Abstract
G protein-coupled receptors (GPCRs) can initiate unique functional responses depending on the subcellular site of activation. Efforts to uncover the mechanistic basis of compartmentalized GPCR signaling have concentrated on the biochemical aspect of this regulation. Here, we assess the biophysical positioning of receptor-containing endosomes as an alternative salient mechanism. We devise a strategy to rapidly and selectively redistribute receptor-containing endosomes ‘on command’ in intact cells without perturbing their biochemical composition. Next, we present two complementary optical readouts that enable robust measurements of bulk- and gene-specific GPCR/cAMP-dependent transcriptional signaling with single-cell resolution. With these, we establish that disruption of native endosome positioning inhibits the initiation of the endosome-dependent transcriptional responses. Lastly, we demonstrate a prominent mechanistic role of phosphodiesterase-mediated cAMP hydrolysis and local PKA activity in this process. Our study, therefore, illuminates a novel mechanism regulating GPCR function by identifying endosome positioning as the principal mediator of spatially-selective receptor signaling.
Introduction
G protein-coupled receptors (GPCRs) orchestrate essential aspects of mammalian physiology. GPCR function is tightly controlled by endocytic trafficking, where the ligand-activated receptor engages arrestins and clathrin machinery and is subsequently internalized into endosomal compartments1. While the endosome-associated receptor pool was classically presumed to be functionally inactive, it is now clear that receptors can also signal from endosomes2–4. Moreover, endosomal receptors can initiate cellular responses that are distinct from those activated at the plasma membrane. Transcriptional reprogramming was one of the first location-biased GPCR responses to be identified and shown to be stimulated from intracellular receptors5, 6. Since then, compartmentalized signaling has been implicated in the transduction of distinct phosphosignaling7, 8 and in the coordination of unique physiologies and drug actions8–17. Yet, how the endosome selectively facilitates these responses compared to other subcellular compartments remains unclear.
Efforts to mechanistically understand compartmentalized GPCR signaling have focused on the biochemical aspect of this regulation through identification and characterization of relevant molecular factors. Studies have identified regulatory roles for beta and alpha arrestins16, 18, 19 as well as trafficking proteins, including sorting nexins and members of the retromer complex18, 20, 21, in fine-tuning the activity of endosomal GPCRs. In contrast, the biophysical properties of endosomes that contribute to this phenomenon have been unexplored. Here, we investigate the mechanistic salience of one biophysical property of endosomes - their subcellular localization. Early endosomes are highly dynamic organelles that bring their cargo toward the cell core. Therefore, their three-dimensional positioning within the cell could play an active role in coordinating GPCR signaling, especially in the context of spatially-biased nuclear responses, such as transcriptional signaling. The challenge in testing this model lies in the ability to selectively manipulate the positioning of early endosomes without perturbing their biochemical composition.
We address this question by focusing on the beta2-adrenergic receptor (β2-AR), one of the best-characterized GPCRs, which controls cardiovascular and pulmonary physiologies in response to adrenaline and noradrenaline22, 23. The β2-AR signals via Gas-dependent stimulation of cyclic AMP (cAMP) to induce protein kinase A (PKA) activation. PKA translocates into the nucleus, where it phosphorylates the cAMP Response Element-Binding Protein (CREB) to regulate gene transcription6, 24–26. It is now well-established that the β2-AR can signal from the plasma membrane and endosomes24. Notably, adrenoceptor activation and cAMP production on early endosomes uniquely drive GPCR-mediated gene transcription, while the plasma membrane fraction of signaling receptors has a negligible contribution to this response6. Therefore, we choose to focus on β2-AR-mediated transcription as a pertinent endosome-biased signaling readout and interrogate the role of organelle positioning in initiating these responses. We devise a strategy that enables the rapid and selective redistribution of receptor-containing endosomes without disrupting the biochemical complexes relevant to receptor trafficking and signaling. We combine this approach with novel cAMP and PKA biosensors and transcriptional readouts to establish the biophysical positioning of endosomes as a primary mediator of location-encoded GPCR signal transduction.
Results
A dimerization strategy rapidly redistributes early endosomes
To assess the significance of endosome positioning in the activation of the spatially-biased GPCR transcriptional responses, we began by devising a strategy for rapid redistribution of endosomes. The positioning of endosomes is tightly regulated by the cytoskeletal framework of dynein and kinesin motors that move endosomes along microtubules toward the nucleus or the cell periphery, respectively27. Early endosomes are bound by dynein motors that transport these organelles toward the minus end of microtubules and in proximity to the cell core28. For that reason, the most commonly used ways to manipulate organelle positioning involve either pharmacologic or genetic perturbations of the cytoskeleton, such as microtubule-depolarizing drugs (e.g., nocodazole), dynein inhibitors (e.g., ciliobrevin D), or depletion/overexpression of motor proteins29, 30. However, these approaches are not selectively tailored toward endosomes. In addition, they can lead to cell toxicity and/or may occur over the course of several days (e.g., in the case of gene depletion/overexpression) and could potentially prompt compensatory genetic mechanisms.
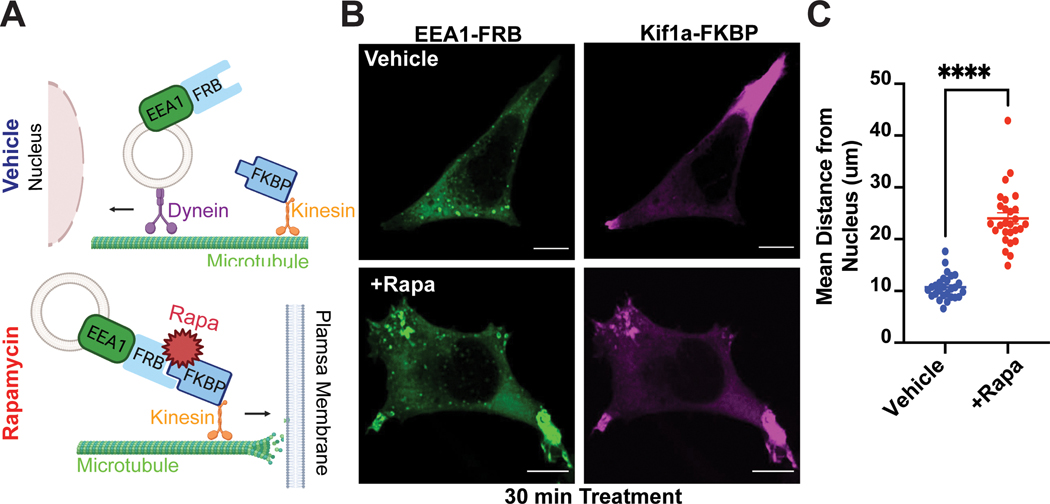
To overcome these limitations, we set up a chemically inducible dimerization (CID) strategy. We fused the early endosome antigen 1 (EEA1) protein to the FKBP-rapamycin-binding (FRB) domain and EGFP to generate EGFP-FRB-EEA1, and the plus-end-directed kinesin motor Kif1a to the FK506 binding protein (FKBP) and tandem dTomato to generate Kif1a-FKBP-tdTomato. Upon addition of rapamycin, FRB dimerizes with FKBP to facilitate ‘on command’ redistribution of endosomes toward the cell periphery (Fig. 1A). As an experimental model system, we chose HEK293 cells, because they express β2-ARs at native levels31 and are an appropriate model to examine receptor signaling under endogenous setting. We transiently transfected HEK293 cells with EGFP-FRB-EEA1 and Kif1a-FKBP-tdTomato and used live- and fixed-cell microscopy to evaluate endosome distribution. In comparison to vehicle-treated cells, which exhibited typical endosomal puncta scattered across the cytosol, rapamycin-treated cells showed rapid and significant recruitment of endosomes toward the cell periphery (Fig. 1B–C, Supplementary Fig. 1A, Supplementary Video 1). In contrast, the dimerization approach did not impact the subcellular distribution of another organelle, the trans-Golgi network (Supplementary Fig. 1B). Thus, our strategy successfully enables selective and rapid redistribution of early endosomes in intact cells.
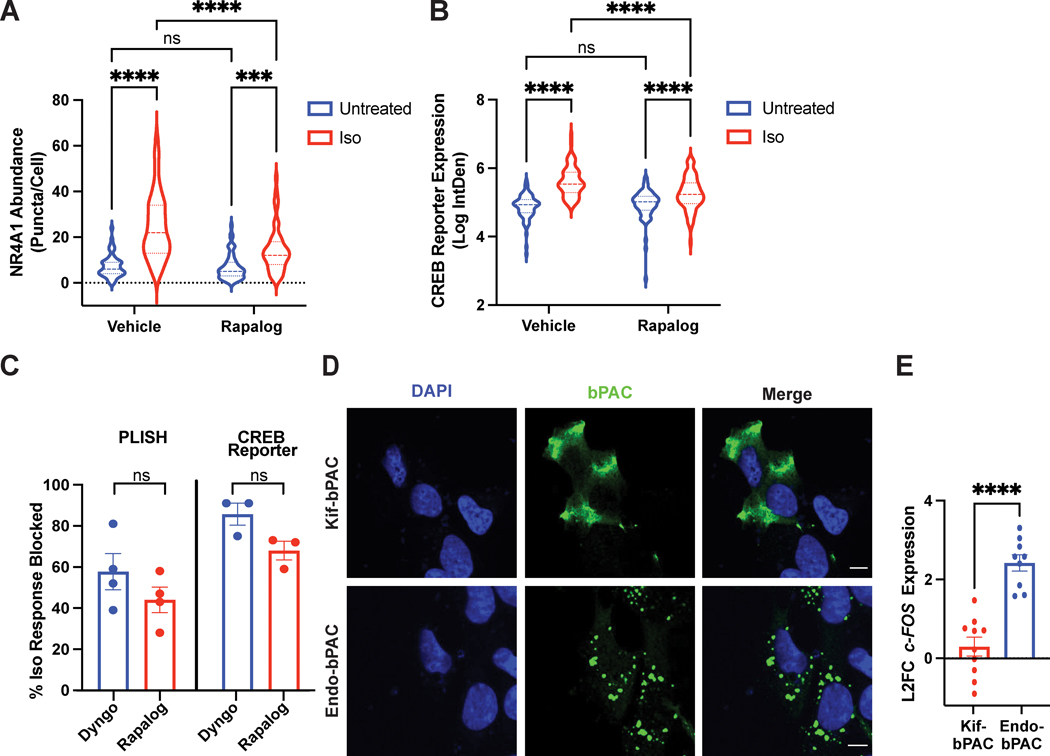
Figure 1. CID approach for redirecting endosomes.
A. Schematic of the approach. B-C. Rapid redistribution of early endosomes by rapamycin treatment. B. FRB-EEA1 (green), localized at early endosomes, and Kif1a-FKBP (magenta), localized near the cell periphery, visualized by immunofluorescence microscopy in fixed cells. In ethanol-treated cells (vehicle, top), early endosomes are dispersed in the cell. After 1μM rapamycin treatment for 30 min (bottom), early endosomes redistribute to the cell periphery. C. Increase in mean endosome distance from the nucleus in rapamycin-treated cells. Mean endosome distance per cell = 10.75μm ± 0.46 (EtOH) and 24.04μm ± 1.08 (rapamycin). Data are average from n = 3 biologically independent replicates ± s.e.m.; 27 cells total/condition; **** = p < 0.0001 by unpaired two-tailed Student’s t-test. Scale bar = 10μm.
β2-AR traffics efficiently through redistributed endosomes
To facilitate the evaluation of endosome positioning in GPCR signaling per se, the dimerization approach should not alter the biochemical composition of endosomes. Hence, we next asked whether the strategy impacted endosomal integrity by assessing β2-AR trafficking and signaling.
