Abstract
The retroviral attachment (att) sites at viral DNA ends are cis-acting regions essential for proviral integration. To investigate the sequence features of att important for human immunodeficiency virus type 1 (HIV-1) integration in vivo, we generated a series of 25 att mutants of HIV-1 by mutagenesis of the U3, U5, or both boundaries of att. Our results indicated that the terminal 11 or 12 bp of viral DNA are sufficient for specific recognition by HIV-1 integrase (IN) and suggested that IN might recognize each att site independently in vivo.
Retroviral integration (22, 30, 55), which is catalyzed by the viral enzyme integrase (IN), is an essential step in the establishment of the provirus and subsequent viral gene expression (7, 13, 20, 32, 38, 43, 46, 47). The reaction catalyzed by IN can be modeled in an in vitro cell-free assay system in which purified IN and a synthetic oligonucleotide mimicking the viral U3 or U5 end are used (3, 8, 10, 18, 26, 35, 49, 54). Subsequently, cis- and trans-acting regions of the human immunodeficiency virus type 1 (HIV-1) genome, including the attachment (att) site (4, 10, 27, 31, 41–43, 49, 54) and the conserved domains within the IN protein (3, 18, 26, 31, 35), have been identified as important for integration. Mutational analyses of att by use of in vitro assays defines the region of the long terminal repeat (LTR) att sequence necessary for efficient interaction with IN (31, 35, 49, 54). In HIV-1, at least 7 and as many as 13 bp adjacent to the highly conserved CA dinucleotide were shown to be required for efficient and specific interaction of the LTR with IN in vitro (31, 35, 49). Importantly, alteration of the conserved CA dinucleotide significantly impairs the in vitro IN activities, i.e., 3′ processing, strand transfer, and disintegration reactions (8, 10, 31, 35, 49, 54). In addition to the CA dinucleotide, the importance of the subterminal LTR sequence for specific interaction has also been reported (2, 3, 27, 31, 35, 49, 54, 56). However, interpretation of these in vitro assays is limited since they examine the action of IN on only a single viral DNA end. In the natural course of retroviral infection, both the U3 and U5 termini are coordinately recognized by IN (concerted integration). Recently, a new series of in vitro assays using modified oligonucleotide substrates that mimic the integration intermediate have provided a more detailed recognition model of IN in which IN (i) recognizes the att and target DNAs asymmetrically (9) and (ii) can juxtapose two viral DNA ends by active-multimer formation (8, 39). Mutational analyses of HIV-1 IN and complementation studies of mutant INs also suggest that HIV-1 IN is active as a multimer (17, 52). These in vitro studies have greatly increased our understanding of the molecular mechanism of retroviral integration. However, recent studies (14, 19, 34, 38, 45) have indicated some differences or discrepancies between results of in vitro and in vivo assays. These discrepancies may reflect differences due to experimental conditions between in vitro and in vivo conditions. Alternatively, involvement of cellular (21) or viral cofactors that may participate in integration in vivo could be also one explanation for the different results. These arguments emphasize the need to check the phenotype of att mutants in vivo, since the sequence features of att important for retroviral integration and replication are not well defined. On the other hand, the terminal U5 region is important for the packaging of viral RNA and initiation of reverse transcription (11, 23, 24, 41). Recent studies using in vitro assays have provided evidence that the A-rich loop in the HIV-1 U5 region 13 to 15 bp upstream of the primer binding site, consisting of a four-adenosine tract (nucleotides [nt] +169 to +172), binds to the four uridine residues in the anticodon loop of tRNA3Lys (23, 24, 36). Thus, it is possible that the function of the att sequence overlaps those of viral replication steps other than integration.
In the present study, we investigated the role of the terminal sequences of viral DNA in the context of an infectious HIV-1 molecular clone by mutagenesis of the U3 or U5 terminal att region or both. Furthermore, we addressed the att site specificity of HIV-1 IN by examining an att site chimeric virus in which the terminal sequences were replaced with the corresponding sequences of murine leukemia virus (MuLV) or feline immunodeficiency virus (FIV). Finally, we analyzed att exchange mutants in which the U3 att region (11 bp) was exchanged with the U5 att region (11 bp) and/or the U5 att region was exchanged with the U3 att region to examine the adequacy and recognition mode of the terminal 11 bp for integration by HIV-1 IN in vivo.
Construction of att site mutants.
