Abstract
Lysine dimethylation (Kme2) is a crucial post-translational modification (PTM) that regulates biological processes and is implicated in diseases. There is significant interest in globally identifying these methylation marks. Unfortunately, this remains challenging due to the lack of robust technologies for selectively labeling Kme2. To address this, we present a chemical method named tertiary amine coupling by oxidation (TACO). This method selectively modifies Kme2 to aldehydes using Selectfluor and a base. The resulting aldehydes from Kme2 were then functionalized using reductive amination, thiolamine, and oxime chemistry. We successfully demonstrated the versatility of TACO in selectively labeling Kme2 peptides and proteins in complex cell lysate mixtures with varying payloads, including affinity tags and fluorophores. We further showed the application of TACO chemistry for the identification of Kme2 sites at a single-molecule level by fluorosequencing. We discovered novel 30 Kme2 sites, in addition to previously known 5 Kme2 sites, by proteomics analysis of TACO-modified nuclear extracts. Our work establishes a unique strategy for covalently modifying Kme2, facilitating the global identification of low-abundance Kme2-PTMs and their sites within complex cell lysate mixtures.
Introduction
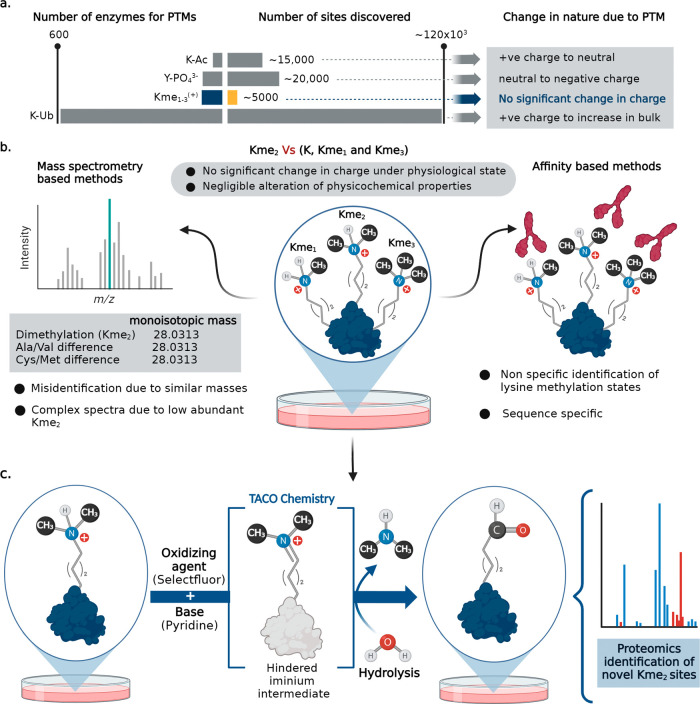
Lysine dimethylation (Kme2) dynamics constitute a crucial epigenetic event heavily involved in gene expression regulation and various biological processes.1−3 Despite increasing interest in this post-translational modification (PTM) and its pivotal role in diverse biological events and diseases beyond chromatin organization, its global identification remains elusive (Figure 1a). This is largely due to the lack of pan-specific chemical tools for labeling dimethyllysine (Kme2). Attempts to identify and characterize lysine dimethylation Kme2 have relied heavily on the use of affinity reagents such as antibodies and methyl-binding domains, MBD.4−7 However, these reagents fail to globally detect Kme2 sites due to their sequence-specific nature and limited specificity for different lysine methylation states. Antibodies are further constrained by the unequal abundance of lysine methylation states, reducing the fidelity in measuring methylation levels (Figure 1b). These challenges can be partially attributed to no significant change in the charge and negligible change in the bulk, hydrophobicity, and physicochemical properties of lysine upon the addition of methyl groups (Figure 1b).4,8−10 Furthermore, affinity-based methods are unable to detect trypsin-digested Kme2 fragments because they require flanking amino acids on either side of Kme2 for efficient recognition.11,12 Alternatively, mass spectrometry (MS)-based proteomics for detecting these low-abundant Kme2 PTMs require additional modification to reduce sample complexity and minimize false positive due to misassignment resulting from amino acid combinations with similar masses to two methyl groups (e.g., Ala vs Val and Cys vs Met) (Figure 1b).10,13,14 Collectively, none of the existing methods can selectively label Kme2 in a specific manner, underscoring the necessity for robust chemical methods for this purpose. Herein, we introduce a pioneering chemical strategy, termed tertiary amine coupling by oxidation (TACO), designed for the selective labeling of tertiary amines (Kme2) to aldehydes regardless of the surrounding amino acid sequence (Figure 1c).15−18 We demonstrated the broad applicability of TACO chemistry by selectively labeling Kme2 on peptides, proteins, nucleosomes, and whole cell lysates using various payloads like affinity tags and fluorophores, independent of the sequence, adjacent PTMs, or multiple Kme2 modifications.
Figure 1.
Different lysine methylation states and methods to profile dimethyl lysine. (a) Number of lysine methylation states discovered so far are much lower than enzymes responsible for such PTMs. The major challenge in identifying lysine methylation is due to the negligible change in the charge and physicochemical properties. (b) Limitations of the affinity-based method and MS in identifying lysine methylation. (c) This work: Development of the oxidative coupling chemical reaction for the selective modification of Kme2 by hydrolysis of iminium ions generating clickable aldehyde sites for identification by proteomics and fluorosequencing.
We demonstrated the potential of TACO chemistry in identifying Kme2 sites at the single molecule level through fluorosequencing. Additionally, we applied TACO chemistry to label and enrich Kme2 proteins, subsequently conducting a proteomic analysis on the labeled samples. This led to the discovery of 30 novel Kme2 sites. Notably, there are no chemical methods available in the literature for the site-selective labeling of Kme2. The capability to selectively label Kme2 sites using various tags in a complex mixture renders this technology exceptionally valuable for identifying and quantifying Kme2 biomarkers across diverse biological samples.
