Summary
Despite the importance of gene-environment interactions (GxEs) in improving and operationalizing genetic discovery, interpretation of any GxEs that are discovered can be surprisingly difficult. There are many potential biological and statistical explanations for a statistically significant finding and, likewise, it is not always clear what can be claimed based on a null result. A better understanding of the possible underlying mechanisms leading to a detected GxE can help investigators decide which are and which are not relevant to their hypothesis. Here, we provide a detailed explanation of five “phenomena,” or data-generating mechanisms, that can lead to nonzero interaction estimates, as well as a discussion of specific instances in which they might be relevant. We hope that, given this framework, investigators can design more targeted experiments and provide cleaner interpretations of the associated results.
Graphical abstract
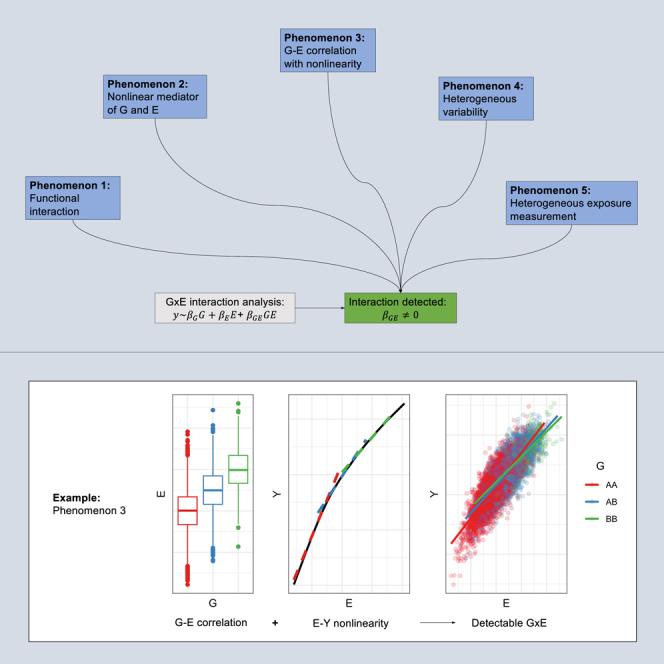
Various data-generating mechanisms may lead to the detection of gene-environment interactions (GxE). We describe five key phenomena, both “real” and spurious, that can produce a detectable GxE and discuss how researchers may plan experiments and interpret GxE findings and their clinical relevance in light of these phenomena.
Background
Gene-environment interactions (GxEs) are of increasing interest for improving genetic discovery, explaining missing heritability and population heterogeneity, and facilitating precision medicine.1 In general, the term describes any departure from a model with pure main effects for genetic and environmental terms, implying differences in the estimated genetic effect depending on the environment or vice versa (i.e., effect modification). They are typically estimated using a product term in a regression setting but can also be derived through comparison of stratified models or other approaches. Associated statistical tests are often underpowered, but increasing sample sizes and associated computationally efficient software options are beginning to enable large-scale discovery efforts.2,3,4,5
Investigators often conduct a GxE analysis to estimate and test such statistical interaction effects without specifying the underlying phenomenon, or data-generating mechanism, being sought or establishing potential explanations leading to any observed GxEs. In this review, we describe five patterns occurring at the biological level that can result in the detection of a statistical interaction as modeled via an interaction term. We hope that this framework will allow investigators to state explicit hypotheses, design better analytical frameworks, and think through possible explanations for their findings.
The organization of this review is as follows. We describe each of five data-generating models (“phenomena”) that can lead to the detection of a statistical interaction as typically tested with a regression product term. For each, we give a general description, provide examples from the literature, and discuss links with other known statistical issues. We provide visualizations based on simulated data, with specifics of the simulation setup and quantitative results provided in the supplemental note. We also discuss potential conditions and limitations related to data availability and domain knowledge. Finally, we provide a series of recommendations for using these observations to inform modeling decisions. Notes on terminology and simplifying assumptions for the purposes of this review are provided in Box 1.
Box 1. Notes on terminology and simplifying assumptions.
First, we make a few notes on terminology for the purposes of this review.
-
•
We use the term “functional” to refer to molecular-level interactions in which specific metabolites, proteins, or other molecular quantities physically interact, such that the activity of one pathway or process is modulated by the activity of another. Though we believe that “mechanistic” may be an equally appropriate term for such a phenomenon, we limit its usage here to avoid confusion with the public health-relevant sense in which it is used by VanderWeele and Knol (related to the sufficient cause framework).6
-
•
We use the term “pathway” in a biological sense (i.e., a set of enzymes and molecules involved in a particular biological process), rather than a statistical sense (i.e., a “causal pathway”), unless otherwise noted.
-
•
The letters G, E, and Y will be used to reference the genotype, exposure, and outcome of interest, respectively.
In laying out these patterns, we also make a few technical, simplifying assumptions.
-
•
We assume continuous outcomes unless otherwise noted.
-
•
We also assume that there are no confounders of the E-Y relationship.
-
•
We primarily focus on genotype as a modifier of exposure-outcome associations. However, all concepts also apply for the converse (i.e., the exposure as a modifier of the genotype-outcome relationship)—we will point out some well-recognized examples.