The U5 or U3 region of viral DNA, synthesized de novo with reverse transcriptase after infection, was derived from the 3′ or 5′ LTR of proviral DNA, respectively. Therefore, to generate U5 or U3 att mutants, we introduced mutations into the 3′-terminal region of the 5′ LTR or the 5′-terminal region of the 3′ LTR of an HIV-1 clone, respectively. Series of deletion or point mutations were introduced into each or both att sites through site-directed mutagenesis of subgenomic fragments (Fig. 1), followed by reconstruction of the mutations in the HIVNLluc-env vector (1, 38, 44), in which the env gene is defective, to allow formation of HIV-1 (amphotropic) pseudotypes, and the nef gene was replaced with the firefly luciferase (Luc) gene (12) to allow efficient monitoring of HIV-1 expression. Briefly, DNA fragments for the mutagenesis of HIV-1 LTR termini were derived from a HIVNLluc-env vector. For mutagenesis of the U5 terminal region, the NcoI-SpeI fragment of HIVNLluc-env (nt 10615 to 14267 and nt 1 to 1508) containing the entire U5 region of the 5′ LTR was subcloned into pGEM5Zf(+) (Promega), forming pGEMU5NS. To introduce mutations at the U5 terminal region, two mutagenic primers were designed to span the HindIII-BssHII (nt 531 to 712) U5 att region. PCR was performed with the mutagenic primers using HIVNLluc-env as template DNA. The amplified products were digested with HindIII and BssHII and ligated to HindIII/BssHII-digested pGEMU5NS. After confirmation of designed mutations by DNA sequence of the entire amplified region, the HindIII-BssHII fragment containing each mutation at the U5 terminal region was inserted back into the HIVNLluc-env vector, replacing the corresponding HindIII-BssHII region of the vector. For mutagenesis of the U3 terminal region, the XhoI-SspI fragment of HIVNLluc-env (nt 8207 to 12677) containing the entire 3′ LTR was subcloned into the XhoI and EcoRV sites of pBluescript II SK(+), forming pSKU3. To introduce mutations at the U3 terminal region, two mutagenic primers were designed to span the MluI-PmaCI (nt 9876 to 10240) region containing the 5′ half of U3 in the 3′ LTR. PCR was performed with the U3 att mutagenic primers as described above. The amplified products were digested with PmaCI and MluI and ligated to PmaCI/MluI-digested vector pSKU3. After confirmation of designed mutations by DNA sequencing of the entire amplified region, the XhoI-NcoI fragment containing each mutation at the U3 terminal region was inserted back into the HIVNLluc-env vector, replacing the corresponding XhoI-NcoI region of the vector. To remove the U3 terminal region in the 5′ LTR of the HIVNLluc-env vector, pGEMU5NS was digested with EcoRV (nt 112) and StuI (site in the 5′ flanking region). The large fragment was purified by agarose gel electrophoresis, followed by self-ligation of the EcoRV/StuI-digested pGEMU5NS vector (pGEMU5NSdelS-RV). The NcoI-SpeI fragment of pGEMU5NSdelS-RV was inserted back into the HIVNLluc-env vector carrying a U3 CA>TG mutation in the 3′ LTR by replacing the corresponding XhoI-SpeI region of the vector (U3 CA>TGX). For certain att mutations, we introduced mutations at both U3 and U5 att sites simultaneously (U3&U5CA>TG, U3&U5DEL8, U3&U5DEL10, U3&U5MuLV, U3&U5FIV, and U3vsU5). To prepare each U3&U5 att mutant, the Xho-NcoI fragment (nt 8207 to 10615) of the HIVNLluc-env vector carrying each U3 att mutation in the 3′ LTR was replaced with the corresponding region of the HIVNLluc-env vector carrying each U5 att mutation.
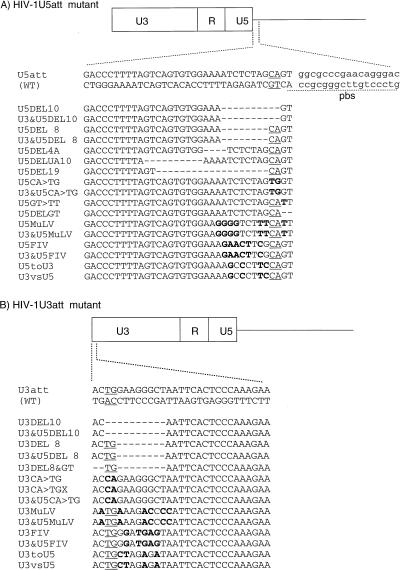
FIG. 1.
Mutation of HIV-1 U3 and U5 att sites. The boundary and nucleotide sequences of the U5 (A) and U3 (B) att sites are shown. Both strands of the WT DNA sequences are shown at the top. The highly conserved CA and TG dinucleotides in each att site are underlined. For att site mutants, only positive-strand DNA sequences are shown. A deleted or altered nucleotide is indicated by a dash or a bold letter, respectively. In MuLV or FIV att chimeric mutants, the terminal 13 bp of each HIV-1 att sequence were replaced with the corresponding terminal 13 bp of MuLV or FIV. The boundary of the primer binding site (pbs) for tRNA3Lys is shown by dashed underlining (A).
We previously showed that the single-round infection system with HIV-1 (amphotropic) pseudotypes was useful for estimation of integration efficiency in vivo by monitoring the level of synthesized viral DNA and Luc activity expressed in infected cells (38). We prepared a pseudotype virus of each att site mutant by cotransfection of COS cells with the HIVNLluc-env vector carrying each att mutation and an amphotropic MuLV envelope expression vector (pJD-1). We also prepared an HIV-1 IN mutant, D116G, carrying an amino acid substitution at the catalytic site of IN (38) for use as an integration-defective control virus. All of the mutants had comparable levels of p24 and Luc activity within lysates of transfected COS cells and comparable levels of p24 in culture supernatants harvested from the transfected cells (data not shown). Thus, none of the mutations had any effect on transfected proviral gene expression.
Effect of each mutation on integration efficiency.
We assessed the ability of att mutants with deletions or point mutations at either the U5 or the U3 terminus or both to form viral DNA following infection of susceptible RD cells (Fig. 2A). For this purpose, PCR was used to monitor the formation of complete or nearly complete viral DNA (R/gag) with primers M667 and M661 (58). For each virus, RD cells (106) were infected by inoculating an aliquot (1 ml) of DNase-treated COS cell virus-containing supernatant. The amount of p24 in each 1-ml aliquot was around 50 ng. At 24 h or 10 days, as indicated on the left of Fig. 2, the entire cell culture was harvested. Total DNA was extracted from infected RD cells by the urea lysis method as previously described (38, 58). Each DNA sample was subjected to quantitative PCR analysis with primer pairs specific for the R/gag region of HIV-1 (38, 58) by 30 cycles of amplification (94°C for 1 min, 65°C for 2 min, and 72°C for 2 min) in a reaction mixture containing 20 mM Tris-HCl (pH 8.4), 50 mM KCl, 0.2 mM deoxynucleoside triphosphates, 1 mM MgCl2, and 2 U of Taq DNA polymerase (GIBCO BRL). For an internal control, total DNA from 4 × 102 to 5 × 104 RD cells was amplified in parallel as human DNA standard primer pairs specific for the human β-globin gene (33) (data not shown). All of the mutants produced comparable levels of complete or nearly complete virus DNA at 24 h postinfection, except the U5DEL GT and U3DEL 8> mutants (Fig. 2A). It is noteworthy that the amplified product (R/gag) of each U5 att deletion mutant (Fig. 1A, U5DEL10, U5DEL 8, U5DELUA10, or U5DEL19) showed a single band with slower mobility than that of the wild type (WT) (200 bp), corresponding to the size of its deletion (Fig. 2A, lanes 2, 3, 5, and 6), indicating that each att mutation was indeed introduced into the de novo-synthesized viral DNA after infection and that a revertant was not generated during the transfection and infection procedures. Replacement of the terminal GT with TT in the U5 att region (Fig. 1, U5GT>TT) did not affect viral DNA synthesis (Fig. 2A, lane 8). Thus, the low levels of viral DNA produced by U5DEL GT and U3DEL 8> suggested that deletion of the terminal nucleotide (T/A) in the U5 and U3 att regions might affect efficient priming of reverse transcription by tRNA3Lys or selection and processing of the polypurine tract to initiate plus-strand DNA synthesis. Deletion of the A-rich loop consisting of a four-adenosine tract in the U5 terminal region (nt +169 to +172) (Fig. 1A, U5DEL4A), which was previously reported to be important for interaction with the tRNA3Lys primer and subsequent negative-strand DNA synthesis in vitro (23, 24), did not affect viral DNA synthesis in our infection system (Fig. 2A, lane 4). Although we cannot exclude the possibility that minor effects of the mutation might accumulate during repeated cycles of virus replication, our observation was consistent with a previous in vivo experiment showing no apparent function of the A-rich loop in viral replication (53). We also examined the stability of viral DNA by monitoring its levels over time in infected cells. The IN catalytic site mutant, D116G, showed low stability, less than 0.5% of the level of the WT (Fig. 2A, compare lanes 1 and 18). Although all of the mutants, except U5DEL GT and U3DEL 8>, produced equivalent levels of viral DNA at 24 h postinfection, the levels at 10 days varied with integration efficiency.
