Abstract
The wide-ranging collection of malignancies arising at the upper aerodigestive tract is categorized as head and neck cancer (HNC), the sixth most prevalent cancer worldwide. Infection with human papillomavirus (HPV) or exposure to carcinogens is the leading causes of HPV+ and HPV− HNCs development, respectively. HPV+ and HPV− HNCs are different in clinical and molecular aspects. Specifically, HPV− HNCs tightly associate with missense mutants of the TP53 gene (encoding for the p53 protein), suggesting a central role for mutant p53 gain-of-function (GOF) in driving tumorigenesis. In contrast, in HPV + HNC, the sequence of TP53 typically remains intact, while the protein is degraded. In tumor cells, the status of the TP53 gene affects the cargo of secreted exosomes. In this review, we describe the accumulated knowledge regarding the involvement of exosomes and p53 on cellular interactions between HPV+ and HPV− HNC cells, and the surrounding tumor microenvironment (TME). Moreover, we envision how TP53 status may determine exosomes cargo in HNC, and, consequently, modify the TME. The potential roles of exosomes described herein are based on both our studies and the studies of others on mutant p53-derived exosomes. Specifically, we showed how exosomes are shed by cancer cells harboring mutant p53 communicate with tumor-associated macrophages in the colon as well as with cancer-associated fibroblasts in the lung, creating immunosuppressive conditions and promoting invasiveness. Altogether, exosomes in HNC in the context of TP53 status are understudied and extensive research is required to shed light on the biology of HPV+ and HPV− HNC.
Keywords: Head and neck, Exosomes, Mutant p53, Human papillomavirus, Tumor microenvironment, CITIM 2019
Introduction
Head and neck cancers (HNC) represent the sixth most common malignancies worldwide and the sixth most common cause of cancer-related deaths. 600,000 new cases of HNC are diagnosed every year, worldwide [1]. HNC is developed primarily via transformation of the squamous epithelium layer of the oral cavity, throat, and larynx. Among these sites, over 90% of the cancers are squamous-cell carcinomas, as reviewed in Warnakulasuriya et al. [2]. Two major risk factors for the development of HNC are exposure to carcinogens and viral infections. The chronic exposure to tobacco (smoking), together with alcohol consumption, is known to be the most common cause of HNC. Viral infections of Epstein–Barr virus (EBV) are known to induce nasopharynx cancer, while the Human Papilloma Virus (HPV) is more prevalent among the oropharyngeal [3]. Moreover, recent studies have identified various types of HPV that are associated with both benign and malignant lesions in the oral cavity [4]. In the last 2 decades, a dramatic increase has been observed in young (age < 60) patients diagnosed with HPV positive (HPV+) cancer. HPV + HNC incidence in the United States (U.S) has increased over the years and has become an epidemic, as reviewed in Khalid et al. [5]. The most recent estimate reports of HPV in HNC indicate that there are about 38,000 cases every year globally [6]. Moreover, in the US, HPV infection has been associated with 60–70% of all patients diagnosed with oropharyngeal cancer [7]. HPV + HNC is more prevalent in younger individuals with a history of multiple sexual partners. In addition, this type of cancer is usually associated with a lower risk of recurrence and death compared with HPV negative (HPV−) HNC [5]. HPV+ and HPV− HNC have diverse and unique molecular and clinical aspects [8]. This significant difference is related to the etiology that differently affects gene expression and the accumulation of genomic alterations (mutations, amplifications, and deletions) [9]. One of the most significant differences between HPV+ and HPV− HNC is reflected by the genomic alterations in the TP53 gene encoding for the hallmark tumor suppressor protein p53.
p53 functions as a central molecular hub, maintaining cellular homeostasis, regulating cell proliferation, cell survival, genome integrity, and a plethora of additional functions [reviewed in 10]. As a transcriptional regulator, p53 integrates stress signals and, accordingly, promotes cellular decisions involving cycle arrest, senescence, and apoptosis to prevent the propagation of damaged cells [11]. The last decade of p53 studies has seen the revelation of additional roles beyond transcription regulation and protection of genome integrity. Evidence has shown that p53 is a major regulator of cellular metabolism, stemness, autophagy, invasion, metastasis, microenvironment control, and inflammation. Notably, mutations in the TP53 gene are considered the most frequent genetic alteration in human cancers [10] (IARC p53 database—http://p53.iarc.fr/). In HNC patients, mutations in TP53 are significantly associated with shorter survival rates. The mutations in TP53 usually occur at an early phase in the malignancy, resulting in dysregulated cell-cycle progression and cellular proliferation [3, 12]. While loss-of-function of the wild-type (WT) form of p53 is enough to expose the cell to a higher risk of transformation, there are additional tumorigenic mechanisms directly governed by the neomorphic mutant p53 (mutp53) proteins [13]. One direct cancer-promoting aspect of mutp53 is through transcriptional blockage of the WT activity mediated via the tetramerization complex in a dominant-negative manner. While this mechanism, in effect, attenuates the WTp53 activity, the most aggressive phenotype driven by mutp53 is referred to as gain-of-function (GOF) [12–17]. Cancer cells gain selective advantages by retaining only the mutant form of p53 (due to loss-of-heterozygosity). This gaining of oncogenic functions is attributed to the ability of several missense mutp53 in reshaping the entire transcription and proteomic profiles of the tumor cell, establishing de-novo interactions with transcription regulators and other cellular proteins. To that end, specific missense mutations in p53 have been reported to undermine central cellular pathways and to drive cancer cell proliferation and survival, promote invasion, migration, metastasis, and chemoresistance, as reviewed in [18]. Furthermore, recent studies, including ours, show that mutp53 can foster a non-cell-autonomous effect over immune and stromal cells in the adjacent tumor microenvironment (TME), via the secretion of extracellular vesicles (exosomes) [19].