Phenomena leading to statistical interactions
Phenomenon 1. Functional
Description
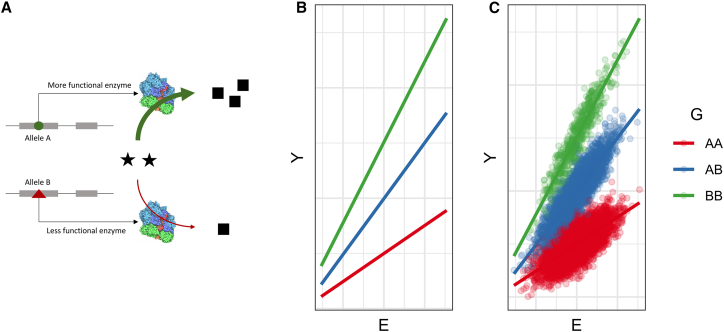
This first category of interaction arises from underlying molecular pathways that physically intersect or modify each other’s function; this is perhaps the most intuitive explanation for a GxE. Here, we might imagine that a genotype, via a causal association with the activity or expression level of some enzyme, modifies the activity of a pathway that mediates the E-Y relationship (Figure 1). Most exposures used in GxE analysis, including human behaviors, environmental exposures, and physiological states (such as BMI and biological sex), ultimately impact health outcomes and biomarkers through molecular mediators such as gene expression changes, protein function, or metabolite concentrations. We can imagine the effect modification by genotype occurring either upstream or downstream of this mediating quantity, as exemplified in the next section (see Preacher and colleagues for more detail on relevant terminology7).
Figure 1.
Simulation of a functional interaction
(A) Two genotypes in the protein coding sequence of a gene (alleles A and B, indicated by the green circle and red triangle) lead to differences in the function of a given enzyme, leading to differential throughput of a biological pathway (squares, representing the outcome Y) despite similar amounts of the input compound (stars, representing the environmental exposure E).
(B) Conceptual visualization of the E-Y effect modification.
(C) Results from simulation of a functional interaction.
Examples
Countless functional GxE effects have been reported in the literature, but we highlight two illustrative examples here. Variants in ERCC2, involved in DNA damage repair, appear to modify the effect of smoking on lung cancer.8 This is “downstream” moderation, in which smoking induces lung DNA damage (the molecular mediator), which then impacts lung cancer risk differentially based on ERCC2-related DNA repair capacity (the functional interaction). In another domain, phenylketonuria is a classic diet-related GxE example, where polymorphisms in PAH reduce or abolish the ability of its protein product to metabolize the amino acid phenylalanine in the liver.9 This is an example of “upstream” moderation, in which the functional interaction involves the exposure itself (the dietary amino acid phenylalanine).
Unlike dietary phenylalanine, for which the relevant metabolic pathways are well characterized, many more complex exposures (such as behavioral traits like physical activity levels) likely act through a series of parallel pathways. Approaches incorporating molecular-level GxE tests (e.g., transcriptomic exposures10 and their multi-exposure extensions11) may be helpful in resolving the relevant biological mechanisms in these cases. Still, such pleiotropic exposures require additional care in considering alternative explanations for any observed GxE.
Phenomenon 2. Nonlinear mediator of genes and environment
Description
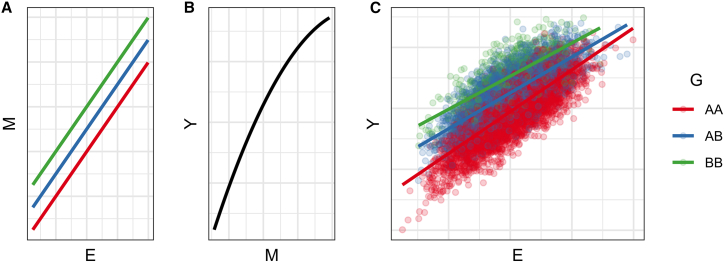
Suppose that both G and E independently impact the expression of the same pathway or a specific mediator M, which itself has a nonlinear relationship with Y. Because G affects the mediator independently of E, i.e., there is a genetic main effect in the statistical model, groups of individuals defined by their genotypes will differ in their mean value of M. Due to the nonlinearity of the M-Y relationship, any nonzero main effect (independently of genotype value) on M of another variable (in this case, E) will translate into changes in Y to a different degree (Figure 2).
Figure 2.
Simulation of a nonlinear mediator
(A) Both G and E have positive effects on the mediator M, but without interaction.
(B) M is nonlinearly related to Y, with a decreasing effect at higher levels of M.
(C) Results from simulation of a nonlinear mediator-based interaction. A GxE interaction with respect to Y is produced despite no interaction with respect to M.
This type of nonlinearity can arise for any number of technical or biological reasons, but two are of particular note. First, floor or ceiling effects are common in continuous biological quantities. Second, many binary outcomes of clinical interest can be thought of as a sharply nonlinear manifestation of an underlying continuous factor (sufficiency of a nutrient, toxicity of a toxin, or surpassing of a disease liability threshold). Eaves describes this phenomenon in more depth as it relates to GxE for psychiatric outcomes defined by specific diagnosis thresholds,12 and Domingue and colleagues explore concerns about interaction with binary and other non-continuous outcomes in-depth outside of the genetic realm.13
Examples
One example of ceiling effects comes from preventive cardiology: statins are very effective at producing reductions in LDL cholesterol (the continuous outcome) via inhibition of HMGCR enzyme function (the mediating quantity), but this relationship is nonlinear, with larger increases in dosage required to produce each additional increment of LDL-C reduction.14 In fact, this nonlinearity is implied in the typical description of statin effects on LDL-C in terms of percentage change, rather than absolute concentrations. Thus, HMGCR inhibition reaches a ceiling in its effect on LDL-C (at which point it may be necessary to target additional biological pathways, such as PCSK9, to achieve further reductions). We might then expect to find interactions between genetic variants affecting the expression of HMGCR and statin usage impacting LDL-C reduction.
