Abstract
Introduction
Colorectal cancer is the third most common cause of cancer death. Rectal cancer makes up a third of all colorectal cases. Treatment for locally advanced rectal cancer includes chemoradiation followed by surgery. We have previously identified ST6GAL1 as a cause of resistance to chemoradiation in vitro and hypothesized that it would be correlated with poor response in human derived models and human tissues.
Methods
Five organoid models were created from primary human rectal cancers and ST6GAL1 was knocked down via lentivirus transduction in one model. ST6GAL1 and Cleaved Caspase-3 (CC3) were assessed after chemoradiation via immunostaining. A tissue microarray (TMA) was created from twenty-six patients who underwent chemoradiation and had pre- and post-treatment specimens of rectal adenocarcinoma available at our institution. Immunohistochemistry was performed for ST6GAL1 and percent positive cancer cell staining was assessed and correlation with pathological grade of response was measured.
Results
Organoid models were treated with chemoradiation and both ST6GAL1 mRNA and protein significantly increased after treatment. The organoid model targeted with ST6GAL1 knockdown was found to have increased CC3 after treatment. In the tissue microarray, 42 percent of patient samples had an increase in percent tumor cell staining for ST6GAL1 after treatment. Post-treatment percent staining was associated with a worse grade of treatment response (p = 0.01) and increased staining post-treatment compared to pre-treatment was also associated with a worse response (p = 0.01).
Conclusion
ST6GAL1 is associated with resistance to treatment in human rectal cancer and knockdown in an organoid model abrogated resistance to apoptosis caused by chemoradiation.
Keywords: ST6GAL1, Radiation resistance, Sialylation, Rectal cancer
Introduction
Colorectal cancer is the third most common cause of cancer death in both men and women. Rectal cancer makes up a third of all colorectal cases and is rising [1]. Treatment for locally advanced rectal cancer includes chemoradiation followed by surgery but response to treatment is variable. About 20 percent of patients have a complete response to chemoradiation, likely do not need surgery, and have been found to exhibit higher 5-year disease-free survival rates [2,3]. While this is promising, we have little understanding of predictors of response.
Altered glycosylation, including hypersialylation, has been identified as one of the hallmarks of cancer [4,5]. ST6GAL1 is the primary enzyme responsible for α2-6 sialylation of N-glycans on select glycoproteins and is increased relative to normal tissues in multiple types of cancer including gastric, pancreatic and colon cancers [6,7]. ST6GAL1 has been associated with tumor progression and therapeutic resistance in pancreatic cancer models [8]. We recently showed that knockdown (KD) of ST6GAL1 decreases resistance to chemoradiation in vitro in colorectal adenocarcinoma using multiple cell lines [9]. We found that the mechanism of sialylation-mediated resistance is likely via TNFR1 sialylation after treatment with chemoradiation and that this sialylation decreases apoptosis.
To determine how relevant these findings are to patients with rectal cancer, we wanted to investigate ST6GAL1 in human organoid models and human rectal cancer samples and assess whether the role we identified in vitro as a mediator of treatment resistance would be validated in vivo. We hypothesized that increased ST6GAL1 would mediate resistance in organoid models and be associated with poor pathologic grade of response in human samples. We found that ST6GAL1 mRNA and protein increased in primary human rectal cancer organoids following treatment. Organoids with knockdown of ST6GAL1 had increased evidence of apoptosis after treatment compared to control organoids. Additionally, in a tissue microarray (TMA) made from a matched cohort of pre- and post-treatment human tumor samples, we found that ST6GAL1 staining is increased in tumor cells in post-treatment surgical specimens. Increased ST6GAL1 staining comparing pre- to post- treatment matched specimens was correlated with worse grade of response. Taken together, these data indicate that ST6GAL1 is not only a mediator of resistance to therapy in vitro but also appears to cause resistance to therapy in vivo. Improved understanding of mechanisms of therapeutic resistance and clinical predictors of treatment response are sorely needed for rectal cancer patients as we move toward consideration of omitting surgery [10].
