Abstract
Functional rescue of NK-cells in solid tumors represents a central aim for new immunotherapeutic strategies. We have conducted a genomic, phenotypic and functional analysis of circulating NK-cells from patients with HCV-related liver cirrhosis and hepatocellular carcinoma. NK-cells were sorted from patients with HCC or liver cirrhosis and from healthy donors. Comparative mRNA gene expression profiling by whole-human-genome microarrays of sorted NK-cells was followed by phenotypic and functional characterization. To further identify possible mediators of NK-cell dysfunction, an in vitro model using media conditioned with patients’ and controls’ plasma was set up. Metabolic and cell motility defects were identified at the genomic level. Dysregulated gene expression profile has been translated into reduced cytokine production and degranulation despite a prevalent phenotype of terminally differentiated NK-cells. NKG2D-downregulation, high SMAD2 phosphorylation and other phenotypic and molecular alterations suggested TGF-β as possible mediator of this dysfunction. Blocking TGF-β could partially restore functional defects of NK-cells from healthy donors, exposed to TGF-β rich HCC patients’ plasma, suggesting that TGF-β among other molecules may represent a suitable target for immunotherapeutic intervention aimed at NK-cell functional restoration. By an unbiased approach, we have identified energy metabolism and cell motility defects of circulating NK-cells as main mechanisms responsible for functional NK-cell impairment in patients with hepatocellular carcinoma. This opens the way to test different approaches to restore NK-cell response in these patients.
Electronic supplementary material
The online version of this article (10.1007/s00262-020-02561-4) contains supplementary material, which is available to authorized users.
Keywords: NK-cells, Hepatocellular carcinoma, Immunometabolism, Cell motility, TGF-β
Introduction
Natural killer (NK) cells are innate lymphoid cells that represent a first-line defense against viral infections and tumors. Besides their direct cytotoxic- and cytokine-producing activities, NK-cells exert a multifaceted role modulating the function of multiple cell types in anti-tumor immunity [1].
The recognition of target cells by NK-cells relies on the integration of multiple signals from a complex array of inhibitory and activating receptors. Inhibitory NK receptors recognize major histocompatibility complex class I molecules that are usually downregulated in tumors. However, NK-cell activation also requires stimulation by the stress-induced ligands of the activating receptors, including NKG2D, NKp46 and NKp30. Additional mechanisms involved in the regulation of NK-cell function are CD16-mediated antibody-dependent cellular cytotoxicity, interaction with cytokines and Toll-like receptor ligands [2].
NK-cell anti-tumor efficacy is progressively dampened during tumor progression. The tumor immune microenvironment by the release of soluble factors or other regulatory mechanisms may be responsible for decreased expression of activating receptors as well as upregulation of inhibitory ones, inefficient trafficking to the tumor site or alteration of intracellular pathways. Moreover, intratumor hypoxic conditions or depletion of fundamental nutrients contributes to NK-cell dysfunction. Thus, the understanding of the metabolic demands of NK-cells is an important goal for cancer-related immune modulatory strategies [3].
Hepatocellular carcinoma (HCC) is the most common type of liver cancer with increasing incidence in Western countries [4]. HCC develops in a context of liver inflammation due to chronic infection with hepatitis B virus (HBV) or hepatitis C virus (HCV), metabolic diseases and chronic alcohol abuse [5]. Liver inflammation triggers a cycle of hepatocyte necrosis and regeneration evolving toward liver cirrhosis that represents a risk factor for the development of HCC. Several lines of evidence support the role played by NK-cells on the evolution of HCC. The multiplicity and phenotype of tumor-infiltrating NK-cells have been associated with disease prognosis [6, 7]; however, the HCC microenvironment has been reported to exert an inhibitory effect on NK-cell function [6–9].
Curative tumor ablation is limited to early HCC, and treatment with multikinase inhibitors is only partially effective in advanced stage. Recently, therapeutic interventions aimed at restoring anti-tumor immune response have shown promising results [10]. NK-cells might represent an attractive immunotherapeutic target due to their rapid activation and lack of antigen specificity. However, even if several studies have described NK-cell functional defects in HCC [6–9, 11], the molecular basis underlying NK-cell functional impairment in HCC is still incompletely understood.
In the present study, we have employed a combined approach including genomic and phenotypic characterization of circulating NK-cells from HCC patients. This strategy allowed the identification of metabolic and functional alterations, together with some clues to possible interventions aimed at restoring NK-cell function as a novel rationale for immune modulatory therapies.
Materials and methods
Patients and biological samples
Peripheral blood mononuclear cells (PBMCs) were isolated from fresh heparinized blood of patients with HCC arising in HCV-related liver cirrhosis (HCC; n. 15) or liver cirrhosis with HCV infection (LC; n. 10) and of healthy controls (HD; n. 14) by Ficoll–Hypaque density-gradient centrifugation. Cells were cryopreserved in liquid nitrogen until the day of analysis. Plasma samples were maintained at − 80 °C until used.
HCC diagnosis had been made by ultrasonography and computed tomography or magnetic resonance imaging in selected cases. Hepatitis B surface antigen (HBsAg) and anti-human immunodeficiency virus were negative in all cases. All patients with LC and HCC were HCV infected (HCV-Ab and HCV-RNA positive) in Child–Pugh class A. HCC patients were in early stage (Barcelona Clinic Liver Cancer stage A). Mean age was similar in patients with LC (72.2 ± 7.4) and HCC (75.1 ± 2.3) and in HD (54.2 ± 5.3). The study was approved by the local ethical committee (Comitato Etico Indipendente of the Azienda Ospedaliero-Universitaria of Parma, Parma, Italy). All participants gave written informed consent to participate in the study.
Gene expression profiling
After thawing, PBMCs from 15 patients with HCC, 10 patients with LC and 7 HD were stained with anti-CD3-PeCy7 (Biolegend), anti-CD16-FITC (BD), anti-CD56-Pe-CF594 (BD) and then 7AAD (7-amino-actinomycin D)-PerCP-Cy5.5 vitality dye. NK-cells identified as CD3−, CD56+, CD16+ or CD16− were sorted by FACSAria III Cell Sorter (BD). RNA was purified from sorted NK-cells with the RNase-Free DNase I Kit (NorgenBiotek), according to the manufacturer’s instructions. Total RNA concentration was determined using a Nanodrop spectrophotometer, and RNA integrity was evaluated with a Bioanalyzer 2100 traces system (Agilent Technologies). RNA was used to synthesize cRNA that was labeled with Cy3 and hybridized to 60-mer oligonucleotide whole-human-genome microarrays (Human GE 8x60K v2 SurePrint G3, Agilent Technologies), following the manufacturer’s protocol. Microarray slides were scanned with an Agilent dual-laser DNA microarray scanner. The Agilent Feature Extraction software version 7.5 was used with default settings to obtain normalized expression values from the raw scans.
Phenotypic analysis of NK-cells
PBMCs from 15 patients with HCC, 8 patients with LC and 14 HD were suspended in RPMI-1640 containing 8% human serum and stained with monoclonal antibodies specific for CD3-Alexa Fluor 700 (BioLegend), CD16-FITC (BD) and CD56-PE-Vio615 (MiltenyiBiotec) on a FACS Canto II (BD) flow cytometer using the FACS Diva Software (BD). Frequency of CD56DIM and CD56BRIGHT NK-cells was defined for each patient evaluating different fluorescence intensities of CD3−CD56+ cells, and expression of each marker was considered on total, CD56DIM or CD56BRIGHT cell populations, both as percentage and as Median Fluorescence Intensity (MFI). Peripheral blood NK-cells (PBNK) was also characterized by the combination of monoclonal antibodies identifying activating and inhibitory receptors, such as: NKG2A (CD94)-PerCP-Cy5.5 (BD Bioscience), NKG2D-PE (eBioscience), NKp30-Alexa Fluor 488 (R&D System), NKp46-APC-Fire750 (BioLegend) and NKp44-PEVio770 (MiltenyiBiotec). Specific differentiation markers, including CD27-FITC (BD) and CD11b-APC-Cy7 (BioLegend), and TGFβR2-Alexa Fluor647 (MiltenyiBiotec) were also evaluated.
