Abstract
Background
Lung adenocarcinoma (LUAD) has become the most frequent histologic type of lung cancer in the past several decades. Recent successes with immune checkpoint blockade therapy have demonstrated that the manipulation of the immune system is a very potent treatment for LUAD. This study aims to explore the role of immune-related genes in the development of LUAD and establish a signature that can predict overall survival for LUAD patients.
Methods
To identify the differential expression genes (DEGs) between normal and tumor tissues, we developed an analysis strategy to combine an independent-sample design and a paired-sample design using RNA-seq transcriptomic profiling data of The Cancer Genome Atlas LUAD samples. Further, we selected prognostic markers from DEGs and evaluated their prognostic value in a prediction model.
Results
We identified and validated PD1, PDL1 and CTLA4 genes as prognostic markers, which are well-known immune checkpoints, and revealed two new potential prognostic immune checkpoints for LUAD, HHLA2 (logFC = 2.55, FDR = 1.89 × 10–6) and VTCN1 (logFC = −2.86, FDR = 1.72 × 10–11). Furthermore, we identified an 18-gene LUAD prognostic biomarker panel and observed that the classified high-risk group presented a significantly shorter overall survival time (HR = 3.57, p value = 4.07 × 10–10). The prediction model was validated in five independent high-throughput gene expression datasets.
Conclusions
The identified DEG features may serve as potential biomarkers for prognosis prediction of LUAD patients and immunotherapy. Based on that assumption, we identified a gene expression-based immune signature for lung adenocarcinoma prognosis.
Electronic supplementary material
The online version of this article (10.1007/s00262-020-02595-8) contains supplementary material, which is available to authorized users.
Keywords: Lung adenocarcinoma, Immunotherapy, Differential expression, Pathway enrichment, Prognosis
Introduction
Lung cancer is a common and severe disease which is the leading cause of cancer mortality worldwide for both men and women [1]. More than half of patients diagnosed with lung cancer have less than a 1-year survival rate, and the 5-year survival rate is around 18% [2]. Among all subtypes of lung cancer, lung adenocarcinoma (LUAD) is at present the most common, and it comprises around 40% of all lung cancer cases. Compared to other subtypes, LUAD tends to grow more slowly but has a greater chance to be found after metastasis [3]. LUAD is the most common type of lung cancer in smokers and nonsmokers in both men and women.
Depending on the stage of LUAD, traditional treatment options usually include surgery, chemotherapy and radiotherapy as well as targeted therapy. In the past few years, advances in molecular-targeted therapy have led to a major paradigm shift in the treatment for LUAD, showing a high positive response rate compared to traditional cytotoxic chemotherapies [4]. Specifically, immunotherapies targeting immune checkpoints for LUAD have shown significantly positive efficacy in clinical trials [5, 6]. Immune checkpoints are primarily initiated through T cell inhibitory receptors, and their ligands, including programmed death 1 (PD1) with PDL1 or PDL2 and cytotoxic T lymphocyte-associated protein 4 (CTLA4) with B7-1 or B7-2, inhibit multiple pathways involved in T cell-mediated immunity to maintain self-tolerance and minimize collateral tissue damage [7]. Recent studies have revealed that the bindings of PD1 and CTLA4 to their ligands could negatively regulate the proliferation and cytokine production of T cells, providing a foundation for the development of PD1, PDL1 and CTLA4 antibodies-based cancer immunotherapy [8, 9]. Furthermore, current clinical trials have demonstrated promising effects of anti-PD1 (e.g., BMS-936558/nivolumab), anti-PDL1 (e.g., BMS-936559/nivolumab) and anti-CTLA4 (e.g., BMS-734016/ipilimumab) monoclonal antibodies in the treatment for advanced cancers, including LUAD [10–12].
Despite the curative potential of cancer immunotherapy, the majority of cancer patients do not benefit from the treatment, and some responders relapse after a period of treatment [13]. Considering the resistance mechanisms to cancer immunotherapy, recent studies have found that neoantigens originating from gene mutations may serve as attractive targets of immunotherapy [14, 15]. However, given that mutation frequency varies among populations and heterogeneity exists among patients, a simple marker may not predict immunotherapy efficacy, and therefore developing a clinical immunotherapy strategy using multiple immune-related biomarkers is essential.
For biomarker development, gene expression profiling provides an essential approach to detect the nature of interactions between the immune system and tumor components in patients, capturing information useful for cancer diagnosis, prognosis or new therapy development (e.g., cancer immunotherapy). For example, Showe MK et al. showed that a diagnostic gene expression signature of 29 genes measured in peripheral blood mononuclear cells was able to distinguish early-stage nonsmall cell lung cancer patients from control patients that had nonmalignant lung disease with 86% accuracy [16]. Furthermore, Kossenkov AV et al. found that the removal of malignant tumors significantly changed the expression of more than 3000 protein-coding genes, especially genes in pathways associated with suppression of the innate immune response, indicating the important role of immune genes in tumor development [17].
In this study, we carried out a transcriptome investigation aiming to examine potential gene expression biomarkers of immune-related genes for LUAD development and to further evaluate their associations with the overall survival outcome of the disease. With the samples of RNA-seq data from The Cancer Genome Atlas (TCGA), the prognostic value of immune-related genes was evaluated, and an 18-gene biomarker panel was identified with a prognosis prediction potential for LUAD. Five microarray datasets from the Gene Expression Omnibus (GEO) database were used for validation of the signature. Our study provides valuable insights into the immunotherapy and prognosis of LUAD by revealing the preliminary evidence of the possible underlying biological mechanisms of immune-related genes in tumor immune evasion.