Today, the involvement of the TME in HNC progression is well acknowledged (reviewed in [20]). Non-cancerous subpopulations of cells are being recruited by cancer cells in an interacted web of communication mechanisms that include secreted cytokines, growth factors, and exosomes. While exosomes had been observed over 50 years ago [21], their significant involvement in intercellular communication has been unraveled only in recent years [22, 23]. Exosomes are small (~ 30–150 nm) lipid bilayer-membrane vesicles assembled in endosomes, formed from multivesicular bodies. The exosomal transfer has been shown to deliver bio-molecules between cells, consequently leading to modulation of signal transduction [24], by delivering proteins, lipids, and nucleic acids which can regulate gene expression in recipient cells. Such cell-to-cell communication by exosomes was reported to affect recipient cells in various physiological but also pathological conditions, including cancer [19]. Transformed cells typically release excessive amounts of exosomes that contribute to malignancy initiation, tumor cell progression [25], invasion, metastasis, angiogenesis, and resistance to therapy [26, 27], and can also determine drug efficacy [28]. In addition to the direct effect of exosomes on neighboring tumor cells, the exosomal cargo has been proven to affect the tumor microenvironment by mediating immune escape and plays a vital role in the formation of the pre-metastatic niche [29].
In this review article, we will ‘zoom-in’ on the potential role played by exosomes in the malignant process of HNC. Depending on our studies, we will analyze the possible link between GOF mutp53, exosomes, and aggressive phenotypes of HNC.
Genomic alterations of TP53, and exosomes in head and neck cancer
HNC biology and etiology More than 90% of HNC are squamous-cell carcinomas. Most patients present with locally advanced disease, involving regional nodes or with distant metastases [30]. HNC standard of care involves surgery, radiotherapy, chemotherapy, targeted therapy (Erbitux), and, most recently, immunotherapy (anti-PD1) [17]. For recurrent or metastatic disease, the standard first-line treatment is currently platinum-based chemotherapy, with Pembrolizumab. Overall survival for patients progressing after the failure of platinum-based chemotherapy and immunotherapy remains unacceptably poor [31] (also reviewed in [32]). The HPV virus can be present as episomal or integrated forms, or both [33]. However, to promote cancer development, viral DNA integration into the host cell genome is essential [9]. HPV is known to drive tumorigenesis through the actions of its two major viral oncoproteins, namely E6 and E7. These proteins determine stable episomal maintenance of HPV [34], and target two major cellular pathways of p53 and retinoblastoma (Rb), respectively. Specifically, E6 enhances the degradation of p53 via ubiquitinylation and proteasome-dependent degradation [35], and E7 releases Rb protein and enables E2F transcription activation [34].
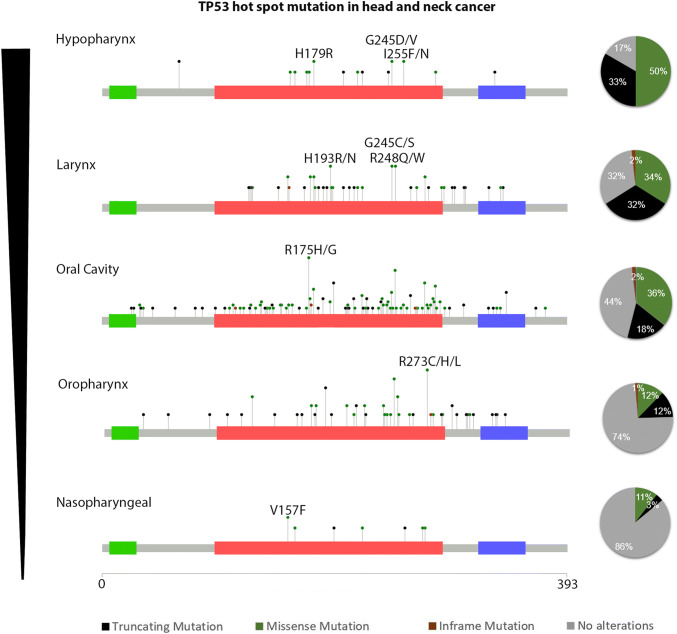
TP53 mutations in HNC The genomic characterization of human malignancy indicates the landscape of the genomic alterations in cancers. Specifically, according to The Cancer Genome Atlas Program (TCGA) analysis, TP53 is mutated in 84% of the cases in HPV− HNC, while in HPV+, the frequency drops to 3% [36]. Previous data analyzed from TCGA by Zhou et al. show that TP53 mutation rate in HNC tumors is most common in the larynx and hypopharynx, less common in the tongue and oral cavity, and least common in the tumors of the oropharynx, including the tonsils, and base of the tongue (83.5%, 75.6%, and 28.6%, respectively) [37]. Among the HPV− HNC patients, the frequent mutations of TP53 are in codons R248, R273, G245, R175, R282, and H179. Our analysis of the GENIE data set, which includes over 1200 HNC samples, shows TP53 mutation distribution in different tumor locations of the HNC: Hypopharynx 83%, Larynx 68%, Oral cavity 55%, Oropharynx 26%, and Nasopharyngeal 14% (Fig. 1).