For the detection of intracellular antigens, cells were fixed and permeabilized with Fixation Permeabilization Concentrate and Diluent and Permeabilization Buffer (eBioscience) following the manufacturer’s instructions and stained with Granzyme-B-AlexaFluor647 (BD), Perforin-eFluor710 (eBioscience), EOMES-PE (Invitrogen) and Phospho-SMAD2-PECF594 (Ser465/467) (BD).
Functional analysis of NK-cells
IFN-γ and TNF-α production by NK-cells was evaluated in all samples available for phenotypic analysis after stimulation with PMA (phorbol 12-myristate 13-acetate) (50 ng/ml) and ionomycin (1 μg/ml) for 4 h. After 1 h, monensin (10 μg/ml) was added. Surface staining with anti-CD3-PE (BD), anti-CD56-PE CF594 (BD) and anti-CD16-FITC (BD) was performed, and then, cells were fixed with the medium A reagent and then permeabilized with medium B reagent (Nordic Mubio) in accordance with manufacturer’s instructions. Cytokine determinations were performed by intracellular cytokine staining (ICS) with anti-IFN-γ-PerCp-Cy5.5 (Biolegend) and anti-TNF-α-APC (Biolegend) monoclonal antibodies and analyzed by flow cytometry. The CD107a degranulation assay was performed to assess the cytotoxic potential. Cells were stimulated with PMA and ionomycin and then incubated with anti-CD107a-PE-Cy7 (BD) and monensin (10 μg/ml) for 4 h. Data were expressed as the difference between the percentage of cytokine- or CD107a-positive NK-cells in the stimulated and unstimulated samples.
Cytotoxic potential of NK-cells
The killing potential of purified PBNKs from HD (n. 7), LC (n. 7) and HCC (n. 7) was evaluated by both bioluminescence and flow-cytometry-based assays. NK-cells were purified by positive selection using CD56 beads (MiltenyiBiotec) following the manufacturer’s instructions and then stimulated o/n with IL-12 + IL-18 (5 ng/ml).
For bioluminescence-based aCella-TOX kit (Cell Technology), target K562 cells were washed, resuspended in RPMI + 10% low IgG FBS (GIBCO), counted and plated in a 96-well U bottom plate (3000 cells/well). Purified and stimulated NK-cells were then counted and resuspended at effector to target (E:T) ratios of 2.5, 7.5, and 15:1. Each condition was seeded in duplicate and incubated for 4 h at 37 °C. The assay was carried out following the manufacturer’s instructions, and the results read by a luminometer (Perkin-Elmer).
NK-cells cytotoxicity toward K562 target cells was also evaluated by flow cytometry. K562 cells were labeled with 0.5 μM CFSE (Carboxyfluorescein succinimidyl ester) and then seeded at 3000 cells/well. Purified NK-cells were added to K562 cells at 7.5:1 E:T ratio. K562 cells without effectors were used as a negative control to establish the spontaneous death rate of target cells. After 4 h incubation, cells were collected, stained with the vitality probe 7AAD (Becton–Dickinson) and analyzed by flow cytometry. Data were expressed as the difference between the percentage of CFSE/7AAD double positive K562 in the experimental and control samples.
Evaluation of NK-cell metabolic status, migration, and conjugation
Mitochondrial membrane potential was measured on NK-cells by the potentiometric probe JC-1 (Molecular Probes, Life Technologies). After surface staining with anti-CD3 and anti-CD56, PBMCs were incubated with JC-1 (2.5 μg/ml) for 10 min at RT and protected from light before flow cytometry analysis. Samples were then stained with the viability probe 7-AAD and finally acquired on FACS Canto II. Depolarized NK-cells were quantified by the percentage of FL1high/FL2low cells (JC-1 staining) detected in the samples in different conditions [12].
Glucose uptake assay was performed under different experimental conditions. PBMCs were washed with PBS 1X to eliminate endogenous glucose and stained for 30 min at 37 °C with the glucose analog 2-NBDG (2-deoxy-2-[(7-nitro-2,1,3-benzoxadiazol-4-yl) amino]-d-glucose, ThermoFisher; 40 μM) in glucose-free RPMI with 10% dialyzed FBS and finally stained with anti-CD3, anti-CD56 and the viability probe 7-AAD. Frequency of 2-NBDG NK-positive cells was measured.
PBMCs were isolated from five healthy controls and were cultured overnight in basal conditions (8% human serum in RPMI1640) or after addition of 30% plasma pools obtained from the 15 patients with HCC or the 10 patients with LC. Cells were incubated with or without 10 μg/ml anti-TGF-β 1,2,3 antibodies (R&D Systems).
The metabolic status of plasma-treated cells was investigated as described above. To test cell migration, PBMCs (5 × 105) were plated in the upper chamber of a 3 µm Transwell Permeable Support (Corning). After 2 h, NK-cells that had migrated to the lower chamber were stained and counted by flow cytometry using six-color TBNK Reagent with BD Trucount™ Tubes (BD). Data were expressed as migration index, obtained as ratio between the absolute number of NK-cells migrating in the lower chamber with respect to those seeded in the upper chamber.
A flow cytometry-based method was used to evaluate adhesion of NK-cells to target cells (K562) in different experimental conditions. The two cell types were labeled with distinct fluorescent dyes (K562 cells with 0.5 μM CFSE, NK-cells with anti CD3-APC-Cy7 and anti-CD56-APC-R700). The CFSE staining was stopped by addition of two volumes of cold fetal bovine serum and washed three times with HBSS. Target cells were added to NK-cells in 5:1 ratio, mixed by gentle vortexing and centrifuged at 4 °C for 3 min at 300 rpm [13]. Samples were incubated at 37 °C for different times (0 min, 15 min and 30 min), then fixed by adding 1 ml of Fixation Permeabilization Concentrate and Diluent (eBioscience) and analyzed on a FACS Canto II (BD) flow cytometer. NK-cells firmly adherent to target cells were quantified, and data were expressed as MFI of NK-cells population gated on target cells.
TGF-β ELISA assay
TGF-β1 levels were quantified in plasma from 15 HCC and eight LC patients and from 7 HD using the Quantikine Human TGF-β1 Immunoassay (R&D). Briefly, 40-µl plasma samples were activated by acidification with 20 µl of 1 N HCl for 10 min at RT and neutralization with an equal volume of 1.2 N NaOH/0.5 M HEPES. Activated samples were then used to perform the ELISA assay, following the manufacturer’s instructions.
Statistical analysis
Statistical analysis was carried out by GraphPad Prism (version 7), JASP 0.10.2 (https://jasp-stats.org/) and Jamovi 1.0.7 (https://www.jamovi.org) softwares. After analysis of variance (F test) and of normality (Kolmogorov–Smirnov test), Mann–Whitney U test, Wilcoxon-matched-paired test or paired t test were applied as appropriate. Differences between groups were evaluated by Kruskal–Wallis test, and p values were corrected for pairwise multiple comparisons, according to the method of Dunn. Moreover, in the experiment where plasma patients were used, the Friedman test was applied and corrected for pairwise multiple comparisons, according to the Conover test. A p value ≤ 0.05 (two tailed) was considered significant.
GeneSpring software package GX13.1.1 (Agilent Technologies) was used for quality control checks, data normalization by the 75th percentile method and initial microarray data analysis. Probes detected in at least four replicates for each condition were retained for further analysis. ANOVA with the Benjamini–Hochberg correction for multiple testing (FDR, ≤ 0.05) was used to track genes differentially expressed between HD, LC and HCC patients. Genes differentially expressed between the three groups were identified by Student–Newman–Keuls post hoc analysis.
