Abstract
Prenylated indole alkaloids (PIAs) possess great structural diversity and biological activities. Despite lots of efforts in investigating the biosynthetic mechanism, the key step of transforming the 2,5-diazabicyclo[2.2.2]octane-containing PIAs to a distinct class of pentacyclic compounds remains unknown. Here, using a combination of gene deletion, heterologous expression, and biochemical characterization, we show that a unique fungal P450 enzyme CtdY catalyzes the cleavage of the amide bond in the 2,5-diazabicyclo[2.2.2]octane system, followed by a decarboxylation step to form the 6/5/5/6/6 pentacyclic ring in 21R-citrinadin A. We also demonstrate the function of a subsequent cascade of stereospecific oxygenases to further modify the 6/5/5/6/6 pentacyclic intermediate en route to the complete 21R-citrinadin A biosynthesis. Our findings reveal a key enzyme CtdY for the pathway divergence in the biosynthesis of PIAs and uncover the complex late-stage post-translational modifications in 21R-citrinadin A biosynthesis.
Introduction
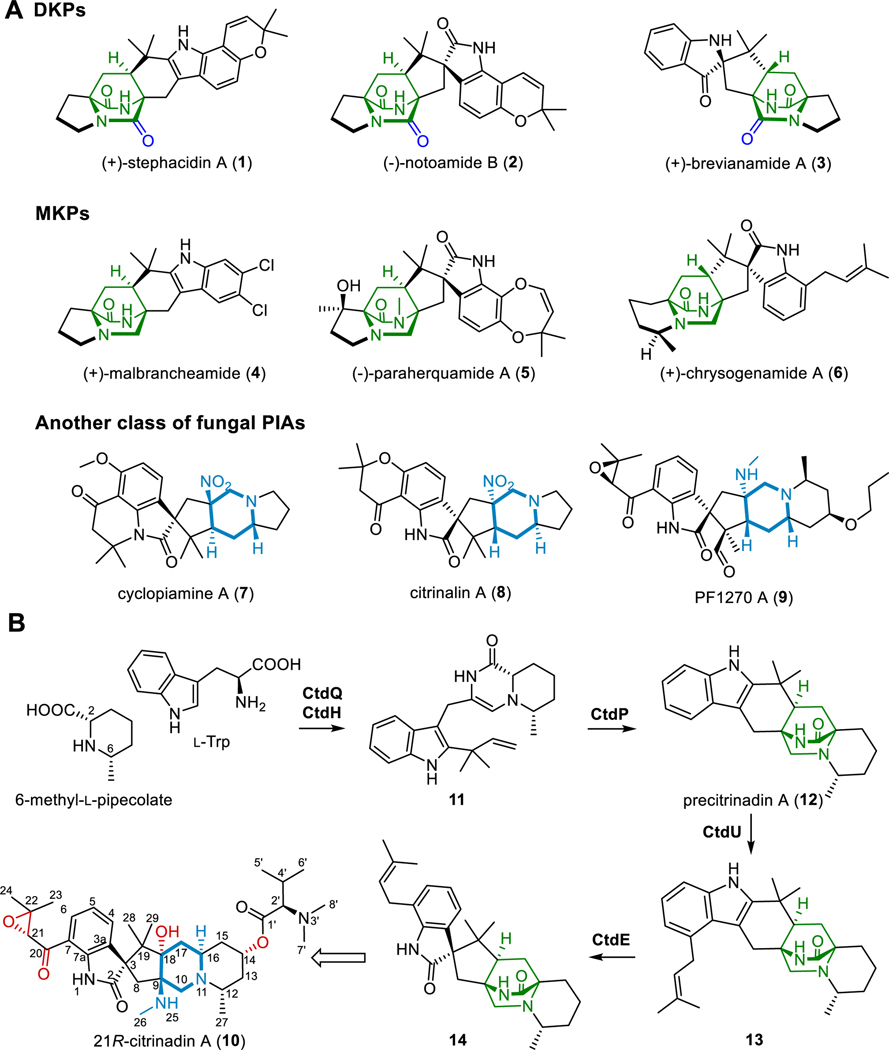
Fungal prenylated indole alkaloids (PIAs) bearing 2,5-diazabicyclo[2.2.2]octane display diverse structural complexity and biological activities, making them attractive synthetic targets and raising broad interests in studying their biosynthesis.1,2 Fungal PIAs are comprised of primary two types: diketopiperazines (DKPs), such as stephacidin A (1), notoamide B (2), and brevianamide A (3); and monoketopiperazines (MKPs), including malbrancheamide (4), paraherquamide A (5), and chrysogenamide A (6) (Figure 1A).3–11 The dipeptide precursors of these PIAs are biosynthesized by nonribosomal peptide synthetases (NRPSs) containing the terminal condensation or reductase domains with different release mechanisms (Figure S1). Followed by the reverse prenylation of the indole moiety in the cyclodipeptide precursor, the azadiene moiety and the dienophile isoprene undergo an intramolecular [4+2] hetero-Diels-Alder (DA) reaction to construct the 2,5-diazabicyclo[2.2.2]octane system.12 Recently, several different classes of Diels-Alderases (DAases) were reported to facilitate the DA reaction and strictly control the specific stereochemistry of bridged bicyclic adducts.13–15 In addition to the PIAs containing the 2,5-diazabicyclo[2.2.2]octane ring structure, another distinct class of fungal PIAs including cyclopiamine A (7), citrinalin A (8), PF1270 A (9), and citrinadin A (10), possess unique 6/6/5/5/6/5 hexacyclic or 6/5/5/6/6 pentacyclic spiroindolinone scaffolds (Figure 1A).16–19
Figure 1.
Chemical structures of representative diketopiperazines (DKPs), monoketopiperazines (MKPs), and another class of PIAs (A); and proposed biosynthetic pathway of 21R-citrinadin A (B).
