Abstract
The human gut is home to trillions of microorganisms that are responsible for the modification of many orally administered drugs, leading to a wide range of therapeutic outcomes. Prodrugs bearing an azo bond are designed to treat inflammatory bowel disease and colorectal cancer via microbial azo reduction, allowing for topical application of therapeutic moieties to the diseased tissue in the intestines. Despite the inextricable link between microbial azo reduction and the efficacy of azo prodrugs, the prevalence, abundance, and distribution of azoreductases have not been systematically examined across the gut microbiome. Here, we curated and clustered amino acid sequences of experimentally confirmed bacterial azoreductases and conducted a hidden Markov model–driven homolog search for these enzymes across 4644 genome sequences present in the representative Unified Human Gastrointestinal Genomes collection. We identified 1958 putative azo-reducing species, corroborating previous findings that azo reduction appears to be a ubiquitous function of the gut microbiome. However, through a systematic comparison of predicted and confirmed azo-reducing strains, we hypothesize the presence of uncharacterized azoreductases in 25 prominent strains of the human gut microbiome. Finally, we confirmed the azo reduction of Acid Orange 7 by multiple strains of Fusobacterium nucleatum, Bacteroides fragilis, and Clostridium clostridioforme. Together, these results suggest the presence and activity of many uncharacterized azoreductases in the human gut microbiome and motivate future studies aimed at characterizing azoreductase genes in prominent members of the human gut microbiome.
SIGNIFICANCE STATEMENT
This work systematically examined the prevalence, abundance, and distribution of azoreductases across the healthy and inflammatory bowel disease human gut microbiome, revealing potentially uncharacterized azoreductase genes. It also confirmed the reduction of Acid Orange 7 by strains of Fusobacterium nucleatum, Bacteroides fragilis, and Clostridium clostridioforme.
Introduction
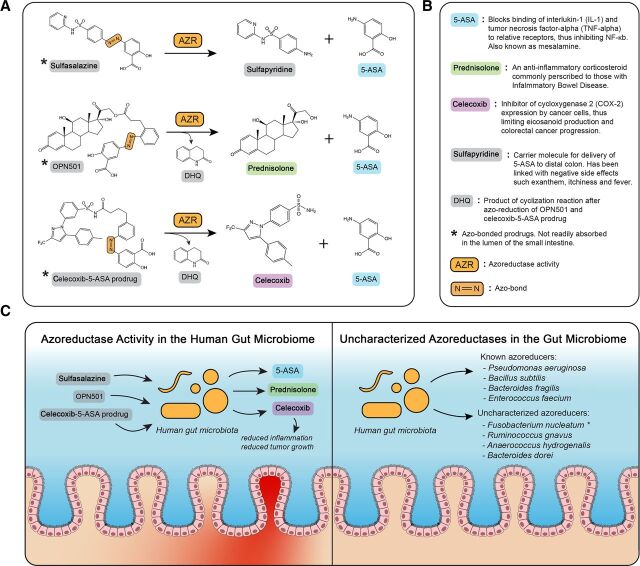
Orally administered drugs are an attractive, noninvasive mode of delivery of pharmaceuticals to the intestines. The human gut microbiome plays an important role in drug metabolism (Spanogiannopoulos et al., 2016) and is capable of activating (Peppercorn and Goldman, 1972; Morrison et al., 2012; Sousa et al., 2014), inactivating (Peters, 1978; Saha et al., 1983; Haiser et al., 2013), and even toxifying (Wallace et al., 2010) pharmaceutical drugs. Prodrugs containing an azo bond actually require bacterial azoreductase activity to release biologically active compounds (Peppercorn and Goldman, 1972). For conditions such as inflammatory bowel disease (IBD) and colorectal cancer (CRC), bacterial azoreductases have been used to deliver therapeutics such as 5-aminosalicylic acid (5-ASA), prednisolone (Ruiz et al., 2011), and celecoxib (Ruiz et al., 2011) topically to diseased intestinal tissues (Fig. 1). Following oral administration of sulfasalazine, bacterial azoreductases in the gut reduce azo bonds, liberating 5-ASA and allowing it to confer its anti-inflammatory properties (Mahida et al., 1991; Rachmilewitz et al., 1992; Weber et al., 2000) topically on inflamed intestinal tissue. Direct oral administration of 5-ASA is nonoptimal because the majority of the drug is absorbed in the small intestine and is sent through systemic circulation (Peppercorn and Goldman, 1973; Tozaki et al., 2002; Friend, 2005; Perrotta et al., 2015; Foppoli et al., 2019). Other examples of azo-bonded prodrugs are OPN501 and celecoxib-5-ASA. OPN501 is made of up prednisolone, 5-ASA, and an inert cyclization product, dihydroquinolone (DHQ). Upon azo reduction, 5-ASA is released and is able to act topically upon the target tissue. Following a spontaneous cyclization reaction, prednisolone and DHQ are released where prednisolone can act upon the target tissue (Ruiz et al., 2011). Celecoxib-5-ASA is made up of celecoxib, 5-ASA, and DHQ, which are all released upon azo reduction and cyclization in a similar mechanistic fashion to OPN501 (Ruiz et al., 2011).
Fig. 1.
Azo reduction by gut microbiota. (A) Pathways of azo-bonded drug activation by bacterial azoreductase activity. DHQ is produced via cyclization of an intermediate of OPN501 and celecoxib-5-ASA prodrug following intramolecular lactamization (Ruiz et al., 2011). (B) Description of downstream metabolites of bacterial azo reduction and the mechanisms of action in IBD and CRC. References for each molecular function described in this subfigure: 5-ASA (Mahida et al., 1991; Cominelli et al., 1992; Rachmilewitz et al., 1992), prednisolone (Cohen et al., 2000), celecoxib (Wu et al., 2003, ; Wu et al., 2010; Gustafsson et al., 2010), sulfapyridine (Nielsen, 1982), and DHQ (Ruiz et al., 2011). (C) Presence of azoreductase-containing bacteria is required for prodrug activation in the IBD gut and CRC gut (left). Many azo-reducing bacteria have been characterized; however, some species have shown experimental evidence of azo-reduction without the full characterization of the genes responsible for azo reduction (right).
A recent review article by Suzuki (2019) collected and curated experimentally confirmed azoreductases and described their preferred flavin cofactors and electron donors. They found that bacterial azoreductases qualitatively cluster into four main clades, which harbor a preference for either flavin mononucleotide (FMN) or flavin adenine dinucleotide as the flavin cofactor and either NADH or NADPH as the preferred electron donor. Clades I, II, and III are flavoproteins, whereas clade IV proteins are flavin free. Clade I azoreductases prefer NADPH as the electron donor, clade II prefers NADH as its electron donor, and clades III and IV generally use both. Of the 37 enzymes examined by Suzuki (2019), eight enzymes formed no distinct phylogenetic clade and featured differences in primary sequence length, flavin cofactor, and preferred electron donor. In addition to their relevance in drug delivery and efficacy, azoreductases are involved in nitroreduction (Brown 1981; Rafii et al., 1990; Rafii and Cerniglia, 1995; Liu et al., 2007; Mercier et al., 2013; Chalansonnet et al., 2017), quinone oxidoreduction (Liu et al., 2008; Leelakriangsak et al., 2008; Ryan et al., 2010a; Ryan et al., 2010b; Ryan et al., 2014), and azo dye reduction. Azo dyes such as Allura Red and Brilliant Black are commonly used in the food and textile industries, and waste from their production and usage pollutes the environment. This has led to a plethora of manuscripts characterizing the activity of azoreductases across the bacterial kingdom (Cerniglia et al., 1982; Zimmermann et al., 1982; Nakanishi et al., 2001; Suzuki et al., 2001; Blümel et al., 2002; Blümel and Stolz, 2003; Chen et al., 2004; Chen et al., 2005; Nachiyar and Rajakumar, 2005; Ooi et al., 2007; Matsumoto et al., 2010; Misal et al., 2011; Feng et al., 2012; Gonçalves et al., 2013; Lang et al., 2013; Misal et al., 2014; Eslami et al., 2016; Zhang et al., 2016), many of which exhibit nonnegligible sequence similarity to gut microbial azoreductases (Suzuki 2019).
There is a growing body of literature suggesting that azo reduction is a ubiquitous function of the human gut microbiome (Javdan et al., 2020), with many prominent bacterial strains showing significant reduction of sulfasalazine in vitro (Zimmermann et al., 2019). Javdan et al. (2020) showed that among 20 different individuals, sulfasalazine reduction was one of the only ubiquitous functions of the gut microbiome. Zimmermann et al. (2019) tested the reduction of sulfasalazine by 76 prominent strains of the gut microbiome and reported a significant [false discover rate (FDR) adjusted P value < 0.05] reduction of sulfasalazine by 62 of these strains. Interestingly, some of these experimentally confirmed azo-reducing strains reported by Zimmermann et al. (2019) have no prior evidence of azo-reduction and, thus, may encode novel or uncharacterized azoreductase genes (Fig. 1C). Identification of known azoreductases in newly reported sulfasalazine-reducing species can help narrow down strains to target for identification of uncharacterized azoreductases.
The prevalence, abundance, expression, and distribution of azoreductase enzymes in the human gut microbiome have implications for the efficacy of existing prodrugs mentioned above, as well as for the development of future azo prodrugs. Although azoreductases have been identified and characterized in many gut bacteria (Supplemental Table 1), the distribution of azoreductases has not been systematically explored across current gut bacterial reference genomes. To address this gap, we conducted a homolog search of known azoreductases across the Unified Human Gastrointestinal Genomes (UHGG) collection (Almeida et al., 2020) to identify putative azoreductases and azo-reducing species in the human gut microbiome. We then assessed the relative abundance and expression of known azoreductases in healthy, ulcerative colitis (UC), and Crohn disease (CD) participants of the Integrative Human Microbiome Project (HMP2) (Proctor and Huttenhower, 2019), the Prospective Registry of IBD Patients at MGH (PRISM) (Franzosa et al., 2019), and the Health Professionals Follow-Up Study (HPFS) (Abu-Ali et al., 2018). Finally, we tested the in vitro azo reduction of Acid Orange 7 by three strains of Fusobacterium nucleatum along with two strains of Bacteroides fragilis and two strains of Clostridium clostridioforme.