Materials and methods
Datasets and immune-related genes
Gene expression data and clinical information were downloaded from the TCGA Web site (https://cancergenome.nih.gov/). The RNA-seq data were generated from Illumina HiSeq 3.1.12.0. Study samples consisted of 457 subjects in total; 57 comprised paired tumor and normal tissues, and 400 had tumor tissue only. Clinical variables included overall survival, race, gender, smoking status, age and tumor stage. For validation of the developed prognosis model, five independent datasets were downloaded from the GEO database, including GSE31210 (N = 246), GSE8894 (N = 138), GSE50081 (N = 181), GSE3141 (N = 111) and GSE30219 (N = 307). These five datasets were generated from the Affymetrix Human Genome U133 Plus 2.0 Array (GPL570). The clinical features, including tumor stage and smoking status of the patients from these validation datasets, are summarized in Supplementary Table 1. A comprehensive list of immune-related genes containing a total of 7052 genes was downloaded from the GeneCards database, which was searched using the keyword “immune” (https://www.genecards.org/).
Differential expressed immune genes
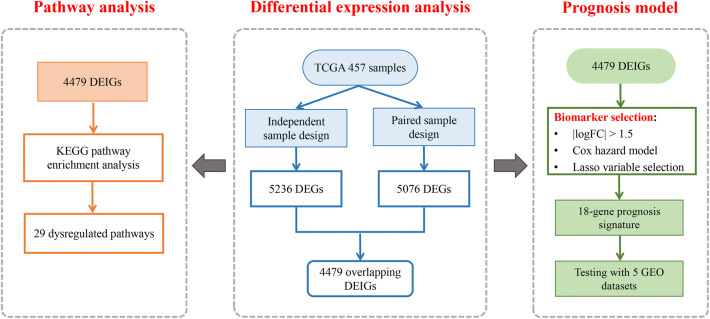
To identify the differential expressed genes (DEGs) in TCGA LUAD tumor tissues compared to normal tissues, we developed a strategy by combining two analysis designs. First, we used an independent-sample design to include all samples in the analysis and enable the confounder adjustment. We analyzed the normal tissues from the paired tumor–normal subjects and the tumor tissues from the tumor-only subjects, guaranteeing that only independent samples were included. Potential confounders, including age, gender, race and smoking status, were adjusted. The RNA-seq raw count data were analyzed using the R package edgeR [18]. Second, we used a paired-sample design with the paired tumor–normal subjects only to eliminate influences from subject-specific random effects and all other confounders. Multivariable linear regression models were used for the detection of DEGs. The Benjamin and Hochberg (BH) method was applied to control the false discovery rate, and adjusted p values < 0.05 were considered to be statistically significant. From the 7052 immune-related genes, significant DEGs were identified from the two designs separately, and the overlapping genes were selected as candidate differential expressed immune genes (DEIGs). The detailed study design of our analysis is displayed in Fig. 1.
Fig. 1.
A flowchart of the study design. First, we developed a novel analysis strategy by combining two different study designs to detect differential expression of immune genes between tumor and normal tissues in TCGA LUAD patients. Next, the significant DEGs were used to perform a pathway enrichment analysis. Further, we constructed a prognosis model with the identified DEGs aiming to distinguish high-risk populations and guide early treatment. The predictive model was validated in five independent lung cancer gene expression datasets
Pathway enrichment analysis of DEIGs
To further explore the immune-related biological processes involved in the detected DEIGs, we conducted a pathway enrichment analysis using GSEA software [19]. The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways were used as reference for understanding the signal transduction, cellular processes and biological pathways. A false discovery rate (FDR) < 0.05 was considered statistically significant.
Development of the immune signature
To develop a multimarker prognosis signature, we used the candidate DEIGs from the gene expression analysis for further biomarker selection. We selected those DEIGs with |logFC| (|log2(fold change)|) > 1.5 as candidate prognostic biomarkers. It is worth noting that the calculation of logFC was normal versus tumor for the whole manuscript. Reads per kilobase per million (RPKM) mapped reads values were calculated from the RNA-seq read counts and were normalized by log2 transformation. Using all 457 tumor samples, we performed univariate Cox proportional hazards regression to evaluate the association between expressions of these DEIGs with the overall survival outcome of the patients. The BH method-corrected FDR < 0.05 indicated significant associations. We then performed a LASSO variable selection using the R package “glmnet,” in which a fivefold cross-validation strategy was used to determine the tuning parameters. Then, we analyzed selected biomarkers in a multivariate Cox proportional hazards model. The best gene model was used to establish the immune signature.
Survival analysis
With the identified immune signature model, the estimated regression coefficients were used to compute a risk score for survival outcome [20]. Patients were then classified into a high-risk group and a low-risk group utilizing the median of risk scores in the sample as the cutoff value. The Kaplan–Meier (K–M) survival curves were generated to graphically demonstrate the overall survival of the high-risk and low-risk groups. The difference between survival curves was evaluated using the log-rank test. The R package “survival” was utilized to perform the survival analysis, and p values < 0.05 were considered to be statistically significant. The performance of the proposed survival prediction model was evaluated separately in five independent GEO datasets. Then, to assess whether the immune signature could be utilized in addition to existing clinicopathologic factors in evaluating risk, we conducted multivariate Cox analysis by adjusting age and stage in independent validation datasets mentioned above.