Unsupervised hierarchical clustering was performed after a median baseline transformation of all the samples and using Ward’s linkage and Euclidean distance. Principal Component Analysis (PCA) was applied to diminish dimensionality of the data with an orthogonal transformation to visualize the clustering of sample populations.
Gene Set Enrichment Analysis (GSEA) was performed on all detected probes to identify molecular pathways significantly overrepresented among the upregulated and downregulated genes and to compare the expression profiles with other published studies. To this end, we used the Molecular Signature Database v 6.0 (C2, canonical pathways; C5, GO gene sets) with the permutation type set to ‘gene set’ to calculate statistical significance, as suggested for fewer than seven replicates; default settings were applied to all other options. Gene sets passing this filter were considered as significantly enriched if the false discovery rate (FDR), calculated using Signal2Noise as metric and 1000 permutations of gene sets, was < 0.25.
Differentially expressed genes (DEGs) with an absolute fold change ≥ |1.5| and p < 0.05 were uploaded into ingenuity pathway analysis software (Qiagen, Redwood City, CA, USA) for core analysis and identification of the most relevant canonical pathways involving DEGs. The diseases and function analysis was also performed to analyze the relationship between DEGs in the dataset and known disease states or biological functions that might be affected by the observed expression alterations. A p value < 0.05 was used as a cutoff for significance. The Z score (number of standard deviations from the mean) positive or negative value was used to predict activation or inhibition, respectively. A Z score > |2| was considered biologically significant.
Results
Gene expression profiling of NK-cells
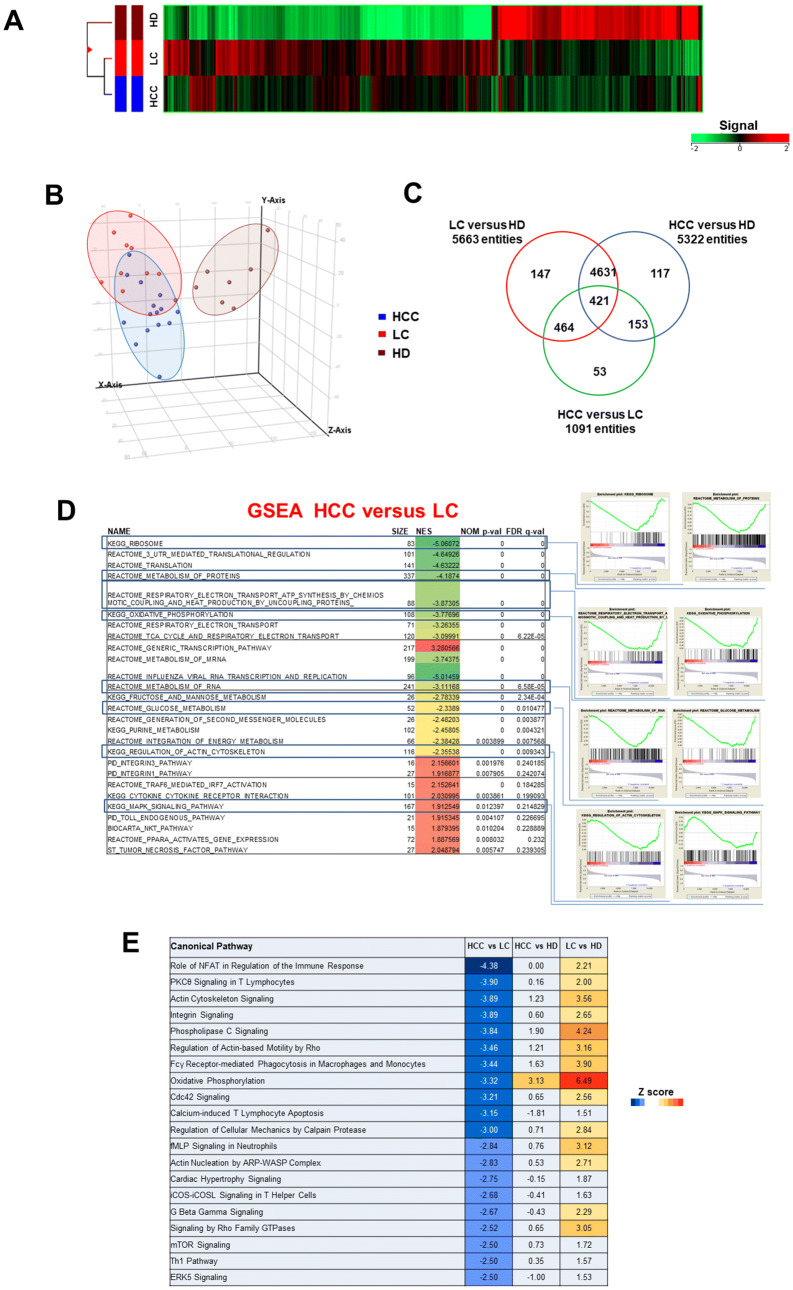
NK-cells (CD3−, CD56+) were sorted from peripheral blood of patients with HCV-related liver cirrhosis (LC; n. 10) and HCC arising in HCV-related LC (HCC; n. 15) and from healthy donors (HD; n. 7) (Supplementary Fig. 1A). Analysis of variance was carried out on genome-wide expression profiling data on detected probes from genome-wide expression profiling data (19,716 genes) identified a subset of 5986 differentially expressed genes (DEGs) in NK-cells from patient subgroups and controls. Hierarchical clustering of differentially expressed genes is depicted in Fig. 1a. PCA analysis showed partial segregation among groups of patients and controls (Fig. 1b). Comparison between LC and HCC identified 1091 DEGs, whereas 5322 and 5663 genes were differentially expressed between HCC and HD and between LC and HD, respectively. Figure 1c shows the overlap between the three comparisons.
Fig. 1.
Gene expression pattern of peripheral blood NK-cells from patients with HCV-related liver cirrhosis (LC) or HCC and healthy donors (HD). a Hierarchical-clustering representation of the 5986 genes identified as differentially expressed in NK-cells from LC (n. 10), HCC (n. 15) and HD (n. 7) by ANOVA with Benjamini–Hochberg correction (p ≤ 0.05). Data were median-normalized before clustering; upregulated and downregulated genes are shown in red and green, respectively. b Patient groups analyzed by Principal Component Analysis (PCA) of ANOVA-filtered data. c Venn diagram representation of differentially expressed genes among the three NK-cell subsets (HCC vs LC, HCC vs HD and LC vs HD), determined by post hoc Student–Newman–Keuls test analysis. d List of enriched gene sets in NK-cells from patients with HCC and LC, identified by GSEA (MSigDB, C2 canonical pathways set). Representative enrichment plots are shown on the right. NES: Normalized enrichment score; FDR: false discovery rate. e IPA canonical pathways (CPs) associated with genes differentially expressed in PBNK. Left: top 20 IPA CPs based on Z score values. Orange indicates upregulation, blue downregulation. The color intensity is proportional to Z score values
We then focused on the comparison between NK-cells from patients with HCC and LC. Analysis by moderated t test with Benjamini–Hochberg correction (p ≤ 0.05, Fold Change threshold 1.5) identified 1435 DEGs, 1114 of which downregulated in HCC (Supplementary Fig. 1B).
Gene Set Enrichment Analysis (GSEA) was performed to deepen the understanding of NK expression modifications related to the carcinogenic process. Gene sets related to cell metabolism (protein synthesis, glucose metabolism, oxidative phosphorylation) and to cell motility (actin cytoskeleton) appeared to be enriched and downregulated in HCC compared to LC (Fig. 1d). A detailed analysis of gene ontology is reported in Supplementary Fig. 2. GSEA was also applied to the comparison between HCC and HD transcriptome profiles allowing to identify an enrichment in downregulated genes related to PI3K (Phosphoinositide 3-kinase-/RAS-mediated intracellular signaling pathways), to molecular interaction between extracellular matrices (integrins pathway) and to actin-associated cell motility. Conversely, comparison between LC and HD transcriptome profiles showed an enrichment in upregulated pathways, such as glycolysis and mitochondrial metabolic functions, proteasome-mediated protein degradation and cell cycle regulation (Supplementary Tables 1 A and B).