Materials and Methods
Description of Publicly Available Shotgun Metagenomic Sequencing Data
Shotgun metagenomic sequencing data obtained from the HMP2 [N (samples) = 703, N (individuals) = 103] (Proctor and Huttenhower, 2019), the PRISM [N (samples) = 218, N (individuals) = 218] (Franzosa et al., 2019), and the HPFS [N (samples) = 220, N (individuals) = 220] (Abu-Ali et al., 2018) are used throughout this work. Note that all samples referred to throughout this work are human stool samples.
Curation of Hidden Markov Models Representing Azoreductase Enzymes
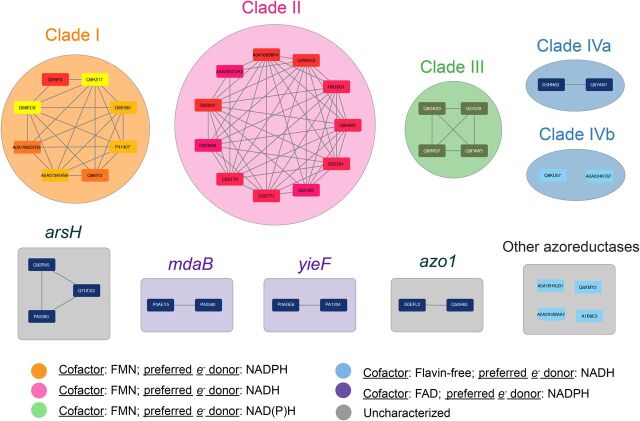
We searched the literature for known and experimentally validated species of bacteria that have azoreductase activity. The list of gene sequences collected, along with the relevant metadata (organism, functional annotation, length, etc.) is available in Supplemental Table 1. Preliminary evidence for azoreductase gene sequence clustering is shown in Suzuki (2019), where sequences were aligned and phylogenetically compared. Next, after collecting 40 sequences of experimentally validated azoreductases, we generated a sequence similarity network using the Enzyme Function Initiative-Enzyme Similarity Tool (EFI-EST) (Gerlt et al., 2015) at a 35% amino acid sequence identity threshold for identifying similar clusters of azoreductase genes. This threshold corroborates the preliminary evidence for a diversity of azoreductase sequences put forth by Suzuki (2019). With the exception of clade IVb sequences, which reached 31% sequence identity, all other clusters of genes had at least 35% sequence identity. The groups shown in Fig. 2 were pressed into profile hidden Markov models (HMMs) using HMMER version 3.1b2 (Finn et al., 2015). Other genes that did not fall into the clade I–clade IV clusters (arsH, yieF, mdaB, azo1, azoR, etc.) were pressed into singular HMMs and included in the homolog search.
Fig. 2.
Bacterial azoreductases cluster by cofactor and electron donor preferences. Following an extensive literature search for experimentally confirmed bacterial azoreductases, amino acid sequences of 40 azoreductase enzymes were collected and clustered with EFI-EST (Gerlt et al., 2015) at 35% sequence identity. Each node in the figure above is a single azoreductase gene, and the edges between nodes indicate at least 35% sequence identity between the two amino acid sequences. The colored clusters, clade I through clade IVa and IVb, are groups of azoreductases previously described by Suzuki (2019) as mechanistically similar groups based on cofactor and electron donor preferences. Clusters labeled with gene names (mdaB, yieF, etc.) represent homologous gene sequences found in two or more organisms. Each mechanistically characterized group of azoreductases were subsequently pressed into profile HMMs using HMMER v3.1b2 (Finn et al., 2015), which formed the basis of the homolog search. The group labeled “other azoreductases” contains sequences that did not fall into any cluster at the 35% identity threshold and were pressed into singular HMMs prior to the homolog search.
Search for Azoreductase Genes across Human Gut Microbial Genomes
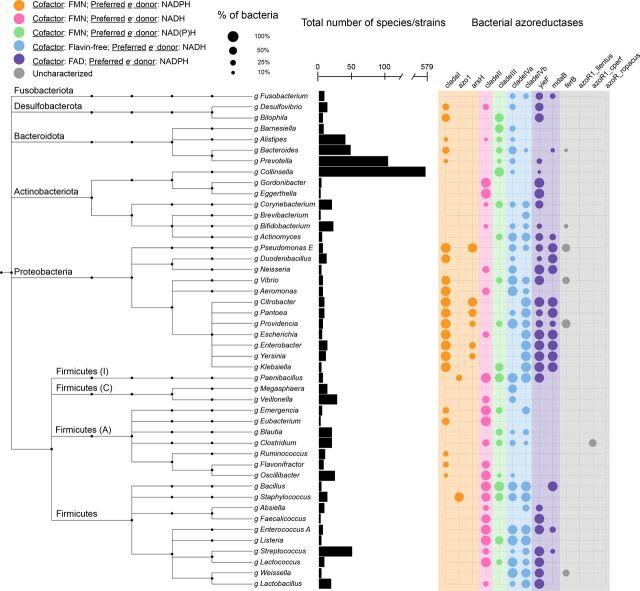
We searched HMMs of known, experimentally validated azoreductases across 4644 nonredundant genomes contained in the UHGG collection (Almeida et al., 2020) using HMMER v3.1b2 (Finn et al., 2015). Alignments to queried HMMs with E-value < 1 × 10−10 and 60% coverage of the query sequence were labeled as putative azoreductase gene sequences. Putative azo-reducing bacterial species with experimentally confirmed azoreductase activity (Supplemental Table 2) were then labeled as “known” azo-reducing species and classified separately from the putative azo-reducing species. Putative azo-reducing species across the bacterial taxonomy were visualized using the iTOL web interface (Letunic and Bork, 2019), and prominent phyla of the gut microbiome were subsetted and presented in Fig. 3.
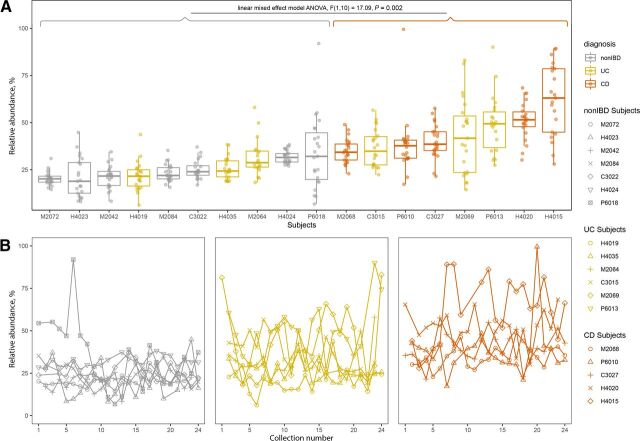
Fig. 3.
Azoreductases are widely distributed across gut bacterial taxonomy. Presence/absence of azoreductases across prominent phyla of the human gut microbiome. The taxonomic tree is obtained from the UHGG (Almeida et al., 2020) which is built on the Genome Taxonomy Database (Chaumeil et al., 2019). Phyla names are annotated on the left side. Phyla names followed by a capital letter, e.g., Firmicutes (A), indicate a novel phylum classified by the Genome Taxonomy Database toolkit. The bar chart in the center indicates the number of species contained in each genus shown in the tree. The size of the circles indicates the number of species that contain hits to the azoreductase genes specified. The color of the circles indicates the cofactor and preferred electron (e-) donor of the enzyme.
Relative Abundance and Azoreductase Gene Abundance and Expression Estimation
Raw sequencing reads for samples from HMP2 (Proctor and Huttenhower, 2019), PRISM (Franzosa et al., 2019), and HPFS (Abu-Ali et al., 2018) were downloaded and extracted with the National Center for Biotechnology Information’s SRA toolkit v2.10.9 (http://ncbi.github.io/sra-tools/). Quality control and adapter trimming of the fastq sequence files were done with the Trim Galore wrapper v0.6.6 (https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/). To remove potential human contaminants, quality trimmed reads were screened against the human genome (hg19) with Bowtie2 v2.4.2 (Langmead and Salzberg, 2012). Putative azoreductase sequences were extracted from UHGG genomes via custom shell and python scripts. Putative azoreductase gene sequences (HMP2, PRISM) and expression levels (HPFS) were quantified using salmon v1.4.0 (Patro et al., 2017) and were normalized and aggregated in R v4.1.1 and were subsequently visualized using the R package ggplot2 (Wickham, 2011) (Fig. 4). Taxonomy profiling of the cleaned metagenomic reads from HMP2 samples was performed using Kraken 2 v2.0.8-beta (Wood et al., 2019) to estimate the relative abundance of bacterial species present in each dataset. These relative abundances were then processed in R v4.1.1 and plotted using ggplot2 (Fig. 5). All computational and bioinformatic procedures are open source and are provided at https://github.com/dombraccia/Azoreductases.
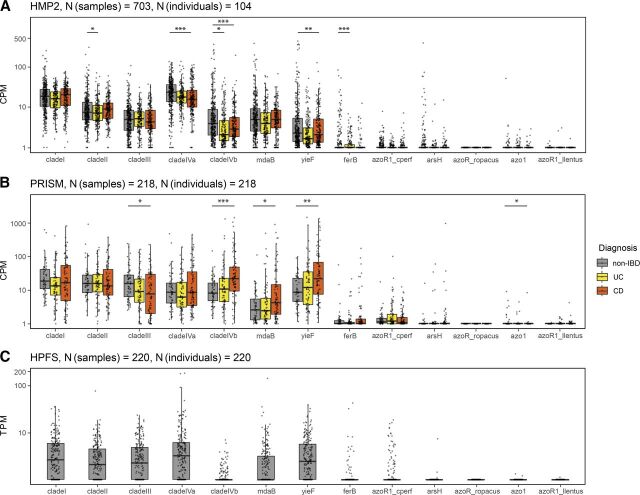
Fig. 4.
