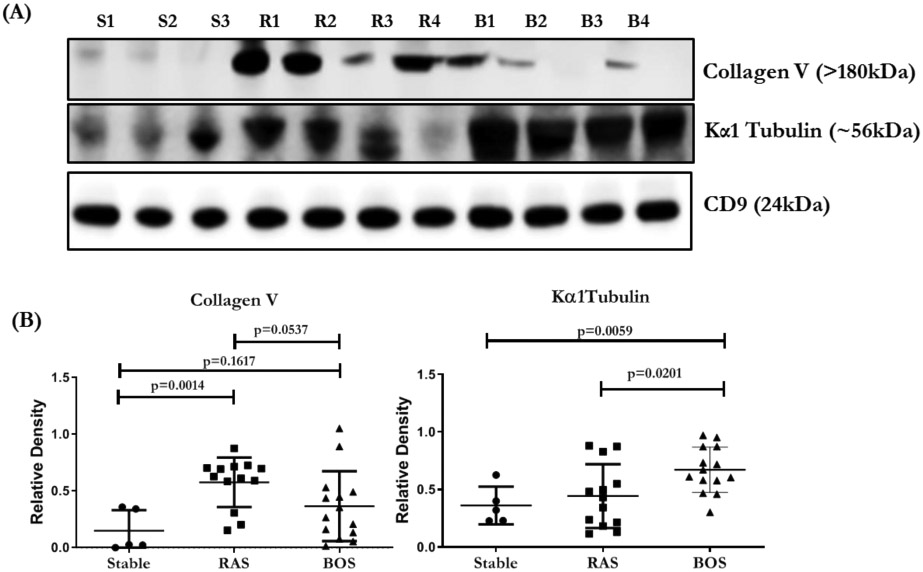
(A) Western blot of exosomal proteins, lung SAgs (Col-V, Kα1T), from LTxRs with stable LTxRs (S1-S3), or RAS (R1-R4) and BOS (B1-B4). (B) Densitometry and statistical analysis of western blots for stable n=5, RAS n=13 and BOS n=14. CD9 given in the figure is used to normalize the blots for Col-V and Kα1 Tubulin. All the graphs are represented as dot plots with Mean (Longest horizontal line) and SD (Vertical Bar Line).*p<0.05, **p<0.01 and ***p<0.001. BOS, bronchiolitis obliterans syndrome; RAS, restrictive allograft syndrome.
All proteins (HLA-DQ, HLA-DR, class I (W6/32), NFkB, 20S peoteasome, PIGR, CIITA, Kα1T and Col-V) have shown mean change in expression after correcting for multiple testing at FDR 5% using Benjamini-Hochberg correction. The adjusted p values after multiple testing for BOS vs RAS are given in Table 2.
Increased levels of proinflammatory cytokines in the circulation of LTxRs with RAS
Plasma from LTxRs with RAS demonstrated significantly higher levels of IL-2R and MIG than plasma from LTxRs with BOS (RAS vs BOS IL-2R: 138.88±33.87 vs 13.05±3.60 pg/ml, p=0.0001; RAS vs BOS MIG: 6.29±3.26 vs 3.45+1.94 pg/ml, p = 0.0104) (Figure 4). In addition, higher concentrations, though not statistically significant, of several proinflammatory cytokines (MCP1, IL-17F, Eotaxin, GMCF, HGF, RANTES) along with lower concentrations of several anti-inflammatory cytokines (IL1RA, IL-10, IL-6, IL-8 and IL-12) were also detected in LTxRs with RAS (Table S2).