Fig. 1.
p53 mutation rate and frequency in HNC. Analysis of the AACR Project GENIE Consortium: Powering Precision Medicine Through An International Consortium [38], showing the frequency and mutation types of TP53 in HNC sites. Left panel: rates of TP53 mutation in different anatomic sites. Middle panel: mutational distribution in the TP53 gene. Right panel: pie analysis of the TP53 mutation types
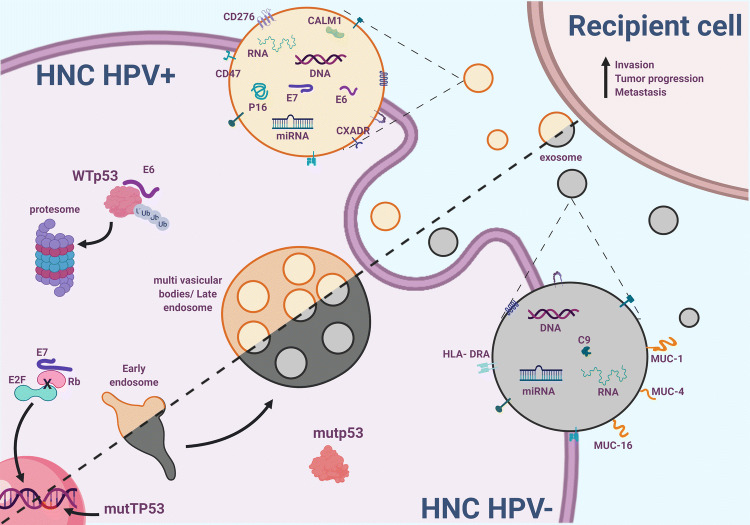
Exosomes in HNC and immunity In the last decade, an extensive research effort described the role of exosomes in cell-to-cell interactions. In different malignancies, including in HNC, tumor cells release exosomes containing immunomodulatory factors, interacting with cells of the TME to facilitate immunosuppressive conditions [39, 40]. Exosomes that are secreted by HNC cells are enriched with suppressive compounds such as cyclooxygenase-2 (COX2), TGF-b, programmed death 1 (PD-1) [41], and cytotoxic T lymphocyte antigen 4 (CTLA-4). These exosomal cargoes promoted CD8+ T-cell apoptosis, inhibited CD4+ T-cell proliferation, upregulated Tregs, and damaged NK-cell function [42]. Furthermore, it was suggested in additional studies that such immunosuppression was mediated by exosomal cytokines, which presented exosomes containing IL-10 and IL-6 secreted by HNC [43]. On the same note, the IL-6-dependent inflammatory stimulation also resulted in increased angiogenesis. Furthermore, the presence of JAG1, a Notch ligand in HNC, released exosomes, has also been noted to affect immune cells negatively [42]. In studies focusing on the plasma of HNC patients, protein cargo of exosomes was indicative of tumor stage, immunosuppressive tumor profile, and disease activity [28]. In addition, HNC-derived plasma exosomes induced apoptosis of activated CD8+ T cells by engaging the extrinsic and intrinsic apoptosis pathways, and thereby modulated the immune response and drove the tumorigenic process. Specifically, plasma-derived exosomes from HNC patients contained TGF-β, FasL, OX40, OX40L, and HSP70 [44]. Notably, exosomes obtained from HPV+ tumor cells carried the viral oncoproteins E6 and E7 [45], in addition to p16 and survivin. In comparison, exosomes derived by HPV− tumor cells did not contain p16, as the encoding gene (CDKN2A) is frequently mutated or deleted [46]. The majority of the detected proteins in exosomes from both HPV(+) and HPV(−) cell lines are cytoplasmic proteins such as integrin family members, proteins mediating proteoglycan-syndecan signaling EGFR [47], and VEGF/VEGFR signaling networks. Furthermore, a study by Pietrowska et al. suggested a list of membrane–surface proteins putatively involved in exosome-mediated cross-talk between the tumor and immune cells. From this list, 172 proteins were mutually expressed in exosomes obtained from HPV+ and HPV− tumor cells. However, there were 43 proteins like CD47, CD276, and CALM1, which were present only in exosomes secreted by HPV + HNC cell lines. Similarly, five proteins were detected only in exosomes from HPV− HNCs, such as C9, MUC-1, HLA-DRA [48].