Abundance and expression of azoreductase genes by human gut microbiota. (A and B) Visualization of shotgun metagenomic sequencing data from the Human Microbiome Project 2, also known as the HMP2 and the PRISM. We used salmon 1.4.0 (Patro et al., 2017) to quantify the abundance of azoreductase genes from hundreds of stool samples across healthy controls (non-IBD), UC, and CD participant cohorts. Raw DNA read alignment counts were normalized to counts per million (CPM), analogous to transcripts-per-million (TPM) normalization. Asterisks above each boxplot indicate statistical significance (*P < 0.05; **P < 0.01; ***P < 0.001, Wilcoxon rank sum test, all FDR adjusted using Benjamini-Hochberg method). (C) Visualization of high-throughput metatranscriptomic data obtained from the HPFS. We quantified the expression of bacterial azoreductases using salmon v1.4.1 (Patro et al., 2017) and normalized the raw read alignment statistics to transcripts per million (TPM). Please see Materials and Methods for a more detailed description of the computational and statistical methods employed.
Fig. 5.
Known and putative azo-reducing species are more abundant in the IBD gut. Relative abundance of known and putative azo-reducing species in HMP2 subjects with more than 20 total stool collections. (A) Relative abundance of known and putative azo-reducing species for qualifying participants across non-IBD, UC, and CD populations. Subjects with CD have significantly higher relative abundances of known+putative azo-reducing species than healthy subjects (non-IBD) per linear mixed-effects model ANOVA, F(1,10) = 17.09, P < 0.003. (B) Relative abundance of putative azo-reducing species over time for healthy (non-IBD), UC, and CD participants. Each line represents a single participant, and each point is the summed relative abundance of known+putative azo-reducing species at that collection point. The key on the right links relative abundance distributions for each subject with the same data point shown over collection numbers. Collections were taken approximately every 14 days.
Statistical Analysis of Relative Metagenomic Sequence Data from HMP2 and PRISM Datasets
Statistical analyses described in Fig. 4 were performed in R 4.1.1 with the Wilcoxon test method using default parameterization. Next, we compared the relative abundances of known and putative azo-reducing species for HMP2 subjects with 20 or more stool samples taken over the course of the study (Fig. 5). A linear mixed-effects model ANOVA was performed on this subset of HMP2 data to determine any statistically significant differences in relative abundance values across non-IBD, UC, and CD subjects. The R package lme4 (Bates et al., 2014) was used to fit the model to the data, and the package lmerTest (Kuznetsova et al., 2017) was used to perform the ANOVA on the model. All statistical analyses were performed in R version 4.1.1 and are provided at https://github.com/dombraccia/Azoreductases.
Acid Orange 7 Azo Reduction Assay
Biologic triplicates were grown in a 10 ml tube containing 10 ml of Yeast Casitone Fatty Acids (YCFA) broth for B. fragilis and C. clostridioforme strains and 10 ml of Brain Heart Infusion (BHI) broth for F. nucleatum strains. Each tube was inoculated with 10 μL of bacteria from glycerol stocks. The final concentrations of each substrate in the bacterial cultures were 50 μg/ml of FMN, 50 μg/ml of NADH, and 50 μmol/ml of Acid Orange 7. The bacterial cultures were left to grow in an anaerobic chamber for 72 hours, and Acid Orange 7 decolorization was measured once every 24 hours since inoculation. The decolorization of Acid Orange 7 was measured by aliquoting triplicates of 200 μL media aliquants to a 96-well plate for each bacterial culture, and absorbance of 550 nm light was measured using a spectrophotometer. The raw absorbance values for each biologic and technical replicate are reported in Supplemental Table 5.
Results
Primary Amino Acid Sequences of Bacterial Azoreductases Group by Mechanistic Preferences
To begin identifying putative azo-reducing species of the gut microbiome, we searched the literature for experimentally verified azoreductase enzymes, collected amino acid sequences, and metadata for these azoreductases (Supplemental Table 1) and compared their primary sequences using the EFI-EST (Gerlt et al., 2015). The resulting sequence similarity network captured mechanistic preferences such as flavin dependence and electron donor preference for each azoreductase reported by Suzuki (2019). We saw near-complete concordance between the clade I–IV azoreductases and the sequence similarity clusters at a 35% amino acid identity edge threshold, with the exception of clade IV, which was split into two separate clusters (Fig. 2). The gene families labeled arsH, mdaB, yieF, and azo1 clustered separately from clade I–IV azoreductases, and we consider each of these clusters as separate subfamilies of azoreductases. Multiple sequence alignments were generated for each cluster shown in Fig. 2 (with the exception of the “Other Azoreductases” group) using MUSCLE v3.8.425 (Edgar, 2004). HMMs were then trained on the multiple sequence alignments from the previous step using the HMMER v3.1b2 method hmmpress (Finn et al., 2015) and were queried against the UHGG collection (Almeida et al., 2020).
Homolog Search for Azoreductases Supports Evidence for Ubiquity of Azo Reduction by the Human Gut Microbiome
We searched for homologs of azoreductases across 4644 representative genomes in the UHGG collection (Almeida et al., 2020) using HMMs generated from sequences of experimentally validated azoreductase enzymes (Fig. 3). This collection contains 204,938 genome sequences of bacteria known to inhabit the human gut, of which 4644 are included in the representative collection (Supplemental Table 4). For the remainder of this work, we refer to species receiving statistically significant (E-value < 1 × 10−10) hits to azoreductase genes as “putative azo-reducing species” or “putative azo-reducing bacteria.” Of the 4644 genomes in the UHGG collection, there are 1443 (31.1%) with one putative azoreductase gene, 343 (7.4%) with two or more putative azoreductases, and 372 (8.0%) with three or more putative azoreductases, indicating the extensive potential of the gut microbiome to reduce azo bonds. Most notably, 364 genomes contain hits to the clade I profile, 452 contain hits to the clade II profile, 793 genomes contain hits to the clade III profile, 568 contain hits to the clade IVa profile, 410 contain hits to the clade IVb profile, 285 contain hits to the mdaB profile, and 477 contain hits to the yieF profile. Prominent phyla of the gut microbiome, such as Proteobacteria and Firmicutes, appear particularly rich with clade I, clade II, clade III, clade IVab, and flavin adenine dinucleotide utilizing azoreductases (purple columns in Fig. 3).
Systematic Evaluation of Predicted Azo-Reducing Species
We next sought to evaluate the results of our azoreductase homolog search with recent findings by Zimmermann et al. (2019) regarding sulfasalazine reduction. Zimmermann et al. (2019) tested the degradation of sulfasalazine by 76 prominent gut bacterial strains, 67 of which had corresponding reference genomes present in the UHGG collection. This provided an excellent source of data to compare our bioinformatic predictions against. We determined the sulfasalazine-reducing status as either sulfasalazine reducing (SR) or non–sulfasalazine reducing for each of the 67 strains based on the significant (FDR adjusted P value < 0.05) reduction of sulfasalazine in vitro reported by Zimmermann et al. (2019) (Table 1). We also determined the predicted reducing status as either a predicted reducer (PR) or a non–predicted reducer (Non-PR) for each strain based on the presence of a putative azoreductase identified from the homolog search step. For each strain, the sulfasalazine-reducing status and predicted reducing status were systematically compared to validate the results of the azoreductase homolog search (Table 1, columns 7–9). We correctly predicted the sulfasalazine-reducing status for 47.8% (32/67) of strains and we incorrectly predicted the sulfasalazine reducing status for 52.2% (35/67) of strains (Table 2). The vast majority (77.1%, 27/35) of incorrectly predicted strains are false negatives, meaning the strain does reduce sulfasalazine in vitro, but we did not identify an azoreductase in the homolog search step (Table 2). Interestingly, the majority of false positives (75%, 6/8) are members of the Proteobacteria phylum, which we previously noted to be particularly rich in azoreductase gene sequences (Fig. 3).
TABLE 1.
Systematic comparison with Zimmermann et al. (2019) sulfasalazine consumption results Complete results from the systematic comparison of predicted sulfasalazine-reducing bacteria to actual sulfasalazine-reducing bacteria. The columns labeled FC (fold change), FC_STD (standard deviation in fold change), P_FDR (FDR adjusted P value), Pct_Consumed (percent consumed), and Pct_Consumed_STD (standard deviation of the percent consumed) were all obtained directly from Zimmermann et al. (2019) (Supplemental Table 3). The SR_Status column contains values SR and non-SR, which were determined based on significant (p_FDR < 0.05) or nonsignificant (p_FDR ≥ 0.05) sulfasalazine reduction. The PR_Status column contains the values PR and non-PR, which were determined based on the presence or absence of one or more azoreductase homologs determined from the homolog search step. The final column, Result, contains the values TP (true positive), TN (true negative), FP (false positive), and FN (false negative). Correctly predicted SR strains have a result of TP and correctly predicted Non-SR strains have a result of TN whereas incorrectly predicted SR strains have a result of FP and incorrectly predicted Non-SR strains have a result of FN.