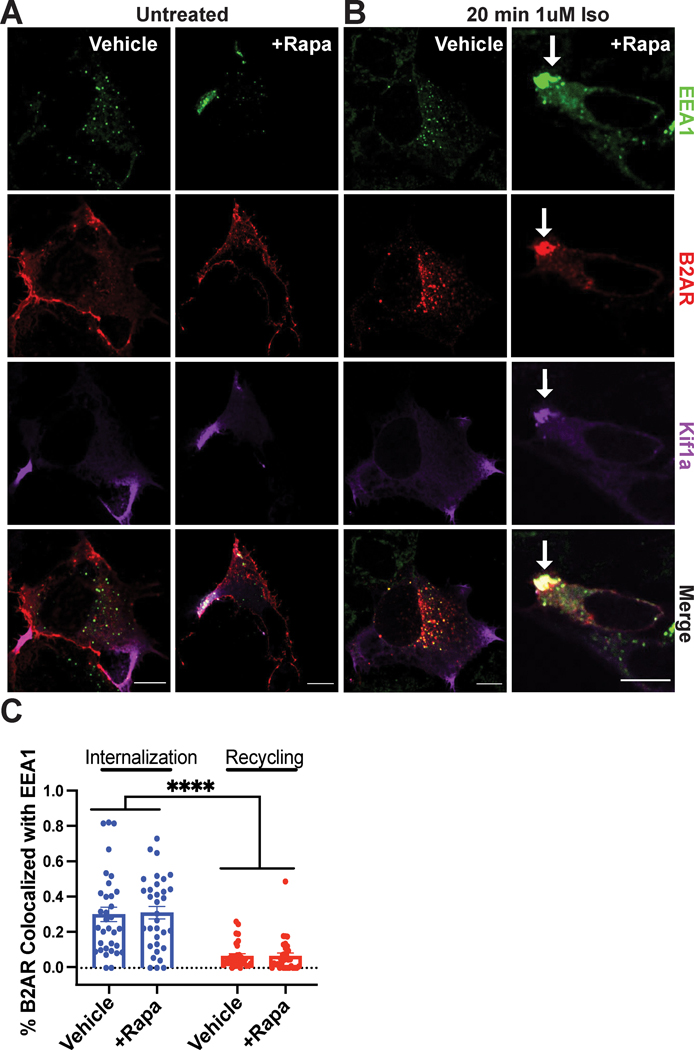
To examine β2-AR trafficking, we co-expressed N-terminally Flag-tagged β2-AR and the dimerization constructs, and stimulated receptor internalization using the synthetic agonist isoproterenol. In the absence of agonist, the β2-AR localized on the plasma membrane in both vehicle and rapamycin conditions (Fig. 2A). In control cells, we observed robust β2-AR endocytosis into EEA1-positive vesicles within 20 minutes of stimulation (Fig. 2B, left). The β2-AR internalized similarly in cells pre-treated with rapamycin, despite the dramatic change in endosome positioning (Fig. 2B, right and Fig. 2C, blue bars). Therefore, rapamycin-induced endosome redistribution does not interfere with proper receptor internalization.
Figure 2. Intact β2-AR trafficking into and out of repositioned endosomes.
A-D. HEK293 cells were pre-treated with ethanol (“Vehicle”) or 1μM rapamycin (“+Rapa”) for 30 min. Cells were fixed and imaged by immunofluorescence microscopy. A. HEK293 cells co-expressing the CID machinery and Flag-β2-AR visualized by immunofluorescence microscopy. β2-AR (red) is present at the cell surface in the absence of β2-AR stimulation. B. β2-AR colocalizes with early endosomes (green) after 20 min of activation with 1μM isoproterenol (“Iso”). β2-AR-containing endosomes selectively redistribute toward the cell periphery upon rapamycin application. Scale bar = 10μm. White arrow indicates repositioned endosomes. C. Endosome redistribution with rapamycin does not impact β2-AR internalization or recycling compared to vehicle-treated cells. Internalization was induced as described in B. Recycling was induced following internalization by application of 10μM alprenolol for 1 h (see also Supplementary Fig. 3). Object-based colocalization analysis was used to calculate internalized or recycled β2-ARs as the ratio of [number of β2-AR puncta colocalized with EEA1]/[total β2-AR puncta] (see “Methods” for details). Data are mean from n = 3 biologically independent replicates ± s.e.m.; 33 cells total/condition; **** = p < 0.0001 by two-way ANOVA test with Tukey.
During the initial hours of agonist stimulation, β2-ARs rapidly recycle from early endosomes back to the plasma membrane. This recycling process is mediated by the retromer complex, which is composed of sorting nexins (SNX) and vacuolar protein sorting (VPS) protein32. To establish whether β2-AR recycling is impacted by endosome redistribution, we first examined the localization of the endosome-associated retromer component SNX27. Both endogenous and overexpressed SNX27 localized similarly to EEA1-positive endosomal compartments in control and rapamycin-treated cells (Supplementary Fig. 2). Next, we visualized and quantified recycled β2-ARs in cells stimulated with isoproterenol, then treated with the antagonist alprenolol to induce synchronized recycling (Supplementary Fig. 3A). We observed very efficient receptor recycling from redistributed endosomes, which paralleled that seen in normal cells (Fig. 2C, red bars). In addition, we examined whether repositioned endosomes can properly mature into recycling endosomes by evaluating the localization of the recycling endosome marker RAB11. RAB11 co-localized with internalized β2-AR in both native and redistributed endosomes following application of alprenolol (Supplementary Fig. 3B). Further, pre-incubation with rapamycin alone did not impact overall trafficking as measured by internalization and recycling of the transferrin receptor (TFR), a constitutively trafficked cargo that is sorted largely by bulk flow (Supplementary Fig. 3C). Hence, neither the localization of necessary sorting components nor the internalization and recycling of β2-ARs is altered by our approach. Next, we investigated whether the receptor stimulates signaling from redistributed endosomes.
β2-AR/Gas signaling is intact in redistributed endosomes
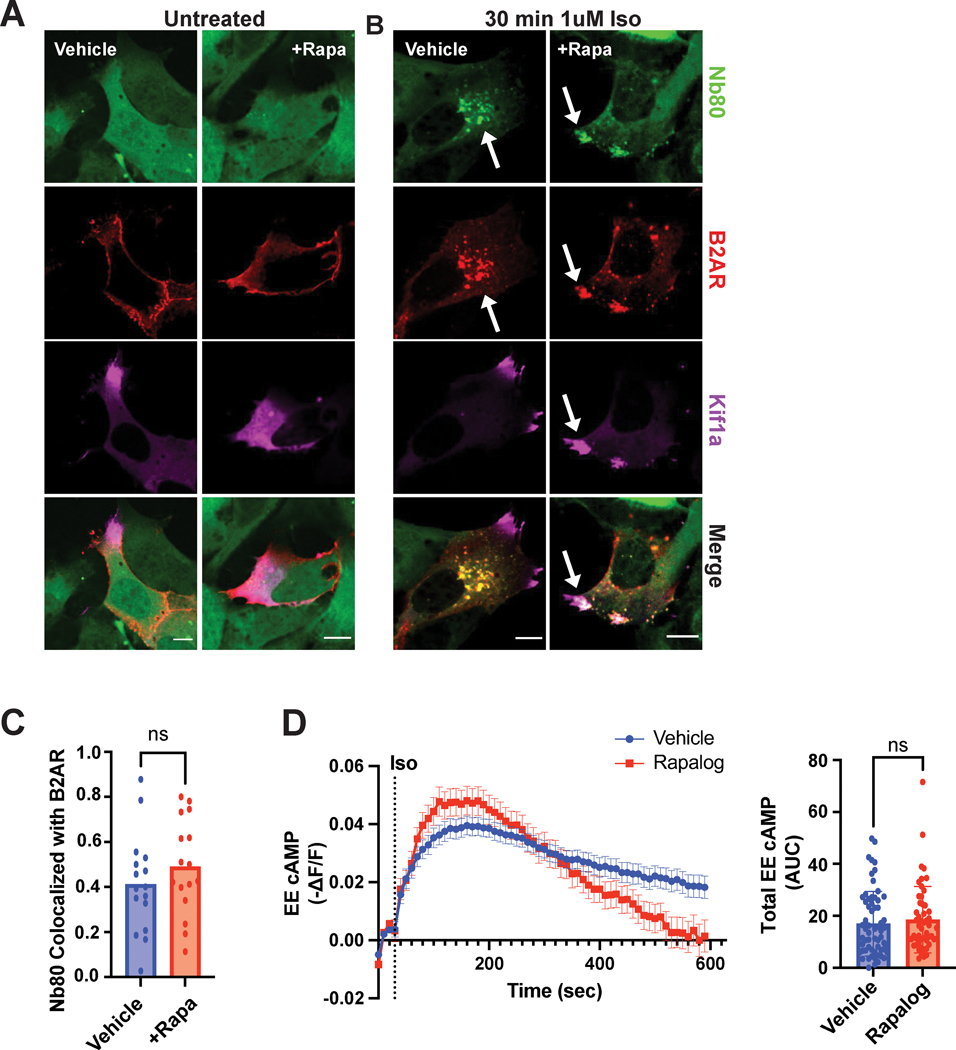
We began by utilizing two conformation-selective nanobody biosensors, Nb80 and Nb37, that enable the in-cell visualization of active β2-ARs and Gas, respectively24. Each nanobody fused to GFP was co-expressed with Flag-tagged β2-AR and the CID constructs. In the absence of receptor stimulation, both Nb80 and Nb37 localized diffusively in the cytosol in control and CID-redistributed cells (Fig. 3A and Supplementary Fig. 4A). Upon isoproterenol stimulation of control cells, we observed nanobody redistribution to receptor-containing endosomes indicating β2-AR and Gas activation in these compartments (Fig. 3B, left, Fig. 3C, blue bar, and Supplementary Fig. 4B). We similarly observed co-localization of both biosensors with endosomal β2-ARs in CID redistributed cells (Fig. 3B, right, Fig. 3C, red bar, and Supplementary Fig. 4C). This demonstrates that the β2-AR can adopt an active conformation and stimulate Gas, despite the change in endosome position.
Figure 3. β2-AR on repositioned endosomes adopts an active conformation and stimulates Gas/cAMP signaling.
A-C. HEK293 cells were pre-treated with ethanol (“Vehicle”) or 1μM rapamycin (“+Rapa”) for 30 min. Flag-β2-AR (red) and Nb80(green) localization in untreated (A) and agonist-stimulated CID-expressing cells (B). A. Nb80 (green) is cytosolic in the absence of β2-AR stimulation. B. Redistributed endosomes containing Flag-β2-AR colocalize with Nb80 following 1μM isoproterenol treatment for 30 min. White arrows indicate examples of colocalization between Nb80 with β2-AR. Scale bar = 10μm. C. Fraction of co-localized β2-AR and Nb80 in control and rapamycin-treated cells. Data are mean from n = 2 biologically independent replicates; 16 cells total/condition. ns by unpaired two-tailed Student’s t-test. D. Cells expressing Flamindo2-FRB-EEA1 and kinesin-FKBP were pre-treated with ethanol (“Vehicle”) or 1μM AP21967 (“Rapalog”) for 30 min before 1μM isoproterenol stimulation. The delta F/F (ΔF/F) ratio was measured every 10 sec and transformed by −1 (left). Total cAMP produced was quantified by calculating the area under the curve (AUC, right). Data are mean from n = 12 (“Vehicle”) and n = 19 (“Rapalog”) biologically independent replicates ± s.e.m.; 52–63 cells total/condition. ns by unpaired two-tailed Student’s t-test.
To directly assess endosomal cAMP in cells with redistributed endosomes, we sought to develop a biosensor that enables local measurements of the second messenger based on several considerations. The heterodimerization strategy necessitated the expression of multiple fluorescent proteins, therefore limiting the available wavelengths for live-cell imaging. In addition, we noted that the accumulation of endosomes in the cell periphery in the presence of rapamycin would lead to enhanced fluorescence intensity of an endosomal sensor confounding the interpretation of the effects of cAMP concentrations on an ‘up’ sensor, i.e. a sensor with an increase in fluorescence upon binding to cAMP. Thus, we focused on validated single-fluorophore ‘down’ sensors that we could target to the endosome. Due to its high dynamic range, we chose Flamindo2, which is composed of the cAMP-binding domain of Epac1 and a yellow fluorescent protein variant, Citrine33. cAMP binding to Flamindo2 leads to a decrease in its fluorescence intensity. We fused Flamindo2 to FRB-EEA1 to generate a Flamindo2-FRB-EEA1 sensor, and confirmed it localized to endosomes (Supplementary Fig. 5A). We next determined that stimulation of endogenous β2-ARs with a saturating dose of isoproterenol yielded responses within the dynamic range of the sensor (Supplementary Fig. 5B). To test the ability of the sensor to detect local cAMP generation, we sought to inhibit β2-AR internalization by overexpression of the dynamin dominant negative mutant, DynK44E (Supplementary Fig. 5C–D)34. Blockade of β2-AR internalization delayed the onset and reduced the total amount of cAMP accumulation, consistent with the endosomal sensor detecting diffusion of cAMP originating from receptor activation at the plasma membrane (Supplementary Fig. 5E). Further, we verified that the construct can be successfully recruited to Kif1a-FKBP-tdTomato in the presence of heterodimerizer ligand (Supplementary Video 2). For these and all subsequent experiments that measure signaling outputs, we used the compound AP21967 (‘Rapalog’) instead of rapamycin. Rapalog permits FRB binding without mTOR crossreaction35. Similar to rapamycin, rapalog application alone did not impact cellular trafficking (Supplementary Fig. 3C) or β2-AR-dependent cAMP production detection by Flamindo2-FRB-EEA1 (Supplementary Fig. 5F). Next, we tested the ability of redistributed endosomes to produce cAMP in the presence of the CID system. We observed the same total amounts of isoproterenol-induced endosomal cAMP in cells with natively localized and repositioned endosomes (Fig. 3D). Therefore, redistributed endosomes support β2-AR/Gas activation and cAMP signaling.
Optical readouts report GPCR-dependent transcription
Endosomal β2-ARs selectively stimulate transcriptional signaling, and we therefore chose to focus on transcription as a representative spatially-biased GPCR response. Because the dimerization strategy requires optimal co-expression of EGFP-FRB-EEA1 and Kif1a-FKBP-tdTomato for efficient endosome relocalization, conventional population-based assays would not be suitable to assess gene transcription. Instead, our experiments necessitated transcriptional readouts compatible with microscopy to allow evaluation of transcriptional signaling in cells that exhibited co-expressed dimerization constructs and efficiently redistributed endosomes.