As an example related to binary outcomes, a series of studies has explored the joint contribution of genetic and dietary effects on choline sufficiency in postmenopausal women.15 Choline sufficiency functions as a roughly binary variable in the sense that, once an individual has a sufficient choline supply, additional choline will not be beneficial in preventing choline-related organ dysfunction. Though dietary choline absorption and endogenous choline production occur via separate pathways, they both contribute to the same biological pool. In postmenopausal women with certain variation in PEMT (required for endogenous choline production), choline production is substantially reduced, such that these women require additional choline from diet or supplementation to avoid deficiency-associated organ dysfunction. A GxE ultimately results, in which exogenous choline reduces organ dysfunction in postmenopausal women with PEMT-reducing alleles but has little effect in others.
Related phenomena
When using binary outcomes, all effects (of genotype, exposure, and mediator) are inherently nonlinear. This makes the choice of scale critical in assessing interactions: investigators can test for departures from additivity affecting raw outcome probabilities (interactions on the “additive scale”) or transformed probabilities (e.g., logistic in logistic regression; interactions on the “multiplicative scale”). Additive interaction analysis is particularly useful for detecting instances in which G and E contribute additively to an underlying liability that manifests as a binary outcome due to thresholding (this threshold could be biological or due to clinical cutoffs). Thus, additive interaction may be relevant for public health applications even when there is no functional interaction of the type that might produce a multiplicative interaction (i.e., of the product term in a logistic regression).6 We note that this question of additive versus multiplicative interaction is most relevant for binary outcomes, where a product term in (for example) logistic regression serves to test for multiplicative interaction. Linear regression product terms for continuous outcomes, our primary focus in this review, test for additive interactions due to the linear covariate-outcome relationship. Phenomenon 2 also has a natural extension to gene-gene interactions, where G and E are replaced by two genetic variants (see Box 2 for additional discussion of connections between outlined GxE concepts and gene-gene interactions).
Box 2. Connections with gene-gene interactions.
Exploration of gene-gene interactions (GxG) is not a goal of this review, and we refer readers to more comprehensive discussions of the topic and the additional considerations it requires.16,17 However, we highlight a few connections between GxE concepts and previously reported GxG phenomena.
-
•
Phenomenon 2 (nonlinear mediator) has an intuitive extension to GxG, where G and E are replaced by two genetic variants, G1 and G2, that are independently associated with the mediator.
-
•
A GxG may manifest as a GxE when one of the genetic variants is associated with E (i.e., G1xG2 combined with a G2-E correlation produces G1xE). This E can be a mediator of the genetic effect, resulting in a complex scenario of interaction plus mediation that has been explored in the statistical7,18 and genetic epidemiology19 literature. Alternatively, E may simply be a non-causal marker for G2. Resolving causal mechanisms in this context may require research designs leveraging environmental variation that is independent of genetic factors.20
-
•
Phenomenon 5 (heterogeneous measurement) has conceptual links to “phantom epistasis,” in which an apparent interaction effect between two non-causal SNPs appears due to a combination of a true causal additive genetic main effect(s) of an unobserved genotype and imperfect linkage disequilibrium of the causal genotype with the marker variants.21
Phenomenon 3. G-E correlation with nonlinearity
Description
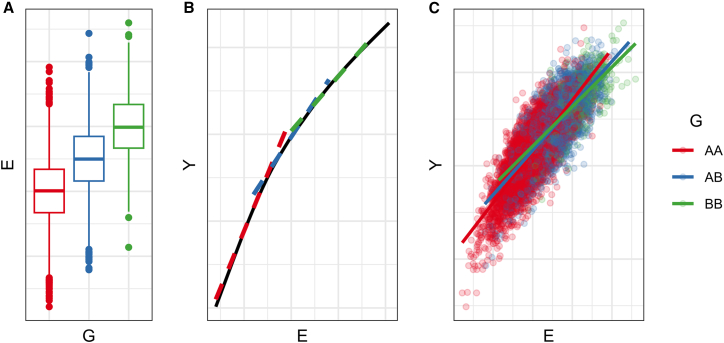
Statistical interactions can also appear when an exposure is (1) correlated with G and (2) related nonlinearly with Y. In this case, the mean E varies across genotype groups and the nonlinearity of the E-Y relationship leads to different E effect estimates by genotype group (Figure 3). This concept is similar to that of the nonlinear mediator described above, but in this case it is E itself, rather than a downstream mediator, that has the nonlinear relationship with Y. Importantly, this G-E correlation doesn’t need to be causal; for example, it can appear systematically across the genome in the presence of population stratification.
Figure 3.
Simulation of G-E correlation with nonlinearity
(A) The mean of E depends on G.