| Strain_Name | FC | FC_STD | p_FDR | Pct_Consumed | Pct_Consumed_STD | SR_Status | PR_Status | Result |
|---|---|---|---|---|---|---|---|---|
| Akkermansia muciniphila ATCCBAA-835 | −0.419 | 0.361 | 0.19 | 25.222 | 18.707 | Non-SR | Non-PR | TN |
| Alistipes indistinctus DSM 22520 | −9.01 | 0.11 | 0.003 | 99.806 | 0.015 | SR | PR | TP |
| Anaerococcus hydrogenalis DSM7454 | −8.826 | 0.198 | 0.008 | 99.78 | 0.03 | SR | Non-PR | FN |
| Anaerotruncus colihominis DSM17241 | −8.088 | 1.575 | 0.016 | 99.633 | 0.401 | SR | Non-PR | FN |
| Bacteroides caccae ATCC43185 | −9.247 | 0.262 | 0.008 | 99.835 | 0.03 | SR | PR | TP |
| Bacteroides cellulosilyticus DSM14838 | −1.502 | 0.308 | 0.002 | 64.691 | 7.532 | SR | PR | TP |
| Bacteroides coprophilus DSM18228 | −0.006 | 0.216 | 0.991 | 0.386 | 14.918 | Non-SR | Non-PR | TN |
| Bacteroides dorei DSM17855 | −4.631 | 0.748 | 0.013 | 95.965 | 2.091 | SR | Non-PR | FN |
| Bacteroides eggerthii DSM20697 | −9.59 | 0.104 | 0.002 | 99.87 | 0.009 | SR | Non-PR | FN |
| Bacteroides finegoldii DSM17565 | −0.79 | 0.296 | 0.042 | 42.176 | 11.877 | SR | Non-PR | FN |
| B. fragilis 3397 T10 | −0.974 | 0.348 | 0.009 | 49.084 | 12.282 | SR | PR | TP |
| B. fragilis ATCC43859 | −10.576 | 0.088 | 0.003 | 99.934 | 0.004 | SR | PR | TP |
| B. fragilis DS-208 | −9.327 | 0.57 | 0.006 | 99.844 | 0.062 | SR | PR | TP |
| B. fragilis HMW610 | −10.522 | 0.159 | 0.004 | 99.932 | 0.007 | SR | PR | TP |
| B. fragilis HMW615 | −10.398 | 0.256 | 0.013 | 99.926 | 0.013 | SR | PR | TP |
| B. fragilis NCTC9343 | −6.184 | 0.835 | 0.006 | 98.625 | 0.796 | SR | PR | TP |
| B. fragilis T(B)9 | −9.252 | 0.144 | 0.005 | 99.836 | 0.016 | SR | PR | TP |
| Bacteroides intestinalis DSM17393 | −1.296 | 0.398 | 0.005 | 59.267 | 11.246 | SR | PR | TP |
| Bacteroides ovatus ATCC8483 | −0.285 | 0.473 | 0.546 | 17.951 | 26.915 | Non-SR | PR | FP |
| Bacteroides pectinophilus ATCC43243 | −0.249 | 0.241 | 0.268 | 15.832 | 14.042 | Non-SR | Non-PR | TN |
| Bacteroides stercoris ATCC43183 | −0.588 | 0.326 | 0.053 | 33.46 | 15.033 | Non-SR | Non-PR | TN |
| Bacteroides thetaiotaomicron 3731 | −1.303 | 0.248 | 0.005 | 59.461 | 6.961 | SR | PR | TP |
| Bacteroides thetaiotaomicron 7330 | −1.032 | 0.237 | 0.003 | 51.082 | 8.044 | SR | PR | TP |
| Bacteroides thetaiotaomicron VPI-5482 | −1.252 | 0.155 | 0.006 | 58.006 | 4.511 | SR | PR | TP |
| Bacteroides uniformis ATCC8492 | −2.605 | 0.389 | 0.001 | 83.558 | 4.436 | SR | Non-PR | FN |
| Bacteroides xylanisolvens DSM18836 | −9.663 | 0.115 | 0.002 | 99.877 | 0.01 | SR | PR | TP |
| Bifidobacterium adolescentis ATCC15703 | −0.7 | 0.271 | 0.014 | 38.442 | 11.584 | SR | Non-PR | FN |
| Bifidobacterium breve DSM20213 | −9.241 | 0.182 | 0.008 | 99.835 | 0.021 | SR | Non-PR | FN |
| Blautia hansenii DSM20583 | −9.234 | 0.155 | 0.005 | 99.834 | 0.018 | SR | Non-PR | FN |
| Bryantia formatexigens DSM14469 | −0.722 | 0.432 | 0.113 | 39.359 | 18.171 | Non-SR | PR | FP |
| Clostridium asparagiforme DSM15981 | −9.184 | 0.203 | 0.01 | 99.828 | 0.024 | SR | Non-PR | FN |
| Clostridium bolteae ATCCBAA-613 | −7.113 | 0.611 | 0.005 | 99.277 | 0.306 | SR | Non-PR | FN |
| Clostridium difficile 120 | −6.039 | 0.703 | 0.013 | 98.479 | 0.741 | SR | PR | TP |
| Clostridium ramosum DSM1402 | −9.046 | 0.169 | 0.006 | 99.811 | 0.022 | SR | PR | TP |
| Clostridium scindens ATCC35704 | −9.064 | 0.324 | 0.004 | 99.813 | 0.042 | SR | Non-PR | FN |
| Clostridium spiroforme DSM1552 | −2.478 | 0.572 | 0.001 | 82.055 | 7.119 | SR | Non-PR | FN |
| Clostridium sporogenes ATCC15579 | −9.072 | 0.158 | 0.006 | 99.814 | 0.02 | SR | PR | TP |
| Clostridium symbiosum ATCC14940 | −9.256 | 0.209 | 0.009 | 99.836 | 0.024 | SR | Non-PR | FN |
| Collinsella aerofaciens ATCC25986 | −6.629 | 0.628 | 0.046 | 98.99 | 0.44 | SR | PR | TP |
| Collinsella intestinalis DSM13280 | −8.916 | 0.118 | 0.003 | 99.793 | 0.017 | SR | Non-PR | FN |
| Coprococcus comes ATCC27758 | −9.182 | 0.199 | 0.009 | 99.828 | 0.024 | SR | Non-PR | FN |
| Dorea formicigenerans ATCC27755 | −4.071 | 0.57 | 0.015 | 94.051 | 2.351 | SR | Non-PR | FN |
| Edwardsiella tarda ATCC23685 | −0.107 | 0.287 | 0.722 | 7.125 | 18.483 | Non-SR | PR | FP |
| Eggerthella lenta ATCC25559 | −0.435 | 0.217 | 0.038 | 26.042 | 11.107 | SR | PR | TP |
| Enterobacter cancerogenus ATCC35316 | −0.853 | 0.579 | 0.076 | 44.621 | 22.214 | Non-SR | PR | FP |
| Enterococcus faecalis V583 | −8.61 | 0.228 | 0.01 | 99.744 | 0.04 | SR | PR | TP |
| Escherichia coli K-12 | −0.714 | 0.331 | 0.038 | 39.057 | 13.995 | SR | PR | TP |
| Eubacterium biforme DSM3989 | −8.927 | 0.098 | 0.008 | 99.795 | 0.014 | SR | Non-PR | FN |
| Eubacterium hallii DSM3353 | −9.031 | 0.381 | 0.028 | 99.809 | 0.05 | SR | Non-PR | FN |
| Eubacterium rectale ATCC33656 | −9.002 | 0.322 | 0.022 | 99.805 | 0.044 | SR | PR | TP |
| Eubacterium ventriosum ATCC27560 | −4.653 | 0.827 | 0.051 | 96.024 | 2.278 | Non-SR | Non-PR | TN |
| Odoribacter splanchnius | −7.892 | 0.744 | 0.004 | 99.579 | 0.217 | SR | PR | TP |
| Parabacteroides distasonis ATCC8503 | −1.007 | 0.298 | 0.022 | 50.253 | 10.272 | SR | Non-PR | FN |
| Parabacteroides johnsonii DSM18315 | −0.529 | 0.281 | 0.123 | 30.702 | 13.51 | Non-SR | Non-PR | TN |
| Parabacteroides merdae ATCC43184 | −0.749 | 0.208 | 0.006 | 40.508 | 8.593 | SR | Non-PR | FN |
| Pretovella copri DSM18205 | −8.693 | 0.41 | 0.015 | 99.758 | 0.069 | SR | Non-PR | FN |
| Proteus penneri ATCC35198 | −1.657 | 0.313 | 0.009 | 68.289 | 6.884 | SR | PR | TP |
| Providencia alcalifaciens DSM30120 | −0.379 | 0.247 | 0.108 | 23.094 | 13.183 | Non-SR | PR | FP |
| Providencia rettgeri DSM1131 | −0.205 | 0.204 | 0.25 | 13.245 | 12.273 | Non-SR | PR | FP |
| Providencia stuartii ATCC25827 | −0.072 | 0.246 | 0.807 | 4.854 | 16.212 | Non-SR | PR | FP |
| Roseburia intestinalis L1-82 | −8.876 | 0.156 | 0.006 | 99.787 | 0.023 | SR | Non-PR | FN |
| Ruminococcus gnavus ATCC29149 | −8.639 | 0.045 | 0 | 99.749 | 0.008 | SR | Non-PR | FN |
| Ruminococcus lactaris ATCC29176 | −9.522 | 0.145 | 0.005 | 99.864 | 0.014 | SR | Non-PR | FN |
| Ruminococcus torques ATCC27756 | −2.384 | 0.298 | 0.016 | 80.841 | 3.961 | SR | PR | TP |
| Salmonella Typhimurium LT2 | −0.673 | 0.712 | 0.252 | 37.287 | 30.936 | Non-SR | PR | FP |
| Subdoligranulum variabile DSM15176 | −8.854 | 0.189 | 0.008 | 99.784 | 0.028 | SR | Non-PR | FN |
| Victivallis vadensis ATCC BAA-548 | −1.844 | 0.392 | 0.004 | 72.146 | 7.574 | SR | Non-PR | FN |
TABLE 2.