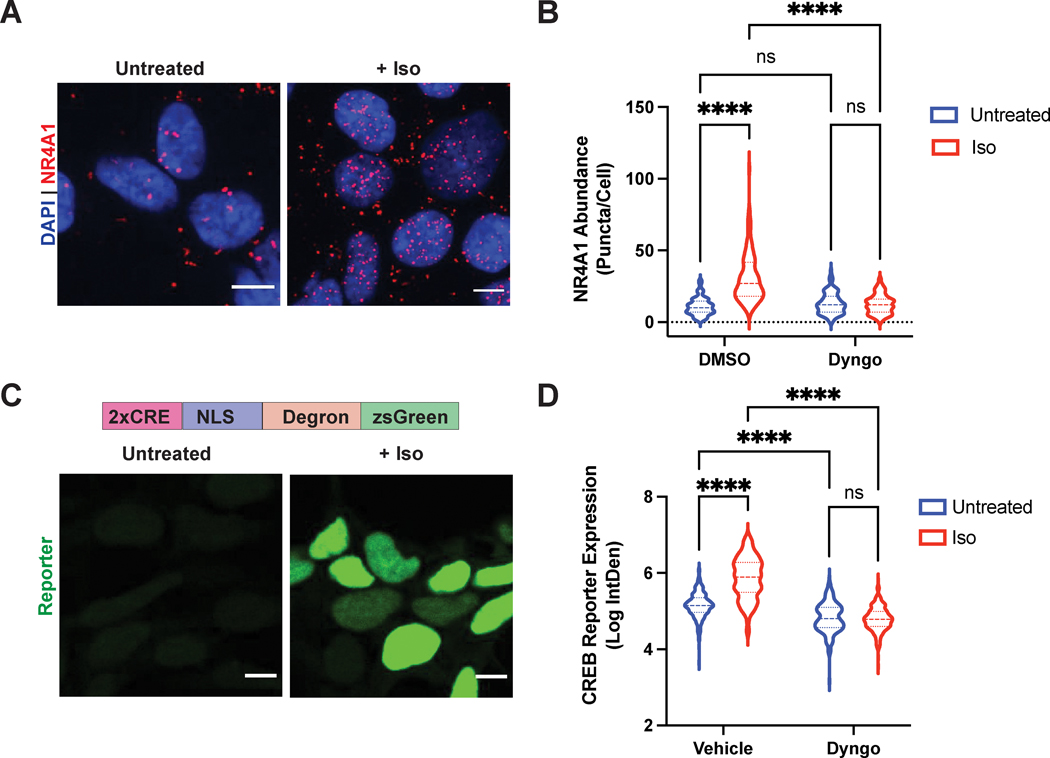
We began by optimizing an optical readout to monitor the expression of endogenous β2-AR/cAMP target genes. To this end, we utilized proximity ligation in situ hybridization (PLISH), a recently developed approach for single-molecule RNA detection in cells with high sensitivity and specificity36. For this analysis, we picked the immediate-early response gene NR4A1 (Nuclear Receptor Subfamily 4 Group A Member 1), a robust transcriptional target gene of β2-AR activation6. NR4A1 expression was induced by isoproterenol treatment, and this induction was blunted when we inhibited dynamin and β2-AR internalization with the drug Dyngo-4a (Fig. 4A–B, Supplementary Fig. 6), as expected for an endosome-dependent response based on previous reports using population-based transcriptional readouts6. Therefore, single-molecule RNA PLISH analysis of NR4A1 successfully captures endosomal β2-AR-mediated transcriptional signaling in cells by microscopy.
Figure 4. Single-cell optical readouts of GPCR-dependent transcriptional signaling.
A. Representative images of NR4A1 mRNA expression by PLISH analysis. Left: untreated; right: treated with 1μM isoproterenol for 2 h. B. Quantification of NR4A1 induction in cells pre-treated with DMSO (“Vehicle”) or 30μM Dyngo-4A for 20 min, then stimulated with 1μM isoproterenol for 2 h. Data are mean from n = 4 biologically independent replicates ± s.e.m.; 132 cells total/condition. C. Representative images of CREB reporter expression in pCRE-NLS-DD-zsGreen1 cells. Schematic of the reporter is shown on top. CRE = cAMP response element; NLS = nuclear localization sequence. Left: untreated in the presence of 1μM Shield-1 for 4 h; right: treated with 1μM isoproterenol in the presence of 1μM Shield for 4 h. D. Quantification of CREB reporter induction in cells pre-treated with DMSO (“Vehicle”) or 30μM Dyngo-4A for 20 min, then stimulated with 1μM isoproterenol for 4 h in the presence of 1μM Shield. Data are mean from n = 3 biologically independent replicates ± s.e.m.; 225 cells total/condition. **** = p < 0.0001 by two-way ANOVA test with Tukey. Scale bar = 10μm.
Next, we established a complementary optical readout of bulk β2-AR/cAMP-dependent transcription. We recently published a fluorescent transcriptional reporter (pCRE-DD-zsGreen1) for the cAMP response element-binding protein (CREB) that we measured using flow cytometry37. We reasoned that we could apply this readout in combination with microscopy. We reengineered the construct to include a nuclear localization sequence (NLS) for ease of quantification, and generated HEK293 cells stably expressing pCRE-NLS-DD-zsGreen1 by lentiviral transduction. Stimulation with isoproterenol induced robust reporter accumulation after 4 hours (>4.5-fold relative to vehicle-treated cells in the presence of stabilizing ligand, Fig. 4C–D). Similar to the NR4A1 analysis, this upregulation was abolished by acute blockade of β2-AR internalization with Dyngo-4a (>6-fold inhibition, Fig. 4D). Thus, these complementary microscopy-based readouts enable respective measurements of bulk and gene-specific cAMP-dependent transcription with single-cell resolution and can be applied in conjunction with the dimerization system to investigate the role of endosome positioning in spatially-biased GPCR signaling.
Endosome redistribution inhibits GPCR-mediated transcription
First, we confirmed that pretreatment of cells with rapalog did not impact basal transcription (log-transformed basal levels of 6.9 versus 7.2 and 4.9 versus 4.9 for rapalog- vs control-treated cells in PLISH and CREB reporter assays, respectively; Fig. 5A and 5B). Therefore, rapalog treatment alone does not affect basal NR4A1 levels measured by RNA in-situ hybridization and CREB signaling measured with the fluorescent reporter.
Figure 5. Endosome positioning is critical for site-selective signaling.
A-C. Cells expressing the CID system were pre-treated with ethanol (“Vehicle”) or 1μM AP21967 (“Rapalog”) for 30 min before agonist stimulation. A. NR4A1 mRNA expression by PLISH analysis of untreated or isoproterenol-stimulated cells (“Iso”, 1μM, 2 h). Data are mean from n = 4 biologically independent replicates ± s.e.m.; 55 cells total/condition. **** = p < 0.0001, *** = p < 0.001 by two-way ANOVA test with Tukey. B. CREB reporter expression in pCRE-NLS-DD-zsGreen1 cells untreated in the presence of 1μM Shield or stimulated with 1μM isoproterenol (“Iso”) in the presence of 1μM Shield for 4 h. Data are mean from n = 3–4 biologically independent replicates ± s.e.m.; 70–77 cells total/condition. **** = p < 0.0001 by two-way ANOVA test with Tukey. C. Endosome positioning regulates the initiation of cAMP-dependent gene transcription to a comparable extent as β2-AR endocytosis. Shown are % inhibition of the isoproterenol-induced transcriptional response from PLISH (left) and CREB reporter (right) analyses. Data are mean from n = 3 (“CREB Reporter”) and n = 4 (“PLISH”) biologically independent replicates ± s.e.m. and analyzed by unpaired two-tailed Student’s t-test. D. Representative images of bPACs (green) localized to the periphery by fusion to a kinesin motor (“Kif-bPAC”, top) or to the early endosome by fusion to a 2X-FYVE domain (“Endo-bPAC”, bottom). Cells were visualized by immunofluorescence microscopy following staining with Alexa 647-conjugated anti-myc antibody. Representative images are selected from n = 2 biologically independent experiments. E. c-FOS induction measured by RT-qPCR analysis after bPAC photostimulation for 3 min. Data represent mean from n = 9 (“Endo-bPAC”) and n = 10 (“Kif-bPAC”) biologically independent replicates ± s.e.m. **** = p < 0.0001 by unpaired two-tailed Student’s t-test. Scale bar = 10μm.
Next, we analyzed the impact of endosome relocalization on the endogenous receptor-induced transcriptional responses using PLISH. In control cells transfected with the CID constructs, we observed robust isoproterenol-dependent NR4A1 accumulation (3.4-fold, Fig. 5A, Supplementary Fig. 7A, left). Remarkably, endosome redistribution significantly blunted this response (1.7-fold decrease, Fig. 5A and Supplementary Fig. 7A, right). At the same time, the expression of the housekeeping gene GAPDH, which is not regulated via the β2-AR cascade, was not impacted by the manipulation (Supplementary Fig. 7B).
Then, we examined transcriptional signaling in pCRE-NLS-DD-zsGreen1 cells. In agreement with the results from the PLISH analysis, here we saw greater than 5-fold induction of CREB reporter accumulation following isoproterenol treatment in control cells expressing the dimerization system, which in turn was dampened by pre-application of rapalog (>2-fold decrease, Fig. 5B and Supplementary Fig. 7C). On the other hand, we observed comparable reporter induction following direct stimulation of CREB with a cell-permeable cAMP analog, which bypasses the need for receptor and adenylyl cyclase activation (Supplementary Fig. 7D). Therefore, the CID-dependent inhibition was specific for the GPCR pathway.
Based on these outcomes, we next quantified the extent to which endosome positioning contributes to the spatial bias of GPCR signaling. Since Dyngo-4a completely blocked endocytosis of β2-ARs (Supplementary Fig. 6B), we defined % loss of isoproterenol-induced signaling in Dyngo-4a-treated cells to represent the endosome-dependent aspect of the transcriptional response. We observed ~60% and ~85% Dyngo-dependent blockade of endosome-dependent transcriptional signaling measured using PLISH and the CREB reporter, respectively (Fig. 5C, blue bars). Strikingly, redistribution of β2-AR-containing endosomes mirrored very closely the extent of inhibition observed with Dyngo-4a (~45% and ~70% by PLISH and CREB reporter analysis, respectively; Fig. 5C, red bars). Hence, endosome positioning is the major mechanistic determinant of endocytosis-dependent transcriptional signaling in response to GPCR stimulation.
The cAMP accumulation site coordinates signaling
As an orthogonal approach to investigate the significance of the biophysical location of signaling, we employed a previously described optogenetic strategy for localized cAMP production6, 7. In this approach, the bacteria-derived photoactivatable adenylyl cyclase, bPAC, is fused to localization-specific targeting sequences to enable spatiotemporal control of second messenger accumulation. We reasoned that bPAC fused to the FYVE domain derived from Hrs38 (“Endo-bPAC”) would mimic signals originating from natively localized endosomes, while bPAC fused to a Kif1a-derived targeting sequence (“Kif-bPAC”) would mirror signals from the subcellular site of repositioned endosome accumulation. We confirmed proper construct expression and localization (Fig. 5D, Supplementary Fig. 8A–C). Then, we measured cAMP concentrations downstream of Kif-bPAC and Endo-bPAC with a colorimetric immunoassay. This analysis showed that comparable cAMP was generated from each site following photostimulation with matched light doses (Supplementary Fig. 8D). Next, we examined transcriptional signaling by quantitative PCR analysis of an established β2-AR target gene, c-FOS6. Photostimulation of Endo-bPAC activated robust signaling, while Kif-bPAC activated a very weak transcriptional response (Fig. 5E). More importantly, this trend held across a range of light doses (Supplementary Fig. 8E). These results are consistent with the outcome from our analysis of endosome positioning and support cAMP signals originating at a distance from natively localized endosomes as effectively uncoupled from downstream transcriptional control.
Local PKA activity is dependent on endosome positioning
To determine how endosome localization coordinates the receptor-dependent transcriptional responses, we began by interrogating the primary cAMP effector kinase, PKA. PKA signaling is highly compartmentalized through the selective localization of its catalytic and regulatory subunits, PKAcat and PKAR, respectively. Specifically, PKAcat, has been found enriched on intracellular puncta and in the perinuclear region5, 26, 39. Utilizing a published functionally validated PKA catalytic Cα construct, mTagBFP2-PKAcatα39, we first verified that this construct co-localized with PKARIIB-mCherry (Supplementary Fig. 9A). Consistent with previous reports5, 10, 26, the complex distributed on puncta in the perinuclear region that overlapped with the Golgi and were proximal to natively localized endosomes (Supplementary Fig. 9B). Notably, the PKAcatα perinuclear localization pattern did not change when endosomes were recruited to the cell periphery by CID (Supplementary Fig. 9C–E). The microscopy analysis of this overexpressed system did not detect significant nuclear translocation of mTagBFP2-PKAcatα upon stimulation of endogenous β2-ARs. Based on other studies, we speculated that this may arise from an overall modest amount of PKAcatα dissociating from the regulatory subunits and a minor fraction of that subsequently entering the nucleus, and/or could result from the larger size of the BFP fluorescent tag precluding passive diffusion through the nuclear pores26, 40, 41. To ensure that the observed PKA localization patterns reflect the native enzyme, in parallel we also examined the distribution of endogenous PKAcatα and PKARIIB under normal and CID-redistributed regimes using immunofluorescence with specific antibodies. In agreement with the results from the overexpressed mTagBFP2-PKAcatα experiments, native PKAcatα and PKARIIB subunits remained enriched in the perinuclear space regardless of endosome positioning (Supplementary Fig. 10).