Results and Discussion
Development of TACO
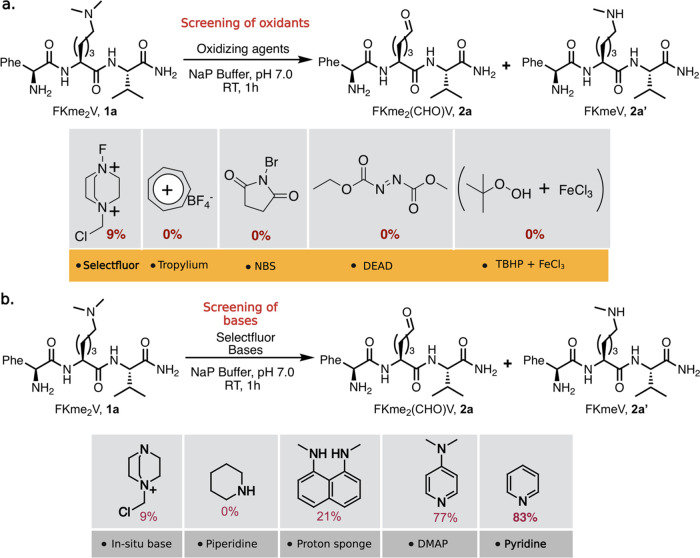
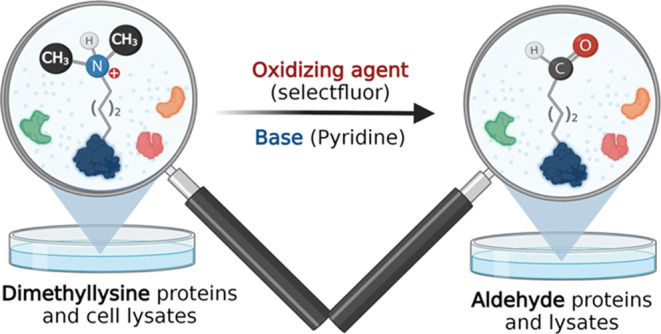
The tertiary amine coupling by oxidation (TACO) reaction capitalizes on the tendency of tertiary amines to generate highly electrophilic iminium ions under oxidative conditions. These ions undergo rapid hydrolysis, leading to the robust conversion of tertiary amines to aldehydes under physiological conditions (Figure 1c).15,19, We explored a variety of oxidizing reagents such as Selectfluor, tropylium tetrafluoroborate, N-bromo succinimide (NBS), diethyl azodicarboxylate (DEAD), and tBuOOH (TBHP) with FeCl3 under hydrolytic conditions on a model peptide FKme2V 1a (Figures 2a and Supporting Figure 1). Of all the reactions, Selectfluor (2 equiv) resulted in the formation of an aldehyde product, FKme2(CHO)V 2a, with 9% conversion in 1 h in sodium phosphate buffer (10 mM, pH 7) at room temperature. However, this reaction also generated small amounts of monomethyllysine FKme1V 2a’, which coeluted with the starting peptide FKme2V 1a as determined by LCMS (Supporting Figure 1). We postulated that the formation of aldehyde 2a and monomethyllysine 2a’ resulted from the nucleophilic attack of water on the more hindered and less hindered iminium ion, respectively (Supporting Figure 2). We did not observe the formation of any aldehyde product in the presence of other oxidants such as tropylium, NBS, DEAD, and (TBHP + FeCl3), due to their poor water compatibility and instability in aqueous conditions. To enhance the conversion of the dimethyl lysine peptide to the peptide aldehyde, we introduced bases stronger than those generated in situ by Selectfluor. Screening various bases such as piperidine, proton sponge, DMAP, and pyridine, we observed high conversion to the peptide aldehyde product FKme2(CHO)V 2a with DMAP (77%) and pyridine (83%) (Figures 2b and Supporting Figure 3). We attribute the high conversion to the planarity of pyridine and DMAP that enables the abstraction of the hindered methylene proton of dimethyl lysine, resulting in the formation of iminium ions followed by hydrolysis to generate an aldehyde. Other bases such as piperidine, DABCO-like base from Selectfluor, and proton sponge are too bulky to remove the proton from the hindered carbon of dimethyl lysine. Next, we generated high amounts of peptide aldehyde (OAc)FKme2(CHO)V 2b from peptide (OAc)FKme2V 1b under optimized reaction conditions and characterized it by NMR (Supporting Figure 4).
Figure 2.
Optimization of the TACO chemistry. (a) Development of the TACO reaction for modification of dimethyllysine Kme2 in peptide FKme2V 1a by exploring varying oxidizing reagents. (b) Evaluation of bases and optimization to the peptide aldehyde FKme2(CHO)V 2a. Reaction conditions: To 6–10 mM of dimethyllysine peptide in 300 μL of 10 mM sodium phosphate buffer (NaP, pH 7.0) were added a base (5 equiv) and oxidizing reagent (2 equiv), and the reaction mixture was stirred for 1 h at room temperature.
Chemoselectivity Studies for Peptide Aldehyde
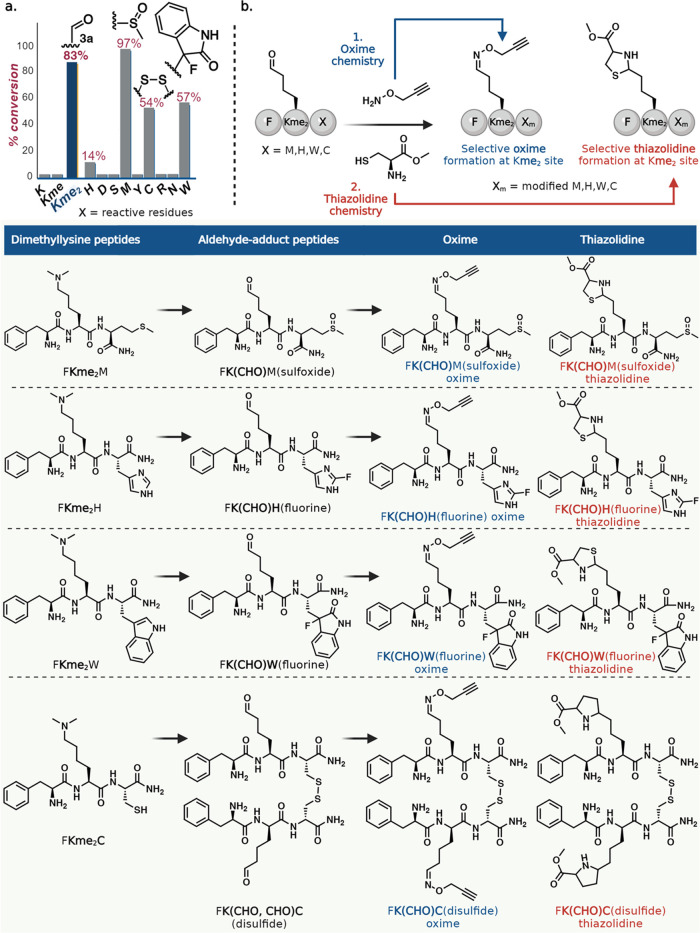
Chemoselectivity studies for peptide aldehyde revealed the specificity of the TACO reaction for generating aldehyde products with Kme2 only, as demonstrated by varying peptides FXV with different lysine methylation states (where X = K, Kme, and Kme2) and reactive amino acids (where X = H, D, S, M, Y, C, R, N, W) (Figures 3a and Supporting Figure 5). Our observations indicated the oxidation of Met and Cys, fluorination of Trp and His, and no formation of quinone and the ipso-fluorinated product with tyrosine under the reaction conditions.21−24 To further evaluate the effect of these side adducts on the enrichment of Kme2-generated aldehyde sites, we synthesized four dimethyllysine tripeptides with methionine (FKme2M), histidine (FKme2H), tryptophan (FKme2W), and cysteine (FKme2C) and subjected them to the optimized TACO reaction conditions. The reactions generated the corresponding aldehyde adducts, FK(CHO)M(sulfoxide), FK(CHO)H(fluorine), FK(CHO)W(fluorine), and FK(CHO)C(disulfide) in high conversions (Figures 3b and Supporting Figure 6). Further, treatment of aldehyde adducts with hydroxylamine and amino-thiol (cysteine methyl ester) generated near-quantitative conversions to the oxime and thiazolidine products. These data strongly validate the negligible effects of TACO-generated adducts on downstream chemoproteomics application on Kme2 in complex mixtures (Figures 3b and Supporting Figure 6).