Recently, we and others have elucidated the early steps of 21R-citrinadin A biosynthesis. Briefly, CtdK, CtdL, and CtdM sharing high homology to CndG, CndF, and CndE might be involved in the biosynthesis of the key building block (2S,6S)-6-methyl-L-pipecolate (Figure S2),20 which is incorporated into the production of the MKP precursor by the NRPS CtdQ.21 A subsequent reverse prenyltransferase (PT) CtdH21 and a specific α-anti-selective DAase CtdP produce the 2,5-diazabicyclo[2.2.2]octane-containing intermediate 12, which is then further prenylated by the other PT CtdU and oxidized to 14 with the 3S-spirooxindole moiety via semipinacol rearrangement by a flavoprotein monooxygenase (FPMO) CtdE21 (Figure 1B). Unlike 21R-citrinadin A possessing the 6/5/5/6/6 pentacyclic structure, the intermediate 14 has the 2,5-diazabicyclo[2.2.2]octane bridged ring system (Figure 1A and 1B). Similarly, the 2,5-diazabicyclo[2.2.2]octane-containing compound citrinalin C was identified from P. citrinum F53, the producer of citrinalin A (8)2 in isotope feeding studies, suggesting that the pentacyclic structure could be derived from the [2.2.2] bridged ring-containing MKP progenitors (Figure S1). However, the biosynthetic steps of such a transformation remain unclear.
The synthesis of the pentacyclic system from the 2,5-diazabicyclo[2.2.2]octane system could undergo the cleavage of the amide bond, however, typical amide bonds featuring high resonance energy (nN→π*C=O conjugation, 15–20 kcal/mol) are pretty stable.22 Synthetically, multiple approaches were developed to break amide bonds, including geometric alteration of the amide bond by twisting or pyramidalization, and electronic delocalization to unlock the ground-state destabilization of amides.23,24 Meanwhile, most reported enzymes cleave amide bonds by hydrolysis, of which proteases, peptidases, amidases, amidohydrolases, and acylases are the most common types.25 Moreover, resistance-related β-lactamases specifically hydrolyze the amide bond in β-lactam antibiotics, thereby inactivating the drug.26 A glycosyltransferase OGT as well as a YcaO enzyme MusD were recently reported to facilitate the hydrolysis of the peptide amide bond.27,28 Distinctively, a Mn(II)-dependent toxoflavin lyase TflA enables the oxidative degradation of the amide in toxoflavin (Figure S3).29 However, after detailed bioinformatic analysis, none of enzymes encoded by the reported biosynthetic gene cluster (BGC) of 21R-citrinadin A belong to the families mentioned above.21 Therefore, uncovering enzymes for deconstructing the 2,5-diazabicyclo[2.2.2]octane might reveal a new biosynthetic strategy to break the amide bond. In addition to the distinct 6/5/5/6/6 pentacyclic scaffold, 21R-citrinadin A features an N, N-dimethylvaline ester unit, an α, β-epoxy-carbonyl moiety, and a C-9 hydroxyl group,18,30 which indicates a variety of enzymatic modification are involved in the late-stage biosynthesis of 21R-citrinadin A, intriguing our further investigation.
Herein, we studied the biosynthetic pathway of 21R-citrinadin A via a combination of gene inactivation, heterologous expression, precursor incorporation, and in vitro biochemical analysis. Our results reveal a unique cytochrome P450 oxygenase CtdY catalyzing the amide bond cleavage and decarboxylation to form the key 6/5/5/6/6 pentacyclic scaffold with an unprecedented mechanism. Moreover, we demonstrate that a set of oxidoreductases consisting of CtdG, CtdR, CtdV, and CtdJ perform the multistep redox reactions on the core structure with strict regio- and stereo-selectivity to install the complete 21R-citrinadin A structure.
Results and Discussion
Probing related enzymes for deconstructing 2,5-diazabicyclo[2.2.2]octane system
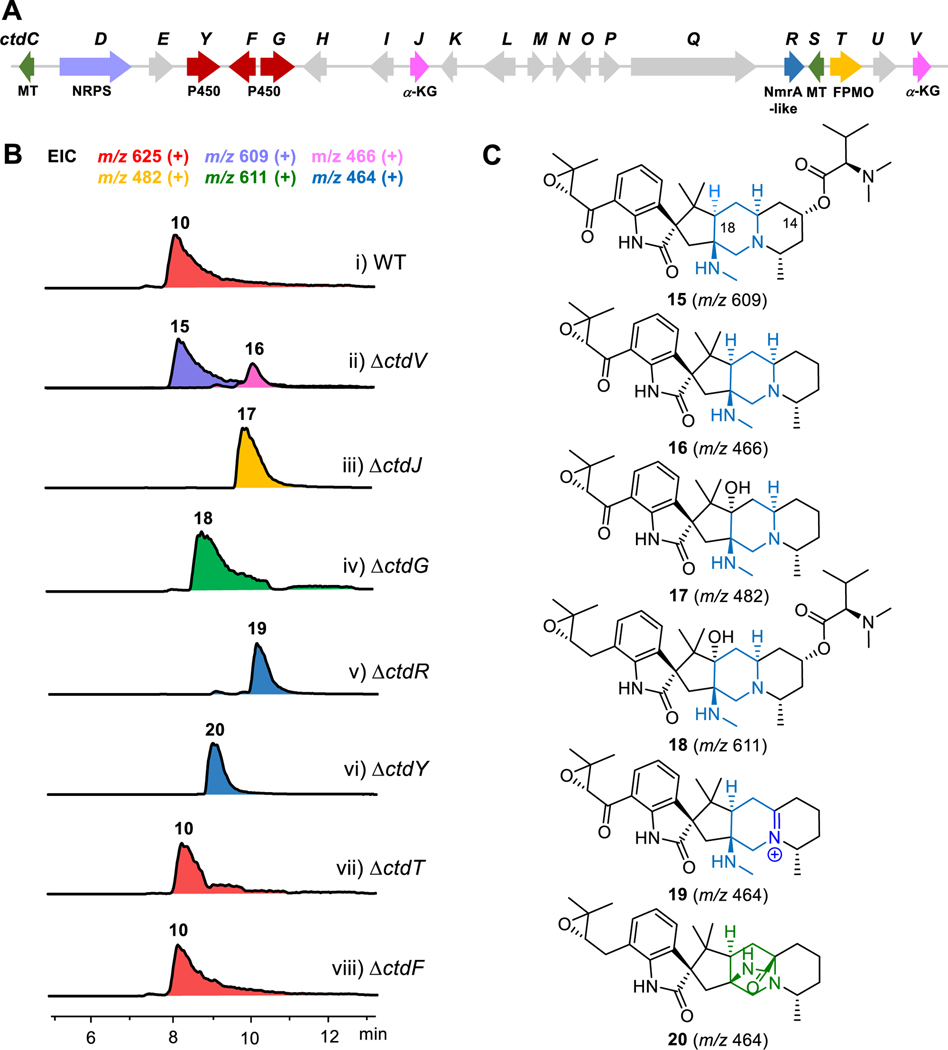
21R-citrinadin A (10) is one of the main metabolites discovered in P. citrinum ATCC 9849 and its putative BGC ctd contains 21 genes based on previous studies and bioinformatic analysis (Figure 2A).15,21 To pinpoint genes related to the deconstruction of 2,5-diazabicyclo[2.2.2]octane, we individually knocked out uncharacterized genes in ctd gene cluster that might have a putative redox function, including four P450-encoding genes (ctdT, ctdF, ctdG, and ctdY), two α-ketoglutarate (α-KG)-dependent oxygenase-encoding genes (ctdV and ctdJ), and one NrmA-like enzyme-encoding gene (ctdR) by the split marker recombination approach (Figure S4).31 As revealed by LC-MS, ΔctdT and ΔctdF mutants still produce 