The involvement of exosomes derived from mutp53 tumor cells in affecting the TME
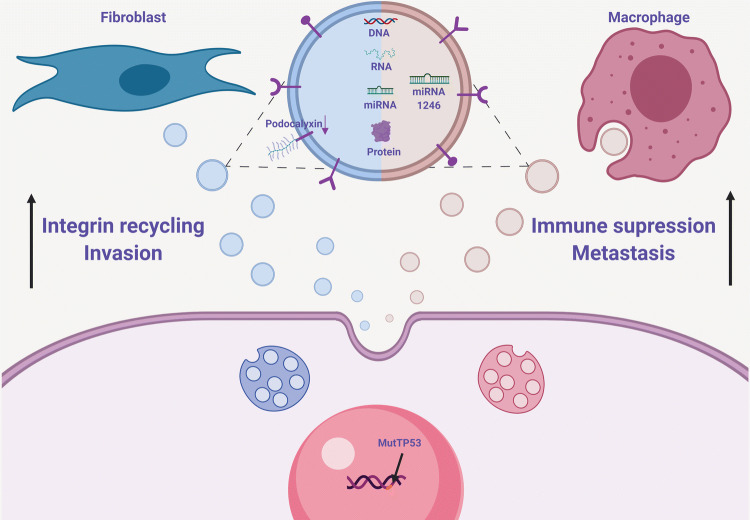
There is now ample evidence linking genetic alterations to the direct effect on the TME [49, 50]. Oncogenic mutations might give rise to a clonal selection imposing a more aggressive tumor progression based on the ability of the mutated clone to communicate with surrounding stromal and immune cells. These dynamics are also reported to take place via subsets of exosomes uniquely released by cells carrying the mutation. To that end, cells harboring mutations in oncogenes such as KRAS, c-Myc, and MET were shown to transfer TME-modulating exosomes taken up by neighboring cells, thus recruiting those normal cells towards the tumorigenic agenda [51–53]. Notably, two recent studies have highlighted mutp53 as another oncogenic event utilizing the exosomal machinery to subordinate the tissue and allow the tumor to progress. Since mutations in p53 are considered a frequent and late event in sporadic colorectal cancer (CRC) [54], exacerbating the disease outcome, we were intrigued to explore the direct mechanisms related to the TME, driven by mutp53 GOF. In particular, we focused on the non-cell-autonomous role played by mutp53 in reprogramming tumor-associated macrophages (TAMs) [19]. Significantly, CRC cells harboring mutp53 were skewing neighboring macrophages to an anti-inflammatory M2-like phenotype not observed when macrophages were cultured together with cells that did not carry a GOF mutant in p53. We showed that this intercellular interaction is exosome-related, as mutp53 CRC cells produced and shed excessive amounts of exosomes, which were engulfed by the TAM. Importantly, we identified a specific microRNA (miR) signature dominated by miR-1246 in the released vesicles. These findings allow us to suggest a direct molecular mechanism through which mutp53 CRC cells can modulate the microenvironment and create an immunosuppressive milieu that would result in an increased metastatic burden and reduced survival rates (Fig. 2).
Fig. 2.
Exosomes originating from mutp53 cancer cells promote cancer progression. Cancer cells harboring mutp53 interact with TME cells via unique subsets of exosomes. In the colon, mutp53 cancer cells release exosomes enriched with miR-1246, taken up by TAMs, reprogramming their inflammatory phenotype, thus promoting immunosuppression and metastasis. In the lung, mutp53 exosomes drive invasiveness by targeting CAFs. Increased integrin recycling governed by low levels of Podocalyxin in exosomes was associated with the modulation of the invasive front
Another recent study sheds light on the mechanistic contribution of mutp53-derived exosomes to metastases developed in lung carcinomas. In accordance with several reports indicating that exosomes have the potential to shape the metastatic niche [55], the authors revealed that lung tumor cells harboring specific GOF mutants of p53 interact with cancer-associated-fibroblasts (CAFs) via exosomes. mutp53 tumor cells shed exosomes carrying integrin receptors remodeling the extracellular matrix (ECM). Integrin receptors directed to ECM were shown to be trafficked through the endosomal pathway and returned, or recycled, to the plasma membrane, thus facilitating the migratory potential of the tumor and immune cells. Interestingly, mutp53 regulated the expression levels of Podocalyxin, a factor frequently dysregulated in various cancer types [56]. mutp53 was suggested to control exosomal levels of Podocalyxin, which consequently led to elevated integrin recycling rates (Fig. 2). Since integrin recycling is a key to metastatic niche priming, this study describes a pathway through which mutp53 operates via a well-orchestrated GOF mechanism to modify the ECM. Such microenvironmental promotion of invasiveness was observed to be directly dependent on mutp53 and Rab35, which join forces and control the content of released exosomes. mutp53-expressing tumors affected collagen organization in the tumor stroma, resulting in ECM cross-linking and assembly of parallel arrays of collagen fibers. This type of ECM organization, detected in the lungs of mutp53 tumor-bearing animals, is more conducive to the metastatic seeding of tumor cells [57].