Figure 3.
Evaluation of chemoselectivity of the TACO reaction. (a) Chemoselectivity studies of the TACO reaction with varying tripeptides FXV, where X = K, Kme, Kme2, H, D, S, M, Y, C, R, N, and W. Only the Kme2 residue modified to an aldehyde under the reaction conditions. Side adducts were observed with histidine (fluorination), methionine (sulfoxide), tryptophan (fluorination and oxygenation), and cysteine (disulfide). (b) Selective derivatization of Kme2-generated aldehydes on tripeptides FKme2X, where X = M, H, W, and C, in the presence of side adducts on histidine, methionine, cysteine, and tryptophan using oxime and thiazolidine chemistry in NaP buffer. The presence of TACO-generated adducts did not interfere with subsequent modification of dimethyl lysine sites, thus indicating the negligible effect of the adducts on downstream application of TACO for profiling of lysine dimethylome.
Application of TACO for Diversification of Dimethyllysine on Histone Peptides
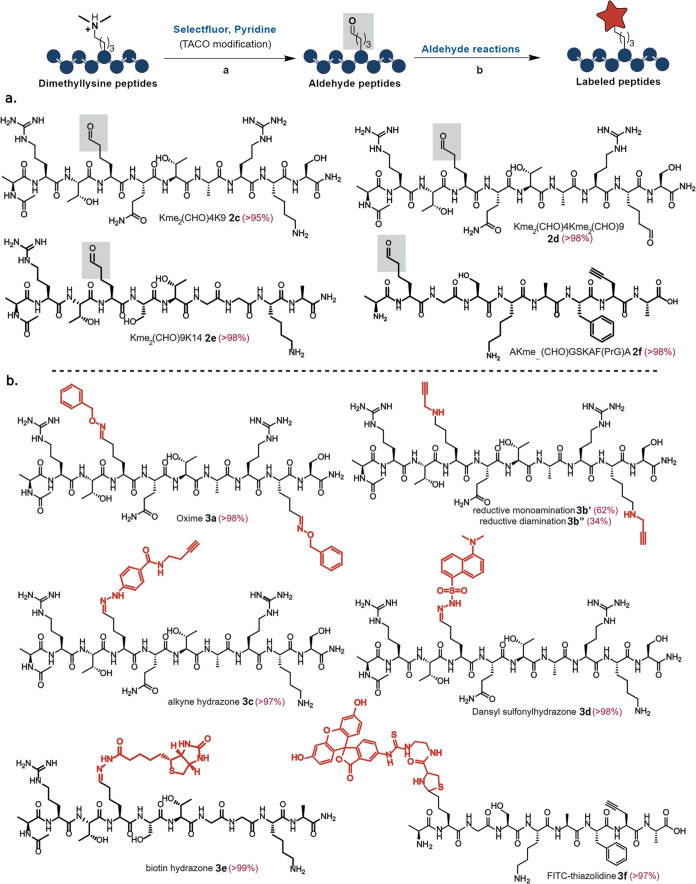
To demonstrate the application of TACO for diversification of histone peptides, we conducted reactions on various peptides of different sizes and amino acid compositions, including histone H3.3 peptide fragments frequently methylated at K4, K9, K27, and K36, known to regulate biological processes and implicated in disease states.25−27 Using solid-phase peptide synthesis, we synthesized the H3.3 peptide fragments Kme24K9 (ARTKme2QTARKS) 1c, Kme24Kme29 (ARTKme2QTARKme2S) 1d, Kme29K14 (ARTKme2STGGKA) 1e, propargyl amine containing peptide AKme2(CHO)GSKAF(PrG)A (where PrG = propargyl glycine) 1f, and a negative control peptide K4K9 (ARTKSTGGKA) 1g. Under optimized conditions, all Kme2-containing peptides underwent modification to aldehyde peptide products (Figures 4a and Supporting Figure 7). Intriguingly, the H3.3 peptide fragment Kme24Kme29 (ARTKme2QTARKme2S) 1d, containing two Kme2, produced the double aldehyde product ARTKme2(CHO)QTARKme2(CHO)S 2d with >98% conversion (Supporting Figure 7). Conversely, the negative control peptide K4K9, ARTKSTGGKA 1g without Kme2, remained unmodified (Figures 4a and Supporting Figure 7). These results unequivocally confirmed the application of TACO toward dimethyllysine Kme2, marking the first chemical method developed for Kme2 modification.
Figure 4.
Application of the TACO reaction for Kme2 and further derivitization with varying payloads. (a) TACO-mediated modification of histone H3.3 peptide (1c–1f) fragments with Kme2 to aldehydes (2c–2f). Reaction conditions: To 6–10 mM of dimethyllysine peptide in 300 μL of 10 mM sodium phosphate buffer (NaP, pH 7.0) were added pyridine (5 equiv) and Selectfluor (2 equiv) and the reaction mixture was stirred for 1 h at room temperature. (b) Late-stage diversification of peptide aldehydes with varying chemistries and payloads.