Methods
Organoid Studies
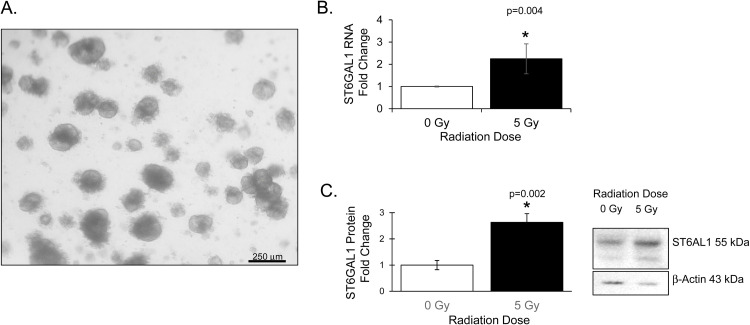
Organoid models were created from surgical samples from rectal cancer patients according to our Institutional Review Board (IRB) approved prospective protocol. Patients were approached in colorectal surgery clinic and consented to allow use of left-over tissue at the time of surgical resection. After resection, samples were given to our research team by the UAB Tissue Biorepository. Samples were washed 3 times in Phosphate Buffered Saline (PBS, Corning) with 100 μg/ml Gentamicin (Gibco) and necrotic tissue excised. Tissue was minced into 0.5 mm3 pieces and allowed to digest in 2 mg/ml Type 1 Collagenase (Gibco) for 45–60 minutes at 37C. Dulbecco's Modified Eagle Medium/Nutrient Mixture F12 Medium (DMEM/F12, Corning) wash buffer containing 10 % Fetal Bovine Serum (FBS, Sigma), 100 units/ml penicillin and 0.1 mg/ml streptomycin (1 % Pen/Strep, Gibco) and 10 μg/ml gentamicin was added and collected digested tissue was filtered through a 70 μm strainer. The filtrate was centrifuged at 250xg for 5 min at room temperature. The resulting pellet was resuspended in an equal volume of Matrigel (Corning). Once Matrigel solidified, cells were cultured with media containing 50 % Advanced Dulbecco's Modified Eagle's Medium (Adv. DMEM), 50 % L-WRN conditioned media (American Type Culture Collection [ATCC] CRL-3276) supplemented with 10 % FBS, 2mM GLUTamax (Gibco), 1 % Pen/Strep, 10 μM SB431542 (Fisher Scientific), 10 μM Y-27632 (Selleck Chemicals), and 50 μg/ml gentamicin. Media was changed every other day. Groups of organoids were harvested at 3 days post treatment for protein and RNA. For ST6GAL1 knockdown, NP26 organoids were plated at low density in Matrigel and transduced using shRNA from Mission Lentiviral Particles targeting ST6GAL1 or a non-mammalian control vector (Sigma). An MOI of 5 was used and antibiotic selection was done using puromycin. Once organoids were visibly growing, clones were pooled and ST6GAL1 mRNA and protein levels were checked to determine success of knockdown. Organoids were treated with 3 μM 5-fluorouracil (5FU, Selleck Chemicals) and irradiated at 5 Gy using an X-RAD 320 (Precision X-Ray, Inc) 48 h after plating. A brightfield image of an organoid model is shown in Fig. 1A.
Fig. 1.
ST6GAL1 mRNA levels and protein expression increase after chemoradiation in human organoid models. A. Brightfield image of a primary rectal cancer organoid model B. ST6GAL1 mRNA increases after chemoradiation. Representative graph of organoid model. Standard deviation error bars C. ST6GAL1 protein increases after chemoradiation. Representative graph of organoid model. Standard deviation error bars. Representative western blot image of organoid model. 0 Gy no chemoradiation, 5 Gy chemoradiation (5FU+5 Gy).
PCR and western blotting
PCR and Western blotting were performed on organoid models. Organoids were lysed with Radioimmunoprecipitation assay buffer (RIPA) containing Halt protease and phosphatase inhibitors (Fisher Scientific) or lysed for RNA using RNeasy Kit (Qiagen) following manufacturer's protocol. Protein concentration was quantified using BioRad DC Protein Assay. Samples were resolved by electrophoresis followed by protein transfer to Immobilon-P membranes (Millipore). Membranes were blocked in 5 % Nonfat dry milk (Fisher Scientific) or Bovine Serum Albumin (BSA, Sigma) in 1X Tris buffered saline (TBS, BioRad Laboratories) and 0.1 % Tween-20 (TBST). Blots were probed with 1:250 antibody against ST6GAL-1 (R&D Systems, #AF5924). Protein loading was verified using 1:5,000 β-Actin (Abcam). Membranes were incubated with anti-goat secondary antibody for ST6GAL-1 and imaged with a BioRad ChemiDoc with Image Lab Software using Immobilon Forte Western HRP Substrate (Millpore). RNA concentration was determined using a NanoDrop One (Thermo Fisher) and equal amounts of RNA were used to generate complementary DNA with the High-Capacity cDNA Reverse Transcription Kit (Fisher Scientific). qPCR was performed using a QuantStudio6 Flex (Applied Biosystems) instrument. TaqMan gene expression assay primers to ST6GAL1 and GAPDH and TaqMan Fast Advanced Master Mix (all Fisher Scientific) were used to generate reactions.
Cleaved caspase-3 immunofluorescent staining and assessment
Organoids were plated in Matrigel on sterile coverslips and allowed to grow 48 h prior to treatment with 3 μM 5FU and 5Gy. Twenty-four hours after chemoradiation, organoids were fixed by adding 10 % volume of well of 4 % paraformaldehyde (Fisher Scientific) directly to media and allowed to incubate at room temperature for 10 min. Media was aspirated and another 1.5 ml of 4 % paraformaldehyde added to each well and allowed to incubate for 20 min. Fixed organoids on coverslips were washed 3 times in 1XPBS to remove any remaining fixative and stored at 4C with fresh 1XPBS until ready for staining. For staining, organoids were washed with PBST (0.1 % Tween-20 in 1XPBS) 3 times for 5 min each and permeabilized for 15 minutes using PBSTX (PBST + 0.5 % Triton X-100). Blocking was for 1 hour with 3 % BSA in PBST and incubated overnight at 4C with 1:400 primary antibody against Cleaved Caspase-3 (CC3, Cell Signaling). Following overnight incubation organoids were washed 3 times with PBST for 5 minutes and incubated with Alexa Fluor 555 donkey anti-rabbit secondary antibody at 1:500 (Invitrogen) in 3 % BSA for 1 h at room temperature. Organoids were washed 2 times with PBST and counterstained with 4′, 6-diamidino-2-phenylindole (DAPI) 1:500 (Invitrogen) in 3 % BSA in PBST for 30 minutes. After 2 washes with PBST for 5 min, coverslips with organoids were washed one more time in 1XPBS. Coverslips were mounted using ProLong Diamond Antifade Mountant (Invitrogen) and imaged with support from UAB High Resolution Imaging Facility at a magnification of 60X on a Nikon A1R-HD confocal microscope or 4X using a Lionheart FX (Biotek). Fluorescence was quantified using Fiji - Image J (v2.14.0/1.54f) from NIH by obtaining amount of DAPI and red CC3 in each image. A minimum of 5 high powered (60X) images for each organoid model and treatment were quantified and experiments were repeated 3 times. The amount of nuclear DAPI (blue) and the amount of cleaved caspase 3 (red) was quantified per high powered field and each was added up for all fields assessed. The total cleaved caspase 3 staining was divided by the total nuclear DAPI staining and multiplied by 100 to get caspase relative to nuclei percent staining.