(B) The primary black line illustrates the true, nonlinear E-Y relationship. Dashed lines illustrate the different linear E-Y relationship at specific E values, with colors corresponding to hypothetical genotypes with different mean E values.
(C) Results from simulation of an interaction arising from G-E correlation with nonlinearity.
Example
This sort of interaction is most likely to appear for exposures with plausibly strong genetic effects. For example, body mass index (BMI) is under strong genetic control. It also shows strong nonlinear relationships with disease risk factors; for example, its effect on LDL-C is much stronger in the lean range compared to the overweight range.22 In a hypothesis-free study examining many genetic variants and exposures, we have previously found many instances of GxBMI interactions impacting cardiometabolic risk factors.23 Though this specific hypothesis wasn’t tested directly, it is likely that a subset of these interactions can be explained by the simultaneous presence of G-BMI associations and a nonlinear BMI-risk factor relationship.
Related phenomena
This general phenomenon leads to the inflation often observed in genome-wide interaction studies when regression models are misspecified with respect to the E-Y relationship.24 In such a case, random G-E correlations genome-wide combine with the nonlinear E-Y relationship to produce statistical evidence of GxE. Though these apparent interaction effects are random and typically small, in aggregate they produce a systematic departure of genome-wide interaction p values from the uniform distribution expected under the null hypothesis.24
Phenomenon 4. Heterogeneous variability
Description
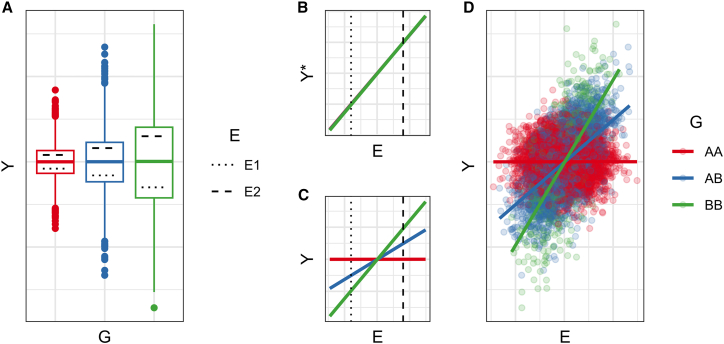
Here, some G or E directly modifies the variability, rather than the mean, of Y (one can imagine decreasing the “friction” in outcome fluctuations or allowing for a wider range of values). When the variability modifier is genetic, it is sometimes referred to as a variance-quantitative trait locus (vQTL). For a genotype that raises variability in Y, the same stimulus (E) might result in a larger absolute change in Y. More rigorously, this scenario assumes that E affects the quantile of Y (the location within its distribution), resulting in a larger linear effect estimate in the more variable genotype group (Figure 4).
Figure 4.
Simulation of heterogeneous variability
(A) G directly affects the variability, but not the mean, of Y. E (with two specific values indicated by the dotted and dashed lines) affects the location of Y within its distribution (i.e., its percentile) in a way that is consistent across G.
(B) E has a consistent effect on Y∗ (Y's percentile) across G (lines for genotypes AA and AB are hidden but identical).
(C) E has differential effects on the actual value of Y across G, due to the simultaneous differential variability.
(D) Results from simulation of a heterogeneous variability-based interaction.
We note an important difference between this concept and the more common usage of the term vQTL to describe any difference in detected statistical variance between genotypes. Often, statistical scans for vQTL effects are conducted to find genetic variants that produce differential variance secondary to a GxE interaction with a specific environmental factor (i.e., the vQTL is a statistical consequence of an underlying functional GxE).25 In contrast, “true” vQTLs have a direct effect on the variability of the phenotype. The relevance of such variants will depend on the research question; detection of a true vQTL might be helpful in predicting an individual’s response to a change in any arbitrary exposure but without adding mechanistic insight into relevant biological pathways for that specific exposure.
Example
Domingue and colleagues describe this scenario as a scaling model.26 Using the UK Biobank dataset, they find a set of genetic variants that associate more strongly with BMI in individuals born later in time. They proceed to show evidence that these variants may confer a general sensitivity to environmental influences (and thus a greater BMI variability) rather than a birth year-specific modifying effect.
Related phenomena
The concept of a “phenotypic capacitor” has been invoked to describe a true vQTL that buffers the effects of cryptic genetic variation, rather than environmental variation. In such a case, the genetic variant would be expected to associate more strongly with phenotypic variance in dizygotic as compared to monozygotic twins due to greater cryptic genetic variation.27 Other, similar non-specific interaction effects have also been described. For example, quantile-specific heritability is a phenomenon in which genetic effects on a phenotype differ across that phenotype’s distribution.28,29 Such quantile-specific genetic effects will produce a “non-specific” GxE, i.e., lead to identification of an additive interaction in standard statistical model, for any exposure having a substantial main effect on the outcome (and thus shifting the location of that phenotype within its distribution).