We further diversified peptide aldehydes using aldehyde-specific reactions, such as oxime, hydrazone, thiazolidine chemistry, and reductive amination (Figure 4b). For instance, the histone peptide ARTKme2(CHO)QTARKme2(CHO)S 2d with double aldehyde groups underwent modifications with benzylhydroxylamine, generating the oxime product 3a in >98% yield (Figures 4b and Supporting Figure 8). Additionally, this double-aldehyde peptide was treated with propragylamine and sodium cyanoborohydride, resulting in reductive amination products, single modification 3b’ (62%) and double modification 3b″ (34%) in high conversions (96%) (Figures 4b and Supporting Figure 9).
Next, we functionalized Kme24K9-aldehyde (ARTKme2(CHO)QTARKS) 2c with alkyne- and dansyl-tagged hydrazines to generate hydrazone products 3c and 3d in >97% and >98% conversions, respectively (Figure 4b and Supporting Figures 10 and 11). Biotin hydrazine treatment of the peptide aldehyde (ARTKme2(CHO)STGGKA) 2e resulted in 99% conversion to biotin peptide hydrazone 3e (Figures 4b and Supporting Figure 12). Concurrently, introducing both an affinity handle and a fluorescent dye into a peptide (AKme2(CHO)GSKAF(PrG)A, where PrG = propargyl glycine) 2f via derivatization with FITC-cysteine yielded the thiazolidine conjugation product 3f in >97% conversion (Figures 4b and Supporting Figure 13). Moreover, treatment of peptide aldehyde FKme2(CHO)V 2a with cysteine resulted in thiazolidine conjugation with high conversion (>99%, Supporting Figure 13). These outcomes underscore the broad scope and efficiency of TACO chemistry in selectively modifying and diversifying Kme2-containing peptides with various payloads.
Selective Labeling of Kme2 Peptides by TACO in a Complex Cell Lysate Mixture
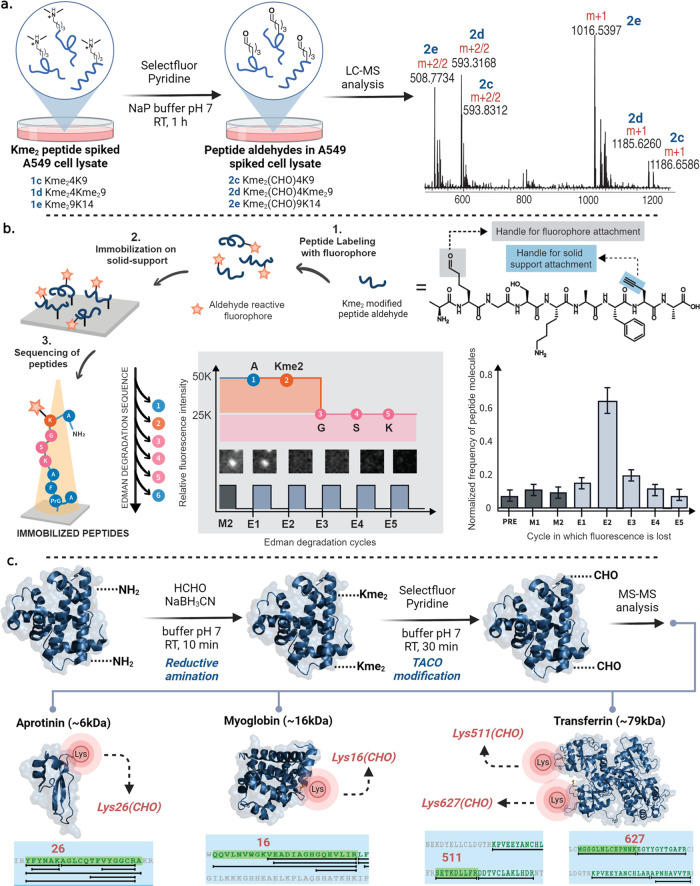
To evaluate the robustness of our TACO technique in labeling Kme2 peptides within a complex mixture, we spiked cell lysates from epithelial carcinoma A549 and H1792 lung cancer cell lines (100 μg) with three different histone H3.3 peptides (0.1 mg each): Kme24K9 (ARTKme2QTARKS) 1c, Kme24Kme29 (ARTKme2QTARKme2S) 1d, and Kme29K14 (ARTKme2STGGKA) 1e and 1c, 1d, and 1g, respectively. Both complex mixtures were incubated with Selectfluor and pyridine for 1 h, followed by LC-MS analysis (Figure 5a, A549 Supporting Figures 14 and 15). All peptides containing Kme2 (1c–1e) were successfully converted to the corresponding peptide aldehydes (2c–2e) without any detectable traces of the unmodified starting peptides in the complex cell lysate mixtures (Figure 5a, A549 Supporting Figures 14 and 15). Notably, peptide 1g, K4K9 (ARTKSTGGKA), lacking Kme2, showed no modification in the H1792 cell lysate (Supporting Figure 15).
Figure 5.
Labeling and identification of Kme2 peptides and proteins in a complex mixture. (a) Selective modification of Kme2 peptides in a complex mixture of A549 lung cancer cell lysate to peptide aldehydes in a quantitative manner. Reaction conditions: To 100 μg of A549 whole cell lysate in 200 μL of NaP buffer pH 7.0 was added 0.1 mg of each histone peptide followed by addition of pyridine (30 equiv with respect to peptides) and selectfluor (10 equiv with respect to peptides). The reaction mixture was stirred for 1 h. (b) Application of TACO in the identification of Kme2 sites on a peptide by single molecule fluorosequencing. The left panel provides an exemplary view of the single fluorescent peptide molecule through cycles of Edman degradation. The right panel indicates the normalized counts of the labeled Kme2 peptide where its fluorescence intensity was lost. (c) Selective modification of chemically generated Kme2 proteins by TACO chemistry followed by the identification of Kme2 sites on proteins by LC-MS/MS analysis.