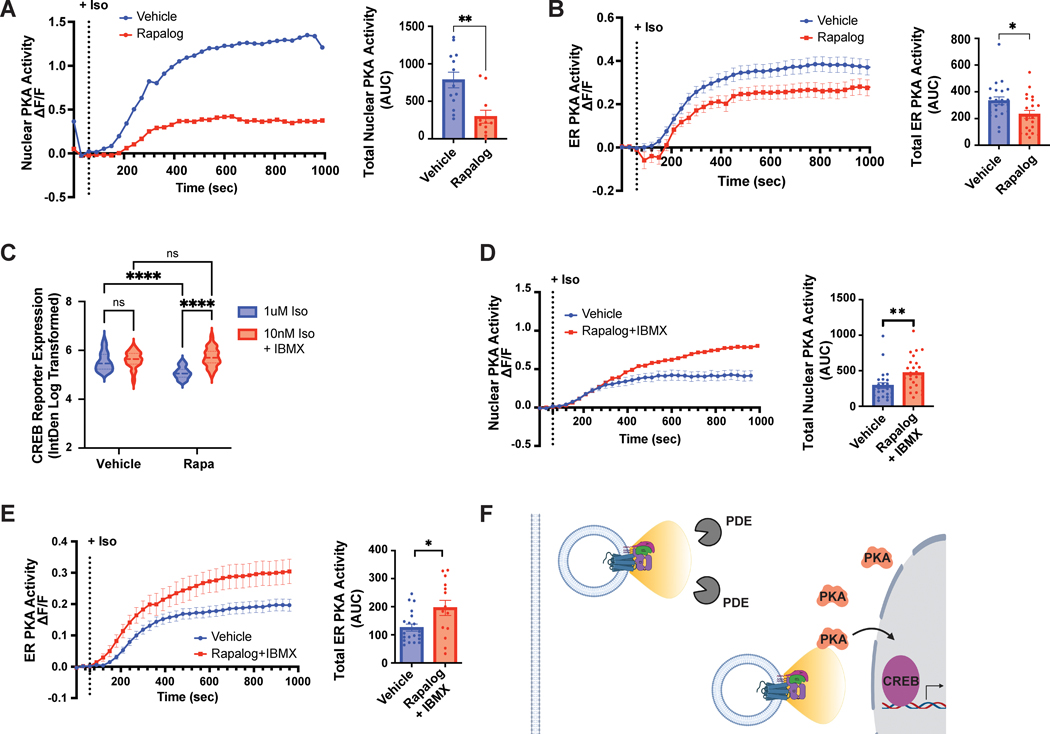
Based on this increased distance between relocalized endosomes and PKAcatα, we hypothesized that endosome positioning may impact local PKA activity. In order to probe local PKA activity, we turned to the highly sensitive single-fluorophore biosensor, ExRai-AKAR2. This sensor utilizes a circularly permuted EGFP (cpEGFP) fused to a PKA activity-dependent molecular switch, consisting of a PKA substrate sequence and the phospho amino acid-binding domain of FHA142. Phosphorylation of the biosensor and binding of the FHA1 domain to the phosphorylated PKA substrate sequence leads to a robust increase in fluorescence emission with 480/488nm excitation and a minor decrease with 400nm excitation. Given the magnitude of the phosphorylation-induced change in fluorescence emission with 480/488nm excitation, ExRai-AKAR2 can be used as an intensiometric sensor to simplify image acquisition relative to excitation ratiometric sensors. Importantly, ExRai-AKAR2 can be targeted to distinct subcellular compartments to measure PKA activity with high spatiotemporal precision42. To specifically examine nuclear PKA activity, which is required for CREB phosphorylation and cAMP-mediated transcription25, we generated ExRai-AKAR2-NLS by fusing an NLS sequence to the C-terminus of ExRai-AKAR2, and verified its localization and specificity (Supplementary Fig. 11). We further observed a dose-dependent increase in ExRai-AKAR2-NLS fluorescence excited at 488 nm following isoproterenol stimulation (Supplementary Fig. 12A). As expected based on previous reports6, 26, this activity was dependent on β2-AR internalization, as DynK44E overexpression eliminated the response (Supplementary Fig. 12B). Next, we evaluated the impact of β2-AR activation from natively localized and redistributed endosomes. Stimulation of β2-AR yielded a robust response in vehicle-treated cells (Fig. 6A). Rapalog treatment alone without co-transfection of the heterodimerization components did not impact these responses (Supplementary Fig. 12C). In contrast, ExRai-AKAR2-NLS induction was almost completely abolished in the presence of both rapalog and CID (Fig. 6A), supporting a critical role for endosome positioning in regulating nuclear PKA activity.
Figure 6. Endosomes precisely position receptors to enable signal compartmentalization and spatially-biased GPCR signaling.
A-B. Cells expressing a Nuclear- (A) or ER- (B) localized ExRai-AKAR2 sensor and the CID system were pre-treated with ethanol (“Vehicle”) or 1μM AP21967 (“Rapalog”) for 30 min before 10nM isoproterenol stimulation. The delta F/F (ΔF/F) ratio was quantified every 30 sec for 1000 sec (left) and the total nuclear (A) or ER (B) PKA activity was quantified by calculating the area under the curve (AUC, right). A. Data are mean from n = 2 biologically independent replicates; 11–13 cells total/condition. ** = p = 0.0019 by unpaired two-tailed Student’s t-test. B. Data are mean from n = 4 (“Vehicle”) and n = 5 (“Rapalog”) biologically independent replicates ± s.e.m.; 21–24 cells total/condition. * = p = 0.0193 by unpaired two-tailed Student’s t-test. C. CREB reporter expression in pCRE-NLS-DD-zsGreen1 cells stimulated with 1μM isoproterenol or 10nM isoproterenol/100μM IBMX in the presence of 1μM Shield for 4 h. Data are mean from n = 3 biologically independent replicates ± s.e.m.; 45 cells total/condition. **** = p < 0.0001, ** = p < 0.01 by two-way ANOVA test with Tukey. D-E. Cells expressing a Nuclear- (D) or ER- (E) localized ExRai-AKAR2 sensor and the CID system were pre-treated with ethanol (“Vehicle”) or 1μM AP21967 (“Rapalog”) for 30 min before isoproterenol stimulation. Vehicle- and Rapalog-treated cells were stimulated with 10nM isoproterenol and 5nM isoproterenol/100μM IBMX, respectively. The delta F/F (ΔF/F) ratio was quantified every 30 sec for 1000 sec (left) and the total nuclear (D) or ER (E) PKA activity was quantified by calculating the area under the curve (AUC, right). D. Data are mean from n = 3 (“Vehicle”) and n = 2 (“Rapalog+IBMX”) biologically independent replicates ± s.e.m.; 25 cells total/condition. E. Data are mean from n = 3 biologically independent replicates ± s.e.m.; 14–20 cells total/condition. ** = p = 0.0059 and * = p = 0.0141 by unpaired two-tailed Student’s t-test. F. Model: Endosomes enable site-selective outputs by functioning as vehicles to deliver the receptor in proximity to PKA and the nucleus and away from phosphodiesterases.
Given the dramatic impact on nuclear PKA signaling, we next sought to examine whether endosome redistribution may similarly shape local kinase activity outside of the nucleus. To this end, we used erExRai-AKAR2, a previously published ExRai-AKAR2 biosensor localized at the outer surface of endoplasmic reticulum (ER) membrane42. We verified that ER-proximal PKA activity increased in a dose-dependent manner following isoproterenol stimulation of endogenous β2-ARs (Supplementary Fig. 12D), and that rapalog alone did not impact the isoproterenol-induced signal (Supplementary Fig. 12E). In the presence of CID, we observed robust ER-proximal PKA activity in vehicle-treated cells. This activity was significantly reduced following endosome redistribution with rapalog (Fig. 6B), albeit to a lesser extent compared to the CID impact on nuclear PKA signaling (Fig. 6A). Markedly, the CID/rapalog-dependent effect fully recapitulated the inhibition of β2-AR-dependent ER-proximal PKA activity following over-expression of DynK44E (Supplementary Fig. 12F). Therefore, the native localization of endosomes mediates local PKA responses both inside and outside of the nucleus.
PDE activity buffers signaling from repositioned endosomes
Activation of β2-AR led to comparable levels of cAMP production at endosomes in vehicle- and rapalog-treated cells (Fig. 3D), yet the downstream PKA and transcriptional responses were dependent on native endosome positioning (Fig. 5 and 6A–B). We thus hypothesized that the propagation of cAMP from endosomes may be impacted by organelle redistribution. To test this, we measured cAMP accumulation in the nucleus using the targeted cAMP biosensor nlsFlamindo233. While rapalog treatment alone did not impact isoproterenol-dependent nuclear cAMP accumulation, we observed a significant CID/rapalog-dependent inhibition of that process (Supplementary Fig. 13). Phosphodiesterases (PDEs) are a well-established class of GPCR/cAMP signaling effectors that hydrolyze cyclic nucleotides and play a critical role in limiting cAMP propagation43. Therefore, we next tested whether PDE hydrolysis may contribute to the selective ability of natively localized endosomes to give rise to spatially-biased GPCR responses.
We began by testing the impact of PDE inhibition on CREB reporter expression in cells with natively localized versus repositioned endosomes. We carefully titrated the stimulation doses to achieve comparable cAMP levels in the presence and absence of PDE inhibition (Supplementary Fig. 14A–B, blue and red curves). Under these matched cAMP induction conditions, blockade of PDE hydrolysis fully reversed the impact of endosome redistribution on CREB reporter levels (Fig. 6C). Further, β2-AR induction in the presence of IBMX also stimulated robust local PKA activity inside and outside of the nucleus, irrespective of endosome positioning (Fig. 6D–E and Supplementary Fig. 14C–D). These results suggest that PDE-dependent cAMP hydrolysis prevents the propagation of receptor signals originating beyond the sites of natively localized endosomes.
Discussion
Recent studies have demonstrated a prominent role of endomembrane GPCRs in nuclear PKA translocation26 and in transcriptional signaling5, 6, 21. Blockade of clathrin or dynamin-dependent endocytosis or collapse of the relevant endomembrane compartment precludes these responses5, 6, 21. Despite this emergent recognition of the functional role of endosomal GPCRs, how intracellular receptors transduce distinct responses has remained an open question. In principle, endosomes could selectively enable transcription by presenting a unique biochemical environment or due to their three-dimensional positioning, which provides optimal localization with respect to relevant signaling effectors. In the present study, we devised an approach to unambiguously distinguish between these models. A rapamycin-dependent dimerization strategy dramatically redistributed endosomes toward the cell periphery without perturbing the complexes required for proper receptor trafficking and signaling, including the localization of an early endosome-resident sorting protein and a recycling Rab GTPase (Fig. 1–3 and Supplementary Fig. 1–5). Coupled with complementary approaches to measure cAMP, PKA and transcriptional signaling with single-cell resolution and high reproducibility, this system allowed us to rigorously demonstrate endosome positioning as the salient mechanism orchestrating location-selective GPCR responses.
We report that rapid endosome redistribution drastically blunted transcriptional signaling downstream of the β2-AR (Fig. 5). We propose that the positioning of endosomal receptors plays an essential regulatory role because of the highly compartmentalized nature of the cAMP cascade, which limits the initiation of downstream responses. PKA signaling is reported to be restricted to nano-sized ‘hotspots’44. This compartmentalization happens through a number of mechanisms. First, the PKA tetramer is ‘trapped’ on organelle membranes via the interactions of the regulatory subunits, PKARI and PKARII, with distinct A-kinase anchoring proteins45. PKA enzymes specifically containing PKARIɑ were shown to concentrate in phase-separated biomolecular condensates39, and PKARIIβ localized the complex to the perinuclear region26, 46. Further, signaling is buffered by PDEs, which efficiently hydrolyze cAMP within a narrow radius43, 47. In addition, PKA requires a high in-cell cAMP activation threshold, recently reported to be in the micromolar range48, which renders local PDE actions and PKA localization especially relevant. Collectively, these properties of the cascade dictate precise, spatially-controlled cAMP- and PKA-dependent responses. Based on this and our data, we propose a model according to which endosomes regulate the propagation of the GPCR signal by providing the optimal biophysical location for cAMP signaling events in two ways. First, they allow the receptor/signaling complex to evade PDE hydrolysis, which we recently showed to be higher at the cell periphery7. Indeed, we detect decreased cAMP accumulation in the nucleus in cells with redistributed endosomes (Supplementary Fig. 13). Consistent with this, inhibition of phosphodiesterase activity rescued the inability of receptor signals originating from relocalized endosomes to activate PKA and gene transcription, even under stimulation conditions that were matched based on total cAMP production (Fig. 6C–E and Supplementary Fig. 14). Second, native endosomes position the receptor in direct proximity to the nucleus and to the relevant downstream effector PKA. In agreement with previous reports,10, 26, 39 we observe the catalytic alpha subunit of PKA, PKAcatα, on intracellular puncta proximal to natively localized endosomes when co-expressed with PKARIIβ (Supplementary Fig. 9–10). Notably, the localization of PKAcatα-containing puncta does not change upon endosome redistribution (Supplementary Fig. 9–10). Hence, cAMP produced via the receptor/Gas on repositioned endosomes is simultaneously buffered by active PDE hydrolysis, while also having to propagate longer distances to reach its spatially-confined downstream effector. We propose that these events ultimately synergize to preclude the initiation of transcriptional signaling (Fig. 6F). In support of this, we reveal that local PKA signaling is strictly dependent on endosome positioning. Downstream of active β2-ARs, redistribution of endosomes blunts PKA activity even at the ER, a continuous organelle that spreads from the perinuclear region throughout the cytosol, making contact sites with all other organelles and the plasma membrane49 (Fig. 6B). The effect is yet more pronounced at greater distances from the cell periphery, with endosome repositioning completely abolishing nuclear PKA signaling (Fig. 6A), which is required for CREB-dependent transcription.