Tissue microarray
Twenty-six Stage 2 and 3 rectal cancer patients were identified on retrospective chart review who had a pre-treatment biopsy of rectal adenocarcinoma at our institution and underwent long course chemoradiation prior to surgical resection and had a post-treatment surgical resection at our institution. This treatment consisted of radiation treatment with 2 Gy of radiation 5 days per week for 5 weeks followed by a boost to the tumor bed the last week. Patients took 5-Fluorouracil (5FU) via infusion pump or oral capecitabine during radiation. Review of the medical record for gender, age, clinical stage, treatment, pathological stage, and pathological grade of response determined at the time the surgical pathology was originally assessed in accordance with the standard grading system [11]. Three TMAs were created by University of Alabama at Birmingham (UAB) Tissue Biorepository utilizing matched pre-treatment biopsies and post-treatment surgical specimens taking care to place samples from individual patients on the same TMA to optimize comparisons within patients. This retrospective study was approved by our Institutional Review Board (IRB).
ST6GAL1 Immunostaining
Human rectal adenocarcinoma TMA tissue sections were deparaffinized and rehydrated by washing in xylene and ethanol. Slides were placed in boiling Citric Acid Based antigen unmasking solution (Vector). The slides were washed, covered in BLOXALL (Vector Labs) for 5 min and 0.5 % Triton-X in PBS for 25 min. Slides were blocked with 2.5 % horse serum and incubated with primary ST6GAL1 antibody 1:100 overnight (R&D Systems, #AF5924). ImmPRESS horse anti-goat immunoglobulin G (IgG, Vector Labs) was used as secondary, with Vector Labs NovaRED peroxidase substrate solution used for primary stain for two minutes. TMA slides were counterstained with hematoxylin for 30 s, dehydrated and mounted with coverslips.
Histopathologic assessment of ST6GAL1
We assessed immunostaining in 26 human rectal adenocarcinoma specimens pre- and post- treatment. Each piece of tissue was assessed for cancer cells and ST6GAL1 staining by a GI pathologist (S.A.) blinded to tissue identification. In each piece, up to 3 fields were identified where ST6GAL1 staining was present. In each field all cancer cells were marked and cancer cells positive for ST6GAL1 staining were marked. Each were counted and a percent positive staining was generated for each sample ((ST5GAL1 positive cells/Total cancer cells)*100). At least 3 areas of tumor were assessed per tumor sample if available. For patients where more than one sample was available on the TMA, an average of the percent positive staining was used. For 5 patients where tumor was not identified in the specimen in the TMA, sections were cut from their original FFPE blocks and used for immunostaining and assessment.
Statistical analysis
Western blot images and photos were quantified using image J and analyzed using ANOVA and Student's t-test for statistical comparison. The pathologic grade of response from the original pathologic assessment in the medical record of the entire surgical resection specimen was used for the analysis. Percent positive staining for pre- and post-treatment samples as well as change in staining between pre- and post-treatment (matched post-treatment percent positive staining minus pre-treatment percent positive staining) samples was assessed for correlation with grade of response using the Kruskal Wallis test. Because Grade 0 is defined as complete pathologic response and thus no tumor cells are present in these post-treatment specimen, Post-treatment grade 0 samples were excluded from assessment of correlation of staining for ST6GAL1.
Results
ST6GAL1 Increases after chemoradiation in rectal cancer organoids
In our prior studies we grew and assessed a limited number of organoids grown from a rectal cancer PDX model [9]. For this study, we grew, and tested further organoids grown from primary human colorectal cancer surgical resection specimens (example organoid model, Fig. 1A). The tumor and patient characteristics are summarized in Table 1. The organoids were treated with radiation and 5FU and harvested 3 days post treatment. RNA and protein were isolated and assessed via qPCR and Western blotting. ST6GAL1 mRNA was significantly increased in 4 of 5 tested models and ST6GAL1 protein was significantly increased in 4 of 5 tested models (Table 1). The mRNA of a representative organoid model was increased by 2.20-fold (p = 0.004) and the protein by 2.5-fold (p = 0.002) as compared to vehicle treatment (Fig. 1B and C).
Table 1.