Figure 4:
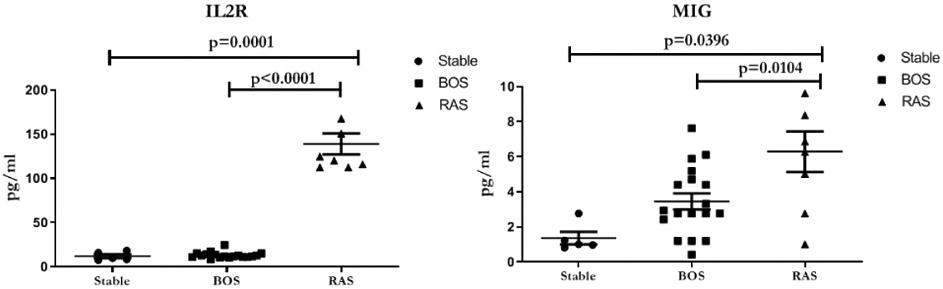
Plasma cytokine levels from LTxRs without CLAD (stable) or diagnosed with BOS or RAS. LTxRs with RAS have higher levels of inflammatory cytokines IL-2R (p=0.0001) and MIG (p=0.0104) compared to LTxRs diagnosed with BOS. Analyzed by Luminex assay. . All the graphs are represented as dot plots with Mean (Longest horizontal line) and SD (Vertical Bar Line). *p<0.05, **p<0.01 and ***p<0.001. BOS, bronchiolitis obliterans syndrome; RAS, restrictive allograft syndrome.
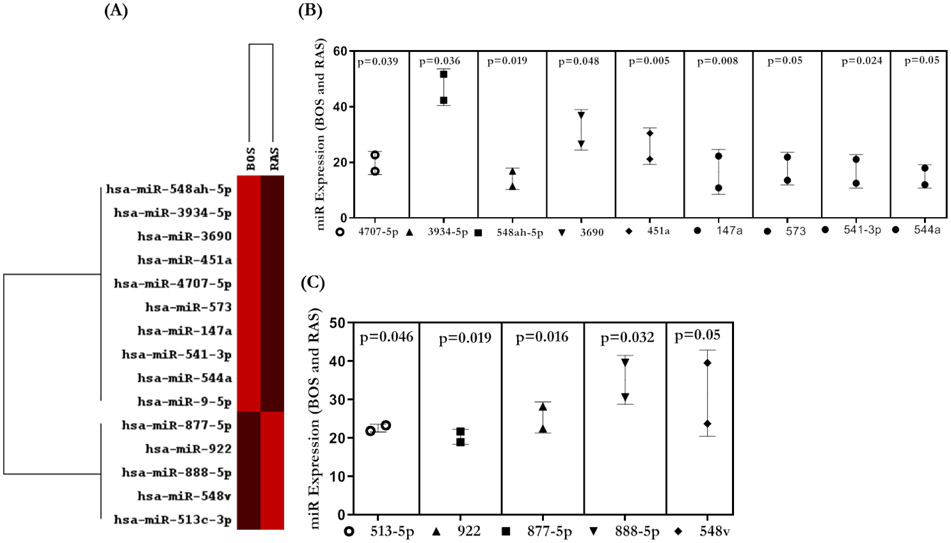
Distinct miRNA in RAS and BOS exosomes
The combined raw read count revealed 798 distinct miRNAs in exosomes isolated from LTxRs with CLAD. We identified 14 miRNAs that were significantly different (≥2-fold) between LTxRs with RAS and LTxRs with BOS (Figure 5). Nine miRNAs were significantly higher in RAS exosomes (548ah-5p, 3934-5p, 3690, 451a, 4707-5p, 573, 147a, 541-3p, and 544a, p<0.05) and 5 miRNAs were significantly higher in BOS exosomes (877-5p, 922, 888-5p, 548v, and 513c-3p, p<0.05). Pathway analysis demonstrated multiple signaling pathways including TGF beta signaling (Supplement).
Figure 5:
(A) Heat map for miRNA profiling from RAS and BOS exosomes. (B) Nested plot for miRNAs higher in BOS samples. (C) Plot for miRNAs present in higher amounts in RAS samples. Only significantly differentially expressed miRNAs are shown. All the graphs represented as Nested Plots with Mean and SD (Vertical Bar Line). *p<0.05, **p<0.01 and ***p<0.001. *p<0.05, **p<0.01 and ***p<0.001. BOS, bronchiolitis obliterans syndrome; miRNA, micro RNA; RAS, restrictive allograft syndrome.