10 (Figure 2A–vii, viii), indicating CtdT and CtdF are not involved in the biosynthesis of 10, while ΔctdV, ΔctdJ, ΔctdG, ΔctdR, and ΔctdY mutants completely abolished the production of 10 (Figures 2B, S5A, and S5B). The ΔctdV mutant accumulated a major product 15 with m/z 609 [M+H]+, along with one minor product 16 with m/z 466 [M+H]+ (Figure 2B–ii). These compounds were isolated and characterized by 1D and 2D NMR spectroscopy. Both compounds 15 and 16 lacked the C-18 hydroxyl group but the N, N-dimethylvaline ester group at C-14 in 10 was absent in 16 (Figure 2C and Table S5–S6). In addition, compound 17 produced by ΔctdJ mutant was similar to 16 while it retained the hydroxyl group at C-18 (Figure 2B–iii and Table S7). These results implicated CtdV and CtdJ as C-18 and C-14 hydroxylases respectively. CtdG shares 45% sequence identity with CloA which transforms the hydroxyl group of elymoclavine into the carboxyl group to provide paspalic acid in the biosynthetic pathway of ergoline alkaloid, suggesting that CtdG may catalyze successive oxidation at the same site.32 Compound 18 with m/z 611 [M+H]+ accumulated in the ΔctdG mutant was isolated and identified by NMR (Figure 2B–iv, 2C, and Table S8). The missing δC 194.9 ppm signal and the appearance of δC 32.7 ppm in 13C NMR spectrum indicated the methylene group rather than the carbonyl group in C-20 of 18. Therefore, CtdG might be responsible for the oxidation of the methylene in the epoxyprenyl side chain to form the α, β-epoxy-carbonyl moiety. Furthermore, the ΔctdR mutant generated an iminium intermediate 19 identified in previous studies (Figure 2B–v and 2C).15 All intermediates purified from the above mutants still have the 6/5/5/6/6 pentacyclic scaffold, indicating there is another enzyme responsible for the deconstruction of the 2,5-diazabicyclo[2.2.2]octane ring in citrinadins. Finally, we isolated a new intermediate 20 from the ΔctdY mutant, which was proven to maintain the 2,5-diazabicyclo[2.2.2]octane moiety, as supported by the HMBC correlation from H-25 to C-30 and other 1D and 2D NMR spectra (Figures 2–Bvi, 2C, and Table S9). Thus, CtdY was proposed to be the key enzyme for the deconstruction of the 2,5-diazabicyclo[2.2.2]octane.
Figure 2.
Identification of genes related to deconstruction of 2,5-diazabicyclo[2.2.2]octane system. (A) The putative ctd gene cluster from P. citrinum ATCC 9849. (B) LC-MS analysis (extracted ion chromatogram, EIC) of P. citrinum wildtype (WT) and ctd knockout mutants. (i) P. citrinum WT, (ii) ΔctdV, (iii) ΔctdJ, (iv) ΔctdG, (v) ΔctdR, (vi) ΔctdY, (vii) ΔctdT, (viii) ΔctdF. EIC (+) traces of m/z=625 (red), m/z=609 (purple), m/z=466 (pink), m/z=482 (yellow), m/z=611 (green), and m/z=464 (blue) are in different colors. (C) Chemical structures of 15-20 that were confirmed by NMR data. NRPS, nonribosomal peptide synthetase; MT, methyltransferase; α-KG, α-ketoglutarate-dependent enzyme; FPMO, flavoprotein monooxygenase.
Functional characterization of CtdY
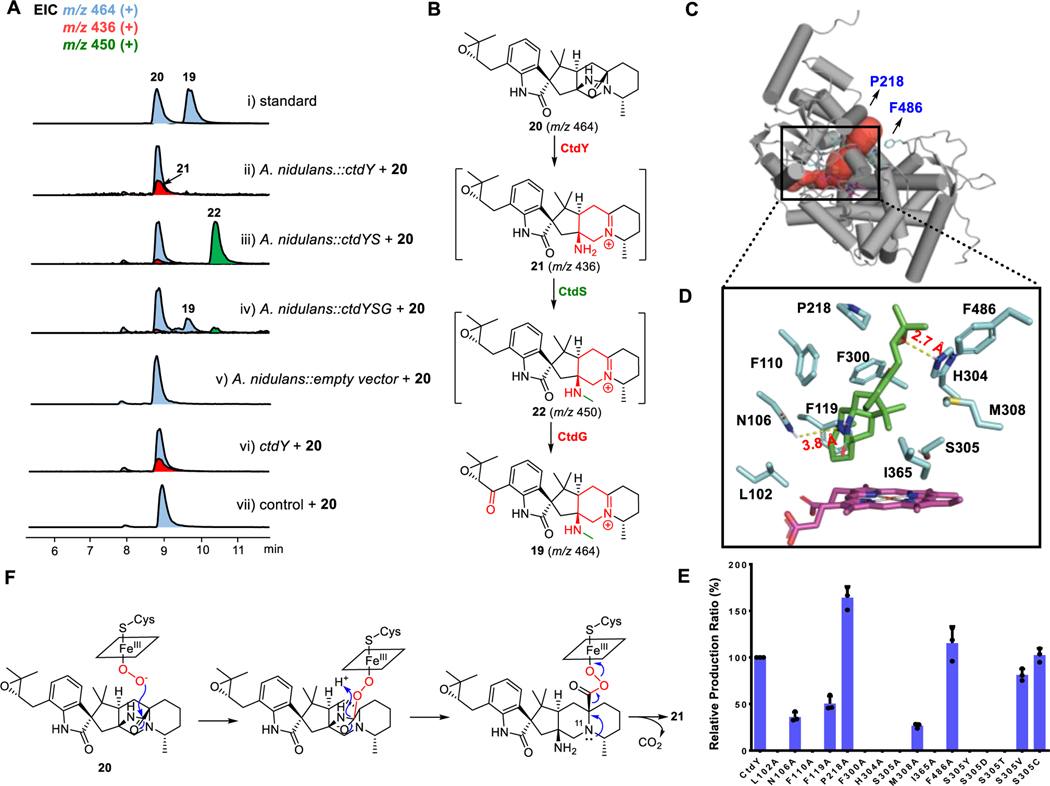
To further explore the function of CtdY, we reconstituted CtdY in A. nidulans A114533, which contains three auxotrophic markers and is widely used as a heterologous host for gene expression. The feeding of 20 to CtdY-containing A. nidulans led to the production of a new compound 21 (Figure 3A–ii). Based on its chemical formula was determined to be C27H38N3O2+ (m/z 436.2949 [M]+, calculated for C27H38N3O2+, 436.2959) deduced by HR-ESI-MS data (Figure S6A) indicating the loss of one carbon and one oxygen compared with 20, CtdY might remove a carbonyl group to produce a putative iminium cation intermediate 21 (Figure 3B). However, the structure of 21 is difficult to be characterized by NMR data due to its instability. Alternatively, we found that 19 purified from ΔctdR mutant shares the same iminium cation moiety as 21 but with an additional carbonyl group in the isopentenyl side chain and an extra methyl at N-25, so we hypothesized that 21 might undergo further methylation and oxidation to synthesize 19. Additionally, the abolishment of the methyl group at N-25 in an off-pathway shunt compound 25 produced by the ΔctdS mutant suggested that CtdS catalyzed the N-methylation at N-25 (Figures S5C, S7, and Table S10). Meanwhile, CtdG was demonstrated to oxidize the CH2 to C=O based on gene knockout results, which was further verified by the biotransformation of 18 to 10 accomplished by CtdG-expressing A. nidulans strain (Figure S8).