Fig. 2.
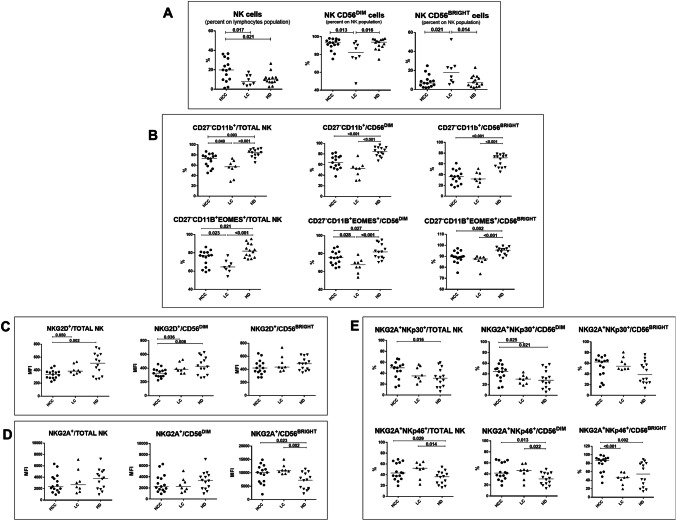
Frequency and phenotype of NK-cells from patients with HCC or HCV-related liver cirrhosis (LC) and healthy donors (HD). a Frequency of total, CD56DIM and CD56BRIGHT NK-cells in HCC (n. 15), LC (n. 8) and HD (n. 14). b Upper panels: percentage of CD11b+CD27− cells in NK subsets from HCC, LC and HD. Lower panels: expression of the transcription factor Eomesodermin (EOMES) in CD11b+CD27− NK-cell subsets from HCC, LC and HD. c, d Median Fluorescence Intensity (MFI) of NKG2D (C) and NKG2A (D) expression in NK-cell subsets from HCC, LC and HD. e Co-expression of NKG2A/NKp30 (upper panels) and NKG2A/NKp46 (lower panels) in NK-cell subsets from HCC, LC and HD. Horizontal lines represent median values. Differences between multiple groups were evaluated by the Kruskal–Wallis test; p values were corrected for pairwise multiple comparisons by the Dunn’s test
To identify the most significant canonical pathways associated with DEGs, data were analyzed by ingenuity pathway analysis (IPA). The results showed differential regulation of pathways involved in NK-cell function, cell motility and energy metabolism. Z score values (Fig. 1e) and significance (number of DEGs in each pathway) values (Supplementary Fig. 1, C) indicated predicted inhibition in the comparison of HCC versus LC, and activation in the comparison of LC versus HD. Interestingly, the HCC-HD comparison only predicted the modest activation of the oxidative phosphorylation pathway. Diseases and function analysis showed that cellular movement and cell-to-cell signaling and interaction were the most represented categories and were both predicted to be downregulated in the comparison of HCC versus LC. In addition, cell death/apoptosis was predicted to be upregulated, whereas cell survival/viability was predicted to be downregulated (Supplementary Fig. 1, D).
NK-cell phenotypes
PBNK analysis by flow cytometry detected a significantly increased percentage of NK-cells on total lymphocytes in HCC (n. 15) compared to LC (n. 8) patients and to HD (n. 14). Phenotypic characterization showed that terminally differentiated NK-cells were more represented in HD than in HCC and LC patients and in HCC compared to LC as shown by higher percentage of CD56DIM NK-cells (Fig. 2a) and CD11b single positive (SP) (CD27 negative) CD56DIM-NK-cells (Fig. 2b). Moreover, staining with Eomesodermin (EOMES), marker of mature NK-cells, showed similar results with an enrichment of CD27−CD11b+EOMES+ NK-cells in HD compared to HCC and LC patients and in HCC compared to LC (Fig. 2b lower panels). NK-cells were also analyzed for the intensity of NKG2A (CD94) expression within CD56BRIGHT and CD56DIM subpopulations, confirming a higher frequency of NKG2ALOW CD56DIM cells in HCC patients compared to LC (Supplementary Fig. 3) and suggesting an enrichment of cytotoxic differentiated NK-cells.
Further analysis showed lower expression of NKG2D in HCC compared to LC and HD and similar levels of NKG2A expression in the three groups (Fig. 2c, d). NKp46 expression was lower in HD compared to both patient groups, and NKp30 expression was significantly higher in HCC compared to LC in CD56 DIM and to HD in the total NK-cell population (not shown). However, significantly higher percentage of NKG2A/NKp30 co-expression was observed in HCC versus LC in the CD56DIM subset, whereas NKG2A/NKp46 co-expression was higher in HCC compared to HD and also to LC in the CD56BRIGHT subset (Fig. 2e).
Representative dot plots of phenotypic analysis are shown in Supplementary Fig. 4.
NK-cell functional defects and TGF-β as possible mediator for dysfunction
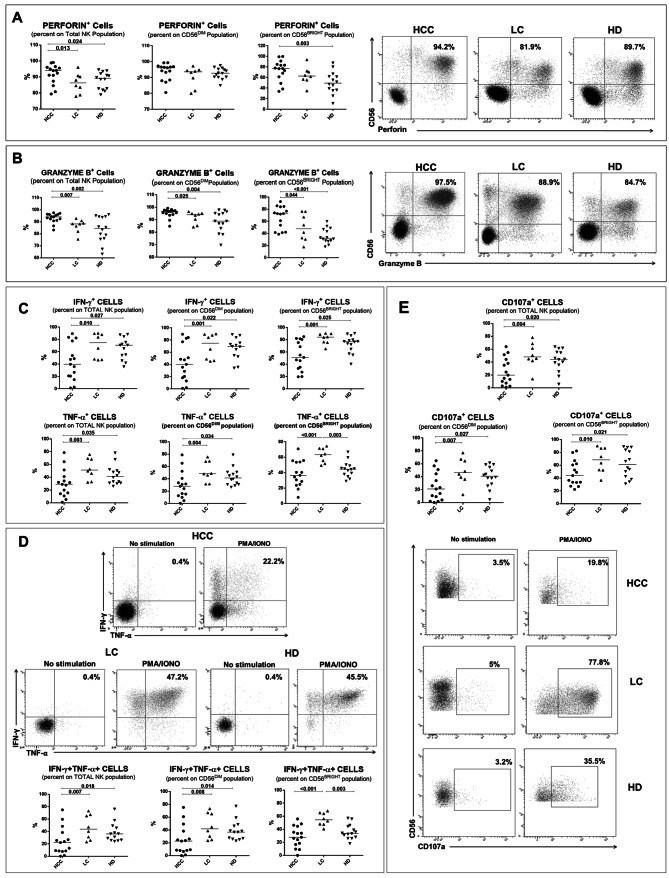
Molecules associated with cytotoxic response were detected in NK-cells from HCC patients at higher levels in total NK-cells from HCC patients compared to LC and HD, and granzyme B was differentially expressed in both the CD56BRIGHT and CD56DIM subsets, while perforin positive cells only in CD56BRIGHT from HCC compared to HD (Fig. 3a, b). However, cytokine production (Fig. 3c, d) and degranulation (CD107a expression) (Fig. 3e) were significantly lower in NK-cells from HCC patients compared to LC patients and, limited to TNF-α, to HD. Polyfunctional NK-cells were significantly more represented in HD and LC compared to HCC (Fig. 3d).
Fig. 3.