Immunization with exosomes from LTxRs with RAS induced higher levels of Abs to lung SAgs with faster kinetics
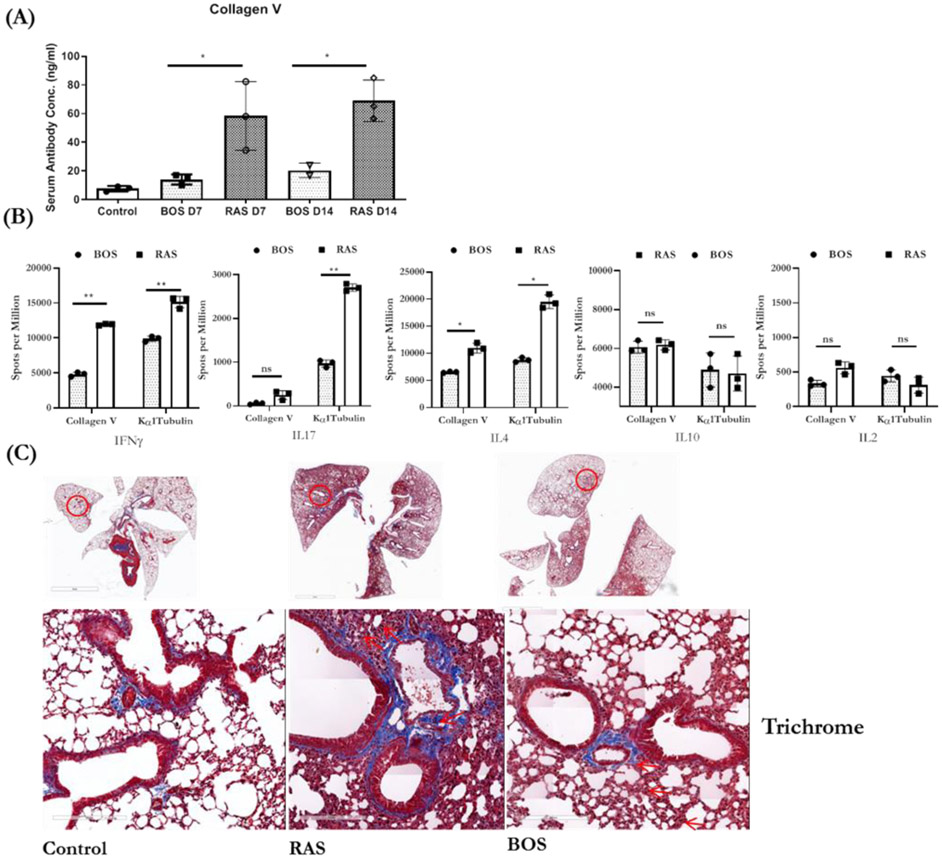
We immunized C57BL/6 mice with exosomes isolated from LTxRs with BOS or RAS to determine the kinetics and the strength of immune responses. All mice immunized with RAS or BOS exosomes developed Abs to lung SAgs.12, 16 However, mice immunized with RAS exosomes developed significantly higher Ab concentrations to Col-V at D7 (BOS vs RAS: 14.08±3.57 vs 58.35±23.91 ng/ml, p=0.0338) and at D14 (BOS vs RAS: 20.35±5.06 vs 68.93±14.51 ng/ml, p=0.0223) (Figure 6A). These results demonstrate that RAS exosomes induce stronger Ab responses with faster kinetics.
Figure 6:
(A) ELISA of serum lung SAg Col-V at D7 and D14 from mice immunized with exosomes isolated from LTxRs with RAS or BOS. (B) EliSPOT values for cytokine producing cells. Graphs in A and B are represented as dot plot with bar, dots as individual values with Mean (represented by Bar height) and SD (Vertical Bar Line).*p<0.05, **p<0.01 and ***p<0.001. (C Trichrome staining of lungs from mice immunized with exosomes isolated from LTxRs with BOS or RAS. Interstitial and inflammatory infiltrates and fibrosis was more prominent in mice injected with LTxRs with RAS compared to mice injected with exosomes from BOS LTxRs. Specific lesions in lungs are marked with red arrows. Images were obtained on a Leica microscope at 40X, and morphometric analysis was performed using Aperio ImageScope software (Leica). Scale bar = 60μm. Lungs from mice immunized with RAS exosomes have inflammation which is predominantly mononuclear and not acute. Based on morphology inflammation is predominantly due to macrophages and lymphocytes and not mediated by neutrophils. BOS, bronchiolitis obliterans syndrome; RAS, restrictive allograft syndrome.