Results
Dysregulated immune genes in LUAD
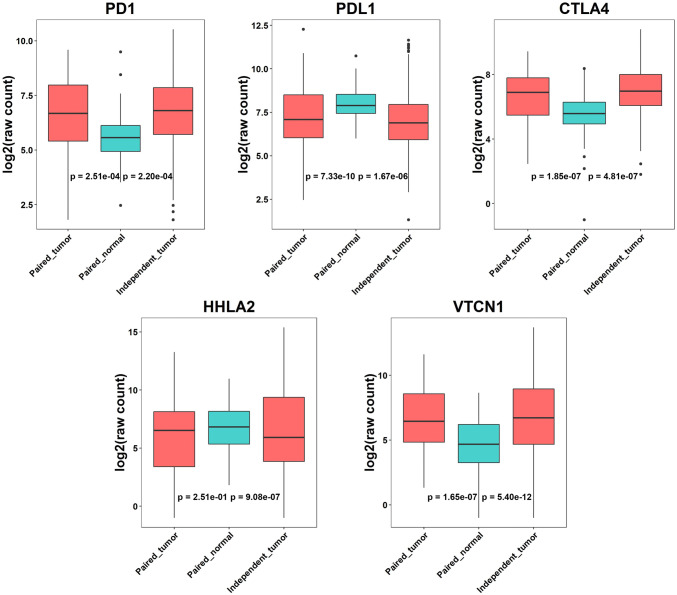
First, we conducted differential expression analysis for the 7052 immune genes obtained from GeneCards database using the discovery TCGA dataset. We performed an independent-sample analysis on 328 tumor tissues and 52 unrelated normal tissues and a paired-sample analysis on 57 paired tumor–normal tissues (see Materials and methods). The basic demographic information of the LUAD patients is summarized in Table 1. Most of these samples (> 90%) were from Caucasians. The independent-sample design analysis and the paired-sample design identified 5,236 and 5,076 significant DEGs (adjusted p value < 0.05), respectively. The overlapping 4479 DEGs with the same direction of regulation from the two analyses were classified as DEIGS. (Supplementary Table 2). Among these DEIGs, the gene expressions of PD1 and CTLA4 were found to be increased in tumor tissues compared to normal tissues, while the expression of PDL1 decreased (logFCs = −1.00, − 0.97 and 0.74, respectively), which was consistent with previous reports on dysregulation of PD1, PDL1 and CTLA4 gene expression in tumors [7, 21, 22], indicating the deactivated tumor infiltrating T cells. We also found another two potential immune differential expression genes, HHLA2 and VTCN1, dysregulated in tumor tissues (|logFC|> 2). From the independent-sample design analysis, HHLA2 showed significantly decreased gene expression in tumor tissues (logFC = 2.55, FDR = 1.89 × 10–6). It was not significant in the paired-sample design analysis, which might be due to the small sample size (logFC = 0.40, FDR = 0.29). Both analyses detected a significantly higher expression of the VTCN1 gene in tumor tissues (independent-sample design: logFC = -2.86, FDR = 1.72 × 10–11; paired-sample design: logFC = -2.14, FDR = 3.73 × 10–7). Boxplots of gene expression for these five candidate immune checkpoints are shown in Fig. 2.
Table 1.
Demographic characteristics of lung adenocarcinoma patients in the discovery dataset. Independent-sample analysis involved 328 tumor tissues and 52 unrelated normal tissues; paired-sample analysis was performed on 57 paired tumor and normal tissues. Age, gender, race and smoking status were adjusted to detect DEGs in the independent-sample design
| Characteristics | Independent-sample design (N = 380) | Paired-sample design (N = 57) |
|---|---|---|
| Median age (years) | 66 | 66 |
| Gender, n (%) | ||
| Male | 167 (43.9) | 33 (57.9) |
| Female | 213 (56.1) | 24 (42.1) |
| Race, n (%) | ||
| Caucasian | 346 (91.1) | 53 (93.0) |
| Asian | 7 (1.8) | / |
| African | 26 (6.8) | 4 (7.0) |
| Indian | 1 (0.3) | / |
| Clinical stage, n (%) | ||
| I | 213 (56.1) | 29 (50.9) |
| II | 89 (23.4) | 13 (22.8) |
| III | 60 (15.8) | 13 (22.8) |
| IV | 18 (4.7) | 2 (3.5) |
| Smoking status, n (%) | ||
| Nona | 60 (15.8) | 7 (12.3) |
| Ref-ndb | 2 (0.5) | / |
| Ref < 15c | 134 (35.3) | 21 (36.8) |
| Ref > 15d | 92 (24.2) | 16 (28.1) |
| Cure | 92 (24.2) | 7 (12.3) |
| Missing information | / | 6 (10.5) |
aLifelong Nonsmoker
bCurrent reformed smoker, duration not specified
cCurrent reformed smoker for < or = 15 years
dCurrent reformed smoker for > 15 years
eCurrent smoker
Fig. 2.
Boxplots for gene expression of five immune checkpoints in tumor and normal tissues
Immune-related gene-involved pathways
To study the dysregulated pathways involved with immune-related genes in LUAD, KEGG pathway enrichment analysis identified 29 significantly dysregulated pathways enriched by the above-identified DEIGs (Table 2). We found that the cell cycle pathway was the most significantly activated pathway in tumor tissues (FDR = 6.54 × 10–4). Genes such as cyclin B1 (CCNB1), pituitary tumor transforming gene 1 (PTTG1), cell division cycle 25c (CDC25C) and proliferating cell nuclear antigen (PCNA) were highly expressed in this pathway. Other pathways such as DNA replication, proteasome, spliceosome and pyrimidine metabolism were also activated in LUAD tumor tissues, which was consistent with previous studies [23, 24]. Importantly, we found immune-related pathways such as cell adhesion molecules (CAMs), intestinal immune network for immunoglobulin A (IgA) production and cytokine–cytokine receptor interaction were downregulated in tumor tissues. Our findings suggest the involvement of tumor immunosuppression in the development of LUAD and implicate the potential of immunotherapy in LUAD treatment [25, 26].