Expression of molecules associated with cytotoxic response in NK-cells and NK-cell functional analysis. a, b Left panels: percentage of Perforin + (a) and Granzyme B + (b) in NK-cell subsets from HCC, LC and HD. Right panels: representative dot plots of Perforin (a) and Granzyme B (b) staining in HCC, LC and HD. C. IFN-γ and TNF-α production was evaluated with or without PMA (phorbol 12-myristate 13-acetate) (50 ng/ml) and ionomycin (1 μg/ml) for 4 h, in NK-cell subsets. After addition of monensin (10 μg/ml), cytokine determinations were performed by intracellular cytokine staining (ICS). d Upper panels: representative dot plots of IFN-γ and TNF-α staining upon stimulation with PMA/Ionomycin in HCC, LC and HD. Lower panels: frequency of polyfunctional IFN-γ+TNF-α+ NK-cells in HCC, LC and HD. e The cytotoxic potential of NK-cells evaluated by the CD107a degranulation assay. After stimulation with PMA and ionomycin, cells were incubated with anti-CD107a, as in c. Upper panels: percentage of CD107a+ cells in NK-cell subsets from HCC, LC and HD. Lower panels: Representative dot plots showing CD107a expression in NK-cells from HCC, LC and HD upon PMA/Ionomycin stimulation. c–e Data are expressed as the difference between the percentage of cytokine+ or CD107a+ NK-cells in the stimulated and unstimulated samples. Horizontal lines represent median values. Differences between multiple groups were evaluated by the Kruskal–Wallis test; p values were corrected for pairwise multiple comparisons by the Dunn’s test
In order to confirm the cytotoxic dysfunction of NK-cells from HCC patients, two different killing assays were used and K562 cells were employed as target cells. Supplementary Fig. 5, A shows decreased cytotoxic capacity of NK-cells from HCC patients compared to LC and HD at different effectors to target (E: T) ratios by a bioluminescence-based assay. In addition, Supplementary Fig. 5, B highlights the cytotoxic dysfunction of NK-cells from HCC patients evaluated by a flow cytometry-based assay. The percentage of CFSE/7-AAD double positive K562 cells (dead target cells) was significantly less abundant when NK-cells from HCC patients were employed as effectors.
NK-cells from a subset of samples from different study groups were tested to confirm the metabolic defects shown by bioinformatic analysis. Mitochondrial membrane depolarization and glucose metabolism (glucose uptake) were measured. As shown in Supplementary Fig. 6, A NK-cells from HCC patients displayed a defective mitochondrial functionality, shown by the increase in depolarized mitochondria compared to LC patients and HD. Glucose uptake capacity was not enhanced in NK-cells from HCC patients as a compensation mechanism for oxidative phosphorylation defect, confirming the existence of metabolic impairment (Supplementary Fig. 6, B).
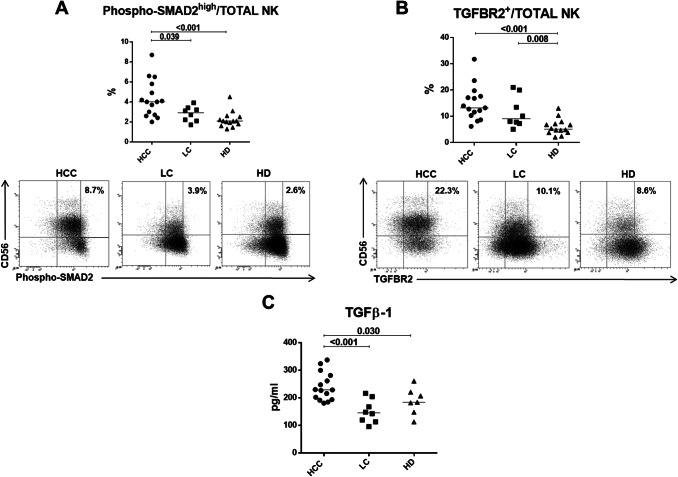
A major determinant of NK effector function is the activating receptor NKG2D that was found to be downregulated in HCC both at mRNA and protein level (data not shown and Fig. 2c). Since TGF-β has been implicated in NKG2D downregulation in the tumor environment [14], we investigated the possible role of TGF-β on the functional impairment of NK-cells in HCC. The phosphorylation of the TGF-β downstream signaling target SMAD2, detected as p-SMAD2high in NK-cells, was significantly more represented in HCC patients compared to LC and HD (Fig. 4a), supporting the activation of TGF-β signaling in HCC. Interestingly, a higher frequency of TGF-βR2-positive NK-cells was also present in HCC and LC patients compared to HD (p <0.05) (Fig. 4b). The higher percentage of phospho-SMAD2high NK-cells suggests that NK-cells from HCC patients may have been more profoundly affected by TGF-β than NK-cells from patients with liver cirrhosis. In agreement with these findings, TGF-β plasma concentration was significantly higher in HCC patients compared to LC and HD (Fig. 4c).
Fig. 4.
SMAD2 activation, TGF-βR2 expression and TGF-β plasma levels. a Upper panel: percentage of carboxy-termini (Ser465/467)-phosphorylated SMAD2 positive NK-cells from HCC, LC and HD. Lower panels: representative flow cytometry dot plots showing Phospho-SMAD2high analysis in study groups. b Upper panel: percentage of NK-cells expressing TGF-βR2 in HCC, LC and HD and Lower panels: representative dot plots of TGF-βR2 staining in HCC, LC and HD. c TGF-β plasma levels in HCC (n. 15), LC (n. 8) and HD (n. 7). Horizontal lines represent median values. Differences between multiple groups were evaluated by the Kruskal–Wallis test; p values were corrected for pairwise multiple comparisons by the Dunn’s test
An in vitro model to recapitulate NK-cell functional defects
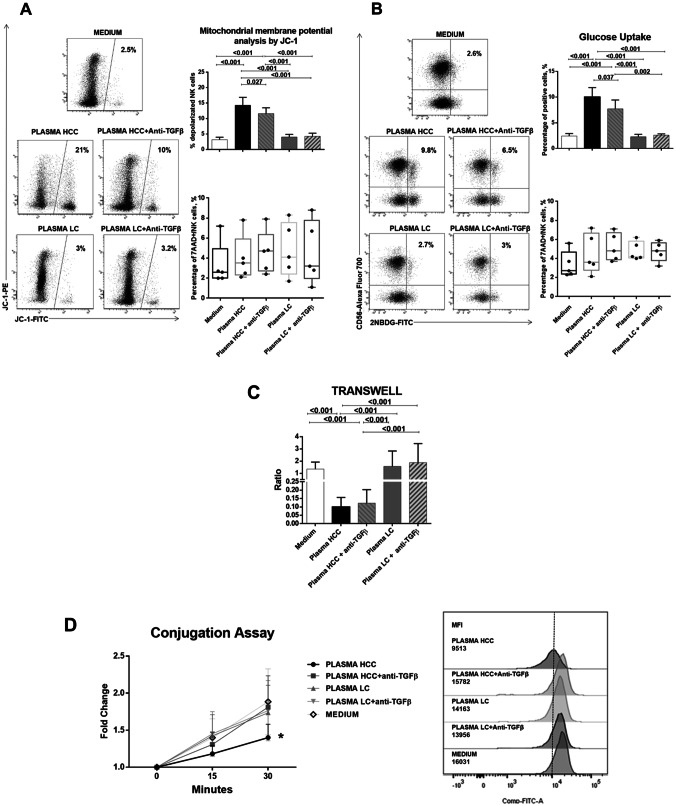
In the attempt to reproduce the metabolic and functional defects shown by immunophenotypic and bioinformatic analysis, NK-cells from healthy donors were tested in basal conditions and after incubation with plasma obtained from HCC or LC patients, with or without anti-TGF-β antibodies. Mitochondrial membrane depolarization, glucose metabolism (glucose uptake), cytokine production, NK-cell degranulation, migration and adhesion to target cells were tested.