Identification of Kme2 Sites by TACO at a Single-Molecule Level Using Fluorosequencing
To determine the site of Kme2 in a peptide at a single-molecule level, we modified a model peptide, AKme2GSKAF(PrG)A (where PrG = propargyl glycine), by TACO chemistry to peptide aldehyde, AKme2(CHO)GSKAF(PrG)A. Next, we functionalized the Kme2-aldehyde group with an Atto647N fluorophore by using dithiolane chemistry under acidic conditions and subjected it to a fluorosequencing workflow including immobilization on the azide-functionalized microscopic slide using PrG on a peptide, AKme2Atto647NGSKAF(PrG)A, by click chemistry (Figures 5b and Supporting Figure 16). In the case of biologically derived peptides, the C-terminus will be used for immobilization using a photoredox catalyst that can differentiate oxidation potential of the C-terminus from the side chains of Asp and Glu, leading to C-terminal immobilization exclusively.28 Next, the fluorophore-labeled immobilized peptide was subjected to several rounds of Edman’s degradation including one prerun and two rounds of mock Edman’s degradation (M1-M2) with all the reagents except phenylisothiocyanate followed by analysis using a total internal reflection fluorescence (TIRF) microscope (Figure 5b).29 No decrease in the fluorescence was observed under the mock conditions, suggesting the high stability of the Kme2-modified fluorophore under Edman’s degradation conditions requiring TFA and pyridine. Next, Edman’s degradation was performed on a peptide with phenylisothiocyanate followed by the analysis of fluorescence by TIRF after each round. A significant decrease in the fluorescence was observed after the second round of Edman’s degradation, confirming the site of Kme2 at a single-molecule level. Subsequent rounds of Edman’s degradation did not lead to a change in the fluorescence. There are no other chemical methods for the identification of sites of Kme2 PTM by any other single-molecule protein sequencing SMPS techniques.30 Future studies will be focused on discovering proteins containing Kme2 and the sites of Kme2 on a proteome-wide scale by fluorosequencing. The work under this direction is currently underway in our laboratory.
Modification of Proteins and Whole Cell Lysate by TACO
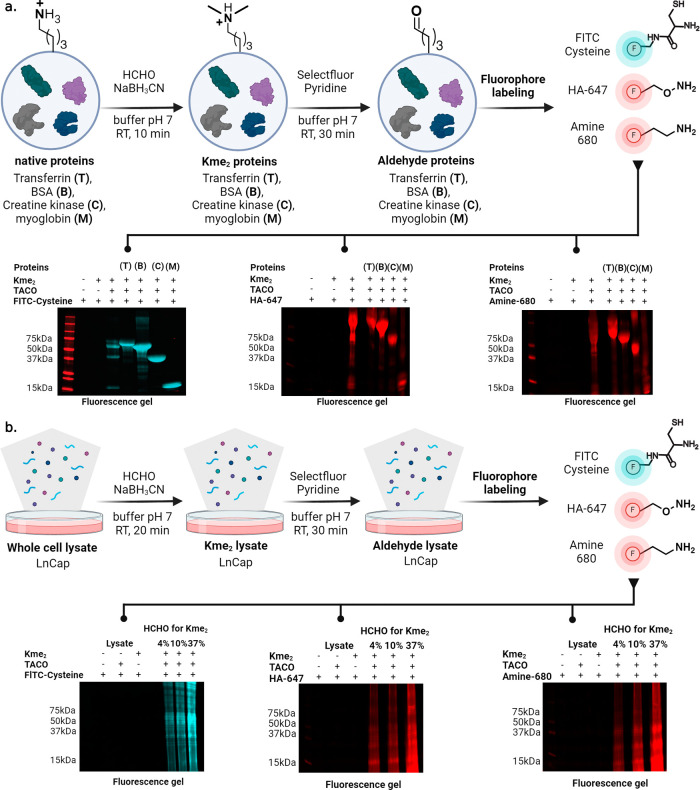
To demonstrate the compatibility of TACO chemistry with proteins, we chemically modified lysines (K) to dimethyl lysine (Kme2) on proteins of varying molecular weights, aprotinin (A, 5 kDa), myoglobin (M, 16 kDa), and transferrin (T, 79 kDa) using reductive amination. Proteins were treated with a solution of formaldehyde and sodium cyanoborohydride in citrate buffer for 10 min. Subsequently, Kme2 proteins were subjected to TACO chemistry to generate aldehyde proteins. The MS/MS analysis of digested aldehyde proteins identified K26 (aprotinin), K16 (myoglobin), K511 (transferrin), and K627 (transferrin) as Kme2 modification sites (Figures 5c and Supporting Figure 17). Moreover, four proteins of varying molecular weights and 3 D structures, transferrin (T), BSA (B), creatine kinase (C), and myoglobin (M), were subjected to reductive amination followed by TACO chemistry to produce aldehyde proteins. These aldehyde proteins were incubated with various fluorophores containing diverse reactive aldehyde functional groups (FITC-cysteine, hydroxylamine (HA)-647 and amine-680) and analyzed via in-gel fluorescence (Figure 6a). The results showed the selective labeling with aldehyde proteins only (lanes 3–7, lane 3 = mixture of all four proteins, Figures 6a and Supporting Figure 18). No fluorophore labeling was observed with native proteins and Kme2 proteins (lanes 1, 2, Figures 6a and Supporting Figure 18). This result demonstrated the ability of TACO chemistry to selectively modify Kme2 sites on proteins to aldehydes, which can then be further modified with varying payloads.
Figure 6.
Selective labeling of Kme2 proteins by TACO chemistry using varying fluorophores. (a) Selective modification of chemically generated Kme2 proteins to aldehyde proteins by TACO chemistry followed by labeling with varying fluorophores using different aldehyde chemistries as analyzed by running SDS PAGE. No fluorophore labeling was observed without the TACO reaction. (b) Selective modification of chemically generated Kme2 proteins in cell lysate to aldehyde proteins by TACO followed by fluorophore labeling and in-gel analysis. High fluorescence was observed in cell lysate in a dose-dependent manner, indicating high amounts of Kme2-proteins. No fluorophore labeling was observed on cell lysate without the TACO reaction or without Kme2-modified cell lysate.
To further demonstrate the robustness of TACO chemistry to modify proteins within cell lysates, we treated prostate cancer cell lysate (LnCap) with reductive amination reagents (formaldehyde and sodium cyanoborohydride) in a dose-dependent manner (4, 10, and 37%), followed by TACO chemistry and subsequent labeling with FITC-cysteine, hydroxylamine (HA)-647, and amine-680 fluorophores (Figures 6b and Supporting Figure 19). Analysis and quantification of in-gel fluorescence results clearly showed selective fluorophore labeling of aldehyde-modified cell lysate and a dose-dependent increase in the fluorescence intensity from 4 to 37% of reductive amination reagents (with Cys-FITC, 37% lane = 1.0, 10% lane = 0.582, 4% lane = 0.420; with HA-647, 37% lane = 1.0, 10% lane = 0.513, 4% lane = 0.459; with amine-680, 37% lane = 1.0, 10% lane = 0.672, 4% lane = 0.373) (lanes 4–6, Figures 6b and Supporting Figure 19). No fluorescence was observed with untreated LnCap cell lysate, Kme2-modified cell lysate, and TACO-treated cell lysate without Kme2 modification (lanes 1–3, Figure 6b and Supporting Figure 19), confirming the high selectivity of TACO chemistry for Kme2.