Table 2.
Dysregulated biological pathways in tumor tissues
| Direction | KEGG pathway | Size | ESa | NESb | p value | FDRc |
|---|---|---|---|---|---|---|
| Upregulated | Cell cycle | 73 | 0.50 | 2.10 | < 0.001 | 6.54E−04 |
| Pyrimidine metabolism | 34 | 0.55 | 2.01 | < 0.001 | 8.94E−04 | |
| Spliceosome | 39 | 0.52 | 2.01 | < 0.001 | 1.01E−03 | |
| Base excision repair | 16 | 0.65 | 2.01 | < 0.001 | 1.07E−03 | |
| Proteasome | 35 | 0.55 | 2.04 | < 0.001 | 1.12E−03 | |
| Aminoacyl TRNA biosynthesis | 18 | 0.62 | 1.95 | < 0.001 | 1.34E−03 | |
| Ubiquitin-mediated proteolysis | 87 | 0.47 | 2.06 | < 0.001 | 1.35E−03 | |
| DNA replication | 17 | 0.61 | 1.92 | < 0.001 | 1.93E−03 | |
| RNA degradation | 22 | 0.54 | 1.84 | 2.41E−03 | 4.65E−03 | |
| Homologous recombination | 15 | 0.59 | 1.76 | 6.51E−03 | 1.32E−02 | |
| Nucleotide excision repair | 22 | 0.50 | 1.69 | 9.64E−03 | 2.51E−02 | |
| N glycan biosynthesis | 19 | 0.51 | 1.69 | 7.17E−03 | 2.66E−02 | |
| Downregulated | Hematopoietic cell lineage | 47 | − 0.51 | − 2.77 | < 0.001 | < 0.0001 |
| Graft versus host disease | 24 | − 0.61 | − 2.72 | < 0.001 | < 0.0001 | |
| Complement and coagulation cascades | 36 | − 0.50 | − 2.54 | < 0.001 | < 0.0001 | |
| Allograft rejection | 23 | − 0.40 | − 2.29 | < 0.001 | 6.10E−04 | |
| Asthma | 20 | − 0.35 | − 2.37 | < 0.001 | 7.12E−04 | |
| Neuroactive ligand receptor interaction | 88 | − 0.56 | − 2.44 | < 0.001 | 8.55E−04 | |
| Vascular smooth muscle contraction | 61 | − 0.55 | − 2.47 | < 0.001 | 1.07E−03 | |
| Autoimmune thyroid disease | 23 | − 0.46 | − 2.11 | < 0.001 | 4.96E−03 | |
| Cell adhesion molecules cams | 78 | − 0.33 | − 2.03 | < 0.001 | 7.04E−03 | |
| Calcium signaling pathway | 76 | − 0.31 | − 2.01 | < 0.001 | 7.55E−03 | |
| Dilated cardiomyopathy | 43 | − 0.35 | − 1.90 | 9.26E−03 | 9.91E−03 | |
| Viral myocarditis | 46 | − 0.32 | − 1.79 | 9.90E−03 | 2.28E−02 | |
| Leukocyte transendothelial migration | 65 | − 0.29 | − 1.77 | < 0.001 | 2.50E−02 | |
| PPAR signaling pathway | 21 | − 0.34 | − 1.67 | 2.90E−02 | 3.85E−02 | |
| Intestinal immune network for IGA production | 31 | − 0.39 | − 1.69 | 2.45E−02 | 3.86E−02 | |
| Cytokine cytokine receptor interaction | 142 | − 0.29 | − 1.64 | 1.85E−02 | 4.21E−02 | |
| Leishmania infection | 53 | − 0.24 | − 1.65 | < 0.001 | 4.36E−02 |
aEnrichment score for the gene set;
bnormalized enrichment score, the enrichment score for the gene set after it has been normalized across analyzed gene sets;
cfalse discovery rate of NES
Immune signature for LUAD prognosis
We then performed survival analysis to identify DEIGs as a prognostic signature for LUAD. From the 4479 DEIGs, 945 genes were selected as candidate signatures with |logFC|> 1.5. Further, we identified 41 genes significantly associated with survival outcome using univariate Cox proportional hazards regression (Supplementary Table 3). Among these, 18 prognostic markers were selected by the LASSO variable selection model (Supplementary Fig. 1). Most of these 18 biomarkers showed discriminative power on the survival curves (Supplementary Fig. 2).