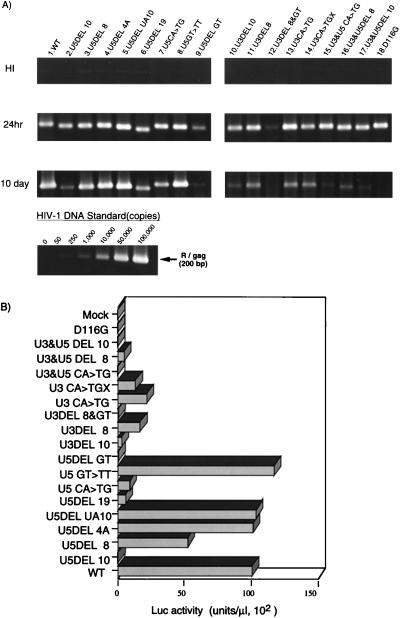
FIG. 2.
Analysis of att mutants. (A) For each virus, 106 RD cells were infected by inoculating a 1-ml aliquot of DNase-treated, virus-containing COS cell supernatant. After 24 h or 10 days, as indicated on the left, the entire cell culture was harvested. Total DNA was extracted from infected RD cells and subjected to quantitative PCR analysis with primer pairs specific for the R/gag region of HIV-1 (38, 58). For HIV-1 DNA standards, 50 to 100,000 copies of linearized HIVNLluc-env were amplified in parallel. Amplified products were resolved on a 2% agar gel and visualized by Sybr Green staining (FMC Bio Product, Rockland, Maine). Quantitative analysis of amplified products was performed by the Epi-Light UV FA1100 system with a Luminous Imager (Aisin Cosmos R&D Co.). An aliquot of each virus preparation was incubated at 65°C for 30 min and used as a heat-inactivated control (HI). (B) At 3 days postinfection, the entire culture was harvested and washed twice with phosphate-buffered saline. The cell pellet was resuspended with 200 μl of cell lysate buffer (Promega Corp.). Ten microliters of each cell lysate was subjected to a Luc assay. Luc activity was determined after subtraction of the background level and normalization to 1 μl of lysate, corresponding to about 103 cells. Luc activity was measured in duplicate for each virus inoculate, and mean values are shown. The standard error for each replicate was less than 10% of the mean. This experiment was performed three times with independently prepared virus. The results of a representative experiment are shown. A virus prepared with a parental (WT) vector or an IN catalytic mutant (D116G) vector was infected in parallel as a WT or integration-defective control, respectively. Mock, mock-transfected COS cells (no virus).
All of the att site mutants showed comparable levels of Luc activity following transfection of COS cells and produced comparable levels of viral DNA after infection, with the exception of U5DEL GT and U3DEL 8> (Fig. 2). Thus, we could estimate relative integration efficiency directly by measuring Luc activity following infection. The WT construct yielded 1.1 × 104 light units (U) of Luc activity per μl of cell lysate (corresponding to about 103 cells) at 3 days postinfection (Fig. 2B, WT). On the other hand, the IN catalytic-site mutant, D116G, yielded only 200 U/μl of cell lysate (Fig. 2B, D116G), indicating that the relative integration efficiency of D116G with respect to the WT level was estimated to be less than 0.2%. In att site mutants, by deletion at each terminus of 10 nt (Fig. 1A, U5DEL10, and B, U3DEL10) which span the highly conserved CA dinucleotide and its 5′ internal region, the relative integration efficiency was reduced to less than 5% of the WT level (Fig. 2B, U5DEL 10 and U3DEL10). When we kept the conserved terminal 4 nt (CAGT) intact and deleted the 8 nt 5′ internal to the conserved CA (Fig. 1A, U5DEL 8; Fig. 1B, U3DEL 8), efficiency was reduced to 20 to 50% of the WT level (Fig. 2B, U5DEL 8 and U3DEL 8). Deletion of the four-adenosine tract in the U5 terminal region (Fig. 1A, U5DEL4A) or 10 nt of its upstream region (Fig. 1A, U5DELUA10) had no drastic effect on integration efficiency (Fig. 2B, U5DEL 4A and U5DEL UA10). These results indicated that the boundary for interaction with IN lies within 11 to 12 nt of each viral DNA terminus. Alteration of the highly conserved CA dinucleotide in each att site (Fig. 1A, U5CA>TG, and B, U3CA>TG) reduced integration efficiency to 10 to 30% of the WT level (Fig. 2B, U5 CA>TG and U3 CA>TG). Similar alterations of the conserved CA dinucleotide resulted in deterioration of the effect on IN activity in vitro (9, 10, 31, 35, 49, 54). Importantly, when we altered the CA dinucleotides of both the U3 and U5 att regions at same time (Fig. 1A and B, U3&U5CA>TG), integration efficiency was synergistically reduced to as low as 1% of the WT level. Thus, consistent with previous in vitro IN studies (8, 10, 31, 35), the results of our in vivo experiment also showed the importance of the CA dinucleotide for integration. However, if nucleotide substitution of the conserved CA was introduced into either the U3 or the U5 att region, the effect was tolerated, as shown above and in our previous report (38). To rule out the possibility that genetic rearrangement of a mutated fragment with the WT sequence at another LTR on the plasmid DNA might generate revertant DNA during the transfection procedure, we generated a U3 CA>TG mutant plasmid with a deletion of another U3 terminal region (StuI-EcoRV) at the 5′ LTR (U3 CA>TGX). U3 CA>TGX mutants showed a