Characteristics of Tumors and Patients for Colorectal Cancer Organoids.
| ID | Patient Age at diagnosis/Sex | Tumor location | Tumor clinic/pathologic stage | Treatment prior to organoid creation | ST6GAL1 protein fold change after treatment | ST6GAL1 mRNA fold change after treatment |
|---|---|---|---|---|---|---|
| *NP26 | 45 years/M | Rectum | cT3N2/ypT3N2 | No | 2.2 | 2.5 |
| NP33 | 50 years/F | Rectum | cT3N2/ypT3N2 | No | 1.9 | 1.8 |
| UAB32 | 26 years/F | Rectum | cT3N1/ypT1N0 | Yes | 2.0 | 2.3 |
| UAB36 | 41 years/M | Ascending colon | pT4N2M1 | No | 2.2 | -0.47 |
| UAB65 | 48 years/F | Ascending colon | T2N0 | No | No Change | 5.0 |
indicates this organoid used for ST6GAL1 knockdown
Rectal cancer organoids with ST6GAL1 knockdown are less resistant to chemoradiation
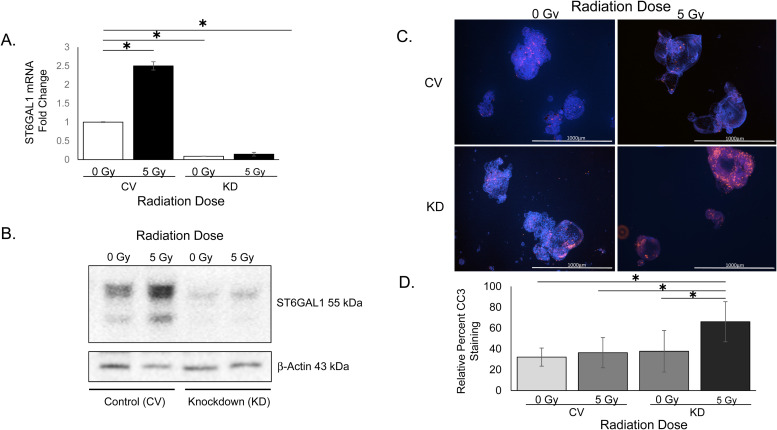
To determine if ST6GAL1 plays a role in chemoradiation resistance in our organoid models we knocked down ST6GAL1 in an organoid line (NP26). Organoids transduced with shRNA targeting ST6GAL1 were analyzed after being treated with 5 Gy radiation and 5FU and compared to control vector transduced organoids. ST6GAL1 KD was confirmed via quantitative PCR and western blot (Fig. 2A and B). To determine if ST6GAL1 KD influenced apoptosis after treatment, we used an antibody to cleaved caspase-3 (CC3) which is indicative of apoptosis. Organoids were cultured on coverslips and treated with 5 Gy radiation and 5FU and 24 hours later immunostaining was performed. Image staining quantified for each treatment group included 6.2 +/– 3.5 organoids per field to 7 +/– 1.6 organoids per field. Cleaved caspase 3 staining quantification revealed a significant increase in CC3 relative to DAPI (nuclei) in ST6GAL1 knockdown organoid models 24 hours after treatment compared to the control vector organoids (Fig. 2C and 2D, n = 3 separate experiments, p = 2.89×10−8).
Fig. 2.
Organoids transduced with shRNA for ST6GAL1 have decreased ST6GAL1 protein expression and are less resistant to apoptosis after chemoradiation. A. ST6GAL1 mRNA expression is decreased with targeted knockdown. B. ST6GAL1 protein is decreased with knock down and this persists even after chemoradiation. Representative western blot shown of organoid model. C. Representative images of one organoid model with Cleaved Caspase-3 staining for apoptosis. Images are 4x, scale bar 1mm. C. Representative analysis of 3 experiments of staining an organoid model for percent Cleaved Caspase-3. Minimum of 5 images were taken at high power (60X) for analysis. Standard deviation error bars. One-way ANOVA p < 0.05. CV control vector, KD ST6GAL1 knockdown organoid model.
Patient characteristics for TMA samples
In order to assess whether ST6GAL1 is associated with poor treatment response, we identified 26 patients who underwent chemoradiation prior to surgery and had a pre- and post-treatment specimen of stage 2 or 3 rectal adenocarcinoma available at our institution. Three TMAs were created with both pre-treatment biopsies and post-treatment surgical specimens. The average age of our cohort was 60.2 (+/−12.0) years old, with 69.2 % being male (Table 2). Abdominoperineal resection (APR) was the most common operation (57.7 %), with the remaining patients undergoing low anterior resection (LAR), indicating tumors were low in the rectum. Clinical stage was stage 3 for 73.1 % of patients and the rest were stage 2. On post-treatment surgical pathology, 7.7 % had a complete response (stage 0), 38.5 % were stage 1, 34.6 % were stage 2, and 19.0 % were stage 3. Pathologic grade of response was 7.7 % grade 0 (complete response), 23.1 % grade 1 (near complete response), 50.0 % grade 2 (partial response) and 19.2 % grade 3 (poor response) (Table 2).
Table 2.