Phenomenon 5. Heterogeneous exposure measurement
Description
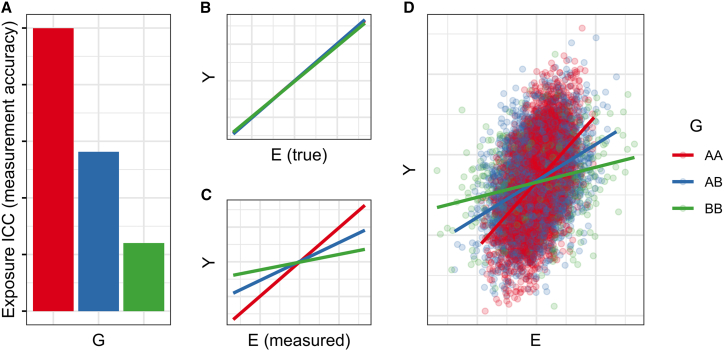
Measurement error in E decreases the estimated magnitude of a true association with Y. So, if G associates with the precision of E measurement, then any nonzero E-Y association will appear stronger in the genotype group with better measurement, inducing a statistical GxE (Figure 5). Importantly, this issue arises most clearly when G associates with the degree of noise in measurement; a G-associated directional bias may or may not manifest as a GxE depending on its nature.
Figure 5.
Simulation of heterogeneous measurement
(A) G directly affects the measurement accuracy of E.
(B) The true E-Y relationship is consistent across G.
(C) The measured E-Y relationship differs across G.
(D) Results from simulation of a heterogeneous measurement-based interaction.
Example
There are many examples of genotype-associated differences in the measurement of biological factors. Studies investigating vitamin D-binding protein (DBP) have established that the chemistry of some immunoassays leads to bias in DBP measurement.30 In another example, self-reported race- and ethnicity-associated differences in skin pigmentation (related to underlying genetic factors) have been shown to lead to differential pulse oximeter performance in measuring blood oxygen saturation.31 Meanwhile, genetic influences on self-report-related quantities such as questionnaire missingness have also been described,32 with implications for differential measurement error.
Related phenomena
For some outcomes of interest, there may be floor or ceiling effects that affect measurement of Y, such as a measurement tool that maxes out at some value. Here, a strong genetic main effect could indirectly increase measurement error by pushing the mean of Y to more extreme values.33,34 Additionally, this phenomenon can be relevant even when measurement of a (non-E) covariate depends on G. It has been recognized that confounder adjustment in interaction models often requires inclusion of G-by-confounder terms, not just their main effects.35 Genotype-associated measurement error in a confounder can affect the “quality” of this adjustment and thus E effect estimates across genotype groups, as has been previously observed in epidemiological models of main effects.36,37
Another related phenomenon combines this measurement concern with the prior discussion of G-E correlation. As observed by Dubridge and Fletcher, spurious GxEs can appear when there is both (1) dependence between a causal genetic variant and the exposure and (2) a non-causal tag variant in imperfect linkage disequilibrium with the causal variant.38 In this case, the interaction appears due to a combination of G-E correlation and measurement error in the genotype.
Recommendations
Given the outlined phenomena, are GxEs “real”? Clearly yes, in the sense that the observed association of G and Y depends on levels of E, and vice versa. However, in some cases the GxE is merely a model- or measurement-related statistical inevitability, without an underlying biological mechanism. For example, phenomenon 5 will be spurious in almost all cases, as measurement error is a practical issue that will rarely be a direct component of biological hypotheses. In contrast, phenomena 1 and 4 are both fundamentally driven by biological processes, with genotypes mechanistically modifying the relationship between the outcome and a specific exposure (phenomenon 1) or all associated exposures (phenomenon 4). Phenomena 2 and 3 represent a middle ground in that they may be considered spurious in a study focused on biological mechanisms, but important for clinical prediction models.
Table S1 provides specific notes on statistical approaches and software tools relevant to each of the phenomena described. Below, we expand on some specific modeling considerations in light of the prior discussion.
-
•
Clinically relevant cutoffs on the raw scale (phenomena 1, 2, 3): Is a cutoff defined on a continuous measure for making clinical decisions, typically to prevent a downstream outcome? If so, any GxE interaction associated with this outcome on the scale of its measurement will be important. For example, it is likely that there is at least some nonlinearity in the increased cardiovascular disease risk (downstream outcome) associated with LDL cholesterol (continuous measure). Nonetheless, specific LDL-C cutoffs are established to guide the use of cholesterol-lowering medication. Therefore, linear interactions of genetic variants and medications affecting LDL-C (as tested by a regression product term) will be clinically important even if they do not correspond to a functional interaction.
-
•
Choice of outcome scale (phenomena 2, 3, 4): Related to the above, the choice of outcome measurement scale requires careful consideration. Choices like log transformation or coding intervention effects in terms of percentage changes are nonlinear transformations of the raw outcome. This consideration is already important in studies of main effects and is amplified for interaction studies due to the phenomena described here. For example, nonlinear mediators, G-E correlation with nonlinearity, and heterogeneous variability are all defined fundamentally in terms of the outcome scale. This choice may also differ according to analysis stage: such nonlinear transformations may be appropriate during hypothesis-free scans (such as genome-wide interaction studies) for which the purpose is mapping loci of interest, with follow-up analyses using the raw outcome scale in order to understand effect sizes and distinguish between some of the phenomena described above.