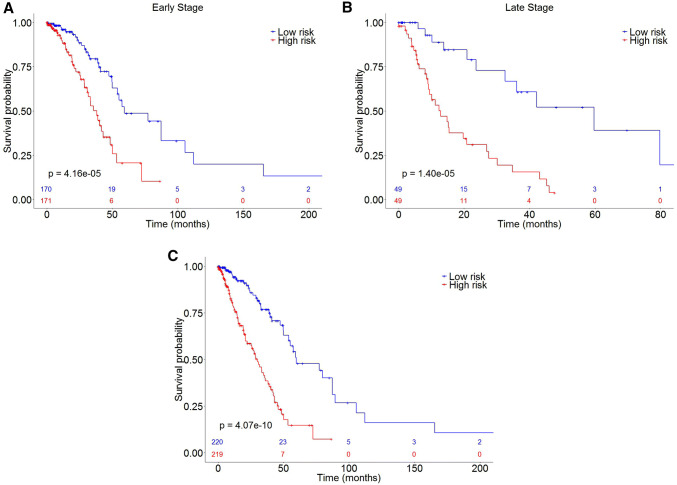
The final prediction model was fit with the 18 biomarkers using a multivariate Cox proportional hazards regression. Risk scores were computed as linear combinations of the gene expression values, and the regression coefficients were estimated from the model (Table 3). Subjects were classified as high-risk group and low-risk group based on the median score, respectively. As expected, the high-risk group had significantly shorter survival times than the low-risk group (HR = 3.57, p value = 4.07 × 10–10, Fig. 3c). Specifically, the risk score of the signature presented slightly better prognosis performance for stage III/IV lung adenocarcinoma (HR = 4.57, p value = 1.40 × 10–5, Fig. 3b) compared to stage I/II (HR = 2.84, p value = 4.16 × 10–5, Fig. 3a). The heatmap showed consistency between gene expression of the 18 biomarkers and tumor stage among LUAD patients (Supplementary Fig. 3).
Table 3.
Univariate and multivariate Cox proportional hazard regression analyses of 18 biomarkers
| Gene | Univariate | Multivariate (18 markers) | ||
|---|---|---|---|---|
| HR (95% CI) | p value | HR (95% CI) | p value | |
| ARNTL2 | 1.35(1.19–1.53) | 4.45E−06 | 1.01(0.83–1.24) | 0.89 |
| PLA2G4F | 0.83(0.77–0.90) | 7.82E−06 | 0.94(0.84–1.04) | 0.24 |
| CYP17A1 | 0.91(0.86–0.95) | 5.49E−05 | 0.94(0.89–1.00) | 3.54E−02 |
| LOXL2 | 1.31(1.15–1.51) | 8.39E−05 | 1.13(0.94–1.35) | 0.20 |
| HMMR | 1.32(1.14–1.53) | 2.13E−04 | 1.03(0.83–1.28) | 0.80 |
| NKX2-5 | 1.09(1.04–1.15) | 3.23E−04 | 1.04(0.98–1.10) | 0.21 |
| GJB2 | 1.20(1.08–1.32) | 3.86E−04 | 1.02(0.90–1.17) | 0.73 |
| DSG3 | 1.10(1.04–1.16) | 4.09E−04 | 1.00(0.95–1.07) | 0.88 |
| SIX1 | 0.84(0.77–0.93) | 4.24E−04 | 0.86(0.76–0.98) | 2.38E−02 |
| MC4R | 0.91(0.86–0.96) | 8.00E−04 | 0.95(0.89–1.00) | 6.65E−02 |
| TRPA1 | 1.13(1.05–1.21) | 9.70E−04 | 1.00(0.91–1.10) | 0.94 |
| RAET1L | 1.07(1.03–1.12) | 1.12E−03 | 1.00(0.95–1.06) | 0.87 |
| KLF4 | 1.28(1.10–1.49) | 1.47E−03 | 1.15(0.96–1.39) | 0.13 |
| HSF2BP | 1.30(1.11–1.53) | 1.48E−03 | 1.04(0.88–1.25) | 0.63 |
| GRIP1 | 1.21(1.08–1.37) | 1.69E−03 | 1.19(1.05–1.35) | 8.53E−03 |
| PTPRN | 1.10(1.03–1.16) | 1.69E−03 | 1.02(0.94–1.09) | 0.66 |
| MAPK4 | 1.10(1.04–1.17) | 2.01E−03 | 1.05(0.99–1.12) | 0.11 |
| PLOD2 | 1.23(1.08–1.40) | 2.14E−03 | 1.06(0.87–1.29) | 0.57 |
Fig. 3.
Kaplan–Meier curves of high-risk and low-risk groups of LUAD patients from TCGA database. a Tumor stage I/II patients. b Tumor stage III/IV patients. c Overall LUAD patients
These 18 immune genes play important roles in cancer progression. Five (KLF4, SIX1, NKX2-5, MAPK4 and TRPA1) are related to cell proliferation, differentiation and metabolism. KLF4 controls the transition of the cell cycle following DNA damage by mediating the tumor suppressor gene p53 [27]. SIX1 is involved in the regulation of cell proliferation and apoptosis and is widely expressed in many cancer types [28, 29]. NKX2-5 is a transcription factor that plays a role in organ formation and development [30]. MAPK4 promotes cell migration in the cell cycle [31]. The function of TRPA1 may play a role in signal transduction and growth control [32]. Other genes, such as HMMR, form a complex with BRCA1 and BRCA2 and are potentially associated with a higher risk of breast cancer [33]. Over-expression of LOXL2 is found in a number of cancers and may play a role in tumor progression [34, 35]. RAET1L plays an important role in antipathogen and anticancer immune responses [36].
Validation of the prognosis signature
To validate the identified immune prognosis signature, we applied the proposed prognostic model to five independent lung cancer transcriptome expression datasets. Four datasets (GSE50081, GSE3141, GSE8894 and GSE30219) showed that patients at high risk had significantly poorer overall survival compared to those at low risk (Supplementary Fig. 4A–D,p values = 9.19 × 10–3, 0.037, 0.018, 9.10 × 10–3, respectively). The GSE31210 dataset analysis showed a p value = 0.051 (Supplementary Fig. 4E), which may be due to its small sample size compared to the other datasets. In addition, multivariate Cox analysis adjusted by age and stage found that the risk score from the identified molecular signature dominated the prediction with weak or moderate additional contributions from tumor stage and age in the TCGA cohort (HR = 3.08, p value = 5.40 × 10–7), GSE50081 (HR = 2.34, p value = 0.0041), GSE8894 (HR = 2.13, p value = 0.042) and GSE30219 (HR = 2.31, p value = 0.0081) datasets, indicating the strong prognostic ability of our signature (Supplementary Fig. 5).