Characteristics of Rectal cancer patients included in TMA.
| Overall (N=26) |
|
|---|---|
| Demographics | |
| Age, mean (stdev) | 60. 2 (12.0) |
| Male sex, N (%) | 18 (69.2 %) |
| Operation, N (%) | |
| APR | 15 (57.7 %) |
| LAR | 11 (42.3 %) |
| Cancer Stage Clinical, N (%) | |
| Stage 2 | 7 (26.9 %) |
| Stage 3 | 19 (73.1 %) |
| Cancer Stage Pathological, N (%) | |
| Stage 0 | 2 (7.7 %) |
| Stage 1 | 10 (38.5 %) |
| Stage 2 | 11 (42.3 %) |
| Stage 3 | 4 (15.4 %) |
| Grade of Response, N (%) | |
| Complete (0) | 2 (7.6 %) |
| Near complete (Grade 1) | 6 (23.1 %) |
| Partial (Grade 2) | 13 (50.0 %) |
| Poor (Grade 3) | 5 (19.2 %) |
Stage 0 = pathologic complete response
Association between ST6GAL1 and grade of response
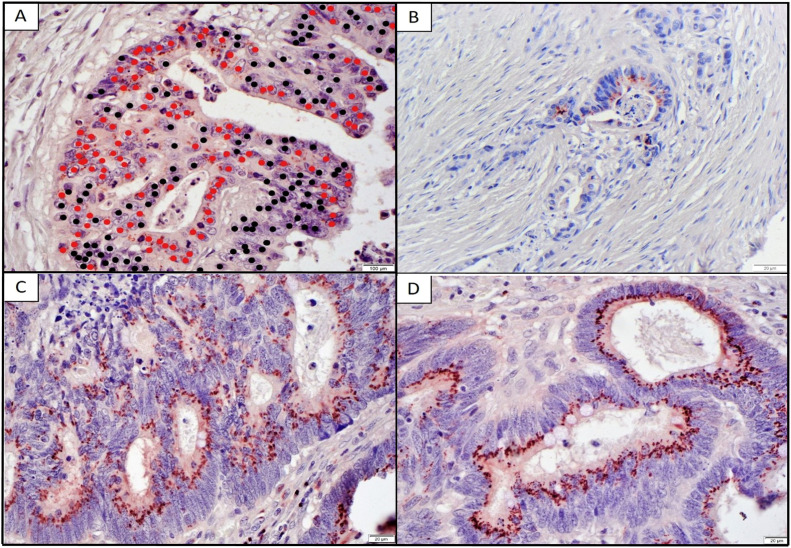
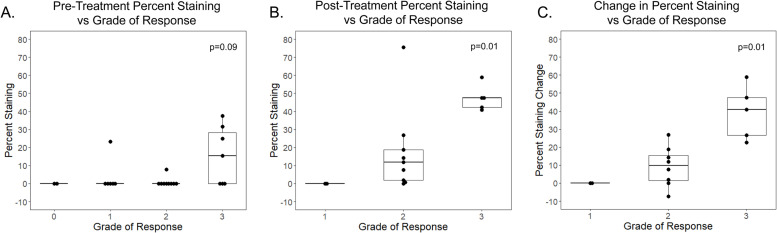
The TMAs were stained for ST6GAL1 using immunohistochemistry and assessed by a blinded gastrointestinal (GI) trained pathologist for percent tumor cells staining for ST6GAL1 by marking all cancer cells in 3 fields and cancer cells staining for ST6GAL1 (Fig. 3A). Representative examples of low percent tumor staining in a patient with a grade 1 (good) response and high percent staining in a grade 3 (poor) tumor response are shown in Fig. 3 B-D. Overall, 42 % of patients had an increase in percent ST6GAL1 staining comparing matched pre-vs-post treatment staining. Additionally, there was a significantly higher percentage of cancer cell staining after treatment (5.9 % pre-treatment vs 22.0 % post-treatment, p < 0.01). We assessed whether pre-ST6GAL1 percent staining, post-treatment percent staining, or change in percent ST6GAL1 staining pre-vs-post treatment were associated with pathologic grade of response to treatment (Fig. 4). For the assessments that included post-treatment staining, we excluded Grade 0 patients (complete pathologic response) because, by definition, these patients had no post-treatment tumor to assess. We found there was a trend toward an association between percent ST6GAL1 staining in pre-treatment specimens and grade of response, however, this was not significant (Fig. 4A, p = 0.09). Additionally, we found that change in ST6GAL1 staining between pre-and post-treatment time points and post-treatment percent staining were both significantly associated with higher (worse) grade of response (p=0.01 and 0.01 respectively, Fig. 4B and C).
Fig. 3.
ST6GAL-1 is increased in higher grade rectal cancer tumors on TMA. A. Example of cancer cells marked by red dots that are positive for ST6GAL-1 and black dots that are negative. Scale bar is 100 μm. B. Post-treatment specimen Grade 1 response. Scale bar is 20 μm. C. & D. Post-treatment specimens Grade 3 response. Scale bar is 20 μm. Magnification 40X. Figure photos are from tissue microarrays.
Fig. 4.
ST6GAL1 is increased in higher grade rectal cancer tumors on TMA. A. Percent ST6GAL1 stained pre-treatment patient samples compared to grade of response. B. Percent ST6GAL1 staining in chemoradiation post-treatment patients compared to grade of response. C. Difference between pre- and post-treatment patient grade of response.