Future perspectives
The concept of mutp53 GOF was first introduced in the early 1990s following the realization that p53 is a tumor suppressor, rather than an oncogene [15, 17, 58]. The fact that several groups worldwide were unknowingly working with mutp53 clones that were yielding oncogenic effects led to the GOF hypothesis. Ever since, a plethora of studies have been published, supporting this notion of oncogenic activities governed directly in the mutp53 cellular setting [59–62]. While the vast majority of these mechanisms are associated with driving tumorigenesis through the cancer cell itself in a cell-autonomous manner, several research groups, including ours, now suggest that mutp53 can also directly modulate other cells of the TME in a non-cell-autonomous manner [19, 57]. In HNC, mutations in the TP53 gene are a central part of the mutational landscape, particularly in the HPV− tumors. While the viral E6 oncoprotein facilitates the degradation of the WTp53, in cases not diagnosed with HPV infection, it is the mutant form of p53 leading the charge [63]. Indeed, several different molecular mechanisms specific to various HNC sites have been suggested in the previous studies [36, 64]. Notwithstanding, and given the new findings described in this review, we hypothesize that in HNC patients, the tumor cells that harbor ‘hotspot’ mutations in p53 might utilize the exosomal machinery to produce and release oncogenic subsets of exosomes serving as a TME-modifying force. Various oncogenic alterations have been shown to both intensify the number of released exosomes and to change the molecular cargo encapsulated into the vesicles. The interaction between the tumor cells and the supporting stromal and immune cells such as CAFs, TAMs, neutrophils, and T cells, might be reinforced by a continuous uptake of exosomes enriched with reprogramming factors (Fig. 3). Hence, it would be intriguing to explore whether exosomes shed by mutp53 tumor cells can exacerbate the pathogenesis of different HNC sites. As we presented here, in HNC, the mutational spectrum of p53 is typically dictated both by HPV infection status as well as the anatomical location of the disease. It is plausible to hypothesize that the involvement of mutp53 derived exosomes will be mutant-specific, site-specific, and negatively correlated with HPV. For example, in the Oropharynx, where we frequently observe lesions harboring mutp53 in residue 273, we can expect to discover a unique GOF mechanism that would not be detected in sites such as the Larynx, where p53 mutations are seldom found. Nevertheless, in HPV+ cases of HNC, exosome-related mechanisms might still be a central avenue through which E6 and E7 are being transported between cells.
Fig. 3.
Prospected oncogenic mechanisms of exosomes released by HNC cells, dependent on HPV infection and p53 status. Exosomes shed by HPV+ cancer cells carry viral oncogenic proteins (E6 and E7), while exosomes from HNC HPV−, mostly p53 mutated, carry different molecular cargo, including complement proteins and mucosal proteins
Finally, since exosomes were reported to be involved in different aspects of tumor progression, from initiation to the metastatic niche, overarching studies covering various facets of tumorigenic mechanisms should be embarked. Isogenic models, both in vitro and in vivo, differing by HPV positivity, by site and by their tumorigenic and metastatic potential, may be used to decipher changes in the molecular cargo carried by exosomes. Potential molecular candidates could be further validated as effectors that modulate signaling pathways that contribute to the malignant process. Unraveling the molecular pathways controlled by exosomes shed by HNC cells will strengthen our understanding of disease pattern by site, by p53 status, and by the non-cancerous compartment involved in the process.
Abbreviations
- AACR
American Association for Cancer Research
- CAFs
Cancer-associated fibroblasts
- COX2
Cyclooxygenase-2
- CRC
Colorectal cancer
- CTLA-4
Cytolytic T lymphocyte-associated antigen 4
- EBV
Epstein–Barr virus
- ECM
Extracellular matrix
- GENIE
Genomics Evidence Neoplasia Information Exchange [AACR Project]
- GOF
Gain-of-function
- HNC
Head and neck cancer
- HPV−
HPV negative
- HPV
Human papillomavirus
- HPV+
HPV positive
- IARC
International Agency for Research on Cancer
- miR
microRNA
- mutp53
Mutant protein 53
- PD-1
Programmed death 1
- Rb
Retinoblastoma
- TAMs
Tumor-associated macrophages
- TCGA
The Cancer Genome Atlas Program
- TME
Tumor microenvironment
- TP53
Tumor protein 53 gene
- WT
Wild type
- WTp53
Wild-type protein 53
Author contributions
EEA surveyed the literature, summarized the published data, and wrote the backbone of the manuscript as well as performed the analysis displayed in Fig. 1, and created all figures presented in this review. TC and ME outlined the sections, edited the text, and finalized it.
Funding
This work was funded by the Israel Science Foundation (ISF, 700/16), the Concern Foundation (#7895), the United States—Israel Binational Science Foundation (BSF, #2017323), and the Israel Cancer Research Foundation (ICRF, 17-1693-RCDA) to Moshe Elkabets. Moshe Elkabets is supported by an Alon Fellowship for outstanding young researchers.
Compliance with ethical standards
Conflict of interest
The authors declare that no potential conflicts of interest exist.