Discussion
In this investigation, we for the first time utilized a novel study design (combining an independent study design and a paired study design) to identify a robust prognostic signature on the basis of a tumor immune microenvironment and its prognostic potential, which was validated in five independent GEO datasets. The identified differential immune signature, including PD1, PDL1, CTLA4 HHLA2 and VTCN1 and the 18-gene panel, may provide valuable potential biomarkers with prognosis potential and possibly serve as targets of immunotherapy for LUAD patients. Enriched pathways of these immune-related differential expression genes between tumor and normal tissues were also discovered, which may shed light on the important role of a tumor-immune microenvironment in the development and progression of LUAD.
In this study, we developed a novel analysis strategy by combining two different designs to identify DEGs between tumor and normal tissues. The independent-sample design was developed to eliminate heterogeneity between patients by adjusting for covariates such as age, gender, race and smoking status. The paired-sample design aimed to avoid baseline differences in complex biological systems among patients. Thus, combining these two study designs can utilize information from the whole dataset and adjust confounding factors. Such a design is novel, and this is the first report of its use.
Among the identified DEIGs, we validated the dysregulation of PD1, PDL1 and CTLA4 immune checkpoints in tumors. We also discovered two potential immune checkpoints, HHLA2 and VTCN1, which may serve as potential immune checkpoints and biomarkers for the selection of patients who may benefit from immunotherapy of LUAD. HHLA2 is a newly identified B7 family member that regulates human T-cell functions through inhibiting the proliferation of both CD4 and CD8 T cells in the presence of T-cell receptor signaling [37, 38]. Closely related to our finding, Cheng et al. found that HHLA2 gene expression was downregulated in human non-small cell lung carcinoma, and its expression was associated with EGFR mutation status [39]. VTCN1 is another important member of the B7 family, and its expression on the cell surface of tumor macrophages negatively regulates T-cell-mediated immune response by inhibiting T-cell activation, proliferation, cytokine production and development of cytotoxicity [40, 41]. Zhang et al. demonstrated a strong role of VTCN1 in tumor growth and lung tumor metastatic progression [42]. Chen et al. observed that VTCN1 was highly expressed in lung cancer cells and promoted apoptotic death of activated tumor antigen-specific T cells [43].
Our pathway enrichment analysis results revealed that immune-related pathways such as CAMs, intestinal immune network for IgA production and cytokine–cytokine receptor interaction were downregulated in tumor tissues, consistent with the previous finding that tumor-originated proinflammatory activity weakens host-mediated antitumor immunity and thereby undergoes immune escape [44, 45]. Thus, further exploration of the underlying mechanisms involved in these dysregulated pathways might provide new targets for LUAD immunotherapy.
In this study, we found that an 18-gene signature was significantly associated with LUAD patients’ overall survival, which was validated in independent datasets. We also found that the performance of the prognosis model was associated with tumor stages, which indicates that this signature is able to provide a robust, global prognostic tool for subgroups of LUAD patients. For the prognosis of LUAD, the prognosis model was more powerful in distinguishing risk groups among patients with late stage (III/IV), which indicated that these markers might be involved in tumor metastasis. Since LUAD patients may have a favorable prognosis after surgery treatment, our model might serve as a potential tool to identify high-risk populations to guide early treatment, which could thereby increase the overall survival rate. Still, our investigation has several limitations. First, the discovery study has a limited sample size, which may lead to limited power of the prognostic model. Second, other clinical prognosis factors (e.g., smoking history, tumor stage) and other pathological factors might be confounders to the signature. We did not take them into consideration as this information is missing from some of the validation datasets. Third, further studies are needed to confirm the promise of our immune signature in guiding immunotherapy with LUAD patients.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We thank the study participants and research staff for their contributions and commitment to this study.
Abbreviations
- BH
Benjamin and Hochberg
- CAMS
Cell adhesion molecules
- DEG
Differential expression genes
- DEIGs
Differential expression immune genes
- FDR
False discovery rate
- GEO
Gene expression omnibus
- HR
Hazard ratio
- IgA
Immunoglobulin A
- KEGG
Kyoto encyclopedia of genes and genomes
- LUAD
Lung adenocarcinoma
- RPKM
Reads per kilobase per million
- TCGA
The Cancer Genome Atlas
Authors' contributions
FX and GC made substantial contributions to the conception and design of the study. LW and XL performed all analyses. All authors made substantial contributions to the acquisition, analysis or interpretation of data for the work. LW wrote the first draft of the manuscript. FX, GC, CC and CIA revised the manuscript for important intellectual content. All authors approved the manuscript.
Funding
The preparation of this manuscript was supported by internal funding from the University of South Carolina for Dr. Feifei Xiao and Dr. Guoshuai Cai.