Figure 3.
Functional characterization of CtdY. (A) LC-MS analysis (EIC) of metabolites from feeding experiments and microsomal in vitro assays of CtdY. (i) standards of compound 19 and 20; feeding of 20 to A. nidulans with (ii) ctdY, (iii) ctdY and ctdS, (iv) ctdY, ctdS, and ctdG, (v) empty vector; (vi) CtdY-containing microsome + 20; (vii) control microsome + 20. (B) The proposed biosynthetic pathway from 20 to 19. EIC (+) traces of m/z=464 (blue grey), m/z=436 (red), m/z=450 (green) are in different colors. (C) The structure model of CtdY-heme was generated by AlphaFold2 and I-TASSER and the enzyme channel (colored in red) of CtdY was predicted by Caver. (D) Overlay of substrate-binding site view of CtdY-heme-20 (20 carbons colored in green, residue carbons colored in light blue, and heme colored in rose red). Molecular docking was performed using AutoDock vina. (E) Analysis of the relative production rate ± standard deviation (SD, three biological replicates) of 20 incubated with microsomal CtdY mutants. (F) Proposed enzymatic mechanism of CtdY.
To further illustrate CtdY function, we then performed feeding experiments of 20 to A. nidulans expressing the different combination of CtdY, CtdS, and CtdG. A. nidulans strain with CtdY and CtdS can transform 20 to a new compound 22 with C28H40N3O2+ (m/z 450.3112 [M]+, calculated for C28H40N3O2+, 450.3115) revealed by HR-ESI-MS data (Figures 3A–iii, 3B, and S6B). We further characterized a stable derivative 22R by the reduction of C=N bond of 22 with NaBH4 by HR-ESI-MS data and NMR spectra (Figure S6 and Table S13). Interestingly, feeding of 20 to A. nidulans expressing CtdY, CtdG, and CtdS led to the production of 19 verified by LC-MS analysis (Figure 3A–iv and 3B). The dramatically decreased biotransformation efficiency of 20 to the demethylated-19 by the A. nidulans strain containing CtdY and CtdG suggested that the methylation of CtdS might be required prior to the oxidation of CtdG (Figure S9). These results reveal that CtdG may catalyze successive oxidation to provide the α, β-epoxy-carbonyl moiety and CtdS is responsible for the methylation at N-25. The identification of 22R and the biotransformation of 20 to 19 strongly support the proposed chemical structure of 21 with the iminium cation produced by CtdY (Figure 3B).
We subsequently cloned ctdY from P. citrinum cDNA and expressed it in Saccharomyces cerevisiae RC01, which contains an integrated copy of cytochrome P450 reductase from A. terreus.34 Since P450 CtdY was predicted to contain a transmembrane domain of 27 amino acids by the TMHMM program (Figure S10),35 we prepared microsomes containing CtdY and then performed the in vitro assays. Compared with the control assay, the CtdY-containing microsomes successfully transformed 20 to 21, which further confirmed the function of CtdY (Figure 3A–vi, vii).
To obtain further insight into the active-site architecture of CtdY, we constructed the enzyme model with heme by AlphaFold2 and I-TASSER (Figure 3C).36,37 Molecular docking with substrate 20 identified an amino acid residues (Figure 3D). The site-directed mutagenesis of these amino acids led to the generation of CtdY variants which were performed by microsomal in vitro assays to reveal their catalytic residue functions (Figure 3E). Notably, the catalytic efficiencies of the P218A and F486A mutants were increased by 50% and 30%, respectively. Since the access channel of CtdY predicted by Caver revealed that P218 and F486 were at the gate of the catalytic channel (Figure 3C),38 we hypothesize that the smaller alanine substitutions might facilitate the substrate to enter the catalytic sites.39 The mutation of non-polar amino acids L102, F110, F300, and I365 into alanine completely eliminated the catalytic activity while the activity of F119A and M308A was reduced by 50% and 70% respectively. These non-polar amino acids might play a critical role in substrate binding through hydrophobic interactions. The catalytic activity of the N106A mutation decreased by 50% indicating N106 might form a hydrogen bond with the amide bond to stabilize the intermediate. S305A also completely abolished the enzymatic activity of CtdY while the S305C and S305V mutations exhibited comparable catalytic activity to wild type indicating S305 may play a steric effect rather than a catalytic role. Furthermore, the catalytic activity of the H304A mutation was also completely abolished. Considering that the distance between H304 and the epoxy of 19 was only 2.3 Å, we proposed H304 might form a hydrogen bond with the epoxy to fix the position of the substrate in the catalytic center for substrate reorganization.