Discussion
Guideline concordant care for locally advanced rectal cancer is chemoradiotherapy followed by surgical resection [12]. Patients who have a better grade of response (0 or 1) have improved short and long term outcomes with lower rates of local recurrence and future metastasis compared to patients who have worse grades of response (grade 2 or 3) [11,13,14]. Therefore, there is a critical need to identify mechanisms of therapeutic resistance as targets for future therapies. We previously identified the glycosyltransferase, ST6GAL1, as a mediator of treatment resistance in vitro via knockdown studies in cell lines and here we validate its role in vivo using human samples and human rectal cancer models. We found that post-treatment tumor samples with increased percent staining of cancer cells for ST6GAL1 had worse grade of response and that patients who had tumors that had an increase in ST6GAL1 comparing their post-treatment to their pre-treatment tumor samples also had worse outcomes. In correlative studies in human-derived organoid models, we found that rectal cancer organoids treated with chemoradiation also had increased ST6GAL1 mRNA and protein. We knocked down ST6GAL1 in a patient derived rectal cancer organoid model and found that following treatment, knock-down cells had increased CC3 immunostaining compared with controls, indicating enhanced apoptosis. Through a combination of the current work and our prior work, we have shown that ST6GAL1 is likely an important cause of radiation resistance in human rectal cancer.
Multiple studies have identified potential predictors of therapeutic resistance in vitro but few have been validated in vivo [15], [16], [17]. Rectal cancers are quite heterogeneous, a trait likely leading to multiple potential paths to resistance [18]. ST6GAL1 is a Golgi sialyltransferase that adds the bulky, negatively-charged sugar, sialic acid, in an α2-6 linkage to select N-glycosylated proteins. The addition of this sugar modulates the structure and function of various cell surface glycoproteins, leading to changes in intracellular signal transduction and gene expression. Prior research on ST6GAL1 by our group in vitro has shown that it causes resistance to 5FU and radiation through decreased apoptosis [9]. The mechanism is likely via sialylation of the TNFR1 and Fas death receptors, as sialylation of TNFR1 and Fas prevents receptor internalization and apoptotic signaling [19,20]. ST6GAL1 has also been shown to mediate resistance to gemcitabine in pancreatic cancer but this mechanism is via abrogation of DNA damage [8]. ST6GAL1 is known to be increased in multiple cancer types and its presence is linked with poor outcome [6,7]. ST6GAL1 is rarely mutated in cancers but is commonly found to have gene amplification and is primarily regulated epigenetically and transcriptionally [6]. Its regulation in rectal cancer has not been assessed.
There are very few biomarkers of outcome in rectal cancer and none have been validated prospectively. High carcinoma embryonic antigen (CEA) at diagnosis was found to be a predictor of poor response to neoadjuvant therapy in rectal cancer but there are no therapies specifically directed at this protein [21]. In a retrospective analysis, KRAS mutation and combined TP53 and KRAS mutations have also been associated with poor response to neoadjuvant treatment [22]. COASY is another potential predictor of response identified in a retrospective cohort and further assessed mechanistically in vitro which was shown to mediate response via PI3K [15]. IGF2 and L1CAM were identified as potential pretreatment biomarkers of poor response in a recent sequencing study of rectal tumor collected as part of multiple randomized trials of neoadjuvant treatment in rectal cancer but neither have been validated prospectively [23]
Our current study has multiple limitations. First, we were only able to identify 26 patients at our institution with matched pre-and-post treatment samples available for assessment. Because we are a referral center, many rectal cancer patients we treat have their initial diagnostic biopsy outside our institution. Additionally, although post-treatment and change in percent ST6GAL1 tumor staining were associated with grade of response to treatment, pre-treatment percent tumor ST6GAL1 staining was not significantly associated with grade of response. While this may mean that ST6GAL1 is not a pre-treatment biomarker of therapeutic response, it is also possible that an association between response and pre-treatment ST6GAL1 could be identified in a larger cohort since our study was limited by small samples size. It is also possible that ST6GAL1 may mediate response to treatment but require stimulation (by treatment) to cause ST6GAL1 upregulation, with the ability for a tumor to increase expression being dependent upon some yet to be discovered factor. Our organoid models all expressed ST6GAL1 and a higher percentage of them had an increase after treatment than we identified in the TMA (80 % vs 42 %). This is not necesarily surprising given that not all tumors can be grown as organoids and that this process likely selects for more robust tumors.
Taken together, our findings indicate that ST6GAL1 is likely an important mediator of treatment resistance in vivo and further study of its regulation and potential targets for treatment may improve therapeutic outcome.
Conflicts of interest and source of funding
No conflicts of interest. KH funded by National Institutes of Health K08CA190645 and 1R01CA265981; MS was supported by the National Human Genome Research Institute of the National Institutes of Health under Award Number 1T32HG008961. SLB funded by U01CA233581 and R01CA225177.