Compliance with ethical standards
Conflict of interest
No potential conflicts of interest were disclosed.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. Cancer J Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2.Institute. NC. SEER Cancer Statistics Review, 1975–2011. Available online: https://seer.cancer.gov/csr/1975_2011/
- 3.Zappa C, Mousa SA. Non-small cell lung cancer: current treatment and future advances. Transl Lung Cancer Res. 2016;5:288–300. doi: 10.21037/tlcr.2016.06.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saito M, Suzuki H, Kono K, Takenoshita S, Kohno T. Treatment of lung adenocarcinoma by molecular-targeted therapy and immunotherapy. Surg Today. 2018;48:1–8. doi: 10.1007/s00595-017-1497-7. [DOI] [PubMed] [Google Scholar]
- 5.Cha YJ, Kim HR, Lee CY, Cho BC, Shim HS. Clinicopathological and prognostic significance of programmed cell death ligand-1 expression in lung adenocarcinoma and its relationship with p53 status. Lung cancer. 2016;97:73–80. doi: 10.1016/j.lungcan.2016.05.001. [DOI] [PubMed] [Google Scholar]
- 6.Teglasi V, Reiniger L, Fabian K, et al. Evaluating the significance of density, localization, and PD-1/PD-L1 immunopositivity of mononuclear cells in the clinical course of lung adenocarcinoma patients with brain metastasis. Neuro-oncology. 2017;19:1058–1067. doi: 10.1093/neuonc/now309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer. 2012;12:252–264. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Mello RA, Veloso AF, Esrom Catarina P, Nadine S, Antoniou G. Potential role of immunotherapy in advanced non-small-cell lung cancer. OncoTargets Therapy. 2017;10:21–30. doi: 10.2147/OTT.S90459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mahoney KM, Rennert PD, Freeman GJ. Combination cancer immunotherapy and new immunomodulatory targets. Nat Rev Drug Discov. 2015;14:561–584. doi: 10.1038/nrd4591. [DOI] [PubMed] [Google Scholar]
- 10.Rizvi NA, Mazieres J, Planchard D, et al. Activity and safety of nivolumab, an anti-PD-1 immune checkpoint inhibitor, for patients with advanced, refractory squamous non-small-cell lung cancer (CheckMate 063): a phase 2, single-arm trial. Lancet Oncol. 2015;16:257–265. doi: 10.1016/S1470-2045(15)70054-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamazaki N, Uhara H, Fukushima S, et al. Phase II study of the immune-checkpoint inhibitor ipilimumab plus dacarbazine in Japanese patients with previously untreated, unresectable or metastatic melanoma. Cancer Chemother Pharmacol. 2015;76:969–975. doi: 10.1007/s00280-015-2870-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brahmer JR, Drake CG, Wollner I, et al. Phase I study of single-agent anti-programmed death-1 (MDX-1106) in refractory solid tumors: safety, clinical activity, pharmacodynamics, and immunologic correlates. J Clin Oncol Off J Am Soc Clin Oncol. 2010;28:3167–3175. doi: 10.1200/JCO.2009.26.7609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sharma P, Hu-Lieskovan S, Wargo JA, Ribas A. Primary, adaptive, and acquired resistance to cancer immunotherapy. Cell. 2017;168:707–723. doi: 10.1016/j.cell.2017.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lennerz V, Fatho M, Gentilini C, Frye RA, Lifke A, Ferel D, Wolfel C, Huber C, Wolfel T. The response of autologous T cells to a human melanoma is dominated by mutated neoantigens. Proc Natl Acad Sci USA. 2005;102:16013–16018. doi: 10.1073/pnas.0500090102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matsushita H, Vesely MD, Koboldt DC, et al. Cancer exome analysis reveals a T-cell-dependent mechanism of cancer immunoediting. Nature. 2012;482:400–404. doi: 10.1038/nature10755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Showe MK, Vachani A, Kossenkov AV, et al. Gene expression profiles in peripheral blood mononuclear cells can distinguish patients with non-small cell lung cancer from patients with nonmalignant lung disease. Can Res. 2009;69:9202–9210. doi: 10.1158/0008-5472.CAN-09-1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kossenkov AV, Vachani A, Chang C, et al. Resection of non-small cell lung cancers reverses tumor-induced gene expression changes in the peripheral immune system. Clin Cancer Res Off J Am Assoc Cancer Res. 2011;17:5867–5877. doi: 10.1158/1078-0432.CCR-11-0737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fehringer G, Liu G, Briollais L, et al. Comparison of pathway analysis approaches using lung cancer GWAS data sets. PLoS ONE. 2012;7:e31816. doi: 10.1371/journal.pone.0031816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cai G, Xiao F, Cheng C, Li Y, Amos CI, Whitfield ML. Population effect model identifies gene expression predictors of survival outcomes in lung adenocarcinoma for both Caucasian and Asian patients. PLoS ONE. 2017;12:e0175850. doi: 10.1371/journal.pone.0175850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Topalian SL, Hodi FS, Brahmer JR, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med. 2012;366:2443–2454. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Patel SP, Kurzrock R. PD-L1 expression as a predictive biomarker in cancer immunotherapy. Mol Cancer Ther. 2015;14:847–856. doi: 10.1158/1535-7163.MCT-14-0983. [DOI] [PubMed] [Google Scholar]