Higher levels of proinflammatory cytokines (IL4, IFN-γ, and IL17) in splenic lymphocytes from mice immunized with RAS exosomes
Splenic lymphocytes from mice immunized with RAS exosomes, compared to those immunized with BOS exosomes, demonstrated more proinflammatory cytokine–secreting cells in response to the lung SAg Col-V: IL4 (BOS vs RAS: 6614.2±61.56 vs 10043.27±1276.75, p=0.0146), IFN-γ (BOS vs RAS: 4776.2±425.11 vs 11920.86±161.69, p=0.0041), and IL17 (BOS vs RAS: 55.66±31.48 vs 228.23±133.83, p=0.0042) spots per million and in response to the lung SAg Kα1T: IL4 (BOS vs RAS: 8819.59±585.38 vs 19663.03±1776.95, p=0.0392) and IFN-γ (BOS vs RAS: 9844.86±523.82 vs 15620.07±47.23, p=0.0020) spots per million. Immunization did not lead to a significant change in IL10, IL17 and IL2 (Figure 6B).
Distinct histopathological lesions in lungs of mice immunized with RAS or BOS exosomes
Lungs were harvested on day 30 after immunization with exosomes isolated from LTxRs with BOS or RAS. Trichrome staining results are given in Figure 6C. Immunization with RAS exosomes resulted in moderate to high inflammation with peribronchial fibrosis. Lesions in airways, pleura, alveoli, and vasculature suggested that RAS exosomes are highly immunogenic and to an extent induce pathology similar to that seen in LTxRs with RAS. The inflammation is predominantly mononuclear and not acute. Based on morphology it is predominantly macrophages and lymphocytes and not neutrophils. In contrast, immunization with BOS exosomes resulted mainly in lesions in small airways, and peripheral lung tissue remained relatively intact with mild to patchy inflammation with fibrosis.
Discussion
De novo development of DSA has been associated with risk for developing CLAD.20 Recent data suggests that not only de novo DSA but also antibody-mediated rejection are potential risk factors for the RAS phenotype of CLAD.21, 22 LTxRs with persistent DSAs are also at higher risk for the RAS phenotype of CLAD.23 Studies have demonstrated an association of RAS with DSAs against mismatched donor HLA class II molecules, especially HLA-DQ.24, 25 In this study, we demonstrated that exosomes isolated from LTxRs with RAS or BOS contain HLA class I and II antigens. However, the concentration of HLA-DR and HLA-DQ molecules are significantly higher in RAS exosomes compared to BOS exosomes. This finding suggests that these exosomes with HLA class II molecules on their surface may induce persistent immune responses leading to DSA to HLA-DR and HLA-DQ in RAS-phenotype CLAD which requires further analysis.
Twelve of the 13 LTxRs with RAS analyzed in this study developed DSA directed to HLA class II with HLA-DR, HLA-DQ and HLA-DPα1 specificities at diagnosis Possible role of exosomes with HLA class II molecules in eliciting immune responses needs further study. It is of interest that only 2 of 18 LTxRs with BOS had detectable DSA with specificities for HLA class II mismatches. However, exosomes from BOS also demonstrated HLA-DR and HLA-DQ, in lower levels than in RAS (Figure 2).