Fungal P450s are structurally and functionally diverse enzymes that usually perform rate-limiting reactions in the biosynthesis of natural products.40 They usually mediate a typical monooxygenation reaction via the canonical ‘oxygen rebound’ mechanism to insert a single oxygen atom into C-H, C=C, N-H, or S-H bonds.41 With an increasing number of characterized P450s, the substrate radical generated by the high active oxoiron(IV) species can initiate more complex oxidation such as C–C bond formation and cleavage.40,42 Notably, the nucleophilic attack of the heme-FeIII hydroperoxide complex contribute to realizing some uncommon reactions, such as epoxidation catalyzed by Fma-P45043,44, and C–C bond cleavage mediated by Baeyer-Villiger rearrangement catalyzed by CYP 1745, JCM 146, and SdnB47. Accordingly, we propose the CtdY catalysis may proceed through a mechanism that the carbonyl of the amide bond in 20 might be attacked by heme-FeIII hydroperoxide species resulting in the cleavage of the C-N bond. The subsequent decarboxylation initiated by the attack of N-11 provided the iminium intermediate 21 (Figure 3F). Moreover, the carbon dioxide detection assay with the screening kit detected the production of CO2 in CtdY assays which further supports the proposed catalytic mechanism (Figure S11). To our best knowledge, it is distinct that a P450 can achieve the cleavage of the amide bond accompanied by the carbonyl elimination. In the sequence similarity network, the node representing CtdY only established a connection with PaxQ, a P450 required for the hydroxylation in paxilline biosynthesis,48 indicating CtdY is distinct from other fungal P450s (Figure S12).
Uncover the cascaded late-stage modification in the biosynthetic pathway
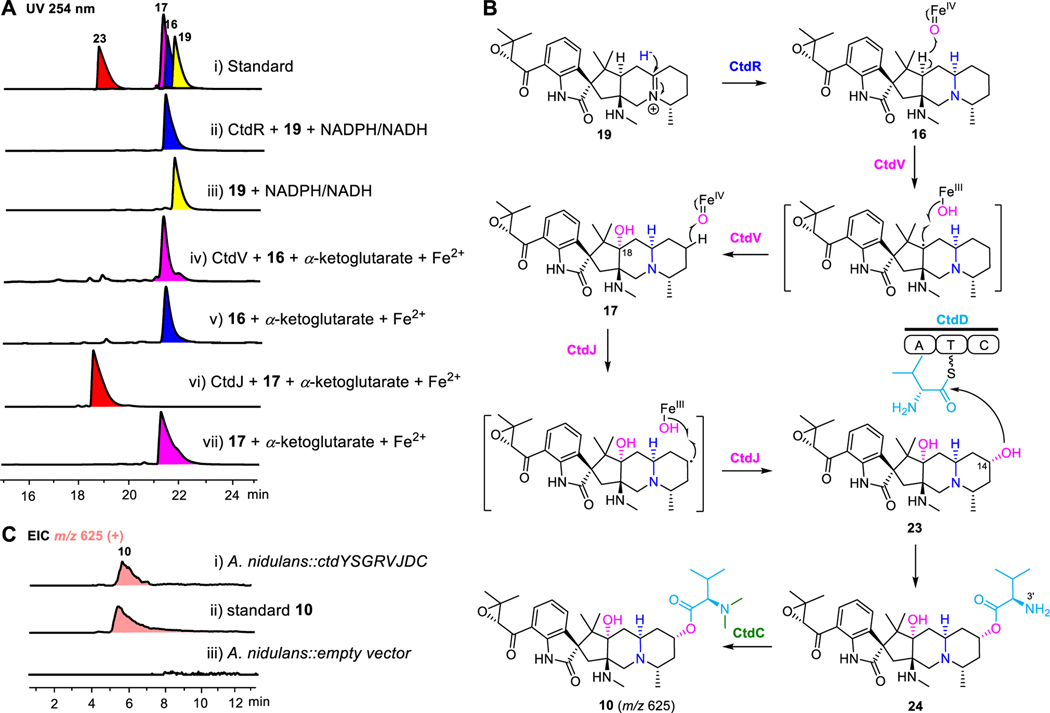
Having elucidated the key enzyme for breaking the bridged bicyclic system, we then focused on the remaining essential enzymes for post-modification of 10. NmrA-like proteins were previously regarded as the transcriptional nitrogen metabolism repressor or redox-sensing regulators, and usually show no catalytic function.49–51 However, CtdP belonging to this family has been reported to participate in the DA reaction and reduce the iminium in a redox cycle with NADP+. Considering the high sequence identity (42.5%) between CtdR and CtdP and the accumulation of the iminium-containing compound 19 in ΔctdR mutants, CtdR was proposed as the key enzyme catalyzing iminium reduction. To further verify the hypothesis, we overexpressed and purified the C-His-tagged form of the CtdR protein from Escherichia coli BL21(DE3) (Figure S13) and carried out the in vitro assays. As a result, 19 was successfully converted to the product 16 of the ΔctdV mutant when incubated with CtdR and NADPH/NADH (Figure 4A–i–iii). And the reduction of the C=N provided a newly formed R-configuration stereo center at C-16 in 16, as confirmed by the NOE correlation between H-16 and H-27 (Table S6). Our results reveal that CtdR can serve as an example of reduction activity achieved by the NmrA-like enzyme (Figure 4B).
Figure 4.
Elucidation of late-stage biosynthesis of 21R-citrinadin A. (A) HPLC analysis of CtdR, CtdJ, and CtdV in vitro assays with UV detected at 254 nm. (i) standards of 16, 17, 19, and 23, (ii) CtdR + 19 + NADPH/NADH, (iii) 19 + NADPH/NADH, (iv) CtdV + 16 + α-ketoglutarate + Fe2+, (v) 16 + α-ketoglutarate + Fe2+, (vi) CtdJ + 17 + α-ketoglutarate + Fe2+, (vii) 17 + α-ketoglutarate + Fe2+. 16, 17, 19, and 23 are colored in blue, rose red, yellow, and red, respectively. (B) Proposed biosynthetic pathway of 10 from 19. (C) Mass filter analysis of 10 (m/z=625 [M+H]+, colored in pink orange) in heterologous host. (i) feeding of 20 to A. nidulans expressing ctdY, ctdS, ctdG, ctdR, ctdV, ctdJ, ctdD, and ctdC. (ii) standard of 10, (iii) feeding of 20 to A. nidulans with empty vector. A, adenylation; T, thiolation; C, condensation.