Alterations of oxidative phosphorylation suggested by bioinformatic and flow cytometry analysis could be reproduced in vitro in NK-cells treated with HCC patients’ plasma. Mitochondrial dysfunction, as shown by high level of electron chain depolarization, was significantly increased by HCC plasma, while partial but significant restoration of mitochondrial function was achieved after TGF-β blocking. LC plasma did not alter NK-cell mitochondrial membrane potential (Fig. 5a). Glucose uptake, augmented as a regulatory mechanism in case of defects in oxidative phosphorylation, was indeed higher upon incubation with HCC plasma and was significantly diminished by anti-TGF-β (Fig. 5b).
Fig. 5.
Analysis of mitochondrial membrane potential, glucose metabolism, cell migration and cell adhesion in NK-cells from healthy controls after exposure to HCC or LC plasma. PBMCs from healthy donors (n. 5) were cultured overnight in basal conditions or with addition of 30% plasma pools derived from HCC or LC patients, with or without anti-TGF-β antibody (10 µg/ml). a Mitochondrial membrane potential was measured on NK-cells by the potentiometric probe JC-1. After surface staining with anti-CD3 and anti-CD56, PBMCs were incubated with JC-1. Samples were then stained with the viability probe 7-AAD and finally acquired on flow cytometer. Depolarized NK-cells (right upper panel) were quantified by the percentage of FL1high/FL2low cells (JC-1 staining) detected in the samples in different conditions. Percentages of 7AAD positive NK-cells (right lower panel) are shown by whiskers plots. Representative dot plots (left panels) show modulation of depolarized NK-cells in different experimental conditions. b Glucose uptake assay was performed on NK-cells. PBMCs were stained with the glucose analog 2-NBDG (2-deoxy-2-[(7-nitro-2,1,3-benzoxadiazol-4-yl) amino]-d-glucose, 40 μM). Frequency of 2-NBDG-positive NK-cells was measured (right upper panel). Percentages of 7AAD positive NK-cells (right lower panel) are shown by whiskers plots. Representative dot plots (right panels) show glucose uptake in NK-cells from different experimental conditions. c 5 × 105 PBMCs from each healthy control were plated in the upper chamber of a 3 µm Transwell Permeable Support, after overnight incubation in the experimental conditions. After additional 2 h, NK-cells that had migrated to the lower chamber were stained and quantified by flow cytometry. Data were expressed as migration index, obtained as ratio between the absolute number of NK-cells migrating in the lower chamber with respect to those seeded in the upper chamber. d Adhesion of NK-cells to target cells (K562) was tested by a flow cytometry-based method in different experimental conditions. The two cell types were labeled with distinct fluorescent dyes (K562 cells with 0.5 μM CFSE, NK-cells with anti-CD3 and anti-CD56). Target cells were added to NK-cells in 5:1 ratio and incubated at 37 °C for different times (0 min, 15 min and 30 min) and then fixed. Data are expressed as the ratio of MFI measured in the 15 and 30 min versus 0 min (left panel). Expression of cell conjugation is represented as MFI values of CD56 + cells in CFSE-FITC K562 fluorescence, comparing NK-cells from healthy subjects in different experimental conditions at 30 min (right panel). Graphs show mean ± SEM values. Statistical analysis was performed by the Friedman test; p values were corrected for pairwise multiple comparisons by the Dunn’s test
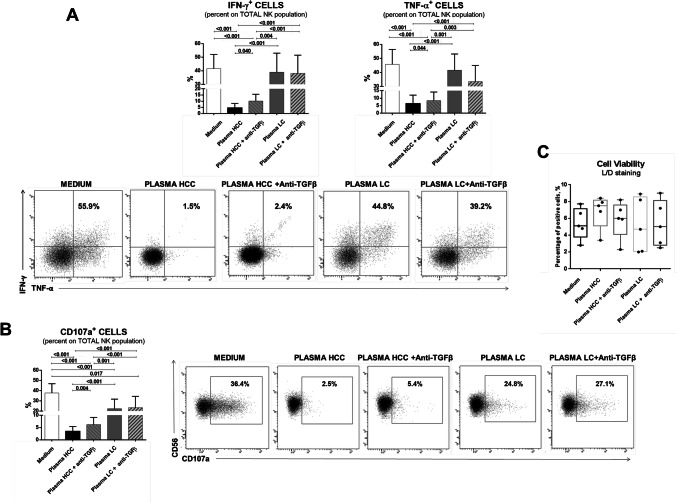
Migration and target cell adhesion were profoundly impaired in the model. While migration could not be restored blocking TGF-β (Fig. 5c), target cells adhesion could be clearly restored by anti-TGF-β (Fig. 5d), suggesting that for this functional defect, TGF-β may play central role. Finally, NK-cell functional defect in terms of cytokine production and degranulation could also be reproduced in this in vitro model and also in this case blocking TGF-β could partially restore NK-cell function (Fig. 6).
Fig. 6.
Functional analysis of NK-cells from healthy controls after exposure to HCC or LC plasma. a Upper panels: percentage of IFN-γ-positive, TNF-α-positive NK-cells obtained by PMA/Ionomycin stimulation of PBMCs from HD (n. 5) in the presence of HCC or LC plasma, with or without TGF-β blocking. Lower panels: representative dot plots illustrate cytokine production in the different experimental conditions b Upper panels: degranulation capacity (CD107a expression). Cells were stimulated with PMA and ionomycin and then incubated with anti-CD107a and monensin. Lower panels: dot plots showed representative CD107a expression in all experimental conditions. c NK-cell viability is shown by whiskers plots in the different experimental conditions. Graphs show mean ± SEM values. Statistical analysis was performed by the Friedman test; p values were corrected for pairwise multiple comparisons by the Dunn’s test
We tested NK-cell viability upon co-culture with HCC and LC patients’ plasma in order to exclude a toxic effect. 7-AAD, and live and dead staining showed no significant differences in all experimental conditions (Figs. 5a, b, 6c).
Discussion
In this study, we have performed a comparison of mRNA expression in NK-cells from HCC patients to patients with the pre-neoplastic condition of liver cirrhosis and to healthy donors. The bioinformatic analysis of gene expression data has been completed by an immunophenotypic study and by the analysis of the main NK-cell functions. It is known that NK-cells may present functional defects in HCC patients; however, previous studies have not systematically addressed the molecular mechanisms associated with these defects by an unbiased gene expression study. We could identify altered cellular pathways and hypothesize possible mechanisms that could be targeted for restoring an immune-protective NK-cell response.
RNA profiling of peripheral blood NK-cells from patients with HCV-related LC and HCC showed alterations of gene expression between the two groups and in comparison with healthy controls. Comparative evaluation indicated that differential regulation mainly involved pathways regarding energy metabolism and cell motility/adhesion. Interestingly, all relevant pathways appeared to be upregulated in cirrhotic patients compared to healthy controls and downregulated in HCC compared to cirrhotic patients. Further analysis confirmed that biological functions involving cellular movement, cell adhesion and gene expression were potentially downregulated in HCC, whereas upregulation of apoptosis was predicted. Metabolic reprogramming is increasingly recognized as a major driver of immune cell activation [3, 15]. The metabolic defects predicted by bioinformatic analysis were evaluated in a subset of patients and controls, supporting the existence of impaired oxidative phosphorylation in NK-cells from HCC patients. Oxidative phosphorylation is required to support both cytotoxic function and IFN-γ production by NK-cells, particularly after cytokine stimulation [16]. Analysis of PBNK from HCC patients showed a higher frequency of NK-cells and in particular of more differentiated phenotypes compared to LC, while HD showed the highest quote of differentiated cells. High frequency of CD56BRIGHT cells is typical of HCV-infected patients [17]. Interestingly, even if HCC patients were also HCV infected, CD56BRIGHT NK-cell subset was less abundant compared LC and similar to HD, suggesting a profound phenotypic change due to the tumor. Differentiated CD11b + CD27- NK-cells, known to be mature NK-cells with the highest cytolytic function [18], in our HCC patients displayed a reduced cytotoxic activity [19, 20]. In addition, the cutting-edge phenotype characterization with transcriptional factor EOMES corroborates the finding that circulating NK-cells from HCC patients were more differentiated to mature state [21] and enriched of NKG2ALOW CD56DIM NK-cells that represent an advanced stage of maturation [22]. Moreover, NK-cells from HCC patients were characterized by decreased cytokine production associated with an impaired degranulation and cytotoxic capacity, despite conserved expression of cytotoxic molecules. These apparently contradictory results might be linked to impaired formation of the immune synapse and to altered cellular motility, compared to LC and HD, as suggested by gene expression profiling. NK-cells from obese mice or humans were reported to have decreased rates of glycolysis and oxidative phosphorylation together with defective killing of tumor cells, partly linked to ineffective immune synapse formation [23]. Indeed, a human study exhibited that tumor-infiltrating liver-resident NK-cells had an increased production of glycolytic-derived lactate with a reduction in intracellular pH, leading to mitochondrial dysfunction and apoptosis [24]. Our transcriptional analysis highlights a putative similar molecular mechanism as shown by the enrichment of dysregulated pathways involved in oxidative phosphorylation and cell death.