phenotype indistinguishable from that of the U3 CA>TG mutant (Fig. 2, U3 CA>TG and U3 CA>TGX). Although the exact mechanism of the observed tolerance of a mutation at one of the att sites in vivo is not clear, similar tolerance effects were also observed when a 10-bp deletion (Fig. 2, U3DEL10, U5DEL 10, and U3&U5DEL 10) and an 8-bp deletion (Fig. 2, U3DEL 8, U5DEL 8, and U3&U5 DEL 8) were introduced into only one of the two att sites. Since the in vitro integration assays analyze the integration events at one att site, compensation of a critical mutation at one of the att sites might happen during the concerted integration of both att sites in vivo. Thus, we hypothesized that binding of IN or the 3′ cleavage product at one of the att sites might help another att processing step, probably by lowering the stringency of the att sequence’s specificity for IN. Recent in vitro studies suggest that IN facilitates the flipping of the terminal bases during catalysis (48). Alteration of the terminal GT to TT (Fig. 1A, U5GT>TT) failed to change viral DNA synthesis (Fig. 2A, U5GT>TT) or integration efficiency (Fig. 2B, U5 GT>TT), indicating that sequence of the G nucleotide at the 3′ end of the U5 att region might not be rigidly required for efficient integration in vivo.
Analysis of att site chimeric virus.
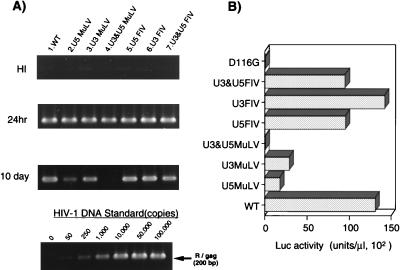
The above analysis of att site mutants showed that the boundary required for interaction with IN lies within 11 or 12 nt of each terminus of the viral DNA. We next examined the att specificity of HIV-1 IN. To address this point, we generated an att site chimeric HIV-1 mutant in which the 13 bp of each terminal sequence of the U3 and/or U5 att region were replaced with the corresponding region of MuLV (Fig. 1, U5MuLV, U3MuLV, and U3&U5MuLV) (35, 46) or FIV (Fig. 1, U5FIV, U3FIV, and U3&U5FIV) (50, 51, 53). None of the replacement mutations had any effect on gene expression of proviral DNA nor virus release after transfection of COS cells (data not shown). Following infection of RD cells, each att chimeric mutant produced a level of viral DNA equivalent to that of the WT (Fig. 3A), indicating no effects of each mutation on reverse transcription after infection. The relative integration efficiencies of the U5 MuLV, U3 MuLV, U5 FIV, and U3 FIV mutants, estimated by Luc activity, were calculated to be 15.6, 28.3, 74.0, and 86.2%, respectively. The MuLV att chimeric mutant showed an integration efficiency as low as that of the corresponding 8-bp deletion mutants (U3DEL8 and U5DEL8), indicating that the 8 nt internal to the conserved CA dinucleotide might be important in determining the att specificity of HIV-1 IN. In contrast, exchange of HIV-1 att with FIV att had less of an effect on integration by HIV-1 IN in vivo. When both the 3′ and 5′ regions of att were replaced at the same time with those of MuLV (U3&U5MuLV) or FIV (U3&U5FIV), U3&U5FIV maintained a high integration efficiency of around 70% of the WT level, while U3&U5MuLV showed a markedly lower integration efficiency of less than 0.5% of the WT level (Fig. 3). Here again, we observed a tolerance for mutations in the U3 or U5 att region in the MuLV att chimeric virus. Thus, our results clearly demonstrated the att site specificity of HIV-1 IN in vivo, consistent with previous in vitro studies (27, 28, 35). On the other hand, replacement of the att sequence with FIV att showed less of an effect on integration in vivo. Similar compatibility of FIV U5 att site usage for HIV-1 IN was reported in vitro (50). In conclusion, we showed that the terminal 11-bp sequence of the U5 att region and the terminal 12-bp sequence of the U3 att region are the minimum cis elements required for specific interaction with HIV-1 IN. Although we did not address the importance of individual residues, the results of a comparison of the att sequences suggested certain key nucleotides at positions 5, 6, 10, and 11 from the end of the U5 region of att and at positions 5, 9, 10, and 12 from the end of the U3 region of att (Fig. 5, indicated by asterisks). Our conclusion was compatible, at least in part, with previous in vitro studies by (i) Katzman and Sudol (27) that indicated the importance of nucleotides at positions 5 and 6 from the end of the U3 region of att and (ii) Reicin et al. (45) that showed the importance of three Gs at positions 6 to 8 from the end of U3. It is noteworthy that the important role of the nucleotides at positions 5 and 6 from the end of the U5 region of att for specific binding of HIV-1 IN has been indicated by an in vitro UV cross-linking study with mismatched att U5 mutagenic oligonucleotides (57).
FIG. 3.