Based on the gene deletion results, both α-KG-dependent oxygenases CtdV and CtdJ were proposed to introduce the hydroxyl group at C-18 and C-14, respectively. To further demonstrate their biochemical function in vitro, CtdV and CtdJ were purified from E. coli BL21(DE3) (Figure S13). We found that incubation of CtdV with FeSO4 and α-ketoglutarate completely converted 16 to C-18 hydroxylated 17 within 1h (Figures 4A–iv, v, and S14), whereas CtdV was unable to hydroxylate 15 which only features an additional N, N-dimethylvaline ester group at C-14 (Figure S15). It indicates that CtdV exhibits a narrow substrate specificity and 16 is its natural substrate. Furthermore, in the appearance of FeSO4 and α-ketoglutarate, CtdJ could convert 17 to a new compound 23 (Figures 4A–vi, vii, and S14), which was demonstrated to be the same intermediate when we knocked out ctdD encoding a single-module NRPS (Figure S5 and Table S11). Furthermore, the remaining methyltransferase-encoding gene ctdC was deleted to yield compound 24 without the dimethyl group at N-3’ suggesting that N-methylation of valine was catalyzed by CtdC and it might be the final step in the biosynthesis of 21R-citrinadin A (Figure S5 and Table S12). Collectively, these results unambiguously elucidated the late-stage modification in the biosynthesis of 10 that CtdR reduced the moiety to produce 16; then CtdV and CtdJ facilitated the continuous oxidation of 16 with strict regioselectivity and stereoselectivity to provide 23; the NRPS CtdD further assemble 23 with L-valine to yield 24; and finally, the methylation of 24 at N-3’ by CtdC lead to the production of 10 (Figure S16).
Eventually, we introduced eight genes including ctdY, ctdS, ctdG, ctdR, ctdV, ctdJ, ctdD, and ctdC simultaneously in A. nidulans accompanied by feeding substrate 20, resulting in the production of final compound 10 in the heterologous strain (Figure 4C). These results reveal that eight enzymes are indispensable and sufficient for the post-translational modification pathway from 20 to 10. In addition, we also found that most of the intermediates we isolated from the knockout strains constructed in this work contain an epoxy group in the isopentenyl side chain. Thus, we hypothesize that the deconstruction of the 2,5-diazabicyclo[2.2.2]octane happens after the epoxidation. This was also verified by incubation of an intermediate 14 without the epoxy group with CtdY-containing microsome and no conversion was observed (Figure S17). Thus, the functions of all the biosynthesis-related genes in ctd have been attributed, but we did not find any genes in ctd involved in the epoxidation of precursor 14 to form 20. The formation of the epoxy group may be related to the genes outside ctd BGC.
Conclusion
In this work, we characterized a distinct P450 CtdY by gene deletion, heterologous expression, and in vitro assays. We propose CtdY can specifically disrupt the amide bond and remove the carbonyl group in the 2,5-diazabicyclo[2.2.2]octane system to provide the 6/5/5/6/6 pentacyclic scaffold with the iminium cation moiety. Moreover, seven enzymes consisting of CtdS, CtdG, CtdR, CtdV, CtdJ, CtdD, and CtdC are verified to complete the subsequent post-translational modification to produce the final compound 21R-citrinadin A. The elucidation of CtdY provides a new enzymatic machinery for cleavage of amide bond, expands the scope of reactions catalyzed by fungal P450s, and lays the foundation for genome mining of indole alkaloids with the pentacyclic structure. The deep insight into the redox enzymes exhibits their potential as biocatalysts and highlights the late-stage diversification of fungal indole alkaloid biosynthesis.
Supplementary Material
Acknowledgments
We thank Prof. László Kürti and Young-Do Kwon for their assistance with HRMS experiments. This work was supported by the NIH grant R35GM138207 and the Robert A. Welch Foundation (C-1952) to X.G.
Footnotes
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/. General procedures, experimental details, and NMR spectra (PDF).
References
- (1).Klas KR; Kato H; Frisvad JC; Yu F; Newmister SA; Fraley AE; Sherman DH; Tsukamoto S; Williams RM Structural and Stereochemical Diversity in Prenylated Indole Alkaloids Containing the Bicyclo[2.2.2]Diazaoctane Ring System from Marine and Terrestrial Fungi. Nat. Prod. Rep. 2018, 35 (6), 532–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Mercado-Marin EV; Garcia-Reynaga P; Romminger S; Pimenta Eli. F.; Romney DK; Lodewyk MW; Williams DE; Andersen RJ; Miller SJ; Tantillo DJ; Berlinck RGS; Sarpong R. Total Synthesis and Isolation of Citrinalin and Cyclopiamine Congeners. Nature 2014, 509 (7500), 318–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Ding Y; Wet de JR; Cavalcoli J; Li S; Greshock TJ; Miller KA; Finefield JM; Sunderhaus JD; McAfoos TJ; Tsukamoto S; Williams RM; Sherman DH Genome-Based Characterization of Two Prenylation Steps in the Assembly of the Stephacidin and Notoamide Anticancer Agents in a Marine-Derived Aspergillus Sp. J. Am. Chem. Soc. 2010, 132 (36), 12733–12740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Li S; Finefield JM; Sunderhaus JD; McAfoos TJ; Williams RM; Sherman DH Biochemical Characterization of NotB as an FAD-Dependent Oxidase in the Biosynthesis of Notoamide Indole Alkaloids. J. Am. Chem. Soc. 2012, 134 (2), 788–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Kato H; Yoshida T; Tokue T; Nojiri Y; Hirota H; Ohta T; Williams RM; Tsukamoto S. Notoamides A–D: Prenylated Indole Alkaloids Isolated from a Marine-Derived Fungus,Aspergillus Sp. Angew. Chem. Int. Ed. 2007, 46 (13), 2254–2256. [DOI] [PubMed] [Google Scholar]