The activating natural cytotoxicity receptors are important mediators of NK-cell cytotoxicity; however, we did not find significantly decreased expression of NKp46 and NKp30 in NK-cells from HCC patients, despite their defective killing capacity. By contrast, NKp30-expressing NK-cells were more frequent in HCC than in LC patients in the CD56DIM subset, different from a recent study [25] showing decreased frequency and expression of NKp30 in HCC. A less advanced disease stage of our patients may account for this discrepancy, consistent with the progressive functional deterioration of NK-cell function during tumor progression [1]. In addition, the higher percentage of NK-cells co-expressing NKp30 and inhibitory NKG2A receptor that we observed in HCC patients might translate into decreased effector function.
The interaction of NKG2D and its ligands is recognized as a major mechanism regulating NK-cell function in the tumor environment [26]. Remarkably, our results showed downregulation of NKG2D in NK-cells from HCC patients, in agreement with a recent study [25]. It is tempting to speculate that soluble factor(s) secreted by HCC may lead to NKG2D downregulation and consequent impairment in cytotoxicity and cytokine secretion, altogether reducing the migratory capacity of NK-cells in patients with HCC. Delivery of TGF-β1 by exosomes from cancer cells is known to downregulate NKG2D [14] and to inhibit oxidative phosphorylation in NK-cells [27] leading to impaired interferon production and NK degranulation [16]. In addition, tumor-derived TGF-β1 was recently reported to alter the migratory capacity of NK-cells by affecting their chemokine receptor repertoire [28]. Phosphorylated SMAD2-positive NK-cells were significantly higher in HCC compared to LC and HD, suggestive of active TGF-β signaling in NK-cells from HCC patients. Plasma from HCC patients contained increased TGF-β1 levels, consistent with previous data [29, 30].
TGF-β1 levels may be even higher at the tumor site where TGF-β1 can contribute to the immunosuppressive tumor microenvironment [31]. Increased TGF-β1 plasma levels are most likely a reflection of local production inhibiting NK-cells function also in the peripheral compartment. Incubation of healthy donor NK-cells with HCC plasma induced a metabolic reprogramming and functional impairment similar to the one predicted in HCC patients, and TGF-β blockade could partially recover NK-cell alterations, further supporting the involvement of TGF-β in NK-cell dysfunction. It is most likely that other soluble mediators beside TGF-β are responsible for the functional defects observed in our model and that the dysfunction of circulating NK-cells may only partially reflect the tumor microenvironment; however, the mechanisms here described allow us to conclude that TGF-β may represent a valuable target to investigate at the clinical level in HCC patients, extending the ongoing studies with TGF-β inhibitors that may find their application as monotherapy or in combination with checkpoint inhibitors.
Previous studies described that liver-resident NK-cells present peculiar characteristics [32, 33]; thus, our results, based on circulating NK-cells, may only partially represent the tumor immune microenvironment. However, it was observed that, in HCC, liver-resident NK-cells are less represented compared to the non-tumorous counterpart [33], thus is tempting to speculate that nonresident NK-cells recruited from the periphery may significantly contribute to the tumor-immune microenvironment.
In conclusion, our results show a metabolic and functional defect in circulating NK-cells in patients with HCC. Incubation of NK-cells from healthy donors with TGF-β-rich plasma from HCC patients can recapitulate the metabolic and functional impairment predicted by gene expression analysis and confirmed by phenotypic characterization of NK-cells from HCC patients. The possibility of partially antagonizing NK-cell dysfunction by anti-TGF-β antibodies corroborates the causal role of TGF-β and suggests a possible line of therapeutic intervention.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Authors’ contribution
AZ was involved in study concept and design, analysis and interpretation of data, drafting of the manuscript, critical revision of the manuscript for important intellectual content and statistical analysis. VB was involved in study concept and design, analysis and interpretation of data, drafting of the manuscript, critical revision of the manuscript for important intellectual content and statistical analysis Diana Canetti was involved in statistical analysis and technical support; VR was involved in statistical analysis and technical support; AO was involved in acquisition of data and analysis and interpretation of data; CC was involved in statistical analysis. VC was involved in technical support; BZ was involved in technical support; TT was involved in funding contribution and critical revision of the manuscript. RDV was involved in acquisition of data and analysis and interpretation of data; CF was involved in critical revision of the manuscript for important intellectual content; EC was involved in study concept and design, analysis and interpretation of data, drafting of the manuscript, critical revision of the manuscript for important intellectual content, obtained funding and statistical analysis. GM was involved in study concept and design, analysis and interpretation of data, drafting of the manuscript, critical revision of the manuscript for important intellectual content, obtained funding and study supervision.
Funding
Italian Association for Cancer Research (AIRC) IG 15485; Accelerator Award 22794 (AIRC, CRUK, AECC).