- 23.Xu H, Ma J, Wu J, Chen L, Sun F, Qu C, Zheng D, Xu S. Gene expression profiling analysis of lung adenocarcinoma. Braz J Med Biol Res Revista brasileira de pesquisas medicas e biologicas. 2016 doi: 10.1590/1414-431X20154861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qian L, Luo Q, Zhao X, Huang J. Pathways enrichment analysis for differentially expressed genes in squamous lung cancer. Pathol Oncol Res POR. 2014;20:197–202. doi: 10.1007/s12253-013-9685-2. [DOI] [PubMed] [Google Scholar]
- 25.Dong ZY, Zhang C, Li YF, et al. Genetic and immune profiles of solid predominant lung adenocarcinoma reveal potential immunotherapeutic strategies. J Thorac Oncol Off Publ Int Assoc Study Lung Cancer. 2018;13:85–96. doi: 10.1016/j.jtho.2017.10.020. [DOI] [PubMed] [Google Scholar]
- 26.Kadara H, Choi M, Zhang J, et al. Whole-exome sequencing and immune profiling of early-stage lung adenocarcinoma with fully annotated clinical follow-up. Ann Oncol Off J Eur Soc Med Oncol. 2017;28:75–82. doi: 10.1093/annonc/mdw436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoon HS, Chen X, Yang VW. Kruppel-like factor 4 mediates p53-dependent G1/S cell cycle arrest in response to DNA damage. J Biol Chem. 2003;278:2101–2105. doi: 10.1074/jbc.M211027200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Behbakht K, Qamar L, Aldridge CS, Coletta RD, Davidson SA, Thorburn A, Ford HL. Six1 overexpression in ovarian carcinoma causes resistance to TRAIL-mediated apoptosis and is associated with poor survival. Can Res. 2007;67:3036–3042. doi: 10.1158/0008-5472.CAN-06-3755. [DOI] [PubMed] [Google Scholar]
- 29.Reichenberger KJ, Coletta RD, Schulte AP, Varella-Garcia M, Ford HL. Gene amplification is a mechanism of Six1 overexpression in breast cancer. Can Res. 2005;65:2668–2675. doi: 10.1158/0008-5472.CAN-04-4286. [DOI] [PubMed] [Google Scholar]
- 30.Fu Y, Yan W, Mohun TJ, Evans SM. Vertebrate tinman homologues XNkx2-3 and XNkx2-5 are required for heart formation in a functionally redundant manner. Development. 1998;125:4439–4449. doi: 10.1242/dev.125.22.4439. [DOI] [PubMed] [Google Scholar]
- 31.Stohr N, Kohn M, Lederer M, Glass M, Reinke C, Singer RH, Huttelmaier S. IGF2BP1 promotes cell migration by regulating MK5 and PTEN signaling. Genes Dev. 2012;26:176–189. doi: 10.1101/gad.177642.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stokes A, Wakano C, Koblan-Huberson M, Adra CN, Fleig A, Turner H. TRPA1 is a substrate for de-ubiquitination by the tumor suppressor CYLD. Cell Signal. 2006;18:1584–1594. doi: 10.1016/j.cellsig.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 33.Blanco I, Kuchenbaecker K, Cuadras D, et al. Assessing associations between the AURKA-HMMR-TPX2-TUBG1 functional module and breast cancer risk in BRCA1/2 mutation carriers. PLoS ONE. 2015;10:e0120020. doi: 10.1371/journal.pone.0120020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.De Craene B, Berx G. Regulatory networks defining EMT during cancer initiation and progression. Nat Rev Cancer. 2013;13:97–110. doi: 10.1038/nrc3447. [DOI] [PubMed] [Google Scholar]
- 35.Barker HE, Cox TR, Erler JT. The rationale for targeting the LOX family in cancer. Nat Rev Cancer. 2012;12:540–552. doi: 10.1038/nrc3319. [DOI] [PubMed] [Google Scholar]
- 36.Raulet DH. Roles of the NKG2D immunoreceptor and its ligands. Nat Rev Immunol. 2003;3:781–790. doi: 10.1038/nri1199. [DOI] [PubMed] [Google Scholar]
- 37.Xiao Y, Freeman GJ. A New B7:CD28 family checkpoint target for cancer immunotherapy: HHLA2. Clin Cancer Res Off J Am Assoc Cancer Res. 2015;21:2201–2203. doi: 10.1158/1078-0432.CCR-14-2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao R, Chinai JM, Buhl S, et al. HHLA2 is a member of the B7 family and inhibits human CD4 and CD8 T-cell function. Proc Natl Acad Sci USA. 2013;110:9879–9884. doi: 10.1073/pnas.1303524110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cheng H, Janakiram M, Borczuk A, et al. HHLA2, a new immune checkpoint member of the B7 family, is widely expressed in human lung cancer and associated with EGFR Mutational Status. Clin Cancer Res Off J Am Assoc Cancer Res. 2017;23:825–832. doi: 10.1158/1078-0432.CCR-15-3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Prasad DV, Richards S, Mai XM, Dong C. B7S1, a novel B7 family member that negatively regulates T cell activation. Immunity. 2003;18:863–873. doi: 10.1016/s1074-7613(03)00147-x. [DOI] [PubMed] [Google Scholar]
- 41.Chen C, Zhu YB, Shen Y, Zhu YH, Zhang XG, Huang JA. Increase of circulating B7–H4-expressing CD68+ macrophage correlated with clinical stage of lung carcinomas. J Immunother. 2012;35:354–358. doi: 10.1097/CJI.0b013e31824212c4. [DOI] [PubMed] [Google Scholar]
- 42.Zhang X, Cai L, Zhang G, Shen Y, Huang J. B7–H4 promotes tumor growth and metastatic progression in lung cancer by impacting cell proliferation and survival. Oncotarget. 2017;8:18861–18871. doi: 10.18632/oncotarget.14475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen C, Qu QX, Shen Y, Mu CY, Zhu YB, Zhang XG, Huang JA. Induced expression of B7–H4 on the surface of lung cancer cell by the tumor-associated macrophages: a potential mechanism of immune escape. Cancer Lett. 2012;317:99–105. doi: 10.1016/j.canlet.2011.11.017. [DOI] [PubMed] [Google Scholar]
- 44.Lin WW, Karin M. A cytokine-mediated link between innate immunity, inflammation, and cancer. J Clin Investig. 2007;117:1175–1183. doi: 10.1172/JCI31537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420:860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.