Analysis of att chimeric mutants. A 1-ml aliquot of each virus containing about 50 ng of p24 was inoculated into 106 RD cells. (A) At 24 h or 10 days after infection, as indicated on the left, total DNA was extracted and subjected to quantitative PCR analyses of de novo-synthesized viral DNA as described in the legend to Fig. 2. (B) For each virus, at 3 days postinfection of the same virus preparation used for the PCR experiment in panel A, the entire culture was harvested. The cell pellet fraction was subjected to a Luc assay as described in the legend to Fig. 2. Luc activity was measured in duplicate for each virus inoculate, and mean values are shown. The standard error for each replicate was less than 10% of the mean. A virus prepared with a parental (WT) or an IN catalytic mutant (D116G) vector was infected in parallel as a WT or integration-defective control, respectively. This experiment was performed three times with independently prepared virus. The results of a representative experiment are shown.
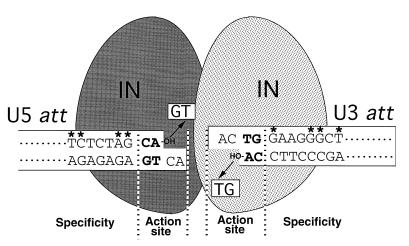
FIG. 5.
Specific att site recognition by HIV-1 IN. The U3 and U5 att regions at both termini of the viral DNA (shown as open boxes) are recognized and subjected to an integration reaction by IN. Removal of the terminal GT dinucleotide by IN (illustrated with arrows) resulted in exposure of the conserved dinucleotide CA-OH (shown in bold letters) at the 3′ ends of both strands. The U3 (12-bp) and U5 (11-bp) att sequences of HIV-1, which are required for efficient interaction with IN, are shown in the boxes. The nucleotides suggested to be important for specific interaction with HIV-1 IN are indicated by asterisks. The independent recognition of each att site by each IN promoter by dimer formation is speculatively illustrated.
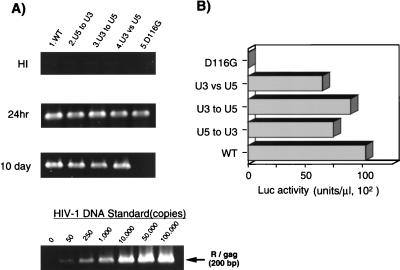
Analysis of att exchange mutants.
There is little apparent nucleotide sequence homology between the U3 and U5 regions of HIV-1 att, except for the terminal 4 bp (Fig. 1). Importantly, the critical nucleotides in each att region that might determine the substrate specificity of HIV-1 IN, as indicated in the chimeric att mutant experiment and previous in vitro studies (27, 56), were not conserved between the two att regions. Thus, it is suggested that IN might recognize U3 and U5 of att independently. Alternatively, IN may recognize both att sites sequentially as a unit. To distinguish these two possibilities, we generated att site exchange mutants (Fig. 1, U5toU3, U3toU5, and U3vsU5) and examined their effects on integration efficiency in vivo. In the U5toU3 mutant, the 11-bp terminal region of U5 of att was replaced with the 11-bp terminal sequence of the U3 att site to produce virus DNA with the 11-bp U3 terminal sequence at both LTR termini. In the second mutant, U3toU5, we replaced the 11-bp terminal sequence of U3 of att with the corresponding U5 att region, resulting in virus DNA having 11-bp U5 att sequences at both termini. In the last mutant, U3vsU5, we exchanged the 11-bp terminal sequences of the U3 and U5 att regions to produce a virus DNA with the U5 att sequence in the U3 terminal region and the U3 att sequence in the U5 terminal region. By analyzing the U3vsU5 mutant, we also addressed the sufficiency of the terminal 11 bp as att, since there is not homology between the U3 and U5 termini outside the terminal 11 bp. All three att exchange mutants produced comparable level of viral DNA (Fig. 4A) with a high integration efficiency of around 70 to 90% of the WT level (Fig. 4B). Thus, our results demonstrated that the terminal 11 bp is almost sufficient for interaction with HIV-1 IN and that HIV-1 IN might recognize the U3 and U5 att sequences not as a unit but, rather, independently. The latter conclusion suggests the importance of dimerization or multimerization of HIV-1 IN for concerted integration of both att sites in vivo.
FIG. 4.
Analysis of att exchange mutants. Each virus was inoculated into 106 RD cells as described in the legend to Fig. 2. (A) At 24 h or 10 days after infection, as indicated on the left, total DNA was extracted and subjected to quantitative PCR analyses of de novo-synthesized viral DNA as described in the legend to Fig. 2. (B) For each virus, at 3 days postinfection with the same virus preparation used for the PCR experiment in panel A, the entire culture was harvested and subjected to a Luc assay as described in the legend to Fig. 2. Luc activity was measured in duplicate for each virus inoculate, and mean values are shown. The standard error for each replicate was less than 10% of the mean. A virus prepared with a parental (WT) vector or an IN catalytic mutant (D116G) vector was infected in parallel as a WT or integration-defective control, respectively. This experiment was performed three times with independently prepared virus. The results of a representative experiment are shown.
As summarized in Fig. 5, the major findings of the present study are the following. (i) The terminal 11 or 12 bp of the U5 and U3 att sites is the minimum cis element required and is almost sufficient for integration or efficient interaction with HIV-1 IN in vivo. (ii) The highly conserved dinucleotides CA and TG are essential probably as an att substrate feature common to all retroviruses for IN-mediated action or 3′ processing, as previously indicated by in vitro studies (10). (iii) The subterminal region 7 to 8 bp 3′ internal to the highly conserved CA might determine the att specificity of HIV-1 IN. (iv) Analysis of att exchange mutants suggested that HIV-1 IN might recognize each att site independently but act in a concerted manner, probably by forming dimers or multimers.
Mutational and complementation analyses of HIV-1 IN in vitro studies of HIV-1 IN showed that IN can be divided into three distinct function domains, the N-terminal, central, and C-terminal domains, that complement each other for full enzymatic activity (17, 52). The central core domain contains the conserved D, D 35E motif, which is essential for the catalytic activity of IN (5, 18, 29, 32). In addition, both the N- and C-terminal regions are required for full IN activities in vitro (17, 52, 59). These in vitro studies indicated that IN might function as a dimer or multimer form (17, 52). The three-dimensional structure of the central domain of HIV-1 has been solved by X-ray crystallography (15). Recently, the structures of the N-terminal (6) and C-terminal (16, 37) domains of HIV-1 IN were also analyzed by nuclear magnetic resonance spectroscopy, which showed that each domain forms a homodimer or a tetramer. Considered together with the results of these in vitro studies, our results obtained with att exchange mutants suggest an important role in dimer or multimer formation for each IN promoter that recognizes the U3 or U5 att site independently for the concerted integration of both att sites in vivo.