Strikingly, the magnitude of inhibition of both PKA and transcriptional signaling following endosome relocalization closely recapitulated the phenotypes observed upon complete blockade of receptor internalization (Fig. 5C, Fig. 6A–B, Supplementary Fig. 12B,F). Therefore, these data argue that the biophysical localization of endosomes per se is the primary mechanistic determinant underlying the ability of these organelles to propagate spatially-biased GPCR responses. We further note that Dyngo-4a and DynK44E manipulations preclude not only β2-AR endocytosis but also subsequent receptor resensitization through recycling. In contrast, β2-ARs recycle efficiently out of repositioned endosomes (Fig. 2, Supplementary Fig. 3A–B), enabling subsequent rounds of cascade activation. Hence, our analysis may in fact underestimate the contribution of endosome positioning to PKA and transcriptional signaling. Our findings, in turn, fit a broader appreciation for the significance of organelle positioning in cellular and human physiology. For example, lysosome positioning was found to regulate mTORC1 and mTORC2 signaling and impact autophagosome formation 29, 50. Similarly, retrograde endosome movement was required for neurotrophic factor signal transmission from distal axons to the nucleus to stimulate various responses, including CREB-dependent transcription51. Conversely, dysregulation of endolysosomal positioning has been linked to a number of human pathologies, including lysosome storage disorders (LSD) and cancer52. Therefore, while elucidation of the biochemical mediators of intracellular receptor activity and their dysregulation in diseases is central in the concerted effort to assess the physiological and pathophysiological implications of intracellular GPCR signaling, the cellular mechanisms that ensure the proper positioning of the intracellular signaling organelle itself may be equally important for disease etiology and should be careful interrogated in parallel.
Lastly, the experimental platform described here could be applied in future analyses to determine how endosome positioning impacts other important outputs of the GPCR cascade. Both G protein-dependent and G protein-independent responses can be initiated from endosomal compartments2, 53. Based on the model proposed above we speculate that, in the case of Gas-coupled receptors which signal from endosomal compartment, any G protein/cAMP-stimulated responses would be preferentially driven by endosomal GPCR activation and coordinated by endosome positioning. Indeed, it was recently shown that phosphosignaling is driven primarily by endosomal cAMP and requires receptor internalization7. However, it was not tested whether these processes are dependent on the precise localization of receptor-containing endosomes. Further, it would be particularly interesting to examine whether endosome positioning plays a role in the recently discovered GPCR-mediated ERK signaling at the endosome54, which is not orchestrated through the actions of PKA, cAMP and PDEs. The novel cAMP and PKA biosensors and the complementary approaches to measure GPCR-dependent transcription by microscopy would also be valuable to further dissect the regulatory logic of the receptor cascade. These functional readouts can be applied to interrogate factors important for endosome positioning or receptor trafficking in and out of endosomes. Because of the single-cell nature of the readouts, we anticipate that these would also offer the unprecedented ability to examine pathway regulation in parallel across different cell types in a heterogeneous setting.
In summary, our study defines a novel mechanism underlying spatially-biased GPCR signaling by demonstrating a central role for endosome positioning in the propagation of site-selective transcriptional signaling. Our findings were corroborated by multiple complementary approaches, including specific readouts for β2-AR, cAMP, PKA and CREB activity, and also by a receptor-independent optogenetic method for cAMP generation. Therefore, these results likely represent a principle of broader significance that would apply to an ever-growing list of Gas/cAMP-coupled GPCRs found to signal from endosomes8, 9, 16, 54–56.
Methods
Chemicals
(−)-Isoproterenol hydrochloride was purchased from Sigma-Aldrich (Cat #I6504), dissolved in water/100mM ascorbic acid to 10mM stock, and used at 1μM final concentration. Alprenolol hydrochloride was purchased from Sigma-Aldrich (Cat # A0360000), dissolved in DMSO to 10mM stock, and used at 10μM final concentration. DD stabilizing compound, Shield-1, purchased from Takara Bio (Cat #632189) and used at 1μM final concentration. 8-(4-Chlorophenylthio)adenosine 3’,5’-cyclic monophosphate (8-CPT-cAMP) was purchased from Abcam (Cat #ab120424), dissolved in water to 150mM stock, and used at 150μM final concentration. 3-isobutyl-1-methylxanthine (IBMX) was purchased from Sigma (Cat #I5879–1G and Cat #I7018), dissolved in DMSO to 100mM stock, and used at 100μM final concentration. Rapamycin was purchased from Sigma-Aldrich (Cat #553210–1MG), dissolved in ethanol to 10mM stock, and used at 1μM final concentration. Rapalog (A/C Heterodimerizer) was purchased from Takara (Cat #635056) and used at 1μM final concentration. Dyngo-4a was purchased from Abcam (Cat #ab120689), dissolved in DMSO to 30mM stock, and used at 30μM final concentration in serum-free medium. Luciferin-D was purchased from Goldbio (Cat # LUCNA1G), dissolved in water + 10mM HEPES to 100mM, and used at 1.6μM final concentration. Coelenterazine H was purchased from Cayman Chemical Company (Cat #16894), dissolved in ethanol to 10mM, and used at 2μM final concentration. Forskolin (Fsk) was purchased from Calbiochem (Cat # 344281) and used at 50μM final concentration. H89 was purchased from Cayman Chemical (Cat #10010556) and used at 20μM final concentration.
Construct cloning
A previously described lentiviral plasmid encoding a transcriptional reporter for CREB activity37 was used as the backbone for the CREB transcriptional reporter in this study. To express this construct in the nucleus, a nuclear localization sequence (NLS) was amplified from dCas9-BFP-KRAB (a gift from Martin Kampmann, UCSF) and inserted into the BamHI-digested reporter backbone using in-fusion cloning (Takara Bio, Cat #638911). pBa-Kif1a(1–396)-tdTomato-FKBP was generated by Asc1/Hpa1 restriction digest of pBa.Kif1a 1–396.GFP (backbone; Addgene, Cat #45058) and of pBa-KIF5C 559-tdTomato-FKBP (insert; Addgene, Cat #64211) followed by ligation. CMV-EGFP-FRB-EEA1 was generated by site-directed mutagenesis to remove the stop codon upstream of FRB from pEGFP-FRB (Addgene, Cat #25919). Next, EEA1 was amplified from GFP-EEA1 (Addgene, Cat #42307) and in-fusion cloning was used to insert the product into the BsiWI-digested pEGFP-FRB backbone. FRB-EEA1 was generated by site-directed mutagenesis to insert an AgeI cut site upstream of EGFP in CMV-EGFP-FRB-EEA1. AgeI digestion was used to remove EGFP, followed by ligation into the CMV-FRB-EEA1 backbone. To clone mCherry-FRB-EEA1, mCherry was amplified from pmCherry-N1-GalT (Addgene, Cat #87327) and inserted into the AgeI/XhoI-digested EGFP-FRB-EEA1 backbone by in-fusion cloning. CMV-Kif1a-bPAC was generated by PCR amplification of Kif1a(1–396) and in-fusion cloning into the Xbal/EcoRV-digested CMV-bPAC backbone described previously6. To clone Nb37 into a lentivirus backbone, Nb37-GFP24 was amplified and inserted into the NheI/XbaI-digested pMK1200 backbone (a gift from Martin Kampmann, UCSF) by in-fusion cloning. Similarly, Nb80-GFP24 was amplified and inserted into the NheI/XbaI-digested pMK1200 backbone by in-fusion cloning to generate Nb80 for lentivirus production. The endosomal cAMP fluorescent sensor was cloned by digestion of the EGFP-FRB-EEA1 (AgeI-HF/XhoI), PCR amplification of Flamindo2 (Addgene, Cat #73938), and in-fusion cloning to generate Flamindo2-FRB-EEA1. ExRai-AKAR2-NLS was generated by subcloning ExRai AKAR2 (Addgene, Cat #161753) into a pcDNA3.1(+) backbone containing a nuclear localization signal (PKKKRKVEDA). To clone GalT-eGFP, GalT was amplified from mCherry-N1-GalT (Addgene, Cat #87327) and inserted into the NheI-digested EGFP-C1 backbone by in-fusion cloning.
Cell culture and transcriptional reporter cell line generation
HEK293, HEK293T, and HeLa cells were obtained from ATCC and grown at 37°C/5% CO2 in Dulbecco’s Modified Eagle Medium (4.5 g/L glucose and L-glutamine, no sodium pyruvate; Thermo Fisher Scientific Cat # 11965118) and supplemented with 10% fetal bovine serum. For live-cell experiments, cells were incubated in imaging media (DMEM - 4.5 g/L glucose and L-glutamine, no sodium pyruvate, no phenol red; Thermo Fisher Scientific Cat #31053–028) + 30mM HEPES (GenClone Cat#25–534). HEK293 cells stably expressing pCRE-NLS-DD-zsGreen1 were generated by lentiviral transduction and fluorescence activated cell sorting for zsGreen1+ in the 488 channel on a BD DIVA (BD Biosciences). Ten GFP-positive cells were sorted per well in a 96-well plate and expanded. Populations were then tested by flow cytometry (BD FACSCanto II) and the flow cytometry data were analyzed using FlowJo software for cAMP-induced GFP expression following stimulation with 1μM isoproterenol in media containing 1μM Shield-1 for 4 h. We selected the population with the highest signal dynamic range for our follow-up experiments.
Lentivirus production
Lentivirus was used to generate the pCRE-NLS-DD-zsGreen1 cell line and to express Nb37, Nb80, and Flag-β2-AR. To make lentivirus, HEK293T cells were transfected with pCRE-NLS-DD-zsGreen1, EF1alpha-Nb37-GFP, EF1alpha-Nb80-GFP, or pFUGW-Flag-β2-AR (a gift from Mark von Zastrow, UCSF) lentivirus vectors and standard packaging vectors (VSVG and psPAX2) using Lipofectamine 2000 (ThermoFisher, Cat #1668027) following recommended protocols. Virus-containing supernatants were collected 72 h after transfection, filtered through a 0.45μ SFCA filter, and either used on the same day or concentrated using Lenti-X concentrator solution (Takara Bio, Cat #631231) and snap frozen at −80C.
Spinning disk confocal imaging
Live- and fixed-cell imaging were performed on Andor Dragonfly Spinning Disk Confocal microscope with 405 nm, 488 nm, 561 nm, 637 nm diode lasers (Andor). The microscope was fitted with 40x/1.3 HC PL APO CS2 (Leica, Cat #11506358), Oil, WD: 0.24mm; 63x/1.47 TIRF HC PL APO CORR (Leica, Cat # 11506319), Oil, WD: 0.10mm; and 100x/1.40–0.70 HCX PL APO (Leica, Cat # 11506210), Oil, WD: 0.09mm objectives. The Andor iXon Life 888 1024×1024 EMCCD, pixel size: 13μm camera was used with and without 2X zoom optics. Live-cell imaging was carried out in a heated (37°C) chamber (Okolab). Data were collected using Fusion software.
Imaging in HeLa cells was performed on a Zeiss AxioObserver Z1 microscope (Carl Zeiss) equipped with a Definite Focus system (Carl Zeiss), a 40×/1.3 NA oil objective and a Photometrics Evolve 512 EMCCD (Photometrics) and controlled by METAFLUOR 7.7 software (Molecular Devices). Dual GFP excitation-ratio imaging was performed using 480DF30 and 405DF40 excitation filters, a 505DRLP dichroic mirror and a 535DF45 emission filter. All filter sets were alternated using a Lambda 10–2 filter changer (Sutter Instruments).