-
•
Specificity of the exposure (phenomenon 4): Is it important that an identified interaction be specific to the exposure in question? A “true” vQTL will amplify the effect of any exposure with a non-zero main effect on the outcome, but this may not be a concern for all studies. For example, such a variant demonstrating a gene-diet interaction question will be useful for precision nutrition (it will truly modify the expected response to dietary changes), but perhaps not for understanding the biological mechanisms mediating the effects of that dietary factor. The answer to this question can guide sensitivity analysis: upon finding a GxE, statistical tools are available to test for evidence of general vQTL effects independent of a specific GxE.26 Furthermore, databases of vQTLs are beginning to appear, in which investigators can look for prior evidence of variance modification at genetic variants of interest,23 though as noted above, these may appear secondary to a specific GxE rather than indicating a general variance modulation effect.
-
•
Shape of the E-Y relationship (phenomena 2, 3): It is almost always helpful to understand the shape of E-Y associations across a substantial dynamic range of E prior to conducting an interaction analysis. Nonlinearities in this relationship may manifest as interactions via these phenomena (e.g., G-E correlation with nonlinearity), and intentional nonlinear transformations of E can be tested for interaction in some cases. This concern also has direct implications for replication across cohorts, a known challenge for the GxE field.1 If two populations have substantially non-overlapping dynamic ranges of an E, their E main effects and GxE interactions will likely be different, impacting both analytical choices (such as nonlinear transformations of E) and population selection (matching populations with similar E distributions for discovery and replication).
-
•
Measurement of E (phenomena 3, 5): Investigators should consider the nuances and limitations of the measurement of the E variable used in GxE tests. Is it a continuous variable, and if so, are there any threshold effects in its ability to capture the underlying trait of interest? Is its value or measurement precision plausibly under genetic control by the variant(s) being tested? Fortunately, some of these questions can be evaluated directly to some extent: G-E correlation can be tested straightforwardly, while measurement heterogeneity can be evaluated by comparing genotype-stratified variances or, ideally, intraclass correlation coefficients from repeated measures.
-
•
Polygenic scores (all phenomena): Polygenic scores can improve statistical power and increase the strength of the genetic instruments being tested for interaction. However, they also render interpretation (and the considerations above) more difficult. For example, the aggregation across multiple biological pathways makes it harder to reason about whether a polygenic score acts as a “true” vQTL or whether it directly influences exposure measurement quality. One strategy that may be helpful is the use of clustered or pathway-specific polygenic scores, which partially reduces the complexity in interpretation.39,40 The accumulation of signal over many variants in polygenic scores also makes the phenomena described here more likely, such as the manifestation of phenomenon P3 due to a stronger potential association with some E.
Conclusions
For any experiment exploring potential GxEs, the underlying hypothesized mechanism is a critical element to understand, describe, and use in guiding experimental choices. Functional interactions, involving direct and specific molecular-level interactions, are often assumed. However, statistical GxEs can appear due to the use of linear models to test relationships that are truly nonlinear (whether the G or E themselves, or a mediator, have nonlinear association with the outcome Y). They may also show up secondary to unaccounted-for genetic effects on phenotype variability or measurement. The extent to which these various explanations are part of the hypothesis in question, versus alternative possibilities that would “explain away” the sought-after effect, will depend on the specific hypothesis being tested. In presenting this framework of data-generating mechanisms for observed GxEs, we hope to facilitate greater clarity and better experimental design in the genetics community.
Data and code availability
The code used for simulation and figure creation is available at https://github.com/kwesterman/roads-to-gxe-review.
Acknowledgments
The authors thank Elliot M. Tucker-Drob, Robert C. Kaplan, and Nicholas L. Smith for helpful feedback on the manuscript. K.E.W. was supported by NIH grant K01DK133637. T.S. was supported by NIH grant R01HL161012.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2024.03.002.
Supplemental information
References