There are some important aspects of retroviral integration that remain to be determined. These include, for example, the exact feature of IN that determines att specificity. Examination of this point was hampered by the nonspecific DNA binding ability of IN, as well as its specific binding ability. Results of recent in vitro studies using chimeric IN of HIV-1 and FIV INs (50) indicate that the determinant of IN for att specificity might be located in the core central domain. Furthermore, lysine residues in the central domain of HIV-1 IN located at position 136 (40) or positions 156 and 159 (25) have been suggested as key sites for specific att binding. Combined with these in vitro studies, further experiments using the in vivo system are necessary for new insight into retroviral integration. Interestingly, the recent studies of Du et al. (14) showed that a mutation in IN can compensate for mutations in simian immunodeficiency virus att.
Acknowledgments
We thank Samson A. Chow, Yoshio Shibagaki, Mari Kannagi, and Irvin S. Y. Chen for the helpful discussion and A. Katayama and J. Minami for technical assistance.
This work was supported by a Grant-in-Aid for Scientific Research on Priority Areas, by a Grant-in-Aid for Scientific Research (C) from the Ministry of Education, Science, Sports and Culture, and by an International AIDS Research Program grant from the Japan Health Scienc Foundation.
REFERENCES
- 1.Adachi A, Gendelman H E, Koenig S, Folks T, Willey R, Rabson A, Martin M A. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balakrishnan M, Jonsson C B. Functional identification of nucleotides conferring substrate specificity to retroviral integrase reactions. J Virol. 1997;71:1025–1035. doi: 10.1128/jvi.71.2.1025-1035.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bushman F D, Craigie R. Activities of human immunodeficiency virus (HIV) integration protein in vitro: specific cleavage and integration of HIV DNA. Proc Natl Acad Sci USA. 1991;88:1339–1343. doi: 10.1073/pnas.88.4.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bushman F D, Craigie R. Sequence requirements for integration of Moloney murine leukemia virus DNA in vitro. J Virol. 1990;64:5645–5648. doi: 10.1128/jvi.64.11.5645-5648.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bushman F D, Engelman A, Palmer I, Wingfield P, Craigie R. Domains of the integrase protein of human immunodeficiency virus type 1 responsible for polynucleotidyl transfer and zinc binding. Proc Natl Acad Sci USA. 1993;90:3428–3432. doi: 10.1073/pnas.90.8.3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cai M, Zheng R, Caffrey M, Craigie R, Clore G M, Gronenborn A M. Solution structure of the N-terminal zinc binding domain of HIV-1 integrase. Nat Struct Biol. 1997;4:567–577. doi: 10.1038/nsb0797-567. [DOI] [PubMed] [Google Scholar]
- 7.Cannon P M, Wilson W, Byles E, Kingsman S M, Kingsman A J. Human immunodeficiency virus type 1 integrase: effect on viral replication of mutations at highly conserved residues. J Virol. 1994;68:4768–4775. doi: 10.1128/jvi.68.8.4768-4775.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chow S A, Brown P O. Juxtaposition of two viral DNA ends in a bimolecular disintegration reaction mediated by multimers of human immunodeficiency virus type 1 or murine leukemia virus integrase. J Virol. 1994;68:7869–7878. doi: 10.1128/jvi.68.12.7869-7878.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chow S A, Brown P O. Substrate features important for recognition and catalysis by human immunodeficiency virus type 1 integrase identified by using novel DNA substrates. J Virol. 1994;68:3896–3907. doi: 10.1128/jvi.68.6.3896-3907.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chow S A, Vincent K A, Ellison V, Brown P O. Reversal of integration and DNA splicing mediated by integrase of human immunodeficiency virus. Science. 1992;255:723–726. doi: 10.1126/science.1738845. [DOI] [PubMed] [Google Scholar]
- 11.Cobrinik D, Soskey L, Leis J. A retroviral RNA secondary structure required for efficient initiation of reverse transcription. J Virol. 1988;62:3622–3630. doi: 10.1128/jvi.62.10.3622-3630.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Wet J R, Wood K V, DeLuca M, Helinski D R, Subramani S. Firefly luciferase gene: structure and expression in mammalian cells. Mol Cell Biol. 1987;7:725–737. doi: 10.1128/mcb.7.2.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Donehower L A, Varmus H E. A mutant murine leukemia virus with a single missense codon in pol is defective in a function affecting integration. Proc Natl Acad Sci USA. 1984;81:6461–6465. doi: 10.1073/pnas.81.20.6461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Du Z, Ilyinskii P O, Lally K, Desrosiers R C, Engelman A. A mutation in integrase can compensate for mutations in the simian immunodeficiency virus att site. J Virol. 1997;71:8124–8132. doi: 10.1128/jvi.71.11.8124-8132.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dyda F, Hickman A B, Jenkins T M, Engelman A, Craigie R, Davies D R. Crystal structure of the catalytic domain of HIV-1 integrase: similarity to other polynucleotidyl transferases. Science. 1994;266:1981–1986. doi: 10.1126/science.7801124. [DOI] [PubMed] [Google Scholar]
- 16.Eijkelenboom A P, Lutzke R A, Boelens R, Plasterk R H, Kaptein R, Hard K. The DNA-binding domain of HIV-1 integrase has an SH3-like fold. Nat Struct Biol. 1995;2:807–810. doi: 10.1038/nsb0995-807. [DOI] [PubMed] [Google Scholar]