Summarized results of systematic Zimmermann et al. (2019) comparison This table displays the summarized results of the systematic comparison of predicted sulfasalazine reducers to experimentally confirmed sulfasalazine reducers reported by Zimmermann et al. (2019).
| PR | Non-PR | |
|---|---|---|
| SR | 38.8% (26/67)a | 40.3% (27/67)b |
| Non-SR | 11.9% (8/67)c | 9.0% (6/67)d |
aThe number of true positives.
bThe number of false negatives.
cThe number of true false positives.
dThe number of true negatives.
Exploratory Analysis of Azoreductase Abundance and Expression Levels in the Human Gut Microbiome
After identifying putative azo-reducing species of the human gut microbiome, we next sought to examine the abundance and expression of putative azoreductases using publicly available metagenomic and metatranscriptomic datasets. We used shotgun metagenomic sequence data from the HMP2 (Proctor and Huttenhower, 2019) and the PRISM (Franzosa et al., 2019) to quantify azoreductase gene abundance. We also used high-throughput metatranscriptomic sequence data from the HPFS (Abu-Ali et al., 2018) to quantify the expression of azoreductases by the human gut microbiota (Fig. 4, A–C). Briefly, raw genomic and transcriptomic reads were filtered and processed using fastp (Chen et al., 2018), and azoreductases were quantified using salmon v1.4.0 (Patro et al., 2017). Please see Materials and Methods for more details on the computational and statistical procedures used.
Significant differences in azoreductase gene abundances between disease conditions are displayed in Fig. 4, A and B with asterisks. We find that clade I, clade II, clade III, clade IVa, clade IVb, mdaB, and yieF genes are considerably higher in abundance than ferB, azoR1_cperf, arsH, azoR_ropacus, azo1, and azoR1_llentus within all three disease conditions for both HMP2 (all FDR adjusted P < 2.2 × 10−16) and PRISM (all FDR adjusted P < 2.6 × 10−7) (Fig. 4, A and B). Clade IVa is higher in abundance than all other azoreductases across healthy, UC, and CD cohorts from HMP2 (Fig. 4A), but the same statistically significant difference was not observed in the PRISM study (Fig. 4B).
The expression of clade I, clade II, clade III, clade IVa, mdaB, and yieF azoreductases are significantly higher than those of clade IVb, ferB, azoR1_cperf, arsH, azoR_ropacus, azo1, and azoR1_llentus azoreductases in healthy individuals (minimum FDR adjusted P value < 1 × 10−7 between mdaB and azoR1_cperf) (Fig. 4C). Although clade IVb abundance levels are comparable to those of clades I, II, III, and IVa, the expression levels of clade IVb azoreductases are significantly lower in vivo than the expression clade I, II, III, and IVa azoreductases (all FDR adjusted P < 2.2 × 10−16).
The Relative Abundance of Putative Azo-Reducing Species Fluctuates over Time
We next sought to examine whether relative abundance levels of combined known and putative azo-reducing species are stable or fluctuate over time. The HMP2 dataset provides a unique opportunity to examine the stability of individuals’ gut microbiomes over time as there are 18 individuals across healthy, UC, and CD cohorts, with at least 20 stool samples taken once every 2 weeks over a 6-month period. To examine the stability of azo reduction in the human gut, we compared the relative abundance of known and putative azo-reducing species from these participants (Fig. 5). The median relative abundance of combined azo-reducing species ranges from 20.3 ± 3.58% to 33.9 ± 19.2% for non-IBD, 21.7 ± 7.6% to 49.0 ± 15.0% for UC, and 34.9 ± 6.39 to 62.3 ± 18.8 for CD subjects. Using linear mixed-effects model ANOVA, we found that combined azo-reducing species are significantly more abundant in CD subjects than in non-IBD subjects (P = 0.002) and are not significantly more abundant in UC subjects than in non-IBD subjects (P = 0.064) (Fig. 5A). Note that Fig. 5B shows the same relative abundance values displayed in Fig. 5A but over the course of the study, from collection 1 to collection 24.
Multiple Strains of F. Nucleatum, B. Fragilis, and C. Clostridioforme Reduce Acid Orange 7 in Vitro
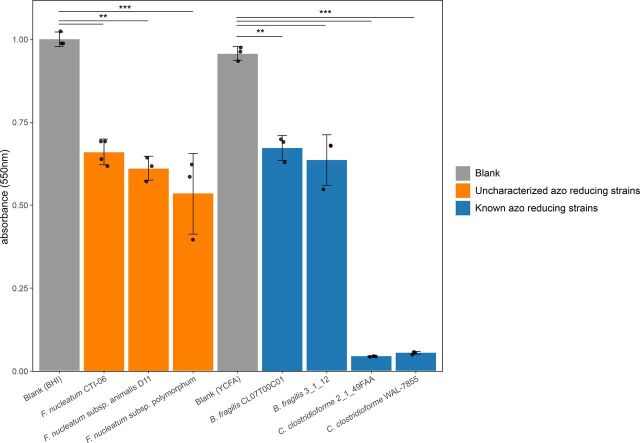
Finally, we sought to test the azo reduction of Acid Orange 7 by three strains of the health-relevant (Castellarin et al., 2012; Kostic et al., 2012; Bashir et al., 2015; Abed et al., 2016) microbe F. nucleatum. As well, we tested the azo reduction of Acid Orange 7 by two positive control species, B. fragilis and C. clostridioforme. Acid Orange 7 is an azo-bonded dye commonly used in the food and textile industries (Bay et al., 2014), and the decolorization of azo-bonded dyes is commonly used to test azo reduction by bacteria in vitro (Feng et al., 2012). F. nucleatum CTI-06, F. nucleatum subsp. animalis D11, and F. nucleatum subsp. polymorphum were grown in BHI media, and Acid Orange 7 was added to the culture after 4 days of growth (Materials and Methods). We also tested the azo reduction of Acid Orange 7 by the known azo-reducing species B. fragilis and Clostridium clostridiforme. B. fragilis strains 3_1_12 and CL07T00C01 and C. clostridioforme strains 2_1_49_FAA and WAL-7855 were grown in YCFA broth and served as positive controls. We found that all strains examined in this assay significantly decolorized Acid Orange 7 in vitro (Fig. 6). To our knowledge, this is the first reporting of azo reduction by F. nucleatum CTI-06, F. nucleatum subsp. animalis D11, and F. nucleatum subsp. polymorphum.
Fig. 6.
Three putative azo-reducing strains of F. nucleatum degrade Acid Orange 7 in vitro. The absorbance of light at 550 nm (corresponding to the absorbance spectra of Acid Orange 7) was measured in cultures of F. nucleatum, B. fragilis, and C. clostridiforme isolate cultures. F. nucleatum strains were grown in BHI media and were compared with BHI-blank control mixture, whereas B. fragilis and C. clostridiforme strains were grown in YCFA media and were thus compared with a YCFA-blank. Each strain was grown and tested in biologic and technical triplicates. Each data point on the plot above is the average of three technical replicates from a single biologic replicate per strain. Please see Materials and Methods for more details regarding our experimental methodology. Asterisks indicate statistical significance calculated via two-sided t tests (*P < 0.05; **P < 0.01; ***P < 0.001).
Discussion
The presence of azo-reducing bacteria in the human gut is necessary for the effective delivery and activation of azo-bonded prodrugs. Although azoreductase activity has been identified in several prominent phyla of the human gut microbiota (Zimmermann et al., 2019) and appears to be ubiquitous across healthy individuals (Javdan et al., 2020), the prevalence, abundance, and distribution of azoreductases have not been systematically examined in the human gut microbiome of healthy individuals nor in individuals living with IBD. In this work, we curated and compiled known azoreductase genes (Fig. 2), searched for azoreductase gene families across a nonredundant set of 4644 human gut bacterial genomes (Almeida et al., 2020), and identified 1958 putative azo-reducing species (Fig. 3). The systematic comparison of our search results to recent experimental evidence of sulfasalazine reduction by prominent gut bacteria (Table 1, Table 2) indicates a disconnect between the current state of azoreductase annotation and experimental evidence of sulfasalazine reduction. Interestingly, the majority (77.1%, 27/35) of incorrectly predicted sulfasalazine-reducing strains are false negatives, meaning these strains did not return a significant hit to an azoreductase gene from the homolog search step but do, in fact, reduce sulfasalazine in vitro. This inconsistency between annotated azoreductases and experimental evidence of azo reduction suggests that many prominent bacterial strains of the human gut microbiome may encode and express previously uncharacterized azoreductase genes. These genes likely serve other endogenous roles such as nitro reduction (Liu et al., 2007; Chalansonnet et al., 2017) and quinone oxidoreduction (Leelakriangsak et al., 2008; Liu et al., 2008; Ryan et al., 2010a; Ryan et al. 2010b; Ryan et al., 2014), with the azo reduction being a side mechanism that these enzymes crossfunctionally participate in.
We next sought to report the relative abundance and expression of azoreductases in the human gut microbiome for healthy controls and IBD patients. Our analysis of 1558 metagenomic samples from 326 individuals across healthy, UC, and CD patient cohorts showed that clade I, II, III, IVa, IVb, mdaB, and yieF azoreductases are significantly more abundant in the gut microbiome compared with the other azoreductases examined in this study (Fig. 4, A and B). We also examined the expression of azoreductases by the human gut microbiota and found that, with the exception of clade IVb, expression levels of azoreductases roughly match with their corresponding genomic abundance (Fig. 4C). The incongruence of clade IVb abundance and expression levels suggests that, when feasible, shotgun metagenomic sequencing of stool samples should be performed in parallel with metatranscriptomic sequencing to better understand the functional landscape of the gut microbiome and the relative contributions of different azoreductases to overall azo reduction. We also sought to examine the relative abundance of known and putative azo-reducing bacteria in healthy, UC, and CD patients over time. We found that the relative abundance of known and putative azo-reducing bacteria is significantly (P = 0.002) higher in individuals with CD and is modestly (P = 0.06) higher in individuals with UC compared with healthy controls (Fig. 5). This bodes well for the future of azo-bonded prodrug development because these therapies are intended to treat individuals afflicted with UC and CD. However, the cumulative relative abundance of known and putative azo-reducing bacteria fluctuates over time (Fig. 5B), and future studies should explore whether there exists some minimum necessary abundance of azo-reducing species for adequate prodrug metabolism and activation.