Early endosome redistribution assay and quantification
Cells plated on Poly-L-lysine (Sigma, Cat #P8920–100ML) coated coverslips (Thomas Scientific, Cat #217N81) were transfected using Lipofectamine 2000 (ThermoFisher, Cat #11668027) with Kif1a-tdTomato-FKBP + EGFP-FRB-EEA1 or mCherry-FRB-EEA1 and EGFP-SNX27 or Rab11a-GFP, where applicable. The following day, the cells were treated with rapamycin or ethanol vehicle for 20 min. Following treatment, cells were fixed using 3.7% formaldehyde (ThermoFisher, Cat #28908) in modified BRB80 buffer (80mM PIPES pH 6.8, 1mM MgCl2, 1mM CaCl2) for 20 min. Fixation was quenched and cells were permeabilized using 2.5%milk/0.1% Triton X-100 (Sigma, Cat #X100–100ML) in TBS for 20 min. Primary antibodies [Anti-EEA1 (Cell Signaling, Cat #3288S), Anti-SNX27 (Abcam, Cat #ab77799), Anti-TGN46 (Abcam, Cat #ab50595)] were diluted 1:500 in permeabilization solution and incubated for 1 h at room temperature. Secondary antibodies [Donkey anti-Mouse IgG (H+L) Secondary Antibody, Alexa Fluor 647 (Invitrogen, Cat #A-315710); Donkey anti-Rabbit IgG (H+L) Secondary Antibody, Alexa Fluor 647 (Invitrogen, Cat #A-31573)] were diluted 1:1000 in permeabilization solution and incubated for 30 min at room temperature, protected from light. Following two washes in permeabilization solution and one wash in PBS, coverslips were mounted using ProlongGold with DAPI (Thermo Scientific, Cat #P36931). Slides were imaged on an Andor Dragonfly Spinning Disk Confocal microscope. Images were imported into FIJI. Single cells were isolated by drawing ROIs around the cell periphery seen in the Kif1A-containing channel and nuclei were identified by drawing ROIs of DAPI-stained nuclei. Clusters of EEA1-containing endosomes were segmented using the Morphological Segmentation plugin (MorphoLibJ)57. Using the distance formula, the distance from segmented EEA1 particles to the nucleus center of mass was calculated based on particle and nuclear x,y coordinates. Distances of all endosomes to the nucleus were averaged per cell. To quantify how endosome redistribution impacted colocalization of early endosomes with SNX27, the Squassh plugin (MosaicSuite)58 was used to remove background, apply a joint deconvolution-segmentation procedure, and calculate object-based colocalization using information about the shapes and intensities of all objects in both channels. From the supplied R script, the C(number) was used to quantify SNX27 colocalization as the ratio of (number of SNX27 puncta colocalized with EEA1)/(total SNX27 puncta) in RStudio.
β2-AR trafficking and quantification
Trafficking was quantified by two methods. For microscopy-based analysis, HEK293 cells plated on Poly-L-lysine-coated coverslips were transfected with Kif1a-tdTomato-FKBP, EGFP-FRB-EEA1, and Flag-β2-AR using Lipofectamine 2000. Cells were first treated with either rapamycin or vehicle (ethanol) for 20 min to induce redistribution, and then supplemented with Alexa 647-conjugated M1 antibody (1:1000) for 10 min to label cell-surface Flag-β2-ARs. To reduce non-specific antibody binding, the media was changed with fresh pre-equilibrated medium containing rapamycin or ethanol and either isoproterenol or vehicle for 20 min to induce β2-AR internalization. Following receptor internalization, media was changed with rapamycin or ethanol control with or without alprenolol for 1 h to induce β2-AR recycling. Cells were fixed as described above and slides were imaged on Andor Dragonfly Spinning Disk Confocal microscope. Images were imported into FIJI and individual channels were saved as TIFF files. The Squassh plugin (MosaicSuite)58 was used to remove background, apply a joint deconvolution-segmentation procedure, and calculate object-based colocalization using information about the shapes and intensities of all objects in both channels. From the supplied R script, the C(number) was used to estimate internalized or recycled β2-ARs as the ratio of (number of β2-AR puncta colocalized with EEA1)/(total β2-AR puncta) in RStudio. To visualize the effect of the dominant-negative dynamin mutant (K44E), HEK293 cells plated on Poly-L-lysine-coated 35mm imaging dishes and transfected with Flag-β2-AR and either DynK44E-mCherry or empty pcDNA3.0 vector using Lipofectamine 2000. The following day, media was changed to pre-warmed imaging media and Alexa 647-conjugated M1 antibody (1:1000) was added for 10 min to label cell-surface Flag-β2-ARs. Images were taken in the absence of β2-AR stimulation or following 20 min of isoproterenol treatment. Cells were imaged live on the Andor Dragonfly Spinning Disk Confocal microscope. Images were imported into FIJI and individual channels were saved as TIFF files.
For flow cytometry-based analysis, we generated HEK293 cells stably expressing Flag-β2-AR and treated with 30μM Dyngo-4a or DMSO in serum-free DMEM for 20 min. Next, cells were treated either isoproterenol or vehicle for 30 min to induce β2-AR internalization. Cells were washed with PBS and then incubated with Alexa 647-conjugated M1 antibody (1:1000) for 1 h at 4°C, shaking at 200 rpm. Then, cells were lifted, placed in tubes for flow cytometry (BD FACS Canto2), and 10,000 cells were analyzed per sample. The gated Alexa 647 mean of the singlet population was used as the amount of receptor surface expression. The % internalized receptors was calculated by 100%-(# surface receptors after 30 min isoproterenol)/(# surface receptors after vehicle) × 100%.
Transferrin receptor trafficking and quantification
HEK293 cells were plated in 12-well plates. The following day, cells were washed with serum-free DMEM, and treated with either ethanol (vehicle), 1μM rapamycin, or 1μM rapalog for 30 min. Then, samples were incubated with 5μg/ml Alexa 488-conjugated diferric transferrin (Tfn-488, Invitrogen, Cat #T133342) for 20 min. To measure recycled TFR, cells were switched into DMEM + 10% FBS for 30 min, then lifted with cold PBS-EDTA, spun down, and resuspended in PBS. To measure internalized TFR, cells were lifted with cold PBS-EDTA, spun down, resuspended in PBS. To measure total TFR, cells were lifted with PBS. Samples were analyzed by flow cytometry (BD FACS Canto2), where 10,000 cells were analyzed per sample. Data analysis was conducted in FlowJo. The gated FITC mean of the singlet population was used, and % Internalized receptors was calculated for each drug treatment as (# of internalized TFR)/(# of total TFR)*100%, while % Recycled receptors was calculated as 100-(# of recycled TFR)/(# internalized TFR)*100%.
Nanobody colocalization in repositioned endosomes
For Nb80 colocalization experiments, HEK293 cells were infected with EF1alpha-Nb80-GFP. Three days later, cells were plated on Poly-L-lysine-coated coverslips. The following day cells were transfected with Kif1A-tdTomato-FKBP, FRB-EEA1, and Flag-β2-AR using Lipofectamine 2000. Cells were first treated with either rapamycin or vehicle (ethanol) for 30 min to induce redistribution, and then supplemented with Alexa 647-conjugated M1 antibody (1:1000) for 10 min to label cell-surface Flag-β2-ARs. To reduce non-specific antibody binding, the medium was replaced with fresh pre-equilibrated medium containing rapamycin or ethanol and either isoproterenol or vehicle for 30 min to induce β2-AR internalization. Cells were fixed as described above and imaged on Andor Dragonfly Spinning Disk Confocal microscope. For Nb37 colocalization experiments, HEK293 cells were infected with EF1alpha-Nb37-GFP. Three days later, cells were plated on Poly-L-lysine-coated 35mm imaging dishes. The following day cells were transfected with Kif1a-tdTomato-FKBP, mCherry-FRB-EEA1, and Flag-β2-AR using Lipofectamine 2000. Cells were first treated with either rapalog or vehicle (ethanol) for 30 min to induce redistribution, and then supplemented with Alexa 647-conjugated M1 antibody (1:1000) for 10 min to label cell-surface Flag-β2-ARs. Cells were imaged live on Andor Dragonfly Spinning Disk Confocal microscope prior to and following isoproterenol stimulation.
Images were imported into FIJI and individual channels were saved as TIFF files. Since these experiments necessitated the transfection of multiple constructs, we observed reduced expression of the dimerization components and, consequently, decreased efficiency of endosome redistribution in cells pre-treated with rapamycin/rapalog. Nevertheless, a sufficient fraction of endosomes relocated to the cell edge in rapamycin-dependent manner to allow evaluation of Nb80 and Nb37 recruitment and demonstration of functional β2-AR signaling. To quantify the Nb80 images, the Squassh plugin (MosaicSuite)58 was used to remove background, apply a joint deconvolution-segmentation procedure, and calculate object-based colocalization using information about the shapes and intensities of all objects in both channels. From the supplied R script, the C(number) was used to quantify Nb80 colocalization as the ratio of (number of Nb80 puncta colocalized with β2-AR)/(total Nb80 puncta). Nb37 recognizes a very transient Gas active state, and therefore Nb37/β2-AR co-localization experiments have intrinsically low dynamic range24. For this reason, we had to utilize line scan analysis to evaluate the colocalization of Nb37, β2-AR, and the CID system. Line scan analysis was performed across 10μm. The fluorescence was measured every 0.2μm and normalized to the maximum fluorescence for each channel. This was done across multiple replicates to show the reproducibility of the colocalization following endosome redistribution (Supplementary Fig. 4).
NR4A1 expression PLISH analysis
HEK293 cells were plated on Poly-L-lysine-coated coverslips and transfected with Kif1A-tdTomato-FKBP and EGFP-FRB-EEA1 using Lipofectamine 2000. When inhibiting receptor internalization, cells were first treated with 30μM Dyngo-4a or DMSO in serum-free DMEM for 20 min. When relocalizing endosomes, cells were first treated with either rapalog or ethanol control in serum-free DMEM media for 20 min. Next, cells were treated with isoproterenol or vehicle control for 2 h. Cells were fixed using 3.7% formaldehyde in PBS/0.1%DEPC (Sigma-Aldrich, Cat #D5758–5ML) for 20 min. Coverslips were washed three times with PBS, dehydrated by an ethanol dilution series, air dried for 10 min, and enclosed by application of a seal chamber (Grace Bio Labs, Cat #621505) as previously described36.
We designed four sets of detection H probes spanning the length of the transcript and tested them individually and in combination. We achieved the best signal dynamic range when we pooled all four H probe sets (isoproterenol versus untreated mean puncta per cell ratio of ~2.9, Fig. 4A–B). The following H-probe oligonucleotides were used for NR4A1:
HL4X-NR4A1–1879 (TTAGTAGGCGAACTTACGTCGTTATGTTGTCAATGATGGGTGGAGG), HR4X-NR4A1–1899 (GCAGCGTGTCCATGAAGATCTTATACGTCGAGTTGAACATAAGTGCG), HL4X-NR4A1–1009 (TTAGTAGGCGAACTTACGTCGTTATGTTGGCGTTTTTCTGCACTGT), HR4X-NR4A1–1029, (CTTGTTAGCCAGGCAGATGTACTTATACGTCGAGTTGAACATAAGTGCG), HL4X-NR4A1–1486 (TTAGTAGGCGAACTTACGTCGTTATGTCGCCTGGCTTAGACCTGTA), HR4X-NR4A1–1506 (AGCAGAAGATGAGCTTGCCCTTATACGTCGAGTTGAACATAAGTGCG), HL4X-NR4A1–421 (TTAGTAGGCGAACTTACGTCGTTATGTCGAACTTGAAGGAGGCAGA), HR4X-NR4A1–441 (AGCCGTACACCTGGAAGTCCTTATACGTCGAGTTGAACATAAGTGCG)
The following H-probe oligonucleotides were used for GAPDH:
HR2X-GAPDH-1107 (GATGATCTTGAGGCTGTTGTCATTATACGTCGAGTTGAACATAAGTGCG), HL2X-GAPDH-1085 (TTAGTAGGCGAACTTACGTCGTTATGTACTTCTCATGGTTCACACCCA).
The 25 nmol H-probe oligonucleotides were ordered from IDT with standard desalting. The 100 nmol B and C oligonucleotides were ordered from IDT with HPLC purification. B and C oligonucleotides were phosphorylated with T4 polynucleotide kinase (NEB, Cat #M0201L) according to the manufacturer recommendations. Imager oligonucleotide was ordered from IDT with HPLC purification and Cy5 conjugation.