Selective Enrichment of Kme2 Proteins from Nucleosomes by TACO
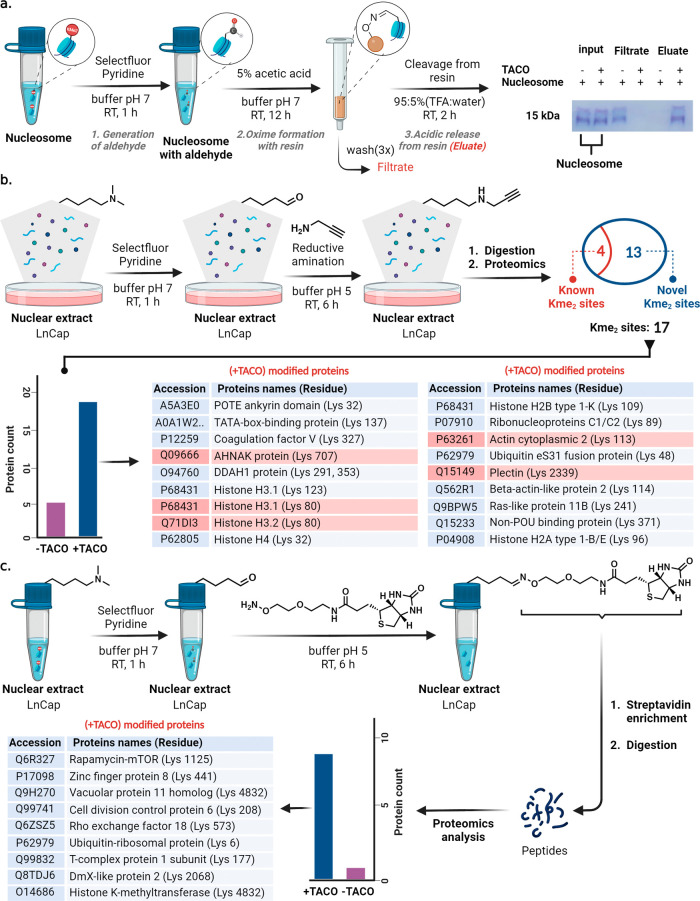
To explore the capability of TACO chemistry in the selective modification and enrichment of naturally occurring Kme2 on histones, we exposed nucleosomes from prostate cancer cell lysate (LnCap) to Selectfluor and pyridine for 1 h, followed by the enrichment of aldehyde proteins via oxime chemistry using hydroxylamine-functionalized resin in pull-down experiments. The resin was thoroughly washed with solvents to remove noncovalently bound proteins (filtrate). Subsequently, enriched proteins were released from the resin under acidic conditions (95% TFA in water).
SDS-PAGE analysis of the cell lysate (filtrate) postenrichment and protein release from the solid support (eluate) distinctly demonstrated the efficient capture and release of Kme2-modified protein aldehydes (Figure 7a, lanes 4 and 6, Supporting Figures 20 and 21). Repetition of this reaction in duplicate showed consistent gel profiles across all replicates, confirming the reproducibility, selectivity, and robustness of the TACO approach in tagging and enriching naturally occurring Kme2 proteins from a complex nucleosome mixture. Enrichment of nucleosomes by hydroxylamine resin without the TACO reaction failed to capture proteins (lane 3 (filtrate) and lane 5 (eluate), Figure 7a). Lanes 1 and 2 represent total nucleosomes with and without TACO reagents, respectively (Figure 7a).
Figure 7.
Discovery of naturally occurring Kme2 proteins and Kme2-sites from a complex cell lysate was carried out by TACO chemistry. (a) Selective modification of naturally occurring dimethyl lysine in nucleosomes using TACO chemistry followed by the enrichment of Kme2-generated aldehyde proteins using hydroxylamine resin and release of trapped proteins under acidic conditions. The filtrate contains proteins remaining after the enrichment process with and without TACO chemistry on nucleosomes, while the eluate comprises proteins enriched from the nucleosomes with and without TACO chemistry using a solid support. The filtrate and eluate were analyzed by SDS PAGE analysis. The gels showed the enrichment of Kme2 proteins from nucleosomes under TACO reaction conditions. (b) Selective modification of Kme2 proteins by TACO chemistry followed by trapping of Kme2-generated aldehyde proteins by reductive amination and proteomic analysis. Twelve novel Kme2 proteins and thirteen new Kme2 sites were identified (novel = blue, known = red). In the negative control, five proteins were identified, which vary from the TACO-treated samples due to the presence of natural occurring allysine on proteins. (c) Enrichment of TACO-modified Kme2 proteins followed by proteomic analysis. Nine low-abundance Kme2 proteins were discovered with nine new Kme2 sites.
Identification and Discovery of Kme2 Proteins and Kme2 Sites by TACO from a Complex Cell Lysate
To demonstrate an exciting application of TACO chemistry for identifying and discovering naturally occurring Kme2 proteins, we treated nuclear extracts obtained from LnCap cells with TACO chemistry followed by labeling of Kme2-generated aldehyde proteins with propargylamine using reductive amination. We performed this reaction to increase the stability of the adducts under proteomics conditions and to add a tag for identification. By digesting and analyzing the cellular mixture via proteomics, we discovered 13 Kme2 sites that had not been previously identified, along with four Kme2-sites previously reported by other methods31 (Figures 7b and Supporting Figure 22). The treatment of nuclear extract of MCF-10A, a preadenocarcinoma cell, with TACO chemistry led to the identification of nine Kme2 sites with one known Kme2 site, K240 of actin, α cardiac muscle 1 (Supporting Figure 22). Notably, none of the identified Kme2 proteins in MCF-10A cells match with Kme2 proteins observed in LnCap cancer cells, thus suggesting the potential of this approach in discovering dimethyllysine biomarkers. Moreover, proteomics analysis of nuclear extracts without TACO modification (negative control) identified five proteins and sites, differing from those observed in the TACO-treated sample. We attribute this modification to the presence of allysine, an endogenous aerobic oxidation aldehyde product of lysine.32
Furthermore, we developed an enrichment protocol for trapping Kme2-generated aldehyde proteins obtained through TACO chemistry to identify low-abundant Kme2 proteins from a complex nuclear extract mixture of LnCap cells. Employing biotin-derivatized alkoxyamine probes to label aldehyde proteins, followed by streptavidin enrichment and digestion of enriched proteins, led to the discovery of nine novel low-abundant Kme2 proteins and identification of nine new Kme2 sites (Figures 7c and Supporting Figure 23). Taken together, these results demonstrate the potential of TACO chemistry to serve as a chemical platform for the identification and discovery of the lysine dimethylome.