- 1.Gauderman W.J., Mukherjee B., Aschard H., Hsu L., Lewinger J.P., Patel C.J., Witte J.S., Amos C., Tai C.G., Conti D., et al. Update on the State of the Science for Analytical Methods for Gene-Environment Interactions. Am. J. Epidemiol. 2017;186:762–770. doi: 10.1093/aje/kwx228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rao D.C., Sung Y.J., Winkler T.W., Schwander K., Borecki I., Cupples L.A., Gauderman W.J., Rice K., Munroe P.B., Psaty B.M., CHARGE Gene-Lifestyle Interactions Working Group∗ Multiancestry Study of Gene-Lifestyle Interactions for Cardiovascular Traits in 610 475 Individuals from 124 Cohorts: Design and Rationale. Circ. Cardiovasc. Genet. 2017;10 doi: 10.1161/CIRCGENETICS.116.001649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bi W., Zhao Z., Dey R., Fritsche L.G., Mukherjee B., Lee S. A Fast and Accurate Method for Genome-wide Scale Phenome-wide G × E Analysis and Its Application to UK Biobank. Am. J. Hum. Genet. 2019;105:1182–1192. doi: 10.1016/j.ajhg.2019.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Westerman K.E., Pham D.T., Hong L., Chen Y., Sevilla-González M., Sung Y.J., Sun Y.V., Morrison A.C., Chen H., Manning A.K. GEM: scalable and flexible gene–environment interaction analysis in millions of samples. Bioinformatics. 2021;37:3514–3520. doi: 10.1093/bioinformatics/btab223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhong W., Chhibber A., Luo L., Mehrotra D.V., Shen J. A fast and powerful linear mixed model approach for genotype-environment interaction tests in large-scale GWAS. Briefings Bioinf. 2023;24 doi: 10.1093/bib/bbac547. [DOI] [PubMed] [Google Scholar]
- 6.VanderWeele T.J., Knol M.J. A tutorial on interaction. Epidemiol. Methods. 2014;3 doi: 10.1515/em-2013-0005. [DOI] [Google Scholar]
- 7.Preacher K.J., Rucker D.D., Hayes A.F. Addressing moderated mediation hypotheses: Theory, methods, and prescriptions. Multivariate Behav. Res. 2007;42:185–227. doi: 10.1080/00273170701341316. [DOI] [PubMed] [Google Scholar]
- 8.Zhou W., Liu G., Miller D.P., Thurston S.W., Xu L.L., Wain J.C., Lynch T.J., Su L., Christiani D.C. Gene-environment interaction for the ERCC2 polymorphisms and cumulative cigarette smoking exposure in lung cancer. Cancer Res. 2002;62:1377–1381. [PubMed] [Google Scholar]
- 9.Williams R.A., Mamotte C.D.S., Burnett J.R. Phenylketonuria: an inborn error of phenylalanine metabolism. Clin. Biochem. Rev. 2008;29:31–41. [PMC free article] [PubMed] [Google Scholar]
- 10.Zhernakova D.V., Deelen P., Vermaat M., Van Iterson M., Van Galen M., Arindrarto W., Van’t Hof P., Mei H., Van Dijk F., Westra H.J., et al. Identification of context-dependent expression quantitative trait loci in whole blood. Nat. Genet. 2017;49:139–145. doi: 10.1038/ng.3737. [DOI] [PubMed] [Google Scholar]
- 11.Moore R., Casale F.P., Jan Bonder M., Horta D., BIOS Consortium. Franke L., Barroso I., Stegle O. A linear mixed-model approach to study multivariate gene–environment interactions. Nat. Genet. 2019;51:180–186. doi: 10.1038/s41588-018-0271-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eaves L.J. Genotype × Environment Interaction in Psychopathology: Fact or Artifact? Twin Res. Hum. Genet. 2006;9 doi: 10.1375/twin.9.1.1. [DOI] [PubMed] [Google Scholar]
- 13.Domingue B.W., Kanopka K., Trejo S., Rhemtulla M., Tucker-Drob E.M. Ubiquitous Bias and False Discovery Due to Model Misspecification in Analysis of Statistical Interactions: The Role of the Outcome’s Distribution and Metric Properties. Psychol. Methods. 2022 doi: 10.1037/met0000532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oni-Orisan A., Hoffmann T.J., Ranatunga D., Medina M.W., Jorgenson E., Schaefer C., Krauss R.M., Iribarren C., Risch N. Characterization of Statin Low-Density Lipoprotein Cholesterol Dose-Response Using Electronic Health Records in a Large Population-Based Cohort. Circ. Genom. Precis. Med. 2018;11:e002043. doi: 10.1161/CIRCGEN.117.002043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fischer L.M., Da Costa K.A., Kwock L., Galanko J., Zeisel S.H. Dietary choline requirements of women: Effects of estrogen and genetic variation. Am. J. Clin. Nutr. 2010;92:1113–1119. doi: 10.3945/ajcn.2010.30064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cordell H.J. Detecting gene-gene interactions that underlie human diseases. Nat. Rev. Genet. 2009;10:392–404. doi: 10.1038/nrg2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ritchie M.D., Van Steen K. The search for gene-gene interactions in genome-wide association studies: challenges in abundance of methods, practical considerations, and biological interpretation. Ann. Transl. Med. 2018;6 doi: 10.21037/atm.2018.04.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kwan J.L.Y., Chan W. Variable system: An alternative approach for the analysis of mediated moderation. Psychol. Methods. 2018;23:262–277. doi: 10.1037/met0000160. [DOI] [PubMed] [Google Scholar]
- 19.Kasela S., Aguet F., Kim-Hellmuth S., Brown B.C., Nachun D.C., Tracy R.P., Durda P., Liu Y., Taylor K.D., Johnson W.C., et al. Interaction molecular QTL mapping discovers cellular and environmental modifiers of genetic regulatory effects. Am. J. Hum. Genet. 2024;111:133–149. doi: 10.1016/j.ajhg.2023.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fletcher J.M., Conley D. The challenge of causal inference in gene-environment interaction research: Leveraging research designs from the social sciences. Am. J. Publ. Health. 2013;103:S42–S45. doi: 10.2105/AJPH.2013.301290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de los Campos G., Sorensen D.A., Toro M.A. Imperfect linkage disequilibrium generates phantom epistasis (& perils of big data) G3 (Bethesda). 2019;9:1429–1436. doi: 10.1534/g3.119.400101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laclaustra M., Lopez-Garcia E., Civeira F., Garcia-Esquinas E., Graciani A., Guallar-Castillon P., Banegas J.R., Rodriguez-Artalejo F. LDL cholesterol rises with BMI only in lean individuals: Cross-sectional U.S. And Spanish representative data. Diabetes Care. 2018;41:2195–2201. doi: 10.2337/dc18-0372. [DOI] [PubMed] [Google Scholar]