In addition, RAS exosomes contained higher levels of PIGR, NFkB a proinflammatory transcription factor and higher levels of CIITA, a transcription factor involved in MHC class II activation. Together, these results strongly suggest that the exosomes from LTxRs with RAS can be highly immunogenic and can increase immune responses leading to DSA as seen in this study. Further, PIGR can bind and transport polymeric immunoglobulins across the epithelial barrier to mucosal surfaces,26 leading to endothelial activation and subsequent damage to transplanted lungs. Since RAS exosomes also contained significantly higher levels of 20S proteasome than BOS exosomes, we propose that the RAS exosomes are highly immunogenic, resulting in humoral immunity to mismatched HLA, especially HLA-DR and HLA-DQ molecules as noted in this study.
Plasma from LTxRs with RAS contained significantly higher levels of the proinflammatory cytokines MIG and IL2R than plasma from LTxRs with BOS. MIG is a member of the CXC subfamily of chemokines and is an inflammatory chemokine capable of recruiting activated T cells.27 MIG is also known to enhance Th1 and Th2 polarization, attracting Th1 cells and inhibiting Th2 migration. The presence of higher levels of IL2R seen in RAS plasma suggests that it can induce proliferation of T cells, which can injure transplanted lungs.
We also identified comparable amounts of several miRNAs that are involved in TGF beta signaling pathways and target the genes related to TGF beta receptors in both the RAS and BOS exosomes.28, 29 Two other studies have also proposed increased levels of TGF beta in BAL fluid from RAS and BOS patients which is in agreement with our results.28, 30 Therefore, we propose that these miRNAs, present in exosomes, may play an important role in the development of both RAS and BOS. Further studies are needed to determine the role of these exosomal miRNAs and how they may induce fibrosis.
Immunization of mice with exosomes from LTxRs diagnosed with RAS or BOS led to the development of Abs to lung SAg (Col-V).Histopathology of the lungs of mice immunized with RAS exosomes demonstrated moderate to severe lesions along with peribronchial fibrosis. In contrast, immunization with BOS exosomes resulted in mild and patchy inflammation with fibrosis.
This study has some limitations: (1) Our analysis includes a small number of LTxRs diagnosed with RAS. However, the results clearly demonstrated a role of DSA specific to HLA class II molecules. Further, RAS exosomes demonstrated significant quantitative differences in the proinflammatory transcription factor NFkB and the MHC class II activating transcription factor CIITA, along with increased 20S proteasome. (2) Our analysis showed significant differences in only two proinflammatory cytokines (MIG and IL2R) in the circulation LTxRs with RAS. However, several other cytokines (MCP1, Rantes, IL1RA, IL6, and IL12) are also increased, but the difference did not reach statistical significance, likely due to the limited number of LTxRs with RAS. (3) Finally, although miRNA analysis of RAS and BOS exosomes revealed differential expression of some miRNAs, additional experiments are required to determine their functions. Despite these limitations, our findings clearly demonstrated a role for de novo development of DSA to HLA class II in the development of RAS and an important role for exosomes in the development of immune responses, especially to HLA class II since the exosomes contained HLA-DR and HLA-DQ along with increased levels of NFkB, CIITA, and 20S proteasome. It was not surprising that we detected only quantitative differences in several immunological and molecular markers analyzed between RAS and BOS subtypes of CLAD since both of these subtypes likely have similar immunopathogenesis. As expected, lung SAgs (Kα1T, Col V) were seen in exosomes isolated from LTxRs with either RAS or BOS since the exosomes likely originated due to injury and stress to the transplanted organ. In conclusion, our results demonstrated that persistent DSA to HLA class II along with increased levels of circulating exosomes containing HLA-DR and HLA-DQ, NFkB, CIITA, 20S proteasome, and PIGR can differentiate between the RAS and BOS phenotype of CLAD.
Supplementary Material
Acknowledgements
We would like to thank Kristina Sanborn for processing the blood samples. We also would like to acknowledge Billie Glasscock and Kristina Nally for their assistance in preparing this manuscript. We would like to thank Kiran Girdhar, Icahn School of Medicine, for helping with Statistical analysis.
Funding sources
This work was supported by grants from the National Institutes of Health AI123034, HL056643, HL092514 (TM).