CRediT authorship contribution statement
Mary Smithson: Investigation, Methodology, Writing – original draft, Funding acquisition. Sameer Al Diffalha: Formal analysis, Investigation, Methodology, Writing – original draft. Regina K. Irwin: Investigation, Methodology, Writing – original draft, Writing – review & editing. Gregory Williams: Investigation, Methodology, Writing – original draft. M. Chandler McLeod: Formal analysis, Methodology. Vivek Somasundaram: Investigation, Methodology. Susan L. Bellis: Investigation, Funding acquisition. Karin M. Hardiman: Conceptualization, Data curation, Funding acquisition, Project administration, Supervision, Visualization, Writing – original draft, Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We would like to acknowledge Arundhati Sawant who screened a list of patients generated by our institutional tumor registry to identify patients who had a pre-treatment biopsy, underwent chemoradiotherapy, and then underwent surgery post-treatment all at our institution. This research was supported by the High Resolution Imaging Facility (HRIF) Service Center which is an institutional core at University of Alabama at Birmingham. HRIF is supported by the Office of the Vice President of Research and Development and the following grant: Cancer Center Support Grant P30 CA013148. Also supporting this research was University of Alabama at Birmingham Tissue Procurement Shared Resource (TPRO) and Tissue Biorepository (TBR) for providing tissue and histology services under award 3P30CA013148-49 from UAB Comprehensive Cancer Center Support Grant.
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2019. CA Cancer J. Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2.Maas M., Nelemans P.J., Valentini V., Das P., Rödel C., Kuo L.-J., Calvo F.A., García-Aguilar J., Glynne-Jones R., Haustermans K., Mohiuddin M., Pucciarelli S., Small W., Suárez J., Theodoropoulos G., Biondo S., Beets-Tan R.G.H., Beets G.L. Long-term outcome in patients with a pathological complete response after chemoradiation for rectal cancer: a pooled analysis of individual patient data. Lancet Oncol. 2010;11:835–844. doi: 10.1016/S1470-2045(10)70172-8. [DOI] [PubMed] [Google Scholar]
- 3.Smith J.J., Chow O.S., Gollub M.J., Nash G.M., Temple L.K., Weiser M.R., Guillem J.G., Paty P.B., Avila K., Garcia-Aguilar J., Rectal Cancer C. Organ Preservation in Rectal Adenocarcinoma: a phase II randomized controlled trial evaluating 3-year disease-free survival in patients with locally advanced rectal cancer treated with chemoradiation plus induction or consolidation chemotherapy, and total mesorectal excision or nonoperative management. BMC. Cancer. 2015;15:767. doi: 10.1186/s12885-015-1632-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Munkley J., Elliott D.J. Hallmarks of glycosylation in cancer. Oncotarget. 2016;7:35478–35489. doi: 10.18632/oncotarget.8155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bellis S.L., Reis C.A., Varki A., Kannagi R., Stanley P. In: Essentials of Glycobiology. 4th Ed. Varki A., Cummings R.D., Esko J.D., Stanley P., Hart G.W., Aebi M., Mohnen D., Kinoshita T., Packer N.H., Prestegard J.H., Schnaar R.L., Seeberger P.H., editors. Cold Spring Harbor; NY: 2022. Glycosylation Changes in Cancer; pp. 631–644. [Google Scholar]
- 6.Dorsett K.A., Marciel M.P., Hwang J., Ankenbauer K.E., Bhalerao N., Bellis S.L. Regulation of ST6GAL1 sialyltransferase expression in cancer cells. Glycobiology. 2021;31:530–539. doi: 10.1093/glycob/cwaa110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garnham R., Scott E., Livermore K.E., Munkley J. ST6GAL1: a key player in cancer. Oncol. Lett. 2019;18:983–989. doi: 10.3892/ol.2019.10458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chakraborty A., Dorsett K.A., Trummell H.Q., Yang E.S., Oliver P.G., Bonner J.A., Buchsbaum D.J., Bellis S.L. ST6Gal-I sialyltransferase promotes chemoresistance in pancreatic ductal adenocarcinoma by abrogating gemcitabine-mediated DNA damage. J. Biol. Chem. 2018;293:984–994. doi: 10.1074/jbc.M117.808584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smithson M., Irwin R., Williams G., Alexander K.L., Smythies L.E., Nearing M., McLeod M.C., Al Diffalha S., Bellis S.L., Hardiman K.M. Sialyltransferase ST6GAL-1 mediates resistance to chemoradiation in rectal cancer. J. Biol. Chem. 2022 doi: 10.1016/j.jbc.2022.101594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garcia-Aguilar J., Patil S., Gollub M.J., Kim J.K., Yuval J.B., Thompson H.M., Verheij F.S., Omer D.M., Lee M., Dunne R.F., Marcet J., Cataldo P., Polite B., Herzig D.O., Liska D., Oommen S., Friel C.M., Ternent C., Coveler A.L., Hunt S., Gregory A., Varma M.G., Bello B.L., Carmichael J.C., Krauss J., Gleisner A., Paty P.B., Weiser M.R., Nash G.M., Pappou E., Guillem J.G., Temple L., Wei I.H., Widmar M., Lin S., Segal N.H., Cercek A., Yaeger R., Smith J.J., Goodman K.A., Wu A.J., Saltz L.B. Organ preservation in patients with rectal adenocarcinoma treated with total neoadjuvant therapy. J. Clin. Oncol. 2022;40:2546–2556. doi: 10.1200/JCO.22.00032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beddy D., Hyland J.M., Winter D.C., Lim C., White A., Moriarty M., Armstrong J., Fennelly D., Gibbons D., Sheahan K. A simplified tumor regression grade correlates with survival in locally advanced rectal carcinoma treated with neoadjuvant chemoradiotherapy. Ann. Surg. Oncol. 2008;15:3471–3477. doi: 10.1245/s10434-008-0149-y. [DOI] [PubMed] [Google Scholar]
- 12.NCCN Clinical Practice Guidelines in Oncology: Rectal Cancer. Version 3.2022 Ed.