- 23.Westerman K.E., Majarian T.D., Giulianini F., Jang D.-K., Miao J., Florez J.C., Chen H., Chasman D.I., Udler M.S., Manning A.K., Cole J.B. Variance-quantitative trait loci enable systematic discovery of gene-environment interactions for cardiometabolic serum biomarkers. Nat. Commun. 2022;13:3993. doi: 10.1038/s41467-022-31625-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Voorman A., Lumley T., McKnight B., Rice K. Behavior of QQ-Plots and Genomic Control in Studies of Gene-Environment Interaction. PLoS One. 2011;6 doi: 10.1371/journal.pone.0019416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paré G., Cook N.R., Ridker P.M., Chasman D.I. On the Use of Variance per Genotype as a Tool to Identify Quantitative Trait Interaction Effects: A Report from the Women’s Genome Health Study. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1000981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Domingue B.W., Kanopka K., Mallard T.T., Trejo S., Tucker-Drob E.M. Modeling Interaction and Dispersion Effects in the Analysis of Gene-by-Environment Interaction. Behav. Genet. 2022;52:56–64. doi: 10.1007/s10519-021-10090-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Conley D., Johnson R., Domingue B., Dawes C., Boardman J., Siegal M. A sibling method for identifying vQTLs. PLoS One. 2018;13 doi: 10.1371/journal.pone.0194541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Williams P.T. Quantile-specific penetrance of genes affecting lipoproteins, adiposity and height. PLoS One. 2012;7 doi: 10.1371/journal.pone.0028764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Williams P.T. Gene-environment interactions due to quantile-specific heritability of triglyceride and VLDL concentrations. Sci. Rep. 2020;10 doi: 10.1038/s41598-020-60965-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nielson C.M., Jones K.S., Chun R.F., Jacobs J.M., Wang Y., Hewison M., Adams J.S., Swanson C.M., Lee C.G., Vanderschueren D., et al. Free 25-hydroxyvitamin D: Impact of vitamin D binding protein assays on racial-genotypic associations. J. Clin. Endocrinol. Metab. 2016;101:2226–2234. doi: 10.1210/jc.2016-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gottlieb E.R., Ziegler J., Morley K., Rush B., Celi L.A. Assessment of Racial and Ethnic Differences in Oxygen Supplementation Among Patients in the Intensive Care Unit. JAMA Intern. Med. 2022;182:849–858. doi: 10.1001/jamainternmed.2022.2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mignogna G., Carey C.E., Wedow R., Baya N., Cordioli M., Pirastu N., Bellocco R., Malerbi K.F., Nivard M.G., Neale B.M., et al. Patterns of item nonresponse behaviour to survey questionnaires are systematic and associated with genetic loci. Nat. Human Behav. 2023;7:1371–1387. doi: 10.1038/s41562-023-01632-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Molenaar D., Van Der Sluis S., Boomsma D.I., Haworth C.M.A., Hewitt J.K., Martin N.G., Plomin R., Wright M.J., Dolan C.V. Genotype by environment interactions in cognitive ability: A survey of 14 studies from four countries covering four age groups. Behav. Genet. 2013;43:208–219. doi: 10.1007/s10519-012-9581-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tucker-Drob E.M. Differentiation of Cognitive Abilities Across the Life Span. Dev. Psychol. 2009;45:1097–1118. doi: 10.1037/a0015864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Keller M.C. Gene × Environment Interaction Studies Have Not Properly Controlled for Potential Confounders: The Problem and the (Simple) Solution. Biol. Psychiatr. 2014;75:18–24. doi: 10.1016/j.biopsych.2013.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spiegelman D., McDermott A., Rosner B. Regression calibration method for correcting measurement-error bias in nutritional epidemiology. Am. J. Clin. Nutr. 1997;65:1179S–1186S. doi: 10.1093/ajcn/65.4.1179S. [DOI] [PubMed] [Google Scholar]
- 37.Thomas D., Stram D., Dwyer J. Exposure Measurement Error: Influence on Exposure-Disease Relationships and Methods of Correction. Annu. Rev. Publ. Health. 1993;14:69–93. doi: 10.1146/annurev.pu.14.050193.000441. [DOI] [PubMed] [Google Scholar]
- 38.Dudbridge F., Fletcher O. Gene-environment dependence creates spurious gene-environment interaction. Am. J. Hum. Genet. 2014;95:301–307. doi: 10.1016/j.ajhg.2014.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim H., Westerman K.E., Smith K., Chiou J., Cole J.B., Majarian T., von Grotthuss M., Kwak S.H., Kim J., Mercader J.M., et al. High-throughput genetic clustering of type 2 diabetes loci reveals heterogeneous mechanistic pathways of metabolic disease. Diabetologia. 2023;66:495–507. doi: 10.1007/s00125-022-05848-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goodman M.O., Cade B.E., Shah N.A., Huang T., Dashti H.S., Saxena R., Rutter M.K., Libby P., Sofer T., Redline S. Pathway-Specific Polygenic Risk Scores Identify Obstructive Sleep Apnea-Related Pathways Differentially Moderating Genetic Susceptibility to Coronary Artery Disease. Circ. Genom. Precis. Med. 2022;15:e003535. doi: 10.1161/CIRCGEN.121.003535. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The code used for simulation and figure creation is available at https://github.com/kwesterman/roads-to-gxe-review.