- 17.Engelman A, Bushman F D, Craigie R. Identification of discrete functional domains of HIV-1 integrase and their organization within an active multimeric complex. EMBO J. 1993;12:3269–3275. doi: 10.1002/j.1460-2075.1993.tb05996.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Engelman A, Craigie R. Identification of conserved amino acid residues critical for human immunodeficiency virus type 1 integrase function in vitro. J Virol. 1992;66:6361–6369. doi: 10.1128/jvi.66.11.6361-6369.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Engelman A, Liu Y, Chen H, Farzan M, Dyda F. Structure-based mutagenesis of the catalytic domain of human immunodeficiency virus type 1 integrase. J Virol. 1997;71:3507–3514. doi: 10.1128/jvi.71.5.3507-3514.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Englund G, Theodore T S, Freed E O, Engleman A, Martin M A. Integration is required for productive infection of monocyte-derived macrophages by human immunodeficiency virus type 1. J Virol. 1995;69:3216–3219. doi: 10.1128/jvi.69.5.3216-3219.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Farnet C M, Bushman F D. HIV-1 cDNA integration: requirement of HMG I(Y) protein for function of preintegration complexes in vitro. Cell. 1997;88:483–492. doi: 10.1016/s0092-8674(00)81888-7. [DOI] [PubMed] [Google Scholar]
- 22.Goff S P. Genetics of retroviral integration. Annu Rev Genet. 1992;26:527–544. doi: 10.1146/annurev.ge.26.120192.002523. [DOI] [PubMed] [Google Scholar]
- 23.Isel C, Ehresmann C, Keith G, Ehresmann B, Marquet R. Initiation of reverse transcription of HIV-1: secondary structure of the HIV-1 RNA/tRNA(3Lys) (template/primer) J Mol Biol. 1995;247:236–250. doi: 10.1006/jmbi.1994.0136. [DOI] [PubMed] [Google Scholar]
- 24.Isel C, Marquet R, Keith G, Ehresmann C, Ehresmann B. Modified nucleotides of tRNA(3Lys) modulate primer/template loop-loop interaction in the initiation complex of HIV-1 reverse transcription. J Biol Chem. 1993;268:25269–25272. [PubMed] [Google Scholar]
- 25.Jenkins T M, Esposito D, Engelman A, Craigie R. Critical contacts between HIV-1 integrase and viral DNA identified by structure-based analysis and photo-crosslinking. EMBO J. 1997;16:6849–6859. doi: 10.1093/emboj/16.22.6849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Katz R A, Merkel G, Kulkosky J, Leis J, Skalka A M. The avian retroviral IN protein is both necessary and sufficient for integrative recombination in vitro. Cell. 1990;63:87–95. doi: 10.1016/0092-8674(90)90290-u. [DOI] [PubMed] [Google Scholar]
- 27.Katzman M, Sudol M. Influence of subterminal viral DNA nucleotides on differential susceptibility to cleavage by human immunodeficiency virus type 1 and visna virus integrases. J Virol. 1996;70:9069–9073. doi: 10.1128/jvi.70.12.9069-9073.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Katzman M, Sudol M. Mapping domains of retroviral integrase responsible for viral DNA specificity and target site selection by analysis of chimeras between human immunodeficiency virus type 1 and visna virus integrases. J Virol. 1995;69:5687–5696. doi: 10.1128/jvi.69.9.5687-5696.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kulkosky J, Jones K S, Katz R A, Mack J P G, Skalka A M. Residues critical for retroviral integrative recombination in a region that is highly conserved among retroviral/retrotransposon integrases and bacterial insertion sequence transposases. Mol Cell Biol. 1992;12:2331–2338. doi: 10.1128/mcb.12.5.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kulkosky J, Skalka A M. Molecular mechanism of retroviral DNA integration. Pharmacol Ther. 1994;61:185–203. doi: 10.1016/0163-7258(94)90062-0. [DOI] [PubMed] [Google Scholar]
- 31.LaFemina R L, Callahan P L, Cordingley M G. Substrate specificity of recombinant human immunodeficiency virus integrase protein. J Virol. 1991;65:5624–5630. doi: 10.1128/jvi.65.10.5624-5630.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.LaFemina R L, Schneider C L, Robbins H L, Callahan P L, LeGrow K, Roth E, Schleif W A, Emini E A. Requirement of active human immunodeficiency virus type 1 integrase enzyme for productive infection of human T-lymphoid cells. J Virol. 1992;66:7414–7419. doi: 10.1128/jvi.66.12.7414-7419.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lawn R M, Efstratiadis A, O’Connell C, Maniatis T. The nucleotide sequence of the human beta-globin gene. Cell. 1980;21:647–651. doi: 10.1016/0092-8674(80)90428-6. [DOI] [PubMed] [Google Scholar]
- 34.Leavitt A D, Robles G, Alesandro N, Varmus H E. Human immunodeficiency virus type 1 integrase mutants retain in vitro integrase activity yet fail to integrate viral DNA efficiently during infection. J Virol. 1996;70:721–728. doi: 10.1128/jvi.70.2.721-728.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leavitt A D, Rose R B, Varmus H E. Both substrate and target oligonucleotide sequences affect in vitro integration mediated by human immunodeficiency virus type 1 integrase protein produced in Saccharomyces cerevisiae. J Virol. 1992;66:2359–2368. doi: 10.1128/jvi.66.4.2359-2368.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liang C, Li X, Rong L, Inouye P, Quan Y, Kleiman L, Wainberg M A. The importance of the A-rich loop in human immunodeficiency virus type 1 reverse transcription and infectivity. J Virol. 1997;71:5750–5757. doi: 10.1128/jvi.71.8.5750-5757.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lodi P J, Ernst J A, Kuszewski J, Hickman A B, Engelman A, Craigie R, Clore G M, Gronenborn A M. Solution structure of the DNA binding domain of HIV-1 integrase. Biochemistry. 1995;34:9826–9833. doi: 10.1021/bi00031a002. [DOI] [PubMed] [Google Scholar]