Footnotes
This paper is a Focussed Research Review based on a presentation given at the Sixth International Conference on Cancer Immunotherapy and Immunomonitoring (CITIM 2019), held in Tbilisi, Georgia, 29th April–2nd May 2019. It is part of a series of CITIM 2019 papers in Cancer Immunology, Immunotherapy.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Tomer Cooks and Moshe Elkabets have contributed equally.
Contributor Information
Tomer Cooks, Email: cooks@bgu.ac.il.
Moshe Elkabets, Email: moshee@bgu.ac.il.
References
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics. CA Cancer J Clin. 2002;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 2.Warnakulasuriya S. Global epidemiology of oral and oropharyngeal cancer. Oral Oncol. 2009;45:309–316. doi: 10.1016/j.oraloncology.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 3.Deng Z, Uehara T, Maeda H, et al. Epstein-barr virus and human papillomavirus infections and genotype distribution in head and neck cancers. PLoS One. 2014;9:e113702. doi: 10.1371/journal.pone.0113702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ritchie JM, Smith EM, Summersgill KF, et al. Human papillomavirus infection as a prognostic factor in carcinomas of the oral cavity and oropharynx. Int J Cancer. 2003;104:336–344. doi: 10.1002/ijc.10960. [DOI] [PubMed] [Google Scholar]
- 5.Khalid MB, Ting P, Pai A, et al. Initial presentation of human papillomavirus-related head and neck cancer: a retrospective review. Laryngoscope. 2019;129:877–882. doi: 10.1002/lary.27296. [DOI] [PubMed] [Google Scholar]
- 6.Forman D, de Martel C, Lacey CJ, et al. Global burden of human papillomavirus and related diseases. Vaccine. 2012;30:F12–F23. doi: 10.1016/J.VACCINE.2012.07.055. [DOI] [PubMed] [Google Scholar]
- 7.Marur S, D’Souza G, Westra WH, Forastiere AA. HPV-associated head and neck cancer: a virus-related cancer epidemic. Lancet Oncol. 2010;11:781–789. doi: 10.1016/S1470-2045(10)70017-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Madeo M, Colbert PL, Vermeer DW, et al. Cancer exosomes induce tumor innervation. Nat Commun. 2018;9:4284. doi: 10.1038/s41467-018-06640-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parfenov M, Pedamallu CS, Gehlenborg N, et al. Characterization of HPV and host genome interactions in primary head and neck cancers. Proc Natl Acad Sci. 2014;111:15544–15549. doi: 10.1073/pnas.1416074111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hollstein M, Sidransky D, Vogelstein B, Harris C. p53 mutations in human cancers. Science (80-) 1991;253:49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- 11.Liu C, Mann D, Sinha UK, Kokot NC. The molecular mechanisms of increased radiosensitivity of HPV-positive oropharyngeal squamous cell carcinoma (OPSCC): an extensive review. J Otolaryngol Head Neck Surg. 2018;47:59. doi: 10.1186/s40463-018-0302-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sabapathy K, Lane DP. Therapeutic targeting of p53: all mutants are equal, but some mutants are more equal than others. Nat Rev Clin Oncol. 2018;15:13–30. doi: 10.1038/nrclinonc.2017.151. [DOI] [PubMed] [Google Scholar]
- 13.Liu Y, Zhang X, Han C, et al. TP53 loss creates therapeutic vulnerability in colorectal cancer. Nature. 2015;520:697–701. doi: 10.1038/nature14418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greathouse KL, White JR, Vargas AJ, et al. Interaction between the microbiome and TP53 in human lung cancer. Genome Biol. 2018;19:123. doi: 10.1186/s13059-018-1501-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim MP, Lozano G. Mutant p53 partners in crime. Cell Death Differ. 2018;25:161–168. doi: 10.1038/cdd.2017.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pfister NT, Prives C. Transcriptional regulation by wild-type and cancer-related mutant forms of p53. Cold Spring Harb Perspect Med. 2017 doi: 10.1101/cshperspect.a026054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oren M, Rotter V. Mutant p53 gain-of-function in cancer. Cold Spring Harb Perspect Biol. 2010;2:a001107. doi: 10.1101/cshperspect.a001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Y, Zhang M-C, Xu X-K, et al. Functional diversity of p53 in human and wild animals. Front Endocrinol (Lausanne) 2019;10:152. doi: 10.3389/fendo.2019.00152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cooks T, Pateras IS, Jenkins LM, et al. Mutant p53 cancers reprogram macrophages to tumor supporting macrophages via exosomal miR-1246. Nat Commun. 2018;9:771. doi: 10.1038/s41467-018-03224-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peltanova B, Raudenska M, Masarik M. Effect of tumor microenvironment on pathogenesis of the head and neck squamous cell carcinoma: a systematic review. Mol Cancer. 2019;18:63. doi: 10.1186/s12943-019-0983-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wolf P. The nature and significance of platelet products in human plasma. Br J Haematol. 1967;13:269–288. doi: 10.1111/j.1365-2141.1967.tb08741.x. [DOI] [PubMed] [Google Scholar]