Conclusions
In summary, we have developed TACO, a versatile chemical method for modifying dimethyllysine Kme2 to an aldehyde group within peptides, proteins, and cell lysates under physiological conditions. The TACO chemistry involves the oxidation of dimethyl lysine/Kme2 by Selectfluor, followed by hydrolysis of the resulting iminium ions to generate aldehydes. Our study demonstrates the robustness of TACO chemistry for selectively labeling of Kme2-containing peptides, proteins, and cell lysates with diverse payloads, including affinity tags such as alkyne, biotin, and various fluorophores, by diverse aldehyde chemistries in high conversions (>90%). Moreover, we showcased the compatibility of TACO chemistry with fluorosequencing, thus demonstrating the potential of identifying Kme2 sites at a single-molecule level. Applying TACO chemistry to the nuclear extract of cancer and preadenocarcinoma cells, we discovered 30 novel Kme2 sites, in addition to 5 previously reported sites using MS/MS analysis. This demonstrates the capability of TACO chemistry to enable the identification of Kme2 proteins and their sites on a proteome-wide scale, through either MS-based proteomics or single molecule fluorosequencing. We compared proteome profiling of diseased cells to healthy controls, and none of the identified Kme2 proteins in MCF-10A cells match with Kme2 proteins observed in LnCap cancer cells, thus suggesting the potential of this approach in discovering Kme2 proteins and Kme2 sites as disease biomarkers. The work in these directions is currently underway in our laboratory. We believe that our chemical approach will complement existing detection methods, significantly expanding the chemical toolbox available for epigenetics research.
Acknowledgments
This research was supported by the NIH (No. 1R35GM133719-01) and NSF (Grant No. CHE-2108774) to M.R. and Research Scholar Grant, RSG-22-025-01-CDP, from the American Cancer Society to M.R. The research was supported by the NIH (No. R35 GM149308-01) to E.V.A. J.S. work was supported through a sponsored research agreement from Erisyon to University of Texas (E.V.A.). The authors thank Heather D Stout for running duplicates of the fluorosequencing experiment. All the images are created with biorender.com.
Glossary
Abbreviations
- Kme2
lysine dimethylation
- PTM
post-translational modification
- TACO
tertiary amine coupling by oxidation
- Kme1
monomethyl lysine
- MS
mass spectrometry
- TIRF
total internal reflection fluorescence
- MBD
methyl-binding domains
- PrG
propargyl glycine
- DEAD
diethyl azodicarboxylate
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/jacs.4c00253.
List of proteins (TACO) (XLSX)
Optimization of the TACO reaction with varying oxidizing reagents and nucleophiles; reaction procedures; procedure of TACO reaction with peptides; proteins; cell lysates; nucleosomes; fluorosequencing; enrichment protocol; proteomic analysis and product characterization by NMR; HPLC; HRMS and computational data; and raw images for fluorosequencing uploaded to Zenodo (PDF)
Author Contributions
The article was written through contributions of all authors. All authors have given approval to the final version of the article.
The authors declare no competing financial interest.
Notes
B.E. and M.R. are the inventors of a United States patent application (PCT/US2022/076782) of Emory University covering the methodology of the TACO reaction for modification of Kme2 and its application in global profiling and identification of Kme2 sites from a complex biological system. J.S. and E.V.A. are cofounders and shareholders of Erisyon, Inc. E.V.A. serves on the scientific advisory board of Erisyon.
Supplementary Material
References
- Moss T. J.; Wallrath L. L. Connections between epigenetic gene silencing and human disease. Mutat. Res., Fundam. Mol. Mech. Mutagen. 2007, 618, 163–174. 10.1016/j.mrfmmm.2006.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egorova K. S.; Olenkina O. M.; Olenina L. V. Lysine methylation of nonhistone proteins is a way to regulate their stability and function. Biochemistry 2010, 75, 535–548. 10.1134/S0006297910050019. [DOI] [PubMed] [Google Scholar]
- Yao X.; Shen W. Crucial function of histone lysine methylation in plant reproduction. Chin. Sci. Bull. 2011, 56, 3493–3499. 10.1007/s11434-011-4814-3. [DOI] [Google Scholar]
- Moore K. E.; Carlson S. M.; Camp N. D.; et al. A general molecular affinity strategy for global detection and proteomic analysis of lysine methylation. Mol. Cell 2013, 50, 444–456. 10.1016/j.molcel.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison E. E.; Carpenter B. A.; Louis L. E. St.; Mullins A. G.; Waters M. L. Development of “Imprint-and-Report” dynamic combinatorial libraries for differential sensing applications. J. Am. Chem. Soc. 2021, 143, 14845–14854. 10.1021/jacs.1c07068. [DOI] [PubMed] [Google Scholar]
- Guo A.; Gu H.; Zhou J.; Mulhern D.; Wang Y.; Lee K. A.; et al. Immunoaffinity enrichment and mass spectrometry analysis of protein methylation. Mol. Cell. Proteomics 2014, 13, 372–387. 10.1074/mcp.O113.027870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emenike B.; Nwajiobi O.; Raj M. Covalent chemical tools for profiling post-translational modifications. Front. Chem. 2022, 10, 868773 10.3389/fchem.2022.868773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polevoda B.; Sherman F. Methylation of proteins involved in translation. Mol. Microbiol. 2007, 65, 590–606. 10.1111/j.1365-2958.2007.05831.x. [DOI] [PubMed] [Google Scholar]
- Bremang M.; Cuomo A.; Agresta A. M.; Stugiewicz M.; Spadotto V.; Bonaldi T. Mass spectrometry-based identification and characterisation of lysine and arginine methylation in the human proteome. Mol. BioSyst. 2013, 9, 2231–2247. 10.1039/c3mb00009e. [DOI] [PubMed] [Google Scholar]