PLISH barcoding buffers and H cocktail for NR4A1 and GAPDH were used as previously described36. BC cocktail was prepared by mixing phosphorylated B and C oligonucleotides in labeling buffer (2X SSC, 20% formamide) at a final concentration of 6μM each. PLISH barcoding was performed in sealed chambers. The workflow was as follows: (1) samples were incubated in the H cocktail at 37°C for 1.5 h, then washed 3× 5 min with H-probe buffer at RT, and then incubated in a high salt buffer (250mM NaCl, 50mM Tris pH 7.4, 2mM EDTA) at 37°C for 10 min; (2) samples were incubated in the BC cocktail + 0.2 mg/mL Heparin at 37°C for 1 h, followed by 2 × 2 min washes with labeling buffer; (3) samples were incubated in ligation buffer at 37°C for 2 h, followed by 2 × 2 min washes with labeling buffer; (4) sample was washed with 1X Nxgen phi29 polymerase buffer at RT for 2 min, then incubated in RCA buffer at 37°C for 2 h, followed by 2 × 2 min washes with labeling buffer; (5) sample was incubated in labeling buffer + 0.2 mg/mL heparin + 100nM Cy5-imaging probe at 37°C for 30 min, followed by 2 × 3 min washes with PBST, and mounted in ProlongGold with DAPI (Thermo Scientific, P36931). The following day, samples were imaged on the Andor Dragonfly Spinning Disk Confocal microscope. Images were taken with 63x objective (Rapalog experiments) or 40x objective (Dyngo-4a experiments). Z stacks were taken with 1.5μm step size and max/min of the stack was determined per image by identifying the height of cells. Images were imported into FIJI and the maximum intensity projections of the channels were saved and exported as TIFF files. DAPI and Cy5 TIFF files were imported into CellProfiler59 and a custom pipeline was created to measure RNA signal intensities at the single-cell level. Briefly, nuclear boundaries were assigned by a shape algorithm, and then expanded by ~10–12μm to define sampling areas. The following data were then recorded: (i) number of nuclei and corresponding sampling areas and (ii) the coordinates of the sampling areas. For the RNA species, puncta were identified using Otsu thresholding and distinguished using an intensity algorithm. The RNA puncta were counted within determined sampling areas to calculate the number of RNA puncta per cell. For calculating % inhibition of Isoproterenol-induced response, the fold changes in samples pre-treated with Dyngo-4a versus DMSO vehicle (Fig. 4B) were compared to samples pre-treated with rapalog versus ethanol vehicle.
pCRE-NLS-DD-zsGreen1 transcriptional reporter quantification
Because the pCRE-NLS-DD-zsGreen1 reporter contains a degron, expression of the construct was stabilized by addition of the ligand Shield-1 at the time of agonist stimulation as described below. Transcriptional reporter cells were plated on Poly-L-lysine-coated coverslips and transfected with Kif1a-tdTomato-FKBP and EGFP-FRB-EEA1 using Lipofectamine 2000. When inhibiting receptor internalization, cells were first treated with 30μM Dyngo-4a or DMSO in serum-free DMEM for 20 min. When relocalizing endosomes, cells were first treated with either rapalog or ethanol for 20 min. Next, cells were then treated with Shield-1 and vehicle, isoproterenol, or isoproterenol + IBMX for 4 h. Cells were fixed with as described above and imaged on Andor Dragonfly Spinning Disk Confocal microscope. Images were imported into FIJI and individual channels were saved as TIFF files. Nuclei were identified by drawing ROIs of DAPI-stained nuclei. Cells were confirmed to have EEA1 redistribution by visual examination of the GFP channel, Kif1a-containing channel, and anti-EEA1-containing channel. In cells with efficient EEA1 redistribution, the nucleus ROI was measured for the integrated density in the GFP (reporter) channel. For calculating % inhibition of Isoproterenol-induced response, the fold changes in samples pre-treated with Dyngo-4a versus DMSO vehicle (Fig. 4D) were compared to samples pre-treated with rapalog versus ethanol vehicle.
cAMP production
cAMP production was measured either with fluorescent Flamindo2-based cAMP sensors and live cell microscopy, or with a luminescent cAMP reporter on a plate reader. To measure endosomal or nuclear cAMP production, cells were plated on Poly-L-lysine-coated 35mm live cell imaging dishes. The following day, cells were transfected with the sensor (Flamindo2-FRB-EEA1 or nlsFlamindo2) and CID components as indicated using Lipofectamine 2000. For DynK44E experiments, cells were transfected with Flamindo2-FRB-EEA1 and either DynK44E-mCherry or empty vector using Lipofectamine 2000. For experiments involving the endosome redistribution system, cells were pretreated with ethanol (vehicle) or 1μM rapalog for 30 min, and then stimulated with indicated doses of isoproterenol. Live-cell images were taken on the Andor Dragonfly Spinning Disk Confocal microscope every 10 sec for the endosomal sensor and every 20 sec for the nuclear sensor, including three baseline frames prior to isoproterenol addition. Images were exported to FIJI. Images were quantified by drawing ROIs around individual cells, measuring the 488-channel integrated density in every frame, and calculating the delta F/F (488-fluorescence–baseline 488-fluorescence)/(baseline 488-fluorescence).
To measure bulk cAMP production, the luminescent cAMP reporter (pGlo-RLuc) was used as previously described37. Cells were transfected with pGlo-RLuc using Lipofectamine 2000. The following day, cells were lifted, resuspended in imaging media + 1.6μM luciferin-D, and incubated in a 96-well plate for 40 min. Plates were analyzed using a Hamamatsu μCell plate reader at 37°C, 5 baseline reads were taken, then drug was added using the automated dispenser, and measurements were taken every 10 sec. After 30 min, cells were lysed in 5mM HEPES, 2% glycerol, 1mM EDTA, 400μM DTT, and 0.2% Triton and incubated with coelenterazine to measure renilla luciferase activity. Data were collected using FDSS software. cAMP production was quantified as luciferin luminescence/renilla luminescence to normalize wells to the expression of the sensor.
Quantitative PCR assay
For bPAC experiments, cells were transfected with each construct using the Lipofectamine 2000. On experiment day, media was changed to serum-free DMEM for 4–6 h. “Light-treated” bPACs were activated using 10% power of a 405-LED array (Sumbulbs) for indicated durations. In parallel, “no light” control samples were included. Following light activation cells, were placed in the incubator for 30 min. For Dyngo-4a experiments, HEK293 cells were incubated with DMSO or Dyngo-4a in serum-free medium for 20 min, then treated with vehicle, isoproterenol or isoproterenol/IBMX for 1 h. Total RNA was extracted from the cells using Zymo Quick-RNA MiniPrep Kit (Genesee Scientific, Cat #11–328). Reverse transcription was performed using iScript RT supermix (Biorad, Cat #1708841) following recommended manufacturer protocols. Power SYBR Green PCR MasterMix (ThermoFisher Scientific, Cat #4367659) and the following primers were used for the qPCR reactions- GAPDH: F 5’-CAATGACCCCTTCATTGACC-3’ and R 5’-GACAAGCTTCCCGTTCTCAG-3’; c-fos: F 5’-GCCTCTCTTACTACCACTCACC-3’ and R 5’-AGATGGCAGTGACCGTGGGAAT-3’. Quantitative PCR was performed using CFX-384 Touch Real-Time PCR System equipped with CFXMaestro software (Biorad). All gene expression levels were normalized to the levels of the housekeeping gene, GAPDH.
Western blotting
HEK293 cells were transfected with bPAC constructs using Lipofectamine 2000. The next day, cells were lysed on ice in ice-cold RIPA buffer (Sigma, Cat #R0278–500mL) with protease inhibitors cocktail (Sigma-Aldrich, Cat #P8340–5ML). Lysates were quantified by Pierce BCA Protein Assay Kit (ThermoScientific, Cat #23225), equal amounts of total protein were boiled in Laemilli dye buffer (Biorad, Cat #1610747) at 70°C for 10 min, and loaded on Mini-PROTEAN TGX Stain-Free 4–15% gels (Biorad, Cat #4568083). Gels were transferred onto nitrocellulose membranes, blocked in Tris-buffered saline (TBS)/0.05% Tween 20/5% milk, and incubated with 1:500 anti–β-actin (Santa Cruz, Cat #sc-69879) and 1:500 anti–myc (Thermo Scientific, Cat #PA1–981) in TBS/0.05% Tween 20/5% milk overnight at 4°C on a shaker. Membranes were washed, incubated with secondary antibody, 1:2500 diluted donkey anti-mouse-680 (LICOR Biosciences, Cat #926–68072) and 1:2500 diluted donkey anti-rabbit-800 (LICOR Biosciences, Cat #926–32213), in LICOR blocking buffer (LICOR Biosciences, Cat #927–40000) for 1 h at room temperature, washed, and visualized Odyssey imager system equipped with ImageStudio software (LICOR). Bands were quantified using ImageStudioLite software. Expression of bPACs were normalized by dividing the intensity of the myc band by the intensity of the beta-actin band.
cAMP ELISA
HEK293 cells transfected with bPAC constructs were stimulated with light or left in the dark (no light control). cAMP was measured using Cayman ELISA cAMP assay (VWR, Cat #75817–364) following manufacturer recommendations. Values were normalized to total protein concentration for each sample.
PKA localization quantification
For experiments with overexpressed PKAcatα and PKARIIB, cells on Poly-L-lysine-coated coverslips were co-transfected with mTagBFP2-PKAcatα39 and PKARIIB-mCherry (a gift from Roshanak Irannejad, UCSF) in the presence or absence of Kif1a-tdTomato-FKBP, EGFP-FRB-EEA1, and GalT-eGFP using Lipofectamine 2000. Cells were pre-treated with either rapalog or ethanol control for 30 min, then stimulated with 1μM isoproterenol, and imaged live on the Andor Dragonfly Spinning Disk Confocal microscope. For colocalization experiments, Z stacks were taken with 0.3μm step size and max/min of the stack was determined per image by identifying the height of cells. Images were imported into FIJI and the maximum intensity projections of the channels were saved and exported as TIFF files. Line scan analysis was performed to evaluate the colocalization of PKAcatα, PKARIIB, and/or GalT. The fluorescence was measured every 0.2μm and normalized to the maximum fluorescence for each channel. To quantify PKAcatα distance to the nucleus, the optimal z frame was determined by visualization of endosomes, kinesin motor, and PKAcatα. Images were taken on the Andor Dragonfly Spinning Disk Confocal microscope and exported to FIJI. ROIs were drawn around the cell periphery and nuclei identified in the Kif1a-containing channel. Clusters of PKAcatα-containing puncta were segmented using the Morphological Segmentation plugin (MorphoLibJ)57. Using the distance formula, the distance from PKA puncta to the nucleus center of mass was calculated based on particle and nuclear x,y coordinates.
For experiments interrogating endogenous PKAcatα and PKARIIB, cells were grown on Poly-L-lysine-coated coverslips. In CID experiments, cells were transfected with Kif1a-tdTomato-FKBP and EGFP-FRB-EEA1 using Lipofectamine 2000 and treated with either rapalog or ethanol vehicle for 30 min. Following treatment, cells were fixed using 3.7% formaldehyde (ThermoFisher, Cat #28908) in modified BRB80 buffer (80mM PIPES pH 6.8, 1mM MgCl2, 1mM CaCl2) for 20 min. Fixation was quenched and cells were permeabilized using 2.5%BSA/0.1% Triton X-100 (Sigma, Cat #X100–100ML) in TBS for 20 min. Primary antibodies [Anti-PKAcatα (BD Biosciences, Cat #610980) or Anti-PKA-RIIB (BD Biosciences, Cat #610625)] were diluted 1:100 in permeabilization solution and incubated for overnight at 4°C. Secondary antibodies [Donkey anti-Mouse IgG (H+L) Secondary Antibody, Alexa Fluor 647 (Invitrogen, Cat #A-315710)] were diluted 1:1000 in permeabilization solution and incubated for 30 min at room temperature, protected from light. Following two washes in permeabilization solution and one wash in PBS, coverslips were mounted using ProlongGold with DAPI (Thermo Scientific, Cat #P36931). Slides were imaged on an Andor Dragonfly Spinning Disk Confocal microscope. Z stacks were taken with 0.3μm step size and max/min of the stack was determined per image by identifying the height of cells. Images were imported into FIJI and the maximum intensity projections of the channels were saved and exported as TIFF files. Line scan analysis was performed to evaluate the colocalization of PKAcatα or PKARIIB with EEA1. The fluorescence was measured every 0.2μm and normalized to the maximum fluorescence for each channel.
PKA activity quantification
Characterization experiments involving the nuclear ExRai-AKAR2 sensor were performed in HeLa cells to be consistent with previously developed localized ExRai-AKAR2 sensors42. Cells were transfected using Lipofectamine 2000 with ExRai-AKAR2-NLS alone or with PKI-NLS-mCherry. The follow day, samples were imaged in pre-equilibrated HBSS and were analyzed by live-cell imaging on Zeiss AxioObserver Z1 microscope. Cells were treated with 50μM forskolin and 100μM IBMX for 10 min. For experiments without the inhibitory PKI construct, cells were then washed with HBSS and treated with 20μM H89 for another 10 min. Images were taken every 30 sec, including ten baseline frames prior to forskolin and IBMX treatment. Images were quantified by drawing ROIs around individual cells, measuring the 480- and 400- channel fluorescence in every frame, and calculating the delta F/F. The ratio of 480-/400- responses was also recorded.
For PKA activity measurements, cells were transfected using Lipofectamine 2000 with either the ER42 or nuclear ExRai-AKAR2 sensor in the presence of Kif1a-tdTomato-FKBP + mCherry-FRB-EEA1 (CID), and either DynK44E-mCherry or empty pcDNA3.0 vector, where applicable. The following day, samples were analyzed by live-cell imaging on the Andor Dragonfly Spinning Disk Confocal microscope. Images were taken every 30 sec, including three baseline frames prior to isoproterenol treatment. Images were quantified by drawing ROIs around individual cells, measuring the 488-channel fluorescence in every frame, and calculating the delta F/F. For dose-response experiments, the indicated doses of isoproterenol were added. For DynK44E experiments, cells were stimulated with 10nM isoproterenol. For experiments involving the endosome redistribution system, cells were pretreated with ethanol (vehicle) or 1μM rapalog for 30 min, and then stimulated with 10nM isoproterenol.