- 22.Skog J, Würdinger T, van Rijn S, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10:1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Valadi H, Ekström K, Bossios A, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 24.Zhang X, Yuan X, Shi H, et al. Exosomes in cancer: small particle, big player. J Hematol Oncol. 2015;8:83. doi: 10.1186/s13045-015-0181-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xie C, Ji N, Tang Z, et al. The role of extracellular vesicles from different origin in the microenvironment of head and neck cancers. Mol Cancer. 2019;18:83. doi: 10.1186/s12943-019-0985-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Theodoraki M-N, Yerneni S, Gooding WE, et al. Circulating exosomes measure responses to therapy in head and neck cancer patients treated with cetuximab, ipilimumab, and IMRT. Oncoimmunology. 2019;8:1593805. doi: 10.1080/2162402X.2019.1593805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ludwig N, Yerneni SS, Razzo BM, Whiteside TL. Exosomes from HNSCC promote angiogenesis through reprogramming of endothelial cells. Mol Cancer Res. 2018;16:1798–1808. doi: 10.1158/1541-7786.MCR-18-0358. [DOI] [PubMed] [Google Scholar]
- 28.Whiteside TL. Tumor-derived exosomes and their role in cancer progression. Adv Clin Chem. 2016;74:103–141. doi: 10.1016/bs.acc.2015.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fong MY, Zhou W, Liu L, et al. Breast-cancer-secreted miR-122 reprograms glucose metabolism in premetastatic niche to promote metastasis. Nat Cell Biol. 2015;17:183–194. doi: 10.1038/ncb3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kannan A, Hertweck KL, Philley JV, et al. Genetic mutation and exosome signature of human papilloma virus associated oropharyngeal cancer. Sci Rep. 2017;7:46102. doi: 10.1038/srep46102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rivera F, García-Castaño A, Vega N, et al. Cetuximab in metastatic or recurrent head and neck cancer: the EXTREME trial. Expert Rev Anticancer Ther. 2009;9:1421–1428. doi: 10.1586/era.09.113. [DOI] [PubMed] [Google Scholar]
- 32.Muratori L, La Salvia A, Sperone P, Di Maio M. Target therapies in recurrent or metastatic head and neck cancer: state of the art and novel perspectives. A systematic review. Crit Rev Oncol Hematol. 2019;139:41–52. doi: 10.1016/J.CRITREVONC.2019.05.002. [DOI] [PubMed] [Google Scholar]
- 33.Marongiu L, Godi A, Parry JV, Beddows S. Human Papillomavirus 16, 18, 31 and 45 viral load, integration and methylation status stratified by cervical disease stage. BMC Cancer. 2014;14:384. doi: 10.1186/1471-2407-14-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yim E-K, Park J-S. The role of HPV E6 and E7 oncoproteins in HPV-associated cervical carcinogenesis. Cancer Res Treat. 2005;37:319. doi: 10.4143/CRT.2005.37.6.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bernard X, Robinson P, Nominé Y, et al. Proteasomal degradation of p53 by human papillomavirus E6 oncoprotein relies on the structural integrity of p53 core domain. PLoS One. 2011;6:e25981. doi: 10.1371/journal.pone.0025981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tanaka N, Zhao M, Tang L, et al. Gain-of-function mutant p53 promotes the oncogenic potential of head and neck squamous cell carcinoma cells by targeting the transcription factors FOXO3a and FOXM1. Oncogene. 2018;37:1279–1292. doi: 10.1038/s41388-017-0032-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou G, Liu Z, Myers JN. TP53 mutations in head and neck squamous cell carcinoma and their impact on disease progression and treatment response. J Cell Biochem. 2016;117:2682–2692. doi: 10.1002/jcb.25592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.cBioPortal for GENIE. http://genie.cbioportal.org/. Accessed 10 Jul 2019
- 39.Atay S, Godwin AK. Tumor-derived exosomes. Commun Integr Biol. 2014;7:e28231. doi: 10.4161/cib.28231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Principe S, Hui AB-Y, Bruce J, et al. Tumor-derived exosomes and microvesicles in head and neck cancer: implications for tumor biology and biomarker discovery. Proteomics. 2013;13:1608–1623. doi: 10.1002/pmic.201200533. [DOI] [PubMed] [Google Scholar]
- 41.Theodoraki M-N, Yerneni SS, Hoffmann TK, et al. Clinical significance of PD-L1 + exosomes in plasma of head and neck cancer patients. Clin Cancer Res. 2018;24:896–905. doi: 10.1158/1078-0432.CCR-17-2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Horton JD, Knochelmann HM, Day TA, et al. Immune evasion by head and neck cancer: foundations for combination therapy. Trends in Cancer. 2019;5:208–232. doi: 10.1016/J.TRECAN.2019.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ferris RL. Immunology and immunotherapy of head and neck cancer. J Clin Oncol. 2015;33:3293–3304. doi: 10.1200/JCO.2015.61.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ludwig S, Floros T, Theodoraki M-N, et al. Suppression of lymphocyte functions by plasma exosomes correlates with disease activity in patients with head and neck cancer. Clin Cancer Res. 2017;23:4843–4854. doi: 10.1158/1078-0432.CCR-16-2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Honegger A, Schilling D, Sültmann H et al (2018) Identification of E6/E7-dependent microRNAs in HPV-positive cancer cells. In: Methods in molecular biology (Clifton, N.J.). pp 119–134 [DOI] [PubMed]