Compliance with ethical standards
Conflict of interest
CF: Consultant for Gilead, Abbvie, Arrowhead.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Chiossone L, Dumas PY, Vienne M, Vivier E. Natural killer cells and other innate lymphoid cells in cancer. Nat Rev Immunol. 2018;18:671–688. doi: 10.1038/s41577-018-0061-z. [DOI] [PubMed] [Google Scholar]
- 2.Guillerey C, Chow MT, Miles K, Olver S, Sceneay J, Takeda K, et al. Toll-like receptor 3 regulates NK cell responses to cytokines and controls experimental metastasis. Oncoimmunology. 2015;4:e1027468. doi: 10.1080/2162402X.2015.1027468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O’Brien KL, Finlay DK. Immunometabolism and natural killer cell responses. Nat Rev Immunol. 2019 doi: 10.1038/s41577-019-0139-2. [DOI] [PubMed] [Google Scholar]
- 4.Kulik L, El-Serag HB. Epidemiology and management of hepatocellular carcinoma. Gastroenterology. 2019;156:477–491.e1. doi: 10.1053/j.gastro.2018.08.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Singal AG, El-Serag HB. Hepatocellular carcinoma from epidemiology to prevention: translating knowledge into practice. Clin Gastroenterol Hepatol. 2015;13:2140–2151. doi: 10.1016/j.cgh.2015.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taketomi A, Shimada M, Shirabe K, Kajiyama K, Gion T, Sugimachi K. Natural killer cell activity in patients with hepatocellular carcinoma: a new prognostic indicator after hepatectomy. Cancer. 1998;83:58–63. doi: 10.1002/(SICI)1097-0142(19980701)83:1<58::AID-CNCR8>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 7.Hoechst B, Voigtlaender T, Ormandy L, Gamrekelashvili J, Zhao F, Wedemeyer H, et al. Myeloid derived suppressor cells inhibit natural killer cells in patients with hepatocellular carcinoma via the NKp30 receptor. Hepatology. 2009;50:799–807. doi: 10.1002/hep.23054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cai L, Zhang Z, Zhou L, Wang H, Fu J, Zhang S, et al. Functional impairment in circulating and intrahepatic NK-cells and relative mechanism in hepatocellular carcinoma patients. Clin Immunol. 2008;129:428–437. doi: 10.1016/j.clim.2008.08.012. [DOI] [PubMed] [Google Scholar]
- 9.Wu Y, Kuang DM, Pan WD, Wan YL, Lao XM, Wang D, Li XF, Zheng L. Monocyte/macrophage-elicited natural killer cell dysfunction in hepatocellular carcinoma is mediated by CD48/2B4 interactions. Hepatology. 2013;57:1107–1116. doi: 10.1002/hep.26192. [DOI] [PubMed] [Google Scholar]
- 10.Iñarrairaegui M, Melero I, Sangro B. Immunotherapy of hepatocellular carcinoma: facts and hopes. Clin Cancer Res. 2018;24:1518–1524. doi: 10.1158/1078-0432.CCR-17-0289. [DOI] [PubMed] [Google Scholar]
- 11.Cariani E, Pilli M, Barili V, Porro E, Biasini E, Olivani A, et al. Natural killer cells phenotypic characterization as an outcome predictor of HCV-linked HCC after curative treatments. Oncoimmunology. 2018;5:e1154249. doi: 10.1080/2162402X.2016.1154249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burshtyn BN, Shin J, Stebbins C, Long EO. Adhesion to target cells is disrupted by killer cell inhibitory receptor. Curr Biol. 2000;10:700–780. doi: 10.1016/S0960-9822(00)00568-6. [DOI] [PubMed] [Google Scholar]
- 13.Troiano L, Ferraresi R, Lugli E, Nemes E, Roat E, Nasi M, et al. Multiparametric analysis of cells with different mitochondrial membrane potential during apoptosis by polychromatic flow cytometry. Nat Protoc. 2007;2:2719–2727. doi: 10.1038/nprot.2007.405. [DOI] [PubMed] [Google Scholar]
- 14.Clayton A, Mitchell JP, Court J, Linnane S, Mason MD, Tabi Z. Human tumor-derived exosomes down-modulate NKG2D expression. J Immunol. 2008;180:7249–7258. doi: 10.4049/jimmunol.180.11.7249. [DOI] [PubMed] [Google Scholar]
- 15.Gardiner CM, Finlay DK. What fuels natural killers? Metabolism and NK Cell responses. Front Immunol. 2017;8:367. doi: 10.3389/fimmu.2017.00367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keating SE, Zaiatz-Bittencourt V, Loftus RM, Keane C, Brennan K, Finlay DK, et al. Metabolic reprogramming supports IFN-γ production by CD56bright NK cells. J Immunol. 2016;196:2552–2560. doi: 10.4049/jimmunol.1501783. [DOI] [PubMed] [Google Scholar]
- 17.Golden-Mason L, Madrigal-Estebas L, McGrath E, Conroy MJ, Ryan EJ, Hegarty JE, et al. Altered natural killer cell subset distributions in resolved and persistent hepatitis C virus infection following single source exposure. Gut. 2008;57:1121–1128. doi: 10.1136/gut.2007.130963. [DOI] [PubMed] [Google Scholar]
- 18.Fu B, Wang F, Sun R, Ling B, Tian Z, Wei H. CD11b and CD27 reflect distinct population and functional specialization in human natural killer cells. Immunology. 2011;133:350–359. doi: 10.1111/j.1365-2567.2011.03446.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang QF, Yin WW, Xia Y, Yi YY, He QF, Wang X, et al. Liver-infiltrating CD11b(-)CD27(-) NK subsets account for NK-cell dysfunction in patients with hepatocellular carcinoma and are associated with tumor progression. Cell Mol Immunol. 2017;14:819–829. doi: 10.1038/cmi.2016.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chiossone L, Chaix J, Fuseri N, Roth C, Vivier E, Walzer T. Maturation of mouse NK cells is a 4-stage developmental program. Blood. 2009;113:5488–5496. doi: 10.1182/blood-2008-10-187179. [DOI] [PubMed] [Google Scholar]
- 21.Simonetta F, Pradier A, Roosnek E. T-bet and eomesodermin in NK cell development, maturation, and function. Front Immunol. 2016;7:241. doi: 10.3389/fimmu.2016.00241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu J, Mao HC, Wei M, Hughes T, Zhang J, Park IK, et al. CD94 surface density identifies a functional intermediary between the CD56bright and CD56dim human NK-cell subsets. Blood. 2010;115:274–281. doi: 10.1182/blood-2009-04-215491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Michelet X, Dyck L, Hogan A, Loftus RM, Duquette D, Wei K, et al. Metabolic reprogramming of natural killer cells in obesity limits antitumor responses. Nat Immunol. 2018;19:1330–1340. doi: 10.1038/s41590-018-0251-7. [DOI] [PubMed] [Google Scholar]
- 24.Harmon C, Robinson MW, Hand F, Almuaili D, Mentor K, Houlihan DD, et al. Lactate-mediated acidification of tumor microenvironment induces apoptosis of liver-resident NK cells in colorectal liver metastasis. Cancer Immunol Res. 2019;7:335–346. doi: 10.1158/2326-6066.CIR-18-0481. [DOI] [PubMed] [Google Scholar]
- 25.Mantovani S, Oliviero B, Lombardi A, Varchetta S, Mele D, Sangiovanni A, et al. Deficient natural killer cell NKp30-mediated function and altered NCR3 splice variants in hepatocellular carcinoma. Hepatology. 2019;69:1165–1179. doi: 10.1002/hep.30235. [DOI] [PubMed] [Google Scholar]
- 26.Duan S, Guo W, Xu Z, He Y, Liang C, Mo Y, et al. Natural killer group 2D receptor and its ligands in cancer immune escape. Mol Cancer. 2019;18:29. doi: 10.1186/s12943-019-0956-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zaiatz-Bittencourt V, Finlay DK, Gardiner CM. Canonical TGF-β signaling pathway represses human NK cell metabolism. J Immunol. 2018;200:3934–3941. doi: 10.4049/jimmunol.1701461. [DOI] [PubMed] [Google Scholar]
- 28.Castriconi R, Dondero A, Bellora F, Moretta L, Castellano A, Locatelli F, et al. Neuroblastoma-derived TGF-β1 modulates the chemokine receptor repertoire of human resting NK cells. J Immunol. 2013;190:5321–5328. doi: 10.4049/jimmunol.1202693. [DOI] [PubMed] [Google Scholar]
- 29.Shirai Y, Kawata S, Tamura S, Ito N, Tsushima H, Takaishi K, et al. Plasma transforming growth factor-beta 1 in patients with hepatocellular carcinoma. Comparison with chronic liver diseases. Cancer. 1994;73:2275–2279. doi: 10.1002/1097-0142(19940501)73:9<2275::AID-CNCR2820730907>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 30.Sacco R, Leuci D, Tortorella C, Fiore G, Marinosci F, Schiraldi O, et al. Transforming growth factor beta1 and soluble Fas serum levels in hepatocellular carcinoma. Cytokine. 2000;12:811–814. doi: 10.1006/cyto.1999.0650. [DOI] [PubMed] [Google Scholar]
- 31.Dituri F, Mancarella S, Cigliano A, Chieti A, Giannelli G. TGF-β as multifaceted orchestrator in HCC progression: signaling, EMT, immune iicroenvironment, and novel therapeutic perspectives. Semin Liver Dis. 2019;39:53–69. doi: 10.1055/s-0038-1676121. [DOI] [PubMed] [Google Scholar]
- 32.Peng H, Tian Z. Diversity of tissue-resident NK cells. SeminImmunol. 2017;31:3–10. doi: 10.1016/j.smim.2017.07.006. [DOI] [PubMed] [Google Scholar]
- 33.Easom NJW, Stegmann KA, Swadling L, Pallett LJ, Burton AR, Odera D, et al. IL-15 overcomes hepatocellular carcinoma-induced NK cell dysfunction. Front Immunol. 2018;9:1009. doi: 10.3389/fimmu.2018.01009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.