- (6).Birch AJ; Wright JJ The Brevianamides: A New Class of Fungal Alkaloid. J. Chem. Soc. D 1969, No. 12, 644b. [Google Scholar]
- (7).Qian-Cutrone J; Huang S; Shu Y-Z; Vyas D; Fairchild C; Menendez A; Krampitz K; Dalterio R; Klohr SE; Gao Q. Stephacidin A and B: Two Structurally Novel, Selective Inhibitors of the Testosterone-Dependent Prostate LNCaP Cells. J. Am. Chem. Soc. 2002, 124 (49), 14556–14557. [DOI] [PubMed] [Google Scholar]
- (8).Yamazaki M; Okuyama E; Kobayashi M; Inoue H. The Structure of Paraherquamide, a Toxic Metabolite From. Tetrahedron Lett. 1981, 22 (2), 135–136. [Google Scholar]
- (9).Martínez-Luis S; Rodríguez R; Acevedo L; González MC; Lira-Rocha A; Mata R. Malbrancheamide, a New Calmodulin Inhibitor from the Fungus Malbranchea Aurantiaca. Tetrahedron 2006, 62 (8), 1817–1822. [Google Scholar]
- (10).Lin Z; Wen J; Zhu T; Fang Y; Gu Q; Zhu W. Chrysogenamide A from an Endophytic Fungus Associated with Cistanche Deserticola and Its Neuroprotective Effect on SH-SY5Y Cells. J. Antibiot. 2008, 61 (2), 81–85. [DOI] [PubMed] [Google Scholar]
- (11).Fraley AE; Garcia-Borràs M; Tripathi A; Khare D; Mercado-Marin EV; Tran H; Dan Q; Webb GP; Watts KR; Crews P; Sarpong R; Williams RM; Smith JL; Houk KN; Sherman DH Function and Structure of MalA/MalA′, Iterative Halogenases for Late-Stage C–H Functionalization of Indole Alkaloids. J. Am. Chem. Soc. 2017, 139 (34), 12060–12068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Porter AEA; Sammes PG A Diels–Alder Reaction of Possible Biosynthetic Importance. J. Chem. Soc. D 1970, 0 (17), 1103a–1103a. [Google Scholar]
- (13).Dan Q; Newmister SA; Klas KR; Fraley AE; McAfoos TJ; Somoza AD; Sunderhaus JD; Ye Y; Shende VV; Yu F; Sanders JN; Brown WC; Zhao L; Paton RS; Houk KN; Smith JL; Sherman DH; Williams RM Fungal Indole Alkaloid Biogenesis through Evolution of a Bifunctional Reductase/Diels–Alderase. Nat. Chem. 2019, 11 (11), 972–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Ye Y; Du L; Zhang X; Newmister SA; McCauley M; Alegre-Requena JV; Zhang W; Mu S; Minami A; Fraley AE; Adrover-Castellano ML; Carney NA; Shende VV; Qi F; Oikawa H; Kato H; Tsukamoto S; Paton RS; Williams RM; Sherman DH; Li S. Fungal-Derived Brevianamide Assembly by a Stereoselective Semipinacolase. Nat. Catal. 2020, 3 (6), 497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Liu Z; Rivera S; Newmister SA; Sanders JN; Nie Q; Liu S; Zhao F; Ferrara JD; Shih H-W; Patil S; Xu W; Miller MD; Phillips GN; Houk KN; Sherman DH; Gao X. An NmrA-like Enzyme-Catalysed Redox-Mediated Diels–Alder Cycloaddition with Anti-Selectivity. Nat. Chem. 2023, 15 (4), 526–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Bond RF; Boeyens JCA; Holzapfel CW; Steyn PS Cyclopiamines A and B, Novel Oxindole Metabolites of Penicillium Cyclopium Westling. J. Chem. Soc., Perkin Trans. 1 1979, 1751. [Google Scholar]
- (17).Kushida N; Watanabe N; Okuda T; Yokoyama F; Gyobu Y; Yaguchi T. PF1270A, B and C, Novel Histamine H3 Receptor Ligands Produced by Penicillium Waksmanii PF1270. J. Antibiot. 2007, 60 (11), 667–673. [DOI] [PubMed] [Google Scholar]
- (18).Tsuda M; Kasai Y; Komatsu K; Sone T; Tanaka M; Mikami Y; Kobayashi J. Citrinadin A, a Novel Pentacyclic Alkaloid from Marine-Derived Fungus Penicillium Citrinum. Org. Lett. 2004, 6 (18), 3087–3089. [DOI] [PubMed] [Google Scholar]
- (19).Pimenta EF; Vita-Marques AM; Tininis A; Seleghim MHR; Sette LD; Veloso K; Ferreira AG; Williams DE; Patrick BO; Dalisay DS; Andersen RJ; Berlinck RGS Use of Experimental Design for the Optimization of the Production of New Secondary Metabolites by Two Penicillium Species. J. Nat. Prod. 2010, 73 (11), 1821–1832 [DOI] [PubMed] [Google Scholar]
- (20).Chen M; Liu C-T; Tang Y. Discovery and Biocatalytic Application of a PLP-Dependent Amino Acid γ-Substitution Enzyme That Catalyzes C–C Bond Formation. J. Am. Chem. Soc. 2020, 142 (23), 10506–10515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Liu Z; Zhao F; Zhao B; Yang J; Ferrara J; Sankaran B; Venkataram Prasad BV; Kundu BB; Phillips GN; Gao Y; Hu L; Zhu T; Gao X. Structural Basis of the Stereoselective Formation of the Spirooxindole Ring in the Biosynthesis of Citrinadins. Nat. Commun. 2021, 12 (1), 4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Gao P; Rahman Md. M.; Zamalloa A; Feliciano J; Szostak M. Classes of Amides That Undergo Selective N–C Amide Bond Activation: The Emergence of Ground-State Destabilization. J. Org. Chem. 2022, DOI: 10.1021/acs.joc.2c01094. [DOI] [PubMed] [Google Scholar]
- (23).Meng G; Zhang J; Szostak M. Acyclic Twisted Amides. Chem. Rev. 2021, 121 (20), 12746–12783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Szostak M; Aubé J. Chemistry of Bridged Lactams and Related Heterocycles. Chem. Rev. 2013, 113 (8), 5701–5765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Sonke T; Kaptein B. Hydrolysis of Amides. In Enzyme Catalysis in Organic Synthesis; Drauz K, Gröger H, May O, Eds.; Wiley, 2012; pp 561–650. [Google Scholar]
- (26).Ghuysen J-M Molecular Structures of Penicillin-Binding Proteins and β-Lactamases. Trends Microbiol. 1994, 2 (10), 372–380. [DOI] [PubMed] [Google Scholar]
- (27).Janetzko J; Trauger SA; Lazarus MB; Walker S. How the Glycosyltransferase OGT Catalyzes Amide Bond Cleavage. Nat. Chem. Biol. 2016, 12 (11), 899–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Zheng Y; Nair SK YcaO-Mediated ATP-Dependent Peptidase Activity in Ribosomal Peptide Biosynthesis. Nat. Chem. Biol. 2023, 19 (1), 111–119. [DOI] [PubMed] [Google Scholar]