- 46.Ludwig S, Sharma P, Theodoraki M-N, et al. Molecular and functional profiles of exosomes From HPV(+) and HPV(−) head and neck cancer cell lines. Front Oncol. 2018;8:445. doi: 10.3389/fonc.2018.00445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Raulf N, Lucarelli P, Thavaraj S, et al. Annexin A1 regulates EGFR activity and alters EGFR-containing tumour-derived exosomes in head and neck cancers. Eur J Cancer. 2018;102:52–68. doi: 10.1016/j.ejca.2018.07.123. [DOI] [PubMed] [Google Scholar]
- 48.Ludwig S, Marczak L, Sharma P, et al. Proteomes of exosomes from HPV(+) or HPV(−) head and neck cancer cells: differential enrichment in immunoregulatory proteins. Oncoimmunology. 2019;8:1593808. doi: 10.1080/2162402X.2019.1593808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dias Carvalho P, Guimarães CF, Cardoso AP, et al. KRAS oncogenic signaling extends beyond cancer cells to orchestrate the microenvironment. Cancer Res. 2018;78:7–14. doi: 10.1158/0008-5472.CAN-17-2084. [DOI] [PubMed] [Google Scholar]
- 50.Bedognetti D, Hendrickx W, Ceccarelli M, et al. Disentangling the relationship between tumor genetic programs and immune responsiveness. Curr Opin Immunol. 2016;39:150–158. doi: 10.1016/j.coi.2016.02.001. [DOI] [PubMed] [Google Scholar]
- 51.Demory Beckler M, Higginbotham JN, Franklin JL, et al. Proteomic analysis of exosomes from mutant KRAS colon cancer cells identifies intercellular transfer of mutant KRAS. Mol Cell Proteomics. 2013;12:343–355. doi: 10.1074/mcp.M112.022806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fonseka P, Liem M, Ozcitti C, et al. Exosomes from N-Myc amplified neuroblastoma cells induce migration and confer chemoresistance to non-N-Myc amplified cells: implications of intra-tumour heterogeneity. J Extracell vesicles. 2019;8:1597614. doi: 10.1080/20013078.2019.1597614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lesnik J, Antes T, Kim J, et al. Registered report: melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Elife. 2016;5:e07383. doi: 10.7554/eLife.07383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li X-L, Zhou J, Chen Z-R, Chng W-J. p53 mutations in colorectal cancer-molecular pathogenesis and pharmacological reactivation. World J Gastroenterol. 2015;21:84. doi: 10.3748/WJG.V21.I1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wortzel I, Dror S, Kenific CM, Lyden D. Exosome-mediated metastasis: communication from a distance. Dev Cell. 2019;49:347–360. doi: 10.1016/j.devcel.2019.04.011. [DOI] [PubMed] [Google Scholar]
- 56.Wang J, Zhao Y, Qi R, et al. Prognostic role of podocalyxin-like protein expression in various cancers: a systematic review and meta-analysis. Oncotarget. 2017;8:52457–52464. doi: 10.18632/oncotarget.14199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Novo D, Heath N, Mitchell L, et al. Mutant p53s generate pro-invasive niches by influencing exosome podocalyxin levels. Nat Commun. 2018;9:5069. doi: 10.1038/s41467-018-07339-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mantovani F, Collavin L, Del Sal G. Mutant p53 as a guardian of the cancer cell. Cell Death Differ. 2019;26:199–212. doi: 10.1038/s41418-018-0246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Solomon H, Dinowitz N, Pateras IS, et al. Mutant p53 gain of function underlies high expression levels of colorectal cancer stem cells markers. Oncogene. 2018;37:1669–1684. doi: 10.1038/s41388-017-0060-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cooks T, Pateras IS, Tarcic O, et al. Mutant p53 prolongs NF-κB activation and promotes chronic inflammation and inflammation-associated colorectal cancer. Cancer Cell. 2013;23:634–646. doi: 10.1016/j.ccr.2013.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Muller PAJ, Caswell PT, Doyle B, et al. Mutant p53 drives invasion by promoting integrin recycling. Cell. 2009;139:1327–1341. doi: 10.1016/j.cell.2009.11.026. [DOI] [PubMed] [Google Scholar]
- 62.Freed-Pastor WA, Mizuno H, Zhao X, et al. Mutant p53 disrupts mammary tissue architecture via the mevalonate pathway. Cell. 2012;148:244–258. doi: 10.1016/j.cell.2011.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Seiwert TY, Zuo Z, Keck MK, et al. Integrative and comparative genomic analysis of HPV-positive and HPV-negative head and neck squamous cell carcinomas. Clin Cancer Res. 2015;21:632–641. doi: 10.1158/1078-0432.CCR-13-3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhou G, Wang J, Zhao M, et al. Gain-of-function mutant p53 promotes cell growth and cancer cell metabolism via inhibition of AMPK activation. Mol Cell. 2014;54:960–974. doi: 10.1016/j.molcel.2014.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]