- Wang Q.; Wang K.; Ye M. Strategies for large-scale analysis of non-histone protein methylation by LC-MS/MS. Analyst 2017, 142, 3536–3548. 10.1039/C7AN00954B. [DOI] [PubMed] [Google Scholar]
- Zoghbi H. Y.; Beaudet A. L. Epigenetics and human disease. Cold Spring Harbor Perspect. Biol. 2016, 8, a019497 10.1101/cshperspect.a019497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busby M.; Xue C.; Li C.; Farjoun Y.; Gienger E.; Yofe I.; et al. Systematic comparison of monoclonal versus polyclonal antibodies for mapping histone modifications by ChIP-seq. Epigenet. Chromatin 2016, 9, 49 10.1186/s13072-016-0100-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young N. L.; DiMaggio P. A.; Plazas-Mayorca M. D.; Baliban R. C.; Floudas C. A.; Garcia B. A. High throughput characterization of combinatorial histone codes. Mol. Cell Proteomics 2009, 8, 2266–2284. 10.1074/mcp.M900238-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nwajiobi O.; Mahesh S.; Streety X.; Raj M. Selective triazenation reaction (STaR) of secondary amines for tagging monomethyl lysine post-translational modifications. Angew. Chem., Int. Ed. 2021, 60, 7344–7352. 10.1002/anie.202013997. [DOI] [PubMed] [Google Scholar]
- Daniels M. H.; Hubbs J. The development of a Selectfluor-mediated oxidative Mannich reaction. Tetrahedron Lett. 2011, 52, 3543–3546. 10.1016/j.tetlet.2011.05.012. [DOI] [Google Scholar]
- Kumaraswamy G.; Murthy A. N.; Pitchaiah A. FeCl3-catalyzed oxidative allylation of sp2 and sp3 C-H bond adjacent to a nitrogen atom: easy access to homoallyl tertiary amines. J. Org. Chem. 2010, 75, 3916–3919. 10.1021/jo1005813. [DOI] [PubMed] [Google Scholar]
- Niu M.; Yin Z.; Fu H.; Jiang Y.; Zhao Y. Copper-catalyzed coupling of tertiary aliphatic amines with terminal alkynes to propargylamines via C– H Activation. J. Org. Chem. 2008, 73, 3961–3963. 10.1021/jo800279j. [DOI] [PubMed] [Google Scholar]
- Xu X.; Li X. Copper/diethyl azodicarboxylate mediated regioselective alkynylation of unactivated aliphatic tertiary methylamine with terminal alkyne. Org. Lett. 2009, 11, 1027–1029. 10.1021/ol802974b. [DOI] [PubMed] [Google Scholar]
- Hauser A.; Bohlmann R. Preparation of aldehydes by oxidation of benzylic amines with selectfluor (F-TEDA-BF4). Synlett 2016, 27, 1870–1872. 10.1055/s-0035-1561642. [DOI] [Google Scholar]
- Seki T.; Fujiwara T.; Takeuchi Y. A facile procedure for synthesis of 3-[2-(N,N-dialkylamino)ethyl]-3-fluorooxindoles by direct fluorination of N,N-dialkyltryptamines. J. Fluorine Chem. 2011, 132, 181–185. 10.1016/j.jfluchem.2010.12.014. [DOI] [Google Scholar]
- Emenike B.; Donovan J.; Raj M. Multicomponent oxidative nitrile thiazolidination reaction for selective modification of N-terminal dimethylation posttranslational modification. J. Am. Chem. Soc. 2023, 145, 16417–16428. 10.1021/jacs.3c02369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karam O.; Martin-Mingot A.; Jouannetaud M.-P.; Jacquesy J.-C.; Cousson A. Efficient oxidative ipso-fluorination of para-substituted phenols using pyridinium polyhydrogen fluoride in combination with hypervalent iodine(III) reagents. Tetrahedron 2004, 60, 6629–6638. 10.1016/j.tet.2004.05.083. [DOI] [Google Scholar]
- Pravst I.; Stavber S. Fluorination of 4-alkyl-substituted phenols and aromatic ethers with fluoroxy and N-F reagents: Cesium fluoroxysulfate and N-fluoro-1,4-diazonia-bicyclo[2.2.2]octane dication salts case. J. Fluorine Chem. 2013, 156, 276–282. 10.1016/j.jfluchem.2013.07.002. [DOI] [Google Scholar]
- Jiang Z.; Ni T.; Wei C.; Tian S.; Li Y.; Dai L.; Liu H.; Zhang D. Selectfluor-promoted direct fluorination at the 4- or 5-position of imidazole derivatives. Synlett 2013, 24, 215–218. 10.1055/s-0032-1317934. [DOI] [Google Scholar]
- Lowe B. R.; Maxham L. A.; Hamey J. J.; Wilkins M. R.; Partridge J. F. Histone H3 mutations: An updated view of their role in chromatin deregulation and cancer. Cancers 2019, 11, 660 10.3390/cancers11050660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjerke L.; Mackay A.; Nandhabalan M.; Burford A.; Jury A.; Popov S.; et al. Histone H3.3 mutations drive pediatric glioblastoma through upregulation of MYCN. Cancer Discovery 2013, 3, 512–519. 10.1158/2159-8290.CD-12-0426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papillon-Cavanagh S.; Lu C.; Gayden T.; Mikael L. G.; Bechet D.; Karamboulas C.; et al. Impaired H3K36 methylation defines a subset of head and neck squamous cell carcinomas. Nat. Genet. 2017, 49, 180–185. 10.1038/ng.3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L.; Floyd B. M.; Chilamari M.; Mapes J.; Swaminathan J.; Bloom S.; Marcotte E. M.; Anslyn E. V. Photoredox-catalyzed decarboxylative C-terminal differentiation for bulk-and single-molecule proteomics. ACS Chem. Biol. 2021, 16, 2595–2603. 10.1021/acschembio.1c00631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaminathan J.; Boulgakov A. A.; Hernandez E. T.; Bardo A. M.; Bachman J. L.; Marotta J.; et al. Highly parallel single-molecule identification of proteins in zeptomole-scale mixtures. Nat. Biotechnol. 2018, 36, 1076–1082. 10.1038/nbt.4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Restrepo-Pérez L.; Joo C.; Dekker C. Paving the way to single-molecule protein sequencing. Nat. Nanotechnol. 2018, 13, 786–796. 10.1038/s41565-018-0236-6. [DOI] [PubMed] [Google Scholar]
- Kapell S.; Jakobsson M. E. Large-scale identification of protein histidine methylation in human cells. NAR: Genomics Bioinf. 2021, 3, lqab045 10.1093/nargab/lqab045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra-Bardenys G.; Peiro S. Enzymatic lysine oxidation as a posttranslational modification. FEBS J. 2022, 289, 8020–8031. 10.1111/febs.16205. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.