- 38.Masuda T, Planelles V, Krogstad P, Chen I S Y. Genetic analysis of human immunodeficiency virus type 1 integrase and the U3 att site: unusual phenotype of mutants in the zinc finger-like domain. J Virol. 1995;69:6687–6696. doi: 10.1128/jvi.69.11.6687-6696.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mazumder A, Gupta M, Pommier Y. Methylphosphonodiester substitution near the conserved CA dinucleotide in the HIV LTR alters both extent of 3′-processing and choice of nucleophile by HIV-1 integrase. Nucleic Acids Res. 1994;22:4441–4448. doi: 10.1093/nar/22.21.4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mazumder A, Neamati N, Ojwang J O, Sunder S, Rando R F, Pommier Y. Inhibition of the human immunodeficiency virus type 1 integrase by guanosine quartet structures. Biochemistry. 1996;35:13762–13771. doi: 10.1021/bi960541u. [DOI] [PubMed] [Google Scholar]
- 41.Murphy J E, Goff S P. Construction and analysis of deletion mutations in the U5 region of Moloney murine leukemia virus: effects on RNA packaging and reverse transcription. J Virol. 1989;63:319–327. doi: 10.1128/jvi.63.1.319-327.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Murphy J E, Goff S P. A mutation at one end of Moloney murine leukemia virus DNA blocks cleavage of both ends by the viral integrase in vivo. J Virol. 1992;66:5092–5095. doi: 10.1128/jvi.66.8.5092-5095.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Panganiban A T, Temin H M. The terminal nucleotides of retrovirus DNA are required for integration but not virus production. Nature. 1983;306:155–160. doi: 10.1038/306155a0. [DOI] [PubMed] [Google Scholar]
- 44.Planelles V, Bachelerie F, Jowett J B M, Haislip A, Xie Y, Banooni P, Masuda T, Chen I S Y. Fate of the human immunodeficiency virus type 1 provirus in infected cells: a role for vpr. J Virol. 1995;69:5883–5889. doi: 10.1128/jvi.69.9.5883-5889.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reicin A S, Kalpana G, Paik S, Marmon S, Goff S. Sequences in the human immunodeficiency virus type 1 U3 region required for in vivo and in vitro integration. J Virol. 1995;69:5904–5907. doi: 10.1128/jvi.69.9.5904-5907.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roth M J, Schwartzberg P L, Goff S P. Structure of the termini of DNA intermediates in the integration of retroviral DNA: dependence on IN function and terminal DNA sequence. Cell. 1989;58:47–54. doi: 10.1016/0092-8674(89)90401-7. [DOI] [PubMed] [Google Scholar]
- 47.Schwartzberg P, Colicelli J, Goff S P. Construction and analysis of deletion mutations in the pol gene of Moloney murine leukemia virus: a new viral function required for productive infection. Cell. 1984;37:1043–1052. doi: 10.1016/0092-8674(84)90439-2. [DOI] [PubMed] [Google Scholar]
- 48.Scottoline B P, Chow S, Ellison V, Brown P O. Disruption of the terminal base pairs of retroviral DNA during integration. Genes Dev. 1997;11:371–382. doi: 10.1101/gad.11.3.371. [DOI] [PubMed] [Google Scholar]
- 49.Sherman P A, Fyfe J A. Human immunodeficiency virus integration protein expressed in Escherichia coli possesses selective DNA cleaving activity. Proc Natl Acad Sci USA. 1990;87:5119–5123. doi: 10.1073/pnas.87.13.5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shibagaki Y, Chow S A. Central core domain of retroviral integrase is responsible for target site selection. J Biol Chem. 1997;272:8361–8369. doi: 10.1074/jbc.272.13.8361. [DOI] [PubMed] [Google Scholar]
- 51.Shibagaki Y, Holmes M L, Appa R S, Chow S A. Characterization of feline immunodeficiency virus integrase and analysis of functional domains. Virology. 1997;230:1–10. doi: 10.1006/viro.1997.8466. [DOI] [PubMed] [Google Scholar]
- 52.van Gent D C, Vink C, Groeneger A A, Plasterk R H. Complementation between HIV integrase proteins mutated in different domains. EMBO J. 1993;12:3261–3267. doi: 10.1002/j.1460-2075.1993.tb05995.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vicenzi E, Dimitrov D S, Engelman A, Migone T-S, Purcell D F J, Leonard J, Englund G, Martin M A. An integration-defective U5 deletion mutant of human immunodeficiency virus type 1 reverts by eliminating additional long terminal repeat sequences. J Virol. 1994;68:7879–7890. doi: 10.1128/jvi.68.12.7879-7890.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vink C, van Gent D C, Elgersma Y, Plasterk R H. Human immunodeficiency virus integrase protein requires a subterminal position of its viral DNA recognition sequence for efficient cleavage. J Virol. 1991;65:4636–4644. doi: 10.1128/jvi.65.9.4636-4644.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Whitcomb J M, Hughes S H. Retroviral reverse transcription and integration: progress and problems. Annu Rev Cell Biol. 1992;8:275–306. doi: 10.1146/annurev.cb.08.110192.001423. [DOI] [PubMed] [Google Scholar]
- 56.Yoshinaga T, Fujiwara T. Different roles of bases within the integration signal sequence of human immunodeficiency virus type 1 in vitro. J Virol. 1995;69:3233–3236. doi: 10.1128/jvi.69.5.3233-3236.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yoshinaga T, Kimura-Ohtani Y, Fujiwara T. Detection and characterization of a functional complex of human immunodeficiency virus type 1 integrase and its DNA substrate by UV cross-linking. J Virol. 1994;68:5690–5697. doi: 10.1128/jvi.68.9.5690-5697.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zack J A, Arrigo S J, Weitsman S R, Go A S, Haislip A, Chen I S Y. HIV-1 entry into quiescent primary lymphocytes: molecular analysis reveals a labile, latent viral structure. Cell. 1990;61:213–222. doi: 10.1016/0092-8674(90)90802-l. [DOI] [PubMed] [Google Scholar]
- 59.Zheng R, Jenkins T M, Craigie R. Zinc folds the N-terminal domain of HIV-1 integrase, promotes multimerization, and enhances catalytic activity. Proc Natl Acad Sci USA. 1996;93:13659–13664. doi: 10.1073/pnas.93.24.13659. [DOI] [PMC free article] [PubMed] [Google Scholar]