- (29).Philmus B; Abdelwahed S; Williams HJ; Fenwick MK; Ealick SE; Begley TP Identification of the Product of Toxoflavin Lyase: Degradation via a Baeyer–Villiger Oxidation. J. Am. Chem. Soc. 2012, 134 (11), 5326–5330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Bian Z; Marvin CC; Martin SF Enantioselective Total Synthesis of (−)-Citrinadin A and Revision of Its Stereochemical Structure. J. Am. Chem. Soc. 2013, 135 (30), 10886–10889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Catlett NL; Lee B-N; Yoder OC; Turgeon BG Split-Marker Recombination for Efficient Targeted Deletion of Fungal Genes. Fungal Genet. Rep. 2003, 50 (1), 9–11. [Google Scholar]
- (32).Gerhards N; Neubauer L; Tudzynski P; Li S-M Biosynthetic Pathways of Ergot Alkaloids. Toxins 2014, 6 (12), 3281–3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Chiang C-Y; Ohashi M; Tang Y. Deciphering Chemical Logic of Fungal Natural Product Biosynthesis through Heterologous Expression and Genome Mining. Nat. Prod. Rep. 2023, 40 (1), 89–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Tang M-C; Lin H-C; Li D; Zou Y; Li J; Xu W; Cacho RA; Hillenmeyer ME; Garg NK; Tang Y. Discovery of Unclustered Fungal Indole Diterpene Biosynthetic Pathways through Combinatorial Pathway Reassembly in Engineered Yeast. J. Am. Chem. Soc. 2015, 137 (43), 13724–13727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Krogh A; Larsson B; von Heijne G; Sonnhammer ELL Predicting Transmembrane Protein Topology with a Hidden Markov Model: Application to Complete Genomes11Edited by F. Cohen. J. Mol. Biol. 2001, 305 (3), 567–580. [DOI] [PubMed] [Google Scholar]
- (36).Jumper J; Evans R; Pritzel A; Green T; Figurnov M; Ronneberger O; Tunyasuvunakool K; Bates R; Žídek A; Potapenko A; Bridgland A; Meyer C; Kohl SAA; Ballard AJ; Cowie A; Romera-Paredes B; Nikolov S; Jain R; Adler J; Back T; Petersen S; Reiman D; Clancy E; Zielinski M; Steinegger M; Pacholska M; Berghammer T; Bodenstein S; Silver D; Vinyals O; Senior AW; Kavukcuoglu K; Kohli P; Hassabis D. Highly Accurate Protein Structure Prediction with AlphaFold. Nature 2021, 596 (7873), 583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Yang J; Yan R; Roy A; Xu D; Poisson J; Zhang Y. The I-TASSER Suite: Protein Structure and Function Prediction. Nat. Methods 2015, 12 (1), 7–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Stourac J; Vavra O; Kokkonen P; Filipovic J; Pinto G; Brezovsky J; Damborsky J; Bednar D. Caver Web 1.0: Identification of Tunnels and Channels in Proteins and Analysis of Ligand Transport. Fungal Genet. Rep. 2019, 47 (W1), W414–W422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Gora A; Brezovsky J; Damborsky J. Gates of Enzymes. Chem. Rev. 2013, 113 (8), 5871–5923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Zhang X; Guo J; Cheng F; Li S. Cytochrome P450 Enzymes in Fungal Natural Product Biosynthesis. Nat. Prod. Rep. 2021, 38 (6), 1072–1099. [DOI] [PubMed] [Google Scholar]
- (41).Guengerich FP Mechanisms of Cytochrome P450-Catalyzed Oxidations. ACS Catal. 2018, 8 (12), 10964–10976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Guengerich FP; Yoshimoto FK Formation and Cleavage of C–C Bonds by Enzymatic Oxidation–Reduction Reactions. Chem. Rev. 2018, 118 (14), 6573–6655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Lin H-C; Tsunematsu Y; Dhingra S; Xu W; Fukutomi M; Chooi Y-H; Cane DE; Calvo AM; Watanabe K; Tang Y. Generation of Complexity in Fungal Terpene Biosynthesis: Discovery of a Multifunctional Cytochrome P450 in the Fumagillin Pathway. J. Am. Chem. Soc. 2014, 136 (11), 4426–4436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Zhang X; Li S. Expansion of Chemical Space for Natural Products by Uncommon P450 Reactions. Nat. Prod. Rep. 2017, 34 (9), 1061–1089. [DOI] [PubMed] [Google Scholar]
- (45).Swinney DC; Mak AY Androgen Formation by Cytochrome P450 CYP17. Solvent Isotope Effect and PL Studies Suggest a Role for Protons in the Regulation of Oxene versus Peroxide Chemistry. Biochemistry 1994, 33 (8), 2185–2190. [DOI] [PubMed] [Google Scholar]
- (46).Miller JC; Lee JHZ; Mclean MA; Chao RR; Stone ISJ; Pukala TL; Bruning JB; De Voss JJ; Schuler MA; Sligar SG; Bell SG Engineering C–C Bond Cleavage Activity into a P450 Monooxygenase Enzyme. J. Am. Chem. Soc. 2023, 145 (16), 9207–9222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Chen Q; Yuan G; Yuan T; Zeng H; Zou Z-R; Tu Z; Gao J; Zou Y. Set of Cytochrome P450s Cooperatively Catalyzes the Synthesis of a Highly Oxidized and Rearranged Diterpene-Class Sordarinane Architecture. J. Am. Chem. Soc. 2022, 144 (8), 3580–3589. [DOI] [PubMed] [Google Scholar]
- (48).Saikia S; Parker EJ; Koulman A; Scott B. Defining Paxilline Biosynthesis in Penicillium Paxilli. J. Biol. Chem. 2007, 282 (23), 16829–16837. [DOI] [PubMed] [Google Scholar]
- (49).Zang W; Zheng X. Structure and Functions of Cellular Redox Sensor HSCARG/NMRAL1, a Linkage among Redox Status, Innate Immunity, DNA Damage Response, and Cancer. Free Radic. Biol. Med. 2020, 160, 768–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Andrianopoulos A; Kourambas S; Sharp JA; Davis MA; Hynes MJ Characterization of the Aspergillus Nidulans NmrA Gene Involved in Nitrogen Metabolite Repression. J. Bacteriol. 1998, 180 (7), 1973–1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Zheng X; Dai X; Zhao Y; Chen Q; Lu F; Yao D; Yu Q; Liu X; Zhang C; Gu X; Luo M. Restructuring of the Dinucleotide-Binding Fold in an NADP(H) Sensor Protein. Proc. Natl. Acad. Sci. U.S.A. 2007, 104 (21), 8809–8814. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.