Finally, we tested the reduction of the azo-bonded dye Acid Orange 7 by three strains of F. nucleatum alongside positive control strains of B. fragilis and C. clostridioforme (Fig. 6). F. nucleatum is positively correlated with colorectal cancer (Marchesi et al., 2011; Kostic et al., 2012), is present in and on cancerous tissue (Castellarin et al., 2012), and possibly contributes to the etiology of the disease (McCoy et al., 2013; Rubinstein et al., 2013; Han, 2015). We found that F. nucleatum CTI-06, F. nucleatum subsp. animalis D11, and F. nucleatum subsp. polymorphum all significantly reduce Acid Orange 7 in vitro, indicating the encoding and activity of azoreductases in these strains of F. nucleatum. The F. nucleatum reference strain present in UHGG received significant hits to the mdaB (E-value = 4.20 × 10−13) and yieF (E-value = 1.40 × 10−23) HMMs and, thus, represents an accurately predicted azo-reducing bacteria (Supplemental Table 3). The identification and characterization of these, and possibly other, F. nucleatum azoreductases could lead to the eventual development of an azo-bonded colorectal cancer therapeutic designed specifically to activate in the presence of F. nucleatum on the surface of colonic tumors.
B. fragilis is a known reducer of azo dyes including Acid Orange 7 (this study), Amaranth, Orange II, and Tartrazine (Bragger et al., 1997), as well as of the quinone menadione (Ito et al., 2020). Additionally, many B. fragilis strains have been shown to be potent reducers of sulfasalazine in vitro (Zimmermann et al., 2019). Although we did identify a significant (E-value < 1 × 10−45) hit to the clade IVa HMM in the two B. fragilis reference strains present in UHGG (Supplemental Table 3), there may be other B. fragilis genes or operons that exhibit azoreductase activity. Ito et al. (2020) described two NADH:quinone oxidoreductase operons, NQR and NUO, and one NADH:quinone oxidoreductase gene, ndh2, capable of reducing the quinone menadione. Recall that bacterial quinone oxidoreductases are often crossreactive with azo compounds and have even been proposed to be a part of the same FMN-dependent superfamily of NAD(P)H utilizing oxidoreductase enzymes (Ryan et al., 2014). Future studies are required to confirm or deny that NQR, NUO, and ndh2 are hitherto uncharacterized azoreductases contributing to the complete azo reduction of sulfasalazine by B. fragilis shown in Zimmerman et al. (2019).
Of the seven bacterial strains tested for reduction of Acid Orange 7, the two C. clostridioforme strains exhibited by far the most effective reduction of Acid Orange 7 (Fig. 6). Although C. clostridioforme is a known azo dye reducer (Raffi and Cerniglia, 1990; Nakamura et al., 2002; Xu et al., 2010), neither of the two reference strains present in UHGG recruited significant alignments to known azoreductase gene families curated in the homolog search step of this work (Supplemental Table 3). This could be the result of either 1) strain-level variation between the reference strains and those tested with Acid Orange 7 in this study or 2) the presence and activity of one or more uncharacterized azoreductases in C. clostridioforme. In either case, further research including a comparative genomics analysis and gene knockout experiment on various strains of C. clostridioforme could lead to an improved understanding of gut microbial azo reduction.
This study has two primary limitations. 1) The E-value and percent of alignment thresholds for determining a putative azoreductase in the homolog search step are not absolute but rather are designed to strike a balance between identifying spurious homologs and missing the identification of true azoreductase homologs. This is an inherent limitation of studies requiring hard cutoffs for homolog classification and, thus, is very difficult to avoid. 2) Bacterial azoreductases exhibit different substrate specificities (Bin et al., 2004; Deller et al., 2006; Sugiura et al., 2006; Joshi et al., 2008; Ryan et al., 2010a, ; Mendes et al., 2011; Lang et al., 2013) and, thus, have varying affinities for different azo prodrugs as well as azo dyes. Though we show a significant reduction of Acid Orange 7 by three strains of F. nucleatum in this work, future experiments showing the reduction of azo drugs such as sulfasalazine would further bolster the hypothesis that F. nucleatum encodes and expresses one or more uncharacterized azoreductases.
In conclusion, we show that known azoreductases are widely distributed in the human gut microbiome and that there are likely many more uncharacterized azoreductases encoded and expressed in the human gut microbiome. These results both 1) bolster previous findings suggesting the ubiquity of azo-reduction in the gut microbiome (Javdan et al., 2020) and 2) suggest the presence and activity of many hitherto uncharacterized azoreductases in the human gut microbiome. The list of false negative strains identified in our systematic comparison analysis can serve as a resource for future studies focused on identifying azoreductases encoded by the human gut microbiome (Table 1). Overall, this work describes the abundance and distribution of known azoreductases in the human gut microbiome and motivates the need for future studies focused on annotating hitherto uncharacterized azoreductases encoded in the human gut microbiome. Further validation and annotation of putative azoreductases encoded by prominent members of the gut flora such as B. fragilis, F. nucleatum, and C. clostridioforme, are important for functional characterization of azo reduction by the human gut microbiome and for the future of azo prodrug development.
Abbreviations
- 5-ASA
5-aminosalicylic acid
- BHI
Brain Heart Infusion
- CD
Crohn disease
- CRC
colorectal cancer
- DHQ
dihydroquinolone
- EFI-EST
Enzyme Function Initiative-Enzyme Similarity Tool
- FDR
false discover rate
- FMN
flavin mononucleotide
- FN
false negative
- FP
false positive
- HMM
hidden Markov model
- HMP2
Integrative Human Microbiome Project
- HPFS
Health Professionals Follow-Up Study
- IBD
inflammatory bowel disease
- PR
predicted reducer
- PRISM
Prospective Registry of IBD Patients at MGH
- SR
sulfasalazine reducing
- TN
true negative
- TP
true positive
- UC
ulcerative colitis
- UHGG
Unified Human Gastrointestinal Genomes
- YCFA
Yeast Casitone Fatty Acid
Authorship Contributions
Participated in research design: Braccia, Minabou Ndjite, Jiang, Pop, Hall.
Conducted experiments: Minabou Ndjite, Weiss, Levy, Abeysinghe.
Performed data analysis: Braccia, Minabou Ndjite.
Wrote or contributed to the writing of the manuscript: Braccia, Minabou Ndjite, Weiss, Levy, Abeysinghe, Jiang, Pop, Hall.
Footnotes
D.B. was supported in part by the National Science Foundation [DGE-1632976] and in part by B.H. startup funding from the University of Maryland. X.J. was supported in part by the Intramural Research Program of National Institutes of Health National Library of Medicine. M.P. was supported by National Institutes of Health [Grant R01-AI-100947]. B.H., G.M.N., A.W., S.L., and S.A. were supported by startup funding from the University of Maryland.
No author has an actual or perceived conflict of interest with the contents of this article.
 This article has supplemental material available at dmd.aspetjournals.org.
This article has supplemental material available at dmd.aspetjournals.org.