Statistics and Reproducibility
Statistical analyses were performed using Prism software (GraphPad), and all relevant information is included in the Figure legends. For instances, in which a representative image is displayed, the experiment was performed in 2–4 independent biological replicates, and at least 10 cells were analyzed per condition.
Supplementary Material
Acknowledgements
We thank members of the Tsvetanova lab and Dr. Roshanak Irannejad (UCSF) for valuable discussions of the project and feedback on the manuscript. Immunofluorescence microscopy imaging was performed using resources from the Duke Light Microscopy Core Facility, with specific training under Lisa Cameron, Yasheng Gao, and Benjamin Carlson. PKARegIIB-mCherry and EGFP-SNX27 were gifts from Roshanak Irannejad (UCSF, CA). SSF- β2-AR, RAB11a-GFP, DynK44E-mCherry, pcDNA3.0, eGFP-C1 and Endo-bPAC were gifts from Mark Von Zastrow (UCSF, CA). MK1200 and dCas9-BFP-KRAB were gifts from Martin Kampmann (UCSF, CA). pBa.Kif1a 1–396.GFP was a gift from Gary Banker & Marvin Bentley (Addgene plasmid # 45058; http://n2t.net/addgene:45058; RRID:Addgene_45058). pBa-KIF5C 559-tdTomato-FKBP was a gift from Gary Banker & Marvin Bentley (Addgene plasmid # 64211; http://n2t.net/addgene:64211; RRID:Addgene_64211). pEGFP-FRB was a gift from Klaus Hahn (Addgene plasmid # 25919; http://n2t.net/addgene:25919; RRID:Addgene_25919). GFP-EEA1 wt was a gift from Silvia Corvera (Addgene plasmid # 42307; http://n2t.net/addgene:42307; RRID:Addgene_42307). pmCherry-N1-GalT was a gift from Lei Lu (Addgene plasmid # 87327; http://n2t.net/addgene:87327; RRID:Addgene_87327). Flamindo2 and nlsFlamindo2 were gifts from Tetsuya Kitaguchi (Addgene plasmid # 73938; http://n2t.net/addgene:73938; RRID:Addgene_73938 and Addgene plasmid # 73939; http://n2t.net/addgene:73939; RRID:Addgene_73939). Figure 1A and 6F were created using BioRender. Research reported in this publication was supported by the National Institutes of Health (R01NS127847 and R35GM142640 to N.G.T., R01DK073368 to J.Z., F31NS120567 to B.K.A.W.) and the American Heart Association (Predoctoral Fellowship 834472 to J.F.Z.).
Footnotes
Competing Interests:
The authors declare no competing interests.
Data availability
All relevant data supporting the findings are available within the paper and the Supplementary data. Source data are provided with the manuscript. Additional information and reagents are available from the corresponding author upon reasonable request.
References:
- 1.Sorkin A. & von Zastrow M. Endocytosis and signalling: intertwining molecular networks. Nat Rev Mol Cell Biol 10, 609–622 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thomsen ARB, Jensen DD, Hicks GA & Bunnett NW Therapeutic Targeting of Endosomal G-Protein-Coupled Receptors. Trends Pharmacol Sci 39, 879–891 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tsvetanova NG, Irannejad R. & von Zastrow M. G protein-coupled receptor (GPCR) signaling via heterotrimeric G proteins from endosomes. J Biol Chem 290, 6689–6696 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vilardaga JP, Jean-Alphonse FG & Gardella TJ Endosomal generation of cAMP in GPCR signaling. Nat Chem Biol 10, 700–706 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Godbole A, Lyga S, Lohse MJ & Calebiro D. Internalized TSH receptors en route to the TGN induce local Gs-protein signaling and gene transcription. Nat Commun 8, 443 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsvetanova NG & von Zastrow M. Spatial encoding of cyclic AMP signaling specificity by GPCR endocytosis. Nat Chem Biol 10, 1061–1065 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsvetanova NG et al. Endosomal cAMP production broadly impacts the cellular phosphoproteome. J Biol Chem 297, 100907 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calebiro D. et al. Persistent cAMP-signals triggered by internalized G-protein-coupled receptors. PLoS Biol 7, e1000172 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferrandon S. et al. Sustained cyclic AMP production by parathyroid hormone receptor endocytosis. Nat Chem Biol 5, 734–742 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Irannejad R. et al. Functional selectivity of GPCR-directed drug action through location bias. Nat Chem Biol 13, 799–806 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nash CA, Wei W, Irannejad R. & Smrcka AV Golgi localized beta1-adrenergic receptors stimulate Golgi PI4P hydrolysis by PLCepsilon to regulate cardiac hypertrophy. Elife 8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Purgert CA et al. Intracellular mGluR5 can mediate synaptic plasticity in the hippocampus. J Neurosci 34, 4589–4598 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vincent K. et al. Intracellular mGluR5 plays a critical role in neuropathic pain. Nat Commun 7, 10604 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.White AD et al. Spatial bias in cAMP generation determines biological responses to PTH type 1 receptor activation. Sci Signal 14, eabc5944 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Okazaki M. et al. Prolonged signaling at the parathyroid hormone receptor by peptide ligands targeted to a specific receptor conformation. Proceedings of the National Academy of Sciences 105, 16525–16530 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feinstein TN et al. Noncanonical control of vasopressin receptor type 2 signaling by retromer and arrestin. J Biol Chem 288, 27849–27860 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuna RS et al. Glucagon-like peptide-1 receptor-mediated endosomal cAMP generation promotes glucose-stimulated insulin secretion in pancreatic β-cells. American journal of physiology-endocrinology and metabolism 305, E161–E170 (2013). [DOI] [PubMed] [Google Scholar]
- 18.Tian X. et al. The α-arrestin ARRDC3 regulates the endosomal residence time and intracellular signaling of the β2-adrenergic receptor. Journal of Biological Chemistry 291, 14510–14525 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thomsen AR et al. GPCR-G protein-β-arrestin super-complex mediates sustained G protein signaling. Cell 166, 907–919 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Varandas KC, Irannejad R. & von Zastrow M. Retromer endosome exit domains serve multiple trafficking destinations and regulate local G protein activation by GPCRs. Current Biology 26, 3129–3142 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bowman SL, Shiwarski DJ & Puthenveedu MA Distinct G protein–coupled receptor recycling pathways allow spatial control of downstream G protein signaling. Journal of Cell Biology 214, 797–806 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Madamanchi A. β-Adrenergic receptor signaling in cardiac function and heart failure. McGill Journal of Medicine: MJM 10, 99 (2007). [PMC free article] [PubMed] [Google Scholar]
- 23.Mutlu GM & Factor P. Alveolar epithelial β2-adrenergic receptors. American journal of respiratory cell and molecular biology 38, 127–134 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Irannejad R. et al. Conformational biosensors reveal GPCR signalling from endosomes. Nature 495, 534–538 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mayr B. & Montminy M. Transcriptional regulation by the phosphorylation-dependent factor CREB. Nature Reviews Molecular Cell Biology 2, 599–609 (2001). [DOI] [PubMed] [Google Scholar]
- 26.Peng GE, Pessino V, Huang B. & von Zastrow M. Spatial decoding of endosomal cAMP signals by a metastable cytoplasmic PKA network. Nat Chem Biol (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schuster M, Lipowsky R, Assmann M-A, Lenz P. & Steinberg G. Transient binding of dynein controls bidirectional long-range motility of early endosomes. Proceedings of the National Academy of Sciences 108, 3618–3623 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reck-Peterson SL, Redwine WB, Vale RD & Carter AP The cytoplasmic dynein transport machinery and its many cargoes. Nature Reviews Molecular Cell Biology 19, 382–398 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Korolchuk VI et al. Lysosomal positioning coordinates cellular nutrient responses. Nat Cell Biol 13, 453–460 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sainath R. & Gallo G. The dynein inhibitor Ciliobrevin D inhibits the bidirectional transport of organelles along sensory axons and impairs NGF-mediated regulation of growth cones and axon branches. Dev Neurobiol 75, 757–777 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Violin JD et al. beta2-adrenergic receptor signaling and desensitization elucidated by quantitative modeling of real time cAMP dynamics. J Biol Chem 283, 2949–2961 (2008). [DOI] [PubMed] [Google Scholar]
- 32.Temkin P. et al. SNX27 mediates retromer tubule entry and endosome-to-plasma membrane trafficking of signalling receptors. Nat Cell Biol 13, 715–721 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Odaka H, Arai S, Inoue T. & Kitaguchi T. Genetically-encoded yellow fluorescent cAMP indicator with an expanded dynamic range for dual-color imaging. PloS one 9, e100252 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Herskovits JS, Burgess CC, Obar RA & Vallee RB Effects of mutant rat dynamin on endocytosis. The Journal of cell biology 122, 565–578 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bayle JH et al. Rapamycin analogs with differential binding specificity permit orthogonal control of protein activity. Chem Biol 13, 99–107 (2006). [DOI] [PubMed] [Google Scholar]
- 36.Nagendran M, Riordan DP, Harbury PB & Desai TJ Automated cell-type classification in intact tissues by single-cell molecular profiling. Elife 7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Semesta KM, Tian R, Kampmann M, von Zastrow M. & Tsvetanova NG A high-throughput CRISPR interference screen for dissecting functional regulators of GPCR/cAMP signaling. PLoS Genet 16, e1009103 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gillooly DJ et al. Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. EMBO J 19, 4577–4588 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang JZ et al. Phase Separation of a PKA Regulatory Subunit Controls cAMP Compartmentation and Oncogenic Signaling. Cell 182, 1531–1544 e1515 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harootunian AT et al. Movement of the free catalytic subunit of cAMP-dependent protein kinase into and out of the nucleus can be explained by diffusion. Molecular Biology of the Cell 4, 993–1002 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith FD et al. Local protein kinase A action proceeds through intact holoenzymes. Science 356, 1288–1293 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang J-F et al. An ultrasensitive biosensor for high-resolution kinase activity imaging in awake mice. Nature chemical biology 17, 39–46 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bock A. et al. Optical Mapping of cAMP Signaling at the Nanometer Scale. Cell 182, 1519–1530 e1517 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mo GC et al. Genetically encoded biosensors for visualizing live-cell biochemical activity at super-resolution. Nat Methods 14, 427–434 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bers DM, Xiang YK & Zaccolo M. Whole-Cell cAMP and PKA Activity are Epiphenomena, Nanodomain Signaling Matters. Physiology (Bethesda) 34, 240–249 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nigg E, Hilz H, Eppenberger H. & Dutly F. Rapid and reversible translocation of the catalytic subunit of cAMP-dependent protein kinase type II from the Golgi complex to the nucleus. The EMBO Journal 4, 2801–2806 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Anton SE et al. Receptor-associated independent cAMP nanodomains mediate spatiotemporal specificity of GPCR signaling. Cell 185, 1130–1142.e1111 (2022). [DOI] [PubMed] [Google Scholar]
- 48.Koschinski A. & Zaccolo M. Activation of PKA in cell requires higher concentration of cAMP than in vitro: implications for compartmentalization of cAMP signalling. Sci Rep 7, 14090 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.English AR & Voeltz GK Endoplasmic reticulum structure and interconnections with other organelles. Cold Spring Harbor perspectives in biology 5, a013227 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jia R. & Bonifacino JS Lysosome Positioning Influences mTORC2 and AKT Signaling. Mol Cell 75, 26–38 e23 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Riccio A, Pierchala BA, Ciarallo CL & Ginty DD An NGF-TrkA-mediated retrograde signal to transcription factor CREB in sympathetic neurons. Science 277, 1097–1100 (1997). [DOI] [PubMed] [Google Scholar]
- 52.Scerra G. et al. Lysosomal positioning diseases: beyond substrate storage. Open Biology 12, 220155 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Murphy JE, Padilla BE, Hasdemir B, Cottrell GS & Bunnett NW Endosomes: a legitimate platform for the signaling train. Proc Natl Acad Sci U S A 106, 17615–17622 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kwon Y. et al. Non-canonical β-adrenergic activation of ERK at endosomes. Nature, 1–7 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Inda C. et al. Different cAMP sources are critically involved in G protein-coupled receptor CRHR1 signaling. J Cell Biol 214, 181–195 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kotowski SJ, Hopf FW, Seif T, Bonci A. & von Zastrow M. Endocytosis promotes rapid dopaminergic signaling. Neuron 71, 278–290 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Legland D, Arganda-Carreras I. & Andrey P. MorphoLibJ: integrated library and plugins for mathematical morphology with ImageJ. Bioinformatics 32, 3532–3534 (2016). [DOI] [PubMed] [Google Scholar]
- 58.Rizk A. et al. Segmentation and quantification of subcellular structures in fluorescence microscopy images using Squassh. Nat Protoc 9, 586–596 (2014). [DOI] [PubMed] [Google Scholar]
- 59.Kamentsky L. et al. Improved structure, function and compatibility for CellProfiler: modular high-throughput image analysis software. Bioinformatics 27, 1179–1180 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data supporting the findings are available within the paper and the Supplementary data. Source data are provided with the manuscript. Additional information and reagents are available from the corresponding author upon reasonable request.