- 13.Mace A.G., Pai R.K., Stocchi L., Kalady M.F. American joint committee on cancer and college of American pathologists regression grade: a new prognostic factor in rectal cancer. Dis. Colon. Rectum. 2015;58:32–44. doi: 10.1097/DCR.0000000000000266. [DOI] [PubMed] [Google Scholar]
- 14.Vecchio F.M., Valentini V., Minsky B.D., Padula G.D., Venkatraman E.S., Balducci M., Micciche F., Ricci R., Morganti A.G., Gambacorta M.A., Maurizi F., Coco C. The relationship of pathologic tumor regression grade (TRG) and outcomes after preoperative therapy in rectal cancer. Int. J. Radiat. Oncol. Biol. Phys. 2005;62:752–760. doi: 10.1016/j.ijrobp.2004.11.017. [DOI] [PubMed] [Google Scholar]
- 15.Ferrandon S., DeVecchio J., Duraes L., Chouhan H., Karagkounis G., Davenport J., Orloff M., Liska D., Kalady M.F. CoA synthase (COASY) mediates radiation resistance via PI3K signaling in rectal cancer. Cancer Res. 2020;80:334–346. doi: 10.1158/0008-5472.CAN-19-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kamran S.C., Lennerz J.K., Margolis C.A., Liu D., Reardon B., Wankowicz S.A., Van Seventer E.E., Tracy A., Wo J.Y., Carter S.L., Willers H., Corcoran R.B., Hong T.S., Van Allen E.M. Integrative molecular characterization of resistance to neoadjuvant chemoradiation in rectal cancer. Clin. Cancer Res. 2019;25:5561–5571. [Google Scholar]
- 17.Wanigasooriya K., Barros-Silva J.D., Tee L., El-Asrag M.E., Stodolna A., Pickles O.J., Stockton J., Bryer C., Hoare R., Whalley C.M., Tyler R., Sillo T., Yau C., Ismail T., Beggs A.D. Patient derived organoids confirm that PI3K/AKT signalling is an escape pathway for radioresistance and a target for therapy in rectal cancer. Front. Oncol. 2022;12 doi: 10.3389/fonc.2022.920444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frydrych L.M., Ulintz P., Bankhead A., Sifuentes C., Greenson J., Maguire L., Irwin R., Fearon E.R., Hardiman K.M. Rectal cancer sub-clones respond differentially to neoadjuvant therapy. Neoplasia. 2019;21:1051–1062. doi: 10.1016/j.neo.2019.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holdbrooks A.T., Britain C.M., Bellis S.L. ST6Gal-I sialyltransferase promotes tumor necrosis factor (TNF)-mediated cancer cell survival via sialylation of the TNF receptor 1 (TNFR1) death receptor. J. Biol. Chem. 2018;293:1610–1622. doi: 10.1074/jbc.M117.801480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Swindall A.F., Bellis S.L. Sialylation of the Fas death receptor by ST6Gal-I provides protection against Fas-mediated apoptosis in colon carcinoma cells. J. Biol. Chem. 2011;286:22982–22990. doi: 10.1074/jbc.M110.211375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park Y.A., Sohn S.K., Seong J., Baik S.H., Lee K.Y., Kim N.K., Cho C.W. Serum CEA as a predictor for the response to preoperative chemoradiation in rectal cancer. J. Surg. Oncol. 2006;93:145–150. doi: 10.1002/jso.20320. [DOI] [PubMed] [Google Scholar]
- 22.Chow O.S., Kuk D., Keskin M., Smith J.J., Camacho N., Pelossof R., Chen C.T., Chen Z., Avila K., Weiser M.R., Berger M.F., Patil S., Bergsland E., Garcia-Aguilar J. KRAS and combined KRAS/TP53 mutations in locally advanced rectal cancer are independently associated with decreased response to neoadjuvant therapy. Ann. Surg. Oncol. 2016;23:2548–2555. doi: 10.1245/s10434-016-5205-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chatila W.K., Kim J.K., Walch H., Marco M.R., Chen C.T., Wu F., Omer D.M., Khalil D.N., Ganesh K., Qu X., Luthra A., Choi S.H., Ho Y.J., Kundra R., Groves K.I., Chow O.S., Cercek A., Weiser M.R., Widmar M., Wei I.H., Pappou E.P., Nash G.M., Paty P.B., Shi Q., Vakiani E., Duygu Selcuklu S., Donoghue M.T.A., Solit D.B., Berger M.F., Shia J., Pelossof R., Romesser P.B., Yaeger R., Smith J.J., Schultz N., Sanchez-Vega F., Garcia-Aguilar J. Genomic and transcriptomic determinants of response to neoadjuvant therapy in rectal cancer. Nat. Med. 2022;28:1646–1655. doi: 10.1038/s41591-022-01930-z. [DOI] [PMC free article] [PubMed] [Google Scholar]