References
- Abed JEmgård JEMZamir GFaroja MAlmogy GGrenov ASol ANaor RPikarsky EAtlan KA, et al. (2016) Fap2 Mediates Fusobacterium nucleatum Colorectal Adenocarcinoma Enrichment by Binding to Tumor-Expressed Gal-GalNAc. Cell Host Microbe 20:215–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abu-Ali GSMehta RSLloyd-Price JMallick HBranck TIvey KLDrew DADuLong CRimm EIzard J, et al. (2018) Metatranscriptome of human faecal microbial communities in a cohort of adult men. Nat Microbiol 3:356–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida ANayfach SBoland MStrozzi FBeracochea MShi ZJPollard KSSakharova EParks DHHugenholtz P, et al. (2021) A unified catalog of 204,938 reference genomes from the human gut microbiome. Nat Biotechnol 39:105–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashir A, Miskeen AY, Bhat A, Fazili KM, Ganai BA (2015) Fusobacterium nucleatum: an emerging bug in colorectal tumorigenesis. Eur J Cancer Prev 24:373–385. [DOI] [PubMed] [Google Scholar]
- Bates D, Mächler M, Bolker B, Walker S (2014) Fitting Linear Mixed-Effects Models Using Lme4. J Stat Softw 67:1–48. DOI: http://arxiv.org/abs/1406.5823. [Google Scholar]
- Bay HH, Lim CK, Kee TC, Ware I, Chan GF, Shahir S, Ibrahim Z (2014) Decolourisation of Acid Orange 7 recalcitrant auto-oxidation coloured by-products using an acclimatised mixed bacterial culture. Environ Sci Pollut Res Int 21:3891–3906. [DOI] [PubMed] [Google Scholar]
- Bin Y, Jiti Z, Jing W, Cuihong D, Hongman H, Zhiyong S, Yongming B (2004) Expression and characteristics of the gene encoding azoreductase from Rhodobacter sphaeroides AS1.1737. FEMS Microbiol Lett 236:129–136. [DOI] [PubMed] [Google Scholar]
- Blümel S, Stolz A (2003) Cloning and characterization of the gene coding for the aerobic azoreductase from Pigmentiphaga kullae K24. Appl Microbiol Biotechnol 62:186–190. [DOI] [PubMed] [Google Scholar]
- Blümel S, Knackmuss H-J, Stolz A (2002) Molecular cloning and characterization of the gene coding for the aerobic azoreductase from Xenophilus azovorans KF46F. Appl Environ Microbiol 68:3948–3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bragger JL, Lloyd AW, Soozandehfar SH, Bloomfield SF, Marriott C, Martin GP (1997) Investigations into the Azo Reducing Activity of a Common Colonic Microorganism. Int J Pharm 157:61–71 DOI: 10.1016/S0378-5173(97)00214-7. [DOI] [Google Scholar]
- Brown JP (1981) Reduction of polymeric azo and nitro dyes by intestinal bacteria. Appl Environ Microbiol 41:1283–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellarin MWarren RLFreeman JDDreolini LKrzywinski MStrauss JBarnes RWatson PAllen-Vercoe EMoore RA, et al. (2012) Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res 22:299–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerniglia CE, Freeman JP, Franklin W, Pack LD (1982) Metabolism of azo dyes derived from benzidine, 3,3′-dimethyl-benzidine and 3,3′-dimethoxybenzidine to potentially carcinogenic aromatic amines by intestinal bacteria. Carcinogenesis 3:1255–1260. [DOI] [PubMed] [Google Scholar]
- Chalansonnet V, Mercier C, Orenga S, Gilbert C (2017) Identification of Enterococcus faecalis enzymes with azoreductases and/or nitroreductase activity. BMC Microbiol 17:126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaumeil PA, Mussig AJ, Hugenholtz P, Parks DH (2019) GTDB-Tk: a toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics 36:1925–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Hopper SL, Cerniglia CE (2005) Biochemical and molecular characterization of an azoreductase from Staphylococcus aureus, a tetrameric NADPH-dependent flavoprotein. Microbiology (Reading) 151:1433–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Wang RF, Cerniglia CE (2004) Molecular cloning, overexpression, purification, and characterization of an aerobic FMN-dependent azoreductase from Enterococcus faecalis. Protein Expr Purif 34:302–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Zhou Y, Chen Y, Gu J (2018) fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34:i884–i890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen RD, Woseth DM, Thisted RA, Hanauer SB (2000) A meta-analysis and overview of the literature on treatment options for left-sided ulcerative colitis and ulcerative proctitis. Am J Gastroenterol 95:1263–1276. [DOI] [PubMed] [Google Scholar]
- Cominelli F, Nast CC, Duchini A, Lee M (1992) Recombinant interleukin-1 receptor antagonist blocks the proinflammatory activity of endogenous interleukin-1 in rabbit immune colitis. Gastroenterology 103:65–71. [DOI] [PubMed] [Google Scholar]
- Deller S, Sollner S, Trenker-El-Toukhy R, Jelesarov I, Gübitz GM, Macheroux P (2006) Characterization of a thermostable NADPH:FMN oxidoreductase from the mesophilic bacterium Bacillus subtilis. Biochemistry 45:7083–7091. [DOI] [PubMed] [Google Scholar]
- Saha JR, Butler VP Jr, Neu HC, Lindenbaum J (1983) Digoxin-inactivating bacteria: identification in human gut flora. Science 220:325–327. [DOI] [PubMed] [Google Scholar]
- Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eslami M, Amoozegar MA, Asad S (2016) Isolation, cloning and characterization of an azoreductase from the halophilic bacterium Halomonas elongata. Int J Biol Macromol 85:111–116. [DOI] [PubMed] [Google Scholar]
- Feng J, Cerniglia CE, Chen H (2012) Toxicological significance of azo dye metabolism by human intestinal microbiota. Front Biosci (Elite Ed) 4:568–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Clements J, Arndt W, Miller BL, Wheeler TJ, Schreiber F, Bateman A, Eddy SR (2015) HMMER web server: 2015 update. Nucleic Acids Res 43 (W1):W30–W38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foppoli A, Maroni A, Moutaharrik S, Melocchi A, Zema L, Palugan L, Cerea M, Gazzaniga A (2019) In vitro and human pharmacoscintigraphic evaluation of an oral 5-ASA delivery system for colonic release. Int J Pharm 572:118723. [DOI] [PubMed] [Google Scholar]
- Franzosa EASirota-Madi AAvila-Pacheco JFornelos NHaiser HJReinker SVatanen THall ABMallick HMcIver LJ, et al. (2019) Gut microbiome structure and metabolic activity in inflammatory bowel disease. Nat Microbiol 4:293–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friend DR (2005) New oral delivery systems for treatment of inflammatory bowel disease. Adv Drug Deliv Rev 57:247–265. [DOI] [PubMed] [Google Scholar]
- Gerlt JA, Bouvier JT, Davidson DB, Imker HJ, Sadkhin B, Slater DR, Whalen KL (2015) Enzyme Function Initiative-Enzyme Similarity Tool (EFI-EST): A web tool for generating protein sequence similarity networks. Biochim Biophys Acta 1854:1019–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonçalves AMD, Mendes S, de Sanctis D, Martins LO, Bento I (2013) The crystal structure of Pseudomonas putida azoreductase - the active site revisited. FEBS J 280:6643–6657. [DOI] [PubMed] [Google Scholar]
- Gustafsson A, Andersson M, Lagerstedt K, Lönnroth C, Nordgren S, Lundholm K (2010) Receptor and enzyme expression for prostanoid metabolism in colorectal cancer related to tumor tissue PGE2. Int J Oncol 36:469–478. [PubMed] [Google Scholar]
- Haiser HJ, Gootenberg DB, Chatman K, Sirasani G, Balskus EP, Turnbaugh PJ (2013) Predicting and manipulating cardiac drug inactivation by the human gut bacterium Eggerthella lenta. Science 341:295–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, YW (2015) Fusobacterium nucleatum: a commensal-turned pathogen. Curr Opin Microbiol 23:141–147. DOI: 10.1016/j.mib.2014.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Gallegos R, Matano LM, Butler NL, Hantman N, Kaili M, Coyne MJ, Comstock LE, Malamy MH, Barquera B (2020) Genetic and Biochemical Analysis of Anaerobic Respiration in Bacteroides Fragilis and Its Importance In Vivo. mBio 11:e03238–19. DOI: 10.1128/mBio.03238-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javdan B, Lopez JG, Chankhamjon P, Lee YJ, Hull R, Wu Q, Wang X, Chatterjee S, Donia MS (2020) Personalized Mapping of Drug Metabolism by the Human Gut Microbiome. Cell 181:1661–1679.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi T, Iyengar L, Singh K, Garg S (2008) Isolation, identification and application of novel bacterial consortium TJ-1 for the decolourization of structurally different azo dyes. Bioresour Technol 99:7115–7121. [DOI] [PubMed] [Google Scholar]
- Peters U, Falk LC, Kalman SM (1978) Digoxin metabolism in patients. Arch Intern Med 137:1074–1076. [PubMed] [Google Scholar]
- Kostic ADGevers DPedamallu CSMichaud MDuke FEarl AMOjesina AIJung JBass AJTabernero J, et al. (2012) Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res 22:292–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznetsova A, Brockhoff PB, Christensen RHB (2017) LmerTest Package: Tests in Linear Mixed Effects Models. J Stat Softw 82:1–26 DOI: 10.18637/jss.v082.i13. [DOI] [Google Scholar]
- Lang W, Sirisansaneeyakul S, Ngiwsara L, Mendes S, Martins LO, Okuyama M, Kimura A (2013) Characterization of a new oxygen-insensitive azoreductase from Brevibacillus laterosporus TISTR1911: toward dye decolorization using a packed-bed metal affinity reactor. Bioresour Technol 150:298–306. [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL (2012) Fast gapped-read alignment with bowtie 2. Nat Methods 9:357–359. DOI: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leelakriangsak M, Huyen NT, Töwe S, van Duy N, Becher D, Hecker M, Antelmann H, Zuber P (2008) Regulation of quinone detoxification by the thiol stress sensing DUF24/MarR-like repressor, YodB in Bacillus subtilis. Mol Microbiol 67:1108–1124. [DOI] [PubMed] [Google Scholar]
- Letunic I, Bork P (2019) Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res 47 (W1):W256–W259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Zhou J, Jin R, Zhou M, Wang J, Lu H, Qu Y (2008) Enhancing survival of Escherichia coli by expression of azoreductase AZR possessing quinone reductase activity. Appl Microbiol Biotechnol 80:409–416. [DOI] [PubMed] [Google Scholar]
- Liu G, Zhou J, Lv H, Xiang X, Wang J, Zhou M, Qv Y (2007) Azoreductase from Rhodobacter sphaeroides AS1.1737 is a flavodoxin that also functions as nitroreductase and flavin mononucleotide reductase. Appl Microbiol Biotechnol 76:1271–1279. [DOI] [PubMed] [Google Scholar]