List of non-standard abbreviations:
- Abs
antibodies
- BOS
bronchiolitis obliterans syndrome
- CLAD
chronic lung allograft dysfunction
- Col-V
collagen V
- DSA
donor specific antibodies
- EliSPOT
Enzyme-linked immunospot
- HLA
human leukocyte antigen
- Kα1T
Kα1 tubulin
- LTx
lung transplantation
- LTxRs
lung transplant recipients
- MHC
major histocompatibility complex
- miRNA
micro RNA
- NFκB
nuclear factor-κB
- PIGR
polymeric immunoglobulin receptor protein
- RAS
restrictive allograft syndrome
- SAgs
self-antigens
Footnotes
Declaration of Competing Interest
The authors declare no conflict of interest. All authors have reviewed and approved the manuscript and have contributed in a substantial and intellectual manner to the work.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Verleden GM, Raghu G, Meyer KC, et al. A new classification system for chronic lung allograft dysfunction. J Heart Lung Transplant 2014;33:127–33. [DOI] [PubMed] [Google Scholar]
- 2.Verleden GM, Glanville AR, Lease ED, et al. Chronic lung allograft dysfunction: Definition, diagnostic criteria, and approaches to treatment-A consensus report from the Pulmonary Council of the ISHLT. J Heart Lung Transplant 2019;38:493–503. [DOI] [PubMed] [Google Scholar]
- 3.Barker AF, Bergeron A, Rom WN, et al. Obliterative bronchiolitis. N Engl J Med 2014;370:1820–8. [DOI] [PubMed] [Google Scholar]
- 4.Chamberlain D, Maurer J, Chaparro C, et al. Evaluation of transbronchial lung biopsy specimens in the diagnosis of bronchiolitis obliterans after lung transplantation. J Heart Lung Transplant 1994;13:963–71. [PubMed] [Google Scholar]
- 5.Sato M, Waddell TK, Wagnetz U, et al. Restrictive allograft syndrome (RAS): a novel form of chronic lung allograft dysfunction. J Heart Lung Transplant 2011;30:735–42. [DOI] [PubMed] [Google Scholar]
- 6.Glanville AR, Verleden GM, Todd JL, et al. Chronic lung allograft dysfunction: Definition and update of restrictive allograft syndrome-A consensus report from the Pulmonary Council of the ISHLT. J Heart Lung Transplant 2019;38:483–92. [DOI] [PubMed] [Google Scholar]
- 7.Willms E, Johansson HJ, Mager I, Lee Y, Blomberg KE, Sadik M, Alaarg A, Smith CI, Lehtio J, ElAndaloussi S, Wood MJ, Vader P Cells release subpopulations of exosomes with distinct molecular and biological properties. Sci Rep 2016;6:22519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rashed MH, Bayraktar E, Helal GK, Abd-Ellah MF, Amero P, Chavez-Reyes A, Rodriguez-Aguayo C Exosomes: from garbage bins to promising therapeutic targets. Int J Mol Sci 2017;18:538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thery C, Zitvogel L, Amigorena S Exosomes: composition, biogenesis and function. Nat Rev Immunol 2002;2(8):569–79. [DOI] [PubMed] [Google Scholar]
- 10.Gunasekaran M, Sharma M, Hachem R, et al. Circulating Exosomes with Distinct Properties during Chronic Lung Allograft Rejection. J Immunol 2018;200:2535–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sharma M, Gunasekaran M, Ravichandran R, et al. Circulating exosomes with lung self-antigens as a biomarker for chronic lung allograft dysfunction: A retrospective analysis. J Heart Lung Transplant 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gunasekaran M, Xu Z, Nayak DK, et al. Donor-Derived Exosomes With Lung Self-Antigens in Human Lung Allograft Rejection. Am J Transplant 2017;17:474–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bansal S, McGilvrey M, Garcia-Mansfield K, et al. Global Proteomics Analysis of Circulating Extracellular Vesicles Isolated from Lung Transplant Recipients. ACS Omega 2020;5:14360–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Foye C, Yan IK, David W, et al. Comparison of miRNA quantitation by Nanostring in serum and plasma samples. PLoS One 2017;12:e0189165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fisher CE, Kapnadak SG, Lease ED, Edelman JD, Limaye AP Interrater agreement in the diagnosis