- Mahida YR, Lamming CE, Gallagher A, Hawthorne AB, Hawkey CJ (1991) 5-Aminosalicylic acid is a potent inhibitor of interleukin 1 beta production in organ culture of colonic biopsy specimens from patients with inflammatory bowel disease. Gut 32:50–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchesi JR, Dutilh BE, Hall N, Peters WHM, Roelofs R, Boleij A, Tjalsma H (2011) Towards the human colorectal cancer microbiome. PLoS One 6:e20447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz JFM, Kedziora K, Keogh B, Maguire J, Reilly M, Windle H, Kelleher DP, Gilmer JF (2011) A double prodrug system for colon targeting of benzenesulfonamide COX-2 inhibitors. Bioorg Med Chem Lett 21:6636–6640. [DOI] [PubMed] [Google Scholar]
- Matsumoto K, Mukai Y, Ogata D, Shozui F, Nduko JM, Taguchi S, Ooi T (2010) Characterization of thermostable FMN-dependent NADH azoreductase from the moderate thermophile Geobacillus stearothermophilus. Appl Microbiol Biotechnol 86:1431–1438. DOI: 10.1007/s00253-009-2351-7. [DOI] [PubMed] [Google Scholar]
- McCoy AN, Araújo-Pérez F, Azcárate-Peril A, Yeh JJ, Sandler RS, Keku TO (2013) Fusobacterium is associated with colorectal adenomas. PLoS One 8:e53653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes S, Pereira L, Batista C, Martins LO (2011) Molecular determinants of azo reduction activity in the strain Pseudomonas putida MET94. Appl Microbiol Biotechnol 92:393–405. [DOI] [PubMed] [Google Scholar]
- Mercier C, Chalansonnet V, Orenga S, Gilbert C (2013) Characteristics of major Escherichia coli reductases involved in aerobic nitro and azo reduction. J Appl Microbiol 115:1012–1022. [DOI] [PubMed] [Google Scholar]
- Misal SA, Lingojwar DP, Lokhande MN, Lokhande PD, Gawai KR (2014) Enzymatic transformation of nitro-aromatic compounds by a flavin-free NADH azoreductase from Lysinibacillus sphaericus. Biotechnol Lett 36:127–131. [DOI] [PubMed] [Google Scholar]
- Misal SA, Lingojwar DP, Shinde RM, Gawai KR (2011) Purification and characterization of azoreductase from alkaliphilic strain Bacillus badius. Process Biochem 46:1264–1269. DOI: 10.1016/j.procbio.2011.02.013. [DOI] [Google Scholar]
- Morrison JM, Wright CM, John GH (2012) Identification, Isolation and Characterization of a Novel Azoreductase from Clostridium Perfringens. Anaerobe 18:229–234. DOI: 10.1016/j.anaerobe.2011.12.006. [DOI] [PubMed] [Google Scholar]
- Nachiyar CV, Rajakumar GS (2005) Purification and Characterization of an Oxygen Insensitive Azoreductase from Pseudomonas Aeruginosa. Enzyme Microb Technol 36:503–509 DOI: 10.1016/j.enzmictec.2004.11.015. [DOI] [Google Scholar]
- Nakamura J, Kubota Y, Miyaoka M, Saitoh T, Mizuno F, Benno Y (2002) Comparison of four microbial enzymes in Clostridia and Bacteroides isolated from human feces. Microbiol Immunol 46:487–490. [DOI] [PubMed] [Google Scholar]
- Nakanishi M, Yatome C, Ishida N, Kitade Y (2001) Putative ACP phosphodiesterase gene (acpD) encodes an azoreductase. J Biol Chem 276:46394–46399. [DOI] [PubMed] [Google Scholar]
- Nielsen OH (1982) Sulfasalazine intolerance. A retrospective survey of the reasons for discontinuing treatment with sulfasalazine in patients with chronic inflammatory bowel disease. Scand J Gastroenterol 17:389–393. [DOI] [PubMed] [Google Scholar]
- Ooi T, Shibata T, Sato R, Ohno H, Kinoshita S, Thuoc TL, Taguchi S (2007) An azoreductase, aerobic NADH-dependent flavoprotein discovered from Bacillus sp.: functional expression and enzymatic characterization. Appl Microbiol Biotechnol 75:377–386. [DOI] [PubMed] [Google Scholar]
- Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C (2017) Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods 14:417–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peppercorn MA, Goldman P (1972) The role of intestinal bacteria in the metabolism of salicylazosulfapyridine. J Pharmacol Exp Ther 181:555–562. [PubMed] [Google Scholar]
- Peppercorn MA, Goldman P (1973) Distribution studies of salicylazosulfapyridine and its metabolites. Gastroenterology 64:240–245. [PubMed] [Google Scholar]
- Perrotta C, Pellegrino P, Moroni E, De Palma C, Cervia D, Danelli P, Clementi E (2015) Five-aminosalicylic Acid: an update for the reappraisal of an old drug. Gastroenterol Res Pract 2015:456895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proctor L, Huttenhower C; Integrative HMP (iHMP) Research Network Consortium (2019) The Integrative Human Microbiome Project. Nature 569:641–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rachmilewitz D, Karmeli F, Schwartz LW, Simon PL (1992) Effect of aminophenols (5-ASA and 4-ASA) on colonic interleukin-1 generation. Gut 33:929–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffi F, Cerniglia CE (1990) An Anaerobic Nondenaturing Gel Assay for the Detection of Azoreductase from Anaerobic Bacteria. J Microbiol Methods 12:139–148. DOI: 10.1016/0167-7012(90)90024-Z. [DOI] [Google Scholar]
- Rafii F, Cerniglia CE (1995) Reduction of azo dyes and nitroaromatic compounds by bacterial enzymes from the human intestinal tract. Environ Health Perspect 103 (Suppl 5):17–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafii F, Franklin W, Cerniglia CE (1990) Azoreductase activity of anaerobic bacteria isolated from human intestinal microflora. Appl Environ Microbiol 56:2146–2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, Han YW (2013) Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/β-catenin signaling via its FadA adhesin. Cell Host Microbe 14:195–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz JFM, Kedziora K, Windle H, Kelleher DP, Gilmer JF (2011) Investigation into drug release from colon-specific azoreductase-activated steroid prodrugs using in-vitro models. J Pharm Pharmacol 63:806–816. DOI: 10.1111/j.2042-7158.2011.01289.x. [DOI] [PubMed] [Google Scholar]
- Ryan A, Kaplan E, Nebel J-C, Polycarpou E, Crescente V, Lowe E, Preston GM, Sim E (2014) Identification of NAD(P)H quinone oxidoreductase activity in azoreductases from P. aeruginosa: azoreductases and NAD(P)H quinone oxidoreductases belong to the same FMN-dependent superfamily of enzymes. PLoS One 9:e98551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan A, Laurieri N, Westwood I, Wang C-J, Lowe E, Sim E (2010a) A novel mechanism for azoreduction. J Mol Biol 400:24–37. [DOI] [PubMed] [Google Scholar]
- Ryan A, Wang C-J, Laurieri N, Westwood I, Sim E (2010b) Reaction mechanism of azoreductases suggests convergent evolution with quinone oxidoreductases. Protein Cell 1:780–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa T, Yadav V, Zann V, Borde A, Abrahamsson B, Basit AW (2014) On the colonic bacterial metabolism of azo-bonded prodrugsof 5-aminosalicylic acid. J Pharm Sci 103:3171–3175. [DOI] [PubMed] [Google Scholar]
- Spanogiannopoulos P, Bess EN, Carmody RN, Turnbaugh PJ (2016) The microbial pharmacists within us: a metagenomic view of xenobiotic metabolism. Nat Rev Microbiol 14:273–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiura W, Yoda T, Matsuba T, Tanaka Y, Suzuki Y (2006) Expression and characterization of the genes encoding azoreductases from Bacillus subtilis and Geobacillus stearothermophilus. Biosci Biotechnol Biochem 70:1655–1665. [DOI] [PubMed] [Google Scholar]
- Suzuki H (2019) Remarkable diversification of bacterial azoreductases: primary sequences, structures, substrates, physiological roles, and biotechnological applications. Appl Microbiol Biotechnol 103:3965–3978. [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Yoda T, Ruhul A, Sugiura W (2001) Molecular cloning and characterization of the gene coding for azoreductase from Bacillus sp. OY1-2 isolated from soil. J Biol Chem 276:9059–9065. [DOI] [PubMed] [Google Scholar]
- Tozaki H, Odoriba T, Okada N, Fujita T, Terabe A, Suzuki T, Okabe S, Muranishi S, Yamamoto A (2002) Chitosan capsules for colon-specific drug delivery: enhanced localization of 5-aminosalicylic acid in the large intestine accelerates healing of TNBS-induced colitis in rats. J Control Release 82:51–61. [DOI] [PubMed] [Google Scholar]
- Wallace BDWang HLane KTScott JEOrans JKoo JSVenkatesh M, et al. (2010) Alleviating cancer drug toxicity by inhibiting a bacterial enzyme. Science 330:831–835. DOI: 10.1126/science.1191175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber CK, Liptay S, Wirth T, Adler G, Schmid RM (2000) Suppression of NF-kappaB activity by sulfasalazine is mediated by direct inhibition of IkappaB kinases α and β. Gastroenterology 119:1209–1218. [DOI] [PubMed] [Google Scholar]
- Wickham H (2011) Ggplot2. Wiley Interdiscip Rev Comput Stat 3:180–185 DOI: 10.1002/wics.147. [DOI] [Google Scholar]
- Wood DE, Lu J, Langmead B (2019) Improved metagenomic analysis with Kraken 2. Genome Biol 20:257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu A-W, Gu J, Ji J-F, Li Z-F, Xu G-W (2003) Role of COX-2 in carcinogenesis of colorectal cancer and its relationship with tumor biological characteristics and patients’ prognosis. World J Gastroenterol 9:1990–1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu WKK, Sung JJ, Lee CW, Yu J, Cho CH (2010) Cyclooxygenase-2 in tumorigenesis of gastrointestinal cancers: an update on the molecular mechanisms. Cancer Lett 295:7–16. [DOI] [PubMed] [Google Scholar]
- Xu H, Heinze TM, Paine DD, Cerniglia CE, Chen H (2010) Sudan azo dyes and Para Red degradation by prevalent bacteria of the human gastrointestinal tract. Anaerobe 16:114–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Ng IS, Chang JS (2016) Cloning and characterization of a robust recombinant azoreductase from Shewanella xiamenensis BC01. J Taiwan Inst Chem Eng 61:97–105. DOI: 10.1016/j.jtice.2016.01.002. [DOI] [Google Scholar]
- Zimmermann M, Zimmermann-Kogadeeva M, Wegmann R, Goodman AL (2019) Mapping human microbiome drug metabolism by gut bacteria and their genes. Nature 570:462–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann T, Kulla HG, Leisinger T (1982) Properties of purified Orange II azoreductase, the enzyme initiating azo dye degradation by Pseudomonas KF46. Eur J Biochem 129:197–203. [DOI] [PubMed] [Google Scholar]