of chronic lung allograft dysfunction after lung transplantation. J Heart and Lung Transplantation 2019;38(3):327–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bansal S, Itabashi Y, Perincheri S, et al. The role of miRNA-155 in the immunopathogenesis of obliterative airway disease in mice induced by circulating exosomes from human lung transplant recipients with chronic lung allograft dysfunction. Cell Immunol 2020;355:104172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saini D, Weber J, Ramachandran S, et al. Alloimmunity-induced autoimmunity as a potential mechanism in the pathogenesis of chronic rejection of human lung allografts. J Heart Lung T ransplant 2011;30:624–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Subramanian V, Ramachandran S, Banan B, et al. Immune response to tissue-restricted self-antigens induces airway inflammation and fibrosis following murine lung transplantation. Am J Transplant 2014;14:2359–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thery C, Witwer KW, Aikawa E, et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles 2018;7:1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roux A, Bendib Le Lan I, Holifanjaniaina S, et al. Antibody-Mediated Rejection in Lung Transplantation: Clinical Outcomes and Donor-Specific Antibody Characteristics. Am J Transplant 2016;16:1216–28. [DOI] [PubMed] [Google Scholar]
- 21.LePavec J, Suberbielle C, Lamrani L, Feuillet S, Savale L, Dorfmuller P, Stephan F, Mussot S, Mercier O, Fadel E De-novo donor-specific anti-HLA antibodies 30 days after lung transplantation are associated with a worse outcome. J Heart and Lung Transplantation 2016;35:1067–77. [DOI] [PubMed] [Google Scholar]
- 22.Todd JL, Jain R, Pavlisko EN, et al. Impact of forced vital capacity loss on survival after the onset of chronic lung allograft dysfunction. Am J Respir Crit Care Med 2014;189:159–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Verleden SE, Vanaudenaerde BM, Emonds MP, et al. Donor-specific and -nonspecific HLA antibodies and outcome post lung transplantation. Eur Respir J 2017;50. [DOI] [PubMed] [Google Scholar]
- 24.Tikkanen JM, Singer LG, Kim SJ, et al. De Novo DQ Donor-Specific Antibodies Are Associated with Chronic Lung Allograft Dysfunction after Lung Transplantation. Am J Respir Crit Care Med 2016;194:596–606. [DOI] [PubMed] [Google Scholar]
- 25.Hachem RR, Kamoun M, Budev MM, et al. Human leukocyte antigens antibodies after lung transplantation: Primary results of the HALT study. Am J Transplant 2018;18:2285–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaetzel CS, Robinson JK, Chintalacharuvu KR, et al. The polymeric immunoglobulin receptor (secretory component) mediates transport of immune complexes across epithelial cells: a local defense function for IgA. Proc Natl Acad Sci U S A 1991;88:8796–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walton DC, Hiho SJ, Cantwell LS, et al. HLA Matching at the Eplet Level Protects Against Chronic Lung Allograft Dysfunction. Am J Transplant 2016;16:2695–703. [DOI] [PubMed] [Google Scholar]
- 28.DerHovanessian A, Weigt SS, Palchevskiy V, et al. The Role of TGF-beta in the Association Between Primary Graft Dysfunction and Bronchiolitis Obliterans Syndrome. Am J Transplant 2016;16:640–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu Z, Ramachandran S, Gunasekaran M, et al. MicroRNA-144 dysregulates the transforming growth factor-beta signaling cascade and contributes to the development of bronchiolitis obliterans syndrome after human lung transplantation. J Heart Lung Transplant 2015;34:1154–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sacreas A, von der Thusen JH, van den Bosch TPP, et al. The pleural mesothelium and transforming growth factor-beta1 pathways in restrictive allograft syndrome: A pre-clinical investigation. J Heart Lung Transplant 2019;38:570–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.